Abstract
Background
Quantification of circulating tumor DNA (ctDNA) levels is a reliable prognostic tool in several malignancies. Dynamic changes in ctDNA levels in response to treatment may also provide prognostic information. Here, we explore the value of changes in ctDNA levels in response to immune checkpoint inhibitors (ICIs).
Methods
We searched MEDLINE (host: PubMed) for trials of ICIs in advanced solid tumors in which outcomes were reported based on change in ctDNA levels. ctDNA reduction was defined as reported in individual trials. Typically, this was either >50% reduction or a reduction to undetectable levels. We extracted HRs and related 95% CIs and/or p values comparing ctDNA reduction versus no reduction for progression-free survival (PFS) and/or overall survival (OS). Data were then pooled in a meta-analysis. Variation in effect size was examined using subgroup analyses.
Results
Eighteen trials were included in the meta-analysis. ctDNA levels were detectable in all participants in all studies prior to initiation of ICIs. A reduction in ctDNA measured 6–16 weeks after starting treatment was associated with significantly better PFS (HR 0.20; 95% CI, 0.14 to 0.28; p<0.001). Similarly, OS was superior in patients with reduced ctDNA levels (HR 0.18; 95% CI, 0.12 to 0.26; p<0.001). The results were consistent across all disease sites, lines of treatment, magnitude of change (to undetectable vs >50% reduction) and whether treatment exposure comprised single or combination ICIs.
Conclusions
In advanced solid tumors, a reduction in ctDNA levels in response to ICIs is associated with substantial improvements in outcome. ctDNA change is an early response biomarker which may allow for de-escalation of cross-sectional imaging in patients receiving ICIs or support treatment de-escalation strategies.
Keywords: Immunotherapy; Biomarkers, Tumor; Neoplastic Cells, Circulating; Tumor Biomarkers
WHAT IS ALREADY KNOWN ON THIS TOPIC
There is a need to validate dynamic markers that provide early and reliable insights about disease response. The predictive role of circulating tumor DNA (ctDNA) change in response to treatment is unclear, especially in unselected advanced solid tumors treated with immune checkpoint inhibitors (ICIs).
WHAT THIS STUDY ADDS
The utilization of a systematic review provides an increased level of certainty regarding the association between ctDNA reduction and improved outcomes. This meta-analysis observed substantially improved progression-free survival (PFS) and overall survival (OS), with a reduction in ctDNA levels in response to ICIs.
These results support the use of ctDNA change as a clinically valid early response biomarker which may allow for de-escalation of cross-sectional imaging in patients receiving ICIs or support treatment de-escalation strategies.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Further research is needed to assess the clinical utility of changes in ctDNA changes. Additional data are also required to standardize detection methods which will improve reproducibility results.
Background
Monitoring response to treatment is critical to the management of advanced solid tumors, where treatment decisions are made typically based on clinical, biochemical and radiological assessment. While in some patients, clinical and/or biochemical changes can be useful in evaluating response early during treatment, in others, there may not be meaningful markers which can be utilized. In addition, symptom assessment can be confounded by toxicity of cancer therapy. Therefore, in most solid tumors, response to treatment is assessed using cross-sectional imaging at set intervals.1
In patients with solid tumors treated with immune checkpoint inhibitors (ICIs), imaging can be difficult to interpret during the initial phase of treatment. Specifically, a phenomenon of pseudoprogression has been described,2 3 where an initial increase in tumor size is observed that is followed typically by tumor regression and does not reflect true disease progression.4 The ability to incorporate complementary parameters in order to accurately evaluate patients’ response to treatment with ICIs is therefore valuable.
Multiple biomarkers can be found in the circulation including circulating tumor cells, circulating cell-free DNA (especially circulating tumor DNA, (ctDNA)), circulating cell-free RNA or extracellular vesicles ((EVs) exosomes) and their cargo (including nucleic acids and proteins).5 ctDNA is released by the tumor cells into the blood and thus harbors the mutational burden of the original tumor.6 ctDNA is considered as a non-invasive ‘real-time’ biomarker that can provide diagnostic and prognostic information before treatment, during treatment and at progression.7 ctDNA platforms differentiate tumor-related DNA from other cell-free DNA by identifying DNA mutations, epigenetic alterations and other forms of tumor-specific abnormalities that act as prominent drivers for cancer development and evolution.8 Quantification of ctDNA levels can be a reliable prognostic tool in several malignancies,9 is being studied as an early cancer diagnostic tool and can be used to guide therapy and surveillance. More recently, detection of genomic alterations in ctDNA has been validated as a predictive biomarker to guide treatment planning especially for targeted agents and ICIs.7
The clinical significance of changes in ctDNA levels over time has been studied in solid tumors. Data show that responses to different treatment modalities are associated with different patterns of ctDNA change. For example, responders to chemotherapy or definitive surgery showed an initial and consistent drop in ctDNA levels, while responders to radiation therapy and targeted therapy (eg, EGFR tyrosine kinase inhibitors) exhibited a slightly different pattern with an initial increase in ctDNA levels, followed by a steady drop. Dynamic changes in ctDNA levels over time and in response to treatment may also provide prognostic information, especially in melanoma.10 11 However, despite these data, there is a paucity of data of the significance of ctDNA in response to immunotherapy in unselected patients with cancer.12 In this study, we aimed to explore whether changes to ctDNA levels in response to ICIs are prognostic in unselected solid tumors focusing on longer term outcomes such as survival. We also sought to provide a more precise estimate of the association between change in ctDNA and outcome.
Methods
A systematic search of the published literature was performed in June 2021 using MEDLINE (host: PubMed). Inclusion criteria comprised clinical trials of any phase in patients with advanced solid tumors receiving ICIs (including programmed cell death (PD-1) inhibitors, programmed cell death ligand-1 (PD-L1) inhibitors, and cytotoxic T-lymphocyte-associated protein 4 (CTLA4) inhibitors) and in whom ctDNA levels were measured before starting ICIs and again early during treatment. Only trials in which survival outcomes were reported based on changes in ctDNA levels were included in the analysis. Reported outcomes needed to include progression-free survival (PFS) and/or overall survival (OS). Each article was screened by two independent reviewers (LA-S and BW). Discrepancy in inclusion criteria was addressed by consensus or with discussion with a third reviewer (EA) if initial consensus could not be reached.
Data extraction was performed separately by two reviewers. Extracted data included: name of the first author, year of publication, disease site, disease stage, number of participants, treatment line, the type of utilized ICIs, method used to measure ctDNA, timing of ctDNA measurement after initiation of treatment, the predetermined definition of ctDNA response, the proportion of patients with radiologically stable disease, the HRs for PFS and/or OS values and their related 95% CIs and/or p values and when reported we also collected data on tumor mutation burden (TMB), microsatellite instability (MSI) status and expression of PD-L1. In articles where HRs for PFS or OS were not reported clearly, Kaplan-Meier curves were utilized to calculate PFS and/or OS results using the Parmar Toolkit.13
Data synthesis and statistical analysis
Data for PFS and OS were pooled in separate meta-analyses utilizing the generic inverse variance. In light of substantial clinical heterogeneity, we utilized random effects modeling irrespective of statistical heterogeneity. Analyses were performed using Review Manager V.5.4 (The Cochrane Collaboration, Copenhagen, Denmark). For both PFS and OS, subgroup analysis was performed based on disease site, treatment type (single vs multiple agents), treatment line (first line vs later lines), the level of ctDNA change (≥50% reduction vs undetectable levels) and timing of second ctDNA sampling (≥ 10 weeks vs <10 weeks). Heterogeneity was reported using the I2 and Cochran Q statistics. Statistical significance was defined as p<0.05. No corrections were applied for multiple significance testing.
Results
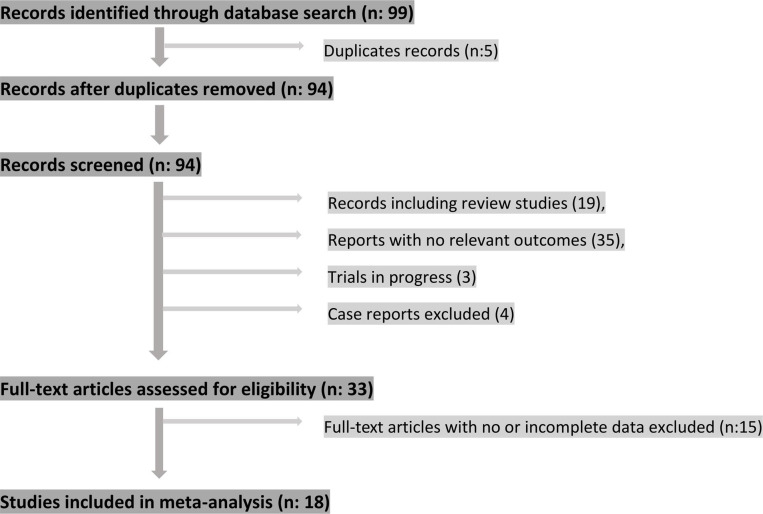
A total of 18 clinical trials comprising 552 participants were included in the meta-analysis (see figure 1 for study selection schema). All trials recruited patients with advanced solid tumors (locally advanced unresectable and/or metastatic). Involved trials included multiple disease sites: melanoma (n:8), lung cancer (n:6), gastric cancer (n:2) and urothelial cancer (n:1). One trial included patients from multiple disease sites (lung cancer, uveal melanoma and microsatellite instability-high (MSI-H) colorectal cancer). ctDNA levels were detectable in all participants in all studies prior to initiation of ICIs. ctDNA clearance was defined as reported in individual trials. Typically, this was defined as either a greater than 50% reduction in ctDNA levels (n=3) or a reduction to undetectable levels (n=14), with one trial comparing relative decrease versus relative increase in ctDNA levels between baseline and the second sampling time point.14 Method of detection included next generation sequencing (NGS) and/or droplet digital PCR assays (ddPCR); the second measurement of ctDNA levels was performed between 6 and 16 weeks after treatment initiation. PFS values were reported in 17 clinical trials while OS values were reported in 11 trials (see table 1).
Figure 1.
Study selection schema.
Table 1.
Characteristics of included clinical trials
| Author | Year of publication | Disease site | Disease stage | Treatment setting | Number of participants | Line of therapy | ICI used | Method used to measure DNA | Expected ctDNA response | Timing of second ctDNA collection |
| Anagnostou et al15 | 2019 | Lung cancer | Metastatic NSCLC | Pallitative | 19 | First line | Nivolimab | TEC-Seq NGS | Undetectable | 6 |
| Cabel et al38 | 2017 | Lung cancer, uveal melanoma and MSI-H CRC | Metastatic | Pallitative | 10 | First line | Mixed | Bidirectional pyrophosphorolysis-activated polymerization, droplet digital PCR or next-generation sequencing depending on the mutation type | Undetectable | 8 |
| Garassino et al39 | 2018 | Lung cancer | Locally advanced unresectable or metastatic | Palliative | 65 | Third or more line | Durvalumab | ctDNA using Guardant360 | Undetectable | 6 |
| Goldberg et al16 | 2018 | Lung cancer | Metastatic NSCLC | Pallitative | 28 | First line | Mixed | Ultra-deep next-generation sequencing/error-suppressed deep sequencing method | 50% reduction | 6 |
| Herbreteau et al40 | 2018 | Melanoma | Locally advanced unresectable or metastatic | Palliative | 53 | First or second line | Nivolumab +- ipilimumab | Droplet digital PCR | Undetectable | 16 |
| Jin et al19 | 2020 | Gastric cancer | Metastatic gastric cancer | Pallitative | 40 | Second or more line | Mixed/multiple | HiSeq4000 NGS platform (Illumina) | Undetectable | 8 |
| Kim et al22 | 2018 | Gastric cancer | Metastatic | Palliative | 23 | Second or more line | Pembrolizumab | ctDNA using Guardant360 | 50% reduction | 6 |
| Lee et al41 | 2017 | Melanoma | Metastatic | Palliative | 40 | First line | Pembrolizumab or nivolumab +- ipilimumab | Droplet digital PCR | Undetectable | 12 |
| Lee et al42 | 2020 | Melanoma | Metastatic melanoma to brain | Pallitative | 72 | First line | Pembrolizumab or nivolumab +- ipilimumab | Droplet digital PCR | Undetectable | 12 |
| Giroux Leprieur et al21 | 2018 | Lung cancer | Metastatic | Pallitative | 15 | Second or more line | Nivolumab | NGS | Undetectable | 8 |
| Marsavela et al43 | 2020 | Melanoma | Metastatic | Palliative | 32 | First line | Variable | Droplet digital PCR | Undetectable | N/A |
| Marsavela et al18 | 2020 | Melanoma | Metastatic | Palliative | 15 | Second or more line | Variable | Droplet digital PCR | Undetectable | 12 |
| Moding et al44 | 2020 | Lung cancer | Locally advanced unresectable NSCLC | Consolidation ICIs following chemoradiation | 15 | Consolidation | Nivolumab | Gene panel sequencing by NGS | 50% reduction | 8 |
| Pedersen et al45 | 2020 | Melanoma | Locally advanced unresectable or Metastatic | Pallitative | 16 | First line | Pembrolizumab or nivolumab/ipilimumab | Droplet digital PCR | Undetectable | 7 |
| Raja et al20 | 2018 | Lung cancer | Metastatic | Palliative | 26 | Second or more line | Durvalumab | ctDNA using Gaurdant360 | Undetectable | 6 |
| Varaljai et al14 | 2020 | Melanoma | Metastatic BRAF V600E positive | Palliative | 18 | First line | Pembrolizumab or nivolumab/ipilimumab | Droplet digital PCR | Decrease vs increase | 10 |
| Warburton et al17 | 2020 | Melanoma | Metastatic | Palliative | 38 | First or second line | Pembrolizumab | Droplet digital PCR | Undetectable | 16 |
| Zheng et al46 | 2018 | Urothelial cancer | Metastatic | Palliative | 27 | Second or more line | Durvalumab | ctDNA using Gaurdant360 | Undetectable vs detectable | 6 |
CRC, colorectal cancer; ICI, immune checkpoint inhibitor; MSI-H, microsatellite instability-high; NGS, next generation sequencing; NSCLC, non-small cell lung cancer.
Progression-free survival
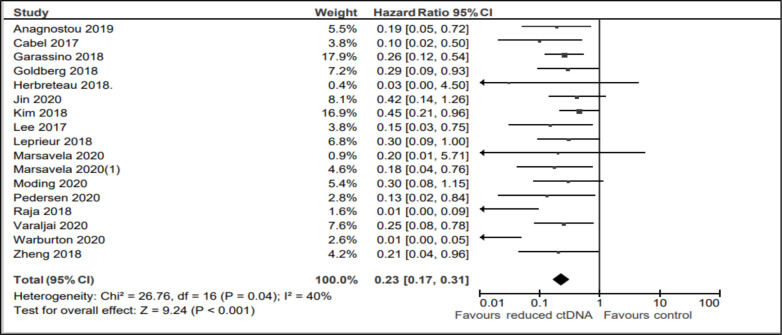
A reduction in ctDNA levels, while on treatment with ICIs, was associated with significantly improved PFS (HR 0.20; 95% CI, 0.14 to 0.28; p<0.001, see figure 2). There was borderline magnitude, but statistically significant heterogeneity (I2 40%, Cochran Q p=0.04). Table 2 shows the subgroup analyses. Overall, results were consistent across all disease sites (melanoma, lung cancer and other solid tumors including gastric cancer, urothelial cancer and MSI-H colorectal cancer), lines of treatment, magnitude of change (to undetectable vs >50% reduction), different intervals of the second ctDNA sampling and whether treatment exposure comprised single or multiple ICIs.
Figure 2.
Forest plot showing the effect of ctDNA clearance on progression-free survival.
Table 2.
Survival outcome data (OS and PFS) correlated with ctDNA clearance, and subgroup analysis based on disease site, treatment type, line of treatment, level of ctDNA change and timing of second ctDNA sampling
| Subgroups/outcome | HR (OS) | 95% CI | Subgroup difference p | HR (PFS) | 95% CI | Subgroup difference p |
| All patients | 0.18 | 0.12 to 0.26 | NA | 0.23 | 0.17 to 0.31 | NA |
| Disease site | ||||||
| Melanoma | 0.20 | 0.12 to 0.33 | 0.27 | 0.12 | 0.06 to 0.25 | 0.15 |
| Lung | 0.16 | 0.08 to 0.31 | 0.23 | 0.14 to 0.37 | ||
| Others | 0.05 | 0.01 to 0.26 | 0.34 | 0.20 to 0.58 | ||
| Treatment | ||||||
| Single agent | 0.14 | 0.07 to 0.31 | 0.45 | 0.23 | 0.16 to 0.33 | 0.004 |
| Multi-agent | 0.20 | 0.12 to 0.32 | 0.23 | 0.13 to 0.41 | ||
| Treatment line | ||||||
| First line | 0.22 | 0.13 to 0.36 | 0.31 | 0.19 | 0.11 to 0.34 | 0.002 |
| Later line | 0.14 | 0.07 to 0.28 | 0.29 | 0.19 to 0.42 | ||
| Level of ctDNA change | ||||||
| ≥50% reduction | 0.17 | 0.05 to 0.62 | 0.93 | 0.29 | 0.12 to 0.71 | 0.21 |
| Undetectable levels | 0.18 | 0.12 to 0.27 | 0.22 | 0.15 to 0.31 | ||
| Interval of second ctDNA sampling | ||||||
| 10–16 weeks | 0.22 | 0.13 to 38 | 0.18 | 0.10 to 0.32 | 0.004 | |
| <10 weeks | 0.13 | 0.06 to 26 | 0.25 | 0.17 to 0.36 |
OS, overall survival; PFS, progression-free survival.
Overall survival
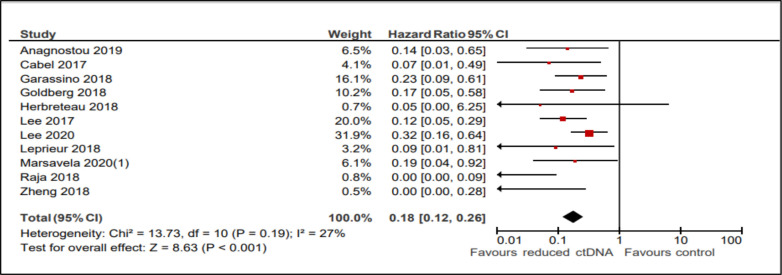
Similar to the PFS analysis, OS was improved in patients with reduced or undetectable ctDNA levels after receiving ICIs (HR 0.18; 95% CI, 0.12 to 0.26; p<0.001, see figure 3). There was no evidence of statistical heterogeneity (I2 27%, Cochran Q p=0.19). Similar to results seen for PFS, there was consistency between subgroups such as disease sites, lines of treatment, magnitude of change (to undetectable vs >50% reduction), different intervals of the second ctDNA sampling and whether treatment exposure comprised single or multiple ICIs.
Figure 3.
Forest plot showing the effect of ctDNA clearance on overall survival.
ctDNA change and radiologic stable disease
The association between ctDNA change and radiologic stable disease was reported in only three studies. Due to the very small number of patients in this analysis and the inconsistent reporting of outcomes, results are reported descriptively as data synthesis was not possible. It appeared that the association between ctDNA reduction and improved outcome also applied to patients with radiologic stable disease. In Anagnostou et al,15 12 out of the 19 patients with NSCLC had stable disease, on initial assessment. Among this group, patients who had ctDNA clearance (n=5) had improved PFS than those who showed detectable ctDNA levels (n=7). As expected from a cohort with study a small sample size, this effect was not statistically significant (HR 0.35; 95% CI, 0.05 to 2.25: p value=0.10). Similar trends were seen in other trials. In Goldberg et al,16 8 out of the 24 patients with NSCLC had stable disease as per RECIST criteria, with a median duration of treatment (mDOR) of 157 days. Among these patients, ctDNA reduction was associated with durable clinical benefit to ICIs, with an mDOR of 218 days (n=3) compared with non-responders who had an mDOR of 120 days (n=5). In Warburton et al,17 70 patients with advanced melanoma received anti-PD-1 therapy for approximately 1 year. Of these, 3 patients had stable disease and among those with fall in ctDNA, authors reported an unspecified longer disease control compared with those whose ctDNA did not fall.
Associations between ctDNA change and other biomarkers
Similar to the assessment of ctDNA change and radiologic stable disease, reporting of data on other biomarkers was variable precluding data synthesis. As such, data were again reported descriptively. In Anagnostou et al,15 patients with NSCLC with high TMB had more favorable survival outcomes than patients with low TMB; however, despite an association between TMB and ctDNA clearance at a study summary level, the latter marker appeared as a more sensitive predictor of PFS. Patients who had ctDNA clearance had better outcomes regardless of TMB status. Also, patients with continued elevation of ctDNA levels, while receiving ICIs, had worse outcomes even the setting of detecting high TMB levels. In Marsavela et al,18 there was no statistically significant difference in clinical benefit between patients with high TMB versus patients with low TMB and no reported association between TMB and ctDNA clearance.
In Jin et al,19 26 out of 32 patients with gastric cancer had known PD-L1 status. There was no significant difference in PFS between PD-L1 high (combined positive score (CPS≥10)) and PD-L1 low patients, and no clear association between PD-L1 expression and ctDNA clearance at a study summary level. In the first-line setting, PFS was not different among patients with combined prognostic score CPS≥10 (7.8 months) vs CPS<10 (7.4 months). The same results were observed in patients who received treatment as second or later lines, where PFS was about 3.4 months, in patients with CPS≥10 and 4.9 months in patients with a CPS<10 (p value>0.05). However, in, patients who had a more than 25% drop in ctDNA levels, during treatment, an improved PFS was observed (7.3 months) compared with those without ctDNA reduction (3.6 months). In Raja et al,20 there was no significant correlation between pretreatment PD-L1 status and differential ctDNA level, among patients with NSCLC and urothelial cancer (p value>0.1). Finally, in Giroux Leprieur et al,21 among patients with NSCLC who received nivolumab, changes in ctDNA levels were not different according to PD-L1 expression (p value=0.695).
In Kim et al,22 there appeared to be study level association between improved outcomes with ctDNA reduction and with MSI-H status. Objective response rates (ORR) were higher in patients with MSI-H tumors (85.7%) and in patients who had a significant drop on ctDNA levels after initiating treatment (ORR: 58%). PFS data were not reported for MSI status.
Discussion
While ICIs have emerged as important therapeutic tools for patients with advanced solid tumors, beyond traditional cross-sectional imaging, there are no standardized real-time markers that can assist treatment decisions.23 Imaging is only of value if done periodically over a relatively long time interval (at least every 2–3 months).24 Furthermore, imaging has other limitations including cost, radiation exposure, availability and, with ICI, there can be difficulties in interpretation due to the phenomenon of pseudoprogression.1 25
Measurement of PD-L1 expression in tissue or blood can provide prediction of benefit from ICIs in some tumors, but it has not been shown to be a robust real-time indicator able to guide treatment decisions after initiation of therapy.26 For example, previous studies showed that 40% of PD-L1-negative patients respond to ICIs.27 Other biomarkers (eg, TMB or MSI) have been reported to have associations with outcome in cancers treated with ICIs, however, factors such as intratumoral and intertumoral heterogeneity or lack of specificity may undermine the predictive role of these biomarker. Data suggest that TMB alone might not be able to identify the group of patients who might benefit from ICIs.18 Therefore, there is a need to validate dynamic markers that can provide clinicians with early and reliable insights about disease response in patient with advanced malignancies.
Dynamic changes in ctDNA levels may provide early assessment of disease response and can be used as a biomarker in this setting. In this study, we evaluated clinical trials in which PFS and OS were reported based on changes in ctDNA levels among patients with advanced solid tumors receiving ICIs. Results showed substantially improved outcome with a reduction in ctDNA levels in response to ICIs. The majority of included trials involved patients with melanoma and lung cancer (14 out of 18 studies). However, there was limited evidence for heterogeneity among studies especially for the OS analysis. Furthermore, there was comparable magnitude of association between change in ctDNA and both PFS and OS among subgroups comprised patients with melanoma, lung cancer and other solid tumors treated with ICIs. Overall, there was a four to five times lower hazard of progression or death with ctDNA reduction and a five to six times lower risk of death from any cause. The utilization of a systematic review and meta-analysis in this study provides increased level of certainty regarding the association between ctDNA reduction and improved outcomes, provides a more robust and precise estimate for the magnitude of effect and provides an assessment of consistency between different studies and different tumor types.28–31
ctDNA is a marker of burden of malignancy. Therefore, it is reasonable to assume that a reduction in ctDNA would be a marker of response to any type of cancer treatment, not just ICIs. In some of the individual trials included in the current analysis (especially those in lung cancer), outcomes in response to ctDNA changes were assessed in patients treated with both ICIs and with cytotoxic chemotherapy. The results showed better outcome in patients who had lower on-treatment ctDNA levels regardless of the type of treatment. However, with summary data on outcomes favoring ICIs over chemotherapy, the absolute impact of a reduction in ctDNA was greater among patients receiving ICI than those receiving chemotherapy.32
These data support the use of ctDNA change as an early response biomarker which could have a complementary role to clinical, radiologic and biochemical parameters. Such information could also allow for de-escalation of cross-sectional imaging in patients receiving ICIs. Furthermore, in a setting exclusive to the use of ICIs, changes in ctDNA could presumably guide clinicians to identify progression from pseudoprogression (ie, if ctDNA levels fall shortly after starting therapy, increase in target tumor measurement would be more consistent with pseudoprogression). Early identification of ineffective therapy can also save patients from being unnecessarily exposed to agents with known toxicity which are unlikely to provide additional benefit. There would also be benefits to this approach from a health economic perspective. Additionally, we attempted to explore the impact of ctDNA change in patients with stable disease on radiological assessment. This analysis was limited by inconsistent reporting of outcome and a small sample size; however, in patients who had stable disease, changes in ctDNA levels appeared to differentiate between those with longer and shorter PFS. This supports the validity of dynamic changes in ctDNA levels as predictive real-time biomarkers both in patients who show response and in those with stable disease with cancer immunotherapy.
Our exploration of association between other biomarkers and change in ctDNA was also limited by inconsistent reporting and small sample size. There did not appear to be a consistent association between TMB or PD-L1 expression and ctDNA change at a study summary level. However, in a single study,22 there did appear to be an association between the effects of MSI-high and ctDNA reduction on response rate. Data on PFS were not reported. This is an interesting observation which warrants further study.
Preliminary data also support role of ctDNA changes in predicting the risk of eventual progression in long-term responders to ICIs. ctDNA clearance has been shown to be associated with significantly better outcomes even if measured more than 2 years after initiation of ICIs.33 In addition to ctDNA clearance, other biomarkers have been studied and have shown promising results as real-time biomarkers for evaluation of response. These included PD-L1 expression in EV. In this setting, increases in EV PD-L1 levels were associated with worse survival.34 Whether a combination of ctDNA and other markers of response would improve on the clinical utility of one of these markers in isolation warrants further research.
This study has limitations. First, to date, there has been no standardized method to measure the ctDNA.35 In all included trials, ctDNA levels were assessed either by ddPCR or multiple variations of NGS (TEC-Seq, HiSeq, ultradeep NGS, Guardiant360, etc), which may increase heterogeneity of the summary data. That said, from a statistical perspective, there did not appear to be large magnitude interstudy heterogeneity in either the PFS or the OS analyses. Second, ctDNA levels were measured at different intervals (6–16 weeks) after treatment initiation. While subgroup analysis did not identify a difference in outcome based on the timing of second sampling, early (before 10 weeks) or late (between 10 and 16 weeks), there may be residual bias which may also affect data interpretation. Third, even though data were consistent among all trials, disease sites, lines of treatment and the level of ctDNA change, the relatively small number of included trials in our meta-analysis will lead to some uncertainty. Fourth, the clinical utility of ctDNA reduction remains unclear. Specifically, it is uncertain to what degree ctDNA measurement would improve clinical decision making and therefore outcomes in comparison to imaging and clinical assessment alone. This should be explored in adequately designed prospective studies. Finally, in this study, we only included studies in which patients had measurable ctDNA at baseline. Data suggest that absence of ctDNA at baseline (so called non-shedders) may have better outcomes than those with measurable ctDNA. The analysis in this article was unable to provide a comparative analysis of ctDNA reduction compared with a cohort without detectable ctDNA at baseline. This limits generalizability.36 37
In summary, in advanced solid tumors, a reduction in ctDNA levels in response to ICIs is associated with substantial improvements in outcome. ctDNA change is a clinically valid early response biomarker which may allow for de-escalation of cross-sectional imaging in patients receiving ICIs or support treatment de-escalation strategies. Further research is needed to assess the clinical utility of changes in ctDNA especially whether such technology provides additive or synergistic information which allows better clinical decision making. Additional data are also required to standardize detection methods which will improve reproducibility results. This should include quantification of variations in sensitivity between available assays, as well as differences in discovery range between assay platforms.
Footnotes
Presented at: A part of this study has been previously published as a meeting abstract at the 2022 ASCO Annual Meeting.1
Contributors: LA-S: conceptualization, methodology, validation, resources, formal analysis, writing—original draft, writing - review and editing; BW: methodology, validation, resources, writing—review and editing; FT: writing - review and editing; CM: visualization, writing—review and editing; AM: writing—review and editing; DWC: conceptualization, writing—review and editing; EA: conceptualization, methodology, validation, data curation, supervision, formal analysis, writing—original draft, writing—review and editing--- Guarantor.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: DWC: Consulting or Advisory Role - AstraZeneca; Eisai; Exact Sciences; Gilead Sciences; GlaxoSmithKline; Merck; Novartis; Pfizer; Roche/GenentechResearch Funding - AstraZeneca (Inst); GlaxoSmithKline (Inst); Inivata (Inst); Merck (Inst); Pfizer (Inst); Roche/Genentech (Inst)Patents, Royalties, Other Intellectual Property - Patent (US62/675,228) for methods of treating cancers characterized by a high expression level of spindle and kinetochore-associated complex subunit 3 (ska3) gene. EA: Honoraria - Exact Sciences; Novartis; Sandoz. LA-S, BW, FT, CM and AM have nothing to disclose.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
Data are available in a public, open access repository. Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Not applicable.
References
- 1.Galldiks N, Kocher M, Ceccon G, et al. Imaging challenges of immunotherapy and targeted therapy in patients with brain metastases: response, progression, and pseudoprogression. Neuro Oncol 2020;22:17–30. 10.1093/neuonc/noz147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park HJ, Kim KW, Pyo J, et al. Incidence of pseudoprogression during immune checkpoint inhibitor therapy for solid tumors: a systematic review and meta-analysis. Radiology 2020;297:87–96. 10.1148/radiol.2020200443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seymour L, Bogaerts J, Perrone A, et al. IRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol 2017;18:e143–52. 10.1016/S1470-2045(17)30074-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia W, Gao Q, Han A, et al. The potential mechanism, recognition and clinical significance of tumor pseudoprogression after immunotherapy. Cancer Biol Med 2019;16:655–70. 10.20892/j.issn.2095-3941.2019.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann MHD, Bender S, Krahn T, et al. Ctdna and ctcs in liquid biopsy-current status and where we need to progress. Comput Struct Biotechnol J 2018;16:190–5. 10.1016/j.csbj.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget 2016;7:48832–41. 10.18632/oncotarget.9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Luo J, Wu S, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov 2020;10:1842–53. 10.1158/2159-8290.CD-20-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma M, Zhu H, Zhang C, et al. “ Liquid biopsy” -ctdna detection with great potential and challenges. Ann Transl Med 2015;3:235. 10.3978/j.issn.2305-5839.2015.09.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ocana A, et al. Circulating DNA and survival in solid tumorscirculating DNA and survival in cancer. Cancer Epidemiology, Biomarkers & Prevention 2016;25:399–406. [DOI] [PubMed] [Google Scholar]
- 10.Larribère L, Martens UM. Advantages and challenges of using ctdna NGS to assess the presence of minimal residual disease (MRD) in solid tumors. Cancers (Basel) 2021;13:5698. 10.3390/cancers13225698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis AA, Iams WT, Chan D, et al. Early assessment of molecular progression and response by whole-genome circulating tumor DNA in advanced solid tumors. Mol Cancer Ther 2020;19:1486–96. 10.1158/1535-7163.MCT-19-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanz-Garcia E, Zhao E, Bratman SV, et al. Monitoring and adapting cancer treatment using circulating tumor DNA kinetics: current research, opportunities, and challenges. Sci Adv 2022;8:eabi8618. 10.1126/sciadv.abi8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815–34. [DOI] [PubMed] [Google Scholar]
- 14.Váraljai R, Wistuba-Hamprecht K, Seremet T, et al. Application of circulating cell-free tumor DNA profiles for therapeutic monitoring and outcome prediction in genetically heterogeneous metastatic melanoma. JCO Precis Oncol 2020;3:PO.18.00229. 10.1200/PO.18.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anagnostou V, Forde PM, White JR, et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res 2019;79:1214–25. 10.1158/0008-5472.CAN-18-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldberg SB, Narayan A, Kole AJ, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin Cancer Res 2018;24:1872–80. 10.1158/1078-0432.CCR-17-1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warburton L, Calapre L, Pereira MR, et al. Circulating tumour DNA in advanced melanoma patients ceasing PD1 inhibition in the absence of disease progression. Cancers (Basel) 2020;12:3486. 10.3390/cancers12113486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsavela G, Johansson PA, Pereira MR, et al. The prognostic impact of circulating tumour DNA in melanoma patients treated with systemic therapies-beyond BRAF mutant detection. Cancers (Basel) 2020;12:3793. 10.3390/cancers12123793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Y, Chen D-L, Wang F, et al. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol Cancer 2020;19:154. 10.1186/s12943-020-01274-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raja R, Kuziora M, Brohawn PZ, et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res 2018;24:6212–22. 10.1158/1078-0432.CCR-18-0386 [DOI] [PubMed] [Google Scholar]
- 21.Giroux Leprieur E, Herbretau G, Dumenil C, et al. Circulating tumor DNA evaluated by next-generation sequencing is predictive of tumor response and prolonged clinical benefit with nivolumab in advanced non-small cell lung cancer. Oncoimmunology 2018;7:e1424675. 10.1080/2162402X.2018.1424675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–58. 10.1038/s41591-018-0101-z [DOI] [PubMed] [Google Scholar]
- 23.An HJ, Chon HJ, Kim C. Peripheral blood-based biomarkers for immune checkpoint inhibitors. Int J Mol Sci 2021;22:17.:9414. 10.3390/ijms22179414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehta S, Shelling A, Muthukaruppan A, et al. Predictive and prognostic molecular markers for cancer medicine. Ther Adv Med Oncol 2010;2:125–48. 10.1177/1758834009360519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y, Wang Q, Dong Q, et al. How to differentiate pseudoprogression from true progression in cancer patients treated with immunotherapy. Am J Cancer Res 2019;9:1546–53. [PMC free article] [PubMed] [Google Scholar]
- 26.Mahoney KM, Atkins MB. Prognostic and predictive markers for the new immunotherapies. Oncology (Williston Park), 2014: 39–48. [PubMed] [Google Scholar]
- 27.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 2015;373:23–34. 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoushtari AN, Collins L, Espinosa E, et al. 1757O early reduction in ctDNA, regardless of best RECIST response, is associated with overall survival (OS) on tebentafusp in previously treated metastatic uveal melanoma (mum) patients. Annals of Oncology 2021;32:S1210. 10.1016/j.annonc.2021.08.1702 [DOI] [Google Scholar]
- 29.Tan L, Sandhu S, Lee RJ, et al. Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann Oncol 2019;30:804–14. 10.1093/annonc/mdz048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sacco A, Forgione L, Carotenuto M, et al. Circulating tumor DNA testing opens new perspectives in melanoma management. Cancers (Basel) 2020;12:2914. 10.3390/cancers12102914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipson EJ, Velculescu VE, Pritchard TS, et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J Immunother Cancer 2014;2:42. 10.1186/s40425-014-0042-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou W, Yaung SJ, Fuhlbrück F, et al. Ctdna predicts overall survival in patients with NSCLC treated with PD-L1 blockade or with chemotherapy. JCO Precis Oncol 2021;5:827–38. 10.1200/PO.21.00057 [DOI] [PubMed] [Google Scholar]
- 33.Hellmann MD, et al. Circulating tumor DNA analysis to assess risk of progression after long-term response to PD-(L) 1 blockade in nsclcctdna analysis in long-term responders to PD-(L) 1 blockade. Clin Cancer Res 2020;26:2849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Miguel-Perez D, Russo A, Arrieta O, et al. Extracellular vesicle PD-L1 dynamics predict durable response to immune-checkpoint inhibitors and survival in patients with non-small cell lung cancer. J Exp Clin Cancer Res 2022;41:186. 10.1186/s13046-022-02379-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saha S, Araf Y, Promon SK. Circulating tumor DNA in cancer diagnosis, monitoring, and prognosis. J Egypt Natl Canc Inst 2022;34:8. 10.1186/s43046-022-00109-4 [DOI] [PubMed] [Google Scholar]
- 36.Kim ES, Velcheti V, Mekhail T, et al. Blood-Based tumor mutational burden as a biomarker for atezolizumab in non-small cell lung cancer: the phase 2 B-F1RST trial. Nat Med 2022;28:939–45. 10.1038/s41591-022-01754-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolfo C, Cardona AF, Cristofanilli M, et al. Corrigendum to “ challenges and opportunities of cfdna analysis implementation in clinical practice: perspective of the international society of liquid biopsy (ISLB)” [ CRIT. rev. oncol. hematol. 151 (july) (2020) 102978 ]. Crit Rev Oncol Hematol 2020;154:103058. 10.1016/j.critrevonc.2020.103058 [DOI] [PubMed] [Google Scholar]
- 38.Cabel L, Riva F, Servois V, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol 2017;28:1996–2001. 10.1093/annonc/mdx212 [DOI] [PubMed] [Google Scholar]
- 39.Garassino MC, Cho B-C, Kim J-H, et al. Durvalumab as third-line or later treatment for advanced non-small-cell lung cancer (atlantic): an open-label, single-arm, phase 2 study. Lancet Oncol 2018;19:521–36. 10.1016/S1470-2045(18)30144-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herbreteau G, Vallée A, Knol A-C, et al. Quantitative monitoring of circulating tumor DNA predicts response of cutaneous metastatic melanoma to anti-PD1 immunotherapy. Oncotarget 2018;9:25265–76. 10.18632/oncotarget.25404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JH, Long GV, Boyd S, et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann Oncol 2017;28:1130–6. 10.1093/annonc/mdx026 [DOI] [PubMed] [Google Scholar]
- 42.Lee JH, Menzies AM, Carlino MS, et al. Longitudinal monitoring of ctDNA in patients with melanoma and brain metastases treated with immune checkpoint inhibitors. Clin Cancer Res 2020;26:4064–71. 10.1158/1078-0432.CCR-19-3926 [DOI] [PubMed] [Google Scholar]
- 43.Marsavela G, Lee J, Calapre L, et al. Circulating tumor DNA predicts outcome from first-, but not second-line treatment and identifies melanoma patients who may benefit from combination immunotherapy. Clin Cancer Res 2020;26:5926–33. 10.1158/1078-0432.CCR-20-2251 [DOI] [PubMed] [Google Scholar]
- 44.Moding EJ, Liu Y, Nabet BY, et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small cell lung cancer. Nat Cancer 2020;1:176–83. 10.1038/s43018-019-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedersen JG, Madsen AT, Gammelgaard KR, et al. Inflammatory cytokines and ctDNA are biomarkers for progression in advanced-stage melanoma patients receiving checkpoint inhibitors. Cancers (Basel) 2020;12:1414. 10.3390/cancers12061414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Y, Narwal R, Jin C, et al. Population modeling of tumor kinetics and overall survival to identify prognostic and predictive biomarkers of efficacy for durvalumab in patients with urothelial carcinoma. Clin Pharmacol Ther 2018;103:643–52. 10.1002/cpt.986 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available in a public, open access repository. Data are available upon reasonable request.