Abstract
COVID-19 has caused significant morbidity and mortality worldwide but also accelerated the clinical use of emerging vaccine formulations. To address the current shortcomings in the prevention and treatment of SARS-CoV-2 infection, this study developed a novel vaccine platform that closely mimics dendritic cells (DCs) in antigen presentation and T-cell stimulation in a cell-free and tunable manner. Genetically engineered DCs that express the SARS-CoV-2 spike protein (S) were chemically converted into extracellular blebs (EBs). The resulting EBs elicited potentially protective humoral immunity in vivo, indicated by the production of antibodies that potently neutralized S-pseudotyped virus, presenting EBs as a promising and safe vaccine.
Keywords: Controlled antigen presentation, Dendritic cell-derived vesicles, Extracellular blebs, COVID-19, Vaccines
1. Introduction
The high morbidity and mortality rate of COVID-19 has necessitated the rapid development of vaccines against severe acute respiratory syndrome coronavirus (SARS-CoV-2). Vaccines based on mRNA, viral vectors, and recombinant protein have been rapidly introduced to a large population and have demonstrated effective protection [1], [2]. However, these vaccines are associated with mild to severe side effects and must be frequently given at a high dose. Moreover, the protection they elicit is of short duration [3], [4], [5], and the vaccines are less effective against emerging strains of SARS-CoV-2 [3].
Dendritic cells (DCs) are professional antigen-presenting cells (APCs) that are capable of activating T cells and inducing adaptive and humoral immune responses after internalizing antigens and presenting antigenic peptides on MHC molecules [6]. DCs also migrate between lymphoid and non-lymphoid tissues and modulate cytokine and chemokine gradients for inducing a durable immune response [7]. The effective use of DCs for vaccination has not been clinically demonstrated due to inefficient antigen presentation, limited migratory capacity, and unsustainable immune stimulation in vivo, also called DC exhaustion [8], [9]. In overcoming the technological shortcomings of employing live DCs for vaccination, DC-derived extracellular vesicles (EVs) have demonstrated the possibility of eliciting antigen-specific neutralization of SARS-CoV-2 [10], [11]. However, vaccine development is challenged by the contradictory immune-privileged property of EVs and by difficulties in their characterization and manufacturing arising from the high structural and functional heterogeneity [12], necessitating an alternative DC-mimicking vaccine.
In an attempt to develop a new vaccine against COVID-19 while avoiding the limitations of live DCs and DC-derived EVs, DCs were genetically engineered to efficiently express the spike protein (S) of SARS-CoV-2 and converted to a cell-free, DC-mimicking vaccine via chemical blebbing [13]. Chemical blebbing creates cell-mimicking vesicles in a highly efficient, rapid, and scalable fashion, and the resulting extracellular blebs (EBs) are homogenous in both structure and function [13], [14]. The S-expressing DC-derived EBs were used to vaccinate mice, and the collected plasma were tested for SARS-CoV-2 neutralization. This study demonstrated the feasibility of developing a novel vaccine platform that is capable of bypassing the hurdles in vaccination, including targeted uptake by APCs and antigen processing in an APC for desired antigen presentation.
2. Materials and methods
2.1. Preparation of SARS-CoV-2 spike protein-expressing cells
The mouse dendritic cell (DC) line DC2.4 (ATCC, Manassas, VA), which was derived from C57BL/6, was cultured in high glucose DMEM supplemented with 10 % (v/v) FBS and penicillin–streptomycin (100 U/mL), all purchased from Thermo Fisher Scientific (Waltham, MA), at 37 °C with 5 % CO2 and passaged every other day. The cells were plated at a density of 5 × 105/well in a 6-well plate for 24 h and then transduced with 2 × 106 TU/well of SARS-CoV-2 spike protein (full-length S alpha)-encoding lentivirus (BPS bioscience, San Diego, CA) in the presence of 5 µg/ml of polybrene (Thermo Fisher Scientific). After 72 h of transduction, transduced cells were selected in the media containing puromycin at 0.5 µg/mL (Thermo Fisher Scientific) for an additional 2 weeks. The S expression in the resulting DC2.4 S cells was analyzed by flow cytometry after staining with anti-S1 primary antibody (BPS Bioscience, San Diego, CA) and FITC-conjugated goat anti-human IgG secondary antibody (Thermo Fisher Scientific).
2.2. Production, collection, and characterization of DC- derived extracellular blebs (EBs)
NEM stock solution was prepared by dissolving N-Ethylmaleimide (NEM, Thermo Fisher Scientific) at 37 °C in 10 mL DI water to a concentration of 2 mM, and sterile filtered using a 0.22 μM syringe filter. Blebbing buffer was prepared immediately before use by adding 90 µL of NEM stock solution to 10 mL DPBS, making it at 0.22 mM NEM. DC2.4 or DC2.4 S cells (5 × 106 cells) were washed 3 times in warm DPBS and incubated in the blebbing buffer for 6 h at 37 °C and 5 % CO2 to produce micro-sized EBs. The supernatant was collected and centrifuged at 1,000 × g for 5 min to pellet cells and cell debris, followed by another centrifugation of the supernatant at 16,100 × g for 15 min. The collected DC2.4 and DC2.4 S EBs were further washed 3 times using 1 × DPBS via repeated centrifugation at 16,100 × g for 10 min to remove any residual blebbing reagent. The DC2.4 and DC2.4 S EB pellets were finally resuspended in 1 × DPBS and confirmed to be free of cells and cell debris under a microscope. The S expression on the surface of the EBs was analyzed by flow cytometry as described for DC2.4 S cells earlier. The surface areas of the EBs were compared with those of the corresponding DC2.4 cells. Briefly, the membrane of an equal number of DC2.4 and DC2.4 S cells was stained with PKH26 according to the manufacturer’s instructions (Thermo Fisher Scientific), followed by blebbing to EBs as described earlier. The PKH26-stained DC2.4 or DC2.4 S cells and DC2.4 or DC2.4 S EBs were lysed in RIPA buffer with gentle vortexing to obtain homogenized membranes. The resulting lysates were analyzed for fluorescence at 550 nm Ex/570 nm Em using a BioTek Synergy plate reader (Agilent, Santa Clara, CA), and their fluorescence was compared using an equal amount by surface area.
2.3. In vivo vaccination, antibody quantification, and virus neutralization
All animal work was reviewed and approved by the UCI Institutional Animal Care and Use Committee (IACUC protocol #AUP-20–116). Female 7–10-week-old C57BL/6 mice (Charles River Laboratories, Wilmington, MA) were subcutaneously injected with 100 μL of 1 × DPBS, S protein (15 ng or 10 μg), 2.5 × 105 DC2.4 or DC2.4 S cells, and DC2.4 or DC2.4 S EBs at an equivalent surface area to DC2.4 and DC2.4 S cells along with an IVAX adjuvant containing 1 nmole MPLA and 3 nmole CpG-1018 in sterile PBS, mixed with an equal volume of AddaVax™ [15]. The mice received a priming injection on Day 0 and a booster injection on Day 14. Immediately before and 10 days after the booster injection (Days 14 and 24), blood was collected into heparinized microcapillary tubes from the saphenous vein. After centrifugation at 6,000 × g for 15 min, the resulting plasma was tested for specific binding to SARS-CoV-2 spike proteins by ELISA. Briefly, recombinant SARS-CoV-2 spike protein (Raybiotech, Peachtree Corners, GA) was coated at 2 μg in 100 μL coating buffer (Thermo Fisher Scientific) per well in a 96-well plate overnight using an orbital shaker. The plates were blocked by adding 100 µL per well of blocking buffer consisting of 5 % (w/v) non-fat dry milk and 0.05 % (w/v) Tween-20 in DPBS and incubated at room temperature (RT) for 2 h. The plate was then rinsed three times using an ELISA wash buffer (Thermo Fisher Scientific). The plasma samples were diluted 20-fold in 150 μL 1 × ELISA diluent buffer, added to the wells and incubated for 2 h at RT. The plates were washed three times using an ELISA wash buffer, and the detection antibody (mouse IgG-HRP [H + L], Waltham, MA) was added according to the manufacturer’s recommended concentration, followed by an additional 1 h incubation at RT. After the plate was washed five times, 100 μL of TMB substrate solution (Thermo Fisher Scientific) was added to each well before incubation for 15 min at RT. The reaction was stopped by addition of 100 μL ELISA stop solution (Thermo Fisher Scientific) per well, and the absorbance was measured at 450 nm and 570 nm using a SpectraMax Plus plate reader (Molecular Devices, San Jose, CA, USA). The absorbance reading at 570 nm was subtracted by that at of 450 nm for optical correction. The antibody concentration in the plasma samples was quantified by comparing the calibration curve of the standard. For the virus neutralization assay, 100 μL of serially diluted plasma was incubated with 100 μL of 1 × 104 GFP-expressing lentivirus pseudotyped with SARS-CoV-2 spike alpha protein (BPS bioscience, San Diego, CA) in DMEM media supplemented with 10 % FBS and 1 % penicillin–streptomycin for 1 h at 37 °C, followed by the addition of 1 × 104 HEK 293 T cells expressing human angiotensin-converting enzyme 2 (hACE2) (BEI Resources [NR-52511], NIAID/NIH) and further incubated for 48 h. For reference, anti-S antibodies, named COVA1-18, gifted from Marit J. van Gils (University of Amsterdam) were used after a series of dilutions from the highest concentration of 1 μg/mL. The cells were analyzed for GFP expression using flow cytometry, and the relative transduction inhibition was determined by mean GFP fluorescence intensity.
2.4. Statistical analysis
For all in vitro studies, triplicate data were analyzed. To achieve statistical significance in in vivo studies, 5 animals per treatment group (n = 5) were used. Two-tailed Student's t-test (GraphPad Prism Ver. 8) was used to calculate the statistical significance of comparisons between two groups, and p-values<0.05 were considered significant. S antibody quantification by ELISA and S-pseudotyped lentivirus neutralization assays were analyzed using one-way analysis of variance (ANOVA) with Tukey’s post hoc test for multiple comparisons between subgroups.
3. Results
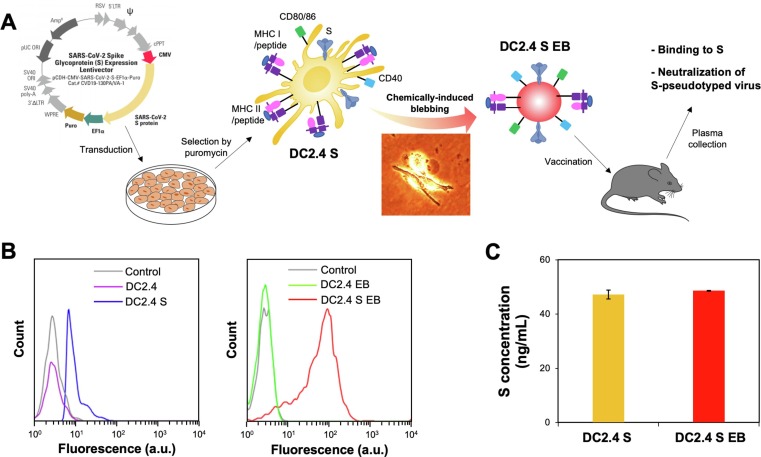
DC2.4 cells derived from C57BL/6 mice were lentivirally transduced to express a SARS-CoV-2 spike protein (S) and further chemically blebbed to prepare a DC-mimicking vaccine (Fig. 1 A). The moderate S expression by DC2.4 S cells, which was preserved on DC2.4 S EBs, was confirmed by flow cytometry (Fig. 1 B). Notably, S expression on DC2.4 S EBs was higher than that on DC2.4 S cells, possibly due to non-specific optical background signal of the smaller DC2.4 S EBs than DC2.4 S cells for the same S density on the surface, as evidenced by the equivalent S contents in the lysates (Fig. 1 C). The highly efficient conversion of DC2.4 and DC2.4 S cells to the corresponding EBs was confirmed by comparing the fluorescent lysates of PKH26-labeled cells and the resulting EBs (Fig. S1). In addition to the S expression, DC surface and maturation markers, CD11c, MHC I, CD40, CD80, and CD86, were found to be comparable between DC2.4 and DC2.4 S cells and their corresponding EBs when characterized by flow cytometry (Fig. S2), although EBs showed slightly higher signals than the cells for CD11c, CD80, and CD86, likely for the reason described for S expression quantification. The results in Fig. 1 confirmed highly efficient preparation of S-expressing DCs from which EBs closely mimicking their molecular profiles were produced for the vaccination of animals against SARS-CoV-2 virus.
Fig. 1.
Preparation and characterization of EBs derived from SARS-CoV-2 spike protein (S)-expressing DC2.4 cells. A) Lentiviral transduction of DC2.4 cells for S expression, followed by blebbing and vaccination of the syngeneic mice. Briefly, DC2.4 cells were transduced with S-expressing lentivirus prior to puromycin selection. DC2.4 S cells were then treated with blebbing buffer to produce EBs. The micro-sized EBs were isolated by centrifugation and used to vaccinate the animals, followed by assays for antibody binding to S and neutralization of S-pseudotyped virus. B) DC2.4 cells, DC2.4 S cells, DC2.4 EBs, and DC2.4 S EBs were analyzed for S expression by flow cytometry. C) 2.5 × 105 DC2.4 S cells and DC2.4 S EBs at a surface area equivalent to 2.5 × 105 DC2.4 S cells were lysed, and their S contents were quantified by ELISA.
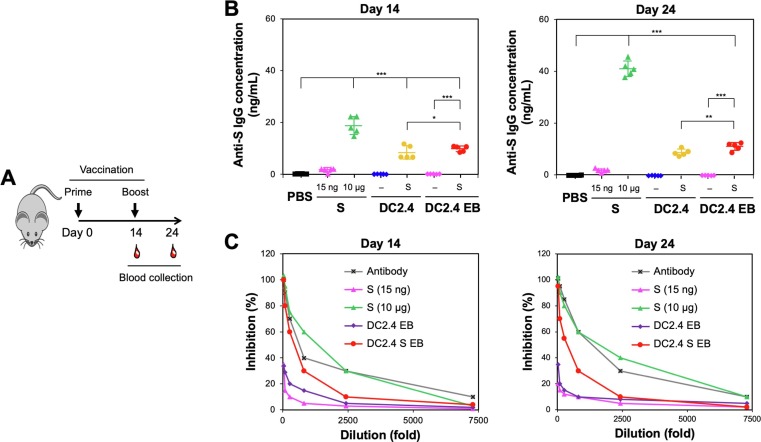
To investigate the DC2.4 S EBs’ ability to induce neutralizing antibodies against SARS-CoV-2 spike-expressing virus, C57BL/6 mice were vaccinated twice with PBS, S (10 μg as a conventional vaccination dose), S (15 ng; equivalent to the S amount in 2.5 × 105 DC2.4 S and DC2.4 S EBs), 2.5 × 105 DC2.4 cells or DC2.4 S cells, and DC2.4 EBs or DC2.4 S EBs an equivalent surface area of 2.5 × 105 of their corresponding cells. The EBs were subcutaneously injected 14 days apart at an equivalent amount of their corresponding cells according to the surface area. Plasma for anti-S IgG and virus neutralization assays were collected at the time of the prime and booster as well as 10 days after the booster (Fig. 2 A). DC2.4 S and DC2.4 S EBs generated similar levels of anti-S antibodies, while no antibody production was observed by S-free DC2.4 cells and their EBs (Fig. 2 B). Notably, the antibody production by DC2.4 S EBs was slightly higher than that of DC2.4 S cells, especially 10 days after the booster, indicating efficient activation of humoral immunity by DC2.4 S EBs in vivo. Compared to an equivalent amount of vaccinated S (15 ng per mouse), DC2.4 S EBs were substantially more efficient (∼350 fold) than the free S in driving antibody production. The booster increased IgG production by ∼ 2-fold with S (10 μg per mouse) but not with DC2.4 S EBs, implying the feasibility of achieving single-shot vaccination. The use of an adjuvant, IVAX-1 increased antibody production by S vaccines (15 ng per mouse) by 5-fold (Fig. S3). In contrast, IVAX-1 marginally affected the antibody production by DC2.4 S and DC2.4 S EBs, likely because these vaccines are already equipped for T cell activation.
Fig. 2.
Animal vaccination by DC2.4 S EBs, measured by antibody generation and virus neutralization. A) Vaccination of C57/BL6 mice, the syngeneic strain from which DCs used to create EBs were derived, with IVAX and blood draw regimen (n = 5). B) anti-S antibody in the plasma from the vaccinated mice with PBS, S (10 μg per mouse; a conventional protein vaccination dose), S (15 ng/mouse; an equivalent S dose to that of DC2.4 S and DC2.4 S EBs), DC2.4 and DC 2.4 S cells, and DC2.4 and DC2.4 S EBs (an equivalent amount by surface area of 2.5 × 105 corresponding cells) was quantified by ELISA. C) The neutralization capability of the plasma harvested from the vaccinated mice were incubated with S-pseudotyped, GFP-expressing lentivirus before transducing 293 T ACE2 cells. No antibodies were quantitated in the plasma collected at the time of prime injection (Day 0) (data now shown). * p < 0.05, p < 0.001, *** p < 0.001 by one-way ANOVA with Tukey’s post hoc test.
In contrast to a wild-type SARS-CoV-2, a pseudotyped lentivirus can only infect cells in a single round, has broad range of host cells, prepared at a high titer, and is not easily inactivated by serum complement [16]. To determine the virus neutralization, plasma samples obtained from mice vaccinated with S (10 μg per mouse) and DC2.4 S EBs were observed by incubation with S-pseudotyped lentivirus (Fig. 2 C). Despite the ∼ 670-fold difference in dose of S, the animals vaccinated with 10 μg had similar, though slightly higher neutralizing antibody responses as the animals vaccinated with DC2.4 S EBs, indicating the high neutralizing efficacy by DC2.4 S EBs. Vaccination with either DC2.4 EBs or S at 15 ng resulted in little if any neutralizing antibody response. These results demonstrate that S-presenting DC-derived EBs given at a very low dose were efficient in producing anti-S antibody that is capable of neutralizing S-pseudotyped virus.
4. Discussion
Traditional forms of vaccines consisting of antigenic proteins have proven safety and efficacy in generating antibodies that target multiple epitopes, while inactivated and attenuated viruses are often regarded as the most potent in generating both cellular and humoral immunity [17]. The shortcomings of the rapidly developed, currently approved mRNA and viral-vector COVID-19 vaccines include mild to critical side effects, limited efficacy and durability against variants, and challenging storage and distribution [4], [5]. In this study, EBs derived from S-expressing DCs (Fig. 1) substantially reduced the dose of S required to elicit a neutralizing antibody response (Fig. 2). In addition, adjuvants were not required with the EB-based vaccine (Fig. S3 and S4), lowering adjuvants-caused side effects [18] and bars for regulatory approval with simpler vaccine formulation, altogether promising for clinical translation. In this study, a novel protein-based vaccination strategy of using DC-derived EBs loaded with SARS-CoV-2 spike proteins to present their antigenic peptides was compared with vaccination by direct administration of the antigenic proteins. The longevity of the antigen-loaded, EB-derived vaccines for sustainable immune activation, especially in comparison with antigen-encoding mRNA vaccines, needs to be evaluated in a subsequent study. EB-based vaccines also offer the possibility for biologically tunable immune activation based on the maturation status of the parent DCs. In this proof-of-concept study, an established cell line of DC2.4 incapable of changing maturation status was used. However, the use of EBs that are derived from primary DCs or bone marrow-derived DCs with varying co-stimulatory signals could allow controlled levels of immune activation. In order to vaccinate a large population of people, allogeneic DCs should be used to manufacture EB vaccines while autologous DC-derived EBs can be used for personalized vaccination.
While DC-derived, S-expressing EBs generated neutralizing antibodies against the S-pseudotyped virus, they might also have been able to induce S-specific cytotoxic CD8 T cells (CTLs). According to recent evidence, cellular immunity plays a crucial role in recovering from COVID-19 [19]. Thus, it will be of interest in future studies to evaluate DC-derived EBs as a COVID-19-treatment strategy. DC2.4 S EBs not only present antigenic peptides on MHC but also carry the protein inside. A study reported that MHC I-loaded exosomes were poorly immunogenic, while the exosomes loaded with a full-length protein elicited strong CD8 T cell responses in vivo [20]. Like exosomes, the S-encapsulating EBs could have been taken up, processed, and presented by APCs, possibly resulting in directed cellular immunity. It is uncertain how much the EBs contributed to neutralizing antibody generation via direct antigen presentation to T cells vs antigenic protein delivery, which could be further investigated by hollow EBs depleted of proteins inside. Ready and affordable access to vaccines is essential in developing community protection.
In conclusion, a new vaccine platform that mimics DCs’ antigen presentation to T cells, likely along with direct antigen delivery, was developed to address the limitations of the current COVID-19 vaccines. The resulting EBs derived from DCs transduced to express the SARS-CoV-2 spike protein generated neutralizing antibodies comparable to levels elicited by a protein vaccine but at a much lower dose. EB-derived vaccines, which are highly stable and tolerate lyophilization (data not shown), unlike currently approved COVID-19 vaccines, may prove to be a useful strategy for preventing COVID-19 in locations where cold chain transportation and storage are unavailable.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
This work was financially supported by UCI COVID-19 CRAFT grant and J.Y.C. was supported by the Korean Health Technology R&D Project of the Korea Health Industry Development Institute (KHIDI) Grant (HI19C0753, the Korea Ministry of Health and Welfare, Republic of Korea). The adjuvants used in this study, IVAX-1, was prepared by Jiin Felgner (UCI Vaccine R&D Center). NIH/NIAID (AI160397) and DTRA (HDTRA1-18-1-0036) supported the work by J.E.D. and D.H.D.
Competing interests
None.
Data and materials availability
All data is available is available from the authors upon request.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellimm.2023.104691.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Teijaro J.R., Farber D.L. COVID-19 vaccines: modes of immune activation and future challenges. Nature Review Immunology. 2021;21:195–197. doi: 10.1038/s41577-021-00526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M.J., Lai L., Edara V.V., Röltgen K., Rogers K., Shirreff L., Ferrell D.E., Wrenn S., Pettie D., Kraft J.C., Miranda M.C., Kepl E., Sydeman C., Brunette N., Murphy M., Fiala B., Carter L., White A.G., Trisal M., Hsieh C.L., Russell-Lodrigue K., Monjure C., Dufour J., Spencer S., Doyle-Meyers L., Bohm R.P., Maness N.J., Roy C., Plante J.A., Plante K.S., Zhu A., Gorman M.J., Shin S., Shen X., Fontenot J., Gupta S., O'Hagan D.T., Van Der Most R., Rappuoli R., Coffman R.L., Novack D., McLellan J.S., Subramaniam S., Montefiori D., Boyd S.D., Flynn J.L., Alter G., Villinger F., Kleanthous H., Rappaport J., Suthar M.S., King N.P., Veesler D., Pulendran B. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–258. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- 3.N. Andrews, J. Stowe, F. Kirsebom, S. Toffa, T. Rickeard, E. Gallagher, C. Gower, M. Kall, N. Groves, A.M. O'Connell, D. Simons, P.B. Blomquist, A. Zaidi, S. Nash, N. Iwani Binti Abdul Aziz, S. Thelwall, G. Dabrera, R. Myers, G. Amirthalingam, S. Gharbia, J.C. Barrett, R. Elson, S.N. Ladhani, N. Ferguson, M. Zambon, C.N.J. Campbell, K. Brown, S. Hopkins, M. Chand, M. Ramsay, J. Lopez Bernal, Covid-19 vaccine effectiveness against the Omicron (B. 1.1. 529) variant, New England Journal of Medicine, 386 (2022) 1532-1546. [DOI] [PMC free article] [PubMed]
- 4.Bruxvoort K.J., Sy L.S., Qian L., Ackerson B.K., Luo Y., Lee G.S., Tian Y., Florea A., Aragones M., Tubert J.E., Takhar H.S., Ku J.H., Paila Y.D., Talarico C.A., Tseng H.F. Effectiveness of mRNA-1273 against delta, mu, and other emerging variants of SARS-CoV-2: test negative case-control study, Effectiveness of mRNA-1273 against delta, mu, and other emerging variants of SARS-CoV-2: test negative case-control study. BMJ. 2021;375 doi: 10.1136/bmj-2021-068848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebinger J.E., Fert-Bober J., Printsev I., Wu M., Sun N., Prostko J.C., Frias E.C., Stewart J.L., Van Eyk J.E., Braun J.G., Cheng S., Sobhani K. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nature Medicine. 2021;27:981–984. doi: 10.1038/s41591-021-01325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabado R.L., Balan S., Bhardwaj N. Dendritic cell-based immunotherapy. Cell Research. 2017;27:74–95. doi: 10.1038/cr.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez M.-I., Heuzé M.L., Martinez-Cingolani C., Volpe E., Donnadieu M.-H., Piel M., Homey B., Lennon-Duménil A.-M., Soumelis V. The human cytokine TSLP triggers a cell-autonomous dendritic cell migration in confined environments. Blood. 2011;118:3862–3869. doi: 10.1182/blood-2010-12-323089. [DOI] [PubMed] [Google Scholar]
- 8.Choi Y., Sunkara V., Lee Y., Cho Y.-K. Exhausted mature dendritic cells exhibit a slower and less persistent random motility but retain chemotaxis against CCL19. Lab on a Chip. 2022;22:377–386. doi: 10.1039/d1lc00876e. [DOI] [PubMed] [Google Scholar]
- 9.Abdi K., Singh N.J., Matzinger P. Lipopolysaccharide-activated dendritic cells: “Exhausted” or alert and waiting? Journal of Immunology. 2012;188:5981–5989. doi: 10.4049/jimmunol.1102868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyashita Y., Yoshida T., Takagi Y., Tsukamoto H., Takashima K., Kouwaki T., Makino K., Fukushima S., Nakamura K., Oshiumi H. Circulating extracellular vesicle microRNAs associated with adverse reactions, proinflammatory cytokine, and antibody production after COVID-19 vaccination. NPJ Vaccines. 2022;7:16. doi: 10.1038/s41541-022-00439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott T.A., Supramaniam A., Idris A., Cardoso A.A., Shrivastava S., Kelly G., Grepo N.A., Soemardy C., Ray R.M., McMillan N.A.J., Morries K.V. Engineered extracellular vesicles directed to the spike protein inhibit SARS-CoV-2. Molecular Therapy Methods & Clinical Development. 2022;24:355–366. doi: 10.1016/j.omtm.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos P., Almeida F. Exosome-based vaccines: History, current state, and clinical trials. Frontiers in Immunology. 2021;12:711565. doi: 10.3389/fimmu.2021.711565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingato D., Edson J.A., Zakharian M., Kwon Y.J. Cancer cell-derived, drug-loaded nanovesicles induced by sulfhydryl-blocking for effective and safe cancer therapy. ACS Nano. 2018;12:9568–9577. doi: 10.1021/acsnano.8b05377. [DOI] [PubMed] [Google Scholar]
- 14.Thone M.N., Kwon Y.J. Extracellular blebs: Artificially-induced extracellular vesicles for facile production and clinical translation. Methods. 2020;177:135–145. doi: 10.1016/j.ymeth.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Davies J.E., Felgner J., Strohmeier S., Pone E.J., Jain A., Jan S., Nakajima R., Jasinskas A., Strahsburger E., Krammer F.J.F.i.I., Felgner P.L., Davies D.H. Administration of multivalent influenza virus recombinant hemagglutinin vaccine in combination-adjuvant elicits broad reactivity beyond the vaccine components. Frontiers in Immunology. 2021;12:692151. doi: 10.3389/fimmu.2021.692151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dautzenberg I.J., Rabelink M.J., Hoeben R.C. The stability of envelope-pseudotyped lentiviral vectors. Gene Therapy. 2021;28:89–104. doi: 10.1038/s41434-020-00193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard A.J., Bijker E.M. A guide to vaccinology: from basic principles to new developments. Nature Reviews Immunology. 2021;21:83–100. doi: 10.1038/s41577-020-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villarreal R., Casale T.B. Commonly used adjuvant human vaccines: advantages and side effects, Journal of Allergy and Clinical Immunology in Practice. 2020;8:2953–2957. doi: 10.1016/j.jaip.2020.04.045. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Yang X., Wang H., Li Z., Deng H., Liu J., Xiong S., He J., Feng X., Guo C., Wang W., Zelinskyy G., Trilling M., Sutter K., Senff T., Menne C., Timm J., Zhang Y., Deng F., Lu Y., Wu J., Lu M., Yang D., Dittmer U., Wang B., Zheng X. Analysis of the long-term impact on cellular immunity in COVID-19-recovered individuals reveals a profound NKT cell impairment. mBio12. 2021:e00085–e121. doi: 10.1128/mBio.00085-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong S., Ruan S., Greenberg Z., He M., McGill J.L. Development of surface engineered antigenic exosomes as vaccines for respiratory syncytial virus. Scientific Reports. 2021;11:21358. doi: 10.1038/s41598-021-00765-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.