Summary
Background
BGB-DXP593, a neutralising monoclonal antibody against SARS-CoV-2, has demonstrated strong activity in reducing viral RNA copy number in SARS-CoV-2-infected animal models. We aimed to examine the efficacy and safety of BGB-DXP593 in ambulatory patients with mild-to-moderate COVID-19.
Methods
This global, randomised, double-blind, phase 2 study (ClinicalTrials.govNCT04551898) screened patients from 20 sites in Australia, Brazil, Mexico, South Africa, and the USA from December 2, 2020, through January 25, 2021. Patients with a first-positive SARS-CoV-2 test (positive reverse transcription–polymerase chain reaction test or authorised antigen test) ≤3 days before screening and mild-to-moderate COVID-19 symptoms for ≤7 days before treatment were randomised 1:1:1:1 to receive a single intravenous infusion of BGB-DXP593 5, 15, or 30 mg/kg, or placebo. The primary endpoint was change from baseline to Day 8 in viral RNA copies/mL as measured in nasopharyngeal swabs. Secondary endpoints were hospitalisation rate due to worsening COVID-19 and treatment-emergent adverse events (TEAEs). A prespecified exploratory endpoint was change in viral RNA copy number in saliva.
Findings
Relative to the natural rate of clearance as assessed in placebo-exposed patients (−3.12 log10 copies/mL), no significant differences in nasopharygneal viral RNA copy number changes were observed (−2.93 to −3.63 log10 copies/mL) by Day 8 in BGB-DXP593-treated patients. Reductions from baseline to Day 8 in saliva viral RNA copy number were larger with BGB-DXP593 5 mg/kg (−1.37 log10 copies/mL [90% confidence interval −2.14, −0.61]; nominal p = 0.003) and 15 mg/kg (−1.26 [−2.06, −0.46]; nominal p = 0.01) vs placebo, and differences favoring BGB-DXP593 were observed by Day 3, although not statistically significant; no difference from placebo was observed for BGB-DXP593 30 mg/kg (−0.71 [−1.45, 0.04]; nominal p = 0.12). Hospitalisation rate due to COVID-19 was numerically lower with BGB-DXP593 (pooled: 2/134 patients; 1.5%) vs placebo (2/47 patients; 4.3%), although not statistically significant. Incidence of TEAEs was similar across treatment groups. No TEAE led to treatment discontinuation. Five serious TEAEs occurred, all attributed to COVID-19 pneumonia.
Interpretation
BGB-DXP593 was well tolerated. Although nasopharyngeal swab SARS-CoV-2 viral RNA copy number was not significantly decreased compared with placebo, viral RNA copy number was inconsistently reduced by Day 8 in saliva at some doses as low as 5 mg/kg.
Funding
BeiGene, Ltd.
Keywords: SARS-CoV-2, Neutralising monoclonal antibody, BGB-DXP593, Viral load or viral RNA copy number, COVID-19, Phase 2 trial
Research in context.
Evidence before this study
We searched PubMed and Google Scholar and reviewed the National Institutes of Health COVID-19 Treatment Guidelines for clinical trials up to March 9, 2022, that reported the efficacy and safety of anti-SARS-CoV-2–neutralising antibodies for the treatment of mild-to-moderate COVID-19. We identified five pivotal trials for five currently recommended neutralising antibodies or antibody combinations: bamlanivimab plus etesevimab, casirivimab plus imdevimab, sotrovimab, bebtelovimab, and regdanvimab (appendix p 1). Mutations in SARS-CoV-2 may confer resistance to antibody therapies. In the face of the ongoing COVID-19 pandemic, development of new neutralising antibodies will provide high-risk patients additional therapeutic options to minimise virulence, and potentially reduce the morbidity and mortality associated with COVID-19.
Added value of this study
BGB-DXP593 is a highly potent, human neutralising immunoglobulin G1 monoclonal antibody against SARS-CoV-2 and has demonstrated strong activity in reducing viral RNA copy number in SARS-CoV-2-infected animal models. The present multicentre, randomised, double-blind, phase 2 study in patients with recently diagnosed mild-to-moderate COVID-19 showed that no significant differences in viral RNA copy number reduction were observed for BGB-DXP593 vs placebo in nasopharyngeal swabs. Exploratory analysis of salivary viral RNA copy number response, with inconsistent dose effects at 2 of 3 dosing levels, was noted. We suggest that additional investigations of response of SARS-CoV-2 salivary viral RNA copy number to treatment with appropriately selected neutralising monoclonal antibodies may be considered.
Implications of all the available evidence
BGB-DXP593 was well tolerated. Nasopharyngeal swab SARS-CoV-2 viral RNA copy number was not significantly decreased by BGB-DXP593 compared with placebo, but saliva SARS-CoV-2 viral RNA copy number was reduced by Day 8 by BGB-DXP593 at doses as low as 5 mg/kg. Although BGB-DXP593 has been escaped by SARS-CoV-2 viral evolution, as is the case for most, if not all, therapeutic and prophylactic antibodies commercially available, conceptually, these results contribute to our understanding of the virologic response to antispike neutralising monoclonal antibodies appropriately selected against susceptible variants at the time of this study.
Introduction
COVID-19 is an acute respiratory infection caused by SARS-CoV-2 that continues to spread rapidly worldwide.1 Patients infected with SARS-CoV-2 display a wide range of disease severity, with most patients asymptomatic or showing only mild symptoms of fever, cough, shortness of breath, or gastrointestinal symptoms. Some patients can rapidly progress from asymptomatic or mild infection to acute respiratory distress requiring hospitalisation.2 Patients with advanced age and chronic health conditions such as cardiovascular disease, diabetes mellitus, immunosuppression, and obesity are more likely to become critically ill, which is associated with a high mortality rate.3
Several treatment options have been recommended for hospitalised patients with COVID-19, including the antiviral agent remdesivir, dexamethasone (recommended only for patients with hypoxaemia), and other immunomodulators (for selected patients with hypoxaemia, as an adjunct to dexamethasone).4 For mild-to-moderate COVID-19, anti-SARS-CoV-2 antibodies including bamlanivimab plus etesevimab, casirivimab plus imdevimab, sotrovimab, and bebtelovimab were initially recommended by the National Institutes of Health for patients at high risk for progressing to severe COVID-19. Mutations in SARS-CoV-2 have, however, been shown to confer resistance to antibody therapies.5,6 The emergence of Omicron variants has reduced or eliminated the efficacy of all of the aforementioned treatment options except bebtelovimab. As a result, bebtelovimab is the only anti-SARS-CoV-2 antibody treatment currently recommended by the National Institutes of Health, but its use is restricted to situations where ritonavir-boosted nirmatrelvir and remdesivir are not available, feasible to use, or clinically appropriate (eg, due to potential drug interactions), and to settings where recipients can be monitored/treated for hypersensitivity reactions.4,7 In vitro studies have shed light on Omicron's emergence, revealing that the B.1.1.529 variant contains ≥15 mutations and escapes neutralisation by most existing SARS-CoV-2 antibodies,7 although T-cell immunity against Omicron induced on vaccination or natural infection has been largely retained.8,9 In the face of the ongoing COVID-19 pandemic with potential new SARS-CoV-2 variants circulating, there is a need for safe and effective therapeutic antibodies in high-risk patients, especially in areas with low or ineffective vaccination rates.
The pathogenesis of COVID-19 is thought to be primarily driven by replication of SARS-CoV-2 early in the clinical course (later disease stages and long-term sequelae appear to be driven by a dysregulated immune/inflammatory response to SARS-CoV-2). Antiviral therapies are, therefore, expected to provide the greatest potential for clinical benefit and limiting disease progression during the early stages of infection.4,10 Many patients with COVID-19 produce endogenous neutralising antibodies (nAbs) to SARS-CoV-2 about 10 days after disease onset.11 Convalescent plasma therapy has been used to treat COVID-19 by passively transferring nAbs,4 but dose-finding data are lacking and its use has not been widely accepted. Exogenously provided neutralising monoclonal antibodies with high affinity to the virus may produce more specific and consistent treatment outcomes. SARS-CoV-2 infects host cells through binding of its spike (S) protein receptor-binding domain (RBD) to angiotensin-converting enzyme-2 (ACE2) expressed on host cells, resulting in virus-host cell membrane fusion and viral invasion.12,13 To date, recommended anti-SARS-CoV-2 antibody-based therapies are all neutralising monoclonal antibodies that target different epitopes in the RBD of the SARS-CoV-2 S protein.4
BGB-DXP593 is a highly potent, human neutralising immunoglobulin G1 monoclonal antibody identified from convalescent COVID-19 patients’ B cells, which is specific for the SARS-CoV-2 S protein.14 It has two identical κ light chains with 2 identical heavy chains covalently interlinked by 4 pairs of disulfide bonds. In vitro assays demonstrated that BGB-DXP593 binds to the RBD of the SARS-CoV-2 S protein with high specificity and affinity, efficiently blocking the binding of RBD to ACE2 and preventing virus from entering host cells. In vivo studies showed strong therapeutic antiviral activity of BGB-DXP593 in reducing viral RNA copy number in SARS-CoV-2-infected humanised ACE2-transgenic mice. A first-in-human phase 1 study of BGB-DXP593 in healthy patients has been completed and no serious treatment-emergent adverse events (TEAEs) were observed (ClinicalTrials.gov Identifier: NCT04532294).
Recent data have shown that higher prevalence of detectable SARS-CoV-2 plasma viral load is associated with worse respiratory disease severity in hospitalised patients and increased risk of mortality,10,15,16 suggesting that complications and death may be partially due to high viral loads. The present global phase 2 study was conducted to evaluate the efficacy of BGB-DXP593 in reducing SARS-CoV-2 viral RNA copy number and its safety profiles in conventional nasopharyngeal swab and noninvasive saliva samples of patients with recently diagnosed mild-to-moderate COVID-19.
Methods
Study design and oversight
This phase 2, randomised, double-blind, placebo-controlled study was conducted at 20 study sites in Australia, Brazil, Mexico, South Africa, and the USA (ClinicalTrials.gov NCT04551898). The trial was conducted in accordance with the International Conference on Harmonization Guidelines for Good Clinical Practice, the Declaration of Helsinki, and applicable local regulations. The protocol was reviewed and approved by the institutional review boards and ethics committees of participating centres, and all patients provided written informed consent before study enrolment.
All patients were randomised in a 1:1:1:1 ratio to receive placebo, or BGB-DXP593 5, 15, or 30 mg/kg. The doses of BGB-DXP593 were based on an overall assessment of in vitro virus neutralisation activity and toxicology data (appendix p 2). Treatment was administered within 7 days of symptom onset. Patients received a single intravenous infusion of BGB-DXP593 or matched placebo over 30–90 min and were followed to the end of the study on Day 113 (±7 days) or within 7 days of premature discontinuation. The present analysis used data extracted at the database lock date of July 13, 2021, when all randomised patients had completed the study or discontinued the study earlier.
Randomisation and masking
Randomisation was generated by site personnel using an interactive response technology system, and stratified by country and disease severity (mild vs moderate COVID-19 symptoms). Except for the pharmacist who dispensed the study drug, treatment assignment in the study was blinded to the investigators, patients, medical staff, and sponsor.
Patients
Eligible patients were aged ≥18 years, had laboratory-confirmed SARS-CoV-2 infection (positive reverse transcription–polymerase chain reaction [RT-PCR] test or authorised antigen testing methods) ≤3 days prior to screening, and mild-to-moderate COVID-19 symptoms for ≤7 days prior to treatment assignments (appendix p 2). Mild-to-moderate COVID-19 symptoms were defined per the National Institutes of Health COVID-19 Treatment Guidelines, which include symptoms such as fever, cough, shortness of breath, sore throat, diarrhea, vomiting, and dysgeusia.4 Patients who had severe COVID-19 and oxygen saturation ≤93% on room air, ratio of arterial oxygen partial pressure to fractional inspired oxygen <300 mm Hg, respiratory rate ≥30/min, heart rate ≥125 beats/min, or history of a positive SARS-CoV-2 test (RT-PCR or other testing authorised by local regulatory authorities) prior to the one serving as eligibility for this study were excluded (appendix p 2).
Study endpoints and assessments
The primary endpoint was change from baseline to Day 8 (±1 day) in SARS-CoV-2 as measured by RT-quantitative PCR (RT-qPCR) in nasopharyngeal swab samples. Secondary endpoints included change in SARS-CoV-2 as measured by RT-qPCR in nasopharyngeal swab samples from baseline to Day 15 (±1 day); time to negative RT-qPCR in all tested samples with no subsequent positive RT-qPCR; proportion of patients requiring hospitalisation due to worsening COVID-19 infection; time to resolution of all COVID-19-related symptoms; incidence and severity of TEAEs and serious TEAEs; and pharmacokinetics (PK) and immunogenicity of BGB-DXP593. Prespecified exploratory endpoints included changes in SARS-CoV-2 levels as measured by RT-qPCR in saliva samples from baseline to Days 8 and 15.
A prespecified subgroup analysis of changes in SARS-CoV-2 levels as measured by RT-qPCR in nasopharyngeal swab samples from baseline to Days 3, 8, and 15 was conducted according to risk status at baseline (high and low risk). High risk was defined as patients who met ≥1 of the following criteria: body mass index ≥35 kg/m2; chronic kidney disease; diabetes mellitus; immunosuppressive disease; currently receiving immunosuppressive treatment; aged ≥65 years; and aged ≥55 years with cardiovascular disease, hypertension, chronic obstructive pulmonary disease, or other chronic respiratory disease.
Nasopharyngeal swab and saliva samples were collected on Day 1 pre-dose (baseline), and Days 3, 8, and 15. Testing was performed at a central laboratory for the quantitative detection of nucleic acid from SARS-CoV-2 using the MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit on the KingFisher system and reagents from the TaqPath™ COVID-19 Combo Kit, which includes the TaqPath RT-PCR COVID-19 Kit (ThermoFisher Scientific Inc., Waltham, Massachusetts, USA). Safety was determined by monitoring TEAEs, laboratory values, vital signs, physical examinations, and electrocardiographic findings. Blood was collected on Day 1 pre- and post-dose, Days 3, 8, 15, and 29, and end of study to characterise the serum PK profile of BGB-DXP593 using a validated electrochemiluminescence assay, with biotinylated anti-DXP593 monoclonal antibody as capture reagent, and anti-DXP593 rabbit polyclonal antibody and MSD® SULFO-TAG Labeled Anti-Rabbit Antibody (Goat; Meso Scale Discovery) as detection reagents. Blood was also collected on Day 1 pre-dose, Days 15 and 29, and end of study for evaluating antidrug antibodies (ADAs) against BGB-DXP593 using validated electrochemiluminescence assays to confirm the presence of anti-DXP593 antibodies.
Statistical analysis
Efficacy analyses were performed in the intent-to-treat population, including all randomised patients. Safety was assessed in the safety analysis set, including all patients who received BGB-DXP593 or placebo. Pharmacokinetics was assessed in the PK analysis set, including all patients who received the study drug and had postdose PK data available. Antidrug antibodies were evaluated in the ADA analysis set, including all patients who received the study drug and had both baseline ADA and ≥1 post-baseline ADA result available.
The statistical hypotheses tested for the primary endpoint were as follows.
-
•
H0: a flat dose–response curve comparing change from baseline to Day 8 in SARS-CoV-2 levels in the placebo and BGB-DXP593 dose groups.
-
•
H1: a nonflat dose–response curve indicating a benefit of BGB-DXP593 over placebo.
A mixed model repeated measures (MMRM) analysis was used to analyse the primary endpoint of least-squares (LS) mean change from baseline to Day 8 in SARS-CoV-2 levels. The primary analysis used methodology employing both multiple comparison procedures and modelling techniques (MCPMod17) to simultaneously evaluate different predefined dose–response patterns for dose finding, while protecting the overall probability of Type I error (1-sided α: 5%; appendix p 3: Fig. S1). The null hypothesis of a flat dose–response relationship over viral level change at Day 8 would be rejected if ≥ 1 model was statistically significant. Target doses could be estimated from the average model by incorporating information on the minimal clinically relevant effect and accounting for safety.
Sample size calculation was based on the assumption that the maximal treatment effect size of BGB-DXP593 vs placebo in LS mean change from baseline to Day 8 in SARS-CoV-2 levels was −1.5 log10 copies/mL in nasopharyngeal swab samples. Forty-one patients were required with primary endpoint data per arm to provide ≥82% power to detect a nonflat dose–response relationship at a 1-sided significance level of 5%. At least 43 patients/arm were required for randomisation to account for a potential 5% dropout rate.
Differences between BGB-DXP593 and placebo—along with 90% confidence intervals (CIs) and p-values—in LS mean changes from baseline to Day 8 in SARS-CoV-2 levels from nasopharyngeal swab samples were estimated using the MMRM model as supportive analyses of the primary endpoint to supplement the MCPMod analysis. The MMRM analysis was also used to analyse LS mean changes in SARS-CoV-2 levels from baseline to Day 15 in nasopharyngeal swab samples and to Day 8 or 15 in saliva samples. Differences between BGB-DXP593 and placebo were estimated, and corresponding 90% CIs and p-values were calculated. The Kaplan–Meier method was used to assess time to negative RT-qPCR and time to resolution of all COVID-19-related symptoms. The proportion of patients requiring hospitalisation due to worsened COVID-19 was presented with a 2-sided binomial exact 90% CI. Since the study enrolled patients with mild-to-moderate COVID-19, it was anticipated that a low number of patients would be hospitalised due to COVID-19 and thus no formal statistical testing was planned for comparison of hospitalisation rates between treatment groups. Adverse events and immunogenicity were summarised descriptively. Pharmacokinetics was assessed by noncompartmental analysis of serial serum concentrations using Phoenix® WinNonlin® v 7.0 or higher (Certara, Princeton, NJ, USA).
Role of the funding source
Beigene, Ltd. developed the protocol, and was involved in the collection, analysis, and interpretation of data, writing the report, and the decision to submit the paper for publication. All authors had full access to all of the data in the study and accepted responsibility for the decision to submit the final manuscript for publication.
Results
Patients
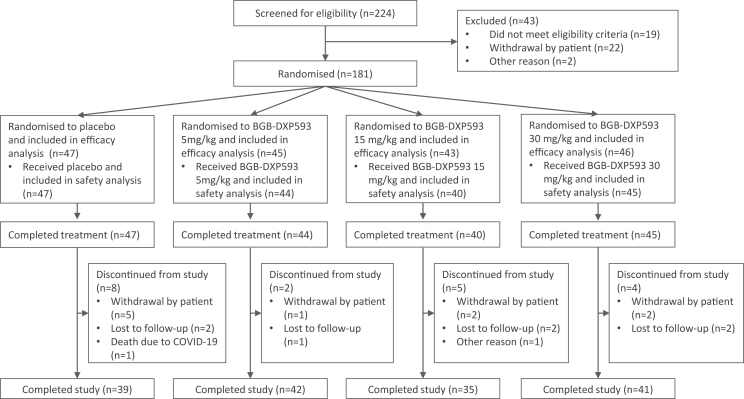
From December 2, 2020 up until January 25, 2021, 224 patients from Australia, Brazil, Mexico, South Africa, and the USA were screened; 181 patients were randomised to receive placebo (n = 47), or BGB-DXP593 5 mg/kg (n = 45), 15 mg/kg (n = 43), or 30 mg/kg (n = 46), and were included in the efficacy analysis (Fig. 1). Five patients were randomised but not treated; 176 patients received and completed the assigned treatment and were included in the safety analysis. Nineteen patients discontinued from the study, with the most common reasons being withdrawal by the patient (n = 10) and lost to follow-up (n = 7); 157 patients (87%) completed the Day 113 visit.
Fig. 1.
Patient disposition.
Treatment groups were generally well balanced at enrollment (Table 1). Median age of patients was 43 years; 93% were <65 years. About half of patients (48%) were women and most (81%) were White. At baseline, 79% of patients had mild COVID-19 symptoms, 35% had a viral RNA copy number >105 copies/mL in nasopharyngeal swab samples, and 27% were at high risk for progressing to severe COVID-19 symptoms. When patients were screened for presence of preexisting immunoglobulin-G antibodies against the SARS-CoV-2 S1 and S2 epitope domains of the spike protein, 82% were serum antibody negative at study entry. Patients received an infusion of placebo or BGB-DXP593 within a median of 5 days after the first COVID-19 symptom.
Table 1.
Demographic and baseline characteristics.
| Placebo (n = 47) | BGB-DXP593 |
Total (N = 181) | |||
|---|---|---|---|---|---|
| 5 mg/kg (n = 45) | 15 mg/kg (n = 43) | 30 mg/kg (n = 46) | |||
| Age | |||||
| Median, y (range) | 42 (18–77) | 46 (18–92) | 43 (21–77) | 39 (18–77) | 43 (18–92) |
| ≥65 y, n (%) | 4 (9) | 6 (13) | 1 (2) | 1 (2) | 12 (7) |
| Sex, n (%) | |||||
| Men | 27 (57) | 28 (62) | 21 (49) | 19 (41) | 95 (52) |
| Women | 20 (43) | 17 (38) | 22 (51) | 27 (59) | 86 (48) |
| Race, n (%) | |||||
| White | 40 (85) | 35 (78) | 39 (91) | 33 (72) | 147 (81) |
| Black or African-American | 3 (6) | 5 (11) | 2 (5) | 5 (11) | 15 (8) |
| Asian, American Indian, or Alaska Native | 3 (6) | 3 (7) | 1 (2) | 2 (4) | 9 (5) |
| Unknown or other | 1 (2) | 2 (4) | 1 (2) | 6 (13) | 10 (6) |
| Median weight, kg (range)a | (n = 47) 77.0 (55.5–137.0) |
(n = 44) 83.7 (54.8–107.0) |
(n = 40) 77.1 (55.6–156.0) |
(n = 45) 81.6 (48.5–127.0) |
(n = 176) 79.5 (48.5–156.0) |
| Country, n (%) | |||||
| Brazil | 5 (11) | 5 (11) | 4 (9) | 5 (11) | 19 (10) |
| Mexico | 3 (6) | 4 (9) | 3 (7) | 4 (9) | 14 (8) |
| South Africa | 4 (9) | 3 (7) | 3 (7) | 3 (7) | 13 (7) |
| USA | 35 (74) | 33 (73) | 33 (77) | 34 (74) | 135 (75) |
| Baseline disease severity, n (%) | |||||
| Mild COVID-19 | 35 (74) | 35 (78) | 35 (81) | 38 (83) | 143 (79) |
| Moderate COVID-19 | 12 (26) | 9 (20) | 5 (12) | 7 (15) | 33 (18) |
| Missing | 0 | 1 (2) | 3 (7) | 1 (2) | 5 (3) |
| Median time from 1st COVID-19 symptom to study drug administration, d (range) | (n = 47) 6 (2–10) |
(n = 44) 5 (2–9) |
(n = 40) 4 (2–8) |
(n = 45) 5 (3–11) |
(n = 176) 5 (2–11) |
| Baseline anti-SARS-CoV-2 IgG, n (%) | |||||
| Negative | 40 (85) | 37 (82) | 32 (74) | 40 (87) | 149 (82) |
| Positive | 7 (15) | 5 (11) | 7 (16) | 5 (11) | 24 (13) |
| Missing | 0 | 3 (7) | 4 (9) | 1 (2) | 8 (4) |
| Baseline SARS-CoV-2 viral RNA copy numberb | |||||
| Viral RNA copy number in nasopharyngeal swab samples >105 copies/mL, n (%) | 15 (32) | 16 (36) | 17 (40) | 15 (33) | 63 (35) |
| Mean in nasopharyngeal swab samples, log10 copies/mL (SD) | (n = 46) 4.74 (2.46) |
(n = 43) 5.20 (2.36) |
(n = 37) 5.04 (2.98) |
(n = 44) 4.81 (2.72) |
(n = 170) 4.94 (2.61) |
| Mean in saliva samples, log10 copies/mL | (n = 42) 4.49 (2.24) |
(n = 40) 4.99 (2.08) |
(n = 33) 4.11 (2.66) |
(n = 44) 4.21 (2.72) |
(n = 159) 4.46 (2.44) |
| High risk for progression to severe COVID-19, n (%)c | 13 (28) | 15 (33) | 9 (21) | 11 (24) | 48 (27) |
Weight for determining drug administration.
Viral RNA copy number value that was below the lower limit of detection was imputed as log10 (1) or 0, while viral RNA copy number value that was above the upper limit of detection was imputed as log10 (5.5∗108).
Defined as patients who met ≥1 of the following criteria: body mass index ≥35 kg/m2; chronic kidney disease; diabetes mellitus; immunosuppressive disease; currently receiving immunosuppressive treatment; ≥65 years of age; or ≥55 years of age with cardiovascular disease, hypertension, chronic obstructive pulmonary disease, or other chronic respiratory disease. IgG, immunoglobulin g; SD, standard deviation.
Viral RNA copy number in nasopharyngeal swab samples
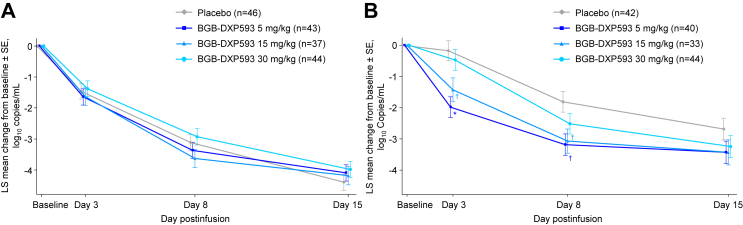
Patients in both the BGB-DXP593 and placebo groups exhibited reductions in viral RNA copy number in nasopharyngeal swab samples by Day 8. The LS mean reductions from baseline to Day 8 in viral RNA copy number were −3.12 log10 copies/mL for the placebo group, and −3.37, −3.63, and −2.93 log10 copies/mL for the BGB-DXP593 5, 15, and 30 mg/kg groups, respectively (Table 2; Fig. 2). Compared with the placebo group, however, changes from baseline to Day 8 in viral RNA copy number were not significantly different for any of the BGB-DXP593 dose groups. By Day 15, viral RNA copy number was decreased even more for the entire population, but there were no statistically significant differences between BGB-DXP593 and placebo in the reduction of viral RNA copy number from baseline.
Table 2.
Efficacy of BGB-DXP593 on SARS-CoV-2.
| Placebo (n = 47) | BGB-DXP593 |
|||
|---|---|---|---|---|
| 5 mg/kg (n = 45) | 15 mg/kg (n = 43) | 30 mg/kg (n = 46) | ||
| Primary endpoint | ||||
| Change in viral RNA copy number from baseline to Day 8 in nasopharyngeal swab samples, log10 copies/mL | ||||
| n | 41 | 42 | 34 | 42 |
| LS mean (SE)a | −3.12 (0.26) | −3.37 (0.26) | −3.63 (0.29) | −2.93 (0.26) |
| Difference vs placebo, (90% CI)a | .. | −0.25 (−0.84, 0.34) | −0.51 (−1.13, 0.11) | 0.19 (−0.40, 0.77) |
| p-valuea | .. | 0.48 | 0.17 | 0.60 |
| Secondary endpoints | ||||
| Change in viral RNA copy number from baseline to Day 15 in nasopharyngeal swab samples, log10 copies/mL | ||||
| n | 41 | 41 | 32 | 42 |
| LS mean (SE)a | −4.39 (0.26) | −4.09 (0.26) | −4.18 (0.30) | −3.98 (0.26) |
| Difference vs placebo, (90% CI)a | .. | 0.30 (−0.29, 0.89) | 0.22 (−0.41, 0.84) | 0.41 (−0.18, 1.00) |
| Nominal p-valuea | .. | 0.40 | 0.57 | 0.25 |
| Median time to negative RT-qPCR in all tested samples, d (90% CI) | 17 (15, 19) | 15 (13, 17) | 10 (8, 16) | 17 (15, 17) |
| Proportion of patients requiring hospitalisation due to worsened COVID-19 | ||||
| Patients hospitalised due to worsened COVID-19, n | 2 | 1 | 1 | 0 |
| Hospitalisation rate, % (90% CI)b | 4.3 (0.8, 12.8) | 2.2 (0.1, 10.1) | 2.3 (0.1, 10.6) | 0.0 (0.0, 6.3) |
| Exploratory endpoints | ||||
| Change in viral RNA copy number from baseline to Day 8 in saliva samples, log10 copies/mL | ||||
| n | 37 | 36 | 30 | 40 |
| LS mean (SE)a | −1.81 (0.33) | −3.19 (0.35) | −3.07 (0.38) | −2.52 (0.33) |
| Difference vs placebo, (90% CI)a | .. | −1.37 (−2.14, −0.61) | −1.26 (−2.06, −0.46) | −0.71 (−1.45, 0.04) |
| Nominal p-valuea | .. | 0.003 | 0.010 | 0.12 |
| Change in viral RNA copy number from baseline to Day 15 in saliva samples, log10 copies/mL | ||||
| n | 33 | 32 | 26 | 35 |
| LS mean (SE)a | −2.68 (0.35) | −3.43 (0.36) | −3.43 (0.41) | −3.25 (0.35) |
| Difference vs placebo, (90% CI)a | .. | −0.74 (−1.54, 0.06) | −0.75 (−1.60, 0.10) | −0.56 (−1.34, 0.22) |
| Nominal p-valuea | .. | 0.13 | 0.15 | 0.24 |
Because the primary endpoint of the study was not met, any a priori statistical testing of secondary and exploratory endpoints could only be considered hypothesis generating and a p-value <0.05 nominally significant.
Mixed model repeated measures analysis was used to estimate least-squares (LS) means, differences vs placebo, and corresponding standard errors (SEs), confidence intervals (CIs), and p-values.
Clopper-Pearson 2-sided 90% CI. RT-qPCR, reverse transcription–quantitative polymerase chain reaction.
Fig. 2.
Least-squares (LS) mean change from baseline in SARS-CoV-2 log10viral RNA copy number over time.A: Nasopharyngeal swab samples: there were no statistically significant differences between BGB-DXP593 and placebo in the reductions of log viral RNA copy numbers from baseline to any of the evaluated time points in nasopharyngeal swab samples. B: Saliva samples: the differences from placebo in the reductions of log viral RNA copy numbers from baseline to Days 3 and 8 in saliva samples were nominally significantly larger for the BGB-DXP593 5 and 15 mg/kg groups, and numerically larger for the 30 mg/kg group. Similar trends were shown by Day 15, but none of the differences between BGB-DXP593 and placebo reached nominal significance. ∗Nominal p < 0.001; †nominal p ≤ 0.01 vs placebo. N corresponds to number of patients at baseline.
When SARS-CoV-2 viral RNA copy number was analysed by baseline risk status, numerical differences from the placebo group were observed for the BGB-DXP593 5 and 15 mg/kg groups in mean reductions of viral RNA copy number from baseline to Day 8 in the subgroup of patients at high risk for progressing to severe COVID-19 (−2.86 log10 copies/mL for the placebo group vs −3.64 and −5.12 log10 copies/mL for the BGB-DXP593 5 and 15 mg/kg groups, respectively; appendix p 4: Fig. S2); there was a smaller difference in the BGB-DXP593 30 mg/kg group (−3.26 log10 copies/mL). No numerical differences between BGB-DXP593 and placebo were observed in the subgroup of patients who were not at high risk for severe COVID-19 at baseline.
Viral RNA copy number in saliva samples
Although all enrolled patients had laboratory-confirmed SARS-CoV-2 infection (positive RT-PCR test or other antigen test) within 3 days of screening per eligibility criteria, 26 patients (14% [7, 4, 7, and 8 patients with placebo, and BGB-DXP593 5, 15, and 30 mg/kg, respectively]) had negative RT-qPCR results from nasopharyngeal swab samples at baseline. Of the 26 patients, 12 were from the same study site, accounting for 57% of patients enrolled at the site and indicating potential issues with nasopharyngeal swab sampling technique. Fortunately, as an exploratory endpoint, saliva samples were analysed in this study in addition to nasopharyngeal swab samples. Because the primary endpoint based on nasopharyngeal swab samples was not met, however, any a priori statistical testing of study endpoints based on saliva samples could only be considered hypothesis generating and a p-value <0.05 nominally significant.
The LS mean reductions in viral RNA copy number in saliva samples from baseline to Day 8 were −1.81, −3.19. −3.07, and −2.52 log10 copies/mL for the placebo, and BGB-DXP593 5, 15, and 30 mg/kg groups, respectively (Table 2; Fig. 2). The differences from placebo in the reduction of viral RNA copy number from baseline to Day 8 in saliva samples were larger for the BGB-DXP593 5 mg/kg group (−1.37 log10 copies/mL; 90% CI −2.14, −0.61; nominal p = 0.003) and 15 mg/kg group (−1.26; 90% CI −2.06, −0.46; nominal p = 0.01); no difference from placebo was observed for the BGB-DXP593 30 mg/kg group (−0.71; 90% CI −1.45, 0.04; nominal p = 0.12). The treatment effect was observed as early as Day 3 for both the 5 and 15 mg/kg group (5 mg/kg: −1.81 log10 copies/mL; 90% CI −2.54, −1.08; nominal p < 0.001; 15 mg/kg, −1.26; 90% CI −2.04, −0.47; nominal p = 0.01; and 30 mg/kg: −0.30; 90% CI −1.02, 0.42; nominal p = 0.50). Similar trends were shown for the differences between the BGB-DXP593 and placebo group in LS mean reductions of viral RNA copy number from baseline to Day 15 in saliva samples, although none of the differences were nominally significant (BGB-DXP593 5 mg/kg group: −0.74 log10 copies/mL; 90% CI −1.54, 0.06; nominal p = 0.13; 15 mg/kg group: −0.75; 90% CI −1.60, 0.10; nominal p = 0.15; and 30 mg/kg group: −0.56; 90% CI −1.34, 0.22; nominal p = 0.24).
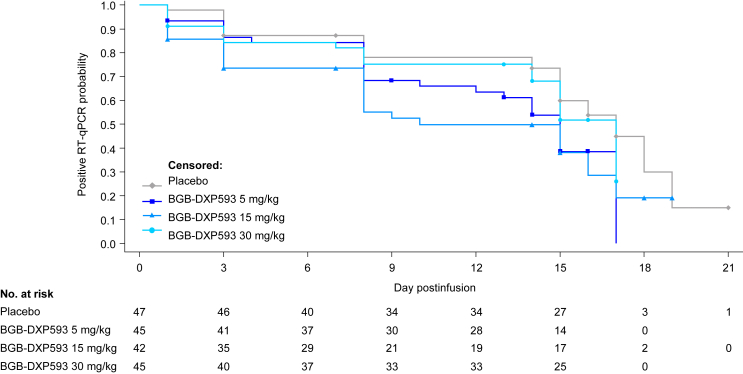
Median times to negative RT-qPCR in all tested samples (nasopharyngeal swab and saliva) were numerically lower with BGB-DXP593 5 mg/kg (15 days) and 15 mg/kg (10 days) vs placebo (17 days; Table 2; Fig. 3), but did not reach statistical significance.
Fig. 3.
Kaplan–Meier plot of time to negative reverse transcription–quantitative polymerase chain reaction (RT-qPCR) in any of the tested samples, including both nasopharyngeal swab and saliva. Median times to negative RT-qPCR were numerically lower with the BGB-DXP593 5 and 15 mg/kg groups vs the placebo group. No formal statistical testing was conducted for comparison between BGB-DXP593 and placebo since this was a phase 2 exploratory study designed primarily for dose finding rather than confirming efficacy.
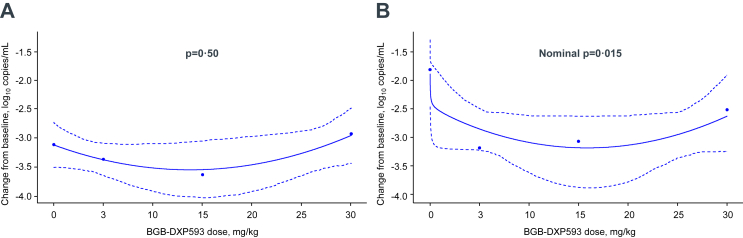
Dose response
Consistent with the viral RNA copy number reduction results, the null hypothesis of a flat dose–response relationship over viral level change at Day 8 could not be rejected in nasopharyngeal swab samples (1-sided p = 0.50); however, post-hoc analysis of saliva samples showed that there was a nominally significant testing result for a dose response in change from baseline to Day 8 in SARS-CoV-2 viral RNA copy number (1-sided nominal p = 0.015), indicating a benefit of BGB-DXP593 over placebo (Fig. 4). Based on analysis of the dose–response curves in saliva samples, BGB-DXP593 5.58 mg/kg was estimated to be the target dose to achieve a treatment difference of −1 log10 copies/mL vs placebo.
Fig. 4.
Estimated dose–response curves in changes from baseline to Day 8 in SARS-CoV-2 viral RNA copy number.A: Nasopharyngeal swab samples: with a 1-sided p-value of 0.50, the null hypothesis of a flat dose–response relationship over SARS-CoV-2 viral RNA copy number change from baseline to Day 8 could not be rejected in nasopharyngeal swab samples. B: Saliva samples: a 1-sided nominal p-value of 0.015 indicates a nominally significant testing result for dose–response curve in change from baseline to Day 8 in SARS-CoV-2 viral RNA copy number in saliva samples. Estimated changes from baseline in viral RNA copy number are plotted in dots. Solid lines represent the fitted dose–response relationship and its 90% confidence bands are plotted in dotted lines.
Clinical outcomes
Median times to resolution of all COVID-19-related symptoms ranged from 14 to 19 days, but there were no substantial differences between the BGB-DXP593 and placebo groups. The proportions of patients who required hospitalisation due to worsening COVID-19 were numerically lower with BGB-DXP593 vs placebo: 1.5% (2/134 patients) for pooled BGB-DXP593 groups (2.2% [1/45] for the 5 mg/kg group, 2.3% [1/43] for the 15 mg/kg group, 0% for the 30 mg/kg group) vs 4.3% (2/47) for the placebo group (Table 2).
Safety
The percentages of patients with ≥1 TEAE were similar across treatment groups (Table 3). The most frequently reported TEAEs were nausea (0%, 5%, 0%, and 7% in the placebo, and BGB-DXP593 5, 15, and 30 mg/kg groups, respectively) and COVID-19 pneumonia (4%, 5%, 3%, and 0%). A mild hypersensitivity reaction was reported in 1 patient treated with placebo. No TEAE led to treatment discontinuation. One patient in the BGB-DXP593 30 mg/kg group had infusion interruption due to TEAEs (ear pruritus and nausea), but restarted and completed the infusion.
Table 3.
Adverse events.
| TEAEs, n (%) | Placebo (n = 47) | BGB-DXP593 |
Total (N = 176) | ||
|---|---|---|---|---|---|
| 5 mg/kg (n = 44) | 15 mg/kg (n = 40) | 30 mg/kg (n = 45) | |||
| ≥1 TEAE | 6 (13) | 6 (14) | 4 (10) | 7 (16) | 23 (13) |
| Grade ≥3 TEAE | 1 (2) | 1 (2) | 1 (3) | 0 (0.0) | 3 (2) |
| Serious TEAEa | 2 (4) | 2 (5) | 1 (3) | 0 (0.0) | 5 (3) |
| TEAE leading to death | 1 (2) | 0 | 0 | 0 | 1 (1) |
| TEAE leading to drug interruption | 0 | 0 | 0 | 1 (2) | 1 (1) |
| TEAE leading to treatment discontinuation or decreased infusion rate | 0 | 0 | 0 | 0 | 0 |
| TEAE according to type | |||||
| Nausea | 0 | 2 (5) | 0 | 3 (7) | 5 (3) |
| COVID-19 pneumonia | 2 (4) | 2 (5) | 1 (3) | 0 | 5 (3) |
| Headache | 0 | 0 | 1 (3) | 1 (2) | 2 (1) |
| Tachycardia | 1 (2) | 1 (2) | 0 | 0 | 2 (1) |
| Diarrhea | 0 | 0 | 0 | 1 (2) | 1 (1) |
| Gastritis | 0 | 1 (2) | 0 | 0 | 1 (1) |
| Vomiting | 0 | 0 | 0 | 1 (2) | 1 (1) |
| Urinary tract infection | 0 | 0 | 1 (3) | 0 | 1 (1) |
| Dizziness | 0 | 0 | 1 (2.5) | 0 | 1 (1) |
| Dysgeusia | 0 | 0 | 0 | 1 (2) | 1 (1) |
| Alanine aminotransferase increased | 0 | 0 | 1 (2.5) | 0 | 1 (1) |
| Aspartate aminotransferase increased | 0 | 0 | 1 (2.5) | 0 | 1 (1) |
| Fibrin D dimer increased | 1 (2) | 0 | 0 | 0 | 1 (1) |
| Ear pruritus | 0 | 0 | 0 | 1 (2) | 1 (1) |
| Medical device site hypersensitivity | 1 (2) | 0 | 0 | 0 | 1 (1) |
| Toxicity to various agents | 0 | 0 | 0 | 1 (2) | 1 (1) |
| Hypoglycaemia | 1 (2) | 0 | 0 | 0 | 1 (1) |
| Myalgia | 1 (2) | 0 | 0 | 0 | 1 (1) |
| Erythema multiforme | 0 | 0 | 0 | 1 (2) | 1 (1) |
| Treatment-related TEAE | 0 | 1 (2) | 0 | 4 (9) | 5 (3) |
| Grade ≥3 treatment-related TEAE | 0 | 0 | 0 | 0 | 0 |
| Serious treatment-related TEAE | 0 | 0 | 0 | 0 | 0 |
| Deathb | 1 (2) | 0 | 0 | 0 | 1 (1) |
Serious treatment-emergent adverse events (TEAEs) were COVID-19 pneumonia in the placebo (n = 2), and BGB-DXP593 5 mg/kg (n = 2), and 15 mg/kg (n = 1) groups.
One death due to COVID-19 pneumonia occurred in the placebo group.
Five serious TEAEs (two with placebo; two with BGB-DXP593 5 mg/kg and one with 15 mg/kg) occurred in the study: all were due to COVID-19 pneumonia and none were attributed by investigators to BGB-DXP593 therapy (Table 3). One patient in the placebo group died due to COVID-19 pneumonia; the patient had multiple risk factors for severe COVID-19, including older age and chronic heart disease.
Pharmacokinetics
The preliminary PK of BGB-DXP593 after single intravenous administration was characterised by a biphasic concentration–time profile and a mean half-life of ∼21–22 days. Dose-proportional increases in maximal concentration and area under the curve from time 0 to ∞ were observed in the 5–30 mg/kg dose range (appendix p 5,6: Fig. S3; Table S1).
Immunogenicity
Antidrug antibodies were evaluable in 126 patients treated with BGB-DXP593. Treatment-emergent ADAs against BGB-DXP593 were detected in 1 patient (<1%) in the 5 mg/kg dose group and were found to be non-neutralising.
Discussion
In this study, nasopharyngeal swab SARS-CoV-2 viral RNA copy number was not significantly decreased by BGB-DXP593 compared with placebo at Day 8; however, viral RNA copy number in saliva was inconsistently reduced at this time point at some doses as low as 5 mg/kg. Hospitalisation rates due to COVID-19 were numerically lower in patients treated with BGB-DXP593 vs placebo.
Baseline characteristics of patients in this study were mostly comparable to those in other studies exploring nAbs in COVID-19.18, 19, 20 In comparison with those studies, patients in this study appeared to have slightly lower viral RNA copy numbers at study entry and slightly longer times to randomisation or treatment after first COVID-19 symptoms. The major differences were that fewer patients in this study were at high risk for progression to severe COVID-19 (27% vs 60%–70%), and the other studies were conducted almost exclusively in the USA, while this trial enrolled patients from both US and non-US countries, including Brazil and South Africa, where SARS-CoV-2 variants may affect neutralisation of the virus.7 Except for a few South African patients who carried SARS-CoV-2 Beta variants, most patients in this trial did not carry SARS-CoV-2 variants of concern or variants with E484K mutations due to the enrollment regions and period of the study.
This study's primary endpoint using nasopharyngeal swab samples was not met. A technical failure of nasopharyngeal swab sampling at one of our study's sites may have led to almost 60% of patients at that site having negative RT-qPCR results despite laboratory-confirmed SARS-CoV-2 infection within 3 days of screening. The suspected nasopharyngeal swab sampling errors, along with the reported high variance of SARS-CoV-2 RNA in the nasopharyngeal swab specimens,21 may have contributed to the lack of significance in the primary endpoint.
Nasopharyngeal swabs and saliva samples are currently the recommended methods for the detection of SARS-CoV-2 RNA. It is, however, well documented that nasopharyngeal swab samples are invasive to collect and may be associated with false-negative results due to inadequate collection of secretions or improper swab technique not reaching the target site of the nasopharynx.22,23
Saliva samples are increasingly accepted as an option for the detection of SARS-CoV-2. Saliva offers advantages of noninvasive collection with viral loads comparable to, or higher than, those in nasopharyngeal swabs after the onset of symptoms and less variation in SARS-CoV-2 RNA levels.21,23,24 There are, however, some testing issues with saliva samples; for example, the dilution of saliva for isolating RNA has not been standardised in clinical practice, which makes it challenging to use saliva samples for quantitative, serial monitoring of viral load. Properly controlled comparative studies vs matched nasopharyngeal swabs and large-scale, longitudinal studies to correlate viral titers in saliva with early clinical manifestations are needed to confirm the role of saliva in the diagnosis and monitoring of COVID-19.23,24
Previous studies of nAbs were conducted using nasopharyngeal swabs.18, 19, 20 The present study explored the effect of BGB-DXP593 on SARS-CoV-2 viral RNA copy number in saliva samples, as well. Investigators in this study were instructed to follow the same laboratory manual for saliva sample collection, dilution, and processing. All except 26 patients had positive RT-qPCR results from saliva samples at baseline. The negative baseline saliva samples were generally evenly distributed across treatment groups (5, 3, 8, and 10 patients in the placebo, and BGB-DXP593 5, 15, and 30 mg/kg groups, respectively), and not concentrated in 1 study site (17 patients scattered in 9 sites and 9 patients from 1 site) as were the negative baseline nasopharyngeal swab samples. The results showed a nominally significant reduction of viral RNA copy number by BGB-DXP593 5 and 15 mg/kg vs placebo at Day 8 post-treatment in saliva samples, with a rapid effect occurring within 3 days and a similar trend of reduction vs placebo observed at Day 15. The differences from placebo in the reduction of viral RNA copy number were larger with lower BGB-DXP593 doses (5 and 15 mg/kg) than with the higher dose (30 mg/kg). A similar observation was reported for bamlanivimab single antibody: the 2800 mg dose was more effective than the 7000 mg dose in reducing viral RNA copy number at Days 3, 7, and 11.18 The reason for the greater response with a lower dose of nAbs, such as BGB-DXP593 and bamlanivimab, is unclear and warrants further investigation. Nevertheless, the nominally significant testing result for dose–response curve in viral RNA copy number reduction at Day 8 in saliva samples was consistent with the clinical benefits of BGB-DXP593 over placebo. These results from saliva samples support the effectiveness of BGB-DXP593 in lowering SARS-CoV-2 viral RNA copy number in patients with mild-to-moderate COVID-19.
Higher SARS-CoV-2 viral loads have been correlated with worse clinical outcomes, including complications and death among hospitalised patients.10,15,16 In the present trial, BGB-DXP593-treated patients had a numerically lower hospitalisation rate due to COVID-19 than placebo-treated patients, which may be related to the better clearance of virus by BGB-DXP593 than placebo. These results should, however, be interpreted with caution due to the small sample size, overlapping 90% CIs in hospitalisation rates between treatment groups, and lack of prespecified statistical testing. The hospitalisation rate in the placebo group (4.3%) was slightly lower than that in other studies of nAbs (5.8%–6.3%),18, 19, 20 which may be due to the small sample size of this study and the low number of patients at high risk for progression to severe COVID-19 at baseline. The safety profile of patients who received BGB-DXP593 was similar to that of placebo-treated patients, indicating that the treatment was well tolerated. All serious TEAEs that occurred in the study were due to COVID-19. The PK profile of BGB-DXP593 was typical of monoclonal antibodies as a class.
This study had several limitations. First, its sample size was smaller than that of other studies of nAbs, although the trial was sufficiently powered to evaluate dose–response relationship over viral level change at Day 8. Second, presumed technical failures with nasopharyngeal swabs at one of the study sites, combined with the high variance of SARS-CoV-2 RNA in nasopharyngeal samples, may have impacted the primary endpoint results. Saliva samples have, however, been shown to detect viral loads that are comparable to or higher than nasopharyngeal swabs,24 and in the present study, nominally significant reductions of SARS-CoV-2 viral RNA copy number were observed with BGB-DXP593 compared with placebo. Third, this study evaluated the efficacy of a single antibody. Previous studies have shown that emergence of treatment-resistant or escape mutant virus may be a concern if a single nAb is used to target a virus.25 However, the present study only collected SARS-CoV-2 sequencing data at baseline. Follow-up sequencing data to document and characterise treatment-emergent resistance were not collected. Despite the lack of data, there is a possibility that BGB-DXP593 may have been supplanted due to the viral evolution and escape of SARS-CoV-2 since the end of this trial in 2021, and further studies of BGB-DXP593 in COVID-19 may not be warranted with its current RBD epitope targeting.
In summary, among patients with mild-to-moderate COVID-19 symptoms, BGB-DXP593 was well tolerated at all doses. No significant difference in viral RNA copy number reduction was observed for BGB-DXP593 and placebo in nasopharyngeal swab samples, possibly due to swab technical issues in some patients. BGB-DXP593 5 and 15 mg/kg reduced SARS-CoV-2 viral RNA copy number at Day 8 in saliva samples compared with placebo. A trend towards a lower hospitalisation rate was observed with BGB-DXP593 vs placebo.
Contributors
All authors reviewed the final manuscript and accepted responsibility for the decision to submit for publication, had full access to all of the data in the study, were involved with the acquisition, analysis, or interpretation of data, take responsibility for the integrity of the data and the accuracy of the data analysis, and provided critical revision of the manuscript for important intellectual content. CP, MM, WZ, and JEJR were involved with the concept and design. CP, MM, FX, WZ, AR, and JEJR were involved with drafting of the manuscript. CP, FX, and WZ provided statistical analysis and verified the underlying data. CP, FX, and JEJR were involved with supervision and project administration.
Data sharing statement
On request, and subject to certain criteria, conditions, and exceptions, BeiGene, Ltd., will provide access to individual de-identified participant data from this study. BeiGene will also consider requests for the protocol, data dictionary, and statistical analysis plan. Data requests may be submitted to DataDisclosure@beigene.com.
Declaration of interests
RV and CP have nothing to disclose. MA has received consulting fees or honoraria from Abbott, AbbVie, Aché, Angion, AstraZeneca, Beigene, Biomedica, Chiesi, EMS, Eurofarma, GSK, Humanigen, IPI-ASAC Brasil, Janssen, Novartis, Rigel, Sanofi Genzyme, and VERU. MM has performed clinical trial contract work done at industry related remuneration with no further incentives for Aspen Pharmacare, AstraZeneca, Boehringer Ingelheim, Cipla Medpro, GSK, MSD, Pharm-Olam, and RedHill. FX and WZ are employed by, and own stock and stock options in BeiGene Co., Ltd. AR is employed by, and owns stock and stock options in BeiGene, Ltd. ZY is employed by, and owns stock and stock options in BeiGene (Beijing) Co., Ltd. JEJR serves or has served on the Australian Cancer Research Foundation Scientific Advisory Board, Cure the Future Board of Directors/Advisory Committee, Board of Directors for FSHD Global Research Foundation, Gene Technology Technical Advisory Committee, National Health and Medical Research Council Mitochondrial Donation Expert Working Committee, Advisory Committee on Biologics of the Therapeutic Goods Administration of the Australian Government; has equity ownership in Genea; has served as a consultant for Imago; has shareholdings with Rarecyte; has consulted with and/or received funding from Athersys, AVI Biopharma, Avigen, bluebird bio, Celgene, Cynata, Elastagen, Gilead, GSK, Imago, Jones Day, Miltenyi, MSD, Novartis, Pfizer, Rarecyte, Roche, SPARK, Takeda, Virax, Wrays; has received various fellowships and research/education grants from the Cancer Council NSW, Cancer Institute NSW, National Health and Medical Research Council, Therapeutic Innovation Australia, and various philanthropic foundations; and serves on the board of the International Society for Cell and Gene Therapy.
Acknowledgments
This study was funded by BeiGene, Ltd., San Mateo, CA, USA. Medical writing and editing assistance was provided by Jinling Wu and Geoff Marx of BioScience Communications, New York, NY, USA, funded by BeiGene, Ltd. We would like to thank Steven D. Nathan, MD, and Camille Kotton, MD, for serving on this trial's data monitoring committee. JEJR wishes to acknowledge the following colleagues for contributing advice, coordination, and logistics: Aimei Lee, Andrew McLachlan, Antony Basten, Stephen Adelstein, David Gattas, Miranda Shaw and RPA Virtual Hospital, Charles Bailey, and Joseph Jewitt. The authors also wish to thank everyone who participated in the study as a researcher.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.101832.
Appendix A. Supplementary data
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlin D.A., Gulick R.M., Martinez F.J. Severe Covid-19. N Engl J Med. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 3.Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease Control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 4.COVID-19 Treatment Guidelines Panel . National Institutes of Health; Bethesda, MD, USA: 2022. Coronavirus disease 2019 (COVID-19) treatment Guidelines.https://www.covid19treatmentguidelines.nih.gov/ [PubMed] [Google Scholar]
- 5.Wang P., Nair M.S., Liu L., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 6.Weisblum Y., Schmidt F., Zhang F., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y., Wang J., Jian F., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y., Cai C., Grifoni A., et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat Med. 2022;28(3):472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeton R., Tincho M.B., Ngomti A., et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022;603(7901):488–492. doi: 10.1038/s41586-022-04460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wölfel R., Corman V.M., Guggemos W., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Zhang L., Sang L., et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130:5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y., Su B., Guo X., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fajnzylber J., Regan J., Coxen K., et al. Massachusetts Consortium for Pathogen Readiness. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pujadas E., Chaudhry F., McBride R., et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8:e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bornkamp B., Pinheiro J., Bretz F. MCPMod: an R package for the design and analysis of dose-finding studies. R package version 1.0-10.1. 2017. https://CRAN.R-project.org/package=MCPMod
- 18.Chen P., Nirula A., Heller B., et al. BLAZE-1 Investigators. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlieb R.L., Nirula A., Chen P., et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinreich D.M., Sivapalasingam S., Norton T., et al. Trial Investigators. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyllie A.L., Fournier J., Casanovas-Massana A., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins T.S., Wu A.W., Ting J.Y. SARS-CoV-2 nasopharyngeal swab testing-false-negative results from a pervasive anatomical misconception. JAMA Otolaryngol Head Neck Surg. 2020;146:993–994. doi: 10.1001/jamaoto.2020.2946. [DOI] [PubMed] [Google Scholar]
- 23.Warsi I., Khurshid Z., Shazam H., et al. Saliva exhibits high sensitivity and specificity for the detection of SARS-COV-2. Diseases. 2021;9:38. doi: 10.3390/diseases9020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khurshid Z., Zohaib S., Joshi C., et al. Saliva as a non-invasive sample for the detection of SARSCoV-2: a systematic review. medRxiv. 2020 doi: 10.1101/2020.05.09.20096354. [DOI] [Google Scholar]
- 25.Simões E.A.F., Forleo-Neto E., Geba G.P., et al. Suptavumab for the prevention of medically attended respiratory syncytial virus infection in preterm infants. Clin Infect Dis. 2021;73:e4400–e4408. doi: 10.1093/cid/ciaa951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.