ABSTRACT
Phenotypic heterogeneity is crucial to bacterial survival and could provide insights into the mechanism of action (MOA) of antibiotics, especially those with polypharmacological actions. Although phenotypic changes among individual cells could be detected by existing profiling methods, due to the data complexity, only population average data were commonly used, thereby overlooking the heterogeneity. In this study, we developed a high-resolution bacterial cytological profiling method that can capture morphological variations of bacteria upon antibiotic treatment. With an unprecedented single-cell resolution, this method classifies morphological changes of individual cells into known MOAs with an overall accuracy above 90%. We next showed that combinations of two antibiotics induce altered cell morphologies that are either unique or similar to that of an antibiotic in the combinations. With these combinatorial profiles, this method successfully revealed multiple cytological changes caused by a natural product-derived compound that, by itself, is inactive against Acinetobacter baumannii but synergistically exerts its multiple antibacterial activities in the presence of colistin. The findings have paved the way for future single-cell profiling in bacteria and have highlighted previously underappreciated intrapopulation variations caused by antibiotic perturbation.
KEYWORDS: antibiotic combinations, phenotypic heterogeneity, high-content analysis, single-cell profiling
INTRODUCTION
Since the discovery of antibiotics, humans have enjoyed decades of success in controlling bacterial infections. However, the overuse and misuse of antibiotics have caused multidrug-resistant (MDR) pathogens to emerge at an alarming rate. Consequently, it is now well recognized by researchers around the globe that the postantibiotic era might arrive sooner than previously assumed (1, 2). In recent years, bacteria have become resistant to almost all antibiotics used clinically, and the number is rapidly rising (3–5). The widespread incidences of MDR Enterococcus spp., Staphylococcus aureus, Klebsiella spp., Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp., collectively called “ESKAPE,” are among the most worrisome. A. baumannii in particular was listed as one of the critical priority pathogens worldwide by WHO in 2017 (5). Many of A. baumannii clinical isolates are resistant to routinely used antibiotics and, carbapenem-resistant A. baumannii (CRAB) is particularly concerning. The outbreak of CRAB has been reported worldwide and recognized as a serious public health issue due to the limitation of treatment available, high cost, and high mortality rate (6–9). Moreover, some strains are also now resistant to the last line of antibiotics for Gram-negative pathogens, including colistin (10–12). Thus, novel antibacterial molecules that are effective against the bacteria are urgently needed to keep them at bay.
In search of novel antibiotics, we recently applied a fluorescence microscopy-based method called bacterial cytological profiling (BCP) (13, 14) to study the mechanism of action (MOA) of antibiotics that are active against A. baumannii (15). BCP identified different MOAs of known antibiotics by analyzing cell morphological profiles upon antibiotic treatment, and it also correctly identified the MOA of unknown compounds, without prior knowledge of its MOA or structure. In principle, a cytological profiling method such as BCP provides morphological data at a single-cell level. However, in order to accurately determine the MOA of compounds, a simple population averaging method had been used (13, 15, 16), thereby losing the ability to capture heterogeneity. It has long been known that phenotypic variations among individual cells are observable even in isogenic populations of bacteria growing under the same environmental conditions (17, 18). The physiological heterogeneity, in an evolutionary perspective, plays a crucial role in ensuring survival of the microorganism under various environmental stresses, including antibiotic treatment (19, 20). Thus, quantitation of heterogeneity in response to treatment can be used to better understand how antibiotics orchestrate their activity in individual cells and provides insight into antibiotic MOA and resistance.
The ability to dissect an individual cell’s response not only provides insight into the MOA of antibiotics but also possibly reveals multiple MOAs simultaneously. Even though population-based BCP analysis was successful in dissecting morphological changes caused by two MOAs at once, it overlooked phenotypic heterogeneity, thereby reporting multiple detected cell profiles as a single MOA (21–23). For example, aminoglycosides are known to both inhibit protein translation and perturb the membrane (24). Using population averages with BCP, aminoglycoside-treated cells were categorized into a single MOA profile of protein translation inhibition, profile P2 (13, 15), despite presenting clear heterogeneity at a single-cell level. This suggests that upon exposure to a single antibiotic with multiple MOAs or multiple antibiotics, the phenotypic heterogeneity was not properly captured by population-based BCP. Single-cell analysis has played a vital role in examining heterogeneity of cellular response in other omics-based profiling methods, leading to an insight into not only disease pathogenesis but also the MOA of drugs (25–28). Recently, various single-cell microbiology studies have provided a fundamental understanding of the heterogenous response of the individual bacterial cell to antibiotics, which is crucial in advancing antibiotic discovery (29–31). Therefore, enhancing BCP resolution to a single-cell level is particularly promising in order to provide a clearer picture of heterogenous bacterial cell profiles and to accurately identify multiple MOAs simultaneously.
In the past decade, the advancement of applied mathematics, engineering, and computer science has accelerated the development of single-cell technologies, delivering both single-cell microscopy and an exceptional quantitative analysis of individual cell profiles (32, 33). In order to identify phenotypic variations, marginal in most cases, among individual cells, machine learning strategies are often introduced into the analysis pipeline to unbiasedly and accurately capture meaningful morphological data patterns that are related to the perturbation of interest (34, 35). This is especially crucial in bacterial cell morphology analysis, where the small size of bacteria limits human visual perception. In this study, we first implemented machine learning strategies in combination with BCP to improve the method’s resolution down to a single-cell level. Then, we showed that this machine learning-assisted single-cell BCP method can identify subprofiles of bacterial cells when treated with antibiotics. Next, we demonstrated that the method can be used to reveal combinatorial effects of dual antibiotic treatment on morphological changes and provide insight into relationships between each morphological profile. Finally, we used the method and the knowledge of dual-treatment combinatorial effects to reveal multiple MOAs of a natural product-derived compound that synergistically kills A. baumannii together with colistin. Altogether, we envision that our findings will enhance the use of BCP as a high-resolution MOA-defining method, improving the screening of compounds and assisting the process of discovery of much-needed antibacterial molecules.
RESULTS
Extended bacterial morphological features were used for MOA classification via high-resolution BCP.
Our previous study indicated that by analyzing 36 bacterial morphological parameters upon antibiotic treatment, BCP in A. baumannii can distinguish different MOAs of antibiotics in spite of inhibiting the same cellular pathway (15). As a result, 8 different MOA profiles, from antibiotics targeting 6 cellular pathways, were reported: protein translation (P1 and P2), cell wall synthesis (C1 and C2), DNA replication (D1), RNA transcription (R1), lipid synthesis (L1), and membrane integrity (M1) (see Table S1 in the supplemental material). Although morphological differences between each profile can easily be detected by mean population data based on morphological features that were selected by experienced researchers (15), subtle variations at the single-cell level that are crucial for differentiating MOA pose a major challenge, thus rendering human-oriented morphological feature selection unsuitable for single-cell variation analysis.
High dimensionality of data sets obtained from extracting hundreds of morphological features from images is known to be one of the major challenges affecting classification accuracy of the analysis (36, 37). Thus, feature selection is necessary to reduce the dimension of the data set by selecting a subset of features that is most relevant to the analysis. In this study, 1,000 individual cell profiles were first randomly selected from three independent experiments for each antibiotic treatment as detailed in Materials and Methods and used as a data set for feature selection. Then, we used the recursive feature elimination with cross-validation based on support vector machine (RFECV-SVM) method (38), which was shown to effectively handle high-dimensional biological data sets (39–41), to identify optimal morphological features that can classify A. baumannii cells treated with different antibiotics into distinct MOA profiles. Of all 156 features from image analysis, 62 features were selected for MOA profile classification analysis with an overall model accuracy at 92.97% (Fig. S1A and B and Table S2). Unsurprisingly, piperacillin (PIP), meropenem (MER), and ciprofloxacin (CIP) were among the top ranks in accuracy (>95%). The previous study found that these antibiotics cause noticeable morphological changes such as elongated cells from PIP and CIP and ovoid cells from MER (15). In contrast, triclosan (TCS)- and colistin (CST)-treated cells resulted in precisions below 90% possibly due to their subtle morphological changes. These results are in agreement with our previous finding that cells treated with these antibiotics clustered closely together and centered around untreated cells (15). Amikacin (AMI), which was previously shown to have more than one morphology, also resulted in a marginally lower precision, 88%. Collectively, the results show that high-resolution BCP can classify individual antibiotic-treated cells according to MOAs of antibiotics with an overall accuracy above 90%.
High-resolution BCP reveals intrapopulation variation in A. baumannii cells upon antibiotic treatment.
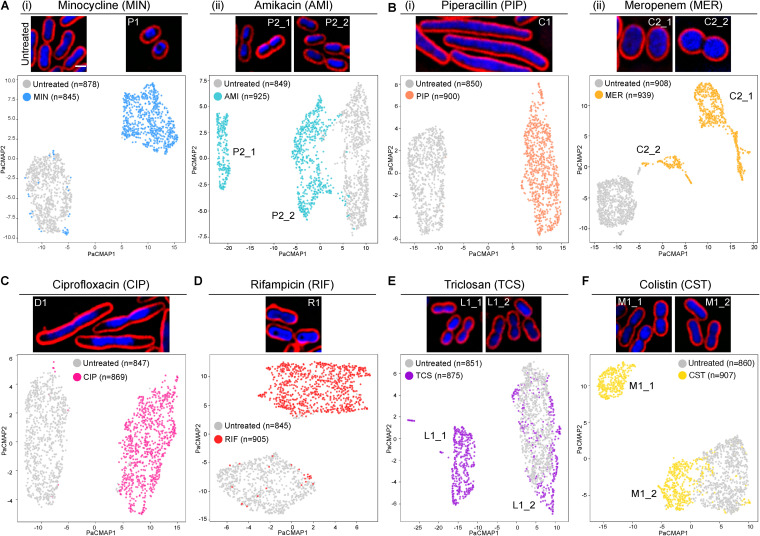
Once the single-cell analysis pipeline had been obtained, we next determined if, at a single-cell resolution, BCP could provide more information regarding the heterogenous response of the cell for each of the reported MOA profiles. Minocycline (MIN)-treated cells, which were known to display a signature toroidal DNA, were grouped into only one cluster, P1_1 (Fig. 1A, panel i), while AMI-treated cells were separated into two subprofiles (Fig. 1A, panel ii). Cells in subprofile P2_1 consisted of doublet cells containing two separated condensed DNA, while cells in P2_2 showed single, slightly condensed nucleoids. Due to the strain difference, A. baumannii ATCC 17978 used in this study, when treated with AMI, did not result in signature toroidal DNA as observed previously in A. baumannii strain ATCC 19606. This is not unexpected since the difference in morphological response to antibiotics between the two strains was previously reported (15). A cell wall synthesis inhibitor, MER, produced ovoid cells that were divided into two subprofiles: C2_1 and C2_2 (Fig. 1B, panel ii). The majority of the cells were in C2_1, i.e., were ovoid, and the rest were clustered into profile C2_2, which consisted of doublet ovoid cells whose cell division was incomplete. These findings are probably due to the meropenem affinity toward different penicillin binding proteins (PBPs) of the bacteria (42). It is possible that cells in which the drug mostly affected PBP2 showed the ovoid shape and clustered in C2_1 group, while cells whose PBP2 and PBP3 were both affected might have shown both ovoid and incomplete cell division and hence clustered into C2_2.
FIG 1.
High-resolution BCP analysis reveals intrapopulation variation of A. baumannii upon antibiotic treatment. Representative cell images and PaCMAP plots are shown. (A) Untreated A. baumannii cells; (i) MIN-treated and (ii) AMI-treated cells; (B) (i) PIP-treated and (ii) MER-treated cells; (C) CIP-treated cells, (D) RIF-treated cells, (E) TCS-treated cells; (F) CST-treated cells. Bacterial cells were treated with 1× MIC of each antibiotic for 1 h and stained with fluorescent dyes. Scale bar represents 1 μm.
Unlike those antibiotics that resulted in multiple subprofiles, PIP (Fig. 1B, panel i), CIP (Fig. 1C) and rifampicin (RIF) (Fig. 1D) displayed a single cluster. Even though TCS (Fig. 1E) and CST (Fig. 1F) were shown to yield two separate subprofiles, one subprofile from each (L1_2 and M1_2) was positioned in close proximity to the untreated cluster. This indistinguishability from the untreated cells explains why the previous study of average-based analysis of these two antibiotics saw them closely clustered to the untreated cells (15). Altogether, we showed that high-resolution BCP is not only able to classify MOA profiles of antibiotics with more than 90% accuracy but also capture morphological variations within the population upon antibiotic treatment.
Combinatorial effects of two antibiotics caused either unique or overlapping morphologies with one of the two antibiotics in the combination.
The ability of the BCP to dissect multiple subprofiles of antibiotic-treated cells urged us to determine if the method can reveal the effect of two antibiotics on bacterial cell morphology. A method that can determine MOAs of mixed compounds simultaneously or compounds possessing multiple MOAs could play an important role in accelerating antibiotic discovery, such as screening for active compounds for combination therapy (43–46). Although previous studies have shown that the combined effect of two antibiotics could be detected using population-based BCP (21, 23) or analysis of representative morphological changes of imaging fields (47), a comprehensive framework of single-cell BCP on antibiotic combinations is still lacking.
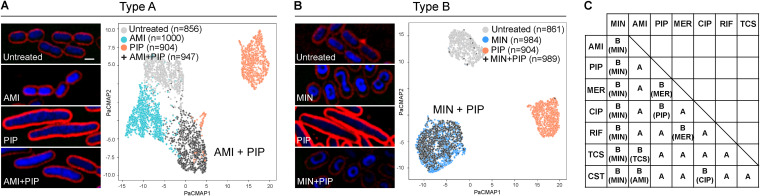
To test if the effect of two antibiotics could be detected, all combinations of the previous 8 MOA profiles (total of 28 combinations) were studied. For each combination, cell features were selected using the RFECV-SVM model based on each single drug in the combination to accurately select relevant features for combinatorial effect analysis (Fig. S2A). By using a similarity index cutoff of 0.75, which was calculated as the lowest similarity index between two randomly divided groups of single antibiotic-treated cells (see Materials and Methods), the 28 combinations were divided into unique (type A) and overlapping (type B) morphological responses (Fig. 2). For type A, the combined effect of the two drugs on the morphology of bacteria resulted in morphological profiles that were clearly separated from the profiles of the single antibiotic treatments (similarity index < 0.75). For example, cells treated with amikacin plus piperacillin (AMI+PIP) exhibited a brighter 4′,6-diamidino-2-phenylindole (DAPI) signal and were longer, which are the morphological changes that are influenced by AMI and PIP, respectively. Thus, the combined morphology gave rise to separate cell profiles that did not significantly overlap individual profiles of either AMI or PIP (Fig. 2A). On the contrary, for type B, the effect of the drug combination resulted in a morphological profiles that was similar to that of either one of the two in the combination (similarity index ≥ 0.75). For instance, treatment with MIN+PIP resulted in a cell profile that was indistinguishable from that of cells treated with MIN alone (Fig. 2B). In summary, from all the 28 combinations (Fig. 2C and Fig. S2B), we found 15 antibiotic pairs that gave rise to morphological changes that were separate from those of single antibiotic-treated cells (type A), suggesting the ability of the method to identify unique morphological changes caused by two antibiotics. However, the combinatorial effect of the other 13 pairs on morphological profiles overlapped with one of the two antibiotics in the combination and hence could not be separated by the method (type B).
FIG 2.
High-resolution BCP analysis of A. baumannii treated with antibiotic combination reveals unique (type A) or overlapping (type B) cytological profiles. (A) Representative cell images and PaCMAP plots of AMI+PIP-treated cells showing unique profiles compared with cells treated with a single antibiotic or untreated cells. (B) Representative cell images and PaCMAP plots of PaCMAP plots of MIN+PIP-treated cells showing overlapped profiles with cells treated with a single antibiotic. Cells were treated with 1× MIC of each antibiotic for 1 h, for both single-drug and antibiotic combination conditions. Scale bar represents 1 μm. (C) Summary of combined morphology types of all 28 combinations. For type B, combined morphology overlapped with the profile of antibiotics shown in parentheses.
It is interesting that antibiotic combinations that resulted in type A are likely to be from the antibiotics causing prominent cell morphological changes such as PIP, CIP, and MER (Fig. 1B and C). Cell length and width have been elucidated to be the major features that contributed to the morphology variation in Escherichia coli (48); therefore, it is possible that the extreme alteration of cell area-related features caused by one of these antibiotics offers more morphological space for the additional antibiotic to show its effect that would, in turn, appear as a “mixed morphology.” In contrast, when MIN was introduced into the combination, its effect on cell morphology dominated all other antibiotics and thus the combined treatments were always found to be largely overlapped with MIN, resulting in type B (Fig. 2C).
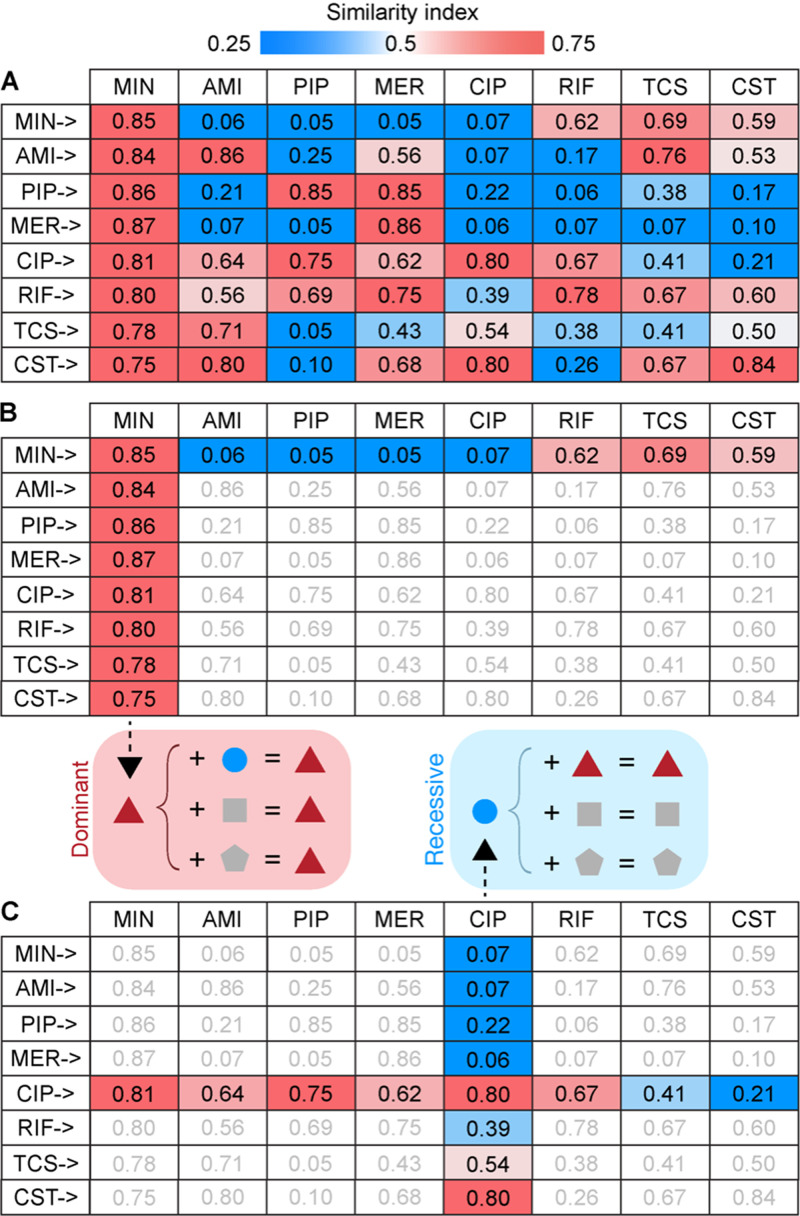
In order to understand the likelihood of morphological changes caused by each antibiotic to become dominant or recessive in combination treatments as observed in type B, the similarity index of each drug toward the other, for each combination, was analyzed (Fig. 3A). In the case of MIN, every combination yielded a higher index of similarity to the profile of MIN (Fig. 3B; similarity index ≥ 0.75 in all cases) and lower similarity to the profile of the other antibiotic, suggesting that morphological changes caused by MIN are likely to be dominant and resilient against those caused by the other antibiotics in the combination. This trend was also observed in MER, although to a lesser degree of dominance (Fig. 3A). In contrast, the presence of CIP in combination with other antibiotics yielded a lower index of similarity to its own profile (Fig. 3C) but a higher index of similarity to the others, suggesting that morphological changes caused by CIP are likely to be masked by the effect of the others. Therefore, dominant or recessive traits of antibiotics present in the combination play an important role in understanding combinatorial effects of two antibiotics on bacterial morphological changes.
FIG 3.
Combinatorial effects of two antibiotics on bacterial morphological changes influenced by dominant or recessive profiling traits of antibiotics. (A) Summary of the indices of similarity of all antibiotic combinations to the antibiotics indicated in the top row. (B) Profiles of cells treated with antibiotic combination containing MIN showed a higher index of similarity to the MIN profile and a lower index of similarity to the other. Antibiotics with a dominant profiling trait suppress morphological changes caused by other antibiotics. (C) Profiles of cells treated with antibiotic combination containing CIP showed a lower index of similarity to the CIP profile and a higher index of similarity to the other. Morphological changes caused by antibiotics with a recessive profiling trait were overshadowed by the other.
Morphological changes caused by antibiotic combination could not accurately indicate antibiotic interaction type.
Based on the finding of unique profiles of cells in the antibiotic combination (type A), we next sought to test if the morphological changes caused by two antibiotics could be used as a predictor of synergism between the two antibiotics. A previous study (48) showed that E. coli strains, whose genetic alteration causes an increase in cell width, are more susceptible to compound A22—which also causes the same morphological outcome, highlighting the synergistic relationship between morphological changes and level of antibiotic susceptibility. To test if this is the case, we examined if type A combination could indicate synergism and type B combination could indicate nonsynergistic interaction by comparing with drug interaction results from checkerboard analysis in all 28 combinations. We found that while some type A combinations corresponded with their synergistic interaction, such as PIP+AMI and MER+CST (type A and synergistic), some gave opposite results, such as PIP+TCS and CIP+RIF (type A but nonsynergistic) (Fig. S3). In contrast, some type B combinations, including PIP+MER, PIP+MIN, and PIP+RIF, exhibited synergistic interaction in the checkerboard assay (type B but synergistic) (Fig. S3). Thus, we did not find the relationship between type of combination profile and type of antibiotic interaction in our study. Our findings were consistent with the previous study in E. coli in which combinatorial effect of two antibiotics on representative morphological changes could not be used to indicate the interaction type of the two antibiotics (47).
Combinatorial profile-based predictive model classifies A. baumannii cells treated with usnic acid (USN) into multiple profiles.
Even though the combinatorial profiles could not be used as indicators of antibiotic interaction type, we sought to determine if the profiles could be used to reveal the possible hidden MOA of a compound that can synergistically work with antibiotics. Finding antibiotics that are active against Gram-negative bacteria, especially A. baumannii, is notoriously challenging (4, 49–51). This is mostly due to the existence of the bacterial outer membrane, which diminishes drug influx, leading to low intracellular concentrations of the drug (52). Thus, potential compounds that target cytosolic components but cannot enter the cell often give negative readouts during screening. In this study, we also encountered the same challenges when we examined a collection of natural product-derived compounds for their antibacterial activity and found that while some of the compounds inhibited the growth of the Gram-positive bacterium Bacillus subtilis, none of them inhibited the growth of A. baumannii at concentrations below 500 μM (Table S3). We hypothesized that in the presence of a membrane-permeabilizing antibiotic that would potentially increase drug influx, the compounds that were previously deemed inactive could enter into the cell and exert their activity, thereby resulting in detectable morphological changes.
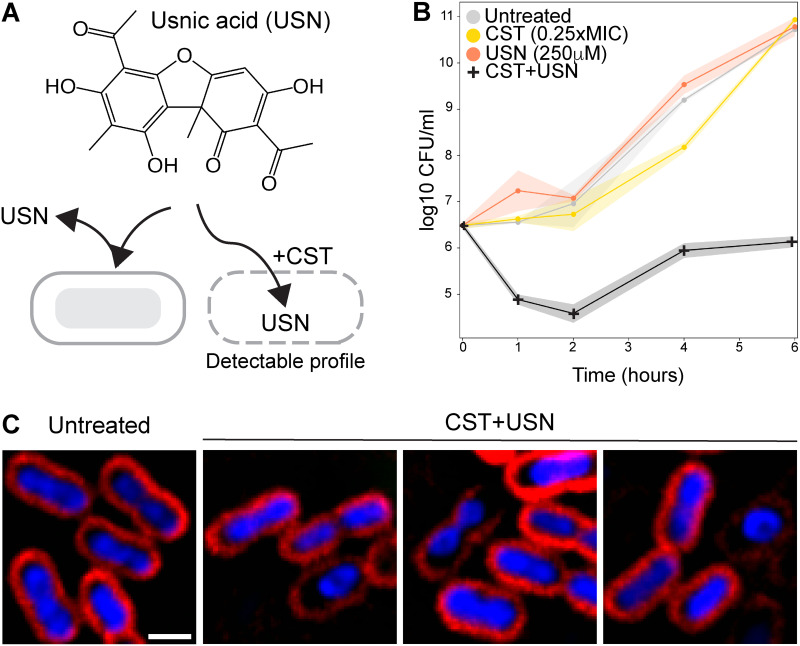
To test this hypothesis, we selected CST as the membrane-permeabilizing antibiotic for a synergy study. In order to test if the compounds synergized with CST, we evaluated the fractional inhibitory concentration (FIC) of each compound in the presence of 0.25× MIC of CST. Out of the compounds tested, 250 μM USN inhibited the growth of the bacteria in the presence of CST, suggesting that CST promotes USN activity against A. baumannii (Fig. 4A and Table S3). To confirm a synergistic effect of CST+USN, a time-kill assay (TKA) was performed; it showed that the combination of 250 μM USN and 0.25× MIC CST significantly decreased the number of bacteria compared to the case with CST or USN alone (Fig. 4B). BCP results showed that even though not all CST+USN-treated cells exhibited morphological changes, various morphological changes were observed in the rest of the population. These morphological changes were similar to various profiles shown earlier, such as toroidal DNA and small cells with a bright DAPI signal (Fig. 4C), suggesting that USN might exert multiple activities against A. baumannii in combination with CST.
FIG 4.
Usnic acid in combination with colistin inhibits the growth of A. baumannii resulted in multiple morphological changes. (A) Chemical structure of USN and a hypothesis diagram showing that CST facilitates the entry of USN into the A. baumannii cell, resulting in detectable cell profile. (B) Time-kill assay of A. baumannii treated with 0.25× MIC CST alone, 250 μM USN alone and in combination, compared with untreated cells. (C) Representative cell images of CST+USN-treated cells showing multiple morphological changes compared with untreated cells. Cells were treated with 0.25× MIC CST and 250 μM USN for 1 h. Scale bar represents 1 μm.
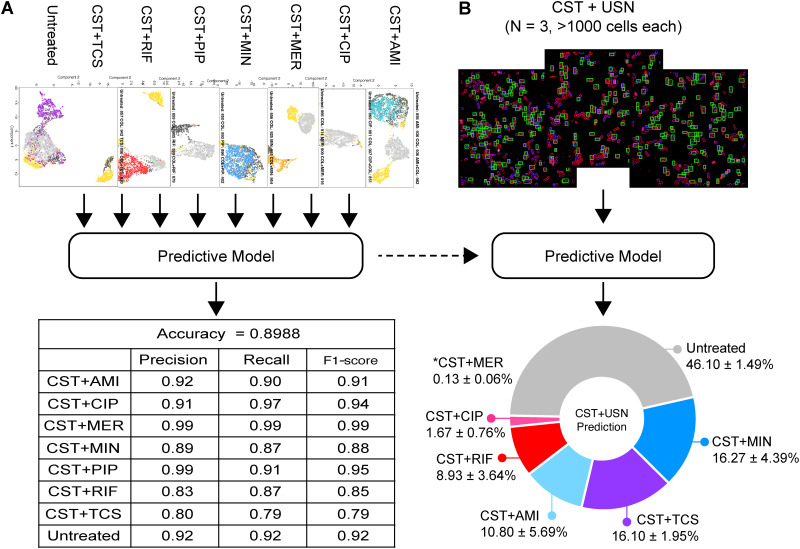
Since multiple morphological changes were observed with CST+USN, we asked if the method could classify CST+USN-treated cells into any profiles of A. baumannii treated with all other antibiotics in combination with CST to relate the morphological change caused by USN to any known MOAs. The predictive model was able to classify the cell profile of each CST combination at an overall accuracy level of 89.88% (Fig. 5A) and was then used to classify individual cell profiles treated with CST+USN (Fig. 5B). The results showed that the majority of cells treated with CST+USN were predicted to be untreated (46.10% ± 1.49%), followed by significant portions of cells treated with CST+MIN (16.27% ± 4.39%), CST+TCS (16.10% ± 1.95%), CST+AMI (10.80% ± 5.69%), and CST+RIF (8.93% ± 3.64%). This finding is in agreement with the previous study in which, despite its lack of activity against Gram-negative bacteria, USN was shown to inhibit the growth of Gram-positive bacteria, S. aureus and B. subtilis, via multiple pathways, including RNA transcription and DNA replication as a primary target and protein translation as a secondary target (53). To rule out the concerns regarding the USN quality in our natural product library, we also performed experiments on commercial usnic acid (cUSN) and obtained similar results (Fig. S4). cUSN alone could not inhibit the growth of A. baumannii but synergistically inhibited the growth of the bacterium at 250 μM with CST (Table S3). Unlike for CST+USN, where the majority of cells were similar to untreated cells, cells treated with CST+cUSN showed various observable morphological changes (Fig. S4A) and were predicted be CST+MIN (35.3%), CST+TCS (21.4%), CST+AMI (11.5%), CST+RIF (9.3%), and CST+MER (8.5%), and followed by untreated (8.1%) (Fig. S4B). Notably, the orders of detectable profiles of CST+cUSN and CST+USN were the same (CST+MIN > CST+TCS > CST+AMI > CST+RIF). Collectively, we demonstrated that high-resolution BCP can be used to reveal multiple profiles of CST+USN-treated cells based on different morphological changes among bacteria population.
FIG 5.
A. baumannii cells treated with usnic acid and colistin were classified into multiple antibiotic combinations. (A) CST combination data set used for predictive model training (Fig. S2B) and the classification accuracy of the resulting model. (B) CST+USN-treated cells were classified into different profiles by the predictive model (*CST+MER not shown in the chart).
DISCUSSION
This study highlights three complementary perspectives in bacterial phenotypic screen for antibiotic discovery: (i) morphological variation of bacteria upon antibiotic treatment, (ii) combinatorial effect of two antibiotics on morphological changes, and (iii) detection of multiple cytological profiles in accordance with the MOAs of antibacterial molecules. In an aspect of morphological variation, we demonstrated that some antibiotics provoked intrapopulation variation and later resulted in cells clustering into different groups based on morphological changes, such as those in AMI-, MER-, and CST+USN-treated populations (Fig. 1 and 4). It is possible that treatment at 1× MIC could be responsible for the intrapopulation variation found in this study. The previous study on antibiotic combination and bacterial morphology found that higher concentrations of antibiotics tends to, but not always, yield lower heterogeneity of bacterial morphology (47). Thus, to minimize heterogeneity noise, treatment with a high concentration of antibiotic is probably suitable for the study that focuses on MOA identification where average-based or representative morphological changes data sets are preferable (16). It is also possible that the findings of cytological variation presented here are limited to A. baumannii ATCC 17978. However, it has been shown in a previous study of BCP using A. baumannii ATCC 19606 that some heterogenous responses of the bacterium to antibiotics were also seen (15). Thus, single-cell studies with other strains would be beneficial in testing if cytological intrapopulation variation is limited to particular strains. Moreover, clinical isolates, especially multidrug-resistant and extensively drug-resistant strains, were not included in the study. Thus, the presence of cytological variation upon antibiotic perturbation in these strains remains unexplored. It has been elucidated that clinical strains have high genetic variations, resulting in heterogenous behavioral and physiological responses to environmental stresses (54, 55). Hence, one would expect cytological variation among these clinical isolates. Whether or not cytological variations upon antibiotic exposure could be detected in other strains of A. baumannii or in other important pathogens is an interesting question worth investigating in the future. Another finding worth noting is that in our study, regardless of the MOA of the antibiotics being used, a small number of treated cells whose morphological profiles were unaltered by treatment were detected in every treatment (Fig. 1). This corresponds with a previous discovery illustrating that under antibiotic or environmental stress, a small fraction of bacteria persist without responding to the stress, thus ensuring the survival of the species (18, 56, 57). A temporal study that can track individual cells before, during, and after antibiotic treatment would provide the key to understanding bacterial survival under antibiotic stress.
In search of antibacterial molecules with novel MOAs, information of novel morphological profiles may expand the border of morphological space that hints at possible new MOAs. In our antibiotic combination study, we found that 15 combinations (type A) gave rise to profiles that are separated from those of the individual antibiotics within the combination, suggesting probable morphological uniqueness. Our finding is slightly in contrast with a previous study in which only four “unexpected morphological phenotypes” were categorized from the higher-throughput and more comprehensive combination study (47). We speculate that this difference is partly from the fact that our study focused on examining single-cell morphological variation rather than detecting a representative morphology of the major population from each imaging well. Whether or not these unique patterns of cytological variation could provide complementary perspective for novel MOA discovery needs further investigation. Another limitation worth noting is that in an attempt to find the relationship between the combination profile and drug interaction type in our study, only one representative antibiotic at one concentration from each previously reported profile was included in the analysis. Since it has been shown that the physiological outcome from drug-drug interaction is highly complex (58) and determined not solely by the MOA of the compound but rather by the characteristics of the compound itself (52, 59), more antibiotics representing each cytological profile at various concentrations are needed to investigate if there is any correlation between cytological profile and drug-drug interaction.
Lastly, in an aspect of detecting multiple cytological profiles that match the MOAs, a platform that can identify multiple MOAs simultaneously will be beneficial to directly screen crude or partially purified extracts from natural products, where mixtures full of compounds are commonly presented (60, 61). In this study, we showed that cells treated with CST+USN were classified into multiple subprofiles that are in accordance with previously reported MOAs of USN. However, it remains unclear whether this phenomenon is limited to the compounds with various activities, such as USN (i.e., one compound with many MOAs). Further investigations are needed to test if this phenomenon can be observed in the case of multitreatment (i.e., multiple compounds with different MOAs). Apart from the ability of the method to reveal multiple subprofiles, the finding of dominant traits of some antibiotics in the study may lead to a better understanding of how multiple active compounds affect individual cells during screens. For instance, antibacterial molecules in the extract whose profiles were masked by another might have been overlooked. This might also be the case with CST+USN treatment in A. baumannii in this study, where we observed significant amount of cells predicted to have profiles similar to those treated with protein translation inhibitors (CST+MIN and CST+AMI), while RNA transcription and DNA replication inhibition activity of USN were also previously reported for S. aureus and B. subtilis (53). It is possible that the dominant trait of protein translation inhibition profiles overshadowed all the effects of the others in A. baumannii as demonstrated in this study. A previous study on transcriptomic analysis of bacteria treated with antibiotics also showed that in some cases, the secondary effect or MOA of the antibiotic might be more pronounced than its primary MOA (62). Moreover, it is likely that the target preferences of antibiotics are different in different bacteria (63), thereby, resulting in different cytological changes in different bacteria. Solving the riddle of the effect of dominant and recessive traits of antibiotics on bacterial morphology will play an important role in developing a phenotypic screening platform that can effectively sift through a large collection of chemical libraries to identify novel antibacterial molecules.
MATERIALS AND METHODS
Bacteria strain and growth.
Acinetobacter baumannii strain ATCC 17978 was used throughout this study. The bacterium was grown in LB medium or LB agar at 30°C.
MIC determination.
The MICs of antibiotics were determined by microdilution method as previously described (15), and the results are shown in Table S1. Briefly, overnight cultures of A. baumannii were diluted 1:100 in LB and grown on a roller at 30°C to an optical density at 600 nm (OD600) of 0.2. The bacterial cultures were diluted 1:100 into each well of a 96-well plate containing antibiotics in LB medium at different concentrations. The bacterial cultures were incubated at 30°C for 24 h. The lowest antibiotic concentration resulting in no visible growth was designated the MIC.
Fluorescence microscopy.
Overnight cultures of A. baumannii were diluted 1:100 in LB and grown at 30°C to an OD600 of 0.2 prior to the addition of antibiotics at the MIC level. After 1 h of incubation with the antibiotic, cultures were stained with 2 μg/mL FM4-64, 2 μg/mL DAPI, and 0.5 μM SYTOX green. Bacterial cells were harvested by centrifugation at 6,000 × g for 1 min and resuspended in 1/10 of the original volume. A few microliters of concentrated bacterial cultures were added onto an agarose pad (1.2% agarose in 10% LB broth) on concave glass slides for microscopy. Experimental setting and imaging parameters were constant throughout every experiment included in the statistical analysis of all training sets of antibiotics.
Image data analysis.
Prior to downstream image analysis, raw images from the microscope were preprocessed into an appropriate image format on ImageJ software (64). Then, machine learning-based image analysis software Ilastik (65) was used to determine individual cells on the images using the bacterial cell membrane to identify each cell. Once all individual cell locations were identified, cell membranes and nuclei were segmented and subsequently analyzed on CellProfiler 4.0 software (66) for cell feature extraction. After image data extraction, statistical calculations and machine learning were carried out using the scikit-learn library (67) in python. Briefly, 1,000 individual cell profiles of each antibiotic treatment from three independent experiments were randomly selected and transformed with QuantileTransformer (68). The outlier data, such as cell debris, abnormal image segmentation, and incomplete cell along the image edge, which amounted to about 10% of the detected objects, were removed with hierarchical density-based spatial clustering of applications with noise (HDBSCAN) (69) with a 10% cutoff. The optimal morphological features were identified by the recursive feature elimination with cross-validation based on support vector machine (RFECV-SVM) model (70). At each iteration, three cross-validations were conducted to determine the best feature set. Then, the performance of the SVM model was evaluated with the data set containing only the selected features to ensure that these selected features were relevant in distinguishing cytological profiles of each antibiotic. The data set was randomly divided into an 80% training and 20% test set on which the model was trained and evaluated, respectively. Accuracy score, f1, recall and confusion matrix were used as indicators for model performance as shown in Fig. S1. Finally, the dimension of the data set was reduced and then visualized with an unsupervised data dimension reduction method, pairwise controlled manifold approximation (PaCMAP) (71).
Similarity index calculation.
The similarity index was calculated based on Euclidean distance between PaCMAP coordinates of the main clusters of single-antibiotic-treated cells and 200 randomly selected dual-antibiotic-treated cells. Due to the scatter coordinate of cell profiles, an unsupervised learning mean shift clustering algorithm (72) was used to determine the main cluster (high density cluster) of single-antibiotic treatment, each of which contained more than 50 cells. Then, only cells in the main cluster of single-antibiotic treatment were used to calculate similarity index as follows:
where is a minimum Euclidean distance between coordinate of dual-antibiotic-treated cell i and single antibiotic-treated cell j.
To examine the degree of overlapping between single- and dual-antibiotic-treated clusters, a similarity index of 0.75 was used as a cutoff, which was calculated from the lowest similarity index of single-antibiotic-treated cells shown in Fig. 1. Briefly, the single-antibiotic-treated cells were randomly divided into two groups and the similarity index between these two groups was calculated. The lowest similarity index between the two groups was selected as the cutoff threshold. For type A or B classification, if the index of similarity of the dual antibiotic to both the single antibiotics in the combination was lower than 0.75, it indicated that the dual antibiotic provided a unique morphological cluster and separated from the single-antibiotic treatments and thus was classified as type A. In contrast, a similarity index higher than 0.75 indicated that dual-antibiotic treatment provided an overlapping morphological cluster with single-antibiotic treatments and thus was classified as type B.
Screening natural product-derived compounds that synergize with colistin in killing A. baumannii.
The collection of natural product-derived compounds was a gift from Warinthorn Chavasiri, Center of Excellence in Natural Products Chemistry (CENP), Chulalongkorn University. MICs of all compounds against A. baumannii and B. subtilis were determined by microdilution method as mentioned above, and the results are shown in Table S3. For the primary synergistic test, compounds that were shown to inhibit the growth of A. baumannii at 500 μM in the presence of 0.25× MIC CST were chosen for the fractional inhibitory concentration (FIC) assay. For FIC testing, overnight cultures of A. baumannii were diluted 1:100 in LB and grown on a roller at 30°C to an OD600 of 0.2. Cultures were added into LB medium containing 0.25× MIC CST and different concentrations of compound and then subjected to 24 h of incubation at 30°C. The FIC of the compound that could synergize with 0.25× MIC CST was determined as the lowest concentration of the compound that resulted in no visible growth of the bacteria.
TKA.
Time-kill assay (TKA) was performed as previously described (73). Briefly, overnight cultures of A. baumannii were diluted 1:100 in LB and grown at 30°C to an OD600 of 0.2. Then, day cultures of A. baumannii were diluted 1:100 into each tube containing 0.25× MIC CST, 250 μM USN, or the combination prior to incubation at 30°C. At each time point (0, 1, 2, 4, and 6 h), 100 μL each of the untreated and treated bacterial culture was collected, serially diluted in LB medium, plated on an LB agar plate, and incubated overnight at 30°C. The CFU were determined, calculated as log CFU per milliliter, and plotted against time.
Classification of USN-induced cytological profiles.
A supervised learning model, support vector machine (SVM) (74), was used to classify cell profiles caused by USN. The SVM model was trained on the data set of untreated cells and treated cell profiles of every colistin-based combination: CST+AMI, CST+CIP, CST+MER, CST+MIN, CST+PIP, CST+RIF, and CST+TCS. Similar to the previously mentioned analysis pipeline, the raw data set was transformed with QuantileTransformer, the outlier was removed with HDBSCAM, and relevant morphological features were selected by RFECV-SVM. The data set was then randomly divided into 80% training and 20% test sets. Finally, the model performance was determined by accuracy score, f1, and recall. The trained SVM model was used to classify each bacterial cell profile from CST+USN (unknown), resulting in a percentage of CST+USN-treated cells distributed over the previously trained categories. The percentage of each predicted category from three independent experiments was averaged and included in a chart (Fig. 3D).
Fractional inhibitory concentration (FIC) determination via checkerboard assay.
An overnight culture of A. baumannii was diluted 1:100 in LB and grown on a roller at 30°C to an OD600 of 0.2. Prior to addition of bacterial culture, two different compounds were serially diluted, in a perpendicular manner to each other, in a 96-well plate to obtain different concentrations of the two antibiotics in the combination. Then, the bacterial cultures were diluted 1:100 into each well and incubated for 24 h at 30°C.
The Σ FICs were calculated as
where A and B are the MICs of each concentration in combination and MICA and MICB are the MICs of each drug individually. Drug interaction is synergistic when the Σ FICs is ≤0.5 and otherwise is nonsynergistic (75).
ACKNOWLEDGMENTS
This research project is supported by Mahidol University under the New Discovery and Frontier Research Grant. This project is also funded by National Research Council of Thailand (NRCT) and Mahidol University (N42A650368). V.C. was supported by NRCT and Chulalongkorn University (N41A640136). P.N. also thanks the Coordinating Center for Thai Government Science and Technology Scholarship Students (CSTS)—the National Science and Technology Development Agency (NSTDA) for grant no. JRA-CO-2563-13729-TH and the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA), Science and Technology Research Partnership for Sustainable Development, SATREPS JPMJSA1806, for chemicals and instruments. T.S. was also supported by NRCT and CSTS.
We thank Mahidol University Frontier Research Facility (MU-FRF) for instrumentation support for DeltaVision Ultra and the Advanced Cell Imaging Center, Institute of Molecular Biosciences, Mahidol University, for the fluorescent microscopy imaging, operated by Patporn Chankong and Naraporn Sirinonthanawech.
Conceptualization, T.S., V.C., and P.N.; Methodology, T.S. and P.N.; Software, T.S.; Validation, T.S., H.H.H., V.C., and P.N.; Formal Analysis, T.S. and P.N.; Investigation, T.S. and H.H.H.; Resources, W.C., V.C., and P.N.; Data Curation, T.S. and H.H.H.; Writing – Original Draft, T.S., H.H.H., V.C., and P.N.; Writing – Review & Editing, H.H.H., J.S., J.P., V.C., and P.N.; Visualization, T.S., H.H.H., and P.N.; Supervision, V.C. and P.N.; Project Administration, T.S., V.C., and P.N.; Funding acquisition, V.C. and P.N.
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roope LSJ, Smith RD, Pouwels KB, Buchanan J, Abel L, Eibich P, Butler CC, Tan PS, Walker AS, Robotham JV, Wordsworth S. 2019. The challenge of antimicrobial resistance: what economics can contribute. Science 364:eaau4679. 10.1126/science.aau4679. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson DL, Walker MJ. 2020. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 33:e00181-19. 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y. 2017. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 6.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins PG, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:233–238. 10.1093/jac/dkp428. [DOI] [PubMed] [Google Scholar]
- 8.Hamidian M, Nigro SJ. 2019. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom 5. 10.1099/mgen.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayobami O, Willrich N, Suwono B, Eckmanns T, Markwart R. 2020. The epidemiology of carbapenem-non-susceptible Acinetobacter species in Europe: analysis of EARS-Net data from 2013 to 2017. Antimicrob Resist Infect Control 9:89. 10.1186/s13756-020-00750-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beceiro A, Moreno A, Fernandez N, Vallejo JA, Aranda J, Adler B, Harper M, Boyce JD, Bou G. 2014. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother 58:518–526. 10.1128/AAC.01597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolain J-M, Diene SM, Kempf M, Gimenez G, Robert C, Raoult D. 2013. Real-time sequencing to decipher the molecular mechanism of resistance of a clinical pan-drug-resistant Acinetobacter baumannii isolate from Marseille, France. Antimicrob Agents Chemother 57:592–596. 10.1128/AAC.01314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doi Y, Murray GL, Peleg AY. 2015. Acinetobacter baumannii: evolution of antimicrobial resistance—treatment options. Semin Respir Crit Care Med 36:85–98. 10.1055/s-0034-1398388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nonejuie P, Burkart M, Pogliano K, Pogliano J. 2013. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc Natl Acad Sci USA 110:16169–16174. 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamsa A, Lopez-Garrido J, Quach D, Riley EP, Pogliano J, Pogliano K. 2016. Rapid inhibition profiling in Bacillus subtilis to identify the mechanism of action of new antimicrobials. ACS Chem Biol 11:2222–2231. 10.1021/acschembio.5b01050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Htoo HH, Brumage L, Chaikeeratisak V, Tsunemoto H, Sugie J, Tribuddharat C, Pogliano J, Nonejuie P. 2019. Bacterial cytological profiling (BCP) as a tool to study mechanism of action of antibiotics that are active against Acinetobacter baumannii. Antimicrob Agents Chemother 63:e02310-18. 10.1128/AAC.02310-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ljosa V, Caie PD, ter Horst R, Sokolnicki KL, Jenkins EL, Daya S, Roberts ME, Jones TR, Singh S, Genovesio A, Clemons PA, Carragher NO, Carpenter AE. 2013. Comparison of methods for image-based profiling of cellular morphological responses to small-molecule treatment. J Biomol Screen 18:1321–1329. 10.1177/1087057113503553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lidstrom ME, Konopka MC. 2010. The role of physiological heterogeneity in microbial population behavior. Nat Chem Biol 6:705–712. 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- 18.Booth IR. 2002. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int J Food Microbiol 78:19–30. 10.1016/S0168-1605(02)00239-8. [DOI] [PubMed] [Google Scholar]
- 19.Dewachter L, Fauvart M, Michiels J. 2019. Bacterial heterogeneity and antibiotic survival: understanding and combatting persistence and heteroresistance. Mol Cell 76:255–267. 10.1016/j.molcel.2019.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Ackermann M. 2015. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13:497–508. 10.1038/nrmicro3491. [DOI] [PubMed] [Google Scholar]
- 21.Lin L, Nonejuie P, Munguia J, Hollands A, Olson J, Dam Q, Kumaraswamy M, Rivera H, Corriden R, Rohde M, Hensler ME, Burkart MD, Pogliano J, Sakoulas G, Nizet V. 2015. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant Gram-negative bacterial pathogens. EBioMedicine 2:690–698. 10.1016/j.ebiom.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nonejuie P, Trial RM, Newton GL, Lamsa A, Perera VR, Aguilar J, Liu W-T, Dorrestein PC, Pogliano J, Pogliano K. 2016. Application of bacterial cytological profiling to crude natural product extracts reveals the antibacterial arsenal of Bacillus subtilis. J Antibiot (Tokyo) 69:353–361. 10.1038/ja.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin JK, Sheehan JP, Bratton BP, Moore GM, Mateus A, Li SH-J, Kim H, Rabinowitz JD, Typas A, Savitski MM, Wilson MZ, Gitai Z. 2020. A dual-mechanism antibiotic kills Gram-negative bacteria and avoids drug resistance. Cell 181:1518–1532.e14. 10.1016/j.cell.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jana S, Deb JK. 2006. Molecular understanding of aminoglycoside action and resistance. Appl Microbiol Biotechnol 70:140–150. 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Bodovitz S. 2010. Single cell analysis: the new frontier in ‘omics.’ Trends Biotechnol 28:281–290. 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomura S. 2021. Single-cell genomics to understand disease pathogenesis. J Hum Genet 66:75–84. 10.1038/s10038-020-00844-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labib M, Kelley SO. 2020. Single-cell analysis targeting the proteome. Nat Rev Chem 4:143–158. 10.1038/s41570-020-0162-7. [DOI] [PubMed] [Google Scholar]
- 28.Heath JR, Ribas A, Mischel PS. 2016. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov 15:204–216. 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hare PJ, LaGree TJ, Byrd BA, DeMarco AM, Mok WWK. 2021. Single-cell technologies to study phenotypic heterogeneity and bacterial persisters. Microorganisms 9:2277. 10.3390/microorganisms9112277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sridhar S, Forrest S, Warne B, Maes M, Baker S, Dougan G, Bartholdson Scott J. 2021. High-content imaging to phenotype antimicrobial effects on individual bacteria at scale. mSystems 6:e00028-21. 10.1128/mSystems.00028-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma P, Amemiya HM, He LL, Gandhi SJ, Nicol R, Bhattacharyya RP, Smillie CS, Hung DT. 2022. Bacterial droplet-based single-cell RNA-seq reveals heterogeneity in bacterial populations and in response to antibiotic perturbation. bioRxiv. 10.1101/2022.08.01.502326. [DOI] [PMC free article] [PubMed]
- 32.Mattiazzi Usaj M, Yeung CHL, Friesen H, Boone C, Andrews BJ. 2021. Single-cell image analysis to explore cell-to-cell heterogeneity in isogenic populations. Cell Syst 12:608–621. 10.1016/j.cels.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, Hyeon DY, Hwang D. 2020. Single-cell multiomics: technologies and data analysis methods. Exp Mol Med 52:1428–1442. 10.1038/s12276-020-0420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji Y, Lotfollahi M, Wolf FA, Theis FJ. 2021. Machine learning for perturbational single-cell omics. Cell Syst 12:522–537. 10.1016/j.cels.2021.05.016. [DOI] [PubMed] [Google Scholar]
- 35.Scheeder C, Heigwer F, Boutros M. 2018. Machine learning and image-based profiling in drug discovery. Curr Opin Syst Biol 10:43–52. 10.1016/j.coisb.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aggarwal CC (ed). 2014. Data classification: algorithms and applications. CRC Press/Chapman & Hall, Boca Raton, FL. [Google Scholar]
- 37.Bellman R. 1966. Dynamic programming. Science 153:34–37. 10.1126/science.153.3731.34. [DOI] [PubMed] [Google Scholar]
- 38.Jeon H, Oh S. 2020. Hybrid-recursive feature elimination for efficient feature selection. Appl Sci 10:3211. 10.3390/app10093211. [DOI] [Google Scholar]
- 39.Zhang Y, Deng Q, Liang W, Zou X. 2018. An efficient feature selection strategy based on multiple support vector machine technology with gene expression data. Biomed Res Int 2018:7538204–7538211. 10.1155/2018/7538204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Xie W, Liu T. 2018. Efficient feature selection and classification for microarray data. PLoS One 13:e0202167. 10.1371/journal.pone.0202167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X, Li C, Zhang Y, Su B, Fan M, Wei H. 2017. Selecting feature subsets based on SVM-RFE and the overlapping ratio with applications in bioinformatics. Molecules 23:52. 10.3390/molecules23010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penwell WF, Shapiro AB, Giacobbe RA, Gu R-F, Gao N, Thresher J, McLaughlin RE, Huband MD, DeJonge BLM, Ehmann DE, Miller AA. 2015. Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob Agents Chemother 59:1680–1689. 10.1128/AAC.04808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doern CD. 2014. When does 2 plus 2 equal 5? A review of antimicrobial synergy testing. J Clin Microbiol 52:4124–4128. 10.1128/JCM.01121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevereau G, Bollenbach T. 2015. Systematic discovery of drug interaction mechanisms. Mol Syst Biol 11:807. 10.15252/msb.20156098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wildenhain J, Spitzer M, Dolma S, Jarvik N, White R, Roy M, Griffiths E, Bellows DS, Wright GD, Tyers M. 2015. Prediction of synergism from chemical-genetic interactions by machine learning. Cell Syst 1:383–395. 10.1016/j.cels.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng W, Sun W, Simeonov A. 2018. Drug repurposing screens and synergistic drug-combinations for infectious diseases. Br J Pharmacol 175:181–191. 10.1111/bph.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coram MA, Wang L, Godinez WJ, Barkan DT, Armstrong Z, Ando DM, Feng BY. 2022. Morphological characterization of antibiotic combinations. ACS Infect Dis 8:66–77. 10.1021/acsinfecdis.1c00312. [DOI] [PubMed] [Google Scholar]
- 48.Ursell T, Lee TK, Shiomi D, Shi H, Tropini C, Monds RD, Colavin A, Billings G, Bhaya-Grossman I, Broxton M, Huang BE, Niki H, Huang KC. 2017. Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. BMC Biol 15:17. 10.1186/s12915-017-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silver LL. 2011. Challenges of antibacterial discovery. Clin Microbiol Rev 24:71–109. 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis K. 2013. Platforms for antibiotic discovery. Nat Rev Drug Discov 12:371–387. 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 51.Lewis K. 2020. The science of antibiotic discovery. Cell 181:29–45. 10.1016/j.cell.2020.02.056. [DOI] [PubMed] [Google Scholar]
- 52.Brochado AR, Telzerow A, Bobonis J, Banzhaf M, Mateus A, Selkrig J, Huth E, Bassler S, Zamarreño Beas J, Zietek M, Ng N, Foerster S, Ezraty B, Py B, Barras F, Savitski MM, Bork P, Göttig S, Typas A. 2018. Species-specific activity of antibacterial drug combinations. Nature 559:259–263. 10.1038/s41586-018-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maciąg-Dorszyńska M, Węgrzyn G, Guzow-Krzemińska B. 2014. Antibacterial activity of lichen secondary metabolite usnic acid is primarily caused by inhibition of RNA and DNA synthesis. FEMS Microbiol Lett 353:57–62. 10.1111/1574-6968.12409. [DOI] [PubMed] [Google Scholar]
- 54.Kamuyu G, Ercoli G, Ramos-Sevillano E, Willcocks S, Kewcharoenwong C, Kiratisin P, Taylor PW, Wren BW, Lertmemongkolchai G, Stabler RA, Brown JS. 2022. Strain specific variations in Acinetobacter baumannii complement sensitivity. Front Immunol 13:853690. 10.3389/fimmu.2022.853690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chin CY, Tipton KA, Farokhyfar M, Burd EM, Weiss DS, Rather PN. 2018. A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat Microbiol 3:563–569. 10.1038/s41564-018-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joers A, Kaldalu N, Tenson T. 2010. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J Bacteriol 192:3379–3384. 10.1128/JB.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fraser D, Kaern M. 2009. A chance at survival: gene expression noise and phenotypic diversification strategies. Mol Microbiol 71:1333–1340. 10.1111/j.1365-2958.2009.06605.x. [DOI] [PubMed] [Google Scholar]
- 58.Roemhild R, Bollenbach T, Andersson DI. 2022. The physiology and genetics of bacterial responses to antibiotic combinations. Nat Rev Microbiol 20:478–490. 10.1038/s41579-022-00700-5. [DOI] [PubMed] [Google Scholar]
- 59.Yeh P, Tschumi AI, Kishony R. 2006. Functional classification of drugs by properties of their pairwise interactions. Nat Genet 38:489–494. 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- 60.Farha MA, Brown ED. 2016. Strategies for target identification of antimicrobial natural products. Nat Prod Rep 33:668–680. 10.1039/c5np00127g. [DOI] [PubMed] [Google Scholar]
- 61.Genilloud O. 2019. Natural products discovery and potential for new antibiotics. Curr Opin Microbiol 51:81–87. 10.1016/j.mib.2019.10.012. [DOI] [PubMed] [Google Scholar]
- 62.O’Rourke A, Beyhan S, Choi Y, Morales P, Chan AP, Espinoza JL, Dupont CL, Meyer KJ, Spoering A, Lewis K, Nierman WC, Nelson KE. 2020. Mechanism-of-action classification of antibiotics by global transcriptome profiling. Antimicrob Agents Chemother 64:e01207-19. 10.1128/AAC.01207-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zapun A, Macheboeuf P, Vernet T. 2017. Penicillin-binding proteins and β-lactam resistance, p 177–211. In Mayers DL, Sobel JD, Ouellette M, Kaye KS, Marchaim D (ed), Antimicrobial drug resistance. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 64.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, Eren K, Cervantes JI, Xu B, Beuttenmueller F, Wolny A, Zhang C, Koethe U, Hamprecht FA, Kreshuk A. 2019. ilastik: interactive machine learning for (bio)image analysis. Nat Methods 16:1226–1232. 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 66.McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, Wiegraebe W, Singh S, Becker T, Caicedo JC, Carpenter AE. 2018. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol 16:e2005970. 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Müller A, Nothman J, Louppe G, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. 2018. Scikit-learn: machine learning in Python. arXiv 12010490 [cs]. 10.48550/arXiv.1201.0490. [DOI]
- 68.Jiang C, Zhao P, Li W, Tang Y, Liu G. 2020. In silico prediction of chemical neurotoxicity using machine learning. Toxicol Res (Camb) 9:164–172. 10.1093/toxres/tfaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Campello RJGB, Moulavi D, Zimek A, Sander J. 2015. Hierarchical density estimates for data clustering, visualization, and outlier detection. ACM Trans Knowl Discov Data 10:1–51. 10.1145/2733381. [DOI] [Google Scholar]
- 70.Caicedo JC, Cooper S, Heigwer F, Warchal S, Qiu P, Molnar C, Vasilevich AS, Barry JD, Bansal HS, Kraus O, Wawer M, Paavolainen L, Herrmann MD, Rohban M, Hung J, Hennig H, Concannon J, Smith I, Clemons PA, Singh S, Rees P, Horvath P, Linington RG, Carpenter AE. 2017. Data-analysis strategies for image-based cell profiling. Nat Methods 14:849–863. 10.1038/nmeth.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Huang H, Rudin C, Shaposhnik Y. 2021. Understanding how dimension reduction tools work: an empirical approach to deciphering t-SNE, UMAP, TriMAP, and PaCMAP for data visualization. arXiv 201204456 [cs, stat]. 10.48550/arXiv.2012.04456. [DOI]
- 72.Carreira-Perpiñán MÁ. 2015. A review of mean-shift algorithms for clustering. arXiv 150300687 [cs, stat]. 10.48550/arXiv.1503.00687. [DOI]
- 73.Pankey G, Ashcraft D, Kahn H, Ismail A. 2014. Time-kill assay and Etest evaluation for synergy with polymyxin B and fluconazole against Candida glabrata. Antimicrob Agents Chemother 58:5795–5800. 10.1128/AAC.03035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. 2018. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics 15:41–51. 10.21873/cgp.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orhan G, Bayram A, Zer Y, Balci I. 2005. Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J Clin Microbiol 43:140–143. 10.1128/JCM.43.1.140-143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.01307-22-s0001.pdf, PDF file, 1.34 MB (1.3MB, pdf)