Abstract
Live-cell imaging is extremely common in synthetic biology research, but its ability to be applied reproducibly across laboratories can be hindered by a lack of standardized image analysis. Here, we introduce a novel cell segmentation method developed as part of a broader Independent Verification & Validation (IV&V) program aimed at characterizing engineered Dictyostelium cells. Standardizing image analysis was found to be highly challenging: the amount of human judgment required for parameter optimization, algorithm tweaking, training and data pre-processing steps forms serious challenges for reproducibility. To bring automation and help remove bias from live-cell image analysis, we developed a self-supervised learning (SSL) method that recursively trains itself directly from motion in live-cell microscopy images without any end-user input, thus providing objective cell segmentation. Here, we highlight this SSL method applied to characterizing the engineered Dictyostelium cells of the original IV&V program. This approach is highly generalizable, accepting images from any cell type or optical modality without the need for manual training or parameter optimization. This method represents an important step toward automated bioimage analysis software and reflects broader efforts to design accessible measurement technologies to enhance reproducibility in synthetic biology research.
Keywords: cell segmentation, self-supervised learning, automated image analysis, reproducibility
Graphical Abstract
1. Introduction
Synthetic biology has the potential to address crucial issues that society will face in the 21st century and beyond, ranging from cell-based therapies (1) to climate change (2). However, for synthetic biology to have a measurable effect on society, basic research results must first be established as reproducible to transition into viable technologies. Reproducibility of key results is increasingly seen as a benchmark of the quality and importance of the research, with research institutions and communities beginning to stress the need for reproducibility efforts and open dialogs as to how to effectively implement them (3–9). Crucial to this aim is the availability of tools that function identically across laboratories in biological research.
As participants in a real-time reproducibility project administered by the U.S. Defense Advanced Research Projects Agency (DARPA), referred to as Independent Verification & Validation (IV&V), the aim was to apply engineering principles of design and control to synthetic biology. At every step in conducting an experiment, if a human judgment is required in the process, it is a potential risk to successful replication. Thus, general ‘tools’ that reduce or eliminate human judgment from a given step are extremely valuable for meaningful biological research. Much of the bandwidth of replication dialogs focuses on the methodology involved in the ‘front end’ of an experiment (10–13). However, less attention has been given to reproducibility at the ‘back end’ of an experiment: how the data are collected, processed and analyzed has significant impact on the ultimate interpretation of an experimental result (6, 14).
Here, we demonstrate the use of a novel back-end tool created in our IV&V program for reproducible cell segmentation through our efforts to replicate key findings from the Iglesias Laboratory in their program with DARPA’s Biological Technology Office. The original experiments tasked for replication (15), an overview of the IV&V program’s scope and broad lessons learned (11) and a detailed description and validation of the self-supervised learning (SSL) segmentation algorithm (16) are published elsewhere.
Cell segmentation poses significant technical challenges in biological research, with many recent efforts to aid the process. Broadly speaking, the segmentation algorithms are often classified as model-based (e.g. CellProfiler) (17) or machine learning (e.g. U-Net) (18) approaches, but neither is completely autonomous. Model-based approaches require manual tuning of multiple parameters in processing steps (e.g. intensity thresholding), sometimes upwards of dozens. Along similar lines, machine learning requires the user to provide annotations on the curated data from which the algorithm is trained—a data-hungry process known as supervised learning (SL). In either approach, there is human involvement, which poses a problem for reproducibility efforts. Algorithms that are tuned or trained at the onset can problematically miss relevant features as the cellular phenotypes or background characteristics evolve, inadvertently skewing the analysis (e.g. variations in label intensity from photobleaching, changes in morphology from blebbing). This requires intervention as experiments or experimental conditions shift, with the end user imparting judgment of both ‘when’ to retune/retrain and ‘how’ to. Especially concerning in SL is that the training process is inherently subjective in nature with no established way to measure bias embedded in training.
This communication focuses on the novelty of SSL segmentation as applied to the engineered Dictyostelium cells, which proposes unique challenges due to the range of morphological and dynamic phenotypes Dictyostelium cells exhibit. The goal was the development of software that enabled blind image segmentation by means of complete automation, thereby achieving best practice in image analysis reproducibility. General segmentation strategies are of enormous value to the biological research community, as evidenced by the significant efforts invested toward developing accurate and accessible segmentation tools (19). SSL segmentation is a natural extension of these efforts—expanding toward a general, configuration-free, single-cell segmentation method. The crux behind this advance is harnessing the dynamic nature of cells (motion) to distinguish what parts of images are cells versus the background. The result is a method that is robust across laboratories and experimental configurations and does not require any input from the end user, showcasing a new approach to reproducible bioimage analysis.
2. Materials and methods
2.1. Cells and microscopy
In their original study, Miao et al. utilized Dictyostelium as a model system to investigate the role of signal transduction excitable networks (STENs) in cell migration, with perturbations to phosphatidylinositol-4,5-bisphosphate (INP54P) levels or Ras/Rap-related activities controlling the migration modes of individual cells between amoeboid and keratocyte-like/oscillatory phenotypes. Details on the engineered Dictyostelium cells and experiments are in the original publication (15), with no significant deviations to report in their replication. A systematic approach, based on open communication channels and site visits, was taken to transfer experimental methodologies and their accurate execution by the IV&V team and performer laboratory (11), a laborious process that is increasingly being mitigated in part by platforms like protocols.io (20). Replicated experiments were conducted on Zeiss Axio Observer microscopes with either 10× phase contrast (0.45 NA), 10× transmitted light (TL, 0.3 NA) or 40× TL (1.4 NA) and imaged with a Zeiss Axiocam 702 CMOS or Hamamatsu ORCA R2 CCD camera. All the data presented here are from three biological replicates—meaning experiments were conducted on different days, on different batches of cells/reagents and on two different microscopes/experimental setups. The Dictyostelium cells were observed for periods of ∼30 min both before and after the addition of 5 µM rapamycin (Rap− and Rap+, respectively).
2.2. Cell segmentation
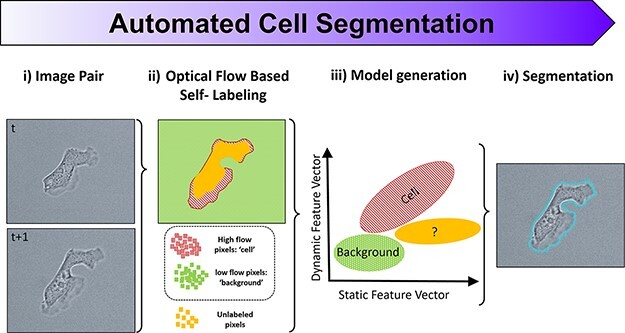
The SSL algorithm is described and validated in detail elsewhere (16), but the overarching concept is outlined in Figure 1A. The crux of SSL is that the relative motion between consecutive images is leveraged to automatically label which parts of an image belong to a cell versus the background. Two consecutive images from a dataset are recursively used to self-train, from t to t + 1. The optical flow between images is calculated by the Farnebäck method (21), which is utilized as a dynamic feature vector to self-label pixels for cell or background classification. A threshold is automatically determined, above which pixels are labeled ‘cell’ (red, hashed), and a similarly lower threshold is used to label pixels as ‘background’ (green). Pixels with intermediate optical flow are left unclassified (solid yellow ‘unlabeled’ pixels). Additional static feature vectors (intensity gradients and entropy) are then generated for each of these self-labeled training pixels. These additional feature vectors are then used to train and generate a naive Bayesian classifier model that is applied to the entire image in a pixel-wise fashion, allowing for the unlabeled pixels by optical flow to be classified and cell-segmented. The entire self-training and reclassification process begins from scratch on the next consecutive images, t + 1 to t + 2, and so on, in a completely automated fashion. The resulting advantage of SSL is that neither parameter tuning nor training images are required, as the self-supervised training data are updated for every image pair automatically. For estimating reproducibility on manual segmentation, three support scientists on the IV&V program were tasked to manually segment over 50 individual Dictyostelium cells on representative 10× TL images, and the ground truth (GT) was established by the lead scientist/liaison with the performer laboratory. Each user’s F1 score was calculated, and the resulting dissimilarity measurement is a simple percentage difference of F1 scores.
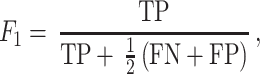 |
(1) |
Figure 1.
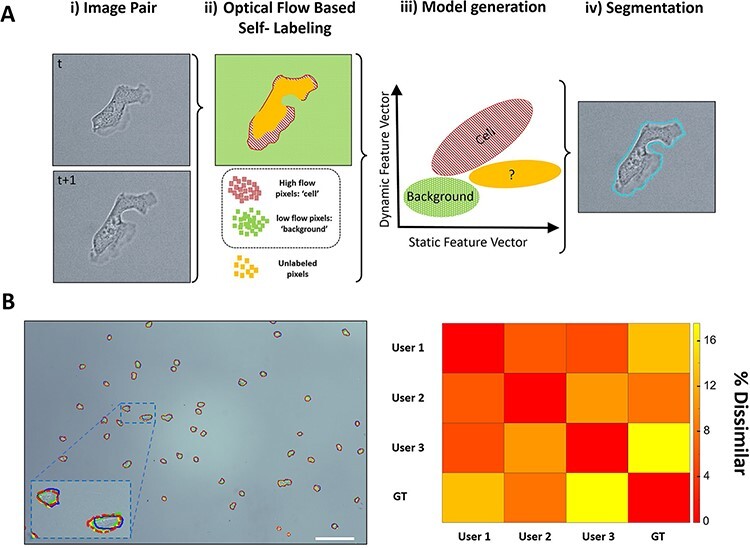
SSL and bias in cell segmentation. (A) The underlying concept of SSL segmentation is that the relative motion between consecutive images (i) is automatically calculated via optical flow, after which pixels with high flow are automatically labeled as ‘cell’, and pixels with low flow are automatically labeled as ‘background’ (ii). Optical flow acts as a dynamic feature vector for automatically labeling training pixels, which then incorporate additional static feature vectors (intensity gradients and entropy) used to train and generate a naive Bayesian classifier model (iii). This model is then applied to the entire image in a pixel-wise fashion, allowing the unlabeled pixels by optical flow to be classified and cell-segmented (iv). (B) Three scientists were tasked to manually segment over 50 individual Dictyostelium cells on standard TL images of the replicated experiments similar to the performer’s microscope set-up (green (solid), red (dashed) and blue (dark solid) outlines). Their results were compared with one another/GT, and the accuracy (F1 score) is tabulated in the above dissimilarity matrix (right). Whether it is manual segmentation, tuning parameters or selecting and labeling data for training, human judgment is always required for current segmentation techniques and thus varies from person to person and laboratory to laboratory. Scale bar 50 µm.
where true positives (TP), false positives (FP) and false negatives (FN) are calculated in a pixel-wise fashion.
2.3. Phenotype classification
Cell segmentation was achieved via SSL, while phenotype analysis and classification strictly followed the methodology of Miao et al. as to not introduce potential discrepancies in results from differing types of analysis and are detailed under the ‘image analysis’ and ‘assignment of migratory modes’ of (15). Briefly, segmented cells that did not leave the field of view and did not interact with other cells were assembled into tracks of a 10-min time window. The segmented area was analyzed over the time of the track, and the coefficient of variation (CoV) of the cell area was calculated. A threshold of COVth = 0.12 was used to designate oscillator phenotypes that exhibit dynamic spreading and contracting behavior. For cells with a COV < COVth, the migration behavior was used to distinguish between amoeboid and fan phenotypes—phenotypes that exhibit highly persistent migration with the direction of movement perpendicularly relative to the long axis of the cell (cell polarity) are deemed fan phenotypes, and the rest are classified as amoeboid.
3. Results and discussion
The replicated experiments tested the hypothesis that the spectrum of migratory modes observed in cells arises from different thresholds of a STEN. Since components in the STEN undergo highly coordinated transient changes during network activation, ‘clamping’ one component near the level it achieves during activation might alter the excitability of the entire network, offering an opportunity to test the idea. The Iglesias team has demonstrated the use of a chemically inducible dimerization system in Dictyostelium to clamp INP54P at low levels, as would be expected to transiently occur during STEN activation (15). In their system, the addition of rapamycin to the extracellular media of INP54P-transfected cells initiated a causal chain of events: the threshold for network activation was lowered, the speed and range of propagating waves of signal transduction activity increased, actin-driven cellular protrusions expanded and, consequently, the cell migratory mode transitions ensued from amoeboid to either an oscillatory or migratory (fan-shaped) phenotype. The resulting data from the replicated experiments are in the form of time-lapse images of individual Dictyostelium cells, which need to be first segmented to extract their dynamic morphology and, ultimately, phenotype to be classified.
The original study utilized a combination of manual and model-based approaches to segment cells and track their migration (15). For the analysis of the replicated experimental data, both manual and conventional SL segmentation methods were deemed to include too much end-user judgment, bias and variability (Figure 1B). Blinded analysis of data is the best practice, but for practical reasons, images were analyzed by the same researchers who designed and conducted the experiments. For a segmentation algorithm to be an effective tool for synthetic biology research that can be applied identically across different laboratories, we sought to abrogate the required human supervision present in cell segmentation by exploiting the motion present in all time-lapse, live-cell microscopy. The SSL method uses optical flow (21) to calculate the relative motion between pairs of time-lapse images of cells (22). Pixels that undergo higher levels of optical flow between consecutive images are automatically self-labeled as ‘cell’, and those that do not are self-labeled as ‘background’ (Figure 1A). These labels are then used to extract the additional feature vectors (entropy and gradients) to train a classification model to fully segment cells. This self-training occurs recursively on every pair of consecutive images, an appealing strategy to account for experimental drifts that lead to changes in image characteristics. By leveraging the structure of time-lapse, live-cell data, this SSL approach achieves accurate single-cell segmentation across different cell types, optical modalities and microscope setups and does so in an entirely automated manner—encapsulating the requirements of a reproducible segmentation method (16).
The SSL approach readily segments Dictyostelium regardless of whether optical objective or imaging modality is used, achieving satisfactory accuracy (F1 score of 0.71), with performance (16) comparable to contemporary segmentation techniques like CellPose (23) and could be compiled into tracks for analysis. Corroborating Miao et al., we observed three distinct phenotypes, which are shown in Figure 2. The amoeboid phenotype has the characteristics of a rounded morphology, small fluctuations in the spread area and low migration speeds/distances (Figure 2A). The fan phenotype exhibits a spread-out morphology with a broad lamellipodia typically at its leading edge. Fan phenotypes also exhibit extremely fast and highly persistent motion with moderate fluctuations in their spread area (Figure 2B). Oscillator phenotypes are characterized by large fluctuations in the spread area and fast motion with low persistence/directionality (Figure 2C). The measured average naive speed of cells approximately doubled from 3.5 ± 1.8 to 6.9 ± 4 µm/min (n > 50 cells taken from three biological replicates, ±standard deviation) 30 min after the addition of rapamycin, in good agreement with the original results (4.2 and 7.3 µm/min, respectively) (15).
Figure 2.
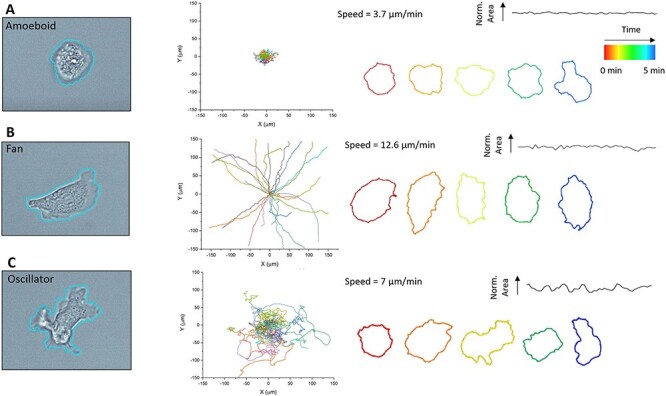
SSL segmentation and tracking of distinct Dictyostelium phenotypes. (Left) Representative SSL segmentation overlays of (A) amoeboid, (B) fan and (C) oscillator cells. (Middle) The centroid tracks of each phenotype of migrations (10 min in the field of view), with each track a different color and reset to the same origin (n = 25 cells for ease of visualization). (Right) Temporal morphological profiles of individual cells highlight the distinct morphological characteristics of each phenotype, with normalized spread area versus time profile and migration speed shown. Source: See the original results in Figure 1C and D in Ref. (15) for comparison.
The heterogeneity within cell populations before and after rapamycin exposure is highlighted in Figure 3, showing the shift in phenotypes that arise from different STEN thresholds. The population of INP54P-transfected cells exhibited 25% oscillator phenotypes (SD 7%) based on a CoVth = 0.12 applied to SSL segmented cells, higher than that reported by Miao et al. (∼7%). However, after exposure to rapamycin, the population shifted to 23% fans and 43% oscillators (SD 5%), in good agreement with that given by Miao et al. (∼20 and 50%, respectively) (15).
Figure 3.
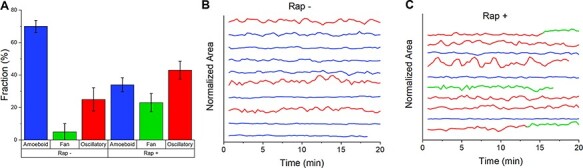
SSL-enabled phenotype classification. (A) Fractions of amoeboid (blue), fan (green) and oscillator (red) phenotypes before and 30 min after the addition of 5 µM rapamycin (n > 50 cells, three biological replicates; error bars indicate ±1 standard deviation). (B) Temporal profiles of normalized areas of 10 representative cells before rapamycin and (C) 30 min after, with each cell track initialized to zero. Source: See the original results in Figure 1E and F in Ref. (15) for comparison.
This IV&V pilot program was unique in that it was built into the grants awarded by DARPA’s Biological Technologies Office (11), but it remains unclear if such efforts will be adopted more widely. The sheer cost in both resources and time, as well as the unclear recognition of conducting replication studies, creates a high barrier-to-entry for IV&V efforts (24, 25). Thus, to encourage replication studies, the field should strive for the creation of automated tools that can be easily implemented by different research groups. We leveraged our opportunity as participants in an IV&V program to create such a tool to address the problem of cell segmentation, which is an important measurement in most biological research. Here, we highlight a configuration-free SSL segmentation method and demonstrate that it can aid in replicating research on the back end by removing human judgment from the process of cell segmentation. Our replicated experiments analyzed via automated SSL segmentation agreed well with the original results, despite the performing laboratory’s use of manual and model-based approaches for cell segmentation.
The automation accompanied by SSL segmentation removes the need for manual labor, saving time and reducing potential bias—ensuring every laboratory is using the same tool in the same manner. However, automation has the trade-off of potentially becoming a ‘black box’ for end users. We tried to avoid this by focusing on motion as the main feature vector used for self-labeling, which is more intuitive and interpretable compared to many machine learning methods. Future work will focus on simulating images from fixed immunofluorescence or confluent cell data in order to enable SSL’s application to static images. To the best of our knowledge, this SSL segmentation represents the first of its kind method and an important step toward the development of general automated bioimage analysis software.
Acknowledgments
The authors would like to thank Pablo Iglesias, Douglas Robinson and Peter Devreotes at Johns Hopkins University. They also thank Xiaoguang Li and Hideaki Matsubayashi for their time and effort spent on protocol transfer.
Contributor Information
Michael C Robitaille, Materials Science and Technology Division, U.S. Naval Research Laboratory, Washington, DC, USA.
Jeff M Byers, Materials Science and Technology Division, U.S. Naval Research Laboratory, Washington, DC, USA.
Joseph A Christodoulides, Materials Science and Technology Division, U.S. Naval Research Laboratory, Washington, DC, USA.
Marc P Raphael, Materials Science and Technology Division, U.S. Naval Research Laboratory, Washington, DC, USA.
Data availability
The SSL application is available for download at Zenodo as (1) a stand-alone Graphical User Interface download for Windows, Mac and Linux operating systems and (2) SSL Matlab source code with user interface application (https://zenodo.org/record/7108601).
The raw data sets used in this replication study are available at https://zenodo.org/record/7429795#.Y5ockXbMIuU.
Funding
National Research Council Research Associateship Program; Jerome and Isabella Karle Distinguished Scholar Fellowship Program; Biological Technology Office of the Defense Advanced Research Program Agency.
Conflict of interest statement.
The authors declare no conflict of interest.
References
- 1. Auslander S., Auslander D. and Fussenegger M. (2017) Synthetic biology—the synthesis of biology. Angew. Chem., Int. Ed., 56, 6396–6419. [DOI] [PubMed] [Google Scholar]
- 2. Georgianna D.R. and Mayfield S.P. (2012) Exploiting diversity and synthetic biology for the production of algal biofuels. Nature, 488, 329–335. [DOI] [PubMed] [Google Scholar]
- 3. Baker M. (2016) Dutch agency launches first grants programme dedicated to replication. Nature, 20. [Google Scholar]
- 4. Begley C.G. and Ellis L.M. (2012) Raise standards for preclinical cancer research. Nature, 483, 531–533. [DOI] [PubMed] [Google Scholar]
- 5. Collins F.S. and Tabak L.A. (2014) Policy: NIH plans to enhance reproducibility. Nature, 505, 612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ioannidis J.P.A. (2005) Why most published research findings are false. PLoS Med., 2, 696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Renshaw A.A. and Gould E.W. (2007) Measuring errors in surgical pathology in real-life practice—defining what does and does not matter. Am. J. Clin. Pathol., 127, 144–152. [DOI] [PubMed] [Google Scholar]
- 8. Deagle R.C., Wee T.L. and Brown C.M. (2017) Reproducibility in light microscopy: maintenance, standards and SOPs. Int. J. Biochem. Cell Biol., 89, 120–124. [DOI] [PubMed] [Google Scholar]
- 9. Redish A.D., Kummerfeld E., Morris R.L. and Love A.C. (2018) Reproducibility failures are essential to scientific inquiry. Proc. Natl. Acad. Sci. U.S.A., 115, 5042–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Errington T.M., Denis A., Perfito N., Iorns E. and Nosek B.A. (2021) Reproducibility in cancer biology: challenges for assessing replicability in preclinical cancer biology. elife, 10, e67995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raphael M.P., Sheehan P.E. and Vora G.J. (2020) A controlled trial for reproducibility. Nature, 579, 190–192. [DOI] [PubMed] [Google Scholar]
- 12. Baker M. (2021) How to write a reproducible lab protocol. Nature, 597, 293–295. [DOI] [PubMed] [Google Scholar]
- 13. Munafo M.R., Nosek B.A., Bishop D.V.M., Button K.S., Chambers C.D., du Sert N.P., Simonsohn U., Wagenmakers E.-J., Ware J.J., Ioannidis J.P.A. (2017) A manifesto for reproducible science. Nat. Hum. Behav., 1, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ioannidis J.P.A. (2019) What have we (not) learnt from millions of scientific papers with P values? Am. Stat., 73, 20–25. [Google Scholar]
- 15. Miao Y.C., Bhattacharya S., Edwards M., Cai H.Q., Inoue T., Iglesias P.A., Devreotes P.N. (2017) Altering the threshold of an excitable signal transduction network changes cell migratory modes. Nat. Cell Biol., 19, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robitaille M.C., Byers J.M., Christodoulides J.A. and Raphael M.P. (2022) A self-supervised machine learning approach for objective live cell segmentation and analysis. Commun. Bio, 5, 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J., Lindquist R.A., Moffat J. et al. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol., 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Falk T., Mai D., Bensch R., Cicek O., Abdulkadir A., Marrakchi Y., Böhm A., Deubner J., Jäckel Z., Seiwald K. et al. (2019) U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods, 16, 67–70. [DOI] [PubMed] [Google Scholar]
- 19. Caicedo J.C., Goodman A., Karhohs K.W., Cimini B.A., Ackerman J., Haghighi M., Heng C., Becker T., Doan M., McQuin C. et al. (2019) Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl. Nat. Methods, 16, 1247–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Teytelman L., Stoliartchouk A., Kindler L. and Hurwitz B.L. (2016) Protocols.io: virtual communities for protocol development and discussion. PLoS Biol., 14, e1002538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farneback G. (2003) Two-frame motion estimation based on polynomial expansion. In: Bigun J, Gustavsson T (eds). Image Analysis, Proceedings. Lecture Notes in Computer Science. Vol. 2749, Springer Berlin Heidelberg, Halmstad, Sweden, pp. 363–370. [Google Scholar]
- 22. Robitaille M.C., Byers J.M., Christodoulides J.A. and Raphael M.P. (2022) Robust optical flow algorithm for general single cell segmentation. PLoS One, 17, e0261763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stringer C., Wang T., Michaelos M. and Pachitariu M. (2021) Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods, 18, 100–106. [DOI] [PubMed] [Google Scholar]
- 24. Amaral O.B. and Neves K. (2021) Reproducibility: expect less of the scientific paper comment. Nature, 597, 329–331. [DOI] [PubMed] [Google Scholar]
- 25. Flier J. (2017) Faculty promotion must assess reproducibility. Nature, 549, 133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The SSL application is available for download at Zenodo as (1) a stand-alone Graphical User Interface download for Windows, Mac and Linux operating systems and (2) SSL Matlab source code with user interface application (https://zenodo.org/record/7108601).
The raw data sets used in this replication study are available at https://zenodo.org/record/7429795#.Y5ockXbMIuU.


