Significance
This study developed a positron emission tomography (PET) imaging agent that detects pulmonary inflammation during acute lung injury. The PET radiotracer uptake was highest in lung regions with high macrophage influx, and quantification of radiotracer uptake strongly correlated with the expression of established inflammatory markers. Further, this PET tracer was able to monitor treatment response to a clinically relevant treatment, providing information beyond what would be obtained by other imaging modalities, like computed tomography, alone. This knowledge will enhance our ability to noninvasively characterize and monitor molecular patterns of inflammation to help develop precision medicine approaches to treat the acutely injured lung.
Keywords: acute lung injury, positron emission tomography, CMKLR1, macrophage, inflammation
Abstract
The lack of techniques for noninvasive imaging of inflammation has challenged precision medicine management of acute respiratory distress syndrome (ARDS). Here, we determined the potential of positron emission tomography (PET) of chemokine-like receptor-1 (CMKLR1) to monitor lung inflammation in a murine model of lipopolysaccharide-induced injury. Lung uptake of a CMKLR1-targeting radiotracer, [64Cu]NODAGA-CG34, was significantly increased in lipopolysaccharide-induced injury, correlated with the expression of multiple inflammatory markers, and reduced by dexamethasone treatment. Monocyte-derived macrophages, followed by interstitial macrophages and monocytes were the major CMKLR1-expressing leukocytes contributing to the increased tracer uptake throughout the first week of lipopolysaccharide-induced injury. The clinical relevance of CMKLR1 as a biomarker of lung inflammation in ARDS was confirmed using single-nuclei RNA-sequencing datasets which showed significant increases in CMKLR1 expression among transcriptionally distinct subsets of lung monocytes and macrophages in COVID-19 patients vs. controls. CMKLR1-targeted PET is a promising strategy to monitor the dynamics of lung inflammation and response to anti-inflammatory treatment in ARDS.
Acute lung injury (ALI) is a heterogeneous life-threatening condition which is caused by diffuse alveolar damage and manifests as acute hypoxemic respiratory failure and non-cardiogenic pulmonary edema, clinically referred to as the acute respiratory distress syndrome (ARDS). With an annual incidence of ~75 per 100,000 individuals and a mortality rate of 27 to 45%, ARDS has long been a major healthcare issue (1). This has been drastically aggravated during the recent pandemic, as ARDS occurs in ~5% of patients with COVID-19 and accounts for most of its fatalities (2).
The disruption of the alveolar-capillary barrier in ALI may occur following direct exposure to various biological, chemical, and physical hazards, or indirect injuries, e.g., sepsis (3–5). Despite the etiological heterogeneity of the triggering events, dysregulated inflammation is a critical driver of ALI/ARDS and its progression to the fibroproliferative phase (5). Therefore, detection of the inter-individual heterogeneity of the immune response plays a crucial role in understanding the pathophysiology of ALI/ARDS and promoting a precision medicine approach through predicting the clinical trajectory of disease in individual patients and monitoring their responses to various anti-inflammatory therapies (6–8).
The current clinical methods to characterize lung inflammation have significant limitations. Invasive techniques, like lung biopsy, pose significant risks to critically ill patients and are subject to sampling bias (9). Additionally, the available plasma biomarkers of inflammation are non-specific, and their alterations may be related to extra-pulmonary processes (10). Molecular imaging, by contrast, can noninvasively provide spatially-resolved information about pathological processes contributing to lung inflammation (11). Early molecular imaging studies of lung inflammation were mostly focused on [18F]fluorodeoxyglucose ([18F]FDG) positron emission tomography (PET), which relies on the detection of enhanced glucose utilization by activated leukocytes (11). However, the non-specificity of [18F]FDG uptake has prompted the development of tracers targeting more specific aspects of the immune response (12, 13). Notably, preclinical studies have shown the feasibility of targeted imaging of C-C motif chemokine receptor-2 (CCR2) (14), very late antigen-4 (VLA4) (15), CD11b (16), and folate receptor-β (17) in ALI, each depicting a different aspect of lung inflammation. However, no single molecular imaging strategy can capture the complexity of the immune response and the substantial heterogeneity of ARDS, which represents major barriers to clinical implementation of therapies targeting different aspects of disease pathogenesis, e.g., immunomodulatory/immunosuppressive or anti-fibrotic drugs, in individual patients (18). For example, while targeted imaging of CCR2 allows for the detection of ongoing influx of monocytes, CCR2 downregulation upon monocyte-to-macrophage differentiation (19, 20) may limit the utility of this approach in inflammatory conditions driven by local proliferation or sustained activation of the accumulated macrophages. Therefore, continued development and validation of tracers that target diverse aspects of the immune response are critical to establish a multipronged approach for characterizing disease endotypes, ultimately leading to the precision management of ALI/ARDS.
Chemokine-like receptor 1 (CMKLR1) is a G-protein-coupled receptor for chemerin and resolvin E1, which plays key roles in recruitment and activation of macrophages, natural killer (NK) cells, and plasmacytoid dendritic cells in inflammatory diseases across multiple organs, including atherosclerosis (21, 22), rheumatoid arthritis (23), and inflammatory bowel disease (24). With respect to the lungs, CMKLR1 has been shown to contribute to the inflammation induced by cigarette smoke (25), traffic-related particles (26), viral pneumonia (27), and ALI (28) in preclinical models. Moreover, a comprehensive single-cell transcriptomic study has recently shown the expression of CMKLR1 by a subset of monocyte-derived macrophages in patients with COVID-19 ARDS (29), supporting the clinical relevance of CMKLR1 as a biomarker of lung inflammation in ARDS.
In this study, we hypothesized that molecular imaging of CMKLR1, as a biomarker of lung inflammation, is feasible and allows for noninvasive quantitative detection of ongoing lung inflammation in an experimental model of ARDS. We describe the development and validation of a CMKLR1-targeting radiotracer, based on a recently discovered high-affinity and proteolytically stable peptidomimetic analog of the carboxyl terminus of chemerin (referred to as CG34) (30), for PET imaging of lung inflammation in a murine model of lipopolysaccharide (LPS)-induced ALI and monitoring the therapeutic response to dexamethasone, as a clinically relevant intervention in ARDS. Additionally, we establish the biological correlates of this imaging approach by determining the relationship between tracer uptake and the expression of multiple markers of lung inflammation, and by immunophenotyping of CMKLR1-expressing leukocytes contributing to tracer uptake in experimental ALI. We further characterize the kinetics of CMKLR1 expression over the course of LPS-induced ALI and compare its pattern to that of CCR2. Finally, we address the clinical relevance of CMKLR1 as a biomarker in ARDS by determining the expression of CMKLR1 in lungs of patients with COVID-19 vs. controls by secondary analysis of a single-nuclei RNA-sequencing (snRNA-seq) dataset (31).
Results
NODAGA-CG34 Is a Potent CMKLR1 Agonist.
To achieve site-specific [64Cu]Cu+2 radiolabeling of CG34, a high-affinity (i.e., low nanomolar) human CMKLR1 agonist (30), we conjugated the N-terminus of CG34 with NODAGA, a chelator with favorable in vivo pharmacokinetics, through a 6-aminohexanoic acid linker (SI Appendix, Figs. S1 and S2). To confirm that NODAGA-6-aminohexanoic acid conjugation did not impair CG34 functionality, calcium flux assays were performed in HeLa cells expressing mouse CMKLR1 (mCMKLR1) and G-protein alpha-15 (Gα15) vs. Gα15 alone (SI Appendix, Figs. S1B and S3). The potency of NODAGA-CG34 in eliciting CMKLR1-specific response was comparable to that of a high-affinity peptide derived from amino acids 145 to 157 of chemerin (Chem145–157, SI Appendix, Fig. S4) with EC50 of 45.7 vs. 42.2 nM, respectively (SI Appendix, Fig. S1C).
[64 Cu]NODAGA-CG34 Exhibits a Favorable In Vitro Profile for CMKLR1-Targeted PET.
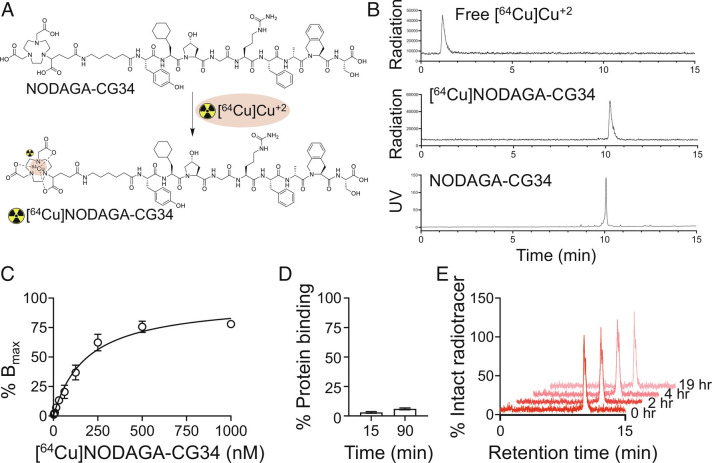
Radiolabeling of NODAGA-CG34 with [64Cu]Cu+2 consistently provided [64Cu]NODAGA-CG34 with radiochemical yields >95% at a molar activity of ~37 MBq/nmol (Fig. 1 A and B). Radioligand binding assays demonstrated a high affinity of [64Cu]NODAGA-CG34 to mCMKLR1 with a dissociation constant (Kd) of 192.1 nM (Fig. 1C), which is further enhanced by the cellular internalization of CMKLR1 agonists (32).
Fig. 1.
Radiolabeling of NODAGA-CG34 and select in vitro pharmacologic properties of [64Cu]NODAGA-CG34. (A) Radiolabeling of NODAGA-CG34 was performed by its incubation with [64Cu]CuCl2 in sodium acetate buffer containing gentisic acid at 40 °C for 30 min. (B) Radiolabeling efficiency >95% was consistently achieved as confirmed by radio-HPLC as a quality control threshold for proceeding to downstream experiments. Radio-HPLC of free [64Cu]Cu+2 (Top) has a retention time of ~1.2 min vs. [64Cu]NODAGA-CG34 (Middle) with a retention time of 10.2 min. Unlabeled NODAGA-CG34 (Lower) demonstrates a similar retention time by HPLC, as measured by UV detection, to that of [64Cu]NODAGA-CG34. (C) A saturation binding curve demonstrates specific (total minus non-specific) binding of [64Cu]NODAGA-CG34 to mCMKLR1 in HeLa cells transiently transfected with mCMKLR1 with a Kd of 192.1 ± 28.6 nM. Total and non-specific binding were conducted in the absence or presence of 2.5 µM Chem145-157, respectively. (D) The plasma protein binding of [64Cu]NODAGA-CG34 was determined by separation of bound and free [64Cu]NODAGA-CG34 using a size-exclusion column followed by γ-counting at specified timepoints. (E) Representative radio-HPLC chromatograms demonstrate high (~100%) in vitro stability of [64Cu]NODAGA-CG34 in mouse plasma at different time-points over a 19-h period. N = 3 independent experiments for all experiments, except for radiolabeling which has been performed >10 times. Data are expressed as the mean ± SEM.
[64Cu]NODAGA-CG34 is highly hydrophilic with an 1-octanol/PBS partition coefficient (logD) of −2.86 ± 0.08. Additionally, [64Cu]NODAGA-CG34 demonstrated low plasma protein binding both at 15 min (3.0% ± 0.7) and 90 min (6.0% ± 0.8) (Fig. 1D).
Radio-HPLC of [64Cu]NODAGA-CG34 after overnight incubation in radiolabeling buffer under ambient conditions did not reveal any additional radioactive peaks (SI Appendix, Fig. S5), confirming its stability to both radiolysis and radiometal chelation. Additionally, radio-HPLC of [64Cu]NODAGA-CG34 after extended incubation in mouse plasma revealed no detectable radiometabolites, indicating its excellent plasma stability (Fig. 1E).
[64Cu]NODAGA-CG34 PET Quantitatively Detects Lung Inflammation.
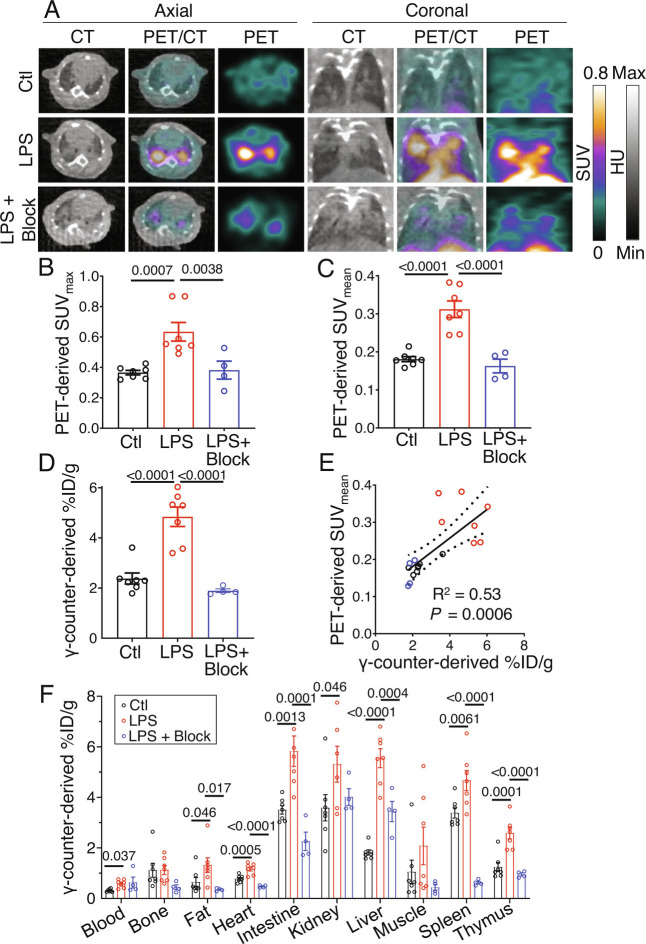
Visual assessment of PET/CT demonstrated both diffuse and distinct focal lung uptake of [64Cu]NODAGA-CG34 in regions of airspace opacification on day 2 post-induction of ALI, which were blocked by co-administration of unlabeled NODAGA-CG34 (Fig. 2A and SI Appendix, Fig. S6). Consistent with this qualitative assessment, the maximum and mean standardized uptake values (SUVmax and SUVmean) of [64Cu]NODAGA-CG34 were ~1.7-fold higher in ALI compared to control lungs (Fig. 2 B and C). A marked reduction in SUVmax and SUVmean in mice co-injected with excess unlabeled NODAGA-CG34 confirmed the in vivo specificity of [64Cu]NODAGA-CG34 uptake (Fig. 2 B and C).
Fig. 2.
[64Cu]NODAGA-CG34 PET/CT in LPS-induced lung injury. (A) Representative axial (Left) or coronal (Right) CT, PET, and co-registered PET/CT images acquired ~90 min after intravenous tracer injection from a control mouse (Top row) vs. a mouse 2 d after intratracheal LPS administration (Middle row) demonstrate markedly increased lung uptake of [64Cu]NODAGA-CG34 mostly localized to regions of airspace opacities detected by CT. Co-injection of 100-fold molar excess of non-radiolabeled NODAGA-CG34 blocks the tracer uptake in inflamed lungs (Bottom row), confirming the specificity of tracer uptake. In vivo PET-derived quantification of tracer uptake demonstrates ~73% increase in lung SUVmax (B) and ~72% increase SUVmean (C) in LPS-injured mice compared to control mice (N = three males and four females per group), which is completely blocked by co-injection of excess non-labeled NODAGA-CG34 despite CT evidence of lung injury (N = two males and two females in the blocked group). (D) Quantification of tracer uptake by γ-counting confirms a similar pattern with 103% increased [64Cu]NODAGA-CG34 uptake in experimental lung injury, compared to control lungs, which is completely blocked by co-injection of unlabeled NODAGA-CG34. (E) In vivo PET-derived (SUVmean) and ex vivo γ-counting-derived (%ID/g) measurements of [64Cu]NODAGA-CG34 uptake correlate significantly (R2 = 0.53, P = 0.0006), confirming the accuracy of noninvasive quantification of tracer uptake in lungs. (F) Biodistribution of [64Cu]NODAGA-CG34, determined by ex vivo γ-counting (%ID/g) in select organs, demonstrated systemic inflammation upon LPS-induced lung injury as evidenced by multi-organ increases in [64Cu]NODAGA-CG34 uptake. SUVmax, SUVmean, and %ID/g data in panels B–E represent the average values of the left and right lungs for each mouse. PBS = phosphate-buffered saline; LPS = lipopolysaccharide. Data are expressed as the mean ± SEM. Linear regressions are shown along with 95% CIs. Statistical significance between groups was calculated using a one-sided ANOVA with a post hoc two-tailed Fisher’s exact test. Linear correlations were determined by calculating the Pearson correlation coefficient.
Similarly, ex vivo γ-counting determined that lung uptake of [64Cu]NODAGA-CG34 was ~twofold higher in ALI mice as compared to controls and was blocked by co-injection of unlabeled NODAGA-CG34 (Fig. 2D). We verified the accuracy of PET-derived quantification of [64Cu]NODAGA-CG34 uptake by demonstrating a strong correlation between SUVmean and γ-counting measurements of [64Cu]NODAGA-CG34 uptake (R2 = 0.53, P = 0.0006) (Fig. 2E).
Consistent with prior reports of systemic inflammation associated with intratracheal LPS-induced ALI (14–16), significant increases in [64Cu]NODAGA-CG34 uptake were present in several organs (particularly liver, spleen, thymus, and intestine) in ALI mice compared to controls (Fig. 2F).
To validate the accuracy of [64Cu]NODAGA-CG34 PET as an imaging biomarker of lung inflammation, we determined the correlations between lung SUVmean and mRNA expression of inflammatory markers. ALI substantially increased the expression of multiple inflammation-associated genes, including chemokines, interleukins, and cytokines, in the lungs (SI Appendix, Fig. S7A), many of which significantly correlated with SUVmean (SI Appendix, Fig. S7B).
[64Cu]NODAGA-CG34 PET Monitors the Therapeutic Response to Dexamethasone.
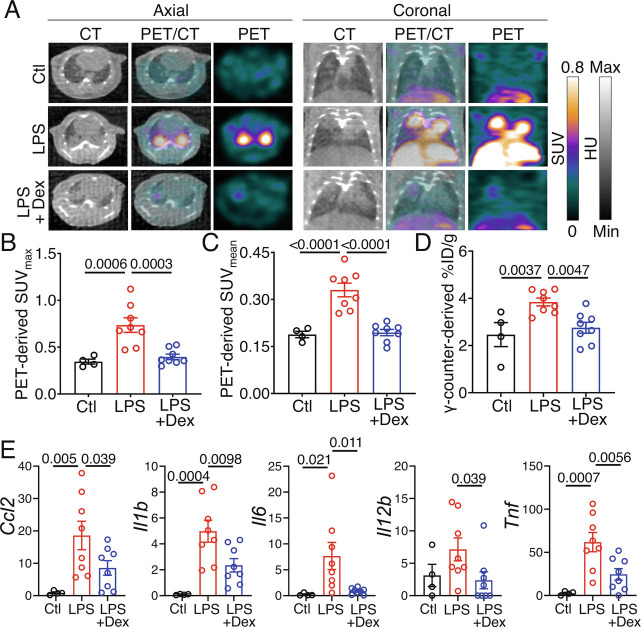
Considering the promise and accuracy of [64Cu]NODAGA-CG34 PET to quantify lung inflammation, we next determined its potential in monitoring the response to anti-inflammatory treatment by dexamethasone. Similar to our prior experiment, lung uptake of [64Cu]NODAGA-CG34 was significantly increased on day 2 post-LPS-induced ALI compared to controls, as determined by in vivo PET and ex vivo γ-counting (Fig. 3 A–D). Dexamethasone treatment (1 and 24 h post-LPS) significantly reduced lung uptake of [64Cu]NODAGA-CG34 in LPS-injured mice, as measured both globally throughout the lungs (SUVmean and %ID/g) and in the most inflamed regions (SUVmax) (Fig. 3 A–D). Notably, dexamethasone-treated ALI mice, when compared to untreated ALI mice, frequently demonstrated consolidated regions with minimal tracer uptake, highlighting the potential of [64Cu]NODAGA-CG34 PET to quantify the inflammatory burden of consolidated lungs, hence supplementing the structural information obtained by CT (Fig. 3A). Consistent with PET findings, dexamethasone significantly reduced the expression of inflammatory biomarkers in LPS-injured lungs (Fig. 3E).
Fig. 3.
Effect of dexamethasone treatment on the lung uptake of [64Cu]NODAGA-CG34 in LPS-induced experimental lung injury. (A) Representative axial (Left) and coronal (Right) CT, PET, and co-registered PET/CT on day 2 post-PBS or LPS treatment. Images were acquired ~90 min after intravenous [64Cu]NODAGA-CG34 injection in control (Top row) and LPS-treated mice after receiving (Bottom row) or not receiving (Middle row) dexamethasone. Corticosteroid treatment reduced radiotracer uptake by PET, although areas of airspace opacities were still frequently observed on CT. (B and C) In vivo PET-derived quantification of tracer uptake demonstrates ~46% decrease in lung SUVmax and ~41% decrease in SUVmean in LPS-injured mice receiving dexamethasone compared to mice treated with LPS only (N = four males and four females per group). The lung radiotracer uptake in dexamethasone-treated mice approached that of control mice (N = two males and two females for control mice). (D) Quantification of lung radiotracer uptake by γ-counting confirms a similar pattern with 28% decreased [64Cu]NODAGA-CG34 uptake in LPS-injured mice treated with dexamethasone, when compared to mice receiving only LPS. (E) The mRNA expression of select inflammatory markers is significantly reduced in dexamethasone-treated, compared to untreated, LPS-injured lungs. mRNA transcript levels are normalized to the geometric mean of Rn18s, the housekeeping gene. SUVmax and SUVmean, and %ID/g values represent the average values of the left and right lungs for each mouse. PBS = phosphate-buffered saline; LPS = lipopolysaccharide; Dex = dexamethasone. Data are expressed as the mean ± SEM. Statistical significance between groups was calculated using a one-sided ANOVA with a post hoc two-tailed Fisher’s exact test.
Increased Uptake of CMKLR1-Targeting Peptide in ALI Predominantly Represents the Expansion of CMKLR1-Expressing Macrophages and Monocytes.
We identified the major immune cells in control and LPS-injured lungs using the gating strategy provided in SI Appendix, Fig. S8. For flow cytometric identification of immune cells contributing to [64Cu]NODAGA-CG34 uptake, we utilized a fluorescently-conjugated peptide derived from an established high-affinity CMKLR1 ligand (33) and analog of CG34 derived from the natural amino acids 145 to 157 of chemerin, 6CF-Chem145–157 (SI Appendix, Fig. S9A). We first determined the high potency and specificity of 6CF-Chem145–157 to mCMKLR1 in murine peritoneal macrophages (SI Appendix, Fig. S9B). Then, we confirmed 6CF-Chem145–157 binding may serve as a proxy for [64Cu]NODAGA-CG34 uptake by demonstrating that NODAGA-CG34 displaced 6CF-Chem145–157 binding on murine peritoneal macrophages (SI Appendix, Fig. S9C). Finally, we confirmed that 6CF-Chem145–157 binding is restricted to CMKLR1-expressing cells by co-staining of peritoneal cells with 6CF-Chem145–157 and an anti-CMKLR1 antibody, which demonstrated 6CF-Chem145–157 binding is only detectable in CMKLR1-expressing macrophages (SI Appendix, Fig. S9D).
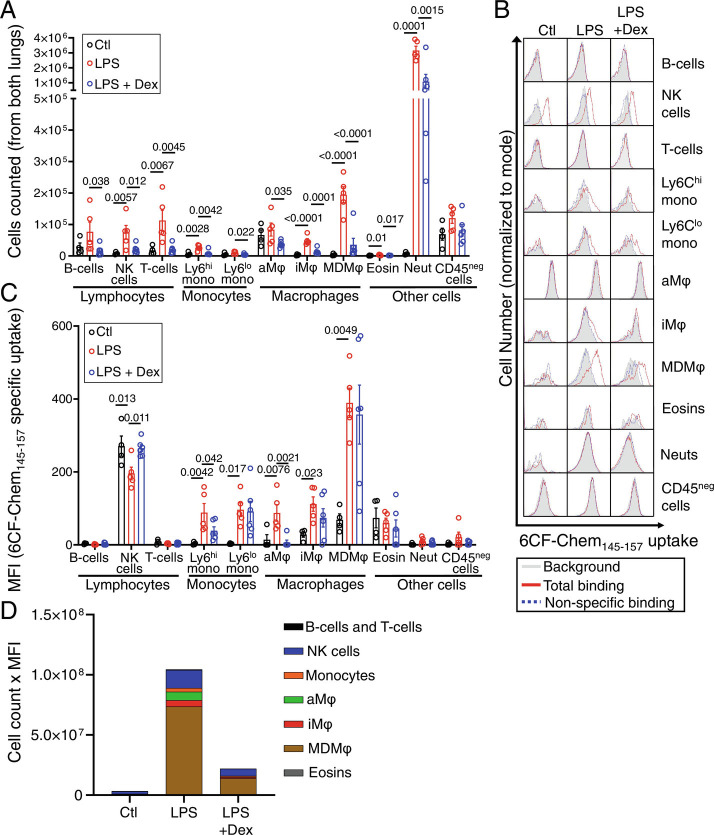
Flow cytometry of mechanically-dissociated lungs demonstrated that 2 d after LPS-induced injury most leukocyte populations, most pronouncedly neutrophils and monocyte-derived macrophages, followed by NK cells, interstitial macrophages, Ly6Chi monocytes, and lymphocytes, were increased (Fig. 4A). However, the LPS-induced increase in 6CF-Chem145–157 uptake at this timepoint was predominantly restricted to monocyte-derived macrophages, followed by modest increases among interstitial macrophages, Ly6Chi monocytes, Ly6Clo monocytes, and alveolar macrophages (Fig. 4 B and C). NK cells were the only population that demonstrated significant 6CF-Chem145–157 uptake both at steady state and following LPS-induced injury. There was no significant uptake of 6CF-Chem145–157 by neutrophils, B-cells, T-cells, and CD45neg cells. Treatment with dexamethasone significantly reduced the number of CMKLR1-expressing cells (e.g., macrophages, monocytes, and NK cells) within the LPS-injured lungs, while only decreasing 6CF-Chem145–157 uptake per cell in alveolar macrophages and Ly6Chi monocytes (Fig. 4 B and C).
Fig. 4.
Flow cytometry identification of CMKLR1 expressing cells in healthy and LPS-injured lungs. (A) The absolute number of most immune cells within the lungs is significantly increased at day 2 post-intratracheal instillation of LPS. (B) Representative histograms showing specific uptake of a CMKLR1-targeted fluorescent ligand, 6CF-Chem145–157 (100 nM), by various immune cells in the lungs of mice treated with PBS, LPS, or LPS plus dexamethasone (gray: background/autofluorescence; red: total-binding of 6CF-Chem145–157 in the absence of Chem145–157; blue: non-specific binding of 6CF-Chem145–157 in the presence of 10 µM Chem145–157). (C) Specific 6CF-Chem145–157 uptake (total minus non-specific/blocked) in different cell subsets was quantified by mean fluorescent intensity (MFI), as a proxy for the uptake of CMKLR1-targeted tracer ([64Cu]NODAGA-CG34), in LPS-injured vs. control mice. Monocyte-derived macrophages, interstitial macrophages, and monocytes (Ly6Chi and Ly6Clo) demonstrated the largest increases in 6CF-Chem145–157 uptake in LPS-induced experimental lung injury. NK cells had significant 6CF-Chem145–157 uptake during steady state, but the uptake was not further induced by lung injury. (D) A stacked-bar graph summary of the cell count multiplied by the cellular uptake (MFI) of 6CF-Chem145–157 highlights that monocyte-derived macrophages are the major contributors to the tracer uptake (~70%) due to a marked increase in their abundance and significant induction of 6CF-Chem145–157 uptake per cell in mice with LPS-induced ALI compared to control or dexamethasone-treated mice. CD45-negative cells and neutrophils are omitted from this graph due to their negligible specific 6CF-Chem145–157 uptake. N for PBS group = two male and two female mice; N for LPS group: three male and two female mice; N for LPS + dexamethasone group: three male and three female mice. aMφ: alveolar macrophages; Eosins: eosinophils; iMφ: interstitial macrophages; MDMφ: monocyte-derived macrophages; NK cells: natural killer cells; Neut: neutrophils. Data are expressed as the mean ± SEM. Statistical significance between groups was calculated using a one-sided ANOVA with a post hoc two-tailed Fisher’s exact test.
To estimate the contribution of individual CMKLR1 expressing cell types to the total lung uptake of 6CF-Chem145–157, as a proxy for the uptake of [64Cu]NODAGA-CG34 in PET imaging studies, we multiplied the number of counted cells per type by their 6CF-Chem145–157 uptake measured by specific mean fluorescence intensity. Using this measure, we confirmed that LPS-treated mice had the highest total lung uptake of 6CF-Chem145–157, while dexamethasone treatment markedly reduced 6CF-Chem145–157 uptake (Fig. 4D). By contrast, there was minimal 6CF-Chem145–157 uptake in PBS-treated mice. Importantly, we determined that monocytes and macrophages represented the majority (~85%) of total 6CF-Chem145–157 uptake on day 2 post-LPS-induced ALI which was largely driven by monocyte-derived macrophages (contributing to ~70% of the uptake) with smaller contributions from alveolar macrophages (~7%), interstitial macrophages (~5%), and monocytes (~3%). NK cells represented only ~14% of 6CF-Chem145–157 uptake in LPS-injured lungs. A similar pattern of 6CF-Chem145–157 uptake by cell type was found in dexamethasone-treated ALI mice though at a significantly reduced total uptake of 6CF-Chem145–157. By contrast, 6CF-Chem145–157 uptake was markedly lower in PBS-treated mice and was largely driven by NK cells (~60%) and alveolar macrophages (~21%), while monocyte-derived macrophages (~6%), interstitial macrophages (~2%), and monocytes (~1%) contributed minimally to the total uptake.
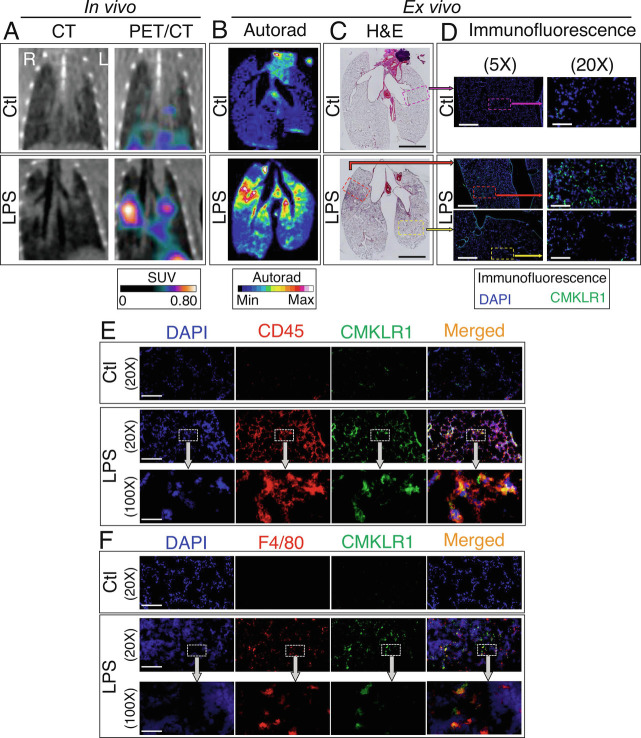
We used combined high-resolution autoradiography and histology to confirm the spatial colocalization of [64Cu]NODAGA-CG34 uptake to regions of lung inflammation and increased CMKLR1 expression. As demonstrated in Fig. 5 A and B, autoradiography revealed minimal diffuse [64Cu]NODAGA-CG34 uptake throughout the lungs in control mice. However, a marked increase in [64Cu]NODAGA-CG34 uptake was present in LPS-injured lungs manifesting as discrete foci of tracer accumulation, mostly in a peri-bronchial distribution as expected by the intratracheal delivery route of LPS, superimposed on a diffusely increased tracer uptake in the remaining parts of the lungs. Notably, the foci of high [64Cu]NODAGA-CG34 uptake colocalized with inflamed regions of lungs as determined by hematoxylin and eosin staining (Fig. 5C) and increased CMKLR1 expression (Fig. 5D). Further, we confirmed that the abundant expression of CMKLR1 in inflamed regions of LPS-injured lungs mostly co-localized with CD45 (Fig. 5E) and F4/80 (Fig. 5F), consistent with flow cytometric finding of 6CF-Chem145–157 uptake in macrophages.
Fig. 5.
Co-localization of [64Cu]NODAGA-CG34 uptake and CMKLR1 expression via PET/CT, autoradiography, and immunohistology in murine lungs. Representative PET/CT images (A) from a control mouse (Top) and day 2 post-LPS (Bottom) reformatted according to the plane of ex vivo autoradiography (B) and hematoxylin and eosin-stained tissues (C) confirm excellent colocalization of [64Cu]NODAGA-CG34 uptake with inflamed regions of the lungs. Representative low- and high-magnification immunofluorescent staining images (D) from adjacent tissue sections demonstrate the abundant expression of CMKLR1 in a highly inflamed region of the lungs in an LPS-treated mouse (red box in C) while there is minimal CMKLR1 expression in a less inflamed (yellow box in C) or non-inflamed (purple box in C) regions of lungs in the LPS- and PBS-treated mice (Scale bar in hematoxylin and eosin imaging = 5 mm, in 5× immunofluorescence images = 800 µM, and in 20× immunofluorescence images = 200 µM. N for PBS and LPS groups = two and three male mice, respectively.) Increased CMKLR1 expression in LPS-induced lung injury mostly colocalizes with (E) CD45+ leukocytes, and in particular (F) F4/80+ macrophages. (Scale bar 20× immunofluorescence images = 200 µM, and in 100× immunofluorescence images = 40 µM.) N for PBS and LPS groups = two female and one male mouse per group.
Distinct Kinetics of CMKLR1 and CCR2 Expression by Monocytes and Macrophages in ALI Allow for Monitoring of Different Aspects of Lung Inflammation by PET.
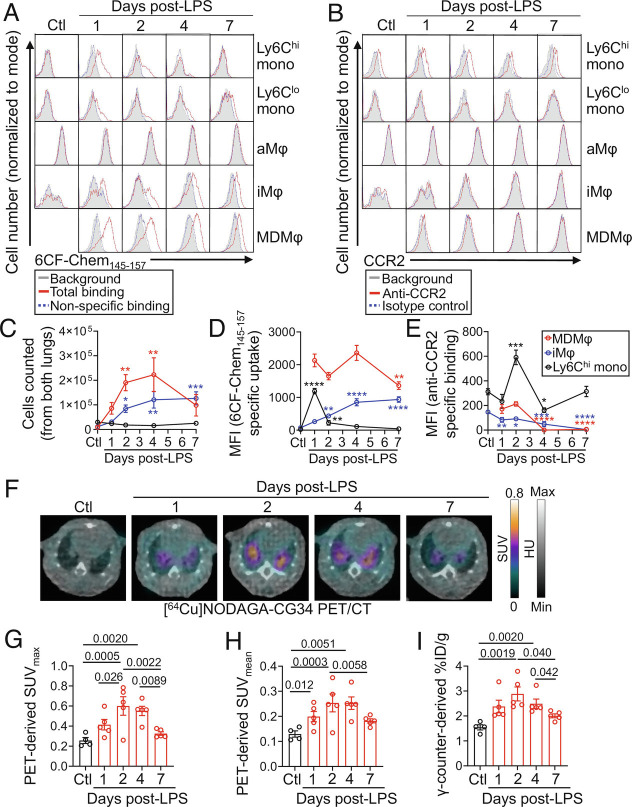
CCR2-targeted PET using [64Cu]DOTA-ECL1i has been reported to detect the early phase of lung inflammation at 24 h post-LPS administration in the murine model of ALI (14). Notably, the lung uptake of [64Cu]DOTA-ECL1i returned to the level of control lungs on days 2 and 6 post-LPS (14). To determine the potential of [64Cu]NODAGA-CG34 PET in detecting a distinct aspect of inflammation captured by [64Cu]DOTA-ECL1i PET (i.e., increased burden of recruited monocytes and macrophages vs. ongoing monocyte flux), we compared the kinetics of CCR2 and CMKLR1 expression at different timepoints after LPS administration by flow cytometry (Fig. 6 A–E). CCR2 expression was highest among Ly6Chi monocytes while it was markedly lower in interstitial and monocyte-derived macrophages particularly on days 4 and 7 post-LPS, consistent with the rapid downregulation of CCR2 upon monocyte-to-macrophage differentiation (34, 35). In contrast, 6CF-Chem145–157 uptake, as a surrogate for CMKLR1 expression, remained high in monocyte-derived macrophages throughout the first week after LPS administration with only a modest decrease on day 7 coinciding with the resolving phase of inflammation as detected by the reduced number of lung inflammatory cells (SI Appendix, Fig. S10). A progressive increase in 6CF-Chem145–157 uptake by interstitial macrophage was also noted throughout the 7-d period, while its uptake by Ly6Chi monocytes markedly dropped from day 1 to day 2 post-LPS. Notably, CCR2 and CMKLR1 expression by SiglecFhi alveolar macrophages remained negligible in control mice and throughout the first week after LPS administration. These data support distinct kinetics of CCR2 and CMKLR1 expression with CCR2 primarily expressed by undifferentiated Ly6Chi monocytes while CMKLR1 predominantly expressed upon differentiation of monocytes to macrophages.
Fig. 6.
Kinetics of CMKLR1 and CCR2 expression by monocytes and macrophages during LPS-induced ALI and time course of [64Cu]NODAGA-CG34 PET/CT. (A and B) Representative histograms showing the specific uptake of a CMKLR1-targeted fluorescent ligand, 6CF-Chem145–157 (100 nm), or expression of CCR2 in lung monocytes and macrophages throughout the course of LPS-induced ALI (gray: background/autofluorescence; red: total-binding of 6CF-Chem145–157 in the absence of Chem145–157 or binding of a CCR2 antibody; blue: non-specific binding of 6CF-Chem145–157 in the presence of 10 µM Chem145–157 or binding of isotype control antibody). Data for monocyte-derived macrophages are omitted for the control group as the cell number was very low for accurate quantification. (C) The absolute number of Ly6Chi monocytes, interstitial macrophages, and monocyte-derived macrophages at different timepoints following LPS instillation. The number of monocyte-derived macrophages peaks on days 2 and 4, whereas the number of interstitial macrophages increases through day 7. By contrast, the number of Ly6Chi monocytes remains consistently low. (D and E) Specific 6CF-Chem145–157 uptake (total minus non-specific/blocked) or anti-CCR2 binding (anti-CCR2 minus isotype control) in Ly6Chi monocytes, interstitial macrophages, and monocyte-derived macrophages following LPS-induced lung injury. Monocyte-derived macrophages maintain high 6CF-Chem145–157 uptake throughout the course of ALI, whereas interstitial macrophages demonstrate increasing 6CF-Chem145–157 uptake through day 7. Ly6Chi monocytes show high 6CF-Chem145–157 uptake only on day 1. By contrast, CCR2 expression is primarily restricted to Ly6Chi monocytes in ALI with only a transient low-level CCR2 expression by monocyte-derived macrophages during days 1 and 2 after LPS administration. (F) Representative axial co-registered [64Cu]NODAGA-CG34 PET/CT images of mice at 1, 2, 4, and 7 d following LPS-treatment or non-treated controls. (G and H) In vivo PET-derived quantification of [64Cu]NODAGA-CG34 uptake demonstrates lung SUVmax and SUVmean peaking around 2 to 4 d post-LPS followed by a decrease on day 7. (I) Quantification of lung radiotracer uptake by γ-counting confirms a similar pattern of tracer uptake to that obtained by in vivo PET. SUVmax and SUVmean, and %ID/g values represent the average values of the left and right lungs for each mouse. Ctl = untreated mice; LPS = lipopolysaccharide; mono = monocytes; aMφ = alveolar macrophages; iMφ = interstitial macrophages; MDMφ = monocyte-derived macrophages. N for panels A–E: three male and two female mice except for day 2 with N = two male and three female mice; N for panels F–I: three male and two female mice except for control group with N = two male and two female mice. Data are expressed as the mean ± SEM. For panels C–E, comparisons were made between the various treatment timepoints to the control group for Ly6Chi monocytes and interstitial macrophages or to the 1-d treatment group for monocyte-derived macrophages (as the control group is omitted). P-values: * < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001. Statistical significance was calculated using a one-sided ANOVA with a post hoc two-tailed Fisher’s exact test.
To confirm that the persistent expression of CMKLR1 by the accumulated burden of lung macrophages allows for the imaging of lung inflammation over an extended period after the induction of ALI, we performed a time course analysis of [64Cu]NODAGA-CG34 PET over 1 wk following LPS administration (Fig. 6 F–I). Interestingly, the lung uptake of [64Cu]NODAGA-CG34 closely paralleled the kinetic changes in CMKLR1 expression by monocytes and macrophages detected by flow cytometry, i.e., a persistent increase in [64Cu]NODAGA-CG34 uptake throughout the first week after LPS administration with a modest decrease on day 7. Together, our data suggest that CMKLR1- and CCR2-targeted PET detect distinct aspects of monocyte/macrophage biology in ALI in which the uptake of [64Cu]NODAGA-CG34 is primarily reflective of the accumulated burden of recruited monocyte-derived and interstitial macrophages, while [64Cu]DOTA-ECL1i uptake is largely driven by ongoing flux of monocytes during the early stage of ALI irrespective of the total burden of macrophage accumulation.
CMKLR1 Expression Is Increased in Lung Monocytes and Macrophages in COVID-19.
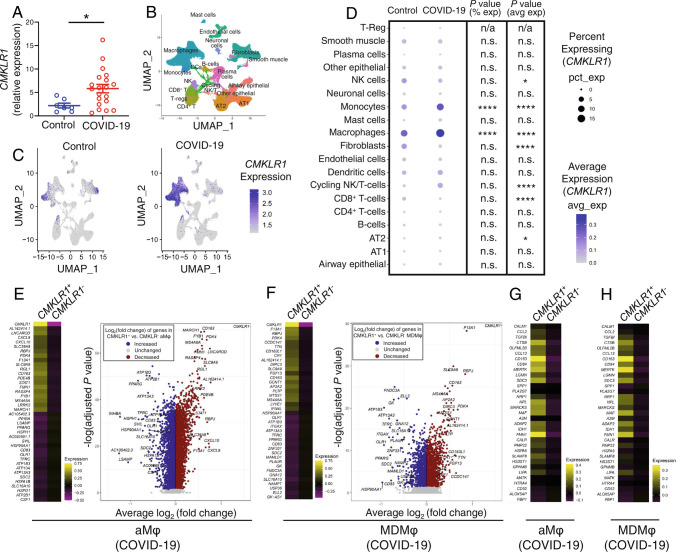
Our preclinical results support the premise of CMKLR1 as an imaging biomarker of monocyte- and macrophage-driven inflammation in an experimental model of ALI. To establish the clinical relevance of CMKLR1 as a biomarker of inflammation in ARDS, we performed a secondary analysis of a snRNA-seq dataset (31) to determine the total and cell-specific expression of CMKLR1 in autopsied lungs of patients with fatal COVID-19-induced ARDS vs. pre-pandemic controls who underwent lung resection or biopsy. CMKLR1 expression was increased nearly threefold on average in COVID-19 compared to control lungs, though there was a significant heterogeneity in CMKLR1 expression among the individual patients (Fig. 7A), supporting the presence of different inflammatory endotypes of COVID-19. Consistent with our data in the murine model, CMKLR1-expressing cells were markedly more abundant in COVID-19 compared to control lungs and were mostly clustered with monocytes and macrophages (Fig. 7 B and C). Detailed analysis across various cell types (Fig. 7D) demonstrated significant increases both in the abundance of CMKLR1-expressing monocytes and macrophages, and their expression level of CMKLR1 in COVID-19 compared to controls.
Fig. 7.
CMKLR1 expression in the lungs of patients with COVID-19-induced ARDS. (A) Pooled-cell analysis of a previous snRNA-seq dataset demonstrates a significant increase in CMKLR1 expression in the lungs of patients with COVID-19-induced ARDS as compared to non-COVID-19 controls. (B) A UMAP projection of the snRNA-seq data allowing for the identification of major lung cell subsets. (C) CMKLR1 expression clusters predominantly among the monocytes and macrophages subsets, and CMKLR1 is significantly increased in abundance and expression (log-normalized, per cell) in COVID-19 (Right) patients vs. controls (Left). (D) Dot plot representation of the average CMKLR1 expression and the percentage of CMKLR1-expressing cells in selected cell subsets. Monocytes and macrophages demonstrate the highest expression level (avg exp) of CMKLR1 and frequency (% exp) of cells expressing (avg exp) CMKLR1 both in control and COVID-19 lungs. Notably, both the frequency of CMKLR1-expressing cells and the expression level of CMKLR1 by monocytes and macrophages are increased in COVID-19 ARDS compared to control lungs. By contrast, CMKLR1 expression is negligible by other cell types, compared to that of monocytes and macrophages, and is only minimally affected by COVID-19 (E and F). Heat maps and volcano plots of the most differentially expressed genes demonstrate distinct transcriptional profiles of CMKLR1-positive and CMKLR1-negative cells among both alveolar macrophages (E) and monocyte-derived macrophages (F) in COVID-19 patients (CMKLR1 data points are excluded from the volcano plots). (G and H) Heat map representations support the transcriptional resemblance of CMKLR1-expressing alveolar macrophages (G) and monocyte-derived macrophages (H) with the previously reported profibrotic CD163/LGMN-expressing profibrotic macrophages in COVID-19 patients. The majority of transcripts reported to be upregulated in CD163/LGMN profibrotic macrophages are also overexpressed in CMKLR1-expressing cells. aMφ: alveolar macrophages; AT1: type 1 alveolar cells; AT2: type 2 alveolar cells; MDMφ: monocyte-derived macrophages; NK: natural killer cells. n.a: not applicable; n.s: not significant. Data are expressed as the mean ± SEM. P-values: * < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001. Statistical significance between two groups was calculated using a two-sided Student’s t test. The percentage of cells positive for CMKLR1 and the average CMKLR1 expression in different cell subsets were compared between COVID-19 and control groups using Fisher’s exact and Wilcoxon rank-sum tests, respectively.
A focused analysis of alveolar macrophages and monocyte-derived macrophages (the two populations with most abundant CMKLR1 expression) in COVID-19 lungs demonstrated distinct transcriptional profiles of CMKLR1-positive and CMKLR1-negative cells, as summarized by heat maps and volcano plots of the most differentially expressed genes (Fig. 7 E and F). Interestingly, the transcriptional profiles of CMKLR1-expressing alveolar macrophages and monocyte-derived macrophages closely resembled those of a recently discovered subset of profibrotic CD163/LGMN-expressing lung macrophages in COVID-19 and idiopathic pulmonary fibrosis (29), as confirmed by the overexpression of a significant number of genes upregulated in these cells (Fig. 7 G and H).
To confirm these results, we analyzed an independent publicly available scRNA-seq dataset which demonstrated the increased expression of CMKLR1 in macrophages obtained from three different sites within the respiratory tract (bronchoalveolar lavage fluid, nasopharynx, and airways) of patients with COVID-19 vs. control subjects (SI Appendix, Fig. S11) (36–38). Similarly, using another publicly available scRNA-seq dataset (38, 39), we demonstrated increased expression of CMKLR1 in macrophages obtained from bronchoalveolar fluid of patients with cystic fibrosis. The expression of CMKLR1 was mostly restricted to two clusters of FOLR2+ and SPP1+ interstitial macrophages which were expanded in cystic fibrosis patients compared to control subjects (SI Appendix, Fig. S12). Finally, we sought to explore CMKLR1 as a macrophage marker in non-pulmonary tissues. Using a fourth independent scRNA-seq dataset (38, 40), we confirmed that CMKLR1 expression in other tissues, including spleen, thymus, liver, and fat, was largely restricted to macrophages (SI Appendix, Fig. S13). These results collectively suggest a broader clinical relevance of CMKLR1 as an imaging biomarker of inflammatory diseases of the lung and other organs.
Discussion
In this study, we demonstrated the accuracy of CMKLR1-targeted PET with [64Cu]NODAGA-CG34 to quantify lung inflammation in a preclinical model of ALI, as validated by strong correlations between radiotracer uptake and expression of biologically relevant inflammatory biomarkers (5), and to monitor the therapeutic response to a clinically relevant anti-inflammatory intervention, i.e., dexamethasone. Additionally, we established the biological basis of [64Cu]NODAGA-CG34 uptake by identifying a marked increase in the abundance of CMKLR1-expressing immune cells in inflamed lungs, which were mostly comprised of monocyte-derived macrophages and to a lesser extent interstitial macrophages and monocytes. Further, we showed that the sustained expression of CMKLR1 on monocyte-derived and interstitial macrophages over the course of LPS-induced ALI represented a distinct pattern than that of CCR2, which was transiently expressed by undifferentiated monocytes in the early stages of ALI. Finally, we confirmed the clinical relevance of CMKLR1, as a biomarker of lung inflammation and a potential target for molecular imaging, by demonstrating significant increases in the abundance and expression of CMKLR1 in transcriptionally distinct subsets of lung monocytes and macrophages in patients with COVID-19, as an example of ARDS associated with a major healthcare burden, and in patients with cystic fibrosis.
Disease endotyping is critical for precision medicine-driven management of ARDS (18, 41). In other inflammatory lung diseases, e.g., asthma and chronic obstructive pulmonary disease, identification of clinically measurable endotypes has enabled successful clinical trials and subsequent approval of treatments specifically for patients with certain molecular patterns of inflammation (42, 43). Although ARDS endotypes exist (41, 44), our limited mechanistic understanding of relevant ARDS biomarkers and molecular pathways along with limited noninvasive approaches for assessment of lung inflammation remain major barriers to endotype ARDS and evaluate targeted immunomodulatory therapies in the clinical setting and clinical trials (18, 41).
Molecular imaging is a promising approach for ARDS endotyping through noninvasive characterization of specific patterns of inflammation. The most extensively studied radiotracer, [18F]FDG, detects pulmonary inflammation in both preclinical and clinical settings (13, 45), but poorly predicts disease course or treatment response (45–47). Considering the non-specificity of [18F]FDG (13), its use is limited as a precision medicine tool in inflammatory lung diseases, and there is a growing interest in developing radiotracers for specific inflammatory biomarkers. Previously tested tracers targeting CCR2 (14), VLA4 (15), CD11b (16), and folate receptor-β (17) have successfully imaged lung inflammation in preclinical models of ALI, though each delineates a selected aspect of the immune response. For example, CCR2 is a promising imaging biomarker for the detection of ongoing influx of classical monocytes to inflamed tissues (14, 48, 49). However, CCR2-targeted PET may not accurately reflect the total burden of tissue macrophages in inflammatory conditions driven by prolonged survival and/or proliferation of tissue macrophages rather than monocyte influx, as CCR2 expression rapidly downregulates upon differentiation of monocytes to macrophages (19, 20). Supporting this scenario, the uptake of a 64Cu-DOTA-ECL1i, a CCR2-targeting tracer, in a murine model of LPS-induced ALI is only detectable in the first 24 h after induction of ALI and returns to the uptake of uninjured lung by days 2 and 6 post-injury (14), the timeframe which corresponds to the maximal accumulation of monocytes and macrophages in this model. On the other hand, folate receptor-β expression is mostly restricted to lung interstitial macrophages compared to monocytes and alveolar macrophages (50, 51). Unlike CCR2 and folate receptor-β, VLA-4 (52) and CD11b (53) have a broader expression across various leukocyte subsets, hence their targeted imaging likely reflects broader, though less specific, aspects of the inflammatory response. Therefore, the complexity and heterogeneity of the inflammatory processes underlying ALI cannot be captured through a single molecular imaging approach, which necessitates the development of a multi-pronged diagnostic approach to delineate various complementary aspects of lung inflammation, paralleling similar efforts for development of therapeutics affecting different inflammatory pathways.
The chemerin-CMKLR1 axis is emerging as a major contributor to monocyte and macrophage recruitment in multiple preclinical models of lung inflammation. For example, the activation of the chemerin-CMKLR1 axis in LPS-induced lung injury increased macrophage migration, while decreasing neutrophil influx and pro-inflammatory cytokine production in the lungs (28). Also, functional CMKLR1 signaling provided a survival benefit along with decreased complications in a murine viral pneumonia model (27). Furthermore, CMKLR1 activation improved viral clearance and production of anti-viral antibodies and decreased the expression of pro-inflammatory mediators (27, 28). CMKLR1 may also influence the innate and adaptive immune responses upon environmental exposures and its deficiency markedly reduces leukocyte recruitment to inflamed lungs (25, 26). While clinical data are still lacking, these preclinical findings highlight the potential of CMKLR1 as a mechanistically relevant biomarker of lung inflammation in a variety of pulmonary diseases.
We are unaware of prior reports using CMKLR1-targeted imaging in inflammatory diseases. To address this gap, we developed [64Cu]NODAGA-CG34 and demonstrated its favorable pharmacodynamics and pharmacokinetic properties for PET, including high affinity at CMKLR1 and receptor-mediated internalization (32), facile site-specific radiolabeling at high molar activity, low plasma protein binding, excellent plasma stability, and fast blood clearance. Consistent with the prior report of a DOTA-conjugated CG34, the incorporation of unnatural amino acids in the CG34 peptide (30) provided excellent stability against proteolytic degradation compared to natural chemerin-derived peptides. To take advantage of its superior radiometal chelation properties for radiolabeling with [64Cu]Cu+2, lower susceptibility to trans-chelation, and higher in vivo stability compared to DOTA (54–56), we used NODAGA as the radiotracer chelating moiety instead of a recently reported DOTA-conjugate (30). NODAGA-based PET tracers, in particular, are readily radiolabeled under mild conditions without the need for post-synthesis radiochemical purification (5, 54, 57). Further, we selected copper-64 as the radiolabeling isotope due to its advantages over other radiometals, such as gallium-68, including its higher spatial resolution and its longer physical half-life which obviates the need for on-site production (58), allowing for its cost-effective nationwide availability (59).
PET/CT showed spatially heterogeneous [64Cu]NODA GA-CG34 uptake in experimental lung injury with focal regions of intense uptake, corresponding to areas of airspace opacites determined by CT, superimposed on relatively diffusely increased uptake, when compared to control lungs. Therefore, we quantified SUVmax and SUVmean to better capture the extent of tracer uptake in the most inflamed regions and the global burden of inflammation, respectively, which both revealed highly reproducible increased tracer uptake in LPS-injured vs. control lungs. The lung uptake of [64Cu]NODAGA-CG34, as measured with γ-counting-derived %ID/g, was quantitatively comparable to that of other pulmonary molecular imaging agents (14, 16, 49). Despite the known difficulties of quantification of tracer uptake in the lungs (13), strong correlations between in vivo (PET-derived SUVmean) and ex vivo (γ-counting-derived %ID/g) measures of [64Cu]NODAGA-CG34 uptake support the accuracy of our approach to noninvasively measure tracer uptake. Encouragingly, in vivo radiotracer uptake also correlated with lung expression of multiple pathogenically relevant and therapeutically targetable biomarkers of lung injury (5, 8), such as Il1b, Il6, and Tnf, highlighting the accuracy of CMKLR1-targeted PET to serve as a surrogate noninvasive marker of lung inflammation.
To highlight the translational potential of CMKLR1-targeted PET to noninvasively monitor lung inflammation during treatment, we performed [64Cu]NODAGA-CG34 PET/CT after dexamethasone treatment in LPS-injured mice, a clinically relevant therapy for ARDS and other inflammatory conditions (8). Dexamethasone treatment significantly reduced lung radiotracer uptake (SUVmax and SUVmean) almost to the control levels, paralleling a significant downregulation of several key pro-inflammatory mediators. Interestingly, PET/CT of LPS-injured mice treated with dexamethasone frequently showed focal airspace opacities (i.e., consolidation) by CT with little [64Cu]NODAGA-CG34 uptake compared to mice not treated with dexamethasone. This finding highlights the potential of [64Cu]NODAGA-CG34 PET to provide biological information beyond what can be achieved by traditional radiographic methods such as CT, which are limited to the detection of anatomical/structural changes.
A key biological validation for [64Cu]NODAGA-CG34 PET was establishing the cells contributing to its uptake in LPS-induced lung injury. Little is known about leukocyte subtypes that express CMKLR1 in steady-state and inflamed lungs, although data from other organs, including spleen (28) and blood (24), have shown the expression of CMKLR1 by monocytes/macrophages, NK cells, and dendritic cells. We performed immunophenotyping of lungs cells which bind to 6CF-Chem145–157, as a proxy for [64Cu]NODAGA-CG34 uptake and CMKLR1 expression, after establishing its competitive receptor-binding profile with NODAGA-CG34 uptake and CMKLR1-specificity. 6CF-Chem145–157 uptake was predominantly restricted to macrophages, monocytes, and NK cells, with minimal binding to other immune and non-immune cells. Notably, recruited monocyte-derived macrophages accounted for nearly 70% of the observed 6CF-Chem145–157 uptake in the lungs on day 2 post-LPS-induced injury driven by a combination of increased cell count and per cell 6CF-Chem145–157 uptake. By contrast, NK cells contributed only 14% to the total lung uptake of 6CF-Chem145–157 in LPS injury. Interestingly, 6CF-Chem145–157 flow cytometry data support that dexamethasone-induced decline in [64Cu]NODAGA-CG34 uptake in lung injury is primarily driven by reducing the influx of CMKLR1-expressing leukocytes with smaller contributions of reduced CMKLR1 expression per cell in alveolar macrophages and Ly6Chi monocytes.
To establish the potential of [64Cu]NODAGA-CG34 PET to capture a distinct aspect of monocyte/macrophage-driven inflammation, we compared the kinetic changes in CMKLR1 and CCR2 expression by monocytes and macrophages along with a time course analysis of [64Cu]NODAGA-CG34 uptake over 1 wk after the induction of ALI. We demonstrated that CCR2 expression by Ly6Chi monocytes is downregulated upon monocyte-to-macrophage differentiation, which could explain the previously reported rapid decrease in the lung uptake of [64Cu]DOTA-ECL1i as early as 2 d after LPS-induced ALI (14). In contrast, CMKLR1 expression and [64Cu]NODAGA-CG34 uptake remained high in monocyte-derived and interstitial macrophages during the first 4 d after the induction of ALI and only modestly declined by monocyte-derived macrophages on day 7, which coincides with the resolution phases of inflammation as detected by the decreased number of infiltrated lung leukocytes. Collectively, our data support that distinct kinetics of CMKLR1 and CCR2 expression by monocytes and macrophages allows for monitoring different aspects of lung inflammation by [64Cu]NODAGA-CG34 PET (total burden of recruited macrophages and monocytes) vs. [64Cu]DOTA-ECL1i PET (ongoing flux of monocytes).
Despite strong preclinical evidence demonstrating the importance of CMKLR1 to the pathophysiology of inflammation, little is known clinically about its role or utility as a biomarker in ARDS (24). Here, we showed a significant increase in CMKLR1 expression in COVID-19 ARDS, which consistent with our findings in the mouse model was mostly due to the accumulation of CMKLR1-overexpressing macrophages and monocytes. The replacement of homeostatic-resident macrophages by monocyte-derived macrophages, which retain their pro-inflammatory phenotype long after the resolution of injury, is a critical pathophysiological process which may drive long-lasting pathogenic remodeling following the resolution of the initial lung injury (60, 61). Therefore, developing tools to monitor the accumulation of hematopoietic-derived lung macrophages may play a major role in ARDS precision medicine by providing insights into its long-term consequences, particularly through molecular imaging of lung macrophage ontogeny via CMKLR1.
Interestingly, a recent snRNA-seq study in an independent dataset has reported that COVID-19 is associated with the expansion of a subset of profibrotic CD163/LGMN-expressing monocyte-derived macrophages which overexpress CMKLR1 and transcriptionally resemble a similar subset of profibrotic macrophages expanded in interstitial lung diseases (29). Consistent with this report, CMKLR1-expressing monocyte-derived and alveolar macrophages in our dataset shared significant transcriptional similarities with CD163/LGMN-expressing macrophages, suggesting the potential of our imaging approach to selectively target a population of macrophages previously identified in fibroproliferative ARDS and other fibrotic interstitial lung diseases. We therefore propose that CMKLR1 serves as a promising biomarker for COVID-19 as a clinical example of viral pneumonia-induced ARDS with a major healthcare concern. Given the heterogenous immunologic response and clinical course of patients with COVID-19 or other causes of ARDS, future studies should explore CMKLR1-targed PET for molecular endotyping of lung inflammation.
There are several limitations of this study. First, no experimental model fully recapitulates the pathogenesis of ARDS in humans. However, LPS-induced lung injury is the most extensively validated experimental model of ALI and shares important features with ARDS (62). Moreover, we established the potential clinical relevance of our data using three independent COVID-19 RNA-seq datasets. Second, while our data indicated that CMKLR1 expression in the murine model of ALI was predominantly restricted to monocyte-derived macrophages and interstitial macrophages with only a low level of expression by alveolar macrophages, the human datasets demonstrated increased CMKLR1 expression by the airspace macrophages of patients with COVID-19. Recent work has highlighted that distinct subsets of monocyte-like cells and resident macrophages contribute to the diversity of the alveolar macrophage pool in both health and disease in humans (63–65). However, our study could not discern the ontogeny of the expanded pool of CMKLR1-expressing alveolar macrophages in COVID-19. Therefore, it remains to be determined if CMKLR1-expressing alveolar macrophages in COVID-19 represent monocyte-derived macrophages which have acquired a close transcriptional resemblance to tissue-resident alveolar macrophages after an extended period of residence in the lungs, as reported in mice (60), and/or embryonically-derived alveolar macrophages which are induced to express CMKLR1 upon lung inflammation. Third, it remains to be determined if CMKLR1 expression plays a mechanistic role in the pathogenesis of lung injury in addition to its role as a biomarker of lung inflammation. However, the primary aim of this study was to validate the utility of CMKLR1 as an imaging biomarker for precision management of ALI by monitoring lung inflammation and response to anti-inflammatory therapy irrespective of its mechanistical contribution to the pathogenesis of the disease. Finally, the imaging components of this study were only performed in animals, as no FDA approval currently exists for this radiotracer. However, the promising results of the current study open the possibility for future New Drug Application (NDA) and conduct of first-in-human PET studies.
In summary, our study demonstrates CMKLR1-targeted PET with [64Cu]NODAGA-CG34 as a promising approach for molecular endotyping of lung inflammation and monitoring the therapeutic response through detection of a distinct subset of monocytes and macrophages. We speculate that [64Cu]NODAGA-CG34 PET may provide similar promising results in the detection of monocyte- and macrophage-driven inflammation in other lung diseases and monitoring response to selective immunomodulatory therapeutics. We envision CMKLR1-targeted PET as an emerging precision medicine tool facilitating the endotyping of inflammatory lung diseases. Finally, considering the growing recognition of the pathogenic roles of macrophages in many inflammatory processes (66) and relative specificity of CMKLR1 expression by macrophages across different organs, we anticipate that CMKLR1 may emerge as a potential biomarker of inflammation beyond diseases of the respiratory system.
Materials and Methods
Additional details on materials and methods are provided in SI Appendix.
Chemicals and Reagents.
The major chemicals and reagents (SI Appendix, Table S1), plasmids (SI Appendix, Table S2), HPLC methods (SI Appendix, Table S3), flow cytometry reagents (SI Appendix, Table S4), histology antibodies (SI Appendix, Table S5), Taqman primers (SI Appendix, Table S6), and details of publicly available single-cell RNA sequencing datasets (SI Appendix, Table S7) are listed in the Supplementary Information. Chemical characterization of NODAGA-CG34 is shown in SI Appendix, Fig. S2.
Cell Culture.
HeLa cells transiently transfected with mouse CMKLR1 and/or Gα15 cDNA plasmids were used for calcium flux or radioligand binding assays. Resident peritoneal cells harvested from C57BL/6J mice via peritoneal lavage (67) were used to determine the binding/uptake of 6CF-Chem145–157 by flow cytometry.
Radiolabeling.
Radiolabeling of NODAGA-CG34 was performed in 0.5 M NaOAc buffer (pH = 6.9 and 0.8 mM gentisic acid dissolved in water) by adding [64Cu]CuCl2 in 0.1 M HCl (37 MBq per 1.0 nmol of NODAGA-CG34) and incubation at 40 °C for 30 min. The radiochemical purity of [64Cu]NODAGA-CG34 was determined by radio-HPLC to confirm a minimum purity of >95% prior to use in all in vitro (measurement of octanol/water partition coefficient (logD), radiolysis and plasma stability assays, and plasma protein binding assay) and in vivo (PET/CT) experiments. Relevant radio-HPLC methods are described in SI Appendix, Table S3.
Mouse Model of Experimental Lung Injury.
Animal experiments were performed on C57BL/6J mice under a protocol approved by the University of Pittsburgh Institutional Animal Care and Use Committee. Adult 9- to 12-wk-old C57BL/6J mice were used throughout the study. Mice were administered intratracheally with 2.5 µg/g LPS from Escherichia coli O111:B4 in 60 µL of phosphate-buffered saline (PBS) to induce lung injury (15). Control mice were intratracheally injected with 60 µL of PBS. In experiments involving anti-inflammatory treatment, mice receiving dexamethasone were given two doses of 10 mg/kg (in 500 µL in PBS) via intraperitoneal injections at 1 and 24 h following LPS instillation, whereas control mice were treated with intraperitoneal PBS (500 µL) injections.
PET/CT and Quantification of [64Cu]NODAGA-CG34 Uptake.
Mice were injected intravenously with [64Cu]NODAGA-CG34 (6.41 ± 0.05 MBq). Tracer specificity was addressed by co-injection of [64Cu]NODAGA-CG34 and 100-fold molar excess of non-radiolabeled NODAGA-CG34. Static PET (~10-min) and CT (180 projections, 140-ms exposure, 180° rotation, 80-kVp, 500-µA, field-of-view: 78.5 × 100-mm) were performed, according to our previous experiments (15) (Inveon, Siemens), 90 min after [64Cu]NODAGA-CG34 injection.
Regions of interest were drawn over the left and right lungs, and the uptake in the left and right lungs of [64Cu]NODAGA-CG34 was averaged and quantified as the mean and maximal standardized uptake value (SUVmean and SUVmax) (IRW software). Biodistribution was performed by γ-counting (Wizard2, PerkinElmer) of harvested organs. Data are reported as percentage of injected dose per gram tissue (%ID/g) after decay correction. Multiplanar reformats of PET/CT images were performed (Vivoquant software) to reconstruct planes matching the autoradiography and histology images.
Gene Expression Assays.
Following PET/CT, the lungs of mice were harvested and stored frozen for >10 half-lives of [64Cu]NODAGA-CG34 prior to use for gene expression assays using TaqMan primers according to standard methods (15). All transcript levels were normalized to the expression level of 18S ribosomal RNA (Rn18s).
Flow Cytometric Immunophenotyping of Murine Lungs.
Mechanically dissociated cells from murine lungs were incubated in the absence or presence of 6CF-Chem145–157 (100 nM) with or without co-incubation with Chem145–157 (10 µM) at 37 °C for 1 h. After washing with PBS and blocking of non-specific binding using 1% BSA and Fc block, a mixture of antibodies and DAPI was added for 0.5 h at 4 °C. Cells were then washed and fixed for flow cytometry.
Lung Immunostaining.
Murine lung cryosections were incubated with primary antibodies (1:200, 4 °C, overnight) prior to incubation with fluorescently-conjugated secondary antibodies (1:200, 4 °C, 2 h) and mounting.
snRNA-Seq of COVID-19 and Control Lungs.
A secondary analysis of a previously published snRNA-seq dataset from the autopsied lungs of 19 patients with lethal COVID-19 and lung tissue (biopsy or lung resection) from seven pre-pandemic control patients without COVID-19 was performed to determine the expression of CMKLR1 among different cell types in the lungs. The RNA sequencing data are available in the Gene Expression Omnibus (GEO) database under accession number GSE171524. Data collection, analysis, and UMAP identification of major cell populations were conducted as previously reported (31). Differential gene expression was performed using the Seurat (v4.1.0) (68).
Analysis of Additional Publicly Available Human scRNA-Seq Datasets.
CMKLR1 expression by different immune cells were determined across different organs through secondary analysis of separate independently published scRNA-seq datasets (GEO accession numbers GSE145926, GSE193782, and GSE201333, or European Genome-Phenome Archive accession number EGAS00001004481) using the UCSC Cell Browser software (available at: https://cells.ucsc.edu/) (36–40).
Statistical Analysis.
Statistical analysis was performed using Prism 9 (GraphPad). Data are presented as mean ± SEM. A Student’s t test was performed to compare the means values between two groups. One-way analysis of variance, followed by Fisher's exact post hoc test, was used to compare mean values in >2 groups. Pearson’s test was used to determine the correlations between continuous variables. For snRNA-seq data, the percentage of cells positive for CMKLR1 and the average CMKLR1 expression in different cell subsets were compared between COVID-19 and control groups using Fisher’s exact and Wilcoxon rank-sum tests, respectively. Statistical significance was considered as P < 0.05.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
This study was supported by the following grants: National Institute of Biomedical Imaging and Bioengineering (NIBIB, R21EB027871) and National Heart, Lung, and Blood Institute (NHLBI, K08HL144911) to ST; NHLBI (F30HL158038) to PZM; NHLBI (R01HL136143, P01HL114453, R01HL142084, and K24HL143285) to JSL; and National Cancer Institute (NCI, K08CA222663, R21CA263381, and R37CA258829), Burroughs Wellcome Fund (Career Award for Medical Scientists), and Fast Grant for COVID-19 research to BI. This work utilized the Hillman Cancer Center In Vivo Imaging Facility, a shared resource at the University of Pittsburgh (supported by P30CA047904). We would like to thank Raymond Yurko and Kazi Islam in the University of Pittsburgh Peptide and Peptoid Synthesis Core for their contributions synthesizing and characterizing the peptides used in this paper. Additionally, we would like to thank the Mass Spectrometry Laboratory (the Department of Chemistry), Flow Cytometry Core Facility, and the Center for Biologic Imaging (funded by S10MH126905) at the University of Pittsburgh.
Author contributions
P.Z.M., F.S.V., J.S.L., and S.T. designed research; P.Z.M., C.E.B., J.D.L., K.E.D., Q.Z., and Z.X. performed research; B.I. contributed new reagents/analytic tools; P.Z.M., C.E.B., J.B., M.T., J.R.N., C.J.A., J.S.L., and S.T. analyzed data; and P.Z.M., C.E.B., J.B., B.I., C.J.A., J.S.L., and S.T. wrote the paper.
Competing interest
The authors declare competing interest. The authors have additional information to disclose, B.I. has received consulting fees from Volastra Therapeutics Inc, Merck, AstraZeneca and Janssen Pharmaceuticals and has received research funding to Columbia University from Alkermes, Arcus Biosciences, Checkmate Pharmaceuticals, Compugen, Immunocore, and Synthekine. J.S.L. is a consultant for Johnson & Johnson/Janssen. None of these represents a conflict of interest pertaining to the presented work. All other authors declare they have no competing interests.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
COVID-19 snRNA-seq data were previously deposited in GEO with the accession number GSE171524 and are also available through the single-cell portal: https://singlecell.broadinstitute.org/single_cell/study/SCP1219. Additional scRNA-seq datasets in COVID-19 and cystic fibrosis patients were previously deposited (GEO accession numbers GSE145926 and GSE193782) and publicly available through UCSC cell browser (available at: https://cells.ucsc.edu/). The other data needed to evaluate the conclusions of the paper are present in the paper and/or the SI Appendix.
Supporting Information
References
- 1.Villar J., Blanco J., Kacmarek R. M., Current incidence and outcome of the acute respiratory distress syndrome. Curr. Opin. Crit. Care 22, 1–6 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Pfortmueller C. A., Spinetti T., Urman R. D., Luedi M. M., Schefold J. C., COVID-19-associated acute respiratory distress syndrome (CARDS): Current knowledge on pathophysiology and ICU treatment - A narrative review. Best Pract. Res. Clin. Anaesthesiol. 35, 351–368 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson B. T., Chambers R. C., Liu K. D., Acute respiratory distress syndrome. N Engl. J. Med. 377, 562–572 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Rubenfeld G. D., et al. , Incidence and outcomes of acute lung injury. N Engl. J. Med. 353, 1685–1693 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Matthay M. A., et al. , Acute respiratory distress syndrome. Nat. Rev. Dis. Primers 5, 18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beitler J. R., et al. , Advancing precision medicine for acute respiratory distress syndrome. Lancet Respir. Med. 10, 107–120 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Famous K. R., et al. , Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am. J. Respir. Crit. Care Med. 195, 331–338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calfee C. S., et al. , Subphenotypes in acute respiratory distress syndrome: Latent class analysis of data from two randomised controlled trials. Lancet Respir. Med. 2, 611–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnabel R. M., et al. , Clinical course and complications following diagnostic bronchoalveolar lavage in critically ill mechanically ventilated patients. BMC Pulm. Med. 15, 107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Laorden M. I., Lorente J. A., Flores C., Slutsky A. S., Villar J., Biomarkers for the acute respiratory distress syndrome: How to make the diagnosis more precise. Ann. Transl. Med. 5, 283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pourfathi M., Kadlecek S. J., Chatterjee S., Rizi R. R., Metabolic imaging and biological assessment: Platforms to evaluate acute lung injury and inflammation. Front. Physiol. 11, 937 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mannes P. Z., Tavakoli S., Imaging immunometabolism in atherosclerosis. J. Nucl. Med. 62, 896–902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D. L., et al. , Quantification of lung PET images: Challenges and opportunities. J. Nucl. Med. 58, 201–207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., et al. , PET-based imaging of chemokine receptor 2 in experimental and disease-related lung inflammation. Radiology 283, 758–768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haddad J., et al. , Molecular imaging of very late antigen-4 in acute lung injury. J. Nucl. Med. 62, 280–286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Q., Huang Q., Mohan C., Li C., Small-animal PET/CT imaging of local and systemic immune response using (64)Cu-alphaCD11b. J. Nucl. Med. 60, 1317–1324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han W., et al. , Molecular imaging of folate receptor beta-positive macrophages during acute lung inflammation. Am. J. Respir. Cell Mol. Biol. 53, 50–59 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bos L. D. J., et al. , Precision medicine in acute respiratory distress syndrome: Workshop report and recommendations for future research. Eur. Respir. Rev. 30, 200317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fantuzzi L., et al. , Loss of CCR2 expression and functional response to monocyte chemotactic protein (MCP-1) during the differentiation of human monocytes: Role of secreted MCP-1 in the regulation of the chemotactic response. Blood 94, 875–883 (1999). [PubMed] [Google Scholar]
- 20.Phillips R. J., Lutz M., Premack B., Differential signaling mechanisms regulate expression of CC chemokine receptor-2 during monocyte maturation. J. Inflamm. 2, 14 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Vorst E. P. C., et al. , Hematopoietic chemR23 (chemerin receptor 23) fuels atherosclerosis by sustaining an M1 macrophage-phenotype and guidance of plasmacytoid dendritic cells to murine lesions-brief report. Arterioscler. Thromb. Vasc. Biol. 39, 685–693 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Zylla S., et al. , Association of circulating chemerin with subclinical parameters of atherosclerosis: Results of a population-based study. Arterioscler. Thromb. Vasc. Biol. 38, 1656–1664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko K., et al. , Chemerin activates fibroblast-like synoviocytes in patients with rheumatoid arthritis. Arthritis Res. Ther. 13, R158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trilleaud C., et al. , Agonist anti-ChemR23 mAb reduces tissue neutrophil accumulation and triggers chronic inflammation resolution. Sci. Adv. 7, 1453 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demoor T., et al. , The role of ChemR23 in the induction and resolution of cigarette smoke-induced inflammation. J. Immunol. 186, 5457–5467 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Provoost S., et al. , Pro- and anti-inflammatory role of chemR23 signaling in pollutant-induced inflammatory lung responses. J. Immunol. 196, 1882–1890 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Bondue B., et al. , ChemR23 dampens lung inflammation and enhances anti-viral immunity in a mouse model of acute viral pneumonia. PLoS Pathog. 7, e1002358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luangsay S., et al. , Mouse ChemR23 is expressed in dendritic cell subsets and macrophages, and mediates an anti-inflammatory activity of chemerin in a lung disease model. J. Immunol. 183, 6489–6499 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Wendisch D., et al. , SARS-CoV-2 infection triggers profibrotic macrophage responses and lung fibrosis. Cell 184, 6243–6261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erdmann S., et al. , CMKLR1-targeting peptide tracers for PET/MR imaging of breast cancer. Theranostics 9, 6719–6733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melms J. C., et al. , A molecular single-cell lung atlas of lethal COVID-19. Nature 595, 114–119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J. X., et al. , Chemerin C9 peptide induces receptor internalization through a clathrin-independent pathway. Acta Pharmacol. Sin. 35, 653–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy A. J., et al. , Chemerin elicits potent constrictor actions via chemokine-like receptor 1 (CMKLR1), not G-protein-coupled receptor 1 (GPR1), in human and rat vasculature. J. Am. Heart Assoc. 5, e004421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Opalek J. M., Ali N. A., Lobb J. M., Hunter M. G., Marsh C. B., Alveolar macrophages lack CCR2 expression and do not migrate to CCL2. J. Inflamm. 4, 19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y. R., et al. , A protocol for the comprehensive flow cytometric analysis of immune cells in normal and inflamed murine non-lymphoid tissues. PLoS One 11, e0150606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao M., et al. , Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Chua R. L., et al. , COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 38, 970–979 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Speir M. L., et al. , UCSC cell browser: Visualize your single-cell data. Bioinformatics 37, 4578–4580 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X., et al. , ScRNA-seq expression of IFI27 and APOC2 identifies four alveolar macrophage superclusters in healthy BALF. Life Sci. Alliance 5, e202201458 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabula Sapiens C., et al. , The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reilly J. P., Calfee C. S., Christie J. D., Acute respiratory distress syndrome phenotypes. Semin Respir. Crit. Care Med. 40, 19–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuruvilla M. E., Lee F. E., Lee G. B., Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin. Rev. Allergy Immunol. 56, 219–233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnes P. J., Inflammatory endotypes in COPD. Allergy 74, 1249–1256 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Ackermann M., et al. , Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl. J. Med. 383, 120–128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D. L., et al. , [18F]fluorodeoxyglucose positron emission tomography for lung antiinflammatory response evaluation. Am. J. Respir. Crit. Care Med. 180, 533–539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scherer P. M., Chen D. L., Imaging pulmonary inflammation. J. Nucl. Med. 57, 1764–1770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones H. A., Marino P. S., Shakur B. H., Morrell N. W., In vivo assessment of lung inflammatory cell activity in patients with COPD and asthma. Eur. Respir. J. 21, 567–573 (2003). [DOI] [PubMed] [Google Scholar]
- 48.English S. J., et al. , CCR2 Positron emission tomography for the assessment of abdominal aortic aneurysm inflammation and rupture prediction. Circ. Cardiovasc. Imaging 13, e009889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brody S. L., et al. , Chemokine receptor 2-targeted molecular imaging in pulmonary fibrosis. A clinical trial. Am. J. Respir. Crit. Care Med. 203, 78–89 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schyns J., et al. , Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat. Commun. 10, 3964 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bain C. C., MacDonald A. S., The impact of the lung environment on macrophage development, activation and function: Diversity in the face of adversity. Mucosal Immunol. 15, 223–234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lobb R. R., Hemler M. E., The pathophysiologic role of alpha 4 integrins in vivo. J. Clin. Invest. 94, 1722–1728 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duan M., et al. , CD11b immunophenotyping identifies inflammatory profiles in the mouse and human lungs. Mucosal Immunol. 9, 550–563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu N., et al. , Promising bifunctional chelators for copper 64-PET imaging: Practical (64)Cu radiolabeling and high in vitro and in vivo complex stability. J. Biol. Inorg. Chem. 21, 177–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wadas T. J., Wong E. H., Weisman G. R., Anderson C. J., Copper chelation chemistry and its role in copper radiopharmaceuticals. Curr. Pharm. Des. 13, 3–16 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Farkas R., et al. , 64)Cu- and (68)Ga-based PET imaging of folate receptor-positive tumors: Development and evaluation of an albumin-binding NODAGA-folate. Mol. Pharm. 13, 1979–1987 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Brasse D., Nonat A., Radiometals: Towards a new success story in nuclear imaging? Dalton Trans. 44, 4845–4858 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Pham T., Rubenfeld G. D., Fifty years of research in ARDS. The epidemiology of acute respiratory distress syndrome. A 50th birthday review. Am. J. Respir. Crit. Care Med. 195, 860–870 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Hendrickson K. W., Peltan I. D., Brown S. M., The epidemiology of acute respiratory distress syndrome before and after coronavirus disease 2019. Crit. Care Clin. 37, 703–716 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Misharin A. V., et al. , Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J. Exp. Med. 214, 2387–2404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McQuattie-Pimentel A. C., Budinger G. R. S., Ballinger M. N., Monocyte-derived alveolar macrophages: The dark side of lung repair? Am. J. Respir. Cell Mol. Biol. 58, 5–16 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Matute-Bello G., Frevert C. W., Martin T. R., Animal models of acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 295, L379–L399 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tabary M., Lee J. S., Alveolar macrophage heterogeneity goes up in smoke? Am. J. Respir. Cell Mol. Biol. 67, 150–152 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mould K. J., et al. , Airspace macrophages and monocytes exist in transcriptionally distinct subsets in healthy adults. Am. J. Respir. Crit. Care Med. 203, 946–956 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liegeois M., et al. , Airway macrophages encompass transcriptionally and functionally distinct subsets altered by smoking. Am. J. Respir. Cell Mol. Biol. 67, 241–252 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watanabe S., Alexander M., Misharin A. V., Budinger G. R. S., The role of macrophages in the resolution of inflammation. J. Clin. Invest. 129, 2619–2628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tavakoli S., et al. , Differential regulation of macrophage glucose metabolism by macrophage colony-stimulating factor and granulocyte-macrophage colony-stimulating factor: Implications for (18)F FDG PET imaging of vessel wall inflammation. Radiology 283, 87–97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hao Y., et al. , Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
COVID-19 snRNA-seq data were previously deposited in GEO with the accession number GSE171524 and are also available through the single-cell portal: https://singlecell.broadinstitute.org/single_cell/study/SCP1219. Additional scRNA-seq datasets in COVID-19 and cystic fibrosis patients were previously deposited (GEO accession numbers GSE145926 and GSE193782) and publicly available through UCSC cell browser (available at: https://cells.ucsc.edu/). The other data needed to evaluate the conclusions of the paper are present in the paper and/or the SI Appendix.