ABSTRACT
Ampicillin-ceftriaxone has become a first-line therapy for Enterococcus faecalis endocarditis. We characterized the penicillin-binding protein (PBP) profiles of various E. faecalis strains and tested for synergy to better inform beta-lactam options for the treatment of E. faecalis infections. We assessed the affinity of PBP2B from elevated-MIC strain E. faecalis LS4828 compared to type strain JH2-2 using the fluorescent beta-lactam Bocillin FL. We also characterized pbp4 and pbpA structures and PBP4 and PBP2B expression and used deletion and complementation studies to assess the impact of PBP2B on the levels of resistance. We tested penicillin-susceptible and -resistant E. faecalis isolates against ceftriaxone or ceftaroline combinations with other beta-lactams in 24-h time-kill studies. Two penicillin-susceptible strains (JH2-2 and L2052) had identical pbp sequences and similar PBP expression levels. One reduced-penicillin-susceptibility strain (L2068) had pbp sequences identical to those of the susceptible strains but expressed more PBP4. The second decreased-penicillin-susceptibility strain (LS4828) had amino acid substitutions in both PBP4 and PBP2B and expressed increased quantities of both proteins. PBP2B did not appear to contribute significantly to the elevated beta-lactam MICs. No synergy was demonstrable against the strains with both mutated PBPs and increased expression (L2068 and LS4828). Meropenem plus ceftriaxone or ertapenem plus ceftriaxone demonstrated the most consistent synergistic activity. PBP2B of strain LS4828 does not contribute significantly to reduced penicillin susceptibility. Neither the MIC nor the level of PBP expression correlated directly with the identified synergistic combinations when tested at static subinhibitory concentrations.
KEYWORDS: Enterococcus faecalis, dual beta-lactams, penicillin-binding protein
INTRODUCTION
Enterococcus faecalis accounts for over 90% of enterococcal infections, and in recent years, concern for resistance is rising (1–6). Features that distinguish E. faecalis are its reduced susceptibility to penicillins, its resistance to cephalosporins, and its tolerance to the bactericidal activity of these antibiotics. The mechanisms underlying enterococcal tolerance remain undefined, but bactericidal activity can be achieved by combining penicillins (or other cell wall-acting agents) with aminoglycosides. Combining penicillin or ampicillin with an aminoglycoside in the clinical setting significantly improved the cure rates for E. faecalis endocarditis and for years was the standard of care for this illness (7). More recently, the spread of high-level aminoglycoside resistance (8, 9) and concerns of aminoglycoside toxicity (10, 11) have prompted the search for suitable alternative therapies.
The two-beta-lactam combination of ampicillin and ceftriaxone has become a standard therapy for endocarditis caused by E. faecalis (10–13). Original reporting on this combination suggested that the in vitro activity could be due to complementary activity against different penicillin-binding proteins (PBPs), resulting in synergy (14). Two E. faecalis PBPs shown to have an impact on beta-lactam susceptibility are PBP4 (encoded by pbp4), where amino acid substitutions within and increased expression have been correlated with decreased beta-lactam susceptibilities (1), and PBP2B (encoded by pbpA), where its deletion in two separate E. faecalis strains was recently reported to result in susceptibility to ceftriaxone (15). Limited studies assessing the impact of PBP2B are available.
While ampicillin resistance in E. faecalis is rare, structural and expression changes of PBP4 may result in penicillin-resistant and ampicillin-susceptible E. faecalis (PRASEF) isolates (1–3, 16). There are increasing reports of PRASEF isolates (1–3, 5, 16, 17) as well as a prospective observational study showing higher mortality rates among patients with PRASEF bloodstream infections who were treated with ampicillin- or piperacillin-based regimens (18). The clinical utility of the combination of ampicillin plus ceftriaxone is also limited by the instability of reconstituted ampicillin, the uncertainty of PBP binding, and its inconvenient dosing schedule (15). Morbidity and mortality rates with ampicillin-ceftriaxone are similar (between 29 and 42%) to those observed with beta-lactam–aminoglycoside regimens (19–21). Given the variety of PBP changes that may underlie the reduced susceptibility of E. faecalis, we elected to examine the in vitro activities of a variety of beta-lactam combinations against a group of 4 E. faecalis isolates, 2 fully susceptible and 2 with elevated MICs, whose resistance mechanisms we fully characterized.
RESULTS
Susceptibility testing.
E. faecalis MICs and minimum bactericidal concentrations (MBCs) are reported in Table 1. All isolates were considered penicillin and ampicillin susceptible according to CLSI breakpoints (susceptible at ≤8 μg/mL) (22). However, isolates L2068 and LS4828 had elevated penicillin MICs of 8 μg/mL and MBCs of 64 and >1,024 μg/mL, respectively. Both JH2-2 and L2052 had a penicillin MIC of 2 μg/mL and an MBC of 4 μg/mL. Susceptibility breakpoints have not been published for the other tested beta-lactams.
TABLE 1.
Enterococcus faecalis isolate MICs and MBCsa
| Drug | MIC (mcg/mL) (MBC [mcg/mL]) |
|||
|---|---|---|---|---|
| Penicillin susceptible |
Penicillin elevated |
|||
| JH2-2 | L2052 | L2068 | LS4828 | |
| Penicillins | ||||
| Penicillin | 2 (4) | 2 (4) | 8 (64) | 8b (>1,024) |
| Ampicillin | 2 (2) | 0.5 (1–2) | 1 (2–4) | 8 (>1,024) |
| Piperacillin-tazobactam | 2 (2) | 2 (4) | 8 (16) | 16 (>1,024) |
| Carbapenems | ||||
| Ertapenem | 4 (16) | 4 (8) | 16 (>64) | 64 (>2,048) |
| Imipenem-cilastatin | 0.5 (1) | 0.5 (1) | 2 (4) | 4–8 (>512) |
| Meropenem | 2 (8) | 2 (8–32) | 8 (64) | 32–64 (>2,048) |
| Cephalosporins | ||||
| Ceftaroline | 1–2 (8) | 1 (8–16) | 16 (>64) | 8 (16) |
| Ceftriaxone | 512 (2,048) | 256 (1,024) | 2,048 (>2,048) | 2,048 (>2,048) |
MICs and MBCs were obtained via broth microdilution according to CLSI guidelines.
The published penicillin MIC for LS4828 was 16 mcg/mL.
Previously reported susceptibilities of JH2-2 to ampicillin, penicillin, imipenem, meropenem, and ceftriaxone were similar to our findings (Table 2) (1, 23). Piperacillin-tazobactam, ertapenem, and ceftaroline MICs have not been previously published for JH2-2. Previously reported LS4828 susceptibilities to ampicillin, penicillin, ceftriaxone, imipenem, and meropenem were similar to our findings (1). Piperacillin-tazobactam, ertapenem, and ceftaroline MICs have not been previously published for LS4828. The MBCs for LS4828, except for ceftaroline, were unattainable due to higher than chemically stable drug concentrations.
TABLE 2.
Impact of PBP2B deletion on susceptibilities of LS4828 determined in BHI broth
| Straina | MIC (mcg/mL) |
|||
|---|---|---|---|---|
| Ampicillin | Penicillin | Ceftriaxone | Imipenem | |
| JH2-2 | 0.78 | 1.56 | >1,000 | 1.56 |
| LS4828 | 12.5 | 6.25 | >1,000 | 12.5 |
| LS4828 ΔPBP2B | 12.5 | 3.13 | >1,000 | 6.25 |
| PBP2B::pRIH310 | 12.5 | 6.25 | >1,000 | 12.5 |
| PBP2B::pRIH311 | 12.5 | 6.25 | >1,000 | 12.5 |
| PBP2B::pBSU100 | 12.5 | 6.25 | >1,000 | 6.25 |
pRIH310, LS4828 pbpA in pBSU100; pRIH311, JH2-2 pbpA in pBSU100.
Genomic analysis.
The genome sizes for L2068 and L2052 are 3,113,293 bp and 2,982,145 bp, respectively.
E. faecalis LS4828 (elevated beta-lactam MICs) expresses increased quantities of PBP4 through increased pbp4 transcription due to a single adenine deletion 8 bases upstream of a putative −35 region of the pbp4 gene (1). LS4828 also has two amino acid substitutions in the translated protein, one of which is associated with a reduced affinity for penicillin (1). Finally, it has a series of amino acid substitutions in its pbpA gene whose impacts on penicillin susceptibility are unknown (see Fig. S1 in the supplemental material). Sequence analysis of the pbp genes of L2068 and L2052 revealed that L2068 shared the adenine deletion found in the pbp4 promoter region of LS4828 (Fig. S2). Both L2068 and L2052 had pbp4 and pbpA amino acid sequences identical to those of JH2-2, with no amino acid substitutions. We did not identify differences in other pbp genes.
PBP experiments.
In order to determine whether the amino acid substitutions in the PBP2B protein contributed to elevated MICs expressed by LS4828, we cloned the pbpA (minus the transmembrane portion) genes from LS4828 and JH2-2 into the expression vector pET-RP1B and introduced these constructs into Escherichia coli BL21 Star(DE3). Purified proteins were used in experiments with Bocillin FL to determine affinities. The Bocillin FL-binding affinities for the two proteins were indistinguishable over a range of Bocillin FL concentrations from 5 to 80 μM (Fig. S3). We then performed a competition experiment with ampicillin and used Bocillin FL to measure the unbound PBP2B proteins. The affinity to ampicillin of LS4828 PBP2B was indistinguishable from that of PBP2B from JH2-2 (Fig. S4). The 50% inhibitory concentration (IC50) for JH2-2 PBP2B was 3.06 μg/mL, and the IC50 for LS4828 PBP2B was 3.93 μg/mL. These data suggest that the numerous amino acid substitutions found in LS4828 PBP2B did not result in a changed penicillin affinity and therefore did not contribute to the elevated beta-lactam MICs.
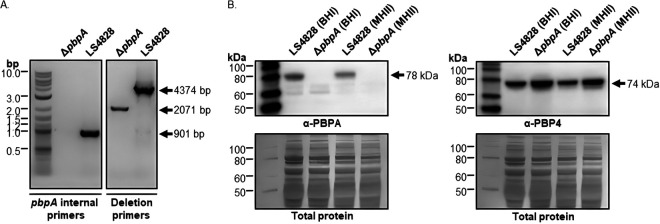
To further assess the contribution of PBP2B to the expression of elevated MICs in our strains, we attempted to delete the pbpA genes from both LS4828 and JH2-2. We successfully deleted pbpA from LS4828, which was confirmed by PCR amplification, whole-genome sequencing, and Western blot experiments (Fig. 1). The whole-genome sequence also confirmed that the deletion did not lead to any other changes to the genome (data not shown). We were unable to delete the pbpA gene from JH2-2 despite multiple attempts with the successful integration and removal of the knockout plasmid in the JH2-2 genome. This suggests that in JH2-2, pbpA serves as an essential gene.
FIG 1.
The LS4828 ΔpbpA mutant is confirmed by PCR and Western blot analysis. (A) pbpA internal primers amplify only if the pbpA gene is present, confirming its absence in the ΔpbpA strain. The deletion primers confirm the expected fragment size of 2,071 bp in the ΔpbpA strain. (B) LS4828 and the ΔpbpA mutant were grown in BHI or MHII broth to mid-log phase, processed for SDS-PAGE, and run on 10% NuPAGE gels. PVDF membranes were probed with a custom PBPA or PBP4 polyclonal antibody for immunodetection. The blots were then stained with Coomassie R-250 for confirmation of equal protein transfer.
After the deletion of pbpA, LS4828 ΔpbpA grew more slowly and to a lower density than the parent strain (Fig. S5). This growth defect was more pronounced in brain heart infusion (BHI) broth than in Mueller-Hinton II (MHII) broth and resulted in a visible change in the morphology of the cells (Fig. S6). The susceptibilities of LS4828 ΔpbpA are shown in Table 2. The deletion did not impact the MIC susceptibilities to the beta lactams tested when compared to the parent strain. Disc susceptibility assay results were consistent with the broth susceptibility findings. The introduction of plasmid-borne JH2-2 or LS4828 pbpA into LS4828 ΔpbpA had a minimal impact on the susceptibilities to beta-lactams (Table 2).
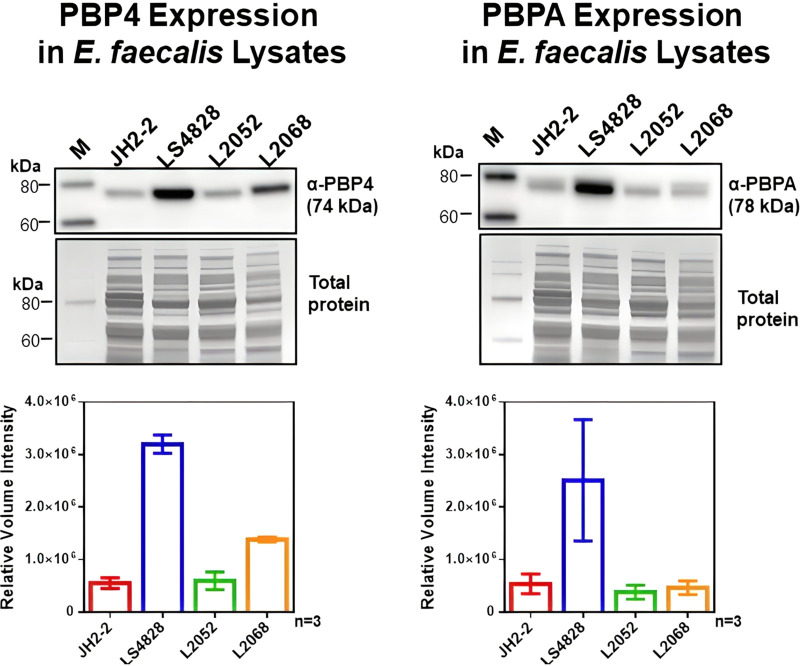
To determine whether the reduced susceptibility of LS4828 to penicillin and ampicillin could be related to the quantity of PBP2B, we used anti-PBP2B and anti-PBP4 antibodies to assess the quantities of PBP2B and PBP4 produced by LS4828, JH2-2, L2052, and L2068 (Fig. 2). The two strains with reduced penicillin susceptibilities produced increased quantities of low-affinity PBP4, with LS4828 producing roughly twice as much as L2068 and roughly six times as much as the susceptible strains. LS4828 also produced PBP2B at levels that exceeded those of all other strains (including L2068) by nearly 5-fold. These data suggested that increased quantities of PBP2B could play a role in the elevated MICs of ampicillin and penicillin expressed by LS4828 relative to the more susceptible strains.
FIG 2.
Quantitation of PBP4 and PBPA expression by Western blot analysis. E. faecalis cells grown in BHI broth to exponential phase were processed for SDS-PAGE and transferred to PVDF membranes for immunoblot detection of PBP4 or PBPA using a custom polyclonal antibody. The graphs below each blot represent expression levels from 3 biological replicates. These data were from normalized densitometry analysis after Coomassie blue R-250 staining of total proteins in the blots. M, molecular weight marker.
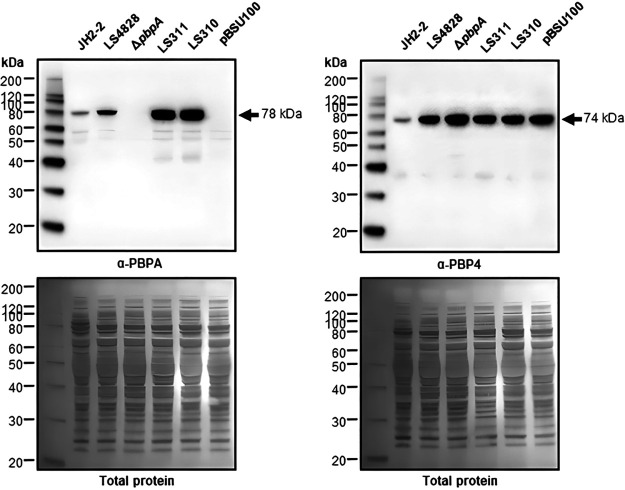
The quantities of PBP4 were compared among JH2-2, LS4828, and LS4828 ΔpbpA. As noted in our previous publication, the level of expression of PBP4 from LS4828 was dramatically higher than that in JH2-2 due to a mutation upstream of its promoter (1). We were, however, surprised to note that the expression level of PBP4 in LS4828 ΔpbpA was approximately 50% higher than that in LS4828. When pbpA was reinserted into this strain on a plasmid, the quantities of PBP4 returned to those seen in LS4828 (Fig. 3). The full Bocillin PBP-binding profiles for all strains are included in Fig. S7.
FIG 3.
Complementation of pbpA from JH2-2 (LS311), LS4828 (LS310), or the pBSU100 vector alone in the ΔpbpA strain is confirmed by Western blotting of E. faecalis strains grown in BHI broth to exponential phase. Cell lysates were prepared for SDS-PAGE, and immunoblots were processed with a custom PBPA or PBP4 polyclonal antibody. The detection of pbp4 in the cell lysates shows increased PBP4 expression in the ΔpbpA strain over that of its parent LS4828 strain. These blots are representative of data from 2 biological replicates. The blots were then stained with Coomassie R-250 for confirmation of equal protein transfer.
In summary, for the four E. faecalis strains used in the following time-kill experiments, LS4828 has the most elevated MICs due to a combination of PBP4 amino acid changes and increased expression of PBP4, L2068 has decreased susceptibility due to increased expression of PBP4 without additional amino acid substitutions, and L2052 and JH2-2 represent wild-type E. faecalis with no PBP4 amino acid substitutions and a baseline level of expression.
Time-kill experiments.
The 24-h combination time-kill assay results are reported in Table 3 and show the change in growth from the initial inoculum. All monotherapy regimens demonstrated inactivity against E. faecalis strains, except for the elevated-penicillin-MIC strain L2068, which demonstrated bacteriostatic activity with ceftaroline (−1.95 ± 0.18 log10 CFU/mL) at 0.5× the MIC, which is why the decrease in growth is not reported as synergistic (Table 4).
TABLE 3.
Dual-combination 24-h time-kill results for changes in the log10 CFU per milliliter of Enterococcus faecalis from the initial inoculuma
| Drug at 0.0625× MIC | Mean change in log10 CFU/mL of strain ± SD |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillin susceptible |
Penicillin elevated |
|||||||||||||||
| JH2-2 (ampicillin MIC of 2 mcg/mL; penicillin MIC of 2 mcg/mL) |
L2052 (ampicillin MIC of 0.5 mcg/mL; penicillin MIC of 2 mcg/mL) |
L2068 (ampicillin MIC of 1 mcg/mL; penicillin MIC of 8 mcg/mL) |
LS4828 (ampicillin MIC of 8 mcg/mL; penicillin MIC of 8 mcg/mL) |
|||||||||||||
| Ceftriaxone |
Ceftaroline |
Ceftriaxone |
Ceftaroline |
Ceftriaxone |
Ceftaroline |
Ceftriaxone |
Ceftaroline |
|||||||||
| 0.25× MIC | 0.5× MIC | 0.25× MIC | 0.5× MIC | 0.25× MIC | 0.5× MIC | 0.25× MIC | 0.5× MIC | 0.25× MIC | 0.5× MIC | 0.25× MIC | 0.5× MIC | 0.25× MIC | 0.5× MIC | 0.25× MIC | 0.5× MIC | |
| Penicillin | 0.68 ± 0.39 | −0.66 ± 0.79 | −1.90 ± 0.10 | −2.32 ± 0.06 | 0.31 ± 1.45 | 0.85 ± 0.87 | 1.59 ± 0.21 | 1.01 ± 0.35 | 0.40 ± 0.08 | 0.02 ± 0.08 | −0.12 ± 0.13 | −2.34 ± 0.09 | 0.54 ± 0.12 | 0.22 ± 0.32 | 0.63 ± 0.00 | 0.86 ± 0.02 |
| Ampicillin | −3.44 ± 0.70 | −3.81 ± 0.26 | −3.18 ± 0.56 | −3.64 ± 0.36 | 1.24 ± 0.56 | 1.10 ± 0.75 | 1.44 ± 0.07 | 0.30 ± 0.05 | 1.99 ± 0.12 | 1.65 ± 0.19 | −0.27 ± 0.01 | −2.57 ± 0.44 | 0.59 ± 0.00 | 0.56 ± 0.32 | 0.71 ± 0.06 | 0.41 ± 0.06 |
| Piperacillin-tazobactam | 0.08 ± 0.13 | −1.69 ± 0.08 | 1.31 ± 0.03 | 0.14 ± 0.05 | 0.56 ± 0.25 | 0.30 ± 0.05 | 1.82 ± 0.14 | 1.41 ± 0.04 | 1.30 ± 0.15 | 2.23 ± 0.10 | 0.07 ± 0.07 | −2.97 ± 0.30 | 0.42 ± 0.12 | 0.48 ± 0.11 | 0.76 ± 0.12 | 0.32 ± 0.49 |
| Ertapenem | −1.96 ± 0.75 | −2.88 ± 0.09 | −0.32 ± 0.10 | −2.39 ± 0.00 | 1.39 ± 0.02 | 1.42 ± 0.10 | 0.48 ± 0.18 | 1.40 ± 0.07 | 1.02 ± 0.06 | −1.03 ± 0.45 | −0.92 ± 0.11 | −3.49 ± 0.49 | 0.48 ± 0.20 | 0.43 ± 0.04 | 0.35 ± 0.19 | 0.22 ± 0.00 |
| Imipenem-cilastatin | 0.10 ± 0.18 | −1.44 ± 0.08 | 1.48 ± 0.06 | 0.44 ± 0.30 | 0.04 ± 0.32 | 1.18 ± 0.55 | 0.26 ± 0.25 | 1.44 ± 0.07 | 0.08 ± 0.15 | −1.47 ± 0.48 | −1.84 ± 0.04 | −3.88 ± 0.00 | 0.54 ± 0.03 | 0.53 ± 0.10 | 0.42 ± 0.04 | 0.36 ± 0.16 |
| Meropenem | −3.04 ± 0.00 | −3.32 ± 0.71 | −2.95 ± 0.04 | −3.43 ± 0.13 | 1.05 ± 0.72 | 0.43 ± 0.95 | 1.42 ± 0.02 | 0.72 ± 1.13 | −0.57 ± 0.02 | −1.29 ± 1.04 | −1.96 ± 0.04 | −3.54 ± 0.43 | 0.41 ± 0.12 | 0.31 ± 0.20 | 0.37 ± 0.21 | 0.10 ± 0.01 |
Antibacterial activity was defined as bactericidal ≥3 log10 CFU/mL or bacteriostatic <3 log10 CFU/mL decrease from initial inoculum at 24 h.
TABLE 4.
Dual-combination 24-h time-kill synergy results for 0.5× MICa
| Drug at 0.0625× MIC | Change in log10 CFU/mL of strain |
|||||||
|---|---|---|---|---|---|---|---|---|
| Penicillin susceptible |
Penicillin elevated |
|||||||
| JH2-2 (ampicillin MIC of 2 mcg/mL; penicillin MIC of 2 mcg/mL) |
L2052 (ampicillin MIC of 0.5 mcg/mL; penicillin MIC of 2 mcg/mL) |
L2068 (ampicillin MIC of 1 mcg/mL; penicillin MIC of 8 mcg/mL) |
LS4828 (ampicillin MIC of 8 mcg/mL; penicillin MIC of 8 mcg/mL) |
|||||
| Ceftriaxone | Ceftaroline | Ceftriaxone | Ceftaroline | Ceftriaxone | Ceftaroline | Ceftriaxone | Ceftaroline | |
| Penicillin | −0.93 | −3.04 | −0.49 | −0.17 | −1.65 | 0.19 | −0.49 | 0.10 |
| Ampicillin | −4.21 | −4.51 | −0.24 | −0.88 | −0.56 | −0.04 | −0.04 | −0.35 |
| Piperacillin-tazobactam | −2.14 | −0.78 | −1.14 | 0.23 | 0.20 | −0.44 | −0.12 | −0.44 |
| Ertapenem | −3.26 | −3.25 | 0.30 | 0.53 | −3.31 | −0.78 | −0.10 | −0.31 |
| Imipenem-cilastatin | −1.84 | −0.42 | −0.26 | 0.26 | −2.47 | −1.35 | −0.07 | −0.40 |
| Meropenem | −3.63 | −4.20 | −0.78 | −0.15 | −3.56 | −0.83 | −0.22 | −0.43 |
Shown are additional decreases in the log10 CFU/mL of the combination compared to its most active single agent at 24 h. Synergy was defined as a ≥2-log10 CFU/mL decrease of the combination compared to tits most active single agent (dark shading). Indifference was defined as a 1- to 2-log10 CFU/mL decrease of the combination compared to its most active single agent (light shading).
(i) Penicillin-susceptible strains (JH2-2 and L2052).
Antibiotics had isolated specific synergistic activity, as reported in Table 4. JH2-2 ceftriaxone synergy was seen with ampicillin, piperacillin-tazobactam, ertapenem, and meropenem, while ceftaroline synergy against JH2-2 occurred in combination with penicillin, ampicillin, ertapenem, and meropenem. Bactericidal activity was noted against JH2-2 for only ampicillin and meropenem plus either ceftriaxone or ceftaroline. Synergistic and bactericidal activities of antibiotic combinations against the clinical L2052 strain were not seen.
(ii) Elevated-penicillin-MIC strains (L2068 and LS4828).
Ceftriaxone in combination with ertapenem, imipenem-cilastatin, and meropenem was synergistic against L2068; however, the activity was not considered bactericidal. Conversely, ceftaroline combinations against L2068 did not demonstrate synergy, but bactericidal activity was observed in combination with ertapenem, imipenem, and meropenem. All dual-beta-lactam combinations demonstrated were inactive against LS4828.
DISCUSSION
Since beta-lactam antibiotics inhibit the penicillin-binding proteins, the nature of the interactions between beta-lactams and these proteins will have an impact on the in vitro MIC and, presumably, the clinical effectiveness of therapy. Two penicillin-binding proteins in E. faecalis have been implicated in the susceptibility of this species to beta-lactams, PBP4 and PBP2B. Resistance in this species has been most commonly attributed to either increased expression of PBP4 or amino acid substitutions that reduce the affinity for beta-lactam antibiotics. We previously documented the contributions of both PBP4 amino acid substitutions and increased expression of PBP4 to the reduced susceptibility expressed by E. faecalis LS4828 (1). In the present study, we defined the contribution of amino acid substitutions in LS4828 PBP4 and PBP2B to elevated beta-lactam MICs and defined the responses to different beta-lactam combinations of LS4828 and several other E. faecalis strains with different PBP4 and PBP2B sequences and expression profiles.
Our data establish that PBP2B does not appreciably contribute to elevated beta-lactam MICs in LS4828. We were unable to demonstrate any significant differences between the affinities for the fluorescent beta-lactam Bocillin FL of the PBP2Bs from LS4828 and the susceptible strain JH2-2. Competition experiments between Bocillin FL and ampicillin also revealed no difference. It was curious that LS4828 PBP2B was produced in larger quantities than in the other strains examined. However, overexpression seems to have no impact on the elevated MICs. We were able to delete the pbpA gene from LS4828, with minimal consequences for in vitro susceptibility to beta-lactams, leading us to conclude that PBP2B does not contribute to the elevated MICs observed in LS4828.
Interestingly, we found that the deletion of pbpA from LS4828 did not result in increased susceptibility to ceftriaxone, which had been previously observed by Djorić et al. in two strains with pbpA deletions (15). Djorić et al. observed in two different wild-type strains, OG1 and CK221, decreases in the ceftriaxone MICs from 64 to 1 mg/L and 512 to 0.25 mg/L, respectively (15). We did note that the deletion of pbpA (and the consequent absence of PBP2B) resulted in a growth defect in the strains as well as an increase in the observed quantities of PBP4 (also observed by Djorić et al.), which were reduced when complemented by plasmid-encoded PBP2B. The regulatory relationship between PBP4 and PBP2B is interesting and worthy of further study.
Observed synergy with dual beta-lactams against E. faecalis was first described by Mainardi and colleagues with amoxicillin plus cefotaxime against two strains, including JH2-2 (14). Our results confirm synergy for ampicillin and ceftriaxone therapy against this strain. We have also determined alternative synergistic beta-lactam combinations with ceftriaxone plus piperacillin-tazobactam, ertapenem, imipenem, and meropenem against this strain. Additionally, we have shown that the following beta-lactams have synergy with ceftaroline against JH2-2: penicillin, ampicillin, ertapenem, and meropenem. Unlike ertapenem and meropenem combinations, the imipenem-including regimens did not demonstrate consistent synergy, which we do not have an explanation for and which requires further research. This is noteworthy since the Infectious Diseases Society of America (IDSA) infective endocarditis guidelines state that imipenem is the most active carbapenem against E. faecalis (13). The lack of penicillin-ceftriaxone synergy for all strains is concerning as penicillin often replaces ampicillin in clinical practice due to the instability of ampicillin once reconstituted (15, 24, 25). For JH2-2, penicillin demonstrated synergy with ceftaroline, but this was not observed for the other strains. Bacteriostatic activity was observed for penicillin plus ceftaroline against L2068 for 0.5× MIC, which was due to the activity of ceftaroline alone. The observed repeated synergies of ceftriaxone or ceftaroline with either meropenem or ertapenem against a penicillin-susceptible strain and an elevated-penicillin-MIC strain warrant further exploration.
The absence of synergy among other dual-beta-lactam combinations against our second penicillin-susceptible strain (L2052) raises questions regarding the synergy testing methodology as well as the role of PBPs. While L2052 was selected for comparison to JH2-2 due to their similar susceptibility profiles (Table 2), their synergy results were not congruent despite their indistinguishable PBP expression profiles and PBP gene sequences. The inconsistency in synergy could be attributed to the lower ampicillin MIC for L2052 than for JH2-2 (0.5 versus 2 μg/mL, respectively), resulting in lower ampicillin exposure at 0.0625× MIC. This lower ampicillin exposure may have impacted PBP saturation and, thus, the synergism ability. However, a previously published time-kill assay study also demonstrated inconsistency in the synergy of ampicillin plus ceftriaxone, with four out of seven E. faecalis bloodstream isolates demonstrating synergy (26). Subinhibitory concentrations were also utilized, but specific concentrations were not mentioned; thus, future studies are warranted to explore the extent of the concentrations required for PBP saturation.
The least susceptible E. faecalis strain, LS4828, showed no synergism with any combinations. Additionally, the MBCs for this strain, except for ceftaroline, were unattainable due to higher than chemically stable drug concentrations. The elevated MICs of LS4828 may be attributed to it producing roughly six times the amount of PBP4 and nearly five times the amount of PBP2B compared to the susceptible strains as well as having two point mutations with amino acid substitutions (V223I and A617T) and a deletion of an adenine located in the predicted pbp4 promoter sequence compared to JH2-2 (1). Enterococcal tolerance to the bactericidal activity of beta-lactam antibiotics remains a largely unexplained phenomenon, but it often has little, if any, correlation with susceptibility. The tolerance of LS4828 to dual-beta-lactam combinations despite relatively modest increases in MICs (depending on the assay) is of concern in clinical practice if tolerance is a predictor of clinical failure. A recent prospective study found that patients with PRASEF bacteremia treated with ampicillin- or piperacillin-based regimens were observed to have higher mortality rates than patients with penicillin-susceptible E. faecalis bacteremia (18).
E. faecalis L2068 was selected for comparison to LS4828 due to the elevated penicillin MICs and overall MICs of other beta-lactams. The elevated penicillin MICs of this strain were likely due to the increased expression of low-affinity PBP4 and may have been driven, such as in LS4828, by the adenine deletion upstream of the pbp4 promoter. Despite higher MICs, synergy was demonstrated among the carbapenems plus ceftriaxone. For ceftaroline combinations, synergy was not demonstrated. However, the synergy capability was likely impacted by the bacteriostatic activity of ceftaroline monotherapy. This observed activity may be due to higher ceftaroline exposures in the time-kill assay given that the MIC against L2068 was 16 μg/mL. The calculated ceftaroline maximum free, unbound concentration (fCmax) of 27.1 μg/mL and fCmin of 3.5 μg/mL from population pharmacokinetic data, however, show that the concentrations utilized would be physiologically achievable (e.g., 0.5× MIC = 8 μg/mL) (27). Another study assessed subinhibitory concentrations (exact concentrations were not specified) of ampicillin plus ceftaroline against E. faecalis strains with ceftaroline MICs of 8 μg/mL (n = 3) and 16 μg/mL (n = 1) and found that one isolate demonstrated bacteriostatic activity against ceftaroline alone (26). This variation in ceftaroline activity may be due to differences in PBP and warrants further exploration. Ceftaroline-based combinations therefore should be explored further, especially for isolates that have elevated penicillin MICs. Ceftaroline, unlike ceftriaxone, does not promote vancomycin-resistant enterococcus (VRE) colonization, likely due to lack of biliary excretion (ceftriaxone, 65%; ceftaroline, <10%) (28–33). Ceftaroline is also not as commonly found to be an independent predictor of Clostridioides difficile infections due to minimal anaerobic activity (34, 35). Previous in vitro pharmacodynamic models have also demonstrated that ampicillin-ceftaroline has kill that is similar or superior to that of ampicillin-ceftriaxone (26, 36).
A potential limitation of this study is the small number of strains examined and that the amino acid substitution found in LS4828 is not one commonly found in PRASEF strains. This limitation is mitigated by the promoter region deletion found in both LS4828 and L2068, which is quite common among E. faecalis strains that are reported to be resistant and are confirmed to impact MICs (37). Moreover, the literature involving amino acid substitutions in PBP4 is largely associative, with very few instances where the mutation associated with resistance is causative (38). Finally, by drawing an analogy to beta-lactamases, which evolved from PBPs, it is not unreasonable to assume that the therapeutic effect of an antibiotic will correlate with MICs more than with the specifics of amino acid substitutions. As such, we believe that our data provide a reasonable approximation of the likely response of other E. faecalis strains with similar MICs.
Overall, despite the lack of the utilization of standardized drug concentrations against each organism, we have identified potential alternative dual-beta-lactam combinations for serious E. faecalis infections. Our findings also suggest that there is a need for clinical laboratories to report both the ampicillin and penicillin MICs, as elevated penicillin MICs, despite being susceptible, appear to impact the efficacy of the currently utilized dual-beta-lactam combination ampicillin-ceftriaxone. Although isolate dependent, carbapenem-based dual beta-lactams appear to be a viable alternative to ampicillin plus ceftriaxone and require further research.
MATERIALS AND METHODS
Strains.
We studied four strains in these experiments. E. faecalis JH2-2 is a laboratory type strain (23) that has been well described in the literature and was among the strains first demonstrating synergy between amoxicillin and cefotaxime (14). E. faecalis LS4828 is a clinical isolate (1) with reduced susceptibilities to penicillin and ampicillin whose resistance mechanisms have been well characterized. E. faecalis L2052 and L2068 are two clinical blood isolates from the Providence Veterans Affairs Medical Center (VAMC) that were chosen because of their differing susceptibilities to penicillins. Details of whole-genome analysis and PBP deletion and complementation experiments can be found in the supplemental material.
Western blot analysis.
Cell lysates were prepared from mid-log-phase cells grown in 50-mL culture volumes in BHI broth as previously described (1). After centrifugation at 4,200 × g for 30 min, cell pellets were suspended in a solution containing 50 mM Tris-HCl (pH 7.5) and 50 mM NaCl and transferred to a 1.5-mL Eppendorf tube. The cells were processed with a second centrifugation step at 13,000 × g for 2 min, the buffer was discarded, and the cell pellet was stored at −20°C. The cell pellet wet weight was measured for each sample to determine the volume of lysis buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.1% NP-40, and Halt protease inhibitor cocktail [Thermo Fisher Scientific]) added to approximate similar protein concentrations (0.4- to 0.9-mL volumes). Cells were lysed by bead beating with the following modifications. Cells were processed by bead beating in lysing matrix B tubes using the MiniBeadBeater-16 instrument (BioSpec Products) four times for 30 s each with 5-min intervals on ice. Protein concentrations were determined by the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher), and 10 μg of protein was used per lane for SDS-PAGE and Western blot analyses. Samples were prepared in LDS (lithium dodecyl sulfate) sample buffer (Thermo Fisher) with 50 mM dithiothreitol (DTT), heated for 10 min at 70°C, and loaded onto 10% NuPAGE gels (Thermo Fisher). Electrophoresis was performed for 10 min at 170 V and then for 40 min at 190 V using chilled 2-(N-morpholino)ethanesulfonic acid (MES) running buffer (Thermo Fisher). Protein transfer to polyvinylidene difluoride (PVDF) membranes using the Invitrogen iBlot gel transfer system and immunodetection of PBP4 and PBP2B were performed according to methods described previously by Rice et al. (1).
PBP2B polyclonal antibody from rabbit was outsourced to New England Peptide (NEP) (Gardner, MA) using purified PBP2B from E. faecalis JH2-2 (according to the PBP4 protein purification protocol described previously by Rice et al. [1]) as the antigen. This custom antibody resulted in a 0.686-mg/mL concentration of affinity-purified antibody. Specificity and selectivity were determined using bacterial lysates, membrane proteins, and purified expressed proteins for optimal antibody dilutions for the Western blot assays (data not shown). The optimal dilution of the PBP2B antibody for cell lysates, membrane proteins, and purified proteins was determined to be 1/1,000, and that of the horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody was 1/10,000 (Thermo Fisher) (data not shown). The isolation of membrane proteins was performed according to a previously described method used for the E. faecalis JH2-2 and LS4828 strains (1).
To determine the amount of protein transferred, Coomassie blue R-250 was used as a total protein stain for the PVDF membranes after immunoblotting according to a previously published protocol (39). The membrane was washed three times in water, followed by staining for 5 min with Coomassie blue stain containing 0.025% (wt/vol) Coomassie brilliant blue R-250 (Millipore-Sigma) in 40% (vol/vol) methanol–7% (vol/vol) acetic acid. The blot was destained with 50% methanol–7% acetic acid for 5 min, followed by two 10-min washes in water. After air drying, the membrane was imaged with the Bio-Rad Chemidoc XRS+ imager, and the volume intensity data for an equally expressed protein band for the protein loading control were used to normalize the expression values for the respective PBP4 and PBP2B Western blots. Densitometry graphs were generated for comparison of the endogenous expression data for the E. faecalis strains.
Bocillin FL binding of purified PBP2Bs.
All Bocillin FL-binding studies were performed at 37°C in phosphate-buffered saline. Bocillin FL titrations with the purified PBP2Bs derived from the JH2-2 or LS4828 E. faecalis strain were performed with 2.78 μg (3.3 μM) of purified protein in 15-μL reaction mixture volumes containing 0 to 80 μM Bocillin FL. These reaction mixtures were run for 20 min at 37°C, stopped with NuPAGE LDS sample buffer (Thermo Fisher) and 45 mM DTT (Sigma), and then incubated at 70°C for 10 min. Sample volumes of 10 μL were loaded onto 10% Bis-Tris NuPAGE gels and electrophoresed for 60 min at 200 V using chilled MES running buffer (Thermo Fisher). Prior to imaging, gels were washed for 5 min twice with distilled water. The Bio-Rad Chemidoc XRS+ imager was used with the trans-UV setting (302-nm filter), and the relative fluorescence intensities were captured using the autoexposure mode. Analysis was performed using the Lane/Band tools of ImageLab software, and backgrounds were automatically subtracted for each gel. The protein affinity for Bocillin FL was determined by the SDS-PAGE gel-based relative fluorescence units from 2 independent experiments. SimplyBlue SafeStain (Thermo Fisher) was used to stain the gels for confirmation of equal protein loading after the fluorescence imaging of all gels. Ampicillin competition with Bocillin FL was performed with 2-fold serial dilutions of ampicillin added to the reaction mixtures with 3.3 μM PBP2B for 10 min prior to the addition of Bocillin FL and then treated as described above for the SDS-PAGE gel assay. Ampicillin IC50 calculations were performed using the percent inhibition of the relative fluorescence intensity values from ImageLab software.
Time-kill experiments.
All isolates were streaked fresh from −80°C stored culture stocks and incubated for 18 to 24 h at 35°C to 37°C before use in time-kill assays. Ampicillin and penicillin were selected as the primary beta-lactams active against E. faecalis. We additionally tested piperacillin-tazobactam, ertapenem, meropenem, and imipenem as these agents are utilized in clinical practice with limited evidence (13). The above-mentioned agents were combined with ceftriaxone given guideline recommendations and previous success with ceftaroline (13, 26, 36). Ampicillin (National Drug Code [NDC] 25021-136-10; Sagent Pharmaceuticals), penicillin G potassium (USP reference standard catalog number 1502508; Sigma-Aldrich, St. Louis, MO), piperacillin-tazobactam (NDC 63323-309-20; Fresenius Kabi USA, LLC), ertapenem (NDC 0006-3843-71; Merck & Co., Inc.), meropenem (NDC 63323-508-30; Fresenius Kabi USA, LLC), imipenem-cilastatin (NDC 0006-3516-59; Merck & Co., Inc.), ceftriaxone (NDC 25021-106-10; Sagent Pharmaceuticals), and ceftaroline dihydrochloride (Allergan, Dublin, Ireland) stock solutions were prepared each week, according to the package inserts, and stored at −20°C.
According to Clinical and Laboratory Standards Institute (CLSI) guidance, Mueller-Hinton broth (MHB; Becton, Dickinson, Sparks, MD, USA), adjusted to 25 mg/L calcium and 12.5 mg/L magnesium, was used for all susceptibility testing and time-kill assays, as previously described (22, 36, 40, 41). Streaks and colonies were plated onto brain heart infusion agar (BHA; Difco, Becton, Dickinson) due to better growth (1, 14, 42).
MIC and minimum bactericidal concentration (MBC) testing was performed by broth microdilution, in duplicate from independent cultures grown overnight that were set up from a single streak overnight, according to CLSI guidelines (22, 41).
In vitro time-kill assay.
Traditional 24-h time-kill assays were performed in duplicate from independent cultures grown overnight that were set up from a single streak overnight as previously described (26, 43, 44). Subinhibitory concentrations of 0.0625× the respective MICs of ampicillin, penicillin, piperacillin-tazobactam, ertapenem, imipenem-cilastatin, and meropenem were tested as monotherapies and in combination with 0.25× and 0.5× the respective MICs of ceftriaxone and ceftaroline. We selected 0.0625× MIC based on subinhibitory concentrations utilized previously by Mainardi et al., who first discovered the in vitro activity of dual beta-lactams against E. faecalis (14). Ceftriaxone and ceftaroline at 0.25× and 0.5× the respective MICs were tested as monotherapies for comparison. We tested various concentrations of ampicillin and ceftriaxone against JH2-2 (data not presented) and found similar results among all concentrations, thus supporting our concentrations tested against other isolates. Samples were obtained to assess the CFU per milliliter at 0, 4, and 24 h. The lower limit of detection was 2.0 log10 CFU/mL.
Activity defined.
Combination therapy activity was described at 24 h as synergy or indifference. Synergy was defined as a ≥2-log10 CFU/mL decrease of the combination from its most active single agent. Additivity was defined as a 1- to 2-log10 CFU/mL decrease of the combination from the most active single agent, and indifference was defined as a <1-log10 CFU/mL decrease (45–47). Antibacterial activity was described as bactericidal or bacteriostatic, defined as a ≥3-log10 CFU/mL or a <3-log10 CFU/mL decrease from the initial inoculum at 24 h, respectively.
ACKNOWLEDGMENTS
Ceftaroline research powder was provided by Allergan.
This work was supported in part by the Office of Academic Affiliations (OAA) at the Department of Veterans Affairs and by COIN, the Center of Innovation in Long-Term Services and Supports for Vulnerable Veterans, Providence, RI. The information provided in the manuscript are is that of the authors and does not necessarily reflect the position or policy of the United States Department of Veterans Affairs. This work was supported in part by award R01 AI141522-01A1 from the National Institute of Allergy and Infectious Diseases.
Kerry L. LaPlante receives research funding from Merck, Pfizer, and Shionogi. Jaclyn A. Cusumano, Kathryn E. Daffinee, Mónica García-Solache, and Charlene Desbonnet have no reported financial relationships relevant to this article. Louis B. Rice serves on a clinical trial data safety monitoring board for VenatoRx Pharmaceuticals.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Rice LB, Desbonnet C, Tait-Kamradt A, Garcia-Solache M, Lonks J, Moon TM, D’Andréa ÉD, Page R, Peti W. 2018. Structural and regulatory changes in PBP4 trigger decreased β-lactam susceptibility in Enterococcus faecalis. mBio 9:e00361-18. 10.1128/mBio.00361-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conceicao N, da Silva LE, Darini AL, Pitondo-Silva A, de Oliveira AG. 2014. Penicillin-resistant, ampicillin-susceptible Enterococcus faecalis of hospital origin: pbp4 gene polymorphism and genetic diversity. Infect Genet Evol 28:289–295. 10.1016/j.meegid.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Ono S, Muratani T, Matsumoto T. 2005. Mechanisms of resistance to imipenem and ampicillin in Enterococcus faecalis. Antimicrob Agents Chemother 49:2954–2958. 10.1128/AAC.49.7.2954-2958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan YE, Ng LSY, Tan TY. 2014. Evaluation of Enterococcus faecalis clinical isolates with ‘penicillin-resistant, ampicillin-susceptible’ phenotype as reported by Vitek-2 Compact system. Pathology 46:544–550. 10.1097/PAT.0000000000000146. [DOI] [PubMed] [Google Scholar]
- 5.Metzidie E, Manolis EN, Pournaras S, Sofianou D, Tsakris A. 2006. Spread of an unusual penicillin- and imipenem-resistant but ampicillin-susceptible phenotype among Enterococcus faecalis clinical isolates. J Antimicrob Chemother 57:158–160. 10.1093/jac/dki427. [DOI] [PubMed] [Google Scholar]
- 6.Guardabassi L, Larsen J, Skov R, Schonheyder HC. 2010. Gentamicin-resistant Enterococcus faecalis sequence type 6 with reduced penicillin susceptibility: diagnostic and therapeutic implications. J Clin Microbiol 48:3820–3821. 10.1128/JCM.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jawetz E, Sonne M. 1966. Penicillin-streptomycin treatment of enterococcal endocarditis. A re-evaluation. N Engl J Med 274:710–715. 10.1056/NEJM196603312741304. [DOI] [PubMed] [Google Scholar]
- 8.Deshpande LM, Fritsche TR, Moet GJ, Biedenbach DJ, Jones RN. 2007. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis 58:163–170. 10.1016/j.diagmicrobio.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Araoka H, Kimura M, Yoneyama A. 2011. A surveillance of high-level gentamicin-resistant enterococcal bacteremia. J Infect Chemother 17:433–434. 10.1007/s10156-010-0175-0. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Hidalgo N, Almirante B, Gavalda J, Gurgui M, Pena C, de Alarcon A, Ruiz J, Vilacosta I, Montejo M, Vallejo N, Lopez-Medrano F, Plata A, Lopez J, Hidalgo-Tenorio C, Galvez J, Saez C, Lomas JM, Falcone M, de la Torre J, Martinez-Lacasa X, Pahissa A. 2013. Ampicillin plus ceftriaxone is as effective as ampicillin plus gentamicin for treating Enterococcus faecalis infective endocarditis. Clin Infect Dis 56:1261–1268. 10.1093/cid/cit052. [DOI] [PubMed] [Google Scholar]
- 11.Pericas JM, Cervera C, del Rio A, Moreno A, Garcia de la Maria C, Almela M, Falces C, Ninot S, Castaneda X, Armero Y, Soy D, Gatell JM, Marco F, Mestres CA, Miro JM, Hospital Clinic Endocarditis Study Group . 2014. Changes in the treatment of Enterococcus faecalis infective endocarditis in Spain in the last 15 years: from ampicillin plus gentamicin to ampicillin plus ceftriaxone. Clin Microbiol Infect 20:O1075–O1083. 10.1111/1469-0691.12756. [DOI] [PubMed] [Google Scholar]
- 12.Gavalda J, Len O, Miro JM, Munoz P, Montejo M, Alarcon A, de la Torre-Cisneros J, Pena C, Martinez-Lacasa X, Sarria C, Bou G, Aguado JM, Navas E, Romeu J, Marco F, Torres C, Tornos P, Planes A, Falco V, Almirante B, Pahissa A. 2007. Brief communication: treatment of Enterococcus faecalis endocarditis with ampicillin plus ceftriaxone. Ann Intern Med 146:574–579. 10.7326/0003-4819-146-8-200704170-00008. [DOI] [PubMed] [Google Scholar]
- 13.Baddour LM, Wilson WR, Bayer AS, Fowler VG, Jr, Tleyjeh IM, Rybak MJ, Barsic B, Lockhart PB, Gewitz MH, Levison ME, Bolger AF, Steckelberg JM, Baltimore RS, Fink AM, O’Gara P, Taubert KA, American Heart Association Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and Stroke Council . 2015. Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications. A scientific statement for healthcare professionals from the American Heart Association. Circulation 132:1435–1486. 10.1161/CIR.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 14.Mainardi JL, Gutmann L, Acar JF, Goldstein FW. 1995. Synergistic effect of amoxicillin and cefotaxime against Enterococcus faecalis. Antimicrob Agents Chemother 39:1984–1987. 10.1128/AAC.39.9.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djorić D, Little JL, Kristich CJ. 2020. Multiple low-reactivity class B penicillin-binding proteins are required for cephalosporin resistance in enterococci. Antimicrob Agents Chemother 64:e02273-19. 10.1128/AAC.02273-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Infante VHP, Conceicao N, de Oliveira AG, Darini ALDC. 2016. Evaluation of polymorphisms in pbp4 gene and genetic diversity in penicillin-resistant, ampicillin-susceptible Enterococcus faecalis from hospitals in different states in Brazil. FEMS Microbiol Lett 363:fnw044. 10.1093/femsle/fnw044. [DOI] [PubMed] [Google Scholar]
- 17.Duez C, Zorzi W, Sapunaric F, Amoroso A, Thamm I, Coyette J. 2001. The penicillin resistance of Enterococcus faecalis JH2-2r results from an overproduction of the low-affinity penicillin-binding protein PBP4 and does not involve a psr-like gene. Microbiology (Reading) 147:2561–2569. 10.1099/00221287-147-9-2561. [DOI] [PubMed] [Google Scholar]
- 18.Kim D, Lee H, Yoon E-J, Hong JS, Shin JH, Uh Y, Shin KS, Shin JH, Kim YA, Park YS, Jeong SH. 2019. Prospective observational study of the clinical prognoses of patients with bloodstream infections caused by ampicillin-susceptible but penicillin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 63:e00291-19. 10.1128/AAC.00291-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miro JM, Pericas JM, del Rio A, Hospital Clinic Endocarditis Study Group . 2013. A new era for treating Enterococcus faecalis endocarditis. Ampicillin plus short-course gentamicin or ampicillin plus ceftriaxone: that is the question! Circulation 127:1763–1766. 10.1161/CIRCULATIONAHA.113.002431. [DOI] [PubMed] [Google Scholar]
- 20.Mylonakis E, Calderwood SB. 2001. Infective endocarditis in adults. N Engl J Med 345:1318–1330. 10.1056/NEJMra010082. [DOI] [PubMed] [Google Scholar]
- 21.Megran DW. 1992. Enterococcal endocarditis. Clin Infect Dis 15:63–71. 10.1093/clinids/15.1.63. [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing: 29th informational supplement (M100-ED30). Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.Arbeloa A, Segal H, Hugonnet J-E, Josseaume N, Dubost L, Brouard J-P, Gutmann L, Mengin-Lecreulx D, Arthur M. 2004. Role of class A penicillin-binding proteins in PBP5-mediated β-lactam resistance in Enterococcus faecalis. J Bacteriol 186:1221–1228. 10.1128/JB.186.5.1221-1228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tritle BJ, Timbrook TT, Fisher MA, Spivak ES. 2020. Penicillin as a potential agent for dual β-lactam therapy for enterococcal endocarditis. Clin Infect Dis 70:1263–1264. 10.1093/cid/ciz594. [DOI] [PubMed] [Google Scholar]
- 25.Lines TH, Greene MH, Nesbitt WJ, Stratton CW, Schmitz JE. 2019. Enterococcus faecalis and penicillin susceptibility testing: the challenge of multiple methods and agent-to-agent predictions. J Appl Lab Med 3:730–732. 10.1373/jalm.2018.027821. [DOI] [PubMed] [Google Scholar]
- 26.Werth BJ, Abbott AN. 2015. The combination of ampicillin plus ceftaroline is synergistic against Enterococcus faecalis. J Antimicrob Chemother 70:2414–2417. 10.1093/jac/dkv125. [DOI] [PubMed] [Google Scholar]
- 27.Matzneller P, Lackner E, Lagler H, Wulkersdorfer B, Osterreicher Z, Zeitlinger M. 2016. Single- and repeated-dose pharmacokinetics of ceftaroline in plasma and soft tissues of healthy volunteers for two different dosing regimens of ceftaroline fosamil. Antimicrob Agents Chemother 60:3617–3625. 10.1128/AAC.00097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakticova V, Hutton-Thomas R, Meyer M, Gurkan E, Rice LB. 2006. Antibiotic-induced enterococcal expansion in the mouse intestine occurs throughout the small bowel and correlates poorly with suppression of competing flora. Antimicrob Agents Chemother 50:3117–3123. 10.1128/AAC.00125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panagiotidis G, Backstrom T, Asker-Hagelberg C, Jandourek A, Weintraub A, Nord CE. 2010. Effect of ceftaroline on normal human intestinal microflora. Antimicrob Agents Chemother 54:1811–1814. 10.1128/AAC.01716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arvidsson A, Alvan G, Angelin B, Borga O, Nord CE. 1982. Ceftriaxone: renal and biliary excretion and effect on the colon microflora. J Antimicrob Chemother 10:207–215. 10.1093/jac/10.3.207. [DOI] [PubMed] [Google Scholar]
- 31.Karachalios G, Charalabopoulos K. 2002. Biliary excretion of antimicrobial drugs. Chemotherapy 48:280–297. 10.1159/000069712. [DOI] [PubMed] [Google Scholar]
- 32.Riccobene T, Jakate A, Rank D, Thye D. 2009. An open-label, pharmacokinetic, safety and tolerability study of single-dose intravenous ceftaroline in subjects with end-stage renal disease on intermittent haemodialysis, abstr P1455. Abstr 19th Annu Meet Eur Soc Clin Microbiol Infect Dis (ECCMID), Helsinki, Finland.
- 33.Ge Y, Maynard D, Rickert DE. 2010. Comparative pharmacokinetics of ceftaroline in rats, rabbits, and monkeys following a single intravenous or intramuscular injection. Antimicrob Agents Chemother 54:912–914. 10.1128/AAC.00645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown KA, Khanafer N, Daneman N, Fisman DN. 2013. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob Agents Chemother 57:2326–2332. 10.1128/AAC.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilcox MH, Chalmers JD, Nord CE, Freeman J, Bouza E. 2017. Role of cephalosporins in the era of Clostridium difficile infection. J Antimicrob Chemother 72:1–18. 10.1093/jac/dkw385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luther MK, Rice LB, LaPlante KL. 2016. Ampicillin in combination with ceftaroline, cefepime, or ceftriaxone demonstrates equivalent activities in a high-inoculum Enterococcus faecalis infection model. Antimicrob Agents Chemother 60:3178–3182. 10.1128/AAC.03126-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazzaro LM, Cassisi M, Stefani S, Campanile F. 2021. Impact of PBP4 alterations on β-lactam resistance and ceftobiprole non-susceptibility among Enterococcus faecalis clinical isolates. Front Cell Infect Microbiol 11:816657. 10.3389/fcimb.2021.816657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gawryszewska I, Żabicka D, Hryniewicz W, Sadowy E. 2021. Penicillin-resistant, ampicillin-susceptible Enterococcus faecalis in Polish hospitals. Microb Drug Resist 27:291–300. 10.1089/mdr.2019.0504. [DOI] [PubMed] [Google Scholar]
- 39.Goldman A, Harper S, Speicher DW. 2016. Detection of proteins on blot membranes. Curr Protoc Protein Sci 86:10.8.1–10.8.11. 10.1002/cpps.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luther MK, Arvanitis M, Mylonakis E, LaPlante KL. 2014. Activity of daptomycin or linezolid in combination with rifampin or gentamicin against biofilm-forming Enterococcus faecalis or E. faecium in an in vitro pharmacodynamic model using simulated endocardial vegetations and an in vivo survival assay using Galleria mellonella larvae. Antimicrob Agents Chemother 58:4612–4620. 10.1128/AAC.02790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clinical and Laboratory Standards Institute. 2019. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed (M07-A11). Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 42.Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, Murray BE, Arias CA. 2012. Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob Agents Chemother 56:4354–4359. 10.1128/AAC.00509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaPlante KL, Rybak MJ. 2004. Clinical glycopeptide-intermediate staphylococci tested against arbekacin, daptomycin, and tigecycline. Diagn Microbiol Infect Dis 50:125–130. 10.1016/j.diagmicrobio.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 44.Smith JR, Barber KE, Raut A, Aboutaleb M, Sakoulas G, Rybak MJ. 2015. β-Lactam combinations with daptomycin provide synergy against vancomycin-resistant Enterococcus faecalis and Enterococcus faecium. J Antimicrob Chemother 70:1738–1743. 10.1093/jac/dkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonapace CR, White RL, Friedrich LV, Bosso JA. 2000. Evaluation of antibiotic synergy against Acinetobacter baumannii: a comparison with Etest, time-kill, and checkerboard methods. Diagn Microbiol Infect Dis 38:43–50. 10.1016/s0732-8893(00)00163-2. [DOI] [PubMed] [Google Scholar]
- 46.Greco WR, Bravo G, Parsons JC. 1995. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev 47:331–385. [PubMed] [Google Scholar]
- 47.Asempa TE, Nicolau DP, Kuti JL. 2019. In vitro activity of imipenem-relebactam alone or in combination with amikacin or colistin against Pseudomonas aeruginosa. Antimicrob Agents Chemother 63:e00997-19. 10.1128/AAC.00997-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.00871-22-s0001.pdf, PDF file, 1.46 MB (1.4MB, pdf)
Supplemental materials and methods. Download aac.00871-22-s0002.docx, DOCX file, 0.03 MB (34.7KB, docx)