Summary
Pan-betacoronavirus neutralizing antibodies may hold the key to developing broadly protective vaccines against novel pandemic coronaviruses and to more effectively respond to SARS-CoV-2 variants. The emergence of Omicron and subvariants of SARS-CoV-2 illustrates the limitations of solely targeting the receptor-binding domain (RBD) of the spike (S) protein. Here, we isolated a large panel of broadly neutralizing antibodies (bnAbs) from SARS-CoV-2 recovered-vaccinated donors, which targets a conserved S2 region in the betacoronavirus spike fusion machinery. Select bnAbs showed broad in vivo protection against all three deadly betacoronaviruses, SARS-CoV-1, SARS-CoV-2, and MERS-CoV, which have spilled over into humans in the past two decades. Structural studies of these bnAbs delineated the molecular basis for their broad reactivity and revealed common antibody features targetable by broad vaccination strategies. These bnAbs provide new insights and opportunities for antibody-based interventions and for developing pan-betacoronavirus vaccines.
Keywords: coronaviruses, SARS-CoV-2, SARS-CoV-2 variants of concern, COVID-19, coronavirus spike, S2 stem-helix site, broadly neutralizing antibodies, broad coronavirus protection, pan-betacoronavirus vaccines
Graphical abstract

Highlights
-
•
Isolated a large panel of stem-helix bnAbs from SARS-CoV-2 recovered-vaccinated humans
-
•
Stem-helix bnAbs possess recurrent germline features desired for targeted vaccines
-
•
IGHV1-46-encoded stem-helix bnAbs are public clonotypes with two binding modes
-
•
Stem-helix bnAbs prophylactically protect against SARS-CoV-1, SARS-CoV-2, and MERS-CoV challenge
Pan-betacoronavirus broadly neutralizing antibodies (bnAbs) may hold the key to developing broadly protective vaccines against pandemic coronaviruses and SARS-CoV-2 variants. Zhou et al. isolate a large panel of S2 stem-helix bnAbs to betacoronaviruses and reveal their public features. Select stem-helix bnAbs protect against all three human betacoronaviruses that cause deadly disease.
Introduction
The initial successes of SARS-CoV-2 vaccines are due in part to the relative ease of inducing protective neutralizing antibodies (nAbs) to immunodominant epitopes on the receptor-binding domain (RBD) of the S1 subunit of the spike protein.1,2,3,4,5 The most potent nAbs target epitopes on the RBD that overlap the angiotensin-converting enzyme 2 (ACE2) receptor-binding site (RBS) and require little affinity maturation to neutralize at very low concentrations.3,6,7,8,9,10,11,12,13 However, mutations, particularly within and around the RBS, readily generate viral variants that are resistant to neutralization by commonly induced classes of antibodies14,15,16,17 without substantially impairing viral fitness, resulting in several variants of concern (VOCs).15,18,19,20,21,22,23,24,25,26,27,28,29,30,31 For example, the K417N/T and E484K mutations in the Beta and Gamma VOCs lead to neutralization escape from the vast majority of RBS nAbs.12,14,15 Other sarbecoviruses that use ACE2 as receptor, such as SARS-CoV-1, and betacoronaviruses using receptors other than ACE2, such as MERS-CoV, show even more sequence divergence in the RBS region.32,33,34
Due to the emergence of SARS-CoV-2 VOCs, and for other zoonotic coronaviruses with pandemic potential, research efforts focus on vaccines and antibodies targeting the most conserved regions of the spike protein.35,36,37 The more conserved faces of the RBD are targeted by many nAbs with greater breadth of neutralization against VOCs and diverse sarbecoviruses than, for example, RBS nAbs.38,39,40,41,42,43,44,45,46,47,48,49 However, the Omicron variants demonstrate escape from some nAbs targeting these more conserved regions of the RBD.22,28,50 An alternative relatively conserved target on the coronavirus spike is the S2 region, which does harbor neutralizing epitopes51 and therefore is of interest to generate vaccines effective against SARS-CoV-2 VOCs and, more ambitiously, pan-betacoronaviruses.35,36,37 We previously isolated a nAb, CC40.8, from a COVID-19 convalescent donor, which neutralizes sarbecoviruses from clades 1a and 1b and SARS-CoV-2 VOCs.52,53 CC40.8 targets the conserved spike S2 stem-helix region that forms part of the spike fusion machinery and can protect against the SARS-CoV-2 challenge in human ACE2 mouse and hamster models.53 Several more broadly neutralizing antibodies (bnAbs) targeting this region have been isolated from humans and from vaccinated animals.54,55,56,57,58,59,60,61,62
These stem-helix bnAbs highlight the opportunities that conserved bnAb S2 epitopes may offer for development of broad betacoronavirus vaccines. A large panel of stem-helix bnAbs is still needed to define the common molecular features of antibodies targeting this site. Accordingly, in this study, we isolated the largest panel of β-CoV stem-helix bnAbs to date and revealed the public antibody features and molecular basis for their broad neutralization of coronaviruses. The isolated S2 stem-helix bnAbs were highly enriched in antibody germline features that could be exploited by targeted vaccines. Select bnAbs protected against infection by all three human betacoronaviruses (SARS-CoV-2, SARS-CoV-1, and MERS-CoV) that cause deadly disease. These bnAbs could facilitate the development of rational vaccine strategies for inducing such bnAbs63,64,65,66,67 and would also provide more options for antibody-based prophylaxis and therapeutic strategies.68
Results
Donors for isolation of β-CoV spike stem-helix bnAbs
To identify suitable donors for the isolation of a panel of β-CoV spike stem-helix bnAbs, we screened immune sera from human donors for cross-reactive binding to 25-mer peptides from the spike S2 stem-helix region, which we previously showed to be a target for bnAbs.52,53 We tested sera from four different groups of donors: (1) COVID-19-recovered donors (n = 15), (2) spike mRNA-vaccinated (2×) donors (n = 10), (3) spike mRNA-vaccinated (3×) donors (n = 9), and (4) COVID-19-recovered then spike-vaccinated (1×) donors (n = 15) (Figure 1A). Whereas weak or no binding was observed for sera from COVID-19-recovered or mRNA-vaccinated individuals to human β-CoV spike S2 stem-helix peptides, sera from 80% (12/15) of recovered-vaccinated (“hybrid immunity69”) donors exhibited strong cross-reactive binding to the peptides (Figure 1A). We noted a strong correlation between binding of recovered-vaccinated sera to SARS-CoV-2 stem-helix peptide with binding to other human β-CoV stem-helix peptides, suggesting targeting of common cross-reactive epitopes (Figure 1B). Accordingly, we sought to isolate β-CoV stem-helix-directed bnAbs from 10 SARS-CoV-2 recovered-vaccinated donors who exhibited cross-reactive binding to this spike region.
Figure 1.
Binding and neutralization properties of S2 stem-helix mAbs
(A) Dot plots showing ELISA binding (OD405) reactivity of immune sera from COVID-19-convalescent donors (n = 15), 2× spike mRNA-vaccinated donors (n = 10), 3× spike mRNA-vaccinated donors (n = 9), and SARS-CoV-2 recovered-vaccinated donors (n = 15) to 25-mer peptides corresponding to spike S2 stem-helix regions of human β-(sarbecoviruses: SARS-CoV-1 or 2; merbecovirus: MERS-CoV; embecoviruses: HCoV-HKU1 and HCoV-OC43) and α-(HCoV-NL63 and HCoV-229E) coronaviruses.
(B) Correlation between binding of recovered-vaccinated sera to SARS-CoV-2 stem-helix peptide and other β-CoV (MERS-CoV, HCoV-HKU1, and HCoV-OC43) stem-helix peptides. Responses for binding to two stem-helix peptides were compared by non-parametric Spearman correlation two-tailed test with 95% confidence interval, and the Spearman correlation coefficient (r) and p values are indicated.
(C) A total of 40 S2 stem-helix mAbs were isolated from 9 SARS-CoV-2 recovered-vaccinated donors (CC9, CC24, CC25, CC67, CC68, CC84, CC92, CC95, and CC99). MAbs were isolated by single B cell sorting using SARS-CoV-2 and MERS-CoV S proteins as baits. Heatmap showing IGHV germline gene usage (colored: VH1-46 [green], VH3-23 [plum], and other VH genes [gray]), IGLV germline gene usage (colored: VK3-20 [light blue], VL1-51 [yellow orange], and other VL genes [gray]), lineage information (unique [cyan] and expanded [tangerine] lineages), and V-gene nucleotide somatic hypermutations (SHMs). EC50 ELISA binding titers of mAbs with β- and α-HCoV spike S2 stem-helix region peptides are shown. IC50 neutralization of mAbs against pseudoviruses of clade 1a (SARS-CoV-2 and Pang17), clade 1b (SARS-CoV-1, WIV1, and SHC014) sarbecoviruses, and MERS-CoV. Spike S2 stem-helix bnAbs, CC40.8, S2P6, and CV3-25, were used as controls for binding and neutralization assays.
(D) Out of 40 stem-helix bnAbs isolated, 32 were unique clones. All 32 unique mAbs neutralized all 5 ACE2-utilizing sarbecoviruses tested. 23 out of the 32 unique mAbs exhibited cross-neutralization against MERS-CoV.
See also Figure S1 and Tables S1–S3.
Isolation of a large panel of β-CoV spike stem-helix mAbs
Using SARS-CoV-2 and MERS-CoV S proteins as baits, we sorted antigen-specific single B cells to isolate 40 stem-helix mAbs (Table S1) from 10 COVID-19 convalescent donors who tested positive for SARS-CoV-2 in March or April of 2020 and were vaccinated at the beginning of 2021 with the Pfizer/BioNTech BNT162b2 (n = 4: CC9, CC92, CC95, and CC99), Johnson & Johnson Ad26.CoV2.S (n = 1: CC67), or Moderna mRNA-1273 (n = 5: CC24, CC25, CC26, CC68, and CC84) vaccines (Figures S1A and S1B).2,70,71 Except for two mAbs that failed to bind stem-helix peptide from human coronavirus HKU1 (HCoV-HKU1), all mAbs bound to stem-helix peptides of endemic β-HCoV (HCoV-HKU1 and HCoV-OC43) but not α-HCoVs (HCoV-NL63 and HCoV-229E) (Figure 1C; Table S2). This finding was consistent with the conservation of key bnAb stem-helix contact residues on β-HCoV.53 Notwithstanding, we also tested binding of mAbs to soluble HCoV S proteins and cell surface-expressed spikes and observed consistent binding to SARS-CoV-2/1 and MERS-CoV spikes but reduced binding to β-HCoV spikes (HCoV-HKU1 and HCoV-OC43), especially in the soluble S-protein format (Table S3). Overall, we isolated 40 stem-helix mAbs, of which 32 were encoded by unique immunoglobulin germline gene combinations (V, D, and J), and 7 of 32 were expanded lineages with two or more clonal members (Figure 1C; Table S3).
Spike stem-helix mAbs exhibit broad neutralization against β-CoVs
We next examined neutralization of stem-helix mAbs against clade 1a (SARS-CoV-1, WIV1, and SHC014) and clade 1b (SARS-CoV-2 and Pang17) ACE2-utilizing sarbecoviruses32,33 and MERS-CoV.34 Consistent with conservation of the stem-helix bnAb epitope region across sarbecoviruses, all of the 32 mAb lineages neutralized all five sarbecoviruses tested with widely varying degrees of neutralization potency (Figure 1C). The bnAbs neutralized clade 1a SHC014 and clade 1b SARS-CoV-2 with higher relative potency, compared with the other sarbecoviruses (geometric mean IC50: SHC014 = 0.8 μg/mL and SARS-CoV-2 = 3.4 μg/mL, compared with Pang17 = 17.1 μg/mL) (Figure 1C), but some bnAbs (n = 8) neutralized all sarbecoviruses in the low μg/mL IC50 range (0.2–6 μg/mL). CC95.102 exhibited the most potent neutralizing activity with all ACE2 sarbecoviruses tested. Of 32 unique stem-helix bnAb lineages, 23 (72%) bnAbs exhibited cross-neutralization against MERS-CoV (Figures 1C and 1D). Neutralization potency of bnAbs against MERS-CoV was lower than to the sarbecoviruses, but many bnAb members were consistently effective.
We tested neutralization of SARS-CoV-2 VOCs (Alpha; Beta; Gamma; Delta; and Omicron subvariants BA.1, BA.2, BA.2.12.1, XBB, BA.2.75, BA.2.75.2, BA.4/5, BA.4.6, and BQ.1.1) by select S2 stem-helix bnAbs (Figure 2). The bnAbs were selected based on their high neutralization potency against sarbecoviruses (CC25.112, CC67.105, CC67.130, and CC95.102; Figure 2B), or against sarbecovirus + MERS-CoV (CC9.104, CC25.103, CC25.106, CC68.104, CC95.108, and CC99.103; Figure 2B). Consistent with the conservation of the S2 stem-helix region in SARS-CoV-2 VOCs, these bnAbs were consistently effective against the VOCs tested (Figures 2B and 2C). Of note, a fraction of stem-helix bnAbs (n = 9) showed some degree of polyreactivity or autoreactivity in both HEp2 cell and polyspecificity reagent (PSR) assays,7,72 but the majority were negative (Figures S1C–S1E; Table S3). Overall, we identified 32 stem-helix bnAb lineages that exhibited broad neutralizing activity against phylogenetically diverse β-HCoVs.
Figure 2.
Neutralization of SARS-CoV-2 VOCs by S2 spike stem-helix bnAbs
(A) Schema showing SARS-CoV-2 spike domains and subdomains of S1 and S2 subunits and spike amino acid changes and deletions in VOCs. Spike regions are labeled (NTD, N-terminal domain; RBD, receptor-binding domain; CTD1, C-terminal domain 1; CTD2, C-terminal domain 2; S1/S2, S1/S2 furin cleavage site; S2′, S2′ TMPRSS2 or cathepsin B/L cleavage site; FP, fusion peptide; HR1, heptad repeat 1; CH, central helix region; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane anchor); the amino acid substitutions are indicated on each VOC spike. The symbols for single mutation, insertion, and deletion are indicated. S2 stem helix is unchanged on all SARS-CoV-2 VOCs.
(B) Neutralization of SARS-CoV-2 (WT) and major SARS-CoV-2 variants (Alpha; Beta; Gamma; Delta; and Omicron subvariants BA.1, BA.2, BA.2.12.1, XBB, BA.2.75, BA.2.75.2, BA.4/5, BA.4.6, and BQ.1.1) by 10 select S2 stem-helix bnAbs.
(C) IC50 neutralization titers of select S2 stem-helix bnAbs against SARS-CoV-2 (WT) and the major SARS-CoV-2 variants. The IC50 neutralization fold-change of S2 stem-helix bnAbs with SARS-CoV-2 variants compared with the WT virus. Spike RBD nAb CC12.1 was used as control.
Immunogenetics of stem-helix bnAbs and vaccine targeting
Immunogenetic analysis of S2 stem-helix antibody sequences (all 32 unique S2 mAbs) shows strong enrichment (25-fold) of IGHV1-46 and marginal enrichment (1.7-fold) of IGHV3-23 germline gene families as compared with human baseline germline frequencies (Figures 3A and S2; Table S3).73,74 In our study, the vast majority of the isolated stem-helix bnAbs were encoded by these two heavy-chain germline genes (81%: IGHV1-46 [62.5%], IGHV3-23 [18.7%]). Previously isolated stem-helix human bnAbs, S2P6 and CC40.8, are IGHV1-46 and IGHV3-23 germline encoded, respectively.53,54 The IGHV1-46 germline gene was slightly more enriched (31-fold) in S2 stem-helix bnAbs that exhibited neutralization with both MERS-CoV and sarbecoviruses (Figures 3A and S2A), suggesting a potential role for this VH-germline gene for broader reactivity against diverse β-HCoV spikes. At least one IGHV1-46-encoded stem-helix bnAb was isolated from each of the 9 donors and likely represents a public clonotype for this bnAb site (Figure 1C; Table S3). For light-chain gene usage, we noted enrichment of IGKV3-20 (3.8-fold) and IGLV1-51 (9.4-fold) germline gene families in S2 stem-helix bnAbs as compared with human baseline frequencies (Figures 3B and S2D).74 Combined, these two light-chain gene families encoded 62.6% (IGKV3-20 [46.9%], IGLV1-51 [15.6%]) of the isolated S2 stem-helix bnAbs. The mAbs possessed modest levels of V-gene nucleotide somatic hypermutation (SHM): for VH, median = 7.3% and for VL, median = 4.5% (Table S3).
Figure 3.
Immunogenetic and kinetic properties of S2 β-CoV spike stem-helix bnAbs
(A and B) Pie plots showing IGHV and IGKV/IGLV gene usage distribution of isolated stem-helix mAbs. Enriched heavy (IGHV1-46 [green] and IGHV3-23 [plum]) (A) and light (IGKV3-20 [sky blue] and IGLV1-51 [yellow]) (B) gene families were colored. Dot plots showing % nucleotide mutations (SHMs) in the heavy (VH) or light (VL) chains of mAbs. The mAbs were grouped by neutralization against sarbecoviruses (SARS) or sarbecoviruses + MERS-CoV (SARS + MERS).
(C and D) CDRH3 (C) or CDRL3 (D) length distributions of isolated mAbs across SARS or SARS + MERS bnAb groups, compared with human baseline germline reference. MAbs with 10 and 11 amino acid CDRH3s or mAbs with 11 amino acid CDRL3s were enriched in isolated S2 stem-helix bnAbs, compared with baseline germline reference and are indicated by arrows.
(E) Sequence conservation logos of 11 amino acid-long CDRL3-bearing stem-helix bnAbs (n = 18) showed enrichment of certain V-J-gene-encoded residues, compared with the human baseline reference. Enriched residues (corresponding to a PPxF motif) were indicated by arrows. The PPxF CDRL3 motif was shown to be important for S2 stem epitope recognition by structural studies below.
(F) BLI binding kinetics of S2 stem-helix mature bnAbs and their inferred germline (iGL) versions to SARS-CoV-2 and MERS-CoV stem-helix peptides. Maximum binding responses, dissociation constants (KDApp), and on-rate (kon) and off-rate constants (koff) for each antibody-protein interaction were compared. KDApp, kon, and koff values were calculated only for antibody-antigen interactions where a maximum binding response of 0.2 nm was obtained. Statistical comparisons between two groups were performed using a Mann-Whitney two-tailed test (∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, p > 0.05).
(G) IC50 neutralization of S2 stem-helix bnAb iGLs with SARS-CoV-2 and MERS-CoV pseudoviruses.
See also Figures S2, S3, S6E, and S6F and Table S3.
We examined the CDRH3 loop lengths in the isolated S2 stem-helix bnAbs and observed a strong enrichment for 10-residue (7.2-fold) and 11-residue (4.5-fold) CDRH3s, compared with the human baseline reference (Figure 3C; Table S3).74 Germline D genes, IGHD3-10 and IGHD3-16, were enriched by 1.6- and 3.5-fold, respectively, compared with the baseline reference (Figure S2B).74 IGHJ4, the most common germline J gene utilized in humans, was slightly enriched (1.3-fold), compared with a reference database, and this germline J gene encoded 72% of stem-helix bnAbs isolated in this study (Figure S2C).74
The CDRL3 loop lengths in the stem-helix bnAbs were modestly enriched (2.9-fold) for 11-residue CDRL3s, compared with the human baseline reference (Figure 3D; Table S3).74 The IGKJ3 germline gene was modestly enriched (4.4-fold) in S2 stem-helix bnAbs as compared with a baseline reference (Figure S2E). The 11-residue CDRL3 loops showed enriched germline VJ light-chain gene-encoded motifs (Figure 3E). These light-chain motifs were shown to be critical for S2 stem epitope recognition by structural studies below. Overall, we observed strong enrichment of IGHV and IGLV germline gene features in β-HCoV spike stem-helix bnAbs. Therefore, rational vaccine strategies could exploit these features to generate a protective B cell response.64,65,75
To examine the potential contribution of antibody SHMs to SARS-CoV-2 neutralization efficiency and cross-neutralization with MERS-CoV, we tested binding of select mAbs to SARS-CoV-2 or MERS-CoV monomeric stem-helix peptides and to their S proteins by biolayer interferometry (BLI) (Figures 3F and S3A). The mAbs bound SARS-CoV-2 and MERS-CoV stem-helix peptides with nanomolar to picomolar apparent dissociation constants (KDApp) (Figures 3F and S3A) that were generally lower for SARS-CoV-2 compared with MERS-CoV stem-helix peptides. We found no strong association of heavy- or light-chain SHMs with binding to SARS-CoV-2 or MERS-CoV stem-helix peptides or with neutralization of the corresponding viruses (Figure S3B). However, binding affinity to MERS-CoV stem-helix peptide was associated with neutralization of the corresponding virus (Figure S3C).
To further investigate the role of SHM in binding and neutralization, we generated inferred germline (iGL) versions of stem nAbs by reverting their heavy- and light-chain V, D, and J regions to the corresponding germlines, as described previously,76 and assessed both binding and neutralization. The BLI binding responses and the KDApp values of the bnAb iGLs with SARS-CoV-2 and MERS-CoV stem-helix peptides were reduced around 10 times, compared with mature bnAbs, but were still strong (Figures 3F and S3A). One caveat is that the iGLs are not fully reverted antibody germlines as reversion of the non-templated CDR3 junctional regions is not possible. Therefore, some of the binding by iGLs could be contributed by non-templated CDR3 regions, and indeed, the structural studies below show that these regions contributed to paratope-epitope interactions of S2 stem-helix bnAbs. This may partly explain the unexpectedly high-affinity binding of the tested iGL antibody versions to S2 stem-helix peptides.
Binding of S2 bnAbs to S proteins were generally of higher KDApp (i.e., lower affinity) than to the corresponding peptides for SARS-CoV-2 but more comparable for MERS-CoV (Figure S3A). The KDApp of iGLs was again higher for S protein than for peptides for SARS-CoV-2. For MERS, KDApp was also higher for S protein than the corresponding peptides. Many of the iGL Abs failed to show detectable binding to MERS-CoV S protein (Figure S3A). Based on these binding patterns, we postulate that S2 bnAb precursors would require affinity maturation to effectively interact with spike trimers, and therefore prime-boost immunogens would need careful design.
By contrast, neutralization of SARS-CoV-2 and MERS-CoV by stem-helix bnAb iGLs was absent (Figure 3G). These results suggest that although overall SHM levels did not correlate with binding or neutralization of mature bnAbs, key antibody mutations were critical for iGLs to attain sufficient affinity for neutralization to be observed.
Spike stem-helix bnAbs recognize a common hydrophobic core epitope
To determine the epitope specificities of the isolated stem-helix bnAbs and potential association with antibody immunogenetic properties, we performed binding of all 32 stem bnAbs to alanine scanning mutants of the SARS-CoV-2 stem peptide (Figure 4A; Table S4). Many bnAbs exhibited a dependence on three hydrophobic residues, F1148, L1152, and F1156, that identified a common core epitope, but the relative dependence of bnAb lineages on each of the hydrophobic core residues varied. Many of the IGHV1-46-encoded bnAbs were paired with IGKV3-20 or IGLV1-51 light chains and all, except two bnAbs, possessed a CDRL3 of 11 residues. The IGHV3-23-encoded bnAbs showed dependence on one or two hydrophobic core epitope residues, and some lineages showed dependence on an upstream acidic residue, D1146. All of the IGHV3-23-encoded bnAbs were paired with an IGKV3-20 light chain with a 9-residue CDRL3 loop. The non-IGHV1-46 or -IGHV3-23-encoded stem-helix bnAbs were also dependent on one or more hydrophobic core epitope residues, with one exception. The negatively charged residues E1151 and D1153 also contributed to the binding of certain bnAbs.
Figure 4.
Fine epitope specificities of S2 β-CoV spike stem-helix bnAbs
(A) ELISA-based epitope mapping of S2 stem-helix bnAbs with alanine scan peptides (25-mer) from the SARS-CoV-2 stem helix. Heatmap shows fold-changes in EC50 binding titers of mAb binding to SARS-CoV-2 stem-helix peptide alanine mutants, compared with the WT peptide. SARS-CoV-2 stem-helix residue positions targeted (2-fold or higher decrease in EC50 binding titer compared with WT stem peptide) are indicated in different colors. Three hydrophobic residues, F1148, L1152, and F1156, were commonly targeted by stem-helix bnAbs and form the core of the bnAb epitope. Association of heavy-chain (IGHV1-46 and IGHV3-23) and light-chain (IGKV3-20 and IGLV1-51) gene usage and CDRL3 length are shown for the mAbs.
(B) Heatmap summary of BLI competition epitope binning of S2 stem-helix bnAbs with human S2 bnAbs of known epitope specificities (CC25.106, CC95.108, CC68.109, CC99.103, S2P6, CC48.8, and CV3-25). The BLI competition was performed with SARS-CoV-2 S protein, and the competition levels are indicated as bright red (very strong), red (strong), orange (moderate), light blue (weak), and gray (very weak).
See also Table S4.
To further map the epitope specificities of the S2 stem-helix bnAbs, we performed BLI epitope binning experiments using stem-helix bnAbs whose molecular epitopes were determined previously (S2P6, CC40.8, and CV3-25)53,54,57 or in this study (CC25.106, CC68.109, CC95.108, and CC99.103; see structural studies below). Except for CV3-25 bnAb, all of the S2 stem-helix bnAbs competed strongly with these bnAbs of known epitope specificity, suggesting that they recognize similar S2 stem-helix epitopes (Figure 4B). Overall, all of the S2 stem-helix bnAbs targeted a similar region containing the hydrophobic core residues in the spike fusion machinery, all of which are highly conserved across betacoronaviruses. The hydrophobic core stem epitope residues on the prefusion S trimer are poorly accessible, and partial disruption of the stem-helix region may be needed to favorably expose this site to bnAb recognition.52,53,54,56
Mechanism of neutralization by S2 stem-helix bnAbs
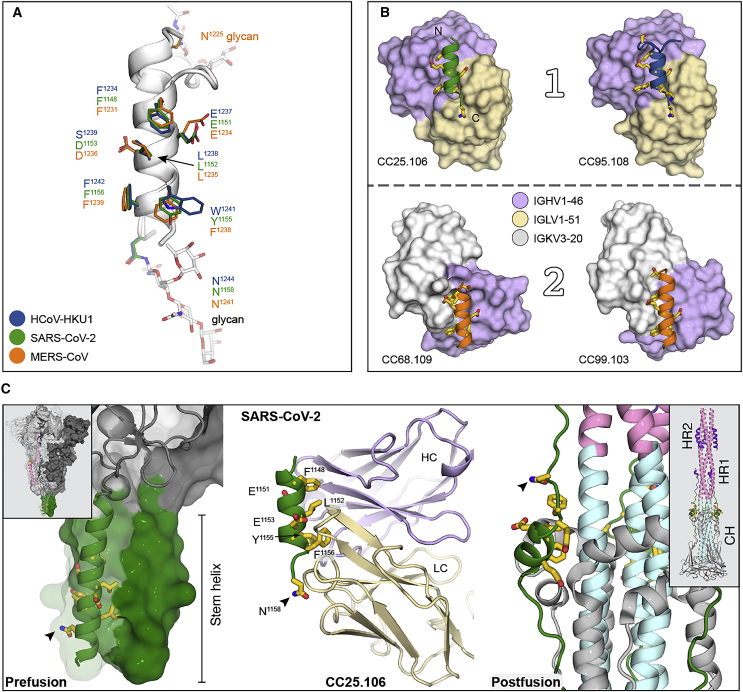
Our data strongly suggested public antibody responses to the betacoronavirus S2 stem-helix region. To understand the structural basis of how these antibodies recognize betacoronaviruses (Figure 1C), we determined crystal structures of four bnAbs (CC25.106, CC95.108, CC68.109, and CC99.103) in complex with betacoronavirus spike stem-helix peptides at resolutions ranging from 1.9 to 2.9 Å (Table S5). All antibodies interacted with a similar epitope site (Figure 5A) that included hydrophobic and aromatic core residues, i.e., F1148, L1152, Y1155, and F1156, on one face of the helix, as well as an acidic residue, E1151, of SARS-CoV-2 and the equivalent positions in MERS-CoV and HCoV-HKU1. These epitope residues are highly conserved across betacoronaviruses (Figure S4A; Table S2), which explained the observed specificities (Figure 1C). This epitope site is located at the interface within a coiled-coil helix bundle at the base of the prefusion SARS-CoV-2 spike and is connected to heptad repeat region 2 (HR2) (Figure 5C). Both helix and heptad repeat undergo dramatic conformational changes between prefusion and postfusion states.77,78 Antibody binding to this stem-helix epitope site may block this transition and hence membrane fusion.53,54,57 Furthermore, all human betacoronaviruses have a glycosylation site at N1158 in SARS-CoV-2 (N1140 in SARS-CoV-1, N1244 in HCoV-HKU1, and N1241 in MERS-CoV) at one end of the epitope, and MERS-CoV has an N-linked glycan at N1225 at the other end (Figures 5A and S4A).79,80 Thus, coronaviruses, including alphacoronaviruses, may use glycans proximal to the stem helix to restrict antibody recognition to this region.
Figure 5.
IGHV1-46 public antibodies target highly conserved residues in betacoronaviruses
(A) Epitope residues in SARS-CoV-2, HCoV-HKU1, and MERS-CoV. Stem helices of these viruses are shown in ribbon representation. Epitope residues involved in interaction with public antibodies are shown as sticks with amino acid positions labeled. Glycan molecules (sticks, white) were modeled (based on structure in PDB: 7LM8) to show potential spatial restrictions at this epitope site. Green, SARS-CoV-2; navy blue, HCoV-HKU1; orange, MERS-CoV.
(B) IGHV1-46 antibodies bind the stem helix in two distinct binding modes, namely, mode 1 and mode 2. The stem helices of betacoronavirus spikes are shown in ribbon mode and aligned in the same orientation. Epitope residues are shown in yellow sticks and antibodies in surface representation colored by their heavy and light chains; lavender, IGHV1-46; beige, IGLV1-51; light gray, IGKV3-20. The antibody approach angle is almost 180° rotated between IGHV1-46 + IGKV3-20 and IGHV1-46 + IGLV1-51 antibodies.
(C) Epitope residues of CC25.106 are involved in the spike fusion activity. Epitope residues are shown as yellow sticks. The epitope location is shown in the prefusion and postfusion structures. Key epitope residues are buried in the stem-helix bundle (green) in prefusion spike (left, PDB: 6XR8) or buried in interaction with the coiled-coil central helices (cyan) in the postfusion spike (right, PDB: 6XRA). Insets in the left and right corners show the overall structure of prefusion and postfusion spike trimer. Ribbon model in the middle shows CC25.106 in complex with SARS-CoV-2 stem helix (green) with antibody in lavender, heavy chain, and beige, light chain. Arrowhead indicates a glycosylation site. CH (cyan), central helix; HR1 (pink), heptad repeat 1; HR2 (magenta), heptad repeat 2.
Two binding modes of IGHV1-46-encoded S2 stem-helix bnAbs
Comparison of the four structures showed that the two public clonotypes recognize the stem helix with two distinct binding modes, even though IGHV1-46 is in common (Figure 5B). The two modes were differentiated by the light-chain residues encoded by different germline genes (Figure 5B). For IGHV1-46 + IGLV1-51 antibodies that adopt binding mode 1, heavy-chain germline residues VH Y33 and I50 interact with hydrophobic epitope residues, i.e., F1148, L1152, and F1156, in SARS-CoV-2 or their equivalent positions in HCoV-HKU1 (Figures 6A, S5A, and S6A). VH N56 forms a π-π interaction with F1148. Residues E1151 and Y1155 hydrogen bond with VH Y33, K52, G95, and H97. Most IGHV1-46+IGLV1-51 antibodies, e.g., CC25.106 and CC95.108, contain VH N52K and S56N somatic mutations (Figure S6D), suggesting convergent mutations that could facilitate interaction with the stem helix. A R/H residue (R98/H97) in CDRH3 of CC25.106 and CC95.108 antibodies interacts with the aromatic ring and backbone carbonyl of Y1155 of SARS-CoV-2 and W1241 of HCoV-HKU1 (Figures 6A, S5A, and S5B). A type II β-turn in CDRH3 of CC25.106 positions H97 at the tip of the turn for interaction with Y1155 of SARS-CoV-2 as well as VL Y32, encoded by IGLV1-51. VL Y32 also interacts with N1158 of SARS-CoV-2, a glycosylation site in the spike protein.79 The R/H residue is therefore structurally constrained in a type II β-turn with a positive phi value, normally accessible only to glycine.
Figure 6.
Antibody germline-encoded residues interact with the stem helix
Key epitope residues and their interacting paratope residues are shown in sticks. Dashed lines represent polar interactions. Antibodies are shown in ribbon representation and stem helices in backbone tubes with side chains as sticks. SARS-CoV-2 stem helix is shown in green and MERS-CoV in orange. ∗indicates somatically hypermutated residue.
(A) CC25.106 interacts with SARS-CoV-2 stem helix. Lavender, heavy chain; beige, light chain. VH Y33, I50, N56, and K52 interact with F1148, E1151, L1152, and Y1155 of the stem helix. IGHV1-46 germline-encoded residues are involved in key interactions with the stem helix.
(B) CC25.106 light chain interacts with SARS-CoV-2 stem helix. VL N51, K66, W91, and Y32 form key interactions with L1152, D1153, F1156, and N1158 of the stem helix. All of these residues are encoded by the IGLV1-51 germline gene.
(C) CC99.103 heavy chain interacts with the MERS-CoV stem helix. The epitope sites are similar between CC25.106 and CC99.103, but their heavy chains bind in opposite directions with respect to the hydrophobic core.
(D) CC99.103 light chain interacts with MERS-CoV stem helix. VL Y32, Y91, S93, and F96 interact with F1231, E1234, and L1235 of MERS-CoV. VL P95 and P95a at the tip of CDRL3 β-turn interact with F1238. The shortest distance between VL P95a and F1238 is 3.5 Å. All of these paratope residues interacting with the stem helix are encoded by IGKV3-20 except for VL F96, which is encoded by IGKJ3 germline.
See also Figures S4B, S4C, S5, and S6 and Table S5.
For IGHV1-46 + IGKV3-20 antibodies CC99.103 and CC68.109 in binding mode 2, the heavy chain interacts with the same epitope site but with an opposite approach angle, compared with IGHV1-46 + IGLV1-51 antibodies (Figure 5B). The side chains of VH Y33, I50, and backbones of VH T57, G95, and I96 from the heavy chain interact with Q1232, L1235, D1236, and F1239 of MERS-CoV stem helix (equivalent to K1149, L1152, D1153, and F1156 of SARS-CoV-2; Figures 5A and S4A). The same heavy chain in the two clonotypes uses VH Y33 and I50 to form a hydrophobic patch that interacts with the hydrophobic core region of the stem helices in four betacoronaviruses, namely, SARS-CoV-2/1, MERS-CoV, and HCoV-HKU1. However, the heavy chain of IGHV1-46 + IGLV1-51 antibodies forms more hydrogen bonds with the stem helix, compared with IGHV1-46 + IGKV3-20 antibodies. In both orientations, most of the key heavy-chain paratope residues are encoded by the IGHV1-46 germline sequence (Figures 6, S5, and S6). In particular, the IGHV1-46 germline-encoded residues, Y33 and I50, form a hydrophobic core interacting with the core S2 stem epitope residues in both binding modes, as revealed by all four S2 bnAbs that were structurally characterized in this study (Figures 6A, 6C, and S6A). We also noted that the CDRH3 junctional residues in S2 bnAbs contribute to epitope recognition (Figure S6A), suggesting that vaccine design strategies may need to take this junctional region into consideration.
The light chains of these IGHV1-46 antibodies were also involved in key interactions with the stem helices. For IGLV1-51 in binding mode 1, VL W91 of CC25.106 and CC95.108 interacts with F1156 and E1153 of SARS-CoV-2 and F1242 of HCoV-HKU1 (Figures 6B and S5C). VL N51 and K66 of CC25.106 and CC95.108 hydrogen bond to N1158 of SARS-CoV-2 or N1244 of HCoV-HKU1. The VL N30 (CC25.106) or VL T30 (CC95.108) main-chain carbonyl oxygen hydrogen bonds to the side-chain amide of N1158 of SARS-CoV-2 or N1244 of HCoV-HKU1 (Figures 6B and S5C). VL Y32 of CC25.106 interacts with N1158 of SARS-CoV-2 and VL F32 of CC95.108 with N1244 of HCoV-HKU1 (Figures 6B and S5C). All of these key paratope residues are encoded by the IGLV1-51 germline sequence. For IGKV3-20 in binding mode 2 (Figure 6D), VL Y32, Y91, and F96 of CC99.103 form a hydrophobic patch for interaction with F1231 and L1235 of MERS-CoV (F1148 and L1152 of SARS-CoV-2, see Figure 5A). F1238 of MERS-CoV (Y1155 of SARS-CoV-2, Y1137 of SARS-CoV-1, and W1241 of HCoV-HKU1) makes hydrophobic interactions with VL P95 and P95a. These paratope residues are also -encoded by IGKV3-20 and IGKJ3/2 germline sequences (Figure S6C).
To address how the glycan at N1158 or its equivalent site in other betacoronaviruses could be accommodated (Figure 6B), we observed that the nitrogen atom on the asparagine amide, where the N-linked glycan is attached, is exposed on the antibody binding surface. In binding mode 1, two hydrogen bonds are formed between VL N51 and K66 and the amide group of SARS-CoV-2 N1158 or its equivalent residue in MERS-CoV and HCoV-HKU1. For binding mode 2, the glycosylation site is over 15 Å from the antibody (Figure 6C). We next tested how the N1158 glycan impacts antibody neutralization. Either N1158A or T1160A mutations (i.e., no glycan at 1,158) showed about 4-fold enhanced antibody neutralization against SARS-CoV-2 pseudovirus variants (Figures S4B and S4C), suggesting that glycan interaction at N1158 had limited impact on the IGHV1-46 public antibodies tested here.
Light-chain-encoded PPxF motif interacts with coronavirus stem helix
The presence of VL P95 and P95a in a β-turn in CDRL3 of CC99.103 and CC68.109 appeared critical for binding to the stem helix. The PP turn left just enough space to accommodate F1238 in MERS-CoV (Y1155 in SARS-CoV-2) stem helix (Figure 6D). We noted that the same PP turn is also present in the S2P6 structure54 in complex with the SARS-CoV-2 stem helix (Figure S5D). Further sequence analysis showed this PP dipeptide is present in all of the IGHV1-46 + IGKV3-20, IGHV1-46 + IGKV1-12, and IGHV1-46 + IGKV1-39 antibodies identified here (Figures S6D–S6F). A bulky hydrophobic residue often follows the PP dipeptide in the fourth position (e.g., PPxF) (Figures 3E and S6F). The PP dipeptide is encoded by the last five nucleotides of the germline sequence of IGKV3-20/IGKV1-12/IGKV1-39 in combination with an N or P nucleotide acquired from VJ recombination (Figure S6E). The F (Y and L in some antibodies in our cohort) is encoded by IGKJ3 (or IGKJ2 for Y) and contributes to a hydrophobic patch along with VL Y32 and Y91; the F is VL F96 in CC99.103 and CC68.109 (Figure 6D). Nevertheless, we did not observe any PPxF motif in CDRL3 of five IGHV1-46 + IGLV1-51 antibodies that we isolated here. The IGHV1-46 + IGLV1-51 antibodies use VL W91 in CDRL3 to interact with L1152, F1156, and D1153 of SARS-CoV-2 stem helix rather than the PPxF motif in IGHV1-46 + IGKV3-20 antibodies (Figure 6B). Hence, the presence of PPxF motif in CDRL3 of these S2 stem binders represents a unique feature for binding mode 2. We surveyed a large human naive database of 1.6 million antibody heavy-light chain pairs and observed ∼270,000 light chains with CDRL3s of 11 amino acids in length.74 Of the 1.6 million heavy-light chain antibody pairs, the 11 AA CDRL3 antibodies that were paired with IGHV1-46-encoded heavy chains, ∼0.005 % possessed a PPxF motif in their CDRL3. In comparison, of the 32 unique S2 bnAbs isolated in our study, 15 (47%) bore a PPxF motif in their CDRL3, suggesting a strong preference for this motif in the S2 stem-helix bnAbs antibodies that could be exploited by targeted vaccines.
In summary, IGHV1-46 + IGLV1-51 antibodies, i.e., CC25.106 and CC95.108, bound SARS-CoV-2 and HCoV-HKU1 stem helices in binding mode 1 (Figures 5B, 6A, 6B, S5B, S5C, and S7). IGHV1-46+IGKV3-20 antibodies, i.e., CC99.103, CC68.109, and S2P6, bound SARS-CoV-2 and MERS-CoV stem helices in binding mode 2 (Figures 5B, 6C, 6D, S5D, and S7). These different binding modes can be accommodated by the highly conserved and quasi-symmetric epitope in the betacoronavirus stem helices (Figures 5A and S4A). Germline-encoded antibody residues, including the PPxF motif in binding mode 2, play a key role in recognizing the stem-helix epitope, providing an explanation as to why IGHV1-46 public clonotypes are naturally favored in targeting the betacoronavirus stem helix (Figures 6 and S5).
Overall, our structural analysis revealed a substantial contribution to S2 stem epitope recognition from both heavy- and light-chain germline-encoded residues, which supported the enrichment of certain antibody germline gene features (Figure 3). These enriched germline features could be exploited by a targeted vaccine approach64,65,75 to induce pan-betacoronavirus bnAbs.
Stem-helix bnAbs protect against challenge with diverse β-CoVs
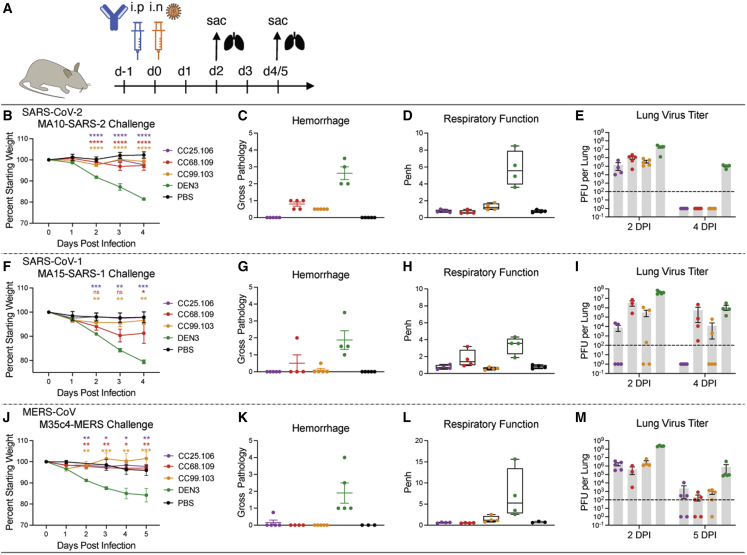
To determine the protective efficacy of the stem-helix bnAbs, we prophylactically treated aged mice81 with individual antibodies followed by virus challenge. We selected three of the broadest and potent stem-helix bnAbs—CC25.106, CC68.109, and CC99.103—and investigated their in vivo protective efficacy against all three deadly human betacoronaviruses—SARS-CoV-2, SARS-CoV-1, and MERS-CoV. Prior to the challenge experiments, we examined neutralization of SARS-CoV-2 and MERS-CoV replication-competent viruses by the three candidate bnAbs and compared with that of pseudoviruses (Table S6). The neutralization IC50s of the stem-helix bnAbs were comparable (less than 10-fold different) for SARS-CoV-2 across the two assay formats, whereas the titers with replication-competent MERS-CoV were higher (lower IC50 values), compared with the pseudovirus format. The three stem-helix bnAbs, individually, or a DEN3 control antibody, were administered intraperitoneally (i.p.) at 300 μg/animal (∼10 mg/kg) into 12 groups of 10 animals (3 groups per antibody; Figure 7A). Also, 12 h prior to the virus challenge, the test antibody in each animal group was administered followed by intranasal (i.n.) challenge with one of three mouse-adapted (MA) betacoronaviruses, MA10-SARS-2 (=SARS-CoV-2), MA15-SARS-1 (=SARS-CoV-1), or M35c4-MERS (=MERS-CoV) (Figure 7A).81,82,83 Post virus challenge, the animals were monitored for signs of clinical disease due to infection, including daily weight changes and pulmonary function. Animals were euthanized at day 2 or day 4 (or day 5 for MERS-CoV challenge group) post infection, and lung tissues were harvested to assess gross pathology and to determine virus titers by plaque assay. Compared with the control antibody DEN3-treated animal groups, the stem-helix bnAb-treated animals in all three betacoronaviruses challenge experiments showed substantially lower weight loss (9.6, 4.6, and 13.1 times less, respectively, for SARS-CoV-2, SARS-CoV-1, and MERS-CoV groups at day 4 or 5) (Figures 7B, 7F, and 7J), reduced hemorrhage (Figures 7C, 7G, and 7K), and normal pulmonary function (6.1, 11.3, and 35.5 times less, respectively, for SARS-CoV-2, SARS-CoV-1, and MERS-CoV groups) (Figures 7D, 7H, and 7L), suggesting a protective role for the bnAbs. We also examined virus load in the lungs harvested at day 2 and day 4/5 post infection, and consistent with the protective role of the bnAbs, the viral titers in stem-helix bnAb-treated animals were substantially reduced compared with the DEN3-treated control group animals (Figures 7E, 7I, and 7M).
Figure 7.
Prophylactic treatment of aged mice with S2 stem-helix bnAbs protected against challenge with diverse betacoronaviruses
(A) Three S2 stem-helix bnAbs (CC25.106, CC68.109, and CC99.103), individually, or a DEN3 control antibody were administered intraperitoneally (i.p.) at 300 μg per animal (∼10 mg/kg) into 12 groups of aged mice (10 animals per group). Each group of animals was challenged intranasally (i.n.) 12 h after antibody infusion with one of 3 mouse-adapted (MA) betacoronaviruses (MA10-SARS-2 = SARS-CoV-2, 1 × 103 plaque-forming units [PFUs] per mouse; MA15-SARS1 = SARS-CoV-1, 1 × 103 PFU per mouse; M35c4-MERS = MERS-CoV, 1 × 105 PFU per mouse). As a control, groups of mice were exposed to PBS in the absence of virus.
(B, F, and J) Percent weight change in S2 stem-helix bnAbs or DEN3 control antibody-treated animals after challenge with mouse-adapted betacoronaviruses. Percent weight change was calculated from day 0 starting weight for all animals. Data are presented as mean values ± SEM. Statistical significance was calculated with Dunnett’s multiple comparisons test between each experimental group and the DEN3 control Ab group. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant (p > 0.05). A one-way ANOVA was used.
(C, G, and K) Day 2 post-infection hemorrhage (Gross Pathology score) scored at tissue harvest in mice prophylactically treated with S2 stem-helix bnAbs or DEN3 control mAb (n = 5 individuals for each group). Data are presented as mean values ± SEM.
(D, H, and L) Day 2 post-infection pulmonary function (shown as Penh score) was measured by whole-body plethysmography in mice prophylactically treated with S2 stem-helix bnAbs or DEN3 control mAb (n = 5 individuals for each group). Data were shown as box-and-whisker plots showing data points from minimum to maximum.
(E, I, and M) Lung virus titers (PFU per lung) were determined by plaque assay of lung tissues collected at days 2 or 4/5 after infection (n = 5 individuals per time point for each group). Data are shown as dot plots with bar heights representing the mean.
See also Table S6.
Overall, all three stem-helix bnAbs protected against severe betacoronavirus disease, with CC25.106 providing more superior protection than the other two bnAbs.
Discussion
In terms of passive antibody treatment, the ability of single mAbs to protect against the two SARS viruses and MERS in the small animal model is encouraging for their potential reagents to stockpile to tackle future outbreaks of viral infection, including novel related betacoronaviruses. Prophylaxis or treatment very early in infection is more likely to be successful than therapy once symptoms are established, based on experience with SARS-CoV-2.84,85 The use of S2 bnAbs, possibly in a cocktail with the most appropriate RBD bnAbs, may be the optimal approach for SARS-CoV-2 prophylaxis, especially as new variants such as Omicron emerge. The dose of S2 bnAbs required to be effective, given the typically lower neutralization potencies of such nAbs compared with RBD nAbs, may be an issue for translation. However, studies in animal models53,54 suggest that S2 bnAbs may protect at lower serum concentrations than would be predicted by their IC50 neutralizing titers—i.e., they may “punch above their weight,” compared with the highly potent RBD site targeting nAbs.53 Therefore, in addition to neutralization, effector functions of the S2 bnAbs may be important for protection,54,86,87,88 and clinical studies will be required to investigate this phenomenon in humans.
We noted that SARS-CoV-2 spike vaccination in COVID-19-recovered donors was effective at generating anti-S2 stem-helix-directed antibody responses, whereas infection or vaccination alone was relatively inefficient and may have implications for designing vaccine strategies. Two hypotheses were readily suggested to explain the paucity of stem-helix bnAb responses in infection or vaccination alone, compared with infection followed by vaccination. First, there could be conformational differences between spike expressed by mRNA vaccines and the spike associated with infectious virions that might influence the exposure of the stem-helix bnAb site and the resulting B cell responses. Our data suggest that the presentation of spike on the virion surface might more favorably prime S2 stem-helix bnAb B cell precursors than the stabilized spike displayed by the mRNA vaccines. Second, T cell responses in infection might be superior and that could help B cell recall responses upon mRNA vaccination.89,90,91 Studies of mixed vaccines that begin priming with inactivated SARS-CoV-2 virus or vector-based vaccines (where the T cell responses are relatively superior) followed by spike mRNA boost vaccination could provide important insights in this context. Overall, hybrid immunity acquired from individuals that are both naturally infected and vaccinated can provide important lessons for broad coronavirus vaccine strategies if the key contributing factors can be determined.
IGHV1-46-encoded betacoronavirus spike stem-helix bnAbs have at least two public clonotypes where the heavy chain can adopt two distinct approach angles depending on the light chain. These two opposing binding modes have interactions with a quasi-symmetric epitope site on the stem helix. For the IGHV1-46 + IGKV3-20 clonotype, a PPxF motif in CDRL3 is positioned next to Y1155 of the SARS-CoV-2 stem helix, F1238 of MERS-CoV, W1241 of HCoV-HKU1, and, presumably, the equivalent position (W1240) of HCoV-OC43. An 11-residue CDRL3 appears ideal for interaction with the stem helix of SARS-CoV-2 in binding mode 2, but we also observed a 9-residue CDRL3 in some antibodies also with a PPxF motif (Figure 3E). Other clonotypes such as IGHV1-46 + IGKV1-12- and IGHV1-46 + IGKV1-39-encoded antibodies also contain a similar PPxF signature in their 11-residue CDRL3, suggesting that even more public clonotypes may target the S2 stem helix.
In terms of vaccine design, a rational strategy is strongly favored by the availability of a panel of bnAbs rather than single mAbs so that the bnAb epitope can be more precisely defined and the qualities of nAbs required for broad neutralization determined.64,65,67,75,92,93 Here, we identified critical hydrophobic S2 residues involved in bnAb binding and showed the prevalence of a IGHV1-46/IGKV3-20 antibody pairing with restricted CDRH3 and CDRL3 lengths in S2 bnAbs. The structural studies revealed substantial contributions of antibody heavy- and light-chain germline-encoded residues for S2 stem-helix epitope recognition. Accordingly, rational vaccine design strategies could take advantage of these germline gene features for vaccine targeting to this site.63,64,65,67,75 Nothwithstanding, accessibility of the S2 stem-helix bnAb site on spike immunogen to effectively engage desired B cell responses might be challenging. Nevertheless, approaches to scaffold immunogen designs that can accommodate such features can now be deployed.55,63,67,94,95,96,97,98 An immunogen that takes advantage of the antibody germline features that we defined in our study may be favored for priming S2 stem bnAb site-specific B cells to generate a memory response. Sequential boost immunogens that further train these memory B cells to recognize the partially occluded S2 bnAb site on native-like spikes in the context of different betacoronaviruses may be required to drive affinity maturation of protective B cell responses toward breadth.
In summary, we isolated the largest panel of β-CoV stem-helix bnAbs to date and revealed the molecular basis for their broad protection. These bnAbs provide a detailed blueprint for rational design of broad coronavirus vaccines and could by themselves be used as reagents to counter betacoronavirus spillovers.
Limitations of the study
Our study has the following limitations. First, we showed that stem-helix bnAbs protected against diverse coronaviruses in a small animal model. Demonstrating protective efficacy in a model that more closely resembles humans, such as non-human primates, would be desirable. Second, despite the relatively low neutralization potency of stem-helix bnAbs, they exhibited apparently robust in vivo protection in our model. This phenomenon needs to be qualitatively evaluated to be firmly established. Third, we noted in our study cohort that S2 stem-helix bnAbs were preferentially elicited in COVID-19 recovered-vaccinated individuals. The reasons for this finding are not clear, but understanding its immunological basis is important for developing vaccine strategies that can efficiently induce S2 stem-helix-directed bnAb responses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-His antibody | Thermo Fisher Scientific | Cat# MA1-21315-1MG; RRID:AB_2536982 |
| Anti-CD3 APC Cy7 | BD Biosciences | Cat# 557757; RRID:AB_396863 |
| Anti-CD4 APC-Cy7 | BioLegend | Cat# 317418; RRID:AB_571947 |
| Anti-CD8 APC-Cy7 | BD Biosciences | Cat# 557760; RRID:AB_396865 |
| Anti-CD14 APC-H7 | BD Biosciences | Cat# 561384; RRID:AB_10611720 |
| Anti-IgM PE | BioLegend | Cat# 314508; RRID:AB_493005 |
| Anti-CD19 PerCP-Cy5.5 | BioLegend | Cat# 302230; RRID:AB_2073119 |
| Anti-CD20 PerCP-Cy5.5 | BioLegend | Cat# 302326; RRID:AB_893283 |
| Anti-IgG BV605 | BD Biosciences | Cat# 563246: RRID:AB_2738092 |
| Streptavidin-BV421 | BD Biosciences | Cat# 563259; RRID:AB_2869475 |
| Streptavidin-AF488 | Thermo Fisher Scientific | Cat# S11223 |
| Streptavidin-AF647 | Thermo Fisher Scientific | Cat# S21374; RRID:AB_2336066 |
| AP-conjugated goat anti-human IgG Fc secondary antibody | Jackson Immuno Research Labs | Cat# 109-055-008; RRID:AB_2337601 |
| PE-conjugated mouse anti-human IgG Fc antibody | SouthernBiotech | Cat# 9040-09; RRID:AB_2796601 |
| Biological samples | ||
| Sera and PBMC samples from convalescent COVID-19 donors, vaccinated donors, and COVID-19 recovered-vaccinated donors | UCSD Human Research Protection Program | UCSD IRB#200236 |
| Chemicals, peptides, and recombinant proteins | ||
| HiFi DNA assembly | New England Biolabs | Cat# E2621L |
| Lipofectamine 2000 | ThermoFisher Scientific | Cat# 11668019 |
| Biotinylated SARS-CoV2 Spike | This paper | N/A |
| Biotinylated MERS-CoV Spike | This paper | N/A |
| SARS-CoV-1 spike | Song et al.52 | N/A |
| SARS-CoV-2 spike | Song et al.52 | N/A |
| HCoV-HKU1 spike | Song et al.52 | N/A |
| HCoV-OC43 spike | Song et al.52 | N/A |
| MERS-CoV spike | Song et al.52 | N/A |
| HCoV-229E spike | Song et al.52 | N/A |
| HCoV-NL63 spike | Song et al.52 | N/A |
| Live/Dead stain | Thermo Fisher Scientific | Cat# L34966 |
| Superscript IV Reverse Transcriptase | Invitrogen | Cat# 18090010 |
| 10mM dNTPs | Invitrogen | Cat# 18427088 |
| Random hexamers | Gene Link | Cat# 26-4000-03 |
| RNAseOUT | Invitrogen | Cat# 10777019 |
| 10% Igepal | Sigma-Aldrich | Cat# 18896 |
| Hot Start DNA Polymerases | Qiagen | Cat# 203643 |
| FectoPRO | Polyplus | Cat# 116-001 |
| Streptavidin | Jackson Immuno Research Labs | Cat# 016-000-084 |
| Phosphatase substrate | Sigma-Aldrich | Cat# S0942-200TAB |
| 25-mer stem helix peptides | A&A Labs | Synthetic Biomolecules |
| Nano-Glo Luciferase Assay System | Promega | Cat# N1130 |
| Bright-Glo Luciferase Assay System | Promega | Cat# E2620 |
| Human insulin | Sigma-Aldrich | Cat# I2643 |
| Single strand DNA | Sigma-Aldrich | Cat# D8899 |
| Solubilized CHO cell membrane protein | This paper | N/A |
| Zombie-NIR viability dye | BioLegend | Cat# 423105 |
| Papain | Sigma-Aldrich | Cat# P3125 |
| Critical commercial assays | ||
| HEp2 slides | Hemagen | Cat# 902360 |
| Deposited data | ||
| Structure of CC25.106 Fab+SARS-CoV-2 stem helix peptide complex | RCSB PDB | PDB: 8DGU |
| Structure of CC68.109 Fab+MERS-CoV stem helix peptide complex | RCSB PDB | PDB: 8DGX |
| Structure of CC95.108 Fab+HCoV-HKU1 stem helix peptide complex | RCSB PDB | PDB: 8DGW |
| Structure of CC99.103 Fab+ MERS-CoV stem helix peptide complex | RCSB PDB | PDB: 8DGV |
| Sequences of 40 stem helix antibodies reported in this paper | Genbank | GenBank: OP699209-OP699288 |
| Experimental models: Cell lines | ||
| FreeStyle293-F cells | Thermo Fisher Scientific | #R79007 |
| Vero E6 cells | ATCC | Cat# CRL-1586; RRID:CVCL_0574 |
| Expi293F cells | Gibco | Cat# A14527 |
| HeLa-ACE2 | This paper | N/A |
| HeLa-DPP4 | This paper | N/A |
| Experimental models: Organisms/strains | ||
| 12-month-old female Balb/c mice | Envigo | stock number 047 |
| C57Bl/6 288/330+/+ mice | Ralph S. Baric Lab | Cockrell et al.82 |
| Recombinant DNA | ||
| pMDL | Addgene | Cat# 12251; RRID:Addgene_12251 |
| pREV | Addgene | Cat# 12253; RRID:Addgene_12253 |
| pVSV-G | Addgene | Cat# 8454; RRID:Addgene_8454 |
| pCMV-dR8.2 dvpr | Addgene | Cat# 8455; RRID:Addgene_8455 |
| pBOB-Luciferase | Addgene | Cat# 170674; RRID:Addgene_170674 |
| phCMV3 | Genlantis | Cat# P003300 |
| pBOB-hACE2 | This paper | N/A |
| pBOB-Hdpp4 | This paper | N/A |
| Software and algorithms | ||
| V-Quest online tool | IMGT | http://www.imgt.org; RRID:SCR_012780 |
| Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| PyMOL V2.4.2 | PyMOL by Schrödinger | https://pymol.org; RRID:SCR_000305 |
| ForteBio Data Analysis software | Sartorius | https://www.sartorius.com/en |
| FlowJo 10 | BD | https://www.flowjo.com/; RRID:SCR_008520 |
| Other | ||
| Expi293 Expression Medium | Gibco | Cat# A1435101 |
| FreeStyl 293 Expression Medium | Gibco | Cat# 12338018 |
| Opti-MEM™ | Thermo Fisher Scientific | Cat# 31985070 |
| HisPur Ni-NTA Resin | Thermo Fisher Scientific | Cat# 88221 |
| 100K Amicon tubes | Millipore | Cat# UFC910024 |
| Superdex 200 Increase10/300 GL column | GE Healthcare | Cat# GE28-9909-44 |
| RPMI1640 medium | Thermo Fisher Scientific | Cat# 11875085 |
| FACS tube with 70-μm mesh cap | Fisher Scientific | Cat# 08-771-23 |
| SPRI beads | Beckman Coulter | Cat# B23318 |
| Protein A Sepharose | GE Healthcare | Cat# 17096302 |
| Protein G Sepharose | GE Healthcare | Cat# 17061805 |
| Econo-Pac columns | BioRad | Cat# 7321010 |
| 30K Amicon tubes | Millipore | Cat# UFC903024 |
| 96-well half-area high binding plates | Corning | Cat# 3690 |
| 96-well plates | Corning | Cat# 3916 |
| Anti-human IgG Fc capture (AHC) biosensors | ForteBio | Cat# 18-5063 |
| Streptavidin biosensors | ForteBio | Cat# 18-5020 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Raiees Andrabi (andrabi@scripps.edu)
Materials availability
Upon specific request and execution of a material transfer agreement (MTA) from The Scripps Research Institute to the lead contact, antibody plasmids will be made available.
Experimental model and subject details
COVID-19 donors
Sera and PBMC samples from convalescent COVID-19 donors, vaccinated donors, and COVID-19 recovered-vaccinated donors, were provided through the “Collection of Biospecimens from Persons Under Investigation for 2019-Novel Coronavirus Infection to Understand Viral Shedding and Immune Response Study” UCSD IRB# 200236 as reported earlier.46 The protocol was approved by the UCSD Human Research Protection Program. Convalescent donor samples were collected based on COVID-19 diagnosis regardless of gender, race, ethnicity, disease severity, or other medical conditions. All human donors were assessed for medical decision-making capacity using a standardized, approved assessment, and voluntarily gave informed consent prior to being enrolled in the study.
Method details
Plasmid construction
To generate soluble S ectodomain proteins from SARS-CoV-1 (residues 1-1190; GenBank: AAP13567),SARS-CoV-2 (residues 1-1208; GenBank: MN908947), HCoV-HKU1 (residue 1-1295; GenBank: YP_173238.1), HCoV-OC43 (residues 1-1300; GenBank: AAX84792.1), MERS-CoV (residues 1-1291; GenBank: APB87319.1), HCoV-229E (residues 1-1110; GenBank: NP_073551.1) and HCoV-NL63 (residues 1-1291; GenBank: YP_003767.1), we synthesized the DNA fragments from GeneArt (Life Technologies) and cloned them into the phCMV3 vector (Genlantis Cat# P003300). In order to produce the stable trimeric prefusion spike proteins, double proline substitutions (2P) were introduced into the S2 subunit: K968/V969 in SARS-CoV-1, K986/V987 in SARS-CoV-2, V1060/L1061 in MERS-CoV, A1071/L1072 in HCoV-HKU1, A1078/L1079 in HCoV-OC43, S1052/I1053 in HCoV-NL63 and T871/I872 in HCoV-229E were replaced by proline. The furin cleavage sites (in SARS-CoV-2 residues 682–685, in SARS-CoV-1 residues 664–667, in HCoV-HKU1 residues 756-760, in HCoV-OC43 residues 762–766, in MERS-CoV residues 748–751, in HCoV-229E residues 564–567 and in HCoV-NL63 residues 745–748) were replaced by a “GSAS” linker; the trimerization T4 fibritin motif was incorporated at the C-terminus of the S proteins. To purify and biotinylate the spike proteins, the HRV-3C protease cleavage site, 6x HisTag, and AviTag spaced by GS-linkers were added to the C-terminus after the trimerization motif. To generate pseudoviruses of MERS-CoV and sarbecoviruses, the DNA fragments encoding the spikes of MERS-CoV and sarbecoviruses without the ER retrieval signal were codon-optimized and synthesized at GeneArt (Life Technologies). The spike encoding genes of Pang17 (residues 1-1249, GenBank: QIA48632.1), WIV1 (residues 1-1238, GenBank: KF367457) and SHC014 (residue 1-1238, GenBank: AGZ48806.1) were constructed into the phCMV3 vector (Genlantis Cat# P003300) using the HiFi DNA assembly (New England Biolabs Cat# E2621L) according to the manufacturer’s instructions.
Cell lines
FreeStyle293-F cells (Thermo Fisher Scientific Cat# R79007) were grown in FreeStyl 293 Expression Medium (Gibco Cat# 12338018), and Expi293F cells (Gibco Cat# A14527) were maintained in Expi293 Expression Medium (Gibco Cat# A1435101). Suspension cells were incubated in the shaker at 150 rpm, 37°C, 8% CO2. Adherent HEK293T cells and HeLa-ACE2 cells were grown in Dulbecco's Modified Eagle Medium (DMEM) with 10% heat-inactivated FBS, 4mM L-Glutamine and 1% P/S, maintaining in the incubator at 37°C, 5% CO2. The stable hACE2 or hDPP4-expressing HeLa cell line was generated using an ACE2 lentivirus protocol previously described.7 Briefly, the pBOB-hACE2 or hDPP4 plasmid and lentiviral packaging plasmids (pMDL, pREV, and pVSV-G (Addgene Cat# 12251, Cat# 12253, Cat# 8454)) were co-transfected into HEK293T cells using Lipofectamine 2000 reagent (ThermoFisher Scientific Cat# 11668019) to generate related lentivirus, then the lentivirus was transduced to HeLa cells.
Expression and purification of HCoV S-proteins
To express the soluble human coronavirus (HCoV) S ectodomain proteins with His-tag or with both His- and Avi-tag at the C-terminus, 350 μg plasmids in 15ml Opti-MEM™ (Thermo Fisher Scientific Cat# 31985070) were filtered and mixed with 1.8 ml 40K PEI (1mg/ml) in 15ml Opti-MEM™, then incubated at room temperature for 30 min and transferred into 1L FreeStyle293-F cells at a density of 1 million cells/ml. Four days after transfection, the cell cultures were centrifuged at 2500xg for 15 min and filtered through a 0.22μm membrane. The His-tagged proteins were purified with the HisPur Ni-NTA Resin (Thermo Fisher Scientific Cat# 88221). After washing by wash buffer (25 mM Imidazole, pH 7.4) for at least 3 bed volumes, the protein was eluted with 25 ml elution buffer (250 mM Imidazole, pH 7.4) at slow gravity speed (∼4 sec/drop), then was buffer exchanged into PBS and concentrated using 100K Amicon tubes (Millipore Cat# UFC910024). After being further purified by size-exclusion chromatography by Superdex 200 Increase 10/300 GL column (GE Healthcare Cat# GE28-9909-44), the protein was pooled and concentrated again for further use.
Flow cytometry B cell profiling and monoclonal antibody isolation
Flow cytometry of PBMC samples from recovered-vaccinated human donors were conducted following methods described previously.7 10ml RPMI1640 medium (Thermo Fisher Scientific Cat# 11875085) with 50% FBS was pre-warmed to 37°C and used to thaw the frozen PBMC samples, followed by centrifugation at 400xg for 5 min. After discarding supernatant, the cells were resuspended in a 5 ml FACS buffer (PBS, 2% FBS, 2 mM EDTA). Fluorescently labeled antibodies specific for cell surface markers were prepared as 1:100 dilution as a master mix in FACS buffer, to stain the PBMC samples for CD3 (APC-Cy7, BD Biosciences Cat# 557757), CD4 (APC-Cy7, BioLegend Cat# 317418), CD8 (APC-Cy7, BD Biosciences Cat# 557760), CD14 (APC-H7, BD Biosciences Cat# 561384), CD19 (PerCP-Cy5.5, BioLegend Cat# 302230), CD20 (PerCP-Cy5.5, BioLegend Cat# 302326), IgG (BV605, BD Biosciences Cat# 563246) and IgM (PE, BioLegend Cat# 314508). Meanwhile, SARS-CoV-2 S protein with Avi-tag was conjugated to streptavidin-BV421 (BD Biosciences Cat# 563259) and streptavidin-AF488 (Thermo Fisher Scientific Cat# S11223), respectively, and the MERS-CoV S protein with Avi-tag was conjugated to streptavidin-AF647 (Thermo Fisher Scientific Cat# S21374). After incubating the cells with Ab mixture for cell surface markers for 15 min in dark, S protein-probes were added to the samples and incubated on ice in the dark for 30 min. FVS510 Live/Dead stain (Thermo Fisher Scientific Cat# L34966) in FACS buffer (1:300) was then added to the samples and incubated on ice in the dark for 15 min. After washing with FACS buffer, the stained cells were resuspended in 500 μl of FACS buffer per 10-20 million cells, filtered through the 70-μm mesh cap into FACS tubes (Fisher Scientific Cat# 08-771-23) and sorted for S protein-specific memory B cells using BD FACSMelody sorter. In brief, after gating of lymphocytes (SSC-A vs. FSC-A) and singlets (SSC-W vs SSC-H and FSC-H vs. FSC-W), live cells were identified by the negative FVS510 live/dead staining phenotype. The CD3- CD4- CD8- CD14- CD19+ CD20+ cells were gated as B cells. By selecting the IgG+ IgM- population, the cells were sequentially gated for SARS-CoV-2-S-BV421+ SARS-CoV-2-S-AF488+ MERS-CoV-S-AF647+ reactivity. Triple positive memory B cells were sorted as single cells into 96-well plates on a cooling platform. Superscript IV Reverse Transcriptase (Invitrogen Cat# 18090010), 10mM dNTPs (Invitrogen Cat# 18427088), random hexamers (Gene Link Cat# 26-4000-03), Ig gene-specific primers, 0.1M DTT, RNAseOUT (Invitrogen Cat# 10777019), and 10% Igepal (Sigma-Aldrich Cat# 18896) were used in the reverse transcription PCR reaction to generate cDNA from the sorted cells right after sorting. Hot Start DNA Polymerases (QIAGEN Cat# 203643) and specific primer sets described previously99,100 were used to perform two rounds of nested PCR reactions to amplify IgG heavy and light chain variable regions using cDNAs as template. After purification with SPRI beads according to manufacturer’s instructions (Beckman Coulter Cat# B23318), PCR products were constructed into expression vectors encoding human IgG1 or Ig kappa/lambda constant domains, respectively, by HiFi DNA assembly (New England Biolabs Cat# E2621L), then transformed into competent E.coli cells. Single colonies were picked for sequencing and analysis on IMGT V-Quest online tool (http://www.imgt.org) and downstream plasmid production.
Expression and purification of monoclonal antibodies
Plasmids of the paired heavy and light chains generated after sorting were co-transfected into Expi293F cells to produce monoclonal antibodies. Briefly, 12μg heavy chain plasmid and 12 μg of light chain plasmid were added into 3ml of Opti-MEM™ (Thermo Fisher Scientific Cat# 31985070), after inverting, 24μl of FectoPRO (Polyplus Cat# 116-001) reagent was added into the mixture and inverted. After incubation at room temperature for 10min, the mixture was added to 30ml of Expi293F cells at 2.8 million cells/ml and incubated in the shaker. 24 hours after transfection, 300μl of 300mM sodium valproic acid solution and 275μl of 45% glucose solution was used to feed each cell culture. Four days post transfection, supernatants of cell cultures were collected by centrifugation at 2500xg for 15 min and filtering through 0.22μm membrane. Protein A Sepharose (GE Healthcare Cat# 17096302) and Protein G Sepharose (GE Healthcare Cat# 17061805) were mixed at 1:1 ratio before adding into the supernatant and rotating overnight at 4°C. The solution was then loaded into Econo-Pac columns (BioRad Cat# 7321010), washed with 1 column volume of PBS, and antibodies were eluted with 10ml of 0.2 M citric acid (pH 2.67). The elution was collected into a tube containing 1ml of 2M Tris Base solution. 30K Amicon centrifugal filters (Millipore Cat# UFC903024) were used for buffer exchange into PBS and further concentrating into smaller volumes.
ELISA using peptides or recombinant proteins
N-terminal biotinylated peptides corresponding to the spike stem-helix regions of SARS-CoV-1/2 (PLQPELDSFKEELDKYFKNHTSPDV), MERS-CoV (PLLGNSTGIDFQDELDEFFKNVSTSIP), HCoV-HKU1 (HSVPKLSDFESELSHWFKNQTSIAP), HCoV-OC43 (TSIPNLPDFKEELDQWFKNQTSVAP), HCoV-229E (TIVPEYIDVNKTLQELSYKLPNYTV) and HCoV-NL63 (TVIPDYVDVNKTLQEFAQNLPKYVK) were synthesized at A&A Labs (Synthetic Biomolecules).53 For peptide ELISA, streptavidin (Jackson Immuno Research Labs Cat# 016-000-084) was coated at 2 μg/ml in PBS onto 96-well half-area high binding plates (Corning Cat# 3690) overnight at 4°C. For recombinant protein ELISA, mouse anti-His antibody (Thermo Fisher Scientific Cat# MA1-21315-1MG) was used at the same concentration to coat the plates. After washing by 0.05% PBST 3 times, 3% BSA was used to block the plates for 2h at 37°C. Then 1 μg/ml of N-terminal biotinylated peptide or 2 μg/ml of His-tagged recombinant spike proteins were applied to plates and incubated for 1h at RT. After washing by 0.05% PBST 3 times, serially diluted serum samples or antibodies were added into plates and incubated for 1h at RT. After another washing, alkaline phosphatase-conjugated goat anti-human IgG Fc secondary antibody (Jackson ImmunoResearch Cat# 109-055-008) was added in 1:1000 dilution and incubated for 1h at RT. After the final wash, phosphatase substrate (Sigma-Aldrich Cat# S0942-200TAB) dissolved in staining buffer was added into each well. Absorption was measured at 405 nm. Fifty percent maximal response concentrations (EC50) were calculated using the Asymmetrical dose-response model of the Richard version in GraphPad Prism 7 (GraphPad Software). To identify critical residues for antibody binding, single alanine mutations were introduced into the 25-mer stem-helix peptide that comprises the linear epitope. These peptides were synthesized at A&A Labs (Synthetic Biomolecules). ELISA as described above was used to test antibody reactivity against peptides with single alanine substitutions.
Pseudovirus production
HIV-based lentivirus backbone plasmid pCMV-dR8.2 dvpr (Addgene Cat# 8455), pBOB-Luciferase (Addgene Cat# 170674) were co-transfected into HEK293T cells along with variously truncated SARS-CoV1, WIV1, SHC014, Pang17, SARS-CoV2, SARS-CoV-2 variants of concern [(B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta) B.1.1.529.1 (Omicron BA.1), B.1.1.529.2 (Omicron BA.2), B.1.1.529.2.12.1 (Omicron BA.2.12.1), Omicron XBB, B.1.1.529.2.75 (Omicron BA.2.75), B.1.1.529.2.75.2 (Omicron BA.2.75.2), B.1.1.529.4/5 (Omicron BA.4/5), B.1.1.529.4.6 (Omicron BA.4.6) and B.1.1.529.5.3.1.1.1.1.1.1 (Omicron BQ.1.1)] and MERS-CoV spike using Lipofectamine 2000 (ThermoFisher Scientific Cat# 11668019) to produce single-round infection-competent pseudoviruses.101 The medium was changed 12-16 hours post transfection. Pseudovirus-containing supernatants were collected 48 hours post transfection and the viral titers were measured by luciferase activity in relative light units (RLU) (Bright-Glo Luciferase Assay System, Promega Cat# E2620). The supernatants were aliquoted and stored at -80°C until further use.
Neutralization assay
Pseudotyped viral neutralization assay was performed as previously reported.7 In brief, neutralization assays were performed by adding 25μl of pseudovirus into 25μl serial dilutions of purified antibodies or plasma from human donors, the mixture was then dispensed into a 96-well plate incubated for one hour at 37°C, then 10,000 HeLa-hACE2 or hDPP4 cells/ well (in 50μl of media containing 20μg/ml Dextran) were directly added to the mixture. After incubation at 37°C for 42-48 h, luciferase activity was measured. Neutralizing activity was measured by reduction in luciferase activity compared to the virus controls. Fifty percent maximal inhibitory concentrations (IC50), the concentrations required to inhibit infection by 50% compared to the controls, were calculated using the dose-response-inhibition model with 5-parameter Hill slope equation in GraphPad Prism 7 (GraphPad Software).
Neutralization assay of replication competent CoVs
Vero E6 cells (ATCCCat# CRL-1586) were seeded at 2x104 cells/well in a black-well, tissue culture treated, 96-well plate (Corning Cat# 3916) 24 h before the assay. Abs were diluted in MEM supplemented with 5% FBS and 1% Pen/Strep media to obtain an 8-point, 3-fold dilution curve with starting concentration at 20 μg/ml. Eight hundred Pfu of SARS2-nLuc and MERS-nLuc replication competent viruses were mixed with Abs at a 1:1 ratio and incubated at 37°C for 1 h. One-hundred microliters of virus and Ab mix was added to each well and incubated at 37°C + 5% CO2 for 20 to 22 h. Luciferase activities were measured by the Nano-Glo Luciferase Assay System (Promega Cat# N1130) following the manufacturer’s protocol using a GloMax luminometer (Promega). Percent inhibition and IC50 were calculated as pseudovirus neutralization assay described above. All experiments were performed as duplicates and independently repeated three times. All the live virus experiments were performed under biosafety level 3 (BSL-3) conditions at negative pressure, by operators in Tyvek suits wearing personal powered-air purifying respirators.
Antibody immunogenetics analysis
We used a previously reported dataset of approximately 1.6 million natively paired human Ab sequences from 4 healthy adult donors.74 All sequences were annotated using the ab[x] toolkit102 and queried using PySpark. To demonstrate enrichment of particular residues within the CDRL3 of the S2-specific bnAbs discovered in this study, we queried our dataset for light chain sequences with CDRL3 length of 11 amino acids (IMGT numbering) and generated CDRL3 sequence logo plots in Python using the logomaker package.103 Comparable plots were also made using the sequences of S2-specific mAbs. To quantify the frequency of antibodies encoding genetic features similar to the isolated S2-specific neutralizing mAbs, we queried the paired sequence dataset for sequences encoding an IGHV1-46 paired with a light chain of CDRL3 of length 11 amino acids, and a CDRL3 sequence matching the regular expression “PP.F..$”.
HEp2 epithelial cell polyreactive assay
HEp2 slides (Hemagen Cat# 902360) were used to determine the reactivity of monoclonal antibodies to human epithelial type 2 (HEp2) by indirect immunofluorescence, according to the manufacturer’s instructions. Briefly, monoclonal antibody was diluted into 50μg/ml by PBS and then added onto immobilized HEp2 slides and incubated for 30 min at RT. After washing by PBS for 3 times, one drop of FITC-conjugated goat anti-human IgG was added onto each well and incubated in the dark for 30 min at RT. After washing, the coverslip was added to HEp2 slide with glycerol and the images were photographed on a Nikon fluorescence microscope for FITC detection.
Polyspecificity reagent (PSR) ELISA
Solubilized CHO cell membrane protein (SMP), human insulin (Sigma-Aldrich Cat# I2643), single strand DNA (Sigma-Aldrich Cat# D8899) were coated onto 96-well half-area high-binding plates (Corning Cat# 3690) at 5μg /ml in PBS overnight at 4°C. After washing with PBST, plates were blocked with 3% BSA for 2h at 37°C. Antibody samples were diluted at 50μg /ml in 1% BSA with 5-fold serial dilution and then added in plates to incubate for 1h at room temperature.7 The assay was performed as described in section “ELISA using peptides or recombinant proteins”.
CELISA binding
Flow cytometry-based Cell-ELISA (CELISA) binding of mAbs with HCoV spikes was performed as described previously.52,104 A total of 4x106 HEK293T cells were seeded into 10cm round cell culture dishes and incubated at 37°C. After 24h, HEK293T cells were transfected with plasmids encoding full-length HCoV spikes and were incubated for 36-48h at 37°C. The cells were harvested and distributed into 96-well round-bottom tissue culture plates for individual staining reactions. For each staining reaction, cells were washed three times with 200μl FACS buffer (1xPBS, 2%FBS, 1mM EDTA). The cells were stained for 1h on ice in 50μl staining buffer with 10μg/ml of primary antibody. After washing three times with 200μl FACS buffer, the cells were stained with 50μl/well of 1:200 diluted R-phycoerythrin (PE)-conjugated mouse anti-human IgG Fc antibody (SouthernBiotech Cat# 9040-09) and 1:1000 dilution of Zombie-NIR viability dye (BioLegend Cat# 423105) on ice in dark for 45min. Following three washes with FACS buffer, the cells were resuspended and analyzed by flow cytometry (BD Lyrics cytometer), and the binding data were generated by calculating the Mean Fluorescence Intensity using FlowJo 10 software. Mock-transfected 293T cells were used as a negative control.
BioLayer Interferometry binding (BLI)
Octet K2 system (ForteBio) was used to determine the monoclonal antibody binding with S-proteins or selected peptides. IgG was first captured for 60s by anti-human IgG Fc capture (AHC) biosensors (ForteBio Cat# 18-5063), then baseline was provided in Octet buffer (PBS with 0.1% Tween) for another 60s. After that, the sensors were transferred into wells containing diluted HCoV S-proteins for 120s for association, and into Octet buffer for disassociation for 240s. Selected peptides that were N-terminal biotinylated were diluted in Octet buffer and first captured for 60s by the hydrated streptavidin biosensors (ForteBio Cat# 18-5020), then unbound peptides were removed by transferring into Octet buffer for 60s to provide the baseline. The sensors were then immersed into the monoclonal antibodies in Octet buffer for 120s for association, followed by transferring into Octet buffer for 240s for dissociation. The data generated were analyzed using the ForteBio Data Analysis software for correction, and the kinetic curves were fit to a 1:1 binding mode. Note that the IgG: spike protomer binding can be a mixed population of 2:1 and 1:1, such that the term ‘apparent affinity’ dissociation constants (KDApp) are shown to reflect the binding affinity between IgGs and spike trimers tested.
Competition BLI
To determine the binding epitopes of the isolated mAbs compared with human stem-helix mAbs of know specificities, we conducted in-tandem epitope binning experiments using the Octet RED384 system. Briefly, 100 nM of biotinylated SARS-CoV-2 Spike in Octet buffer was captured by streptavidin biosensors (FortéBio Cat# 18-5020) for 5 min and then transferred into Octet buffer (PBS plus 0.1% Tween 20) for 30s to remove unbound spike protein and provide the baseline. Then, the protein bound sensors were moved into the saturating antibody in Octet buffer at a concentration of 100 μg/mL for 10 min. The biosensors were then transferred into 100 μg/mL competitor antibody in Octet buffer for 5 min to measure binding in the presence of saturating antibody. As control, biosensors loaded with antigen were directly moved into competitor antibody solution. The percent (%) inhibition in binding was calculated with the formula: [Percent (%) binding inhibition = 1- (competitor antibody binding response in presence of saturating antibody) / (binding response of the competitor antibody without saturating antibody).
Expression and purification of Fabs
To generate Fabs, IgGs were digested by papain (Sigma-Aldrich Cat# P3125) at 37°C for 4 hours and then incubated with protein A beads for 2 hours at 4°C to remove Fc fragments. Each Fab was concentrated afterward and further purified by size exclusion chromatography using a Superdex 200 Increase 10/300 GL column (GE Healthcare Cat# GE28-9909-44). The selected fractions were pooled and concentrated again for further use.
Crystallization and X-ray structure determination
Purified Fabs of antibodies CC25.106, CC95.108, CC99.103, CC68.109 were mixed with betacoronavirus stem-helix peptides in a molar ratio of 1:5-8 (Fab:peptide) and incubated on ice for 1 hour before setting up for crystallization trials. 384 conditions of the JCSG Core Suite (Qiagen) were used for setting-up trays for screening on our robotic CrystalMation system (Rigaku) at Scripps Research. Crystallization trials were set-up by the vapor diffusion method in sitting drops containing 0.1 μl of protein complex and 0.1 μl of reservoir solution. Crystals appeared on day 14, were harvested on day 21, pre-equilibrated in cryoprotectant containing 0–10% ethylene glycol, and then flash cooled and stored in liquid nitrogen until data collection. Diffraction quality crystals were obtained for the following Fab-peptide complexes with indicated crystallization conditions: [CC25.106+SARS-CoV-2: 19% (v/v) isopropanol, 19% (w/v) polyethylene glycol 4000, 5% (v/v) glycerol, 0.095 M sodium citrate pH 5.6]; [CC68.109+MERS-CoV: 0.17 M sodium acetate, 25.5% (w/v) polyethylene glycol 4000, 15% (v/v) glycerol, 0.085 M Tris pH 8.5]; [CC95.108+HCoV-HKU1: 0.2 M ammonium formate, 20% (w/v) polyethylene glycol 3350]; [CC99.103+MERS-CoV: 19% (v/v) isopropanol, 19% (w/v) polyethylene glycol 4000, 5% (v/v) glycerol, 0.095 M sodium citrate pH 5.6]. Diffraction data were collected at cryogenic temperature (100 K) at either beamline 23-ID-D of the Advanced Photon Source (APS) at Argonne National Laboratory (for CC25.106 and CC68.109) or beamlines 12-1 and 12-2 of the Stanford Synchrotron Radiation Lightsource (SSRL) (for CC95.108 and CC99.103). The X-ray data were processed with HKL2000.105 The X-ray structures were initially solved by molecular replacement (MR) using PHASER106 with MR models for the Fabs from PDB:7JMW107 and PDB:7KN4.108 Iterative model building and refinement were then carried out in COOT109 and PHENIX,110 respectively. Epitope and paratope residues, as well as their interactions, were identified by using PISA program111 with buried surface area (BSA > 0 Å2) as the criterion. Antibody and coronavirus spike sequences were aligned for analysis using MUSCLE program112 built in European Bioinformatics Institute (EBI) web services.113 All residues in the antibody structures and analyses were numbered using the Kabat scheme for easy comparison.114
In vivo virus challenge in mouse model
All mouse experiments were performed at the University of North Carolina, NIH/PHS Animal Welfare Assurance Number: D16-00256 (A3410-01), under approved IACUC protocols. The animal manipulation and virus work were performed in a Class 2A biological safety cabinet in a BSL3 approved facility and workers wore PAPRs, tyvek suites and were double gloved. 12-month-old female Balb/c mice (strain 047) were purchased from Envigo for Sarbecovirus challenge experiments.81,115 C57Bl/6 288/330+/+ mice, which encode two human codons in the mouse dipeptidyl peptidase gene, were used for MERS-CoV mouse adapted challenge experiments.82 Mice were housed in individually ventilated Seal-Safe cages, provided food and water ad libitum, and allowed to acclimate at least seven days before experimental use. Twelve hours prior to infection, 300μg antibody was injected into mice intraperitoneally. The mice used in this study were between 25-30g at the start of the experiment, so the antibody dose per mouse was between 10 and 12 mg/kg. Immediately prior to infection, mice were anesthetized by injection of ketamine and xylazine intraperitoneally and weighed. Virus (SARS-CoV MA15, SARS-CoV2 MA10 and mouse adapted MERS-CoV-M35c4) was diluted in 50μl sterile PBS and administered intranasally.81,82,83,115 Mice were weighed daily and observed for signs of disease. The mice were euthanized via isoflurane overdose at the designated timepoint, followed by assessment of gross lung pathology and collection of the inferior lobe for virus titration. Respiratory function was measured at day2 post infection via Buxco whole body plethysmography, as previously described.116 No animals or data points were excluded from the analyses.
Virus titration
SARS-CoV-2-MA10, SARS-CoV-1-MA15 and MERS-CoV-M35c4 were grown and titered using VeroE6 cells as previously described.117 Briefly, lung tissue was homogenized in 1ml sterile PBS, Via Magnalyser (Roche), centrifuged to pellet debris, plated in 10-fold serial dilutions on VeroE6 cells on a 6-well plate and covered with a 1:1 mixture of 1.6% agarose and media. At two (SARS-CoV-1) or three (SARS-CoV-2) days post plating, cells were stained with neutral red and plaques counted.
Quantification and statistical analysis
Statistical analysis was performed using Graph Pad Prism 8, Graph Pad Software, San Diego, California, USA. ID50 or IC50 titers were compared using the non-parametric unpaired Mann-Whitney-U test. The correlation between two groups was determined by Spearman rank test. Groups of data were compared using the Kruskal-Wallis non-parametric test. Dunnett's multiple comparisons test were also performed between experimental groups. Data were considered statistically significant when p < 0.05.
Acknowledgments
We thank all of the human cohort participants for donating samples. This work was supported by NIH CHAVD UM1 AI44462 (D.R.B. and I.A.W.), the IAVI Neutralizing Antibody Center, the Bill and Melinda Gates Foundation INV-004923 (I.A.W. and D.R.B.), the Translational Virology Core of the San Diego Center for AIDS Research (CFAR) grant NIH AI036214 (D.M.S.), NIH 5T32AI007384 (S.A.R.), NIH U54 CA260543 and AI157155 (R.S.B.), NIH R21 AI145372 (L.E.G.), and the John and Mary Tu Foundation and the James B. Pendleton Charitable Trust (D.M.S. and D.R.B.). L.V.T. is supported by Pfizer NCBiotech Distinguished Postdoctoral Fellowship in Gene Therapy. D.R.M. is currently supported by a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award and a Hanna H. Gray Fellowship from the Howard Hughes Medical Institute. We thank Henry Tien for technical support with the crystallization robot and Robyn Stanfield for assistance in X-ray data collection. We are grateful to the staff of the Stanford Synchrotron Radiation Lightsource (SSRL) beamlines 12-1 and 12-2 and Advanced Photon Source (APS) beamline 23ID-D for assistance. This research used resources of the SSRL, SLAC National Accelerator Laboratory, which is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health – National Institute of General Medical Sciences (including P41GM103393). This research also used resources of the APS, a U.S. Department of Energy (DOE) Office of Science User Facility, operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
Author contributions
P.Z., G.S., H.L., M.Y., I.A.W., R.S.B., L.E.G., D.R.B., and R.A. conceived and designed the study. N.B., M.P., E.G., S.A.R., D.M.S., and T.F.R. recruited donors and collected and processed plasma and peripheral blood mononuclear cell (PBMC) samples. P.Z., G.S., W.-t.H., R.M., K.D., S.C., P.Y., L.P., T.C., O.L., and F.A. performed BLI, ELISA, virus preparation, neutralization and isolation, and characterization of monoclonal antibodies. H.L., Y.S., and B.B. performed immunogenetic analysis of antibodies. H.L. crystallized the antibody-antigen complexes and determined the crystal structures. H.L., M.Y., and X.Z. collected X-ray data. H.L., M.Y., and I.A.W. analyzed the structural data. L.V.T. performed live virus neutralizations assays, and D.R.M., A.S., and L.E.G. conducted in vivo animal protection studies. P.Z., G.S., H.L., M.Y., W.-t.H., N.B., X.Z., L.V.T., D.R.M., A.S., F.A., P.Y., L.P., K.D., R.M., S.C., T.C., O.L., M.P., E.G., D.N., J.G.J., I.A.W., Y.S., B.B., T.F.R., R.S.B., L.E.G., D.R.B., and R.A. designed the experiments and/or analyzed the data. P.Z., G.S., H.L., I.A.W., D.R.B., and R.A. wrote the paper, and all authors reviewed and edited the paper.
Declaration of interests
P.Z., G.S., W.-t.H., D.R.B., and R.A. are listed as inventors on pending patent applications describing the betacoronavirus bnAbs isolated in this study. D.R.B. is a consultant for IAVI and for Invivyd. R.S.B. and L.E.G. have ongoing collaborations with Invivyd.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: February 16, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2023.02.005.
Contributor Information
Ian A. Wilson, Email: wilson@scripps.edu.
Ralph S. Baric, Email: rbaric@email.unc.edu.
Lisa E. Gralinski, Email: lgralins@email.unc.edu.
Dennis R. Burton, Email: burton@scripps.edu.
Raiees Andrabi, Email: andrabi@scripps.edu.
Supplemental information
Data and code availability
The data supporting the findings of this study are available within the published article and summarized in the corresponding tables, figures, and supplemental materials. Antibody sequences have been deposited in GenBank under accession numbers OP699209-OP699288. X-ray coordinates and structure factors have been deposited in the RCSB Protein Data Bank under accession codes 8DGU, 8DGV, 8DGW, and 8DGX.
References
- 1.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert P.B., Montefiori D.C., McDermott A., Fong Y., Benkeser D.C., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Title: immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy trial. Science. 2021;375:43–50. doi: 10.1101/2021.08.09.21261290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greaney A.J., Loes A.N., Gentles L.E., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci. Transl. Med. 2021;13:eabi9915. doi: 10.1126/scitranslmed.abi9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H., Wilson I.A. Protective neutralizing epitopes in SARS-CoV-2. Immunol. Rev. 2022;310:76–92. doi: 10.1111/imr.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H.H., Barnes C.O., Cipolla M., Ramos V., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan M., Huang D., Lee C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittal A., Khattri A., Verma V. Structural and antigenic variations in the spike protein of emerging SARS-CoV-2 variants. PLoS Pathog. 2022;18:e1010260. doi: 10.1371/journal.ppat.1010260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox M., Peacock T.P., Harvey W.T., Hughes J., Wright D.W., Willett B.J., Thomson E., Gupta R.K., Peacock S.J., Robertson D.L., et al. SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat. Rev. Microbiol. 2022 doi: 10.1038/s41579-022-00809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 19.Mascola J.R., Graham B.S., Fauci A.S. SARS-CoV-2 viral variants-tackling a moving target. JAMA. 2021;325:1261–1262. doi: 10.1001/jama.2021.2088. [DOI] [PubMed] [Google Scholar]
- 20.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., et al. COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 23.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 25.Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 28.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu P., Evans J.P., Faraone J., Zheng Y.-M., Carlin C., Anghelina M., Stevens P., Fernandez S., Jones D., Lozanski G., et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe. 2023;31:9–17. doi: 10.1016/j.chom.2022.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arora P., Kempf A., Nehlmeier I., Schulz S.R., Jäck H.M., Pöhlmann S., Hoffmann M. Omicron sublineage BQ.1.1 resistance to monoclonal antibodies. Lancet Infect. Dis. 2022;23:22–23. doi: 10.1016/s1473-3099(22)00733-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Planas D., Bruel T., Staropoli I., Guivel-Benhassine F., Porrot F., Maes P., Grzelak L., Prot M., Mougari S., Planchais C., et al. Resistance of Omicron subvariants BA.2.75.2, BA.4.6 and BQ.1.1 to neutralizing antibodies. Nat. Commun. 2023;14:824. doi: 10.1038/s41467-023-36561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menachery V.D., Yount B.L., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 35.Burton D.R., Topol E.J. Variant-proof vaccines - invest now for the next pandemic. Nature. 2021;590:386–388. doi: 10.1038/d41586-021-00340-4. [DOI] [PubMed] [Google Scholar]
- 36.Koff W.C., Berkley S.F. A universal coronavirus vaccine. Science. 2021;371:759. doi: 10.1126/science.abh0447. [DOI] [PubMed] [Google Scholar]
- 37.Morens D.M., Taubenberger J.K., Fauci A.S. Universal coronavirus vaccines - an urgent need. N. Engl. J. Med. 2022;386:297–299. doi: 10.1056/NEJMp2118468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tortorici M.A., Czudnochowski N., Starr T.N., Marzi R., Walls A.C., Zatta F., Bowen J.E., Jaconi S., Di Iulio J., Wang Z., et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 41.Martinez D.R., Schafer A., Gobeil S., Li D., De la Cruz G., Parks R., Lu X., Barr M., Stalls V., Janowska K., et al. A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Sci. Transl. Med. 2021;14:eabj7125. doi: 10.1126/scitranslmed.abj7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He W.-t., Yuan M., Callaghan S., Musharrafieh R., Song G., Silva M., Beutler N., Lee W., Yong P., Torres J., et al. Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques. Sci. Transl. Med. 2022;14:eabl9605. doi: 10.1126/scitranslmed.abl9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D., Edwards R.J., Manne K., Martinez D.R., Schäfer A., Alam S.M., Wiehe K., Lu X., Parks R., Sutherland L.L., et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell. 2021;184:4203–4219.e32. doi: 10.1016/j.cell.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He W.-T., Musharrafieh R., Song G., Dueker K., Tse L.V., Martinez D.R., Schäfer A., Callaghan S., Yong P., Beutler N., et al. Targeted isolation of diverse human protective broadly neutralizing antibodies against SARS-like viruses. Nat. Immunol. 2022;23:960–970. doi: 10.1038/s41590-022-01222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H., Kaku C.I., Song G., Yuan M., Andrabi R., Burton D.R., Walker L.M., Wilson I.A. Human antibodies to SARS-CoV-2 with a recurring YYDRxG motif retain binding and neutralization to variants of concern including Omicron. Commun. Biol. 2022;5:766. doi: 10.1038/s42003-022-03700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan M., Zhu X., He W.T., Zhou P., Kaku C.I., Capozzola T., Zhu C.Y., Yu X., Liu H., Yu W., et al. A broad and potent neutralization epitope in SARS-related coronaviruses. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2205784119. e2205784119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jette C.A., Cohen A.A., Gnanapragasam P.N.P., Muecksch F., Lee Y.E., Huey-Tubman K.E., Schmidt F., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C., et al. Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies. Cell Rep. 2021;36:109760. doi: 10.1016/j.celrep.2021.109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song G., He W.T., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou P., Yuan M., Song G., Beutler N., Shaabani N., Huang D., He W.T., Zhu X., Callaghan S., Yong P., et al. A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection. Sci. Transl. Med. 2022;14:eabi9215. doi: 10.1126/scitranslmed.abi9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinto D., Sauer M.M., Czudnochowski N., Low J.S., Tortorici M.A., Housley M.P., Noack J., Walls A.C., Bowen J.E., Guarino B., et al. Broad Betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsieh C.L., Werner A.P., Leist S.R., Stevens L.J., Falconer E., Goldsmith J.A., Chou C.W., Abiona O.M., West A., Westendorf K., et al. Stabilized coronavirus spike stem elicits a broadly protective antibody. Cell Rep. 2021;37:109929. doi: 10.1016/j.celrep.2021.109929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauer M.M., Tortorici M.A., Park Y.J., Walls A.C., Homad L., Acton O.J., Bowen J.E., Wang C., Xiong X., de van der Schueren W., et al. Structural basis for broad coronavirus neutralization. Nat. Struct. Mol. Biol. 2021;28:478–486. doi: 10.1038/s41594-021-00596-4. [DOI] [PubMed] [Google Scholar]
- 57.Li W., Chen Y., Prévost J., Ullah I., Lu M., Gong S.Y., Tauzin A., Gasser R., Vézina D., Anand S.P., et al. Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern. Cell Rep. 2022;38:110210. doi: 10.1016/j.celrep.2021.110210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C., van Haperen R., Gutiérrez-Álvarez J., Li W., Okba N.M.A., Albulescu I., Widjaja I., van Dieren B., Fernandez-Delgado R., Sola I., et al. A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Nat. Commun. 2021;12:1715. doi: 10.1038/s41467-021-21968-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jennewein M.F., MacCamy A.J., Akins N.R., Feng J., Homad L.J., Hurlburt N.K., Seydoux E., Wan Y.H., Stuart A.B., Edara V.V., et al. Isolation and characterization of cross-neutralizing coronavirus antibodies from COVID-19+ subjects. Cell Rep. 2021;36:109353. doi: 10.1016/j.celrep.2021.109353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hurlburt N.K., Homad L.J., Sinha I., Jennewein M.F., MacCamy A.J., Wan Y.H., Boonyaratanakornkit J., Sholukh A.M., Jackson A.M., Zhou P., et al. Structural definition of a pan-sarbecovirus neutralizing epitope on the spike S2 subunit. Commun. Biol. 2022;5:342. doi: 10.1038/s42003-022-03262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dacon C., Peng L., Lin T.-H., Tucker C., Lee C.-C.D., Cong Y., Wang L., Purser L., Cooper A.J.R., Williams J.K., et al. Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses. Cell Host Microbe. 2023;31:97–111.e12. doi: 10.1016/j.chom.2022.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piepenbrink M.S., Park J.-G., Deshpande A., Loos A., Ye C., Basu M., Sarkar S., Khalil A.M., Chauvin D., Woo J., et al. Potent universal beta-coronavirus therapeutic activity mediated by direct respiratory administration of a Spike S2 domain-specific human neutralizing monoclonal antibody. PLoS Pathog. 2022;18:e1010691. doi: 10.1371/journal.ppat.1010691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andrabi R., Bhiman J.N., Burton D.R. Strategies for a multi-stage neutralizing antibody-based HIV vaccine. Curr. Opin. Immunol. 2018;53:143–151. doi: 10.1016/j.coi.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steichen J.M., Lin Y.C., Havenar-Daughton C., Pecetta S., Ozorowski G., Willis J.R., Toy L., Sok D., Liguori A., Kratochvil S., et al. A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science. 2019;366:aax4380. doi: 10.1126/science.aax4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jardine J., Julien J.P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P.S., MacPherson S., Jones M., et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erbelding E.J., Post D.J., Stemmy E.J., Roberts P.C., Augustine A.D., Ferguson S., Paules C.I., Graham B.S., Fauci A.S. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018;218:347–354. doi: 10.1093/infdis/jiy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burton D.R., Walker L.M. Rational vaccine design in the time of COVID-19. Cell Host Microbe. 2020;27:695–698. doi: 10.1016/j.chom.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hurt A.C., Wheatley A.K. Neutralizing antibody therapeutics for COVID-19. Viruses. 2021;13:628. doi: 10.3390/v13040628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crotty S. Hybrid immunity. Science. 2021;372:1392–1393. [Google Scholar]
- 70.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guthmiller J.J., Lan L.Y.-L., Fernández-Quintero M.L., Han J., Utset H.A., Bitar D.J., Hamel N.J., Stovicek O., Li L., Tepora M., et al. Polyreactive Broadly Neutralizing B cells Are Selected to Provide Defense against Pandemic Threat Influenza Viruses. Immunity. 2020;53:1230–1244.e5. doi: 10.1016/j.immuni.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Briney B., Inderbitzin A., Joyce C., Burton D.R. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature. 2019;566:393–397. doi: 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaffe D.B., Shahi P., Adams B.A., Chrisman A.M., Finnegan P.M., Raman N., Royall A.E., Tsai F., Vollbrecht T., Reyes D.S., et al. Functional antibodies exhibit light chain coherence. Nature. 2022;611:352–357. doi: 10.1038/s41586-022-05371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andrabi R., Voss J.E., Liang C.H., Briney B., McCoy L.E., Wu C.Y., Wong C.H., Poignard P., Burton D.R. Identification of common features in prototype broadly neutralizing antibodies to HIV envelope V2 apex to facilitate vaccine design. Immunity. 2015;43:959–973. doi: 10.1016/j.immuni.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Andrabi R., Pallesen J., Allen J.D., Song G., Zhang J., de Val N., Gegg G., Porter K., Su C.Y., Pauthner M., et al. The chimpanzee SIV envelope trimer: structure and deployment as an HIV vaccine template. Cell Rep. 2019;27:2426–2441.e6. doi: 10.1016/j.celrep.2019.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fan X., Cao D., Kong L., Zhang X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat. Commun. 2020;11:3618. doi: 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watanabe Y., Berndsen Z.T., Raghwani J., Seabright G.E., Allen J.D., Pybus O.G., McLellan J.S., Wilson I.A., Bowden T.A., Ward A.B., Crispin M. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020;11:2688. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cockrell A.S., Yount B.L., Scobey T., Jensen K., Douglas M., Beall A., Tang X.C., Marasco W.A., Heise M.T., Baric R.S. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol. 2016;2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Douglas M.G., Kocher J.F., Scobey T., Baric R.S., Cockrell A.S. Adaptive evolution influences the infectious dose of MERS-CoV necessary to achieve severe respiratory disease. Virology. 2018;517:98–107. doi: 10.1016/j.virol.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cohen M.S. Monoclonal antibodies to disrupt progression of early Covid-19 infection. N. Engl. J. Med. 2021;384:289–291. doi: 10.1056/NEJMe2034495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fischer W.A., 2nd, Eron J.J., Jr., Holman W., Cohen M.S., Fang L., Szewczyk L.J., Sheahan T.P., Baric R., Mollan K.R., Wolfe C.R., et al. A Phase 2a clinical trial of Molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med. 2021;14:eabl7430. doi: 10.1126/scitranslmed.abl7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greenbaum U., Klein K., Martinez F., Song J., Thall P.F., Ramdial J.L., Knape C., Aung F.M., Scroggins J., Knopfelmacher A., et al. High levels of common cold coronavirus antibodies in convalescent plasma are associated with improved survival in COVID-19 patients. Front. Immunol. 2021;12:675679. doi: 10.3389/fimmu.2021.675679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaplonek P., Wang C., Bartsch Y., Fischinger S., Gorman M.J., Bowman K., Kang J., Dayal D., Martin P., Nowak R.P., et al. Early cross-coronavirus reactive signatures of humoral immunity against COVID-19. Sci. Immunol. 2021;6:eabj2901. doi: 10.1126/sciimmunol.abj2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pušnik J., König J., Mai K., Richter E., Zorn J., Proksch H., Schulte B., Alter G., Streeck H. Persistent maintenance of intermediate memory B cells following SARS-CoV-2 infection and vaccination recall response. J. Virol. 2022;96:e0076022. doi: 10.1128/jvi.00760-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reynolds C.J., Pade C., Gibbons J.M., Butler D.K., Otter A.D., Menacho K., Fontana M., Smit A., Sackville-West J.E., Cutino-Moguel T., et al. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science. 2021;372:1418–1423. doi: 10.1126/science.abh1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haynes B.F., Mascola J.R. The quest for an antibody-based HIV vaccine. Immunol. Rev. 2017;275:5–10. doi: 10.1111/imr.12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwong P.D., Mascola J.R., Nabel G.J. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 94.Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O'Connor M.A., Chen C., et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Joyce M.G., King H.A.D., Elakhal-Naouar I., Ahmed A., Peachman K.K., Macedo Cincotta C., Subra C., Chen R.E., Thomas P.V., Chen W.H., et al. A SARS-CoV-2 ferritin nanoparticle vaccine elicits protective immune responses in nonhuman primates. Sci. Transl. Med. 2021;14:eabi5735. doi: 10.1126/scitranslmed.abi5735. [DOI] [PubMed] [Google Scholar]
- 97.Ofek G., Guenaga F.J., Schief W.R., Skinner J., Baker D., Wyatt R., Kwong P.D. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl. Acad. Sci. USA. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Correia B.E., Ban Y.E., Holmes M.A., Xu H., Ellingson K., Kraft Z., Carrico C., Boni E., Sather D.N., Zenobia C., et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 99.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Doria-Rose N.A., Bhiman J.N., Roark R.S., Schramm C.A., Gorman J., Chuang G.Y., Pancera M., Cale E.M., Ernandes M.J., Louder M.K., et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. 2016;90:76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou X., Ma F., Xie J., Yuan M., Li Y., Shaabani N., Zhao F., Huang D., Wu N.C., Lee C.D., et al. Diverse immunoglobulin gene usage and convergent epitope targeting in neutralizing antibody responses to SARS-CoV-2. Cell Rep. 2021;35:109109. doi: 10.1016/j.celrep.2021.109109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Briney B., Burton D.R. 2018. Massively scalable genetic analysis of antibody repertoires. [DOI] [Google Scholar]
- 103.Tareen A., Kinney J.B. Logomaker: beautiful sequence logos in Python. Bioinformatics. 2020;36:2272–2274. doi: 10.1093/bioinformatics/btz921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou P., Wang H., Fang M., Li Y., Wang H., Shi S., Li Z., Wu J., Han X., Shi X., et al. Broadly resistant HIV-1 against CD4-binding site neutralizing antibodies. PLoS Pathog. 2019;15:e1007819. doi: 10.1371/journal.ppat.1007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 106.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., van Schooten J., Zhu X., Lee C.D., Brouwer P.J.M., et al. Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity. 2020;53:1272–1280.e5. doi: 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qiang M., Ma P., Li Y., Liu H., Harding A., Min C., Wang F., Liu L., Yuan M., Ji Q., et al. Neutralizing antibodies to SARS-CoV-2 selected from a human antibody library constructed decades ago. Adv. Sci. (Weinh) 2022;9:e2102181. doi: 10.1002/advs.202102181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 110.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 112.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dunbar J., Deane C.M. ANARCI: antigen receptor numbering and receptor classification. Bioinformatics. 2016;32:298–300. doi: 10.1093/bioinformatics/btv552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86:884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Menachery V.D., Gralinski L.E., Baric R.S., Ferris M.T. New metrics for evaluating viral respiratory pathogenesis. PLoS One. 2015;10:e0131451. doi: 10.1371/journal.pone.0131451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the published article and summarized in the corresponding tables, figures, and supplemental materials. Antibody sequences have been deposited in GenBank under accession numbers OP699209-OP699288. X-ray coordinates and structure factors have been deposited in the RCSB Protein Data Bank under accession codes 8DGU, 8DGV, 8DGW, and 8DGX.