Abstract
Polycomb group (PcG) proteins are critical chromatin regulators required for stable cell fate that maintain repression of lineage-inappropriate genes. Recent advances show that PcG proteins form distinct multi-protein complexes in different cellular environments, such as in early development, adult tissue maintenance, and cancer. This surprising compositional diversity provides the basis for mechanistic diversity. Understanding this complexity deepens and refines the principles of PcG complexes’ recruitment, target gene repression, and inheritance of memory. We review how the core molecular mechanism of Polycomb complexes operates in diverse developmental settings. We propose that context-dependent changes in composition and mechanism are essential for the proper epigenetic regulation in development.
Introduction
Stable maintenance of acquired cell fate is the hallmark of cellular differentiation. Robust development, from making the correct number of digits to producing myriad brain cell types, relies on the memory of cell fate, especially on proper gene expression. To maintain active or inactive status of genes, eukaryotes employ chromatin and associated proteins to mark and sequester DNA elements. A key group of chromatin modifying complexes required for maintaining the repressed state in metazoans are formed by Polycomb group (PcG) proteins.
PcG genes were initially discovered in Drosophila melanogaster by homeotic transformation phenotypes, implicating them in cell fate determination1,2. Subsequent studies showed that PcG proteins are critical for preventing misexpression of Hox genes in inappropriate body segments3–5. Core PcG genes are conserved across many species from flowering plants to vertebrates, and they play similar roles in the maintenance of repression6. This Polycomb function is not only critical for proper embryonic development, but also for the regulation of adult stem cells. As such, PcG genes are often mutated or amplified in cancer.
Polycomb group proteins form stable multi-protein complexes to modify chromatin. Initial biochemical studies identified distinct Polycomb complexes called Polycomb Repressive Complex 1 (PRC1) and PRC2 with different molecular activities. Both PRC1 and PRC2 have subsequently been shown to encompass numerous related complexes with differing auxiliary subunits. PRC1 complexes catalyze mono-ubiquitylation of Lys119 residue of H2A (H2AK119Ub1)7,8 and can compact chromatin9,10. PRC2 complexes catalyze mono- di- and tri-methylation of Lys27 residue of H3 (H3K27me1/2/3)11–14. The genome localization of PRC1 and PRC2 complexes frequently overlap at the promoters of developmentally important transcription factors that need to be repressed in a given cell type15–18. Cooperation between PRC1 and PRC2 activities can induce positive feedback of their recruitment and their biochemical activities11,19–24, which is critical for the maintenance of the silent transcriptional state.
While the core functions of Polycomb complexes are conserved in diverse cell types and species, advances in the past decade have revealed surprisingly unique functions of distinct Polycomb complexes in different cellular contexts25–27. Both PRC1 and PRC2 form sub-complexes either by combinatorial assembly of paralogous components or by inclusion of accessory proteins. These distinct Polycomb complexes possess different molecular activities. Furthermore, the rapid advance of technologies now enables genome-wide chromatin analyses of these complexes in rare and specialized cell types, including adult stem cells and preimplantation embryos. With findings from these diverse contexts, there is a growing appreciation that different cell types might use different Polycomb complexes to meet their unique cellular needs. In this review, we will focus on how the behaviour of distinct Polycomb complexes in diverse contexts has expanded and refined our understanding of the Polycomb system and chromatin-mediated epigenetic regulation. More general principles on molecular mechanisms and developmental roles of PcG proteins can be found in other recent reviews28,29.
Here, we first introduce distinct Polycomb complexes and their molecular functions. Then we will describe context-specific functions of Polycomb complexes, which are often achieved by formation of cell type-specific sub-complexes. We will examine how the genomic distribution of PcG is altered in special cell types and under certain perturbation conditions, and how this plasticity reveals the regulatory principles on the activity and targeting of PcG proteins. Finally, we will discuss how histone modifications and PcG proteins together maintain the memory of repression and stabilize cell fate.
Functions of Polycomb complexes
The composition and molecular function of cPRC1, ncPRC1 and PRC2
Three major Polycomb repressive complexes have been defined based on their distinct biochemical activities: canonical PRC1 (cPRC1), non-canonical PRC1 (ncPRC1, also known as variant PRC1, vPRC1), and PRC2. PRC2 and ncPRC1 are histone modifying enzyme complexes, while cPRC1 modifies chromatin structure and organization primarily through non-enzymatic means (FIG. 1, upper boxes).
Figure 1 |. Composition and molecular functions of Polycomb complexes.
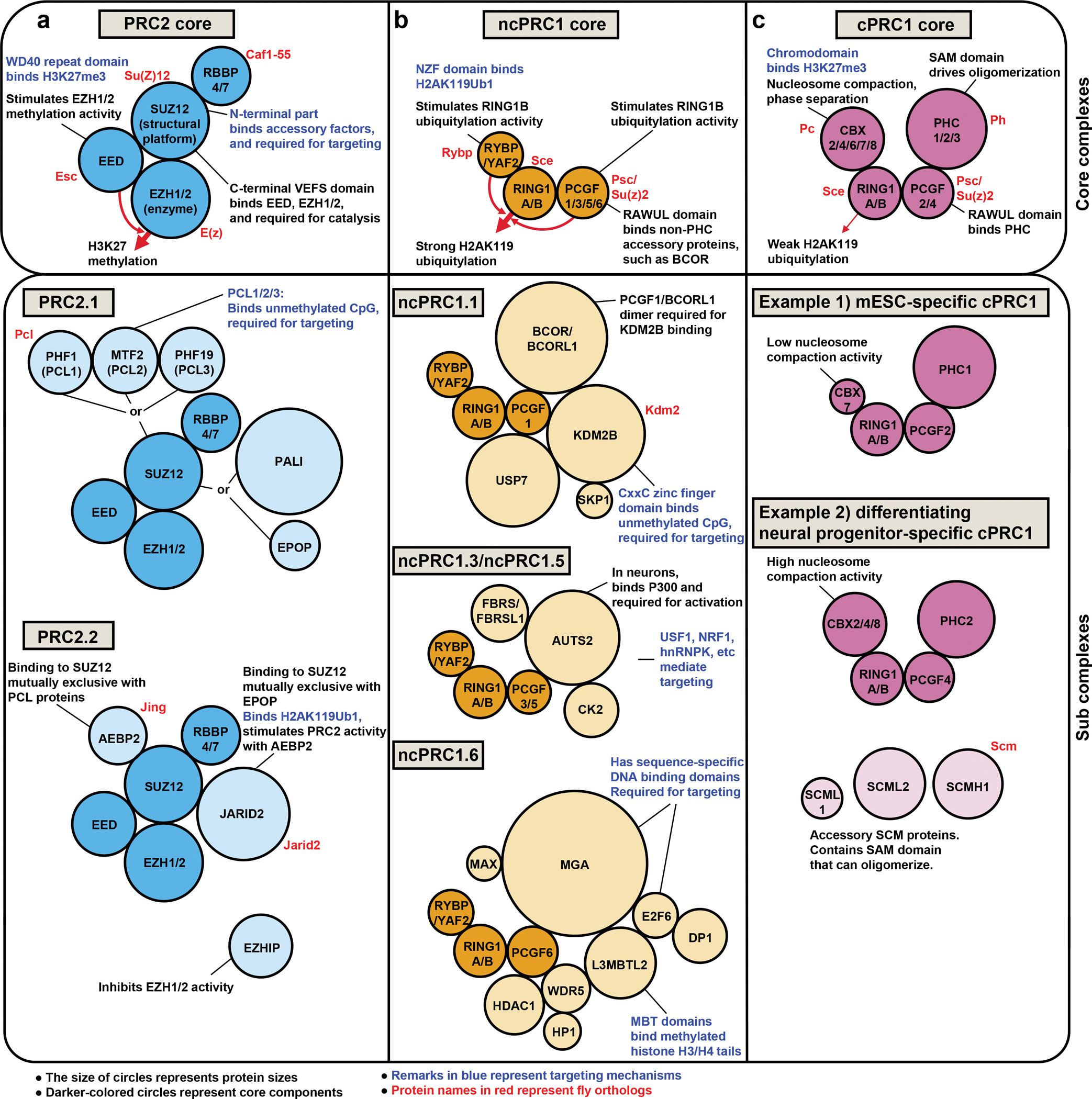
PRC2, ncPRC1 and cPRC1 have core components that form stable complexes with little variation between cell types (upper boxes) except when one paralog is switched with the other or accessory components are included (lower boxes). a | The core PRC2 complex can catalyze H3K27 methylation through the SET domain in EZH proteins. Sub-stoichiometric accessory proteins bind the N-terminal region of SUZ12. They can help PRC2 targeting. For example, PRC2.1 has PCL proteins that bind unmethylated CpG-rich DNA, and PRC2.2 has JARID2 that can bind H2AK119Ub1. Other accessory proteins such as PALI, EPOP and EZHIP modulate PRC2 catalytic activity. b, c | Both cPRC1 and ncPRC1 have RING and PCGF proteins that dimerize through their respective RING domains203,204. RING1A/B has a RAWUL domain that binds either CBX proteins (for cPRC1), or RYBP/YAF2 (for ncPRC1). b | ncPRC1 can be defined by its specific PCGF paralog (for example, ncPRC1.1 contains PCGF1). In addition, each ncPRC1 has diverse accessory components that confer unique functions to the complexes. c | Cell type-specific cPRC1 complexes can be formed by combinatorial assembly of CBX, PHC and PCGF paralogs as shown in two examples of mESC and NPC-specific cPRC1s. In addition, SCM proteins can be incorporated in cPRC1 in a cell type-specific manner.
PRC2 is composed of SUZ12, EED, RBBP4 (or paralog RBBP7) and EZH2 (or paralog EZH1) (FIG. 1a). It catalyzes mono-, di-, and tri-methylation of H3K27 using the EZH proteins’ methyltransferase activity11–14. PRC2 is responsible for di-methylation of the broad intergenic genome30,31, and tri-methylation of H3K27, for example on inactive and unmethylated CpG-rich promoters32–35. While H3K27me3 is strongly correlated with low gene expression36, this histone mark alone is not sufficient for gene repression37–39. Proteins recruited to H3K27me3-enriched regions and the resulting physical changes in the chromatin structure are also required for stable gene silencing9,40.
ncPRC1 is composed of RING1B (or paralog RING1A), PCGF1 (or paralog PCGF3/5/6, less likely PCGF2/4), and RYBP (or paralog YAF2) (FIG. 1b)8,25,41,42. The RING subunit8,43 of ncPRC1 catalyzes mono-ubiquitylation of Lys119 of H2A at promoters44,45 and across the genome31,46. RYBP, a specific component of ncPRC1, stimulates the ubiquitylation activity47. While PRC2 and cPRC1 are preferentially localized at inactive promoters, ncPRC1 is seen at both inactive (H3K27me3-high) and active (H2K27me3-low) promoters when assessed by RYBP, KDM2B, and RING1B binding patterns48,49. In mouse embryonic stem cells (mESCs) and developing skin, loss of ubiquitylation resulted in derepression of hundreds of Polycomb target genes46,50–52. However, a significant reduction of ubiquitylation by catalytic mutation of Ring1b in mice or Sce (Ring1a/b ortholog) in flies did not show patterning defects, indicating the maintenance of Hox gene repression53,54. Even fly larval cells replaced with histone H2A mutant that cannot be ubiquitylated did not derepress Hox genes54. Thus, the role for ubiquitylation in maintenance of a repressed state is likely context-dependent55.
Contrary to PRC2 and ncPRC1, cPRC1 modifies chromatin primarily through non-enzymatic mechanisms. cPRC1 is composed of RING1A/B, PCGF2/4, CBX2 (or paralog CBX4/6/7/8), and PHC1 (or paralog PHC2/3) (FIG. 1c). CBX binding to RING1B is mutually exclusive with RYBP56. cPRC1 compacts nucleosome arrays and blocks chromatin remodeling by the mammalian SWItch/Sucrose NonFermentable (mSWI/SNF) complex (also called BAF) in vitro through its CBX subunit9,10,40. In addition, PHC proteins can bridge distant Polycomb bound sites through the oligomerization activity of its Sterile Alpha Motif (SAM) domain57–60. Both CBX and PHC proteins can phase separate with target chromatin in vitro61–63. These self-association properties might contribute to the formation of exclusive nuclear structures called Polycomb bodies, which have been proposed to be refractory to transcriptional activation64,65. Reflecting cPRC1’s chromatin organization activity, PcG bound regions occupy less space than transcriptionally active regions66, and distant PcG bound sites interact with each other67,68. Perturbing cPRC1 function by mutating CBX or PHC subunits resulted in derepression of Polycomb target genes in mESCs69–71 or homeotic transformation of axial skeletons57,69,72–74, suggesting that the structural function of cPRC1 is critical for maintenance of the repressed state.
Stabilization of repressed state by positive feedback of PcG complexes
One model that is largely consistent with the evidence on interplay between Polycomb complexes invokes hierarchical recruitment (FIG. 2)75,76. First, ncPRC1 is recruited to hypomethylated CpG-rich promoters through its KDM2B subunit42,49,77 and ubiquitylates H2A (FIG. 2a). PRC2 then recognizes the H2AK119Ub1 with its accessory component JARID223,78 and catalyzes tri-methylation of H3K27 (FIG. 2b). The EED subunit of PRC2 binds H3K27me3. This interaction further stimulates PRC2 activity, which promotes methylation of neighboring nucleosomes to spread H3K27me3 away from the initial nucleation site (FIG. 2c)19,79. Similar to H3K27me3 spreading, RYBP in ncPRC1 binds H2AK119Ub1, and this interaction stimulates ncPRC1 activity to spread H2AK119Ub1 to neighboring nucleosomes (FIG. 2c)21,80. The CBX subunit of cPRC1 then binds H3K27me3 through its chromodomain11,22,24. CBX and PHC subunits in cPRC1 further bridge and sequester Polycomb bound loci into Polycomb bodies57,59–63. They maintain a high local concentration of PcG complexes and a repressed state. PRC2 activity has been shown to be stimulated when the template contains densely packed nucleosomes20, and ncPRC1 activity was stimulated when the template was compacted with H121, suggesting a compact local chromatin environment might strengthen the feedback loop in this proposed pathway (FIG. 2d)57.
Figure 2 |. Formation of repressed domains by positive feedback of Polycomb complexes.
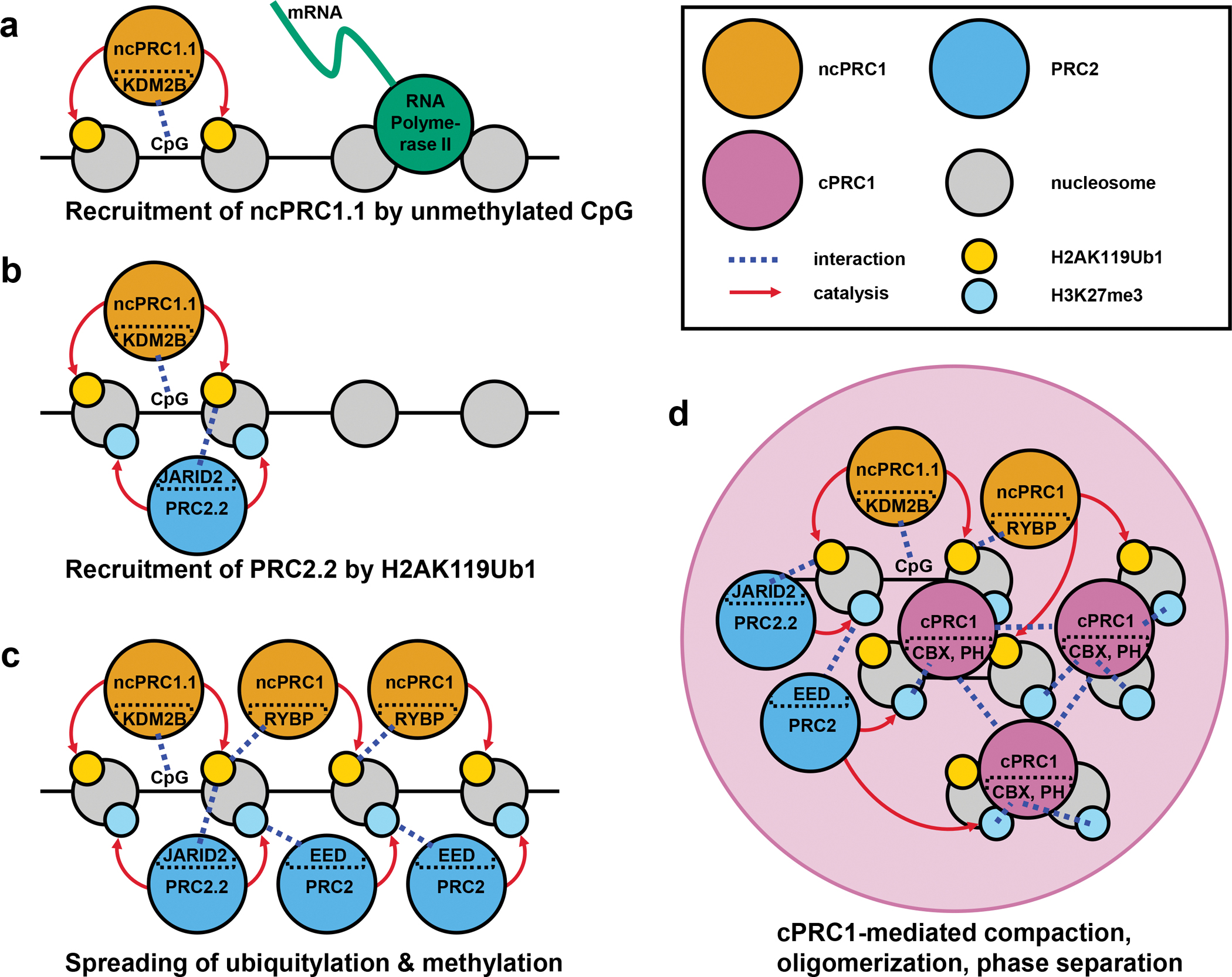
The process of Polycomb repressed domain formation by hierarchical recruitment of PcG complexes is represented. a | ncPRC1.1 can be targeted to hypomethylated CpG-rich promoters through its KDM2B subunit. KDM2B can be targeted to promoters regardless of its transcription status. Note that PRC2.1 can also be targeted to CpG-rich promoters through its PCL subunit, but that is not depicted here for simplicity and to emphasize the interplay between Polycomb complexes. b | PRC2.2 can be recruited to promoters by recognition of H2AK119Ub1 with its JARID2 subunit. Lack of transcription also contributes to PRC2 recruitment. c | H2AK119Ub1 and H3K27me3 modifications can spread beyond the initial recruitment site by 1) RYBP (ncPRC1) interaction with H2AK119Ub1, and 2) EED (PRC2) interaction with H3K27me3. d | The chromodomain in CBX proteins binds H3K27me3 and targets cPRC1 to the H3K27me3 enriched regions. CBX and PH subunits of cPRC1 compact nucleosome targets, bridge distant Polycomb bound regions, and phase separate to form Polycomb bodies. Modification of chromatin structure by cPRC1 can further help maintain high levels of H2AK119Ub1 and H3K27me3 as compacted and dense nucleosomes are better substrates for ncPRC1 and PRC2 enzymatic activities.
This model has been developed based upon functional analyses in vitro, structural analyses, and studies in cell culture done primarily with mESCs. It is not clear to what extent this model accurately depicts PcG function in every cell type. However, the overarching principle is that Polycomb complexes are capable of positive feedback interactions that might facilitate durable maintenance of repression at silenced genes across cell divisions. Thus, we will discuss findings from mESCs and diverse differentiated cells under the framework of this interplay of Polycomb complexes
Diverse Polycomb complexes
Advances over the past decade have identified diverse members of each of the three central Polycomb complexes in different cells, or even in a single cell type. The diversity of sub-complexes in each family is generated by distinct combinatorial assemblies of paralogs of core subunits and/or by inclusion of accessory proteins. How might this diversity impact PcG function?
Multiple cPRC1 complexes can be formed by combinatorial assembly of CBX2/4/6/7/8, PCGF2/4 or PHC1/2/3 proteins. In addition, sub-stoichiometric SCM (SCML1/2, SCMH1) proteins are also found in cPRC1 in different cell types (FIG. 1c). Different paralogs possess distinct molecular activities; for example, CBX4 has SUMOylation activity81, and CBX7 has significantly lower chromatin compaction activity than other CBXs10,82. The genes encoding CBX2/4/8, which are normally expressed in differentiating cells, are found in tandem within a 70kb region of the same chromosome in most mammals. They all have analogous domain structures with similar compaction and phase separation properties suggesting that they might be the result of gene duplications during evolution6,83. However, CBX paralogs have diverged in their binding to H3K27me3 peptides84, chromatin85 and to mitotic chromosomes86. Among the CBX paralogs, only CBX2 has an AT-hook domain and can be targeted to AT-rich pericentromeres in mouse zygotes87. Another component of cPRC1, PHC also normally switches from PHC1 to PHC2 as cells differentiate59,70. As this component mediates long-range chromatin interactions57,59 and is involved in phase separation62, it might contribute to changes in the cPRC1 functions that shape nuclear organization.
ncPRC1 is categorized by the presence of PCGF1/3/5/6 proteins and by specific accessory factors which are associated with each of the PCGF-specific ncPRC1s (FIG. 1b)25. Like CBXs in cPRC1, each PCGF paralog has different molecular characteristics. PCGF1/3/5/6 are more effective in relieving auto-inhibition of RING1B ubiquitylation activity than PCGF2/4 in cPRC143. This difference contributes to the substantively stronger ubiquitylation activity of ncPRC1 compared to cPRC1. Furthermore, RING finger and WD40-associated ubiquitin-like (RAWUL) domains in PCGF2/4 can bind PHC proteins, whereas those of PCGF1/3 cannot88. In fact, sequences of RAWUL domains of PCGF1/3/5/6 are substantially different from PCGF2/4’s, possibly conferring PHC-mediated oligomerization activity only to PCGF2/4 containing cPRC189. Reflecting distinct nature of PCGF2/4, double KO of Pcgf2/4 in developing skin resulted in ectopic formation of Merkel cells, while KO of Pcgf1, Pcgf3/5, or Pcgf6 did not show noticeable defects50. PCGF3/5 are also functionally distinct from other PCGFs. When ectopically recruited to an artificial promoter, PCGF2/4/6 repressed while PCGF3/5 activated luciferase expression in a human cell line90. In addition, PCGF3/5 have longer residence times on the Xist RNA domains of the inactive X-chromosome and are required for ubiquitylation of the inactive X chromosome91. PCGF6 is also unique in that it is required to specifically repress genes involved in meiotic initiation in mESCs92,93.
PRC2 can be divided into PRC2.1 (contains a PCL paralog and mutually exclusive EPOP or PALI) and PRC2.2 (contains AEBP2 and JARID2) (FIG. 1a) (recently reviewed by Yu et al.94). One major difference in these two PRC2 complexes is that accessory subunits provide different targeting mechanisms. PRC2.1 can be targeted to CpG-rich promoters by PCL subunit95,96. PRC2.2 can also be targeted to the CpG-rich promoters but by recognizing H2AK119Ub1 with the JARID2 subunit23,78. PRC2.1 and PRC2.2 act redundantly to deposit H3K27me3 at target genes in mESCs97,98. On the other hand, during neural progenitor cell (NPC) differentiation, PRC2.1 and PRC2.2 repress distinct sets of genes, suggesting context-dependent functions of PRC2 subcomplexes99.
Differences in the composition of subcomplexes in each of the three families can therefore alter both functional characteristics and targeting of the complexes. Paralogs and accessory proteins of Polycomb complexes often show cell type-specific expression patterns (FIG. 3). Do these changes in function impact the cooperative interactions illustrated in the hierarchical model? How might they strengthen the fidelity of PcG function during differentiation processes? We summarize below studies that begin to address these issues, emphasizing that a full characterization of this area is essential to understand maintenance during development but is in its infancy.
Figure 3 |. Examples of cell type-specific Polycomb complexes.
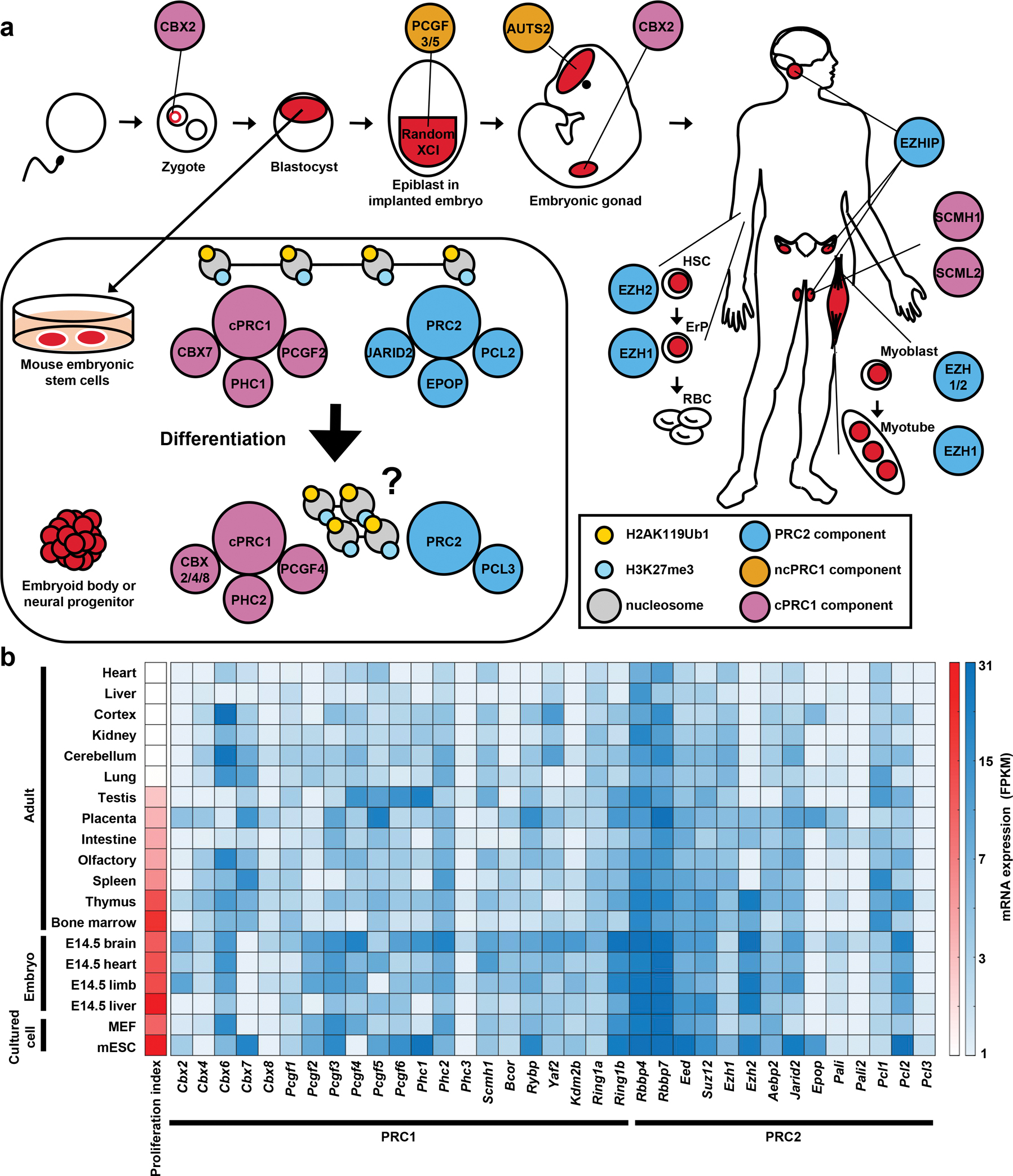
a | A cartoon describing mammalian development shows specific expression or phenotypes of PcG complex components in distinct cell types in embryogenesis or adult tissues. Inset shows paralog switching and accessory component incorporation in the transition from mESCs to differentiated cell types. b | Heat map showing mRNA expression of different PcG genes in the mouse embryo and adult tissues, as well as in cultured cell types, such as MEFs and mESCs. Note that core components of the complexes, such as Ring1b, Eed, show relatively uniform expression, while accessory components or components with many paralogs exhibit more variable or tissue-specific expression. Proliferation index represents how proliferative the cells are in the tissues and is derived from median FPKM of 16 cell cycle genes, including Mcms, Cyclins, Cdks. Data are from Gene Expression Omnibus GSE29278205. XCI, X-chromosome inactivation; HSC, hematopoietic stem cell; ErP, erythroid progenitor; E, embryonic day; MEF, mouse embryonic fibroblast; FPKM, fragments per kilobase million.
Context-specific functions
Polycomb paralogs, overlapping or specific functions?
Paralogous components of Polycomb complexes have some functional redundancy so they can compensate for each other in certain settings. Many PCGF or CBX paralogs possess the same molecular function of supporting ubiquitylation43 or compacting chromatin10, but they vary in efficiency. They are also frequently expressed in the same cell types, as has been shown in mESCs and NPCs by quantitative mass spectrometry100. When these paralogs are expressed in the same cell, the target gene binding patterns largely overlap. A significant fraction of H3K27me3 high-occupancy sites were co-bound by multiple PCGF paralogs in mESCs46,90. In addition, CBX6/7/8 and RING1A/B chromatin binding patterns largely overlapped by ChIP-seq from human fibroblasts101.
These Polycomb complexes with different paralogous components are likely to have redundant functions, as simultaneous knockouts (KOs) of more than one paralog often exacerbate cellular and organismal phenotypes. While Ring1b KO mESCs can maintain the expression of genes associated with pluripotency, Ring1a and Ring1b double KO mESCs spontaneously differentiate and cannot be maintained as stem cells102. Depletion of both Ezh1 and Ezh2 showed stronger derepression of PcG target genes in mESCs103 and stronger defects in hair follicle morphogenesis104 than the Ezh2 single KO. In characterization of genes encoding cPRC1 components, Pcgf2 and Pcgf4 double KO and Phc1 and Phc2 double KO mice showed more severe homeotic transformations of the axial skeletons than the single KOs72,105. ncPRC1s also have redundant functions. Combined KO of Pcgf1/3/5 or Pcgf1/3/5/6 had stronger depletion of H2AK119Ub1 than the single Pcgf KOs in mESCs46. When a PcG protein is absent, sometimes expression of the other paralog is upregulated. RING1A was upregulated post-transcriptionally in Ring1b mutant mESCs102,106 and in oocytes107. However, paralogous protein levels were not always upregulated by the loss of the other as in the examples of some PCGFs46. Often the amount of remaining paralog-containing Polycomb complexes is sufficient to provide compensatory function. Even different classes of Polycomb complexes can provide redundant functions, as combined KO of PRC1 and PRC2 exacerbates cellular and gene expression phenotypes108–110.
Cell type-specific mutant phenotypes were the first indication that the Polycomb paralogs may have unique functions (FIG. 3a). Many viable Polycomb mutant mice show a homeotic transformation phenotype57,69,72–75,92,105,111, which supports their conserved role in the maintenance of Hox gene repression in early development. However, in addition to the axial patterning defect, Cbx2 KO mice showed a male-to-female sex reversal phenotype, suggesting CBX2 has a specialized role in the suppression of female pathway in the embryonic gonad74,112. Furthermore, Pcgf3/5 double KO mice showed that only female embryos were severely degraded at mid-gestation with malformed placentas, while male embryos looked seemingly normal at the same stage91. This female-specific phenotype is likely due to failure of X-inactivation. Indeed, Pcgf3/5 double KO mESCs showed defects in Xist-mediated chromosome-wide gene silencing by RNA-seq91. On the other hand, KO of accessory subunits of cPRC1, Scmh1 or Scml2 showed male-specific sterility111,113,114.
Distinct cellular responses driven by different paralogous components were also shown by overexpression studies. PCGF4 was first isolated as an oncogene to drive B cell lymphomagenesis when overexpressed, even before it was identified as a part of Polycomb complexes115,116. Another cDNA overexpression screen isolated CBX7 to block replicative senescence of human prostate epithelial cells117. CBX7, but not CBX2/4/8 drove proliferation and contributed to stem cell potential and leukemogenesis when overexpressed in hematopoietic stem/progenitor cells118. CBX7 overexpression also maintained mESCs close to the pluripotent state without Xist expression71. These data suggest that CBX7’s major function is to maintain an undifferentiated state, which is consistent with its high expression in mESCs70,71. Other CBX proteins can perform similar roles in different contexts. When CBX4/6/7/8 were each expressed in human primary keratinocytes, only CBX4 induced the cells to remain in a quiescent and undifferentiated state119. On the other hand, CBX8 was unique in its ability to drive MLL-AF9 fusion leukemogenesis or mammary tumorigenesis120,121.
These loss-of-function and overexpression data suggest clear non-overlapping, context-specific functions of different Polycomb paralogous proteins. Distinct complexes containing different paralogs might have 1) different abilities in covalently modifying or altering chromatin structure, 2) possible underappreciated roles in direct gene regulation, 3) differential targeting to specific genes, or 4) a combination of these properties. The challenge will be to understand which of the changes listed above drive diverse phenotypic outcomes. The key conclusion is that mammalian PcG complexes are not only diverse, but that diversity is important for normal development.
Paralog switching and cell type-specific components
In line with cell type-specific paralog functions, there are many cases in which one paralog is replaced with another during cell fate transitions (FIG. 3a). One well studied example is the differentiation of mESCs to embryoid bodies, neural, or mesoderm progenitors. CBX7 is the dominant CBX component in mESCs, and it is replaced by CBX2/4/8 in embryoid bodies70,71. CBX7 is also replaced by CBX2 during the differentiation of mESC to early cardiac mesoderm122. Similarly, PHC1, the major component of cPRC1 in mESCs is replaced with PHC2 in NPCs59,70. PCGF proteins also transition from being PCGF1/6-dominant in mESCs to PCGF1/4-dominant in NPCs100. PRC2 can undergo EZH2 to EZH1 subunit switching in hematopoietic stem/progenitor to erythroid progenitor differentiation123. During skeletal muscle differentiation, both EZH1 and EZH2 are expressed in proliferative myoblasts, while EZH1 expression becomes dominant in post-mitotic myotubes124. In general, EZH2 expression is restricted to embryonic and proliferative tissues, whereas EZH1 expression is more ubiquitous, albeit in lower levels than EZH2 (FIG. 3b)125,126. Consistent with their expression pattern, EZH2 is required for embryonic development, whereas EZH1 is dispensable104,127. Similarly, RING1B but not RING1A is specifically required for embryogenesis128,129. Expression of mESC-specific paralogs, such as CBX7 that cannot compact chromatin10,82, may contribute to the unique characteristics of pluripotency, when chromatin is hypothesized to be more fluid130. As mESCs differentiate into specific lineages, cPRC1 subunits such as compaction-capable CBXs are upregulated, and it has been proposed that this functional change consolidates gene expression changes. Knocking the domain responsible for chromatin compaction from CBX2 into CBX7 disrupted the ability of mESCs to differentiate properly82, suggesting the importance of forming a cPRC1 complex with the appropriate characteristics.
mESCs are also characterized by the strong expression of many accessory components of PRC2 (FIG. 3a and 3b). mESCs have high expression of PCL2 (MTF2), which is replaced by PCL3 (PHF19) during NPC differentiation100. JARID2 is also highly expressed in mESCs or in pluripotent embryonic carcinoma cells and downregulated during embryoid body or neuronal differentiation131,132. EPOP is another PRC2 component with high expression in mESCs and inner cell mass of mouse embryos133. In fact, mESCs are distinct in that they have much higher levels of core PRC2 components than differentiated cells100,134. It has been hypothesized that this higher expression of PRC2 components may be a characteristic of pluripotent cells, which may need higher levels of de novo PRC2 recruitment for their uncommitted state100. Alternatively, the fast cell cycle of mESCs may demand more PRC2 to restore H3K27me3 levels before next replication100.
Cell type-specific subunits also confer unique properties to PcG complexes (FIG. 3a). Like PHC, the SCM proteins have SAM domains, which can form helical polymers in vitro, and are required for PcG target gene repression by bridging different PcG complexes135,136. SCMH1, a sub-stoichiometric component of mammalian cPRC1, showed testis-enriched expression, and is required for preventing mistargeting of cPRC1 to the XY-body (a male germ cell-specific X chromosome silenced compartment) in spermatocytes111. SCML2 is another testis-enriched component of PRC1 required for prevention of somatic gene expression in progenitor spermatogonia113. Interestingly, SCML2 is also involved in blocking H2AK119 ubiquitylation in the XY-body of differentiated spermatocytes113,114. The fly ortholog, Scm, is specifically upregulated in differentiating female germ cells (nurse cells) and is required for the formation of punctated H3K27me3 domains and fertility137.
Cell type-specific components can reduce the repressive activity or even provide an opposing, gene activation role for PcG complexes. AUTS2 is undetectable in mESCs but is upregulated in motor neurons, and forms ncPRC1.3 and ncPRC1.5 with PCGF3/5, P300 and CK2138,139 (FIG. 1b). AUTS2-ncPRC1.3 is required for neuronal gene activation, and a transcription factor, NRF1 is required for AUTS2-ncPRC1.3 chromatin targeting139. ncPRC1.3 in mESCs required a different transcription factor, USF1 for chromatin targeting, suggesting cell type-specific recruitment mechanisms90. In resting B lymphocytes, Aurora B kinase binds RING1B and inhibits H2AK119 ubiquitylation activity at active promoters. Both proteins are required for transcription of active genes140. EZHIP (also called CATACOMB141) is a placental mammal-specific accessory component of PRC2, which can inhibit PRC2 activity141–143. EZHIP expression is enriched in both male and female germ cells143 and is also upregulated in posterior fossa type A (PFA) ependymoma144. Consistent with its gonad-specific expression, Ezhip KO females had a progressive decrease in the number of follicles and showed lower fertility143.
Even without explicit subunit switching, cellular differentiation and the associated chromatin environment changes can demand different Polycomb functions. For example, in mouse neocortex development, ubiquitylation by ncPRC1 was required for gene repression only in the early neurogenic phase, but was dispensable in the late astrogenic phase. The oligomerization activity of PHC2 was instead required for repression of the same set of genes in the astrogenic phase145.
In sum, paralogous Polycomb proteins clearly play redundant roles, providing a critical safeguarding mechanism for Polycomb-mediated gene silencing. At the same time, these paralogs and accessory proteins possess unique molecular functions and are expressed in a cell type-specific manner, contributing to the diversified cellular behaviors in developing mammalian cells. The simplest hypothesis to explain the biological role for these changes in activity is that regulation of cell type-specific genes in distinct lineages has evolved to require specific functions that are only provided by certain PcG paralogs.
Atypical genomic distributions
Atypical H3K27me3 in early development and redistribution of PcG complexes
Dynamic changes in PcG function in different cellular contexts are further highlighted by atypical genomic distributions of PcG proteins and histone modifications early in development. Furthermore, in certain perturbation conditions, PcG proteins are redistributed from the usual sites to ectopic loci, suggesting correct allocation of limited amounts of PcG proteins is a key aspect of PcG regulation.
PcG proteins and their associated histone modifications are usually found at promoters and a subset of strong PcG enrichment sites, spread out across tens of kilobases. However, in mouse preimplantation embryos, the usual promoter H3K27me3 is depleted on both the maternal and paternal genome146. Before activation of zygotic genome transcription, developing fly, zebrafish, and human embryos also showed loss of H3K27me3 enrichment at promoters147–149, indicating that the erasure and then re-establishment of promoter H3K27me3 is conserved across species. Additionally, in mouse oocytes and in preimplantation embryos, H3K27me3 is found in broad intergenic regions with little DNA methylation and transcription (FIG. 4a)146. The broad intergenic maternal H3K27me3-modified loci in mouse oocytes and early embryos form self-interacting domains that are dependent on PRC1 and not on the more commonly observed genome organizing protein cohesin150. H2AK119Ub1 also showed atypical broad distribution that foreshadows H3K27me3 pattern in mouse oocytes and early embryos151,152. Reflecting these events on intergenic regions, H3K27me3 and cPRC1 are enriched at paternal but not maternal pericentromeres in one-cell mouse zygotes, further underscoring plasticity of PcG functions in different regulatory contexts87,153.
Figure 4 |. Redistribution of Polycomb complexes.
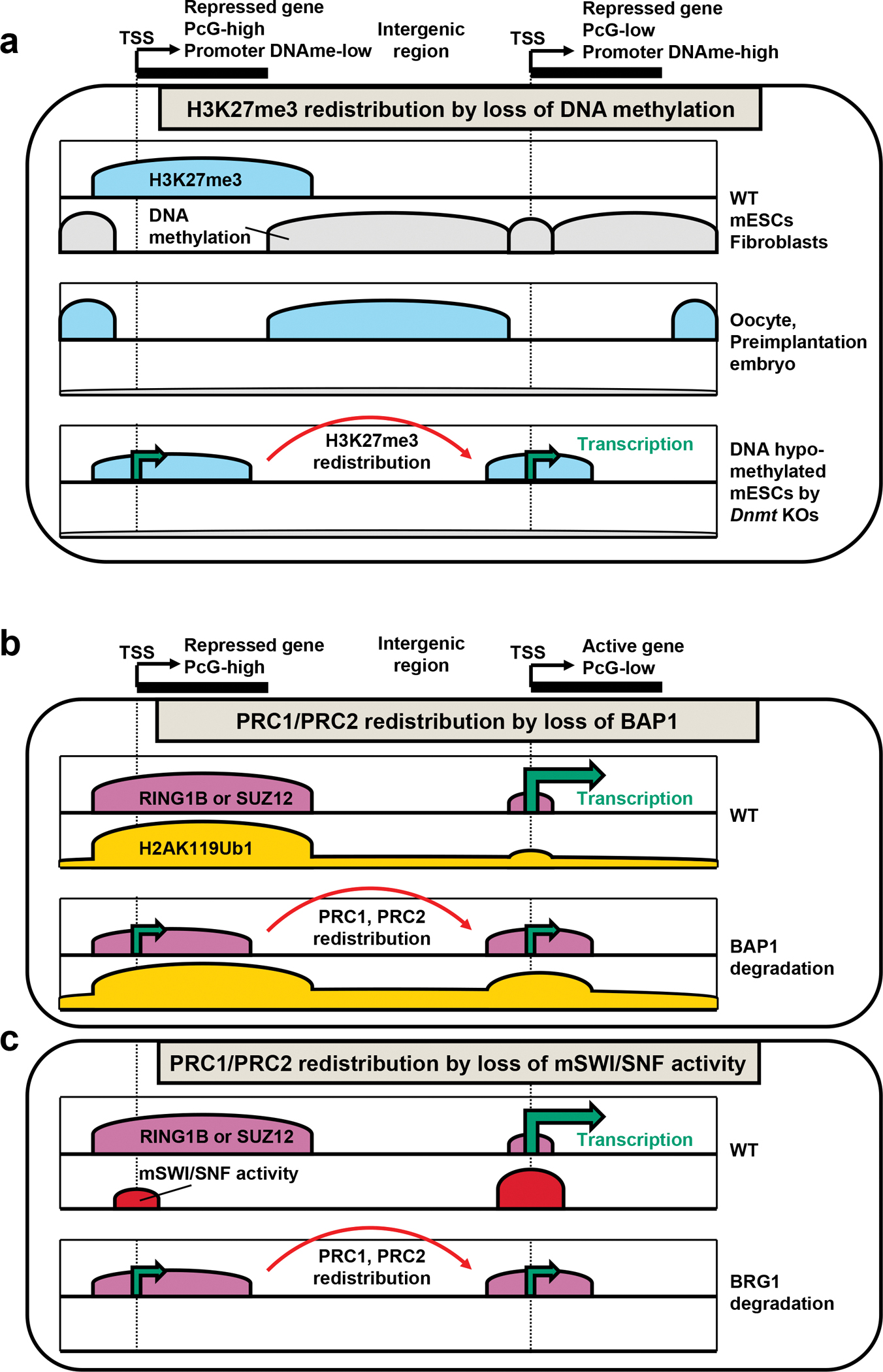
Schematic representation of PcG complexes and related DNA and histone modifications in a genome browser format to show the level and breadth of enrichment. a | Loss of DNA methylation: while one class of promoters (PcG-low, DNA-methyl high) gain H3K27me3 as they lose DNA methylation, another class of promoters (PcG-high, DNA-methyl low) show decrease in H3K27me3, suggesting potential redistribution of PRC2 from PcG-high to PcG-low promoters by global loss of DNA methylation. In oocytes and preimplantation embryos with developmentally regulated genome-wide DNA hypomethylation, H3K27me3 undergoes global remodelling to localize at intergenic regions. b | Loss of H2AK119Ub1 deubiquitylase BAP1: Global increase in H2AK119Ub1 level results in redistribution and decrease of PRC1(RING1B) and PRC2(SUZ12) from promoters normally have high levels of PcG complexes, accompanied by gene derepression. c | Loss of activity of the opposing chromatin modifying complex, mSWI/SNF: degradation of BRG1, the catalytic component of mSWI/SNF results in the increase of PRC1(RING1B) and PRC2(SUZ12) occupancy at usually PcG-low promoters, accompanied by the decrease of PRC1 and PRC2 from normally PcG-high promoters and gene derepression. TSS, transcription start site; DNAme, DNA methylation; Dnmt, DNA methyltransferase.
What is the utility of this atypical organization and how is it regulated? DNA hypomethylation is hypothesized to be one mechanism for the unusual Polycomb domain expansion in intergenic regions in oocytes and preimplantation embryos. DNA methylation antagonizes binding of a targeting component of ncPRC1.1, KDM2B, to the CpG-rich sequences (e.g. CpG islands)42,49,154. Furthermore, the degree of CpG methylation at CpG islands anti-correlates with H3K27me3 levels in human ES cells155. When mESCs or MEFs were severely hypomethylated by knocking out DNA methyltransferases, ectopic H3K27me3 domains appeared in previously CpG-methylated promoters and intergenic regions155–157. The ectopic accumulation of H3K27me3 was accompanied by the decrease of H3K27me3 in normal target sites156,157, suggesting formation of ectopic H3K27me3 domains may dilute away Polycomb complexes from the normal targets (FIG. 4a). However, when PcG proteins are redistributed in developmentally regulated settings, such as in oocytes or in preimplantation embryos, this atypical localization is hypothesized to have a role for compartmentalizing the maternal genome for efficient repression of non-canonical targets, such as transposons150. In addition, this maternally-inherited H3K27me3 was critical for repression of maternal alleles for a small group of genes including Xist, providing a DNA methylation-independent means of imprinting158–161. On the other hand, human embryos appear to lack H3K27me3-mediated imprinting, indicating divergent usage of Polycomb mechanisms even within mammalian species149.
PcG complexes can also be redistributed following disruption of opposing chromatin modifying proteins. Proper levels of H2AK119Ub1 deposition are maintained by the balance between ubiquitin ligase and deubiquitylation activities. BAP1 is an evolutionarily conserved H2AK119Ub1 deubiquitylase, and loss of BAP1 in mESCs resulted in the increase of the genome-wide H2AK119Ub1 level162,163. However, some genes with strong Polycomb enrichment showed a decrease in PRC1(RING1B) and PRC2(SUZ12) occupancy with accompanying derepression of gene expression, potentially because PcG complexes were titrated away from high-occupancy targets when H2AK119Ub1 level increased genome-wide (FIG. 4b)162,163. mSWI/SNF complexes also contribute to concentrating Polycomb complexes at target loci. BRG1 is the enzymatic component of the mSWI/SNF chromatin remodeling complexes, which can counteract Polycomb repression164–166. Acute depletion of BRG1 in mESCs paradoxically resulted in derepression of many highly Polycomb-bound genes including HoxA and HoxD genes167. ChIP-seq revealed that PRC1(RING1B) and PRC2(SUZ12) were redistributed from high-occupancy sites to low-occupancy sites, resulting in the derepression of Polycomb high-occupancy genes (FIG. 4c)167. The effects of both BAP1 and BRG1 depletion underscore the importance of balancing the genomic distribution of the finite amount of PcG complexes to appropriate targets.
These examples of PcG redistribution suggest that PcG proteins’ nuclear concentrations might be maintained at limiting levels to ensure proper regulation. In line with this, PcG genes show dosage sensitive phenotypes when mutated, and overexpression also disrupts PcG functions168–170. In addition, wild-type CBX2, a component of cPRC1, cannot be significantly overexpressed in mESCs, while a compaction and phase separation-deficient mutant CBX2 can171. Since concentration is an important factor in determining genomic distribution, overall enzymatic activity, and non-enzymatic functions such as condensation172, one hypothesis is that regulation of PcG concentration might be critical to its role in genome organization.
Maintenance of Gene Repression
A defining feature of the Polycomb system is that PcG proteins are frequently required to maintain, rather than establish, gene silencing during differentiation. In both flies and mice, expression of Hox boundaries were initially correctly established, but PcG gene mutant embryos failed to maintain boundaries of Hox repression3,105. Recruitment of the PcG proteins is responsive to changes in gene expression, as exemplified by findings that PRC2 can be passively recruited to CpG-rich promoters by transcriptional inactivity34,155,173,174. Active recruitment of the PcG machinery also occurs. For example, ncPRCs can be recruited to chromatin through sequence-specific DNA binding proteins such as USF1 for ncPRC1.3 or MAX/MGA and E2F6 for ncPRC1.6 in mESCs90,92,175. However, findings from many developmental transitions suggest that a key role for the PcG complexes is to stabilize acquired cell fates established by changes in gene expression. Among many examples, when H3K27me3 is depleted, embryoid bodies are more likely to adopt an earlier mESC-like state when cultured in mESC media176, and pancreatic β-cells dedifferentiate to an immature state177. In addition, Ring1a/b double KO astrogenic neural progenitors aberrantly adopt an earlier neurogenic state178, and E(z) (ortholog of Ezh1/2) mutant fly female germ cells cannot acquire oocyte fate and instead transdifferentiate to nurse-like cells179.
Divergent roles of PcG complexes in the maintenance of repression
How might ncPRC1, PRC2 and cPRC1 be involved in maintenance? One straightforward model is that the hierarchical recruitment pathway elucidated in mESCs (FIG. 2) is recapitulated at each cell division in stably differentiated tissues, thereby recreating a PcG repressed pattern on the genomes of daughter cells. By this model, the interplay between these three complexes would remain when pluripotent cells become differentiated, however changes in function of components of these complexes (e.g., changes in CBX or PCGF proteins) would change the balance of activities in this cascade, impacting maintenance. Genetic analysis implies that ncPRC1, PRC2 and cPRC1 might all be required for maintenance although the extent of contribution differs between these three complexes in different cellular contexts. We summarize current data on the roles for each family of complexes and relate them to the hierarchical model. We emphasize the hypothetical nature of any model for maintenance at the current time as this is a developing area in whole organisms.
A role for PRC2 in maintenance is attractive because transmission of the H3K27me3 modification to daughter cells might ensure the transmission of the entire PcG machinery. However, loss of function analyses indicate that cellular division leads to a decrease in H3K27me3 by replicative dilution and ensuing gene derepression even in the presence of functional PRC2. For example, when de novo recruitment of PcG complexes was lost by excision of the Polycomb Response Element (PRE) in fly larvae, after several cell divisions H3K27me3 was depleted, and Hox genes were derepressed in wing discs180,181. In contrast, when proliferation of the PRE-excised cells was blocked by developmental arrest or drug treatment, H3K27me3 levels and gene repression were maintained180,181. The fact that H3K27me3 cannot be maintained without a PRE suggests that the mark alone is not sufficient for self-maintenance over replication and requires additional de novo recruitment mechanisms.
Replicative dilution of H3K27me3 has been observed in mammalian systems, although this has largely been studied in the context of PRC2 disruption or inhibition. In mouse intestinal stem cells, it took several cellular divisions over 21 days to completely lose H3K27me3 and derepress PcG target genes after deletion of PRC2182. Cellular proliferation was also required for derepression in cultured lymphoma cells after inhibition of PRC2 enzymatic activity, and blocking cell cycle significantly delayed derepression182. Importantly, in these studies, the derepression kinetics were distinct in different cell types and for different genes in the same cell type. Basal levels of activating transcription factors180 or promoter H3K4me3 level were proposed to be the determinants of the derepression potential183. Numerous genetic studies, previously reviewed elsewhere94, imply that PRC2 is necessary for maintenance, but the studies above suggest that PRC2 alone is not sufficient to generate memory and likely works with PRC1 family complexes.
The loss of memory can be stochastic. This observation came from studies on cPRC1 components and supports a key role for cPRC1 in faithful memory. The ability of cPRC1 to compact and phase separate increases the concentration of PcG components in the condensates (Polycomb bodies), which might help preserve memory by increasing local concentrations of PcG machinery to allow reformation of repressive structures across cell division. Fly cPRC1 remains bound to replicated chromatin in vitro184, and mammalian cPRC1 component CBX2 associates with mitotic chromosomes in cells86; both of these may assist in the re-formation of condensates during cell division. During vernalization of plants, FLC, a repressor of flowering, is silenced by the Polycomb system. When a chromodomain protein LHP1, a putative functional ortholog of the mammalian CBX protein, was mutated, FLC became de-repressed, but sporadically in a few clusters of distinct cells in the roots (FIG. 5c)185. Even within a single cell, one allele could be derepressed while the other allele remained silent, suggesting the memory is stored in cis185. Mammalian cells also showed stochastic derepression by loss of cPRC1. When CBX7 was ectopically recruited and then released from a fluorescent reporter locus, mESCs showed a bimodal state of the reporter reactivation, with more than half of the cells maintaining full repression after 10–12 cell divisions106. Similar stochastic derepression was also observed in human cells with live-imaging studies that monitored reporter gene reactivation after the release of tethered PRC2186. In addition, certain PRC2 mutant flies showed small patches of posteriorly transformed cells, reminiscent of the sporadic FLC derepression seen in plants187. Even when PcG was functional, transgenic flies carrying PREs occasionally showed variegated silencing of associated reporters in eyes and wing discs188,189. This stochastic, all-or-nothing response of the maintenance of repression reflects the bistable nature of Polycomb-mediated gene silencing by competition between repressors (PcG) and activators (trithorax group) (FIG. 5c and 5d)190,191. Polycomb mutant flies and mice are known to have variable phenotypes73,192–196. In extreme cases, the same Scmh1 mutant mice exhibit phenotypes ranging from infertile by loss of post-meiotic germ cells to fertile and indistinguishable from the wild type111. The stochastic nature of Polycomb-mediated memory provides one possible mechanism for the variable penetrance.
Figure 5 |. Hypothetical models on different roles of Polycomb complexes in the maintenance of gene repression.
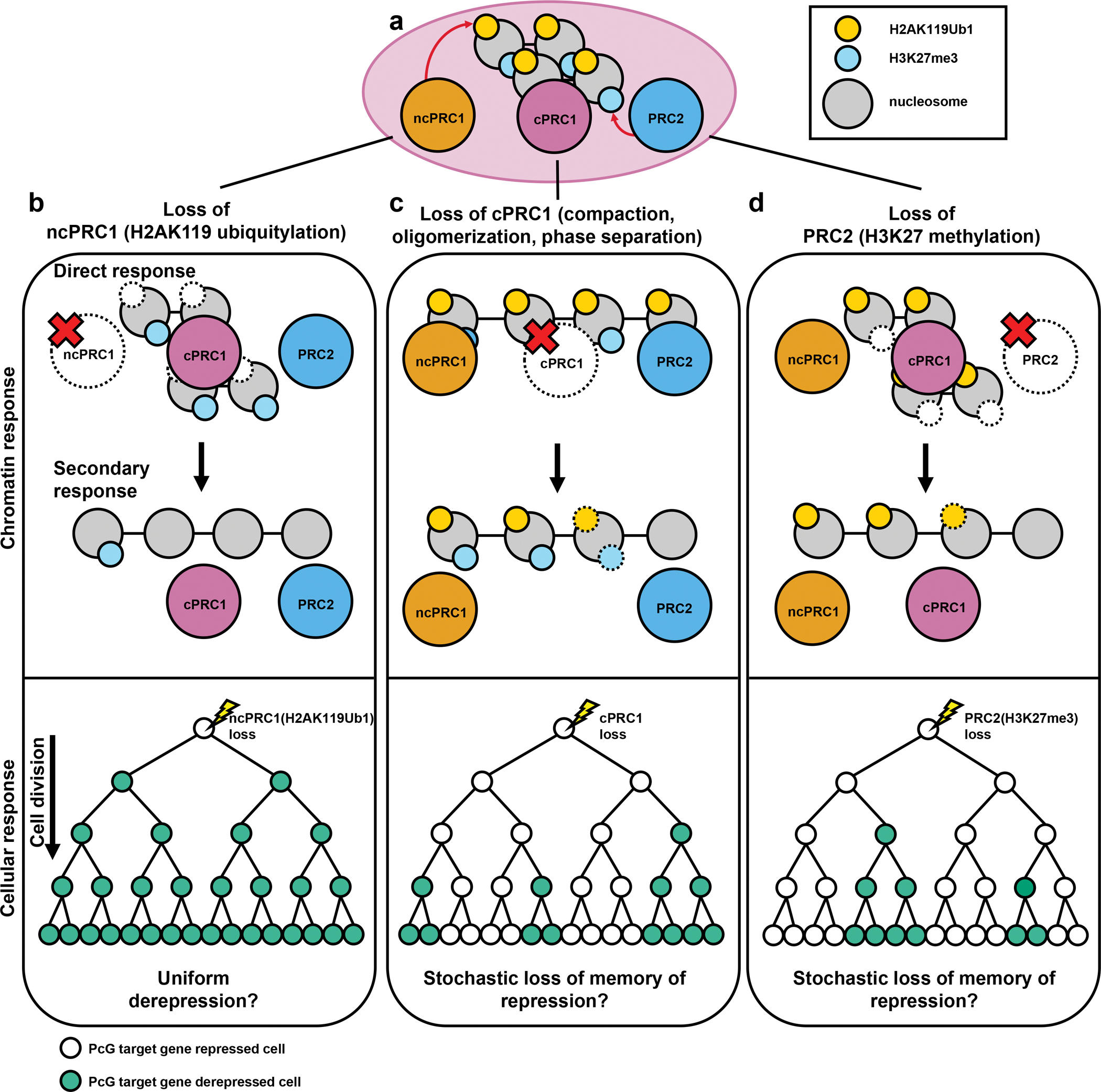
a | A schematic of a repressed Polycomb domain with H2AK119Ub1 and H3K27me3 modifications. b | Chromatin response (upper): When ncPRC1 activity is disrupted, initially H2AK119Ub1 is lost. Because H2AK119Ub1 recruits PRC2.2, over time H3K27me3 deposition and in turn cPRC1 recruitment is decreased. Cellular response (lower): It is possible that ubiquitylation is involved in direct gene repression. Therefore, loss of H2AK119Ub1 may result in fast and uniform derepression of PcG target genes. c | Chromatin response: When cPRC1 activity is lost, chromatin compaction and long-range interactions between PcG bound regions are disrupted. Because compacted and dense nucleosomes are better substrates for ncPRC1 and PRC2, loss of cPRC1 may eventually lead to decrease in H2AK119Ub1 and H3K27me3 levels over long term. Cellular response: Unlike ncPRC1, existing data are consistent with cPRC1 being involved in the memory of repression, resulting in slower and stochastic conversion to the “on” state of PcG target genes by loss of cPRC1 function. d | Chromatin response: When PRC2 activity is disrupted, H3K27me3 level is decreased at first, followed by the decrease in the cPRC1. Resulting disruption of Polycomb domain structure will negatively impact in reaching full H2AK119Ub1 level. Cellular response: Similar to cPRC1, PRC2 and H3K27me3 may also be involved in the memory of repression, resulting in slower and stochastic conversion of PcG targets to the “on” state by loss of PRC2 function.
Not all Polycomb mutants show stochastic, bimodal pattern of target gene derepression. These observations imply that different PcG complexes may contribute to distinct aspects of maintenance. When RING1B was degraded in mESCs using a degron system, most of the cells showed uniform derepression of Polycomb target genes37. Unlike PRE or PRC2 mutants where derepression occurred over the course of multiple cell divisions, RING1B degradation resulted in almost immediate derepression, occurring two hours from the induction of degradation37. This might reflect the fact that RING1B is essential for formation of both ncPRC1 and cPRC1 and eliminating both complexes might generate a severe phenotype in a short period. Alternatively, the fast derepression may indicate that the role of ubiquitylation catalyzed by RING1B (mostly by ncPRC1) is essential for the direct repression of genes (FIG. 5b). Supporting ncPRC1 and H2AK119Ub1’s role in the direct repression, inducible loss of H2AK119Ub1 by conditional catalytic point mutations of RING1B resulted in rapid derepression of PcG target genes within 72 hours52. In addition, release of tethered RYBP (ncPRC1) from an ectopic fluorescent reporter locus showed uniform derepression of the reporter within 6 days106. Studies in flies also support divergent roles of PcG complexes in the maintenance of repression197, while exact molecular functions of orthologs can be different between species10,198. When mutant clones were made in fly larval wing discs, all the clones showed fast, strong derepression in Psc (Pcgf) or Ph (Phc) mutant clones, while slower and variable derepression was observed in Pc (Cbx) or E(z) (Ezh) mutants197.
Overall, the Polycomb system provides maintenance of gene silencing, which can be inherited across cell divisions. The varying kinetics and heterogenous cellular responses by disrupting different Polycomb complexes might reflect the two distinct core features of the Polycomb system, direct repression and memory/maintenance. Mutations that impact direct repression might be expected to show widespread levels of derepression while mutations that impact memory might be expected to show stochastic derepression. For example, if a memory function went from nearly 100% effective in wild-type cells to 90% effective in cPRC1 or PRC2 mutant cells, that decrease to 90% effectiveness would result in 50% of cells with derepression after 6–7 cell divisions and thus disrupt normal development. In contrast, components involved directly in repressing gene expression, such as ncPRC1, might be required for that repression on all cells. Uncoupling direct repression and memory remains a hypothesis that is difficult to test in animals as perturbation of one aspect of PcG complex influences other complexes. Furthermore, it is unlikely that each PcG complex’s role is exclusive to one aspect of gene silencing; rather, it could be that cPRC1 and PRC2 have more pronounced roles in the ‘stabilization’ of silencing than ncPRC1. Nevertheless, it will be critical to investigate the distinct and occasionally stochastic nature of gene derepression by dysregulation of Polycomb components to better understand pathogenesis of diseases associated with Polycomb mutations.
Conclusions and future perspectives
In the past decade, many specialized Polycomb complexes have been identified, and their molecular functions have been characterized. What has emerged from recent work is a defined model for the interactions between the three major complexes central to full repression of gene expression in pluripotent cells. Studies also have uncovered that these complexes change components and modify their function as cells differentiate. These variations in composition and function can be lineage-specific. Understanding the divergent roles of these complexes in direct repression to the memory maintenance in differentiated cells is essential for understanding stabilization of cell fate.
With our expanded understanding of Polycomb mechanisms, there are new challenges and opportunities ahead. How do the mechanisms identified through biochemical or cell culture experiments play out in the development of complex animals? For example, does hierarchical recruitment of ncPRC1 to PRC2 to cPRC1 identified in mESCs happen in every cell type in every cell division? Consistent with the hierarchical recruitment model, ncPRC1.1-mediated H2AK119Ub1 was required for PRC2-mediated H3K27me3 during the differentiation of mESCs to embryoid bodies199. In addition, deposition of H2AK119Ub1 preceded H3K27me3 in mouse preimplantation embryos151,152 and in zebrafish embryos before zygotic genome activation200, and also in an inducible X-inactivation system201. However, in growing oocytes without cell divisions, there were minimal changes in H3K27me3 when H2AK119Ub1 was lost by Pcgf1/6 dKO152. Human hematopoietic stem cells divide every several months, while zebrafish embryonic cells divide every 16 minutes, how do these dramatic differences in timing impact necessary mechanisms? Further studies, in many contexts, will be needed to investigate, generalize, and refine the models discussed here. Finally, little is known about the behaviour of Polycomb bodies in the context of whole animals. These Polycomb domains contain high local concentrations of Polycomb complexes. Which gene targets reside in these bodies? Do these separated structures add an additional dimension in self-reinforcing mechanism and consolidation of memory? Formation and dissolution of Polycomb bodies are developmentally controlled137,202, and there will be many surprises as we further our understanding of their behaviour in their native contexts. In summary, analyses of the different requirements of diverse lineages in distinct settings will be required to understand the full scope of PcG mechanisms that are at play in complex organisms.
Technological advances will enable us to tackle these important questions. PcG complexes positively influence each other, thereby perturbing one component invariably impacts other PcG complexes as well. In addition, compensation by other paralogous proteins makes it difficult to observe the effects clearly in genetic loss-of-function studies. Acute depletion of target proteins using inducible genetic deletion or degron-based approach and evaluating immediate responses will be helpful to resolve these issues. Sequencing and microscopy-based single cell approaches in the whole animal will be helpful to identify critical cell populations and stages that require PcG complexes and to assess stochastic responses by disruption of Polycomb function. Many new low-input chromatin profiling methodologies will be crucial to assess genome-wide chromatin changes in those critical small populations. Importantly, simultaneous advances in the mechanistic understanding based on live-single molecule tracking, biochemistry and structural biology will be the foundation for these in vivo studies.
Biochemistry and genetics have been successfully integrated to advance our knowledge of the Polycomb system. With an even deeper understanding of the molecular mechanisms and by applying the principles to developing cells in the organisms, the Polycomb system will provide fruitful ground for future unexpected discoveries and will continue to serve as a paradigm of epigenetic regulation of cell fate.
Acknowledgements
We thank J. Zhu, M. Mauger, E. Steinson, J. Wucherpfennig, U. Cho and E. Grow for critical reading of the manuscript. This work was supported by NIH grant R35-GM131743 to R.E.K.
Glossary
- Nucleosome array:
In vitro reconstituted chromatin template used to study biochemical properties of chromatin modifying proteins, which is made from DNA with nucleosome positioning sequences and linker mixed with histone octamers.
- mSWI/SNF complex:
A protein complex that can destabilize histone-DNA interactions in an ATP-dependent manner. It can create accessibility to DNA and counteract Polycomb-mediated repression.
- Phase separation:
A phenomenon where proteins transition to another phase with different physicochemical properties, often through multivalent interactions among themselves. Potentially one of the driving forces to form membranelles organelles and condensates in the cell.
- Homeotic transformation:
A class of mutant phenotypes that a body segment transforms into another body segment usually by misregulation of Hox genes, such as fly Antennapedia mutant producing legs instead of antennae.
- Paralog and ortholog:
From an ancestral gene, paralogs are derived by gene duplication events within the same species, whereas orthologs are derived by speciation events (therefore orthologs are present in different species). Paralogous proteins can retain similar functions, but they can also acquire distinct functions. The same is true for orthologs.
- Preimplantation embryo:
Placental animal embryos from zygote to prior to implantation stages. First lineage specification between inner cell mass (gives rise to embryo proper) and trophectoderm (placenta) cells happens during this stage.
- Pericentromere:
A region of chromosome adjacent to the centromere, composed of AT-rich satellite DNA tandem repeats, usually DNA-methylated and decorated with H3K9 methylation.
- CpG islands:
A stretch of ~1kb DNA region in vertebrates with overrepresentation of CpG dinucleotides than the genome average. They are often a site of transcription initiation, and more than half of annotated gene promoters are CpG islands.
- Polycomb Response Element:
Discrete regulatory DNA element that can nucleate Polycomb complexes recruitment and silencing in flies.
- Vernalization:
A process of prolonged exposure to the cold that induces flowering in plants. Genetic screens to find genes required for vernalization uncovered a number of genes later identified to be part of plant Polycomb complexes.
- Zygotic genome activation:
After fertilization, transcription is absent in the zygotic genome, therefore embryos develop with maternally provided transcripts. Zygotic genome activation happens as maternal mRNA decays, and genes are transcribed from the zygotic genome through the process called maternal-to-zygotic transition.
- Trithorax group (trxG):
A group of chromatin regulators that maintains an active state of gene expression that includes mSWI/SNF complex. Genes encoding trxG proteins were originally discovered by a genetic suppression screen in flies to suppress the Polycomb mutant phenotype.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Lewis PH Pc: Polycomb. Drosoph Inf Serv 21, 69 (1947). [Google Scholar]
- 2.Slifer EH A mutant stock of Drosophila with extra sex-combs. Journal of Experimental Zoology 90, 31–40, doi: 10.1002/jez.1400900103 (1942). [DOI] [Google Scholar]
- 3.Struhl G & Akam M Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J 4, 3259–3264 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wedeen C, Harding K & Levine M Spatial regulation of Antennapedia and bithorax gene expression by the Polycomb locus in Drosophila. Cell 44, 739–748, doi: 10.1016/0092-8674(86)90840-8 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Zink B & Paro R In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature 337, 468–471, doi: 10.1038/337468a0 (1989). [DOI] [PubMed] [Google Scholar]
- 6.Whitcomb SJ, Basu A, Allis CD & Bernstein E Polycomb Group proteins: an evolutionary perspective. Trends Genet 23, 494–502, doi: 10.1016/j.tig.2007.08.006 (2007). [DOI] [PubMed] [Google Scholar]
- 7.de Napoles M et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7, 663–676, doi: 10.1016/j.devcel.2004.10.005 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Wang H et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878, doi: 10.1038/nature02985 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Francis NJ, Kingston RE & Woodcock CL Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577, doi: 10.1126/science.1100576 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Grau DJ et al. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev 25, 2210–2221, doi: 10.1101/gad.17288211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao R et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043, doi: 10.1126/science.1076997 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Czermin B et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196, doi: 10.1016/s0092-8674(02)00975-3 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P & Reinberg D Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16, 2893–2905, doi: 10.1101/gad.1035902 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller J et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208, doi: 10.1016/s0092-8674(02)00976-5 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Boyer LA et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353, doi: 10.1038/nature04733 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Bracken AP, Dietrich N, Pasini D, Hansen KH & Helin K Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20, 1123–1136, doi: 10.1101/gad.381706 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz YB et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38, 700–705, doi: 10.1038/ng1817 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Tolhuis B et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38, 694–699, doi: 10.1038/ng1792 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Margueron R et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767, doi: 10.1038/nature08398 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan W et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337, 971–975, doi: 10.1126/science.1225237 (2012). [DOI] [PubMed] [Google Scholar]
- 21. Zhao J et al. RYBP/YAF2-PRC1 complexes and histone H1-dependent chromatin compaction mediate propagation of H2AK119ub1 during cell division. Nat Cell Biol 22, 439–452, doi: 10.1038/s41556-020-0484-1 (2020). This study demonstrates that the interaction between RYBP and H2AK119Ub1 is critical for spreading of H2AK119Ub1 by ncPRC1.
- 22.Fischle W et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17, 1870–1881, doi: 10.1101/gad.1110503 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalb R et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21, 569–571, doi: 10.1038/nsmb.2833 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Min J, Zhang Y & Xu RM Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17, 1823–1828, doi: 10.1101/gad.269603 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao Z et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45, 344–356, doi: 10.1016/j.molcel.2012.01.002 (2012). Through systematic pull-downs and follow up experiments, this study chracterizes that the composition of distinct PRC1 complexes are based on combinations of PCGF, CBX and RYBP.
- 26.Lagarou A et al. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22, 2799–2810, doi: 10.1101/gad.484208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nekrasov M et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J 26, 4078–4088, doi: 10.1038/sj.emboj.7601837 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackledge NP & Klose RJ The molecular principles of gene regulation by Polycomb repressive complexes. Nat Rev Mol Cell Biol, doi: 10.1038/s41580-021-00398-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piunti A & Shilatifard A The roles of Polycomb repressive complexes in mammalian development and cancer. Nat Rev Mol Cell Biol 22, 326–345, doi: 10.1038/s41580-021-00341-1 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Ferrari KJ et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53, 49–62, doi: 10.1016/j.molcel.2013.10.030 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Lee HG, Kahn TG, Simcox A, Schwartz YB & Pirrotta V Genome-wide activities of Polycomb complexes control pervasive transcription. Genome Res 25, 1170–1181, doi: 10.1101/gr.188920.114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ku M et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4, e1000242, doi: 10.1371/journal.pgen.1000242 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee TI et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313, doi: 10.1016/j.cell.2006.02.043 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendenhall EM et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6, e1001244, doi: 10.1371/journal.pgen.1001244 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikkelsen TS et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560, doi: 10.1038/nature06008 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein BE et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326, doi: 10.1016/j.cell.2006.02.041 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Dobrinic P, Szczurek AT & Klose RJ PRC1 drives Polycomb-mediated gene repression by controlling transcription initiation and burst frequency. Nat Struct Mol Biol 28, 811–824, doi: 10.1038/s41594-021-00661-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henikoff S & Shilatifard A Histone modification: cause or cog? Trends Genet 27, 389–396, doi: 10.1016/j.tig.2011.06.006 (2011). [DOI] [PubMed] [Google Scholar]
- 39.O’Geen H et al. dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res 45, 9901–9916, doi: 10.1093/nar/gkx578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao Z et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98, 37–46, doi: 10.1016/S0092-8674(00)80604-2 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Tavares L et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678, doi: 10.1016/j.cell.2011.12.029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farcas AM et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 1, e00205, doi: 10.7554/eLife.00205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taherbhoy AM, Huang OW & Cochran AG BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat Commun 6, 7621, doi: 10.1038/ncomms8621 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Endoh M et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet 8, e1002774, doi: 10.1371/journal.pgen.1002774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kallin EM et al. Genome-wide uH2A localization analysis highlights Bmi1-dependent deposition of the mark at repressed genes. PLoS Genet 5, e1000506, doi: 10.1371/journal.pgen.1000506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fursova NA et al. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Mol Cell 74, 1020–1036 e1028, doi: 10.1016/j.molcel.2019.03.024 (2019). Through systematic deletion of PCGF proteins in mESCs, the authors characterize the synergistic roles of the PCGF proteins in H2A ubiquitylation and gene repression. See also Scelfo et al., (2019)
- 47.Rose NR et al. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. Elife 5, doi: 10.7554/eLife.18591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morey L, Aloia L, Cozzuto L, Benitah SA & Di Croce L RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep 3, 60–69, doi: 10.1016/j.celrep.2012.11.026 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Wu X, Johansen JV & Helin K Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 49, 1134–1146, doi: 10.1016/j.molcel.2013.01.016 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Cohen I et al. PRC1 Fine-tunes Gene Repression and Activation to Safeguard Skin Development and Stem Cell Specification. Cell Stem Cell 22, 726–739 e727, doi: 10.1016/j.stem.2018.04.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamburri S et al. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol Cell 77, 840–856 e845, doi: 10.1016/j.molcel.2019.11.021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blackledge NP et al. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol Cell 77, 857–874 e859, doi: 10.1016/j.molcel.2019.12.001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Illingworth RS et al. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev 29, 1897–1902, doi: 10.1101/gad.268151.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pengelly AR, Kalb R, Finkl K & Muller J Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev 29, 1487–1492, doi: 10.1101/gad.265439.115 (2015). With Illingworth et al. (2015), the authors show that the bulk of ubiquitylation is dispensable for correct patterning of animals by generating ubiquitylation deficient flies and mice.
- 55.Cohen I, Bar C & Ezhkova E Activity of PRC1 and Histone H2AK119 Monoubiquitination: Revising Popular Misconceptions. Bioessays 42, e1900192, doi: 10.1002/bies.201900192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang R et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 18, 966–975, doi: 10.1016/j.str.2010.04.013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Isono K et al. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell 26, 565–577, doi: 10.1016/j.devcel.2013.08.016 (2013). This study demonstrates that the PHC protein’s SAM domain-mediated clustering is critical for Polycomb body formation and proper mouse development.
- 58.Kim CA, Gingery M, Pilpa RM & Bowie JU The SAM domain of polyhomeotic forms a helical polymer. Nat Struct Biol 9, 453–457, doi: 10.1038/nsb802 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Kundu S et al. Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Mol Cell 65, 432–446 e435, doi: 10.1016/j.molcel.2017.01.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wani AH et al. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat Commun 7, 10291, doi: 10.1038/ncomms10291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Plys AJ et al. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev 33, 799–813, doi: 10.1101/gad.326488.119 (2019). With Tatavosian et al. (2019), these studies show that the cPRC1 component CBX2 can phase separate, suggesting another potential way of physically regulating Polycomb domains.
- 62.Seif E et al. Phase separation by the polyhomeotic sterile alpha motif compartmentalizes Polycomb Group proteins and enhances their activity. Nat Commun 11, 5609, doi: 10.1038/s41467-020-19435-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tatavosian R et al. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J Biol Chem 294, 1451–1463, doi: 10.1074/jbc.RA118.006620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Satijn DP et al. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol 17, 4105–4113, doi: 10.1128/MCB.17.7.4105 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saurin AJ et al. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol 142, 887–898, doi: 10.1083/jcb.142.4.887 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Boettiger AN et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422, doi: 10.1038/nature16496 (2016). Using oligopaint FISH and super resolution microscopy, this study is one of the first imaging-based studies to show that Polycomb bound regions are more densely packed than transcriptionally active regions.
- 67. Schoenfelder S et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet 47, 1179–1186, doi: 10.1038/ng.3393 (2015). Through promoter capture Hi-C, this study demonstrates the role of PRC1 in genome organization.
- 68.Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D & Noordermeer D Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci U S A 112, 4672–4677, doi: 10.1073/pnas.1504783112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lau MS et al. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science 355, 1081–1084, doi: 10.1126/science.aah5403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Morey L et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10, 47–62, doi: 10.1016/j.stem.2011.12.006 (2012). With O’Loghlen et al. (2012), these studies show CBX component switching occurs during mESC differentiation, suggesting cPRC1 composition can be dynamically changed depending on the cellular contexts.
- 71.O’Loghlen A et al. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell 10, 33–46, doi: 10.1016/j.stem.2011.12.004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Isono K et al. Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mol Cell Biol 25, 6694–6706, doi: 10.1128/MCB.25.15.6694-6706.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Core N et al. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development 124, 721–729 (1997). [DOI] [PubMed] [Google Scholar]
- 74.Katoh-Fukui Y et al. Male-to-female sex reversal in M33 mutant mice. Nature 393, 688–692, doi: 10.1038/31482 (1998). [DOI] [PubMed] [Google Scholar]
- 75. Blackledge NP et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459, doi: 10.1016/j.cell.2014.05.004 (2014). With Cooper et al. (2014), through ectopic recruitment of various PRC components to artificial loci, the authors demonstrate that ncPRC1 is upstream of PRC2 and cPRC1 in the hierarchical pathway in mESCs.
- 76.Cooper S et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep 7, 1456–1470, doi: 10.1016/j.celrep.2014.04.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He J et al. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 15, 373–384, doi: 10.1038/ncb2702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cooper S et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun 7, 13661, doi: 10.1038/ncomms13661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oksuz O et al. Capturing the Onset of PRC2-Mediated Repressive Domain Formation. Mol Cell 70, 1149–1162 e1145, doi: 10.1016/j.molcel.2018.05.023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arrigoni R et al. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett 580, 6233–6241, doi: 10.1016/j.febslet.2006.10.027 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Kagey MH, Melhuish TA & Wotton D The polycomb protein Pc2 is a SUMO E3. Cell 113, 127–137, doi: 10.1016/s0092-8674(03)00159-4 (2003). [DOI] [PubMed] [Google Scholar]
- 82.Jaensch ES et al. A Polycomb domain found in committed cells impairs differentiation when introduced into PRC1 in pluripotent cells. Mol Cell, doi: 10.1016/j.molcel.2021.09.018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim J & Kingston RE The CBX family of proteins in transcriptional repression and memory. J Biosci 45 (2020). [PubMed] [Google Scholar]
- 84.Bernstein E et al. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26, 2560–2569, doi: 10.1128/MCB.26.7.2560-2569.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhen CY et al. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. Elife 5, doi: 10.7554/eLife.17667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhen CY, Duc HN, Kokotovic M, Phiel CJ & Ren X Cbx2 stably associates with mitotic chromosomes via a PRC2- or PRC1-independent mechanism and is needed for recruiting PRC1 complex to mitotic chromosomes. Mol Biol Cell 25, 3726–3739, doi: 10.1091/mbc.E14-06-1109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tardat M et al. Cbx2 targets PRC1 to constitutive heterochromatin in mouse zygotes in a parent-of-origin-dependent manner. Mol Cell 58, 157–171, doi: 10.1016/j.molcel.2015.02.013 (2015). [DOI] [PubMed] [Google Scholar]
- 88.Junco SE et al. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure 21, 665–671, doi: 10.1016/j.str.2013.02.013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gray F et al. BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization. Nat Commun 7, 13343, doi: 10.1038/ncomms13343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scelfo A et al. Functional Landscape of PCGF Proteins Reveals Both RING1A/B-Dependent-and RING1A/B-Independent-Specific Activities. Mol Cell 74, 1037–1052 e1037, doi: 10.1016/j.molcel.2019.04.002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Almeida M et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356, 1081–1084, doi: 10.1126/science.aal2512 (2017). Using live cell imaging and genetic mouse models, the authors show PCGF3/5’s specific roles in X-inactivation.
- 92.Endoh M et al. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. Elife 6, doi: 10.7554/eLife.21064 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zdzieblo D et al. Pcgf6, a polycomb group protein, regulates mesodermal lineage differentiation in murine ESCs and functions in iPS reprogramming. Stem Cells 32, 3112–3125, doi: 10.1002/stem.1826 (2014). [DOI] [PubMed] [Google Scholar]
- 94.Yu JR, Lee CH, Oksuz O, Stafford JM & Reinberg D PRC2 is high maintenance. Genes Dev 33, 903–935, doi: 10.1101/gad.325050.119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li H et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549, 287–291, doi: 10.1038/nature23881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perino M et al. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat Genet 50, 1002–1010, doi: 10.1038/s41588-018-0134-8 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Healy E et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol Cell 76, 437–452 e436, doi: 10.1016/j.molcel.2019.08.012 (2019). [DOI] [PubMed] [Google Scholar]
- 98.Hojfeldt JW et al. Non-core Subunits of the PRC2 Complex Are Collectively Required for Its Target-Site Specificity. Mol Cell 76, 423–436 e423, doi: 10.1016/j.molcel.2019.07.031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petracovici A & Bonasio R Distinct PRC2 subunits regulate maintenance and establishment of Polycomb repression during differentiation. Mol Cell 81, 2625–2639 e2625, doi: 10.1016/j.molcel.2021.03.038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kloet SL et al. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat Struct Mol Biol 23, 682–690, doi: 10.1038/nsmb.3248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pemberton H et al. Genome-wide co-localization of Polycomb orthologs and their effects on gene expression in human fibroblasts. Genome Biol 15, R23, doi: 10.1186/gb-2014-15-2-r23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Endoh M et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135, 1513–1524, doi: 10.1242/dev.014340 (2008). [DOI] [PubMed] [Google Scholar]
- 103.Shen X et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32, 491–502, doi: 10.1016/j.molcel.2008.10.016 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ezhkova E et al. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25, 485–498, doi: 10.1101/gad.2019811 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Akasaka T et al. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128, 1587–1597 (2001). [DOI] [PubMed] [Google Scholar]
- 106.Moussa HF et al. Canonical PRC1 controls sequence-independent propagation of Polycomb-mediated gene silencing. Nat Commun 10, 1931, doi: 10.1038/s41467-019-09628-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Posfai E et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev 26, 920–932, doi: 10.1101/gad.188094.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leeb M et al. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24, 265–276, doi: 10.1101/gad.544410 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cohen I et al. Polycomb complexes redundantly maintain epidermal stem cell identity during development. Genes Dev 35, 354–366, doi: 10.1101/gad.345363.120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zepeda-Martinez JA et al. Parallel PRC2/cPRC1 and vPRC1 pathways silence lineage-specific genes and maintain self-renewal in mouse embryonic stem cells. Sci Adv 6, eaax5692, doi: 10.1126/sciadv.aax5692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takada Y et al. Mammalian Polycomb Scmh1 mediates exclusion of Polycomb complexes from the XY body in the pachytene spermatocytes. Development 134, 579–590, doi: 10.1242/dev.02747 (2007). [DOI] [PubMed] [Google Scholar]
- 112.Garcia-Moreno SA et al. CBX2 is required to stabilize the testis pathway by repressing Wnt signaling. PLoS Genet 15, e1007895, doi: 10.1371/journal.pgen.1007895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hasegawa K et al. SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev Cell 32, 574–588, doi: 10.1016/j.devcel.2015.01.014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Luo M et al. Polycomb protein SCML2 associates with USP7 and counteracts histone H2A ubiquitination in the XY chromatin during male meiosis. PLoS Genet 11, e1004954, doi: 10.1371/journal.pgen.1004954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haupt Y, Alexander WS, Barri G, Klinken SP & Adams JM Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell 65, 753–763, doi: 10.1016/0092-8674(91)90383-a (1991). [DOI] [PubMed] [Google Scholar]
- 116.van Lohuizen M et al. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65, 737–752, doi: 10.1016/0092-8674(91)90382-9 (1991). [DOI] [PubMed] [Google Scholar]
- 117.Gil J, Bernard D, Martinez D & Beach D Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol 6, 67–72, doi: 10.1038/ncb1077 (2004). [DOI] [PubMed] [Google Scholar]
- 118.Klauke K et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol 15, 353–362, doi: 10.1038/ncb2701 (2013). [DOI] [PubMed] [Google Scholar]
- 119.Luis NM et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell 9, 233–246, doi: 10.1016/j.stem.2011.07.013 (2011). [DOI] [PubMed] [Google Scholar]
- 120.Chung CY et al. Cbx8 Acts Non-canonically with Wdr5 to Promote Mammary Tumorigenesis. Cell Rep 16, 472–486, doi: 10.1016/j.celrep.2016.06.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tan J et al. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell 20, 563–575, doi: 10.1016/j.ccr.2011.09.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Morey L et al. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell 17, 300–315, doi: 10.1016/j.stem.2015.08.009 (2015). [DOI] [PubMed] [Google Scholar]
- 123.Xu J et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol Cell 57, 304–316, doi: 10.1016/j.molcel.2014.12.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stojic L et al. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics Chromatin 4, 16, doi: 10.1186/1756-8935-4-16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Margueron R et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32, 503–518, doi: 10.1016/j.molcel.2008.11.004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Laible G et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J 16, 3219–3232, doi: 10.1093/emboj/16.11.3219 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.O’Carroll D et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21, 4330–4336, doi: 10.1128/MCB.21.13.4330-4336.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Voncken JW et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci U S A 100, 2468–2473, doi: 10.1073/pnas.0434312100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.del Mar Lorente M et al. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127, 5093–5100, doi: 10.1242/dev.127.23.5093 (2000). [DOI] [PubMed] [Google Scholar]
- 130.Bhattacharya D, Talwar S, Mazumder A & Shivashankar GV Spatio-temporal plasticity in chromatin organization in mouse cell differentiation and during Drosophila embryogenesis. Biophys J 96, 3832–3839, doi: 10.1016/j.bpj.2008.11.075 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li G et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24, 368–380, doi: 10.1101/gad.1886410 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Oliviero G et al. Dynamic Protein Interactions of the Polycomb Repressive Complex 2 during Differentiation of Pluripotent Cells. Mol Cell Proteomics 15, 3450–3460, doi: 10.1074/mcp.M116.062240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Beringer M et al. EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol Cell 64, 645–658, doi: 10.1016/j.molcel.2016.10.018 (2016). [DOI] [PubMed] [Google Scholar]
- 134.Silva J et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4, 481–495, doi: 10.1016/s1534-5807(03)00068-6 (2003). [DOI] [PubMed] [Google Scholar]
- 135.Frey F et al. Molecular basis of PRC1 targeting to Polycomb response elements by PhoRC. Genes Dev 30, 1116–1127, doi: 10.1101/gad.279141.116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kang H et al. Sex comb on midleg (Scm) is a functional link between PcG-repressive complexes in Drosophila. Genes Dev 29, 1136–1150, doi: 10.1101/gad.260562.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.DeLuca SZ, Ghildiyal M, Pang LY & Spradling AC Differentiating Drosophila female germ cells initiate Polycomb silencing by regulating PRC2-interacting proteins. Elife 9, doi: 10.7554/eLife.56922 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gao Z et al. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 516, 349–354, doi: 10.1038/nature13921 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu S et al. NRF1 association with AUTS2-Polycomb mediates specific gene activation in the brain. Mol Cell, doi: 10.1016/j.molcel.2021.09.020 (2021). [DOI] [PubMed] [Google Scholar]
- 140.Frangini A et al. The aurora B kinase and the polycomb protein ring1B combine to regulate active promoters in quiescent lymphocytes. Mol Cell 51, 647–661, doi: 10.1016/j.molcel.2013.08.022 (2013). [DOI] [PubMed] [Google Scholar]
- 141.Piunti A et al. CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci Adv 5, eaax2887, doi: 10.1126/sciadv.aax2887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jain SU et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun 10, 2146, doi: 10.1038/s41467-019-09981-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ragazzini R et al. EZHIP constrains Polycomb Repressive Complex 2 activity in germ cells. Nat Commun 10, 3858, doi: 10.1038/s41467-019-11800-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pajtler KW et al. Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol 136, 211–226, doi: 10.1007/s00401-018-1877-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Tsuboi M et al. Ubiquitination-Independent Repression of PRC1 Targets during Neuronal Fate Restriction in the Developing Mouse Neocortex. Dev Cell 47, 758–772 e755, doi: 10.1016/j.devcel.2018.11.018 (2018). This study shows that distinct PRC1 activity (ubiquitylation vs. PHC-mediated clustering) is required for gene silencing in different stages of neural progenitor cells during cortical development.
- 146. Zheng H et al. Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals. Mol Cell 63, 1066–1079, doi: 10.1016/j.molcel.2016.08.032 (2016). Through low-input ChIP-seq experiments, the authors identify atypical H3K27me3 distribution in mouse oocytes and preimplantation embryos.
- 147.Li XY, Harrison MM, Villalta JE, Kaplan T & Eisen MB Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife 3, doi: 10.7554/eLife.03737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vastenhouw NL et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464, 922–926, doi: 10.1038/nature08866 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xia W et al. Resetting histone modifications during human parental-to-zygotic transition. Science 365, 353–360, doi: 10.1126/science.aaw5118 (2019). [DOI] [PubMed] [Google Scholar]
- 150.Du Z et al. Polycomb Group Proteins Regulate Chromatin Architecture in Mouse Oocytes and Early Embryos. Mol Cell 77, 825–839 e827, doi: 10.1016/j.molcel.2019.11.011 (2020). [DOI] [PubMed] [Google Scholar]
- 151.Chen Z, Djekidel MN & Zhang Y Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos. Nat Genet 53, 551–563, doi: 10.1038/s41588-021-00821-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mei H et al. H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Nat Genet 53, 539–550, doi: 10.1038/s41588-021-00820-3 (2021). [DOI] [PubMed] [Google Scholar]
- 153.Puschendorf M et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet 40, 411–420, doi: 10.1038/ng.99 (2008). [DOI] [PubMed] [Google Scholar]
- 154.Koyama-Nasu R, David G & Tanese N The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat Cell Biol 9, 1074–1080, doi: 10.1038/ncb1628 (2007). [DOI] [PubMed] [Google Scholar]
- 155.Lynch MD et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J 31, 317–329, doi: 10.1038/emboj.2011.399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brinkman AB et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res 22, 1128–1138, doi: 10.1101/gr.133728.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Reddington JP et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol 14, R25, doi: 10.1186/gb-2013-14-3-r25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Harris C et al. Conversion of random X-inactivation to imprinted X-inactivation by maternal PRC2. Elife 8, doi: 10.7554/eLife.44258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Inoue A, Chen Z, Yin Q & Zhang Y Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev 32, 1525–1536, doi: 10.1101/gad.318675.118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Inoue A, Jiang L, Lu F, Suzuki T & Zhang Y Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424, doi: 10.1038/nature23262 (2017). This study identifies a set of genes that require maternally inherited H3K27me3 for their allele-specific repression, demonstrating that Polycomb system is involved in imprinting in early mouse embryos.
- 161.Inoue A, Jiang L, Lu F & Zhang Y Genomic imprinting of Xist by maternal H3K27me3. Genes Dev 31, 1927–1932, doi: 10.1101/gad.304113.117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Conway E et al. BAP1 enhances Polycomb repression by counteracting widespread H2AK119ub1 deposition and chromatin condensation. Mol Cell 81, 3526–3541 e3528, doi: 10.1016/j.molcel.2021.06.020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fursova NA et al. BAP1 constrains pervasive H2AK119ub1 to control the transcriptional potential of the genome. Genes Dev 35, 749–770, doi: 10.1101/gad.347005.120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kadoch C et al. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet 49, 213–222, doi: 10.1038/ng.3734 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kennison JA & Tamkun JW Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A 85, 8136–8140, doi: 10.1073/pnas.85.21.8136 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Stanton BZ et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet 49, 282–288, doi: 10.1038/ng.3735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Weber CM et al. mSWI/SNF promotes Polycomb repression both directly and through genome-wide redistribution. Nat Struct Mol Biol 28, 501–511, doi: 10.1038/s41594-021-00604-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sharp EJ, Martin EC & Adler PN Directed overexpression of suppressor 2 of zeste and Posterior Sex Combs results in bristle abnormalities in Drosophila melanogaster. Dev Biol 161, 379–392, doi: 10.1006/dbio.1994.1039 (1994). [DOI] [PubMed] [Google Scholar]
- 169.Bel S et al. Genetic interactions and dosage effects of Polycomb group genes in mice. Development 125, 3543–3551, doi: 10.1242/dev.125.18.3543 (1998). [DOI] [PubMed] [Google Scholar]
- 170.Alkema MJ, van der Lugt NM, Bobeldijk RC, Berns A & van Lohuizen M Transformation of axial skeleton due to overexpression of bmi-1 in transgenic mice. Nature 374, 724–727, doi: 10.1038/374724a0 (1995). [DOI] [PubMed] [Google Scholar]
- 171.Lau MS Mutations in the Charged Domain of CBX2 Disrupt PRC1 Function in Vivo Doctoral thesis, Harvard University, (2016). [Google Scholar]
- 172.Zhu L & Brangwynne CP Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol 34, 23–30, doi: 10.1016/j.ceb.2015.04.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hosogane M, Funayama R, Shirota M & Nakayama K Lack of Transcription Triggers H3K27me3 Accumulation in the Gene Body. Cell Rep 16, 696–706, doi: 10.1016/j.celrep.2016.06.034 (2016). [DOI] [PubMed] [Google Scholar]
- 174.Riising EM et al. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 55, 347–360, doi: 10.1016/j.molcel.2014.06.005 (2014). [DOI] [PubMed] [Google Scholar]
- 175.Stielow B, Finkernagel F, Stiewe T, Nist A & Suske G MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet 14, e1007193, doi: 10.1371/journal.pgen.1007193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Miller SA, Damle M, Kim J & Kingston RE Full methylation of H3K27 by PRC2 is dispensable for initial embryoid body formation but required to maintain differentiated cell identity. Development 148, doi: 10.1242/dev.196329 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lu TT et al. The Polycomb-Dependent Epigenome Controls beta Cell Dysfunction, Dedifferentiation, and Diabetes. Cell Metab 27, 1294–1308 e1297, doi: 10.1016/j.cmet.2018.04.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hirabayashi Y et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600–613, doi: 10.1016/j.neuron.2009.08.021 (2009). [DOI] [PubMed] [Google Scholar]
- 179.Iovino N, Ciabrelli F & Cavalli G PRC2 controls Drosophila oocyte cell fate by repressing cell cycle genes. Dev Cell 26, 431–439, doi: 10.1016/j.devcel.2013.06.021 (2013). [DOI] [PubMed] [Google Scholar]
- 180. Coleman RT & Struhl G Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 356, doi: 10.1126/science.aai8236 (2017). This study, together with Laprell et al. (2017), demonstrates that existing H3K27me3 is not sufficient for self-maintenance and gene repression by inducing the deletion of DNA elements needed for de novo PcG recruitment.
- 181.Laprell F, Finkl K & Muller J Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 356, 85–88, doi: 10.1126/science.aai8266 (2017). [DOI] [PubMed] [Google Scholar]
- 182. Jadhav U et al. Replicational Dilution of H3K27me3 in Mammalian Cells and the Role of Poised Promoters. Mol Cell 78, 141–151 e145, doi: 10.1016/j.molcel.2020.01.017 (2020). This study shows slow replicative dilution and resulting gene derepression in mammalian systems including intestinal stem cells and human lymphoma cells after disrupting PRC2.
- 183.Jadhav U et al. Acquired Tissue-Specific Promoter Bivalency Is a Basis for PRC2 Necessity in Adult Cells. Cell 165, 1389–1400, doi: 10.1016/j.cell.2016.04.031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Francis NJ, Follmer NE, Simon MD, Aghia G & Butler JD Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell 137, 110–122, doi: 10.1016/j.cell.2009.02.017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Yang H et al. Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis. Science 357, 1142–1145, doi: 10.1126/science.aan1121 (2017). This study shows that the loss of Polycomb-mediated memory can be variable between cells within an organism.
- 186.Bintu L et al. Dynamics of epigenetic regulation at the single-cell level. Science 351, 720–724, doi: 10.1126/science.aab2956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Struhl G A gene product required for correct initiation of segmental determination in Drosophila. Nature 293, 36–41, doi: 10.1038/293036a0 (1981). [DOI] [PubMed] [Google Scholar]
- 188.Chan CS, Rastelli L & Pirrotta V A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J 13, 2553–2564 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.De S, Mitra A, Cheng Y, Pfeifer K & Kassis JA Formation of a Polycomb-Domain in the Absence of Strong Polycomb Response Elements. PLoS Genet 12, e1006200, doi: 10.1371/journal.pgen.1006200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mukund A & Bintu L Temporal signaling, population control, and information processing through chromatin-mediated gene regulation. J Theor Biol 535, 110977, doi: 10.1016/j.jtbi.2021.110977 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sneppen K & Ringrose L Theoretical analysis of Polycomb-Trithorax systems predicts that poised chromatin is bistable and not bivalent. Nat Commun 10, 2133, doi: 10.1038/s41467-019-10130-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Akasaka T et al. A role for mel-18, a Polycomb group-related vertebrate gene, during theanteroposterior specification of the axial skeleton. Development 122, 1513–1522 (1996). [DOI] [PubMed] [Google Scholar]
- 193.Capdevila MP, Botas J & Garcia-Bellido A Genetic interactions between the Polycomb locus and the Antennapedia and Bithorax complexes of Drosophila. Rouxs Arch Dev Biol 195, 417–432, doi: 10.1007/BF00375746 (1986). [DOI] [PubMed] [Google Scholar]
- 194.Dura JM, Brock HW & Santamaria P Polyhomeotic: a gene of Drosophila melanogaster required for correct expression of segmental identity. Mol Gen Genet 198, 213–220, doi: 10.1007/BF00382998 (1985). [DOI] [PubMed] [Google Scholar]
- 195.Tokunaga C & Stern C The Developmental Autonomy of Extra Sex Combs in Drosophila Melanogaster. Dev Biol 11, 50–81, doi: 10.1016/0012-1606(65)90037-0 (1965). [DOI] [PubMed] [Google Scholar]
- 196.van der Lugt NM et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev 8, 757–769, doi: 10.1101/gad.8.7.757 (1994). [DOI] [PubMed] [Google Scholar]
- 197.Beuchle D, Struhl G & Muller J Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128, 993–1004 (2001). [DOI] [PubMed] [Google Scholar]
- 198.Beh LY, Colwell LJ & Francis NJ A core subunit of Polycomb repressive complex 1 is broadly conserved in function but not primary sequence. Proc Natl Acad Sci U S A 109, E1063–1071, doi: 10.1073/pnas.1118678109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sugishita H et al. Variant PCGF1-PRC1 links PRC2 recruitment with differentiation-associated transcriptional inactivation at target genes. Nat Commun 12, 5341, doi: 10.1038/s41467-021-24894-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hickey GJ et al. Establishment of developmental gene silencing by ordered polycomb complex recruitment in early zebrafish embryos. Elife 11, doi: 10.7554/eLife.67738 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zylicz JJ et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 176, 182–197 e123, doi: 10.1016/j.cell.2018.11.041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cheutin T & Cavalli G Progressive polycomb assembly on H3K27me3 compartments generates polycomb bodies with developmentally regulated motion. PLoS Genet 8, e1002465, doi: 10.1371/journal.pgen.1002465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Buchwald G et al. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J 25, 2465–2474, doi: 10.1038/sj.emboj.7601144 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li Z et al. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem 281, 20643–20649, doi: 10.1074/jbc.M602461200 (2006). [DOI] [PubMed] [Google Scholar]
- 205.Shen Y et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120, doi: 10.1038/nature11243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]


