Significance
Hydrogen sulfide (H2S) is an endogenous signaling molecule associated with both cytoprotective and cytotoxic properties. Despite its physiological importance, the cellular metabolism of H2S is not well defined. Our results here reveal that H2S is predominantly detoxified by the cytosolic enzyme, SOD1, which is a major determinant in preventing H2S-mediated cytotoxicity. Additionally, SOD1-sulfide oxidase activity limits the formation of reactive sulfur species (RSS), such as per- and polysulfides. Furthermore, while H2S acts as an antioxidant, the interaction of reactive oxygen species with H2S forms thiol oxidizing polysulfide species. This significantly changes our understanding of how H2S is metabolized to prevent its cytotoxic effects and indicates that SOD1 antioxidant properties extend to limiting both H2S and related RSS.
Keywords: hydrogen sulfide, persulfide, polysulfide, SOD1, reactive sulfur species
Abstract
Although hydrogen sulfide (H2S) is an endogenous signaling molecule with antioxidant properties, it is also cytotoxic by potently inhibiting cytochrome c oxidase and mitochondrial respiration. Paradoxically, the primary route of H2S detoxification is thought to occur inside the mitochondrial matrix via a series of relatively slow enzymatic reactions that are unlikely to compete with its rapid inhibition of cytochrome c oxidase. Therefore, alternative or complementary cellular mechanisms of H2S detoxification are predicted to exist. Here, superoxide dismutase [Cu-Zn] (SOD1) is shown to be an efficient H2S oxidase that has an essential role in limiting cytotoxicity from endogenous and exogenous sulfide. Decreased SOD1 expression resulted in increased sensitivity to H2S toxicity in yeast and human cells, while increased SOD1 expression enhanced tolerance to H2S. SOD1 rapidly converted H2S to sulfate under conditions of limiting sulfide; however, when sulfide was in molar excess, SOD1 catalyzed the formation of per- and polysulfides, which induce cellular thiol oxidation. Furthermore, in SOD1-deficient cells, elevated levels of reactive oxygen species catalyzed sulfide oxidation to per- and polysulfides. These data reveal that a fundamental function of SOD1 is to regulate H2S and related reactive sulfur species.
H2S is a toxin that inhibits cellular respiration with a similar potency as cyanide (1, 2). Additionally, excessive cellular H2S is associated with oxidative stress and polysulfide formation (3–7). Despite its potent toxicity, endogenous or exogenous H2S exerts regulatory and beneficial actions in cell and animal models (8–11), consistent with sulfide being efficiently metabolized to avoid its toxicity.
H2S mediates cellular signaling in part by reacting with disulfide bonds to form persulfides (Reaction 1) (12–14).
| [1] |
In x&addition to signaling, persulfide formation is currently considered the primary mechanism of H2S detoxification via sulfide quinone oxidoreductase (SQOR) and the “sulfide oxidizing unit” in the mitochondrial matrix (15–17). The SQOR active site exists as a trisulfide bond between two cysteine residues, and the detoxification of sulfide is thought to proceed as in Reaction 2, forming two cysteine persulfide residues (18).
| [2] |
However, persulfide formation from sulfide reacting with either disulfide (Reaction 1) or trisulfide (Reaction 2) are thermodynamically unfavorable reactions (Keq << 1) (13, 19), and correspondingly, SQOR reduction by H2S has only been described for excessive sulfide concentrations (15, 17, 20). In contrast, complex IV (cytochrome c oxidase) is rapidly and potently inhibited by H2S (1, 21). Due to the incongruities of H2S reactivity described above, together with the lack of SQOR sulfide detoxification in central nervous tissue (22), we challenged the currently accepted mechanism of sulfide detoxification and postulated that a cytosolic H2S oxidase exists to limit mitochondrial inhibition by H2S.
Superoxide dismutase [Cu-Zn] (SOD1) is a highly expressed, ubiquitous copper, and zinc-containing protein that is conserved throughout evolution and across taxonomical kingdoms (23–25). Although it is generally accepted to function as a superoxide oxidoreductase (26), SOD1 can catalyze other chemical reactions (27–30). Here we show that SOD1 rapidly detoxifies endogenous and exogenous sulfide to sulfate. Furthermore, we demonstrate that this sulfide oxidase function of SOD1 protects cells from H2S and limits reactive sulfur species (RSS) (i.e., persulfides and polysulfides) that mediate sulfide signaling and toxicity (31, 32).
Results and Discussion
SOD1 Expression is Critical for Sulfide Tolerance.
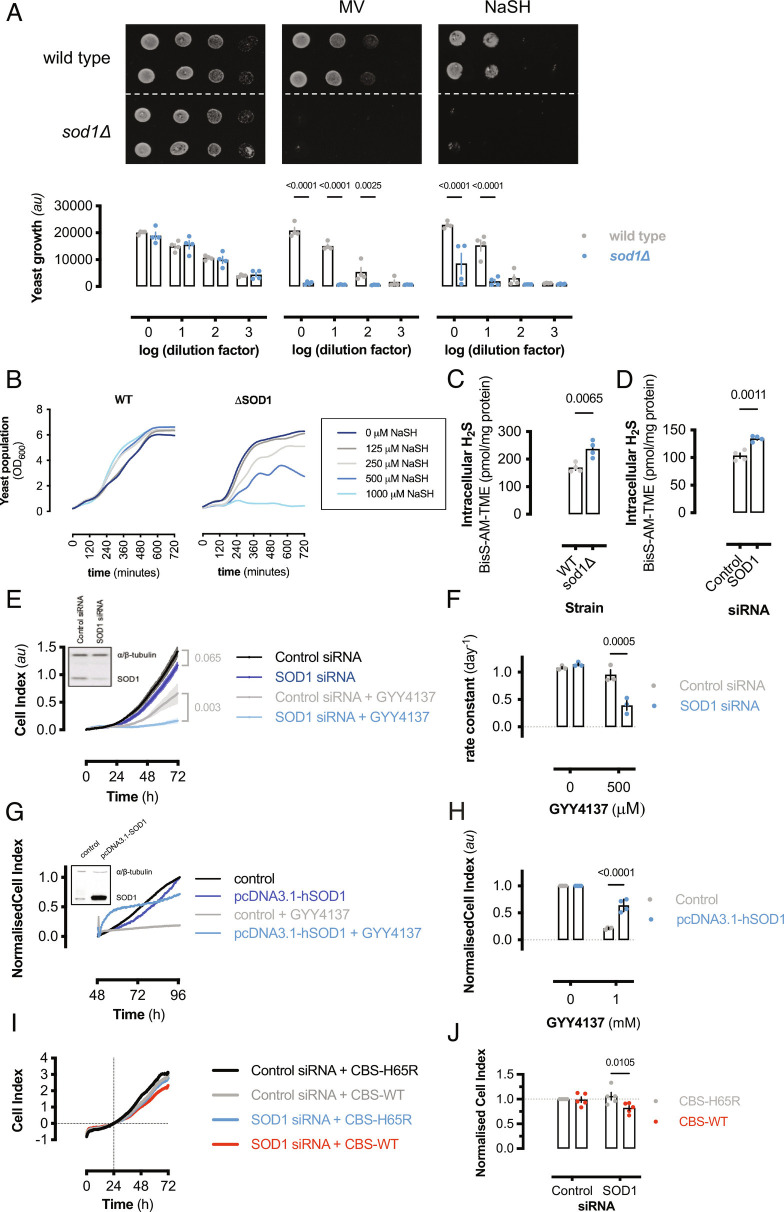
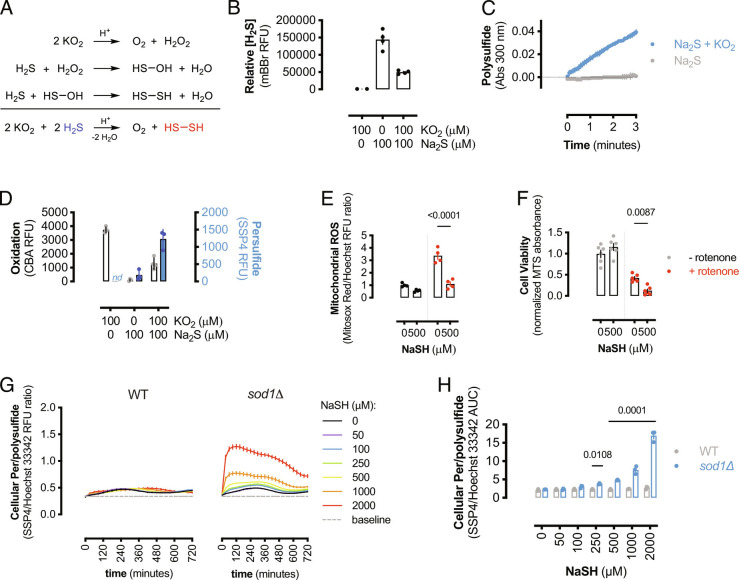
As H2S is a potent toxin present at the origins of life (33), we speculated that a universal mechanism of sulfide detoxification exists. SOD1 is a ubiquitous, highly expressed redox-active protein present in all kingdoms of life; therefore, using a yeast knockout model, the role of SOD1 in limiting H2S toxicity was investigated. Using a serial dilution spot assay, sod1Δ yeast showed inhibited growth on NaSH compared to control agar (Fig. 1A). The toxic effect of NaSH on sod1Δ yeast was similar to methyl viologen (MV), a superoxide-generating compound. Additionally, manganese supplementation, which functions as a superoxide dismutase mimetic (34–37), but not a sulfide oxidase at physiological pH (SI Appendix, Fig. S1C), limited MV-induced cytotoxicity, but did not alter sulfide-mediated toxicity in sod1Δ yeast (SI Appendix, Fig. S1 A and B). These results indicate that sulfide-induced cytotoxicity in sod1Δ yeast is due to limited sulfide metabolism and not due to general oxidative stress associated with sod1 deletion. Furthermore, wild-type or sod1Δ yeast was cultured in complete liquid media with or without NaSH and growth was monitored over time (Fig. 1B). Wild-type yeast growth rates slightly increased in response to NaSH, while sod1Δ yeast growth rates were concentration-dependently inhibited by sulfide (SI Appendix, Fig. S2A). To measure the effect of SOD1 expression on endogenous cellular H2S levels, wild-type or sod1Δ yeast was lysed in the presence of N-iodoacetyl tyrosine methyl ester (TME-IAM), a sulfide and polysulfide labeling and stabilizing reagent (38). TME-IAM adducted hydrogen sulfide/hydrosulfide (Bis-S-AM-TME) was analyzed by liquid chromatography-electrospray ionization–tandem mass spectrometry (LC-ESI-MS/MS) (SI Appendix, Table S1) (38). H2S was significantly elevated in sod1Δ yeast compared to wild-type yeast (Fig. 1C). Similarly, endogenous H2S levels were significantly higher in HEK293 cells transfected with SOD1-specific siRNA compared to control cells (Fig. 1D).
Fig. 1.
SOD1 protects cells from H2S toxicity. (A) (Top) Representative image of yeast spot assay of wild-type or sod1Δ yeast grown on YPD agar media or YPD containing either 100 μM MV2+ or NaSH. (Bottom) Bar graph showing mean values from densitometric analysis of yeast spot assay images (±SEM; n = 4 experiments, n = 2 spots per experiment). Significance calculated by two-way ANOVA with Sidak’s test. (B) Representative growth (OD600) of wild-type or sod1Δ yeast cultured ± NaSH measured over 12 h. (C) Intracellular H2S content measured by LC-ESI-MS/MS (38) from wild-type or sod1Δ yeast or (D) control or SOD1-silenced HEK293 cells. Data represent mean values [±SEM; n = 4 colonies (yeast); n = 4 wells (HEK293)] and significance calculated by unpaired, two-tailed t tests. (E) Proliferation, measured by electrical impedance, of SOD1-silenced or control HEK293 cells cultured with or without 500 μM GYY4137. (Inset: immunoblot showing relative SOD1 expression.) Data shown are mean cell index values (±SD, n = 3 to 4 wells per group) from a representative experiment. Significance at 72-h time point was calculated by two-way ANOVA with Sîdak’s test. (F) Graph showing final cell index values from (E) normalized to control values. Bars represent mean normalized cell index (±SEM; n = 5 experiments, n = 3 to 4 wells per experiment). (G). Proliferation, measured by electrical impedance, of SOD1-overexpressing or control HEK293 cells cultured with or without 1 mM GYY4137. (Inset: immunoblot showing relative SOD1 expression.) (H) Graph showing final cell index values from (G) normalized to control values. Bars represent mean normalized cell index (±SEM; n = 4 experiments, n = 3 to 4 wells per experiment). (I) Cell proliferation of control or SOD1-silenced HEK293 cells expressing either WT or H65R-mutant human CBS. Mean data from a representative experiment are shown. Data normalized to 24-h time point (gridlines). (J) Graph showing 72-h cell index values from (I). Bars represent mean values (±SEM; n = 5 experiments, n = 2 to 3 wells per experiment).
To examine the role of SOD1 expression on sulfide tolerance in mammalian cells, human HEK293 cells were transiently transfected with control or SOD1-specific silencing RNA oligonucleotides and proliferation was measured in cell cultured with the slow-releasing H2S-donor, GYY4137 (500 μM), or vehicle. This concentration of GYY4137 formed a steady-state sulfide concentration of ≈74 μM after 24 h (SI Appendix, Fig. S2B). Sulfide reduced control cell proliferation after 72 h to approximately half of vehicle treated controls, whereas sulfide almost completely inhibited SOD1-silenced cell proliferation (Fig. 1 E and F). To confirm that the inhibition of proliferation was not due to decreased cell adhesion, GYY4137 was added to cells 24 h after seeding. GYY4137 has similar inhibitory effects on adherent cell proliferation (SI Appendix, Fig. S2 C and D). Conversely, SOD1 overexpression significantly increased HEK293 cell tolerance to 1 mM GYY4137 (Fig. 1 G and H). The steady-state concentration of H2S under these conditions was ≈150 μM (SI Appendix, Fig. S1B). Cystathionine β-synthase (CBS) overexpression results in elevated intracellular H2S and/or cysteine persulfide levels (39, 40). To examine the effect of SOD1 on endogenous H2S production, HEK293 cells were transiently transfected with either GFP-tagged, wild-type, or mutant (H65R) CBS expression vector with either control or SOD1-specific siRNA (SI Appendix, Fig. S2 E and F). Wild-type but not mutant CBS overexpression showed decreased proliferation in SOD1-silenced cells; however, wild-type or mutant CBS overexpression had no effect on SOD1-replete cells (Fig. 1 I and J). Despite similar superoxide metabolizing function, SOD2 silencing had no effect on exogenous sulfide-mediated cellular toxicity in HEK293 cells (SI Appendix, Fig. S2 G and H). These data reveal that SOD1 is essential for protecting cells against both endogenous and exogenous H2S-mediated cytotoxicity.
SOD1 Oxidizes Sulfide to Sulfate.
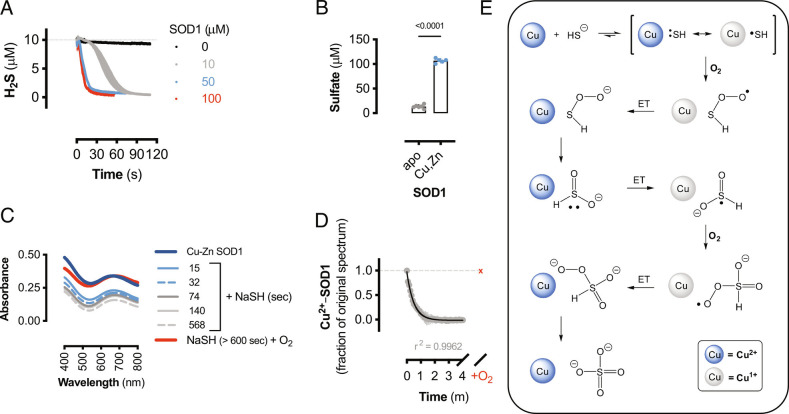
As SOD1 expression appears to be critical for sulfide detoxification, the reaction between SOD1 and sulfide was investigated. Sulfide oxidation by SOD1 has been reported to be a very slow process that forms hydrogen persulfide (H2S2); however, this reaction was only studied under non-physiological conditions of excess H2S compared to SOD1 (41). Cellular and tissue H2S concentrations are estimated to be in the low nanomolar range (42), while cellular SOD1 concentrations are estimated to be between 10 and 100 μM (43). Therefore, the physiologically relevant reaction between excess SOD1 and limiting H2S was examined. H2S was rapidly consumed by the addition of SOD1 in a concentration-dependent manner (Fig. 2A). Sulfate was the major reaction product from SOD1 and NaSH; however, sulfate formation was significantly decreased in apo-SOD1 (Fig. 2B). Under anaerobic conditions, H2S rapidly reacted with SOD1, as the Cu2+ d-d transition centered at 670 nm follows a second-order decay (Fig. 2 C and D), implying that sulfide coordinated to the copper center forming a reduced copper-sulfide species. Upon exposure to air, the d-d band returned to the starting absorbance, signifying that the SOD1-sulfide complex was oxidized by O2 (Fig. 2 C and D).
Fig. 2.
SOD1 is a sulfide oxidase. (A) Amperometric electrode measurements of H2S (10 μM) consumption by the addition of human SOD1 (10 to 100 μM) or buffer (0 μM) at t = 0. Data shown are mean values (±SEM, n = 3 reactions). (B) Sulfate measurements from 100 μM NaSH alone or reacted with 150 μM human Cu-Zn SOD1 or apo-SOD1. Data shown are mean values (±SEM, n = 5 to 6 reactions). Significance calculated by unpaired, two-tailed t test. (C) Visible spectra of bovine SOD1 alone (dark blue) and reacted with NaSH for indicated times under anaerobic conditions and after exposure to air (red line). (D) Change in absorbance (680 nm) from bovine SOD-Cu2+ reduction by NaSH as in (C). Gray circles represent anaerobic spectra, and red “x” represents air-treated samples. Data shown are mean values (±SEM; n = 3 reactions) and fit to a second-order decay curve (black line) with corresponding goodness of fit. (E) Proposed chemical mechanism for the oxidation of sulfide to sulfate by SOD1. Blue circles represent Cu2+-SOD1, gray circles represent Cu1+-SOD1, and ET represents electron transfer reaction.
Copper and nickel (2+) thiolate complexes are in structural resonance with thiyl radical coordinated to a reduced metal center, and the paramagnetic nature of these complexes permits spin-allowed reactions with molecular oxygen to generate sulfinic acids (44, 45). Here we propose that HS− coordination to Cu2+-SOD1, instead of an alkyl thiolate, initially forms sulfhydryl thioperoxide, which is a tautomer of sulfoxylic acid (Fig. 2E). Sulfoxylic acid is thermodynamically susceptible to further oxidation by Cu2+ to form sulfoxylic peroxide radical, which is reduced by Cu1+ and rearranges to form sulfate (46). Thus, the Cu2+ center of SOD1 catalyzes the autoxidation of sulfide by removing the spin-forbidden nature of Reaction 3, which is thermodynamically highly favorable.
| [3] |
Metal-catalyzed sulfide oxidation is an effective route to detoxification, as cobinamide, a vitamin B12 analog, reduces H2S toxicity (3). Therefore, the ubiquitous and highly expressed [Cu-Zn] SOD1 is an efficient sulfide oxidase under physiological conditions that converts toxic hydrogen sulfide into innocuous sulfate.
SOD1 Oxidizes Excess Sulfide to Per/Polysulfides.
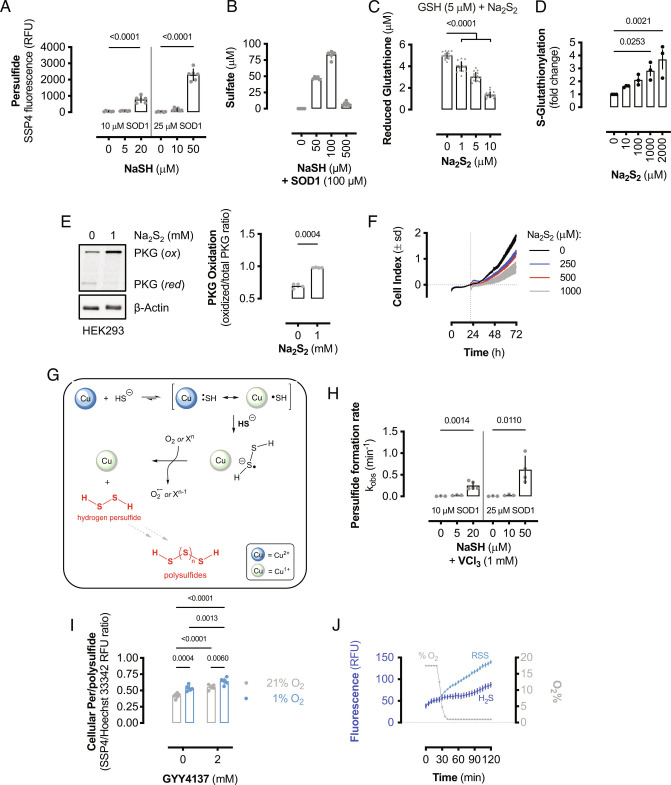
SOD1 is reported to oxidize H2S to hydrogen persulfide (H2S2); however, H2S2 formation was only investigated under conditions of limiting SOD1 and excess sulfide (41). Here we show that H2S2 and RSS formation by SOD1 is limited to conditions where sulfide is in molar excess compared to SOD1. Solutions of NaSH were added to SOD1 in the presence of SSP4, a fluorescent probe that detects persulfides (47). SSP4 fluorescence did not increase when limiting amounts of sulfide were added to SOD1; however, persulfide was detected when sulfide concentrations were greater than SOD1 (Fig. 3A). Similarly, sulfate production was nearly linear with respect to sulfide concentration; however, when the molar concentration of H2S exceeded that of SOD1, sulfate production was significantly reduced (Fig. 3B), suggesting that another chemical process dominates under these conditions. These data indicate that SOD1 does not contribute to persulfide and polysulfide formation under normal physiological H2S levels (42).
Fig. 3.
SOD1 oxidizes excess H2S to RSS. (A) RSS formation from the oxidation of NaSH by human SOD1. Bars represent mean values (±SEM; n = 6 reactions). Significance calculated by one-way ANOVA with Dunnett’s test. (B) Sulfate formation from the oxidation of NaSH by 100 μM human SOD1. Bars represent mean values (±SEM; n = 5 to 6 reactions). (C) Reduced glutathione remaining after reacting GSH (5 μM) with Na2S2 at the indicated concentrations. Bars represent mean values (±SEM; n = 18 wells). Significance calculated by one-way ANOVA with Dunnett’s test. (D) Quantification of protein S-glutathionylation from MCF10A cells treated with Na2S2. Bars represent mean values (±SEM; n = 3 wells) and significance calculated by one-way ANOVA with Dunnett’s test. (E) Immunoblot of relative reduced and oxidized PKG and β-actin expression in HEK293 cells treated with or without 1 mM Na2S2. Bars represent mean oxidized PKG fraction (±SEM; n = 4 wells). (F) Cellular proliferation as measured by electrical impedance (cell index). HEK293 cells were cultured for 24 h before addition of Na2S2 stock solutions and cellular proliferation monitored for 48 h. Data shown are mean cell index values (±SD; n = 3 wells). (G) Proposed chemical mechanism for SOD1-catalyzed RSS formation from excess sulfide. The reaction proceeds via a disulfide radical anion intermediate. (H) Observed rate constants for the formation of persulfide from the anaerobic reaction of limiting or excess sulfide with SOD1 in the presence of 1 mM VCl3. Bars represent mean rate constants (±SEM; n = 3 to 5 reactions). Significance calculated by one-way ANOVA with Dunnett’s test. (I) Relative cellular persulfide and/or polysulfide formation in HEK293 cells cultured with GYY4137 or vehicle in normoxic (21% O2) or anaerobic conditions (1% O2). Bars represent mean values (±SEM; n = 6 wells). Significance calculated by two-way ANOVA with Sidak’s test. (J) Atmospheric O2 content and relative fluorescence of wild-type yeast loaded with either a H2S- or RSS-specific probe cultured at 30 °C. Data shown are mean values (±SEM; n = 9 to 10 wells).
When SOD1 expression or function is decreased, or if high cellular levels of H2S are achieved, H2S2 is predicted to be formed; this is potentially significant as hydrogen persulfide is a cellular oxidant that is isoelectronic with hydrogen peroxide. Additionally, H2S2 disproportionates to regenerate H2S and form H2S3 and longer polysulfide chains; therefore, hydrogen persulfide results in RSS formation via multiple equilibria. To determine if H2S2 is a cellular thiol oxidant like hydrogen peroxide, authentic sodium persulfide (Na2S2) was reacted with reduced glutathione (GSH). Sodium persulfide concentration-dependently oxidized GSH (Fig. 3C) and protein S-glutathionylation significantly increased in MCF10A cells treated with Na2S2 (Fig. 3D and SI Appendix, Fig. S3A). Similarly, Na2S2 resulted in rapid cGMP-dependent protein kinase 1 (PKG) oxidation (Fig. 3E) and reduced HEK293 cell proliferation in a concentration-dependent manner (Fig. 3F). Therefore, SOD1 catalyzes the formation of RSS when sulfide is in molar excess, which has cellular thiol oxidizing effects.
H2S2 from sulfide oxidation has been proposed to proceed via thiyl radical production and recombination (41); however, this is not a thermodynamically favorable reaction (HS• + e− = HS−; Eo = +0.92 V) (48) and chemically unlikely, as O2 would rapidly compete for thiyl radical recombination. We propose that under excess sulfide conditions, the Cu-thiyl complex will react with another equivalent of HS− (instead of O2 under limiting sulfide conditions) to form a cuprous persulfide radical anion (Fig. 3G). Disulfide radical anions are potent one-electron reducing agents (49), and we show here that SOD1-catalyzed persulfide formation proceeds via persulfide radical anion, as the reaction occurs under anaerobic conditions in the presence of electron acceptors of varying reduction potentials. In addition to O2, the SOD1-persulfide complex reduced Fe(CN)63− and V3+ (Fig. 3H and SI Appendix, Fig. S3 B–D) but did not reduce MV2+ to MV•+ (SI Appendix, Fig. S3E). This indicates that the SOD1-persulfide complex reduction potential is bracketed between 0.26 and 0.43 V, consistent with a metal-coordinated disulfide radical anion (50). These results also indicate that SOD1-catalyzed persulfide formation under high relative sulfide conditions may proceed under hypoxic conditions, provided an alternative electron acceptor is present. To measure persulfide formation under hypoxic conditions, HEK293 cells were cultured in 21% or 1% O2-containing atmosphere for 24 h with vehicle or 2 mM GYY4137, which corresponded to ≈270 μM H2S (SI Appendix, Fig. S1B). Hypoxia resulted in increased basal persulfide formation compared to normoxic controls and sulfide donation resulted in elevated persulfide formation in both conditions; however, sulfide donation under hypoxic conditions resulted in significantly more persulfide formation compared to normoxia (Fig. 3I). These data suggest that SOD1 may act as a pro-oxidant under high H2S levels, even in the absence of O2. Thus, cellular hypoxia may be a scenario of elevated H2S and RSS formation and reactivity.
To further examine if endogenous cellular H2S and RSS accumulate in hypoxic conditions, wild-type yeast was loaded with either H2S-specific or RSS-specific chemical probes and monitored over time at variable O2 concentrations. When yeast was cultured under normoxic conditions, H2S and RSS probes remained relatively stable; however, as the O2 concentration decreased to 1%, both H2S and RSS florescent probes gained in signal intensity (Fig. 3J and SI Appendix, Fig. S3 F and G). Overall, these results indicate that cellular hypoxia limits SOD1-mediated sulfide oxidation and detoxification to sulfate and increases per- and polysulfide formation, as observed in hypoxic brains (51). Therefore, as oxygen tension affects H2S and RSS levels, in vitro experiments performed in 21% O2 may artificially limit intracellular H2S levels and therefore suppress RSS signaling.
SOD1 Limits Cellular Per- and Polysulfides.
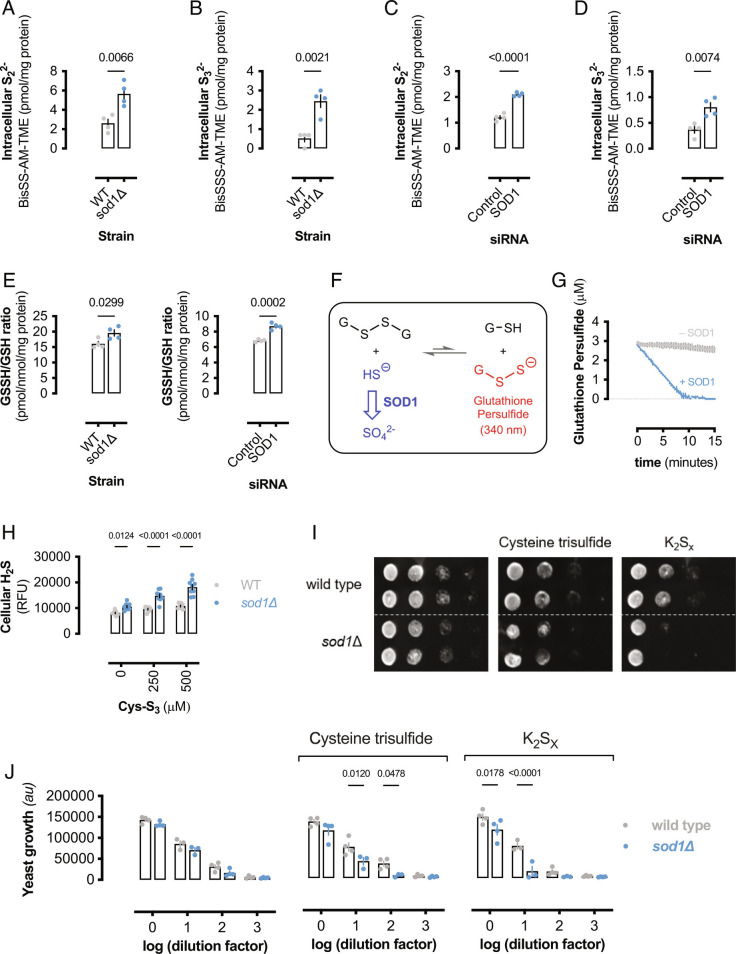
H2S is thought to elicit signaling via formation of persulfides as shown in Reaction 1 (14). As SOD1 deletion or silencing resulted in increased basal H2S levels (Fig. 1 C and D), endogenous RSS was predicted to increase in response to decreased SOD1 expression. TME-IAM adducted persulfide (Bis-SS-AM-TME), trisulfide (Bis-SSS-AM-TME), reduced glutathione (GS-AM-TME), and glutathione hydropersulfide (GSS-AM-TME) were measured by LC-ESI-MS/MS (SI Appendix, Table S1) (38). The inorganic polysulfides S22− and S32− were significantly elevated in sod1Δ yeast compared to wild-type yeast (Fig. 4 A and B). Similarly, S22− and S32− were significantly elevated in SOD1-silenced HEK293 cells compared to control cells (Fig. 4 C and D). Furthermore, basal glutathione hydropersulfide (GSSH/GSH ratio) was also elevated in cells lacking SOD1 (Fig. 4E), suggesting that SOD1 exerts its antioxidant effects partially by limiting H2S and RSS.
Fig. 4.
SOD1 oxidation of sulfide decreases RSS. (A) Intracellular dihydrogen disulfide (S22−) and (B) dihydrogen trisulfide (S32−) content from wild-type or sod1Δ yeast. Bars represent mean polysulfide levels (±SEM; n = 4 colonies) and significance calculated by unpaired, two-tailed t test. (C) Intracellular dihydrogen disulfide (S22−) and (D) dihydrogen trisulfide (S32−) content from control or SOD1-silenced HEK293 cells. Bars represent mean polysulfide levels (±SEM; n = 4 wells) and significance calculated by unpaired, two-tailed t test. (E) Glutathione persulfide (GSSH) content normalized to total glutathione (GSH) and protein from (Left) wild-type or sod1Δ yeast and (Right) control or SOD1-silenced HEK293 cells. Bars represent mean polysulfide levels (±SEM; n = 4) and significance calculated by unpaired, two-tailed t test. (F) Schematic showing SOD1 regulation of glutathione persulfide levels by consuming H2S. H2S reacts with glutathione disulfide and is in equilibrium with glutathione persulfide and GSH. (G) Glutathione persulfide concentration after addition of buffer or bovine SOD1 (100 μM). Data represent mean concentrations (±SEM; n = 3 reactions). (H) Relative cellular H2S in wild-type or sod1Δ yeast treated with Cys-S3 for 1 h. Bars represent mean relative fluorescence units (±SEM; n = 9). Significance calculated by two-way ANOVA with Sidak’s test. (I) Representative image of yeast spot assay of wild-type or sod1Δ yeast grown on YPD agar media containing either 100 μM cysteine trisulfide or potassium polysulfide (K2Sx). (J) Bar graph showing mean values from densitometric analysis of yeast spot assay images (I) (±SEM; n = 4 experiments, n = 2 spots per experiment). Significance calculated by two-way ANOVA with Sidak’s test.
SOD1 consumption of H2S is predicted to shift the equilibrium in Reaction 1 to decrease persulfide concentration (Fig. 4F). To examine if SOD1 limits persulfide formation, glutathione persulfide was formed by reacting glutathione disulfide (GSSG) with Na2S. An equilibrium constant of ≈1 × 10−5 was calculated by reacting increasing concentrations of Na2S (SI Appendix, Fig. S4 A and B), consistent with other reports that Reaction 1 is thermodynamically unfavorable (13, 19). Addition of SOD1 to GSS- in equilibrium with HS− and GSSG caused the persulfide absorbance to rapidly decay, whereas addition of buffer did not significantly alter the persulfide equilibrium (Fig. 4G), demonstrating that SOD1-sulfide oxidase activity limits the cellular pool of RSS.
Cysteine trisulfide is predicted to react with thiols to generate cysteine persulfide (52), which can further react with thiols to form mixed disulfides and H2S (SI Appendix, Fig. S4C). Indeed, reaction of cysteine trisulfide with 2-mercaptoethanol rapidly formed H2S (SI Appendix, Fig. S4D). Therefore, cysteine trisulfide was utilized as an in situ cellular persulfide and H2S-donor (19, 52). To determine if cysteine trisulfide treatment resulted in increased H2S, wild-type or sod1Δ yeast was treated with cysteine trisulfide and relative cellular H2S measured by fluorescent probe (53). Cysteine trisulfide significantly increased H2S in sod1Δ yeast compared to wild-type yeast (Fig. 4H), indicating that alkyl trisulfides promote cellular H2S formation. To determine if SOD1 can detoxify tri- and polysulfides via H2S oxidation, wild-type or sod1Δ yeast growth on agar plates containing either cysteine trisulfide or potassium polysulfide (K2Sx) was determined by a serial dilution spot assay (Fig. 4I). The absence of SOD1 resulted in increased sensitivity to both cysteine trisulfide and K2Sx (Fig. 4J), indicating that SOD1 metabolizes cellular H2S produced from alkyl trisulfides, persulfides, and polysulfides and limits RSS toxicity. While RSS may serve important antioxidant and signaling roles (40, 54), excessive levels of per- and polysulfides will be maladaptive due to the oxidative stress they impart (7, 19, 32) and here we show that SOD1 attenuates these effects via shifting sulfide equilibria away from RSS.
Reactive Oxygen Species (ROS) Oxidizes H2S to RSS.
H2S exhibits antioxidant effects in multiple cellular settings (55). Here, we have examined the role of SOD1 on sulfide oxidation by using SOD1-knockdown or deletion strategies; however, cells lacking SOD1 have elevated levels of ROS (56). Superoxide rapidly disproportionates to molecular oxygen and H2O2, and H2S is predicted to react with H2O2 to form H2S2 (Fig. 5A). Therefore, we investigated if ROS contributed to RSS cytotoxicity, while simultaneously inhibiting ROS-mediated oxidation. To mimic cellular ROS generation in the absence of SOD1, potassium superoxide (KO2) was dissolved in alkaline aqueous media, which rapidly forms O2 and H2O2 (57). Decomposed KO2 solutions containing approximately 50 μM H2O2 were reacted with Na2S, and the remaining sulfide was measured by monobromobimane (mBBr) fluorescence. ROS consumed sulfide (Fig. 5B) to form polysulfides (Fig. 5C). Furthermore, the reaction between sulfide and ROS inhibited ROS-mediated oxidation but increased per- and polysulfide reactivity (Fig. 5D). Additionally, H2S treatment reduced intracellular Ca2+ levels in SOD1-silenced HEK293 cells (SI Appendix, Fig. S5A), consistent with its ROS antioxidant effects (58). Therefore, H2S is a ROS antioxidant; however, this interaction results in the formation of thiol oxidizing RSS. To examine this effect of H2S and ROS in a cellular setting, HEK293 cells were incubated with rotenone or vehicle and mitochondrial ROS formation was measured in response to NaSH. Rotenone treatment increased ROS formation, which was significantly decreased by H2S (Fig. 5E). However, this decrease in ROS production by H2S resulted in increased cytotoxicity after 24 h (Fig. 5F), indicating that the interaction between ROS and H2S limits ROS-mediated oxidation, but forms other cytotoxic molecules. Similarly, NaSH treatment of SOD1-silenced HEK293 cells resulted in decreased mitochondrial respiration and ATP production, while NaSH did not decrease mitochondrial activity in SOD1-replete control cells (SI Appendix, Fig. S5 B and C).
Fig. 5.
ROS oxidizes H2S to RSS. (A) Reactions depicting the oxidation of H2S by KO2 to form hydrogen persulfide (H2S2). (B) Bar graph showing the relative amount of H2S remaining after reaction with KO2 at pH 7.5. Data are mean RFU values (±SEM; n = 3 reactions) and significance calculated by one-way ANOVA with Dunnett’s test. (C) The rate of polysulfide formation from the reaction of Na2S (100 μM) with KO2 (100 μM) compared to Na2S alone. Data shown are mean absorbance values (±SEM; n = 4 reactions). (D) ROS and RSS formation from H2S reacting with KO2 as measured by CBA and SSP4 fluorescence, respectively. Bar graphs show mean RFU (±SEM; n = 3 to 4 reactions). (E) Mitochondrial superoxide (MitoSOX Red) normalized to DNA content (Hoechst 333422) in HEK293 cells treated ± rotenone (100 nM) and ± NaSH (500 μM) for 2 h. Bars represent mean RFU ratios (±SEM; n = 4 experiments) and significance calculated by two-way ANOVA. (F) Cell viability after 24 h in HEK293 cells treated with rotenone and/or NaSH as in (E) as measured by MTS reduction. Bars represent mean absorbance values normalized to untreated control (±SEM; n = 6 experiments), and significance was calculated by two-way ANOVA. (G) Relative cellular persulfide/polysulfide formation in wild-type or sod1Δ yeast cultured with NaSH as in Fig. 1C. Data presented as persulfide normalized to nuclear DNA content and data shown are mean ratios (±SEM; n = 3 experiments, n = 2 wells per replicate). (H) Quantification of yeast persulfide/polysulfide formation over 12 h in wild-type or sod1Δ yeast cultured with NaSH. Bars represent mean AUC (±SEM; n = 3 experiments, n = 2 wells per replicate) from (G).
To measure the cellular RSS formation from ROS and H2S, wild-type or sod1Δ yeast was loaded with SSP4 and Hoechst 33342 and cultured with increasing concentrations of NaSH. In the absence of SOD1, H2S donation resulted in rapid intracellular RSS formation in a concentration-dependent manner (Fig. 5 G and H), whereas endogenous SOD1 expression in wild-type yeast significantly limited RSS formation (Fig. 5H). Furthermore, RSS formation (Fig. 5 G and H) preceded cellular death in sod1Δ yeast (Fig. 1B) and strongly indicates that RSS formation is associated with cellular toxicity. The increased RSS formation in SOD1-knockdown cells suggests that cellular thiol oxidation may occur under conditions of excess sulfide compared to SOD1. Control or SOD1-silenced HEK293 cells were treated with 100 μM NaSH, and cellular thiol oxidation was determined by PKG dimerization and protein S-glutathionylation. In control cells, 100 μM NaSH was not sufficient to induce either PKG oxidation or increased protein S-glutathionylation, consistent with SOD1-mediated detoxification (SI Appendix, Fig. S5 D and E). However, 100 μM NaSH caused both PKG thiol oxidation and global protein S-glutathionylation in SOD1-silenced cells (SI Appendix, Fig. S5 D and E), suggesting that RSS formation induces cellular oxidative stress similar to authentic hydrogen persulfide (Fig. 3 C–G) and cysteine trisulfide (19). Sulfide is known to induce oxidants other than ROS from inhibited mitochondrial electron transport chain (3, 6), and these results indicate that sulfide-mediated RSS formation is linked to cellular oxidation and toxicity. Therefore, we conclude that SOD1 limits persulfide/polysulfide toxicity by consuming H2S as well as preventing the interaction between ROS and H2S.
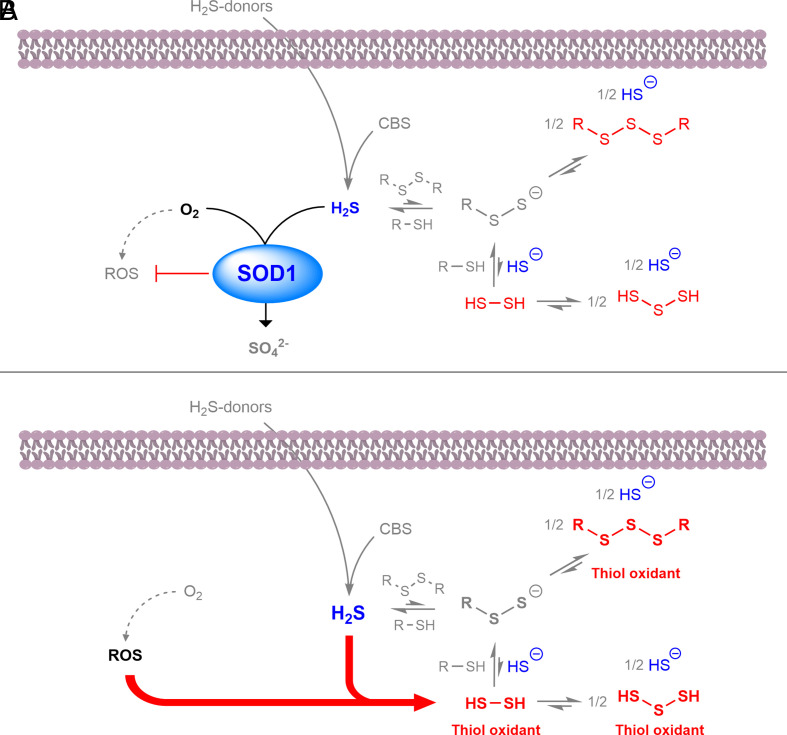
Overall, our data indicate that SOD1 restricts cellular H2S levels and limits cellular RSS by shifting the multiple equilibria associated with H2S (Fig. 6A). In the absence of SOD1, elevated ROS catalyzes the formation of toxic RSS (Fig. 6B). SOD1 has an antioxidant effect in microbes, animals, and plants (23); our results indicate that the antioxidant effect of SOD1 may be expanded to include its role in sulfide and RSS detoxification. In addition to superoxide metabolism, SOD1 has other cellular function; for example, SOD1 regulates cellular NADPH production by inhibiting GAPDH activity (59), as well as functioning as a nuclear transcription factor (60). Our data indicate an alternative role for SOD1 in sulfur metabolism and suggest that RSS such as polysulfides are cellular oxidants limited by SOD1, in addition to ROS. Although ROS are generally accepted to be the principle cellular oxidant, our results indicate that the oxidative role of RSS may be equally important, as RSS have been proposed to be the major cellular oxidant pool (32, 61). Just as the discovery of the superoxide dismutation spurred research into the role of ROS in human disease (26), our results indicate a similar amount of attention may be justified for RSS in cellular dysfunction.
Fig. 6.
SOD1 detoxifies H2S and limits RSS-mediated thiol oxidation. (A) SOD1 catalyzes the oxidation of H2S from endogenous and exogenous sources to sulfate, in addition to limiting cellular ROS formation. The consumption of H2S by SOD1 shifts multiple equilibria away from thiol oxidizing RSS (red). (B) In the absence of SOD1, ROS levels increase and directly react with H2S to form thiol oxidizing RSS. Therefore, SOD1 antioxidant properties include suppression of cellular H2S and related RSS.
Materials and Methods
Detailed methods are accessible in SI Appendix, Supplementary Text. Reagents and resources used in this study are listed in SI Appendix, Table S2. Briefly, yeast strains (WT and sod1Δ) were grown on YPD agar or liquid media supplemented with either NaSH or MV and growth assessed by either spot assay or OD600 measurements. Human embryonic kidney epithelial cell line (HEK293) was transfected with control or SOD1-specific siRNA, and growth was measured by electrical impedance in the presence of a H2S-donor, GYY4137. Alternatively, HEK293 cells were co-transfected with siRNA and expression plasmids encoding for GFP-tagged wild-type or mutant (H65R) CBS and proliferation measured by electrical impedance. Human or bovine [Cu-Zn] SOD1 or apo-SOD1 was reacted with NaSH, and sulfide consumption was measured by amperometric electrode, while sulfate was measured by anion chromatography. Cellular and biochemical persulfide formation was measured by SSP4 fluorescence and LC-ESI-MS/MS. Protein thiol oxidation is measured by biotinylated glutathione ethyl ester incorporation and protein kinase G disulfide formation.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
C.H.S. was supported by British Heart Foundation project grant PG/19/33/34385. P.E. was supported by The Barts Charity Cardiovascular Programme Award G00913 and by program grants from the British Heart Foundation and the Medical Research Council. This work was supported, in part, by a Grant-in-Aid for Scientific Research (C), Challenging Exploratory Research, and Transformative Research Areas “Life Science Innovation Driven by Supersulfide Biology”(AIII) from the Ministry of Education, Sciences, Sports, Technology (MEXT), Japan, to S.K. (22K06148) and to H.I. (21H05263). The authors thank Dr. Natalie Ludgate (QMUL School of Geography) for assistance with ion chromatography methodology and data collection.
Author contributions
C.H.S. designed research; C.H.S. and S.K. performed research; S.K. and H.I. contributed new reagents/analytic tools; C.H.S., S.K., and P.E. analyzed data; and C.H.S. and P.E. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
DNA plasmids data have been deposited in Addgene (182922, 182923, 182924). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Petersen L. C., The effect of inhibitors on the oxygen kinetics of cytochrome c oxidase. Biochim. Biophys. Acta 460, 299–307 (1977). [DOI] [PubMed] [Google Scholar]
- 2.Beauchamp R. O., et al. , A critical review of the literature on hydrogen sulfide toxicity. Crit. Rev. Toxicol. 13, 25–97 (1984). [DOI] [PubMed] [Google Scholar]
- 3.Jiang J., et al. , Hydrogen sulfide—Mechanisms of toxicity and development of an antidote. Sci. Rep. 6, 20831 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stubbert D., et al. , Protein kinase G Iα oxidation paradoxically underlies blood pressure lowering by the reductant hydrogen sulfide. Hypertension 64, 1344–1351 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Ide M., et al. , Excess hydrogen sulfide and polysulfides production underlies a schizophrenia pathophysiology. EMBO Mol. Med. 11, e10695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truong D. H., Eghbal M. A., Hindmarsh W., Roth S. H., O’Brien P. J., Molecular mechanisms of hydrogen sulfide toxicity. Drug Metab. Rev. 38, 733–744 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Greiner R., et al. , Polysulfides link H2S to protein thiol oxidation. Antioxid. Redox Signal. 19, 1749–1765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Q., Ying J., Xiang L., Zhang C., The biologic effect of hydrogen sulfide and its function in various diseases. Medicine 97, e13065 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura H., Shibuya N., Kimura Y., Hydrogen sulfide is a signaling molecule and a cytoprotectant. Antioxid. Redox Signal. 17, 45–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hine C., et al. , Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 160, 132–144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovinazzo D., et al. , Hydrogen sulfide is neuroprotective in Alzheimer’s disease by sulfhydrating GSK3β and inhibiting Tau hyperphosphorylation. Proc. Natl. Acad. Sci. U.S.A. 118, e2017225118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park C.-M., Weerasinghe L., Day J. J., Fukuto J. M., Xian M., Persulfides: Current knowledge and challenges in chemistry and chemical biology. Mol. BioSyst. 11, 1775–1785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuevasanta E., et al. , Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J. Biol. Chem. 290, 26866–26880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul B. D., Snyder S. H., H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13, 499–507 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Libiad M., Yadav P. K., Vitvitsky V., Martinov M., Banerjee R., Organization of the human mitochondrial hydrogen sulfide oxidation pathway*. J. Biol. Chem. 289, 30901–30910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szabo C., et al. , Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br. J. Pharmacol. 171, 2099–2122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson M. R., Melideo S. L., Jorns M. S., Human sulfide: Quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry 51, 6804–6815 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Landry A. P., Ballou D. P., Banerjee R., Hydrogen sulfide oxidation by sulfide quinone oxidoreductase. Chembiochem 22, 949–960 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Switzer C. H., Guttzeit S., Eykyn T. R., Eaton P., Cysteine trisulfide oxidizes protein thiols and induces electrophilic stress in human cells. Redox Biol. 47, 102155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landry A. P., Ballou D. P., Banerjee R., H2S oxidation by nanodisc-embedded human sulfide quinone oxidoreductase. J. Biol. Chem. 292, 11641–11649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill B. C., et al. , Interactions of sulphide and other ligands with cytochrome c oxidase. An electron-paramagnetic-resonance study. Biochem. J. 224, 591–600 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linden D. R., et al. , Sulphide quinone reductase contributes to hydrogen sulphide metabolism in murine peripheral tissues but not in the CNS. Br. J. Pharmacol. 165, 2178–2190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Case A. J., On the origin of superoxide dismutase: An evolutionary perspective of superoxide-mediated redox signaling. Antioxidants 6, 82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culotta V. C., Luk E., "Chaperones for metalloproteins" in Encyclopedia of Biological Chemistry, Lennarz W. J., Lane M. D., Eds. (Elsevier, New York, 2004), pp. 383–386, 10.1016/B0-12-443710-9/00101-0. [DOI] [Google Scholar]
- 25.Pardo C. A., et al. , Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc. Natl. Acad. Sci. U.S.A. 92, 954–958 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCord J. M., Fridovich I., Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244, 6049–6055 (1969). [PubMed] [Google Scholar]
- 27.Bakavayev S., et al. , Cu/Zn-superoxide dismutase and wild-type like fALS SOD1 mutants produce cytotoxic quantities of H(2)O(2) via cysteine-dependent redox short-circuit. Sci. Rep. 9, 10826–10826 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranguelova K., Bonini Marcelo G., Mason Ronald P., (Bi)sulfite oxidation by copper, zinc-superoxide dismutase: Sulfite-derived, radical-initiated protein radical formation. Environ. Health Perspect. 118, 970–975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liochev S. I., Fridovich I., Nitroxyl (NO−): A substrate for superoxide dismutase. Arch. Biochem. Biophys. 402, 166–171 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Hobbs A. J., Fukuto J. M., Ignarro L. J., Formation of free nitric oxide from l-arginine by nitric oxide synthase: Direct enhancement of generation by superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 91, 10992 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishanina T. V., Libiad M., Banerjee R., Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 11, 457–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Switzer C. H., Fukuto J. M., The antioxidant and oxidant properties of hydropersulfides (RSSH) and polysulfide species. Redox Biol. 57, 102486 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson K. R., Straub K. D., The role of hydrogen sulfide in evolution and the evolution of hydrogen sulfide in metabolism and signaling. Physiology 31, 60–72 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Barnese K., Gralla E. B., Cabelli D. E., Valentine J. S., Manganous phosphate acts as a superoxide dismutase. J. Am. Chem. Soc. 130, 4604–4606 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Reddi A. R., et al. , The overlapping roles of manganese and Cu/Zn SOD in oxidative stress protection. Free Radic. Biol. Med. 46, 154–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNaughton R. L., et al. , Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 15335–15339 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnese K., Gralla E. B., Valentine J. S., Cabelli D. E., Biologically relevant mechanism for catalytic superoxide removal by simple manganese compounds. Proc. Natl. Acad. Sci. U.S.A. 109, 6892–6897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasamatsu S., et al. , High-precision sulfur metabolomics innovated by a new specific probe for trapping reactive sulfur species. Antioxid. Redox Signal. 34, 1407–1419 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Chen X., Jhee K.-H., Kruger W. D., Production of the neuromodulator H2S by cystathionineβ-synthase via the condensation of cysteine and homocysteine*. J. Biol. Chem. 279, 52082–52086 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Ida T., et al. , Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 7606–7611 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson K. R., et al. , Metabolism of hydrogen sulfide (H2S) and production of reactive sulfur species (RSS) by superoxide dismutase. Redox Biol. 15, 74–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furne J., Saeed A., Levitt M. D., Whole tissue hydrogen sulfide concentrations are orders of magnitude lower than presently accepted values. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R1479–R1485 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Furukawa Y., O’Halloran T. V., Posttranslational modifications in Cu, Zn-Superoxide dismutase and mutations associated with amyotrophic lateral sclerosis. Antioxid. Redox Signal. 8, 847–867 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farmer P. J., Reibenspies J. H., Lindahl P. A., Darensbourg M. Y., Effects of sulfur site modification on the redox potentials of derivatives of [N, N’-bis(2-mercaptoethyl)-1,5-diazacyclooctanato]nickel(II). J. Am. Chem. Soc. 115, 4665–4674 (1993). [Google Scholar]
- 45.Ording-Wenker E. C. M., Siegler M. A., Lutz M., Bouwman E., Cul thiolate reactivity with dioxygen: The formation of cuII sulfinate and cuII sulfonate species via a cuII thiolate intermediate. Inorg. Chem. 52, 13113–13122 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Kumar M. R., Farmer P. J., Chemical trapping and characterization of small oxoacids of sulfur (SOS) generated in aqueous oxidations of H2S. Redox Biol. 14, 485–491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W., et al. , New fluorescent probes for sulfane sulfurs and the application in bioimaging. Chem. Sci. 4, 2892–2896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das T. N., Huie R. E., Neta P., Padmaja S., Reduction potential of the sulfhydryl radical: Pulse radiolysis and laser flash photolysis studies of the formation and reactions of ·SH and HSSH·- in aqueous solutions. J. Phys. Chem. A 103, 5221–5226 (1999). [Google Scholar]
- 49.Surdhar P. S., Armstrong D. A., Reduction potentials and exchange reactions of thiyl radicals and disulfide anion radicals. J. Phys. Chem. 91, 6532–6537 (1987). [Google Scholar]
- 50.Antonello S., Daasbjerg K., Jensen H., Taddei F., Maran F., Formation and cleavage of aromatic disulfide radical anions. J. Am. Chem. Soc. 125, 14905–14916 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Marutani E., et al. , Sulfide catabolism ameliorates hypoxic brain injury. Nat. Commun. 12, 3108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henderson C. F., et al. , Cysteine trisulfide protects E. coli from electrophile-induced death through the generation of cysteine hydropersulfide. Chem. Res. Toxicol. 33, 678–686 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Zhang J., et al. , o, o-Difluorination of aromatic azide yields a fast-response fluorescent probe for H2S detection and for improved bioorthogonal reactions. Org. Biomol. Chem. 15, 4212–4217 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Zivanovic J., et al. , Selective persulfide detection reveals evolutionarily conserved antiaging effects of S-Sulfhydration. Cell Metab. 30, 1152–1170.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shefa U., Kim M.-S., Jeong N. Y., Jung J., Antioxidant and cell-signaling functions of hydrogen sulfide in the central nervous system. Oxid. Med. Cell. Longev. 2018, 1873962–1873962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., Branicky R., Noë A., Hekimi S., Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 217, 1915–1928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayyan M., Hashim M. A., AlNashef I. M., Superoxide ion: Generation and chemical implications. Chem. Rev. 116, 3029–3085 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Görlach A., Bertram K., Hudecova S., Krizanova O., Calcium and ROS: A mutual interplay. Redox Biol. 6, 260–271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Montllor-Albalate C., et al. , Sod1 integrates oxygen availability to redox regulate NADPH production and the thiol redoxome. Proc. Natl. Acad. Sci. U.S.A. 119, e2023328119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsang C. K., Liu Y., Thomas J., Zhang Y., Zheng X. F. S., Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 5, 3446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeLeon E. R., et al. , A case of mistaken identity: Are reactive oxygen species actually reactive sulfide species? Am. J. Physiol. Regul. Integr. Comp. Physiol. 310, R549–R560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
DNA plasmids data have been deposited in Addgene (182922, 182923, 182924). All study data are included in the article and/or SI Appendix.