Abstract
Trafficking of live mammals is considered a major risk for emergence of zoonotic viruses. SARS-CoV-2-related coronaviruses have previously been identified in pangolins, the world’s most smuggled mammal. A new study identifies a MERS-related coronavirus in trafficked pangolins with broad mammalian tropism and a newly acquired furin cleavage site in Spike.
Trafficking of live mammals is considered a major risk for emergence of zoonotic viruses. SARS-CoV-2-related coronaviruses have previously been identified in pangolins, the world’s most smuggled mammal. A new study identifies a MERS-related coronavirus in trafficked pangolins with broad mammalian tropism and a newly acquired furin cleavage site in Spike.
Main text
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has revealed how vulnerable the human population is to the emergence of new highly infectious respiratory viruses, particularly families of coronaviruses that circulate in bat species. Though interactions between humans and bats in the wild are of concern as environments change, the overspill of bat viruses into intermediary host animals with which people have more contact is an important pandemic risk.1
Over the last 20 years, four beta coronaviruses (βCoVs) have emerged in humans: the sarbecoviruses SARS-CoV-1 and SARS-CoV-2, the embecovirus HKU1-CoV, and the merbecovirus Middle Eastern respiratory syndrome coronavirus (MERS-CoV).1 All have either been shown, or strongly suspected, to have come into humans via intermediary species. MERS-CoV has spilled over multiple times from domesticated camels and causes respiratory infections with high mortality. SARS-CoV-1 entered humans in 2002 via the sale of farmed palm civets in live animal markets in Forshan, China. With SARS-CoV-2, despite ongoing controversies, the extant epidemiological and genetic data implicates a similar emergence in the Huanan Seafood Market in Wuhan where sarbecovirus-susceptible species were on sale.2 , 3
The illicit trade in wild animals and their live sale is thus regarded as a clear and present danger for viral emergence.1 Pangolins, whose “scales” are used in traditional medicine and whose meat is considered a delicacy in parts of Southeast Asia, are the most smuggled mammals worldwide.4 Importantly, sarbecoviruses very closely related to SARS-CoV-2 have been identified in smuggled Malayan pangolins (Manis javanicus),5 , 6 with evidence of wider seropositivity indicating their exposure to bat βCoVs either in the wild or during their trafficking.7
In this issue of Cell, a cross-institutional team led by researchers at the Wuhan Institute of Virology has identified and isolated a novel MERS-related virus in swabs taken from Malayan pangolins seized by Chinese customs officials8 The virus (MjHKU4r-CoV-1) is related to the HKU4 group of merbecoviruses found previously in Tylonycteris species bats. Similar to other merbecoviruses, the Spike glycoprotein can use a broad range of mammalian dipeptidyl peptidase-4 (DPP4) proteins as cell-surface receptors, including orthologs from bats, pangolins, and humans, as well as several other livestock animals. MjHKU4r-CoV-1 replicates robustly in human cells in culture, and in both airway and intestinal organoid tissues. While also sensitive to small molecule inhibitors of the RNA-dependent RNA polymerase shown previously to have efficacy against MERS-CoV, MjHKU4r-CoV-1 is resistant to recombinant neutralizing antibodies directed at MERS-CoV Spike. In the current study, the virus replicated to high titers in the lungs of transgenic mice expressing human DPP4. Although not causing much outward sign of illness in the animals, viral titers were maintained for several days with histological evidence of mild-to-moderate pneumonia. Thus, the virus the researchers isolated is a broad generalist MERS-related CoV with clear potential to cause respiratory infection, and perhaps illness, in humans.
Spike glycoproteins of CoVs are divided into 2 domains—S1, which contains the receptor binding domain, and the membrane-anchored S2, which encodes the fusion mechanism.9 Spike requires proteolytic processing at the S1/S2 junction and a spatially proximal site, S2′, to release the fusion peptide and activate membrane fusion. Although often mediated by either endosomal cathepsins or transmembrane serine proteases (TMPRSS) in the target cell, some CoVs have evolved polybasic cleavage sites to broaden their protease usage to those of the furin family. This allows Spike to be processed as the virus leaves the infected cell—essentially pre-priming it for more efficient target cell entry. Furin cleavage sites (FCS) are a common feature of many enveloped viral glycoproteins, but among respiratory viruses they have been associated with enhanced transmissibility and pathogenesis.
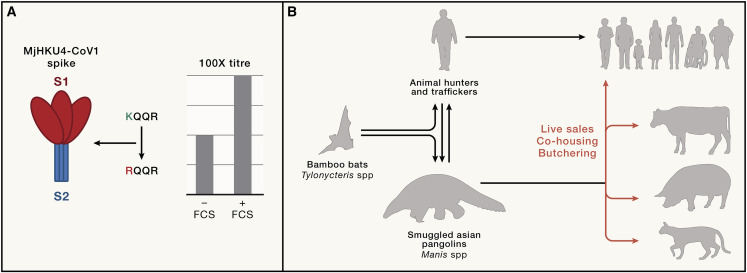
The acquisition of an FCS at the S1/S2 junction of SARS-CoV-2 (so far unique in the sarbecovirus family) was a key determinant of its pandemic emergence, and its deletion impairs respiratory transmission in animal models.9 FCS are more common in the merbecovirus family including in MERS-CoV itself,10 but in the HKU4 group thus far sequenced, no such FCS exists. However, the substitution of a KQQR motif at the S1/S2 junction in the bat HKU4 to the RQQR found in MjHKU4r-CoV-1 results in the generation of a minimal consensus FCS (RXXR) (Figure 1 A). The authors show that although weak in comparison to MERS-CoV, MjHKU4r-CoV-1 Spike is processed by furin. The FCS enhances viral pseudotype entry into human cells by 2–3 orders of magnitude compared to bat HKU4. Similarly, viral production in the presence of a furin inhibitor specifically affects only the wild-type MjHKU4r-CoV-1 Spike. Thus, MjHKU4r-CoV-1 Spike has acquired a weak, but functioning FCS that confers more efficient entry into human cells.
Figure 1.
The Spike protein of MjHKU4-CoV-1 and the risk of its transmission to humans
(A) The Spike glycoprotein from the MjHKU4-CoV-1 virus, like its bat CoV relatives, has a broad tropism for mammalian DPP4 proteins. However, a single amino acid substitution (K-to-R) in at the S1/S2 boundary generates a minimal furin cleavage site (FCS) that can be processed during viral assembly. This site gives the MjHKU4-CoV-1 S a 100-fold higher infectivity in human cells.
(B) The presence of MjHKU4-CoV-1 in smuggled Malayan pangolins raises questions about whether these viruses are endemic in the wild or have been acquired after capture and trafficking. Furthermore, through its broad mammalian tropism, it presents a clear risk of zoonotic emergence either via the traffickers themselves or live sale, co-housing, or butchery of the animals in the presence of the general public or domesticated and farm animals.
This study raises several important questions (Figure 1B). First, how commonly are pangolins exposed to bat coronaviruses? Four seized animals were positive for viral RNA, but a further seven had antibody responses consistent with infection. Second, as mainly solitary animals, where were the pangolins infected—in the wild or after their capture, and did MjHKU4r-CoV-1 come directly from bats or via another species? And last, have human handlers been exposed, and if so, is there any evidence of infection or illness?
Recent studies of live game sales in China have revealed evidence of extensive cross-species transmission of mammalian viruses, including bat coronaviruses, to animals on sale in markets.11 Bat merbecoviruses and sarbecoviruses have wide species tropism defined by receptor usage, raising the question of whether acquisition of expanded protease activation tips the balance between a low risk of infection and the potential for respiratory transmission in humans or other species, which would require further in vivo transmission studies in model animals using MjHKU4r-CoV-1 with the FCS reverted to KQQR. Recent demonstration of bat merbecoviruses with the ability to use the sarbecovirus mammalian entry receptor, ACE2, further expands the potential of these viruses to emerge in humans.12 With smuggled pangolins now shown to harbor two groups of pandemic-potential coronaviruses, this study highlights how the illegal wildlife trade is a key risk area for emerging viruses that requires improved international surveillance and regulation as a priority.
Acknowledgments
Declaration of interests
The author declares no competing interests.
References
- 1.Keusch G.T., Amuasi J.H., Anderson D.E., Daszak P., Eckerle I., Field H., Koopmans M., Lam S.K., Das Neves C.G., Peiris M., et al. Pandemic origins and a One Health approach to preparedness and prevention: Solutions based on SARS-CoV-2 and other RNA viruses. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2202871119. e2202871119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pekar J.E., Magee A., Parker E., Moshiri N., Izhikevich K., Havens J.L., Gangavarapu K., Malpica Serrano L.M., Crits-Christoph A., Matteson N.L., et al. The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2. Science. 2022;377:960–966. doi: 10.1126/science.abp8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worobey M., Levy J.I., Malpica Serrano L., Crits-Christoph A., Pekar J.E., Goldstein S.A., Rasmussen A.L., Kraemer M.U.G., Newman C., Koopmans M.P.G., et al. The Huanan Seafood Wholesale Market in Wuhan was the early epicenter of the COVID-19 pandemic. Science. 2022;377:951–959. doi: 10.1126/science.abp8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinrich S., Wittmann T.A., Ross J.V., Shepherd C.R., Challender D.W.S., Cassey P. 2017. The Global Trafficking of Pangolins: A comprehensive summary of seizures and trafficking routes from 2010–2015. Southeast Asia Regional Office, Petaling Jaya, Selangor, Malaysia., Southeast Asia Regional Office. [Google Scholar]
- 5.Lam T.T.Y., Jia N., Zhang Y.W., Shum M.H.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 6.Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 7.Wacharapluesadee S., Tan C.W., Maneeorn P., Duengkae P., Zhu F., Joyjinda Y., Kaewpom T., Chia W.N., Ampoot W., Lim B.L., et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 2021;12:972. doi: 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J., Yang X., Si H., Gong Q., Que T., Li J., et al. A bat MERS-like coronavirus circulates in pangolin and utilizes human DPP4 and host proteases for cell entry. Cell. 2023;186 doi: 10.1016/j.cell.2023.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stout A.E., Millet J.K., Stanhope M.J., Whittaker G.R. Furin cleavage sites in the spike proteins of bat and rodent coronaviruses: Implications for virus evolution and zoonotic transfer from rodent species. One Health. 2021;13:100282. doi: 10.1016/j.onehlt.2021.100282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He W.T., Hou X., Zhao J., Sun J., He H., Si W., Wang J., Jiang Z., Yan Z., Xing G., et al. Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell. 2022;185:1117–1129.e8. doi: 10.1016/j.cell.2022.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong Q., Cao L., Ma C., Tortorici M.A., Liu C., Si J., Liu P., Gu M., Walls A.C., Wang C., et al. Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. Nature. 2022;612:748–757. doi: 10.1038/s41586-022-05513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]