Significance
OGT is the only enzyme that catalyzes the addition of O-GlcNAc to serine and threonine residues on intracellular proteins. It has been known for more than two decades ago that OGT is essential for mammalian cell viability, but the underlying mechanisms have remained elusive. We show here that the block in cell viability induced by OGT deficiency is primarily due to a large, deleterious increase in mitochondrial oxidative phosphorylation (OXPHOS). Mechanistically, OGT suppresses mTOR activity and maintains mitochondrial fitness by restricting proteasome activity and imposing homeostasis of intracellular amino acid levels. Our studies provide important mechanistic insights into the role of OGT and O-GlcNAc modifications in intracellular homeostasis and cell viability.
Keywords: OGT, genome-wide CRISPR/Cas9 screen, mitochondrion, mTOR, proteasome
Abstract
O-GlcNAc transferase (OGT) modifies serine and threonine residues on nuclear and cytosolic proteins with O-linked N-acetylglucosamine (GlcNAc). OGT is essential for mammalian cell viability, but the underlying mechanisms are still enigmatic. We performed a genome-wide CRISPR–Cas9 screen in mouse embryonic stem cells (mESCs) to identify candidates whose depletion rescued the block in cell proliferation induced by OGT deficiency. We show that the block in cell proliferation in OGT-deficient cells stems from mitochondrial dysfunction secondary to mTOR (mechanistic target of rapamycin) hyperactivation. In normal cells, OGT maintains low mTOR activity and mitochondrial fitness through suppression of proteasome activity; in the absence of OGT, increased proteasome activity results in increased steady-state amino acid levels, which in turn promote mTOR lysosomal translocation and activation, and increased oxidative phosphorylation. mTOR activation in OGT-deficient mESCs was confirmed by an independent phospho-proteomic screen. Our study highlights a unique series of events whereby OGT regulates the proteasome/ mTOR/ mitochondrial axis in a manner that maintains homeostasis of intracellular amino acid levels, mitochondrial fitness, and cell viability. A similar mechanism operates in CD8+ T cells, indicating its generality across mammalian cell types. Manipulating OGT activity may have therapeutic potential in diseases in which this signaling pathway is impaired.
O-GlcNAc transferase (OGT) is an essential X chromosome-encoded enzyme that catalyzes the addition of N-acetylglucosamine (GlcNAc) to the hydroxyl groups of serine and threonine residues on many nuclear and cytosolic proteins (1, 2). This posttranslational modification is reversible and is actively removed by the O-GlcNAcase OGA (1, 2). It has been known for more than two decades that OGT is essential for mammalian cell viability (3), but the underlying mechanisms are still unclear (1). Given the close association between OGT and human diseases such as cancer, diabetes, and cardiovascular disease (4–6), knowledge of the mechanisms by which OGT controls cell viability is essential to understand the role of OGT and the O-GlcNAc modification in cellular function.
A functional interaction of OGT with mitochondria has been suggested by several previous studies, many of which focused on mitochondrial OGT (mOGT), a splice variant of OGT that contains a mitochondrial targeting sequence (7). Detailed analyses of mOGT function have been complicated by difficulties in specifically and efficiently depleting mOGT (8) and the reportedly low levels of O-GlcNAcylated proteins in the mitochondria (9). The relation between O-GlcNAcylation and mitochondrial function depends strongly on the particular cellular system used. Increasing O-GlcNAc levels by treatment with the OGA inhibitor thiamet-G led to increased oxygen consumption and adenosine trisphosphate (ATP) production rates in cardiac mitochondria (10), but decreased basal oxygen consumption rates (OCR) and ATP production without an effect on maximal OCR in SH-SY5Y neuroblastoma or NT2 human embryonal carcinoma cells (11). Partial (50 to 70%) depletion of total OGT with a pan-OGT siRNA, which would be expected to decrease cellular O-GlcNAc levels, resulted in increased basal and maximal OCR in HeLa cells (8), whereas hematopoietic stem cells (HSCs) in which the Ogt gene was deleted by treatment of Ogt-floxed Mx1Cre mice with polyI:polyC had increased mitochondrial mass coupled to decreased copy numbers of mitochondrial DNA and decreased spare respiratory capacity, defined as the difference between maximal and basal OCR (12). However, these findings are complicated by the fact that Ogt deletion resulted in severe reduction of the numbers of HSC and likely promoted the accumulation of cells in which Ogt deletion was incomplete. The effects of OGT deletion on mitochondrial function need to be reevaluated in better-defined cellular systems with efficient OGT deletion.
The mechanistic target of rapamycin (mTOR) is a serine/threonine kinase that regulates fundamental cellular processes including cell growth and metabolism in response to environmental and intracellular signals (13, 14). mTOR is the catalytic subunit of two functionally distinct protein complexes: mTOR complex 1 (mTORC1) defined by the presence of the rapamycin-sensitive subunit Raptor, and mTORC2, which contains the alternate subunit Rictor (15). The mTORC1 complex has been described as a “coincidence detector” because it is activated only when both growth factors and nutrients are present to facilitate cellular growth (13). mTORC1 activation requires two separate steps that occur after translocation of mTOR to the lysosomal membrane (13, 14). Growth factors and cellular stress signals induce activation of the small GTPase Rheb, which is located on the lysosomal membrane and directly stimulates mTOR kinase activity in its GTP-bound form. However, mTORC1 only localizes in the vicinity of GTP-bound Rheb when nutrient signals such as amino acids are available to activate the Rag complex, a heterodimer of the small GTPases, RagA and RagC (or RagB and RagD). When RagA (or RagB) are GTP bound and RagC (or RagD) are bound to GDP, the Rag heterodimers recruit mTORC1 to the lysosomal membrane. The Rag heterodimer is itself activated and anchored to the lysosomal membrane by the pentameric Ragulator complex, a noncanonical guanine nucleotide exchange factor that contains Lamtor1-5 (16, 17). Thus, mTOR indirectly senses amino acid levels through proteins associated with the Rag and Ragulator complexes. Moreover, mTORC1 has been reported to regulate mitochondrial function in at least two ways: through transcriptional control of mitochondrial regulators such as PGC-1α (PPARγ coactivator 1α) (18) and by regulation of protein synthesis of a subset of mitochondrial proteins encoded by nuclear DNA through inhibition of eukaryotic translation initiation factor 4E-binding proteins (4E-BPs) (19).
To investigate why OGT is essential for the survival of proliferating cells (1), we generated mouse embryonic stem cell (mESC) lines with one or two floxed Ogt alleles. The blastocysts from which the ESCs were derived also expressed a Cre–ERT2 fusion protein for inducible deletion of the Ogt gene, and a Rosa26-YFPLSL reporter gene so that cells in which Cre–ERT2 had been activated in the nucleus with 4-hydroxytamoxifen (4-OHT) were marked with YFP. We used these cells to perform genome-wide CRISPR–Cas9 screens for small-guide RNAs (sgRNAs) that rescued the arrested cell proliferation of Ogt-deleted mESCs. The enriched sgRNAs targeted a large number of genes related to mitochondrial function, as well as genes whose products were involved in amino acid sensing by mTOR. We show that OGT deficiency leads to arrested cell proliferation and loss of mESC viability at least partly by increasing proteasome activity and intracellular amino acid levels, which in turn promotes lysosomal translocation and activation of mTORC1. The net result is a massive, deleterious increase in mitochondrial oxidative phosphorylation (OXPHOS), which correlates strongly with decreased cell viability. Genome-wide proteomic and phosphoproteomic analyses show extensive changes in global signaling and confirm our finding of mTOR hyperactivation in OGT-deficient cells. By highlighting the biochemical connection between OGT deficiency and high aberrant activation of proteasome function, the mTOR pathway, and mitochondrial OXPHOS, our studies provide important mechanistic insights into the role of OGT and O-GlcNAc modifications in intracellular homeostasis and cell viability.
Results
Inducible Deletion of the Ogt Gene Impairs Cell Viability in mESCs.
To overcome the lethality of Ogt deletion in mESCs (3), we generated conditional Ogt-floxed mice homozygous for knock-in alleles of Cre–ERT2 or LSL-YFP in the Rosa26 locus. We crossed Ogt floxed Cre–ERT2KI/KI mice with Ogt floxed Rosa26-LSL-YFPKI/KI mice to obtain OgtfloxedCre–ERT2+/KI Rosa26-LSL-YFP+/KI (hereafter termed Ogt fl) blastocysts (embryonic day 3.5), from which we derived Ogt fl mESC lines (Fig. 1A). Subsequent experiments were performed with male Ogt fl mESCs to maximize the likelihood of complete deletion of the X-linked Ogt gene in response to 4-hydroxytamoxifen (4-OHT) treatment.
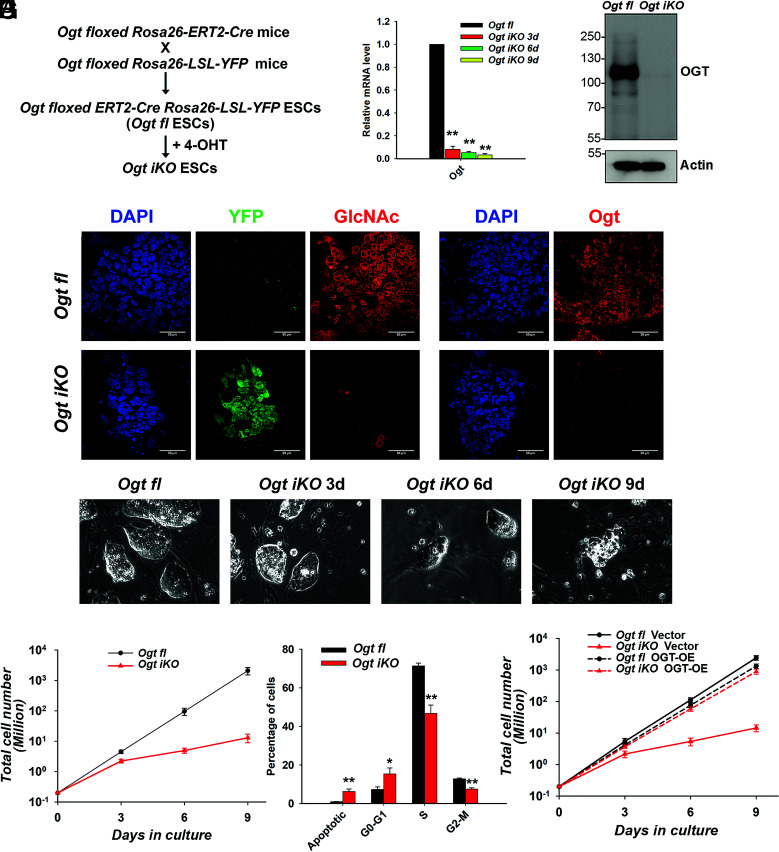
Fig. 1.
Generation of mESCs with inducible Ogt deletion. (A) Scheme used for the generation of inducible Ogt fl Cre–ERT2+/KI Rosa26-YFP+/KI (Ogt fl) mESC lines. (B) Quantitative real-time PCR (qRT-PCR) analysis of Ogt transcript levels in Ogt fl control and Ogt iKO mESCs 3, 6, and 9 d after 4-OHT treatment. The expression level of Ogt mRNA is shown relative to the level in control mESCs. Data are shown as mean ± SD (N = 3). (C) Western blot analysis of OGT protein level in Ogt fl mESCs treated with or without 4-OHT for 6 d. Actin was used as a loading control. (D) Immunohistochemistry of Ogt fl and Ogt iKO mESCs 6 d after 4-OHT treatment, using antibodies against O-GlcNAc and OGT. Nucleus staining: DAPI (blue). (Scale bar: 10 μm.) (E) Phase-contrast images of Ogt fl control and Ogt iKO mESCs 3, 6, and 9 d after 4-OHT treatment. (F) Cumulative growth curves of Ogt fl mESCs treated with or without 4-OHT at day 0 and counted at each passage (every 3 d) thereafter until day 9. Data are shown as mean ± SD (N = 3). Note the logarithmic scale on the Y-axis. (G) Percentage of Ogt fl and Ogt iKO mESCs 6 d after 4-OHT treatment in different phases of the cell cycle. Data are shown as mean ± SD (N = 3). (H) Cumulative growth curves of Ogt fl mESCs stably expressing empty vector or WT OGT and treated with or without 4-OHT. Data are shown as mean ± SD (N = 3). Note the logarithmic scale on the Y-axis.
At day 3 after 4-OHT treatment, the extent of Ogt gene deletion was >90% as assessed by qRT-PCR for Ogt mRNA, and the extent of deletion continued to increase at days 6 and 9 after 4-OHT treatment (Fig. 1B). Deletion at the protein level was nearly complete by 6 d as assessed by western blot (Fig. 1C). Immunostaining confirmed that both OGT and its catalytic product, the O-GlcNAc modification, were almost completely eliminated in Ogt-deleted cells (hereafter termed Ogt iKO mESCs; i for “inducible”) by day 6 (Fig. 1D). Ogt iKO mESCs gradually underwent a significant morphology change, from large, tightly packed colonies in the case of Ogt fl mESCs to small, loosely distributed colonies after Ogt deletion (Fig. 1E). OGT-deficient mESCs displayed a significantly impaired growth rate beginning at ~3 d after 4-OHT addition, resulting in a ~90% decrease in the number of Ogt iKO cells by day 6 and >95% decrease by day 9 (Fig. 1F, please note logarithmic scale of Y-axis). The Ogt iKO mESCs also showed decreased cell viability and decreased expression of the pluripotency markers Oct4 and Nanog (SI Appendix, Fig. S1 A and B). Cell cycle analysis using BrdU, a thymidine analog that is incorporated into newly synthesized DNA during S phase, and 7-AAD, a DNA intercalating dye, showed that Ogt iKO mESCs displayed significant cell cycle arrest at the G0-G1 stage as well as a significant increase of apoptotic cells by day 6 (Fig. 1G and SI Appendix, Fig.S1C). G1-phase arrest of cell cycle progression provides an opportunity for cells to repair DNA and other damage or, if this fails, to undergo apoptosis (20). The impaired growth rate was due specifically to Ogt deletion, since it was rescued by overexpression of wild-type (WT) OGT in Ogt iKO mESCs, concomitantly with restoration of O-GlcNAc staining (Fig. 1H and SI Appendix, Fig. S1D). 4-OHT treatment did not affect the cell growth of WT mESCs (SI Appendix, Fig. S1E).
A Genome-Wide CRISPR–Cas9 Screen Identifies Key Regulators of Cell Survival in OGT-Deficient mESCs.
To identify genes whose deletion could restore cell proliferation and increase the survival of Ogt iKO mESCs, we conducted an unbiased genome-wide CRISPR–Cas9 viability screen. We stably expressed Cas9 in Ogt fl mESCs, and lentivirally transduced them with the sgRNA Brie library (21), which contains a pool of 78,637 sgRNAs targeting 19,647 genes with four sgRNAs per gene and 1,000 control nontargeting sgRNAs. After puromycin treatment for 7 d to select for cells stably expressing the sgRNAs, the cells were split into two groups: an untreated (Ogt fl) group that served as the control and a second group treated with 4-OHT for 6 d to induce Ogt deletion (Ogt iKO). On day 13, YFP-negative cells from the control Ogt fl group and YFP+ cells from the Ogt iKO group were sorted, and the enriched sgRNAs were identified by next-generation deep sequencing (Fig. 2A). The sequencing data were analyzed using the PinAPL-Py platform (22).
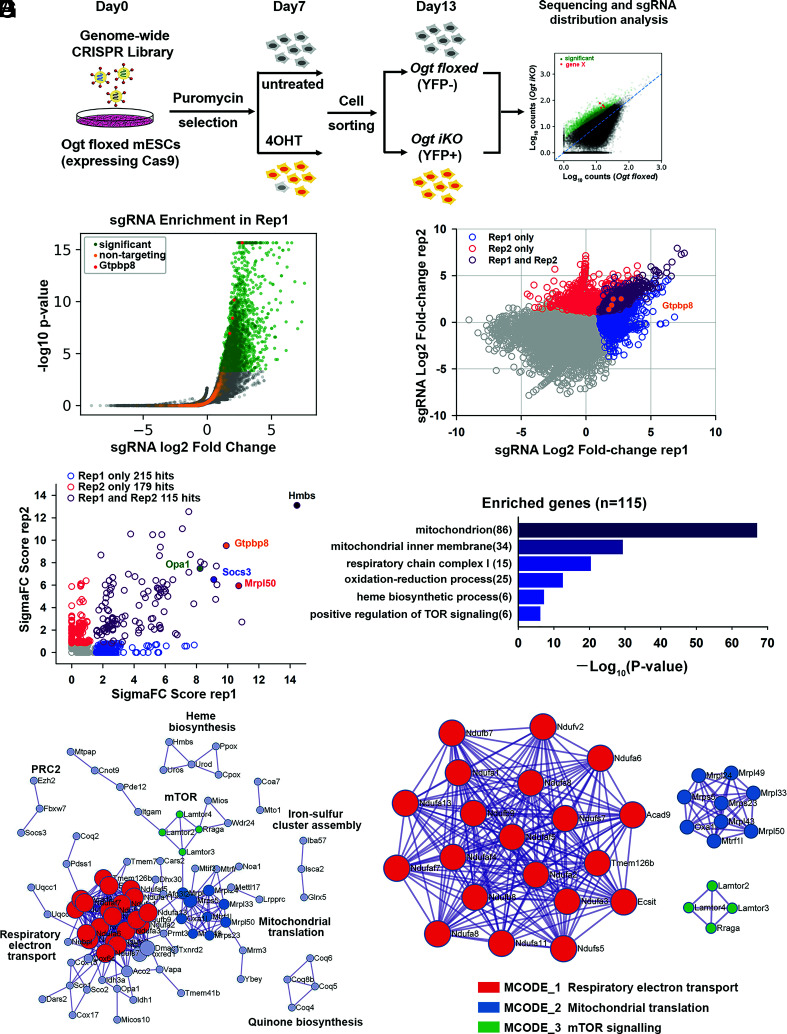
Fig. 2.
Genome-wide CRISPR–Cas9 screen to identify key regulators of cell survival in OGT-deficient mESCs. (A) Strategy for the genome-wide CRISPR–Cas9 viability screen to identify key regulators of cell growth in OGT-deficient mESCs. (B) Volcano plot of P-value versus fold enrichment for each sgRNA in Replicate 1. Significantly enriched sgRNAs are shown in green and nontargeting control sgRNAs in orange. All the four sgRNAs for Gtpbp8 were significantly enriched in the screen (red dots). (C) sgRNAs that were enriched in one or both biological replicates of the CRISPR–Cas9 viability screen. The four sgRNAs targeting Gtpbp8 are highlighted. The correlation between the fold changes in sgRNA abundance has a Pearson’s r of 0.438. (D) Rank of enriched hits by gene score, calculated based on the log fold changes of all sgRNAs targeting each gene and the number of sgRNAs targeting each gene that reached statistical significance. 115 enriched hits in both biological replicates are shown in purple. A few of the most highly scored hits (Hmbs,Gtpbp8, Opa1, Socs3, and Mrpl50) are labeled. (E) Gene ontology analysis of biological pathways of the 115 enriched hits. (F) Metascape visualization of the protein–protein interactome network of the 115 highly scored hits from the CRISPR screen. Each MCODE complex identified by Metascape was assigned a unique color. (G) Three MCODE complexes and their associated functional pathways identified by Metascape.
We performed two independent screens that were sequenced to a depth of >10 M mapped reads per sample; >98% of all genes in the sgRNA library were represented in each screen (SI Appendix, Fig. S2A). The volcano plot showed significantly enriched sgRNAs in green and 1,000 control nontargeting sgRNAs in orange, which allowed us to calculate an empirical false discovery rate of 3.1% for replicate 1 and 1.1% for replicate 2 (Fig. 2B and SI Appendix, Fig.S2B and Dataset S1). The overlap of enriched sgRNAs and their rank in each replicate are shown in Fig. 2C. Gene scores were calculated considering the log fold changes of all sgRNAs targeting each gene and the number of sgRNAs targeting each gene that reached statistical significance (22). In the two replicate experiments, 115 hits were identified by the overlap of sgRNAs enriched in the Ogt iKO groups compared to the control Ogt fl groups (P-value < 0.01) (Fig. 2C and Dataset S2). Hits with the highest scores, including Hmbs, Mrpl50, Gtpbp8, Socs3, and Opa1, are highlighted (Fig. 2D).
Gene scores were calculated using SigmaFC, which considers the sum of the log fold changes of all sgRNAs targeting each gene and multiplies this number by the number of sgRNAs that reached statistically significant enrichment (22). We plotted SigmaFC scores from two independent screen replicates and only investigated those that were significant across both screens. In the two replicate experiments, 115 candidate genes were identified by the overlap of sgRNAs enriched in the Ogt iKO groups compared to the control Ogt fl groups (P-value < 0.01) (SI Appendix, Fig. S2C and Dataset S2). Genes with the highest scores, including Hmbs, Mrpl50, Gtpbp8, Socs3, and Opa1, are highlighted in Fig. 2D.
More than 80 of the 115 gene candidates were annotated by gene ontology analysis as related to the mitochondrion or the mitochondrial respiratory chain complex (Fig. 2E). Metascape, which applies the MCODE (Molecular Complex Detection) algorithm to identify protein complexes (23), identified three MCODE complexes: respiratory electron transport, including NADH:ubiquinone oxidoreductase family (Nduf) proteins in mitochondrial complex I (shown in red); proteins involved in mitochondrial translation, such as mitochondrial ribosomal proteins (Mrpl) (shown in blue); and mTOR signaling, specifically Rraga and Lamtor2-4 (shown in green) (Fig. 2 F and G). Metascape analysis also identified protein complexes involved in heme biosynthesis (Hmbs, Uros, and Urod), quinone biosynthesis (Coq4, Coq5, Coq6, and Coq8b), and iron–sulfur cluster assembly (Glrx5, Isca2, and Iba57) (Fig. 2F). Screen hits related to mitochondria and mTOR signaling are considered in detail below. Other candidate hits are being investigated further as part of a separate project.
OGT Deficiency Results in Mitochondrial Dysfunction.
To follow up on the large number of enriched sgRNAs targeting mitochondrial proteins, we assessed mitochondrial function in control and Ogt-deleted cells by evaluating mitochondrial membrane potential, mitochondrial mass, and mitochondrial OXPHOS. Immunostaining and flow cytometry analysis showed that mitochondrial membrane potential assessed by MitoTracker red CMXRos staining (Fig. 3 A–C) and mitochondrial mass assessed by MitoTracker deep red staining (SI Appendix, Fig. S3 A–C) were both increased in Ogt iKO relative to Ogt fl mESCs. However, OGT deficiency did not affect mitophagy, a process for removal of damaged mitochondria through autophagy, as assessed by using a small-molecule fluorescent probe (24) (Mtphagy Dye; SI Appendix, Fig. S3 D and E). Evaluation of mitochondrial OXPHOS showed that basal and maximal OCR as well as ATP production, the indicator of OXPHOS, were aberrantly increased in Ogt iKO compared to Ogt fl mESCs (Fig. 3D and SI Appendix, Fig. S3F). As expected, overexpression of WT OGT in Ogt iKO mESCs rescued the aberrant increase of mitochondrial OXPHOS (Fig. 3E and SI Appendix, Fig. S3G). OGT deficiency was associated with an approximately twofold increase in the levels of reactive oxygen species (ROS) measured by cellROX deep red, presumably as a consequence of increased OXPHOS (SI Appendix, Fig. S3 H and I).
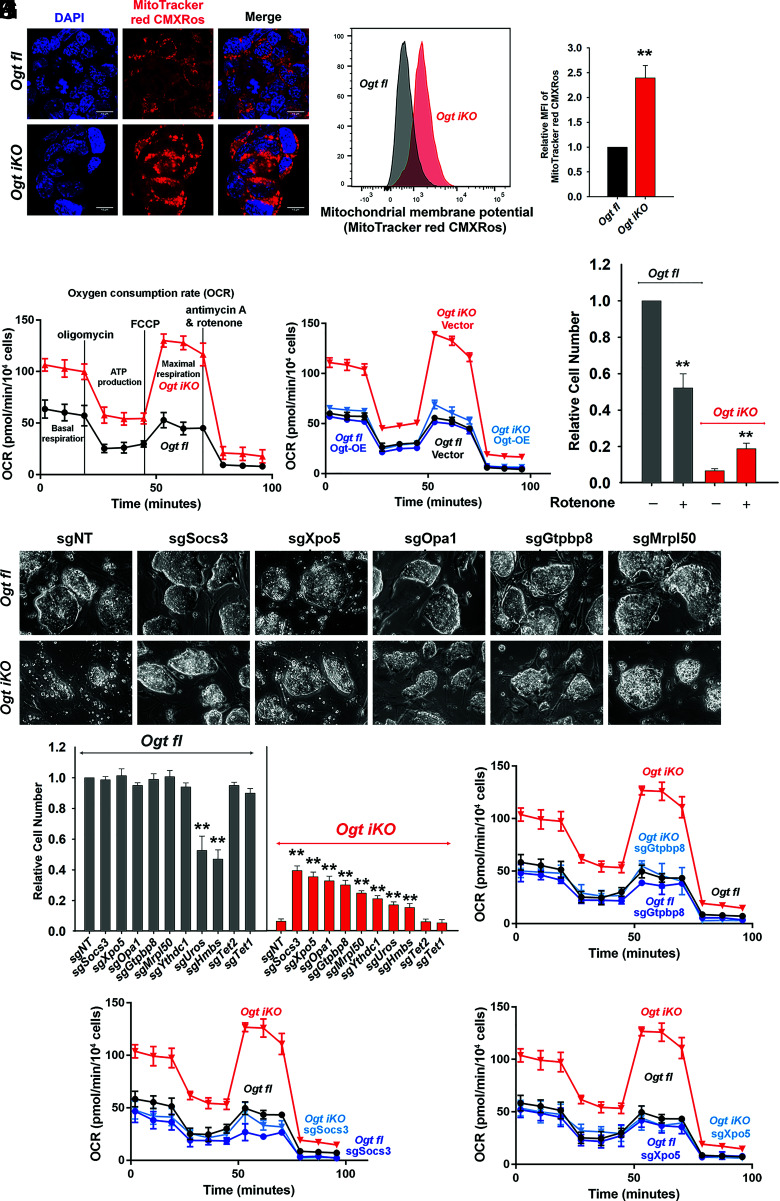
Fig. 3.
OGT deficiency results in mitochondrial dysfunction. (A) Fluorescence images of Ogt fl and Ogt iKO mESCs 6 d after 4-OHT treatment. Cells were stained with MitoTracker red CMXRos. Nucleus staining: DAPI (blue). (Scale bar: 10 μm.) (B) Representative histogram of MitoTracker red CMXRos fluorescence in Ogt fl and Ogt iKO mESCs 6 d after 4-OHT treatment. (C) Relative MFI (mean fluorescent intensity) of MitoTracker red CMXRos shown in Fig. 3B. (D) Analysis of OCR using Seahorse XFe24 in Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, for 6 d. Data are shown as mean ± SD (N = 3). (E) Analysis of OCR using Seahorse XFe24 in Ogt fl mESCs stably expressing empty vector or WT OGT treated with or without 4-OHT for 6 d. Data are shown as mean ± SD (N = 3). (F) Relative cell numbers of Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, and with or without the mitochondrial Complex I inhibitor rotenone (75 nM) for 8 d. Data are shown as mean ± SD (N = 3). (G) Phase-contrast images of Ogt fl and Ogt iKO mESCs expressing nontargeting sgRNA (sgNT) or sgRNAs targeting the indicated genes (Socs3, Xpo5, Opa1, Gtpbp8, and Mrpl50) and treated without or with 4-OHT, respectively, for 8 d. (H) Relative cell numbers of Ogt fl and Ogt iKO mESCs expressing nontargeting sgRNA (sgNT) or sgRNAs targeting the indicated genes (Socs3, Xpo5, Opa1, Gtpbp8, Mrpl50, Ythdc1, Uros, Hmbs, Tet2, or Tet1) and treated without or with 4-OHT, respectively, for 8 d. Data are shown as mean ± SD (N = 3). (I–K), Analysis of OCR using Seahorse XFe24 in Ogt fl and Ogt iKO mESCs expressing sgRNAs targeting Gtpbp8 (I), Socs3 (J), and Xpo5 (K) and treated without or with 4-OHT, respectively, for 6 d. Data are shown as mean ± SD (N = 3).
Metabolome analyses showed that the TCA cycle metabolites such as citrate, alpha-ketoglutarate, succinate, fumarate, and malate, which are tightly associated with OXPHOS (25), were increased, while the glycolytic pathway metabolites such as 3-phosphoglycerate and phosphoenolpyruvate were decreased, in Ogt iKO compared to Ogt fl mESCs (SI Appendix, Fig. S3J). At the transcriptional level, the majority of genes encoded in the mitochondrial genome showed increased expression levels in Ogt iKO mESCs (SI Appendix, Fig. S3K). The mitochondrial complex I inhibitor Rotenone partially rescued the blocked proliferation of Ogt iKO mESCs, despite its substantial toxicity for Ogt fl mESCs (Fig. 3F and SI Appendix, Fig. S4A), supporting the conclusion that mitochondrial function is hyperactivated in OGT-deficient mESCs. As shown above, reconstitution with WT OGT rescued both the cell proliferation defect and the aberrant increase in mitochondrial OXPHOS of Ogt iKO mESCs (Figs. 1H and 3E and SI Appendix, Fig. S3G), and OGT-reconstituted Ogt iKO cells were as sensitive to rotenone as WT cells (50% decrease in cell numbers, compare Fig. 3F and SI Appendix, Fig. S4B).
We used an all-in-one CRISPR/Cas9 vector system to individually interrogate the effects of enriched sgRNAs targeting eight of the top candidate genes identified in our screen (Fig. 2D). The candidate proteins that we selected were involved in diverse cellular pathways – mitochondrial proteins (Opa1, Gtpbp8, Mrpl50), proteins involved in heme biosynthesis (Hmbs, Uros), and proteins involved in other cellular functions (Socs3, Xpo5, Ythdc1). Nontargeting (NT) sgRNAs and sgRNAs targeting Tet1 or Tet2 (which are expressed in ES cells, but whose sgRNAs were not enriched in our screen) were used as controls. Each gene was efficiently deleted as assessed by qRT-PCR (SI Appendix, Fig. S5A). Transduction with Cas9 and sgRNAs targeting all eight genes improved the morphology and growth rate of Ogt iKO mESCs, leading to the formation of large, tightly packed colonies as well as promoting a three to seven fold increase in cell numbers; transduction with control NT and Tet1/Tet2 sgRNAs had no effect (Fig. 3 G and H and SI Appendix, Fig. S5B). Moreover, sgRNAs against Socs3, Xpo5, and Gtpbp8 restored expression levels of the pluripotency markers Oct4 and Nanog (SI Appendix, Fig. S5C). Hmbs and Uros sgRNAs reproducibly rescued the proliferation of Ogt iKO mESCs to the same degree as rotenone (compare (Fig. 3F and 3H) Fig. 3, but were not further considered because they also diminished the proliferation of normal (Ogt fl) mESCs (Fig. 3H). Notably, sgRNAs targeting the top three candidate genes, Socs3, Xpo5, and Gtpbp8, reduced the aberrant increase of OCR in Ogt iKO mESCs to normal levels (Fig. 3 I–K and SI Appendix, Fig. S6 A–C). Our data indicate that sgRNA-mediated depletion of each of these proteins indirectly inhibits the increase in mitochondrial OXPHOS resulting from OGT depletion, although the exact pathways and mechanisms remain to be understood.
mTOR Is Hyperactivated in OGT-Deficient mESCs.
Gene ontology analysis of the 115 candidate genes identified in our sgRNA screen highlighted six genes related to positive regulation of mTOR signaling (Lamtor2, Lamtor3, Lamtor4, Rraga, Wdr24, and Mios) (Fig. 4A). Lamtor2, 3, and 4 are subunits of the Ragulator complex, which controls the activity of the Rag complex, a heterodimer of two guanine nucleotide-binding proteins, RagA (encoded by Rraga) and RagC (or alternately, RagB and RagD) (16). Wdr24 and Mios are subunits of the GATOR2 complex, which indirectly senses amino acid levels and activates mTORC1 by inhibiting the GAP activity of GATOR1 (13, 14, 16). Since mTOR is an established regulator of mitochondrial function at both the transcriptional and translational levels (18, 19), we examined mTOR activity in OGT-deficient cells. Analysis of mTOR activity at a single-cell level by flow cytometry and immunostaining showed that the levels of phosphorylation of S6 ribosomal protein on serine 235/236, a reliable marker for mTOR activity, were strikingly increased in Ogt iKO mESCs, without a change in total S6 ribosomal protein levels; this increase was prevented by the ATP-competitive pan-mTOR inhibitor Torin2 (Fig. 4 B and C and SI Appendix, Fig. S7 A and B). Akt signaling, which is upstream of mTOR (26), was also slightly increased in Ogt iKO mESCs (SI Appendix, Fig. S7 C and D). Torin2 also diminished the high OCR (Fig. 4D and SI Appendix, Fig. S7E), mitochondrial membrane potential, and mitochondrial mass (SI Appendix, Fig. S7 F–I) of Ogt iKO mESCs to nearly normal levels. At the transcriptional level, Torin2 also prevented the increase in expression levels of genes encoded in mitochondrial DNA in Ogt iKO mESCs (SI Appendix, Fig. S7J). In contrast, the mitochondrial complex 1 inhibitor rotenone and an sgRNA targeting Socs3, which inhibited mitochondrial OXPHOS (Fig. 3 J), did not affect mTOR hyperactivation in OGT iKO mESCs (SI Appendix, Fig. S8 A–D). These data indicate that increased mitochondrial OXPHOS activity is largely downstream of mTOR hyperactivation in OGT-deficient mESC. Consistent with our observation in Ogt iKO mESCs, inhibition of OGT catalytic activity by the OGT inhibitor OSMI-1 (27) significantly increased mTOR activity in control Ogt fl mESCs, as assessed by flow cytometry for phosphorylation of S6 ribosomal protein (SI Appendix, Fig. S8 E and F).
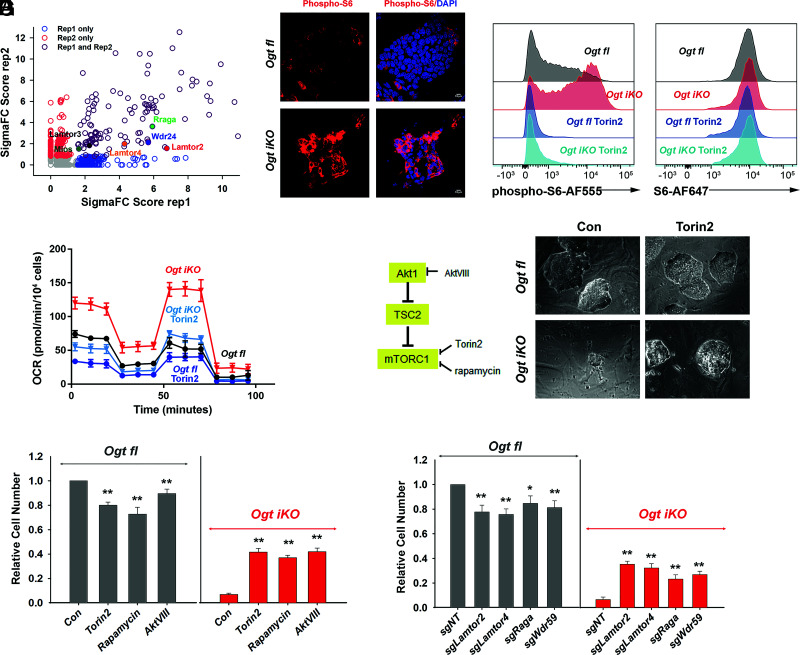
Fig. 4.
mTOR is hyperactivated in OGT-deficient mESCs. (A) Scatterplot of enriched genes by gene score. Genes related to positive regulation of mTOR signaling are highlighted. (B) Immunohistochemistry of Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, for 6 d. Cells were stained with antibody against phospho-S6 ribosomal protein. Nucleus staining: DAPI (blue). Scale bar: 10 μm. (C) Flow cytometry analysis of phospho-S6 (Left) and total S6 (Right) ribosomal protein in Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, for 6 d and then with or without 200 nM Torin2 for 2 h. Data are representative of three biological replicates. (D) Analysis of OCR using Seahorse XFe24 in Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, and with or without Torin2 (25 nM) for 6 d. Data are shown as mean ± SD (N = 3). (E) Left, Simplified schematic diagram of mTOR pathway activation and the proteins targeted by the inhibitors Torin2 (25 nM), rapamycin (50 nM), and AktVIII (7.5 μM). Right, Phase-contrast images of Ogt fl and Ogt iKO mESCs treated with or without Torin2. (F) Relative cell numbers of Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, and with or without Torin2 (25 nM), rapamycin (50 nM), and AktVIII (7.5 μM) for 8 d. Data are shown as mean ± SD (N = 3). (G) Relative cell numbers of Ogt fl and Ogt iKO mESCs expressing gRNAs against the indicated genes (Lamtor2, Lamtor4, Raga, and Wdr59) and treated without or with 4-OHT, respectively, for 8 d. Data are shown as mean ± SD (N = 3).
Since mTOR signaling was strongly up-regulated upon Ogt deletion, we tested whether mTOR inhibition could rescue the viability of OGT-deficient mESCs. Treatment with the mTOR inhibitor Torin2, the mTORC1 inhibitor rapamycin, or the AKT inhibitor AKTVIII partially rescued colony morphology and the decrease in mESC cell numbers observed in Ogt iKO cells (Fig. 4 E and F and SI Appendix, Fig. S9A), although Ogt fl cells showed a slight decrease in numbers upon treatment with these inhibitors. Treatment with Torin2 also rescued the expression of mRNAs encoding the pluripotency markers Oct4 and Nanog in Ogt iKO cells (SI Appendix, Fig. S9B). Similarly, CRISPR–Cas9-mediated disruption of the genes encoding mTOR pathway components such as Lamtor2, Lamtor4, Rraga, and Wdr59 partially rescued cell numbers in Ogt-deleted cells, despite reducing cell numbers somewhat in Ogt fl mESCs (Fig. 4G and SI Appendix, Fig. S9 C and D). Consistent with this observation, expression of sgRNA targeting Lamtor2 inhibited hyperactivated mTOR signaling in Ogt iKO mESCs as judged by immunostaining for phosphorylation of S6 ribosomal protein (SI Appendix, Fig. S9E). The mTOR pathway plays key roles in protein synthesis (28) and has been reported to regulate mitochondrial function by regulating protein synthesis (19). Notably, cycloheximide, an inhibitor of protein synthesis that is toxic for control mESCs, rescued the decreased proliferation of Ogt iKO mESCs (SI Appendix, Fig. S10 A and B). These results point to a functional link between OGT, mTOR activity, and mitochondrial function that is essential for the proliferation and survival of OGT-deficient mESCs.
Extensive Changes in Global Signaling and Protein Abundance in Ogt iKO mESCs.
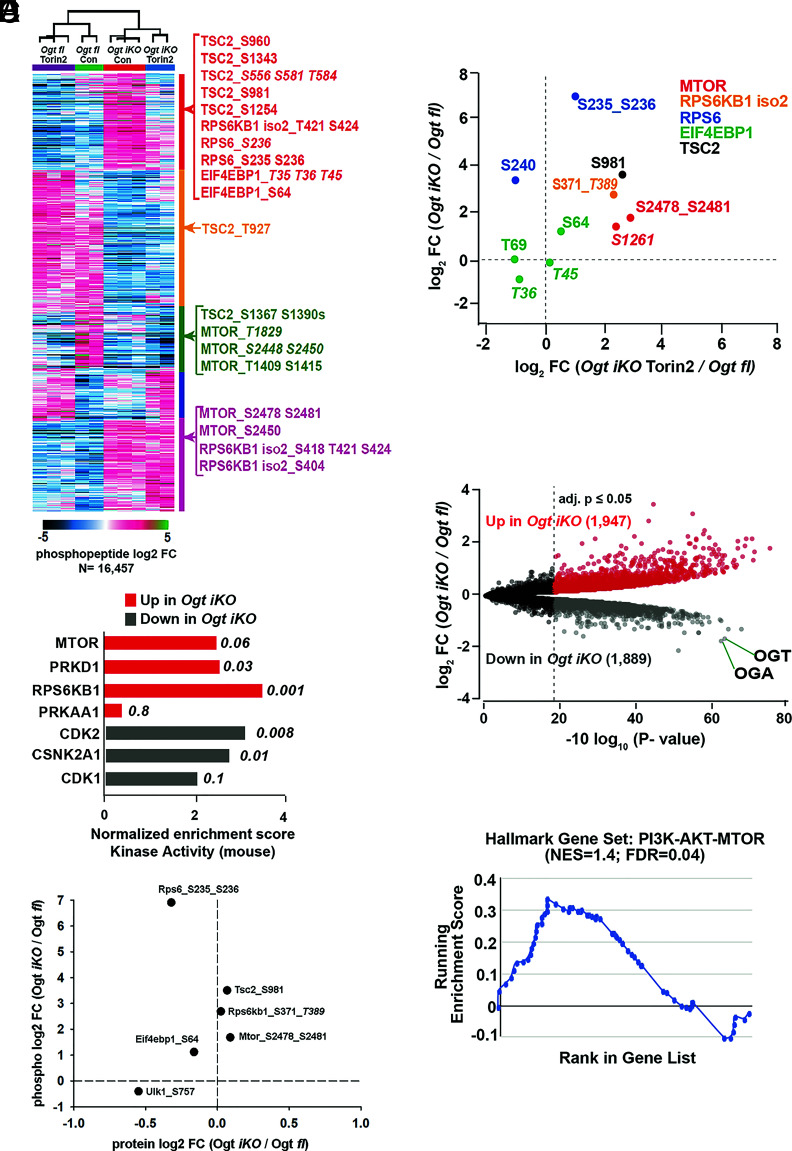
To evaluate the global effects of mTOR activation in OGT-deficient cells, we performed quantitative proteomic and phosphoproteomic analysis of Ogt fl and Ogt iKO mESCs in the presence and absence of the mTOR inhibitor Torin2. Phosphoproteomics revealed extensive changes in signaling in mESCs lacking OGT (Fig. 5A). Assessment of kinase activity by posttranslational modification-set enrichment analysis (PTM-SEA) (29) showed significant upregulation of mTOR, RPS6KB1 (S6 kinase), and PKRD1 signaling pathways in OGT-deficient cells compared to Ogt fl cells (Fig. 5B). As a comparison, AMPKA1 (PRKAA1) kinase, which senses ATP levels in the cell, showed only minor activation upon Ogt deletion. Kinases associated with cell cycle progression, CDK1/2 and CSNK2A1, had diminished activity in OGT-deficient mESCs (Fig. 5B). Comparing changes in phosphorylation levels with changes in protein levels to assess phospho-site stoichiometry of canonical mTOR signaling components, we found that activating phosphorylations on mTOR and Rps6kb1 (an established target for phosphorylation by mTOR) were significantly up-regulated upon Ogt deletion without changes in mTOR or Rps6kb1 protein levels (Fig. 5C). Ribosomal protein S6 (RPS6), a well-established target of S6K, showed a small though significant decrease in protein levels with a concomitant increase in phosphorylation levels at S235 and S236, indicating an increase in stoichiometry (Fig. 5C and SI Appendix, Fig.S11A). TSC2, a negative regulator of mTORC1 signaling, showed significant upregulation of its inhibitory phosphorylation site S981, also without changes in protein levels (Fig. 5C). The stoichiometry of phosphorylation (normalized to protein levels) of two downstream targets of mTOR – EIF4EBP1 at S64 and ULK1 at S757 – was also increased in Ogt iKO mESCs (Fig. 5C and SI Appendix, Fig.S11A).
Fig. 5.
Extensive changes in global signaling and protein abundances in Ogt iKO mESCs. (A) K-means clustering analysis of all regulated (BH-corrected adjusted P-value ≥ 0.05) phosphopeptides as determined by a moderated F-test. Example phosphopeptides for canonical mTOR pathway components are highlighted. Italicized phosphosite numbers indicate phosphosite assignment ambiguity. (B) Normalized enrichment scores for kinase activity as determined by PTM-SEA of phospho-proteomics data performed in Ogt fl and Ogt iKO mESCs. Top three pathways in both directions, as well as their respective P-values, are shown. PRKAA1 (AMPKA1) is included as a comparison. Only mouse-derived PTM sets were used for the analysis. (C) Changes in phosphopeptide and protein levels of mTOR signaling components in Ogt fl and Ogt iKO mESCs as determined by quantitative proteomics. Italicized phosphosite numbers indicate phosphosite assignment ambiguity. (D) Changes in phosphorylation levels on mTOR signaling components upon Ogt deletion alone (y-axis) and Ogt deletion in the presence of Torin2 (x-axis). Both are relative to Ogt fl. Italicized phosphosite numbers indicate phosphosite assignment ambiguity. “Iso 2” refers to isoform 2 of RPS6KB1. (E) Volcano plot of differentially abundant proteins between Ogt-floxed and Ogt iKO mESCs. BH-corrected P-values of less than or equal to 0.05 are highlighted. (F) Leading edge analysis for proteome level-GSEA of proteins involved in the PI3K–AKT–MTOR pathway. Positive enrichment indicates up in Ogt iKO; negative, down in Ogt iKO. Normalized enrichment score and false discovery rate (FDR, q-value) are shown.
Treatment of Ogt iKO cells with Torin2 decreased the abundance of ~20% of the phosphopeptides dysregulated by Ogt deletion and subsequent mTOR activation (Fig. 5A). Among these were several phosphorylation sites known to be downstream of mTOR, including RPS6KB1, RPS6, and EIF4EBP1 (Fig. 5D). Torin2 did not alter a phosphorylation site, S1261, on mTOR that is responsive to amino acids (30), suggesting that the ability of mTOR to sense changes in amino acid levels may remain intact in OGT-deficient cells. The proteome of Ogt iKO mESCs also showed major changes in protein levels (3,836 of the 8,470 measured) compared to Ogt fl cells, where OGT and OGA were among the most down-regulated proteins upon Ogt deletion (Fig. 5E). Gene set enrichment analysis of the proteome-level data also showed increased expression of proteins in the PI3K–AKT–mTOR signaling pathway in Ogt-deficient mESCs (Fig. 5F). Consistent with the increased frequency of apoptotic cells in OGT-deficient cells (Fig. 1G and SI Appendix, Fig.S1C), protein levels of the mitochondrial apoptosis markers CDKN1A (p21), CDKN2A (p16), CASP8, APAF1, and BAX were increased in Ogt iKO cells, and this increase could be rescued by the mTOR inhibitor Torin2 (SI Appendix, Fig.S11B). Together, these data show that Ogt deletion leads to the activation of mTOR/S6K signaling and extensive remodeling of the proteome and phosphoproteome.
OGT Deficiency Promotes mTOR Translocation by Increasing Proteasome Activity.
We investigated the mechanisms underlying aberrant mTOR activation in the absence of OGT. Notably, all six mTOR pathway hits from our CRISPR/Cas9 screen were related to mTOR translocation to the lysosomal membrane in response to amino acid sensing – mediated on the one hand by the Rag and Ragulator complexes (sgRNAs against Rraga, Lamtor 2-4), and on the other hand to amino acid sensing mediated by the GATOR2 complex (sgRNAs against Wdr24 and Mios) (13, 14). Moreover, an amino acid-responsive phosphorylation site on mTOR was induced upon Ogt deletion and was not altered by Torin2 (Fig. 5D). We, therefore, assessed mTOR subcellular localization in Ogt fl and Ogt iKO mESCs by immunocytochemistry and also monitored steady-state amino acid levels in these cells (Fig. 6 A and B). In Ogt fl mESCs, mTOR staining was diffusely localized to the cytoplasm, with little overlap with the lysosomal marker LAMP1, whereas in Ogt iKO mESCs, mTOR showed perinuclear staining that overlapped considerably with LAMP1 (Fig. 6A and SI Appendix, Fig. S12A). sgRNA-mediated disruption of Lamtor2 inhibited mTOR translocation to the lysosome in Ogt iKO mESCs (SI Appendix, Fig. S12B), confirming that mTOR was activated by translocation to the lysosome in the absence of OGT.
Fig. 6.
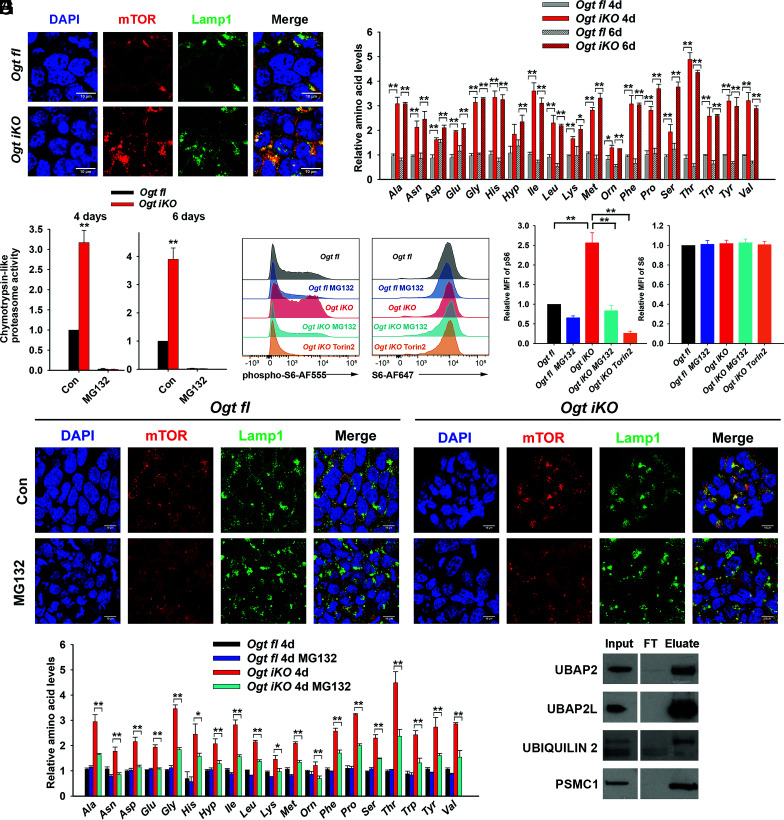
OGT deficiency promotes the translocation of mTOR by increasing proteasome activity. (A) Immunohistochemistry of Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, for 6 d. Cells were stained with antibodies against mTOR and Lamp1. Nucleus staining: DAPI (blue). Scale bar: 10 μm. (B) Relative amino acid levels in Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, for 4 or 6 d. Data are shown as mean ± SD (N = 3). (C) Chymotrypsin-like proteasome activity in Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, for 4 or 6 d. Data are shown as mean ± SD (N = 3). (D) Flow cytometry analysis of phospho-S6 (Left) and total S6 (Right) ribosomal protein in Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, for 6 d and then with or without 10 μM MG132 or 200 nM Torin2 for 2.5 h. (E) Relative mean fluorescence intensity (MFI) of phospho-S6 and S6 ribosomal protein in three independent experiments similar to that shown in (D). (F) Immunohistochemistry of Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, for 6 d and then with or without 10 μM MG132 for 2.5 h. Cells were stained with antibodies against mTOR and Lamp1. Nucleus staining: DAPI (blue). Scale bar: 10 μm. (G) Relative amino acid levels in Ogt fl and Ogt iKO mESCs treated without or with 4-OHT, respectively, for 4 d and then with or without 10 μM MG132 for 2.5 h. (H) Western blot of UBAP2, UBAP2L, ubiquilin 2, and PSMC1 proteins after WGA pull down of cell lysates from Ogt fl mESCs. FT: Flow Through.
The lysosomal translocation and activation of mTOR correlated with a global increase in amino acid levels, including the branched chain amino acids leucine, isoleucine, and valine, which are important regulators of mTOR (31) in Ogt iKO mESCs, apparent as early as 4 d or 6 d after 4-OHT treatment (Fig. 6B). Steady-state amino acid levels are controlled by the balance between protein synthesis, which depends on mRNA translation by ribosomes, and protein degradation, effected by proteasomes, lysosomes, and autophagosomes. We focused on 26S proteasome activity, which was previously reported to be reversibly inhibited by OGT through direct O-GlcNAc modification of proteasome subunits including the ATPase Rpt2 (also known as Psmc1) (32). Indeed, three major proteasome activities (chymotrypsin-like, trypsin-like, and caspase-like) were all significantly increased in Ogt iKO compared to Ogt fl mESCs (Fig. 6C and SI Appendix, Fig. S13 A and B), and this increase in activity was completely inhibited by treatment for short times (1.5 h) with 10 μM of the proteasome inhibitor MG132 (Fig. 6C and SI Appendix, Fig. S13 A and B). Furthermore, an in-gel proteasome assay showed that 26S and 30S proteasome activities were significantly increased, while the activity of the 20S proteasome (which is present at low levels in ES cells) was slightly increased in Ogt iKO mESCs (SI Appendix, Fig. S13 C and D). These data indicate that OGT inhibits the ATPase activity of the capped 26S and 30S proteasomes. However, the mitochondrial complex 1 inhibitor rotenone and the pan-mTOR inhibitor Torin2 could not block the increase of proteasome activity in Ogt iKO cells (SI Appendix, Fig. S13E). Given that Torin2 significantly blocked the increase of mTOR activity (SI Appendix, Fig. S13 F and G) and rotenone significantly blocked the increase of mitochondrial membrane potential in Ogt iKO cells (SI Appendix, Fig. S13 H and I), our data suggest that mTOR hyperactivation occurs downstream of proteasome activity. To confirm the hypothesis that mTOR is hyperactivated at least partly because of increased proteasome activity, we measured mTOR hyperactivation in the presence or absence of proteasome inhibitors in control Ogt fl mESCs and Ogt iKO mESCs. Treatment of the cells for 2.5 h with MG132 or the structurally unrelated proteasome inhibitor bortezomib significantly reduced phospho-S6 staining, assessed by flow cytometry and immunocytochemistry for phospho-S6, both in control cells and after Ogt deletion (Fig. 6 D and E and SI Appendix, Fig.S14 A–C). The levels of total S6 ribosomal protein were unchanged under these conditions (Fig. 6 D and E and SI Appendix, Fig. S14 A–C). MG132 treatment also significantly reduced the lysosomal localization of mTOR in Ogt iKO mESCs, as indicated by the poor overlap of mTOR staining with the lysosome marker LAMP1 (Fig. 6F and SI Appendix, Fig. S14D), and partially rescued the aberrant increase of amino acids in Ogt iKO mESCs (Fig. 6G).
In addition to the direct O-GlcNAc modification of proteasome subunit Psmc1 (32), ubiquilin-2 and ubiquitin-associated proteins Ubap2 (ubiquitin-associated protein 2) and Ubap2l (ubiquitin-associated protein 2-like) have been reported to be directly O-GlcNAcylated (Dataset S3) (33, 34). Ubiquilin-2 links the proteasome and the polyubiquitin chains on targeted proteins through its N-terminal ubiquitin-like domain (UBL) and C-terminal ubiquitin-associated domain (UBA) (35). Both Ubap2 and Ubap2l contain UBA domains and play important roles in the ubiquitin–proteasome pathway (36, 37). To corroborate these findings in mESCs, we precipitated whole cell lysates of Ogt fl mESCs with wheat germ agglutinin (WGA), a lectin that recognizes the O-GlcNAc modification, and immunoblotted for Pmsc1, ubiquilin-2, Ubap2, and Ubap2l. These experiments confirmed previous findings (Dataset S3) (34) showing that Pmsc1/Rpt2, ubiquilin-2, Ubap2, and Ubap2l were O-GlcNAc modified in Ogt fl mESCs (Fig. 6H). In the case of ubiquilin-2, we observed clear enrichment for a slowly migrating, presumably O-GlcNAc-modified protein species. Together, these data suggest that proteasomal subunit and ubiquitin-associated proteins are O-GlcNAc modified by OGT; in the absence of OGT, high proteasome activity and consequent high intracellular amino acid levels result in aberrant mTOR activation and increased mitochondrial OXPHOS, thus causing decreased proliferation and viability of mESCs.
Increased Amino Acid Levels and Increased mTOR Activation Are Also Observed in OGT-Deficient T Cells.
Next, we asked if what we observed in mESCs is a general mechanism across mammalian cell types. To answer this question, we used CD8+ T cells isolated from male Ogt floxedCre–ERT2+/KI Rosa26-LSL-YFP+/KI mice. At day 6 after 4-OHT treatment, the O-GlcNAc modification was completely eliminated in Ogt iKO CD8+ T cells as assessed by flow cytometry (SI Appendix, Fig. S15A). As expected, Ogt iKO CD8+ T cells showed a significant impairment of growth rate (SI Appendix, Fig. S15B; note logarithmic scale of Y-axis). Consistent with our results in mESCs, OGT deficiency in CD8+ T cells was associated with a significant increase in the levels of certain amino acids, most notably the branched chain amino acids leucine, isoleucine, and valine and the aromatic amino acids phenylalanine, tryptophan, and tyrosine (SI Appendix, Fig. S15C), as well as with mTOR activation assessed by flow cytometry for phospho-S6 (SI Appendix, Fig. S15 D and E). The levels of phosphorylation of S6 ribosomal protein on serine 235/236 were significantly increased in Ogt iKO CD8+ T cells, while the levels of total S6 ribosomal protein were slightly decreased as analyzed by flow cytometry (SI Appendix, Fig. S15 D and E). Treatment with the mTOR inhibitor Torin2 and the mitochondrial inhibitor rotenone partially rescued the decrease of cell numbers in Ogt iKO CD8+ T cells, despite a decrease in cell numbers of control Ogt fl CD8+ T cells upon treatment with these inhibitors (SI Appendix, Fig. S15 F and G). Together, these data suggest that OGT controls cell viability through the proteasome/mTOR/mitochondrial axis in both embryonic stem cells and somatic cells.
Discussion
The O-GlcNAc transferase OGT is highly conserved across species (1, 38) and is absolutely required for the proliferation and survival of mammalian cells (3, 39, 40), but the reason for this requirement has been unclear (1). To address this point, we performed an unbiased, genome-wide CRISPR–Cas9 “suppressor” screen in Ogt fl mESCs, in which the Ogt gene could be inducibly deleted by treatment with 4-OHT. The screen provided important insights, revealing that mitochondrial dysfunction is a major cause for the block in cell proliferation and eventual loss of cell viability induced by OGT deficiency. We traced this phenotype to mTOR hyperactivity, resulting from increased steady-state amino acid levels that in turn stemmed from increased proteasomal activity (SI Appendix, Fig. S16). An independent phosphoproteomic screen also emphasized the pronounced activation of the mTOR pathway in OGT-deficient mESCs. The data are generalizable to other cell types, since increased amino acid levels and increased mTOR activation are also observed in OGT-deficient CD8+ T cells. A useful outcome is that we have been able to obtain viable, proliferating, OGT-deficient mESCs by inhibiting mTOR hyperactivation with Torin2, or by depleting candidate gene products from the screen. The availability of cultured OGT-deficient cells will permit a comprehensive analysis of how OGT regulates diverse aspects of cellular function, in the absence of complications posed by cell cycle arrest and ensuing apoptosis.
Our study connects OGT deficiency directly to several aberrations in mitochondrial function, including overactivity of the mitochondrial electron transport chain, abnormally increased mitochondrial mass and mitochondrial membrane potential, and increased expression of genes encoded in the mitochondrial genome. -OGT-deficient mESCs displayed a dramatic increase in mitochondrial OXPHOS, and despite its toxicity to normal cells, the mitochondrial complex one inhibitor rotenone partially rescued the proliferation defect of Ogt-deficient cells. sgRNA-mediated disruption of genes encoding three of the most prominent hits in our screen – Socs3, Xpo5, and Gtpbp8 – returned mitochondrial OXPHOS to almost normal levels while also rescuing the proliferation defect of OGT-deficient cells. These proteins were not previously known to have specific mitochondrial functions. Gtpbp8, a member of the Obg family of P-loop-containing small G proteins, is annotated as mitochondrial (41); a homolog, Gtpbp10, resides in the mitochondrial matrix and is required for proper maturation of the large subunit of the mitochondrial ribosome (42). Gtpbp8 may have a related function, since several Mrpl proteins – components of the mitochondrial large ribosomal subunit – were also hits in our screen. Socs3 is a key negative regulator of Stat3, which is at least partially localized to mitochondria (43). Xpo5 is an exportin that exports pre-miRNAs and tRNAs from the nucleus to the cytoplasm (44). Further analysis of the functional roles of these three proteins in mitochondria, and the mechanism by which they suppress mitochondrial dysfunction in OGT-deficient cells, would be of considerable interest. Notably, rescue by the top sgRNA candidates identified in our whole-genome screen was incomplete; given that OGT has thousands of substrates, other mechanisms (in addition to mTOR hyperactivation and the associated mitochondrial dysfunction) might contribute to the defect in cell proliferation in Ogt-deficient cells.
Despite these unambiguous findings, it is clear from numerous papers in the OGT arena that cellular responses to manipulating OGT, OGA, and O-GlcNAc levels are complex and vary with the exact cell types and approaches used. Parameters that might affect cellular and mitochondrial function include whether the analyzed cells are cycling or quiescent, the deletion efficiencies of Ogt or Oga genes, and the precise level of OGT or OGA activity remaining in different cellular compartments after inhibitor treatment. In our mESC system in which the parental Ogt fl mESCs are highly proliferative and the efficiency of Ogt deletion is >95%, Ogt iKO cells displayed a dramatic increase in mitochondrial OXPHOS and OCR. Likewise, in HeLa cells, even partial (50 to 70%) depletion of OGT with siRNA resulted in increased basal and maximal OCR (8). In contrast, treatment of SH-SY5Y neuroblastoma or NT2 human embryonal carcinoma cells with the OGA inhibitor thiamet-G decreased basal OCR and ATP production (11), but the same inhibitor resulted in increased oxygen consumption and ATP production rates in isolated cardiac mitochondria (10). In a particularly complex example, Ogt deletion in HSC yielded cells with decreased copy numbers of mitochondrial DNA and decreased spare respiratory capacity; a caveat is that the numbers of HSC were greatly decreased, presumably because of loss of HSC viability in the absence of OGT (12). In this context, Ogt iKO CD8+ T cells resembled Ogt iKO mESC in showing decreased proliferation, mTOR hyperactivation, and increased levels of several amino acids including branched-chain and aromatic amino acids, and partial rescue of the cell proliferation defect by Torin2 and rotenone. Whole-genome screens for rescue of cell viability in different cell types might be useful to distinguish cell-specific versus common pathways after Ogt deletion.
Many previous studies of the crosstalk between OGT and mTOR have focused on how mTOR signaling affects OGT function. In breast cancer cells, mTOR activation was associated with increased levels of OGT and O-GlcNAc modification (45). In hepatic cancer cells, pharmacological inhibition of mTOR led to decreased OGT expression concomitant with a global reduction of O-GlcNAc modification (46). Thus, in these settings, mTOR appears to activate OGT. Our focus here is in the opposite direction: i.e., how OGT affects mTOR activity. Because mTOR activity promotes cell proliferation (47), and because of the dramatic decrease in cell proliferation in Ogt iKO cells, it was generally assumed that OGT deficiency would lead to decreased mTOR (48). Consistent with this assumption, treatment of neurons with OGT inhibitors significantly down-regulated mTOR activity (48), suggesting that OGT might activate mTOR. Notably, however, nondividing cells such as neurons – unlike proliferating cells – can survive in the absence of OGT (49), and given the differences in cellular context, the mechanism of crosstalk between OGT and mTOR has not been easy to decipher. However, a previous publication showed that mTOR activity (assessed by immunoblotting for phospho-S6K) was significantly increased in OGT-deficient CD8+ T cells, although the authors pursued a different finding, the downregulation of Myc protein in these cells (40). Thus, our study shows that complete Ogt gene deletion is (perhaps paradoxically) associated with mTOR hyperactivation, leading to increased mitochondrial OXPHOS, proliferation arrest, and eventual loss of viability of Ogt iKO cells (SI Appendix, Fig. S16).
Dysregulated metabolism is one of the hallmarks of cancer, and many of the proteins that play important roles in the proteasome/mTOR/mitochondrial axis (SI Appendix, Fig. S16) are aberrantly expressed in cancer. Increased levels of OGT expression and O-GlcNAc modification are often observed in cancer cells (50–52). Hyperactivation of mTOR signaling is observed in many types of cancers; mTOR signaling is the second most frequently altered signaling pathway in cancer, and multiple mTOR inhibitors have been used in cancer clinical trials (53, 54). Increased expression of proteasome subunits and high proteasome activity are also characteristic of many cancer types (55–57), and proteasome inhibitors have been used in the clinic for the treatment of hematological malignancies (58). We have shown that several proteasome subunits and ubiquitin-associated proteins, including Psmc1, Ubap2, Ubap2l, and Ubiquilin2, are targets for O-GlcNAcylation by OGT and display altered expression in cancer cells compared to normal controls (59–64). Therefore, a detailed investigation of OGT activity in proteasome/mTOR signaling in different cancer cell types may provide novel targets for cancer therapy in the future.
Materials and Methods
Detailed materials and methods are described in SI Appendix, SI Materials and Methods.
Generation and Culture of mESCs.
Ogtfloxed Cre–ERT2KI/KI mice were crossed with Ogtfloxed Rosa26-YFPKI/KI mice to obtain OgtfloxedCre–ERT2+/KI Rosa26-LSL-YFP+/KI blastocysts (embryonic day 3.5) to generate male and female Ogt floxed mESC lines. Ogt deletion was then induced by the addition of 1 μM 4-hydroxytamoxifen (4-OHT) (TOCRIS, catalog number: 3412). Mouse ESCs were maintained on mitomycin C-treated mouse embryonic fibroblasts (MEFs; feeder cells) with LIF in knockout DMEM medium (ThermoFisher Scientific, catalog number: 10829018) supplemented with 15% KOSR (KnockOut Serum Replacement, ThermoFisher Scientific, catalog number: 10828028), 2 mM L-glutamine, 1 X MEM nonessential amino acids, and 50 μM β-mercaptoethanol. Cell numbers were counted by flow cytometry on a BD Accuri C6 (BD Biosciences).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
We thank Dr. Joyce Chen and Dr. Anand Balasubramani for generating Ogtfloxed Cre–ERT2KI/KI mice and Ogtfloxed Rosa26-LSL-YFPKI/KI mice; Dr. Mohit Jain (UCSD), Dr. Lucas Sullivan (Fred Hutchinson Cancer Research Center), and members of the Rao laboratory for suggestions and discussions. We thank Dr. Natasha Zachara from Johns Hopkins University School of Medicine for providing Ogt-floxed mice; Dr. Ronald M. Evans (Salk Institute) for providing WT OGT plasmid; C. Kim, D. Hinz, C. Dillingham, M. Haynes, and S. Ellis at the La Jolla Institute Flow Cytometry facility for help with cell-sorting experiments; J. Day, S. Alarcon, H. Dose, K. Tanguay, and A. Hernandez of the La Jolla Institute Sequencing facility for help with next-generation sequencing; and Olga Zagnitko at Cancer Metabolism Core at Sanford Burnham Prebys Medical Discovery Institute for assistance with GC/MS analyses. This work was supported by NIH R01 grant R35 CA210043 and R01 CA247500 (to A.R.). FACSAria II Cell Sorter was acquired through the Shared Instrumentation Grant (SIG) Program S10 RR027366, and Hiseq 2500 was funded by S10OD016262. The Sanford Burnham Prebys Cancer Metabolism Core is supported by NCI Cancer Center Support Grant P30 CA030199. X.L. was supported by a postdoctoral fellowship from CIRM UCSD Interdisciplinary Stem Cell Research and Training Grant II (TG2-01154). H.S. was supported by Pew Latin American Fellows Program from the Pew Charitable Trusts and from EDUC4-12804 from California Institute for Regenerative Medicine (CIRM).
Author contributions
X.L., X.Y., S.A.M., and A.R. designed research; X.L., X.Y., H.S., R.A.B., and D.A.S. performed research; S.A.C. contributed new reagents/analytic tools; X.L., X.Y., S.A.M. and A.R. analyzed data; and X.L., X.Y., S.A.M., and A.R. wrote the paper.
Competing interest
The authors declare competing interest. The authors have additional information to disclose, co-author Burt and reviewer Slawson are co-authors on a 2022 review together (https://www.frontiersin.org/articles/10.3389/fmolb.2022.920727/full).
Footnotes
Reviewers: M.R.P., University of Southern California; and C.S., University of Kansas Medical Center.
Contributor Information
Samuel A. Myers, Email: sam@lji.org.
Anjana Rao, Email: arao@lji.org.
Data, Materials, and Software Availability
The proteomics and phosphoproteomics data can be accessed at https://massive.ucsd.edu, dataset MSV000088393. All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Levine Z. G., Walker S., The biochemistry of O-GlcNAc transferase: Which functions make it essential in mammalian cells? Annu. Rev. Biochem. 85, 631–657 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Bond M. R., Hanover J. A., A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shafi R., et al. , The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Z., Vosseller K., O-GlcNAc in cancer biology. Amino Acids 45, 719–733 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Yang X., et al. , Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Marsh S. A., Collins H. E., Chatham J. C., Protein O-GlcNAcylation and cardiovascular (patho)physiology. J. Biol. Chem. 289, 34449–34456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanover J. A., et al. , Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409, 287–297 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Sacoman J. L., Dagda R. Y., Burnham-Marusich A. R., Dagda R. K., Berninsone P. M., Mitochondrial O-GlcNAc transferase (mOGT) Regulates mitochondrial structure, function, and survival in HeLa cells. J. Biol. Chem. 292, 4499–4518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Love D. C., Kochan J., Cathey R. L., Shin S. H., Hanover J. A., Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J. Cell Sci. 116, 647–654 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Ma J., et al. , O-GlcNAcomic profiling identifies widespread O-Linked β-N-Acetylglucosamine modification (O-GlcNAcylation) in oxidative phosphorylation system regulating cardiac mitochondrial function. J. Biol. Chem. 290, 29141–29153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan E. P., et al. , Sustained O-GlcNAcylation reprograms mitochondrial function to regulate energy metabolism. J. Biol. Chem. 292, 14940–14962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami K., et al. , OGT Regulates hematopoietic stem cell maintenance via PINK1-dependent mitophagy. Cell Rep. 34, 108579 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Condon K. J., Sabatini D. M., Nutrient regulation of mTORC1 at a glance. J. Cell sci. 132, jcs222570 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G. Y., Sabatini D. M., mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Sancak Y., et al. , The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science, 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L., Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham J. T., et al. , mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Morita M., et al. , mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Murad H., et al. , Induction of G1-phase cell cycle arrest and apoptosis pathway in MDA-MB-231 human breast cancer cells by sulfated polysaccharide extracted from Laurencia papillosa. Cancer Cell Int. 16, 39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spahn P. N., et al. , PinAPL-Py: A comprehensive web-application for the analysis of CRISPR/Cas9 screens. Sci. Rep. 7, 15854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y., Metascape provides a biologist-oriented rerefrefsource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwashita H., et al. , Live cell imaging of mitochondrial autophagy with a novel fluorescent small molecule. ACS Chem. Biol. 12, 2546–2551 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Martínez-Reyes I., Chandel N. S., Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11, 102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paplomata E., O’Regan R., The PI3K/AKT/mTOR pathway in breast cancer: Targets, trials and biomarkers. Thera. Adv. Med. Oncol. 6, 154–166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortiz-Meoz R. F., et al. , A small molecule that inhibits OGT activity in cells. ACS Chem. Biol. 10, 1392–1397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Proud C. G., The mTOR pathway in the control of protein synthesis. Physiology 21, 362–369 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Krug K., et al. , A curated resource for phosphosite-specific signature analysis. Mol. Cell. Proteomics 18, 576–593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acosta-Jaquez H. A., et al. , Site-specific mTOR phosphorylation promotes mTORC1-mediated signaling and cell growth. Mol. Cell. Biol. 29, 4308–4324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy B. K., Lamming D. W., The mechanistic target of rapamycin: The grand ConducTOR of metabolism and aging. Cell Metab. 23, 990–1003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F., et al. , O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–725 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Burt R. A., et al. , Novel antibodies for the simple and efficient enrichment of native O-GlcNAc modified peptides. Mol. Cell. Proteomics 20, 100167 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wulff-Fuentes E., et al. , The human O-GlcNAcome database and meta-analysis. Sci. Data 8, 25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renaud L., Picher-Martel V., Codron P., Julien J. P., Key role of UBQLN2 in pathogenesis of amyotrophic lateral sclerosis and frontotemporal dementia. Acta Neuropathol. Commun. 7, 103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmann K., Bucher P., The UBA domain: A sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21, 172–173 (1996). [PubMed] [Google Scholar]
- 37.Wilde I. B., Brack M., Winget J. M., Mayor T., Proteomic characterization of aggregating proteins after the inhibition of the ubiquitin proteasome system. J. Proteome Res. 10, 1062–1072 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Hart G. W., Nutrient regulation of signaling and transcription. J. Biol. Chem. 294, 2211–2231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Donnell N., Zachara N. E., Hart G. W., Marth J. D., Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swamy M., et al. , Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 17, 712–720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma Y., et al. , MRX8, the conserved mitochondrial YihA GTPase family member, is required for de novo Cox1 synthesis at suboptimal temperatures in Saccharomyces cerevisiae. Mol. Biol. Cell 32, ar16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maiti P., Kim H. J., Tu Y. T., Barrientos A., Human GTPBP10 is required for mitoribosome maturation. Nucleic Acids Res. 46, 11423–11437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szczepanek K., Chen Q., Larner A. C., Lesnefsky E. J., Cytoprotection by the modulation of mitochondrial electron transport chain: The emerging role of mitochondrial STAT3. Mitochondrion 12, 180–189 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu K., He J., Pu W., Peng Y., The role of exportin-5 in microRNA biogenesis and cancer. Genom. Proteom. Bioinform. 16, 120–126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sodi V. L., et al. , mTOR/MYC axis regulates O-GlcNAc transferase expression and O-GlcNAcylation in breast cancer. Mol. Cancer Res. 13, 923–933 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park S., Pak J., Jang I., Cho J. W., Inhibition of mTOR affects protein stability of OGT. Biochem. Biophys. Res. Commun. 453, 208–212 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Morita M., et al. , mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 14, 473–480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahman M. A., Cho Y., Hwang H., Rhim H pharmacological inhibition of O-GlcNAc transferase promotes mTOR-dependent autophagy in rat cortical neurons. Brain Sci. 10, 958 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruan H. B., et al. , O-GlcNAc transferase enables AgRP neurons to suppress browning of white fat. Cell 159, 306–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu D., Jin J., Qiu Z., Liu D., Luo H., Functional analysis of O-GlcNAcylation in cancer metastasis. Front. Oncol. 10, 585288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mi W., et al. , O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochimica. Biophysica. Acta 1812, 514–519 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Wang L., et al. , Suppressed OGT expression inhibits cell proliferation while inducing cell apoptosis in bladder cancer. BMC Cancer 18, 1141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian T., Li X., Zhang J., mTOR signaling in cancer and mTOR inhibitors in solid tumor targeting therapy. Int. J. Mol. Sci. 20, 755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de la Cruz López K. G., Toledo Guzmán M. E., Sánchez E. O., García Carrancá A., mTORC1 as a regulator of mitochondrial functions and a therapeutic target in cancer. Front. Oncol. 9, 1373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumatori A., et al. , Abnormally high expression of proteasomes in human leukemic cells. Proc. Natl. Acad. Sci. U.S.A. 87, 7071–7075 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arlt A., et al. , Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2). Oncogene 28, 3983–3996 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Chen L., Madura K., Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res. 65, 5599–5606 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Manasanch E. E., Orlowski R. Z., Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 14, 417–433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahlman K. B., et al. , Modulators of prostate cancer cell proliferation and viability identified by short-hairpin RNA library screening. PloS one 7, e34414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jantrapirom S., Piccolo L. L., Pruksakorn D., Potikanond S., Nimlamool W., Ubiquilin networking in cancers. Cancers 12, 1586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshida K., et al. , Clinical significance of ubiquitin-associated protein 2-like in patients with uterine cervical cancer. In Vivo 34, 109–116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Latonen L., et al. , Amplification of the 9p13.3 chromosomal region in prostate cancer. Genes Chromosomes Cancer 55, 617–625 (2016). [DOI] [PubMed] [Google Scholar]
- 63.He J., Chen Y., Cai L., Li Z., Guo X., UBAP2L silencing inhibits cell proliferation and G2/M phase transition in breast cancer. Breast Cancer 25, 224–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li D., Huang Y., Knockdown of ubiquitin associated protein 2–like inhibits the growth and migration of prostate cancer cells. Oncol. Rep. 32, 1578–1584 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
The proteomics and phosphoproteomics data can be accessed at https://massive.ucsd.edu, dataset MSV000088393. All study data are included in the article and/or SI Appendix.