Abstract
Spermatogenesis is regulated by several Y chromosome-specific genes located in a specific region of the long arm of the Y chromosome, the azoospermia factor region (AZF). AZF microdeletions are the main structural chromosomal abnormalities that cause male infertility. Assisted reproductive technology (ART) has been used to overcome natural fertilization barriers, allowing infertile couples to have children. However, these techniques increase the risk of vertical transmission of genetic defects. Despite widespread awareness of AZF microdeletions, the occurrence of de novo deletions and overexpression, as well as the expansion of AZF microdeletion vertical transmission, remains unknown. This review summarizes the mechanism of AZF microdeletion and the function of the candidate genes in the AZF region and their corresponding clinical phenotypes. Moreover, vertical transmission cases of AZF microdeletions, the impact of vertical inheritance on male fertility, and the prospective direction of research in this field are also outlined.
Keywords: azoospermia factor, male infertility, microdeletion, vertical transmission
INTRODUCTION
Approximately 10%–15% of couples of child-bearing age suffer from infertility worldwide. Genetic factors play well-recognized roles in male infertility, one of the most prominent of which is the azoospermia factor region (AZF) microdeletion.
The human Y chromosome is a submetacentric chromosome with two pseudoautosomal regions (PARs). The male-specific region (MSY) spans 95% of the chromosome length and is flanked by PAR1 and PAR2. It is subdivided into three discrete classes of sequences: X-transposed, X-degenerate, and ampliconic. The ampliconic sequence contains eight groups of palindromic structures, as they are largely homogeneous with almost identical sequence identity, leading to nonallelic homologous recombination (NAHR), which is the primary cause of AZF microdeletion.1 The AZF regions, which contain genes critical for spermatogenesis and male fertility, include AZFa, AZFb, and AZFc regions.2 Kent-First et al.3 found another region between AZFb and AZFc that also contained genes involved in spermatogenesis and termed it the AZFd region. However, it is currently ignored in the European Academy of Andrology (EAA)/European Molecular Quality Network (EMQN) best practice guidelines.4
AZF microdeletions account for approximately 14% cases of oligozoospermia and azoospermia.5 Owing to the development of ART, a greater number of AZF microdeletion patients have been able to have offspring. Studies have revealed that AZF microdeletions are vertically transmitted, but the extent of AZF microdeletions in progeny remains controversial.6 This article reviews candidate gene functions and AZF microdeletion types. In addition, we specifically discuss the transmission characteristics of AZF microdeletions and outline the future research goals.
AZF GENE PARTITION AND STRUCTURE
Ubiquitin-specific peptidase 9, Y-linked (USP9Y) was the first recognized gene in the AZFa locus, 160 kb long with 46 exons. USP9Y encodes a ubiquitin-specific protease that potentially ensures that meiosis proceeds normally, transforming germ cells into mature spermatozoa.7 Dead box on Y (DBY) is composed of 17 exons and extends for approximately 16 kb, playing a role in the earliest stages of human germ cell development. DBY encodes a potential RNA helicase that may be involved in mRNA translation.8 Ubiquitously transcribed tetratricopeptide repeat gene, Y-linked (UTY) consists of 50 exons and is expressed in the multiple organs of the human body. It encodes a tetratricopeptide repeat gene that may play a function in transcriptional regulation.9 RNA binding motif, Y-linked (RBMY) comprises 6 copies and 12 exons. The N-terminus of the encoded protein has an RNA recognition motif, which edits mRNA precursors into specific mRNAs.10 Eukaryotic translation initiation factor 1A, Y-linked (EIF1AY) contains 7 exons discovered on the nonrecombining region of the Y chromosome. It encodes a translation initiation and elongation factor with homologs in Xq22 and chromosome 1p. This protein may promote the stabilization of the initiator Met-binding transfer RNA to the 40S ribosomal subunit.11 Deleted-in-azoospermia (DAZ) is a multicopy gene that is widely studied in the AZFc locus. DAZ contains 16 exons and is approximately 42 kb long and composed of two clusters of DAZ for four genes. DAZ is specifically expressed in all stages of germ cell development and encodes RNA-binding proteins.12 The encoded RNA-binding protein is located in the germinal epithelium and sperm tail; thus, DAZ is associated with spermatogenesis and sperm motility.13 Chromodomain Protein Y linked (CDY) is a reverse derivative of Chromosome Domain Y-like (CDYL) found on the autosomal chromosome. The protein encoded by CDY specifically recognizes methylate histone H3 lysine 27, enhancing its methyltransferase activity and promoting histone methylation, which is important in spermatogenesis.14
MECHANISM OF AZF MICRODELETION
There are approximately 50 million base pairs on the Y chromosome, approximately one-tenth of which are found in palindromic sequences. These palindromic sequences can cause structural changes such as inversion and deletion of the Y chromosome through NAHR. Human endogenous retroviruses (HERVs) are closely related to AZF microdeletions. A HERV is a defective or inactive original virus, and the region around the sex-determining gene of the human Y chromosome is the hotspot of integration.15
The AZFa locus is located in the proximal region of Yq11 where it spans around 1100 kb. Only single-copy genes are encoded. NAHR does not arise in the absence of HERV intervention. Sun et al.16 discovered that the similarity of sequences at both ends of the AZFa microdeletions area with human endogenous retrovirus 15 (HERV15) is as high as 96% or more, implying that HERV15 might be a substrate for NAHR occurring at both ends of the AZFa microdeletions region. Patients with AZFa microdeletions also contained HERV15yq1 (9747 nucleotides from Yq11 interval D3) and HERV15yq2 (9969 nucleotides from Yq11 interval D6) at both ends of the deletion area, as well as two sets of highly homologous segments, identical sequence domains (ID), including ID1 (1278 bp) and ID2 (1690 bp), which might produce NAHR between them.17,18 AZFa deletion caused by HERV15 nonallelic recombination events is 793 kb in length (Figure 1).
Figure 1.
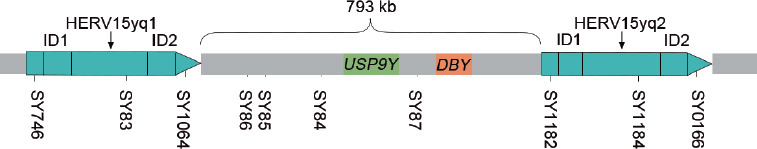
The blue box represents HERV15yq1 and HERV15yq2, and there are two groups of highly homologous fragments ID1 and ID2, with the deletion length of about 793 kb. The green box is USP9Y, the orange box represents DBY, and the position of the STS markers are displayed below. USP9Y: ubiquitin-specific peptidase 9, Y-linked; DBY: dead box on Y; HERV15: human endogenous retrovirus 15; ID: identical sequence domains; STS: sequence tagged sites.
The AZFb locus is located at the center of Yq11 and spans approximately 7 Mb, part of which overlaps with the AZFc locus by 1.5 Mb. On the one hand, owing to the high sequence similarity between the P1.1 (P1 distal) and P1.2 (P1 proximal) palindromes and P5 palindromes P5/P1.1 and P5/P1.2, NAHR can occur at both. The former deletes 32 genes in MSY, covering approximately 6.2 Mb in length. The latter deletes 42 genes, covering approximately 7.6 Mb in length.19 HERV, on the other hand, is associated with testis-specific transcripts to the Y (TTY) deletion in the AZFb region. The two sides of the HERV genome sequences are long terminal repeats (LTRs), coding regulatory elements that act as gene expression promoters or silencers in specific tissues and cell lines.20 NAHR of the 5’LTR and 3’LTR of human endogenous retrovirus K14C results in the deletion of TTY13 in the AZFb locus21 (Figure 2).
Figure 2.
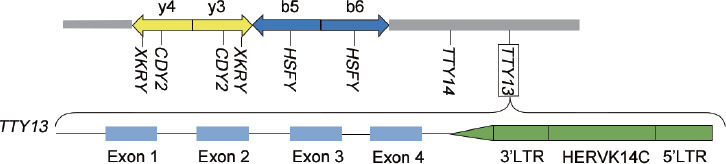
The green box represents HERVK14C, and the 3’ LTR and 5’ LTR at both ends are highly homologous; the blue box represents the exon of TTY13, and TTY13 contains HERVK14C. HERVK14C: human endogenous retrovirus K14C; LTR: long terminal repeat; TTY: testis-specific transcripts to the Y; HSFY: heat shock factors on Y; XKRY: X-kell blood group precursor related Y; CDY: Chromodomain Protein Y linked; y: yellow; b: blue.
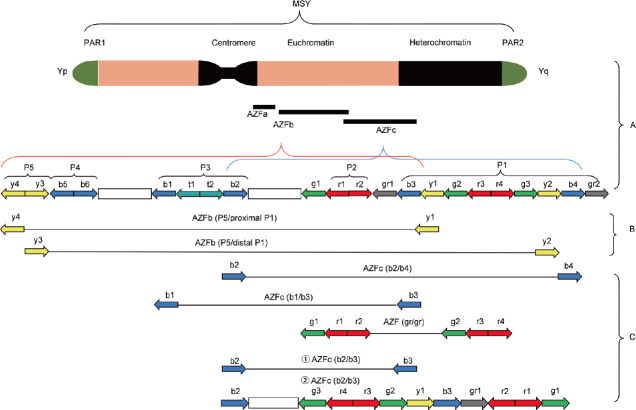
The AZFc locus is approximately 4.5 Mb and is located at the distal end of Yq11. Two complete palindromic structures (P1P2) and the distal portion of palindrome structure P3 are located in the AZFc locus. The AZFc locus has 5 amplicons, named after colors of the fluorescent probe: blue (b), green (g), red (r), yellow (y) and gray (g), and partially overlaps with AZFb. AZFc microdeletions consist of four subtypes: b2/b4, b1/b3, b2/b3 and gr/gr (Figure 3).22
Figure 3.
Part A is Y chromosome structure, PAR1 and PAR2 are shown in light gray, and the autosomal domains in specific regions of Y are shown in dark gray. The black circular areas represent centromeres. AZFc amplicon is divided into five color sequence families: yellow (y), blue (b), green (g), red (r), and gray (g). The direction of the arrow indicates the polarity of the amplicon. Part B describes two recombination situations in which AZFb region is missing. Part C describes the deletion mechanism of four different subtypes in AZFc region. PAR: pseudoautosomal region; MSY: male-specific region; AZF: azoospermia factor; P1–5: palindrome sequence 1–5.
b2/b4 is a two-direct repeat sequence found at both ends of the AZFc locus, with a fragment length of approximately 230 kb, and the incidence of NAHR here is the lowest. Once NAHR occurs, the 3 Mb segment is lost, which corresponds to complete AZFc deletion. Currently, b2/b4 loss is considered a high-risk factor for severe spermatogenesis disorders since it leads to a 145-fold increase in the risk of severe spermatogenesis disorders. This is because this loss results in the deletion of all DAZ gene families (DAZ1–DAZ4), causing the deletion of two important coding genes, CDY1 and basic protein on Y chromosome, 2 gene (BPY2).23 The incidence of b1/b3 deletion is relatively low, and its microdeletion mechanism is similar to that of b2/b4. NAHR occurs in the same sequence direction, resulting in the deletion of nearly 1.6 Mb.
Two mechanisms underlie the b2/b3 deletion type. In the first, gr/rg inversion causes g1-r1-r2 to recombine with r3-r4-g3, followed by b2/b3 deletion. In the second, b2/b3 inversion leads to the reversal of the b3-y1-g2 sequence in the region, which turns the original b2-b3 reverse repeat sequence into a direct repeat sequence, followed by rg/rg deletion.24 Both mechanisms result in the final loss of the same fragment, which is approximately 1.8 Mb in length, including 12 genes and multiple copy number transcripts. b2/b3 deletions occur mainly due to gene inversion, resulting in DAZ3/DAZ4 deletions in the four DAZ copies.25 Worldwide, b2/b3 deletions occur most often in China,26 and b2/b3 deletions have a higher risk of spermatogenic disorders than gr/gr deletions.27 The b2/b3 deletion has not demonstrated a significant effect on spermatogenesis in infertile populations in Northern Europe and Germany. This suggests that the association between b2/b3 deletion and spermatogenesis disorders is population and region dependent.28
The gr/gr region constitutes approximately half of the AZFc region, spans approximately 1.6 Mb and accounts for the highest incidence of AZFc microdeletions. NAHR in a pair of direct repeat sequences (g1-r1-r2 and g2-r3-r4) leads to the deletion of gr/gr, spanning approximately 1.6 Mb in length.29 Deletion of gr/gr results in the deletion of DAZ1/DAZ2 genes as well as the two copies of CDY1a and BPY2. The incidence of gr/gr deficiency in infertile populations exhibits obvious ethnic and geographical differences, accounting for approximately 10% of infertility in Asia and 15% in Africa.26 At the country level, it reaches as high as 20% in Japan,30 approximately 5% in France31 and 3% in Italy.32 Studies show that a lack of the gr/gr region is strongly associated with reduced sperm concentration and male infertility.33 However, the relationship between gr/gr loss and male infertility remains controversial. A meta-analysis of more than 100 000 infertile men by Stouffs et al.34 found that the deletion of gr/gr is only a high-risk factor for male infertility in certain regions and populations, with the strongest association found among Caucasians, while no significant association was found in white Brazilians.35 The incidence of gr/gr deletion and the correlation between male infertility and ethnic and geographic changes may be due to differences in the genetic backgrounds of the haplogroup of the Y chromosome.36
VERTICAL TRANSMISSION OF AZF MICRODELETIONS
Because most individuals with AZF microdeletions are infertile, de novo deletions account for approximately 80% of AZF microdeletions.37 A small percentage of individuals with AZF microdeletions are fertile. AZF microdeletions in infertile individuals whose partners give birth to children through microtesticular sperm extraction (micro-TESE), testicular sperm aspiration (TESA), intracytoplasmic sperm injection (ICSI) or other ARTs are still transferred to the progeny. It has also been observed that the AZF microdeletion range might be enlarged in offspring.38
AZFa deletions are rare and account for approximately 0.5%–4% of AZF deletions. The complete AZFa deletion blocks the production and maturation of spermatozoa in the seminiferous tubules, leading to the appearance of Sertoli cell-only syndrome (SCOS). Consequently, men with complete AZFa deletion have a low probability of having spermatozoa cells.39
Recent reports have indicated that patients who lack USP9Y generally do not produce spermatozoa, but they have been detected in patients with partial USP9Y deletion. Two cases of partial USP9Y deletion vertical inheritance were reported by Krausz et al.40 In the first case, the proband exhibited severe oligozoospermia, and the patient in the second case had azoospermia. Their fathers had the same partial microdeletion but a normal phenotype. The first proband had no amplification of sY84, sY86 or exon 22 of USP9Y, and the second case lacked the 3’ end of the USP9Y transcript. This implies that USP9Y may not be the decisive gene for spermatogenesis. One possible implication of this is that while USP9Y contributes to spermatogenesis, it does not cause SCOS. Rodovalho et al.41 reported the special cases of vertical transmission where the father of the proband lacked sY84 and gave rise to 3 offspring naturally. The proband also lacked sY84 and thus had azoospermia, but one of the brothers had sY84 and sY127 deletions and had normal semen quality. The other brother did not have an AZF deletion but still suffered from azoospermia. It may be that there was a microdeletion upstream or downstream of the paternal detection region, suggesting an expanded range of deletions during gametogenesis that resulted in offspring of different genotypes and phenotypes.41 Jia et al.42 found a fertile man with normal semen missing sY83, sY1064 and sY86 whose father had the same deletions. This was passed to his son (proband). Moreover, Jiang et al.43 found a case of an infertile patient with partial AZFa (sY86) deletion but normal sperm concentration and vitality. Alksere et al.44 described a Caucasian man who was infertile despite having a partial deletion of USP9Y (sY84 and sY1323) and normal sperm; his father was likewise affected by the deletion. It has been suggested that partial AZFa deletions may not always result in azoospermia and that USP9Y may not play a decisive role in spermatogenesis. Given the differences among these cases, the relationship between the deletion of USP9Y and spermatogenesis is not clear (Table 1).
Table 1.
Summary of vertical transmission in azoospermia factor a microdeletion
| Study | Population | Deletion gene of proband | Type of deletion | Type of transmission (natural or ART) | Detection method | Proband phenotype (total sperm count per ml) | Father phenotype | Sibling phenotype | Son phenotype |
|---|---|---|---|---|---|---|---|---|---|
| Krausz et al.40 2006 | Family I: Italian Family II: Romanian | I: sY84, sY86 and exon 22 of USP9Y II: the 3’end of USP9Y transcript | I: USP9Y complete II: USP9Y partial | I: natural II: natural | STS PCR and CGH | I: infertility (140×106) II: azoospermia | S and fertility | ND | ND |
| Rodovalho et al.41 2008 | Mixed | Family I: sY84 Family II: sY84 | I: partial II: partial | I: natural II: ART | STS PCR | I: azoospermia II: severe oligospermia | I: S and fertility II: ND | I: S and severe oligospermia II: ND | ND |
| Luddi et al.45 2009 | Italian | USP9Y | USP9Y complete | Natural | STS PCR and CGH | Mild asthenozoospermia (54×106–660×106) | S and fertility | S and fertility | ND |
| Alksere et al.44 2019 | Latvian | sY84, sY1323 | Partial | Natural | STS PCR | Infertility | S and fertility | ND | ND |
| Tang et al.46 2020 | Han Chinese | sY84, sY86 | Complete | Natural | STS PCR and sanger sequencing | Fertility | S and fertility | ND | ND |
| Jia et al.42 2020 | Han Chinese | sY83, sY1064, sY86 | Complete | Natural | STS PCR and CGH | Fertility | S and fertility | ND | ND |
S: the deletion range is the same as the proband; NS: the deletion range is not the same as the proband; ND: the phenotype is not determined; STS: sequence tagged sites; PCR: polymerase chain reaction; CGH: comparative genomic hybridization; USP9Y: ubiquitin-specific peptidase 9, Y-linked; ART: assisted reproductive technology
Few cases of completely lacking USP9Y with normal semen quality have been described in the literature. An analysis by Luddi et al.45 described a proband with mild asthenozoospermia and complete deletion of USP9Y in the AZFa region. The father and brother of the proband also had this USP9Y deletion, suggesting that the father naturally passed the AZFa deletion to his offspring. USP9Y is thus more likely a factor that improves spermatogenic efficiency rather than one required for spermatogenesis. Similarly, Tang et al.46 reported a case of a vertical genetic family lacking AZFa (sY84 and sY86). The proband’s semen analysis was normal, but both he and his father lacked the hg38Y fragment (Y: 12470437–12690385) in the AZFa region. These results further support the idea that the deletion of sY84 and sY86 may indicate the high probability of complete deletion of AZFa. The type of deletion should be identified through gene sequencing.
The incidence of AZFb microdeletions is modest, accounting for 1%–55% of all AZF deletions. The lack of AZFb causes spermatogenesis to stop at the primary spermatocyte stage; testicular biopsy can reveal spermatogonia and primary spermatocytes but no spermatozoa. As a result, AZFb microdeletions are generally not naturally inherited and only infrequently inherited via ART.47
Although azoospermia comprises the vast majority of clinical phenotypes of AZFb microdeletions, a few individuals have fertility, indicating the presence of natural inheritance. Stouffs et al.48 discovered spermatozoa in two individuals who had severe oligo-astheno-teratozoospermia and cryptozoospermia with AZFb microdeletion, with one of them achieving fertility via ICSI. A specific family of AZFb microdeletions with three generations of natural transmission was found by Plotton et al.49 The proband with oligo-asthenozoospermia had an AZFb microdeletion (sY142, sY143, sY1197, sY1192, and G34984), shared by his father and son. Similarly, Rolf et al.50 reported a family with three generations of natural AZFb deletion transmission (sY143 and sY130). The proband had moderate oligo-astheno-teratozoospermia with natural conception. In addition, Samli et al.51 discovered that an azoospermia proband, his uncle, father, and three brothers all had an AZFb, RNA-binding motif on Y (RBM1), microdeletion, and the proband also had an additional AZFa (sY81) deletion. Despite the fact that the father naturally transmitted the AZFb (RBM1) microdeletion to his four sons, the elder brothers were unable to conceive, but the younger brother was the father of two daughters. This suggests that some AZFb microdeletion patients can be naturally transmitted vertically and that the deletion range can be expanded in offspring, resulting in a variety of clinical phenotypes51 (Table 2).
Table 2.
Summary of vertical transmission in azoospermia factor b microdeletion
| Study | Population | Deletion gene of proband | Type of deletion | Type of transmission (natural or ART) | Detection method | Proband phenotype (total sperm count per ml) | Father phenotype | Sibling phenotype | Son phenotype |
|---|---|---|---|---|---|---|---|---|---|
| Plotton et al.49 2010 | French | sY142, sY143, sY1197, sY1192, G34984 (PRY) | Partial | Natural | STS PCR | Fertility (3.6×106) | S and fertility | ND | S |
| Rolf et al.50 2002 | Italian | sY130, sY143 | Partial | Natural | STS PCR | Fertility (2.4×106–8.3×106) | S and fertility | ND | S |
| Samli et al.51 2006 | Turks | sY84, sY127, RBM1 | Partial | Natural | STS PCR | Azoospermia | NS and fertility | I: NS and azoospermia II: NS and azoospermia III: NS and fertility | ND |
| Zhang et al.52 2017 | Han Chinese | sY121, sY127, sY134, sY143 | Complete | ART | STS PCR | Azoospermia | Fertility | ND | S |
S: the deletion range is the same as the proband; NS: the deletion range is not the same as the proband; ND: the phenotype is not determined; STS: sequence tagged sites; PCR: polymerase chain reaction; ART: assisted reproductive technology; PRY: PTP-BL related on the Y chromosome; RBM1: RNA-binding motif on Y
Zhang et al.52 described a patient with a full AZFb deletion (sY121, sY127, sY134, and sY143) who had severe oligozoospermia and transferred the same deletion type to his offspring by ICSI. There was no significant difference in the clinical pregnancy rate of ICSI when spermatozoa from patients with AZFb microdeletion were compared to spermatozoa from nonaffected individuals. This shows that sperm from patients with AZFb microdeletions can transmit the deletion to their offspring through ART.53
AZFc microdeletions have been associated with a variety of clinical and histological phenotypes, ranging from azoospermia to oligozoospermia. TESE can achieve a 50% spermatozoon retrieval rate, and the success rate of testicular sperm retrieval with micro-TESE ranges from 9% to as high as 80%.54 As a result, the natural and ART inheritance of AZFc microdeletions is more prevalent than those of the other two.
While the majority of AZFc microdeletions result in male sterility, researchers in one study found that the father’s AZFc microdeletions were passed on to three offspring through natural reproduction. Two of them had azoospermia, whereas the other had normal semen quality, demonstrating the variety of AZFc microdeletion phenotypes.55 Saut et al.56 described a unique example of a family in which the father lacked DAZ and CDY yet transmitted the Y chromosome to his three sons through natural reproduction. The children had the same deletion range as their parents, but they developed azoospermia. Chang et al.57 found that the proband’s father was a DAZ gene deletion carrier who produced naturally four DAZ gene-deleted and infertile offspring; however, the proband’s 63-year-old father had been diagnosed with azoospermia. This implies that people with AZFc microdeletions may initially have normal fertility, but their fertility diminishes with age, leading to azoospermia. Pan et al.58 found that 3 infertile patients naturally inherited b2/b3 region deletions from their father, expanding the deletion range. Patient 1, with an AZFc deletion (sY152, sY157, sY254, and sY255), had oligozoospermia; Patient 2, with an AZFa+b+c deletion, had azoospermia; and Patient 3, with an AZFb+c deletion, also had azoospermia. This discovery is consistent with the findings of Calogero et al.59 who discovered that the proband exhibited severe oligozoospermia with additional sY1192 and sY153 deletions when compared with his father.
Since most infertile patients with AZFc microdeletions have a relatively high probability of harboring spermatozoa accessible via micro-TESE or TESA, these deletions are mainly passed to the offspring through ART. Several studies have revealed that AZFc microdeletions are inherited vertically via ICSI, which does not cause their expansion.60–62 In addition, Oates et al.63 concluded that the clinical outcome of ICSI was not affected by the presence of an AZFc deletion; progeny generally inherited AZFc deletions, but the length of the deletion did not increase. Furthermore, Lynch et al.64 found that the gr/gr deletion was inherited vertically through ICSI, and no de novo deletion of gr/gr was detected; hence, ICSI is not a risk factor for the expansion of deletion in the AZFc region. However, Komori et al.65 found three groups of fathers and sons who had inherited AZFc microdeletions vertically through ICSI, one of which had progeny deletion extension; when compared with his father, the proband of this group had azoospermia with additional deletions (sY245, sY255, sY236, and sY267). After reviewing approximately 100 infertile patients with AZFc microdeletions, Kim et al.66 found that the microdeletions had no significant effect on the ICSI results. Nevertheless, the AZFc microdeletions could be inherited vertically by the offspring via ICSI. In short, ICSI might not be a risk factor for AZFc microdeletions and does not affect the clinical pregnancy outcomes from this procedure67 (Table 3).
Table 3.
Summary of vertical transmission in azoospermia factor c microdeletion
| Study | Population | Deletion gene of proband | Type of deletion | Type of transmission (natural or ART) | Detection method | Proband phenotype (total sperm count per ml) | Father phenotype | Sibling phenotype | Son phenotype |
|---|---|---|---|---|---|---|---|---|---|
| Kühnert et al.55 2004 | German | sY1192, sY152, sY157, sY158, sY255, sY254, sY1125, sY1054 | Partial | Natural | STS PCR | Azoospermia | S and fertility | I: S and azoospermia III: ND |
ND |
| Chang et al.57 1999 | American | sY149, sY147, sY145, sY148, sY152, sY154, sY158 | Partial | Natural | STS PCR | Infertility (0.5×106) | S and fertility | I: S and azoospermia II: S and azoospermia III: severe oligospermia |
ND |
| Pan et al.58 2018 | Han Chinese | Family I: sY152, sY157, sY254, sY255 Family II: AZFa+b + c complete deletion Family III: sY127, sY134, sY143, sY152, sY157, sY254, sY255 | I: partial II: partial III: partial | I: natural II: natural III: natural | High-throughput MLPA sequencing | I: infertility (5.6×106–7.2×106) II: azoospermia III: azoospermia | I: B2/b3 subdeletion and fertility II: B2/b3 subdeletion and fertility III: B2/b3 duplication and fertility |
ND | ND |
| Calogero et al.59 2002 | Italian | sY1192, sY153, sY152, sY155, sY158, sY147, sY149, sY220, sY254, sY255, sY243, sY236, sY283, sY202, sY277 | Partial | Natural | STS PCR | Infertility (0.2×106–0.5×106) | NS and fertility | ND | ND |
| Saut et al.56 2000 | French | sY153, sY152, sY155, sY154, sY158, sY148, sY220, sY243, sY269 | Partial | Natural | STS PCR | Azoospermia | S and fertility | I: S and azoospermia II: S and azoospermia | ND |
| Page et al.61 1999 | American | Family I: sY205, sY254, sY624, sY602, sY202, sY158 Family II: sY205, sY254, sY624, sY602, sY202, sY158 Family III: sY205, sY254, sY624, sY602, sY202, sY158 | I: partial II: partial III: partial | I: ART II: ART III: ART | STS PCR | I: azoospermia II: azoospermia III: azoospermia |
S and fertility | ND | S and ND |
| Kleiman et al.60 1999 | Mixed | Y153, sY254, sY255, sY158, DAZ, sY160 | Partial | ART | STS PCR | Azoospermia | S and fertility | ND | S and ND |
| Komori et al.65 2002 | Japanese | Family I: sY240 Family II: sY233, sY240, sY245, sY277, sY254, sY255, sY283, sY236, sY267 Family III: sY233, sY240, sY254, sY283 | I: partial II: partial | I: ART II: ART | STS PCR | I: azoospermia II: azoospermia III: azoospermia |
ND | ND | I: S and ND II: S and ND III: S and ND |
S: the deletion range is the same as the proband; NS: the deletion range is not the same as the proband; ND: the phenotype is not determined; STS: sequence tagged sites; PCR: polymerase chain reaction; MLPA: multiplex ligation-dependent probe amplification; DAZ: deleted-in-azoospermia; ART: assisted reproductive technology
In summary, the clinical phenotype of patients with AZFc deletions is heterogeneous, ranging from normal seminal sperm concentrations to azoospermia, and the natural inheritance of these deletions is not uncommon. With the development of ART, AZFc microdeletions are mainly vertically transmitted through ICSI. Most recent studies show that AZFc microdeletions do not affect the pregnancy outcome of ICSI. Finally, while ICSI does not cause de novo deletions in offspring, the vertical transmission of AZFc microdeletions may increase the risk of deletion expansion.68
PERSPECTIVES
A significant genetic component of male infertility is AZF locus microdeletion. Consequently, each male patient with severe oligospermia should have a Y microdeletion test performed in clinical practice.69 However, because the clinical phenomenology of the various deletions is heterogeneous, it can be determined by several variables, including environment and genetics. This has resulted in the vertical transmission of deletions in the AZF region, resulting in infertility in the children.
One of the limitations of current research is that the 6 sequence tagged sites (STS) of the European Andrology Society’s regular clinical test cannot be thoroughly examined for this kind of deletion and that gene heterogeneity leads to an ambiguous genotype-phenotype association.4 The majority of clinical research on vertical transmission in the AZF region to date has involved case reports using various methodologies. The majority of these methodologies utilized STS polymerase chain reaction (PCR) rather than gene sequencing to determine the type and extent of the deletion; some deletion expansion fragments may be overlooked as a result. With the completion of the human genome sequence, advanced tools and methods for AZF microdeletion molecular research, such rapid and large-scale high-throughput genome sequencing technology and the multiplex ligand probe-dependent amplification (MLPA) method, are becoming more widely available. These techniques will better emphasize the type of AZF microdeletion and the associated clinical phenotype. Furthermore, the function of the candidate gene will be clarified, resulting in a breakthrough in AZF microdeletion research and therapy.70,71
Although ART is beneficial to infertile individuals, it increases the chance of the vertical transmission of AZF microdeletions. To minimize the probability of transmission of AZF microdeletions, genetic analysis and consultation should be performed on both parents when performing ART. The deletion range of offspring with AZF microdeletions often expands with age, resulting in a progressive sperm decrease. Patients with oligozoospermia resulting from an AZFc microdeletion should have their sperm cryopreserved as soon as feasible to avoid the need for ART in future.72
The possible pathological consequences of the vertical transmission of AZF deletions in male offspring should be noted. Lysine (K)-specific demethylase 5D (KDM5D) is located in the AZFb region and encodes a JmjC domain-containing protein. KDM5D inhibits the invasiveness of prostate cancer cells, and the gene is frequently deleted in metastatic prostate cancer.73,74 The findings from these studies suggest that low KDM5D expression may be associated with poor prognosis in prostate cancer patients. In addition, Nathanson et al.75 reported that gr/gr deletion is associated with a two-fold increased risk of testicular germ cell tumors. A recent study showed that normozoospermic gr/gr deletion carriers carry a four-fold increased risk of developing the disease.76 Evidence from studies suggests that gr/gr deletion may be an independent risk factor for testicular germ cell tumors. In addition, there is some evidence to suggest that AZFb+c deletion has been linked to abnormal height (severely short or extremely tall) and neuropsychiatric disorders.77 Notwithstanding the relatively limited sample and the lack of molecular information in tissues, these works offer valuable insights into the relationship between the vertical transmission of AZF deletions in male offspring and genitourinary cancers.
Distinct Y lineages classified by Y chromosome haplogroups (Y-hgs) are linked with numerous vertical transmission processes. In Chilean individuals, Y chromosome haplogroups M and Y chromosome haplogroups H are associated with gr/gr deletion, and Y chromosome haplogroups Q1a3a might enhance susceptibility to AZFb deletion. However, partial AZFc deletion does not appear to produce serious spermatogenic problems.78 In Chinese individuals, Y chromosome haplogroups C and Y chromosome haplogroups DE might contribute to spermatogenic failure in AZFc deficiency, whereas Y-hgQ3 may have the opposite effect.79 Overall, a link has been shown between Y-hg and the AZF deletion mechanisms, and the AZF genes have varying influences on spermatogenic function in different species of Y-hg.80 However, to our knowledge, no research has yet identified a definite link between the expansion of deletions in the vertical transmission of AZF and Y-hg.
AUTHOR CONTRIBUTIONS
CYD collected information from database and writing the manuscript. ZZ helped with the design of charts and tables. WHT and HJ contributed to the design of the review and supervised the research. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
ACKNOWLEDGMENTS
The present study was supported by National Natural Science Foundation of China (No. 81901535 and No. 82071698), the National Key Research & Developmental Program of China (No. 2021YFC2700203), and Natural Science Foundation of Beijing Municipality (No. 7222208).
REFERENCES
- 1.Vogt PH, Bender U, Deibel B, Kiesewetter F, Zimmer J, et al. Human AZFb deletions cause distinct testicular pathologies depending on their extensions in Yq11 and the Y haplogroup: new cases and review of literature. Cell Biosci. 2021;11:60. doi: 10.1186/s13578-021-00551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witherspoon L, Dergham A, Flannigan R. Y-microdeletions: a review of the genetic basis for this common cause of male infertility. Transl Androl Urol. 2021;10:1383–90. doi: 10.21037/tau-19-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kent-First M, Muallem A, Shultz J, Pryor J, Roberts K, et al. Defining regions of the Y-chromosome responsible for male infertility and identification of a fourth AZF region (AZFd) by Y-chromosome microdeletion detection. Mol Reprod Dev. 1999;53:27–41. doi: 10.1002/(SICI)1098-2795(199905)53:1<27::AID-MRD4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 4.Krausz C, Hoefsloot L, Simoni M, Tüttelmann F. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology. 2014;2:5–19. doi: 10.1111/j.2047-2927.2013.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waseem AS, Singh V, Makker GC, Trivedi S, Mishra G, et al. AZF deletions in Indian populations: original study and meta-analyses. J Assist Reprod Genet. 2020;37:459–69. doi: 10.1007/s10815-019-01661-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu X, Wei Z, Jiang Y, Zhang S. Y chromosome azoospermia factor region microdeletions and transmission characteristics in azoospermic and severe oligozoospermic patients. Int J Clin Exp Med. 2015;8:14634–46. [PMC free article] [PubMed] [Google Scholar]
- 7.Lee KH, Song GJ, Kang IS, Kim SW, Paick JS, et al. Ubiquitin-specific protease activity of USP9Y, a male infertility gene on the Y chromosome. Reprod Fertil Dev. 2003;15:129–33. doi: 10.1071/rd03002. [DOI] [PubMed] [Google Scholar]
- 8.Kotov AA, Olenkina OM, Godneeva BK, Adashev VE, Olenina LV. Progress in understanding the molecular functions of DDX3Y (DBY) in male germ cell development and maintenance. Biosci Trends. 2017;11:46–53. doi: 10.5582/bst.2016.01216. [DOI] [PubMed] [Google Scholar]
- 9.Gažová I, Lengeling A, Summers KM. Lysine demethylases KDM6A and UTY: the X and Y of histone demethylation. Mol Genet Metab. 2019;127:31–44. doi: 10.1016/j.ymgme.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Dreumont N, Bourgeois CF, Lejeune F, Liu Y, Ehrmann IE, et al. Human RBMY regulates germline-specific splicing events by modulating the function of the serine/arginine-rich proteins 9G8 and Tra2-β. J Cell Sci. 2010;123:40–50. doi: 10.1242/jcs.055889. [DOI] [PubMed] [Google Scholar]
- 11.Yarahmadi E, Borjian Boroujeni P, Totonchi M, Gourabi H. Genotyping of the EIF1AY gene in Iranian patients with non-obstructive azoospermia. Curr Urol. 2019;13:46–50. doi: 10.1159/000499295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Liang Z, Yang J, Wang D, Wang H, et al. DAZL is a master translational regulator of murine spermatogenesis. Natl Sci Rev. 2019;6:455–68. doi: 10.1093/nsr/nwy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vangompel MJ, Xu EY. The roles of the DAZ family in spermatogenesis: more than just translation?Spermatogenesis. 2011;1:36–46. doi: 10.4161/spmg.1.1.14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Yang X, Gui B, Xie G, Zhang D, et al. Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J Biol Chem. 2011;286:42414–25. doi: 10.1074/jbc.M111.271064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guliyev M, Yilmaz S, Sahin K, Marakli S, Gozukirmizi N. Human endogenous retrovirus-H insertion screening. Mol Med Rep. 2013;7:1305–9. doi: 10.3892/mmr.2013.1295. [DOI] [PubMed] [Google Scholar]
- 16.Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, et al. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum Mol Genet. 2000;9:2291–6. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- 17.Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–72. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- 18.Choi J, Koh E, Matsui F, Sugimoto K, Suzuki H, et al. Study of azoospermia factor-a deletion caused by homologous recombination between the human endogenous retroviral elements and population-specific alleles in Japanese infertile males. Fertil Steril. 2008;89:1177–82. doi: 10.1016/j.fertnstert.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Repping S, Skaletsky H, Lange J, Silber S, Fulco V, et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 2002;71:906–22. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandi N, Cadeddu M, Blomberg J, Tramontano E. Contribution of type W human endogenous retroviruses to the human genome: characterization of HERV-W proviral insertions and processed pseudogenes. Retrovirology. 2016;13:67. doi: 10.1186/s12977-016-0301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sin HS, Koh E, Taya M, Iijima M, Sugimoto K, et al. A novel Y chromosome microdeletion with the loss of an endogenous retrovirus related, testis specific transcript in AZFb region. J Urol. 2011;186:1545–52. doi: 10.1016/j.juro.2011.05.044. [DOI] [PubMed] [Google Scholar]
- 22.Cerván-Martín M, Castilla JA, Palomino-Morales RJ, Carmona FD. Genetic landscape of nonobstructive azoospermia and new perspectives for the clinic. J Clin Med. 2020;9:300. doi: 10.3390/jcm9020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozen SG, Marszalek JD, Irenze K, Skaletsky H, Brown LG, et al. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. Am J Hum Genet. 2012;91:890–6. doi: 10.1016/j.ajhg.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35:247–51. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 25.Ferlin A, Tessari A, Ganz F, Marchina E, Barlati S, et al. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet. 2005;42:209–13. doi: 10.1136/jmg.2004.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu B, Lu NX, Xia YK, Gu AH, Lu CC, et al. A frequent Y chromosome b2/b3 subdeletion shows strong association with male infertility in Han-Chinese population. Hum Reprod. 2007;22:1107–13. doi: 10.1093/humrep/del499. [DOI] [PubMed] [Google Scholar]
- 27.Xiao X, Jiang L, Wang T, Wang Y, Lu L, et al. [Study on the relationship between partial deletions in the azzospermia factor C region of Y chromosome and male infertility. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010;27:294–8. doi: 10.3760/cma.j.issn.1003-9406.2010.0.013. [Article in Chinese] [DOI] [PubMed] [Google Scholar]
- 28.Hucklenbroich K, Gromoll J, Heinrich M, Hohoff C, Nieschlag E, et al. Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod. 2005;20:191–7. doi: 10.1093/humrep/deh558. [DOI] [PubMed] [Google Scholar]
- 29.Repping S, van Daalen SK, Korver CM, Brown LG, Marszalek JD, et al. A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics. 2004;83:1046–52. doi: 10.1016/j.ygeno.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 30.Carvalho C, Zuccherato L, Fujisawa M, Shirakawa T, Ribeiro A, et al. Study of AZFc partial deletion gr/gr in fertile and infertile Japanese males. J Hum Genet. 2006;51:794–9. doi: 10.1007/s10038-006-0024-2. [DOI] [PubMed] [Google Scholar]
- 31.Ravel C, Sandra C, Brahim E, Rouba H, Legendre M, et al. Y-chromosome AZFc structural architecture and relationship to male fertility. Fertil Steril. 2009;92:1924–33. doi: 10.1016/j.fertnstert.2008.08.135. [DOI] [PubMed] [Google Scholar]
- 32.Giachini C, Laface I, Guarducci E, Balercia G, Forti G, et al. Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet. 2008;124:399–410. doi: 10.1007/s00439-008-0561-1. [DOI] [PubMed] [Google Scholar]
- 33.Bansal SK, Jaiswal D, Gupta N, Singh K, Dada R, et al. Gr/gr deletions on Y-chromosome correlate with male infertility: an original study, meta-analyses, and trial sequential analyses. Sci Rep. 2016;6:19798. doi: 10.1038/srep19798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stouffs K, Lissens W, Tournaye H, Haentjens P. What about gr/gr deletions and male infertility?Systematic review and meta-analysis. Hum Reprod Update. 2011;17:197–209. doi: 10.1093/humupd/dmq046. [DOI] [PubMed] [Google Scholar]
- 35.Carvalho C, Zuccherato L, Luciana B, Santos F, Pena S. No association found between gr/gr deletions and infertility in Brazilian males. Mol Hum Reprod. 2006;12:269–73. doi: 10.1093/molehr/gal029. [DOI] [PubMed] [Google Scholar]
- 36.Paulo N, Gonçalves J, Plancha C. The AZFc region of the Y chromosome: at the crossroads between genetic diversity and male infertility. Hum Reprod Update. 2010;16:525–42. doi: 10.1093/humupd/dmq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai RL, Sun LK, Yang X, Li LL, Zhu HB, et al. Expansion and de novo occurrence of Y chromosome microdeletions occurring via natural vertical transmission in northeastern China. J Int Med Res. 2012;40:1182–91. doi: 10.1177/147323001204000339. [DOI] [PubMed] [Google Scholar]
- 38.Lee S, Ahn S, Lee K, Kwack K, Jun H, et al. Intracytoplasmic sperm injection may lead to vertical transmission, expansion, and de novo occurrence of Y-chromosome microdeletions in male fetuses. Ferti Steril. 2006;85:1512–5. doi: 10.1016/j.fertnstert.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 39.Kleiman SE, Almog R, Yogev L, Hauser R, Lehavi O, et al. Screening for partial AZFa microdeletions in the Y chromosome of infertile men: is it of clinical relevance? Fertil Steril. 2012;98:43–7. doi: 10.1016/j.fertnstert.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 40.Krausz C, Degl’Innocenti S, Nuti F, Morelli A, Felici F, et al. Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility. Hum Mol Genet. 2006;15:2673–81. doi: 10.1093/hmg/ddl198. [DOI] [PubMed] [Google Scholar]
- 41.Rodovalho R, Arruda J, Moura K. Tracking microdeletions of the AZF region in a patrilineal line of infertile men. Genet Mol Res. 2008;7:614–22. doi: 10.4238/vol7-3gmr455. [DOI] [PubMed] [Google Scholar]
- 42.Jia Y, Niu Z, Li W, Qin Q, Sun T, et al. A fertile male with a single sY86 deletion on the Y chromosome. Asian J Androl. 2020;22:333–4. doi: 10.4103/aja.aja_94_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang KA, Chen X, Liang YB, Tong KY, Ye H. A case of partial deletion of AZFa region of Y chromosome and review. Nat J Androl. 2019;25:953–5. [Google Scholar]
- 44.Alksere B, Berzina D, Dudorova A, Conka U, Andersone S, et al. Case of inherited partial AZFa deletion without impact on male fertility. Case Rep Genet. 2019;2019:3802613. doi: 10.1155/2019/3802613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luddi A, Margollicci M, Gambera L, Serafini F, Cioni M, et al. Spermatogenesis in a man with complete deletion of USP9Y. N Engl J Med. 2009;360:881–5. doi: 10.1056/NEJMoa0806218. [DOI] [PubMed] [Google Scholar]
- 46.Tang D, Liu W, Li G, He X, Zhang Z, et al. Normal fertility with deletion of sY84 and sY86 in AZFa region. Andrology. 2020;8:332–6. doi: 10.1111/andr.12692. [DOI] [PubMed] [Google Scholar]
- 47.Kleiman SE, Yogev L, Lehavi O, Hauser R, Botchan A, et al. The likelihood of finding mature sperm cells in men with AZFb or AZFb-c deletions: six new cases and a review of the literature (1994-2010) Fertil Steril. 2011;95:2005–12. doi: 10.1016/j.fertnstert.2011.01.162. 12.e1–4. [DOI] [PubMed] [Google Scholar]
- 48.Stouffs K, Vloeberghs V, Gheldof A, Tournaye H, Seneca S. Are AZFb deletions always incompatible with sperm production? Andrology. 2017;5:691–4. doi: 10.1111/andr.12350. [DOI] [PubMed] [Google Scholar]
- 49.Plotton I, Ducros C, Pugeat M, Morel Y, Lejeune H. Transmissible microdeletion of the Y-chromosome encompassing two DAZ copies, four RBMY1 copies, and both PRY copies. Fertil Steril. 2010;94:2770.e11–6. doi: 10.1016/j.fertnstert.2010.04.038. [DOI] [PubMed] [Google Scholar]
- 50.Rolf C, Gromoll J, Simoni M, Nieschlag E. Natural transmission of a partial AZFb deletion of the Y chromosome over three generations: case report. Hum Reprod. 2002;17:2267–71. doi: 10.1093/humrep/17.9.2267. [DOI] [PubMed] [Google Scholar]
- 51.Samli H, Murat S, Solak M. Natural transmission of AZFb Y-chromosomal microdeletion from father to his three sons. Arch Androl. 2006;52:423–6. doi: 10.1080/01485010600822655. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Li L, Xue L, Zhang H, Zhu Y, et al. Complete azoospermia factor b deletion of y chromosome in an infertile male with severe oligoasthenozoospermia: case report and literature review. Urology. 2017;102:111–5. doi: 10.1016/j.urology.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 53.Choi JM, Chung P, Veeck L, Mielnik A, Palermo GD, et al. AZF microdeletions of the Y chromosome and in vitro fertilization outcome. Fertil Steril. 2004;81:337–41. doi: 10.1016/j.fertnstert.2003.06.030. [DOI] [PubMed] [Google Scholar]
- 54.Pavan-Jukic D, Stubljar D, Jukic T, Starc A. Predictive factors for sperm retrieval from males with azoospermia who are eligible for testicular sperm extraction (TESE) Syst Biol Reprod Med. 2020;66:70–5. doi: 10.1080/19396368.2019.1680764. [DOI] [PubMed] [Google Scholar]
- 55.Kühnert B, Gromoll J, Kostova E, Tschanter P, Luetjens CM, et al. Case report: natural transmission of an AZFc Y-chromosomal microdeletion from father to his sons. Hum Reprod. 2004;19:886–8. doi: 10.1093/humrep/deh186. [DOI] [PubMed] [Google Scholar]
- 56.Saut N, Terriou P, Navarro A, Lévy N, Mitchell MJ. The human Y chromosome genes BPY2, CDY1 and DAZ are not essential for sustained fertility. Mol Hum Reprod. 2000;6:789–93. doi: 10.1093/molehr/6.9.789. [DOI] [PubMed] [Google Scholar]
- 57.Chang PL, Sauer MV, Brown S. Y chromosome microdeletion in a father and his four infertile sons. Hum Reprod. 1999;14:2689–94. doi: 10.1093/humrep/14.11.2689. [DOI] [PubMed] [Google Scholar]
- 58.Pan Y, Li L, Yu Y, Jiang Y, Yang X, et al. Natural transmission of b2/b3 subdeletion or duplication to expanded Y chromosome microdeletions. Med Sci Monit. 2018;24:6559–63. doi: 10.12659/MSM.911644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calogero AE, Garofalo MR, Barone N, Longo GA, De Palma A, et al. Spontaneous transmission from a father to his son of a Y chromosome microdeletion involving the deleted in azoospermia (DAZ) gene. J Endocrinol Invest. 2002;25:631–4. doi: 10.1007/BF03345088. [DOI] [PubMed] [Google Scholar]
- 60.Kleiman SE, Yogev L, Gamzu R, Hauser R, Botchan A, et al. Three-generation evaluation of Y-chromosome microdeletion. J Androl. 1999;20:394–8. [PubMed] [Google Scholar]
- 61.Page DC, Silber S, Brown LG. Men with infertility caused by AZFc deletion can produce sons by intracytoplasmic sperm injection, but are likely to transmit the deletion and infertility. Hum Reprod. 1999;14:1722–6. doi: 10.1093/humrep/14.7.1722. [DOI] [PubMed] [Google Scholar]
- 62.Cram DS, Ma K, Bhasin S, Arias J, Pandjaitan M, et al. Y chromosome analysis of infertile men and their sons conceived through intracytoplasmic sperm injection: vertical transmission of deletions and rarity of de novo deletions. Fertil Steril. 2000;74:909–15. doi: 10.1016/s0015-0282(00)01568-5. [DOI] [PubMed] [Google Scholar]
- 63.Oates RD, Silber S, Brown LG, Page DC. Clinical characterization of 42 oligospermic or azoospermic men with microdeletion of the AZFc region of the Y chromosome, and of 18 children conceived via ICSI. Hum Reprod. 2002;17:2813–24. doi: 10.1093/humrep/17.11.2813. [DOI] [PubMed] [Google Scholar]
- 64.Lynch M, Cram DS, Reilly A, O’Bryan MK, Baker HW, et al. The Y chromosome gr/gr subdeletion is associated with male infertility. Mol Hum Reprod. 2005;11:507–12. doi: 10.1093/molehr/gah191. [DOI] [PubMed] [Google Scholar]
- 65.Komori S, Kato H, Kobayashi S, Koyama K, Isojima S. Transmission of Y chromosomal microdeletions from father to son through intracytoplasmic sperm injection. J Hum Genet. 2002;47:465–8. doi: 10.1007/s100380200066. [DOI] [PubMed] [Google Scholar]
- 66.Kim MJ, Choi HW, Park SY, Song IO, Seo JT, et al. Molecular and cytogenetic studies of 101 infertile men with microdeletions of Y chromosome in 1,306 infertile Korean men. J Assist Reprod Genet. 2012;29:539–46. doi: 10.1007/s10815-012-9748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu XY, Wang RX, Fu Y, Luo LL, Guo W, et al. Outcomes of intracytoplasmic sperm injection in oligozoospermic men with Y chromosome AZFb or AZFc microdeletions. Andrologia. 2017;49:e12602. doi: 10.1111/and.12602. [DOI] [PubMed] [Google Scholar]
- 68.Xi Q, Zhang Z, Wang R, Li L, Li L, et al. Obstetric and perinatal outcomes of intracytoplasmic sperm injection for infertile men with Y chromosome microdeletions. Medicine (Baltimore) 2019;98:e17407. doi: 10.1097/MD.0000000000017407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson M, Raheem A, De Luca F, Hallerstrom M, Zainal Y, et al. An analysis of the frequency of Y-chromosome microdeletions and the determination of a threshold sperm concentration for genetic testing in infertile men. BJU Int. 2019;123:367–72. doi: 10.1111/bju.14521. [DOI] [PubMed] [Google Scholar]
- 70.Sun K, Chen XF, Zhu XB, Hu HL, Zhang W, et al. A new molecular diagnostic approach to assess Y chromosome microdeletions in infertile men. J Int Med Res. 2012;40:237–48. doi: 10.1177/147323001204000124. [DOI] [PubMed] [Google Scholar]
- 71.Franchim CS, Soares-Junior JM, Serafini PC, Monteleone PA, Coccuzza MS, et al. Efficacy of MLPA for detection of Y-chromosome microdeletions in infertile Brazilian patients. J Assist Reprod Genet. 2020;37:1251–9. doi: 10.1007/s10815-020-01777-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Colaco S, Modi D. Consequences of Y chromosome microdeletions beyond male infertility. J Assist Reprod Genet. 2019;36:1329–37. doi: 10.1007/s10815-019-01492-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li N, Dhar SS, Chen TY, Kan PY, Wei Y, et al. JARID1D is a suppressor and prognostic marker of prostate cancer invasion and metastasis. Cancer Res. 2016;76:831–43. doi: 10.1158/0008-5472.CAN-15-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jangravi Z, Tabar MS, Mirzaei M, Parsamatin P, Vakilian H, et al. Two splice variants of y chromosome-located lysine-specific demethylase 5D have distinct function in prostate cancer cell line (DU-145) J Proteome Res. 2015;14:3492–502. doi: 10.1021/acs.jproteome.5b00333. [DOI] [PubMed] [Google Scholar]
- 75.Nathanson KL, Kanetsky PA, Hawes R, Vaughn DJ, Letrero R, et al. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am J Hum Genet. 2005;77:1034–43. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moreno-Mendoza D, Casamonti E, Paoli D, Chianese C, Riera-Escamilla A, et al. gr/gr deletion predisposes to testicular germ cell tumour independently from altered spermatogenesis: results from the largest European study. Eur J Hum Genet. 2019;27:1578–88. doi: 10.1038/s41431-019-0420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castro A, Rodríguez F, Flórez M, López P, Curotto B, et al. Pseudoautosomal abnormalities in terminal AZFb+c deletions are associated with isochromosomes Yp and may lead to abnormal growth and neuropsychiatric function. Hum Reprod. 2017;32:465–75. doi: 10.1093/humrep/dew333. [DOI] [PubMed] [Google Scholar]
- 78.Lardone MC, Ortega V, Ortiz E, Flórez M, Piottante A, et al. Partial-AZFc deletions in Chilean men with primary spermatogenic impairment: gene dosage and Y-chromosome haplogroups. J Assist Reprod Genet. 2020;37:3109–19. doi: 10.1007/s10815-020-01957-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang Y, Ma M, Li L, Zhang W, Chen P, et al. Y chromosome haplogroups may confer susceptibility to partial AZFc deletions and deletion effect on spermatogenesis impairment. Hum Reprod. 2008;23:2167–72. doi: 10.1093/humrep/den229. [DOI] [PubMed] [Google Scholar]
- 80.Vogt PH. AZF deletions and Y chromosomal haplogroups: history and update based on sequence. Hum Reprod Update. 2005;11:319–36. doi: 10.1093/humupd/dmi017. [DOI] [PubMed] [Google Scholar]