Abstract
Recent evidence shows that small RNAs are transferred from a species to another through cross-species transmission and exhibit biological activities in the receptor. In this study, we focused on tomato-derived sRNAs play a role of defense against Botrytis cinerea. Bioinformatics method was firstly employed to identify tomato-encoded sRNAs as the cross-species antifungal factors targeting B. cinerea genes. Then the expression levels of some identifed sRNAs were checked in B. cinerea-infected plant using qRT-PCR method. Exogenic RNA-induced gene silences analysis were performed to investigate the antifungal roles of the sRNAs, and the target genes in B. cinerea of antifungal sRNAs would be confirmed by using co-expression analysis. Results showed that a total of 21 B.cinerea-induced sRNAs with high abundance were identified as the cross-kingdom regulator candidates. Among them, three sRNAs containing a miRNA (miR396a-5p) and two siRNA (siR3 and siR14) were selected for experimental validation and bioassay analysis. qRT-PCR confirmed that all of these 3 sRNAs were induced in tomato leaves by B. cinerea infection. Correspondingly, 4 virulence genes of B. cinerea respectively targeted by these 3 sRNAs were down-regulated. Bioassay revealed that all of these 3 cross-species sRNAs could inhibit the virulence and spore gemination of B. cinerea. Correspondingly, the coding genes of B. cinerea targeted by these sRNAs were also down-regulated. Moreover, the virulence inhibition by double strand sRNA was more effective than that by single strand sRNA. The inhibition efficiency of sRNA against B. cinerea increased with the increase of its concentration. Our findings provide new evidence into the coevolution of pathogens and host plants, as well as new directions for the use of plant-derived sRNAs to control pathogens.
Keywords: tomato-derived sRNA, cross-species regulation, RNAi, virulence inhibition, B. cinerea
1. Introduction
Botrytis cinerea represents one of the most predominant and common necrotrophic fungal pathogen promoting postharvest decay of fresh fruit and vegetables (Romanazzi and Feliziani, 2014). B. cinerea has a wide range of hosts and can infect over 200 plant species, causing grey mould disease (Schumacher, 2012; Kumar et al., 2020). Correspondingly, plants have also developed effective strategies against B. cinerea pathogen involving in two internal immune systems: PAMP triggered immunity (PTI) and effector triggered immunity (ETI). PTI and ETI mainly use proteins as action points, in which the microbial associated molecular pattern (MAMP) from pathogens or the damage associated molecular pattern (DMAP) from plants are used as triggers, and plant receptors are used as detectors (Duanis-Assaf et al., 2022). In addition, more and more evidence showed that ncRNA can be used as a mobile immune factor to antagonize the virulence of invasive filamentous pathogens (fungi and oomycetes) in a sequence dependent manner. In 2013, a groundbreaking work showed that the plant fungal pathogen B. cinerea could deliver sRNA to plant cells, and the mobile fungal sRNA could be loaded onto the plant silencing complex containing AGO1 to hijack plant immunity (Weiberg et al., 2013).
In contrary, expressing sRNAs targeting B. cinerea (Bc) DCL genes in plants results in successful silencing of the BcDCL genes, which in turn inhibits the generation of B. cinerea sRNAs that have been proven to be able to hijack plant immunity in a trans-kingdom manner (Wang et al., 2016). Cai et al. (2018) successfully transferred sRNA from Arabidopsis thaliana to B. cinerea through extracellular vesicles and then silenced fungus target genes in vivo. In addition, dsRNAs and small RNAs (sRNAs) targeting DCL1 and DCL2 of B. cinerea have been applied for disease control through RNA spraying (Wang et al., 2016; Wang et al., 2017; Meng et al., 2020; Qiao et al., 2021). Spraying a β2-tubulin dsRNA could conferred plant resistance to B. cinerea (Gu et al., 2019). Nerva et al. (2020) applied dsRNA targeting BcCYP51, Bcchs1, and BcEF2 to suppress B. cinerea infection by high pressure spraying of grapevine leaves and postharvest spraying of grape bunches, and results indicate that RNA-based method to control B.cinerea is effective and environmentally friendly (Wang and Jin, 2017). Therefore, systematic identification of numerous RNAs, which could inhibit B. cinerea virulence infection, is important for understanding the resistance mechanism of plant against B. cinerea and applying RNA-based method to control B. cinerea. In this study, we report the discovery and validation of tomato-derived sRNAs in genome wide, which can inhibit the infection of B. cinerea. Our findings provide new evidence into the coevolution of pathogens and host plants, as well as new directions for the use of plant-derived sRNAs to control pathogens.
2. Materials and methods
2.1. Data sets
Eight sRNA-seq data sets produced from B. cinerea-infected tomato were downloaded from SRA database according to the accession numbers (GSM1101912, GSM1101913, GSM1101914, GSM1101915, GSM1101916, GSM1101917, SRR1482408 and SRR1463412). Among them, GSM1101912, GSM1101914 and GSM1101915 were respectively produced from the B.cinerea-inoculated leaves at 0h, 24 h and 72 h by Weiberg et al. (2013), whereas SRR1482408 and SRR1463412 were produced from the mock- and B.cinerea-inoculated tomato at 7 days (Jin and Wu, 2015). Tomato genome sequences were downloaded from ftp://ftp.solgenomics.net/(version:build_2.40). The datasets of virulence factors in fungal pathogens were downloaded from http://sysbio.unl.edu/DFVF/index.php (Lu et al., 2012).
2.2. Systemic identification of tomato-derived sRNAs targeting B. cinerea
To identify B. cinerea-inducted sRNA from tomato, two sRNA-seq datasets which were sequenced from B. cinerea-inoculated (TD7d) and mock-inoculated (TC7d) tomato leaves at 7 days post-inoculation (dpi) were downloaded from NCBI SRA database with the accession number SRP043615 (Jin and Wu, 2015). The unique sequences in each library were extracted and combined into 1 sRNA library for sRNA identification; all reads in both libraries that were exactly mapped to tomato genomic sequences but unmapped to tRNA, rRNA and B.cinerea genome sequence were extracted as tomato sRNA. The raw read count of these unique sRNAs were retrieved from each sRNA libraries and normalized to reads per millions (RPM). The sRNA sequences with a minimum RPM of 10 in each library were extracted for different expression analysis. The upregulated sRNAs with the Log2(TD7d/TC7d) of >1 and and p-value <0.001 in B. cinerea-infected tomato were considered as the candidates of B. cinerea-induced sRNAs in tomato. Then these candidates were confirmed by sRNA-seq datasets produced from B.cinerea-inoculated tomato at 24 hpi and 72 hpi comparing with mock-inoculated tomato by Weiberg et al. (2013). These B.cinerea-induced sRNAs were considered as miRNAs through sequences alignment with known miRNAs deposited in miRBase database (Kozomara and Griffiths-Jones, 2011). The remaining sRNAs were considered as unknown siRNA. The target genes of the identified sRNA including miRNAs and siRNAs were predicted by using psRNATarget server against cDNA sequences of B. cinerea (Dai and Zhao, 2011). The virulence factors were analyzed for the putative targets by using blastp with the parameters of E-value <1e-5, identity >40% and coverage >70%.
2.3. Preparation of plant materials and inoculation of B. cinere
The seeds of tomatoes cv. Micro Tom and Nicotiana benthamiana were seeded directly into the soil with a 12h:12h photoperiod at ~22°C in a greenhouse. This work used 6-week-old plants. Potato dextrose agar was used for the cultivation of B. cinerea. Conidiospore has collected from 2-week-old infected tomato washed with distilled water two times and adjusted the concentration of 5 × 106 conidiospores/mL for bioassay. Based on the sequence shown in Table S2 , both single strand (ss) sRNAs and double strand (ds) sRNAs were synthesized by GenePharma (Shanghai, China). Each sRNA was added to the conidiospore suspension at a final concentration of 10 µM and immediately drop-inoculated onto five tobacco leaves or tomato leaves for several days. The leaves in control were inoculated with conidiospores in the same manner but with water or negative control RNA (NC RNA, 5′-UUCUCCGAACGUGUCACGUTT-3′. has no target gene in B. cinerea). The experiment was repeated three times. We analyzed the intensity of infection of B. cinerea control and treatment with sRNA by imaging and Trypan blue staining (Woods-Tör et al., 2018).
2.4. Conidiospore germination assays
Conidiospore germination was detected using the cellophane strip method described by Bilir et al. (2019). Briefly, cellophane strips were cut into 1.5 cm2, sterilized by high pressure, and then placed onto MS media in Petri dishes and were added with 10 µL of B. cinerea conidiospore with sRNA or NC-RNA treatment. After incubating for 12 h:12 h photoperiod at 22°C, conidiospores were examined under a light microscope.
2.5. Total RNA extraction and quantitative RT-PCR (qRT-PCR)
Total RNAs were extracted using TRNzol-A+ reagent (TIANGEN, Beijing, China), and a Nano Drop 2000 spectrophotometer was used for RNA quantification. The steps of reverse transcription for the mRNA and sRNA are different. For mRNAs, reverse transcription was performed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China) according to the manufacturer’s recommendations. A similar reaction without reverse transcriptase was performed as a control to confirm the absence of genomic DNA in subsequent steps. For miRNAs, we added poly(A) using E. coli poly(A) polymerase (NEB, Beijing, China). 3’ RT-Primer (Invitrogen) was used as the reverse transcription primer for the following reverse transcription according to the manufacturer’s protocol.
SYBR Green PCR was performed in accordance with the manufacturer’s instructions (NEB, Beijing, China). In brief, 1 μL of cDNA template was added to 10 μL of 2X Master Mix (NEB, Beijing, China), and 10 μM of specific primers and ddH2O were added to a final volume of 20 μL. The reaction was pre-denatured for 3 min at 94 °C, followed by 50 cycles of 94°C for 30 s and 58°C for 30 s. All reactions were performed in triplicate, and controls (no template) were included for each gene. Threshold cycle (Ct) values were automatically determined using the ABI 7500 Real-Time PCR System (USA). The confirmation of amplicon specificity was based on the melt curve at the end of each run. Fold changes were calculated using the 2-ΔΔCt method, where 2-ΔΔCt = (Ct, target − Ct, inner) Treatment − (Ct, target – Ct, inner) Control (Livak and Schmittgen, 2001). All oligos used in this study are listed in Supplementary Table S5 .
2.6. Vector construction and co-expression of the target with miR396a
Pre-miR396a was synthesized and then cloned into the pBIN438 vector with BamH I and Sal I. The cDNA sequences of B. cinerea genes targeted by miR396a were obtained using the RT-PCR method and then inserted into the pEarleyGate 100 expression vector down-stream of the CaMV 35S promoter region. The target sites of miR396a-5p in the target genes were deleted using overlap PCR. Subsequently, the mutated sequences were also inserted into the pEarleyGate 100 vector with expression driven by CaMV 35S promoter.
Transient expression experiments were performed in accordance with the method described by Meng et al. (2020). The Agrobacterium tumefaciens strain GV3101 was transformed with the constructs p35S::MIR396a, p35S::target (normal target site), and p35S::targetmu (target site deletion). Transformed A. tumefaciens samples were harvested and resuspended in infiltration medium [10 mM MgCl2, 10 mM MES (pH 5.6), 200 μM acetosyringone] with the OD600 adjusted to approximately 0.2 for 3-4 h. The A. tumefaciens was infiltrated into 2-week-old N. benthamiana leaves, which were harvested 2 days later.
2.7. Statistical analysis
The statistical analysis was performed with SPSS statistical software 22.0 (United States). All the results were expressed as the Means with SDs from three independent experiments. The t-test was selected and the P-values < 0.05 were considered statistically significant.
3. Results
3.1. Genome-wide identification of tomato-derived sRNAs targeting B. cinerea
Two sRNA-seq datasets were sequenced from B. cinerea-inoculated (TD7d), and mock-inoculated (TC7d) tomato leaves at 7 days post-inoculation (dpi) were downloaded from the NCBI SRA database with the accession number of SRP043615 to identify tomato-derived sRNAs targeting B. cinerea (Jin and Wu, 2015). By using 10 normalized RPM sRNA reads as a cutoff, a total of 1373 sRNAs, ranging from 20 nt to 24 nt, were identified in both libraries ( Table S1 ). Among them, 53 sRNAs were upregulated in B. cinerea-infected leaves compared with mock-infected leaves and then considered as B. cinerea-induced sRNA candidates ( Table S2 ). Moreover, 21 out of the 53 sRNAs were upregulated in B. cinerea-inoculated tomato libraries reported by Weiberg et al. (2013) ( Table S2 , Table 1 ), but the remaining 32 sRNAs could not be confirmed ( Table S2 ). Of these 21 expression-confirmed sRNAs, six had been reported as miRNAs in miRBase, and the remaining 15 were unknown, which were labeled as siR1–siR15 ( Table 1 ). In addition, six B. cinerea-induced sRNAs including 2 miRNAs (miR156d-5p and miR396a-5p) and four siRNAs (siR3, siR10, siR13, and siR14) were confirmed by two or more B. cinerea-treated datasets reported by Weiberg et al. (2013) ( Table 1 ).
Table 1.
Confirmation of B.cinerea-induced sRNA in tomato by sRNA-seq datasets produced from B.cinerea-inoculated tomato at 24 hpi and 72 hpi comparing with mock-inoculated tomato by Weiberg et al. (2013).
| sRNA_id* | annotation | sRNA_Name | Confirmation in different dataset | |||
|---|---|---|---|---|---|---|
| 24hpi_leaf vs. Mock | 72hpi_leaf vs. Mock | 24hpi_fruit vs. Mock | 72hpi_fruit vs. Mock | |||
| s100001 | ath-miR157a-5p | miR157a_5p | Up | |||
| s100006 | sly-miR156d-5p | miR156d_5p | Up | Up | ||
| s100033 | sly-miR5300 | miR5300 | Up | |||
| s100063 | Unknown siRNA | siR1 | Up | |||
| s100120 | Unknown siRNA | siR2 | Up | |||
| s100182 | Unknown siRNA | siR3 | Up | Up | ||
| s100241 | sly-miR396a-5p | miR396a_5p | Up | Up | ||
| s100282 | Unknown siRNA | siR4 | Up | |||
| s100407 | Unknown siRNA | siR5 | Up | |||
| s100439 | sly-miR396a-3p | miR396a_3p | Up | |||
| s100544 | Unknown siRNA | siR6 | Up | |||
| s100841 | sly-miR482b | miR482b | Up | |||
| s100916 | Unknown siRNA | siR7 | Up | |||
| s101071 | Unknown siRNA | siR8 | Up | |||
| s101117 | Unknown siRNA | siR9 | Up | |||
| s101225 | Unknown siRNA | siR10 | Up | Up | ||
| s101249 | Unknown siRNA | siR11 | Up | |||
| s101279 | Unknown siRNA | siR12 | Up | |||
| s101316 | Unknown siRNA | siR13 | Up | Up | Up | |
| s101327 | Unknown siRNA | siR14 | Up | Up | ||
| s101332 | Unknown siRNA | siR15 | Up | |||
We identified potential target genes of these 21 B. cinerea-induced sRNAs in silico and focused on those associated with fungi virulence factors to understand the roles of these sRNAs in cross-kingdom. The results showed that a total of 149 transcripts of B. cinerea were identified as putative targets by psRNAtarget with default parameters (http://plantgrn.noble.org/psRNATarget/) ( Table S3 ). By searching the database of virulence factors in fungal pathogens, we revealed that 82 transcripts targeted by the 21 sRNAs shared high sequence similarity with 63 virulence factors, indicating their potential roles in the pathogenicity of B. cinerea ( Table S4 ). Moreover, 13 transcripts were targeted by nine sRNAs, namely, miR396a-5p, miR482b, siR2, siR3, siR5, siR8, siR9, siR14, and siR15, which were involved in “Grey mould” ( Table 2 ). The transcripts were targeted by the remaining 12 sRNAs, namely, miR157a-5p, miR156d-5p, miR5300, miR396a_3p, siR1, siR4, siR6, siR7, siR10, siR11, siR12, and siR13, which were reported as virulence factors in other fungal pathogens.
Table 2.
The target genes of sRNAs were related with Grey mould.
| sRNA_name | Taget proteins | Annotation | UniProtID | Gene Symbol |
|---|---|---|---|---|
| miR396a_5p | Bcin15p02590.2 | adenylate cyclase | Q9P880_BOTFU | BAC |
| miR396a_5p | Bcin04p04920.1 | thiamine biosynthesis protein | O94101_BOTFU | BCNMT1 |
| miR482b | Bcin09p01310.1 | neutral trehalase protein | Q156F4_BOTFU | NULL |
| miR482b | Bcin09p02390.1 | cmgc mapk protein kinase protein | Q000T6_BOTFU | BMP3 |
| miR482b | Bcin11p04600.1 | mfs transporter protein | Q9P8L8_BOTFU | BCMFS1 |
| siR2 | Bcin05p02840.1 | phosphoinositide-specific phospholipase C (BCPLC1) | P78585_BOTFU | BCPLC1 |
| siR3 | Bcin04p03120.1 | putative chitin synthase g protein | Q8TG14_BOTFU | BCCHSIII |
| siR5 | Bcin08p02970.1 | pectin methylesterase protein | Q9C2Y1_BOTFU | BCPME1 |
| siR8 | Bcin13p01840.1 | Colletotrichum karsti peptidyl-prolyl cis-trans isomerase 10 (CkaCkLH20_04771), partial mRNA/unknown | Q6WP53_BOTFU | BCP1 |
| siR9 | Bcin15p02590.2 | putative adenylate cyclase | Q9P880_BOTFU | BAC |
| siR12 | Bcin01p06260.6 | two-component osmosensing histidine kinase BOS1 | Q8X1E7_BOTFU | BOS1 |
| siR14 | Bcin01p06260.1 | two-component osmosensing histidine kinase BOS1 | Q8X1E7_BOTFU | BOS1 |
| siR15 | Bcin03p06840.1 | nadph oxidase regulator protein | B0BER8_BOTFU | NOXR |
| siR15 | Bcin05p00350.1 | nadph oxidase protein | B0BES1_BOTFU | NOXA |
| siR15 | Bcin09p06130.1 | tetraspanin protein | Q8J0E3_BOTFU | PLS1 |
3.2. Expression patterns of B. cinerea-induced sRNAs and their target genes
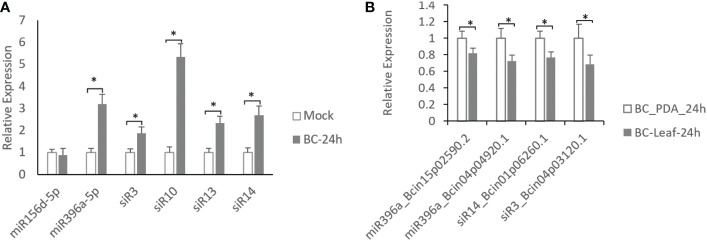
To confirm the expression of these identified sRNAs induced by B. cinerea, 6 sRNAs (miR156d-5p, miR396a-5p, siR3, siR10, siR13 and siR14) supported by two or more other B. cinerea-treated datasets were selected to measure their expression patterns in B. cinerea-inoculated tomato leaves at 24 hours post-inoculation (hpi) using quantitative reverse transcription PCR (qRT-PCR). Results showed that all of these sRNAs were upregulated in B. cinerea-infected tomato leaves compare with mock-treated leaves except for miR156d-5p ( Figure 1A ), showing a consistent expression pattern with sRNA-seq datasets (Weiberg et al., 2013; Jia et al., 2015). Of these 5 B. cinerea-induced sRNAs, only 3 sRNAs might target four virulence factor coding genes of grey mould disease, including Bcin15p02590.2 and Bcin04p04920.1 targeted by miR396a-5p, Bcin04p03120.1 targeted by siR3 and Bcin01p06260.1 targeted by miR14 ( Table 2 ). Therefore, expression patterns of these 4 coding genes were also investigated in B. cinerea inoculated on the potato leaves comparted with that on the the PDA plates the by using qRT-PCR. Results showed that all of these four targets were down-regulated ( Figure 1B ), showing a negative regulation with their sRNAs.
Figure 1.
Expression analysis of B.cinerea-induces sRNAs and their targets. (A) Expression levels of sRNAs in B cinerea-inoculated leaves compared with mock-inoculated leaves in tomato. (B) Expression levels of 4 coding genes targeted by 3 sRNAs in B cinerea grew on tomato leaves comparted with that grew on PDA plates. Asterisks indicate a significant difference (∗P < 0.05).
3.3. Antifungal activities of B. cinerea-inducted sRNAs
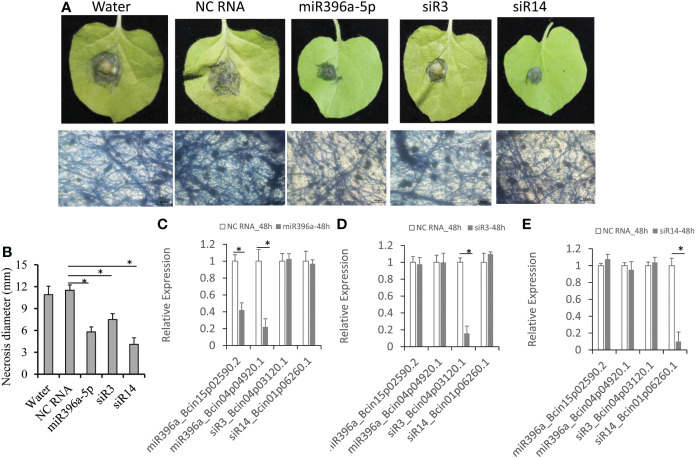
Evidences showed that exogenous RNA-induced gene silence (ERIGS) is an efficient method to control grey mould disease by adding B. cinerea gene-targeted sRNAs (Wang et al., 2016; Wang et al., 2017; Meng et al., 2020; Qiao et al., 2021). Therefore, we used ERIGS to test the antifungal activities of these 3 cross-species sRNAs against B. cinerea. These three cross-species sRNAs (miR396a-5p, siR3 and siR14) were added separately into the B. cinerea conidiospore solution to a final concentration of 10 μM and then drop-inoculated to at least six tobacco leaves for 3 days. Result showed that the average diameter of necrosis reached ~11 mm on B. cinerea-inoculated leaves with water or NC RNA treatments ( Figures 2A, B ). On the leaves of sRNA-treated B. cinerea, three sRNAs had smaller necrosis than those mock treated ( Figure 2A ). Among them, siR14 hardly had a necrotic diameter (4.1 mm), followed by miR396a-5p (5.8 mm), and the remaining siR3 had a similar necrotic diameter (~7.5 mm, Figure 2B ). Correspondingly, the virulence factors coding genes (Bcin15p02590.2 and Bcin04p04920.1) targeted by miR396a-5p were significantly down-regulated in B. cinerea treated by miR396a-5p, but no expression changes in B. cinerea treated by the other two sRNAs ( Figures 2C–E ). Similarity, the expression of Bcin04p03120.1 targeted by siR3 and Bcin01p06260.1 targeted by siR14 were only down-regulated in B. cinerea treated by siR3 and siR14, respectively ( Figures 2C–E ). These results suggesting that these 3 sRNAs control grey mould disease by targeting the virulence genes of B. cinerea.
Figure 2.
Antifungal activities of tomato-derived sRNAs. (A) Tobacco leaves were inoculated with B cinerea conidiospores containing tomato-derived sRNAs. (B) Diameters of the necrosis on tobacco leaves. (C–E) Expression analysis of 4 coding genes targeted by the 3 sRNAs in B cinerea treated with miR396a-5p, siR3 and siR14, respectively. Results are expressed as means ± SD of 3 biological replicates. Asterisks indicate a significant difference (*P<0.05) compared to the corresponding NC-RNA treatment.
3.4. sRNAs inhibits spore germination
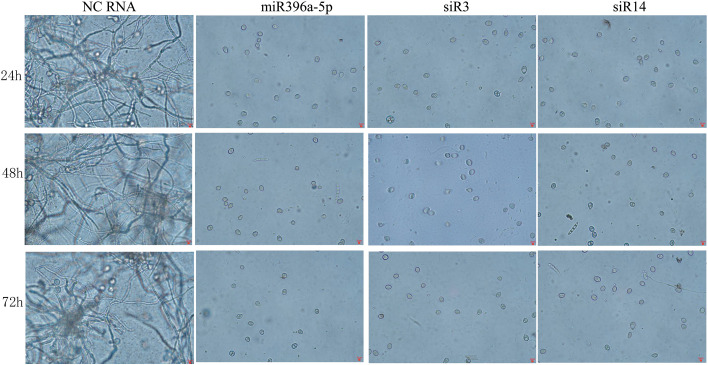
Evidences showed that adding exogenous sRNAs can inhibit the spore germination and hence infection of pathogenic fungi by inducing RNAi pathway (Bilir et al., 2019; Meng et al., 2020). Therefore, Germination assays were performed at 24 h, 48 h and 72 h to understand the effects of the 3 sRNAs on the conidiospore germination of B. cinerea. Results showed that most of the spores incubated with NC-RNA germinated successfully and the mycelia grew well within 24 hours, but the spores treated with each of these 3 sRNAs (miR396a-5p, siR3 and siR14) failed to germinate ( Figure 3 ). These results suggest that RNA has a significant inhibitory effect on spore germination of B. Cinerea.
Figure 3.
Germination assays of B cinerea conidiospores treated by tomato-derived sRNAs.
3.5. Effects of sRNA concentration on antifungal activities
miR396a-5p which has a medium antifungal activity in all of three sRNAs was selected in the subsequent assay for the effects of sRNA concentration. miR396a-5p was added to the conidiospore solution of B. cinerea with a final concentration of 5, 10 and 20 μM, and then inoculated to tobacco leaves for 4 days. For the single stand sRNA, miR396a-5p with 5μM has a largest necrosis diameter (13 mm); followed by 10μM; miR396a-5p treated with 20 μM has a minimal necrosis (8 mm) ( Figures 4A, C ). These results indicated that high concentration of sRNA has better anti-B.cinerea activity. A similar result was observed for the double strand miR396a-5p ( Figure 4B, C ). These results showed that the suppression by double strand miR396a-5p (ds-miR396a-5p) was more efficient than that by single strand miR396a-5p (ss-miR396a-5p) ( Figure 4C ). In addition, the antifungal activity of ds-miR396a-5p were also investigated on tomato leaves through inoculating conidiospore solution of B. cinerea with 10 uM ds-miR396a-5p. Results showed that the necrosis area of B. cinerea were significantly reduced in ds-miR396a-5p-treated leaves compared with that in NC RNA-treated leaves ( Figure 5 ), exhibiting strong antifungal activity.
Figure 4.
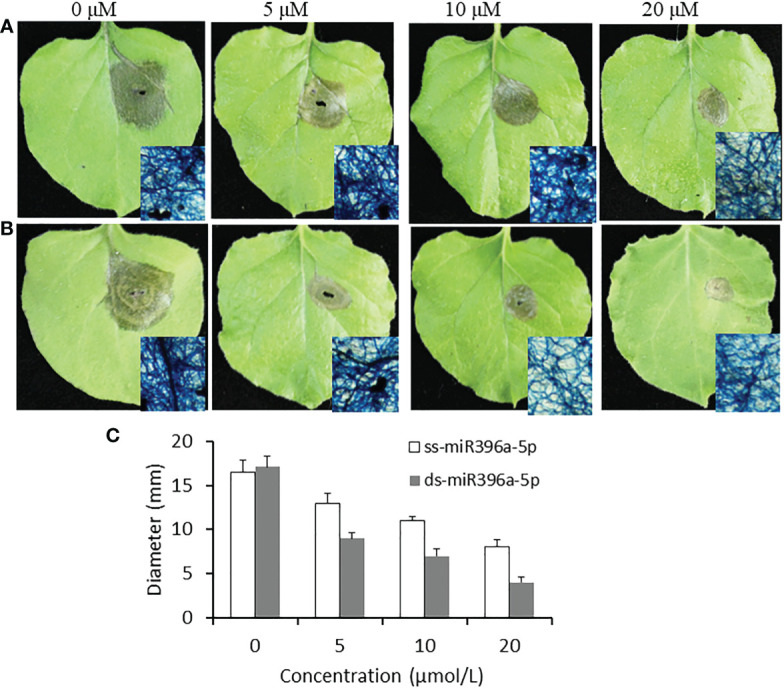
Effect of miR396a-5p concentration on the virulence inhibition. A) Tobacco leaves were inoculated with B cinerea conidiospores containing 5 μM, 10 μM and 20 μM ss-miR396a-5p. Partial necrosis were stained by trypan blue method. B) Tobacco leaves were inoculated with B cinerea conidiospores containing 5 μM, 10 μM and 20 μM ds-miR396a-5p. Partial necrosis were stained by trypan blue method. C) Diameters of the necrosis on tobacco leaves. Results are expressed as means ± SD of 3 biological replicates.
Figure 5.

Antifungal activities of miR396a-5p in tomato leaves with three branches. Tomato leaves were inoculated with B cinerea conidiospores containing 20 μM ds-miR396a-5p and NC RNA.
3.6. Validation of B. cinerea genes targeted by miR396a-5p
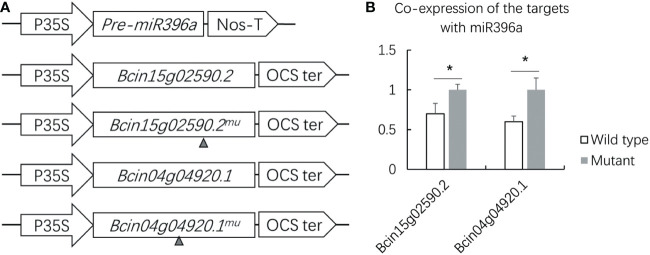
To understand the anti-B. cinerea mechanism, transient co-expression was performed to validate two B. cinerea coding genes targeted by miR396a-5p which was predicted by psRNAtarget ( Table 2 , Table S3 ). We firstly constructed two types of the target gene vectors, which contained either a target site or lacked a target site, to further validate the identified targets of miR396a-5p ( Figure 6A ). target constructs together with the pre-miR396a were transiently co-expressed in N. benthamiana. As the control, each of two wild type target constructs was also transiently expressed in N. benthamiana. Compared with transient expression of the control, the relative expression levels of Bcin15g02590.2 and Bcin04g04920.1 without miR396a-5p target sites were not repressed by co-expression with pre-miR396a ( Figure 6B ). However, the transcript levels of Bcin15g02590.2 and Bcin04g04920.1 containing a target site were significantly decreased when co-expressed with pre-miR396a ( Figure 6B ). The results suggested that both virulence genes of B. cinerea could be negatively regulated by miR396a.
Figure 6.
Validation of target gene of miR396a by transient co expression. (A) Overexpression vector constructs of miR396a-5p and its 2 target genes in B cinerea (Bcin15g02590.2 and Bcin04g04920.1). Gray small triangle: refers to the deletion of the target site. (B) Relative transcription levels of the 2 target genes co-expressed with miR396a-5p in N. benthamiana. Results are expressed as means ± SD of 3 biological replicates. Asterisks indicate a significant difference (*P < 0.05) compared to the corresponding the transient expression of the wild type target gene.
4. Discussion
Emerging evidence supports that RNAs can be transmitted as cross-species effectors from a low trophic level of the food chain and exhibit biological activities at a high trophic level (Jiang et al., 2012; Han and Luan, 2015; Zhang et al., 2012; Zhang H et al., 2016; Zeng et al., 2019). In addition to the study on the sRNAs transferred from a high trophic level to a low trophic level of the food chain and exhibit biological activities, Zhang T et al. (2016) found that cotton-derived miRNAs target V. dahliae virulence genes. The same phenomenon was found in our previous research that tomato-derived miR1001 can cross-species inhibit the growth and virulence of B. cinerea (Meng et al., 2020). In this study, we performed a genome-wide identification to screen the tomato-derived sRNAs acting as the cross-species inhibitors in the plant against B. cinerea. A total of 21 sRNAs were identified as the candidates of cross-species inhibitors. Of them, 3 tomato-derived sRNAs, namely miR396a-5p, siR3 and siR14, showed a role of the cross-species regulation of B. cinerea virulence. For these 3 sRNAs, a total of four virulence genes were predicted as the targets including adenylate cyclase, thiamine biosynthesis protein, chitin synthase and two-component histidine kinase ( Table 2 ). Evidence that the mutants of adenylate cyclase (Liu et al., 2012), thiamine biosynthesis protein (Liu et al., 2022), chitin synthase (Zhang et al., 2016) and two-component histidine kinase (Zhang et al., 2010) would significantly reduce the pathogenicity of the pathogens. Therefore, we proposed that these 3 sRNAs inhibited the pathogenicity by targeting the virulence genes of B. cinerea. In addition, we also investigate the roles of endogenous genes in plant targeted by these 3 antifungal sRNAs ( Table S6 ). Results showed that a large number of the coding genes can be targeted by these 3 sRNA in tomato, and most of these endogenous targets are related to plant growth and development and stress tolerances. For example, a total of 178 coding genes were predicted as the targets of miR396a-5p. previous study showed that miR396a is a plant conserved mRNA, which mainly regulates the growth and development of plant leaves by targeting GRF family members. Moreover, miR396a could also regulate the expression of bHLH74, participating to regulate the development of leaf margin shape and roots (Chen et al., 2015; Chen et al., 2017; Chandran et al., 2019). Interestingly, studies have found that overexpression of miR396a reduced the resistance of plant against to pathogens (Chen et al., 2015). In this study, we found that the exogenous application of miR396a can suppress the virulence of B. cinerea. Therefore, we speculate that the expression of pathogenic sRNAs in plants induced by B. cinerea is to help them successfully infect the host plants. On the contrary, in order to counter this invasion mode of pathogens, host might transport these induced pathogenic sRNAs into pathogens to weaken the role of pathogenic sRNAs in plant and inhibit the virulence of pathogens.
We consider that low-abundance sRNAs hardly play their role in cross-species gene regulation because they are difficult to transport to receptor species or the abundance in the receptor species is excessively low to play a corresponding role. Jia et al. (2015) found that the mulberry miR166b in silkworm is nonfunctional in cross-kingdom gene regulation. Lin et al. (2019) also mentioned that the abundance of cross-kingdom miRNAs detected in receptor species should correlate with the miRNA abundance in the original species unless a particular miRNA is highly stable or preferentially absorbed in animals (Zhang et al., 2012). However, the read count of miR162a is relatively high in H. larvae and shows a significant effect on cross-kingdom gene regulation (Zhu et al., 2017). Basing on this notion and sRNA-seq data, we selected the tomato-derived sRNA candidates with a minimum RPM of 10 in tomato, corresponding to the raw reads of >140.
Bilir et al. (2019) showed that exogenous single-strands sRNAs (ss-sRNAs) and double-strand sRNAs (ds-sRNAs) can be absorbed by spores and combined with target genes to form RNA complexes and inhibit the expression of the target genes. Ds-sRNA is more effective than ss-sRNA. A similar phenomenon was found in this study. When ss-miR396a-5p or ds-miR396a-5p was combined with the spores of B. cinerea, both sRNAs can inhibit the infection of B. cinerea. Moreover, ds-sRNA was more effective than ss-sRNA. The reason may be that ds-sRNA has higher binding efficiency with RISC than ss-sRNA. Martinez et al. (2002) revealed that RISC formation is undetectable at lower concentrations of ss-sRNA but can be detected even at 1 nmol/L concentration of ds-siRNA. Holen et al. (2003) showed that an excess of ds-siRNA can competitively block ss-sRNA. Lima et al. (2009) found that siRNA was more efficient than ssRNA in binding Dicer.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Author contributions
FW and WBJ conceived and designed the research. FW, YH and WQJ organized and performed the experiments. FW and WBJ analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Diao Huang for his technical assistance on bioassay analysis.
Funding Statement
This work was supported by the grant from the Natural Science Foundation of China (32172496), the Natural Science Foundation of Zhejiang Province (LY21C140004), and the 521 Talent Foundation of Zhejiang Sci-Tech University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1072181/full#supplementary-material
References
- Bilir Ö., Telli O., Norman C., Budak H., Hong Y., Tör M. (2019). Small RNA inhibits infection by downy mildew pathogen hyaloperonospora arabidopsidis. Mol. Plant Pathol. 20, 1523–1534. doi: 10.1111/mpp.12863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Qiao L., Wang M., He B., Lin F. M., Palmquist J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. doi: 10.1126/science.aar4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V., Wang H., Gao F., Cao X. L., Chen Y. P., Li G. B., et al. (2019). miR396-OsGRFs module balances growth and rice blast disease-resistance. Front. Plant Sci. 9, 1999. doi: 10.3389/fpls.2018.01999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Luan Y., Zhai J. (2015). Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 34, 2013–2025. doi: 10.1007/s00299-015-1847-0 [DOI] [PubMed] [Google Scholar]
- Chen L., Meng J., Zhai J., Xu P., Luan Y. (2017). MicroRNA396a-5p and -3p induce tomato disease susceptibility by suppressing target genes and upregulating salicylic acid. Plant Sci. 265, 177–187. doi: 10.1016/j.plantsci.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Dai X., Zhao P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanis-Assaf D., Galsurker O., Davydov O., Maurer D., Feygenberg O., Sagi M., et al. (2022). Double-stranded RNA targeting fungal ergosterol biosynthesis pathway controls botrytis cinerea and postharvest grey mould. Plant Biotechnol. J. 20, 226–237. doi: 10.1111/pbi.13708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu K. X., Song X. S., Xiao X. M., Duan X. X., Wang J. X., Duan Y. B., et al. (2019). A β2-tubulin dsRNA derived from fusarium asiaticum confers plant resistance to multiple phytopathogens and reduces fungicide resistance. Pest. Biochem. Physiol. 153, 36–46. doi: 10.1016/j.pestbp.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Han L., Luan Y. S. (2015). Horizontal transfer of small RNAs to and from plants. Front. Plant Sci. 6, 1113. doi: 10.3389/fpls.2015.01113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen T., Amarzguioui M., Babaie E., Prydz H. (2003). Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 31, 2401–2407. doi: 10.1093/nar/gkg338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M. X., Sang X. L., Hong Z. (2012). Beyond nutrients: Food-derived microRNAs provide cross-kingdom regulation. Bioessays 34, 280–284. doi: 10.1002/bies.201100181 [DOI] [PubMed] [Google Scholar]
- Jia L., Zhang D., Xiang Z., He N. (2015). Nonfunctional ingestion of plant miRNAs in silkworm revealed by digital droplet PCR and transcriptome analysis. Sci. Rep. 5, 12290. doi: 10.1038/srep12290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Wu F. (2015). Characterization of miRNAs associated with botrytis cinerea infection of tomato leaves. BMC Plant Biol. 15, 1. doi: 10.1186/s12870-014-0410-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRbase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157. doi: 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Hatan E., Bar E., Davidovich-Rikanati R., Doron-Faigenboim A., Spitzer-Rimon B., et al. (2020). Phenylalanine increases chrysanthemum flower immunity against botrytis cinerea attack. Plant J. 104, 226–240. doi: 10.1111/tpj.14919 [DOI] [PubMed] [Google Scholar]
- Lima W. F., Murray H., Nichols J. G., Wu. H., Sun H., Prakash T. P., et al. (2009). Human dicer binds short single-strand and double-strand RNA with high affinity and interacts with different regions of the nucleic acids. J. Biol. Chem. 284, 2535–2548. doi: 10.1074/jbc.M803748200 [DOI] [PubMed] [Google Scholar]
- Lin Y., Zhu S., Hu C., Wang J., Jiang P., Zhu L., et al. (2019). Cross-species suppression of hepatoma cell growth and migration by a schistosoma japonicum MicroRNA. Mol. Ther. Nucleic Acids 18, 400–412. doi: 10.1016/j.omtn.2019.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Peng G., Xia Y. (2012). The adenylate cyclase gene MaAC is required for virulence and multi-stress tolerance of metarhizium acridum. BMC Microbiol. 12, 163. doi: 10.1186/1471-2180-12-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang X., Deng S., Wang H., Zhao Y. (2022). Thiamine is required for virulence and survival of pseudomonas syringae pv. tomato DC3000 on tomatoes. Front. Microbiol. 13, 903258. doi: 10.3389/fmicb.2022.903258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method . Methos 25, 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu T., Yao B., Zhang C. (2012). DFVF: database of fungal virulence factors. DATABASE (Oxford) 2012, bas032. doi: 10.1093/database/bas032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska A., Urlaub H., Lührmann R., Tuschl T. (2002). Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110, 563–574. doi: 10.1016/S0092-8674(02)00908-X [DOI] [PubMed] [Google Scholar]
- Meng X., Jin W., Wu F. (2020). Novel tomato miRNA miR1001 initiates cross-species regulation to suppress the conidiospore germination and infection virulence of botrytis cinerea in vitro . Gene 759, 145002. doi: 10.1016/j.gene.2020.145002 [DOI] [PubMed] [Google Scholar]
- Nerva L., Sandrini M., Gambino G., Chitarra W. (2020). Double-stranded RNAs (dsRNAs) as a sustainable tool against Gray mold (Botrytis cinerea) in grapevine: Effectiveness of different application methods in an open-air environment. Biomolecules 10, 20. doi: 10.3390/biom10020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L., Lan C., Capriotti L., Ah-Fong A., Nino Sanchez J., Hamby R., et al. (2021). Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 19, 1756–1768. doi: 10.1111/pbi.13589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanazzi G., Feliziani E. (2014). “Botrytis cinerea (gray mold),” in Postharvest decay, Control Strategies; Bautista-Banos Editor: Elsevier Inc. 131–146. doi: 10.1016/B978-0-12-411552-1.00004-1 [DOI] [Google Scholar]
- Schumacher J. (2012). Tools for botrytis cinerea: new expression vectors make the gray mold fungus more accessible to cell biology approaches. Fungal Genet. Biol. 49, 483–497. doi: 10.1016/j.fgb.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Wang M., Jin H. (2017). Spray-induced gene silencing: a powerful innovative strategy for crop protection. Trends Microbiol. 25, 4–6. doi: 10.1016/j.tim.2016.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Weiberg A., Dellota E., Jr., Yamane D., Jin H. (2017). Botrytis small RNA bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 14, 421–428. doi: 10.1080/15476286.2017.1291112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Weiberg A., Lin F. M., Thomma B. P., Huang H. D., Jin H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2, 16151. doi: 10.1038/nplants.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F. M., Zhao H., Zhang Z., Kaloshian I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods-Tör A., Studholme D. J., Çevik V., Telli O., Holub E. B., Tör M. (2018). A suppressor/avirulence gene combination in hyaloperonospora arabidopsidis determines race specificity in arabidopsis thaliana. Front. Plant Sci. 9, 1957 doi: 10.3389/fpls.2018.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Gupta V. K., Jiang Y., Yang B., Gong L., Zhu H. (2019). Cross-kingdom small RNAs among animals, plants and microbes. Cells 8, 371. doi: 10.3390/cells8040371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Z., Chen Q., Liu C. H., Liu Y. B., Yi P., Niu K. X., et al. (2016). Chitin synthase gene FgCHS8 affects virulence and fungal cell wall sensitivity to environmental stress in fusarium graminearum. Fungal Biol. 120 (5), 764–774. doi: 10.1016/j.funbio.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Zhang L., Hou D., Chen X., Li D., Zhu L., Zhang Y., et al. (2012). Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 22, 107–126. doi: 10.1038/cr.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li Y. P., Liu Y. N., Liu H. M., Wang H. Y., Jin W., et al. (2016). Role of plant microRNA in cross-species regulatory networks of humans. BMC Syst. Biol. 10, 60. doi: 10.1186/s12918-016-0292-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu K., Zhang X., Song W., Zhao Q., Dong Y., et al. (2010). A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, magnaporthe oryzae. Curr. Genet. 56 (6), 517–528. doi: 10.1007/s00294-010-0319-x [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhao Y. L., Zhao J. H., Wang S., Jin Y. , Chen Z. Q., et al. (2016). Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen . Nat Plants 2, 16153. doi: 10.1038/nplants.2016.153 [DOI] [PubMed] [Google Scholar]
- Zhu K., Liu M., Fu Z., Zhou Z., Kong Y., Liang H., et al. (2017). Plant microRNAs in larval food regulate honeybee caste development. PloS Genet. 13, e1006946. doi: 10.1371/journal.pgen.1006946 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.