Abstract
Current clinical tools for breast cancer (BC) diagnosis are insufficient but liquid biopsy of different bodily fluids has recently emerged as a minimally invasive strategy that provides a real-time snapshot of tumour biomarkers for early diagnosis, active surveillance of progression, and post-treatment recurrence. Extracellular vesicles (EVs) are nano-sized membranous structures 50–1000 nm in diameter that are released by cells into biological fluids. EVs contain proteins, nucleic acids, and lipids which play pivotal roles in tumourigenesis and metastasis through cell-to-cell communication. Proteins and miRNAs from small EVs (sEV), which range in size from 50–150 nm, are being investigated as a potential source for novel BC biomarkers using mass spectrometry-based proteomics and next-generation sequencing. This review covers recent developments in sEV isolation and single sEV analysis technologies and summarises the sEV protein and miRNA biomarkers identified for BC diagnosis, prognosis, and chemoresistance. The limitations of current sEV biomarker research are discussed along with future perspective applications.
Keywords: Breast cancer, Extracellular vesicles, Liquid biopsy, Biomarker, Diagnosis, Prognosis
Introduction
Breast cancer (BC) is a global public health concern accounting for nearly 30% of female cancers [1]. In 2021, about 284,200 females were newly diagnosed with BC in the US resulting in 43,600 deaths [2, 3]. Early diagnosis of BC is essential for selection of effective treatments and reducing the possibility of cancer metastasis [4]. BC is a heterogenous and dynamic disease with unique somatic mutations accompanied by changes in gene and protein expression. It is classified into distinct subtypes based on estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) expression. Individual biomarker profiles influence tumour recurrence, drug resistance, and mortality and require different therapeutic approaches [5, 6]. Due to the complexity of BC disease, identification of additional clinical biomarkers is necessary to further stratify patients and provide more information on initial diagnosis as well as the monitoring cancer progression, metastasis, and relapse [7].
Currently, the most common tool for detection, staging, and prognosis of cancer is tissue biopsy. However, a tissue biopsy is difficult to obtain, and tumour molecular and genetic information from the biopsy provides limited information for early detection, screening, and monitoring. Mammography is the only clinically proven imaging method for the early diagnosis of BC, with high potential for false-negative diagnosis and low sensitivity for dense breast tissue [8, 9]. Small lesions are frequently missed and may not be visible by mammograms [10, 11]. Therefore, a significant proportion of detected tumours in women undergoing regular screening have already disseminated, and therefore these patients present with invaded lymph nodes or general metastases at the time of diagnosis. Furthermore, one- to two-year regular mammogram screening does not allow the detection of high growth rate tumours. Although the survival of patients has increased over the last few decades due to screening programs and postoperative adjuvant systemic therapies (i.e., hormone therapy and chemotherapy), many patients die from metastatic relapse [12].
Serum levels of cancer antigen 15–3 (CA 15–3) is the biomarker currently used for BC monitoring. However, CA 15–3 measurements are not helpful in diagnosis, especially in patients with early-stage cancers, and not useful in therapeutic decision-making of patients with BC [13]. Traditional prognostic markers (age at diagnosis, tumour size, hormonal receptor status, tumour grade) are not sufficient for precise risk group discrimination in BC. Therefore, there is an unmet need for discovery of accurate and minimally invasive biomarkers for early detection, prognosis, prediction, monitoring of therapy response, and anticipation of drug resistance in BC patients.
To overcome the challenges of techniques that have traditionally been utilised for identifying and validating clinical biomarkers, analyses based on liquid biopsies have been proposed which focus on circulating tumour cells (CTCs), circulating tumour DNA (ctDNA), tumour-educated platelets, and extracellular vesicles (EVs) in bodily fluids including blood, urine, and saliva [14–17]. Liquid biopsies provide several advantages over collection of tissue biopsies: it is less invasive; it is possible to trace the heterogeneity [18], metastasis, and progression of cancer in a real time [19]; and a patient’s response to treatment can be constantly monitored [20]. Pain and side effects are significantly lower than after collection of tissue biopsies [21, 22].
According to the International Society of Extracellular Vesicles (ISEV), the term “extracellular vesicles” is the appropriate terminology for the heterogeneous populations of vesicles isolated from cell culture supernatants or physiological fluids [23]. The EV populations are defined by their size and biogenesis: exosomes are small-sized EVs (50–150 nm) generated from the fusion of plasma membrane and multivesicular bodies; large-sized microvesicles (large EVs) (100–1000 nm) are formed directly from plasma membrane; and apoptotic bodies (500–5000 nm) are the largest EVs created during programmed cell death [23]. However, there are different classification systems and the groups are not necessarily mutually exclusive. For example, in the ISEV 2018 guideline EV subtypes are classified by physical characteristics such as EV size (small EVs < 200 nm, large EVs > 200 nm), density, and biochemical composition (e.g., CD63, CD81, Annexin A5) [24]. Large EVs (lEVs) have a density 1.10–1.15 g/mL and Cav-1, CK18, and GAPDH expression, while small EVs (sEVs) are characterised by CD9, CD63, CD81, TSG101, Alix, Flotilin-1, and heat-shock proteins (HSPs) and have a density range of 1.05–1.2 g/mL [25]. Based on this classification, this review will focus on sEVs and references that refer to exosomes will fall under this group.
EVs are present in all biofluids including blood [26, 27], urine [28], breast milk, saliva [29], ascites [30], and cerebrospinal fluid [31]. In multicellular organisms, sEVs play a significant role in transmitting biological information from cell to cell and within the tumour microenvironment through various RNAs, proteins, and lipids [32–34]. Several studies have reported that cancer cells release more sEVs than normal cells [35, 36] and their biological cargoes reflect the cell of origin and therefore serve as superior biomarker candidates for disease diagnosis, prognosis, and surveillance [27, 37–39]. Furthermore, genomic and proteomic profiling studies have shown that sEV contents and number are different between BC patients and healthy controls, suggesting they may be a unique source of tumour information [40, 41].
In this review, new techniques that have been developed for sEV isolation and single sEV analysis are summarised. sEVs contain numerous types of biomolecules but the focus here will be on protein and miRNA biomarkers for BC diagnosis and prognosis. The limitations of sEV-based biomarker research will be discussed along with future perspectives of sEVs as a source of novel clinical biomarkers.
Advantages of sEVs as a biomarker source in BC liquid biopsy
CTCs and ctDNA within liquid biopsies have been studied as diagnostic and prognostic biomarker candidates in various cancers [42–46]. However, the clinical utility of CTCs and ctDNA is facing several critical challenges that need to be resolved. Firstly, CTCs have a short life span (1–2.4 h) [47] and the process of releasing CTCs into the bloodstream is still unclear [48]. The low concentration, dynamically heterogeneous form, and lack of sensitive detection methods of CTCs in bodily fluids make their isolation and characterisation difficult [49, 50]. Analysis of ctDNA is challenging because of low concentration and short half-life, requiring elaborate detection and isolation methods [51].
Currently, many researchers in the liquid biopsy area have turned their focus on sEVs because of their relatively greater abundance in biofluids, rich biological content from parental cells, stable form [52], and ability to be stored at -80℃ for a long periods of time without significant change in protein contents and morphology [53]. For example, Kirsten rat sarcoma viral oncogene homolog gene (KRAS) and epidermal growth factor receptor (EGFR) DNA mutations detected in plasma sEVs were reported to be sensitive tumour prognostic indicators for pancreatic cancer [54], early-stage non-small-cell lung cancer [55], colorectal cancer, and melanoma [56]. In addition to genetic mutations, the RNAs and proteins from tumour-derived sEVs were shown to have diagnostic value in various cancers such as breast, ovarian, and prostate [38, 57, 58]. For clinical use, the first commercial sEV-based test, ExoDx™ Prostate (IntelliScore), was launched in 2017 and approved by the US Food and Drug Administration (FDA) and uses urinary sEV RNA transcripts. Notably, this test was able to reduce collection of tissue biopsies by 27% in prostate cancer patients [59, 60]. This is evidence that sEVs from liquid biopsy have opened a new pathway for biomarker development which can impact diagnosis, prediction, and treatment of various cancers including BC (Fig. 1).
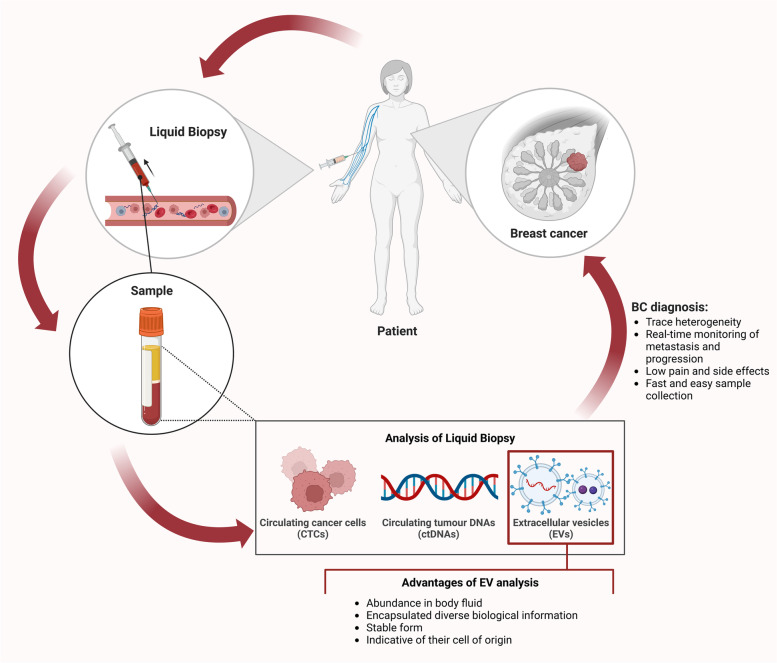
Fig. 1.
Liquid biopsy for breast cancer (BC) diagnosis. Body fluids are collected from BC patients for liquid biopsy analysis. Circulating tumour cells, circulating tumour DNA, and extracellular vesicles can be detected via liquid biopsy. This approach has several advantages in BC diagnosis such as ability to trace heterogeneity, monitoring of tumour metastasis and progression in a real-time, reduced pain and side effects, and it is a faster and easier process to obtain samples compared to tissue biopsy. Among the analytes detected by liquid biopsy, extracellular vesicles have superior characteristics, for example, abundance in various bodily fluids, stable form with encapsulated bioactive molecules, and reflective of their cell of origin. This figure was created with BioRender.com
Advanced techniques for BC sEV isolation and characterisation
EV research relies on precise and efficient isolation which requires accurate quantitation and high purity. However, heterogeneity in size, density, biochemical composition, and morphology of EVs complicate isolation strategies of specific sEV subpopulations [24, 61, 62]. Various isolation methods have been devised by exploiting a unique characteristic of sEVs such as size, density, composition, morphology, surface proteins, and gauge [63]. The classical sEV separation methods include ultracentrifugation (UC), ultrafiltration, size exclusion chromatography (SEC), polymer precipitation, and immunoaffinity. However, low purity and yield, high cost, and damage to sEVs are the main disadvantages of the traditional isolation methods, and in response, researchers have established more reliable and robust isolation methods. The more recent isolation methods that have emerged include immunoaffinity-based microfluidic chips and magnetic beads which provide simpler and faster detection and isolation of sEVs from cancer patients and with higher efficiency and specificity than the conventional methods [64–68]. Some of the limitations of these approaches include a higher cost and low sEV yield following removal of the affinity agents. Recent reviews discussed the strengths and merits of these approaches [63, 69, 70], and the focus here will centre on newly advanced sEV isolation techniques as it pertains to BC research.
Wang et al. [71] reported isolation of nanoscale vesicles with high yield and purity from plasma of BC patients using microfluidic chips with ciliated micropillars [72]. The Sub-ExoProfile chip is a microfluidic nanodevice with three self-assembled 3D nanopillars to capture CD81, EpCAM, and HER2-specific sEVs and tested in BC cell lines and plasma samples where it was capable of distinguishing HER2+ BC and triple-negative BC (TNBC) using a small volume of sample [73]. Additionally, microfluidic chips with filtration approaches such as ExoDIF [74] and ExoID-Chip [75] were validated in BC blood samples, showing high sEV concentration, sensitivity, and selectivity, and they successfully were able to distinguish between BC patients and healthy donors for diagnosis. Most recently, a novel lipid microarray based on supporting lipid membranes carrying antibody CD63 and EpCAM offered a rapid and accurate capture of cancer-specific sEVs in BC cells with minimal sample volume [67]. Despite the effort to develop novel isolation techniques, it is still difficult to apply these methods in a clinical setting due to issues with separation of similar-sized particles, obtaining a high yield of sEVs from a small volume of sample, and high cost.
Because of the high cost of immuno-based isolation methods, researchers have developed alternative approaches that do not rely on antibodies, for example, lipid nanoprobes [76], anion-exchange chromatography [77], and cholesterol-modified magnetic beads [78]. A synthetic peptide, Vn96, was developed that could bind HSPs on the surface of sEVs [79]. Tests showed that this affinity reagent was able to isolate sEVs with reduced background from BC cancer cell lines and plasma samples compared to UC. Moreover, applying Vn96 isolation with TRIzol© extraction in BC cell lines allowed multi-omic analysis of sEV proteins and RNAs on the same samples with a minimal processing time [80]. Zang et al. developed a DNA aptamer-based magnetic technique that targets CD63 on the surface of sEVs [81]. This approach maintained high bioactivity of sEVs and was able to detect increased MUC1+ sEVs in plasma samples of BC patients. However, a recent report showed that the DNA aptamer demonstrated low sEV specificity and reproducibility for sEV proteins in clinical settings [82]. Development of higher quality aptamers would require higher cost and more labour [83]. Therefore, method improvement is essential if clinical affinity-based isolation techniques are to be adopted in the future and this requires efficiencies in development and manufacturing cost.
Currently, standardised sEV isolation methods have not been established. The ideal sEV enrichment methodology would be simple, high throughput, and fast with low cost and high purity. However, none of the current isolation methodologies meet these criteria due to the complexity of biological samples and heterogenous nature of EVs. One possibility to overcome these obstacles may be to conjoin two or more isolation techniques [84]. For example, SEC followed by polyethylene glycol (PEG)-based precipitation in BC cell lines was reported to enrich 20- to 200-fold higher concentrations of sEV proteins and in less time than SEC or UC alone [85]. Also, ExoQuick or Total Exosome Isolation kits followed by a half cycle of UC in the serum of BC patients led to significantly purer yields, higher throughput, and with a less time-consuming process [86]. Tayebi et al. developed EV subpopulation sorting using combined electrical and acoustic forces [87]. The purity of sEVs recovered and recovery rate were higher in the combinational approach than in either individual approach. Lastly, clinical grade sEVs from human adipose mesenchymal stromal cells were isolated by a combination of UC and PEG-based isolation techniques and these sEVs were used for aerosol inhalation in lung injury to decrease inflammation [88]. Taken together, continued development of combined isolation techniques may be a viable option for selecting and isolating sEVs, which could aid in downstream analysis and clinical applications.
Single sEV analysis in BC research
To overcome issues such as heterogeneity, impurities during enrichment, inaccurate quantitation, and damage to sEVs, techniques based on molecular composition of single sEVs have been developed [89–91]. Single particle interferometric reflectance imaging sensor (SP-IRIS) can help overcome limitations such as overestimation of sEV counts and detection size limits, and simultaneously detect and count single sEV particles and analyse relative size distribution [92]. Jung et al. used SP-IRIS to compare several different sEV isolation methods based on protein marker expression such as CD9, CD63, and CD81 and found that Exoquick isolation kits had higher sEV particle concentration compared to exoEasy and UC isolates [93].
Instead of conventional flow cytometry (FCM), researchers have shown that a custom-built high-sensitivity flow cytometer [94], a fluorescence-activated vesicle sorting (FAVS) instrument [95], and a high-resolution flow cytometer (hFCM) [96] improved upon sensitivity and resolution. For example, FAVS was shown to localise EGFR ligand on the outside of sEVs in a BC cell line [97]. Risha et al. used hFCM to verify expression levels of glucose transporter 1 (GLUT-1), glypican 1 (GPC-1), and disintegrin and metalloproteinase domain-containing protein 10 (ADAM10) on the surface of sEVs derived from BC [98].
In addition to these techniques, nano-flow cytometry (nFCM) can analyse single sEVs below diameters of 40 nm with low background signal, enhanced signal integration, and high sensitivity [99, 100]. This method can quantify individual sEVs without a surface anchoring process and only requires small volumes compared to conventional FCM [101]. Recent studies demonstrated that nFCM can capture heterogeneously distributed cargoes across different EV subpopulations in colorectal cancer cell lines and Expi293F cells [102, 103]. In BC plasma samples, Salmond et al. used nFCM to distinguish sEVs derived from platelets and protein aggregations depending on their size, markers, and detergent treatment, showing the relatively high sensitivity of nFCM. Also, the majority of CD9+ and mammaglobin+ sEVs in human BC platelets were successfully identified using nFCM [104]. Expression of tissue factor (TF/F3) and EGFR among EV subpopulations from a BC cell line were mapped by nFCM [105].
A method for super-resolution microscopy, called photoactivated localisation microscopy/stochastic optical reconstruction microscopy (PALM/STORM), has also been utilised for the detection of single sEVs (< 100 nm) in BC cell lines. Results showed that a large portion of sEVs taken up by BC cell lines were localised inside lysosomes, providing evidence that sEVs can be endocytosed by recipient cells and transported into lysosomes for further degradation [106].
The droplet-based single-exosome-counting enzyme-linked immunoassay (droplet digital ExoELISA) was used for absolute quantitation of glypican-1+ sEVs in the plasma of BC patients by targeting protein expression on single sEVs in droplets [107]. Results showed that glypican-1+ sEVs were highly expressed in BC cells compared to benign breast disease and numbers were reduced after surgery, showing good potential of this technique for BC diagnosis. Recently, digital profiling of proteins on individual sEVs (DPPIE) was developed to multiplex detection of proteins on individual sEVs. DPPIE showed higher CD63/EpCAM/MUC1+ sEVs in BC plasma compared to healthy donors [108]. Finally, digital droplet PCR (ddPCR) was able to simultaneously quantify four mRNAs in individual sEVs derived from BC plasma samples with high sensitivity, demonstrating a novel method for profiling PR, ER, and HER2 subtypes [109].
Precise isolation and characterisation of each EV subpopulation is still a formidable challenge [110]. However, advances are ongoing and the advantages of current techniques for analysis of individual sEVs in BC research are summarised in Table 1 and sEV detection, characterisation, and analytical methodologies are shown in Fig. 2. In summary, single sEV detection holds great promise for early diagnosis and progression monitoring of BC and personalised medicine.
Table 1.
Summary of small extracellular vesicle analysis methods in breast cancer research
| Detection technique | Detection material | Advantage | Reference |
|---|---|---|---|
| SP-IRIS | Concentration and size distribution | High throughput and sensitive detection of single EVs | [92] |
| HSFCM | Specific EV population and size distribution | Multiparameter analysis of single EVs as small as 40 nm and analysis 10 000 particles per minute | [94] |
| FAVS | Specific EV population | High sensitivity, speedy detection, and elimination of background noise | [95, 111] |
| hFCM | Specific EV population | Low detection limit and multiparameter qualitative analysis | [96] |
| nFCM | Specific EV population | Analysation of EVs smaller than 40 nm, high throughput, and high resolution with multiparametric scattered light to detect individual EVs | [99, 100, 112] |
| dSTORM | Three-dimensional shape of sEVs | Imaging resolution of approximately 20 nm, inexpensive equipment | [106, 113] |
| Droplet digital ExoELISA | Absolute quantification of sEVs | Detection of maximum 5 sEVs per microliter of samples with high sensitivity and specificity, absolute quantification of targeting sEVs | [107] |
| DPPIE | Multi-quantification of surface protein of sEVs | Analysation of muti-protein expression in individual sEVs with high sensitivity and small volume of samples, and no purification step required | [108] |
| ddPCR | Quantify RNAs from single sEVs | Absolute quantification of rare targets | [114] |
Abbreviations: ddPCR droplet digital PCR, DPPIE Digital profiling of proteins on individual sEVs, Droplet digital ExoELISA Droplet-based single-exosome-counting enzyme-linked immunoassay, dSTORM direct stochastic optical reconstruction microscopy, EV Extracellular vesicle, FAVS Fluorescence-activated vesicle sorting, hFCM high-resolution flow cytometer, HSFCM High-sensitivity flow cytometer, nFCM nano-flow cytometer, SP-IRIS Single particle interferometric reflectance imaging sensor
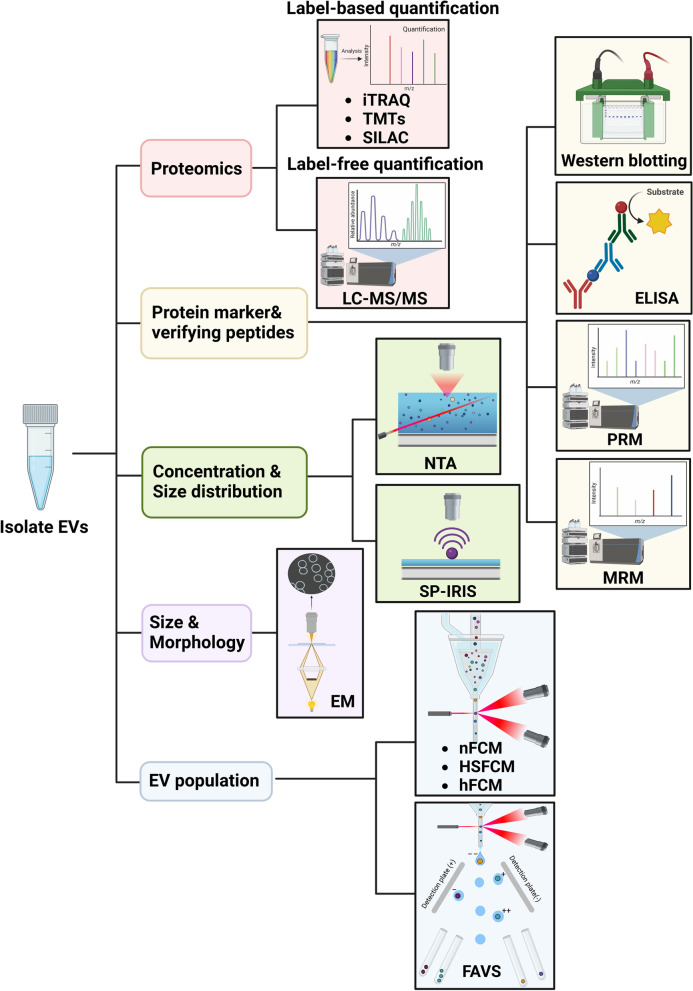
Fig. 2.
Small extracellular vesicle (sEV) detection and characterisation. sEVs isolated from breast cancer (BC) blood samples and cell lines can be characterised by size, morphology, concentration, and sEV markers. sEV size distribution and concentration can be analysed by nanoparticle tracking analysis (NTA) and single particle interferometric reflectance imaging sensor (SP-IRIS). Various types of electron microscopy (EM) can help classify sEVs by size and morphology. Nano flow cytometry (nFCM), high-sensitivity flow cytometry (HSFCM), high-resolution flow cytometry (hFCM), and fluorescence-activated vesicle sorting (FAVS) can sort sEV subpopulations in the isolated samples using various sEV markers. sEV proteins can be profiled by mass spectrometry-based analysis including label-free and label-based quantification. For verifying sEV protein and peptide markers, multiple reaction monitoring (MRM), parallel reaction monitoring (PRM), western blotting (WB), and enzyme-linked immunosorbent assay (ELISA) can be used. This figure was created with BioRender.com
sEV protein biomarkers and post translational modifications (PTMs) in BC diagnosis, prognosis, and chemoresistance
Early proteomic approaches for sEV biomarker research evolved from western blotting (WB) to ELISA, one-dimensional electrophoresis, and liquid chromatography with tandem mass spectroscopy (LC–MS/MS), however, these methods are limited by low acquisition rate, limited throughput, low sensitivity, and high cost [115, 116]. Unlike ELISA and WB which quantify single proteins, improved MS-based technology allows global analysis by detection of thousands of sEV proteins in one experiment [32]. Current LC–MS/MS-based quantitative proteomics are categorised into two approaches: label-free and label-based quantification. Label-free quantification is a commonly used proteomic approach to identify sEV proteins in cells [117] and various body fluids samples such as plasma [118], serum [119], saliva [120], urine [121], and cerebrospinal fluid [122, 123]. However, it is among the lowest for throughput among MS-based methods [124] and has low reproducibility because of multi-step sample processing [125]. One researcher reported improved throughput by using label-free LC–MS/MS combined with the absolute protein expression (APEX) algorithmic tool in cancer-derived sEV protein detection, showing a more time-effective and less laborious detection process for cancer biomarker discovery [126].
For label-based quantification, isobaric tags for absolute and relative quantification (iTRAQ), tandem mass tags (TMTs), and stable isotope labelling of amino acids (SILAC) are examples of label-based quantification that have been applied to sEV research in various diseases including BC [127–132]. However, these techniques have limitations in terms of high cost, low throughput, and lack of specificity during ion selection [133, 134]. To increase throughput and specificity, Ting et al. utilised a multiplex approach using TMT reagents which requires an additional isolation and fragmentation event (MS3) [135]. For the purpose of identifying potential biomarkers in BC diagnosis and monitoring of disease progression, Clark et al. combined TMTs with support vector machine analytics to detect 251 sEV proteins in BC cells [136].
One of the modern techniques used for sEV protein biomarker validation is multiple reaction monitoring (MRM), which provides a more accurate quantification of low abundant peptides than other targeted methods and without the necessity for expensive antibodies [137, 138]. Several studies have used MRM to obtain detailed expression profiles of sEV proteins and to identify potential biomarkers such as HSP90 and syndecan-1 in bladder cancer, and annexin A2 and clusterin in Alzheimer's disease [139–141]. However, the precise measurements made by MRM can only be applied for small peptides, not for multiple peptides in the same protein [142], which may limit its clinical translation as an assay.
The parallel-reaction monitoring (PRM) technique is similar to MRM, but quantitation is performed with MS2 in a high-resolution mass spectrometer. It has higher throughput, high sensitivity, and less preparatory time in peptide identification, and it is adequate for the identification of PTMs [143, 144]. PRM was used to validate selected sEV proteins from a global profiling study [129]. Additionally, it was utilised to detect elevated sEV phosphoproteins with a potential diagnostic value such as GTPase-activating protein subunit alpha-2 (RALGAPA2), cGMP-dependent protein kinase1 (PKG1), and tight junction protein 2 (TJP2) in BC patients [145]. Proteomic techniques used for sEV protein biomarker detection are shown in Fig. 2.
sEV protein biomarkers in BC diagnosis
Proteins contained within sEVs consist of almost half of the human proteome and are reflective of the cell types of origin, suggesting an ideal choice for disease-specificity and biomarker discovery [146]. Accumulating evidence indicates sEV protein biomarkers play an important role in BC diagnosis. Lee et al. detected 270 sEV proteins in invasive BC cell lines using LC–MS/MS and validated EGF-like repeat and discoidin I-like domain containing protein 3 (EDIL3) as a diagnostic biomarker that is correlated with metastasis [147]. Another study profiled 241 uniquely expressed sEV proteins in several BC cell lines, demonstrating fibronectin (FN) as a diagnostic biomarker candidate specifically for distinguishing between ER+ and ER− BC [40, 148]. Using a reverse phase protein microarray, up-regulated focal adhesion kinase (FAK) and mitogen-activated protein kinase 1 (MEK1) in plasma sEVs were identified as candidate biomarkers for BC diagnosis [40]. With micro FCM, CD47 was found to have relatively lower abundance levels in sEVs isolated from serum of BC patients compared to healthy controls [149]. Risha et al. profiled 726 uniquely expressed proteins in TNBC cells by nano LC–MS/MS and identified three potential biomarkers located on the membrane surface of sEVs (GPC-1, glucose GLUT-1, and ADAM10), and up-regulated when compared to a non-tumourigenic epithelial breast cell line [98]. A multiplexed cantilever array was used to profile GPC-1 in sEVs released from BC cell lines, showing high sensitivity and throughput in real-time acquisition [150].
Recently, several novel sEV screening methods including microfluidic chips [151], surface enhanced Raman scattering nanotags [152], and DNA aptamer-mediated microfluidics [153] have demonstrated simple and time-saving ways to profile sEV EpCAM and HER2 proteins for diagnosis of HER2+ BC. Cumulatively, these results demonstrate the diagnostic value of sEV protein biomarkers in BC, however, further validation in large studies of independent clinical samples is required to confirm their clinical significance.
sEV protein biomarkers in BC prognosis
Determination of the prognostic value of sEV protein biomarkers is an important area of BC research. Higher levels of sEV annexin A2 was reported using a combination of two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) MS in BC cell lines and plasma of BC patients [154]. Also, sEV annexin A2 in serum was shown to have a prognostic value with a positive correlation to tumour grade of TNBC and poor survival [155]. In BC patients treated with neoadjuvant chemotherapy, elevated nerve growth factor (NGF) within serum-derived sEVs was related to poor survival outcomes, suggesting NGF in sEVs was an independent prognostic factor for overall survival [156]. The level of insulin-like growth factor receptor β subunit (IGFRβ) in plasma sEVs [40] and CD82 in serum sEVs [157] was found to gradually increase with BC grade, showing the potential of these protein biomarkers for monitoring BC progression or prediction of disease prognosis. In the earliest stage of BC, up-regulated developmental endothelial locus-1 (Del-1) in plasma sEVs was identified as a potential prognostic indicator by LC–MS/MS and ELISA [158]. Survivin was elevated in serum sEVs from early-stage BC and TNBC patients, and its splice variants were inversely correlated to tumour grade in BC [159], suggesting the expression of survivin and its variants might be a potential indicator of BC progression. Interestingly, urinary sEVs from early-stage BC patients had significantly higher levels of matrix metalloproteinase-1 (MMP-1)/CD63 than healthy control subjects, showing a high sensitivity for primary screening of early-stage BC cases [160].
sEV protein biomarkers in predicting BC chemoresistance
Chemoresistance is a major challenge for BC therapy and prediction of its occurrence can help guide treatment and improve prognosis. Serum from HER2+ BC patients was reported to harbour sEVs with elevated levels of HER2 [161]. Furthermore, trastuzumab was found to bind to sEV HER2 in treated patients and likely contributed to resistance to this drug. In addition, unique expression profiles of 51 sEV proteins was reported in HER2+ BC cells treated with trastuzumab and these proteins had functional roles related to organelle organisation, cytokinesis, and response to lipids [162]. This study highlighted the role of nano-ultra-HPLC–MS/MS in identifying potential biomarker candidates for predicting trastuzumab chemoresistance. Durcker et al. observed that trastuzumab treatment led to elevated levels of sEV PERP, ITB1, GNAS2, and GNA13 proteins in HER2+ and trastuzumab-sensitive BC cells, but not in the resistant cells [163]. Further evidence showed that these proteins were also elevated in plasma sEVs of HER2+ BC patients treated with trastuzumab therapy, however, a larger patient cohort is required for verification [163].
Several lines of research suggested that sEV-mediated intercellular cargo delivery from chemoresistant BC cells to sensitive cells might induce a chemoresistant phenotype in the acceptor cells, including GSTP1 [164], UCH-L1 [165], and transient receptor potential channel 5 (TrpC5) [166] proteins in adriamycin-resistant BC cells and serum. In the same way, high abundance of ABCG2/breast cancer resistance protein (BCRP) in plasma sEVs isolated from anthracycline-taxane-based neoadjuvant chemotherapy-resistant BC patients [167], and annexin A6 in gemcitabine-resistant BC cells was found to transfer to chemo-sensitive BC cells via sEVs and induce chemoresistance [168]. During this transfer, annexin A6 derived from gemcitabine-resistant BC cells induced the inhibition of EGFR ubiquitination and degradation [168, 169]. Kavanagh et al. demonstrated that therapeutic-induced senescent (TIS) TNBC cells by paclitaxel (PTX) had 142 significantly elevated sEV proteins such as ATPases, annexins, tubulins, integrins, and Rabs that are associated with cell proliferation, ATP depletion, apoptosis, and insoluble senescence-associated secretory phenotype [170]. This data shows the potential of TNBC TIS as a source of predictive biomarkers as well as support for chemotherapeutic challenge. Thus, sEV protein biomarkers are an important source for predicting BC chemoresistance in the clinical setting and future development of personalised medicine.
PTMs as BC sEV protein biomarkers
Most proteins contain dynamic PTMs which affect function and stability, and PTMs are known to be integral to sEV formation and sEV-related biological processes [171]. For example, Aguilar et al. detected about 10,000 phosphopeptides and 1,500 N-glycopeptides in a small volume of human plasma-derived sEVs [172]. Several studies have addressed the possibility of phosphorylation and glycosylation of sEV proteins as cancer biomarkers, for example, phospho-EGFR in head and neck cancer [173], phospho-AKT and -ERK1/2 in non-small-cell lung cancer [174], and EGFR-specific N-glycan in colorectal cancer [175].
Although recent studies have profiled phosphoproteins in BC, research that specifically targets sEV phosphoproteins in BC is very limited. A few studies showed evidence that sEV phosphoproteins may have diagnostic value for BC. For example, 144 up-regulated phosphoproteins in plasma sEVs of BC patients were identified by LC–MS/MS and phosphorylation of RALGAPA2, PKG1, and TJP2 were validated by PRM, therefore these phosphoproteins are candidate sEV biomarkers for BC diagnosis [145]. Moreover, in TNBC and HER+ BC, sEVs contained hyperphosphorylated receptor tyrosine kinase, non-receptor tyrosine kinase, and MAP kinase, and the downstream signalling pathways of these kinases are related to migration and angiogenesis. It was postulated that BC-derived sEVs can transfer hyperphosphorylated proteins and activate intracellular signalling pathways related to metastasis in recipient cells [176].
For glycosylation PTMs, LC–MS/MS analysis showed the differential expression of 77 glycoproteins in sEVs from plasma of BC patients compared to healthy controls, of which 20 were up-regulated [177]. Elevated sEV glycoproteins include lymphocyte antigen 6 complex locus protein G6f (LY6G6F), von Willebrand factor (VWF), CD147/basigin (BSG), complement C1q subcomponent subunit A (C1QA), and angiopoietin-1 (ANGPT1/Ang1) [177]. Moreover, highly abundant glycosylated sEV proteins glycoprotein 130 in BC cells and CD147 in BC patient serum showed a diagnostic value for BC [178, 179]. Glycoprotein 130 can transfer to macrophages via BC-derived sEVs, leading to phosphorylation of transcription factor STAT3 and induction of tumourigenesis-associated genes IL-6 and IL-10 [179], suggesting the potential of both glycoprotein 130 and CD147 as biomarkers for BC progression monitoring.
sEVs have been reported to enhance the expression of P-glycoprotein (P-gp) in recipient cells. For example, sEVs from adriamycin-resistant BC cells were shown to transfer UCH-L1, activate the MAPK/ERK signalling pathway, and up-regulated P-gp [166]. Similarly, TrpC5 was also delivered to adriamycin-sensitive cells by sEVs but stimulated P-gp expression through the translocation of transcription factor, T-cells isoform c3 (NFATc3) [165]. In PTX-induced TNBC TIS, sEV glycoprotein profiling identified elevated P-gp, CD44, galectin-3, and glycogenin-1, suggesting that altered abundance of glycoproteins in sEVs could be a tool to evaluate the treatment efficacy of TNBC cells that are amenable to PTX chemotherapy [170, 180].
More research is required to verify PTMs as BC biomarkers. Moreover, high resolution methods for profiling cancer-specific and dynamic phosphorylation and glycosylation of sEV proteins as well as development of PTM-specific antibodies are required for further validation. The role of potential sEV protein biomarkers, including PTMs, in BC diagnosis, prognosis, and chemoresistance are summarised in Fig. 3 and Table 2.
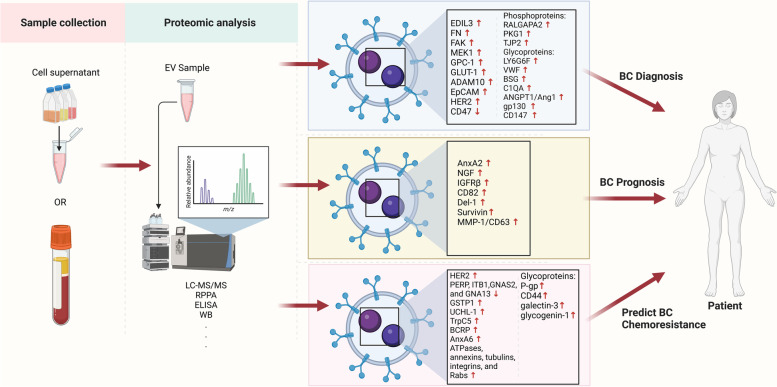
Fig. 3.
Small extracellular vesicle (sEV) protein biomarkers, including post-translational modifications (PTMs), in breast cancer (BC) diagnosis, prognosis, and chemoresistance. sEVs are collected from cell supernatants and blood samples of BC patients. Isolated sEVs are profiled by proteomic strategies such as liquid chromatography with tandem mass spectrometry (LC–MS/MS) and reverse phase protein microarray (RPPA) and validated by sEV protein detection methods including enzyme-linked immunosorbent assay (ELISA) and western blotting (WB). Various up- or down-regulated proteins in BC sEV samples were identified and can be used in BC diagnosis, prognosis, and prediction of BC chemoresistance. This figure was created with BioRender.com
Table 2.
Small extracellular vesicle protein biomarkers, including post-translational modifications, identified in breast cancer diagnosis, prognosis, chemoresistance
| Biomarker | Source | Purpose | Expression | Feature | Technology | Reference |
|---|---|---|---|---|---|---|
| EDIL3 | Cells | Diagnosis | ↑ | Related to invasion ability | LC–MS/MS | [147] |
| FN | Cells/plasma | Diagnosis | ↑ | Highest expression in ER− BC followed by ER+ and healthy control | LC–MS/MS | [40, 148] |
| FAK | Plasma | Diagnosis | ↑ | N/A | RPPA | [40] |
| MEK1 | Plasma | Diagnosis | ↑ | N/A | RPPA | [40] |
| CD47 | Serum | Diagnosis | ↓ | N/A | Micro FCM | [149] |
| GPC-1 | Cells | Diagnosis | ↑ | Located on cell surface, related to proliferation |
Nano-LC–MS/MS Sandwich cantilever assay |
[98, 150] |
| GLUT-1 | Cells | Diagnosis | ↑ | Located on the cell surface related to BC migration | Nano-LC–MS/MS | [98] |
| ADAM10 | Cells | Diagnosis | ↑ | Located on the cell surface | Nano-LC–MS/MS | [98] |
| EpCAM | Cells/plasma | Diagnosis | ↑ |
Overexpressed in various cancers Diagnosis for HER2+ BC |
Microfluidic chips SERS nanotags DNA aptamers mediated- microfluidic |
[151–153] |
| HER2 | Cells/plasma |
Diagnosis Chemoresistance |
↑ |
Diagnosis for HER2+ BC Related to trastuzumab resistance |
Microfluidic chips SERS nanotags DNA aptamers mediated- microfluidic nano-ultra-HPLC–MS/MS |
[151–153, 161, 162] |
| RALGAPA2, PKG1 & TJP2 | Plasma | Diagnosis | ↑ | Phosphoproteins |
LC–MS/MS PRM |
[145] |
| LY6G6F, VWF, BSG, C1QA & ANGPT1/Ang1 | Plasma | Diagnosis | ↑ | Glycoproteins | LC–MS/MS | [177] |
| Glycoprotein 130 | Cells | Diagnosis | ↑ | Glycoprotein | WB | [179] |
| CD147 | Serum | Diagnosis | ↑ | Glycoprotein |
LC–MS/MS WB |
[178] |
| Annexin A2 | Cells/serum | Prognosis | ↑ | Related to poor survival of BC | MALDI-TOF MS | [154, 155] |
| NGF | Serum | Prognosis | ↑ | Related to poor survival of BC who undergo neoadjuvant chemotherapy | ProcartaPlex immune-related panels | [156] |
| IGFRβ | Plasma | Prognosis | ↑ | Higher expression in later-stage BC | RPPA | [40] |
| CD82 | Serum | Prognosis | ↑ | Higher expression in later-stage BC | ELISA | [157] |
| Del-1 | Plasma | Prognosis | ↑ | N/A |
LC–MS/MS ELISA |
[158] |
| Survivin | Serum | Prognosis | ↑ |
High expressed in early-stage BC and TNBC Survivin splice variants negatively correlated with tumour grade |
ELISA WB |
[159] |
| MMP-1/CD63 | Urine | Prognosis | ↑ | Higher expression in early-stage BC | WB | [160] |
| PERP, ITB1, GNAS2 & GNA13 | Cells/plasma | Chemoresistance | ↓ | Higher expressed in HER2+ and trastuzumab-sensitive BC | LC–MS/MS | [163] |
| GSTP1 | Cells/serum | Chemoresistance | ↑ | Higher expressed in adriamycin-resistance BC |
FCM WB |
[164] |
| UCHL-1 | Cells/serum | Chemoresistance | ↑ | Higher expressed in adriamycin-resistance BC |
FCM WB |
[165] |
| TrpC5 | Cells/serum | Chemoresistance | ↑ | Higher expressed in adriamycin-resistance BC |
FCM RT-PCR |
[166] |
| BCRP | Plasma | Chemoresistance | ↑ | Higher expressed in TEC therapy resistance BC |
FCM RT-PCR |
[167] |
| Annexin 6 | Cells | Chemoresistance | ↑ | Higher expressed in gemcitabine resistance BC | Isobaric peptide labelling LC–MS/MS, WB | [168] |
| ATPases, annexins, tubulins, integrins & Rabs | Cells | Chemoresistance | ↑ | Higher expressed in TIS cells |
LC–MS/MS WB |
[170] |
| P-gp, CD44, galectin-3 & glycogenin-1 | Cells | Chemoresistance | ↑ |
Glycoproteins PTX chemotherapy-resistance in TNBC |
LC–MS/MS | [170, 180] |
Abbreviations: BC Breast cancer, ELISA Enzyme-linked immunosorbent assay, ER Estrogen receptor, FCM Flow cytometry, LC–MS/MS Liquid chromatography with tandem mass spectrometry, N/A Not applicable, PRM Parallel-reaction monitoring, PTX Paclitaxel, RPPA Reverse phase protein microarray, RT-PCR Reverse transcriptase-polymerase chain reaction, SERS Surface enhanced Raman scattering, TEC The anthracycline-taxane-based neoadjuvant chemotherapy (docetaxel, epirubicin, and cyclophosphamide), TIS Therapeutic induced senescent, TNBC Triple negative breast cancer, UHPLC Ultra-high performance liquid chromatography, WB Western blotting
Limitations of sEV protein biomarkers in cancer research
MS-based proteomics is the most common method for profiling sEV protein biomarkers in cancer. Although better enrichment strategies for sEVs and more specificity for accurate protein identification by MS are required, current sEV isolation methods attempt to find a balance between purity and abundance which are inversely related. Askeland et al. recently compared three sEV isolation techniques such as UC, SEC, and peptide-affinity precipitation from human plasma, and found all isolates still had contaminations which required additional steps for higher purity but consequently led to a lower yield [82]. In addition, protein contents of sEVs were found to vary depending on the isolation method. For example, when SEC, UC, and PEG-based isolation methods combined with SEC were compared, all showed different relative quantities of sEV proteins [85]. In a comparison study between SEC, PEG-based isolation, and protein organic solvent precipitation (PROSPR), common sEV markers such as CD9, CD63 and CD81 were only detected in sEVs isolated from SEC [181]. Moreover, protein precipitation with acetone in the PROSPR methodology may have led to peptide modification when glycine is the second residue. Since up to 6% of peptides contain glycine as the second residue, this could lead to consequential artifacts in the data [182, 183]. The ISEV guideline has advocated for a universal protocol for obtaining pure sEVs that are compatible with proteomic analysis [184].
Selection of the enzymatic digestion procedure in sample preparation steps which involve contaminant removal for MS-based analysis, including in-gel, in-solution, and filter-aided digestion, affects the identification yield of sEV peptides. When in-solution digestion is used, the profile of sEV proteins varies depending on the choice of sEV isolation method as well as the detergents used for solubilising sEV proteins. For example, using nano LC–MS/MS, Risha et al. identified 986 sEV proteins isolated by UC and digestion in the presence of n-dodecyl β-d-maltoside (DDM) detergent, and this total was higher compared to use of other detergents (Triton X-100, Digitonin) and isolation methods (ultrafiltration combined with UC or ExoQuick®) [98]. In-gel digestion is likely the most popular method but can lead to heavy keratin contamination. Xu et al. suggested the use of an electrostatic eliminator to solve the contamination problem, but it was not completely eliminated [185]. Several studies introduced modified filter-aided digestion protocols with sodium dodecyl sulfate detergents [186] and multiple enzyme-filter aided sample preparation (MED-FASP) [187, 188] for LC–MS/MS analysis, showing the highest yield of sEV peptide and protein identifications and with the cleanest MS spectra from blood samples. However, because of its time-consuming and labour-intensive processes, translation to clinical settings remains a challenge.
Several investigations reported that sEVs isolated from blood samples were contaminated with non-vesicular entities such as lipoproteins and protein aggregates [189, 190]. Sodar et al. showed that lipoprotein morphology was similar to sEVs and there were tenfold more lipoprotein particles compared to sEVs in the sEV isolates from human blood which are likely contributing factors to contamination. The authors further postulated that any of the known sEV isolation methods cannot separate lipoproteins from the isolates [191]. However, combining isolation methods can help to reduce contamination. Utilising SEC with a density cushion significantly reduced lipoprotein particles in human plasma samples [192]. The sequential use of PEG precipitation, linear iodixanol density gradients, and SEC filtered out 90% of lipoproteins in plasma samples, however, significant losses of sEVs and arduous sample preparation were limitations for this approach [193].
Considerations for choosing an appropriate sEV isolation technique include the volume of biofluid and the sEV contaminants known to reside in the biofluid. Development of enhanced sEV sample preparation techniques will lead to higher purity, yield, and throughput, as well as better reproducibility and more accurate protein quantification.
sEV miRNA biomarkers in BC diagnosis, prognosis, and chemoresistance
miRNAs are short single-stranded non-coding RNA molecules about 19–25 nucleotides in length, and can inhibit gene function by directly binding to the 3’-untranslated region of target mRNAs. An RNA profiling study using next-generation sequencing (NGS) found that miRNAs were the most prominent RNA species in sEVs and identified 593 unique miRNAs [194]. In blood, encapsulation of miRNAs in sEVs can provide a stable environment and facilitate long distance cell-to-cell communication. Therefore, tumour-derived sEVs carrying miRNAs that are released into the bloodstream could be a valuable biomarker resource for disease diagnosis, prognosis, and treatment management [116, 195].
Identification of miRNAs and their targets have been studied by quantitative real-time PCR (qRT-PCR), microarray analysis, and NGS [196–198]. NGS is a second-generation sequencing technique that has followed Sanger sequencing methods. Today, it is the popular method for miRNA profiling with several advantages including high sensitivity and throughput, ability to identify miRNA variants, and low cost [199, 200]. The 4 most widely used NGS systems are the 454 pyrosequencing method, Illumina’s sequencing by synthesis (SBS) technique, Ion Torrent technology, and sequencing by oligonucleotide ligation and detection (SOLiD). The 454, Illumina, and SOLiD methods use four fluorescence dyes for sequence detection. Illumina uses fluorescently labelled reversible terminators that are imaged once a dNTP is added to the sequence, and then cleaved to stop addition of the next base. The SOLiD method does not use DNA polymerase like other methods but rather detects fluorescence when an 8-mer oligonucleotide is added to sequence using a DNA ligase. Instead of fluorescence, Ion Torrent detects proton changes during elongation [201]. Among these four systems, most BC sEV miRNA studies have utilised Illumina sequencing to identify metastatic features related to miRNAs, new therapeutic targets, and biomarker candidates [202–205]. The BC sEV miRNAs identified as a potential diagnostic, prognostic, and chemoresistant biomarkers as well as potential therapeutic targets are summarised in Fig. 4. Current sEV miRNA biomarker information including diagnosis, prognosis, and prediction of chemoresistance in BC is shown in Table 3.
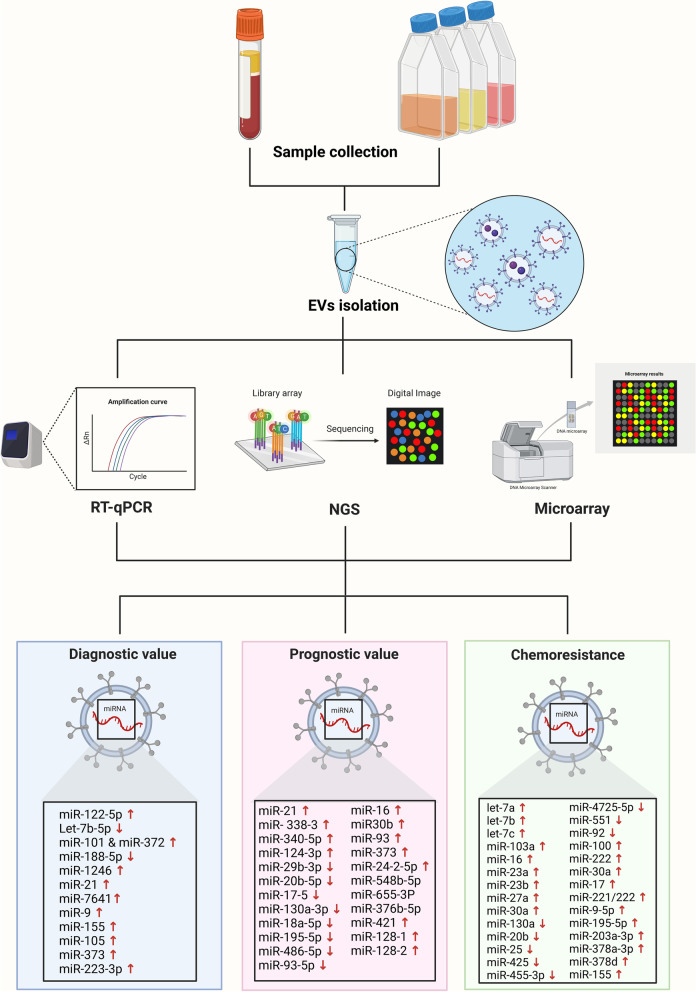
Fig. 4.
Small extracellular vesicle (sEV) miRNA biomarkers in breast cancer (BC) diagnosis, prognosis, chemoresistance. BC plasma and cell supernatant samples are collected, and the isolated sEVs containing miRNAs are analysed by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR), next-generation sequencing (NGS), and microarray analysis. The unique sEV miRNA expression profiles of BC samples have value for diagnosis, prognosis, and predicting chemoresistance of BC. This figure was created with BioRender.com
Table 3.
Small extracellular vesicle miRNA biomarkers in breast cancer diagnosis, prognosis and chemoresistance
| Biomarker | Source | Type of purpose | Expression | Feature | Technology | Reference |
|---|---|---|---|---|---|---|
| miR-122-5p | Plasma | Diagnosis | ↑ | N/A | RT-qPCR | [206] |
| Let-7b-5p | Plasma | Diagnosis | ↓ | N/A | RT-qPCR | [206] |
| miR-101 & miR-372 | Serum | Diagnosis | ↑ | N/A | RT-qPCR | [207] |
| miR-188-5p | Serum | Diagnosis | ↓ | N/A | RT-qPCR | [208] |
| miR-1246 | Plasma | Diagnosis | ↑ | Combination with miR-21 showing better diagnostic value | NGS-Illumina | [209] |
| miR-21 | Plasma |
Diagnosis Prognosis |
↑ | High expression in later-stage BC | NGS-Illumina | [209–211] |
| miR-7641 | Cells/plasma | Diagnosis | ↑ |
Up-regulated in TNBC Related to proliferation & migration |
Microarray/RT-qPCR | [212] |
| miR-9 | Cells | Diagnosis | ↑ |
Up-regulated in TNBC Related to metastasis |
RT-qPCR | [213] |
| miR-155 | Cells |
Diagnosis Chemoresistance |
↑ |
Up-regulated in TNBC Related to DOX and PTX resistance in BC |
RT-qPCR | [213, 214] |
| miR-105 | Cells/Mouse | Diagnosis | ↑ | Up-regulated in TNBC | Solex deep sequencing | [58] |
| miR-373 | Cells/serum | Diagnosis | ↑ |
Up-regulated in TNBC Related to proliferation & migration |
RT-qPCR | [207, 215] |
| miR-223-3p | Plasma | Diagnosis | ↑ | Related to invasive BC | RT-qPCR | [216] |
| miR- 338-3p, miR-340-5p & miR-124-3p | Serum | Prognosis | ↑ | Related to BC recurrence | miRNA PCR array | [217] |
| miR-29b-3p, miR-20b-5p, miR-17-5p, miR-130a-3p, miR-18a-5p, miR-195-5p, miR-486-5p & miR-93-5p | Serum | Prognosis | ↓ | Related to BC recurrence | miRNA PCR array | [217] |
| miR-16 & miR30b | Plasma | Prognosis | ↑ | Up-regulated in BC with recurrence | Microarray | [218] |
| miR-93 | Plasma | Prognosis | ↑ | Up-regulated in DCIS | Microarray | [218] |
| miR-373 & miR-24–2-5p | Plasma | Prognosis | ↑ | Negatively correlated with survival | NGS-Illumina | [219] |
| miR-548b-5p, miR-655-3P & miR-376b-5p | Plasma | Prognosis | ↓ | Positively correlated with survival | NGS-Illumina | [219] |
| miR-421, miR-128–1 &miR-128–2 | Serum | Prognosis | ↑ |
GI-derived three sEV miRNA signature Related to poor prognosis |
NGS-Illumina | [202] |
| miR-100, miR-222, miR-30a & miR-17 | Cells | Chemoresistance | ↑ | Related to DTX resistance in BC | Microarray | [220] |
| miR-221/222 | Cells | Chemoresistance | ↑ | Related to tamoxifen resistance in BC | RT-qPCR | [221] |
| let-7a, let-7b, let-7c, miR-103a, miR-16, miR-23a, miR-23b, miR-27a & miR-30a | Cells | Chemoresistance | ↑ | Related to DTX resistance in BC | Microarray | [222] |
| miR-130a, miR-20b, miR-25, miR-425, miR-455-3p, miR-4725-5p, miR-551, miR-92 | Cells | Chemoresistance | ↓ | Related to DTX resistance in BC | Microarray | [222] |
| miR-9-5p, miR-195-5p & miR-203a-3p | Cells | Chemoresistance | ↑ | Related to DOX and DTX resistance in BC | NGS-Illumina | [223] |
| miR-378a-3p, miR-378d | Cells | Chemoresistance | ↑ | Related to DOX and PTX resistance in BC | RT-qPCR | [224] |
Abbreviations: BC Breast cancer, DCIS Ductal carcinoma in situ, DOX Doxorubicin, DTX Docetaxel, GI Genomic instability, NGS Next-generation sequencing, N/A Not applicable, PTX Paclitaxel, RT-qPCR Reverse transcriptase-quantitative polymerase chain reaction, TNBC Triple- negative breast cancer
sEV miRNA biomarkers in BC diagnosis
Several studies have evaluated distinctive miRNA profiles in sEVs between BC patients and healthy controls to find potential blood-based biomarkers for diagnosis. For example, up-regulated miR-122-5p and down-regulated Let‐7b‐5p were identified [206] in sEVs from BC plasma. In serum sEVs, miR-101 and miR-372 [207] were elevated and miR-188-5p [208] levels were lower in BC patients relative to healthy controls. Hannafon et al. profiled miRNAs in BC plasma-derived sEVs using NGS and found higher levels of miR-1246 and miR-21 in patients compared to healthy controls, and the combination of those miRNAs were a better indicator for BC diagnosis than individual levels [209]. Abundance of miR-7641 [212], miR-9, miR-155 [213], miR-105 [58], and miR-373 [207] was higher in sEVs from TNBC cells compared to human mammary epithelial cells and non-metastatic BC cells, suggesting that sEV miRNA levels may be tissue-type dependent and can be used to differentiate BC subtypes. Elevated miR-7641 and miR-373 was also found in sEVs from TNBC serum and plasma and linked to the proliferation and migration ability of BC cells [207, 212]. Also, miR-223-3p levels were higher in plasma sEVs of invasive ductal carcinoma BC patients when compared to those from ductal carcinoma in situ (DCIS) patients and healthy control subjects, showing potential diagnostic value for invasive BC [216]. All in all, sEV miRNAs are a promising biomarker source for BC early diagnosis and for distinguishing different stages of BC for personalised treatment options.
sEV miRNA biomarkers in BC prognosis
Evaluating prognosis using sEV-based liquid biopsy is an important component for optimising treatment choices for BC patients. An sEV miRNA profiling study indicated that miR-338-3p, miR-340-5p, and miR-124-3p were up-regulated, and miR-29b-3p, miR-20b-5p, miR-17-5p, miR-130a-3p, miR-18a-5p, miR-195-5p, miR-486-5p, and miR-93-5p were down-regulated in the serum of BC patients with recurrence compared to patients without recurrence [217]. Ni et al. profiled sEV miRNAs in plasma of DCIS patients and primary BC patients with recurrence compared to healthy women [218]. Levels of miR-16 and miR30b were found to be relatively higher in recurrent patients, and miR-93 in DCIS patients [218], demonstrating the promise of sEV miRNAs to distinguish recurrent BC from the early stages of the disease. In another study, 35 differentially expressed miRNAs in plasma sEVs from early-stage BC patients were profiled [219]. Among these miRNAs, highly abundant miR-375 and miR-24–2-5p were found to be negatively correlated with patient survival, and significantly down-regulated miR-548b-5p, miR-655-3P, and miR-376b-5p were found to be positively correlated with survival. Recently, Bao et al. used a combination of genomic instability (GI) analysis with sEV miRNA profiling to identify novel BC biomarkers. In their study, three higher GI-derived sEV miRNA signatures (miR-421, miR-128–1, and miR-128–2) in the serum of BC patients were found to be associated with poor prognosis [202]. These findings support sEV-derived miRNAs as a useful tool for BC prognosis.
sEV miRNA biomarkers in predicting BC chemoresistance
Deciphering mechanisms of cancer chemoresistance is a critical step for developing or selecting therapeutic agents that bypass resistance and this can also aid in the identification of biomarkers that can be used as predictors of resistance. Chen et al. showed that miR-100, miR-222, miR-30a, and miR-17 were transferred from BC-resistant to -sensitive cells by sEVs and miR-222 reduced expression of the tumour suppressor gene PTEN in sensitive BC cells [220]. Up-regulation of sEV miR-221/222 was also detected in tamoxifen-resistant BC cells [221]. When miR-211/222 sEV levels were reduced, tamoxifen resistance decreased accordingly, showing the potential of using this miRNA as a predictive tool to monitor the progression of tamoxifen-resistant BC.
In addition to these findings, Chen et al. found that sEVs from doxorubicin (DOX)- and docetaxel (DTX)-resistant BC cells had over 300 differentially expressed miRNAs compared to drug-sensitive BC cells [220]. The most abundant sEV miRNAs in DTX-resistant cells were let-7a, let-7b, let-7c, miR-103a, miR-16, miR-23a, miR-23b, miR-27a, and miR-30a, and the most down-regulated miRNAs were miR-130a, miR-20b, miR-25, miR-425, miR-455-3p, miR-4725-5p, miR-551, and miR-92 [222]. These miRNAs are known to modulate target genes involved in transcriptional regulation, protein phosphorylation, kinase activity, and protein binding. Shen et al. also profiled sEV miRNAs in DOX- and DTX-resistant BC cells and reported up-regulation of miR-9-5p, miR-195-5p, and miR-203a-3p that target the one cut homeobox 2 (ONECUT2) transcription factor [223]. Yang et al. reported that serum sEVs from DOX- and PTX-resistant BC patients had higher levels of miR-378a-3p and miR-378d, and suggested a novel mechanism whereby DOX and PTX chemotherapy activated the EZH2/STAT3 pathway in BC cells and led to higher abundance of these miRNAs in serum sEVs [224]. Both of the sEV miRNAs were delivered to neighbouring BC cells to activate the WNT and NOTCH stemness pathways via suppression of Dickkopf 3 (DKK3) and NUMB expression and could induce chemoresistance. Moreover, relatively higher miR-155 abundance was detected in DOX- and PTX-resistant TNBC cell sEVs and the resistance was found to be mediated by translocation of miR-155 via BC-derived sEVs, suggesting that sEV miRNAs could be used as a biomarker for prediction of DOX and PTX resistance and to help guide therapy [213, 214].
Limitations of sEV miRNA biomarkers in cancer research
It is difficult to orthogonally validate miRNA profiling using microarray and NGS techniques, possibly due to the reduced selectivity of NGS [225]. In addition, miRNA library composition for NGS is dependent on sample library preparation kits. For example, in different individual libraries, the number of detectable sEV miRNAs was found to vary from 380 to 474 [194]. The different miRNA libraries might cause biased NGS data for certain transcripts, but highly abundant miRNAs are less affected by library preparation [226].
Several studies have shown that the choice of methodology for sEV isolation and RNA extraction can significantly affect the abundance of sEV miRNAs and this is also influenced by sample type, resulting in significant differences in miRNA profiling. For example, Park et al. reported that the number of unique miRNA species from urinary sEVs isolated by UC, qEV, and ExoQuick preparations were different, and ExoQuick showed the lowest number of sEV miRNA species compared to the others [227]. However, in blood samples, Kuhlmann et al. demonstrated that ExoQuick provided optimal miRNA yield and consistent small RNA libraries [228]. On the other hand, Buschmann et al. showed that precipitation and membrane affinity-based sEV isolation methods were equally efficient at sEV miRNA profiling, resulting in high absolute numbers of mapped miRNA reads and increased separation of patients group [226]. When six commercial RNA extraction kits were compared, the Norgen Biotek exosome purification and RNA isolation kit had a > fourfold increase of sEV miRNA species in human urine than the other kits [229]. For miRNA biomarker discovery, selection of a high throughput platform, library preparation method, downstream analysis, and sEV isolation and RNA extraction protocol regarding sample type must all be carefully considered.
miRNAs contain heterogenous ends leading to miRNA isoforms (isomiRs) which can affect the precision and accuracy of miRNA profiling and detection [230]. NGS-based sequencing is the only high throughput platform with the ability to detect individual isomiRs but most computational tools do not include isomiR deconvolution by default, so users must consider this option [231, 232]. Some studies have suggested that isomiRs have a role in sorting miRNAs into sEVs and therefore contain discriminatory information which is useful for disease biomarkers [233–235]. Thus, when searching for novel sEV miRNAs and verifying them as cancer biomarkers or therapeutic targets, isomiRs and their functional association with disease should be included the development pipeline.
Conclusions and future perspectives
To improve BC diagnosis and prognosis, researchers have increasingly focused on liquid biopsy due to its limited invasiveness and suitability for cancer monitoring. According to the evidence described here, monitoring changes of sEV-derived proteins and miRNAs holds great potential and promise for BC biomarker translation. Based on the advances in technology and amount of research described in this review, there are some critical considerations pertaining to sEV-related BC research. Firstly, a combination of approaches might be necessary to accurately diagnose or predict BC. Several lines of recent research have focused on BC multi-omic biomarker signatures that have higher sensitivity for diagnosis and prognosis than a single-omic biomarker. For example, the combination of serum sEV differentiation antagonising non-protein coding RNA (DANCR) and traditional tumour markers CA 15–3 and CEA showed 91.4% specificity and 90.8% sensitivity in BC diagnostic value with improved accuracy over individual biomarkers [236]. Moreover, the combination of serum sEV miR-1910-3p with CA 15–3 was more sensitive than individual biomarkers with 96% diagnostic sensitivity in BC [237]. For early BC patients, combined expression of urinary sEV miR-21 and matrix metalloproteinase-1 (MMP-1) showed improved sensitivity (95%) and specificity (79%) [160]. Additional studies to evaluate multi-omic biomarkers are required.
Secondly, extracellular vesicle and particles (EVPs) are heterogenous nanovesicles and particles classified into subpopulations according to size. In 2018, it was discovered that a new EVP subpopulation called exomeres (smaller than 35 nm), had distinct N-glycan and proteomic profiling compared to other EV subpopulations, and exomeres proteins were associated with glycolysis, metabolic pathways, coagulation, and hypoxia [238]. In 2021, the smallest EVPs named supermeres was reported, and these supermeres showed different RNA and protein profiles relative to exomeres and sEVs [239]. Several tumour-associated proteins and EV-related RNAs were enriched in supermeres [239]. Thus, proteins, glycans, lipids, and nucleic acids might be selectively encapsulated into different EVP subpopulations and have unique biomechanical characteristics and functions in terms of tumour biology. More work is required to thoroughly characterise each EVP subpopulation and better techniques for EVP isolation need to be developed to obtain high-quality samples in a cost and time efficient manner.
Progress has been made but there is still a long road ahead to bring sEV biomarkers from the bench to the bedside. A lack of standardisation in isolation and characterisation obstructs progress, resulting in lower reproducibility and significant inter-study variation. In addition, a limited number of study cohorts, heterogeneous clinical samples, and the complexity of biofluids are additional obstacles to sEV biomarker utility in the clinical setting. Despite these challenges, sEV proteins and miRNAs are promising candidate biomarkers for BC, and research and development for clinical usage are moving forward.
Acknowledgements
Editorial assistance, in the form of language editing and correction, was provided by XpertScientific Editing and Consulting Services.
Authors’ contributions
Yujin Lee and Yong Li wrote the main manuscript text and Yujin Lee prepares Figs. 1, 2, 3 and 4. All authors read and approved the final manuscript.
Funding
This work is mainly supported by the St. George Hospital Cancer Research Trust Fund and St. George and Sutherland Medical Research Foundation. Lee YJ is supported by the University International Postgraduate Award (UIPA) Scholarship from UNSW Sydney, Australia.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Group USCSW. U.S. Cancer Statistics Data Visualizations Tool, Based on 2019 Submission Data (1999–2017). US Department of Health and Human Services. 2020.
- 4.Indicators CAsNCC. Relative Survival by Stage at Diagnosis (Female Breast Cancer) 2019. https://ncci.canceraustralia.gov.au/relative-survival-stage-diagnosis-female-breast-cancer. Accessed 20 Sept 2021.
- 5.Clarke R, Tyson JJ, Dixon JM. Endocrine resistance in breast cancer–An overview and update. Mol Cell Endocrinol. 2015;418 Pt 3(0 3):220–34. doi: 10.1016/j.mce.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Koren S, Bentires-Alj M. Breast tumor heterogeneity: source of fitness. Hurdle for Therapy Mol Cell. 2015;60(4):537–546. doi: 10.1016/j.molcel.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 8.Wang L. Early diagnosis of breast cancer. Sensors (Basel) 2017;17(7):1572. doi: 10.3390/s17071572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi KS, Yoon M, Song SH, Suh M, Park B, Jung KW, et al. Effect of mammography screening on stage at breast cancer diagnosis: results from the Korea National Cancer Screening Program. Sci Rep. 2018;8(1):8882. doi: 10.1038/s41598-018-27152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerlikowske K, Grady D, Barclay J, Sickles EA, Ernster V. Likelihood ratios for modern screening mammography. Risk of breast cancer based on age and mammographic interpretation. JAMA. 1996;276(1):39–43. doi: 10.1001/jama.1996.03540010041028. [DOI] [PubMed] [Google Scholar]
- 11.Bae JM, Shin SY, Kim EH, Kim YN, Nam CM. Distribution of dense breasts using screening mammography in Korean women: a retrospective observational study. Epidemiol Health. 2014;36:e2014027. doi: 10.4178/epih/e2014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–717. https://doi-org.wwwproxy1.library.unsw.edu.au/10.1016/S0140-6736(05)66544-0. [DOI] [PubMed]
- 13.Duffy MJ, Evoy D, McDermott EW. CA 15–3: uses and limitation as a biomarker for breast cancer. Clin Chim Acta. 2010;411(23–24):1869–1874. doi: 10.1016/j.cca.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 14.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang B, Zhu Y, Ni J, Thompson J, Malouf D, Bucci J, et al. Extracellular vesicles: the next generation of biomarkers for liquid biopsy-based prostate cancer diagnosis. Theranostics. 2020;10(5):2309–2326. doi: 10.7150/thno.39486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28(5):666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshioka Y, Kosaka N, Konishi Y, Ohta H, Okamoto H, Sonoda H, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. 2014;5:3591. doi: 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Luca F, Rotunno G, Salvianti F, Galardi F, Pestrin M, Gabellini S, et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget. 2016;7(18):26107–26119. doi: 10.18632/oncotarget.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, Han L, Yuan J, Sun Y. Circulating tumor cell free DNA from plasma and urine in the clinical management of colorectal cancer. Cancer Biomark. 2020;27(1):29–37. doi: 10.3233/CBM-182344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilgour E, Rothwell DG, Brady G, Dive C. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell. 2020;37(4):485–495. doi: 10.1016/j.ccell.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Robertson EG, Baxter G. Tumour seeding following percutaneous needle biopsy: the real story! Clin Radiol. 2011;66(11):1007–1014. doi: 10.1016/j.crad.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Marrugo-Ramírez J, Mir M, Samitier J. Blood-based cancer biomarkers in liquid biopsy: a promising non-invasive alternative to tissue biopsy. Int J Mol Sci. 2018;19(10):2877. doi: 10.3390/ijms19102877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witwer KW, Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visan KS, Wu LY, Voss S, Wuethrich A, Möller A. Status quo of Extracellular Vesicle isolation and detection methods for clinical utility. Semin Cancer Biol. 2022;88:157–171. doi: 10.1016/j.semcancer.2022.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 27.Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–281. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raj DA, Fiume I, Capasso G, Pocsfalvi G. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 2012;81(12):1263–1272. doi: 10.1038/ki.2012.25. [DOI] [PubMed] [Google Scholar]
- 29.Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, et al. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16(4):782–790. doi: 10.1038/mt.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med. 2012;10:5. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim DK, Kang B, Kim OY, Choi DS, Lee J, Kim SR, et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013;2:20384. doi: 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH, et al. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics. 2015;31(6):933–939. doi: 10.1093/bioinformatics/btu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17(1):147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36(13):1770–1778. doi: 10.1038/onc.2016.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14(11):1891–1900. doi: 10.1039/C4LC00136B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67(1):33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lou G, Song X, Yang F, Wu S, Wang J, Chen Z, et al. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8:122. doi: 10.1186/s13045-015-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vinik Y, Ortega FG, Mills GB, Lu Y, Jurkowicz M, Halperin S, et al. Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci Adv. 2020;6(40):eaba5714. doi: 10.1126/sciadv.aba5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang JP, Tang YY, Fan CM, Guo C, Zhou YH, Li Z, et al. The role of exosomal non-coding RNAs in cancer metastasis. Oncotarget. 2018;9(15):12487–12502. doi: 10.18632/oncotarget.23552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bidard FC, Peeters DJ, Fehm T, Nolé F, Gisbert-Criado R, Mavroudis D, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15(4):406–414. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 43.Yang C, Zou K, Zheng L, Xiong B. Prognostic and clinicopathological significance of circulating tumor cells detected by RT-PCR in non-metastatic colorectal cancer: a meta-analysis and systematic review. BMC Cancer. 2017;17(1):725. doi: 10.1186/s12885-017-3704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan K, Leong SM, Kee Z, Caramat PV, Teo J, Blanco MVM, et al. Longitudinal monitoring reveals dynamic changes in circulating tumor cells (CTCs) and CTC-associated miRNAs in response to chemotherapy in metastatic colorectal cancer patients. Cancer Lett. 2018;423:1–8. doi: 10.1016/j.canlet.2018.02.039. [DOI] [PubMed] [Google Scholar]
- 45.Pantel K, Hille C, Scher HI. Circulating tumor cells in prostate cancer: from discovery to clinical utility. Clin Chem. 2019;65(1):87–99. doi: 10.1373/clinchem.2018.287102. [DOI] [PubMed] [Google Scholar]
- 46.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10(24):8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 48.Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell. 2013;23(5):573–581. doi: 10.1016/j.ccr.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi E, Fabbri F. CTCs 2020: great expectations or unreasonable dreams. Cells. 2019;8(9):989. doi: 10.3390/cells8090989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh RF, et al. Molecular biomarker analyses using circulating tumor cells. PLoS ONE. 2010;5(9):e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ignatiadis M, Lee M, Jeffrey SS. Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin Cancer Res. 2015;21(21):4786–4800. doi: 10.1158/1078-0432.CCR-14-1190. [DOI] [PubMed] [Google Scholar]
- 52.Cai X, Janku F, Zhan Q, Fan JB. Accessing genetic information with liquid biopsies. Trends Genet. 2015;31(10):564–575. doi: 10.1016/j.tig.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Maroto R, Zhao Y, Jamaluddin M, Popov VL, Wang H, Kalubowilage M, et al. Effects of storage temperature on airway exosome integrity for diagnostic and functional analyses. J Extracell Vesicles. 2017;6(1):1359478. doi: 10.1080/20013078.2017.1359478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allenson K, Castillo J, San Lucas FA, Scelo G, Kim DU, Bernard V, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28(4):741–747. doi: 10.1093/annonc/mdx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan Y, Liu B, Lei H, Zhang B, Wang Y, Huang H, et al. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann Oncol. 2018;29(12):2379–2383. doi: 10.1093/annonc/mdy458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Möhrmann L, Huang HJ, Hong DS, Tsimberidou AM, Fu S, Piha-Paul SA, et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clin Cancer Res. 2018;24(1):181–188. doi: 10.1158/1078-0432.CCR-17-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou J, Li X, Wu X, Zhang T, Zhu Q, Wang X, et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. 2018;6(12):1578–1592. doi: 10.1158/2326-6066.CIR-17-0479. [DOI] [PubMed] [Google Scholar]
- 58.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKiernan J, Donovan MJ, O'Neill V, Bentink S, Noerholm M, Belzer S, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2016;2(7):882–889. doi: 10.1001/jamaoncol.2016.0097. [DOI] [PubMed] [Google Scholar]
- 60.McKiernan J, Donovan MJ, Margolis E, Partin A, Carter B, Brown G, et al. A prospective adaptive utility trial to validate performance of a novel urine exosome gene expression assay to predict high-grade prostate cancer in patients with prostate-specific antigen 2–10ng/ml at initial biopsy. Eur Urol. 2018;74(6):731–738. doi: 10.1016/j.eururo.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 61.Sharma A, Khatun Z, Shiras A. Tumor exosomes: cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine (Lond) 2016;11(4):421–437. doi: 10.2217/nnm.15.210. [DOI] [PubMed] [Google Scholar]
- 62.Ludwig N, Whiteside TL, Reichert TE. Challenges in exosome isolation and analysis in health and disease. Int J Mol Sci. 2019;20(19):4684. doi: 10.3390/ijms20194684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7(3):789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao S, Wu M, Yang S, Wu Y, Gu Y, Chen C, et al. A disposable acoustofluidic chip for nano/microparticle separation using unidirectional acoustic transducers. Lab Chip. 2020;20(7):1298–1308. doi: 10.1039/D0LC00106F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci U S A. 2017;114(40):10584–10589. doi: 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Li Q, Shi H, Tang K, Qiao L, Yu G, et al. Microfluidic Raman biochip detection of exosomes: a promising tool for prostate cancer diagnosis. Lab Chip. 2020;20(24):4632–4637. doi: 10.1039/D0LC00677G. [DOI] [PubMed] [Google Scholar]
- 67.Liu HY, Kumar R, Zhong C, Gorji S, Paniushkina L, Masood R, et al. Rapid capture of cancer extracellular vesicles by lipid patch microarrays. Adv Mater. 2021;33(35):e2008493. doi: 10.1002/adma.202008493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han S, Xu Y, Sun J, Liu Y, Zhao Y, Tao W, et al. Isolation and analysis of extracellular vesicles in a Morpho butterfly wing-integrated microvortex biochip. Biosens Bioelectron. 2020;154:112073. doi: 10.1016/j.bios.2020.112073. [DOI] [PubMed] [Google Scholar]
- 69.Coumans FAW, Brisson AR, Buzas EI, Dignat-George F, Drees EEE, El-Andaloussi S, et al. Methodological guidelines to study extracellular vesicles. Circ Res. 2017;120(10):1632–1648. doi: 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- 70.Xu R, Greening DW, Zhu HJ, Takahashi N, Simpson RJ. Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest. 2016;126(4):1152–1162. doi: 10.1172/JCI81129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, Godin B, et al. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip. 2013;13(15):2879–2882. doi: 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim J, Choi M, Lee H, Kim YH, Han JY, Lee ES, et al. Direct isolation and characterization of circulating exosomes from biological samples using magnetic nanowires. J Nanobiotechnology. 2019;17(1):1. doi: 10.1186/s12951-018-0433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Wang S, Chen A, Wang R, Li L, Fang X. Efficient exosome subpopulation isolation and proteomic profiling using a Sub-ExoProfile chip towards cancer diagnosis and treatment. Analyst. 2022;147(19):4237–4248. doi: 10.1039/D2AN01268E. [DOI] [PubMed] [Google Scholar]
- 74.Kang YT, Kim YJ, Bu J, Cho YH, Han SW, Moon BI. High-purity capture and release of circulating exosomes using an exosome-specific dual-patterned immunofiltration (ExoDIF) device. Nanoscale. 2017;9(36):13495–13505. doi: 10.1039/C7NR04557C. [DOI] [PubMed] [Google Scholar]
- 75.Dong X, Chi J, Zheng L, Ma B, Li Z, Wang S, et al. Efficient isolation and sensitive quantification of extracellular vesicles based on an integrated ExoID-Chip using photonic crystals. Lab Chip. 2019;19(17):2897–2904. doi: 10.1039/C9LC00445A. [DOI] [PubMed] [Google Scholar]
- 76.Wan Y, Cheng G, Liu X, Hao SJ, Nisic M, Zhu CD, et al. Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nat Biomed Eng. 2017;1:0058. doi: 10.1038/s41551-017-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J, Xu Y, Lu Y, Xing W. Isolation and visible detection of tumor-derived exosomes from plasma. Anal Chem. 2018;90(24):14207–14215. doi: 10.1021/acs.analchem.8b03031. [DOI] [PubMed] [Google Scholar]
- 78.He F, Wang J, Yin BC, Ye BC. Quantification of exosome based on a copper-mediated signal amplification strategy. Anal Chem. 2018;90(13):8072–8079. doi: 10.1021/acs.analchem.8b01187. [DOI] [PubMed] [Google Scholar]
- 79.Ghosh A, Davey M, Chute IC, Griffiths SG, Lewis S, Chacko S, et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS ONE. 2014;9(10):e110443. doi: 10.1371/journal.pone.0110443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Joy AP, Ayre DC, Chute IC, Beauregard AP, Wajnberg G, Ghosh A, et al. Proteome profiling of extracellular vesicles captured with the affinity peptide Vn96: comparison of Laemmli and TRIzol© protein-extraction methods. J Extracell Vesicles. 2018;7(1):1438727. doi: 10.1080/20013078.2018.1438727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang K, Yue Y, Wu S, Liu W, Shi J, Zhang Z. Rapid capture and nondestructive release of extracellular vesicles using aptamer-based magnetic isolation. ACS Sens. 2019;4(5):1245–1251. doi: 10.1021/acssensors.9b00060. [DOI] [PubMed] [Google Scholar]
- 82.Askeland A, Borup A, Østergaard O, Olsen JV, Lund SM, Christiansen G, et al. Mass-spectrometry based proteome comparison of extracellular vesicle isolation methods: comparison of ME-kit, size-exclusion chromatography, and high-speed centrifugation. Biomedicines. 2020;8(8):246. doi: 10.3390/biomedicines8080246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu H, Alkhamis O, Canoura J, Liu Y, Xiao Y. Advances and challenges in small-molecule DNA aptamer isolation, characterization, and sensor development. Angew Chem Int Ed Engl. 2021;60(31):16800–16823. doi: 10.1002/anie.202008663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu WZ, Ma ZJ, Kang XW. Current status and outlook of advances in exosome isolation. Anal Bioanal Chem. 2022;414(24):7123–7141. doi: 10.1007/s00216-022-04253-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martínez-Greene JA, Hernández-Ortega K, Quiroz-Baez R, Resendis-Antonio O, Pichardo-Casas I, Sinclair DA, et al. Quantitative proteomic analysis of extracellular vesicle subgroups isolated by an optimized method combining polymer-based precipitation and size exclusion chromatography. J Extracell Vesicles. 2021;10(6):e12087. doi: 10.1002/jev2.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ryu KJ, Lee JY, Park C, Cho D, Kim SJ. Isolation of small extracellular vesicles from human serum using a combination of ultracentrifugation with polymer-based precipitation. Ann Lab Med. 2020;40(3):253–258. doi: 10.3343/alm.2020.40.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tayebi M, Yang D, Collins DJ, Ai Y. Deterministic sorting of submicrometer particles and extracellular vesicles using a combined electric and acoustic field. Nano Lett. 2021;21(16):6835–6842. doi: 10.1021/acs.nanolett.1c01827. [DOI] [PubMed] [Google Scholar]
- 88.Shi MM, Yang QY, Monsel A, Yan JY, Dai CX, Zhao JY, et al. Preclinical efficacy and clinical safety of clinical-grade nebulized allogenic adipose mesenchymal stromal cells-derived extracellular vesicles. J Extracell Vesicles. 2021;10(10):e12134. doi: 10.1002/jev2.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szatanek R, Baj-Krzyworzeka M, Zimoch J, Lekka M, Siedlar M, Baran J. The methods of choice for extracellular vesicles (EVs) characterization. Int J Mol Sci. 2017;18(6):1153. doi: 10.3390/ijms18061153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Erdbrügger U, Rudy CK, Etter ME, Dryden KA, Yeager M, Klibanov AL, et al. Imaging flow cytometry elucidates limitations of microparticle analysis by conventional flow cytometry. Cytometry A. 2014;85(9):756–770. doi: 10.1002/cyto.a.22494. [DOI] [PubMed] [Google Scholar]
- 91.Akers JC, Ramakrishnan V, Nolan JP, Duggan E, Fu CC, Hochberg FH, et al. Comparative analysis of technologies for quantifying extracellular vesicles (EVs) in Clinical Cerebrospinal Fluids (CSF) PLoS ONE. 2016;11(2):e0149866. doi: 10.1371/journal.pone.0149866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Daaboul GG, Gagni P, Benussi L, Bettotti P, Ciani M, Cretich M, et al. Digital detection of exosomes by interferometric imaging. Sci Rep. 2016;6:37246. doi: 10.1038/srep37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jung HH, Kim JY, Lim JE, Im YH. Cytokine profiling in serum-derived exosomes isolated by different methods. Sci Rep. 2020;10(1):14069. doi: 10.1038/s41598-020-70584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W, et al. Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano. 2018;12(1):671–680. doi: 10.1021/acsnano.7b07782. [DOI] [PubMed] [Google Scholar]
- 95.Higginbotham JN, Zhang Q, Jeppesen DK, Scott AM, Manning HC, Ochieng J, et al. Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J Extracell Vesicles. 2016;5:29254. doi: 10.3402/jev.v5.29254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van der Vlist EJ, Nolte-'t Hoen EN, Stoorvogel W, Arkesteijn GJ, Wauben MH. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7(7):1311–1326. doi: 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- 97.Higginbotham JN, Demory Beckler M, Gephart JD, Franklin JL, Bogatcheva G, Kremers GJ, et al. Amphiregulin exosomes increase cancer cell invasion. Curr Biol. 2011;21(9):779–786. doi: 10.1016/j.cub.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Risha Y, Minic Z, Ghobadloo SM, Berezovski MV. The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers. Sci Rep. 2020;10(1):13572. doi: 10.1038/s41598-020-70393-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tian Y, Gong M, Hu Y, Liu H, Zhang W, Zhang M, et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J Extracell Vesicles. 2020;9(1):1697028. doi: 10.1080/20013078.2019.1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arab T, Mallick ER, Huang Y, Dong L, Liao Z, Zhao Z, et al. Characterization of extracellular vesicles and synthetic nanoparticles with four orthogonal single-particle analysis platforms. J Extracell Vesicles. 2021;10(6):e12079. doi: 10.1002/jev2.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Friedrich R, Block S, Alizadehheidari M, Heider S, Fritzsche J, Esbjörner EK, et al. A nano flow cytometer for single lipid vesicle analysis. Lab Chip. 2017;17(5):830–841. doi: 10.1039/C6LC01302C. [DOI] [PubMed] [Google Scholar]
- 102.Liu H, Tian Y, Xue C, Niu Q, Chen C, Yan X. Analysis of extracellular vesicle DNA at the single-vesicle level by nano-flow cytometry. J Extracell Vesicles. 2022;11(4):e12206. doi: 10.1002/jev2.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Silva AM, Lázaro-Ibáñez E, Gunnarsson A, Dhande A, Daaboul G, Peacock B, et al. Quantification of protein cargo loading into engineered extracellular vesicles at single-vesicle and single-molecule resolution. J Extracell Vesicles. 2021;10(10):e12130. doi: 10.1002/jev2.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Salmond N, Khanna K, Owen GR, Williams KC. Nanoscale flow cytometry for immunophenotyping and quantitating extracellular vesicles in blood plasma. Nanoscale. 2021;13(3):2012–2025. doi: 10.1039/D0NR05525E. [DOI] [PubMed] [Google Scholar]
- 105.Choi D, Montermini L, Jeong H, Sharma S, Meehan B, Rak J. Mapping subpopulations of cancer cell-derived extracellular vesicles and particles by nano-flow cytometry. ACS Nano. 2019;13(9):10499–10511. doi: 10.1021/acsnano.9b04480. [DOI] [PubMed] [Google Scholar]
- 106.Chen C, Zong S, Wang Z, Lu J, Zhu D, Zhang Y, et al. Imaging and Intracellular tracking of cancer-derived exosomes using single-molecule localization-based super-resolution microscope. ACS Appl Mater Interfaces. 2016;8(39):25825–25833. doi: 10.1021/acsami.6b09442. [DOI] [PubMed] [Google Scholar]
- 107.Liu C, Xu X, Li B, Situ B, Pan W, Hu Y, et al. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett. 2018;18(7):4226–4232. doi: 10.1021/acs.nanolett.8b01184. [DOI] [PubMed] [Google Scholar]
- 108.Zhang J, Shi J, Zhang H, Zhu Y, Liu W, Zhang K, et al. Localized fluorescent imaging of multiple proteins on individual extracellular vesicles using rolling circle amplification for cancer diagnosis. J Extracell Vesicles. 2020;10(1):e12025. doi: 10.1002/jev2.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu C, Li B, Lin H, Yang C, Guo J, Cui B, et al. Multiplexed analysis of small extracellular vesicle-derived mRNAs by droplet digital PCR and machine learning improves breast cancer diagnosis. Biosens Bioelectron. 2021;194:113615. doi: 10.1016/j.bios.2021.113615. [DOI] [PubMed] [Google Scholar]
- 110.Wang T, Xing Y, Cheng Z, Yu F. Analysis of single extracellular vesicles for biomedical applications with especial emphasis on cancer investigations. TrAC, Trends Anal Chem. 2022;152:116604. doi: 10.1016/j.trac.2022.116604. [DOI] [Google Scholar]
- 111.van der Pol E, van Gemert MJ, Sturk A, Nieuwland R, van Leeuwen TG. Single vs. swarm detection of microparticles and exosomes by flow cytometry. J Thromb Haemost. 2012;10(5):919–30. doi: 10.1111/j.1538-7836.2012.04683.x. [DOI] [PubMed] [Google Scholar]
- 112.Groot Kormelink T, Arkesteijn GJ, Nauwelaers FA, van den Engh G, Nolte-'t Hoen EN, Wauben MH. Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high-resolution flow cytometry. Cytometry A. 2016;89(2):135–147. doi: 10.1002/cyto.a.22644. [DOI] [PubMed] [Google Scholar]
- 113.Jensen E, Crossman DJ. Technical review: types of imaging-direct STORM. Anat Rec (Hoboken) 2014;297(12):2227–2231. doi: 10.1002/ar.22960. [DOI] [PubMed] [Google Scholar]
- 114.Wang C, Ding Q, Plant P, Basheer M, Yang C, Tawedrous E, et al. Droplet digital PCR improves urinary exosomal miRNA detection compared to real-time PCR. Clin Biochem. 2019;67:54–59. doi: 10.1016/j.clinbiochem.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 115.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312(5771):212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 116.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 117.Benedikter BJ, Bouwman FG, Heinzmann ACA, Vajen T, Mariman EC, Wouters EFM, et al. Proteomic analysis reveals procoagulant properties of cigarette smoke-induced extracellular vesicles. J Extracell Vesicles. 2019;8(1):1585163. doi: 10.1080/20013078.2019.1585163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Veerman RE, Teeuwen L, Czarnewski P, Güclüler Akpinar G, Sandberg A, Cao X, et al. Molecular evaluation of five different isolation methods for extracellular vesicles reveals different clinical applicability and subcellular origin. J Extracell Vesicles. 2021;10(9):e12128. doi: 10.1002/jev2.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kennel PJ, Saha A, Maldonado DA, Givens R, Brunjes DL, Castillero E, et al. Serum exosomal protein profiling for the non-invasive detection of cardiac allograft rejection. J Heart Lung Transplant. 2018;37(3):409–417. doi: 10.1016/j.healun.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 120.Sun Y, Liu S, Qiao Z, Shang Z, Xia Z, Niu X, et al. Systematic comparison of exosomal proteomes from human saliva and serum for the detection of lung cancer. Anal Chim Acta. 2017;982:84–95. doi: 10.1016/j.aca.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 121.Bijnsdorp IV, Geldof AA, Lavaei M, Piersma SR, van Moorselaar RJ, Jimenez CR. Exosomal ITGA3 interferes with non-cancerous prostate cell functions and is increased in urine exosomes of metastatic prostate cancer patients. J Extracell Vesicles. 2013;2:22097. doi: 10.3402/jev.v2i0.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guha D, Lorenz DR, Misra V, Chettimada S, Morgello S, Gabuzda D. Proteomic analysis of cerebrospinal fluid extracellular vesicles reveals synaptic injury, inflammation, and stress response markers in HIV patients with cognitive impairment. J Neuroinflammation. 2019;16(1):254. doi: 10.1186/s12974-019-1617-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bachi A, Bonaldi T. Quantitative proteomics as a new piece of the systems biology puzzle. J Proteomics. 2008;71(3):357–367. doi: 10.1016/j.jprot.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 124.Lindemann C, Thomanek N, Hundt F, Lerari T, Meyer HE, Wolters D, et al. Strategies in relative and absolute quantitative mass spectrometry based proteomics. Biol Chem. 2017;398(5–6):687–699. doi: 10.1515/hsz-2017-0104. [DOI] [PubMed] [Google Scholar]
- 125.Xie F, Liu T, Qian WJ, Petyuk VA, Smith RD. Liquid chromatography-mass spectrometry-based quantitative proteomics. J Biol Chem. 2011;286(29):25443–25449. doi: 10.1074/jbc.R110.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Choi DS, Choi DY, Hong BS, Jang SC, Kim DK, Lee J, et al. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J Extracell Vesicles. 2012;1:18704. doi: 10.3402/jev.v1i0.18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kong J, Tian H, Zhang F, Zhang Z, Li J, Liu X, et al. Extracellular vesicles of carcinoma-associated fibroblasts creates a pre-metastatic niche in the lung through activating fibroblasts. Mol Cancer. 2019;18(1):175. doi: 10.1186/s12943-019-1101-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chan YK, Zhang H, Liu P, Tsao SW, Lung ML, Mak NK, et al. Proteomic analysis of exosomes from nasopharyngeal carcinoma cell identifies intercellular transfer of angiogenic proteins. Int J Cancer. 2015;137(8):1830–1841. doi: 10.1002/ijc.29562. [DOI] [PubMed] [Google Scholar]
- 129.Zheng X, Xu K, Zhou B, Chen T, Huang Y, Li Q, et al. A circulating extracellular vesicles-based novel screening tool for colorectal cancer revealed by shotgun and data-independent acquisition mass spectrometry. J Extracell Vesicles. 2020;9(1):1750202. doi: 10.1080/20013078.2020.1750202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao X, Wu Y, Duan J, Ma Y, Shen Z, Wei L, et al. Quantitative proteomic analysis of exosome protein content changes induced by hepatitis B virus in Huh-7 cells using SILAC labeling and LC-MS/MS. J Proteome Res. 2014;13(12):5391–5402. doi: 10.1021/pr5008703. [DOI] [PubMed] [Google Scholar]
- 131.Kalra H, Gangoda L, Fonseka P, Chitti SV, Liem M, Keerthikumar S, et al. Extracellular vesicles containing oncogenic mutant β-catenin activate Wnt signalling pathway in the recipient cells. J Extracell Vesicles. 2019;8(1):1690217. doi: 10.1080/20013078.2019.1690217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Koh YQ, Ng DQ, Ng CC, Boey A, Wei M, Sze SK, et al. Extracellular vesicle proteome of breast cancer patients with and without cognitive impairment following anthracycline-based chemotherapy: an exploratory study. Biomark Insights. 2021;16:11772719211018204. doi: 10.1177/11772719211018204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Savitski MM, Mathieson T, Zinn N, Sweetman G, Doce C, Becher I, et al. Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J Proteome Res. 2013;12(8):3586–3598. doi: 10.1021/pr400098r. [DOI] [PubMed] [Google Scholar]
- 134.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics. 2010;9(9):1885–1897. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8(11):937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Clark DJ, Fondrie WE, Liao Z, Hanson PI, Fulton A, Mao L, et al. Redefining the breast cancer exosome proteome by tandem mass tag quantitative proteomics and multivariate cluster analysis. Anal Chem. 2015;87(20):10462–10469. doi: 10.1021/acs.analchem.5b02586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9(6):555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 138.Rardin MJ, Held JM, Gibson BW. Targeted quantitation of acetylated lysine peptides by selected reaction monitoring mass spectrometry. Methods Mol Biol. 2013;1077:121–131. doi: 10.1007/978-1-62703-637-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Song Z, Xu Y, Zhang L, Zhou L, Zhang Y, Han Y, et al. Comprehensive proteomic profiling of urinary exosomes and identification of potential non-invasive early biomarkers of Alzheimer's disease in 5XFAD mouse model. Front Genet. 2020;11:565479. doi: 10.3389/fgene.2020.565479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang T, Anderson KW, Turko IV. Assessment of extracellular vesicles purity using proteomic standards. Anal Chem. 2017;89(20):11070–11075. doi: 10.1021/acs.analchem.7b03119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tomiyama E, Matsuzaki K, Fujita K, Shiromizu T, Narumi R, Jingushi K, et al. Proteomic analysis of urinary and tissue-exudative extracellular vesicles to discover novel bladder cancer biomarkers. Cancer Sci. 2021;112(5):2033–2045. doi: 10.1111/cas.14881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Elliott MH, Smith DS, Parker CE, Borchers C. Current trends in quantitative proteomics. J Mass Spectrom. 2009;44(12):1637–1660. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- 143.Xu L, Gimple RC, Lau WB, Lau B, Fei F, Shen Q, et al. The present and future of the mass spectrometry-based investigation of the exosome landscape. Mass Spectrom Rev. 2020;39(5–6):745–762. doi: 10.1002/mas.21635. [DOI] [PubMed] [Google Scholar]
- 144.Sowers JL, Mirfattah B, Xu P, Tang H, Park IY, Walker C, et al. Quantification of histone modifications by parallel-reaction monitoring: a method validation. Anal Chem. 2015;87(19):10006–10014. doi: 10.1021/acs.analchem.5b02615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen IH, Xue L, Hsu CC, Paez JS, Pan L, Andaluz H, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A. 2017;114(12):3175–3180. doi: 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015;9(3–4):358–367. doi: 10.1002/prca.201400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee JE, Moon PG, Cho YE, Kim YB, Kim IS, Park H, et al. Identification of EDIL3 on extracellular vesicles involved in breast cancer cell invasion. J Proteomics. 2016;131:17–28. doi: 10.1016/j.jprot.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 148.Moon PG, Lee JE, Cho YE, Lee SJ, Chae YS, Jung JH, et al. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget. 2016;7(26):40189–40199. doi: 10.18632/oncotarget.9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kibria G, Ramos EK, Lee KE, Bedoyan S, Huang S, Samaeekia R, et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci Rep. 2016;6:36502. doi: 10.1038/srep36502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Etayash H, McGee AR, Kaur K, Thundat T. Nanomechanical sandwich assay for multiple cancer biomarkers in breast cancer cell-derived exosomes. Nanoscale. 2016;8(33):15137–15141. doi: 10.1039/C6NR03478K. [DOI] [PubMed] [Google Scholar]
- 151.Fang S, Tian H, Li X, Jin D, Li X, Kong J, et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS ONE. 2017;12(4):e0175050. doi: 10.1371/journal.pone.0175050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kwizera EA, O'Connor R, Vinduska V, Williams M, Butch ER, Snyder SE, et al. Molecular detection and analysis of exosomes using surface-enhanced raman scattering gold nanorods and a miniaturized device. Theranostics. 2018;8(10):2722–2738. doi: 10.7150/thno.21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu C, Zhao J, Tian F, Chang J, Zhang W, Sun J. λ-DNA- and aptamer-mediated sorting and analysis of extracellular vesicles. J Am Chem Soc. 2019;141(9):3817–3821. doi: 10.1021/jacs.9b00007. [DOI] [PubMed] [Google Scholar]
- 154.Jeon YR, Kim SY, Lee EJ, Kim YN, Noh DY, Park SY, et al. Identification of annexin II as a novel secretory biomarker for breast cancer. Proteomics. 2013;13(21):3145–3156. doi: 10.1002/pmic.201300127. [DOI] [PubMed] [Google Scholar]
- 155.Chaudhary P, Gibbs LD, Maji S, Lewis CM, Suzuki S, Vishwanatha JK. Serum exosomal-annexin A2 is associated with African-American triple-negative breast cancer and promotes angiogenesis. Breast Cancer Res. 2020;22(1):11. doi: 10.1186/s13058-020-1251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jung HH, Kim JY, Cho EY, Oh JM, Lee JE, Kim SW, et al. Elevated Level of Nerve Growth Factor (NGF) in Serum-Derived Exosomes Predicts Poor Survival in Patients with Breast Cancer Undergoing Neoadjuvant Chemotherapy. Cancers (Basel) 2021;13(21):5260. doi: 10.3390/cancers13215260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang X, Zhong W, Bu J, Li Y, Li R, Nie R, et al. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol Carcinog. 2019;58(5):674–685. doi: 10.1002/mc.22960. [DOI] [PubMed] [Google Scholar]
- 158.Moon PG, Lee JE, Cho YE, Lee SJ, Jung JH, Chae YS, et al. Identification of Developmental Endothelial Locus-1 on Circulating Extracellular Vesicles as a Novel Biomarker for Early Breast Cancer Detection. Clin Cancer Res. 2016;22(7):1757–1766. doi: 10.1158/1078-0432.CCR-15-0654. [DOI] [PubMed] [Google Scholar]
- 159.Khan S, Bennit HF, Turay D, Perez M, Mirshahidi S, Yuan Y, et al. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer. 2014;14:176. doi: 10.1186/1471-2407-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ando W, Kikuchi K, Uematsu T, Yokomori H, Takaki T, Sogabe M, et al. Novel breast cancer screening: combined expression of miR-21 and MMP-1 in urinary exosomes detects 95% of breast cancer without metastasis. Sci Rep. 2019;9(1):13595. doi: 10.1038/s41598-019-50084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ciravolo V, Huber V, Ghedini GC, Venturelli E, Bianchi F, Campiglio M, et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol. 2012;227(2):658–667. doi: 10.1002/jcp.22773. [DOI] [PubMed] [Google Scholar]
- 162.Marconi S, Santamaria S, Bartolucci M, Stigliani S, Aiello C, Gagliani MC, et al. Trastuzumab Modulates the Protein Cargo of Extracellular Vesicles Released by ERBB2(+) Breast Cancer Cells. Membranes (Basel) 2021;11(3):199. doi: 10.3390/membranes11030199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Drucker A, Yoo BH, Khan IA, Choi D, Montermini L, Liu X, et al. Trastuzumab-induced upregulation of a protein set in extracellular vesicles emitted by ErbB2-positive breast cancer cells correlates with their trastuzumab sensitivity. Breast Cancer Res. 2020;22(1):105. doi: 10.1186/s13058-020-01342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yang SJ, Wang DD, Li J, Xu HZ, Shen HY, Chen X, et al. Predictive role of GSTP1-containing exosomes in chemotherapy-resistant breast cancer. Gene. 2017;623:5–14. doi: 10.1016/j.gene.2017.04.031. [DOI] [PubMed] [Google Scholar]
- 165.Ning K, Wang T, Sun X, Zhang P, Chen Y, Jin J, et al. UCH-L1-containing exosomes mediate chemotherapeutic resistance transfer in breast cancer. J Surg Oncol. 2017;115(8):932–940. doi: 10.1002/jso.24614. [DOI] [PubMed] [Google Scholar]
- 166.Ma X, Chen Z, Hua D, He D, Wang L, Zhang P, et al. Essential role for TrpC5-containing extracellular vesicles in breast cancer with chemotherapeutic resistance. Proc Natl Acad Sci U S A. 2014;111(17):6389–6394. doi: 10.1073/pnas.1400272111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen Y, Wang L, Zhu Y, Chen Z, Qi X, Jin L, et al. Breast cancer resistance protein (BCRP)-containing circulating microvesicles contribute to chemoresistance in breast cancer. Oncol Lett. 2015;10(6):3742–3748. doi: 10.3892/ol.2015.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li T, Tao Z, Zhu Y, Liu X, Wang L, Du Y, et al. Exosomal annexin A6 induces gemcitabine resistance by inhibiting ubiquitination and degradation of EGFR in triple-negative breast cancer. Cell Death Dis. 2021;12(7):684. doi: 10.1038/s41419-021-03963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 170.Kavanagh EL, Lindsay S, Halasz M, Gubbins LC, Weiner-Gorzel K, Guang MHZ, et al. Protein and chemotherapy profiling of extracellular vesicles harvested from therapeutic induced senescent triple negative breast cancer cells. Oncogenesis. 2017;6(10):e388. doi: 10.1038/oncsis.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang Y, Wu X, Andy TW. Characterization and applications of extracellular vesicle proteome with post-translational modifications. Trends Analyt Chem. 2018;107:21–30. doi: 10.1016/j.trac.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Andaluz Aguilar H, Iliuk AB, Chen IH, Tao WA. Sequential phosphoproteomics and N-glycoproteomics of plasma-derived extracellular vesicles. Nat Protoc. 2020;15(1):161–180. doi: 10.1038/s41596-019-0260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Raulf N, Lucarelli P, Thavaraj S, Brown S, Vicencio JM, Sauter T, et al. Annexin A1 regulates EGFR activity and alters EGFR-containing tumour-derived exosomes in head and neck cancers. Eur J Cancer. 2018;102:52–68. doi: 10.1016/j.ejca.2018.07.123. [DOI] [PubMed] [Google Scholar]
- 174.van der Mijn JC, Sol N, Mellema W, Jimenez CR, Piersma SR, Dekker H, et al. Analysis of AKT and ERK1/2 protein kinases in extracellular vesicles isolated from blood of patients with cancer. J Extracell Vesicles. 2014;3:25657. doi: 10.3402/jev.v3.25657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sethi MK, Kim H, Park CK, Baker MS, Paik YK, Packer NH, et al. In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology. 2015;25(10):1064–1078. doi: 10.1093/glycob/cwv042. [DOI] [PubMed] [Google Scholar]
- 176.Rontogianni S, Synadaki E, Li B, Liefaard MC, Lips EH, Wesseling J, et al. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol. 2019;2:325. doi: 10.1038/s42003-019-0570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chen IH, Aguilar HA, Paez Paez JS, Wu X, Pan L, Wendt MK, et al. Analytical pipeline for discovery and verification of glycoproteins from plasma-derived extracellular vesicles as breast cancer biomarkers. Anal Chem. 2018;90(10):6307–6313. doi: 10.1021/acs.analchem.8b01090. [DOI] [PubMed] [Google Scholar]
- 178.Menck K, Scharf C, Bleckmann A, Dyck L, Rost U, Wenzel D, et al. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J Mol Cell Biol. 2015;7(2):143–153. doi: 10.1093/jmcb/mju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ham S, Lima LG, Chai EPZ, Muller A, Lobb RJ, Krumeich S, et al. Breast cancer-derived exosomes alter macrophage polarization via gp130/STAT3 signaling. Front Immunol. 2018;9:871. doi: 10.3389/fimmu.2018.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kavanagh EL, Halasz M, Dowling P, Withers J, Lindsay S, Higgins MJ, et al. N-Linked glycosylation profiles of therapeutic induced senescent (TIS) triple negative breast cancer cells (TNBC) and their extracellular vesicle (EV) progeny. Mol Omics. 2021;17(1):72–85. doi: 10.1039/D0MO00017E. [DOI] [PubMed] [Google Scholar]
- 181.Gámez-Valero A, Monguió-Tortajada M, Carreras-Planella L, Franquesa M, Beyer K, Borràs FE. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles' characteristics compared to precipitating agents. Sci Rep. 2016;6:33641. doi: 10.1038/srep33641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Simpson DM, Beynon RJ. Acetone precipitation of proteins and the modification of peptides. J Proteome Res. 2010;9(1):444–450. doi: 10.1021/pr900806x. [DOI] [PubMed] [Google Scholar]
- 183.Gallart-Palau X, Serra A, Wong AS, Sandin S, Lai MK, Chen CP, et al. Extracellular vesicles are rapidly purified from human plasma by PRotein Organic Solvent PRecipitation (PROSPR) Sci Rep. 2015;5:14664. doi: 10.1038/srep14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Xu B, Zhang Y, Zhao Z, Yoshida Y, Magdeldin S, Fujinaka H, et al. Usage of electrostatic eliminator reduces human keratin contamination significantly in gel-based proteomics analysis. J Proteomics. 2011;74(7):1022–1029. doi: 10.1016/j.jprot.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 186.Schmudlach A, Felton J, Cipolla C, Sun L, Kennedy RT, Dovichi NJ. Sample preparation protocol for bottom-up proteomic analysis of the secretome of the islets of Langerhans. Analyst. 2016;141(5):1700–1706. doi: 10.1039/C5AN02265G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fel A, Lewandowska AE, Petrides PE, Wiśniewski JR. Comparison of proteome composition of serum enriched in extracellular vesicles isolated from polycythemia Vera patients and healthy controls. Proteomes. 2019;7(2):20. doi: 10.3390/proteomes7020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wiśniewski JR. Quantitative Evaluation of Filter Aided Sample Preparation (FASP) and Multienzyme Digestion FASP Protocols. Anal Chem. 2016;88(10):5438–5443. doi: 10.1021/acs.analchem.6b00859. [DOI] [PubMed] [Google Scholar]
- 189.György B, Módos K, Pállinger E, Pálóczi K, Pásztói M, Misják P, et al. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood. 2011;117(4):e39–48. doi: 10.1182/blood-2010-09-307595. [DOI] [PubMed] [Google Scholar]
- 190.Yuana Y, Levels J, Grootemaat A, Sturk A, Nieuwland R. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J Extracell Vesicles. 2014;3:23262. doi: 10.3402/jev.v3.23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sódar BW, Kittel Á, Pálóczi K, Vukman KV, Osteikoetxea X, Szabó-Taylor K, et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep. 2016;6:24316. doi: 10.1038/srep24316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci. 2018;75(15):2873–2886. doi: 10.1007/s00018-018-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang X, Borg EGF, Liaci AM, Vos HR, Stoorvogel W. A novel three step protocol to isolate extracellular vesicles from plasma or cell culture medium with both high yield and purity. J Extracell Vesicles. 2020;9(1):1791450. doi: 10.1080/20013078.2020.1791450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 196.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3(4):563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 197.Gyvyte U, Juzenas S, Salteniene V, Kupcinskas J, Poskiene L, Kucinskas L, et al. MiRNA profiling of gastrointestinal stromal tumors by next-generation sequencing. Oncotarget. 2017;8(23):37225–37238. doi: 10.18632/oncotarget.16664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sørensen SS, Nygaard AB, Nielsen MY, Jensen K, Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with acute ischemic stroke. Transl Stroke Res. 2014;5(6):711–718. doi: 10.1007/s12975-014-0364-8. [DOI] [PubMed] [Google Scholar]
- 199.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chu A, Robertson G, Brooks D, Mungall AJ, Birol I, Coope R, et al. Large-scale profiling of microRNAs for The Cancer Genome Atlas. Nucleic Acids Res. 2016;44(1):e3. doi: 10.1093/nar/gkv808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hu Y, Lan W, Miller D. Next-generation sequencing for MicroRNA expression profile. Methods Mol Biol. 2017;1617:169–177. doi: 10.1007/978-1-4939-7046-9_12. [DOI] [PubMed] [Google Scholar]
- 202.Bao S, Hu T, Liu J, Su J, Sun J, Ming Y, et al. Genomic instability-derived plasma extracellular vesicle-microRNA signature as a minimally invasive predictor of risk and unfavorable prognosis in breast cancer. J Nanobiotechnology. 2021;19(1):22. doi: 10.1186/s12951-020-00767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu X, Cao M, Palomares M, Wu X, Li A, Yan W, et al. Metastatic breast cancer cells overexpress and secrete miR-218 to regulate type I collagen deposition by osteoblasts. Breast Cancer Res. 2018;20(1):127. doi: 10.1186/s13058-018-1059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Buschmann D, González R, Kirchner B, Mazzone C, Pfaffl MW, Schelling G, et al. Glucocorticoid receptor overexpression slightly shifts microRNA expression patterns in triple-negative breast cancer. Int J Oncol. 2018;52(6):1765–1776. doi: 10.3892/ijo.2018.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33–42. doi: 10.1016/j.canlet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 206.Li M, Zou X, Xia T, Wang T, Liu P, Zhou X, et al. A five-miRNA panel in plasma was identified for breast cancer diagnosis. Cancer Med. 2019;8(16):7006–7017. doi: 10.1002/cam4.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Eichelser C, Stückrath I, Müller V, Milde-Langosch K, Wikman H, Pantel K, et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5(20):9650–9663. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang M, Zhang H, Yang F, Qiu R, Zhao X, Gong Z, et al. miR-188-5p suppresses cellular proliferation and migration via IL6ST: A potential noninvasive diagnostic biomarker for breast cancer. J Cell Physiol. 2020;235(5):4890–4901. doi: 10.1002/jcp.29367. [DOI] [PubMed] [Google Scholar]
- 209.Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18(1):90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 211.Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, et al. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117(1):131–140. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 212.Shen S, Song Y, Zhao B, Xu Y, Ren X, Zhou Y, et al. Cancer-derived exosomal miR-7641 promotes breast cancer progression and metastasis. Cell Commun Signal. 2021;19(1):20. doi: 10.1186/s12964-020-00700-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kia V, Paryan M, Mortazavi Y, Biglari A, Mohammadi-Yeganeh S. Evaluation of exosomal miR-9 and miR-155 targeting PTEN and DUSP14 in highly metastatic breast cancer and their effect on low metastatic cells. J Cell Biochem. 2019;120(4):5666–5676. doi: 10.1002/jcb.27850. [DOI] [PubMed] [Google Scholar]
- 214.Santos JC, Lima NDS, Sarian LO, Matheu A, Ribeiro ML, Derchain SFM. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep. 2018;8(1):829. doi: 10.1038/s41598-018-19339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem. 2013;59(10):1489–1496. doi: 10.1373/clinchem.2013.205161. [DOI] [PubMed] [Google Scholar]
- 216.Yoshikawa M, Iinuma H, Umemoto Y, Yanagisawa T, Matsumoto A, Jinno H. Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol Lett. 2018;15(6):9584–9592. doi: 10.3892/ol.2018.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sueta A, Yamamoto Y, Tomiguchi M, Takeshita T, Yamamoto-Ibusuki M, Iwase H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget. 2017;8(41):69934–69944. doi: 10.18632/oncotarget.19482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ni Q, Stevic I, Pan C, Müller V, Oliveira-Ferrer L, Pantel K, et al. Different signatures of miR-16, miR-30b and miR-93 in exosomes from breast cancer and DCIS patients. Sci Rep. 2018;8(1):12974. doi: 10.1038/s41598-018-31108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yan C, Hu J, Yang Y, Hu H, Zhou D, Ma M, et al. Plasma extracellular vesicle-packaged microRNAs as candidate diagnostic biomarkers for early-stage breast cancer. Mol Med Rep. 2019;20(5):3991–4002. doi: 10.3892/mmr.2019.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS ONE. 2014;9(4):e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y, et al. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res Treat. 2014;147(2):423–431. doi: 10.1007/s10549-014-3037-0. [DOI] [PubMed] [Google Scholar]
- 222.Chen WX, Xu LY, Cheng L, Qian Q, He X, Peng WT, et al. Bioinformatics analysis of dysregulated microRNAs in exosomes from docetaxel-resistant and parental human breast cancer cells. Cancer Manag Res. 2019;11:5425–5435. doi: 10.2147/CMAR.S201335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Shen M, Dong C, Ruan X, Yan W, Cao M, Pizzo D, et al. Chemotherapy-induced extracellular vesicle mirnas promote breast cancer stemness by targeting ONECUT2. Cancer Res. 2019;79(14):3608–3621. doi: 10.1158/0008-5472.CAN-18-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yang Q, Zhao S, Shi Z, Cao L, Liu J, Pan T, et al. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J Exp Clin Cancer Res. 2021;40(1):120. doi: 10.1186/s13046-021-01901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Backes C, Sedaghat-Hamedani F, Frese K, Hart M, Ludwig N, Meder B, et al. Bias in high-throughput analysis of miRNAs and Implications for biomarker studies. Anal Chem. 2016;88(4):2088–2095. doi: 10.1021/acs.analchem.5b03376. [DOI] [PubMed] [Google Scholar]
- 226.Buschmann D, Kirchner B, Hermann S, Märte M, Wurmser C, Brandes F, et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J Extracell Vesicles. 2018;7(1):1481321. doi: 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Park S, Lee K, Park IB, Kim NH, Cho S, Rhee WJ, et al. The profiles of microRNAs from urinary extracellular vesicles (EVs) prepared by various isolation methods and their correlation with serum EV microRNAs. Diabetes Res Clin Pract. 2020;160:108010. doi: 10.1016/j.diabres.2020.108010. [DOI] [PubMed] [Google Scholar]
- 228.Kuhlmann JD, Chebouti I, Kimmig R, Buderath P, Reuter M, Puppel SH, et al. Extracellular vesicle-associated miRNAs in ovarian cancer - design of an integrated NGS-based workflow for the identification of blood-based biomarkers for platinum-resistance. Clin Chem Lab Med. 2019;57(7):1053–1062. doi: 10.1515/cclm-2018-1048. [DOI] [PubMed] [Google Scholar]
- 229.Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014;86(2):433–444. doi: 10.1038/ki.2013.502. [DOI] [PubMed] [Google Scholar]
- 230.Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, et al. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16(11):2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Avendaño-Vázquez SE, Flores-Jasso CF. Stumbling on elusive cargo: how isomiRs challenge microRNA detection and quantification, the case of extracellular vesicles. J Extracell Vesicles. 2020;9(1):1784617. doi: 10.1080/20013078.2020.1784617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Backes C, Haas J, Leidinger P, Frese K, Großmann T, Ruprecht K, et al. miFRame: analysis and visualization of miRNA sequencing data in neurological disorders. J Transl Med. 2015;13:224. doi: 10.1186/s12967-015-0594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8(6):1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 234.Koppers-Lalic D, Hackenberg M, de Menezes R, Misovic B, Wachalska M, Geldof A, et al. Non-invasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget. 2016;7(16):22566–22578. doi: 10.18632/oncotarget.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li D, Liu J, Guo B, Liang C, Dang L, Lu C, et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun. 2016;7:10872. doi: 10.1038/ncomms10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shi W, Jin X, Wang Y, Zhang Q, Yang L. High serum exosomal long non-coding RNA DANCR expression confers poor prognosis in patients with breast cancer. J Clin Lab Anal. 2022;36(3):e24186. doi: 10.1002/jcla.24186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wang B, Mao JH, Wang BY, Wang LX, Wen HY, Xu LJ, et al. Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-κB signaling pathway. Cancer Lett. 2020;489:87–99. doi: 10.1016/j.canlet.2020.05.038. [DOI] [PubMed] [Google Scholar]
- 238.Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20(3):332–343. doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhang Q, Jeppesen DK, Higginbotham JN, Graves-Deal R, Trinh VQ, Ramirez MA, et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat Cell Biol. 2021;23(12):1240–1254. doi: 10.1038/s41556-021-00805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.