Significance
It is well known that plasticity-inducing neuronal activity drives de novo synthesis of plasticity-related proteins, which are required for expression of downstream plasticity mechanisms. It is not very well known, however, whether cellular machineries exist that specifically target and regulate production of these activity-induced nascent proteins. The discovery of a neuronal membrane proteasome (NMP), which was found to preferentially degrade activity-induced nascent proteins in neuronal cultures, added a new player to the arena. Here, we report in vivo evidence for functional significance of NMPs. We demonstrated the expression and proteolytic function of NMPs in the vertebrate brain and, importantly, provided in vivo evidence for the roles of NMPs in regulating both neuronal activity and behavioral plasticity in intact neuronal circuits.
Keywords: proteasome, neuron, activity-induced nascent proteins, spontaneous activity, BONCAT
Abstract
Protein degradation is critical for brain function through processes that remain incompletely understood. Here, we investigated the in vivo function of the 20S neuronal membrane proteasome (NMP) in the brain of Xenopus laevis tadpoles. With biochemistry, immunohistochemistry, and electron microscopy, we demonstrated that NMPs are conserved in the tadpole brain and preferentially degrade neuronal activity–induced newly synthesized proteins in vivo. Using in vivo calcium imaging in the optic tectum, we showed that acute NMP inhibition rapidly increased spontaneous neuronal activity, resulting in hypersynchronization across tectal neurons. At the circuit level, inhibiting NMPs abolished learning-dependent improvement in visuomotor behavior in live animals and caused a significant deterioration in basal behavioral performance following visual training with enhanced visual experience. Our data provide in vivo characterization of NMP functions in the vertebrate nervous system and suggest that NMP-mediated degradation of activity-induced nascent proteins may serve as a homeostatic modulatory mechanism in neurons that is critical for regulating neuronal activity and experience-dependent circuit plasticity.
Proteostasis, the collective protein synthesis, folding, and degradation mechanisms that maintain the integrity of the cellular proteome, is pivotal for the health and function of the nervous system (1). Protein synthesis and degradation are tightly controlled and closely coordinated in many cellular processes, as shown in the classical studies of cell cycle progression (2). Proteostasis in neurons is no exception—both protein synthesis and degradation play essential roles in neuronal circuit plasticity and homeostasis (3–8). A key signature of Hebbian-type plasticity mechanisms is activity-dependent upregulation of plasticity-related protein synthesis (9–11). These activity-induced nascent proteins then trigger downstream signaling mechanisms that are important for expression of synaptic plasticity. Recent studies, however, suggest that our understanding of the mechanisms and the cellular machinery that control the degradation of nascent neuronal proteins may be incomplete (12–14).
Proteasomes constitute a major component of the proteostasis network in neurons. Proteasomes are large macromolecular complexes, with 28 subunits, including six catalytic subunits (15). Together, these make up the core 20S particle (CP), which can interact with another set of proteins that make up a 19S cap, forming the 26S (singly capped) or 30S (doubly capped) proteasome. We refer below to the 19S-capped proteasomes as 26S. 26S proteasomes are responsible for the majority of ubiquitin-dependent degradation in the nervous system and require ATP (16). On the other hand, 20S uncapped proteasomes do not require ubiquitin or ATP and operate independently of 26S to degrade intracellular proteins, such as damaged/oxidized or unstructured proteins (17, 18). Emerging evidence suggests that 20S uncapped proteasomes are relatively enriched in the brain and may play critical roles in neuronal physiology (1, 13, 14, 19). Indeed, recent work demonstrated that an uncapped 20S proteasome complex is tightly associated with the neuronal plasma membrane in the mammalian central nervous system (CNS) (13, 14). In prior in vitro studies, neuronal membrane proteasomes (NMPs) appear to preferentially degrade activity-induced nascent polypeptides as they are actively translated, and degradation occurs in a ubiquitin-independent manner (13). Furthermore, blocking NMP activity in cortical neuronal cultures rapidly altered ongoing neuronal activity (14). Together, these studies suggest that NMPs could play a previously undiscovered role in proteostasis of activity-induced nascent proteins and may also be involved in regulating neuronal activity in vitro; however, whether NMPs have similar properties in vivo and the functional significance of NMPs in intact animals are unknown.
Here, we examined the function of NMPs in the brains of live tadpoles, with a focus on the potential role of NMPs in activity-dependent neuronal and behavioral plasticity. The visual circuit of tadpoles is a robust experimental system to investigate mechanisms underlying experience-dependent plasticity through the use of in vivo drug treatments, calcium imaging, and analysis of visually guided behavioral plasticity (20, 21). Prior studies in the Xenopus visual system applied bioorthogonal noncanonical amino acid tagging (BONCAT) to identify visual experience–induced nascent proteins in the living tadpole brain and demonstrated the requirement for newly synthesized proteins in visual experience–dependent behavioral plasticity (11, 22). We optimized the in vivo BONCAT labeling protocol to successfully label nascent proteins produced within a short period of 30 min, providing the temporal resolution to quantitatively evaluate rapid degradation of nascent proteins by NMPs in vivo. With biochemical, immunohistochemical, and electron microscopic evidence, we demonstrated that NMPs are present in the tadpole brain. Using the BONCAT labeling of nascent proteins in the tadpole brain, we showed that inhibiting NMPs in vivo significantly increased the level of activity-induced nascent proteins detected both as the pooled total amount of nascent proteins and as individual activity-induced nascent proteins. Interestingly, NMP inhibition rapidly increased spontaneous neuronal activity in tectal neurons and increased synchronous activity across the tectal circuit in intact animals, suggesting that NMPs play a role in regulating neuronal activity. The effect of NMP inhibition is significantly more prominent under the stimulated condition than under the basal condition. To test whether NMP function is required for experience-dependent neuronal plasticity at the circuit and behavioral levels, we employed a tectally mediated visual avoidance behavioral paradigm and found that inhibiting NMPs abolished visual experience–dependent behavioral plasticity, consistent with extensive evidence that aberrant synchronization in brain circuits disrupts cognitive function and behavior (23–25). Particularly intriguingly, NMP inhibition did not affect basal visual avoidance behavior in the absence of enhanced visual training, suggesting a specific role of NMP function during periods of heightened neuronal activity. Together, these results demonstrate that NMPs are conserved in the vertebrate brain and degrade nascent proteins in vivo. These results also provide in vivo evidence that NMPs play an important role in regulating neuronal activity and are required for neural circuit function in experience-dependent behavioral plasticity.
Results
NMPs Are Present in Neurons in the Tadpole Brain.
To study NMP functions in vivo, we employed a membrane-impermeable 20S core proteasome inhibitor, biotin–epoxomicin (BE). Epoxomicin (Epox) is a highly specific proteasome inhibitor, which covalently binds to catalytic β subunits of the core 20S proteasome (26). Biotinylation renders Epox impermeable to the cell membrane in live cells. When BE is added to neuronal cultures, it specifically binds to and blocks NMPs on the neuronal membrane (13, 14). Therefore, BE can be used both as a marker and as a highly specific inhibitor for NMPs in living neurons. To determine whether NMPs are present in the Xenopus brain, we injected BE (1 mM, note lower concentrations were used for subsequent physiology experiments; see Materials and Methods for details) into brain ventricles of live tadpoles and collected brain tissue either 30 min or 6 h after injection for subcellular fractionation and western blotting. The 20S proteasome core subunits (α1–7) are abundantly expressed in the tadpole brain and are present in the isolated membrane fraction (Fig. 1A). Importantly, the biotin bands unique to the BE-injected samples were only seen in the membrane fraction at the molecular weight corresponding to the 20S β subunits, revealing the BE-bound β subunits (Fig. 1A; the full molecular range of the biotin blot is provided in Fig. 1A (SI Appendix, Fig. S1). In addition, the BE-unique biotin bands were detected exclusively in the membrane fraction at both 30 min and 6 h after the injection, suggesting that BE binding to the 20S β subunits on the membrane (i.e. NMPs) was stable, consistent with prior work demonstrating irreversible covalent binding between Epox and the catalytic β subunits (13, 14, 26). To further examine whether BE binds solely at the epoxomicin-binding site, we preinjected tadpoles with Epox prior to BE injection to test for occlusion of BE binding and used neutravidin-coated resin to pull down biotin-bound proteins from the total brain lysate. Western blots of the neutravidin pull-down samples showed that 20S β5 subunits coprecipitated with biotin only in the BE-injected samples (Fig. 1B). BE-specific biotin bands corresponding to the 20S β subunits were also blocked in the epoxomicin-preinjected samples. The occlusion of BE binding to the 20S β subunits by preinjection of Epox indicates that BE and Epox share binding sites in complex brain tissue.
Fig. 1.
NMPs are present in tadpole neurons. (A) Biochemical evidence for NMPs in the tadpole brain. Tadpole brain tissue was collected at 30 m or 6 h after either BE or Epox injection, and an equivalent amount of protein from the membrane and cytosolic fractions were labeled with indicated antibodies on western blots. The 20S core proteasome subunits (α1–7) are abundantly expressed in the tadpole brain and are enriched in the membrane fraction. Importantly, the biotin signal was recovered at the molecular weight corresponding to the 20S catalytic subunits (run around 20 to 25 kD) bound to the injected BE in (and only in) the membrane preparation both at 30 min and 6 h after the injection. Actin, a cytosolic protein, and GluA1, which is enriched in the neuronal membrane, serve as positive controls for the cell fractionation preparation. (B) Preinjection of Epox occluded BE binding to NMPs. Brain lysates from uninjected, BE-injected, and Epox preinjected (Epox-BE-inj) samples were purified by neutravidin pulldown to coprecipitate BE-bound proteins and blotted with biotin and 20S β5 antibody. (C) Anatomical illustration of cellular organization of the optic tectum (OT) and the ventricle (V). CBL: neuronal cell body layer. NPL: neural progenitor cell layer. N: neuropil. (D) Immunohistological localization of NMPs bound to BE. Vibratome sections of the OT immunostained with Anti-Biotin antibody show punctate biotin signal in neuronal cell body layers and the neuropil in the BE-injected tectum but not uninjected, or Epox-preinjected, or biotin-injected controls. All samples processed in parallel under the same conditions. Gold: biotin; blue: DAPI. (Scale bar, 20 µm.) (E–J) Ultrastructural distribution of the 20S proteasome shown by preembedding immunogold labeling with anti-α1-7 20S proteasome subunit antibody and FluoroNanogold secondary antibody (1.4-nm-diameter gold particles). Plasma membranes are highlighted in light blue. (E and F) Ultramicrographs from the tectal cell body layer showing anti-α1-7 20S immunogold labeling (E) and no primary controls (F). Boxed regions are shown at higher magnification in E’ and F’. Immunogold particles identifying 20S α subunits are associated with the plasma and nuclear membranes and found in the cytoplasm. Asterisks mark nuclei. (Scale bar, 1 µm.) (G–J) Ultramicrographs from the tectal neuropil labeled with anti-α1-7 20S antibodies (G and I) or no primary control (H and J). Numerous immunogold particles identifying 20S α subunits are localized at or near the plasma membrane in G and I. The 20S α subunits are also visible at synaptic sites, which are marked by arrows. (Scale bar, 500 nm.)
Next, we used immunohistochemistry to examine the distribution of BE-bound NMPs in the optic tectum (OT) with Anti-Biotin antibody (Fig. 1 C and D). Strong biotin signal was observed in the neuronal soma layer and the neuropil layer of the OT from BE-injected animals, with little signal seen in the ventricular layer where neuroprogenitor cells reside (Fig. 1D), consistent with previous observations that NMPs are primarily detected in mature neurons (14). No signal was seen in uninjected brains or in the brains preinjected with Epox. In addition, injecting the brain with free biotin did not result in any signal above background, indicating that BE binding in the brain was not due to nonspecific biotin binding. These data suggest that BE-bound 20S CP is present in the tadpole brain and is associated with optic tectal neurons, supporting the presence of NMPs in tectal neurons. To further probe the subcellular and ultrastructural localization of NMPs in the OT, we employed pre-embedding immuno-EM using antibodies against the 20S α subunits. As 20S α subunits are obligatory subunits found in all 26S and free 20S proteasomes, we identified NMPs by 20S α subunit immunoreactivity localized to the plasma membrane (Fig. 1 E, E’, G, and I). Controls with no primary antibody showed low background levels of nonspecific Nanogold labeling (Fig. 1 F, F’, H, and J). We observed 20S α subunit immunolabeling in the nuclear membrane and cytoplasm (Fig. 1 E and E’), which are known to contain both intracellular 26S and free 20S proteasomes (18). Importantly, we observed a subset of 20S α subunits closely associated with the plasma membrane (i.e. NMPs) in both the tectal cell body layer (Fig. 1 E and E’) and neuropil, which includes both axonal and dendritic processes (Fig. 1 G and I). These putative NMPs were also observed at synaptic sites (Fig. 1G, arrows) similar to what has been reported in mouse hippocampal neurons (14). Taken together, these data show that NMPs are expressed in the Xenopus laevis brain, indicating that NMPs are conserved across nonmammalian and mammalian vertebrates.
NMPs Preferentially Degrade Nascent Proteins in the Tadpole Brain.
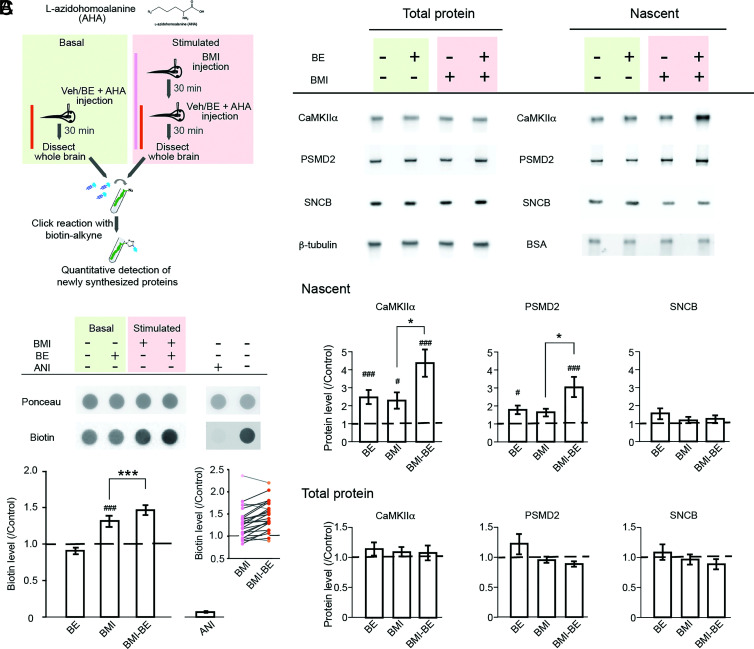
Prior studies in mouse cortical neuronal cultures indicated that NMPs rapidly degrade nascent proteins synthesized in response to increased neuronal activity and that NMPs function independently of ubiquitin (13). To determine whether activity-induced nascent proteins are degraded by NMPs in vivo, we treated animals with intraventricular injection of bicuculline methiodide (BMI), which blocks GABAergic inhibition and significantly increases neuronal activity Fig. 2A, and measured nascent protein production over the next 30 min in live tadpole brains in the presence or absence of the NMP inhibitor BE (Fig. 2A). Animals were injected intraventricularly with the noncanonical amino acid, L-azidohomoalanine (AHA), to label nascent proteins for quantitative analysis. Fifty micromolar BE was injected together with AHA under either basal or BMI-stimulated conditions to inhibit NMPs during the labeling period, after which whole-brain tissue was dissected and processed with click chemistry to specifically tag AHA-labeled nascent proteins with biotin. Dot blots immunolabeled with biotin antibody were used to assess the total level of nascent proteins. Neither endogenous biotin nor biotin signal from the injected BE was detectable by dot blot under the linear exposure range for the biotinylated AHA-tagged nascent proteins, assuring quantitative assessment of the biotinylated nascent proteins (SI Appendix, Fig. S2). Treatment with the protein synthesis inhibitor, anisomycin, blocked all biotin labeling (Fig. 2B, Upper), indicating that AHA-tagged nascent proteins were the sole source for the biotin signal detected. Under the basal condition, AHA-tagged nascent proteins were readily detected in tadpole brain tissue after the 30-min AHA incubation period, indicative of protein synthesis under ambient activity (Fig. 2B, Upper). BMI treatment significantly increased the level of nascent proteins, consistent with our prior reports of rapidly increased protein synthesis in response to heightened neuronal activity (11, 22). Interestingly, inhibiting NMPs with BE in the presence of BMI induced a further significant increase in the level of nascent proteins, shown in representative dot blots (Fig. 2B, Upper) and quantitatively (Fig. 2B, Lower: BMI: 1.32 ± 0.08; BMI–BE: 1.47 ± 0.07; mean ± SEM, normalized to control from the same batch), suggesting that about a third of nascent proteins synthesized in response to increased neuronal activity was rapidly degraded by NMPs in the tadpole brain. By contrast, under basal conditions, no significant difference was observed in the total level of AHA-tagged nascent proteins when NMPs are inhibited with BE (Fig. 2B; BE: 0.91 ± 0.04).
Fig. 2.
NMPs degrade nascent proteins in the tadpole brain. (A) Experimental design and timeline. Animals under basal or stimulated (treated with BMI) conditions were injected in the brain ventricle with either vehicle (Veh) or BE together with AHA to label nascent proteins using the BONCAT. For each batch of animals, 25 to 30 brains were collected for each experimental group and processed in parallel. (B, Top): representative Ponceau staining (Top) and biotin-immunolabeled dot blots (Bottom) under basal and BMI-stimulated conditions. (B, Bottom): summary data of AHA–biotin-labeled total nascent protein level detected by biotin signal in dot blots. Data from different experimental groups were normalized to the corresponding control group (marked by the dashed line) from the same batch of animals run on the same blot. Data shown as mean ± SEM, n = 24 batches. RM one-way ANOVA with Bonferroni’s multiple comparison test. ###P < 0.001, compared with the control group. ***P < 0.001, comparison as marked. Inset shows data from individual batches for the BMI and BMI–BE groups. Data points of the same batch were line-connected for visualization of within-batch comparisons (Cohen’s D = 0.43). Anisomycin (ANI) was coinjected with AHA under control condition in a subset (n = 3 batches) of experiments to confirm that the detected biotin signal was from biotinylated AHA-labeled nascent proteins. (C) Nascent CaMKIIα and PSMD2, but not SNCB are degraded by NMPs in the tadpole brain. Top: experimental conditions for each group are shown with representative western blots for each candidate and loading controls (β-tubulin for total protein and biotinylated BSA for purified nascent proteins). Middle: quantification of nascent proteins (Middle) and total protein (Bottom). All data were normalized to the corresponding control group (dashed line) from the same batch that was run on the same blot. NMP inhibition increased nascent CaMKIIα and PSMD2 under basal and stimulated conditions. Total protein was not affected by NMP inhibition. The Friedman test with Dunn’s multiple comparison test. #P < 0.05, ###P < 0.001, compared with the control group. *P < 0.05, comparison as marked. n = 16 batches.
Data shown in Fig. 2B indicate that NMPs degrade nascent proteins under stimulated conditions in the tadpole brain, suggesting that proteins known to be synthesized in response to increased activity are plausible substrates for NMPs. To identify potential NMP substrates, we purified the biotinylated AHA-labeled nascent proteins with neutravidin-coated resin from brain homogenates. We first verified the specificity of the neutravidin purification of biotinylated nascent proteins with negative controls of AHA-tagged samples without click chemistry for biotinylation (SI Appendix, Fig. S3). Samples from brains treated with or without BE under either basal or stimulated conditions were then prepared for western blot analysis using antibodies against individual protein candidates. We selected three candidate proteins based on prior reports of either their involvement in activity-dependent protein synthesis in the tadpole brain or potential interaction with free 20S proteasomes (11, 13, 18, 22). CaMKIIα plays pivotal roles in activity-dependent plasticity (27), and its synthesis is known to increase in response to activity, including the Xenopus brain (22, 28). PSMD2 (26S proteasome non-ATPase regulatory subunit 2) is a proteasome subunit that we have shown is up-regulated in Xenopus in visual experience–dependent learning paradigms (11). β-synuclein (SNCB) is an intrinsically disordered (ID) protein that is thought to play a role in synaptic function (29) but with no prior report of activity-induced upregulation in synthesis. Proteins with ID domains have been reported to be degraded by free 20S in vitro without ubiquitination (18), which suggests that it is a potential NMP substrate. All three proteins were readily detected in the nascent protein samples under basal control conditions (Fig. 2C, Top). Increasing neuronal activity with BMI treatment significantly increased nascent CaMKIIα. Inhibiting NMPs with BE in BMI-treated animals resulted in a further significant increase in nascent CaMKIIα, suggesting that NMPs rapidly degrade nascent CaMKIIα under stimulated conditions in vivo. Interestingly, inhibiting NMP activity under the basal condition also revealed a significant increase in nascent CaMKIIα, suggesting that ongoing CaMKIIα synthesis occurs under basal activity conditions and that NMPs degrade this nascent CaMKIIα too. Inhibiting NMP activity also significantly increased nascent PSMD2 under both basal and BMI-stimulated conditions. Interestingly, an increased synthesis in response to BMI alone was not detected, but blocking NMPs in the presence of BMI revealed a significant net increase in nascent PSMD2 over the short 30-min time course of our experiments, suggesting that nascent PSMD2 was rapidly degraded by NMPs. Notably, not all nascent proteins examined were degraded by NMPs. The level of nascent SNCB was not affected by NMP inhibition under either basal or stimulated conditions, suggesting that SNCB is not a substrate for NMPs. In addition, the level of nascent SNCB was also not affected by BMI in the presence or absence of BE, suggesting that SNCB synthesis is not up-regulated by increased activity. These data also suggest that NMPs degrade nascent proteins under basal conditions. It should be noted that animals were free swimming and exposed to an ambient light environment with sensory inputs under basal condition. The NMP-degraded subset of nascent proteins in the whole brain homogenate revealed by NMP inhibition could reflect ongoing active neural activity in at least some brain regions under this condition. However, the observation that NMP-mediated degradation of nascent protein was not detectable in the total nascent protein level by dot blot but could only be detected by western blotting with individual protein candidates suggests that the overall level of NMP-mediated degradation of nascent proteins is significantly lower under basal condition compared with that under stimulated condition. No changes were detected in total protein samples of the candidate proteins tested under either basal or stimulated conditions (Fig. 2C, Bottom), suggesting that the nascent proteins synthesized over 30 min in vivo are a minor proportion of these samples.
Prior studies indicate that NMP-mediated protein degradation is ubiquitin-independent (13). To test if this is also the case in vivo, we used ubiquitin dot blots to assess ubiquitin levels in total protein samples from tadpole brains. We observed no change in the ubiquitin level in BE-treated samples compared with the control group under either basal or stimulated conditions (SI Appendix, Fig. S4). As a positive control, Epox treatment, which inhibits all proteasome activities including the ubiquitin-dependent protein degradation, significantly increased the ubiquitin level in the brain. This result corroborated the prior in vitro finding that proteolytic activity of NMP is ubiquitin independent, supporting the interpretation that NMPs do not play a significant role in the degradation of ubiquitinated pre-existing proteins in vivo.
NMPs Regulate Neuronal Activity In Vivo.
Blocking NMPs with BE in pharmacologically stimulated neuronal cultures significantly changed neuronal activity (14). To test whether NMP inhibition affects neuronal and circuit activity in vivo, we used time-lapse 2-photon Ca++ imaging in GCaMP6f-expressing tectal neurons to examine the effects of acute inhibition of NMPs on spontaneous neuronal activity in the brain of awake tadpoles. Ca++ signals recorded from neuronal soma faithfully report neuronal activity in tectal neurons (30–32). The time-lapse imaging protocol allowed us to record the activity of a population of neurons with single-neuron resolution over extended periods in live animals. As shown in representative images in Fig. 3A, tectal neurons can be identified over different time points throughout the time series, enabling us to extract Ca++ activity data from the same individual neurons before and after different in vivo drug treatments for within-cell comparisons (Fig. 3 B and C).
Fig. 3.
Inhibiting NMPs induced a rapid increase in spontaneous neuronal activity and network synchrony in the tadpole brain. (A) Representative time-lapse images of GCaMP fluorescence collected in the OT of a live tadpole throughout an experiment. The animal was recorded for 5 min before and after intraventricular injection of BMI and for 5 min after BE injection. (Scale bar, 50 µM.) (B) Traces of Ca++ activity extracted from the GCaMP fluorescence signal in the same individual tectal neuronal soma (ROIs) during the 5-min recording period following the indicated treatments. (C) Raster plots (Top) of Ca++ events derived from the data in B. (D–K) Summary data of average Ca++ event counts (D–G) and integrated Ca++ responses (H–K) at different time points in animals under different conditions. (L–M) Scatterplots of Ca++ event counts for individual neurons in BMI-treated animals before and after treatment with either vehicle (L) or BE (M). (N–P) The synchrony index calculated from Ca++ activity of all neurons recorded in each animal in animals treated with BE (N), BMI-veh (O), or BMI–BE (P). Data points from the same animal were connected by straight lines. (Q) Magnitude of change in Ca++ event counts following different treatments. *P < 0.05, **P < 0.01, and ***P < 0.001. The Friedman test with Dunn’s multiple comparison post hoc test. Veh: n = 54 neurons and N = 3 animals; BE: n = 214 neurons and N = 6 animals (data pooled from 50 µM and 250 µM BE). BMI-Veh: n = 119 neurons and N = 6 animals; BMI–BE: n = 194 neurons and N = 8 animals.
Under the basal condition, inhibiting NMPs with intraventricular BE injection rapidly increased spontaneous neuronal activity, shown by the increased frequency of Ca++ events (Fig. 3 D and E), increased integrated signal of compound Ca++ transients (Fig. 3 H and I), and increased synchronous activity across the neuronal population (Fig. 3N) compared with control. The effect of NMP inhibition was dose dependent (SI Appendix, Fig. S5). Intraventricular injection of epoxomicin, which blocks both NMPs and intracellular 26S proteasomes, increased neuronal activity similar to BE (SI Appendix, Fig. S5E), suggesting that the rapid increase in neuronal activity may be mostly attributed to inhibition of NMPs.
We next asked how NMP inhibition affects neuronal activity under pharmacologically stimulated conditions. Intraventricular BMI injection increased the frequency of spontaneous firing events and integrated compound Ca++ transients within individual tectal neurons over extended recording periods and across neurons in the OT (Fig. 3 F, G, J, and K). BMI also increased synchronous firing across the population of tectal neurons (Fig. 3 O and P). Importantly, inhibiting NMPs with BE following BMI treatment induced a further increase in neuronal activity to levels significantly above that seen with BMI alone (Fig. 3 G, K, M and P), an effect not seen in vehicle-injected animals (Fig. 3 F, J, L and O). These data suggest a sequence of events in which increased neuronal activity increases protein synthesis; the activity-induced nascent proteins trigger rapid NMP-mediated degradation of these substrates, which in turn limits neuronal activity. This scenario suggests that NMPs play a role in preventing runaway of neuronal activity that could potentially disrupt activity patterns within the tectal network: Under basal activity levels, fewer nascent proteins are synthesized (Fig. 2B); the level of NMP-mediated degradation of nascent proteins remains relatively low, and basal neuronal activity levels are modestly modulated by NMPs; under stimulated conditions with increased protein synthesis, NMP-mediated degradation of nascent proteins is more prominent and may play a larger role in regulating neuronal activity. To assess the feasibility of this idea, we compared the relative change in neuronal activity induced by NMP inhibition under basal and BMI-stimulated conditions (Fig. 3Q). Blocking NMPs results in a significantly greater increase in spontaneous neuronal activity in BMI-treated animals than that under the basal condition (change in Ca++ event counts before and after BE injection: basal condition + BE = 1.42 ± 0.19; BMI + BE = 4.21 ± 0.56, normalized to baseline activity from the same cell), supporting the idea that NMP function plays a significantly larger role in modulating neuronal activity under stimulated conditions in which neurons are firing extensively.
NMP Function Is Required for Learning-Induced Behavioral Plasticity.
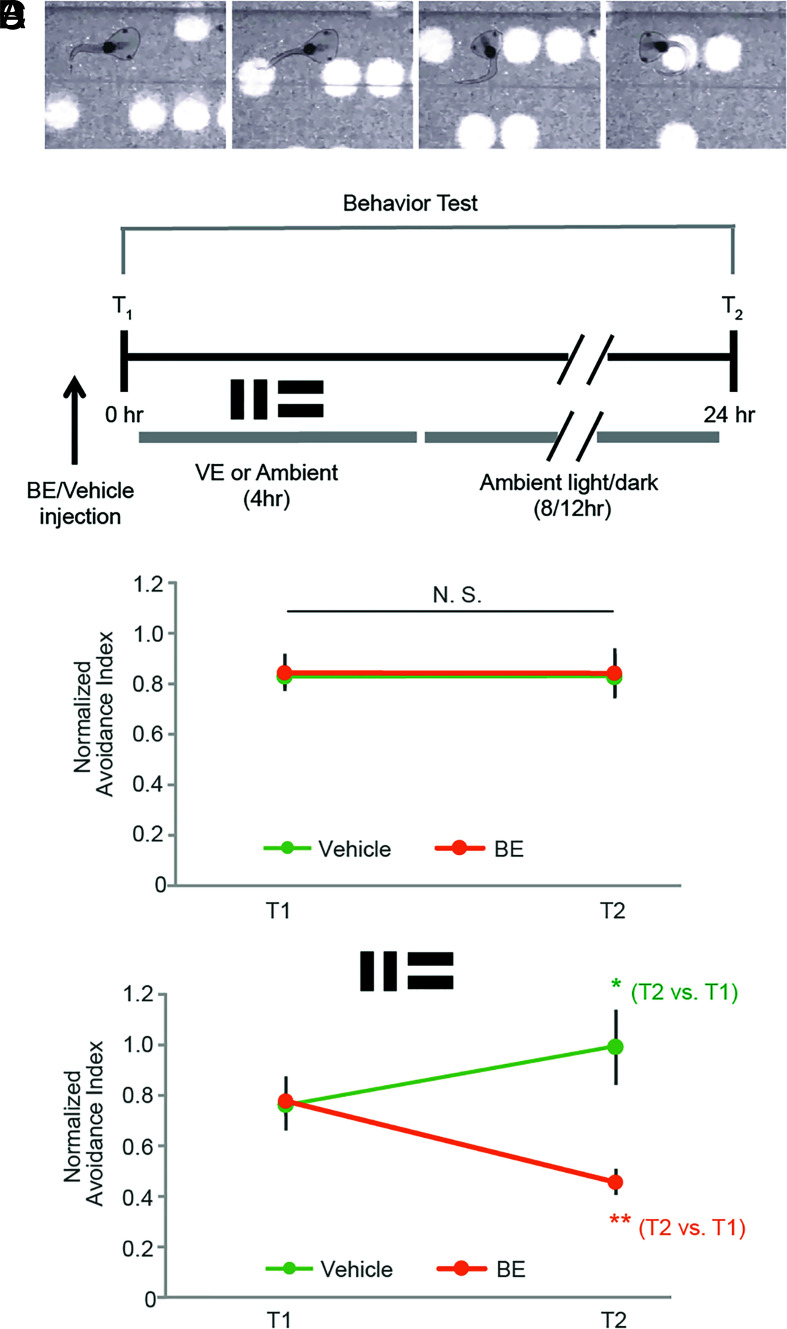
Two important features of the nervous system are the ability to process information and to adapt behavioral responses in accordance with changes in the environment, for instance, through learning. This requires that neurons are capable of detecting signal from noise and changes in the activity pattern in afferent inputs, which in turn requires that neuronal activity is maintained within an intermediate dynamic range (33–35). Consequently, dysregulated neuronal activity is predicted to interfere with both normal circuit function and experience-dependent plasticity. Indeed, studies in several animal models of neurodevelopmental and neurodegenerative diseases have shown that aberrant neuronal activity led to hypersynchronization and learning deficits in neural networks (24, 36). Our data presented above show that inhibiting NMPs abruptly increased neuronal activity and synchrony across the network, particularly under stimulated conditions when network activity was already elevated. This suggests that NMPs may play a role in maintaining neuronal activity within a dynamic range that is essential for normal circuit function and experience-dependent circuit plasticity. To test whether the dysregulated neuronal activity incurred by NMP inhibition impairs circuit function or plasticity, we employed a well-established visual avoidance behavioral paradigm in tadpoles (32, 37, 38) to assay the effect of inhibiting NMPs in the OT on visuomotor behavior and learning-induced behavioral plasticity. The visual avoidance behavior is a tectally mediated behavior in which an animal changes swim trajectory in response to an approaching visual stimulus (Fig. 4A). In addition, the performance of visual avoidance behavior can be significantly improved by exposing animals to enhanced visual training (22, 39, 40), likely through experience-dependent refinement of the tectal visuomotor circuits (38, 41, 42). This behavioral paradigm therefore can be used as a readout for both the basic function and learning-induced behavioral plasticity of the visuomotor circuit. Animals received an intraventricular injection of either BE or vehicle and were tested shortly thereafter for baseline avoidance performance (Fig. 4B). Animals were then exposed to either normal ambient light (control) or 4 h of enhanced visual training (VE) and tested again the next day to evaluate changes in their behavioral performance. Blocking NMPs under control conditions, without visual training, did not affect baseline avoidance behavior performance (Fig. 4C), suggesting that the modest increase in neuronal activity induced by NMP inhibition under basal conditions (Fig. 3 E and I) did not disrupt basic circuit function. By contrast, inhibiting NMPs with BE during visual training (VE) completely blocked the VE-induced behavioral plasticity that was observed in vehicle-injected animals (Fig. 4D). Intriguingly, blocking NMPs not only blocked the learning-induced behavioral enhancement but also led to a significant deterioration of the behavioral performance compared with the baseline performance before VE (Fig. 4D). These data suggest that blocking NMPs does not interfere with baseline neuronal function and that the effect of NMP inhibition on learning-induced behavioral plasticity was specifically related to the visual training process. Taken together, these results indicate that NMPs play an essential role in experience-dependent behavioral plasticity.
Fig. 4.
NMP activity is required for learning-induced behavioral improvement and the maintenance of normal visual avoidance behavior following enhanced visual training in tadpoles. (A) Illustration of the visual avoidance behavior in tadpoles. Animals make a sharp turn in their swimming trajectory when they encounter an approaching visual stimulus. The percentage of avoidance events out of the first 10 encounters is quantified as the avoidance index (AI) to evaluate the behavioral performance of the animals. (B) Experimental timeline for visual training and behavioral test schedule. Animals first received a ventricular injection of either BE or vehicle (Veh) and were tested 30 min later for baseline visual avoidance behavior performance (T1). Then, animals were subjected to 4 h of either visual training (VE) or ambient light and returned to their normal rearing conditions. Animals were tested again for avoidance behavior the next day (T2). (C) In the absence of VE, baseline behavioral performance was not affected by BE, n = 9 animal batches. (D) Following VE, vehicle-injected control animals improved their visual avoidance response. Inhibiting NMPs with BE not only completely blocked learning-induced behavioral improvement following VE but also caused a significant deterioration of the behavioral performance comparing with the pretraining baseline level. *P < 0.05 and **P < 0.01. Paired two-tailed Student’s t test, n = 6 animal batches.
Discussion
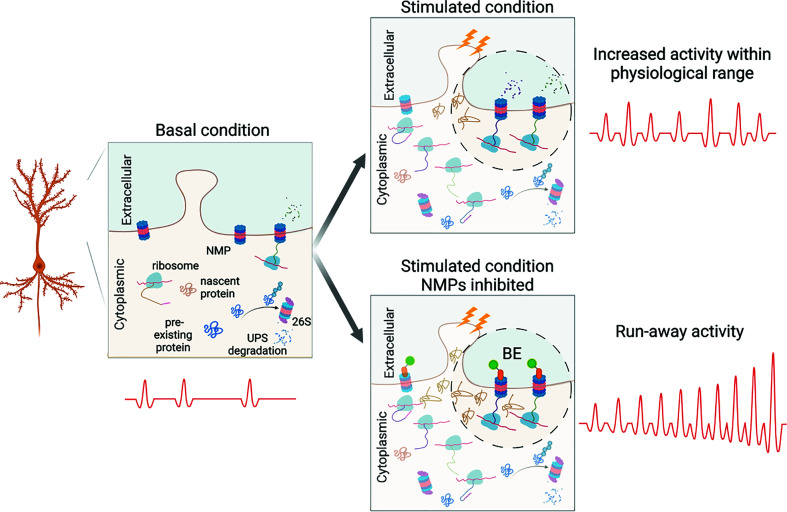
Regulated protein synthesis and degradation in CNS neurons are required for learning and memory, highlighting the critical importance of tight control over proteostasis in neurons. Nevertheless, our understanding of mechanisms engaged by the proteostasis network during neuronal plasticity remains incomplete (35, 43, 44). Here, we took advantage of the Xenopus tadpole visual system to test the potential role of NMPs in visual system function and experience-dependent behavioral plasticity in intact animals. We provide in vivo evidence that NMPs control the accumulation of nascent proteins in the tadpole brain, and NMP function is required for experience-dependent behavioral plasticity. We demonstrated that NMPs are present in the tadpole brain and rapidly degrade nascent proteins in vivo. Pharmacologically increasing brain activity increased nascent protein synthesis, as expected, and in turn increased NMP-mediated degradation of nascent proteins. Conversely, inhibiting NMPs rapidly increased spontaneous neuronal activity in the brain and increased synchronous neuronal activity across the optic tectal circuit, suggesting that NMPs play a role in regulating neuronal activity and help to prevent aberrant synchronous network activity, especially under stimulated conditions. Finally, we show that inhibiting NMPs blocked visual experience–dependent behavioral plasticity in a visual avoidance task and further degraded baseline visual avoidance behavioral performance specifically in animals exposed to enhanced visual experience. Our results indicate that NMPs add to the well-documented mechanisms for protein degradation by UPS and autophagic pathways (1, 43), offering an additional level of regulatory vigilance over the dynamic maintenance of proteostasis in neurons, especially in the face of fluctuations in neuronal activity, when protein synthesis changes rapidly. The contribution of protein homeostasis to neuronal functions has been most studied in the context of the pool of total existing proteins. Relatively less is known about the rapid degradation of newly synthesized proteins (1, 13, 14). Our study suggests that NMPs are poised to detect increases in neuronal activity based on the increased presence of nascent protein substrates and provide negative feedback by degrading a proportion of the activity-induced nascent proteins. Together, these mechanisms may contribute to homeostatic control of overall network activity within a healthy range. In the absence of NMP function, this feedback is blocked, resulting in aberrant synchronous spontaneous activity, which interferes with circuit function and disrupts learning (Fig. 5).
Fig. 5.
Schematic of the proposed function of NMP contribution to the maintenance of homeostasis of neuronal activity in response to elevated neuronal activity. A portion of the neuronal dendrite including a spine is exemplified under three different conditions (boxes): the basal condition (Left), the stimulated condition (Upper Right), and the stimulated condition in the presence of BE, which blocks NMP activity (Lower Right). NMPs are shown in the plasma membrane, ribosomes, nascent proteins (tan), preexisting proteins (blue), and the 26S proteasome, which carries out UPS-mediated protein degradation, are shown in the cytoplasm. Under basal conditions (Left), there is a low level of NMP-mediated degradation of nascent proteins, which are produced by either constitutive protein synthesis or from ongoing neural activity. Upon enhanced stimulation (Upper Right), protein synthesis increases, and the resultant nascent proteins contribute to activity-dependent synaptic plasticity mechanisms that strengthen synaptic connections and increase synaptic transmission. A significant proportion of these activity-induced plasticity-related nascent proteins is degraded by NMPs, which help to maintain neuronal activity within a normal range. Under the stimulated condition, when NMPs are inhibited by BE (Lower Right, BE: green and orange shape bound to NMPs), excess nascent proteins that were not degraded induce aberrant neuronal activity.
We characterize NMPs in the Xenopus brain, showing that they are associated with the plasma membrane of neurons. Our data do not exclude the possibility that NMPs may be present in nonneuronal cell types (such as astrocytes, microglia, and radial glial processes). The ultrastructural localization of NMPs at synaptic sites in the OT, reported here and in mouse hippocampal neurons (14), positions NMPs to be directly involved in the timely degradation of locally translated activity-induced proteins, which can be particularly relevant in modulating activity-dependent plasticity of synaptic transmission (45). Synaptic localization and activity of the UPS pathway have been well documented (46, 47); however, the NMP is unique with respect to its substrate selectivity for acutely synthesized nascent proteins, as shown here and in our previous studies (13, 14).
Increased neuronal activity significantly increases protein synthesis in neurons compared with constitutive levels of protein synthesis (45, 48). Based on the observation that NMPs preferentially degrade nascent proteins (13), one might expect that the total amount of nascent proteins degraded by NMPs would be higher under stimulated conditions than that under the basal condition because of increased substrate availability. Indeed, by quantifying the total amount of BONCAT-labeled acutely synthesized proteins over 30 min, we detected significant NMP-mediated nascent protein degradation in the stimulated brain compared with the basal condition when protein synthesis is relatively low. Estimated from the dot blot data, ~30% of the activity-induced total nascent proteins were degraded by NMPs.
We were interested in whether NMPs degrade a similar proportion of different nascent proteins. While dot blots provide a quantitative measure of population changes in nascent proteins, western blots of individual candidate nascent proteins allowed greater sensitivity to detect changes in individual nacent protein levels. Among the three candidate proteins examined, we find different degrees of NMP-mediated degradation across the candidates. For instance, activity-induced nascent CaMKIIα and activity-induced nascent PSMD2 are both NMP substrates, while nascent SNCB does not appear to be an NMP substrate under these conditions. Western blot analysis also revealed that NMPs degrade nascent proteins generated under the basal condition, which was not detectable with the dot blot assay when all constitutively synthesized nascent proteins were pooled together. The individual proteins we examined were selected based on prior reports of their involvement in activity-dependent protein synthesis. CaMKIIα is a key molecule underlying synaptic plasticity (27). It is well documented that both the mRNA and protein levels of CaMKIIα are up-regulated in neurons in response to activity (11, 22, 28, 49, 50). CaMKIIα mRNA is localized to dendrites and is rapidly recruited to dendritic spines and translated into protein when activity increases (28, 51–53), making it a plausible candidate substrate for NMPs. Indeed, our data clearly demonstrate that CaMKIIα was synthesized under the basal condition, and its synthesis rapidly increased after pharmacological stimulation. Furthermore, NMP inhibition revealed a significantly higher level of nascent CaMKIIα under both basal and stimulated conditions, indicating that nascent CaMKIIα is a substrate of NMPs under both conditions. Synthesis of the proteasome subunit PSMD2 is up-regulated in the tadpole brain following 4 h of enhanced visual stimulation (11). The synthesis and degradation of proteasome components are known to be regulated by neuronal activity in the brain (19, 48). In the brief pharmacological stimulation paradigm we employed here, we observed no change in nascent PSMD2 in the stimulated samples compared with the basal condition with normal NMP function. However, inhibiting NMPs significantly increased nascent PSMD2, indicating that NMPs efficiently degraded newly synthesized PSMD2 and prevented accumulation of nascent PSMD2. Taken together, these data suggest that NMPs may function as a mechanism to rapidly limit accumulation of activity-induced nascent proteins. In addition, our data suggest that NMP-mediated degradation of activity-induced individual nascent proteins differs between substrates, possibly reflecting the amounts of activity-induced nascent proteins and therefore the availability of different NMP substrates. Similarly, the lower level of NMP-mediated degradation of nascent proteins under the basal condition may be due to the lower level of protein synthesis and therefore lower substrate availability.
Our results demonstrated that inhibiting NMP function rapidly increased spontaneous neuronal activity and led to hypersynchronization of activity across the tectal circuit. Inhibiting NMPs not only abolished visual training–induced behavioral plasticity but also disrupted circuit function. Our biochemistry data showed that BE remained bound to the NMP for at least 6 h after injection (Fig. 1C), consistent with the irreversible binding of Epox to the 20S core particle (26) and ensuring that the NMPs remain blocked during the VE training period. The formation of new memories is thought to result from a series of fine-tuned modifications that occur in a highly specific manner at various synapses involved in learning and behavior modification from sensory inputs to motor output (54). The training process engages synaptic plasticity mechanisms that initiate strengthening of some synapses and weakening of others, thus changing the detection threshold to certain sensory inputs and eventually the response manifested by animal behavior. Spaced visual training in the retinotectal circuit increased the correlation in spontaneous activity across tectal neurons (55). An optimal level of correlated spontaneous activity following plasticity-inducing stimulation might be required for synaptic potentiation as either blocking spontaneous activity (56) or increasing it by random visual stimulation (57) following theta burst stimulation (TBS) disrupted persistent retinotectal synaptic potentiation induced by TBS. Interestingly, acute proteasome inhibition rapidly increased neuronal activity in neuronal cultures (58) and enhanced LTP induction but blocked expression of late-phase LTP in hippocampal slice (59). Even though these effects were attributed to the UPS pathway, it is noteworthy that the proteasome inhibitor used in the study, epoxomicin, also inhibits NMPs, suggesting that NMPs may contribute to the reported increase in neuronal activity and effects on plasticity. As we have shown here, Epox rapidly increased spontaneous neuronal activity similar to what we observed with the application of the specific NMP inhibitor, BE (SI Appendix, Fig. S5). The increased activity induced by acute NMP inhibition may lead to a transient further increase in activity that could have been interpreted as enhanced LTP induction. Furthermore, the effect of proteasome inhibition on late-phase LTP was blocked by the protein synthesis inhibitor anisomycin (59), indicating that the underlying mechanism involves newly synthesized proteins. Similarly, blocking protein synthesis in the tadpole brain also blocked the learning-induced visuomotor behavioral plasticity (22) studied here.
It is particularly interesting that inhibiting NMPs specifically disrupted the visual avoidance behavior in animals subjected to visual training, a 4-h period of increased visual experience, which increases neuronal excitability and stimulus detection (41), but inhibiting NMPs had no deleterious effect on visual avoidance behavior in animals that did not receive visual training. This suggests that the increase in neuronal activity seen with NMP inhibition under the basal condition was below a threshold to interfere with circuit function. By contrast, in response to the enhanced visual experience, when neuronal activity was elevated, NMP function then becomes essential to limit neuronal activity to a level that permits circuit function and plasticity. This is consistent with the drastically different magnitude of the effect of NMP inhibition on neuronal activity under basal and stimulated conditions (Fig. 3Q). Whether specific activity-induced nascent proteins are essential NMP substrates for limiting neuronal activity or whether different activity-induced proteins contribute to the differential effects of NMP inhibition on neuronal activity under basal and stimulated conditions remain to be elucidated.
What might be the mechanism underlying the role NMPs play in regulating neuronal activity? The NMP-mediated degradation of nascent proteins may underlie a rapid homeostatic mechanism that helps maintain neuronal activity in check in the face of rising activity. Runaway neuronal activity disrupts the activity patterns in neural networks that are essential to form synapse specific modifications of neuronal connectivity, which are the basis for learning and memory (35). Maintaining neuronal activity in an intermediate range is the core function of homeostatic plasticity mechanisms (35). Activity-induced nascent proteins trigger subsequent signaling pathways for the expression of synaptic plasticity (60–62). Timely degradation of these nascent proteins may hold such activity-induced synaptic changes in check, protecting the network from destabilizing positive feedback loops. The activity-dependent regulation of proteostasis in neurons is likely a highly intricate and delicate process. NMP-mediated degradation of activity-induced nascent proteins may contribute to rapid homeostatic regulation of neuronal activity, particularly in the face of elevated activity levels. For example, NMDAR activation induces proteasomal degradation of MOV10, a synaptic translational repressor, which leads to increased expression of CaMKIIα (63). Activation of CaMKII following increased synaptic activity leads to phosphorylation of serine 120 on Rpt6, which in turn leads to enhanced 26S proteasome activity (64), potentially generating a feedback loop within the signaling cascade. Putting a cap on the amount of newly synthesized CaMKII would help put a brake on the downstream signaling cascade. Likewise, tight control of the newly synthesized proteasome subunits such as PSMD2 could be part of the dynamic regulatory scheme underlying activity-dependent proteostasis. Failure to degrade excessive activity-induced nascent proteins may not only jeopardize the expression of synaptic plasticity but also impair circuit functions such as information processing, as shown by the detrimental effect of blocking NMPs on visual avoidance behavior following enhanced visual training we observed.
Another potential pathway through which NMPs function may affect neuronal activity is by the peptides generated by the 20S catalytic degradation of nascent protein substrates (1, 14), although thus far very little is known about the identities and functional properties of these peptides. Multiple mechanisms may underlie the functional involvement of NMPs in regulating neuronal activity. Emerging data suggest that different activity patterns can induce different compositions of the nascent transcriptome and proteome as a result of the recruitment of different classes of neurons, involvement of a differential neural circuitry, and even induction of different plasticity mechanisms (11, 65–67). The functional contribution of NMP-mediated degradation of activity-induced nascent proteins may also vary under different activity paradigms depending on the nascent proteome induced. Our data provided direct evidence for a functional role of NMPs in regulating neuronal activity in vivo and the requirement of NMP activity for experience-dependent behavioral plasticity. Additional investigations are warranted to further elucidate the cellular and molecular components and mechanisms underlying the homeostatic control of activity-induced nascent proteomes and their contributions to neuronal function.
Materials and Methods
Animals.
Albino X. laevis embryos were obtained from in-house fertilization or Xen Express (Brooksville, FL) and reared at 21 to 22 °C with 12-h dark/12-h light cycle in 0.1×Steinberg solution (in millimolar: 58.0 NaCl, 0.67 KCl, 0.34 Ca(NO3)2, 0.83 MgSO4, and 3.0 HEPES, pH 7.2). Animals were fed from stage 47 (68). All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of The Scripps Research Institute and the Georgetown University. Stage 47 to 48 tadpoles of either sex were used for all experiments.
Intraventricular Injection.
Reagents (BMI, BE, Epox, anisomycin, and AHA) were injected intraventricularly as described before (69) and in Supporting Information. Drug concentrations were listed as the injected concentrations. To maximize direct detection of injected BE in the brain, 1 mM BE was injected for IHC and membrane preparation experiments in Fig. 1, and lower concentrations (5 to 250 µM BE) were injected for biochemical, functional imaging, and behavioral experiments.
Immunohistochemistry.
To visualize BE binding in the tadpole brain, animals were injected intraventricularly with 1 mM BE and fixed in 4% paraformaldehyde for immunocytochemistry (IHC) together with uninjected controls. Sample preparation is described in detail in SI Appendix. Thirty micromolar vibratome sections of the dissected brains were incubated with goat Anti-Biotin polyclonal antibody (Invitrogen; catalog #31852, RRID: AB_228243, 1:500 in PBSTw with 1% normal goat serum and 1% fish gelatin) for 72 h at 4 °C followed by secondary antibody (donkey anti-goat Alexa Fluor 488; Thermo Fisher Scientific; catalog #A-32814, RRID: AB_2762838). Sections were rinsed and mounted on slides in Vectashield Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA). Brains from the BE-injected and uninjected groups were embedded in the same blocks and processed under exactly the same conditions throughout the experiments. Images were collected on a Nikon C2 confocal microscope with a 40× Plan Fluor Oil Objective (NA 1.3).
Electron Microscopy.
Sample preparation for stage 47 tadpole brains is described in detail in SI Appendix. Fifty micromolar vibratome sections were incubated in 1:500 mouse anti-proteasome 20S α1, 2, 3, 5, 6 & 7 subunits (Enzo; BML-PW8195, RRID: AB_10541045) followed by 1:800 anti-mouse FluoroNanogold (Life Technologies; A24920). GoldEnhance EM Plus (Nanoprobes; 2114) was used to enhance Nanogold particle size for visualization. Sections were processed for electron microscopy, and ultrathin sections were examined with a FEI Talos L120C Electron Microscope, and photographs were taken with a Ceta 16M CMOS camera at 8,500× or 22,000× magnification. In electron micrographs, membranes were pseudocolored blue using Photoshop (Adobe) for easier visualization.
Membrane Preparation and Western Blotting.
Tadpoles (40/group) received intraventricular injections of 1 mM BE or 1 mM Epox, and brains were dissected either 30 min or 6 h later. The membrane preparation was performed for biochemical analysis as previously described (14) and summarized in SI Appendix. Equal volume of both membrane and cytosolic extracts was loaded onto the SDS–PAGE gel and processed for western blotting.
BONCAT and Immunoblotting.
1 M Azidohomoalanine (AHA) stock solution was made from powder (Click Chemistry Tools; catalog #1066100) in 1× PBS with pH adjusted to 7 using 1 N NaOH. All drug solutions were prepared to have a final concentration of 350 mM AHA and contain ~0.01% Fast Green dye for injection visualization. BE (synthesized de novo and purchased from the Leiden Institute of Chemistry with purity and verified as described before (14); 50 to 100 µM) and BMI (R&D Systems; catalog #2503, 50 to 100 µM) were injected into the midbrain ventricle of stage 48 tadpoles. For a subset of experiments, extra animal groups were injected with anisomycin (Sigma-Aldrich; catalog #A5862, 25 µM) or Epox (ApexBio; catalog #50-190-4754, 50 to 100 µM), as specified in the results. Thirty minutes after injection, the brains were dissected. About 25 to 30 animal brains were collected for each experimental group and stored at −80 °C to be processed the following day. Tadpole brain tissue was processed as previously described (11). Dot blot with Anti-Biotin (Invitrogen; catalog #31852, RRID: AB_228243, 1:1,000) was used for quantitative evaluation of total nascent proteins. For ubiquitin dot blot, 5 mg of total protein samples was loaded onto the dot blot and immunoblotted with anti-ubiquitin antibody (Invitrogen; catalog #131600, RRID: AB_2533002, 1:1,000), followed by quantitative analysis. For purification of biotinylated AHA-tagged nascent proteins and subsequent western blotting, an internal loading control, 1.5 µL of 0.1 mg/mL biotinylated BSA (BioVision; catalog #7097-25), was added to each sample before processing for click chemistry. Biotinylated nascent proteins were purified by methanol/chloroform/water precipitation and pulled down with neutravidin beads (Pierce High Capacity Neutravidin Agarose Resin; Thermo Fisher Scientific; catalog #29202), followed by western blotting. For quantification, the band intensity of the protein of interest was first normalized to the BSA loading control and then normalized to the value of the control sample from the same batch run on the same blot. For neutravidin pulldown of the BE-injected samples with or without Epox preinjection (no AHA labeling), neutravidin beads were directly added to the total lysate of brain tissues and processed as described above. A detailed description of the BONCAT and sample processing protocol can be found in SI Appendix.
In Vivo Two-Photon Imaging of Spontaneous Ca++ Activities in Tectal Neurons and Data Analysis.
For functional imaging of spontaneous calcium activity, animals were co-electroporated with pGP-CMV-GCaMP6f (2 mg/mL; Addgene plasmid #40755) and CMV-turboRFP (1 mg/mL) at stages 46 to 47. Animals were prescreened 3 to 5 d after electroporation for those with moderate to high number of transfected cells expressing turboRFP. On the day of imaging, the animal was immobilized with pancuronium dibromide (1 mM in 0.1× Steinberg solution; ApexBio; catalog #15500-66-0) for 1 min (55). A time series of spontaneous Ca++ activity was collected at 30-Hz frame rate either on a Scientifica multiphoton resonant microscope (Scientifica, UK) with a 25× water immersion objective (Olympus Ultra 25×MPE, 1.05 NA) or a Bruker Ultima Investigator multiphoton microscope (Bruker, Billerica, MA) in the resonant scanning mode with a 20x water immersion objective (Olympus XLUMPLFLN20XW, 1.0 NA). Wavelength of 940 nm was used to excite GCaMP6f. For each experiment, a 5-min baseline was taken before the animal received intraventricular injections for subsequent treatments, and a 5-min time series was taken for each time point after the injection. The patterns of turboRFP-expressing cells were used as landmarks to help identifying the imaged optical plane to ensure the same population of GCaMP6f-expressing neurons were imaged for all time points.
A time series of calcium response data were processed in ImageJ (NIH) by manually identifying the region of interest (cell soma) and then further analyzed with customized MATLAB scripts. The extracted GCaMP6 fluorescence was used to calculate dF/F based on an exponentially weighted moving average algorithm (70) to remove the slow drifting of baseline signal and fast oscillatory noise that resulted from occasional tissue pulsation. Spontaneous Ca++ events were defined as 3 SD above the mean dF/F in the GCaMP fluorescence during the 5-min baseline recording session immediately before the first drug injection (71). The same threshold (mean dF/F + 3 SD during the baseline session) was applied to dF/F data of all subsequent time points recorded from the same neuron. The total number of spontaneous Ca++ event peaks during each 5-min recording session was designated as the Ca++ event count. The integrated Ca++ response was calculated as the integrated area under the curve above threshold for each Ca++ event. Synchronous Ca++ events were analyzed based on previously published method (24). The synchrony index was a value between 0 and 1, with 0 being no synchrony, and 1 being all cells were synchronously active. For example, a synchrony index value of 0.2 means 20% of the whole time period there were synchronous events going on in the neuronal population.
For BMI–BE data presented in Fig. 3, treatments with 50 to 250 μM BE showed a consistently rapid increase in neuronal activity on top of BMI-induced heightened neuronal activity across animals from different clutches, and thus, we pooled data from all animals injected with 50- to 250-μM BE following the BMI treatment for the BMI–BE dataset.
Visual Avoidance Assay and Visual Training.
The visual avoidance assay and training were conducted as previously described (22) and in SI Appendix. Briefly, stage 47 tadpoles were placed in a testing chamber, and a random pattern of moving dots was presented. Videos of animal behavior were captured and manually analyzed post hoc to score visual avoidance responses. An AI was quantified as the fraction of avoidance responses out of the first ten encounters.
Tadpoles received intraventricular injection of either vehicle or BE (50 μM), and visual avoidance behavior was tested for time point (T1) after 20 to 40 min to allow animals to recover from anesthesia. After T1, a group assigned for enhanced visual training (VE) was exposed to visual stimulus training, as described, for 4 h (37), while controls were exposed to ambient light. Visual avoidance behavior was tested the following day (T2). All data were analyzed post hoc as described above, blind to the experimental groups.
Statistical Tests.
Statistical tests were performed using GraphPad Prism version 9.2.0 for Mac OS (GraphPad Software, San Diego, California USA). The Kolmogorov–Smirnov test was used to determine if the dataset was normally distributed and parametric (for normally distributed dataset), or nonparametric tests were used accordingly. All data are presented as mean ± SEM. Data are considered significantly different when p values are less than 0.05. Post hoc power analysis was performed using JMP Pro 15 (SAS Institute). The statistical test used for each experiment is specified in the results. Experiments and analysis were performed by the experimenter blinded to the experimental conditions.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Acknowledgments
We thank the Microscopy Core at The Scripps Research Institute for their technical support. We thank Dr. Han-Hsuan Liu and Eric Carlson for their help in the initial optimization of the in vivo BONCAT labeling and sample processing protocol. We thank Dr. Ju Lu for generously sharing the MATLAB script for synchrony analysis. This work was funded by the NIH (KL2TR001432 to H.-Y.H.; EY011261, EY031597, and NS114975 to H.T.C.; and NS110754 to S.S.M.).
Author contributions
H.-Y.H., K.V.R., S.S.M. and H.T.C. conceived the project. H.-Y.H. and H.T.C. designed experiments and supervised the research. H.-Y.H., A.A. R.B., N.M., R.F., and K.V.R. conducted experiments and analyzed the data. H.-Y.H., S.S.M. and H.T.C. interpreted the data and wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
Reviewers: C.M.A., New York University; and K.P., University of Wyoming.
Contributor Information
Hai-yan He, Email: haiyan.he@georgetown.edu.
Hollis T. Cline, Email: cline@scripps.edu.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Turker F., Cook E. K., Margolis S. S., The proteasome and its role in the nervous system. Cell Chem. Biol. 28, 903–917 (2021), 10.1016/j.chembiol.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitaker M., Patel R., Calcium and cell cycle control. Development 108, 525–542 (1990), 10.1242/dev.108.4.525. [DOI] [PubMed] [Google Scholar]
- 3.DiAntonio A., et al. , Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature 412, 449–452 (2001), 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- 4.Dorrbaum R., Kochen L., Langer J. D., Schuman E. M., Local and global influences on protein turnover in neurons and glia. Elife 7, e34202 (2018), 10.7554/eLife.34202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fonseca R., Vabulas R. M., Hartl F. U., Bonhoeffer T., Nagerl U. V. A., A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 52, 239–245 (2006), 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Sutton A., Schuman E. M., Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58 (2006), 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Tai C., Schuman E. M., Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9, 826–838 (2008), 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 8.K. Pandey, X. W. Yu, Steinmetz A., C. M. Alberini, Autophagy coupled to translation is required for long-term memory. Autophagy 17, 1614–1635 (2021), 10.1080/15548627.2020.1775393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fonseca R., Nagerl U. V., Bonhoeffer T., Neuronal activity determines the protein synthesis dependence of long-term potentiation. Nat. Neurosci. 9, 478–480 (2006), 10.1038/nn1667. [DOI] [PubMed] [Google Scholar]
- 10.Wang D. O., et al. , Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 324, 1536–1540 (2009), 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H., et al. , Role of the visual experience-dependent nascent proteome in neuronal plasticity. Elife 7, e33420 (2018), 10.7554/eLife.33420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balchin D., Hayer-Hartl M., Hartl F. U., In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016), 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran V., et al. , Activity-dependent degradation of the nascentome by the neuronal membrane proteasome. Mol. Cell 71, 169–177.e166 (2018), 10.1016/j.molcel.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramachandran V., Margolis S. S., A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function. Nat. Struct. Mol. Biol. 24, 419–430 (2017), 10.1038/nsmb.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bard J. A. M., et al. , Structure and function of the 26S proteasome. Annu. Rev. Biochem. 87, 697–724 (2018), 10.1146/annurev-biochem-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coux O., Tanaka K., Goldberg A. L., Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847 (1996), 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 17.Sahu I., et al. , The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag. Nat. Commun. 12, 6173 (2021), 10.1038/s41467-021-26427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Nissan G., Sharon M., Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 4, 862–884 (2014), 10.3390/biom4030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tai H. C., Besche H., Goldberg A. L., Schuman E. M., Characterization of the brain 26S proteasome and its interacting proteins. Front. Mol. Neurosci. 3, 12 (2010), 10.3389/fnmol.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pratt G., Khakhalin A. S., Modeling human neurodevelopmental disorders in the Xenopus tadpole: From mechanisms to therapeutic targets. Dis. Model. Mech. 6, 1057–1065 (2013), 10.1242/dmm.012138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruthazer E. S., Aizenman C. D., Learning to see: Patterned visual activity and the development of visual function. Trends Neurosci. 33, 183–192 (2010), 10.1016/j.tins.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen W., et al. , Acute synthesis of CPEB is required for plasticity of visual avoidance behavior in Xenopus. Cell Rep. 6, 737–747 (2014), 10.1016/j.celrep.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ives-Deliperi V., Butler J. T., Mechanisms of cognitive impairment in temporal lobe epilepsy: A systematic review of resting-state functional connectivity studies. Epilepsy. Behav. 115, 107686 (2021), 10.1016/j.yebeh.2020.107686. [DOI] [PubMed] [Google Scholar]
- 24.Goncalves J. T., Anstey J. E., Golshani P., Portera-Cailliau C., Circuit level defects in the developing neocortex of Fragile X mice. Nat. Neurosci. 16, 903–909 (2013), 10.1038/nn.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Pino E., Gessele N., Koch U., Enhanced excitatory connectivity and disturbed sound processing in the auditory brainstem of fragile X mice. J. Neurosci. 37, 7403–7419 (2017), 10.1523/JNEUROSCI.2310-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng L., et al. , Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. U.S.A. 96, 10403–10408 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lisman J., Schulman H., Cline H., The molecular basis of CaMKII function in synaptic and behavioural memory. Nat. Rev. Neurosci. 3, 175–190 (2002), 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- 28.Ouyang Y., Rosenstein A., Kreiman G., Schuman E. M., Kennedy M. B., Tetanic stimulation leads to increased accumulation of Ca(2+)/calmodulin-dependent protein kinase II via dendritic protein synthesis in hippocampal neurons. J. Neurosci. 19, 7823–7833 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams J. K., Yang X., Baum J., Interactions between the Intrinsically disordered proteins beta-synuclein and alpha-synuclein. Proteomics 18, e1800109 (2018), 10.1002/pmic.201800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunfield D., Haas K., In vivo single-cell excitability probing of neuronal ensembles in the intact and awake developing Xenopus brain. Nat. Protoc. 5, 841–848 (2010), 10.1038/nprot.2010.10. [DOI] [PubMed] [Google Scholar]
- 31.He H. Y., Shen W., Hiramoto M., Cline H. T., Experience-dependent bimodal plasticity of inhibitory neurons in early development. Neuron 90, 1203–1214 (2016), 10.1016/j.neuron.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H., Khakhalin A. S., Nurmikko A. V., Aizenman C. D., Visual experience-dependent maturation of correlated neuronal activity patterns in a developing visual system. J. Neurosci. 31, 8025–8036 (2011), 10.1523/JNEUROSCI.5802-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abraham W. C., Metaplasticity: Tuning synapses and networks for plasticity. Nat. Rev. Neurosci. 9, 387 (2008), 10.1038/nrn2356. [DOI] [PubMed] [Google Scholar]
- 34.Li J., Park E., Zhong L. R., Chen L., Homeostatic synaptic plasticity as a metaplasticity mechanism–A molecular and cellular perspective. Curr. Opin. Neurobiol. 54, 44–53 (2019), 10.1016/j.conb.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turrigiano G. G., The dialectic of Hebb and homeostasis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 372, 20160258 (2017), 10.1098/rstb.2016.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin J., VanDongen A. M., Enhanced neuronal activity and asynchronous calcium transients revealed in a 3D organoid model of alzheimer’s disease. ACS Biomater. Sci. Eng. 7, 254–264 (2021), 10.1021/acsbiomaterials.0c01583. [DOI] [PubMed] [Google Scholar]
- 37.Shen W., McKeown C. R., Demas J. A., Cline H. T., Inhibition to excitation ratio regulates visual system responses and behavior in vivo. J. Neurophysiol. 106, 2285–2302 (2011), 10.1152/jn.00641.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong W., et al. , Visual avoidance in Xenopus tadpoles is correlated with the maturation of visual responses in the optic tectum. J. Neurophysiol. 101, 803–815 (2009), 10.1152/jn.90848.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He H. Y., Shen W., Zheng L., Guo X., Cline H. T., Excitatory synaptic dysfunction cell-autonomously decreases inhibitory inputs and disrupts structural and functional plasticity. Nat. Commun. 9, 2893 (2018), 10.1038/s41467-018-05125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H. H., Cline H. T., Fragile X mental retardation protein is required to maintain visual conditioning-induced behavioral plasticity by limiting local protein synthesis. J. Neurosci. 36, 7325–7339 (2016), 10.1523/JNEUROSCI.4282-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aizenman C. D., Akerman C. J., Jensen K. R., Cline H. T., Visually driven regulation of intrinsic neuronal excitability improves stimulus detection in vivo. Neuron 39, 831–842 (2003), 10.1016/s0896-6273(03)00527-0. [DOI] [PubMed] [Google Scholar]
- 42.Khakhalin A. S., Koren D., Gu J., Xu H., Aizenman C. D., Excitation and inhibition in recurrent networks mediate collision avoidance in Xenopus tadpoles. Eur. J. Neurosci. 40, 2948–2962 (2014), 10.1111/ejn.12664. [DOI] [PubMed] [Google Scholar]
- 43.Hegde A. N., Proteolysis, synaptic plasticity and memory. Neurobiol. Learn. Mem. 138, 98–110 (2017), 10.1016/j.nlm.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holt C. E., Schuman E. M., The central dogma decentralized: New perspectives on RNA function and local translation in neurons. Neuron 80, 648–657 (2013), 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holt C. E., Martin K. C., Schuman E. M., Local translation in neurons: Visualization and function. Nat. Struct. Mol. Biol. 26, 557–566 (2019), 10.1038/s41594-019-0263-5. [DOI] [PubMed] [Google Scholar]
- 46.Bingol B., Schuman E. M., Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature 441, 1144–1148 (2006), 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- 47.Drinjakovic J., et al. , E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron 65, 341–357 (2010), 10.1016/j.neuron.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiapparelli L., et al. , Activity-induced cell type specific nascent proteome. J. Neurosci. 42, 7900–7920 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayford M., Baranes D., Podsypanina K., Kandel E. R., The 3’-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc. Natl. Acad. Sci. U.S.A. 93, 13250–13255 (1996), 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bagni C., Mannucci L., Dotti C. G., Amaldi F., Chemical stimulation of synaptosomes modulates alpha -Ca2+/calmodulin-dependent protein kinase II mRNA association to polysomes. J. Neurosci. 20, RC76 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Havik B., Rokke H., Bardsen K., Davanger S., Bramham C. R., Bursts of high-frequency stimulation trigger rapid delivery of pre-existing alpha-CaMKII mRNA to synapses: A mechanism in dendritic protein synthesis during long-term potentiation in adult awake rats. Eur. J. Neurosci. 17, 2679–2689 (2003), 10.1046/j.1460-9568.2003.02712.x. [DOI] [PubMed] [Google Scholar]
- 52.Sajikumar S., Navakkode S., Frey J. U., Identification of compartment- and process-specific molecules required for "synaptic tagging" during long-term potentiation and long-term depression in hippocampal CA1. J. Neurosci. 27, 5068–5080 (2007), 10.1523/JNEUROSCI.4940-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wells D. G., et al. , A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J. Neurosci. 21, 9541–9548 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeuchi T., Duszkiewicz A. J., Morris R. G., The synaptic plasticity and memory hypothesis: Encoding, storage and persistence. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 369, 20130288 (2014), 10.1098/rstb.2013.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunfield D., Haas K., Metaplasticity governs natural experience-driven plasticity of nascent embryonic brain circuits. Neuron 64, 240–250 (2009), 10.1016/j.neuron.2009.08.034. [DOI] [PubMed] [Google Scholar]
- 56.Gong L. Q., et al. , Postinduction requirement of NMDA receptor activation for late-phase long-term potentiation of developing retinotectal synapses in vivo. J. Neurosci. 31, 3328–3335 (2011), 10.1523/JNEUROSCI.5936-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Q., Tao H. W., Poo M. M., Reversal and stabilization of synaptic modifications in a developing visual system. Science 300, 1953–1957 (2003), 10.1126/science.1082212. [DOI] [PubMed] [Google Scholar]
- 58.Rinetti G. V., Schweizer F. E., Ubiquitination acutely regulates presynaptic neurotransmitter release in mammalian neurons. J. Neurosci. 30, 3157–3166 (2010), 10.1523/JNEUROSCI.3712-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong C., Upadhya S. C., Ding L., Smith T. K., Hegde A. N., Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learn Mem. 15, 335–347 (2008), 10.1101/lm.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coultrap S. J., Bayer K. U., CaMKII regulation in information processing and storage. Trends Neurosci. 35, 607–618 (2012), 10.1016/j.tins.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheng M., Greenberg M. E., The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4, 477–485 (1990), 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 62.Sun X., Lin Y., Npas4: Linking neuronal activity to memory. Trends Neurosci. 39, 264–275 (2016), 10.1016/j.tins.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee S., Neveu P., Kosik K. S., A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 64, 871–884 (2009), 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 64.Djakovic S. N., Schwarz L. A., Barylko B., DeMartino G. N., Patrick G. N., Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 284, 26655–26665 (2009), 10.1074/jbc.M109.021956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schanzenbacher C. T., Sambandan S., Langer J. D., Schuman E. M., Nascent proteome remodeling following homeostatic scaling at hippocampal synapses. Neuron 92, 358–371 (2016), 10.1016/j.neuron.2016.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bowling H., et al. , BONLAC: A combinatorial proteomic technique to measure stimulus-induced translational profiles in brain slices. Neuropharmacology 100, 76–89 (2016), 10.1016/j.neuropharm.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garay P. M., et al. , RAI1 regulates activity-dependent nascent transcription and synaptic scaling. Cell Rep. 32, 108002 (2020), 10.1016/j.celrep.2020.108002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nieuwkoop P. D., Faber J., Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis (Garland Pub, 1994). [Google Scholar]
- 69.Mogi K., Adachi T., Izumi S., Toyoizumi R., Visualisation of cerebrospinal fluid flow patterns in albino Xenopus larvae in vivo. Fluids. Barriers. CNS 9, 9 (2012), 10.1186/2045-8118-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jia H., Rochefort N. L., Chen X., Konnerth A., In vivo two-photon imaging of sensory-evoked dendritic calcium signals in cortical neurons. Nat. Protoc. 6, 28–35 (2011), 10.1038/nprot.2010.169. [DOI] [PubMed] [Google Scholar]
- 71.Hewapathirane D. S., Dunfield D., Yen W., Chen S., Haas K., In vivo imaging of seizure activity in a novel developmental seizure model. Exp. Neurol. 211, 480–488 (2008), 10.1016/j.expneurol.2008.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Data Availability Statement
All study data are included in the article and/or SI Appendix.