Highlights
-
•
Microwells allow combining up to 100 human primary liver spheroids in one system.
-
•
The model is set up in a 96-well (liver-plate) and microfluidic format (liver-chip).
-
•
Cell viability and CYP activity of liver-chips are comparable to liver spheroids.
-
•
Liver-chips can be deployed for multiparametric drug safety testing.
-
•
Liver-chips possess a higher metabolic capacity versus single liver spheroids.
Keywords: MPS, liver, Microwells, Spheroid, DILI, Drug metabolism
Abstract
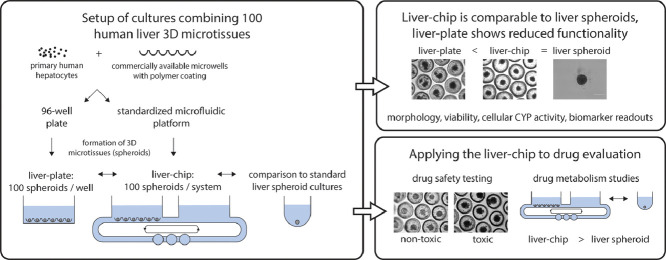
Human 3D liver microtissues/spheroids are powerful in vitro models to study drug-induced liver injury (DILI) but the small number of cells per spheroid limits the models’ usefulness to study drug metabolism. In this work, we scale up the number of spheroids on both a plate and a standardized organ-chip platform by factor 100 using a basic method which requires only limited technical expertise. We successfully generated up to 100 spheroids using polymer-coated microwells in a 96-well plate (= liver-plate) or organ-chip (= liver-chip). Liver-chips display a comparable cellular CYP3A4 activity, viability, and biomarker expression as liver spheroids for at least one week, while liver-plate cultures display an overall reduced hepatic functionality. To prove its applicability to drug discovery and development, the liver-chip was used to test selected reference compounds. The test system could discriminate toxicity of the DILI-positive compound tolcapone from its less hepatotoxic structural analogue entacapone, using biochemical and morphological readouts. Following incubation with diclofenac, the liver-chips had an increased metabolite formation compared to standard spheroid cultures. In summary, we generated a human liver-chip model using a standardized organ-chip platform which combines up to 100 spheroids and can be used for the evaluation of both drug safety and metabolism.
Graphical abstract
1. Introduction
Primary human hepatocytes (PHH) are essential in vitro cultures used in drug discovery and development to allow a relevant and translatable assessment of potential drug candidates for clearance, metabolism, and potential hepatotoxicity. PHH have mainly been utilized in suspension or as two-dimensional (2D) monolayer cultures, however, only short-term incubations are possible as hepatocytes quickly dedifferentiate in vitro [1].
Three-dimensional (3D) cultures of PHH as liver microtissues (= liver spheroids) preserve the hepatocyte phenotype and functionality for several weeks [2,3]. The longevity of the model allows long-term drug incubations with repeated drug administrations to be performed, mimicking the chronic treatment regimen of many marketed compounds and achieving a higher relevance to the clinical setting. Such a long-term drug exposure gives human 3D liver spheroids a superior sensitivity compared to 2D cultures to detect the hepatotoxicity of drugs causing drug-induced liver injury (DILI) in patients (= DILI-positive compounds). Both 2D and 3D models show high specificity and limit the number of false positive signals for drugs with less or no DILI concerns in patients (= DILI-negative compounds) [4,5]. In recent years, human liver spheroids became an increasingly useful tool for pharma companies to screen drug candidates for DILI and to gain mechanistic insights into toxicity [4,6,7].
Recently, human liver spheroids were evaluated to predict the in vivo hepatic clearance of compounds [8,9]. The long-term stability of spheroids allows these cultures to be incubated with one or more compound(s) for multiple days. However, spheroids contain considerably fewer cells than a typical suspension model, which again limits the model's sensitivity to detect the clearance of compounds with high metabolic stabilities. To address this limitation, Kanebratt et al. successfully pooled 3 spheroids in one well [9]. Riede et al. also explored this approach by pooling 5 spheroids in the same well, however, they observed aggregation of the spheroids and a decrease of cytochrome P450 (CYP) enzyme expression over 7 days, leading to an underprediction of intrinsic clearance [8]. While human liver spheroids are highly suitable for long-term incubations, multiple spheroids will need to be combined to increase the total cell number and metabolic capacity in one incubation. Therefore, the development of a method which allows the pooling of multiple spheroids whilst preventing aggregation and fusion is highly desirable.
One potential solution to overcome this challenge is the use of microwell arrays with multiple small cavities that can each contain a spheroid. By introducing a microwell array in the system, spheroids can be contained in separate microwells, thereby preventing their aggregation and fusion, whilst still allowing all spheroids to share the same medium compartment. Recently, microwell plates and inserts became commercially available to combine spheroids of the liver and other tissues [10], [11], [12], [13].
Finally, microphysiological systems (MPS), also known as organ-on-chips are increasingly being implemented to create more physiological and predictive in vitro cultures suitable for long-term culturing experiments. MPS allow circulation of the medium, which emulates the blood flow. Circulation of the medium leads to an improved exchange of nutrients, gasses, waste products and drugs between the medium and the cells resulting in improved longevity and physiological relevance [14,15]. Due to medium flow, the culture will also experience shear stress, which is a key biological signal to some cell types to reach physiological functionality [16,17]. Finally, medium flow allows the interconnection of multiple compartments and thus the setup of multi-organ chips. To properly emulate organ-organ interactions, the cell ratio between organs in vitro must closely approach in vivo values [18,19]. Several liver MPS cultures have been set up in custom and standardized chip platforms [14,15,20]. A hepatic compartment is also implemented in most multi-organ chips, since the liver is the prime site of drug metabolism and has shown physiological interplay with several other organs in vitro [18,[21], [22], [23], [24]].
The combination of MPS with PHH-derived liver spheroids has only been reported in a limited number of articles to our knowledge [21,23,25,26]. Application of human liver spheroids from primary cells on organ-chip models has either been in customized systems by pooling 10-50 spheroids in a gel [23,25] or with 10 spheroids in a standardized model combining flow and hanging-drop technology [21,26]. It would be useful to have an easy-to-use approach that can combine numerous PHH-derived liver spheroids on a standardized organ-chip platform to allow the implementation of such a novel model with good reproducibility of results across different labs. Such a model could have an added value over less complex and physiological models to investigate the safety and metabolism of potential drug candidates during drug discovery and development.
In this work, we describe the setup of a simple method using commercially available microwells to simultaneously form and culture 100 human liver spheroids from primary cells within one system. The model was set up in a 96-well plate and standardized organ-chip model. We termed these models liver-plate and liver-chip, respectively, comparable to other articles describing similar systems [14,27]. Comparison of both systems against conventional liver spheroid cultures demonstrated that the liver-chip had a comparable hepatic functionality as liver spheroids, while the liver-plate did not meet the same standards. Finally, the liver-chip will be shown to have been successfully applied to study drug safety and metabolism profiling, and possesses an added value compared to liver spheroid cultures to study drug metabolism.
2. Materials and methods
2.1. Microwell insert preparation
Dynarray microwells (polycarbonate, porous, 500 µm diameter per microwell) were obtained from 300MICRONS (Karlsruhe, Germany). Air bubbles in microwell cavities were removed by immersing the arrays in an alcohol-water series according to the manufacturer's instructions and finally washed in distilled water (Sigma Aldrich). Microwells were subsequently made cell-repellent by coating with BIOFLOAT FLEX solution (faCellitate, Mannheim, Germany) for 3 min, followed by airdrying the solution for at least 30 min under laminar flow. Finally, 96-well inserts were cut out using a 6 mm skin biopsy puncher (Acuderm, Ford Lauderdale, FL, USA) and transferred to a cell culture plate or chip with the cavities facing upwards.
2.2. Preparation of chips
Microphysiological 2-organ chips were obtained from TissUse (Berlin, Germany). Every circuit was composed of two 96-well compartments (of which one equipped with a reservoir holder) and 3 on-chip micropumps. Every compartment was filled with 300 µl of Lonza hepatocyte complete medium (HCM) (Lonza, Verviers, Belgium) with 10% premium-grade FBS (VWR, Radnor, PA, USA). In the experiments using microwell arrays, a microwell insert was placed at the bottom of every reservoir holder.
2.3. Cell sources
PHH and crude mixed non-parenchymal cells (NPC) were all obtained from BioIVT (Brussels, Belgium). The human biological samples were sourced, and their research use was in accord with the terms of the informed consent. PHH were thawed in human cryopreserved hepatocyte thawing medium (Lonza), NPC were thawed in CP medium (BioIVT). After centrifugation for 10 minutes at 100 g cells were resuspended in Lonza HCM + 10% FBS and counted using a hemocytometer (Nano-entek, Waltham, MA, USA). PHH were used for the model setup (Fig. 1 and 2), PHH + NPC were utilized for further characterization and drug testing (Fig. 3 and 4). PHH and NPC were mixed in a 2:1 ratio, which was previously found as the optimal ratio in spheroids [6].
Fig. 1.
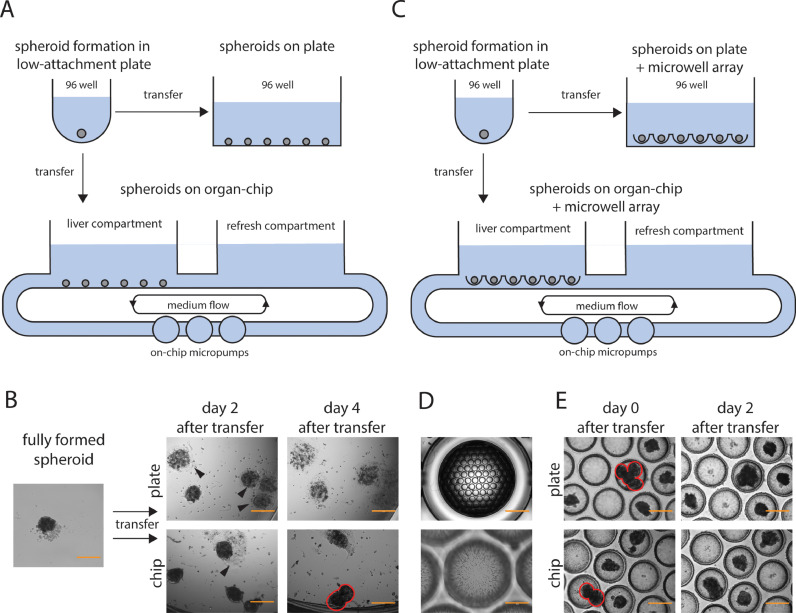
Pooling human liver spheroids in an organ-chip and plate setup.
A: overview figure of the culture setup with spheroid formation in an ultra-low attachment plate, and transfer of several spheroids to a 96-well plate or organ-chip. B: brightfield pictures showing fully formed spheroids (after 7-8 days) before and after transfer to a plate or organ-chip. Arrowheads indicate spheroids adhering to the bottom, red circles indicate fusing spheroids. Scale bar, 400 µm. C: overview figure of the culture setup with spheroid formation in an ultra-low attachment plate, and transfer of several spheroids to a 96-well plate or organ-chip containing microwells. D: overview picture of microwells, showing ∼ 100 cavities per array and a zoomed-in picture of microwells, showing pores in the material. Scale bar, 2000 µm (top picture), 200 µm (bottom picture). E: pictures of plate and organ-chip with microwells after transfer of several spheroids. Red circles indicate fusing spheroids. Scale bar, 400 µm.
Fig. 2.
Human liver spheroid formation in a plate and organ-chip using microwells.
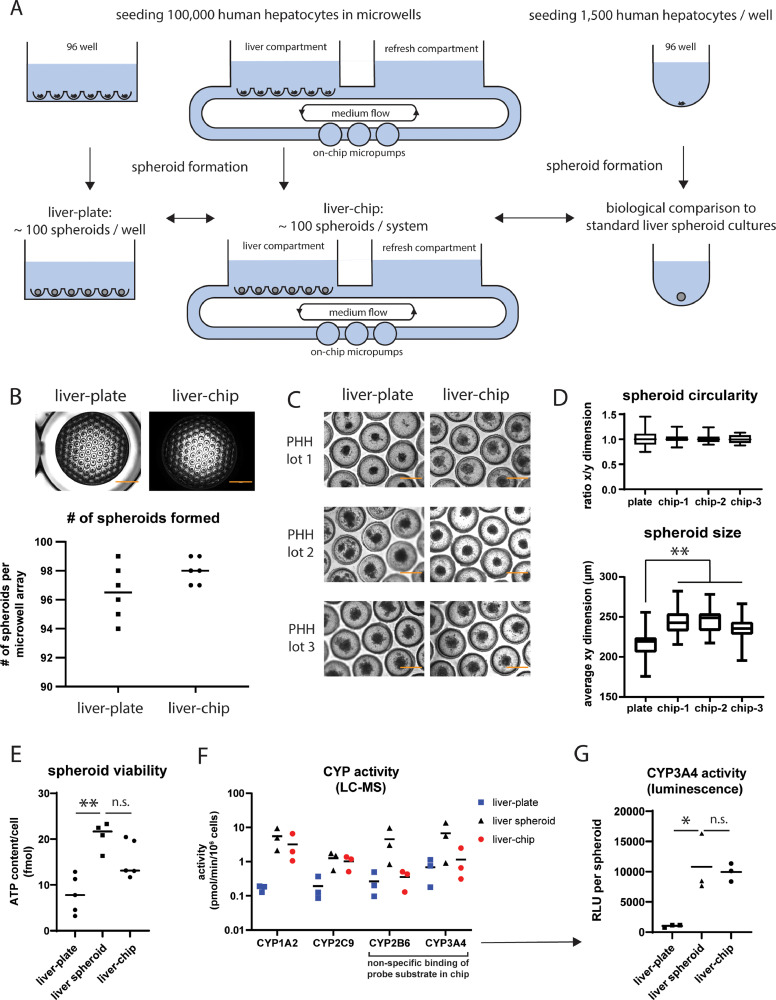
A: overview figure of the culture setup showing immediate seeding of PHH in microwells in a 96-well plate and organ-chip. Both models were biologically compared to conventional liver spheroid cultures, formed by seeding cells in low-attachment wells. B: brightfield pictures showing spheroids formed in a microwell array in a plate and organ-chip after 7 days of culture. The number of spheroids per system was shown in the graph below, which represents the mean + individual values. N = 6 individual cultures. Scale bar, 2000 µm. C: brightfield pictures of spheroids formed after 6-10 days of culture in microwells in a plate and organ-chip. Spheroid formation was successful in all three tested PHH lots. Scale bar, 400 µm. D: spheroid size and circularity were quantified in fully formed spheroid cultures (day 7-8 of culture) by measuring the x- and y-dimensions of spheroids in individual microwell array in both the plate and organ-chip. Each box plot summarizes data from 21-27 individual spheroids within the same microwell array. **, p < 0.01. E: spheroid viability of the liver-plate, liver-chip and liver spheroid culture after 2 weeks of culture. The graph shows the mean and individual values expressed as fmol ATP/cell. **, p < 0.01; n.s., not significant (p ≥ 0.05). F: CYP enzyme activity (determined using LC-MS quantification of primary metabolites) in liver-chip, liver-plate, and liver spheroids after 2 weeks of culture. The graph shows the mean and individual values obtained from 3 different donors. G: CYP3A4 activity per spheroid (as measured by a luminescence-based assay) of the liver-plate, liver-chip and single liver spheroid after 2 weeks of culture. The graph shows the mean and individual values per condition. The luminescent signal is expressed in relative luminescent units (RLU) and corrected for medium volume and number of spheroids per condition. *, p < 0.05; n.s., not significant (p ≥ 0.05).
Fig. 3.
Biomarker characterization of the liver-chip over time.
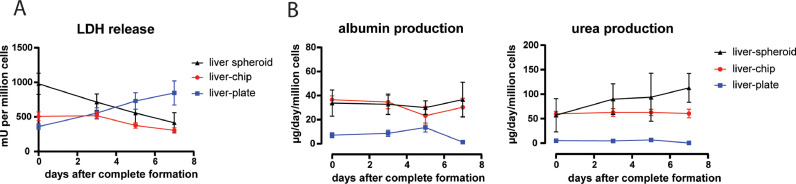
A: LDH release in the medium of the different cultures over time. The data is shown as the amount of LDH present in the medium (milli-units, mU) normalized to the number of cells in each culture. Data represents mean ± standard deviation of the mean. N = 3. B: albumin and urea production in the different cultures over time. Data represents mean ± standard deviation. N = 3-4.
Fig. 4.
Application of the liver-chip to study drug safety and drug metabolism.
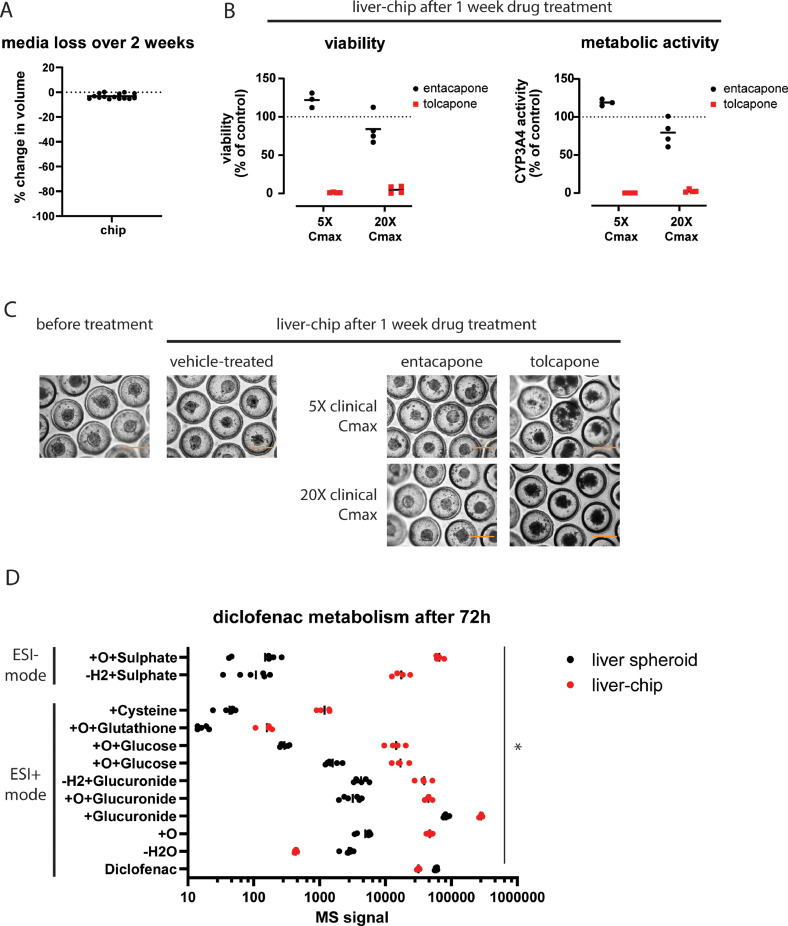
A: media loss in the liver-chip after 2 weeks of culturing. Graph indicates mean with 15 individual data points. B: Liver-chip viability and CYP3A4 activity after being treated for 7 days with entacapone (black circles) or tolcapone (red squares) at 5X or 20X the clinical Cmax. Signal was normalized to DMSO-treated controls. Data represents the mean with individual data points. N = 3-4. C: brightfield pictures of spheroids in the liver-chip before and after treatment. Scale bar, 400 µm. D: metabolite profile following incubation of the liver-chip and single spheroid culture for 72 hours with 50 µM diclofenac. *, p < 0.05 between the liver spheroid and liver-chip for all reported metabolites.
2.4. Liver spheroid formation
Single spheroid culture: 1,500 PHH with or without 750 NPC were seeded in 100 µl Lonza HCM + 10% FBS per well in BIOFLOAT 96-well plates (faCellitate). Plates were centrifuged for 2 min at 100 g and incubated at 37 °C. 2–4 days later, each well was supplemented with 100 µl of serum-free Lonza HCM, giving a total volume of 200 µl per well. Next, 50% media changes with serum-free Lonza HCM were performed for 3 more days. After 7–8 days, spheroids were fully formed and showed a well-defined edge (as seen in Fig. 2C and in line with other reports [3]) and were used for downstream applications (transfer, maintenance, drug treatment).
Loading spheroids in a plate: 16 or 32 fully formed spheroids were collected, pooled in 100 µl Lonza HCM using widebore pipette tips (Thermo Fisher, Waltham, MA, USA) and transferred into one well of a transparent 96-well plate (Corning, Corning, NY, USA) with or without microwell insert containing 100 µl Lonza HCM. Low-adhesive wells were obtained by adding 100 µl BIOFLOAT FLEX solution (faCellitate, Mannheim, Germany) for 3 min, followed by airdrying of the solution for at least 30 min under laminar flow.
Loading spheroids in a chip: 16 fully formed spheroids were pooled in 100 µl Lonza HCM using wide-bore pipette tips (Thermo Fisher). In the 2 organ-chip (TissUse), 100 µl of the medium in the reservoir compartment (total volume per circuit: 600 µl) was replaced by 100 µl Lonza HCM containing 16 spheroids.
Liver-plate formation: 100,000 PHH with or without 50,000 NPC were seeded in 100 µl Lonza medium + 10% FBS per well in a transparent 96-well plate (Corning) previously loaded with Dynarray microwell inserts (300MICRONS) and pre-wetted with 50 µl Lonza HCM + 10% FBS. The seeding medium was carbogen-saturated unless otherwise specified, while the medium for refreshing was not saturated with carbogen. The plate was centrifuged for 2 minutes at 100 g. 2-4 days later, each well was supplemented with 150 µl of serum-free Lonza HCM, giving a total volume of 300 µl. The next day, 50% media changes with serum-free Lonza HCM were performed for 3 more days. On day 7-8, spheroids were fully formed.
Liver-chip formation: In the 2-organ chip (TissUse), 100 µl of the medium in the reservoir compartment with microwells was replaced by 100 µl carbogen-saturated Lonza medium + 10% FBS with 100,000 PHH and with or without 50,000 NPC, giving a total volume of 600 µl per circuit. The chip was centrifuged for 2 min at 100 g. Immediately afterwards, the medium of the other compartment of the chip was replaced by carbogen-saturated Lonza HCM + 10% FBS. The chip was connected to the HUMIMIC starter (TissUse) with a pumping frequency of 0.5 Hz and a pressure of ±200 mbar. After 1 day, 50% of the total medium was replaced with serum-free Lonza HCM (without carbogen saturation) in the cell-free compartment. As the cell-containing compartment does not have to be disturbed directly, the first media refreshment can be performed earlier than for the liver-plate and liver spheroids. Media changes with serum-free Lonza HCM were performed for 4 days. After the last medium refreshment, the pumping parameters were set to a pumping frequency of 0.75 Hz and a pressure of ±300 mbar. On day 7-8, spheroids were fully formed.
Images were taken using the EVOS XL or the EVOS M5000 (Thermo Fisher). Spheroid dimensions were measured using FIJI (Fiji Is Just ImageJ, https://fiji.sc/, [28]).
2.5. Culture maintenance and drug treatment
Following completed spheroid formation, 50% of the culture medium was refreshed every 2-3 days with serum-free Lonza HCM, on a Monday, Wednesday, or Friday. Diclofenac, entacapone, and tolcapone were bought from Merck/Millipore Sigma (Burlington, MA, USA). Drugs were either dissolved as powder in medium or diluted from a DMSO stock to reach a maximal final concentration of 0.1% DMSO. For drug studies, the added medium contained the drug at double the intended final concentration (2X) or vehicle. To study metabolism, diclofenac was added at a final concentration of 50 µM and incubated for 3 days. For safety testing, liver-chips were treated for 7 days with 5X or 20X the total clinical Cmax of tolcapone (Cmax = 47.577 µM) and entacapone (Cmax = 3.588 µM) [29]. Tolcapone is a strongly hepatotoxic compound (most DILI), while entacapone is a less hepatotoxic structural analogue (less DILI) [29,30]. Half of the media was refreshed every 2-3 days (on a Monday, Wednesday, and Friday) with a 2X solution of the drug or vehicle in Lonza HCM for a total of 7 days. For liquid chromatography-mass spectrometry (LC-MS) or biomarker analysis, medium was removed, diluted depending on the downstream application, and stored at -80 °C.
2.6. Biomarker assays
Supernatants (SN) were collected and stored either undiluted (for albumin and urea analysis) or diluted 1:50 (for liver-plate and liver-chip SN) or 1:5 (for liver spheroids) in lactate dehydrogenase (LDH) storage buffer (for LDH analysis, buffer prepared according to manufacturer's instructions) at -80 °C. Samples diluted in LDH storage buffer were thawed once and analyzed for LDH content using the LDH-glo cytotoxicity assay (Promega, Madisone, WI, USA) following the manufacturer's instructions. To determine albumin production, thawed supernatant samples were diluted 1:500 (for liver-plate and liver-chip SN) or 1:50 (for liver spheroids) in dilution buffer C and analyzed using the human albumin ELISA kit E88-129 (Bethyl laboratories, Montgomery, TX, USA). Urea analysis was performed on 20 µl (for liver-plate and liver-chip or 40 µl (for liver spheroids) of SN using the urea assay kit KA1652 (Abnova, Heidelberg, Germany). All assays were read on a Victor Nivo plate reader (Perkin Elmer, Waltham, MA, USA).
2.7. CYP3A4-Glo and viability assay
Fully formed liver models were maintained for 7 days in the presence or absence of drug. To determine CYP3A4 activity using a luminescence-based assay, 50% of the medium was replaced by the same volume of 2X CYP3A4-Glo probe (Promega) in Lonza HCM. Cultures were incubated for 2 h and 25 µl samples were taken and processed according to the manufacturer's instructions. Raw signals were normalized for volume and spheroid number. For viability assessment, liver spheroids were analyzed for ATP content by removing 150 µl of SN and adding 50 µl CellTiter-Glo® (CTG) 3D assay (Promega) and were processed according to manufacturer's instructions. On the liver-plate and liver-chip, 50% of the total volume was replaced with an equal volume of CTG 3D assay and first put on a shaker for 5 min to lyse the cells. Next, a sample of the lysate was diluted 1:5 in Lonza HCM and 50 µl was mixed with 50 µl CTG 3D assay in a 96-well plate. Raw signals were normalized to the number of seeded cells in each culture. For both the CYP3A4-Glo and CTG 3D assay, luminescence was read using a Victor Nivo plate reader (Perkin Elmer).
2.8. CYP probe cocktail incubation
To evaluate the metabolic activity of CYP enzymes in liver cultures, 50% of the media was removed and replaced by a cocktail of 4 probe substrates diluted in Lonza HCM. The 4 substrates were bupropion (CYP2B6 substrate, final concentration: 100 µM), phenacetin (CYP1A2 substrate, final concentration: 10 µM), diclofenac (CYP2C9 substrate, final concentration: 10 µM), midazolam (CYP3A4 substrate, final concentration: 3 µM). After 4 h of incubation, 50 µl samples were taken and frozen until analysis.
2.9. Non-specific binding of compounds to the chip
To determine the impact of non-specific binding on the nominal concentration of compounds in an empty chip, every compartment of a TissUse 2-organ chip was filled with 300 µl Lonza HCM. Next, 300 µl of the refresh compartment was replaced with 300 µl Lonza HCM containing either the CYP probe cocktail at 2x the final concentration or 300 µl Lonza HCM containing a mix of entacapone and tolcapone at 10x the clinical Cmax. The pumping parameters were set to a pumping frequency of 0.75 Hz and a pressure of ±300 mbar. After 4 and 24 h, 50 µl media samples were taken from the receiver compartment and frozen until LC-MS analysis.
2.10. LC-MS analysis
CYP enzyme activity and cocktail parent quantification: Frozen media samples were thawed. A volume of 50 µl of media was mixed with 10 µl of a 70/30 water/acetonitrile mix containing 4 internal standards (0.32 µM 1-OH-midazolam - D4, 0.35 µM 4-OH-diclofenac - 13C6, 0.42 µM OH-bupropion - D6, 0.71 µM Acetaminophen - D4) and 50 µl of a 95/5 water/acetonitrile mix. A volume of 4 µl (for metabolite quantification) or 0.1–1 µl (for parent quantification) was injected on an API 5000 triple quadrupole LC-MS instrument (AB Sciex, Framingham, MA, USA) coupled to a HP1290 INFINITY II ultra-performance liquid chromatography instrument (UPLC; Agilent Technologies, Santa Clara, CA, USA) with an ZORBAX Eclipse plus XDB C18 50 × 2.1 mm column (Agilent Technologies). The flow rate of the mobile phase was 0.412 ml/min. The mobile phase was a mix of solvent A (water + 0.1 % trifluoroacetic acid, pH = 2.4) and solvent B (acetonitrile). The gradient profile was: 0–2.16 min 5% B, 2.16–2.4 min 5-25% B, 2.40–3.86 min 25–40% B, 3.87–4.34 min 50–80% B, 4.34–4.83 min 80% B, 4.84–5.56 min 90% B, 5.57–6.5 min 5% B. Positive electrospray ionization (ESI+) was used in combination with multiple reaction monitoring. Additional instrument parameters are listed in Supplementary Table 1.
Entacapone and tolcapone quantification: Media samples were first diluted with Lonza HCM to a concentration < 10 µM and frozen until further use. Next, 50 µl of the diluted sample was mixed with 100 µl of acetonitrile with 0.5 µM dextromethorphan as internal standard. Samples were centrifuged for 5 minutes at 3000 g, 50 µl of the supernatant was mixed with 150 µl water + 0.1% formic acid. A volume of 5 µl of each sample was injected on an API 5000 triple quadrupole LC-MS instrument (AB Sciex) coupled to a HP1290 INFINITY II UPLC (Agilent Technologies) with an ACE Excel 2 C18-AR 150 × 2.1 mm column (Advanced Chromatography Technologies, Aberdeen, UK). The flow rate of the mobile phase was 0.4 ml/min. The mobile phase was a mix of solvent A (water + 0.1 % formic acid) and solvent B (acetonitrile + 0.1 % formic acid). The gradient profile was: 0–-0.3 min 10% B, 0.4–4 min 10–70% B, 4.01-5 min 95% B, 5.01–-6 min 10% B. Positive electrospray ionization (ESI+) was used in combination with multiple reaction monitoring. Additional analysis parameters are listed in Supplementary Table 1.
Metabolite identification: Supernatants were collected after 72 h of incubation with diclofenac and stored undiluted at -80°C. Thawed media samples (50 µl) were mixed with 1 volume of acetonitrile, vortexed for 30 s, and centrifuged at 3000 g for 10 min at 4°C. Supernatant was diluted with 1 volume of water and 100 µl was transferred to an autosampler plate on which 5 µl of sample was injected. Metabolite identification was performed on an Acquity UPLC coupled to a Vion IMS QTof Mass Spectrometer (MS; Waters, Wilmslow, UK). The mass spectrometer was operated in positive and negative mode with electrospray ionization (ESI+ and ESI-). The MS source settings were the following: capillary voltage of 0.5 kV, sample cone voltage of 35 V, source temperature and desolvation temperature were set at 150°C and 450°C. Nitrogen was used as the cone gas (6 l/h), desolvation gas (600 l/h) and as the collision gas. Diclofenac and its metabolites were separated using an Acquity CSH column (100 × 150µm, particle size 1.7 µm, Waters) maintained at 45°C. The flow rate of the mobile phase was 0.4 ml/min. The mobile phase was a mix of solvent A (10 mM ammonium acetate/0.1% acetic acid/ 5% acetonitrile/water) and solvent B (100% acetonitrile). The gradient profile was: 0-1 min 10% B, 1–9 min 10–90% B, 9-10.5 min 90% B, 10.5–12 min 10% B. Metabolites were screened by using UNIFI software (Waters) based on accurate mass measurement. The structure of diclofenac and its metabolites were elucidated by tandem mass spectrometry data acquisition using alternating low energy and high energy collision-induced dissociation where the former is used to obtain precursor ion accurate mass (mass errors less than 5 ppm) and the latter used to obtain product accurate mass ions (mass errors less than 10 ppm).
2.11. Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 8.1.1). In every analysis, n ≥ 3 with every data point being the average of multiple technical replicates unless otherwise indicated. For comparison of two groups, an unpaired two-tailed t-Student test was applied. In case of multiple comparisons (more than two groups), one-way analysis of variance (ANOVA) was done followed by Tukey's multiple comparisons test. Statistical significance was defined as P < 0.05.
3. Results
3.1. Pooling of human liver spheroids using a microwell approach
To explore upscaling of human liver spheroids in one system, PHH spheroids were first formed by seeding 1500 cells/well in low-attachment 96-well plates. Following complete spheroid formation after 7–8 days of culture, multiple spheroids were pooled into a 96-well plate or one 96-well compartment of a TissUse 2-organ-chip circuit (Fig. 1A). A total of 16 spheroids were pooled per well as a balance between upscaling the number of cells/well and minimizing the number of spheroids in close contact with each other immediately after seeding. When pooling 32 spheroids per well, multiple spheroids were already in very close proximity from the beginning (Suppl. Fig. 1A), which can easily lead to spheroid fusion and loss of functionality. Following the cultures for 2-4 days, spheroids in a regular 96-well attached to the bottom and lost their spherical 3D structure (Fig. 1B). Spheroid attachment to the bottom could be prevented by coating wells with a low-adhesive solution. However, spheroids could freely move within the well during plate handling or media refreshment, which inevitably led to physical contact between spheroids, and eventually spheroid fusion (Suppl. Fig. 1B). In the chip, several spheroids also attached to the bottom, although to a lesser extent compared to the uncoated liver-plate. Other spheroids in the chip gradually moved and aggregated with neighboring spheroids (Fig. 1B). In summary, simply pooling spheroids without any restriction led to spheroid attachment to other spheroids or the well bottom. Therefore, it was concluded that the model in its current form cannot retain its original morphology over time and is not suitable for long-term drug exposure experiments.
To pool spheroids whilst limiting their aggregation and attachment to the bottom, a microwell array approach was evaluated (Fig. 1C). We used a commercially available microwell array containing U-shaped cavities. To fit the microwell array into a well of a plate or organ-chip, we punched out a 96-well-sized array with ∼ 100 cavities (as described in section 2). The material of the microwell array is porous (as shown in Fig. 1D) which is expected to facilitate spheroid perfusion in flow conditions. Pooling 16 pre-formed spheroids and seeding them in different cavities of a microwell array in a 96-well plate or organ-chip successfully separated them. Moreover, spheroids did not appear to attach to the low-adhesive microwells (Fig. 1E). However, when loading multiple spheroids simultaneously, they often ended up in adjacent cavities or even the same cavity. Eventually, spheroids within the same cavity fused (Fig. 1E). In conclusion, the use of a microwell array prevents spheroid aggregation when seeded in separate cavities, but spheroids easily end up in the same cavity when loading them onto the array. Therefore, we explored immediate formation of spheroids in microwells as an option to combine multiple spheroids in a less labor-intensive approach.
3.2. Generation of a functional liver-chip model with 100 human liver spheroids
Spheroid formation in low-adhesive microwells was tested in both a plate setup and an organ-chip setup. Each microwell array (containing ∼ 100 cavities) was loaded with 100,000 PHH, followed by centrifugation (Fig. 2A). Quantification showed that close to 100 spheroids were generated in each microwell array, both in the plate setup (hereafter denoted as ‘liver-plate’; 97 ± 2 spheroids per array) and chip setup (hereafter referred to as ‘liver-chip’; 98 ± 1 spheroids per array) (Fig. 2B). We tested spheroid formation in microwells using PHH from three different donors with spheroid-forming abilities in conventional liver spheroid culture (Suppl. Fig. 2A). All three lots successfully formed spheroids using the microwell approach (as confirmed by visual inspection under a microscope; Fig. 2C). With each tested PHH lot, spheroids in the liver-chip appeared less variable in size and showed a better-defined edge versus the liver-plate (Fig. 2C). To understand if this difference in morphology is evident from the first days of the culture or only arises later, we monitored spheroid formation over time in low-attachment plates, liver-chips, and liver-plates. Following the cultures over time suggests the fastest spheroid compaction occurred in liver-chips compared to the liver-plate and the single spheroid cultures (Suppl. Fig. 2B). To evaluate the uniformity of spheroid shape and size within one microwell array, we measured the dimensions of fully formed spheroids on day 7-8 of culture (Fig. 2D). When comparing both microwell setups, spheroids in the liver-chip are on average larger with less non-circular outliers versus the liver-plate (Fig. 2D). Next, we benchmarked spheroid health and functionality in the liver-chip and liver-plate against conventional single spheroid cultures of 3 different donors after 2 weeks of culture. The cellular ATP content per cell in the liver-plate was lower compared to liver spheroids, while there was no significant difference between the liver-chip and single liver spheroids (Fig. 2E). Next, we analyzed the activity of several CYP enzymes in the cultures by quantification of the primary metabolites of specific CYP substrate compounds by LC-MS. No statistical differences were observed (due to inter-donor variability), however some trends are that 1) the liver-plate culture has lower activity of all analyzed CYP enzymes compared to liver spheroids and 2) the liver-chip has comparable CYP1A2 and CYP2C9 activity as liver spheroids, while CYP2B6 and CYP3A4 activity seems lower (Fig. 2F). However, data on the liver-chip for CYP3A4 and CYP2B6 must be interpreted with caution, as the probe substrates midazolam (CYP3A4 substrate) and bupropion (CYP2B6 substrate) show a poor recovery in empty chips (Suppl. Fig. 2C) which likely leads to an underestimation of CYP2B6 and CYP3A4 activity in the chip. As an alternative to estimate CYP3A4 activity, we deployed a luminescence-based assay. This assay revealed a similar CYP3A4 activity/spheroid in the liver-chip and liver spheroids, while the liver-plate has a significantly lower CYP3A4 activity (Fig. 2G). In conclusion, we successfully set up a human liver-chip in vitro model with a comparable cellular viability and CYP activity as in conventional spheroid cultures, while liver-plate cultures showed lower viability and metabolic activity.
One hypothesis to explain the reduced viability and functionality of the liver-plate is hypoxia during seeding. Therefore, we tested if there was a negative impact on spheroid formation in the liver-plate when the plating medium was not saturated with carbogen before seeding the cells, and if spheroid formation improved when plating the cells in a lower total volume to reduce the distance of the cells from the air-liquid interface. However, none of the alternative approaches resulted in a significant change in viability or CYP3A4 activity of these cultures (Suppl. Fig. 2D). The liver-chip also did not show any difference in viability or CYP3A4 activity when the plating medium was saturated with carbogen or not (Suppl. Fig. 2E).
We further characterized the liver-chip and liver-plate models and benchmarked their functionality over time against single liver spheroid cultures. For this purpose, we included both PHH and NPC in all cell systems. First, we measured the release of several biomarkers in the medium for 7 days following complete spheroid formation. The amount of LDH released in the medium, indicative of cell lysis, showed a decreasing trend in liver-chip and liver spheroid cultures over time, while the liver-plate showed higher levels over time (Fig. 3A). Both albumin and urea production rates, typical markers of hepatic health, were maintained at stable and physiologically relevant levels in the liver-chip and single liver spheroid cultures for 7 days following complete spheroid formation, while the liver-plate showed low production rates (Fig. 3B). We also analyzed the stability of CYP enzyme function over 2 weeks after spheroid formation within the same culture. In the liver-chip and liver spheroids, all analyzed CYP enzymes were still active 2 weeks after spheroid formation, while we could not detect any CYP activity anymore in the liver-plate after 2 weeks (Suppl. Fig. 3). In conclusion, monitoring biomarker release and CYP activity levels show that hepatocytes in the liver-chip and liver spheroids remain healthy and functionally active throughout the in vitro culturing period, while the liver-plate shows a poor hepatic phenotype over time.
3.3. Application of the liver-chip for drug safety and metabolism studies
Following biological characterization, the liver-chip containing PHH and NPC was tested for its suitability to be deployed as a drug testing platform for safety and metabolism studies. First, we estimated the loss of volume of the liver-chip throughout the culturing period of 2 weeks (e.g. evaporation, frequent opening of the compartments and regularly refreshing of 50% of the medium). Weighing chips over time showed a limited weight loss (on average < 5% of the total mass of the medium) between the onset and end of the culturing time (Fig. 4A). Next, we tested the toxicity of catechol-O-methyltransferase (COMT) inhibitors tolcapone (most DILI) and entacapone (less DILI) at 5x and/or 20x the reported clinical Cmax values in human plasma. Recovery of both entacapone and tolcapone in empty chips was close to 100% (Suppl. Fig. 4A), showing that these compounds are not prone to non-specific binding to the chip housing at the used concentrations. The different liver cultures were treated with 3 drug administrations of entacapone, tolcapone or vehicle over a total time course of 7 days and analyzed for viability and metabolic activity. Liver-chips treated with tolcapone, a known hepatotoxic drug, showed a strongly reduced viability and CYP3A4 activity, while its structural analogue entacapone did not provoke a similar response (Fig. 4B). This difference in response was confirmed by monitoring the spheroid morphology at the end of the drug treatment. Tolcapone-treated spheroids presented as dark microtissues with a severely disrupted outer rim whilst both vehicle-treated and entacapone-treated spheroids retain their translucent appearance and smooth outer rim (Fig. 4C). Testing this pair of molecules in liver spheroids gave similar outcomes on spheroid viability and metabolic activity (Suppl. Fig. 4B). The liver-plate could also correctly flag both compounds using viability as a readout, however tolcapone toxicity was not detected at 5X the Cmax using metabolic activity as a readout (Suppl. Fig. 4C).
Finally, we evaluated the metabolic capacity of the liver-chip and benchmarked it against single liver spheroid cultures by metabolite identification in the supernatant. We incubated 50 µM diclofenac in the system for 72h, which is the maximal time we currently applied between 2 media refreshes. Liver-chip cultures showed a higher metabolic turnover of diclofenac versus single spheroid cultures, resulting in a significantly increased signal for nearly all observed metabolites and a lower signal for unconjugated diclofenac and its dehydrated form. Both cultures displayed both phase I (hydroxy-metabolite), phase II (glucuronide- or glucose, or sulphate-conjugated) and glutathione-conjugated metabolites (Fig. 4D).
In conclusion, we describe the setup and optimization of a functionally competent human liver-chip composed of 100 liver microtissues cultured under flow. This model shows a greatly improved metabolic capacity compared to single liver spheroids and can be deployed for both drug safety and metabolism studies.
4. Discussion
In vitro models are commonly used in the early stages of drug development, to allow simple, cost-effective and high-throughput assays for compound screening and selection. However, traditional in vitro models lack complexity to confidently answer questions involving interconnected processes. To allow a higher complexity and relevance of in vitro models to predict in vivo clinically relevant findings, the development and validation of both 3D organ models and organ-on-chip technologies will be key. The technological improvements in the field allow the combination of both approaches to study 3D models under flow in a single- or multi-organ setup [23,31,32]. Since many MPS platforms are still highly customized requiring very specific expertise, their acceptance across laboratories and companies remains limited. Some key requirements for the widespread use of this technology are broad commercialization, ease of use and reproducibility of the generated data.
In this work, we have explored different methods to combine multiple human liver spheroids in a plate and a commercially available organ-chip setup. Next, we benchmarked these models against standard spheroid cultures and applied the appropriate model to study drug metabolism and safety. When pooling liver spheroids, aggregation of multiple 3D structures is undesirable, as it will result in a larger structure in which the center can turn into a necrotic core due to difficulties with nutrient and oxygen supplementation [33]. Recently, Riede et al. demonstrated that pooling multiple liver spheroids results in one large, fused structure with a decreased expression of various CYP genes, like CYP3A4 and CYP1A2 [8]. Our first attempts of pooling spheroids also resulted in 1) spheroid fusion and 2) disintegration of the 3D structure after adhering to the plastic. Low-adhesive microwells already improved the model by restricting physical contact between spheroids and with the plastic of the well. However, when seeding multiple spheroids at the same time, spheroids often ended up in the same microwell, leading again to a large, fused structure. A potential solution could be to load spheroids individually while monitoring the well under a microscope, however, this approach would be labor-intensive and was not further pursued in this study. Alternatively, the microwells could be resized based on the spheroid diameter to only allow one spheroid per cavity.
Eventually, we utilized the microwell array to simultaneously form 100 spheroids within one culture. The approach of using microwells to form multiple liver spheroids per well has been described by other groups [11,13,34,35]. However, this study differentiates from other studies by the combination of 1) using liver spheroids derived from primary human hepatocytes 2) setting up the model with commercially available and easy-to-use consumables in a standardized MPS platform, which facilitates the implementation in other labs and 3) evaluating the model from the perspective of drug evaluation. A similar platform is the liver-chip model developed by the Griffith lab and commercialized by the company CNBio which has been evaluated for drug safety and metabolism testing in multiple reports [15,36,37]. In this model, hepatocytes are also seeded in a scaffold with multiple porous microchannels to form a 3D microtissue. The main difference with our described model is the adherence of cells to the scaffold and the shape of the resulting 3D microtissue. The CNBio model uses a collagen-coated scaffold where cells attach to the scaffold, resulting in an adherent structure which is different from typically described PHH spheroids [3,4]. In this work, we aimed to develop a system which combines liver spheroids while maintaining their typical structure and minimizing the interaction with any scaffold, and therefore treated the microwell array with a low-adhesive coating. The additional advantage of this approach is that spheroids from the same system can easily be collected and divided to use for multiple purposes. Finally, the spheroids in the microwells can be monitored throughout the experiment.
This work reveals some remarkable differences between spheroids in the liver-chip and liver-plate models. While the liver-chip model shows a similar cellular functionality as conventional liver spheroid cultures, liver-plate models show diminished hepatic activity. Already during the first days of culture, the liver-chip shows a faster spheroid formation compared to the liver-plate, suggesting that the culture conditions are more optimal in the liver-chip. Once fully formed, the spheroids in the liver-plate have a lower viability and strongly reduced CYP3A4 activity compared to the liver-chip, despite that the same number of cells are seeded in both systems (i.e. 100,000 PHH per culture or 1,000 PHH per cavity). One hypothesis is that cells in the liver-plate might experience a O2-deficient environment when seeding a high number of hepatocytes. Hepatocytes have a high oxygen demand, especially during the first 24 hours after seeding [38]. In a static system, oxygen exchanged at the air-liquid interface might take too long to sufficiently supply a high number of cells further away from the surface, while oxygen exchange is expected to be more efficient in organ-chips. The microfluidic channels in the chips are made from polydimethylsiloxane (PDMS), a gas-permeable material which allows oxygen exchange between the medium and the air, thereby chips have a larger surface area where oxygen exchange can happen. In addition, the medium flow will allow dissolved oxygen (as well as nutrients) to quickly reach the cells [39]. To increase the oxygen supplementation during the seeding step, the plating medium of the liver-chip and liver-plate was saturated with carbogen before seeding the cell suspension [40]. However, even with this approach the CYP3A4 activity in liver-plates was still significantly lower than in liver spheroids and liver-chips. Notably, a very recent publication shows the setup of a model combining multiple human liver spheroids in a plate with microwells [35]. The model shows albumin production rates and CYP activity levels comparable to liver spheroids. Some key differences are 1) the use of less cells than described in this paper, which likely reduces the total need for oxygen and nutrient supplementation and 2) the use of a 96-well plate with the microwells integrated in the bottom [35], which could potentially allow oxygen supplementation from the bottom of the plate.
The low CYP3A4 activity in the liver-plate versus the liver-chip can potentially be linked with other published reports. A significant decrease in CYP3A4 protein levels and specific CYP gene expression levels (CYP3A4, CYP1A2) was reported in aggregated liver spheroids, possibly due to intra-spheroid hypoxia [8]. Hypoxia might cause a downregulation of specific CYP genes like CYP1A2 and CYP3A4 [38].
Finally, the liver-chip model was assessed for use in drug safety testing and metabolism studies. First, empty chips were evaluated for several basic properties. The chips show low evaporation rates, which comply with specifications recommended by the International Consortium for Innovation and Quality in Pharmaceutical Development (IQ) MPS affiliate [41]. However, many organ-chips are made from PDMS, a hydrophobic material to which several small molecules show non-specific binding [42]. In this study, we showed that free drug levels of midazolam and bupropion rapidly decline in the chip (Suppl. Fig. 2C). To reduce/slow down non-specific binding, albumin was included in the medium used in this study [43]. Second, the liver-chip correctly discriminated the hepatotoxic effects of the DILI-positive compound tolcapone from the less toxic compound entacapone, which highlights the liver-chip's potential applicability to evaluate the safety of a therapy. This pair of structural analogues is often used to evaluate the predictive value of liver in vitro models [4,30]. Given the high number of liver spheroids in each liver-chip, the model is amenable to collect enough cells to apply different endpoint assays to the same system (e.g. immunohistochemical readouts, omics analyses). To further increase the physiological relevance of the model to predict outcomes in human subjects, NPCs were added in the culture. Various reports show that adding other liver cell types in liver in vitro models can modulate the metabolism and toxicity of drugs, e.g. through inflammation and fibrosis [6,44,45]. Whether this liver-chip has a higher predictive value to detect DILI than liver spheroids, needs to be determined by testing an extensive list of known DILI-positive and DILI-negative compounds on this system.
Next, the liver-chip model was evaluated for its utility in drug metabolism studies in comparison to liver spheroid cultures, which have previously been evaluated for this purpose in recent studies [8,9]. Here, we used diclofenac as test compound as it is metabolized in the liver through both phase I and phase II metabolic pathways. We observed an increased metabolic turnover of diclofenac in the liver-chip compared to the liver spheroids, due to the higher cell density in the liver-chip. When looking at the formed metabolites, we detected several in vivo-relevant metabolites resulting from a phase I and phase II metabolic conversion, as well as glutathione- and cysteine-conjugated metabolites [46], [47], [48]. The only metabolite that had a higher signal in the liver spheroid versus the liver-chip was a dehydrated form of diclofenac, potentially resulting from an internal cyclisation in equilibrium with the higher remaining levels of diclofenac in the liver spheroid [48]. The increased metabolic capacity of the liver-chip and other microwell setups could especially be an added value to determine the metabolic turnover and metabolite of compounds with a low hepatic clearance, where single spheroid assays are not sensitive enough [8]. To further build on the liver-chip model as a metabolically competent system, the utilized MPS platform easily allows the liver model to be interconnected with a second organ model to study the interplay of these organs and to assess potential (side) effects of drug metabolites on downstream organs [18,21,22,24].
In conclusion, we demonstrate the setup of a human liver-chip model which allows the generation of 100 liver spheroids per system using commercially available microwell arrays in a standardized chip platform. This model shows promise to assess drug metabolism and safety in increasingly complex and physiologically relevant human liver in vitro models.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
At the time of conducting the work, all authors were employees, and some were stakeholders of UCB Biopharma SRL.
Acknowledgements
We thank Sylvie Dell'Aiera and Kimberley Perkins for technical assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbiosy.2022.100054.
Appendix. Supplementary materials
References
- 1.Vilas-Boas V., Cooreman A., Gijbels E., Van Campenhout R., Gustafson E., Ballet S., et al. In: Drug-induced Liver Injury, Adv Pharmacol. Ramachandran A., Jaeschke H., editors. Elsevier Inc; 2019. Primary hepatocytes and their cultures for the testing of drug-induced liver injury; pp. 1–30. [DOI] [PubMed] [Google Scholar]
- 2.Messner S., Agarkova I., Moritz W., Kelm J.M. Multi-cell type human liver microtissues for hepatotoxicity testing. Arch Toxicol. 2013;87:209–213. doi: 10.1007/s00204-012-0968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell C.C., Hendriks D.F.G., Moro S.M.L., Ellis E., Walsh J., Renblom A., Fredriksson Puigvert L., Dankers A.C.A., Jacobs F., Snoeys J., Sison-Young R.L., Jenkins R.E., Nordling Å., Mkrtchian S., Park B.K., Kitteringham N.R., Goldring C.E.P., Lauschke V.M., Ingelman-Sundberg M. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci Rep. 2016;6:1–13. doi: 10.1038/srep25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proctor W.R., Foster A.J., Vogt J., Summers C., Middleton B., Pilling M.A., Shienson D., Kijanska M., Ströbel S., Kelm J.M., Morgan P., Messner S., Williams D. Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Arch Toxicol. 2017;91:2849–2863. doi: 10.1007/s00204-017-2002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vorrink S.U., Zhou Y., Ingelman-Sundberg M., Lauschke V.M. Prediction of drug-induced hepatotoxicity using long-term stable primary hepatic 3D spheroid cultures in chemically defined conditions. Toxicol Sci. 2018;163:655–665. doi: 10.1093/toxsci/kfy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell C.C., Chouhan B., Andersson L.C., Andersson H., Dear J.W., Williams D.P., Söderberg M. Functionality of primary hepatic non parenchymal cells in a 3D spheroid model and contribution to acetaminophen hepatotoxicity. Arch Toxicol. 2020 doi: 10.1007/s00204-020-02682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schofield C.A., Walker T.M., Taylor M.A., Patel M., Vlachou D.F., Macina J.M., et al. Evaluation of a three-dimensional primary human hepatocyte spheroid model: adoption and industrialization for the enhanced detection of drug-induced liver injury. Chem Res Toxicol. 2021 doi: 10.1021/acs.chemrestox.1c00227. [DOI] [PubMed] [Google Scholar]
- 8.Riede J., Wollmann B.M., Molden E., Ingelman-Sundberg M. Primary human hepatocyte spheroids as an in vitro tool for investigating drug compounds with low clearance. Drug Metab Dispos. 2021 doi: 10.1124/dmd.120.000340. [DOI] [PubMed] [Google Scholar]
- 9.Kanebratt K.P., Janefeldt A., Vilén L., Vildhede A., Samuelsson K., Milton L., Björkbom A., Persson M., Leandersson C., Andersson T.B., Hilgendorf C. Primary human hepatocyte spheroid model as a 3D in vitro platform for metabolism studies. J Pharm Sci. 2021;110:422–431. doi: 10.1016/j.xphs.2020.10.043. [DOI] [PubMed] [Google Scholar]
- 10.Fey S.J., Wrzesinski K. Determination of drug toxicity using 3D spheroids constructed from an immortal human hepatocyte cell line. Toxicol Sci. 2012;127:403–411. doi: 10.1093/toxsci/kfs122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucendo-Villarin B., Meseguer-Ripolles J., Drew J., Fischer L., Ma E., Flint O., Simpson K.J., MacHesky L.M., Mountford J.C., Hay D.C. Development of a cost-effective automated platform to produce human liver spheroids for basic and applied research. Biofabrication. 2020;13 doi: 10.1088/1758-5090/abbdb2. [DOI] [PubMed] [Google Scholar]
- 12.Kakni P., Hueber R., Knoops K., López-Iglesias C., Truckenmüller R., Habibovic P., Giselbrecht S. Intestinal organoid culture in polymer film-based microwell arrays. Adv Biosyst. 2020;4 doi: 10.1002/adbi.202000126. [DOI] [PubMed] [Google Scholar]
- 13.Ogihara T., Iwai H., Inoue Y., Katagi J., Matsumoto N., Motoi-Ohtsuji M., Kakiki M., Kaneda S., Nagao T., Kusumoto K., Ozeki E., Jomura T., Tanaka S., Ueda T., Ohta K., Ohkura T., Arakawa H., Nagai D. Utility of human hepatocyte spheroids for evaluation of hepatotoxicity. Fund Toxicol Sci. 2015;2:41–48. doi: 10.2131/fts.2.41. [DOI] [Google Scholar]
- 14.Foster A.J., Chouhan B., Regan S.L., Rollison H., Amberntsson S., Andersson L.C., Srivastava A., Darnell M., Cairns J., Lazic S.E., Jang K.J., Petropolis D.B., Kodella K., Rubins J.E., Williams D., Hamilton G.A., Ewart L., Morgan P. Integrated in vitro models for hepatic safety and metabolism: evaluation of a human Liver-Chip and liver spheroid. Arch Toxicol. 2019;93:1021–1037. doi: 10.1007/s00204-019-02427-4. [DOI] [PubMed] [Google Scholar]
- 15.Rubiano A., Indapurkar A., Yokosawa R., Miedzik A., Rosenzweig B., Arefin A., Moulin C.M., Dame K., Hartman N., Volpe D.A., Matta M.K., Hughes D.J., Strauss D.G., Kostrzewski T., Ribeiro A.J.S. Characterizing the reproducibility in using a liver microphysiological system for assaying drug toxicity, metabolism and accumulation. Clin Transl Sci. 2020 doi: 10.1111/cts.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang K.J., Mehr A.P., Hamilton G.A., McPartlin L.A., Chung S., Suh K.Y., Ingber D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integrative Biol (United Kingdom) 2013;5:1119–1129. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 17.Cucullo L., Hossain M., Puvenna V., Marchi N., Janigro D. The role of shear stress in Blood-Brain Barrier endothelial physiology. BMC Neurosci. 2011;12:40. doi: 10.1186/1471-2202-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer S., Wennberg Huldt C., Kanebratt K.P., Durieux I., Gunne D., Andersson S., Ewart L., Haynes W.G., Maschmeyer I., Winter A., Ämmälä C., Marx U., Andersson T.B. Functional coupling of human pancreatic islets and liver spheroids on-a-chip: Towards a novel human ex vivo type 2 diabetes model. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-14815-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramme A.P., Koenig L., Hasenberg T., Schwenk C., Magauer C., Faust D., Lorenz A.K., Krebs A.-C., Drewell C., Schirrmann K., Vladetic A., Lin G.-C., Pabinger S., Neuhaus W., Bois F., Lauster R., Marx U., Dehne E.-M. Autologous induced pluripotent stem cell-derived four-organ-chip. Future Sci OA. 2019;5:FSO413. doi: 10.2144/fsoa-2019-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J.-H., Santhosh M., Choi J. In vitro blood–brain barrier-integrated neurological disorder models using a microfluidic device. Micromachines (Basel) 2019;11:21. doi: 10.3390/mi11010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J.Y., Fluri D.A., Marchan R., Boonen K., Mohanty S., Singh P., Hammad S., Landuyt B., Hengstler J.G., Kelm J.M., Hierlemann A., Frey O. 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. J Biotechnol. 2015;205:24–35. doi: 10.1016/j.jbiotec.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 22.McAleer C.W., Pointon A., Long C.J., Brighton R.L., Wilkin B.D., Bridges L.R., Narasimhan Sriram N., Fabre K., McDougall R., Muse V.P., Mettetal J.T., Srivastava A., Williams D., Schnepper M.T., Roles J.L., Shuler M.L., Hickman J.J., Ewart L. On the potential of in vitro organ-chip models to define temporal pharmacokinetic-pharmacodynamic relationships. Sci Rep. 2019;9:9619. doi: 10.1038/s41598-019-45656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajan S.A.P., Aleman J., Wan M., Pourhabibi Zarandi N., Nzou G., Murphy S., Bishop C.E., Sadri-Ardekani H., Shupe T., Atala A., Hall A.R., Skardal A. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomater. 2020:1–12. doi: 10.1016/j.actbio.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W.L.K., Edington C., Suter E., Yu J., Velazquez J.J., Velazquez J.G., Shockley M., Large E.M., Venkataramanan R., Hughes D.J., Stokes C.L., Trumper D.L., Carrier R.L., Cirit M., Griffith L.G., Lauffenburger D.A. Integrated gut/liver microphysiological systems elucidates inflammatory inter-tissue crosstalk. Biotechnol Bioeng. 2017;114:2648–2659. doi: 10.1002/bit.26370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao S., Coppeta J.R., Rogers H.B., Isenberg B.C., Zhu J., Olalekan S.A., McKinnon K.E., Dokic D., Rashedi A.S., Haisenleder D.J., Malpani S.S., Arnold-Murray C.A., Chen K., Jiang M., Bai L., Nguyen C.T., Zhang J., Laronda M.M., Hope T.J., Maniar K.P., Pavone M.E., Avram M.J., Sefton E.C., Getsios S., Burdette J.E., Kim J.J., Borenstein J.T., Woodruff T.K. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun. 2017;8 doi: 10.1038/ncomms14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohasz C., Bonanini F., Hoelting L., Renggli K., Frey O., Hierlemann A. Predicting metabolism-related drug–drug interactions using a microphysiological multitissue system. Adv Biosyst. 2020;4:1–13. doi: 10.1002/adbi.202000079. [DOI] [PubMed] [Google Scholar]
- 27.Docci L., Milani N., Ramp T., Romeo A.A., Godoy P., Franyuti D.O., Krähenbühl S., Gertz M., Galetin A., Parrott N., Fowler S. Exploration and application of a liver-on-a-chip device in combination with modelling and simulation for quantitative drug metabolism studies. Lab Chip. 2022 doi: 10.1039/d1lc01161h. [DOI] [PubMed] [Google Scholar]
- 28.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker P.A., Ryder S., Lavado A., Dilworth C., Riley R.J., Riley R.J. The evolution of strategies to minimise the risk of human drug induced liver injury (DILI) in drug discovery and development. Arch Toxicol. 2020 doi: 10.1007/s00204-020-02763-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baudy A.R., Otieno M.A., Hewitt P., Gan J., Roth A., Keller D., Sura R., van Vleet T.R., Proctor W.R. Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry. Lab Chip. 2020;20:215–225. doi: 10.1039/c9lc00768g. [DOI] [PubMed] [Google Scholar]
- 31.Park J., Lee B.K., Jeong G.S., Hyun J.K., Lee C.J., Lee S.H. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer's disease. Lab Chip. 2015;15:141–150. doi: 10.1039/c4lc00962b. [DOI] [PubMed] [Google Scholar]
- 32.Skardal A., Murphy S.V., Devarasetty M., Mead I., Kang H.W., Seol Y.J., Zhang Y.S., Shin S.R., Zhao L., Aleman J., Hall A.R., Shupe T.D., Kleensang A., Dokmeci M.R., Jin Lee S., Jackson J.D., Yoo J.J., Hartung T., Khademhosseini A., Soker S., Bishop C.E., Atala A. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci Rep. 2017;7:1–16. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin R.-Z., Chang H.-Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J. 2008;3:1172–1184. doi: 10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]
- 34.Choi J.H., Loarca L., De Hoyos-Vega J.M., Dadgar N., Loutherback K., Shah V.H., Stybayeva G., Revzin A. Microfluidic confinement enhances phenotype and function of hepatocyte spheroids. Am J Physiol Cell Physiol. 2020 doi: 10.1152/ajpcell.00094.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preiss L.C., Lauschke V.M., Georgi K., Petersson C. Multi-well array culture of primary human hepatocyte spheroids for clearance extrapolation of slowly metabolized compounds. The AAPS J. 2022;24 doi: 10.1208/s12248-022-00689-y. [DOI] [PubMed] [Google Scholar]
- 36.Tsamandouras N., Kostrzewski T., Stokes C.L., Griffith L.G., Hughes D.J., Cirit M. Quantitative assessment of population variability in hepatic drug metabolism using a perfused three-dimensional human liver microphysiological system. J Pharmacol Exp Ther. 2017;360:95–105. doi: 10.1124/jpet.116.237495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domansky K., Inman W., Serdy J., Dash A., Lim M.H.M., Griffith L.G. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip. 2010;10:51–58. doi: 10.1039/B913221J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kidambi S., Yarmush R.S., Novik E., Chao P., Yarmush M.L., Nahmias Y. Oxygen-mediated enhancement of primary hepatocyte metabolism, functional polarization, gene expression, and drug clearance. Proc Natl Acad Sci U S A. 2009;106:15714–15719. doi: 10.1073/pnas.0906820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wulftange W.J., Rose M.A., Garmendia-Cedillos M., da Silva D., Poprawski J.E., Srinivasachar D., Sullivan T., Lim L., v. Bliskovsky V., Hall M.D., Pohida T.J., Robey R.W., Morgan N.Y., Gottesman M.M. Spatial control of oxygen delivery to three-dimensional cultures alters cancer cell growth and gene expression. J Cell Physiol. 2019;234:20608–20622. doi: 10.1002/jcp.28665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hengstler J.G., Utesch D., Steinberg P., Platt K.L., Diener B., Ringel M., Swales N., Fischer T., Biefang K., Gerl M., Böttger T., Oesch F. Cryopreserved primary hepatocytes as a constantly available in vitro model for the evaluation of human and animal drug metabolism and enzyme induction. Drug Metab Rev. 2000;32:81–118. doi: 10.1081/DMR-100100564. [DOI] [PubMed] [Google Scholar]
- 41.S. Fowler, L. Kelly, D.B. Duignan, A. Gupta, N. Hariparsad, J.R. Kenny, W.G. Lai, J. Liras, J.A. Phillips, J. Gan, Lab on a Chip Microphysiological systems for ADME-related applications : current status and recommendations for system development and characterization, (2020). doi: 10.1039/c9lc00857h. [DOI] [PubMed]
- 42.van Meer B.J., de Vries H., Firth K.S.A., van Weerd J., Tertoolen L.G.J., Karperien H.B.J., Jonkheijm P., Denning C., IJzerman A.P., Mummery C.L. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem Biophys Res Commun. 2017;482:323–328. doi: 10.1016/j.bbrc.2016.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T. Sasserath, J.W. Rumsey, C.W. Mcaleer, L.R. Bridges, C.J. Long, D. Elbrecht, F. Schuler, A. Roth, C. Bertinetti-lapatki, M.L. Shuler, J.J. Hickman, Differential Monocyte Actuation in a Three-Organ Functional Innate Immune System-on-a-Chip, 2000323 (2020) 1–19. doi: 10.1002/advs.202000323. [DOI] [PMC free article] [PubMed]
- 44.Messner S., Agarkova I., Moritz W., Kelm J.M. Multi-cell type human liver microtissues for hepatotoxicity testing. Arch Toxicol. 2013;87:209–213. doi: 10.1007/s00204-012-0968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang K., Otieno M.A., Ronxhi J., Lim H., Ewart L., Kodella K.R., Petropolis D.B., Kulkarni G., Rubins J.E., Conegliano D., Nawroth J., Simic D., Lam W., Singer M., Barale E., Singh B., Sonee M., Streeter A.J., Manthey C., Jones B., Srivastava A., Andersson L.C., Williams D., Park H., Barrile R., Sliz J., Herland A., Haney S., Karalis K., Ingber D.E., Hamilton G.A. Reproducing human and cross-species drug toxicities using a liver-chip. Sci Transl Med. 2019;11:eaax5516. doi: 10.1126/scitranslmed.aax5516. [DOI] [PubMed] [Google Scholar]
- 46.Pickup K., Gavin A., Jones H.B., Karlsson E., Page C., Ratcliffe K., Sarda S., Schulz-Utermoehl T., Wilson I. The hepatic reductase null mouse as a model for exploring hepatic conjugation of xenobiotics: application to the metabolism of diclofenac. Xenobiotica. 2012;42:195–205. doi: 10.3109/00498254.2011.607196. [DOI] [PubMed] [Google Scholar]
- 47.Stierlin H., Faigle J.W., Sallmann A., Kung W., Richter W.J., Kriemler H.P., Alt K.O., Winkler T. Biotransformation of diclofenac sodium (voltaren® in animals and in man: I. Isolation and identification of principal metabolites. Xenobiotica. 1979;9:601–610. doi: 10.3109/00498257909042327. [DOI] [PubMed] [Google Scholar]
- 48.Sarda S., Page C., Pickup K., Schulz-Utermoehl T., Wilson I. Diclofenac metabolism in the mouse: Novel in vivo metabolites identified by high performance liquid chromatography coupled to linear ion trap mass spectrometry. Xenobiotica. 2012;42:179–194. doi: 10.3109/00498254.2011.607865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.