Significance
Plants are continuously challenged by pathogens, such as rhizogenic agrobacteria, which cause the formation of "crazy" or "hairy" roots in susceptible plant species. Despite this pathogenic effect, hairy root platforms are also valuable biotechnological tools and are intensively explored as potential sustainable production systems for bioactive plant-specialized metabolites. However, many of the molecular mechanisms underlying hairy root formation remain unknown. Here, we combined a number of cutting-edge omics technologies to unravel the molecular function of one of the major players in hairy root disease and development, namely the RolB oncoprotein. Our data support a model in which this bacterial protein directs the plant transcription machinery to promote hairy root development.
Keywords: hairy roots, Rol proteins, rhizogenic Agrobacterium, Solanum lycopersicum, TOPLESS
Abstract
Rhizogenic Agrobacterium strains comprise biotrophic pathogens that cause hairy root disease (HRD) on hydroponically grown Solanaceae and Cucurbitaceae crops, besides being widely explored agents for the creation of hairy root cultures for the sustainable production of plant-specialized metabolites. Hairy root formation is mediated through the expression of genes encoded on the T-DNA of the root-inducing (Ri) plasmid, of which several, including root oncogenic locus B (rolB), play a major role in hairy root development. Despite decades of research, the exact molecular function of the proteins encoded by the rol genes remains enigmatic. Here, by means of TurboID-mediated proximity labeling in tomato (Solanum lycopersicum) hairy roots, we identified the repressor proteins TOPLESS (TPL) and Novel Interactor of JAZ (NINJA) as direct interactors of RolB. Although these interactions allow RolB to act as a transcriptional repressor, our data hint at another in planta function of the RolB oncoprotein. Hence, by a series of plant bioassays, transcriptomic and DNA-binding site enrichment analyses, we conclude that RolB can mitigate the TPL functioning so that it leads to a specific and partial reprogramming of phytohormone signaling, immunity, growth, and developmental processes. Our data support a model in which RolB manipulates host transcription, at least in part, through interaction with TPL, to facilitate hairy root development. Thereby, we provide important mechanistic insights into this renowned oncoprotein in HRD.
Many hydroponically grown Solanaceae and Cucurbitaceae crops, including Solanum lycopersicum (tomato), are affected by a root disorder, called “hairy roots.” The hairy root disease (HRD), also termed “crazy root” or “root mat” disease, is characterized by extensive root proliferation, resulting in suppressed water uptake, strong vegetative growth, reduced fruit yield, and, eventually, withering of the plant (1, 2). The causal agents of HRD are rhizogenic Agrobacterium strains harboring a root-inducing (Ri) plasmid that is required for transferring part of the Ri plasmid, namely the transfer DNA (T-DNA) to the plant nucleus, T-DNA integration into the plant genome, and subsequent hairy root development (1). The infection mechanism employed by rhizogenic Agrobacterium strains is similar to that of the well-studied pathogen Agrobacterium tumefaciens, but the molecular mechanisms underlying the symptom development remain largely unknown. This lack of knowledge does not only involve the establishment of pathogenic hairy roots in Solanaceae and Cucurbitaceae crops but also of the well-known plant hairy root cultures that are intensely explored as potential sustainable production systems for numerous bioactive specialized metabolites from a wide variety of medicinal plant species and generated by distinct rhizogenic Agrobacterium strains upon infection (3, 4).
The T-DNA of Ri plasmids encodes several annotated open reading frames (ORFs) of which several, designated root oncogenic locus (rol) genes, play major roles in the hairy root symptom development (5). However, the exact function of these ORFs is still poorly understood. One of these rol genes, i.e., rolB, has been thoroughly studied because its overexpression can promote rapid, adventitious root formation upon plant transformation (6, 7). Until now, no explicit function and molecular mode-of-action have been attributed to the RolB protein yet and no sequence homology with plant proteins has been found (8). Nonetheless, in the past decades, numerous, but remarkably diverse, putative activities have been reported for RolB. Its Ri capacity has been linked to altered auxin sensitivity (8, 9). Further, RolB has been reported to be able to reduce reactive oxygen species (ROS) levels by activation of scavenging enzymes, to increase tolerance against biotic and abiotic stresses, and to modulate photosynthesis. Finally, although other rol genes can also increase the production of specialized metabolites in transformed plants, rolB is the most active and has been described frequently as the major player in specialized metabolite production (8, 9). Previously, two different enzymatic activities have been assigned to RolB, indoxyl-β-glucosidase, and protein tyrosine phosphatase (8, 9). Furthermore, the RolB interaction with a protein of the 14-3-3 family in Nicotiana tabacum (tobacco) was linked with its nuclear localization and enhanced rooting activity (10). Finally, similarly to the 6b oncogene of A. tumefaciens, RolB was suggested to influence posttranscriptional silencing through the upregulation of genes encoding the microRNA (miRNA) processing machinery (9). Notwithstanding this expanding and quite diverse knowledge, an unambiguous allocation of a clear biological function to the rolB-encoded protein remains complicated.
Protein–protein interactions (PPIs) are an essential interface in the communication between a plant and a microbe and eventually determine a successful plant’s defense response or, conversely, the disease emergence. Accordingly, studies on the interactions between proteins from the crown gall-inducing A. tumefaciens and the model plant Arabidopsis thaliana (Arabidopsis) have provided interesting insights into how plant proteins contribute to Agrobacterium-mediated plant transformation (11, 12). To defeat the plant defense responses, pathogens typically secrete various effector proteins. Characterization of effector PPIs often allows the elucidation of their molecular function and gain insights into pathogen infection mechanisms. Frequently, so-called effector hubs, i.e., plant proteins targeted by two or more effectors, are revealed (13).
Notably, recent discoveries have exposed that the corepressor TOPLESS (TPL), which functions in numerous biological processes to regulate growth, development, and response to environmental stresses (14), is an important pathogen susceptibility-promoting target of several fungal effectors (15–17). Multiple TPL repression mechanisms have been described, either by recruitment of chromatin-remodeling enzymes, such as histone deacetylase 19 (18) and the repressive cyclin-dependent kinase eight mediator complex (19), or by direct binding to histone proteins (20) or to components of the core mediator complex (21). TPL consists of three protein domains at the N-terminus, followed by WD40 repeats at the C-terminus that are involved in PPIs (22, 23). At the N-terminus, the LIS homology (LisH) domain that mediates protein dimerization is followed by a C-terminal to the LisH (CTLH) domain that interacts with proteins containing an ethylene response factor (ERF)-associated amphiphilic repression (EAR) motif (14, 24–26). These two N-terminal domains are followed by a CT11-RanBPM (CRA) domain that contributes to TPL dimerization and stabilizes the LisH domain (27, 28). In jasmonate (JA) signaling, the adaptor protein Novel Interactor of JAZ (NINJA) connects TPL to the JA ZIM domain (JAZ) repressor proteins, which themselves target various transcription factors (TFs), ultimately repressing the TF-controlled JA-responsive genes (29). NINJA belongs to the ABI-FIVE BINDING PROTEIN (AFP) family and is characterized by three conserved protein domains (30): the EAR-containing A domain that is essential for binding with TPL, the C domain that is responsible and sufficient for JAZ protein interaction, and a B domain. Accordingly, both TPL and NINJA proteins function as negative regulators of JA signaling (29).
Here, we made use of an innovative interactomics approach to study protein interaction networks in living cells, namely TurboID-mediated proximity labeling (PL) in tomato hairy roots (31). By means of this technology followed by biochemical analysis, we identified the corepressor TPL and the adaptor protein NINJA as direct bona fide interactors of the rhizogenic Agrobacterium protein RolB. We further demonstrate that RolB possesses a C-terminal EAR motif that interacts with the N-terminal domain of TPL and that this interaction is essential for its function in hairy root development.
Results
TurboID-Mediated PL Identifies Tomato TPL Proteins as RolB Interactors.
We recently established a TurboID-mediated PL platform in tomato hairy roots (31), which we here implemented to identify plant protein interactors of the rhizogenic Agrobacterium protein RolB. The engineered promiscuous biotin ligase TurboID was C-terminally fused to the bait (RolB) or a control (enhanced green fluorescent protein, eGFP) protein. These fusion constructs were stably expressed in tomato hairy roots under the control of an estradiol-inducible promoter and a strong constitutive cauliflower mosaic virus (CaMV) 35S promoter, respectively. Intact protein expression and (auto-)biotinylation activity of the TurboID-fusion proteins in tomato hairy roots was confirmed by western blot analysis (SI Appendix, Fig. S1). After streptavidin purification of biotinylated proteins and mass spectrometry analysis, 64 and 17 plant protein interactors were identified as significantly enriched in the RolB samples when compared to the eGFP control samples at a false discovery rate (FDR) of 0.01 and 0.001, respectively (Dataset S1). Among the latter, six were over fivefold enriched (log2), including the bait RolB itself, and, remarkably, four of the tomato TPL proteins (SlTPL1, SlTPL3, SlTPL4, and SlTPL5; Dataset S1 and SI Appendix, Fig. S2).
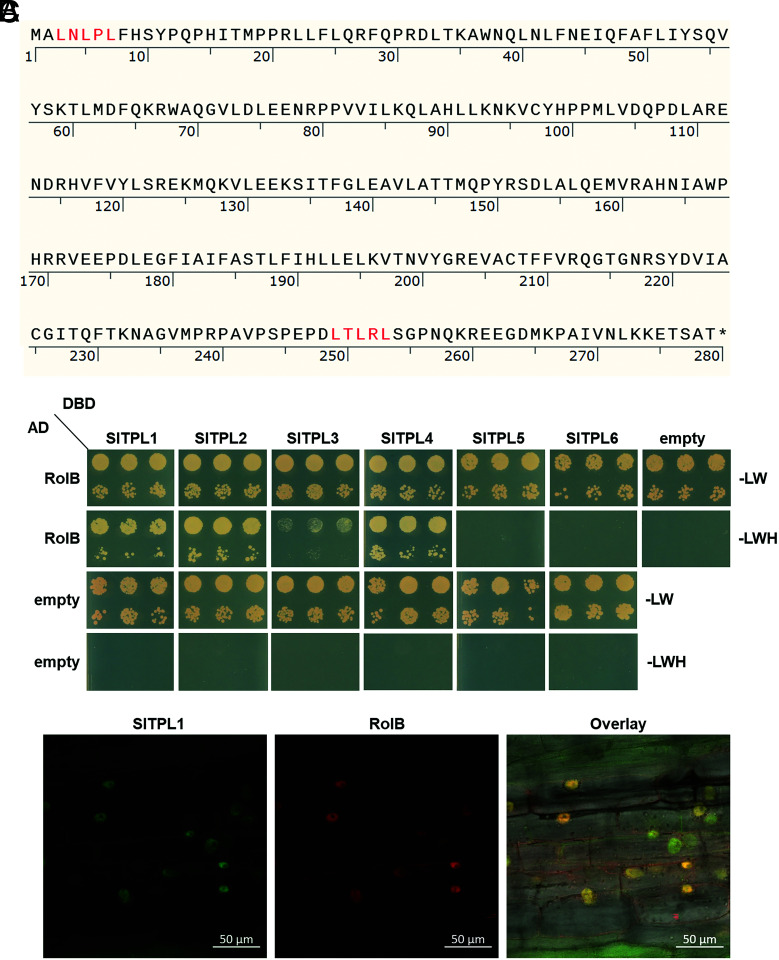
Given that TPL plays a central role in hormone and stress signaling and that it is also reported as the target of fungal effectors, we focused our downstream analysis on the interaction of TPL proteins with RolB. The TPL family in tomato comprises six members, SlTPL1 to SlTPL6 (32). As the TPL proteins can interact with EAR motif-containing proteins, we scanned the RolB sequence and detected two putative EAR motifs, one N-terminal (LNLPL starting at position 3) and one C-terminal (LTLRL starting at position 249) (Fig. 1A), hinting at a direct interaction with TPL proteins. To assess such a direct physical interaction, we performed yeast two-hybrid (Y2H) analyses with RolB as the bait, confirming that SlTPL1, SlTPL2, SlTPL3, and SlTPL4 can directly interact with RolB (Fig. 1B). In our hands, no direct interaction between SlTPL5 and SlTPL6 could be observed, at least not in the Y2H system. Whether the detection of SlTPL5 in the TurboID experiment might be caused by an indirect interaction with RolB, for instance via the reported heterodimerization of TPL proteins in planta (27), was not further assessed.
Fig. 1.
Direct interaction of RolB with the tomato TPL proteins and colocalization with SlTPL1 in the plant nucleus. (A) The protein sequence of RolB with the EAR motifs (LXLXL highlighted in red). Image created by Snapgene. (B) Y2H assay with RolB as bait and the six tomato TPL homologs as preys: SlTPL1 (Solyc03g117360), SlTPL2 (Solyc08g076030), SlTPL3 (Solyc01g100050), SlTPL4 (Solyc03g116750), SlTPL5 (Solyc07g008040), and SlTPL6 (Solyc08g029050). Transformed yeasts were spotted in 10-fold (Top row) and 100-fold (Bottom row) dilutions on control medium (-Leu-Trp [-LW]) or selective medium (-Leu-Trp-His [-LWH]). Controls are yeasts transformed with empty vectors. Gene constructs in the pGBT9 and pGADT7 vectors carry the GAL4 DBD or the transcription activation domain (AD), respectively. (C) Nuclear colocalization of tagged SlTPL1 and RolB in transformed tomato hairy roots. GFP and mCherry fluorescent reporter genes were C-terminally fused to the SlTPL1 and rolB ORFs, respectively, and coexpressed in tomato hairy roots under the control of the constitutive Ubiquitin (UBI) and CaMV35S promoter, respectively.
Previously, both RolB and TPL proteins have been described to localize to the plant nucleus (10, 32). To confirm the nuclear localization of these proteins in tomato hairy roots as well, we C-terminally fused the fluorescent reporter proteins GFP and mCherry to SlTPL1 and RolB, respectively. SlTPL1 was selected as most representative for further biological assays because it is the tomato TPL isoform with the highest expression in most tomato organs, including roots (32). The rolB and SlTPL1 fluorescent reporter constructs were coexpressed in tomato hairy roots under the control of a constitutive promoter. In line with previous publications, RolB and SlTPL1 colocalized in the nucleus of tomato hairy root cells (Fig. 1C). Taken together, these data demonstrate that RolB directly interacts with tomato TPL proteins in the nucleus of tomato hairy root cells.
RolB Interacts with TPL via Its C-Terminal EAR Motif.
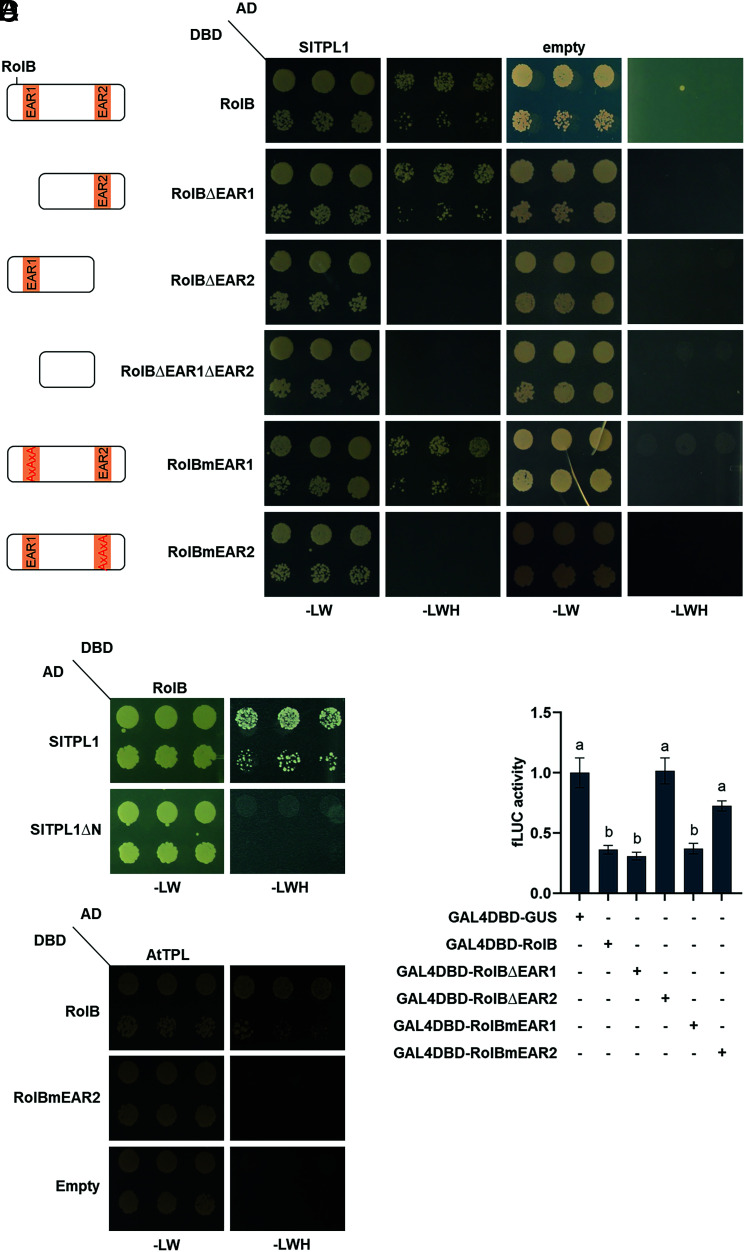
We designated the N-terminal and C-terminal EAR motifs in the RolB protein sequence EAR1 and EAR2, respectively. To determine whether both EAR motifs were required and/or sufficient for the direct interaction with TPL, we generated single- and double-deletion variants of RolB, i.e., RolBΔEAR1, RolBΔEAR2, and RolBΔEAR1ΔEAR2. Y2H analysis revealed that deletion of the C-terminal motif (from amino acid 249 to 279) abolished the interaction with TPL, whereas deletion of the N-terminal motif (from amino acid 1 to 7) did not (Fig. 2A). By means of site-directed mutagenesis, mutant variants of RolB were developed in which the three leucine residues were mutated to alanine residues (LXLXL→AXAXA), i.e., RolBmEAR1 and RolBmEAR2. In agreement with our observations with the truncated versions, interaction was abolished with RolBmEAR2 but remained unaffected with RolBmEAR1 (Fig. 2A). Notably, the presence of the C-terminal EAR2, but not of the N-terminal EAR1 motif, was conserved across the RolB sequences of different rhizogenic Agrobacterium strains (SI Appendix, Fig. S3), further underscoring its potentially functional importance.
Fig. 2.
TPL interaction mediated by the C-terminal EAR motif of RolB. (A) Y2H assay with RolB and RolB derivatives as baits and tomato SlTPL1 as prey. Y2H analysis was done as in Fig. 1. (B) Y2H assay with RolB as bait and SlTPL1 deleted in the N-terminal portion (comprising the LisH and CTLH domains, amino acids 1-37) as prey. (C) Y2H assay with RolB and RolBmEAR2 as baits and Arabidopsis TPL (AtTPL, AT1G15750) as prey. (D) Transactivation activity in tobacco protoplasts transfected with a pUAS–fLUC reporter construct and a construct with RolB or RolB mutants C-terminally fused with the GAL4 DBD. Values represent the mean ± SE (n = 8) relative to a control transfection with a GAL4-DBD-GUS control construct. The different letters indicate significant differences between groups evaluated by one-way ANOVA with Tukey’s multiple comparison test at 5% significance level.
Conversely, to verify whether the TPL CTLH domain, which typically mediates the interaction of TPL proteins with an EAR motif-containing protein, also connects with RolB, we generated an N-terminally truncated version of SlTPL1 (SlTPL1ΔN), lacking the LisH and CTLH domains (from amino acid 1 to 37). Deletion of this fragment abolished the interaction with RolB in yeast (Fig. 2B), implying the probable involvement of the TPL CTLH domain in binding EAR2 of RolB. Finally, to assess whether the interaction between RolB and TPL is conserved among other plant species, we carried out a Y2H analysis with the Arabidopsis TPL protein as prey. RolB and Arabidopsis TPL interacted strongly and, as expected, this interaction was not observed with RolBmEAR2 (Fig. 2C). Together, these results indicate that the C-terminal EAR motif of RolB is necessary and sufficient to mediate interaction with TPL proteins, presumably via the known EAR motif-binding CTLH domain in TPL and with TPL proteins across the plant kingdom.
RolB Can Act as a Transcriptional Repressor.
Considering that RolB localizes to the nucleus of tomato hairy roots, in which it interacts with the corepressor TPL, we assessed whether RolB could also act as a transcriptional repressor by using a tobacco protoplast-based reporter assay. Indeed, a protein fusion of RolB with the DNA-binding domain (DBD) of GAL4 repressed the basal activity of a firefly luciferase (fLUC) reporter construct driven by a promoter containing the GAL4 upstream activation sequence (UAS) elements (Fig. 2D). In agreement with the Y2H RolB-TPL interaction data, deletion or mutation of EAR1 did not affect the RolB repressor capacity, whereas deletion or mutation of EAR2 abolished it (Fig. 2D). Taken together, these results show that RolB can act as a transcriptional repressor, at least when recruited to a promoter by fusion with a DNA-binding protein, and highlight the importance of the C-terminal EAR2 motif in this capacity.
RolB Interacts Directly with SlbZIP11 and the NINJA Protein.
Based on the results of the transcription repressor assays, we first hypothesized that RolB and TPL might function together as a complex to repress still unknown target genes by RolB-mediated recruitment of TPL to yet unknown TF(s). In such a scenario, RolB-interacting TF(s) could have been retrieved with TurboID PL as well, either as direct or indirect interactors. To identify potential protein targets of a RolB-TPL repressor complex, we mined the RolB TurboID list for other putative nuclear-localized TFs. Two such proteins were significantly enriched at an FDR of 0.01, namely bZIP proteins, designated SlbZIP11 and SlbZIP30 (Dataset S1). Tomato encodes 69 bZIP proteins that have been categorized into 11 groups (I to XI) based on alignment and type of amino acid residues present in their basic and hinge regions. SlbZIP11 and SlbZIP30 both belong to group IX (33). The Arabidopsis homolog of SlbZIP11, AtbZIP29, regulates genes involved in cell cycle regulation and cell wall organization (34). Interestingly, the Arabidopsis homolog of SlbZIP30 is the VirE2-interacting protein 1 (AtVIP1), which is involved in the activation of defense response genes upon pathogen attack (35). Dominant negative repression of either AtbZIP29 and AtVIP1 results in a wavy root phenotype reminiscent of the hairy root phenotype (34, 36). Therefore, we speculated that RolB and TPL might function together to repress the activity of the tomato SlbZIP11 and SlbZIP30. Y2H analysis confirmed that SlbZIP11 was a direct RolB interactor and that this interaction did not depend on the EAR motifs of RolB (SI Appendix, Fig. S4A). The interaction between SlbZIP30 and RolB could not be validated by Y2H (SI Appendix, Fig. S4B). However, given that bZIP proteins had previously been reported to form homodimers and heterodimers (37) and that both homodimerization and heterodimerization of SlbZIP11 and SlbZIP30 were confirmed with Y2H (SI Appendix, Fig. S4C); SlbZIP30 might plausibly be an indirect RolB interactor. To verify a possible “adaptor” role for RolB, i.e., as a “bridging” protein between TPL and the bZIP TFs, we applied yeast-3-hybrid (Y3H) experiments. However, because RolB could not link the TPL and bZIP proteins, a scenario in which the activity of bZIPs is inhibited by a RolB-TPL corepressor complex seems improbable (SI Appendix, Fig. S4D). Accordingly, RolB could not attenuate the mild (not significant) transactivation capacity of a GAL4DBD-SlbZIP11 fusion protein in tobacco protoplasts (SI Appendix, Fig. S4E).
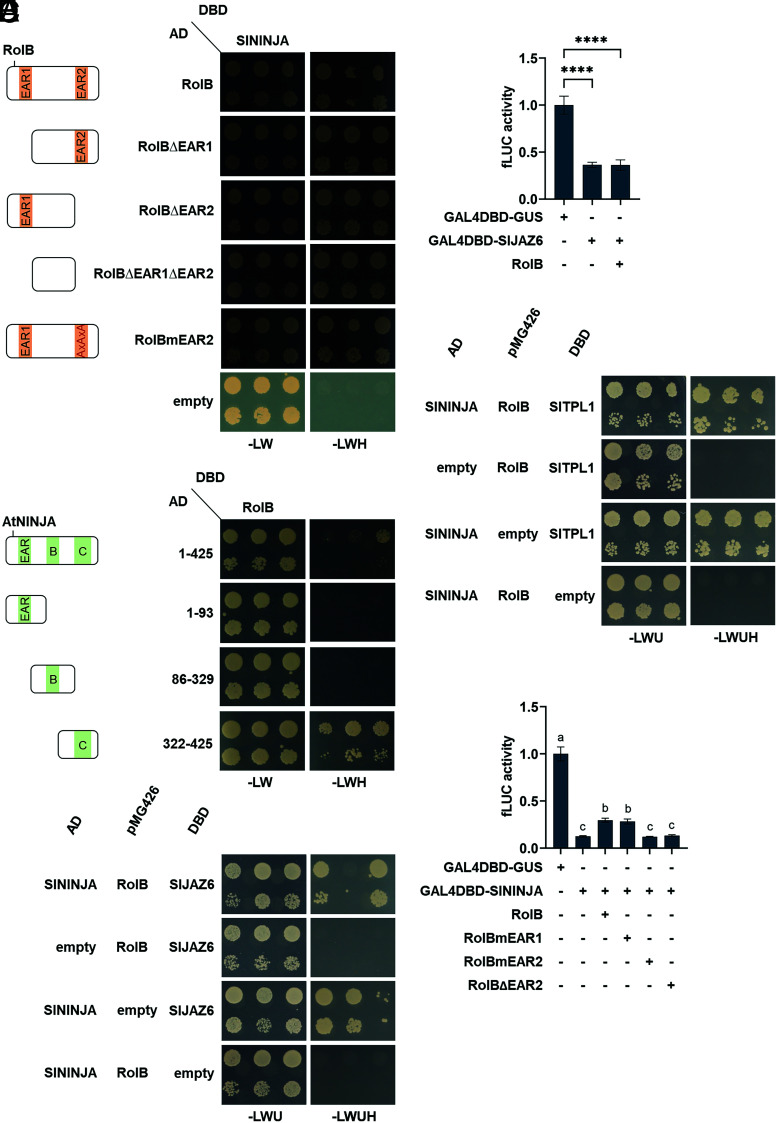
Besides enriched TFs in the RolB TurboID dataset, we found the adaptor protein NINJA among the significantly enriched proteins at an FDR of 0.01 (Dataset S1). As NINJA is a known and well-characterized interactor of TPL proteins and associates them with the JAZ repressor proteins to suppress the action of JAZ-bound TFs, such as MYC2 (29), NINJA could be assumed to be an indirect interactor of RolB. Unexpectedly, Y2H analysis confirmed SlNINJA not only as a direct interactor of SlTPL1 (Fig. 3D) but also of RolB itself (Fig. 3A). Furthermore, this interaction was independent of the presence of the RolB EAR motifs. Arabidopsis NINJA (AtNINJA) consists of three conserved (A–C) protein domains (29, 30). By using previously generated AtNINJA deletion constructs (29), we found that the C-terminal domain of NINJA is responsible for the interaction with RolB (Fig. 3B), which is the same domain mediating interaction with the JAZ proteins (29). Based on this finding, we speculated that RolB might be able to disrupt the interaction between NINJA and the JAZ repressor proteins. However, subsequent Y3H analysis with SlJAZ6 did not support such a physical action (Fig. 3C). Accordingly, RolB could not suppress the repressor capacity of a GAL4DBD-SlJAZ6 fusion protein in tobacco protoplasts (Fig. 3D). Likewise, the interaction between SlNINJA and SlTPL1 was not impeded by RolB (Fig. 3E), which may not be surprising, because RolB interacts directly with both and, thus, principally can act as an adaptor as well as an interferer. Similarly to RolB, a protein fusion of AtNINJA with the DBD of GAL4 was reported to repress the pUAS-fLUC reporter construct in tobacco protoplasts (29). Here, we confirmed that finding with SlNINJA but also found that the GAL4DBD-SlNINJA transcriptional repressor activity could be significantly counteracted by the coexpression with free RolB (Fig. 3F). This negative effect of free RolB on GAL4DBD-SlNINJA depended on the presence of the EAR2 motif, thus on its capacity to recruit TPL proteins (Fig. 3F). This finding was counterintuitive because the repressor activity of GAL4DBD-RolB itself was observed only when it can recruit TPL (Fig. 2D). Taken together, we believe that these results might point to a complex competitive molecular effect, in which RolB can mitigate the repressor functioning of either or both NINJA and TPL rather than act as a repressor itself. We further speculate that this mitigation effect is an essential in planta activity of RolB to facilitate hairy root formation.
Fig. 3.
Direct interaction of RolB with SlNINJA and mitigation of its repressor function. (A) Y2H assay with SlNINJA (Solyc05g018320) as bait and RolB derivatives as preys. Y2H analysis was done as in Fig. 1. (B) Y2H assay with RolB as bait and the Arabidopsis NINJA (AtNINJA; AT4G28910) domains A, B, and C as preys. (C) Y3H analysis with SlNINJA as prey, SlJAZ6 (Solyc01g005440) as bait, and RolB as "bridge" protein. (D) Transactivation activity in tobacco protoplasts transfected with a pUAS–fLUC reporter construct, a SlJAZ6 construct fused with GAL4-DBD, and a RolB construct. Values represent the mean ± SE (n = 8) relative to a control transfection with a GAL4-DBD-GUS control construct. Statistical significance was assessed with one-way ANOVA with Tukey’s honest significance difference (****P ≤ 0.0001). (E) Y3H analysis with SlTPL1 as bait, SlNINJA as prey, and RolB as "bridge" protein. Transformed yeasts were spotted in 10-fold (Top row) and 100-fold (Bottom row) dilutions on a control medium (SD-Leu-Trp-Ura [SD-LWU]) and selective medium (SD-Leu-Trp-Ura-His [SD-LWUH]). Gene constructs in the pGBT9 and pGADT7 vectors carry the GAL4-DBD or the transcription activation domain, respectively, in contrast to constructs expressed in pMG426, which do not carry DNA-binding or transcription activation domains. Controls are yeast transformed with empty vectors. (F) Transactivation activity in tobacco protoplasts transfected with a pUAS–fLUC reporter construct, an SlNINJA construct fused with GAL4-DBD, and a RolB or RolB variant construct. Values represent the mean ± SE (n = 8) relative to a control transfection with a GAL4-DBD-GUS control construct. The different letters indicate significant differences between groups evaluated by one-way ANOVA with Tukey’s honest significance at 5% significance level.
The C-terminal EAR Motif of RolB Is a Major Determinant of a Successful Hairy Root Formation upon Rhizogenic Agrobacterium Infection.
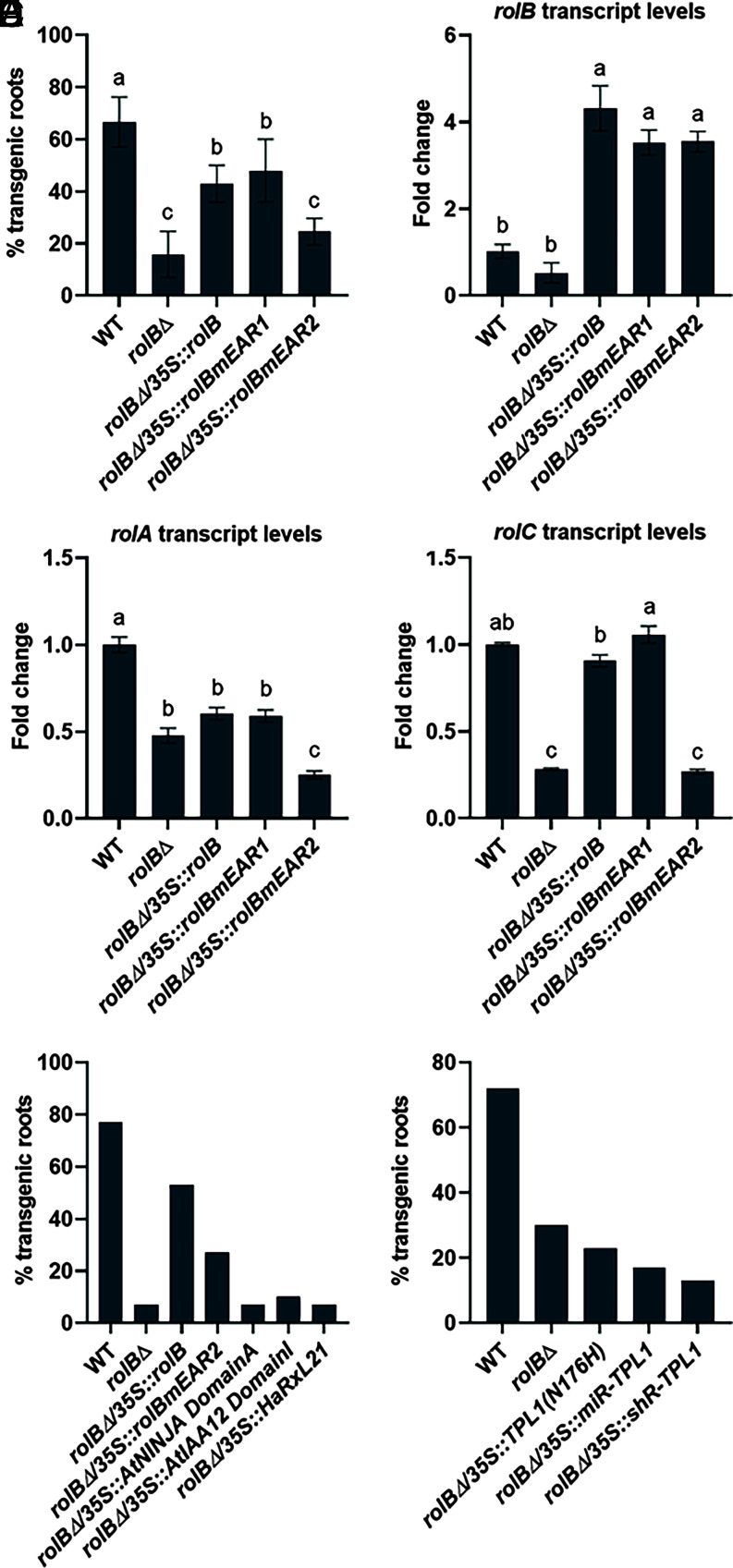
To gain more insights into the importance of the ability of RolB to interact with TPL for tomato hairy root development, we applied in planta bioassays. To this end, we used an adapted version of the tomato composite plant bioassay (38) and the rhizogenic Agrobacterium biovar 1 strain K599 as the causal agent of HRD (39). This strain became our model strain to study the function of the rol genes and to infect tomato seedlings because i) it has an Ri plasmid with a rather small T-DNA that encodes a set of only 11 ORFs, which is far less than other rhizogenic Agrobacterium strains (40), thus facilitating functional analyses, and ii) a K599 strain mutated in the rolB oncogene had been generated through base editing (41). Here, we used the wild-type (WT) K599 and rolB mutant strain (designated rolB∆ hereafter) to compare their infection efficiency in our tomato composite plant bioassay. To allow a more robust scoring of hairy root development and distinction between true hairy roots from “WT” adventitious roots, both strains were transformed with a construct expressing the mCherry fluorescent reporter gene. In agreement with previous reports, the percentage of transformed fluorescent roots upon infection with the rolB∆ strain was significantly lower than that obtained upon infection with the WT strain (Fig. 4A and SI Appendix, Fig. S5 A and B), confirming the key role of RolB in tomato hairy root development as well as the utility and robustness of our assay.
Fig. 4.
Interaction with TPL as important determinant of the RolB activity in the establishment of tomato hairy roots. Tomato seedlings (~30 seedlings per strain) were infected with rhizogenic Agrobacterium K599 strain variants and the percentage of emerging fluorescent transgenic roots was scored 4 wk post-infection. (A) Percentage of transgenic fluorescent tomato roots upon infection with the rhizogenic Agrobacterium K599 WT strain, the rolB∆ mutant strain, and the rolB∆ strain transformed with a construct expressing either the native rolB oncogene, the rolBmEAR1 mutant, or the rolBmEAR2 mutant, each under the control of the constitutive CaMV35S promoter, along with a GFP fluorescent reporter generolB∆/35S::rolBrolB∆/35S::rolBmEAR1, and rolB∆/35S::rolBmEAR2, respectively. The WT and rolB∆ strain were transformed with a construct expressing the mCherry fluorescent reporter gene. Values represent the mean ± SE (n = 5). The different letters indicate significant differences between groups evaluated by one-way ANOVA with Tukey’s honest significance at a 5% significance level. (B–D) qRT-PCR analysis of the fold-changes in rolB (B), rolA (C), and rolC (D) transcript levels upon infection with the different strains 4 wk post-infection. Fold change is relative to the transcript levels in roots upon K599 WT infection and normalized to the SlCAC (Solyc08g006960) transcript levels. Values represent the mean ± SE (n = 3). The different letters indicate significant differences between groups evaluated by ANOVA with Tukey’s honest significance difference at 5% significance level. (E) Percentage of transgenic fluorescent tomato roots upon infection with the rolB∆ strain transformed with a construct expressing the gene that encodes either the N-terminal domain of the Arabidopsis NINJA (Domain A), the N-terminal domain of Arabidopsis IAA12 (Domain I), or the downy mildew effector HaRxL21, each under the control of the constitutive CaMV35S promoter, along with an GFP fluorescent reporter gene. (F) Percentage of transgenic fluorescent tomato roots upon infection with the rolB∆ strain transformed with SlTPL1 silencing constructs, miR-SlTPL1 and short hairpin RNA(shR)-SlTPL1, or a construct carrying a dominant negative mutant version of the SlTPL1 gene, SlTPL1(N176H). Given the lack of complementation with any of the constructs, some of the assays shown (E and F) were not repeated, hence n = 1 without error bars.
Next, to determine whether the interaction with TPL is an essential element of the RolB function, we transformed the rolB∆ strain with a construct expressing either the native rolB oncogene (for complementation), the rolBmEAR1 mutant, or the rolBmEAR2 mutant, each under the control of the constitutive CaMV35S promoter, along with a GFP fluorescent reporter gene. As expected, ectopic expression of the native rolB oncogene largely restored the percentage of transgenic fluorescent roots obtained upon infection (Fig. 4A and SI Appendix, Fig. S5C). A similar effect was observed with the ectopic expression of the rolBmEAR1 mutant, but not of the rolBmEAR2 mutant (Fig. 4A). The latter restored only partially the percentage of transgenic fluorescent roots in comparison to infections with the rolB∆ and WT strains (Fig. 4A). Accordingly, the steady-state levels of the rolA and rolC transcripts were markedly lower in tomato roots upon infection with the rolB∆ strain than those upon infection with the WT strain (Fig. 4 B–D). Conversely, ectopic expression of the native rolB oncogene or the rolBmEAR1 mutant, but not of the rolBmEAR2 mutant, largely restored the rolA and rolC transcript levels upon infection with the rolB∆ strain (Fig. 4 C and D). Together, these data reveal that the C-terminal EAR motif of RolB and, thus, the capacity of RolB to recruit TPL play an important, albeit not indispensable, role in hairy root formation. In accordance, a construct ectopically expressing the genetic fusion of TurboID to the rolB oncogene could complement the rolB∆ strain to the same extent as the construct expressing the nontagged rolB (SI Appendix, Fig. S6), evidencing that the RolB-TurboID fusion protein, which had demonstrated the TPL-binding activity in vivo in tomato root cells, corresponds to a functional RolB protein.
If the RolB mode of action to mitigate the TPL activity would rely on mere binding to TPL, thereby scavenging it in a nonspecific manner from its many potential in planta protein interactors (directly) and/or known target genes (indirectly via its interacting target TFs), we hypothesized that ectopic overexpression of genes encoding proteins that can bind to TPL, but without presumable additional function, might also complement the rolB∆ mutant phenotype. To test this assumption, we chose three proteins, or fragments thereof, including i) the N-terminal domain of the Arabidopsis NINJA that cannot interact with RolB (Fig. 3B), ii) the N-terminal domain of the Arabidopsis indole-3-acetic acid 12 (IAA12), and iii) the downy mildew effector HaR × L21 that had recently been found to interact with TPL upon infection. The common determinant of these protein fragments is that they all contain the EAR motif to which TPL binds (14, 28, 29). However, none of these constructs could complement the rolB∆ strain (Fig. 4E), in terms of the percentage of transgenic fluorescent roots obtained upon infection. As an alternative manner to decrease TPL levels in planta, we silenced the TPL expression. Therefore, we designed miRNA (miR) and short hairpin RNA (shR) constructs that target SlTPL1 and transformed them into the rolB∆ strain. In addition to the silencing constructs, we also generated an overexpression construct carrying a mutant version of the SlTPL1 gene, SlTPL1(N176H), corresponding to a mutant version of AtTPL with dominant negative effects (18). None of these constructs could mediate an increase in the percentage of transgenic fluorescent roots upon infection when compared to the rolB∆ strain (Fig. 4F and SI Appendix, Fig. S7). Noteworthy, although SlTPL1 had the highest expression in roots (32), it was not the only SlTPL gene expressed. Hence, functional redundancy cannot be excluded to have hampered the detection of some degree of possible complementation by one or more of the other five SlTPL homologs. Nonetheless, taken together, our data indicate that the renowned key role of RolB in tomato HRD depends largely on the presence of its C-terminal EAR motif and of its capacity to recruit TPL. This RolB-TPL interaction seems to have some specificity determinants, probably to endow RolB with the ability to target a specific cellular process because this role cannot be taken over by other TPL-interacting proteins.
The RolB Activity Modulates the Plant Transcriptome Depending on Its TPL-Recruiting Capacity.
As TPL is a corepressor protein, we considered that the targeted scavenging of TPL by RolB should ultimately be reflected in the plant transcriptome during the establishment of HRD. Therefore, RNA sequencing of tomato roots was carried out upon infection with the WT K599 strain, the rolB∆ strain, and the rolB∆ strain with the constructs expressing either the native rolB oncogene or the rolBmEAR2 mutant. Unfortunately, principal component analysis (PCA) indicated a large variability in the tomato root samples infected by rolB∆, rolB∆/35S::rolB, and rolB∆/35S::rolBmEAR2 (SI Appendix, Fig. S8). The observed variability was to be expected given that the number of transformed roots emerging after infection with the rolB∆ strain was lower and the growth speed slower than those of roots infected with the WT K599 strain (Fig. 4A), plausibly affecting the transcriptome robustness. Nonetheless, we attempted to look for significantly differentially expressed genes (DEGs) with criteria of log2 fold ≥two and a P value and FDR cutoff of 0.05. This analysis revealed 110 DEGs when tomato root samples infected with rolB∆/35S::rolB vs. WT K599 (83 up- and 27 down-regulated; Dataset S2) were compared, but none with rolB∆ or rolB∆/35S::rolBmEAR2 vs. the WT K599 (Dataset S2). Although this observation does not provide further functional insights, it supports the importance of the C-terminal EAR motif for the RolB function in hairy root formation. Enrichment analysis of Gene Ontology (GO) terms on the 110 DEGs of the root samples infected by rolB∆/35S::rolB vs. WT K599, possibly reflecting effects of ectopic rolB expression, was also not informative (Dataset S2E).
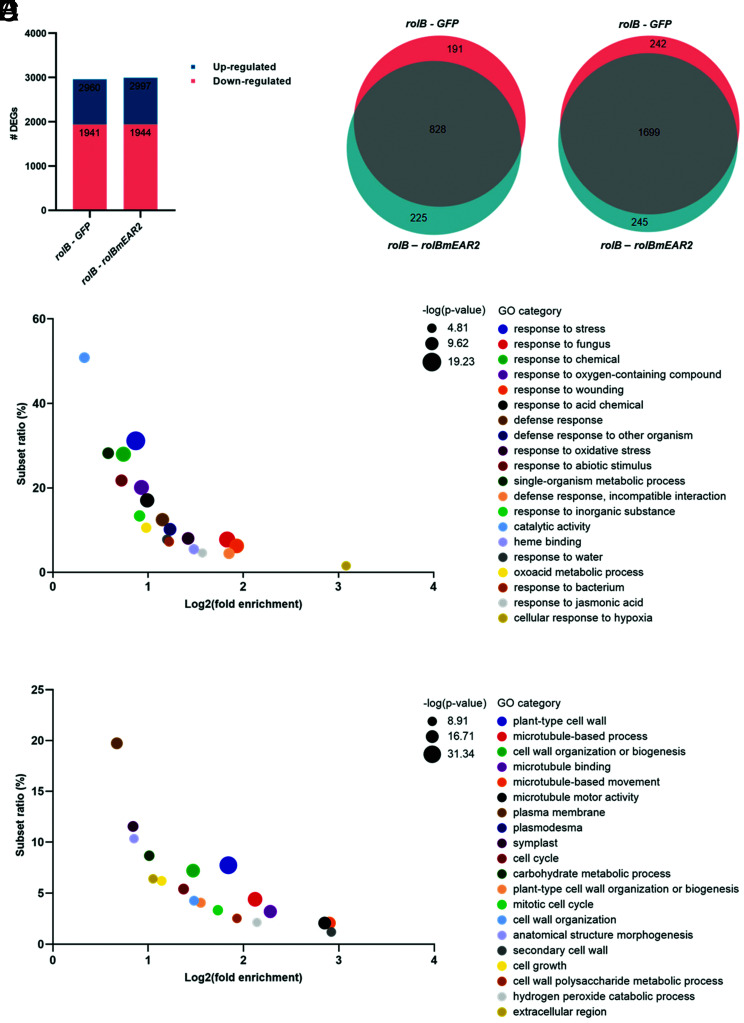
As comparable transcriptomes could not be generated through our in planta bioassays, we turned to the tomato hairy root platform that had successfully been used for the TurboID PL analysis and in which the different relevant rolB variants that we created could be inducibly expressed. More specifically, we compared the gene expression in tomato hairy root lines transformed with inducible rolB, rolBmEAR2, or GFP overexpression constructs, 24 h after mock and β-estradiol treatments (SI Appendix, Fig. S9 A and B). PCA indicated that in this setup, indeed robust transcriptomes could be generated. Further, rolB and rolBmEAR2 as well as rolB and GFP transgenic hairy root lines could be separated after β-estradiol induction (SI Appendix, Fig. S9 C and D), but not the mock-treated samples or the rolBmEAR2 and GFP transgenic hairy root lines after β-estradiol induction (SI Appendix, Fig. S9 C and D). Next, we mined this dataset for up- and down-regulated DEGs. Implementation of the same selection criteria as above did not reveal DEGs when the mock-treated samples or rolBmEAR2 vs. GFP transgenic hairy root lines after β-estradiol induction were compared (Dataset S3B), in agreement with the PCA. In contrast, 2,997 genes were differentially expressed between rolB and rolBmEAR2 transgenic hairy root lines after β-estradiol induction, namely 1,053 up-regulated and 1,944 down-regulated DEGs (Fig. 5A and Dataset S3 B, E, and F). These DEGs highly overlapped (~70%) with the genes differentially expressed between rolB and GFP transgenic hairy root lines, namely 1,019 up-regulated and 1,941 down-regulated DEGs (Fig. 5 A and B and Dataset S3 B–D). Conversely, there was little overlap between the DEGs in rolB vs. rolBmEAR2 (or rolB vs. GFP) transgenic hairy root lines and those in tomato root samples infected by rolB∆/35S::rolB vs. WT K599, 80 up- and 49 down-regulated. Plausibly, the limited overlap might correspond to the long-term (4 wk in the bioassay) vs. short-term (24 h in the β-estradiol induction) effect on the ectopic and/or overexpression of rolB. In the DEGs up-regulated by rolB vs. rolBmEAR2 (as well as rolB vs. GFP), we found a significant enrichment of the GO terms involved in response to stimulus (44%), more specifically, response to stress, response to abiotic and biotic stimulus, and defense response (Fig. 5D and SI Appendix, Dataset S4 A and C), indicating that the RolB induction of plant immune responses in tomato roots depends on its TPL interaction. In addition, we identified four hormone-related GO categories in the up-regulated DEGs: “response to abscisic acid,” “response to jasmonic acid,” “response to ethylene,” and “response to salicylic acid.”
Fig. 5.
DEGs in rolB compared to rolBmEAR2 transgenic tomato hairy roots identified by transcriptome analysis. RNA sequencing was performed of transgenic tomato hairy roots expressing rolBrolBmEAR2, and GFP. (A) The number of up- and down-regulated DEGs between rolBrolBmEAR2, and GFP root lines after 24 h of β-estradiol (100 µM) induction. Genes were significantly differentially expressed in the contrast groups, with P value < 0.05, FDR < 0.05, and log(FC) > 2 (up-regulated DEGs) or <−2 (down-regulated DEGs). Proportional Venn diagrams show DEGs in the contrast groups rolB-GFP and rolB-rolBmEAR2 with an overlap of 828 up-regulated (B) and 1,699 down-regulated (C) genes. GO term enrichment was analyzed on the up-regulated (D) and down-regulated DEGs (E) between rolB and rolBmEAR2 root lines by means of PLAZA 4.5 Dicots. GO enrichment bubble plots (comprising molecular function, biological process, and cellular component ontologies) show the 20 most significant hits. The log2(fold enrichment) was plotted against the subset ratio in percentage.
In the DEGs down-regulated by rolB vs. rolBmEAR2 (as well as rolB vs. GFP), the GO terms found were involved in developmental processes, anatomical structure development, cellular component organization, and root development (Fig. 5E and Dataset S4 B and D). Catalytic activity, such as oxidoreductase activity, was enriched across both up- and down-regulated DEGs. Altogether, our transcriptome data clearly imply that RolB manipulates host transcription through interaction with TPL.
RolB-Activated Genes Are Enriched for TF Targets Involved in the JA/ET Pathway.
Given the possible role for RolB in the deregulation of TPL-mediated repression, we focused on the up-regulated DEGs. Twelve DEGs in the GO category “response to ethylene (ET)” were annotated as APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) family TFs (Dataset S4E); eight of which were identified as orthologs of the Arabidopsis AP2/ERF family TFs ERF1 (AT3G23240), ERF-1 (AT4G17500), and ESE1 (AT3G23220) that belong to the B3 subcluster. Two of these AP2/ERFs also appear as DEGs in the GO category “response to JA.” Many of the AP2/ERFs from this subcluster have been shown to i) act as positive transcriptional regulators of defense-related transcription, ii) have a JA- or pathogen-inducible expression, and iii) be involved in the signaling pathways mediated by the JA and ET hormones to install pathogen defense programs (42). Constitutive overexpression of ERF1, for instance, activates several plant defense-related genes, such as PLANT DEFENSIN1.2 (PDF1.2) and chitinases in Arabidopsis (42, 43). We detected two chitinases, ChiB and ChiC, up-regulated by rolB when compared to rolBmEAR2. rolB also induced two 1-aminocyclopropane-1-carboxylate oxidases (ACOs), two allene oxide synthases (AOSs), and a lipoxygenase (LOX) that are putatively involved in ET and JA biosynthesis (Dataset S4 E and F) (42). Taken together, these results show that the rolB expression induces the expression of several AP2/ERFs, as well as defense-related genes that are likely targets of the AP2/ERFs, suggesting that RolB stimulates the JA/ET signaling pathway.
The DEGs in the GO category “response to JA” also comprise some other TFs often found in stress hormone signaling, such as NAC (NAM, ATAF1/2, and CUC2), WRKY, and MYB TFs (44), but surprisingly no MYC2 or other bHLH TFs. To verify this result and to allow the identification of transcriptional regulators that are possibly specifically affected by the RolB-TPL effect in a nonbiased manner, we performed a TF-binding site enrichment analysis on the rolB vs. rolBmEAR2 up- and down-regulated DEGs. We adapted a previously described method, TF2Network (45), for tomato and used enriched binding sites together with information about the TF binding to detect candidate regulators and their target genes. Confirming our initial findings, we obtained a significant enrichment of the AP2/ERF, NAC, and WRKY binding sites in the up-regulated DEG set (Dataset S5A), whereas Homeodomain, DNA-binding with one finger (DOF), and Teosinte branched1/Cincinnata/proliferating cell factor (TCP) TF-binding sites were significantly enriched in the DEG set down-regulated by rolB (Dataset S5B). Remarkably again, MYC2 and/or other known JA-responsive bHLH TFs did not top these lists. Nonetheless, a large fraction of the transcriptional regulators that excel in the RolB tomato-enriched TF list, as well as their targets, were also responsive to JA in tomato hairy roots, as deduced from another available in-house generated RNA-sequencing dataset (SI Appendix, Fig. S10 A and B and Datasets S5 and S6). Together, these data point to a potential interference with JA signaling, particularly via AP2/ERF family TFs possibly involved in the JA/ET branch of the signaling pathway. Further, induction of these JA/ET TFs was not limited to increased expression but may also involve increased activity because of the strong enrichment of genes with TF-binding sites in the RolB transcriptome. Plausibly, this could be a consequence of the RolB-mitigated recruitment of the TPL corepressors to these TFs. However, a noncomprehensive screening for the presence of EAR motifs in the top-ranked RolB tomato-enriched TF list did not imply many of them as direct TPL interactors. This indication was corroborated by a pilot Y2H analysis, in which no interaction between a subset of these TFs with TPL proteins could be detected (SI Appendix, Fig. S10C). Hence, the precise molecular mechanism by which RolB affects the activity of these TFs in a TPL-dependent manner remains unresolved for now.
Discussion
The rhizogenic Agrobacterium rolB oncogene encoded on the T-DNA of the Ri plasmid plays an essential role in hairy root development, but the exact molecular function of RolB remains enigmatic, despite intensive research over the past decades (8). Here, we demonstrate that RolB contains a C-terminal EAR motif (amino acid sequence LTLRL), conserved in the RolB sequences of different rhizogenic Agrobacterium strains, which is necessary and sufficient to interact with the N-terminal LisH CTLH region of the corepressor TPL in the nucleus of tomato root cells. In planta bioassays revealed that the key role of RolB in hairy root development largely depends on this C-terminal EAR motif and, hence, on its capacity to recruit TPL.
Are TPL Proteins a Hub for Pathogen Effectors to Dampen Immune Responses?.
Through our findings, RolB joined a recently identified small group of fungal effectors that can target plant TPL proteins. As the TPL proteins as general corepressors are recruited for nearly any plant hormone, stress, or developmental signaling pathway (14, 29), they may be an interesting target (or hub) for pathogens to modify the plant hormonal landscape to their benefit (13). More specifically, TPL is already known to be targeted by the effectors Naked1 (Nkd1) and Jsi1 from the gall-inducing smut fungus Ustilago maydis and HaRxL21 from the oomycete pathogen Hyaloperonospora arabidopsidis, the causal agent of downy mildew (15–17). Although the in planta consequences of fungal effector binding to TPL vary, they all seem to modulate the host plant’s immune responses. For instance, HaRxL21 and TPL interact also via an EAR motif (amino acid sequence LMLTL) at the C-terminus of the effector that has been shown to be necessary for the virulence function of the effector. Furthermore, HaRxL21 uses the interaction with TPL proteins to repress Arabidopsis immunity and enhance host susceptibility to both biotrophic and necrotrophic pathogens (15). Nkd1 binds to TPL also via a C-terminal EAR-motif (amino acid sequence LDLSL) and prevents the recruitment of AUX/IAA repressors, resulting in an increase in auxin signaling and subsequent inhibition of pathogen-associated molecular patterns-triggered ROS bursts in maize (Zea mays) (16). Moreover, engineered Nkd1 variants with increased expression or enhanced EAR-mediated TPL binding can not only up-regulate genes involved in auxin signaling and metabolism but also genes involved in JA and salicylic acid (SA) signaling and metabolism, thereby promoting pathogen susceptibility. As such, it has been postulated that the Nkd1-mediated TPL inhibition and ensuing up-regulated hormonal signaling restrains defense and, possibly, also facilitates smut gall development (16). Conversely, the interaction between the other U. maydis effector, Jsi1, and the maize TPL proteins is determined by an N-terminal EAR motif, not of the LXLXL-type but of the DLNXXP-type (amino acid sequence DLNELP) that interacts with the C-terminal WD40 motif of TPL proteins. The Jsi1 function seems to increase biotroph susceptibility by activating the ERF branch of the JA/ET defense signaling pathway that is typically induced upon necrotrophic pathogen infection (17). Jsi1 has been proposed to interfere with the AP2/ERF TF activities involved in the JA/ET defense pathway, but the precise molecular mechanism behind such possible interference remains unknown. Our results provide evidence that the rhizogenic Agrobacterium protein RolB exerts a molecular function similar to that of Jsi1, i.e., activation of the ERF branch of the JA/ET signaling pathway through TPL-mediated repression deregulation.
The JA/ET response pathway consists of two mutually antagonistic branches: the MYC and ERF branches. The MYC branch is mainly induced upon wounding and herbivore attack and controlled by the basic helix-loop-helix leucine zipper (bHLH) TFs, such as MYC2, whereas the ERF branch is triggered upon necrotrophic pathogen attack and regulated by the AP2/ERF TFs, such as ERF1 (46, 47). ERF1 activates the expression of defense-related genes, such as PDF1.2 and chitinases via binding of the ERF domain with the GCC-box motif, and can repress MYC2 target genes (46, 47). In line with our finding that ERF1 is one of the major TF regulators of the target genes induced by rolB, we identified the chitinases ChiB and ChiC along with the JA/ET biosynthesis-involved genes ACO, AOS, and LOX that are up-regulated by rolB. Given the absence of bHLH TFs, such as MYC2 in both our TurboID and transcriptome data, RolB might possibly mitigate the repressor activity of either or both TPL and NINJA to activate the ERF branch of the JA/ET defense signaling. These observations are analogous to those found with HaRxL21 from H. arabidopsidis, also a biotrophic pathogen, like agrobacteria. More specifically, in tomato, rolB activates TFs of the AP2/ERF, NAC, and WRKY families with a JA or necrotrophic pathogen-inducible expression (44). Among the AP2/ERF family TFs, eight TFs were identified as orthologs of the Arabidopsis ERF1, ERF-1, and ESE1 that belong to the B3 subcluster of the AP2/ERF family and act as positive transcriptional regulators of JA/ET-mediated defense responses (42). Similar findings were obtained with Jsi1 in maize (17), but, intriguingly although RolB and Jsi1 both possess an EAR motif, they are of a different type (LXLXL vs. DLNXXP, respectively), located at different positions in the effector (C- vs. N-terminus, respectively), and targeting different domains in TPL proteins (N- vs. C-terminus, respectively). Although the precise molecular mechanisms by which either RolB or Jsi1 interfere with the ERF branch of the JA/ET signaling are still unknown, unraveling how seemingly different molecular modes of physical interaction lead to identical or similar physiological effects would be exciting. In this regard, it would also be interesting to further investigate with which part of TPL Nkd1 and HaRxL21 interact with and whether this has similar molecular and/or physiological consequences.
The RolB Mode of Action, More Than Just Interference with TPL Function to Dampen Immune Responses?.
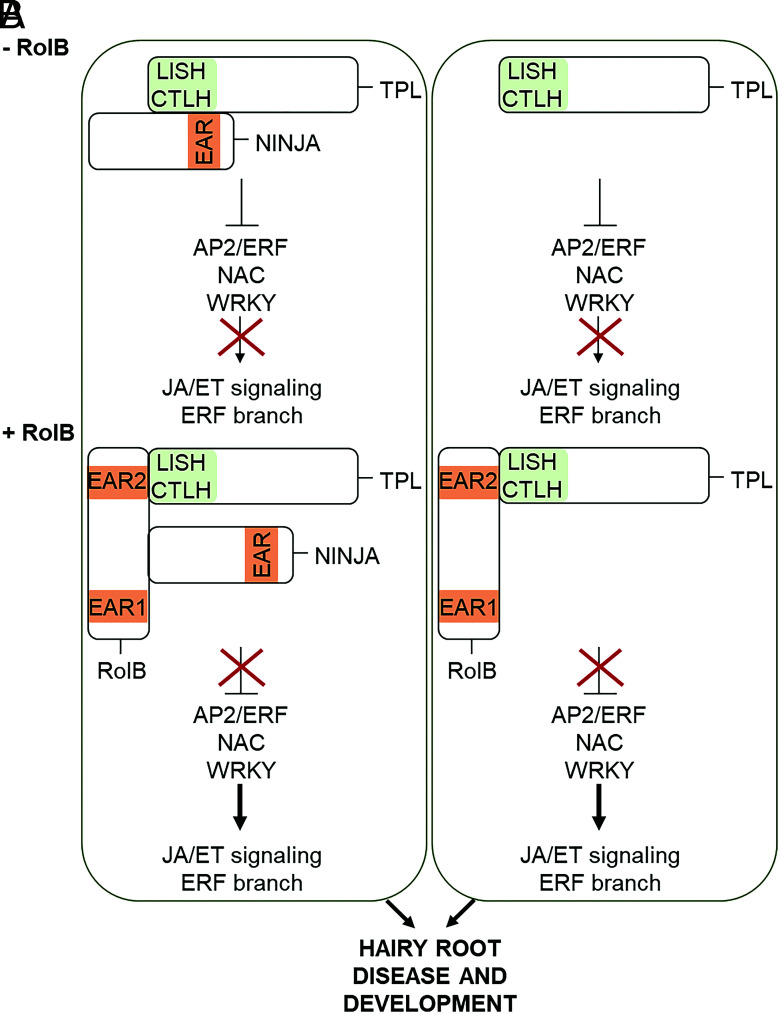
Besides their participation in JA/ET-mediated defense responses, AP2/ERF TFs are also known to play an important role in root development. Although auxin is a key phytohormone in root development (48), other phytohormones, including JA and ET, are integrated into the auxin signaling pathways to regulate the development of primary, lateral, and adventitious roots, largely through TFs that function as crosstalk nodes. For instance, in Arabidopsis, ERF1 and ERF109 connect ET and JA with auxin signaling by modulation of the auxin biosynthesis (48). Furthermore, ERF115 has been identified as a repressor of adventitious root formation in Arabidopsis and to be transcriptionally activated by JA signaling in a NINJA-dependent manner (49). The crucial role of the ERF family TFs as well as of the NINJA adaptor protein in the integration of JA and ET signaling into auxin signaling, thereby regulating the root formation, might attribute a role for RolB, not only in modulating immune responses but also in promoting the hairy root development. Indeed, such a pleiotropic RolB role is supported by both our transcriptome and interactome data. Particularly, our finding that RolB can directly interact with the adaptor protein NINJA and mitigate its repressor activity supports this hypothesis. Therefore, we propose a model in which RolB-mediated derepression of either or both the repressor proteins TPL and NINJA results in the activation of the ERF branch of the JA/ET signaling, facilitating the HRD and development (Fig. 6). As such, RolB might exert multiple roles, perhaps similar to those of the fungal effector Nkd1, which were postulated to promote both pathogen susceptibility and smut gall development (16). Whether Nkd1 has other (direct) interactors besides TPL proteins to mediate such pleiotropic effects is still not known. Our data indicate that the RolB picture is undoubtedly far more complex, even when a mere role as host transcription manipulator is considered. Indeed, our TurboID analysis revealed many more potential RolB interactors and, hence, functions. These potential interactors include several bZIP proteins, at least one of which was shown to be a direct RolB interactor. As we focused on TPL as a RolB target, we did not investigate the physiological relevance of the RolB-bZIP interaction. However, our preliminary data indicate that the interaction did not link these bZIP TFs to TPL to inhibit their action.
Fig. 6.
Hypothetical model of RolB-mediated manipulation of plant transcription. Two possible mechanistic models are proposed that might concur in planta. In the absence of rhizogenic agrobacteria, the repressor complexes consisting of either TPL-NINJA (A) or TPL (B) inhibit downstream AP2/ERF, NAC, and WRKY TFs. Upon rhizogenic agrobacteria infection, RolB mitigates the repressor function of these TPL-containing repressor complexes, with derepression of AP2/ERF, NAC, and WRKY TFs as a consequence. Subsequently, the AP2/ERF, NAC, and WRKY TFs activate the ERF branch of JA signaling, triggering microbial growth and hairy root development.
Finally, it is worthwhile pinpointing that we identified several RNA binding and editing proteins, including an Argonaute (AGO) protein, as potential RolB interaction targets in our TurboID analysis. The tomato AGO protein is a putative homolog of Arabidopsis AGO10, which represses the miR165/166 activity to maintain stem cell homeostasis in the Arabidopsis shoot apical meristem and floral meristems (50). Although direct interaction between tomato AGO10 and RolB could not readily be confirmed by Y2H, interactions with such proteins involved in gene silencing might provide a molecular basis for RolB and its potential action in modulation of posttranscriptional silencing (9, 51).
In conclusion, our results allowed us to unambiguously reveal and establish at least one crucial part of the modus operandi of the RolB oncoprotein. Furthermore, our transcriptome and interactome data provide an exciting foundation to further unravel the pleiotropic actions of RolB, thereby not only providing a molecular rationale for some of its previously reported in planta effects but also uncovering still unknown activities and functions.
Materials and Methods
Generation of Tomato Hairy Root Lines.
For TurboID-mediated PL in tomato hairy roots, the promiscuous engineered biotin ligase TurboID was C-terminally and genetically fused to the bait (rolB) and the control (eGFP) genes by means of Gateway cloning. More specifically, entries carrying the promoter sequence (EN-RPS5α_XVE or pEN-CaMV35S), ORF (pDONR221-rolB or pDONR221-eGFP), and tag of choice (pEN-TurboID-flag) were recombined in the Gateway-compatible binary vector pKCTAP.
For subcellular localizations, GFP and mCherry were C-terminally and genetically fused to SlTPL1 and rolB under the control of the constitutive promoter ubiquitin (UBI) and CaMV35S, respectively, by Golden Gibson cloning. More specifically, expression units were assembled with Golden Gate cloning into the shuttle vectors pGGIB-U1-AG-U2 and pGGIB-U2-AG-U3 to yield pGGIB-U1-pUBI-SlTPL1-GFP-35ST-U2 and pGGIB-U2-pCaMV35S-rolB-mCherry-35ST-U3. In these vectors, unique nucleotide sequences (U sites) of 40 bp are flanked with the I-SceI restriction sites. By means of the Gibson assembly, the fragments were combined into pK-U1-AG-U9 with pGGIB-U3-linker-U9 to yield pK-pUBI-SlTPL1-GFP-35ST-pCaMV35S-rolB-mCherry-35ST.
For transcriptome analysis, entries carrying the estradiol-inducible promoter sequence (pRPS5α_XVE) and the ORFs (rolB, rolBmEAR2, or eGFP) were assembled in the binary vector pGGK-AG by Golden Gate cloning. For the generation of EAR motif mutants of RolB, the Leu residues were mutated to Ala residues by site-directed mutagenesis, of which the primer sequences are presented in SI Appendix, Table S1. The structure and sequence of all used entry and destination vectors are accessible online at https://gatewayvectors.vib.be/.
All destination vectors were transformed into the rhizogenic Agrobacterium ATCC15834 strain (52, 53). Tomato hairy roots were transformed as previously described (54). Briefly, tomato cotyledons cv. Moneymaker were infected with the transformed rhizogenic Agrobacterium ATCC15834 strain by wounding and placed on nonselective Murashige and Skoog medium with 3% (w/v) sucrose (MS+). Three days postinfection, the cotyledons were transferred to a selective MS+ medium (200 µg/mL cefotaxime and 50 µg/mL kanamycin). After 2 wk of incubation in the dark at 22 to 25 °C, adventitious roots were cut from the cotyledons and transferred to a selective MS+ medium. This last step was repeated twice. After 6 wk of growth on a selective MS+ medium, the roots were transferred to a nonselective MS+ medium and used for experimental purposes.
TurboID-Mediated PL.
TurboID-mediated PL involves transformed hairy root cultivation, sample preparation for MS, immunoblot analysis, LC-MS/MS analysis, and MS-data analysis, which are all described in more detail in the SI Appendix, Materials and Methods.
Confocal Imaging.
For subcellular localization of RolB and SlTPL1 in tomato hairy roots, mCherry and GFP were imaged using the Carl Zeiss inverted LSM710 confocal laser microscope, equipped with objective Plan-Apochromat 20x/0.8 M27. GFP fluorescence was observed after excitation using a 488-nm laser and detected using the 505 to 530-nm bandpass emission filter. mCherry fluorescence was observed after excitation using a 587-nm laser and detected using the 610-nm longpass emission filter.
Yeast Two- and Three-Hybrid Assays.
Y2H and Y3H assays were done as previously described (55) and are described in more detail in SI Appendix, Materials and Methods.
Transient Expression Assay.
Transient expression assays were carried out as previously described (56) and are described in detail in SI Appendix, Materials and Methods.
Tomato Composite Plant Bioassay.
Tomato composite plant bioassays were carried out as previously described with some adaptations (38). For details, see SI Appendix, Materials and methods. Three and four weeks post-infection, the percentage of fluorescent transgenic roots was determined (for instance, 50% transgenic roots corresponded to 15 out of 30 tomato seedlings with fluorescent transgenic roots) under a fluorescence microscope (Leica).
RNA-Seq.
Tomato hairy roots were grown in 5 mL liquid MS+ medium for 2 wk at 22 to 25 °C and 150 rpm, in the dark. After 2 wk, hairy roots were treated with mock and β-estradiol (100 µM) for 24 h and subsequently harvested by flash-freezing in liquid nitrogen. For each construct (pRPS5α_XVE::rolB, pRPS5α_XVE::rolBmEAR2, and pRPS5α_XVE::eGFP), three independently transformed tomato hairy root lines were used. RNA was extracted from root tissue with the RNeasy Plant Mini Kit (Qiagen). Of total RNA, 1 µg was sent to the VIB Nucleomics Core for sequencing with the Illumina NovaSeq6000 (20 M; 100 bp single-end reads).
The total number of reads was ~20,000,000 for each sample. Transcript abundances were generated by mapping the reads on the tomato Exome ITAG4.0 by means of the Salmon mapping tool (57). Statistical analysis was done in R with the egdeR package. The three biological replicates were grouped for the analysis. The data were filtered for low expressed genes (Dataset S2A) that did not meet the following criteria: minimum read count in at least two samples >25 and count per million reads >1.5. RNA-Seq read counts were normalized with Trimmed Mean of M values method (58). Models of expression contrasts were fitted with a generalized linear regression model. Genes were significantly differentially expressed in the contrast groups when the P value < 0.05, FDR < 0.05, and log2 fold >2 (up-regulated DEGs) or <−2 (down-regulated DEGs). GO enrichment or biological process, cellular component, and molecular function were analyzed with PLAZA 4.5 Dicots (59).
TF-Binding Site Enrichment Analysis.
For TF-binding site enrichment in tomato, the 2-kb upstream sequence of the translation start site was extracted for each locus of the tomato International Tomato Annotation Group (ITAG) 4.0 genome annotation; the sequence was shortened, when there was an overlap with a neighboring gene within the 2 kb. A tomato motif collection was used derived from the combined Catalog of Inferred Sequence Binding Preferences (CisBP; version 2.00) (60) and JASPAR 2020 (61) databases. Motifs were mapped on the upstream sequences with Cluster-Buster (CB) (62) (compiled on September 22, 2017) and Find Individual Motif Occurrences (FIMO; meme version 4.11.4) (63), both with default parameters. This motif collection contained 1,376 motifs that were associated with 943 tomato TFs. Based on previous results in Arabidopsis (64), the top 7,000 top-scoring motif matches for FIMO and the top 4,000 scoring motif matches for CB were retained. Motifs were linked to target genes based on the presence/absence of a motif match in the promoter sequence. Finally, motif enrichment was done by applying the hypergeometric distribution on the sets of up- and down-regulated genes. FDR correction was done with the Benjamini–Hochberg procedure for multiple testing correction (significance threshold set at adjusted P value < 0.05). Gene identifiers from CisBP and JASPAR TFs were converted from the ITAG2.5 nomenclature to ITAG4.0 using Liftoff (65) (version 1.6.1; default parameters). The child features of the gene models of the ITAG2.5/SL2.5 genome version were also mapped onto the SL4.0 genome assembly by means of Liftoff. The resulting GFF3 file with the coordinates of aligned features was filtered for overlaps (75% alignment coverage) with the ITAG4.0 genome annotation file using BEDTools (66) (version 2.2.28) to generate a gene identifier mapping table between gene models overlapping with at least 90% of their length.
qPCR.
RNA was extracted with the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized with the qScript cDNA synthesis kit (Quantabio). Quantitative real-time PCR reactions were run with the LightCycler 480 System (Roche) and Fast SYBR Green Master Mix (Applied Biosystems). Transcript levels were determined with the 2-ΔΔCt method, normalized to the amount of CAC (Solyc08g006960) transcript (67). Primer sequences are included in SI Appendix, Table S1.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Acknowledgments
We thank Rebecca De Clercq for technical assistance and Lieven Sterck for support with the RNA-Seq analysis. This research was supported by the Research Foundation-Flanders (project no. G051120N to P.V.D.) and ELIXIR Belgium (grant I002819N to K.V.), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (PROPHECY grant agreement no. 803972 to P.V.D.), the Ghent University for the Special Research Fund “Concerted Research Action grant” (BOF18-GOA-013 to A.G.), and a research grant (BOF24Y2019001901 to N.M.P.). E.C. is a predoctoral fellow of the Research Foundation Flanders. Research is supported by KU Leuven Internal Funds. S.D.R. is supported by Research Foundation-Flanders Predoctoral Fellowship 1S43920N.
Author contributions
A.G. designed research; L.G., E.C., N.M.P., J.V.-M., A.C.J.-M., S.D.R., L.D.M., and V.J. performed research; L.G., E.C., N.M.P., M.V.M., B.D.C., K.V., P.V.D., and A.G. analyzed data; and L.G. and A.G. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
Reviewers: M.I.G., Center of Plant Systems Biology and Biotechnology; S.Y.H., Duke University; and R.S., Centro Nacional de Biotecnologia.
Contributor Information
Marc Van Montagu, Email: vanmontagu@ugent.be.
Alain Goossens, Email: alain.goossens@psb.vib-ugent.be.
Data, Materials, and Software Availability
All data are included in the article, SI Appendix and/or public databanks. RNA-Seq data have been deposited in the ArrayExpress database (accessions E-MTAB-11876, E-MTAB-11880, and E-MTAB-12405).
Supporting Information
References
- 1.Weller S. A., Stead D. E., O’Neill T. M., Morley P. S., Root mat of tomato caused by rhizogenic strains of Agrobacterium biovar 1 in the UK. Plant Pathol. 49, 799 (2000). [Google Scholar]
- 2.Bosmans L., et al. , Rhizogenic agrobacteria in hydroponic crops: Epidemics, diagnostics and control. Plant Pathol. 66, 1043–1053 (2017). [Google Scholar]
- 3.Georgiev M. I., Agostini E., Ludwig-Müller J., Xu J., Genetically transformed roots: From plant disease to biotechnological resource. Trends Biotechnol. 30, 528–537 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Gutierrez-Valdes N., et al. , Hairy root cultures—a versatile tool with multiple applications. Front. Plant Sci. 11, 33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White F. F., Taylor B. H., Huffman G. A., Gordon M. P., Nester E. W., Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. J. Bacteriol. 164, 33–44 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delbarre A., et al. , The rolB gene of Agrobacterium rhizogenes does not increase the auxin sensitivity of tobacco protoplasts by modifying the intracellular auxin concentration. Plant Physiol. 105, 563–569 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altamura M. M., Agrobacterium rhizogenes rolB and rolD genes: Regulation and involvement in plant development. Plant Cell Tissue Organ Cult. 77, 89–101 (2004). [Google Scholar]
- 8.Mauro M. L., Bettini P. P., Agrobacterium rhizogenes rolB oncogene: An intriguing player for many roles. Plant Physiol. Biochem. 165, 10–18 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Bulgakov V. P., Vereshchagina Y. V., Bulgakov D. V., Veremeichik G. N., Shkryl Y. N., The rolB plant oncogene affects multiple signaling protein modules related to hormone signaling and plant defense. Sci. Rep. 8, 2285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriuchi H., et al. , Nuclear localization and interaction of RolB with plant 14–3–3 proteins correlates with induction of adventitious roots by the oncogene rolB. Plant J. 38, 260–275 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Gelvin S. B., Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu. Rev. Phytopathol. 48, 45–68 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Lee L.-Y., et al. , Screening a cDNA library for protein-protein interactions directly in planta. Plant Cell 24, 1746–1759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceulemans E., Ibrahim H. M. M., De Coninck B., Goossens A., Pathogen effectors: Exploiting the promiscuity of plant signaling hubs. Trends Plant Sci. 26, 780–795 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Causier B., Ashworth M., Guo W., Davies B., The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 158, 423–438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harvey S., et al. , Downy mildew effector HaRxL21 interacts with the transcriptional repressor TOPLESS to promote pathogen susceptibility. PLoS Pathog. 16, e1008835 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarrete F., et al. , TOPLESS promotes plant immunity by repressing auxin signaling and is targeted by the fungal effector Naked1. Plant Commun. 3, 100269 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darino M., et al. , Ustilago maydis effector Jsi1 interacts with Topless corepressor, hijacking plant jasmonate/ethylene signaling. New Phytol. 229, 3393–3407 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long J. A., Ohno C., Smith Z. R., Meyerowitz E. M., TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Ito J., et al. , Auxin-dependent compositional change in mediator in ARF7- and ARF19-mediated transcription. Proc. Natl. Acad. Sci. U.S.A. 113, 6562–6567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma H., et al. , A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci. Adv. 3, 1601217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leydon A. R., et al. , Repression by the Arabidopsis TOPLESS corepressor requires association with the core mediator complex. Elife 10, e66739 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins J., O’Grady K., Chen S., Gurley W., The C-terminal WD40 repeats on the TOPLESS co-repressor function as a protein–protein interaction surface. Plant Mol. Biol. 100, 47–58 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Liu X., Galli M., Camehl I., Gallavotti A., RAMOSA1 ENHANCER LOCUS2-mediated transcriptional repression regulates vegetative and reproductive architecture. Plant Physiol. 179, 348–363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim M. H., et al. , The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure 12, 987–998 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Kagale S., Links M. G., Rozwadowski K., Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152, 1109–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delto C. F., et al. , The LisH motif of muskelin is crucial for oligomerization and governs intracellular localization. Structure 23, 364–373 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Ke J., et al. , Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 1, e1500107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Arevalillo R., et al. , Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 114, 8107–8112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pauwels L., et al. , NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia M. E., Lynch T., Peeters J., Snowden C., Finkelstein R., A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 67, 643–658 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Arora D., et al. , Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell 32, 3388–3407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao Y., et al. , Genome-wide identification, phylogenetic analysis, expression profiling, and protein-protein interaction properties of TOPLESS gene family members in tomato. J. Exp. Bot. 65, 1013–1023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D., Fu F., Zhang H., Song F., Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genomics 16, 771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Leene J., et al. , Functional characterization of the Arabidopsis transcription factor bZIP29 reveals its role in leaf and root development. J. Exp. Bot. 67, 5825–5840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djamei A., Pitzschke A., Nakagami H., Rajh I., Hirt H., Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 318, 453–456 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Tsugama D., Liu S., Takano T., The bZIP protein VIP1 is involved in touch responses in Arabidopsis roots. Plant Physiol. 171, 1355–1365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deppmann C. D., et al. , Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 32, 3435–3445 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho-Plágaro T., Huertas R., Tamayo-Navarrete M. I., Ocampo J. A., García-Garrido J. M., An improved method for Agrobacterium rhizogenes-mediated transformation of tomato suitable for the study of arbuscular mycorrhizal symbiosis. Plant Methods 14, 34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mankin S. L., et al. , Disarming and sequencing of Agrobacterium rhizogenes strain K599 (NCPPB2659) plasmid pRi2659. In Vitro Cell. Dev. Biol. Plant 43, 521–535 (2007). [Google Scholar]
- 40.Otten L., T-DNA regions from 350 Agrobacterium genomes: Maps and phylogeny. Plant Mol. Biol. 106, 239–258 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Rodrigues S. D., et al. , Efficient CRISPR-mediated base editing in Agrobacterium spp. Proc. Natl. Acad. Sci. U.S.A. 118, e2013338118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N., Han X., Feng D., Yuan D., Huang L.-J., Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 20, 671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Z., Molecular basis for jasmonate and ethylene signal interactions in Arabidopsis. J. Exp. Bot. 65, 5743–5748 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Ruan J., et al. , Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 20, 2479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulkarni S. R., Vaneechoutte D., Van de Velde J., Vandepoele K., TF2Network: Predicting transcription factor regulators and gene regulatory networks in Arabidopsis using publicly available binding site information. Nucleic Acids Res. 46, e31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorenzo O., Piqueras R., Sánchez-Serrano J. J., Solano R., ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berrocal-Lobo M., Molina A., Solano R., Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Xu P., et al. , Integration of jasmonic acid and ethylene into auxin signaling in root development. Front. Plant Sci. 11, 271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakehal A., et al. , ETHYLENE RESPONSE FACTOR 115 integrates jasmonate and cytokinin signaling machineries to repress adventitious rooting in Arabidopsis. New Phytol. 228, 1611–1626 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Zhang C., et al. , Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis. Dev. Cell 55, 603–616 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bulgakov V. P., Veremeichik G. N., Shkryl Y. N., The rolB gene activates the expression of genes encoding microRNA processing machinery. Biotechnol. Lett. 37, 921–925 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Wen-jun S., Forde B. G., Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucleic Acids Res. 17, 8385–8385 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kajala K., Coil D. A., Brady S. M., Draft genome sequence of Rhizobium rhizogenes strain ATCC 15834. Genome Announc. 2, e01108–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ron M., et al. , Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 166, 455–469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pérez Cuéllar A., Pauwels L., De Clercq R., Goossens A., Yeast two-hybrid analysis of jasmonate signaling proteins. Methods Mol. Biol. 1011, 173–185 (2013). [DOI] [PubMed] [Google Scholar]
- 56.De Sutter V., et al. , Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J. 44, 1065–1076 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Bel M., et al. , PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46, D1190–D1196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weirauch M. T., et al. , Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fornes O., et al. , JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frith M. C., Li M. C., Weng Z., Cluster-Buster: Finding dense clusters of motifs in DNA sequences. Nucleic Acids Res. 31, 3666–3668 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulkarni S. R., Jones D. M., Vandepoele K., Enhanced maps of transcription factor binding sites improve regulatory networks learned from accessible chromatin data. Plant Physiol. 181, 412–425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shumate A., Salzberg S. L., Liftoff: Accurate mapping of gene annotations. Bioinformatics 37, 1639–1643 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Data Availability Statement
All data are included in the article, SI Appendix and/or public databanks. RNA-Seq data have been deposited in the ArrayExpress database (accessions E-MTAB-11876, E-MTAB-11880, and E-MTAB-12405).