Abstract

Materials with the ability to change properties can expand the capabilities of in vitro models of biological processes and diseases as it has become increasingly clear that static, stiff materials with smooth surfaces fall short in recapitulating the in vivo cellular microenvironment. Here, we introduce a patterned material that can be rapidly stiffened and softened in situ in response to an external magnetic field through the addition of magnetic inclusions into a soft silicone elastomer with topographic surface patterning. This substrate can be used for cell culture to investigate short-term cellular responses to dynamic stiffening or softening and the interaction with topography that encourages cells to assume a specific morphology. We investigated short-term cellular responses to dynamic stiffening or softening in human ventricular cardiac fibroblasts. Our results indicate that the combination of dynamic changes in stiffness with and without topographic cues induces different effects on the alignment and activation or deactivation of myofibroblasts. Cells cultured on patterned substrates exhibited a more aligned morphology than cells cultured on flat material; moreover, cell alignment was not dependent on substrate stiffness. On a patterned substrate, there was no significant change in the number of activated myofibroblasts when the material was temporally stiffened, but temporal softening caused a significant decrease in myofibroblast activation (50% to 38%), indicating a competing interaction of these characteristics on cell behavior. This material provides a unique in vitro platform to observe the time-dependent dynamics of cells by better mimicking more complex behaviors and realistic microenvironments for investigating biological processes, such as the development of fibrosis.
Introduction
Cardiac tissue, post-myocardial infarction (MI), exhibits a continuously changing local mechanical environment or remodeling of the extracellular matrix (ECM).1−3 During the healing process post-MI, there is an increase in the amount of collagen as a scar forms that alters the mechanical properties of the remodeling tissue.4,5 Additionally, early studies have suggested that the increased stiffness of infarcted tissue can be attributed not only to higher collagen density but also to the straightening of the collagen fibers during systolic contraction.6 In addition to the cell sensing ECM alignment, cardiac fibroblasts migrate and modulate matrix turnover in response to biochemical and biomechanical cues sent by cardiomyocytes during intense or prolonged stress,7 and it is therefore suspected that larger topographic changes such as cardiomyocyte enlargement, cell death, or other cardiac tissue modification will affect fibroblast response and activation during injury. The interplay of stiffness and anisotropy on the cellular level is complex; however, it has been shown that there is a fundamental relationship between the collagen fiber topography and the conversion of cardiac fibroblasts (CFs) into activated myofibroblasts, which in turn precipitates the development of cardiac fibrosis.1,8−10 CFs have the capability to sense the mechanical microenvironment and respond to changes in environmental stiffness, which has also been linked to driving the myofibroblast phenotype. Combining dynamic control of topography and stiffness in an in vitro model of post-MI cardiac tissue allows us to systematically examine how these tissue characteristics affect myofibroblast activation and how mechanical modulation at different activation stages could alter remodeling in unique ways.
Creating a model of post-MI cardiac tissue requires mimicking the spatial topographic properties and time-dependent mechanical properties of the infarcted myocardium. Micropatterned materials enable us to recapitulate various structural organizations in the infarcted heart and examine how the structural alignment cues affect differential cellular behavior in the remodeling process. Dynamic materials provide a window into understanding these complex biological changes by mimicking the continual environmental remodeling, which has driven the development of substrates with tunable stiffness through a variety of approaches, including applying light, changes in temperature, pH, or biomolecules.11−14 However, a key limitation of these approaches is that most provide a unidirectional and permanent change in material properties, limiting the ability to study time-dependent processes. Magnetically tunable materials are a highly innovative solution that overcome the shortcomings of conventional materials for studying biomechanical responses, allowing for rapid and reversible changes in mechanical properties. We have recently introduced magnetorheological elastomers (MREs) for cell culture experiments that allow for substrate stiffness to be temporally manipulated to mimic in vivo processes, thus revealing unique biomechanical responses in cardiac cells.15 This MRE platform provides conditions for a deeper mechanistic understanding of how cells are influenced by their dynamic environments and can guide strategies to control biological processes but is also expandable to consider other critical features of the microenvironment.
In this paper, we demonstrate a robust technique to create MREs of tunable stiffness integrated with defined micropatterns to explore interdependent effects of mechanical properties and topography on the morphology and phenotype of cardiac fibroblasts differentially. Topographic patterning and stiffness of culture substrates have become a mainstay in mechanobiology to control and study various cell phenotypes. Building on our previous work with dynamic culture substrates, we combined functionality to control both stiffness and patterning in an integrated platform to mimic post-myocardial infarction. As an alternative to using Sylgard 527, a polydimethylsiloxane (PDMS)-based elastomer, to fabricate MREs, we chose the commercially available Ecoflex Gel as the elastomer component because of its fast cure time; thus, it is less time-consuming to manufacture and provides greater feature fidelity. This platform allows for temporal investigation of dynamic mechanical alterations, whereby we can pre-condition cells on the same baseline before performing any changes in stiffness, better mimicking in vivo conditions of ECM pre- and post-MI. It has been established that the altered mechanical properties of the myocardium associated with cardiac diseases activate CFs.16 Here, we investigate how dynamically changing the stiffness experienced by cardiac fibroblasts on both un-patterned and micropatterned surfaces affects the activation of myofibroblasts through the expression of α-smooth muscle actin (α-SMA), which has been broadly used as one of many pro-fibrotic biomarkers after cardiac fibrosis, the expression of which characterizes the pro-fibrotic phenotype of myofibroblasts.17 Integrating topographic cues into dynamic materials can ultimately provide an insightful and unique approach to study mechanotransduction in cardiac function and disease.
Methods
Dynamic Micropatterned Substrate Fabrication
MREs were made from Ecoflex Gel (lot no. 2110161, Smooth-On, Inc.), a skin-safe product that has been widely used for various applications, such as prosthetics and orthotic devices, in order to accommodate the micropatterning procedure. To facilitate molding procedures to replicate micropatterns on the dynamic Ecoflex Gel MREs (or Sylgard MREs), which are super hydrophobic and sticky, an agarose molding method previously used in soft lithography was chosen in this study for ease of peel-off, mild preparation conditions, and high reproducibility.18 We follow the approach described by Mayer et al. and used a high-strength, 2 wt % agarose gel to ensure the stability of patterns during transfer as feature integrity is preserved,18 unlike softer gels of lower agarose concentration. Briefly, photolithography was used to prepare masters containing microchannels of SU8 photoresists with width of 40, 60, and 100 μm. Such dimensions are not only comparable to the diameter of collagen fiber bundles comprising the extracellular matrix diameter fibrosis but also the order of magnitude of cardiomyocytes and myocardial fibers. Sylgard 184 PDMS (GMID: 02065622, Corning Inc.) at a 10:1 base to curing agent ratio was replicated on the photolithographic masters. Then, 2 wt % hot agarose solution was prepared and casted onto the Sylgard 184 PDMS mold. The PDMS mold can be easily separated from the solidified agarose gel due to the hydrophilicity of agarose (Figure 1). Magnetorheological elastomers prepared with Ecoflex Gel (EcoGel MREs), 50 wt % carbonyl iron powder (lot no. C8M031, Chemical Store), and thinner (lot no. 2105485, Smooth-On, Inc.) were casted on microchannel featured agarose and cured at 60 °C for 20 min. The agarose gel was then separated from the polymerized EcoGel MREs. After placing the EcoGel MREs into a 35 mm dish, we backfilled 184 Sylgard PDMS (10:1, base to curing agent ratio) into the surrounding area. Features of the micropatterned materials were compared with patterns on MREs created with Sylgard 527 PDMS (GMID: 01696742, Corning Inc.), which we used in previous work,15,19,20 and were prepared utilizing the same hydrogel molding method as described above and cured at 60 °C for 24 h.
Figure 1.
Overview of the micropatterning of magnetorheological elastomers and function. (A) Schematics of the micropatterning procedure using the hydrogel molding method. (B) Composite consisting of the polymer matrix and iron particles that can change stiffness when a magnetic field is applied. (C) Representative images of channels patterned on the substrate and cross section of the channel profile.
Surface Topography and Roughness Characterization
To characterize the surface profile of our transferred features on our patterned MREs, we used white light interferometry (WLI, Veeco Wyko NT9100), a non-contact optical surface profile measurement to preserve the surface structure, which we utilized to characterize our opaque MRE surfaces in previous studies.15,20 A 10× objective lens was used to observe the surface structure. We measured the surface profile under both magnet and no magnet conditions. To measure the surface profile of micropatterned features under a magnetic field, we applied a disc axially magnetized N42 neodymium magnet (1.25″ diameter, 0.25″ thick, K&J Magnetics, Inc.) under the patterned MREs. The center line perpendicular to the channel direction in the scan image was chosen to analyze the z-direction height change profile in Vision software.
We also characterized the roughness of the EcoGel MRE surface using WLI. The mean surface roughness (Ra) of the EcoGel MRE surface with and without a magnetic field applied was calculated over a 50 μm × 50 μm region.
Mechanical Property Measurements
The shear and elastic moduli of the un-patterned EcoGel MREs were characterized both with and without an applied magnetic field. Rheological characterization was performed with a DRH 3 rheometer (TA Instruments) using a 25 mm circular parallel plate geometry to extract shear storage modulus (G′) and loss modulus (G″). The frequency dependence of the MRE properties was characterized with a frequency sweep from 20 to 0.02 Hz at 2% strain, whereas the strain dependence was characterized with a strain sweep from 2 to 22% at 1 rad/s.
The elastic modulus of the un-patterned EcoGel MREs was measured on a custom microindenter like that described by Rennie et al.21 and Schulze et al.22 For the indenting probe, a 2 mm diameter ruby sphere was attached to a calibrated titanium cantilever and indentation force and displacement were measured by a capacitance probe (Capacitec) and optical, linear encoder (Renishaw). Indentations were performed at 5 μm/s with target load at 3 mN to obtain force–displacement curves. Using the classical adhesive contact model by Johnson et al., the unloading curves were fit to extract the elastic modulus and work of adhesion of the MRE–probe contact pair.23
To obtain the elastic modulus of the patterned EcoGel MREs, we performed nanoindentation tests on the top and bottom of microchannels using a Bruker bioscope catalyst atomic force microscope (AFM) equipped with a Bruker SAA-SPH-1UM probe (0.236 N/m and 1 μm diameter spherical tip). A digital microscope (Dino-Lite) was mounted on top of the AFM header to visualize the probing location. Ramp mode was performed with a forward/reverse velocity of 1 μm/s and scan area of 5 μm × 5 μm. Elastic modulus was calculated from the retraction curve fitted by the Hertzian contact model using NanoScope Analysis software.
Cell Culture of Cardiac Fibroblasts
Normal human cardiac fibroblasts from ventricles (NHCF-V) (46 year-old male donor; Lonza, Walkersville, MD) were commercially obtained and cultured in fibroblast basal medium (FBM, catalog no. CC-3131, Lonza) supplemented with fibroblast growth media (FGM-3) SingleQuots (catalog no. CC-4525, Lonza), which comprised fetal bovine serum, rhFGF-B, recombinant human insulin 0.5%, and gentamicin/amphotericin-B. Cells were maintained in a humidified incubator at 37 °C with 5% CO2. Cells with passage number between 3 and 8 were used for all immunofluorescence assays. Prior to seeding cells on EcoGel MREs, the surface of the substrate was sterilized with 70% ethanol for 20 min followed by three times washing with 1× phosphate-buffered saline (PBS). After sterilization, EcoGel MREs were functionalized with fibronectin (10 μg/mL) overnight at 4 °C.
The viability of NHCF-V on EcoGel MREs was measured using a Live/Dead Viability/Cytotoxicity kit (lot no. 2369061, ThermoFisher Scientific). NHCF-V was cultured on EcoGel MREs at a seeding density of 200 cells/mm2 with FGM prepared above for 24 and 48 h. Tested samples were incubated with 0.5 μM of a acetomethoxy derivative of calcein (calcein AM, green; live) and 0.5 μM BOBO-3 iodide (red; dead) for 15 min in 1× PBS per product protocol. The cells were then rinsed twice with 1× PBS, and the samples were immediately imaged on a Zeiss Axio Imager upright microscope with a 5×/0.16 EC Plan-Neofluar objective. Images were used for counting and calculating the densities of cells in the fluorescein isothiocyanate (FITC, green; live) and the Texas Red (red; dead) channels. The ratio of integrated density in the FITC to Texas Red channel defined the cell viability. Ten different fields of view were scanned and assessed to obtain average cell viability.
Immunostaining of Cardiac Fibroblasts
To evaluate stiffness- and topography-dependent structural and phenotypic conversion of NHCF-V on micropatterned EcoGel MREs, we fixed the cells with 4% (v/v) paraformaldehyde (Sigma Aldrich) in 1× PBS for 10 min and permeabilized with 0.1% (v/v) Triton X-100 (Sigma Aldrich) and 1% (w/v) bovine serum albumin (Sigma Aldrich) in 1× PBS for 5 min. After fixation and permeabilization, cells were either stained for α-SMA or f-actin. For α-SMA, cells were incubated with the mouse anti-α-smooth muscle actin antibody (α-SMA, product no. A5228, Sigma-Aldrich, 1:500) at 4 °C fridge overnight followed by two washes with 1× PBS and incubated with goat-anti-mouse Alexa Fluor 488 (lot no. 2420714, Invitrogen, 1:500) at room temperature in the dark for 1 h. For f-actin, cells were incubated with phalloidin 488 (catalog no. A12379, Invitrogen, 1:200) for 10 min. The samples were then washed twice with 1× PBS and incubated with NucBlue (Hoetchst 33342, lot no. 2397762, ThermoFisher Scientific) for 10 min. The final samples were washed twice with 1× PBS and imaged on a Zeiss Axio Imager upright microscope with a 20×/0.5 N-Achroplan dip-in objective, with N ≥ 5 images in the static stiffness culture set and N ≥ 10 for the dynamic platform stiffness set.
Quantification of Cardiac Fibroblast Alignment
Cardiac fibroblast directional alignment was determined relative to the pattern direction. The bottom of microchannels of the material was identified under bright-field microscopy, and the direction of channels was aligned vertically when capturing both bright-field and fluorescence images. Phalloidin was used to stain the f-actin of cardiac fibroblast for analysis. The long axis of actin cytoskeleton was identified, and we defined alignment of individual cells by the angle between the long axis of actin filament and direction of the microchannel (vertically aligned in our study). The angle of alignment was calculated by using ImageJ software.
Quantification of Myofibroblast Activation
Myofibroblast activation was determined through manually identifying the number of activated cells, indicated by α-SMA stress fiber formation, and determining the percent activated from the total cell count, indicated by the nuclear stain (NucBlue). At least five regions of interest were imaged and calculated for each group.
Dynamic Experiment Design
To better understand the temporal changes in cardiac fibroblast cell phenotypes, we performed dynamic experiments through modulating stiffness for both un-patterned and patterned EcoGel MREs with cells pre-cultured on substrates for 48 h. In the matrix-stiffening experiment, cardiac fibroblasts were cultured on soft un-patterned and patterned EcoGel MREs for 48 h followed by in situ stiffening of the substrate by applying external magnets. Cells were fixed and stained after 1 and 3 h on the stiffened substrate to quantify activation. For the matrix-softening experiment, cells were cultured on stiff un-patterned and patterned EcoGel MREs with magnets applied for 48 h followed by in situ softening of the substrate by removing magnets. Cells were fixed and stained after 1 and 3 h on the softened substrate to quantify activation.
Statistical Analysis
All images and data are representative of the results of three independent experiments. Student’s t test was performed to compare the pattern resolution of different devices as well as alignment of the un-patterned substrates. Two-way ANOVAs were used to determine effects of pattern width and magnet conditions on both cell activation and alignment with Tukey’s test used for post hoc pairwise comparison. Significance was determined at p < 0.05. Error is reported in figures as standard deviation of the mean unless otherwise noted.
Results and Discussion
Surface Characterization of Micropatterned EcoGel MREs
To ensure proper transference of topographic features onto our MRE substrates, we fabricated 60 μm-tall microchannels with varying widths of 40, 60, and 100 μm into our MRE devices. The surface topography profile of both Sylgard and EcoGel MREs as well as the master mold was confirmed with white light interferometry. Figure 2 effectively reveals the importance of material selection where we compare the 100 μm microchannels of EcoGel MREs to Sylgard MREs, whereby we observed that the patterned features were far less pronounced with Sylgard 527. By applying a magnet to the micropatterned EcoGel MREs, we detected height and surface roughness changes of the features; specifically, both the depth of microchannels (Figure 2) and surface roughness increased (Figure S1A). These changes in surface topography are associated with increases in magnetic field and stiffness, which are caused by motion of the particles within the matrix with an overall very small effect on elastomer volume.24−26 The microchannel depth of EcoGel MREs with a magnet applied was increased approximately from 60 to 80 μm. This can be explained by the increase in the magnetic force on the bottom of channels caused by the closer proximity to the magnet as opposed to the top of the channels, leading to displacement differences between the top and bottom of the channels. This variation in displacement, with a smaller displacement at the top of the channels compared to the bottom of the channel, gives rise to the channel depth increase. In contrast, lower fidelity of the patterned microchannels on Sylgard MREs results in similar magnetic attraction force between top and bottom surfaces of the channel, thus a less displacement difference and less pronounced height increase. The feature profile change is induced by the application of a magnetic field difference between the top and bottom microchannels driving iron particle motion in the materials,19 which also resulted in the surface roughness increase due to internal deformation of the soft silicone microstructure.20
Figure 2.
Surface characterization by white light interferometry. Surface topography and profiles under both no magnet and magnet conditions for (A) EcoGel MREs and (B) Sylgard 527 MREs.
We quantified the fidelity of pattern features on both EcoGel MREs and Sylgard 527 MREs by analyzing how well they compared to our master mold reference (Figure S2), with resolution defined as the standard deviation of the residuals. We found that there was a significant difference in resolution between the materials with a z resolution of 7.3 ± 0.69 μm for EcoGel as compared to 14.6 ± 1.4 μm for Sylgard 527 (p < 0.0001) (Figure S2C). This difference in resolution can be attributed to an increase in deformation in the agarose mold with Sylgard 527 compared to EcoGel (Figure S3A–C), caused by increased precipitation of iron particles with longer polymerization time required for Sylgard 527 MREs (24 h) compared to EcoGel MREs (20 min). Additionally, the microfeatures on agarose gels are extremely sensitive to hydration at high temperature; the longer the microchannels agarose gels were kept in the oven, the more dehydrated and deformed they became, and resulted in less pronounced features. Therefore, it is necessary to prepare the agarose shortly before usage to preserve and replicate features from agarose gels. We also note that the short curing time for EcoGel MREs did not hit the threshold time for agarose to significantly dry out and shrink;27 hence, the desired features were well transferred. In general, the novel EcoGel MRE materials show higher fidelity of microchannel replication and are more cost effective and less time-consuming to manufacture compared to patterned MREs created with Sylgard 527.
Micropatterned EcoGel MREs Allow for Stiffness Tunability
The EcoGel MRE substrates reproducibly stiffen in response to a magnetic field (Figure 3). The stiffness of the material was confirmed using rheology, microindentation, and nanoindentation with and without an applied magnet. Figure 3A shows the global and local characterizations of mechanical properties of EcoGel MREs. Figure 3B shows both shear storage and loss moduli of un-patterned EcoGel MREs as a function of angular frequency and oscillation strain. The shear storage modulus, G′, ranged from 2.28 kPa with no magnet up to over 20 kPa with the magnet and no spacer at 1 rad/s, whereas the loss modulus, G″, ranged between 0.3 and 10 kPa under the same conditions. There was a minimal dependence of G′ on frequency while G″ increased with angular frequency. G″ of EcoGel MREs with a magnet doubled from 11.28 to 23.40 kPa with the frequency increasing from 0 to 20 rad/s, while G″ without a magnet increased from 0.12 to 1.59 kPa over the same frequency range. For the oscillation strain sweep measurements from 0.2% to 20%, both G′ and G″ have very low dependence on the strain for the no magnet condition and were approximately 2.5 and 0.35 kPa, respectively. However, in the magnet condition, both G′ and G″ decreased with increasing strain (G′: 98.35 to 21.63 kPa; G″ 25.30 to 9.34 kPa), revealing a strain-softening behavior. This indicates that at higher magnetic fields, PDMS-based MREs soften as the strain increases,27 which could potentially be due to the realignment of incorporated iron particles in EcoGel MREs during shear straining. Overall, the EcoGel MREs exhibited the in situ modulation of stiffness by simply applying external magnets.
Figure 3.
Mechanical characterization of the viscoelastic EcoGel MREs to magnetic fields. (A) Schematic overview of all mechanical characterizations performed on un-patterned and patterned substrates: (i) shear rheology on un-patterned surfaces, (ii) microindentation on un-patterned surfaces using a 2 mm ruby sphere tip, and (iii) nanoindentation on micropatterned EcoGel MREs using AFM equipped with a 1 μm tip. (B) Shear storage, G′, and loss moduli, G″, of frequency and strain sweeps for cylindrical EcoGel MREs under no magnet and magnet conditions at 2% strain and 1 rad/s, respectively. (C) Elastic modulus of un-patterned substrate calculated from the microindenter under no magnet and magnet conditions (p = 0.00331). (D) Elastic modulus of the micropatterned substrate calculated from AFM on the top and bottom features for both no magnet (p = 0.44179) and magnet conditions (p = 0.03035). (NM, no magnet; M, magnet).
We also verified the elastic moduli of the un-patterned substrate, where EcoGel MREs without an applied magnet exhibited an elastic modulus of 8.14 kPa, which represents the healthy myocardium (Figure 3C).28 The elastic modulus significantly increased to 82.49 kPa with an applied magnet (p = 0.003), which mimics the pathological stiffness of infarcted myocardial tissue.29 We further measured the local stiffness of microfeatures of the patterned EcoGel MREs. Using AFM, we indented the top and bottom of channels on EcoGel MREs with a micro-sized sphere tip under both no magnet and magnet conditions (Figure 3D). Under the no magnet condition, the elastic moduli of the top and bottom of microchannels were 6.75 ± 7.20 and 4.81 ± 2.49 kPa, respectively, and were not significantly different (p = 0.44). As expected, the elastic modulus increased to approximately 144.17 ± 62.81 and 175.81 ± 83.88 kPa on the top and bottom of channels when a disc magnet applied, where the bottom of the microchannel was significantly stiffer than the top of the channel (p = 0.03). This is caused by the small difference in distance of these two locations to the applied magnet resulting in a higher local magnetic field strength at the bottom of the channel and therefore higher stiffness. Notably, the large variability in the measured elastic modulus on the bottom of the channel might result from the increased roughness on the bottom as presented above, which consequently will increase the contact area of the probe during measurements and result in measurement uncertainty.
Alignment of Cardiac Fibroblasts on Varying Topography and Substrate Stiffnesses
To study the collective alignment response of primary cardiac fibroblasts to topographic and stiffness changes, we cultured the NHCF-V on both un-patterned and patterned substrates under static conditions (no magnet and magnet), with cell alignment analyzed after 48 h in culture. The cells on the patterned substrates became more elongated and oriented in parallel with the aligned patterns as compared to that on the un-patterned substrates (Figure 4A). Rose plots reflect the distribution of cell alignments relative to a given direction, where the 0° orientation represents the direction of the microchannels. The symmetry of the micropatterned device allowed us to consider the cell direction within a 0–90° range. Visual inspection of the soft, no magnet condition of un-patterned substrates suggests a wide distribution of angles between 0 and 90°, indicating no particular collective alignment. However, alignment was observed on stiff, un-patterned substrates, which agrees with previous findings that stiffer substrates drove the actin cytoskeleton into polarized and aligned morphology.30 Cells on patterned substrates exhibit a distribution of alignments around an orientation of 0°, with an increasing preference toward 0° on the patterned substrates as the features increase in width (Figure 4B).
Figure 4.
Morphology alteration of NHCF-V on the EcoGel MRE substrate featured with microchannels. (A) Representative images of cardiac fibroblasts on un-patterned and patterned substrates. Cells were stained for phalloidin (green) and nuclei (blue). (B) Quantification of actin cytoskeleton alignment for cells on the un-patterned and patterned substrate under no magnet and magnet conditions.
Overall, the patterned feature width (the channel dimension) influenced cell alignment whereas substrate stiffness had no influence on directional alignment. Specifically, two-way ANOVA (n = 5) revealed that alignment on larger features is significantly higher (p = 0.005) but was not dependent on substrate stiffness (p = 0.124). Tukey’s post hoc test revealed a significant difference in alignment between the 40 μm channels and the 100 μm channels (p = 0.004). While controlling the alignment of cells is critical for any engineered tissue, topographic cues play a particularly critical role in wound healing where the cells align parallel to microstructure direction.31,32 The recent literature has indicated that the viscous component of a substrate influences cell responses33 and it is possible that not only the stiffness is affecting the cell behavior but that viscosity also plays a role in the results of this study.
Interplay of Anisotropic Topography and Matrix Stiffness Regulates Cardiac Fibroblast Activation
Given that cardiac fibroblasts respond to topography-dependent alterations,1,31 we postulated that cardiac fibroblasts behave differentially on the stiffness-modulated topographic substrates. NHCF-V were seeded on both un-patterned and micropatterned EcoGel MREs of various dimensions either with or without magnets applied for 48 h. Student’s t test showed that under static conditions on the un-patterned substrates for 48 h, the myofibroblast activation significantly increased on the stiff (magnet) condition (n ≥ 5, p < 0.0002), similar to previous studies.34 Two-way ANOVA (n ≥ 5) revealed that under static conditions on micropatterned substrates for 48 h, the percentage of cells with α-SMA stress fibers significantly depended on feature width and substrate stiffness (Figure 5), such that larger features and stiffer substrates with the magnet added both result in an increase in α-SMA positive cells (each p < 0.0001). Finally, we determined that there is a significant pattern × stiffness interaction effect on activation (p = 0.048), indicating that on the largest patterns, the effect of substrate stiffness on activation was further augmented relative to the smaller patterns. It is worth noting from surface profiles by WLI characterization that the height of the microchannels increased in response to the magnetic field and might also play a role in cell activation. Using 100 μm channels (our most responsive width feature), we varied height and stiffness and observed an increase of cardiac fibroblast activation on patterned substrates with taller features compared to shorter features (Figure S5). Combined with the results of microchannel-width dependent-activation, larger geometric sizes of microchannels probably lead to much higher activation of studied human cardiac fibroblasts, and the change of size with applied magnet could influence this effect. One question for future studies is whether there is a specific geometric size of the microchannel for human cardiac fibroblasts to probe and maximize their activation. In this study, we saw the greatest activation effects on the 100 μm-width patterned EcoGel MREs, and as such we will focus this next set of dynamic experiments on those features.
Figure 5.

Percentage of myofibroblast activation on un-patterned and patterned substrates with microchannel widths of 40, 60, and 100 μm under no magnet and magnet conditions for 48 h cell culturing. Significant difference is indicated by # and † compared to 60 and 100 μm, respectively, within the group.
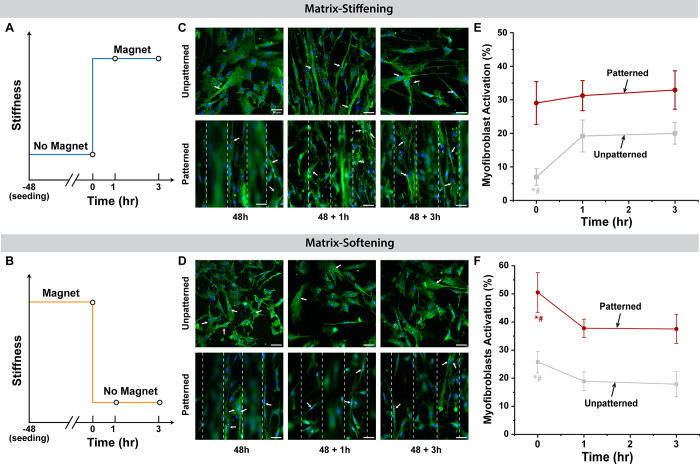
To highlight the dynamic ability of our device, we can mimic the development of acute MI and post-infarction remodeling processes by modulating the stiffness of the microenvironment temporally: matrix stiffening and softening are achieved by means of applying and removing magnets, respectively, to mimic physiological or pathological states. After the addition or removal of the magnets, we observed phenotypic changes in myofibroblast activation at 1 and 3 h (Figure 6A,B), which agrees with our previous work.15 On un-patterned substrates, the percent myofibroblast activation under the matrix stiffening condition increased significantly from 6.9% to 19.1% (p = 0.002) and remained unchanged from 1 to 3 h. Interestingly, matrix stiffening did not significantly upregulate cell activation on the 100 μm patterned substrate after 1 h (p = 0.589) (Figure 6E), which demonstrates that acutely, the pre-conditioned substrate with anisotropic microchannels tended to maintain high myofibroblast activation regardless of matrix stiffness. In Figure 6F, matrix softening downregulated the cell activation on both un-patterned and patterned samples and indicated an apparent plateau after 1 h; however, matrix softening on patterned substrates significantly decreased activation from 50.5% to 37.8% (p < 0.0006), in contrast to the lack of change in activation when the matrix stiffened. We again observed that the percentage of α-SMA-positive cells from the matrix softening condition was significantly higher on the patterned substrate than on the un-patterned substrate.
Figure 6.
Temporal test of matrix-stiffening and matrix-softening effect on NHCF-V phenotype conversion on the un-patterned/patterned substrate. (A, B) Schematic illustration of temporal tests for matrix-stiffening and matrix-softening conditions. (C) Representative fluorescence images of cardiac fibroblast activation under dynamic temporal stiffening at 48 h prior to stiffening followed by 1 and 3 h post stiffening. (D) Representative fluorescence images of cardiac fibroblast activation under dynamic temporal softening at 48 h prior to softening followed by 1 and 3 h post-softening. Immunofluorescence staining for α-SMA (green) and nucleus (blue). White arrows indicate cells with polymerized stress fiber identified as activated myofibroblasts. Scale bars, 50 μm. (E, F) Percentage of myofibroblasts activation under matrix-stiffening and matrix-softening conditions, respectively. Significant difference is indicated by * and # compared with 1 and 3 h time points, respectively.
Noticeably, the activated NHCF-V on both un-patterned and patterned substrates initially seeded under stiff conditions were partially deactivated when the substrate was softened (Figure 6F). However, it should be noted that compared to the soft control, the partial deactivation was still greater, suggesting that these cells exhibit an apparent mechanical memory, meaning that the mechanical conditions prior to the matrix softening event influences the retention of the pre-conditioned phenotype. While this 3 h time course is short, we have previously shown that cardiac fibroblasts can reversibly deactivate within that time course after a prolonged pre-seeding.15 We also recognize that α-SMA expression only partially describes fibroblast activation and inclusion of other markers could more fully characterize the activation response on these substrates.35 Collectively, our results indicate that myofibroblast activation is highly affected by topographic cues along with the mechanical cues in our study and that memory-dependent behavior of NHCF-V limits the complete deactivation during the post-infarction remodeling process.
Conclusions
Micropatterned and stiff substrates have been used as in vitro platforms individually and separately to study effects on cellular morphology and phenotype changes. However, few studies considered dynamic substrates with tunable stiffness, which can be modulated in situ. Our novel substrate further incorporates micropatterns onto the stiffness-tunable substrate in which mechanical cues and topographic cues can be integrated and dynamically controlled for the first time to study the time-dependent interplay of these effects on cellular responses. This substrate enabled us to better recapitulate mechanical and topographic characteristics of healthy and infarcted myocardium tissue as well as the dynamics of infarction development and remodeling processes. The microstructural configurations interplayed with reversible mechanical property modulations pose an intriguing finding on temporal control of cells in an acute manner. While this platform is designed to add mechanical microenvironmental features not previously investigated with in vitro models together, this is a step toward a more representative microenvironment and sheds light on possibilities that incorporate more complex components into one platform. In the future, this biomimetic platform is expected to be broadened to the investigation of various cell types and interactions with their environment for potential use to develop therapeutic strategies.
Acknowledgments
This research was supported in part by the Delaware INBRE program (P20 GM103446), the Institutional Development Award (U54 GM104941), and the Delaware Center for Musculoskeletal Research COBRE (P20 GM139760), with grants from the National Institute of General Medical Science from the National Institutes of Health and the State of Delaware.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c06529.
EcoGel MREs contents: Ecoflex Gel, silicone thinner, carbonyl iron powder; Sylgard 527 MREs contents: Sylgard 527 silicone, carbonyl iron powder (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Bugg D.; Bretherton R.; Kim P.; Olszewski E.; Nagle A.; Schumacher A. E.; Chu N.; Gunaje J.; DeForest C. A.; Stevens K.; Kim D.-H.; Davis J. Infarct Collagen Topography Regulates Fibroblast Fate via P38-Yes-Associated Protein Transcriptional Enhanced Associate Domain Signals. Circ. Res. 2020, 127, 1306–1322. 10.1161/CIRCRESAHA.119.316162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano L. R.; Lee J.-J.; Holmes J. W. Surgical Reinforcement Alters Collagen Alignment and Turnover in Healing Myocardial Infarcts. Am. J. Physiol.: Heart Circ. Physiol. 2018, 315, H1041. 10.1152/AJPHEART.00088.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman P.; Spack Daniel E.; Shumer N. J. N. Physiological Implications of Myocardial Scar Structure. Physiol. Behav. 2017, 176, 139–148. 10.1002/cphy.c140067.Physiological.28363838 [DOI] [Google Scholar]
- McCormick R. J.; Musch T. I.; Bergman B. C.; Thomas D. P. Regional Differences in LV Collagen Accumulation and Mature Cross-Linking after Myocardial Infarction in Rats. Am. J. Physiol.: Heart Circ. Physiol. 1994, 266, H354. 10.1152/AJPHEART.1994.266.1.H354. [DOI] [PubMed] [Google Scholar]
- Capasso J. M.; Robinson T. F.; Anversa P. Alterations in Collagen Cross-Linking Impair Myocardial Contractility in the Mouse Heart. Circ. Res. 1989, 65, 1657–1664. 10.1161/01.RES.65.6.1657. [DOI] [PubMed] [Google Scholar]
- Omens J. H.; Miller T. R.; Covell J. W. Relationship between Passive Tissue Strain and Collagen Uncoiling during Healing of Infarcted Myocardium. Cardiovasc. Res. 1997, 33, 351–358. 10.1016/S0008-6363(96)00206-4/2/33-2-351-FIG7.GIF. [DOI] [PubMed] [Google Scholar]
- Rogers J. D.; Holmes J. W.; Saucerman J. J.; Richardson W. J. Mechano-Chemo Signaling Interactions Modulate Matrix Production by Cardiac Fibroblasts. Matrix Biol. Plus 2021, 10, 100055 10.1016/j.mbplus.2020.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W. J.; Clarke S. A.; Alexander Quinn T.; Holmes J. W. Physiological Implications of Myocardial Scar Structure. Compr. Physiol. 2015, 5, 1877–1909. 10.1002/cphy.c140067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goergen C. J.; Chen H. H.; Sakadžić S.; Srinivasan V. J.; Sosnovik D. E. Microstructural Characterization of Myocardial Infarction with Optical Coherence Tractography and Two-photon Microscopy. Physiol. Rep. 2016, 4, 12894. 10.14814/phy2.12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W. J.; Holmes J. W. Emergence of Collagen Orientation Heterogeneity in Healing Infarcts and an Agent-Based Model. Biophys. J. 2016, 110, 2266–2277. 10.1016/j.bpj.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester C. B.; Pugazenthi A.; Grande-Allen K. J.; Ghanta R. K.; Debakey M. E. Cell-Laden Bioactive Poly(Ethylene Glycol) Hydrogels for Studying Mesenchymal Stem Cell Behavior in Myocardial Infarct-Stiffness Microenvironments. Cardiovasc. Eng. Technol. 2021, 12, 183. 10.1007/s13239-020-00515-6. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Wood N. A.; Fok K.; Yoshizumi T.; Park D. W.; Jiang H.; Schwartzman D. S.; Zenati M. A.; Uchibori T.; Wagner W. R.; Riviere C. N. Design of a Coupled Thermoresponsive Hydrogel and Robotic System for Postinfarct Biomaterial Injection Therapy. Ann. Thorac. Surg. 2016, 102, 780. 10.1016/J.ATHORACSUR.2016.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbern J. C.; Minami E.; Stayton P. S.; Murry C. E. Delivery of Basic Fibroblast Growth Factor with a PH-Responsive, Injectable Hydrogel to Improve Angiogenesis in Infarcted Myocardium. Biomaterials 2011, 32, 2407–2416. 10.1016/J.BIOMATERIALS.2010.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.; Liu W. Functional Hydrogels for the Treatment of Myocardial Infarction. NPG Asia Mater. 2022, 14, 1–15. 10.1038/s41427-021-00330-y. [DOI] [Google Scholar]
- Corbin E. A.; Vite A.; Peyster E. G.; Bhoopalam M.; Brandimarto J.; Wang X.; Bennett A. I.; Clark A. T.; Cheng X.; Turner K. T.; Musunuru K.; Margulies K. B. Tunable and Reversible Substrate Stiffness Reveals a Dynamic Mechanosensitivity of Cardiomyocytes. ACS Appl. Mater. Interfaces 2019, 11, 20603–20614. 10.1021/acsami.9b02446. [DOI] [PubMed] [Google Scholar]
- Van Putten S.; Shafieyan Y.; Hinz B. Mechanical Control of Cardiac Myofibroblasts. J. Mol. Cell. Cardiol. 2016, 10.1016/j.yjmcc.2015.11.025. [DOI] [PubMed] [Google Scholar]
- Herum K. M.; Choppe J.; Kumar A.; Engler A. J.; McCulloch A. D. Mechanical Regulation of Cardiac Fibroblast Profibrotic Phenotypes. Mol. Biol. Cell 2017, 28, 1871–1882. 10.1091/mbc.E17-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.; Yang J.; Gitlin I.; Gracias D. H.; Whitesides G. M. Micropatterned Agarose Gels for Stamping Arrays of Proteins and Gradients of Proteins. Proteomics 2004, 4, 2366–2376. 10.1002/pmic.200300748. [DOI] [PubMed] [Google Scholar]
- Clark A. T.; Marchfield D.; Cao Z.; Dang T.; Tang N.; Gilbert D.; Corbin E. A.; Buchanan K. S.; Cheng X. M. The Effect of Polymer Stiffness on Magnetization Reversal of Magnetorheological Elastomers. APL Mater. 2022, 10, 041106 10.1063/5.0086761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. T.; Bennett A.; Kraus E.; Pogoda K.; Ce̅bers A.; Janmey P.; Turner K.; Corbin E. A.; Cheng X. Magnetic Field Tuning of Mechanical Properties of Ultrasoft PDMS- Based Magnetorheological Elastomers for Biological Applications. Multifunct. Mater. 2021, 035001 10.1088/2399-7532/ac1b7e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie A. C.; Dickrell P. L.; Sawyer W. G. Friction Coefficient of Soft Contact Lenses: Measurements and Modeling. Tribol. Lett. 2005, 18, 499. 10.1007/s11249-005-3610-0. [DOI] [Google Scholar]
- Schulze K. D.; Bennett A. I.; Marshall S.; Rowe K. G.; Dunn A. C. Real Area of Contact in a Soft Transparent Interface by Particle Exclusion Microscopy. J. Tribol. 2016, 138, 041404 10.1115/1.4032822. [DOI] [Google Scholar]
- Johnson K. L.; Kendall K.; Roberts A. D. Surface Energy and the Contact of Elastic Solids. Proc. R. Soc. Lond. A. 1971, 324, 301–313. 10.1098/rspa.1971.0141. [DOI] [Google Scholar]
- Yao J.; Sun Y.; Wang Y.; Fu Q.; Xiong Z.; Liu Y. Magnet-Induced Aligning Magnetorheological Elastomer Based on Ultra-Soft Matrix. Compos. Sci. Technol. 2018, 162, 170–179. 10.1016/j.compscitech.2018.04.036. [DOI] [Google Scholar]
- Pessot G.; Schümann M.; Gundermann T.; Odenbach S.; Löwen H.; Menzel A. M. Tunable Dynamic Moduli of Magnetic Elastomers: From Characterization by x-Ray Micro-Computed Tomography to Mesoscopic Modeling. J. Phys. Condens. Matter 2018, 30, 125101. 10.1088/1361-648X/aaaeaa. [DOI] [PubMed] [Google Scholar]
- Chen S.; Li R.; Li X.; Wang X. Magnetic Field Induced Surface Micro-Deformation of Magnetorheological Elastomers for Roughness Control. Front. Mater. 2018, 5, 1–8. 10.3389/fmats.2018.00076. [DOI] [Google Scholar]
- Kurniawan N. A.; Wong L. H.; Rajagopalan R. Early Stiffening and Softening of Collagen: Interplay of Deformation Mechanisms in Biopolymer Networks. Biomacromolecules 2012, 13, 691–698. 10.1021/BM2015812/SUPPL_FILE/BM2015812_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- Gupta K. B.; Ratcliffe M. B.; Fallert M. A.; Henry Edmunds L.; Bogen D. K.. Changes in Passive Mechanical Stiffness of Myocardial Tissue With Aneurysm Formation. 1994, 2315. [DOI] [PubMed]
- Berry M. F.; Engler A. J.; Woo Y. J.; Pirolli T. J.; Bish L. T.; Jayasankar V.; Morine K. J.; Gardner T. J.; Discher D. E.; Sweeney H. L. Mesenchymal Stem Cell Injection after Myocardial Infarction Improves Myocardial Compliance. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H2196. 10.1152/AJPHEART.01017.2005. [DOI] [PubMed] [Google Scholar]
- Doss B. L.; Pan M.; Gupta M.; Grenci G.; Mège R. M.; Lim C. T.; Sheetz M. P.; Voituriez R.; Ladoux B. Cell Response to Substrate Rigidity Is Regulated by Active and Passive Cytoskeletal Stress. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 12817–12825. 10.1073/pnas.1917555117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. N.; Hong Y.; Kim M. S.; Kim S. M.; Suh K. Y. Effect of Orientation and Density of Nanotopography in Dermal Wound Healing. Biomaterials 2012, 33, 8782–8792. 10.1016/J.BIOMATERIALS.2012.08.038. [DOI] [PubMed] [Google Scholar]
- Kim D. H.; Lipke E. A.; Kim P.; Cheong R.; Thompson S.; Delannoy M.; Suh K. Y.; Tung L.; Levchenko A. Nanoscale Cues Regulate the Structure and Function of Macroscopic Cardiac Tissue Constructs. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 565. 10.1073/PNAS.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier E. E.; Pogoda K.; Wells R. G.; Janmey P. A. Control of Cell Morphology and Differentiation by Substrates with Independently Tunable Elasticity and Viscous Dissipation. Nat. Commun. 2018, 9, 1–13. 10.1038/s41467-018-02906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y. C.; Corbin E. A.; Caliari S. R.; Ouyang L.; Vega S. L.; Truitt R.; Han L.; Margulies K. B.; Burdick J. A. Mechanically Dynamic Pdms Substrates to Investigate Changing Cell Environments. Biomaterials 2017, 145, 23–32. 10.1016/j.biomaterials.2017.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist M. D.; Molkentin J. D. Redefining the Identity of Cardiac Fibroblasts. Nat. Rev. Cardiol. 2017, 14, 484–491. 10.1038/nrcardio.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.