Significance
Several degenerative diseases are caused by expansions of CAG trinucleotide repeats. The repeat-containing RNAs contribute to cellular pathology in two unusual ways. These RNAs may agglomerate in the nucleus as foci or undergo aberrant repeat-associated non-AUG (RAN) translation. We show that RAN translation is preceded by prolonged retention of repeat-containing RNAs at nuclear foci, chronologically connecting these two pathomechanisms. Upon RAN translation, the repeat-containing RNAs coaggregate with the cognate RAN translation products. This cytoplasmic RNA–RAN protein aggregation correlates with the mislocalization of various RNA-binding proteins and cell toxicity, while accumulation of nuclear foci alone does not produce these defects. Our findings suggest that cytoplasmic RNA aggregation may be a common feature of diseases caused by CAG trinucleotide repeat expansions.
Keywords: Repeat expansion diseases, RNA localization, RNA aggregation, RAN translation
Abstract
CAG trinucleotide repeat expansions cause several neurodegenerative diseases, including Huntington's disease and spinocerebellar ataxia. RNAs with expanded CAG repeats contribute to disease in two unusual ways. First, these repeat-containing RNAs may agglomerate in the nucleus as foci that sequester several RNA-binding proteins. Second, these RNAs may undergo aberrant repeat-associated non-AUG (RAN) translation in multiple frames and produce aggregation-prone proteins. The relationship between RAN translation and RNA foci, and their relative contributions to cellular dysfunction, are unclear. Here, we show that CAG repeat-containing RNAs that undergo RAN translation first accumulate at nuclear foci and, over time, are exported to the cytoplasm. In the cytoplasm, these RNAs are initially dispersed but, upon RAN translation, aggregate with the RAN translation products. These RNA–RAN protein agglomerates sequester various RNA-binding proteins and are associated with the disruption of nucleocytoplasmic transport and cell death. In contrast, RNA accumulation at nuclear foci alone does not produce discernable defects in nucleocytoplasmic transport or cell viability. Inhibition of RAN translation prevents cytoplasmic RNA aggregation and alleviates cell toxicity. Our findings demonstrate that RAN translation-induced RNA-protein aggregation correlates with the key pathological hallmarks observed in disease and suggest that cytoplasmic RNA aggregation may be an underappreciated phenomenon in CAG trinucleotide repeat expansion disorders.
Over 40 genetic diseases are traced to aberrant expansions of short tandem repeats (1, 2). This family of diseases, collectively referred to as nucleotide repeat expansion disorders, includes Huntington’s disease (HD), several spinocerebellar ataxias (SCA), myotonic dystrophy, and certain forms of amyotrophic lateral sclerosis/frontotemporal dementia (ALS/FTD). Disease-causing repeats are found in both protein-coding as well as noncoding regions. For example, Huntington’s disease results from a CAG trinucleotide repeat expansion in exon 1 of the huntingtin (HTT) gene, whereas myotonic dystrophy is caused by a CTG repeat expansion in the 3′ untranslated region of dystrophia myotonica protein kinase (DMPK). Disease usually manifests when the number of repeats exceeds a critical threshold (1, 2).
Although repeat expansion diseases are monogenic disorders, the disease mechanisms are often more complex than the simple loss of function of the mutated gene (1, 2). Expanded repeats may contribute to cellular dysfunction and disease via at least three possible routes. First, when repeats are present in the protein-coding parts of the gene, they may produce aggregation-prone homopolymeric proteins, such as ones with expanded polyglutamine stretches. Second, the repeats may also be translated in multiple reading frames without a bona fide AUG start codon: a process referred to as repeat-associated non-AUG (RAN) translation (3). Third, the repeat-containing RNAs form higher order assemblies and accumulate as "foci" in the nucleus. These nuclear RNA foci sequester various RNA-binding proteins and result in transcriptome-wide RNA processing defects (4, 5). These mechanisms of pathology are not mutually exclusive, which makes it challenging to tease out their individual contributions to cellular dysfunction.
In previous work, we showed that pathogenic guanine/cytosine (GC)-rich repeat expansions in RNA (such as expanded CAG repeats) create sites for multivalent intermolecular base-pairing (6). An increase in the number of repeats corresponds to an increased valency for RNA–RNA interaction, and purified RNA with expanded repeats form micron-sized clusters in vitro. In cells, expression of RNA with expanded CAG repeats that do not lie within a canonical protein-coding open reading frame was sufficient to produce nuclear foci. However, expression of these minimal CAG repeat-containing RNAs did not induce noncanonical RAN translation, nor did it produce noticeable cell toxicity (6).
Here, we sought to develop a synthetic system that recapitulates RAN translation. Since prior work has shown that the sequences surrounding the repeats may affect this process (3, 7, 8), we created a small library where the same CAG repeat tract was placed downstream of different flanking sequences. We found that while expression of most CAG repeat-containing RNAs induced nuclear foci, a subset of these sequences were also exported to the cytoplasm and underwent RAN translation. Strikingly, cytoplasmic CAG repeat-containing RNAs coaggregated with the RAN translation products. These cytoplasmic RNA–RAN protein agglomerates recruited RNA-binding proteins such as TDP-43 and FUS, and were associated with nucleocytoplasmic transport defects and pronounced cell death. These toxic effects could be suppressed by inhibiting RAN translation. In contrast, CAG repeat-containing RNAs that accumulated at nuclear foci alone neither produced these abnormalities nor affected cell viability. Our findings suggest that translation of CAG repeat-containing RNA and accompanying cytoplasmic RNA-protein aggregation underlie the key pathological defects observed in disease.
Results
RAN Translation of RNA with Expanded CAG Repeats Depends on the Surrounding Sequence Context.
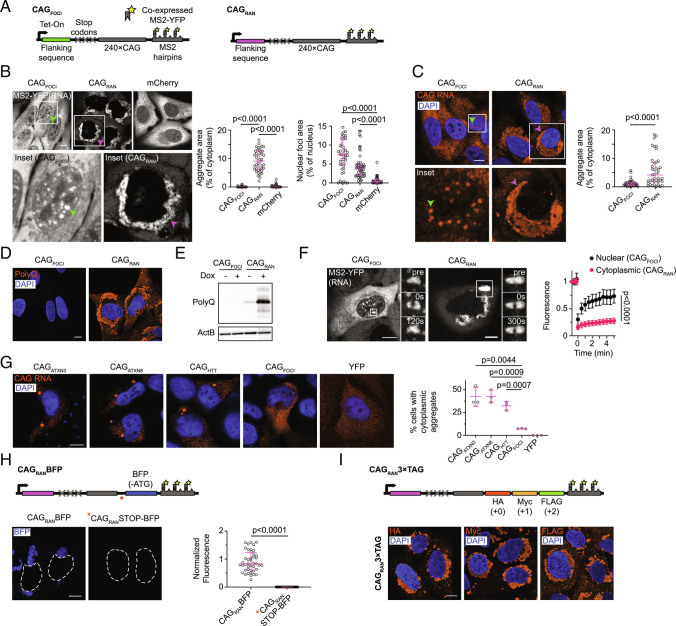
We previously reported that RNA transcripts that primarily consist of expanded CAG repeats cluster at RNA foci in the nucleus, but the expression of these RNAs did not produce RAN translation products (6). In contrast, expression of CAG repeat-containing minigenes, where the repeat tract is flanked by sequences native to disease-associated genes (such as HTT, ATXN3, and ATXN8 associated with HD, SCA3, and SCA8, respectively), recapitulated RAN translation in U-2OS cells (SI Appendix, Fig. S1A), as previously reported in other cell types (3). Several studies have indicated that sequences surrounding the expanded repeat tracts may facilitate RAN translation (3, 7–9). To determine whether adjacent nucleotide stretches could induce RAN translation of our synthetic CAG repeat construct, we appended a small library of 12 different 250-nt sequences upstream of the same 240×CAG repeat tract (sequences in Dataset S1). To preclude canonical translation of the repeat region, we incorporated multiple stop codons immediately upstream of the repeats (Fig. 1A). Downstream of the repeats, we cloned multiple MS2 aptamers, which serve as binding sites for a coexpressed enhanced yellow fluorescent protein-tagged MS2 coat protein (MS2-YFP) and thus allow RNA visualization (10). Upon expression of these constructs in U-2OS cells, we observed two distinct classes of cellular phenotypes. The first class exhibited liquid-like nuclear RNA foci and showed no detectable RAN translation products (representative sequence CAGFOCI in Fig. 1 B–E, other sequences in SI Appendix, Fig. S1 B and C), similar to our earlier report (6).
Fig. 1.
Sequence context modulates the localization and RAN translation of CAG repeat-containing RNAs. A, Schematics for CAGRAN and CAGFOCI constructs. B and C, Representative micrographs of cells expressing the indicated constructs imaged using MS2–YFP (B, Left) or via FISH probes against the MS2 tag (C, Left), and corresponding quantification of the aggregate and foci areas (B and C, Right). Boxes show regions used for Insets. D and E, Immunofluorescence micrographs (D) and immunoblot (E) for indicated samples using polyglutamine (polyQ) antibody. F, Representative micrographs before (pre) and at the indicated timepoints after photobleaching for nuclear foci of CAGFOCI (Left) and cytoplasmic inclusions of CAGRAN (Middle) and corresponding fluorescence recovery after photobleaching (FRAP) trajectories (Right). G, Fluorescence micrographs of cells transfected with the indicated constructs and stained using a FISH probe against the CAG repeats (Left) and corresponding quantification of the percentage of cells with cytoplasmic RNA aggregates (Right). H, Schematic for CAGRANBFP construct. Asterisk denotes the location of stop codons in CAGRANSTOP-BFP construct (Top). Representative fluorescence micrographs 24 h post-induction (Bottom, Left), and quantification of BFP fluorescence, normalized to cell area (Bottom, Right). I, Schematic for CAGRAN3xTag (Top) and representative immunofluorescence micrographs 24 h post-induction staining for the indicated tags (Bottom). Micrographs in F are representative of ≥20 independent photobleaching events; all other micrographs are representative of ≥40 cells across ≥2 independent experiments. Each data point in B, C, and H represents a single cell. Error bars denote median ± interquartile range. Data in G are summarized as mean ± SD over 3 independent experiments. Significance values were calculated using Mann–Whitney U tests in B, C, F, and H and using Student’s t test in G. Micrographs were independently scaled in B, C, F, and G. Where indicated, cell nuclei are stained with DAPI. (Scale bars, 10 µm.)
In the second class, we observed minimal nuclear foci but instead, the repeat-containing RNAs accumulated in perinuclear cytoplasmic inclusions (Fig. 1 B and C and SI Appendix, Fig. S1B). Unlike the round nuclear foci, the cytoplasmic RNA agglomerates had a dendritic mesh-like morphology (Fig. 1 B and C). The repeat-containing RNA at these cytoplasmic inclusions was immobile as evidenced by a lack of fluorescence recovery upon photobleaching (Fig. 1F and Movie S1). Similar RNA localization was observed via fluorescence in situ hybridization (FISH) both with and without the MS2 tag on the RNAs (Fig. 1C and SI Appendix, Fig. S1 D and E, controls for the MS2 tag are further discussed in SI Appendix, Supplemental Discussion). These localization phenotypes could not be accounted for by differences in RNA expression levels (SI Appendix, Fig. S1 F and G), and for a given sequence, changing RNA expression did not noticeably affect its subcellular localization (SI Appendix, Fig. S1 H and I).
Interestingly, constructs that resulted in cytoplasmic RNA aggregates also exhibited pronounced staining for polyglutamine-containing proteins (Fig. 1 D and E and SI Appendix, Fig. S1C), likely arising due to RAN translation of the repeat tract. Likewise, the expression of minigenes that undergo RAN translation (upstream sequences corresponding to HTT, ATXN3, and ATXN8), also produced cytoplasmic aggregates of the cognate repeat-containing RNA (Fig. 1G). We were intrigued by this correlation between cytoplasmic RNA aggregation and RAN translation, and we chose a representative sequence from this class in our synthetic library that produced RAN translation products and cytoplasmic RNA aggregates (CAGRAN) for further analysis.
Two hallmarks of RAN translation are that i) the expanded repeats are translated even though they do not occur in a canonical open reading frame with translation initiating within or possibly upstream of repeat region (3, 8, 11), and ii) the repeats may be translated in multiple reading frames (3, 12). Our constructs contained stop codons in each reading frame immediately upstream of the repeat tract; thus, the CAG repeats in CAGRAN are not a part of a canonical (or near-canonical) open reading frame. To validate that translation initiation occurs within or upstream of the repeats, we incorporated a fluorescent protein, enhanced blue fluorescent protein 2 (EBFP2), lacking an ATG-start codon, immediately downstream of the CAG-repeats (CAGRANBFP, EBFP2 incorporated in the CAG or 0 frame, Fig. 1H). The EBFP2 protein sequence does not contain another in-frame methionine in the first 78 amino acids and translation initiation at this residue should not result in a fluorescent protein product (13). If translation initiates within or upstream of the CAG repeats, the translating ribosomes will read through the downstream EBFP2, resulting in fluorescent protein production. Consistent with this hypothesis, cells transduced with CAGRANBFP exhibited BFP fluorescence upon induction (Fig. 1H). Incorporation of stop codons between the CAG-repeats and EBFP2 (CAGRANSTOP-BFP) led to a near-complete loss of BFP fluorescence (Fig. 1H), reinforcing that EBFP2 in CAGRANBFP initiates in the upstream region and is produced via RAN translation.
Noncanonical RAN translation of CAG repeats may produce polyglutamine (in 0 or CAG frame), polyserine (in +1 or AGC frame), and polyalanine (in +2 or GCA frame). We observe abundant polyglutamine production upon the expression of CAGRAN both with and without the MS2 tag (Fig. 1 D and E and SI Appendix, Fig. S1J). There are no commercial antibodies currently available against polyserine or polyalanine peptides. To test whether CAGRAN is translated in multiple reading frames, we incorporated epitope tags downstream of the CAG repeats (CAGRAN3xTag, HA in the 0 frame, Myc in the +1 frame, and FLAG in the +2 frame).
Immunofluorescence microscopy showed that cells expressing CAGRAN3xTag produced proteins corresponding to all three frames (Fig. 1I). Overall, these results show that CAGRAN recapitulates RAN translation in multiple frames, while a related construct with an identical repeat tract, CAGFOCI, does not and only produces nuclear RNA foci.
Cytoplasmic RNA Aggregation Is Preceded by Accumulation of RNA at Nuclear Foci.
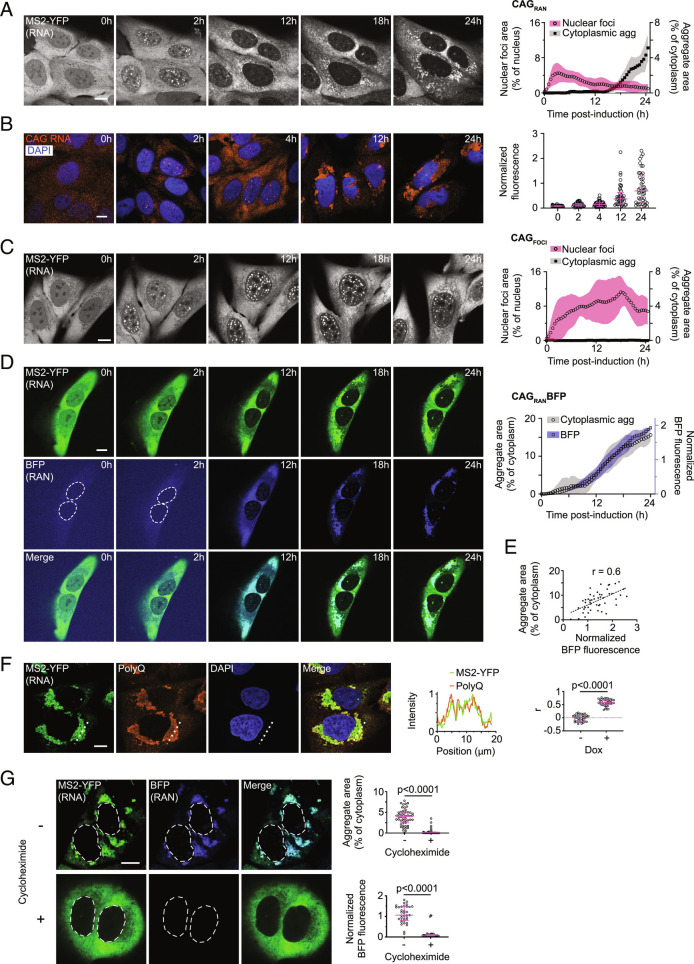
We next examined the localization of CAGRAN RNA in live cells via the MS2 hairpin tag (10). We found that within minutes after induction, CAGRAN RNA formed numerous micron-sized nuclear foci (Fig. 2A and Movie S2). CAGRAN RNA at these nuclear foci was mobile and exhibited rapid fluorescence recovery upon photobleaching (characteristic recovery time, τ ~ 80 s, SI Appendix, Fig. S2A and Movie S3). This recovery rate is similar to that observed for RNA that exclusively form nuclear foci (τ ~ 60 s for CAGFOCI) (Fig. 1F).
Fig. 2.
Cytoplasmic RNA aggregation follows accumulation at foci and coincides with RAN translation. A, Representative micrographs of cells expressing CAGRAN at the stated timepoints after induction (Left) and corresponding quantification (Right) of nuclear (circles, pink shading, left y-axis) and cytoplasmic (squares, gray shading, right y-axis) inclusions. B, Similar to A, imaged via FISH probes against the CAG repeats (Left) and corresponding quantification of the FISH signal (Right). C, Similar to A except for cells expressing CAGFOCI. D, Representative micrographs of cells expressing CAGRANBFP at the stated timepoints after induction imaged in MS2–YFP (Top) and BFP (Middle) channels, and their overlay (Bottom). Plot to the right shows the cytoplasmic aggregate area (circles, gray shading) and BFP fluorescence (squares, blue shading). E, Quantification of BFP fluorescence in relation to cytoplasmic RNA aggregate area. Dashed line represents a linear regression fit. F, Representative immunofluorescence micrographs showing CAGRAN RNA and polyQ-containing proteins. Intensity profile of MS2–YFP and polyQ signal along the dashed line in the micrographs (Middle). Correlation between MS2–YFP and polyQ-immunofluorescence signals (Right). G, Micrographs of cells expressing CAGRANBFP without (−) or with 10 µg/mL cycloheximide (+) treatment for 14 h (Left). Corresponding quantification of RNA aggregate area (Right, Top) and of BFP fluorescence (Right, Bottom). In A, C, and D, each data point is the mean of ≥4 cells and is a rolling average of three successive timepoints, shaded regions depict SD, and data are representative of ≥4 experiments with ≥25 cells. Each data point in B, E, F, and G represents a single cell and micrographs are representative of ≥2 independent experiments with ≥40 cells. Fluorescence in B, D, E, and G is normalized to cell area. Data in B, F, and G are summarized as median ± interquartile range and significance values were calculated by Mann–Whitney U tests. Images in B and D and MS2–YFP images in G are independently scaled. Where indicated, cell nuclei are stained with DAPI. (Scale bars, 10 µm.)
Over time (6 to 12 h post induction), CAGRAN RNA was exported to the cytoplasm and the nuclear foci dissolved or diminished in size (Fig. 2A). Immediately after export, CAGRAN RNA in the cytoplasm was well-dispersed and no obvious aggregates were observed. At ~18 h post-induction, several micron-sized RNA puncta began appearing throughout the cytoplasm. These small aggregates migrated inward toward the nucleus and merged to form a larger perinuclear cluster (2D-projection area 90 ± 40 µm2, Fig. 2A and Movie S2). Fixed timepoint analysis of the localization of CAGRAN RNA by FISH confirmed this same sequence of events (Fig. 2B). In comparison, no appreciable cytoplasmic inclusions were observed in cells expressing CAGFOCI over a comparable 24-h time-course or even up to 6 d after induction (Fig. 2C and Movie S4 and SI Appendix, Fig. S2B). Control RNAs encoding for mCherry or the reverse complement of mCherry (a noncoding RNA) with MS2 tags did not form observable inclusions in the nucleus or the cytoplasm (SI Appendix, Fig. S2 C and D).
RAN Translation Coincides with Cytoplasmic Aggregation of the CAG Repeat-Containing RNA.
To evaluate the timing of RAN translation with respect to RNA localization, we used our reporter cell line with EBFP2 cloned downstream of CAG repeats (CAGRANBFP). The RNA could be detected via the MS2 hairpin tag while EBFP2 fluorescence reported on RAN translation. At early timepoints after transcription induction, when the RNA primarily accumulated at nuclear foci, no appreciable BFP fluorescence was observed (Fig. 2D). After ~12 h, we observed an increase in BFP fluorescence concomitant with a reduction in size of nuclear foci but prior to any apparent cytoplasmic RNA or protein aggregation (Fig. 2D and Movie S5). By 18 h of induction, aggregates of the BFP-tagged RAN translation products appeared throughout the cytoplasm. These BFP aggregates colocalized with CAG-repeat-containing RNA even at the earliest stages of aggregation that we could detect using confocal microscopy. Approximately 24 h post-induction, both BFP-tagged RAN translation products and CAGRAN RNA colocalized at a few large perinuclear aggregates. Quantitative single-cell image analysis showed that the size of RNA aggregates correlated with the expression levels of RAN translation products (Fig. 2E, Pearson’s correlation coefficient, r, between MS2–YFP aggregate area and BFP fluorescence = 0.6). Combined staining for polyglutamine (as a marker for RAN translation products) and CAGRAN RNA (via MS2–YFP) confirmed the results described above that RAN translation products colocalize with the repeat-containing RNA (Fig. 2F, r = 0.58 between MS2–YFP fluorescence and polyglutamine fluorescence compared to r = 0.00 for uninduced controls, P < 0.0001 by Mann–Whitney test, n ≥ 40 cells).
The observation that RAN translation preceded cytoplasmic CAGRAN RNA aggregation suggested that RAN translation could be involved in the RNA aggregation process. To further test whether translation is required for the cytoplasmic aggregation of its template repeat-containing RNA, we treated cells with a translation elongation inhibitor, cycloheximide (10 µg/mL) concomitantly with induction of RNA expression. Cycloheximide treatment reduced the levels of RAN translation products by >90% (Fig. 2G) but did not affect RNA stability (SI Appendix, Fig. S2E). Interestingly, cycloheximide reduced cytoplasmic RNA aggregates by >90% compared with untreated controls (Fig. 2G; P < 0.0001 by Mann–Whitney test, n ≥ 40 cells), while it did not disrupt the nuclear foci formed by the CAGFOCI RNA (SI Appendix, Fig. S2F). Similarly, puromycin, another translation inhibitor, abrogated both protein translation and cytoplasmic RNA aggregation (SI Appendix, Fig. S2G). These results indicate that RAN translation products coaggregate with the template CAGRAN RNA, and that translation is required for cytoplasmic RNA aggregation.
Cytoplasmic RNA–RAN Protein Aggregates Recruit Nuclear RNA Binding Proteins.
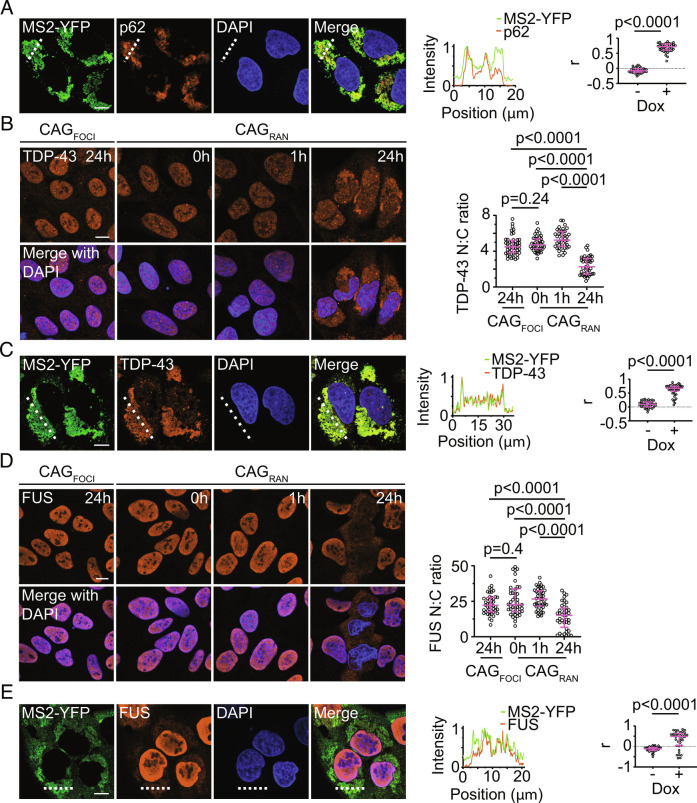
We examined the localization of RNA–RAN protein aggregates with respect to markers associated with cellular stress and neurodegenerative disease. The RNA aggregates did not colocalize with various cytoplasmic RNA granules including stress granules (G3BP1) (14), P-bodies (EDC4) (15), and TIS granules (TIS11b) (16) (SI Appendix, Fig. S3 A−C). In contrast, p62, an autophagy receptor and a commonly used marker for protein aggregation (17), strongly accumulated at the RNA inclusions in cells expressing CAGRAN with or without the MS2 tag (Fig. 3A and SI Appendix, Fig. S3D; r = 0.72, compared with r = -0.08 for uninduced controls, P < 0.0001 by Mann–Whitney test, n ≥ 40 cells).
Fig. 3.
RNA–RAN protein aggregates recruit nuclear RNA-binding proteins. A, Representative immunofluorescence micrographs showing colocalization between CAGRAN RNA and p62 (Left), and intensity profile of the two channels along the dashed line (Middle). Correlation between MS2–YFP and p62 signals with (+) and without (−) RNA induction (Right). B, Representative immunofluorescence micrographs showing TDP-43 in cells expressing CAGFOCI or CAGRAN for the stated times (Left). Quantification of the ratio of nuclear to cytoplasmic TDP-43 (Right). C, Similar to A except staining for TDP-43 instead of p62. D, Similar to B except staining for FUS instead of TDP-43. E, Similar to A except staining for FUS instead of p62. Cell nuclei are stained with DAPI. Data are representative of ≥2 independent experiments with ≥40 cells. Each data point represents a single cell and data are summarized as median ± interquartile range. Significance values were calculated by Mann–Whitney U tests. (Scale bars, 10 µm.)
A common theme in nucleotide repeat expansion disorders is the aberrant cytoplasmic localization of RNA-binding proteins such as TAR DNA-binding protein (TDP-43) and fused in sarcoma (FUS) (18–23). We found that cytoplasmic levels of TDP-43 were significantly higher in cells that express CAGRAN than in uninduced controls (Fig. 3B). TDP-43 was also substantially enriched at the cytoplasmic RNA aggregates (Fig. 3C, r = 0.68, compared with r = 0.10 for uninduced cells, P < 0.0001 by Mann–Whitney test, n ≥ 40 cells). FUS, another RNA-binding protein that forms aggregates in several repeat expansion disorders, also mislocalized to the cytoplasm (Fig. 3D) and accumulated at the cytoplasmic RNA aggregates (Fig. 3E; r = 0.49, compared to r = −0.11 for uninduced controls, P < 0.0001 by Mann–Whitney test, n ≥ 38 cells). Similar results were observed both with and without the MS2 tag on the CAGRAN RNA (SI Appendix, Fig. S3 E and F). Neither TDP-43 nor FUS mislocalized in cells expressing CAGFOCI, and the nuclear-to-cytoplasmic ratio of these proteins in cells with RNA foci only was comparable to that observed in the uninduced controls (Fig. 3 B and D). Likewise, in cells that expressed CAGRAN for 1 h, which exhibited nuclear foci but had not yet accumulated substantial RNA–RAN protein aggregates, the RNA-binding protein localization patterns were similar to those observed in uninduced control cells.
RNA–RAN Peptide Aggregates Disrupt Nucleocytoplasmic Transport and Induce Cell Death.
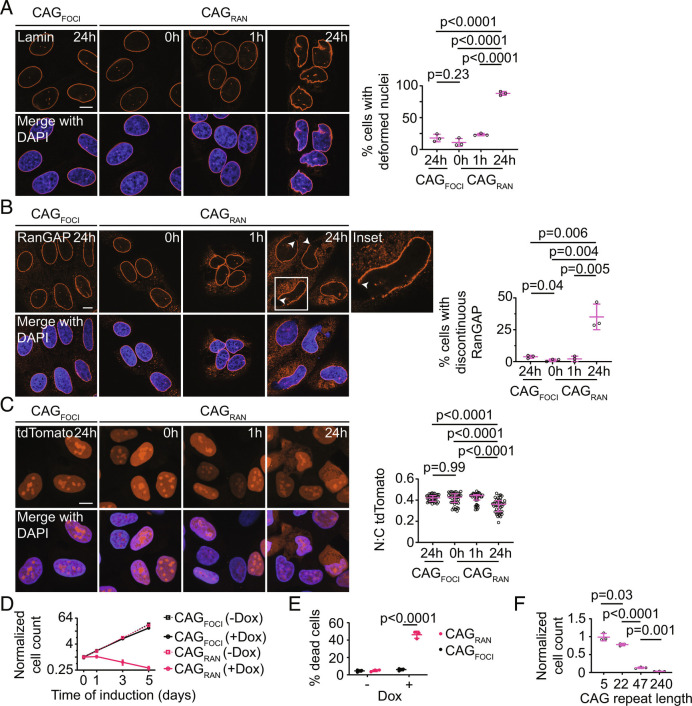
Another common defect observed in nucleotide repeat expansion disorders is the disruption of nucleocytoplasmic transport (24–27). Cells with cytoplasmic RNA aggregates displayed a deformed nuclear morphology (Fig. 4A), and the nuclear envelope was indented in the regions adjoining the RNA aggregates (SI Appendix, Fig. S4A). We examined the distribution of RanGAP1, which typically localizes to the cytoplasmic face of nuclear pore complexes and mediates recycling of importins. Upon prolonged (24 h) expression of CAGRAN, a substantial fraction (35 ± 10%, mean ± SD) of cells exhibited a clear disruption of RanGAP1 along the nuclear envelope (Fig. 4B). This disruption of RanGAP1 localization was not observed in cells expressing CAGFOCI or at early timepoints in cells expressing CAGRAN when only nuclear foci were present (Fig. 4B). Similar features were observed in cells expressing CAGRAN without the MS2 tag (SI Appendix, Fig. S4 B and C).
Fig. 4.
RNA–RAN protein aggregates disrupt nucleocytoplasmic transport and induce cell death. A and B, Representative immunofluorescence micrographs showing Lamin (A) or RanGAP1 (B) in cells expressing CAGRAN or CAGFOCI for the stated times (Left). Quantification of cells with deformed nuclei (A, Right) and discontinuous RanGAP1 staining (B, Right). Each data point represents an independent experiment (≥3 experiments) evaluating ≥40 cells. C, Representative micrographs of NLS-tdTomato-NES nuclear transport reporter in fixed cells expressing CAGRAN or CAGFOCI for the stated times (Left), and corresponding quantification of the nuclear to cytoplasmic tdTomato fluorescence (Right). Each data point represents a single cell and data are summarized as the median ± interquartile range, n ≥ 40 cells. Significance values were calculated by Mann–Whitney U tests. D, Quantification of cell populations expressing CAGRAN or CAGFOCI over 5 d, normalized to population size at day 0 (immediately prior to induction). Each data point is the mean of three biological replicates, and error bars denote SD. E, Quantification of cell death caused by expression of CAGRAN or CAGFOCI over 3 d, as measured by trypan blue staining. F, Quantification of cell populations expressing CAGRAN with varying number of CAG repeats 5 d post-induction. Cell counts are normalized to uninduced controls. Each data point in E and F represents a separate biological replicate, n ≥ 3. In A, B, E, and F, data are summarized as mean ± SD and significance values were calculated by Student’s t tests. Where indicated, cell nuclei are stained with DAPI. (Scale bars, 10 µm.)
We also assessed nuclear import/export dynamics using a fluorescent reporter tagged with both a nuclear localization sequence and a nuclear export sequence that shuttles between the nucleus and the cytoplasm (27). Cells exhibiting RNA foci did not exhibit measurable changes in the nuclear-to-cytoplasmic distribution of this reporter as compared with uninduced controls. In contrast, in cells expressing CAGRAN, a significantly higher fraction of the reporter accumulated in the cytoplasm (Fig. 4C). Again, this change in distribution was not observed at earlier timepoints in cells expressing CAGRAN when we do not observe substantial accumulation of RAN translation products and cytoplasmic RNA aggregates (Fig. 4C).
Long-term expression of CAGRAN caused substantial cell toxicity (69 ± 5% decrease in cell number over 5 d, mean ± SD), while expression of CAGFOCI had no significant effect on cell proliferation compared with uninduced controls (Fig. 4 D and E). A similar effect on cell viability was observed in cells expressing CAGRAN without the MS2 tag (SI Appendix, Fig. S4 D and E). Cell toxicity increased with an increasing number of CAG repeats (Fig. 4F) at comparable expression levels (SI Appendix, Fig. S4F) and a construct with the same upstream flanking sequence and downstream MS2 tag as CAGRAN but only 5×CAG did not induce detectable cell death (Fig. 4F). In summary, these results demonstrate that accumulation of RAN translation products correlates with the disruption of nucleocytoplasmic transport and is substantially more toxic to cells than retention of the CAG repeat-containing RNA at nuclear foci.
Translation Inhibition Prevents Repeat-Containing RNA-Mediated Cell Toxicity.
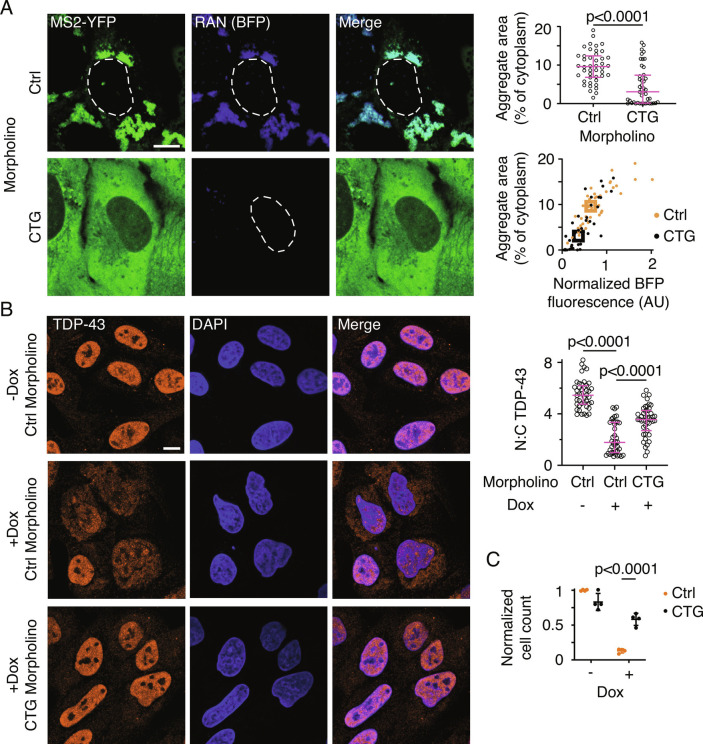
We tested whether targeted translation inhibition of CAG-repeat-containing RNA could rescue cell toxicity. We designed a phosphorodiamidate backbone 8×CTG morpholino that can hybridize with the CAG-repeats. Morpholinos sterically inhibit protein translation without affecting the stability of the target RNA (28). Pretreatment with this 8×CTG morpholino suppressed translation and aggregation of CAGRAN RNA (reduction by 43% and 71% in RAN translation and RNA aggregation respectively compared with treatment with control morpholinos, P < 0.001 by Mann–Whitney test, n ≥ 40 cells) while only modestly affecting RNA levels (Fig. 5A and SI Appendix, Fig. S5A). Morpholino treatment also prevented TDP-43 mislocalization and deformation of the nuclear envelope and significantly reduced cell death (Fig. 5 B and C and SI Appendix, Fig. S5B). The 8×CTG morpholino hybridizes with the CAG repeats and may directly perturb its clustering. Previous studies have shown that targeting morpholinos to regions upstream of the coding sequence occludes the assembly of translation initiation complexes and thus inhibits protein production (29). Since we do not know the precise translation initiation site(s) for CAGRAN, we designed another morpholino that targets a region close to the 5′ end of the transcript. Treatment with this upstream-region targeting morpholino also markedly inhibited RAN translation, cytoplasmic RNA aggregation, and cell toxicity (SI Appendix, Fig. S5 C and D). These results reinforce that RAN translation is substantially more toxic to cells than the expression of CAG repeat-containing RNA alone and that suppressing translation of the repeat-containing RNA is sufficient to mitigate cytoplasmic RNA aggregation and the key cellular defects observed in disease.
Fig. 5.
Inhibition of RAN translation prevents cell toxicity and nuclear transport defects. A, Representative micrographs of cells expressing CAGRANBFP for 24 h and treated with 50 µM of a control (Ctrl) or 8×CTG morpholino (CTG) for 48 h (Left). MS2–YFP images are independently scaled. Quantification of aggregate area (Right, Top) and in relation to BFP fluorescence (Right, Bottom). Data are representative of ≥3 independent experiments and ≥40 cells. B, Representative immunofluorescence micrographs showing TDP-43 localization in cells treated with indicated morpholinos for 48 h, with (+Dox) or without (−Dox) CAGRAN induction (Left), and corresponding quantification of the ratio of nuclear to cytoplasmic TDP-43 (Right). Cell nuclei are stained with DAPI. Data are representative of ≥40 cells. C, Quantification of cell survival 4 d post CAGRAN induction after treatment with the indicated morpholinos. Each data point represents a separate biological replicate, n = 4. Each data point in A and B represents a single cell; bar graph data are summarized as median ± interquartile range and boxes in the scatterplot in A represent the median values for each population. Significance values in A and B were calculated by Mann–Whitney U tests. Data in C are summarized as mean ± SD and the significance value was calculated by Student’s t test. (Scale bars, 10 µm.)
Discussion
Nuclear foci of repeat-containing RNA were first observed in myotonic dystrophy (30) and have been subsequently described in several repeat expansion diseases (5). We demonstrate that, in addition to aggregating in the nucleus, CAG repeat-containing RNAs also aggregate in the cytoplasm. Using two similar constructs, one that accumulates at nuclear foci, and another that is exported and aggregates in the cytoplasm, we have been able to tease apart their effects on cellular pathology. Cytoplasmic localization of a noncoding CAG repeat-containing RNA correlates with its RAN translation. The RAN translation products coaggregate with the repeat-containing RNA, and these cytoplasmic RNA–RAN protein inclusions are the sites for the accumulation of several RNA-binding proteins as well as markers for protein aggregation. Cytoplasmic aggregation of CAG repeat-containing RNA may provide a mechanistic explanation for the aberrant cytoplasmic localization of various RNA-binding proteins such as FUS and TDP-43 in these diseases (18–22).
Why have these cytoplasmic RNA inclusions not been observed in previous studies on RNA localization in CAG repeat expansion diseases? We found that a commonly used chemical cross-linking fixative, formaldehyde, reduced the detection of RNA at cytoplasmic aggregates by FISH. While both nuclear foci and cytoplasmic aggregates of CAGRAN RNA were observed in methanol-fixed cells, only nuclear foci were detected in cells that were fixed with formaldehyde (SI Appendix, Fig. S6). Even in cells where RNA detection by MS2-YFP revealed abundant cytoplasmic inclusions of MS2 hairpin-tagged CAG-repeat-containing RNA, detection of cytoplasmic RNA aggregates by FISH was substantially impeded after formaldehyde fixation (SI Appendix, Fig. S6). Chemical cross-linking of proteins and RNA may prevent probes from accessing the RNA at inclusions. Formaldehyde treatment can also result in the addition of mono-methylol groups to RNA bases—particularly to adenine and cytosine bases—and obscure RNA detection via FISH (31).
Aberrant cytoplasmic localization and aggregation of RNA-binding proteins is commonplace in nucleotide repeat expansion diseases (1, 18–22). Our work raises the possibility that these cytoplasmic inclusions of RNA-binding proteins may also contain the corresponding repeat-containing RNA. Consistent with this notion, cytoplasmic “foci” of repeat-containing RNA have been reported in myotonic dystrophy (32, 33), C9orf72-linked FTD/ALS (34–37), and fragile X-related tremor/ataxia syndrome (FXTAS) (38). In several cases, these cytoplasmic RNA foci colocalize with RNA-binding proteins. Mizielinska et al. reported that in C9orf72 ALS/FTD patients, foci of sense and antisense GGGGCC repeat-containing RNA colocalize with both p62 (marker for RAN peptides) and TDP-43 inclusions, albeit in a small proportion of cells (35). In FXTAS, staining with a G-quadruplex-specific antibody revealed the aggregation of CGG-repeat-containing RNA (which forms G-quadruplexes) with the cognate polyglycine RAN translation products, and these inclusions recruited numerous RNA-binding proteins in an RNA-dependent manner (38). In SCA10, (caused by an intronic ATTCT repeat expansion) the cytoplasmic aggregates of repeat-containing RNA colocalize with hnRNP K (39, 40). Whether such cytoplasmic RNA aggregates form when the repeats are canonically translated remains to be seen. Coaggregation of RNA and proteins may also be a common feature of protein aggregation disorders that do not involve repeat expansions. Early studies on amyloid beta plaques in Alzheimer’s disease showed that these aggregates stain with acridine orange, an RNA-binding dye (41), and a recent study found that pathogenic tau aggregates contain RNA (42). Modifications in tissue storage and fixation methods to retain RNA integrity and advancements in detection technologies may allow one to determine the true prevalence of RNA aggregates in neurodegenerative disease.
Our data also demonstrate that RAN translation of CAG repeat-containing RNAs is more toxic to cells than the accumulation of these RNAs at nuclear foci. RNAs that coalesced exclusively at nuclear foci did not induce substantial cytotoxicity in our cell culture model, and inhibiting RAN translation in cells where repeat-containing RNA was exported to the cytoplasm, was sufficient to mitigate cell death. Cytoplasmic RNA aggregation accompanies RAN translation and currently, we are not able to tease out the relative contributions of RNA and protein aggregates to the cellular pathology and toxicity. Cell toxicity may result from proteotoxic stress induced by aggregation of RAN peptides, RNA aggregation, or a combination of these factors. RNA–RAN protein aggregates could also sequester essential proteins and RNAs and produce indirect deficits that disrupt crucial cellular functions (43). Additionally, translation of repeat-containing RNAs may induce stress via ribosomal collisions (44). Future investigations may uncover the molecular basis of RNA–RAN protein coaggregation and decouple their roles in mediating disease.
Our findings, based upon a tissue culture model, support the notion that sequestration of CAG repeat-containing RNAs in the nucleus at foci may be protective to cells, and if so, may provide a therapeutic intervention strategy to reduce disease burden. These findings are similar to observations in fly models of ALS/FTD where arginine-containing dipeptide repeat products of RAN translation are more toxic than expression of GGGGCC repeat-containing RNA that exclusively forms foci (45, 46). Previous studies showed that overexpression of MBNL1 may increase RNA retention in the nucleus and consequently inhibit RAN translation and cell death (47, 48). Genetic screens have also identified several RNA export-factors such as NXF1 and NXT1 as well as nuclear pore components as potential regulators of repeat expansion induced toxicity (36, 49). However, we cannot exclude the possibility that prolonged accumulation of CAG repeat-containing RNA at nuclear foci also interferes with cell function, as has been observed in myotonic dystrophy where nuclear foci of CUG/CCUG-repeat-containing RNA are associated with various splicing defects (4).
Our results highlight the role of sequence context surrounding the repeat tract as a potential disease modifier. Previous studies have shown that the surrounding sequence context influences CAG-repeat instability (50) and polyglutamine protein aggregation propensity (51). We show that flanking sequences can also influence cytoplasmic RNA localization and RAN translation. CAG repeats in their native sequence context in ATXN3, ATXN8, and HTT genes undergo RAN translation (3). In our small-scale screen, we tested flanking sequences corresponding to the regions upstream of CTG repeats in DMPK (associated with myotonic dystrophy type 1), upstream of CCTG repeats in CNBP (associated with myotonic dystrophy type 2), and downstream of CAG repeats in HTT gene, but these sequences did not produce RAN translation products. In contrast, we identified several nonnative or synthetic sequences that potently induced RAN translation (see Materials and Methods). At this time, we are unable to identify features of the upstream sequences that mediate cytoplasmic localization and RAN translation; expansion of our screening approach to a larger library or mutational studies may help to reveal the responsible sequence motifs. We speculate that the sequences that cause cytoplasmic localization and RAN translation may load certain RNA-binding proteins that facilitate this process. For example, RNA helicases such as DDX3X and serine/arginine-rich-family proteins have been proposed to modulate RNA export and RAN translation (49, 52, 53). Once in the cytoplasm, the repeat-containing RNA may also activate the double-stranded RNA-dependent protein kinase (PKR) pathway, which potently stimulates RAN translation (54). Another possibility, which is not mutually exclusive, is that the upstream sequences together with CAG repeats may form RNA secondary structures that result in read-through of the stop codons adjacent to the CAG repeats (55, 56). The availability of synthetic models that recapitulate RAN translation, such as those described here, may help elucidate the molecular basis for this noncanonical translation process.
Our work also reveals several crucial differences between nuclear and cytoplasmic RNA aggregation. We previously showed that disease-associated repeat expansions in RNA provide sites for multivalent intermolecular base-pairing, which promote RNA phase separation and foci formation in the nucleus (6). The same repeat sequence in the cytoplasm does not appear to form aggregates until it is translated, suggesting that the repeat-containing RNA in the cytoplasm is initially in a state that inhibits extensive intermolecular RNA–RNA interactions. The process of RAN translation may relieve this repression. The RAN translation products themselves are aggregation-prone, and another possibility is that the repeat-containing RNA may cluster cotranslationally with the RAN-translated proteins. The smaller RNA–RAN protein aggregates are deposited, possibly by the minus-end-directed motor dynein, into large perinuclear, aggresome-like inclusions (57). These cytoplasmic inclusions sequester various RNA-binding proteins, disrupt nuclear morphology, and over time, cause cell death. Future work may reveal the factors involved in repeat-RNA export, RAN translation, and the mechanism of RNA–RAN-protein coaggregation.
Materials and Methods
Experimental Model and Subject Details.
U-2OS cells (ATCC, HTB-96) stably expressing a TetOn3G transactivator protein and an MS2 hairpin-binding protein fused to YFP were previously described (6). HEK293T (ATCC, CRL-3216) cells were used for generating lentivirus. ATCC authenticated cell lines by STR profiling. Cells were grown in DMEM (Life Technologies, 11965-126) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, 26140-079) and 1× penicillin, streptomycin, and glutamine (PSG, Gibco, 10378016). Transgene expression, wherever indicated, was induced by adding 1 µg mL−1 doxycycline (Sigma-Aldrich, D9891) for 24 h except where otherwise specified. Cells were maintained in a humidified incubator at 37 °C supplemented with 5% CO2.
Cloning and Plasmid Preparation.
Complete plasmid sequences are provided in SI Appendix, Table S1. Lentiviral transfer plasmids with 47× and 240×CAG-repeats were previously described (6). Lentiviral packaging and envelope constructs were obtained from Addgene: pCMV-VSV-G was a gift from Bob Weinberg (Addgene plasmid # 8454; https://www.addgene.org/8454/; RRID: Addgene_8454) (58); psPAX2 was a gift from Didier Trono (Addgene plasmid # 12260; https://www.addgene.org/12260/; RRID: Addgene_12260). Plasmids containing CAG repeats with endogenous flanking sequences from ATXN3, ATXN8, and HTT were generously provided by Laura Ranum (3).
The library of 240×CAG repeat-containing plasmids with variable flanking sequences was generated as follows. Double-stranded DNA oligonucleotides (~300 bases) were purchased from Quintara Biosciences. These DNA fragments were inserted in plasmids with 47× or 240×CAG repeats between EcoRI and MluI sites using standard restriction digestion and ligation procedures. To construct variants of CAGRAN with 5× or 22× CAG repeats, we purchased CAG repeat-containing single-stranded DNA (Integrated DNA Technologies, IDT), annealed and incorporated them between EcoRI and SgrDI sites downstream of the flanking sequence in CAGRAN. To construct CAGRANBFP, EBFP2 was obtained as a double-stranded DNA fragment (IDT) and was cloned between BamHI and NotI sites in CAGRAN. All cloning and plasmid preparations were performed in Stbl3 Escherichia coli cells (Invitrogen, C7373-03) grown at 30 °C. Since repeat number can spontaneously change during the cloning process, for each construct we verified the repeat tract in two ways. One, we optimized a Sanger sequencing protocol (in collaboration with Quintara Biosciences), which used betaine and 7-deaza-dGTP. This optimized sequencing protocol provided ~800 base long reads from each end. In constructs with 240×CAG repeats, Sanger sequencing did not provide sufficient read length to unambiguously determine the number of repeats, so the repeat number was verified by examining the size of the insert after restriction digestion and gel electrophoresis. Sanger sequencing revealed eight unintended interruptions in the CAG repeat track in our constructs with 240×CAG repeats (sequences in Dataset S1). These sites contained a deletion of G nucleotide and the first interruption occurred at 42 bases from the start of the repeat tract. These interruptions were present in our parent plasmid, and were common to all 240×CAG repeat-containing constructs examined in this study (CAGRAN, CAGFOCI, and related constructs). We observed similar phenotypes (e.g., toxicity, cytoplasmic RNA aggregation, and RAN translation) in constructs containing these repeat interruptions or corresponding constructs with uninterrupted 47×CAG repeats.
Transfections and Lentiviral Transductions.
Lentivirus was generated using second-generation lentiviral packaging system in HEK293T cells. In brief, ~500,000 HEK293T cells were plated in a 6-well plate until they reached 60 to 80% confluence. Each well was transfected with 0.5 µg envelope plasmid (Addgene #8454), 1 µg packaging plasmid (Addgene #12260), and 2 µg of the transfer plasmid with 8 µL Lipofectamine LTX (Invitrogen, 15338-100) in 500 µL of Opti-MEM reduced serum media (Gibco, 31985-070). This solution was incubated for 5 min before pipetting onto HEK293T cells in IMDM (Gibco, 12440-053) + 10% FBS + 1× PSG (Gibco, 10378016). After 3 d of incubation, virus was collected by spinning the supernatant for 5 min at 20,000g. U-2OS cells were transduced with varying titers of virus in 10 µg/mL polybrene (Millipore, TR-1003-G). For transient transfections in U-2OS cells, 500 ng plasmid was mixed with Opti-MEM reduced serum media and 1.5 µL Viafect transfection reagent (Promega, E4981) for a total volume of 50 µL. 10 µL of this mixture was added per well of U-2OS cells grown in a 96-well glass bottom plate (Brooks, MGB096-1-2-LG-L) with 100 µL fresh media.
Transgene Copy Number and Expression Quantification.
Genomic DNA and total RNA were collected from cells using PureLink Genomic DNA mini kits (Invitrogen, K1820-01) and PureLink RNA mini kits (Invitrogen, 12183018A), respectively, according to the manufacturer’s protocols. RNA was converted to cDNA using SuperScript III reverse transcriptase and Vilo Master Mix with random primers (LifeTechnologies, 11754050). Transgene genomic DNA or cDNA copies were quantified using real-time quantitative PCR (RT-qPCR) with SYBR green reagents (Applied Biosystems, 4309155) using primers targeting the TetPromoter for DNA and targeting the WPRE for cDNA (SI Appendix, Table S1). To estimate the transgene copy number, data were normalized to ACTB (SI Appendix, Fig. S1E and Fig. 4C, primers listed in SI Appendix, Table S1).
Toxicity Assays.
Cell toxicity was quantified using two methods: 1) counting the number of adhered cells and 2) assessing the exclusion of trypan blue dye. 1) Cells were plated on a six-well plate at 10,000 cells per well. The following day, cells were induced with 1 µg mL−1 doxycycline or left as uninduced controls in a total volume of 2.5 mL cell culture medium. Each cell line was compared with a corresponding uninduced control to account for any differences in plating density between cell lines. Five days post induction (unless otherwise specified), cells were washed five times with DPBS to remove dead cells from the plate, dissociated from the plate using trypsin, and neutralized with growth medium (DMEM + 10% (v/v) FBS + 1% (v/v) PSG) two times in order to collect all cells from the plate. The cells were then pelleted at 500× g for 3 min and resuspended in growth medium. Cells were counted using a Countess II FL automated cell counter (Invitrogen, AMQAF1000) and disposable cell counting chamber slides (Invitrogen, C10283). At least two technical replicate counts were performed for each of at least three biological replicates.
2) Trypan blue assays were adapted from published protocols (59). Cells were plated on a six-well plate and induced with doxycycline the following day. Five days after induction, the supernatant which contains any dead or detached cells was collected. Cells were washed with DPBS, and this wash solution was added to the supernatant. After trypsinization, cells were collected in growth medium and were added to the supernatant/wash solutions. Cells were pelleted at 500× g for 3 min, and pellets were resuspended in 100 µL DPBS. This resuspension was mixed 1:1 with 0.4% trypan blue (Invitrogen, T10282) for 3 min, and cell counts were performed using a Countess II FL automated cell counter as described above. At least two technical replicate counts were performed for each sample.
Fluorescence Microscopy.
Cells were plated in a glass-bottom 96-well plate (Brooks, MGB096-1-2-LG-L) and imaged using a Dragonfly 505 spinning-disk confocal microscope (Andor Technologies) equipped with a piezo Z-stage (ASI) and an iXon Ultra 888 EMCCD camera. Pin-hole size was kept at 40 μm. Z-stacks were acquired with a step size of 0.3 to 0.5 µm. Live cells were imaged in a humidified chamber (OKO labs) maintained at 37 °C and 5% (v/v) CO2 using a 100× oil immersion objective NA 1.45 (Nikon, MRD01905) (pixel size 121 nm × 121 nm). Fixed cells were imaged at room temperature. BFP and DAPI were excited with a 405-nm laser, and fluorescence was collected using a 445/46 bandpass filter. YFP and Alexa488 were imaged using a 488-nm laser and corresponding 521/38-nm band pass emission filter. Cy3 or mCherry-labeled samples were imaged using a 561-nm excitation and a 594/43 emission filter, and Cy5 or Atto647N-labeled samples were imaged using 640-nm laser line and a 698/77 bandpass emission filter. At least 40 cells were imaged in at least two independent experiments.
Image Analysis and Aggregate Quantification.
All image analysis was performed using FIJI (60). For quantification of fluorescence intensities, background fluorescence was determined from the average signal of 10 regions of interest (approximately 50 μm2) that did not contain any cells. This background signal was determined for each channel in each experiment and subtracted prior to intensity quantification. For characterization of cytoplasmic/nuclear inclusions, cells were manually segmented to nuclei and cytoplasm. To identify inclusions, we empirically determined the intensity and size threshold that adequately recapitulated the features that were observed by manual inspection. We tested several thresholding parameters, and obtained comparable results.
The parameters used for data presented in the paper are described below. Cytoplasmic inclusions for MS2–YFP images were identified as objects with 2× the mean intensity of the cytoplasm and a size threshold of 0.1 μm2. A similar thresholding was used for identifying cytoplasmic inclusions by RNA FISH, after appropriate adjustments for the background fluorescence. We classified cells with at least 2% of their cytoplasm occupied by inclusions as having cytoplasmic aggregates. Likewise, nuclear foci were identified as having intensity 1.5-fold the median nucleoplasm intensity and a size threshold at 0.14 μm2. We classified cells with at least 1% of their nucleoplasm occupied by inclusions as having nuclear foci. In cases where transient transfection was used, we set the size threshold for aggregates as 0.1 μm2 and set an intensity threshold at 4× the mean cytoplasmic intensity after background subtraction. We classified cells with at least 0.2% of their cytoplasm occupied by inclusions as having cytoplasmic aggregates. To quantify total RNA levels by FISH, average background signal was measured in the nucleus and cytoplasm of uninduced cells to account for differences in background staining between nucleus and cytoplasm. The integrated intensity of each compartment after background subtraction was used to estimate the relative transgene RNA levels.
In immunofluorescence experiments to determine the nuclear or cytoplasmic protein ratios, we manually segmented the cells (based on DAPI to identify nuclei and faint background immunofluorescence signal to identify cell body). Nuclear to cytoplasmic intensity ratios were calculated after background subtraction as the ratios between the mean pixel intensities in each compartment or the ratios of total integrated fluorescence intensities of the two compartments. Similar results were observed in both cases. We analyzed cells that exhibited similar fluorescence intensities under the various treatment conditions in order to preclude possible artifacts due to the differences in permeabilization and/or antibody accessibility. Nuclear envelope deformations and defective RanGAP1 localization were scored manually by visual inspection.
For colocalization analysis, line-profiles were obtained using the “Plot profile” tool in FIJI (60). Background signal was subtracted from all points and the intensity of each channel was normalized to the maximum intensity along the line. For calculating correlation coefficients, we empirically determined the intensity threshold to classify a pixel as background or foreground. The thresholded images were used to determine Pearson’s correlation coefficients of the two intensities on a per-pixel basis. Thresholds were determined using the sample image, and identical settings were used for the corresponding controls. Movies were stabilized using the Image Stabilizer plugin in FIJI (61).
FRAP Imaging and Analysis.
Regions approximately one square micron in size within cytoplasmic or nuclear inclusions were photobleached using a MicroPoint FRAP module (Andor Technologies) using a 405-nm wavelength photoablation laser. Cells were imaged for five frames prior to bleaching, immediately after the bleach (within 2 s), and then in 30 s intervals for 5 min after bleaching. After background subtraction, fluorescence of the bleached region at each timepoint was normalized to the fluorescence of the bleached region prior to photobleaching to determine fluorescence recovery. To correct for photobleaching over the course of imaging, fluorescence of the bleached region was also normalized to the fluorescence of an unbleached region of the aggregate at each timepoint. FRAP was performed at 24 h (± 3 h) after induction with doxycycline. At least 10 FRAP events from two independent experiments were analyzed for each case. The characteristic time for fluorescence recovery was determined by fitting the fluorescence to the equation: I = A−I0 exp(−t/τ), where I is the fluorescence intensity, t is the time, τ is the characteristic recovery time, and A and I0 are fitting constants.
RNA FISH.
Cells plated in 96-well glass bottom plates were fixed with a solution of 75% (v/v) methanol, 25% (v/v) acetic acid for 10 min at 4 °C. Fixed cells were washed three times with a wash solution (PBS or nuclease-free water with 300 mM NaCl, 30 mM sodium citrate, 10% (v/v) formamide, and 0.1% (v/v) NP-40 substitute) at room temperature (23 °C). Hybridization was performed at 37 °C for 3 h with 200 nM Cy3-conjugated probes targeting either the CAG repeats or the MS2 hairpin region (probe sequences SI Appendix, Table S1). The probes were dissolved in hybridization buffer (100 mg mL−1 dextran sulfate, 10% formamide, 300 mM NaCl, and 30 mM sodium citrate in nuclease-free water). After hybridization, cells were washed for 30 min with wash solution and counterstained for 30 min with wash solution containing 0.5 µg mL−1 DAPI; both this wash step and DAPI staining were performed at 37 °C. Cells were then washed three times with PBS, and kept in PBS for imaging.
Where indicated, cells were fixed with 2% (w/v) formaldehyde for 10 min, followed by a similar protocol to that described above, except that the probes were hybridized for 16 h. Detection of cytoplasmic inclusions was substantially impeded after formaldehyde treatment (see Discussion).
Immunofluorescence.
Cells were fixed with 2% formaldehyde in PBS for 40 min, washed four times with PBS, and then permeabilized with 0.1% (v/v) Triton-X-100 in PBS for 10 min at room temperature. Cells were then blocked in 0.45-µm filtered 3% (w/v) bovine serum albumin (BSA; Sigma-Aldrich, A7906) in PBS. Primary antibodies (SI Appendix, Table S1) were diluted 1:100 in 1% (w/v) BSA in PBS and incubated with cells for 1 h at room temperature. Cells were washed three times with PBS and then incubated with the appropriate secondary antibody (SI Appendix, Table S1) diluted 1:2,000 in 1% (w/v) BSA (in PBS) for 1 h at room temperature. After three washes with PBS, cells were counterstained with DAPI solution (PBS containing 0.5 µg mL−1 DAPI) for 3 min, washed three times with PBS again, and then imaged as previously described. To enhance MS2-YFP signal after fixation, cells were incubated with an anti-GFP antibody and a subsequent Alexa488-conjugated secondary antibody. These incubations were performed along with other antibodies (i.e., cells were simultaneously incubated with anti-G3BP1 and anti-GFP primary antibodies). At least two independent experiments were performed for all immunofluorescence experiments.
Dual Immunofluorescence-FISH.
Cells were fixed with 3% glyoxal (v/v) (Sigma-Aldrich, 50649) diluted in PBS for 15 min at 4 °C. Fixed cells were washed three times with PBS and then permeabilized with 0.1% Triton-X-100 (v/v) in PBS for 10 min at room temperature. Immunofluorescence was then carried out as described above with an RNase-free BSA (Sigma-Aldrich, 2930-100GM). After secondary staining, cells were refixed with 3.7% glyoxal for 10 min at 4 °C. Cells were then washed with RNA FISH wash buffer (PBS or nuclease-free water with 300 mM NaCl, 30 mM sodium citrate, 10% (v/v) formamide, and 0.1% (v/v) NP-40 substitute) at room temperature (23 °C). Hybridization was carried out with 7×CTG-Atto647 (IDT) oligonucleotide probes at 37 °C overnight in a humidified chamber. After hybridization, cells were washed for 30 min with wash solution and counterstained for 30 min with wash solution containing 0.5 µg mL−1 DAPI; both this wash step and DAPI staining were performed at 37 °C. Cells were then washed three times with PBS, and kept in PBS for imaging.
Western Blots.
Cells were washed with PBS and then lysed with RIPA lysis buffer (25 mM Tris-HCl pH = 7.5, 150 mM NaCl, 1% (v/v) NP-40, 1% (w/v) sodium deoxycholate, 0.1% (w/v) SDS) with protease and phosphatase inhibitors (Thermo Scientific, 78442). The lysate was homogenized by passing through a 22-gauge syringe 10 times and then incubated on ice for 30 min with vortexing every 5 min. Cell debris was removed by centrifugation at 500× g for 10 min at 4 °C and lysates were mixed with 4× Bolt™ lithium dodecyl sulfate (LDS) sample buffer (Invitrogen, B0007) with 50 mM DTT and heated at 70 °C for 10 min. Samples were loaded onto a Bolt™ 4 to 12% Bis-Tris polyacrylamide gel (Invitrogen, NW04122) and transferred to a PVDF membrane using iBlot 2 dry blotting system (Invitrogen, IB21001). Membranes were blocked in 5% (w/v) skim milk in tris-buffered saline with 0.1% (v/v) Tween-20 (TBST) for 1 h at room temperature. Membranes were then incubated with primary antibodies (SI Appendix, Table S1) that were diluted 1:1,000 in 1% (w/v) skim milk in TBST at 4 °C overnight. After three TBST washes, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies at a 1:10,000 dilution in 1% (w/v) skim milk in TBST for 1 h at room temperature (SI Appendix, Table S1). Membranes were washed three times with TBST and chemiluminescence signals were detected using SuperSignal West Femto Maximum sensitivity substrate (Thermo Scientific, 34095) on a ChemiDoc XRS+ imager (Bio-Rad). Band intensities were quantified using FIJI after subtracting background signal.
Translation Inhibition.
Where indicated, cells were treated with 10 µg mL−1 cycloheximide (Sigma-Aldrich, C1998). To induce transgene expression, 1 µg mL−1 doxycycline was concurrently added. Cells were then imaged as previously described 14 h post treatment. Cells transduced with a high viral titer were used in these experiments so that aggregates began to form prior to substantial cell death due to the general toxicity induced by cycloheximide treatment. Two independent experiments were performed.
Targeted translation inhibition was achieved through addition of 8×CTG morpholinos or a morpholino targeting an upstream sequence in CAGRAN (morpholino sequence “CGACGGTGGCCAGGAACCTCATAT,” GeneTools). Control experiments were performed using a standard control morpholino (CCTCTTACCTCAGTTACAATTTATA; GeneTools), targeting a mutated intron of the human beta-globin gene. In each case, 50 µM morpholino was delivered with 6 µM EndoPorter peptide (GeneTools, OT-EP-PEG-1) for 24 h prior to induction with doxycycline. Morpholinos were kept in the media after doxycycline addition giving a total treatment time of 48 h with morpholinos and 24 h of treatment with doxycycline. For toxicity assays, cells were plated at 5,000 cells/well in 24-well plates in a total volume of 300 µL. Cells were treated with 50 µM 8×CTG or control morpholinos for 5 d and with 10 ng/mL doxycycline for 4 d.
Quantification and Statistical Analyses.
Cell viability, qPCR, and quantifications of percent of cells exhibiting a phenotype (e.g., nuclear deformation, RanGAP1 discontinuities, or the presence of inclusions) were analyzed by two-tailed unpaired Student’s t tests. Morpholino rescue of cell viability was analyzed by two-way analysis of variance (ANOVA) in addition to t tests. FRAP experiments were analyzed using Wilcoxon signed-rank test. All other experiments were analyzed by Mann–Whitney U tests. All statistical tests are described in figure legends including the test used, minimum values of n, definitions of center, and precision measurements.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
FRAP of cytoplasmic RNA aggregates. Time-lapse of cytoplasmic RNA aggregate bleached at t = 0 s. Cells transduced with construct CAGRAN were induced for 24 h prior to bleaching. Arrowheads denote photobleached regions. Images are taken at 30 s intervals after bleaching. Scale bar, 10 μm.
Real-time visualization cytoplasmic RNA aggregation. Time-lapse movie of cells transduced with CAGRAN. Expression of CAGRAN construct is induced at time t = 0. Images show MS2-YFP. Images are taken at 30 min intervals after induction. Scale bar, 10 μm.
FRAP of nuclear RNA foci. Time-lapse of cytoplasmic RNA aggregate bleached at t = 0 s. Cells transduced with construct CAGRAN were induced for 2 h prior to bleaching. Arrowheads denote photobleached regions. Images are taken at 30 s intervals after bleaching. Scale bar, 10 μm.
Real-time visualization of RNA foci. Time-lapse movie of cells transduced with CAGFOCI. Expression of CAGFOCI construct is induced at time t = 0. Images show MS2-YFP. Images are taken at 30 min intervals after induction. Scale bar, 10 μm.
Real-time visualization of RAN translation and RNA aggregation. Time-lapse movie of cells transduced with CAGRAN-BFP translation reporter. Expression of CAGRAN-BFP construct is induced at time t = 0. Images show composite (cyan) of MS2-YFP (green) and BFP (blue) channels. Images are taken at 30 min intervals after induction. Scale bar, 10 μm.
Acknowledgments
We thank members of the Jain lab for helpful discussions. Plasmids containing CAG repeats with flanking sequences from ATXN3, ATXN8, and HTT were a gift from Laura Ranum. This work was supported by grants from the NIH (R00AG053434), the David and Lucile Packard Foundation, and the Smith Family Awards Program. Y.C. was supported by a fellowship from the National Research Foundation of Korea. R.D.V. is an investigator with the Howard Hughes Medical Institute.
Author contributions
M.R.D., Y.C., R.A., R.A.S., R.D.V., and A.J. designed research; M.R.D., Y.C., R.A., R.A.S., N.Z., C.P.T., and A.J. performed research; M.R.D. contributed new reagents/analytic tools; M.R.D., Y.C., R.A., and R.A.S. analyzed data; and M.R.D., R.D.V., and A.J. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Some study data available (Some reagents (cell lines, plasmids) use proprietary components and will be shared upon request under appropriate MTA.).
Supporting Information
References
- 1.Malik I., Kelley C. P., Wang E. T., Todd P. K., Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 22, 589–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulson H., “Repeat expansion diseases” in Handbook of Clinical Neurology, (Elsevier, 2018), pp. 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zu T., et al. , Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A. 108, 260–265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang H., Mankodi A., Swanson M. S., Moxley R. T., Thornton C. A., Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 13, 3079–3088 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Wojciechowska M., Krzyzosiak W. J., Cellular toxicity of expanded RNA repeats: Focus on RNA foci. Hum. Mol. Genet. 20, 3811–3821 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain A., Vale R. D., RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kearse M. G., et al. , CGG repeat-associated non-AUG translation utilizes a Cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol. Cell 62, 314–322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabet R., et al. , CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat. Commun. 9, 152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jazurek-Ciesiolka M., et al. , RAN translation of the expanded CAG repeats in the SCA3 disease context. J. Mol. Biol. 432, 166699 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Bertrand E., et al. , Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Todd P. K., et al. , CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen L., Cleary J. D., Ranum L. P. W., Repeat-associated non-ATG translation: Molecular mechanisms and contribution to neurological disease. Annu. Rev. Neurosci. 42, 227–247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heim R., Prasher D. C., Tsien R. Y., Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 91, 12501–12504 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tourrière H., et al. , The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Yu J. H., Yang W.-H., Gulick T., Bloch K. D., Bloch D. B., Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11, 1795–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma W., Mayr C., A membraneless organelle associated with the endoplasmic reticulum enables 3’UTR-mediated protein-protein interactions. Cell 175, 1492–1506.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zatloukal K., et al. , p62 is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 160, 255–263 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwab C., Arai T., Hasegawa M., Yu S., McGeer P. L., Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J. Neuropathol. Exp. Neurol. 67, 1159–1165 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Woulfe J., Gray D. A., Mackenzie I. R. A., Fus-immunoreactive intranuclear inclusions in neurodegenerative disease. Brain Pathol. 20, 589–597 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori F., FUS colocalizes with polyglutamine, but not with TDP-43 in neuronal intranuclear inclusions in spinocerebellar ataxia type 2. Neuropathol. Appl. Neurobiol. 40, 351–355 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Niss F., Zaidi W., Hallberg E., Ström A.-L., Polyglutamine expanded Ataxin-7 induces DNA damage and alters FUS localization and function. Mol. Cell. Neurosci. 110, 103584 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Yokoyama T., Ishiyama M., Hasegawa K., Uchihara T., Yagishita S., Novel neuronal cytoplasmic inclusions in a patient carrying SCA8 expansion mutation: Ribosomal aggregates in neuronal inclusions. Neuropathology 34, 27–31 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Conlon E. G., Manley J. L., RNA-binding proteins in neurodegeneration: Mechanisms in aggregate. Genes Dev. 31, 1509–1528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou C.-C., et al. , TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasset-Rosa F., et al. , Polyglutamine-expanded Huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 94, 48–57.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grima J. C., et al. , Mutant huntingtin disrupts the nuclear pore complex. Neuron 94, 93–107.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K., et al. , The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summerton J. E., Morpholino, siRNA, and S-DNA compared: Impact of structure and mechanism of action on off-target effects and sequence specificity. Curr. Top. Med. Chem. 7, 651–660 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Summerton J., Morpholino antisense oligomers: The case for an RNase H-independent structural type. Biochim. Biophys. Acta BBA -Gene Struct. Expr. 1489, 141–158 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Taneja K. L., McCurrach M., Schalling M., Housman D., Singer R. H., Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 128, 995–1002 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda N., Ohnishi T., Kawamoto S., Monden M., Okubo K., Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 27, 4436–4443 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dansithong W., et al. , Cytoplasmic CUG RNA Foci are insufficient to Elicit Key DM1 features. PLoS One 3, e3968 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones K., et al. , Reduction of toxic RNAs in myotonic dystrophies type 1 and type 2 by the RNA helicase p68/DDX5. Proc. Natl. Acad. Sci. U.S.A. 112, 8041–8045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper-Knock J., et al. , Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol. (Berl.) 130, 63–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizielinska S., et al. , C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. (Berl.) 126, 845–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S., et al. , Nuclear export and translation of circular repeat-containing intronic RNA in C9ORF72-ALS/FTD. Nat. Commun. 12, 4908 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glineburg M. R., et al. , Enhanced detection of expanded repeat mRNA foci with hybridization chain reaction. Acta Neuropathol. Commun. 9, 73 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asamitsu S., et al. , CGG repeat RNA G-quadruplexes interact with FMRpolyG to cause neuronal dysfunction in fragile X-related tremor/ataxia syndrome. Sci. Adv. 7, eabd9440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White M. C., et al. , Inactivation of hnRNP K by expanded intronic AUUCU repeat induces Apoptosis Via translocation of PKCδ to Mitochondria in Spinocerebellar Ataxia 10. PLoS Genet. 6, e1000984 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White M., et al. , Transgenic mice with SCA10 pentanucleotide repeats show motor phenotype and susceptibility to seizure: A toxic RNA gain-of-function model. J. Neurosci. Res. 90, 706–714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ginsberg S. D., Crino P. B., Lee V. M., Eberwine J. H., Trojanowski J. Q., Sequestration of RNA in Alzheimer’s disease neurofibrillary tangles and senile plaques. Ann. Neurol. 41, 200–209 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Lester E., et al. , Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron 109, 1675–1691.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melamed Z., et al. , Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 22, 180–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aviner R., et al. , Ribotoxic collisions on CAG expansions disrupt proteostasis and stress responses in Huntington’s Disease. BiorXiv (2022), 10.1101/2022.05.04.490528. [DOI] [Google Scholar]
- 45.Moens T. G., et al. , Sense and antisense RNA are not toxic in Drosophila models of C9orf72-associated ALS/FTD. Acta Neuropathol. (Berl.) 135, 445–457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran H., et al. , Differential toxicity of nuclear RNA Foci versus dipeptide repeat proteins in a Drosophila model of C9ORF72 FTD/ALS. Neuron 87, 1207–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zu T., et al. , RAN translation regulated by Muscleblind proteins in myotonic dystrophy Type 2. Neuron 95, 1292–1305.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X., et al. , Nuclear retention of full-length HTT RNA is mediated by splicing factors MBNL1 and U2AF65. Sci. Rep. 5, 12521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng W., et al. , CRISPR-Cas9 screens identify the RNA helicase DDX3X as a repressor of C9ORF72 (GGGGCC)n repeat-associated Non-AUG translation. Neuron 104, 885–898.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brock G. J., Anderson N. H., Monckton D. G., Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: Associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet. 8, 1061–1067 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Duennwald M. L., Jagadish S., Muchowski P. J., Lindquist S., Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. U.S.A. 103, 11045–11050 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linsalata A. E., et al. , DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation. EMBO Rep. 20, e47498 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malik I., et al. , SRSF protein kinase 1 modulates RAN translation and suppresses CGG repeat toxicity. EMBO Mol. Med. 13, e14163 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zu T., et al. , Metformin inhibits RAN translation through PKR pathway and mitigates disease in C9orf72 ALS/FTD mice. Proc. Natl. Acad. Sci. U.S.A. 117, 18591–18599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Firth A. E., Wills N. M., Gesteland R. F., Atkins J. F., Stimulation of stop codon readthrough: Frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 39, 6679–6691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wangen J. R., Green R., Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides. eLife 9, e52611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnston J. A., Ward C. L., Kopito R. R., Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart S. A., et al. , Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strober W., Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 21, A.3B.1–A.3B.2. (2001), 10.1002/0471142735.ima03bs21. [DOI] [PubMed] [Google Scholar]
- 60.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li K., "The image stabilizer plugin for ImageJ," http://www.cs.cmu.edu/~kangli/code/Image_Stabilizer.html (2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
FRAP of cytoplasmic RNA aggregates. Time-lapse of cytoplasmic RNA aggregate bleached at t = 0 s. Cells transduced with construct CAGRAN were induced for 24 h prior to bleaching. Arrowheads denote photobleached regions. Images are taken at 30 s intervals after bleaching. Scale bar, 10 μm.
Real-time visualization cytoplasmic RNA aggregation. Time-lapse movie of cells transduced with CAGRAN. Expression of CAGRAN construct is induced at time t = 0. Images show MS2-YFP. Images are taken at 30 min intervals after induction. Scale bar, 10 μm.
FRAP of nuclear RNA foci. Time-lapse of cytoplasmic RNA aggregate bleached at t = 0 s. Cells transduced with construct CAGRAN were induced for 2 h prior to bleaching. Arrowheads denote photobleached regions. Images are taken at 30 s intervals after bleaching. Scale bar, 10 μm.
Real-time visualization of RNA foci. Time-lapse movie of cells transduced with CAGFOCI. Expression of CAGFOCI construct is induced at time t = 0. Images show MS2-YFP. Images are taken at 30 min intervals after induction. Scale bar, 10 μm.
Real-time visualization of RAN translation and RNA aggregation. Time-lapse movie of cells transduced with CAGRAN-BFP translation reporter. Expression of CAGRAN-BFP construct is induced at time t = 0. Images show composite (cyan) of MS2-YFP (green) and BFP (blue) channels. Images are taken at 30 min intervals after induction. Scale bar, 10 μm.
Data Availability Statement
Some study data available (Some reagents (cell lines, plasmids) use proprietary components and will be shared upon request under appropriate MTA.).