Abstract
Gingerols, mainly [6]-gingerol (6G), [8]-gingerol (8G), and [10]-gingerol (10G), are the functional and specific pungent phytochemicals in ginger. However, poor oral bioavailability limits their applications owing to extensive metabolism. The present study aims to characterize the cytochrome P450 (CYP) metabolic characteristics of 6G, 8G, and 10G by using pooled human liver microsomes (HLM), different animal liver microsomes, and the expressed CYP enzymes. It is shown that NADPH-dependent oxidation and hydrogenation metabolisms of gingerols are the main metabolic types in HLM. With the increase of the carbon chain, the polarity of gingerols decreases and the formation of hydrogenated metabolites is more efficient (CLint: 1.41 μL min−1 mg−1 for 6G, 7.79 μL min−1 mg−1 for 8G and 14.11 μL min−1 mg−1 for 10G), indicating that the phase I metabolism of gingerols by HLM varied with the chemical structure of the substrate. The phase I metabolism of gingerols revealed considerable species variations, and compared to HLM, novel metabolites such as (3S,5S)-gingerdiols and demethylated metabolites are generated in some animal liver microsomes. The primary enzymes involved in the oxidized and demethylated metabolism of these gingerols are CYP1A2 and CYP2C19, but their affinities for gingerols are not the same. CYP2D6 and CYP2B6 contributed significantly to the formation of (3R,5S)-[8]-gingerdiol and (3R,5S)-[10]-gingerdiol, respectively; however, the enzyme responsible for the production of (3R,5S)-[6]-gingerediol is yet to be identified. Some metabolites in microsomes cannot be detected by the 12 investigated CYP enzymes, which may be related to the combined effects of multiple enzymes in microsomes, the different affinity of mixed liver microsomes and CYP enzymes, gene polymorphisms, etc. Overall, this work provides a deeper knowledge of the influence of CYP metabolism on the gingerols, as well as the mode of action and the possibility for drug–herbal interactions.
Cytochrome P450 metabolism characteristics of [6]-gingerol, [8]-gingerol, and [10]-gingerol.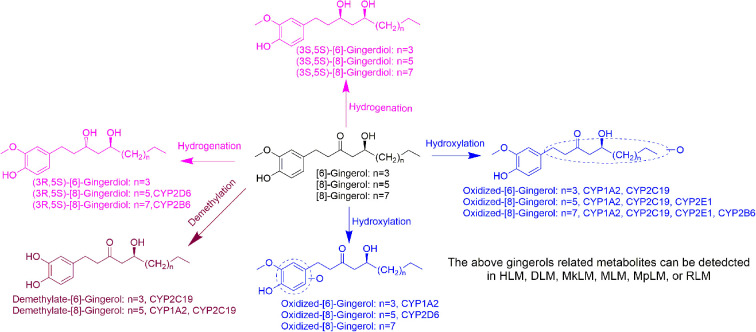
Introduction
Ginger, the rhizome of Zingiber officinale, has been cultivated throughout the world for centuries.1 As one of the most important food medicine homology species, ginger has been extensively applied in the fields of food, agriculture, spices, and traditional or modern medicine worldwide due to its numerous significant health-beneficial effects such as anti-inflammatory, antioxidation, antitumor, and antidiabetic.2 These beneficial effects are believed to be mainly attributed to its major pungent phytochemicals (mainly gingerols and shogaols).3,4 In particular, gingerols, the characteristic pungent constituents and quality control markers of ginger, make up about 25% of its oleoresin, with [6]-gingerol being the most abundant, followed by [8]- and [10]-gingerol, and they are responsible for most of the beneficial effects of ginger,5–8 such as analgesic, antipyretic and anti-inflammatory activities, anticancer activity, antidiabetic activity, and so on.9
Pharmacokinetics of ginger in humans showed that [6]-, [8]-, and [10]-gingerol were rapidly absorbed and cleared, with the highest plasma concentrations just of 42.0 ± 16.3 nmol L−1, 5.3 ± 0.8 nmol L−1, and 4.8 ± 0.08 nmol L−1, respectively.10 These findings indicated that the metabolic characteristics had an important impact on the bioavailability and even health-promoting effects. Our previous study showed that 141 xenobiotics (36 prototypes and 105 metabolites) in rats were identified after oral administration of ginger, and nearly 60% of the xenobiotics in vivo were derived from the pungent compounds, especially gingerols.11 Furthermore, gingerdiols and mono-oxidation products (phase I reaction), as well as glucuronidated conjugates (phase II reaction), were identified as the main metabolites of gingerols,11–16 indicating that gingerols undergo phase I or phase II metabolism by corresponding enzymes.
Glucuronidation is one of the main models of phase II metabolism; previous studies have shown that UGT1A9 and UGT2B7 were the main contributors to the glucuronidated metabolism of [6]-, [8]-, and [10]-gingerol based on the analysis of the relationship between enzyme reaction kinetics and activity. Unfortunately, glucuronidation was a detoxification mechanism because the glucuronide was generally pharmacologically inactive and rapidly eliminated from the body due to its highly polar nature.17 However, the phase I metabolites of gingerols, such as [6]-gingerdiol and [10]-gingerdiol, have been reported to have anticancer activity in H-1299 cells or hematopoietic effects in zebrafish embryos.12,13 And it was noted that the cytochrome P450 superfamily (CYP450s) is a large variety of enzymes, and they are the main enzymes involved in drug metabolism of phase I reaction and biological activation, accounting for about 75% of the total number of metabolic reactions. Additionally, although gingerols were mainly excreted in the form of glucuronidated conjugates, in many cases these conjugates were formed based on phase I metabolites. Hence, it is essential to investigate the metabolic pathway and mechanism of gingerols in CYP subtype enzymes, which allow for a more accurate prediction of their distribution and a greater comprehension of their mode of action in vivo.
Therefore, the metabolic pathways and rates of [6]-, [8]-, and [10]-gingerol (Fig. 1) in HLM were firstly characterized by incubating each compound with nicotinamide adenine dinucleotide phosphate (NADPH) supplemented microsomes. Furthermore, species differences of gingerols in different animal liver microsomes were analysed. In addition, reaction phenotyping was performed by ultrahigh-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry (UHPLC/Q-TOF-MS) to identify the main CYPs contributing to the metabolism of gingerols. Further, kinetic parameters were derived by fitting an appropriate model to the data. With these approaches, the present study is of significant value for better predicting the disposal of gingerols and deeply comprehending their mechanism of action in human tissues, which also helps illustrate their beneficial effects and potential drug–drug interactions.
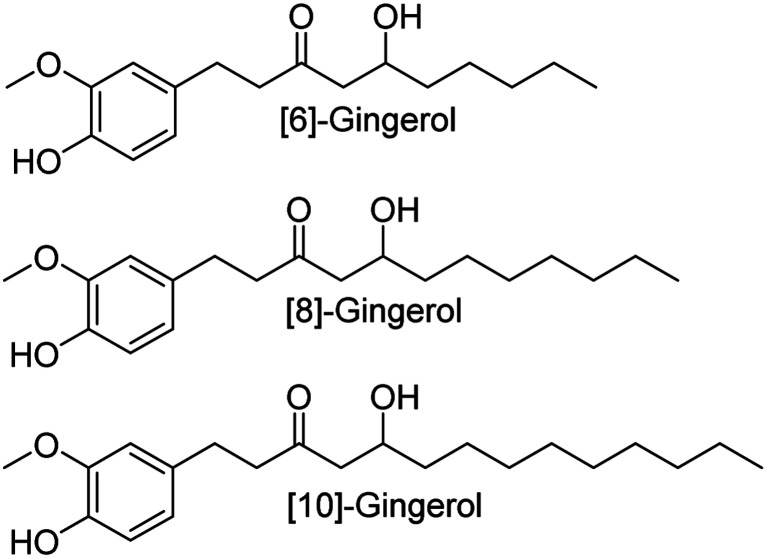
Fig. 1. The chemical structural information of [6]-gingerol, [8]-gingerol and [10]-gingerol.
Experimental
Chemicals and reagents
Pooled human liver microsomes (HLM), rat liver microsomes (RLM), mice liver microsomes (MLM), monkey liver microsomes (MkLM), dog liver microsomes (DLM), mini pig liver microsomes (MpLM) and recombinant expressed human CYP enzymes (CYP1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2 × 101, 3A4 and 3A5) were obtained from Corning Biosciences (New York, USA). Magnesium chloride and NADPH were provided by Sigma-Aldrich (St. Louis, MO, USA). [6]-gingerol (6G), [8]-gingerol (8S), [10]-gingerol (10G), (3R,5S)-[6]-gingerdiol, (3S,5S)-[6]-gingerdiol, (3R,5S)-[8]-gingerdiol, (3S,5S)-[8]-gingerdiol, (3R,5S)-[10]-gingerdiol, (3S,5S)-[10]-gingerdiol with purities over 98% were isolated and identified in our laboratory, and their 13C-NMR and HRMS data were listed in the ESI.† Other chemicals and materials were all analytical grades.
In vitro metabolism assay
As described previously,18 the incubation system (100 μL) for phase I metabolism contained Tris–HCl buffer solution (50 mM, pH 7.4), drug-metabolizing enzyme solutions (0.5 mg ML−1, including pooled HLM, animal liver microsomes and purified CYP enzymes), MgCl2 (5 mM), a series of [6]-, [8]-, and [10]-gingerol solutions and NADPH solution (1 mM). After 2 hours of co-incubation, the reaction was terminated by adding same volume of ice-cold acetonitrile, and the samples were vortexed and centrifuged at 14 000 g for 10 min. Then, the supernatant was subjected to UPLC-Q/TOF-MS or UPLC for analysis. The negative control was constructed by an incubation system without NADPH to confirm that the metabolites produced are NADPH-dependent. All experiments in this study were performed in triplicate. Preliminary experiments were performed to ensure that the rates of metabolism were determined under linear conditions with respect to the incubation time and protein concentration.
UPLC-Q/TOF-MS and UPLC analysis
The UPLC analysis was performed on an Acquity UPLC 1-Class system equipped with a binary solvent system, an automatic sample manager, and a photodiode array detector (Waters Corporation). A BEH RP C18 column (2.1 × 50 mm, 1.7 mm) maintained at 40 °C was used for the separation of gingerols (6G,8G, and 10G) and their NADPH-dependent metabolites. The mobile phases consisted of water (A) and acetonitrile (B), both including 0.1% formic acid (v/v), at a flow rate of 0.3 mL min−1. The gradient elution program was optimized as follows: 5% B from 0–0.5 min, 5–10% B from 1–1.5 min, 10–70% B from 1.5–4.5 min.70–95% B from 4.5–4.5 min. 95–95% B from 4.5–5.5 min, 95–5% B from 5.5–6.5 min, 5% B from 6.5–7 min. The injection volume was set at 4 μL and the detection wavelength was set at 280 nm. The phase I metabolites of gingerols were quantitated by UPLC based on the standard curve of the parent compound using the same method as described before.2,19,20 Calibration curves were constructed by plotting each gingerol peak area ratio (Y) versus its concentration (X) using a 1/X2 weighting factor. For 6G (Y6G = 1174.2X + 120.19), 8G (Y8G = 1248.8X − 100.19), and 10G (Y10G = 1010X + 129.88), an acceptable linear correlation was confirmed by correlation coefficients (r2) of 0.9960, 0.9976, and 0.9998, respectively.
Kinetic evaluation
A series of [6]-, [8]- and [10]-gingerol concentrations (6.25–400 μM for 6G, 6.25–400 μM for 8G, and 3.125–400 μM for 10G) was incubated with pooled HLM, animal liver microsomes and purified CYP enzymes, respectively, to determine the metabolic rates. The kinetic models of Michaelis–Menten (eqn 1) and substrate inhibition model (eqn. (2)) were fitted to the data of phase I metabolism rates versus substrate concentrations. Model selection was based on a visual inspection of the Eadie–Hofstee plot. A straight line in the Eadie–Hofstee plot indicated that the data were best described by the Michaelis–Menten model, whereas a hook in the upper panel suggested that the substrate inhibition model should be used. Model fitting and parameter estimation were performed using the GraphPad Prism V7 software (San Diego, CA, USA).
 |
1 |
 |
2 |
Species difference analysis
In this study, a series of [6]-, [8]- and [10-gingerol solutions were incubated with five animal liver microsomes (RLM, MLM, MkLM, DLM, and MpLM) to determine phase I metabolism rates, respectively. Kinetic parameters were derived from the appropriate model fitting. Additionally, the CLint value for each phase I metabolite by HLM and different animal liver microsomes were used as the evaluation parameters to estimate species diversity, as published previously.18,21
Statistical analysis
Data are expressed as mean ± standard deviation (SD). The two-tailed Student's t-test was used to compare the mean differences, and the prior level of significance was set at p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***).
Results
Structural identification of gingerols metabolites in HLM
After incubation of 6G, 8G, and 10G with NADPH-supplemented HLM, respectively, metabolites were detected by UHPLC/Q-TOF-MS for each gingerol (Table 1. Fig. S1:† HLM). For phase I metabolism of 6G in HLM, four metabolites were identified as oxidized or hydrogenated products. Among them, M6G-1, M6G-2, and M6G-3 showed the same [M + Na]+ at m/z 333.1678 (C17H26O5Na), which was 16 Da higher than that of 6G, indicating that they were oxidized products. In the MS/MS analysis, the daughter ions at m/z 275.1637 and 193.0863 for M6G-2 and M6G-3 suggested that the oxygen atom was conjugated to the benzene ring of 6G. However, the daughter ion at m/z 137.0603 for M6G-1 indicated that the oxygen atom was conjugated to the carbon chain of 6G. M6G-5 exhibited the [M + Na]+ ion at m/z 319.1885 (C17H28O4Na), which was 2 Da higher than that of 6G. Hence, it was identified as [6]-gingerdiol because 6G has only one carbonyl reduction site, and its absolute configuration is defined as (3R, 5S)-[6]-gingerdiol by the reference standard isolated in our laboratory. Similarly, five phase I metabolites of 8G in HLM have been identified as oxidized or hydrogenated products (Table 1. Fig. S2:† HLM). M8G-1 and M8G-2 were characterized as oxidized products on the carbon chain of 8G due to the ion fragments of m/z 177.0905 and m/z 137.0603. M8G-3 and M8G-4 were suggested as oxidized metabolites on the benzene of 8G because of the ions at m/z 193.0863. M8G-6 was also identified as (3R, 5S)-[8]-gingerdiol by the reference standard isolated in our laboratory. Likewise, four 10G related metabolites were characterized in HLM (Table 1. Fig. S3:† HLM). M10G-1 and M10G-2 were oxidized products on the carbon chain, and M10G-3 was the oxidized metabolite on the benzene ring of 10G, whereas M10G-4 was (3R, 5S)-[10]-gingerdiol, the hydrogenated product of 10G.
The identified metabolites of [6]-, [8], [10]-gingerol in different liver microsomes by UPLC-Q/TOF-MSa.
| No. | RT (min) | Formula | [M + Na]+ ion | Error (ppm) | (+) ESI-MS/MS | Identification | Liver microsomes |
|---|---|---|---|---|---|---|---|
| M6G-O | 4.66 | C17H26O4Na | 317.1724 | 1.9 | 277.1804, 137.0604 | 6-G | ALL |
| M6G-1 | 3.42 | C17H26O5Na | 333.1678 | −0.9 | 275.1650, 137.0603 | Mono-oxidized-6G (carbon chain) | H, R |
| M6G-2 | 3.89 | C17H26O5Na | 333.1678 | 0.8 | 275.1637, 193.0863 | Mono-oxidized-6G (benzene) | H |
| M6G-3 | 4.27 | C17H26O5Na | 333.1678 | 3.3 | 275.1637, 193.0863, 167.0714 | Mono-oxidized-6G (benzene) | H, M |
| M6G-4 | 4.30 | C16H24O4Na | 303.1570 | −0.7 | 263.1640, 163.0570, 123.0440 | Demethylated-6G | R |
| M6G-5 | 4.45 | C17H28O4Na | 319.1885 | 0.1 | 279.1949, 261.1862, 163.0758, 137.0598 | [3R,5S]-[6]-Gingerdiol | ALL |
| M6G-6 | 4.57 | C17H28O4Na | 319.1885 | 0.8 | 279.1957, 261.1853, 163.0761, 137.0606 | [3S,5S]-[6]-Gingerdiol | Mk |
| M8G-O | 5.23 | C19H30O4Na | 345.2036 | 2.7 | 305.2115, 177.0915, 137.0602 | 8G | ALL |
| M8G-1 | 3.89 | C19H30O5Na | 361.1991 | 1.7 | 303.1962, 177.0905, 137.0607 | Mono-oxidized-8G (carbon chain) | H, R |
| M8G-2 | 3.95 | C19H30O5Na | 361.1991 | 1.7 | 321.2055, 303.1960, 177.0913, 137.0597 | Mono-oxidized-8G (carbon chain) | H, D, Mk, Mp, R |
| M8G-3 | 4.53 | C19H30O5Na | 361.1991 | −3.9 | 321.2066, 303.1968, 193.0869 | Mono-oxidized-8G (benzene) | H |
| M8G-4 | 4.87 | C19H30O5Na | 361.1991 | 4.2 | 321.2045, 303.2054, 193.0863 | Mono-oxidized-8G (benzene) | H, M |
| M8G-5 | 4.91 | C18H28O4Na | 331.1903 | 5.4 | 291.196, 163.057, 123.0450 | Demethylated-8G | R |
| M8G-6 | 5.01 | C19H32O4Na | 347.2198 | 4.6 | 307.2274, 289.2174, 163.0752, 137.0598 | [3R,5S]-[8]-Gingerdiol | ALL |
| M8G-7 | 5.19 | C19H32O4Na | 347.2198 | 3.5 | 307.2267, 289.2171, 163.0765, 137.0597 | [3S,5S]-[8]-Gingerdiol | D |
| M10G-O | 5.74 | C21H34O4Na | 373.2343 | 3.3 | 333.2426, 177.0913, 137.0602 | 10G | ALL |
| M10G-1 | 4.39 | C21H34O5Na | 389.2304 | 4.4 | 331.2274, 177.0916, 137.0599 | Mono-oxidized-10G (carbon chain) | H, Mk, M, Mp, R |
| M10G-2 | 4.45 | C21H34O5Na | 389.2304 | −4.6 | 349.2384, 331.2274, 177.0913, 137.0599 | Mono-oxidized-10G (carbon chain) | ALL |
| M10G-3 | 5.09 | C21H34O5Na | 389.2304 | −0.3 | 349.2372, 331.2274, 193.0869 | Mono-oxidized-10G (benzene) | H, Mp |
| M10G-4 | 5.57 | C21H36O4Na | 375.2511 | −1.1 | 335.2583, 317.2475, 177.0914, 163.0758 | [3R,5S]-[10]-Gingerdiol | ALL |
| M10G-5 | 5.72 | C21H36O4Na | 375.2511 | 3.6 | 335.2564, 317.2470, 177.0907, 163.0759 | [3S,5S]-[10]-Gingerdiol | R |
6G, 8G, and 10G means [6]-gingerol, [8]-gingerol, and [10]-gingerol, respectively. H, D, Mk, M, Mp, and R represents liver microsomes of human, dog, monkey, mice, mini pig, and rat, respectively.
Phase I metabolism of gingerols in HLM
In this study, compared with the parent compound, the phase I metabolites showed no essential changes in the mother nucleus structure, so their ultraviolet absorption was almost the same. Hence, the phase I metabolites of gingerols were quantitated by UPLC based on the standard curve of the parent compound.
Due to the decrease of the sensitivity of the UPLC compared to LC-MS, some metabolites cannot be detected. The enzyme kinetics model was determined by observing the Eadie–Hofstee plot visually. A straight line in the Eadie–Hofstee plot indicated that the data were best described by the Michaelis–Menten model, while a hook in the upper panel indicated that the substrate inhibition model should be employed. The results (Fig. 2–4, Table S1†) showed that the oxidized products on the carbon chains of M6G-1, M8G-1, M8G-2, M10G-1, and M10G-2 exhibited different enzymatic reaction characteristics. Among them, M6G-1 followed the Michaelis equation, but with carbon chain growth, M8G-1, M8G-2, M10G-1, and M10G-2 all met the inhibition kinetics. The hydrogenated metabolites of 6G (Michaelis equation), 8G (Michaelis equation), and 10G (substrate inhibition equation) in HLM also showed similar kinetic characteristics. Furthermore, the hydrogenated metabolism formation for 6G, 8G, and 10G was 432.82 pmol min−1 mg−1, 848.90 pmol min−1 mg−1, and 1957.00 pmol min−1 mg−1, respectively, which was higher than that of oxidized metabolism formation, indicating that the hydrogenated metabolism was much more efficient of gingerols in HLM. Meanwhile, the corresponding total CLint value of the hydrogenated metabolite was 1.41 μL min−1 mg−1 for 6G, 7.79 μL min−1 mg−1 for 8G, and 14.11 μL min−1 mg−1 for 10G in HLM, suggesting that the hydrogenated activity for gingerols in HLM might be promoted as the length of their alkyl chains increased. The CLint value of gingerol oxidative metabolites also showed selectivity, of which the oxidation product with smaller polarity showed a higher CLint value.
Fig. 2. Enzyme kinetic curve of CYP metabolism of [6]-gingerol in HLM (A), DLM (B), MkLM (C), MLM (D), MpLM (E), and RLM (F), the insert shows the corresponding Eadie–Hofstee plot. HLM, DLM, MkLM, MLM, MpLM, and RLM, represents liver microsomes of Human, Dog, Monkey, Mice, Mini Pig, and Rat, respectively.
Fig. 3. Enzyme kinetic curve of CYP metabolism of [8]-gingerol in HLM (A), DLM (B), MkLM (C), MLM (D), MpLM (E), and RLM (F), the insert shows the corresponding Eadie–Hofstee plot. HLM, DLM, MkLM, MLM, MpLM, and RLM, represents liver microsomes of Human, Dog, Monkey, Mice, Mini Pig, and Rat, respectively.
Fig. 4. Enzyme kinetic curve of CYP metabolism of [10]-gingerol in HLM (A), DLM (B), MkLM (C), MLM (D), MpLM (E), and RLM (F), the insert shows the corresponding Eadie–Hofstee plot. HLM, DLM, MkLM, MLM, MpLM, and RLM, represents liver microsomes of Human, Dog, Monkey, Mice, Mini Pig, and Rat, respectively.
Species differences of phase I metabolism of gingerols
In this study, the liver microsomes of rats, mice, monkeys, dogs, and mini pigs were incubated with 6G, 8G, and 10G, respectively, to explore the phase I metabolic characteristics of gingerols in different species. The results showed that oxidized and hydrogenated products of gingerols were the main phase I metabolites in liver microsomes of different species, whereas the demethylated metabolites were only highly exposed in RLM (Fig. S1†).
As shown in Fig. S1,† hydrogenated products of 6G (M6G-5) were identified in all animal liver microsomes; oxidized metabolites of 6G (M6G-1 and M6G-3) showed a certain exposure in MLM and RLM, and the demethylated metabolite of 6G (M6G-4) was exposed in RLM. Of all metabolites of 6G followed the Michaelis–Menten equation, except for M6G-4 which met the substrate inhibition model (Table S1†). It was important to note that the hydrogenated metabolite (M6G-6, 3S,5S-[6]-gingerdiol), which could be detected in some animal liver microsomes, was not found in HLM. The catalyzation efficiencies (reflected by CLint values, Fig. 2) for M6G-5 of human and animal microsomes followed the order of MkLM (8.71 mL min−1 mg−1) > MLM (8.24 mL min−1 mg−1) > HLM (2.90 mL min−1 mg−1) > DLM (2.47 mL min−1 mg−1) > RLM (1.30 mL min−1 mg−1) > MpLM (0.56 mL min−1 mg−1). Similarly, the CLint values for M6G-1 were RLM (1.41 mL min−1 mg−1) > HLM (0.68 mL min−1 mg−1). Based on the metabolic profile and catalytic efficiency in liver microsomes of various species, rats were likely the best model for human 6G phase I metabolism studies.
The oxidized metabolites (M8G-1, M8G-2, M8G-4) and hydrogenated products (M8G-6 and M8G-7) of 8G also showed high exposure in different animal liver microsomes, while the demethylated metabolite of M8G-5 was exposed in RLM (Fig. S2†). In RLM, MkLM, DLM and MpLM, there were mainly the oxidized metabolites (M8G-1 and M8G-2) on the carbon chain, whereas the oxidized metabolite (M8G-4) on the benzene ring was the main one for 8G in MLM. Among them, as shown in Fig. 3 and Table S1,† the CLint value for M8G-2 (major oxidation product) in RLM was closer to that of HLM, and they (M8G-2 in RLM and HLM) showed the same enzyme kinetic characteristic of the substrate inhibition model rather than Michaelis–Menten equation (M8G-2 in MkLM, DLM and MpLM). For the hydrogenated products (M8G-6: 3R,5S-[8]-gingerdiol and M8G-7: 3S,5S-[8]-gingerdiol), 3R,5S-[8]-gingerdiol with a high Vmax could be detected among all animal liver microsomes, whereas 3S,5S-[8]-gingerdiol was only found in the DLM. The CLint values for M8G-6 of human and animal microsomes followed the order of MkLM (16.35 mL min−1 mg−1) > HLM (7.79 mL min−1 mg−1) > MLM (6.48 mL min−1 mg−1) > MpLM (4.27 mL min−1 mg−1) > DLM (4.21 mL min−1 mg−1) > RLM (0.40 mL min−1 mg−1).
The results showed that oxidized metabolites (M10G-1 and M10G-2) and hydrogenated products (M10G-4 and M10G-5) were the main phase I metabolites of 10G in different animal liver microsomes (Fig. S3†). Kinetic profiling (Fig. 4 and Table S1†) revealed that phase I metabolites of 10G in MkLM, DLM, MLM and MpLM all followed the Michaelis–Menten equation, whereas the products in RLM met the substrate inhibition model. 3R,5S-[10]-gingerol (M10G-4) showed the highest Vmax among the metabolites of 10G in each liver microsomes, indicating that M10G-4 was the main metabolite of 10G. Combined with metabolic products and CLint values in different animal liver microsomes, mini pigs were probably the best model for the phase I metabolism studies of 10G instead of humans.
Reaction phenotyping by CYP enzymes
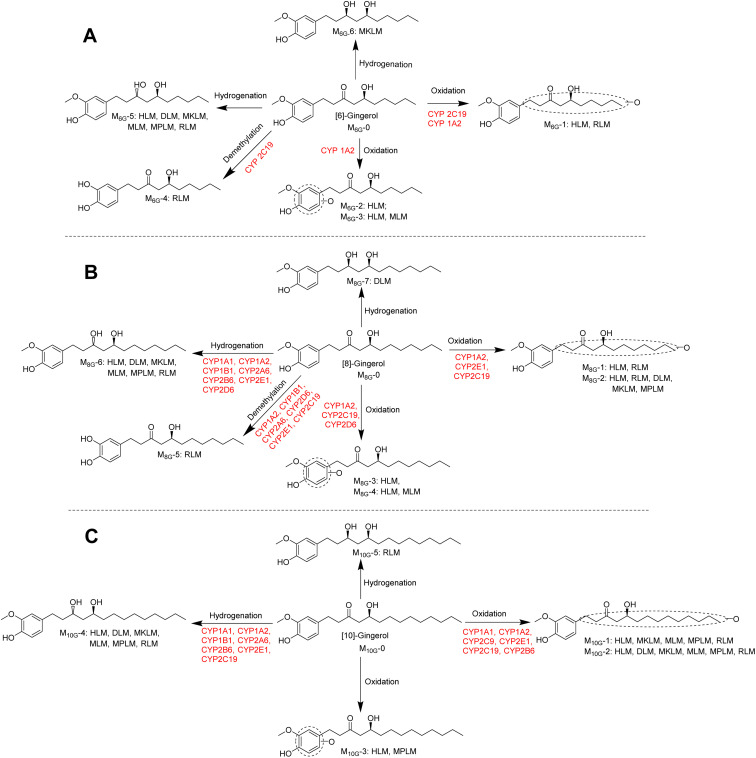
To determine the enzymes involved in the contribution to phase I metabolism, two test concentrations of [6]-, [8]- and [10]-gingerol were incubated with various expressed CYPs enzymes, respectively, which were qualitatively analysed by UPLC-Q-TOF-MS. It showed that CYP1A2 and CYP2C19 were involved in the oxidative metabolism of [6]-gingerol, and CYP2C19 also contributed to forming the demethylated product (Fig. 6). 8G was catalysed by CYP1A2, CYP2C19 and CYP2E1 to form the oxidative metabolism (M8G-1, M8G-2, M8G-3, and M8G-4), while the formation of the demethylated product was contributed by CYP1A2, 1B1, 2A6, 2D6, and 2 × 101, and CYP1A1, 1A2, 1B1, 2A6, 2B6, and 2 × 101 catalysed the production of M8G-6 (Fig. 6). Similarly, CYP1A1, 1A2, 2C9, 2 × 101, and 2C19 exhibited catalysis in the formation of M10G-1, while CYP1A1, 1A2, 1B1, 2A6, 2B6, 2 × 101, and 2C19 catalysed the production of M10G-4 (Fig. 6). Unfortunately, in this study, the contribution of CYPs in the formation of 3S,5S-gingerdiols was still unclear. Furthermore, because the concentration of gingerols-related metabolites was lesser than the limit of quantification, it was unable to determine the complete kinetic parameters of these CYP enzymes.
Fig. 6. Metabolic pathways of [6]-gingerol (A), [8]-gingerol (B), and [10]-gingerol (C) involving CYPs.
Phase I metabolism kinetics of [6]-gingerol by expressed CYP enzymes
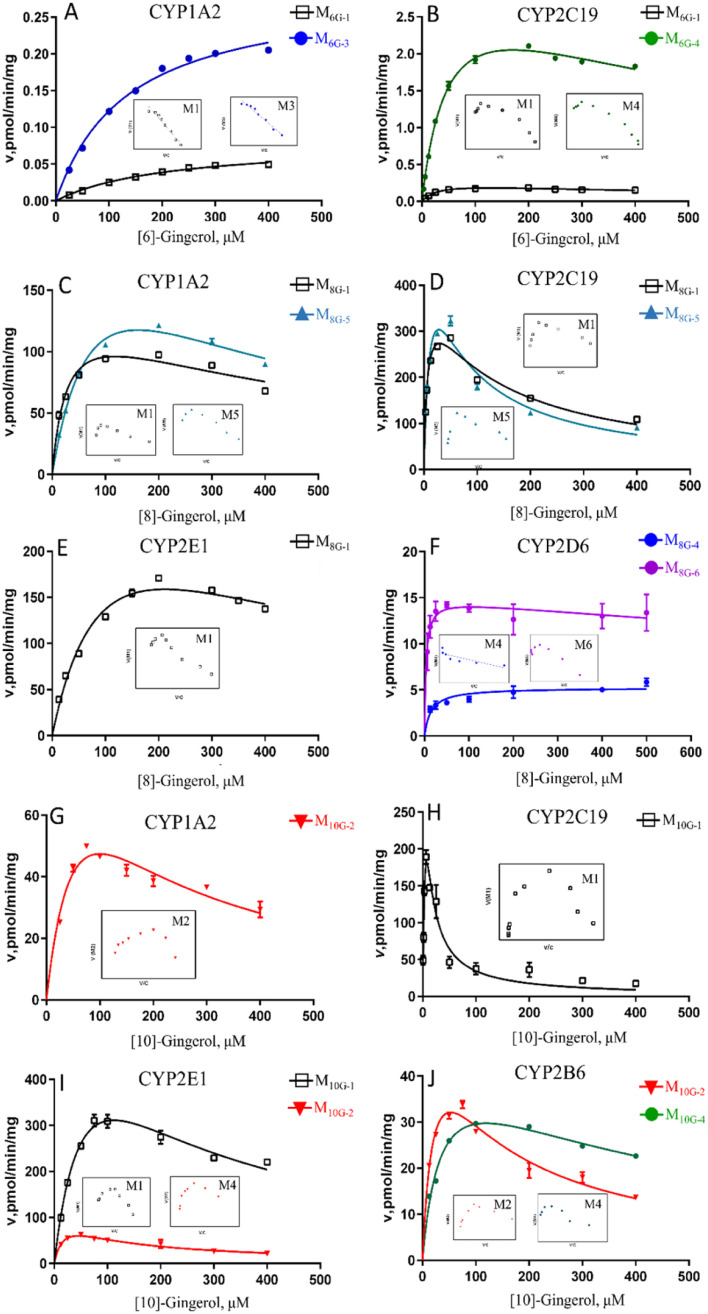
Based on the reaction phenotyping results, CYP1A2 and CYP2C19 were the main enzymes involved in the oxidative metabolism of 6G (Fig. 5, Table S2†). The metabolic rates of M6G-1 and M6G-3 catalysed by CYP1A2 all met classical Michaelis–Menten kinetics, whereas the formation of M6G-1 and M6G-4, which were metabolized by CYP2C19 were modelled by the substrate inhibition equation. According to the literature,18 the kinetic model in CYPs that did not follow the same kinetic as in HLM was a normal phenomenon. The CLint of M6G-1 catalysed by CYP2C19 (8.85 ± 0.86 μL min−1 mg−1) was almost 24 times higher than that of CYP1A2, and CYP2C19 showed the highest CLint of the metabolite of M6G-4 (63.31 ± 5.06 μL min−1 mg−1). In addition, the Km value of 6G metabolized by CYP2C19 (30.21 μM ∼52.22 μM) was less than that of CYP1A2 (135.80 μM ∼212.83 μM), which indicated that CYP2C19 had a good affinity for 6G, and it was also an important factor for the high inherent clearance rate of 6-gingerol in CYP2C19 system.
Fig. 5. Enzyme kinetic curve of CYP metabolism of [6]-, [8]-, [10]-gingerol in CYP subtype enzymes. A–B, [6]-gingerol in CYP1A2 (A), and CYP2C19 (B); C–F, [8]-gingerol in CYP1A2 (C), CYP2C19 (D), CYP2E1 (E), and CYP2D6 (F); G–J, [10]-gingerol in CYP1A2 (G), CYP2C19 (H), CYP2E1 (I), and CYP2B6 (J). The insert shows the corresponding Eadie–Hofstee plot.
Phase I metabolism kinetics of [8]-gingerol by expressed CYP enzymes
CYP1A2, CYP2C19, and CYP2E1 were the primary enzymes involved in the formation of M8G-1 and M8G-5 of 8G (Fig. 5, Table S2†). Enzymatic kinetics and the Eadie–Hofstee plot demonstrated that the metabolic rates of M8G-1 and M8G-5 catalysed by CYP1A2, CYP2C19, and CYP2E1 were all satisfied the conventional substrate inhibition equation. The CLint of M8G-1 and M8G-5 catalyzed by CYP2C19 (CLint of M8G-1: 59.19 ± 7.69 μL min−1 mg−1, CLint of M8G-5: 45.20 ± 12.68 μLmin−1mg) was almost 12–22 times higher than that of CYP1A2 (CLint of M8G-1: 5.02 ± 0.97 μL min−1 mg−1, CLint of M8G-5: 2.63 ± 0.49 μL min−1 mg−1) and CYP2E1 (CLint of M8G-1: 2.81 ± 0.70 μL min−1 mg−1), suggesting that CYP2C19 was important for the bioavailability of 8G. Furthermore, CYP2C19 also showed a smaller Km value than that of CYP1A2 and CYP2E1, indicating that CYP2C19 may be the main contributor to phase I metabolism of 8G. CYP2D6 contributed to the formation of the oxidized metabolite (M8G-4) and hydrogenated product (M8G-6); the metabolic rate of M8G-4 catalysed by CYP2D6 was met classical Michaelis–Menten kinetics, while the formation of M8G-6 was modelled by the substrate inhibition equation.
Phase I metabolism kinetics of [10]-gingerol by expressed CYP enzymes
Formation of phase I metabolites of 10G was mainly observed with CYP1A2, CYP2B6, CYP2C19, and CYP2E1 among the 12 expressed CYP enzymes, and the oxidized products on the carbon chain of M10G-1 and M10G-2 were the main oxidized products incubated by CYP enzymes (Fig. 5, Table S2†). The enzyme kinetics for oxidation of 10G by the above-mentioned CYP enzymes displayed a consistent substrate inhibition profile. CYP1A2, CYP2B6, and CYP2E1 catalysed the formation of M10G-2, of which CYP2E1 showed the highest CLint and lowest Km, suggesting that CYP2E1 was the main enzyme contributing to the formation of M10G-2 from 10G. Likewise, CYP2C19 might be the main active enzyme in the oxidized form of M10G-2 from 10G. Furthermore, CYP2B6 also helped to catalyse the formation of hydrogenated products (M10G-4), whose enzyme kinetic was modelled by the substrate inhibition equation (CLint = 1.31 ± 0.45 μL min−1 mg−1, Km = 36.6 ± 11.09 μM).
Discussion
Gingerols, the major and bioactive pungent ingredients in the rhizome of zingiber officinale, has drawn increasing attention to their pharmacological activities.1–4 However, previous metabolic profile research and pharmacokinetic study of ginger showed that gingerols experienced a series of phase I and phase II reactions that resulted in poor bioavailability.11–16 It has been reported that UGT1A9 and UGT2B7 were the main contributors to the glucuronidation (the main phase II reaction type) of gingerols,17 but the relationship between enzyme reaction kinetics of their phase I reaction (mainly CYPs) and the activity of their metabolism mechanism was still unclear. Hence, in order to better predict the disposal of gingerols and understand their mechanism action in individual tissues, especially in humans, it inspired us to further explore the phase I metabolic pathways of gingerols, which also helped to illustrate the variation in their beneficial effects and potential drug–herbal interactions.
It is notable that the outcome of enzyme kinetics assays could be affected by the drug–protein binding because of the presence of microsomal protein, which will weaken the ability to predict pharmacokinetic properties. Hallifax and Houston model has been proved to accurately predict on free unbound values (fu) for the moderately lipophilic compounds (log P = 2.5–5.0).22 The fu values were estimated to be 98%, 95% and 80% for 6G, 8G, and 10G (0.05 mg mL−1 protein), respectively. As the estimated nonspecific binding of 6G, 8G, and 10G under the incubation conditions used was less than 20%, the incubation system was not corrected for nonspecific protein binding in calculations of kinetic parameters.23
Determination of the activities of CYP isozymes and microsomes was based on the intrinsic clearance values (CLint) obtained from kinetic profiling over a wide range of substrate concentrations. CLint is calculated by Vmax and Km, which represents the catalytic efficiency and is independent of the substrate concentration of enzymes or microsomes. Furthermore, CLint is more relevant in an attempt to predict the clearance activity in vivo compared with other parameters (i.e. Vmax and Km). Consequently, in this study, the human liver showed a wide range of CLint values (0.68 μL min−1 mg−1 ∼202.00 μL min−1 mg−1) for the different types of phase I reactions in the metabolism of gingerols. By comparing the values of CLint, it found that with the increase of gingerols polarity, individuals tended to rapidly increase the polarity of chemicals through metabolic reaction.
Our study demonstrated that oxidized metabolites (oxidation on the carbon chain or benzene ring) and hydrogenated metabolites ([6]-,[8]-,[10]-gingerdiol) were the major phase I products of gingerols in HLM. Interestingly, the absolute configuration of the hydrogenated metabolites in HLM was determined as the R conformer of C-3 and the S conformer of C-5, and only this conformer could be formed in the HLM, whereas in DLM or MkLM, both absolute configurations of “3R,5S” and “3S, 5S” could be found. The results indicated that the hydrogenated metabolism of gingerols by corresponding enzymes showed regioselectivity (position preference). Furthermore, in the HLM system, the formation of 3R,5S-[6]-gingerdiol (CLint = 2.90 μL min−1 mg−1) was more efficient than the oxidation products (CLint = 0.68 μL min−1 mg−1), but with the increase of the carbon chain, the polarity of gingerols decreases and the formation of hydrogenated metabolites was more efficiency, which indicated that the phase I metabolism of gingerols by HLM varied with the chemical structure of the substrate. Hence, the NADPH-dependent phase I reaction activity could be regulated by alteration of the gingerol structure.
As shown Fig. 6, this study found that the phase I metabolism of gingerols in liver microsomes was mainly divided into three categories: oxidation, hydrogenation, and demethylation. In this study, the oxidation metabolites of [6]-, [8]-and [10]-gingerol can be divided into two categories: carbon chain oxidation products or benzene ring oxidation products. It was noted that both CYP1A2 and CYP2C19 are involved in the oxidative metabolism of these three gingerols. Nonetheless, the affinity of these enzymes to gingerols is not the same, and oxidation metabolites on the carbon chain are the main products of these enzymes. The hydrogenated metabolites ([6]-,[8]-,[10]-gingerdiol) are the major phase I products of gingerols in HLM; CYP2D6 and CYP2B6 showed the main contribution to the formation of 3R,5S-[8]-gingerdiol and 3R,5S-[10]-gingerdiol, respectively, but which enzyme produces 3R,5S-[6]-gingerediol remains unclear. It is also noted that all the 12 CYPs tested in this study cannot catalyse gingerols to the formation of 3S,5S-gingerediols, as a class of bioactive chemicals, which indicates that more in-depth research is necessary. [6]-gingerol and [8]-gingerol can be metabolized to the formation of demethylated products by CYP1A2 or CYP2C19, and the CYP2C19 showed a high capability of demethylation with a high value of CLint (63.31 ± 5.06 μL min−1 mg−1 to 6G, 45.20 ± 12.68 μL min−1 mg−1 to 8G). With the increase of gingerols polarity, 10G cannot be metabolized to the corresponding demethylated production in liver microsomes, which indicated that the regioselective action of the CYP enzymes on the metabolism of gingerols.
Glucuronidation metabolism was thought to be a detoxifying process because of the strong polarity of glucuronidated products and their quick elimination from the body. In contrast, phase I metabolism may introduce active groups to the prototype. Hence, the inhibition or activation of the enzyme was of some significance to the exertion of the efficacy of the corresponding chemicals. Previous research revealed that UGT1A9 and UGT2B7 were the main contributors to the formation of the glucuronidated gingerols, and this study indicated that CYP1A2, CYP2C19, CYP2D6 and CYP2B6 involved in the phase I metabolism of gingerols. These results indicated a high possibility of drug–herbal interactions interaction between ginger and the drugs such as zidovudine, naloxone, morphine, and others whose main metabolic pathways were catalysed by the above enzymes.
In this study, it was shown that some metabolites in microsomes cannot be detected in the 12 tested CYP enzymes. Firstly, there may be another phase I metabolic enzymes involved in the metabolism of gingerols beyond the 12 tested CYP enzymes. Second, the metabolic differences may be that the catalysis of gingerols by human liver microsomes is a combination of multiple enzymes. In addition, the different affinity of mixed liver microsomes and CYP subtype enzymes to the binding sites of gingerol metabolites and gene polymorphisms may also be another reason for the differences in the metabolism.
Conclusions
Totally, CYP metabolism characteristics of gingerols ([6]-, [8]-, and [10]-gingerol), the main functional and specific chemicals in ginger, are revealed by microsomes of humans and different species tissues combined with expressed CYP enzymes. Oxidation, hydrogenation, and demethylation metabolisms of gingerols are the main metabolic type in microsomes. However, there are differences in the metabolic types and metabolic kinetics of different species of liver microsomes. CYP1A2 and CYP2C19 are the main enzymes involved in the oxidized and demethylated metabolism of these gingerols, but the affinity of these enzymes to gingerols is not the same. CYP2D6 and CYP2B6 exhibited the primary contribution to the creation of 3R,5S-[8]-gingerdiol and 3R,5S-[10]-gingerdiol, respectively, however which enzyme makes 3R,5S-[6]-gingerediol remains unclear. Some metabolites in microsomes cannot be detected in the 12 tested CYP enzymes, which may be related to the combined effects of multiple enzymes, the different affinity of mixed liver microsomes and CYP enzymes, and gene polymorphisms, and so on. Overall, this study provided a better insight into the impact of CYP metabolism on the functional effects of gingerols, which also gave in-depth data on an understanding of their mechanism action and potential drug–herbal interactions.
Author contributions
Liangliang He: supervision, conceptualization, investigation, methodology, funding acquisition; Xintong Chen, Chanjuan Chen: investigation, data curation; Jie Liu, Qingmei Mo: data curation; Xinsheng Yao: supervision; Xin Di: project administration, supervision; Zifei Qin, Zhihong Yao: project administration, funding acquisition, supervision.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was financially supported by the National Key R&D Program of China (2022YFC0867400), National Natural Science Foundation of China (82003914, 81974519), Natural Science Foundation of Guangdong Province (2019A1515110391), Program of Introducing Talents of Discipline to Universities (B13038), Guangzhou Key Laboratory of Formula-Pattern of Traditional Chinese Medicine (202102010014).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra06184h
References
- Afzal M. Al-Hadidi D. Menon M. Pesek J. Dhami M. S. Drug Metab. Drug Interact. 2001;18:159–190. doi: 10.1515/dmdi.2001.18.3-4.159. [DOI] [PubMed] [Google Scholar]
- He L. Xu J. Wang Q. Zhang Y. Qin Z. Yu Y. Qian Z. Yao Z. Yao X. RSC Adv. 2018;8:41368–41375. doi: 10.1039/c8ra08466a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga M. S. Haniadka R. Pereira M. M. D'Souza J. J. Pallaty P. L. Bhat H. P. Popuri S. Crit. Rev. Food Sci. Nutr. 2011;51:499–523. doi: 10.1080/10408391003698669. [DOI] [PubMed] [Google Scholar]
- Kubra I. R. Rao L. J. M. Crit. Rev. Food Sci. Nutr. 2012;52:651–688. doi: 10.1080/10408398.2010.505689. [DOI] [PubMed] [Google Scholar]
- Schwertner H. A. Rios D. C. J. Chromatogr. B. 2007;856:41–47. doi: 10.1016/j.jchromb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Choi J. G. Kim S. Y. Jeong M. Myung S. O. Pharmacol. Ther. 2018;182:56–69. doi: 10.1016/j.pharmthera.2017.08.010. [DOI] [PubMed] [Google Scholar]
- Torres de Lima R. M. dos Reis A. C. Melo de Menezes A.-A. P. de Oliveira Santos J. V. Gomes de Oliveira Filho J. W. de Oliveira Ferreira J. R. Oliveira Barros de Alencar M. V. Oliveira Ferreira da Mata A. M. Khan I. N. Islam A. Uddin S. J. Ali E. S. Islam M. T. Tripathi S. Mishra S. K. Mubarak M. S. de Carvalho Melo-Cavalcante A. A. Phytother. Res. 2018;32:1885–1907. doi: 10.1002/ptr.6134. [DOI] [PubMed] [Google Scholar]
- Semwal R. B. Semwal D. K. Combrinck S. Viljoen A. M. Phytochemistry. 2015;117:554–568. doi: 10.1016/j.phytochem.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Semwal R. B. Semwal D. K. Combrinck S. Viljoen A. M. Phytochemistry. 2015;117:554–568. doi: 10.1016/j.phytochem.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Schoenknecht C. Andersen G. Schmidts I. Schieberle P. J. Agric. Food Chem. 2016;64:2269–2279. doi: 10.1021/acs.jafc.6b00030. [DOI] [PubMed] [Google Scholar]
- He L. Qin Z. Li M. Chen Z. Zeng C. Yao Z. Yu Y. Dai Y. Yao X. J. Agric. Food Chem. 2018;66:9010–9033. doi: 10.1021/acs.jafc.8b03600. [DOI] [PubMed] [Google Scholar]
- Lv L. Chen H. Soroka D. Chen X. Leung T. Sang S. J. Agric. Food Chem. 2012;60:11372–11377. doi: 10.1021/jf303879b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Soroka D. N. Haider J. Ferri-Lagneau K. F. Leung T. Sang S. J. Agric. Food Chem. 2013;61:5353–5360. doi: 10.1021/jf401501s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer E. Heuschmid F. F. Kranz S. Metzler M. J. Agric. Food Chem. 2006;54:8769–8774. doi: 10.1021/jf062235l. [DOI] [PubMed] [Google Scholar]
- Zick S. M. Djuric Z. Ruffin M. T. Litzinger A. J. Normolle D. P. Alrawi S. Feng M. R. Brenner D. E. Cancer Epidemiol., Biomarkers Prev. 2008;17:1930–1936. doi: 10.1158/1055-9965.EPI-07-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Li C.-Y. Wen X.-D. Li P. Qi L.-W. J. Chromatogr. B. 2009;877:671–679. doi: 10.1016/j.jchromb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- Wu Z. Liu H. Wu B. J. Pharm. Pharmacol. 2015;67:583–596. doi: 10.1111/jphp.12351. [DOI] [PubMed] [Google Scholar]
- Xu J. Li M. Yao Z. Zhang Y. Li S. Hu L. Qin Z. Gonzalez F. J. Yao X. J. Pharm. Biomed. Anal. 2018;158:351–360. doi: 10.1016/j.jpba.2018.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Hong X. Yao Z. Dai Y. Zhao G. Qin Z. Wu B. Gonzalez F. J. Yao X. Xenobiotica. 2018;48:357–367. doi: 10.1080/00498254.2017.1323139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z. Li S. Qin Z. Hong X. Dai Y. Wu B. Ye W. Gonzalez F. J. Yao X. RSC Adv. 2017;7:52661–52671. [Google Scholar]
- Qin Z. Li S. Yao Z. Hong X. Xu J. Lin P. Zhao G. Gonzalez F. J. Yao X. J. Pharm. Biomed. Anal. 2018;155:157–168. doi: 10.1016/j.jpba.2018.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallifax D. Houston J. B. Drug Metab. Dispos. 2006;34:724–726. doi: 10.1124/dmd.105.007658. [DOI] [PubMed] [Google Scholar]
- Zhou J. Tracy T. S. Remmel R. P. Drug Metab. Dispos. 2010;38:431–440. doi: 10.1124/dmd.109.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.