Abstract
Long COVID (LC), a type of post-acute sequelae of SARS-CoV-2 infection (PASC), occurs after at least 10% of SARS-CoV-2 infections, yet its etiology remains poorly understood. Here, we used multiple “omics” assays (CyTOF, RNAseq, Olink) and serology to deeply characterize both global and SARS-CoV-2-specific immunity from blood of individuals with clear LC and non-LC clinical trajectories, 8 months following infection and prior to receipt of any SARS-CoV-2 vaccine. Our analysis focused on deep phenotyping of T cells, which play important roles in immunity against SARS-CoV-2 yet may also contribute to COVID-19 pathogenesis. Our findings demonstrate that individuals with LC exhibit systemic inflammation and immune dysregulation. This is evidenced by global differences in T cell subset distribution in ways that imply ongoing immune responses, as well as by sex-specific perturbations in cytolytic subsets. Individuals with LC harbored increased frequencies of CD4+ T cells poised to migrate to inflamed tissues, and exhausted SARS-CoV-2-specific CD8+ T cells. They also harbored significantly higher levels of SARS-CoV-2 antibodies, and in contrast to non-LC individuals, exhibited a mis-coordination between their SARS-CoV-2-specific T and B cell responses. Collectively, our data suggest that proper crosstalk between the humoral and cellular arms of adaptive immunity has broken down in LC, and that this, perhaps in the context of persistent virus, leads to the immune dysregulation, inflammation, and clinical symptoms associated with this debilitating condition.
Introduction
Intense efforts are underway to determine the pathophysiology of post-acute sequelae of SARS-CoV-2 infection (PASC), a set of conditions affecting at least 10% of individuals recovering from COVID-19 1–3. PASC, which includes the unexplained, debilitating symptoms that characterize LC, remains a major public health challenge despite the availability of SARS-CoV-2 vaccination and treatment 4–8. Although the underlying cause or causes of LC are incompletely understood, multiple mechanisms including microvascular dysregulation 9,10, autoimmune phenomena 11–13, and reactivation of latent human herpesviruses 12,14,15 have been proposed as contributors to inflammatory responses, particularly in tissues, which could in turn drive symptoms that individuals experience. In addition, persistence of SARS-CoV-2 antigen has recently been demonstrated in a subset of immunocompetent individuals with LC 16–18. However, there are currently no accepted therapies for LC, in part due to limited insight into the underlying mechanisms of the condition to date 2.
To try to better understand the molecular underpinnings of LC, multiple “omics”-based approaches have recently been implemented on plasma specimens. Such studies have revealed individuals with LC to more often have elevated levels of inflammatory cytokines such as IFNβ and IL8, but low levels of cortisol 12,19,20. These results are consistent with the ongoing immunologic perturbations that have been consistently observed in individuals experiencing LC 12,20–25. Serological analyses have also found the presence of auto-antibodies during the acute or post-acute phases of infection to be associated with LC 11–13, although this has not been observed consistently 19,26,27. Elevated levels of SARS-CoV-2 antibodies have also been associated with LC 19. “Omics” analyses of immune cells in the form single-cell transcriptomics on PBMCs have likewise been performed, resulting in the classification of LC into multiple endotypes, and uncovering the persistent elevation of select immune subsets – including myeloid and NK subsets – in some phenotypes of LC 12. This study, however, did not examine individuals whose symptoms persisted beyond three months, and did not examine LC resulting from initial mild-to-moderate (non-hospitalized) cases of COVID-19, which comprise the vast majority of those experiencing this condition.
T cells play an important role in SARS-CoV-2 immunity and pathogenesis, yet relatively little is known about their role in LC. A limited set of studies that have examined SARS-CoV-2-specific T cell responses have implicated these cells in LC, albeit with conflicting results. While some studies have found elevated SARS-CoV-2-specific T cell responses in LC as compared to non-LC individuals 24,28, we have observed faster decay of subsets of SARS-CoV-2-specific CD8+ T cells in the context of LC 29. Apart from a transcriptomic/CITE-seq analysis of SARS-CoV-2-specific CD8+ T cells by MIRA, which identified unique features associated with LC two to three months after COVID-19 hospitalization 12, in-depth analyses of the phenotypic and functional features of SARS-CoV-2-specific T cells from individuals with LC are lacking. In particular, the profile of CD4+ T cells, key orchestrators of adaptive immunity, in individuals with LC is currently unknown.
We have previously used deep phenotypic characterization of SARS-CoV-2-specific T cells by CyTOF to identify differentiation states, effector functions, and/or homing properties of SARS-CoV-2-specific T cells associated with long-lived memory responses, fatal COVID-19, vaccination, and hybrid immunity 30–33, and to characterize pulmonary T cell responses in a mouse model of severe COVID-19 34. As these insights into COVID-19 immunity and pathogenesis were obtained using these next-generation T cell characterization assays in ways that would not have been captured using solely more traditional T cell assays, we reasoned that a similar in-depth analysis could identify T cells that protect or contribute to the symptoms of LC.
Therefore, in this study we deeply characterized T cell immunity during the post-acute phase of infection by CyTOF. We then combined these data with standard serological analyses, as well as two additional “omics” techniques: RNAseq and high-dimensional plasma proteomics using the Olink Explore Proximity Extension Assay (PEA), the latter of which enables simultaneous quantitation of 384 analytes from plasma. We leveraged a cohort of LC and non-LC individuals with detailed longitudinal characterization and biospecimen collection prior to SARS-CoV-2 vaccination or reinfection, which could confound interpretation of SARS-CoV-2-specific T cell and antibody responses, to identify clues to the immunologic processes that might drive LC. By performing this holistic, integrative analysis on a well-matched set of LC and non-LC individuals with consistent phenotypes for 8 months after infection, we were able to identify unique immune features associated with LC that inform on the mechanistic underpinnings of this debilitating disease.
Results
LC and non-LC participants
To study the phenotypes and effector functions of immune cells from individuals experiencing Long COVID-19 symptoms, we analyzed blood samples from 27 LC and 16 non-LC individuals from the San Francisco-based Long-term Impact of Infection with Novel Coronavirus (LIINC) cohort (NCT04362150)35. Specimens were collected 8 months following infection, but individuals had been followed since at least 4 months post-infection to characterize LC over time. Individuals with LC were defined as those that consistently met the case definition for LC (at least one COVID-19-attributed symptom that was new or worsened since the time of SARS-CoV-2 infection, and was at least somewhat bothersome) at both 4 and 8 months, while clinically matched non-LC individuals did not experience any lingering symptoms for the entire 8 months after SARS-CoV-2 infection. Importantly, at the time of specimen collection (in 2020–2021), none of the participants had yet received a SARS-CoV-2 vaccine, which would confound the SARS-CoV-2-specific serological and T cell analyses. Overall, the enrolled individuals had a median age of 46 years (range 19 to 71) and 58.1% identified as White (Table 1, Table S1). Individuals with LC were more likely to be female (63% vs 44%) (Fig. S1A) and to have been previously hospitalized during the acute phase of COVID-19 (26% vs 13%) (Fig. S1B). The individuals with LC analyzed herein were all highly symptomatic, with the number of Long COVID symptoms significantly increasing (from mean 6.7 to 9.2) with time (Fig. S1C).
Table 1:
Participants’ demographics, hospitalization status, and sequelae status at month 8 visit
| Participants | All | Gender | Hospitalized | Status | |||
|---|---|---|---|---|---|---|---|
| Female | Male | Yes | No | LC | Non-LC | ||
| N | 43 | 24 | 19 | 9 | 34 | 27 | 16 |
| Age (Median) | 46 | 43 | 53 | 46 | 48 | 46 | 45.5 |
| M41 (Median) | 2.5 | 5 | 0 | 7 | 2 | 4.5 | 0 |
| M82 (Median) | 5 | 7 | 2 | 10 | 3 | 7 | 0 |
| Female (N) | 24 | - | - | 6 | 18 | 17 | 7 |
| Race (N) | |||||||
|
| |||||||
| White | 25 | 13 | 12 | 0 | 25 | 14 | 11 |
| Latinx* | 11 | 6 | 5 | 7 | 4 | 9 | 2 |
| Black | 2 | 2 | 0 | 1 | 1 | 1 | 1 |
| Asian | 3 | 1 | 2 | 1 | 2 | 2 | 1 |
| NA# | 2 | 2 | 0 | 0 | 2 | 1 | 1 |
M4: Symptom counts at month 4 visit.
M8: Symptom counts at month 8 visit.
Latinx: Hispanic or Latino.
NA: Data are not available here.
Experimental design
SARS-CoV-2 serological analysis and four “omics” assays were performed on the same blood specimens from our cohort of LC and non-LC individuals (Fig. 1). Plasma/sera were analyzed for RBD-specific antibody levels, and for the levels of 394 analytes using the Olink platform. PBMCs from the same specimens were subjected to bulk RNAseq, as well as in-depth CD4+ and CD8+ T cell phenotyping using a 39-parameter CyTOF panel designed to simultaneously interrogate the differentiation states, activation states, effector functions, and homing properties of T cells (Table S2). Cells were phenotyped by CyTOF at baseline and following a 6-hour stimulation with SARS-CoV-2 peptides to identify SARS-CoV-2-specific T cells through intracellular cytokine staining. The RNAseq and Olink datasets, as well as the CyTOF datasets corresponding to total and SARS-CoV-2-specific T cells, were visualized and analyzed using a variety of integrative high-dimensional analysis approaches (Fig. 1). In total, we obtained 5 distinct datasets, enabling us to assess humoral response (serology), plasma analytes (Olink), transcriptional signatures (RNAseq), T cell features (CyTOF), and SARS-CoV-2-specific T cell phenotypes and effector functions (CyTOF).
Fig. 1. Study Design.
Schematic of experimental design and data analyses. Plasma and sera from 27 individuals with Long COVID (LC) and 16 individuals without LC (Non-LC) were subjected to Olink and serological analyses. PBMCs from the same individuals were subjected to RNAseq analysis, as well as to CyTOF analysis at baseline, or following a 6-hour stimulation with peptides derived from SARS-CoV-2 spike proteins (see Methods) to analyze T cell responses. The cells for CyTOF were treated with viability marker, fixed, and stained with a 39-parameter panel (Table S2) prior to analysis on a CyTOF instrument. The indicated tools on the right were then used for analyses of the resulting high-dimensional datasets.
SARS-CoV-2-specific T cells exhibit similar frequencies and effector profiles in LC and non-LC individuals
To quantitate total and SARS-CoV-2-specific T cells, normalized events from the CyTOF datasets were gated on intact, live, singlet events, followed by gating for CD4+ and CD8+ T cells (Fig. 2A). The T cells were assessed for expression of all our panel’s effector molecules, which were chosen because of their roles in T-cell immunity and pathogenesis. These consisted of the cytokines IFNγ, TNFα, IL2, IL4, IL6, IL17, and MIP1β, and the cytolytic markers granzyme B, perforin, and LAMP1 (Fig. S2). To determine which T cells were SARS-CoV-2-specific, we established a stringent set of rules based on the frequencies of cells expressing these effectors in samples treated or not with SARS-CoV-2 peptides (details in Methods). This analysis revealed that a combination of IFNγ, TNFα, and/or IL2 specifically identified the vast majority of SARS-CoV-2-specific CD4+ T cells (Fig. 2B, S2A), while a combination of IFNγ, TNFα, and/or MIP1β specifically identified the vast majority of SARS-CoV-2-specific CD8+ T cells (Fig. 2C, S2B).
Fig. 2. Identification and characterization of SARS-CoV-2-specific T cells in individuals from the LIINC cohort.
A. Gating strategy to identify T cells. Intact, live, singlet cells were gated for T cells (CD3+) followed by sub-gating on CD4+ and CD8+ T cells as indicated. B. SARS-CoV-2-specific CD4+ T cells can be identified as those producing IFNγ, TNFα, or IL2 in response to SARS-CoV-2 peptide stimulation. Cells were analyzed by intracellular cytokine staining in the absence (top row) or presence (bottom row) of SARS-CoV-2 peptides. C. SARS-CoV-2-specific CD8+ T cells can be identified as those producing IFNγ, TNFα, or MIP1β in response to SARS-CoV-2 peptide stimulation. Cells were analyzed by intracellular cytokine staining in the absence (top row) or presence (bottom row) of SARS-CoV-2 peptides. D, E. No significant differences in the magnitude of the T cell responses were observed between LC and non-LC individuals within the CD4+ (D) or CD8+ (E) T cell compartments (student’s t-tests). F. Analysis of polyfunctionality of SARS-CoV-2-specific CD4+ T cells. SPICE analysis revealed that polyfunctional SARS-CoV-2-specific CD4+ T cells co-expressing IFNγ, IL2, and TNFα (category 1) trended higher in non-LC than LC individuals albeit insignificantly (permutation test). TNFα single positive cells (category 7) made up the vast majority of SARS-CoV-2-specific CD4+ T cells in both LC and non-LC individuals. G. Analysis of polyfunctionality of SARS-CoV-2-specific CD8+ T cells. SPICE analysis revealed that polyfunctional SARS-CoV-2-specific CD8+ T cells co-expressing IFNγ, MIP1β, and TNFα (category 1) trended higher in non-LC than LC individuals albeit insignificantly (permutation test). TNFα single positive cells (category 7) made up the majority of SARS-CoV-2-specific CD8+ T cells in both LC and non-LC individuals, but to a lesser extent than for SARS-CoV-2-specific CD4+ T cells. Relative to SARS-CoV-2-specific CD4+ T cells, SARS-CoV-2-specific CD8+ T cells more frequently produced IFNγ.
Using Boolean gating, we then compared the frequencies of SARS-CoV-2-specific T cells between LC and non-LC individuals. No significant differences were observed when looking at the total population of SARS-CoV-2-specific T cells (expressing any combination of IFNγ, TNFα, IL2, and/or MIP1β) (Fig. 2D, E), or when looking at SARS-CoV-2-specific T cells producing any of the individual effector cytokines (Fig. S3A, B).
To quantitate polyfunctional cells, we implemented Simulation Program with Integrated Circuit Emphasis (SPICE) analyses. Overall, the distribution of polyfunctional SARS-CoV-2-specific T cells was similar between the LC and non-LC individuals, among both the CD4+ and CD8+ T cell compartments (Fig. 2F, G). The most polyfunctional T cells (IFNγ+TNFα+IL2+ for SARS-CoV-2-specific CD4+ T cells, and IFNγ+TNFα+MIP1β+ for SARS-CoV-2-specific CD8+ T cells) tended to be more abundant in non-LC individuals, but this trend did not reach statistical significance (Fig. 2F, G). TNFα single-positive cells made up the majority of SARS-CoV-2-specific T cells in both LC and non-LC individuals, particularly among CD4+ T cells where >50% of the responding cells singly produced this effector cytokine (Fig. 2F, G). By contrast, SARS-CoV-2-specific CD8+ T cells more frequently produced IFNγ than SARS-CoV-2-specific CD4+ T cells, independent of LC status (Fig. 2F, G). Overall, these analyses suggest that SARS-CoV-2-specific T cells have similar effector profiles in LC and non-LC individuals. However, one interesting exception was found in that IL6 was found to be induced within CD4+ T cells after SARS-CoV-2 peptide stimulation (Fig. S3C) exclusively among individuals with LC, albeit only in a small subset of these people (14%) (Fig. S3D). These results suggest that, although rare, SARS-CoV-2-specific CD4+ T cells producing IL6 may be specifically associated with a subset of individuals with LC.
Individuals with LC exhibit different distributions of T cell subsets including in sex-dimorphic fashion
T cells can be classified not only by the effector molecules they produce, but also by T cell lineage markers. We next took advantage of the deep phenotyping capabilities of CyTOF to compare classical subset distributions among total and SARS-CoV-2-specific T cells. Naïve T cells (Tn), stem cell memory cells (Tscm), central memory T cells (Tcm), effector memory T cells (Tem), transitional memory cells (Ttm), and effector memory RA (Temra) cells were identified from both CD4+ and CD8+ T cells through sequential gating strategies (Fig. S4A, B). In addition, T follicular helper cells (Tfh) and regulatory T cells (Treg) were identified from the CD4+ compartment (Fig. S4A). Among total CD4+ T cells, the Tcm, Tfh, and Treg subsets were all significantly elevated among the individuals with LC (Fig. 3A). The other CD4+ T cell subsets were not different between the LC and non-LC groups, and among SARS-CoV-2-specific CD4+ T cells none of the subsets were significantly different (Fig. 3A, B). Among both total and SARS-CoV-2-specific CD8+ T cells, no subsets were statistically different between the LC and non-LC groups; however, there was a trend (p=0.09) for the Tem subset to be higher among total CD8+ T cells in the LC group (Fig. S5).
Fig 3. Tcm, Tfh, and Treg frequencies differ between LC and Non-LC individuals.
A. CD4+ T cell subset analysis reveals higher proportions of Tcm, Tfh, and Treg in LC vs. non-LC individuals. **p<0.01, *p<0.05 (student’s t-test). B. No significant differences were observed in the proportion of the indicated SARS-CoV-2-specific CD4+ T cell subsets between LC vs. non-LC individuals. Phenotypic definition of subsets were as follows: naïve T cells (Tn): CD45RA+CD45RO-CCR7+CD95-, stem cell memory T cells (Tscm): CD45RA+CD45RO-CCR7+CD95+, central memory T cells (Tcm): CD45RA-CD45RO+CCR7+CD27+, effector memory T cells (Tem): CD45RA-CD45RO+CCR7-CD27-, transitional memory T cells (Ttm): CD45RA-CD45RO+CCR7-CD27+, effector memory RA T cells (Temra): CD45RA+CD45RO-CCR7-, T follicular helper cells (Tfh): CD45RA-CD45RO+PD1+CXCR5+, and regulatory T cells (Treg): CD45RA-CD45RO+CD127-CD25+.
To examine T cell distribution between LC and non-LC individuals using not only the T cell lineage markers, but all the phenotyping and effector markers analyzed in our CyTOF panel, we performed clustering analyses. CD4+ and CD8+ T cells fell into six (Clusters A1-A6) and five (Clusters B1-B5) clusters, respectively (Fig. S6A, S7A), that did not differ significantly between the LC and non-LC groups, except when we stratified the participants by sex. Among CD4+ T cells, cluster A1 was significantly over-represented in LC than non-LC females (but not males), while cluster A4 was significantly over-represented in LC than non-LC males (but not females) (Fig. S6B). Cluster A1 was composed of naïve CD4+ T cells, and expressed low levels of activation markers and inflammatory tissue homing receptors, and high levels of the lymph node homing receptors (Fig. S6C). By contrast, cluster A4 was composed of terminally differentiated effector memory CD4+ T cells and expressed high levels of receptors associated with homing to inflamed tissues but not those associated with homing to lymph nodes (Fig. S6D). They also expressed elevated levels of the cytolytic markers perforin, granzyme B, and LAMP1 (Fig. S6D). Among CD8+ T cells, cluster B1 was significantly under-represented in LC while cluster B2 was significantly over-represented, but only among females (Fig. S7B). Interestingly, the phenotypic features of cluster B1 mirrored those of cluster A1, while the features of cluster B2 mirrored those of cluster A4 (Fig. S7 C, D). These observations, together with the observation that cluster A4 trended higher in the female LC group, suggest that female individuals with LC harbor relatively low frequencies of resting naïve T cells expressing low levels of inflammatory tissue-homing receptors, and high frequencies of terminally differentiated effector memory T cells expressing inflammatory tissue homing receptors and cytolytic markers; this was true among both CD4+ and CD8+ T cells. More broadly, the results suggest that there are sex-dimorphic differences in the subset distribution of T cells between LC and non-LC individuals.
Preferential expression of some tissue-homing receptors on CD4+ T cells from individuals with LC
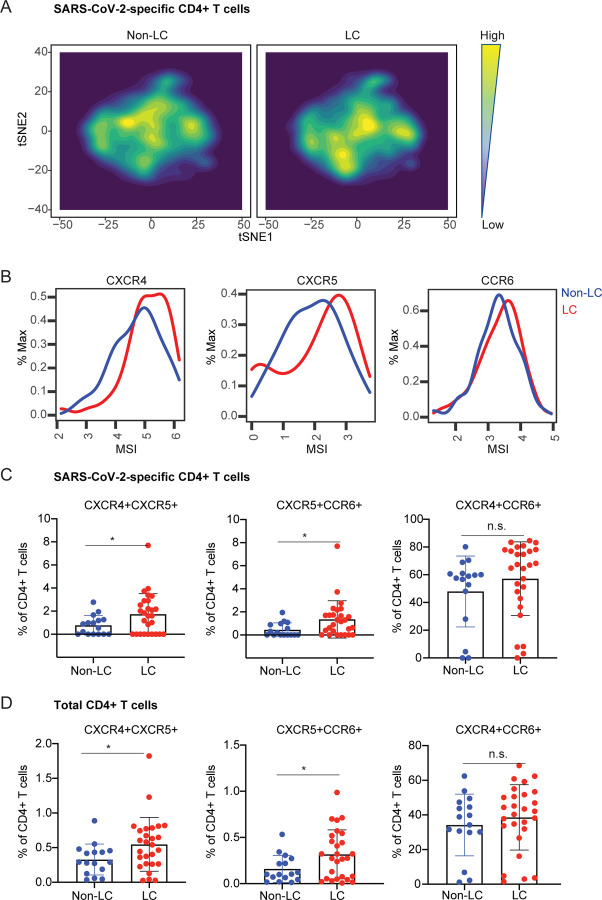
We then focused on the phenotypic features of SARS-CoV-2-specific T cells. Contour-based tSNE visualization of the datasets revealed that cells from the LC vs. non-LC groups tended to concentrate in different areas suggesting some phenotypic differences (Fig. 4A, 5A). Focusing first on the SARS-CoV-2-specific CD4+ T cells, we found that the chemokine receptors CXCR4, CXCR5, and CCR6 were all expressed at higher levels in the cells from the LC as compared to the non-LC individuals (Fig. 4B). Gating on SARS-CoV-2-specific CD4+ T cells co-expressing various pairs of these chemokine receptors revealed that those that were CXCR4+CXCR5+ and CXCR5+CCR6+, but not those that were CXCR4+CCR6+, were significantly increased in individuals with LC (Fig. 4C). Interestingly, this same pattern was observed among total CD4+ T cells (Fig. 4D), suggesting that the preferential expression of these tissue-homing receptors in CD4+ T cells from individuals with LC extended beyond those with specificity for SARS-CoV-2.
Fig. 4. SARS-CoV-2-specific CD4+ T cells from individuals with LC preferentially express homing receptors associated with migration to inflamed tissues.
A. tSNE contour depiction of SARS-CoV-2-specific CD4+ T cells from LC and non-LC individuals, highlighting different distribution of cells from the two groups. B. Expression of the chemokine receptors CXCR4, CXCR5, and CCR6 are elevated in SARS-CoV-2-specific CD4+ T cells from LC as compared to non-LC individuals. MSI corresponds to mean signal intensity of the indicated markers’ expression level. C, D. CXCR4+CXCR5+ and CXCR5+CCR6+ but not CXCR4+CCR6+ SARS-CoV-2-specific (C) and total (D) CD4+ T cells are significantly elevated in LC as compared to non-LC individuals. *p<0.05 (student’s t-test).
Fig. 5. SARS-CoV-2-specific CD8+ T cells from individuals with LC preferentially express exhaustion markers PD1 and CTLA4.
A. tSNE contour depiction of SARS-CoV-2-specific CD8+ T cells from LC and non-LC individuals, highlighting different distribution of cells from the two groups. B. Expression of exhaustion markers PD1 and CTLA4, but not TIGIT, are elevated on SARS-CoV-2-specific CD8+ T cells from LC as compared to non-LC individuals. MSI corresponds to mean signal intensity of the indicated markers’ expression level. C, D. PD1+CTLA4+ cells are significantly enriched among SARS-CoV-2-specific CD8+ T cells (C) but not total CD8+ T cells (D) in LC as compared to non-LC individuals. By contrast, TIGIT+CTLA4+ and PD1+TIGIT+ total and SARS-CoV-2-specific CD8+ T cells were equivalently distributed between LC and non-LC individuals. *p<0.05 (student’s t-test).
SARS-CoV-2-specific CD8+ T cells from individuals with LC preferentially co-express the checkpoint molecules PD1 and CTLA4
A similar manual inspection of CD8+ T cell data revealed the checkpoint/exhaustion markers PD1 and CTLA4 to be expressed at elevated levels on SARS-CoV-2-specific CD8+ T cells from the LC as compared to non-LC individuals, while the exhaustion marker TIGIT was not differentially expressed (Fig. 5B). Consistent with these data, SARS-CoV-2-specific CD8+ T cells that were PD1+CTLA4+, but not those that were TIGIT+CTLA4+ or PD1+TIGIT+, were significantly elevated in individuals with LC (Fig. 5C). Interestingly, however, frequencies of total PD1+CTLA4+ CD8+ T cells were not different between the LC and non-LC individuals (Fig. 5D). These results together suggest that LC-derived CD8+ T cells recognizing SARS-CoV-2 uniquely exhibit features of exhaustion, as reflected by co-expression of PD1 and CTLA4, perhaps due to persistence of SARS-CoV-2 antigen that cannot be eliminated in individuals with LC.
Individuals with LC exhibit a mis-coordinated T and antibody response
Serological analysis revealed significantly higher SARS-CoV-2 RBD antibody levels in the LC group than in the non-LC group (Fig. 6A). Interestingly, the two individuals with LC with the highest antibody levels (green oval) were not those that had the highest frequencies of exhausted (PD1+CTLA4+) SARS-CoV-2-specific CD8+ T cells (purple oval) (Fig. 6B), even though both features are consistent with a persistent SARS-CoV-2 reservoir. Interestingly, however, the individuals with the highest frequencies of exhausted SARS-CoV-2-specific CD8+ T cells did have the lowest frequencies of SARS-CoV-2-specific CD4+ Treg cells, and the frequencies of these two subsets of cells negatively correlated in LC but not non-LC individuals (Fig. 6C). When we assessed the association between antibody levels and SARS-CoV-2-specific T cell frequencies, we found a significant (p=0.0418 for CD4, p=0.0007 for CD8) positive correlation, but only in the non-LC individuals (Fig. 6C, D). These results suggest that a mis-coordinated humoral and cell-mediated response, previously implicated in severe COVID-19 36, may also be a hallmark of LC.
Fig. 6. Dis-coordinated humoral and adaptive immunity in individuals with LC.
A. SARS-CoV-2 spike RBD antibody levels are elevated in LC as compared to non-LC individuals. *p<0.05 (student’s t-test). B. Individuals with LC harboring the highest humoral response (green oval) are not those exhibiting highest levels of exhausted PD1+CTLA4+ SARS-CoV-2-specific CD8+ T cells (purple oval). C. Frequencies of PD1+CTLA4+ SARS-CoV-2-specific CD8+ T cells and SARS-CoV-2-specific CD4+ Treg cells are negatively associated only in individuals with LC. D, E. SARS-CoV-2 spike RBD antibody levels are significantly positively associated with the frequencies of SARS-CoV-2-specific CD4+ (D) and CD8+ (E) T cells in non-LC individuals, but not in individuals with LC.
Individuals with LC exhibit global alterations in PBMCs reflecting immune dysregulation and inflammation
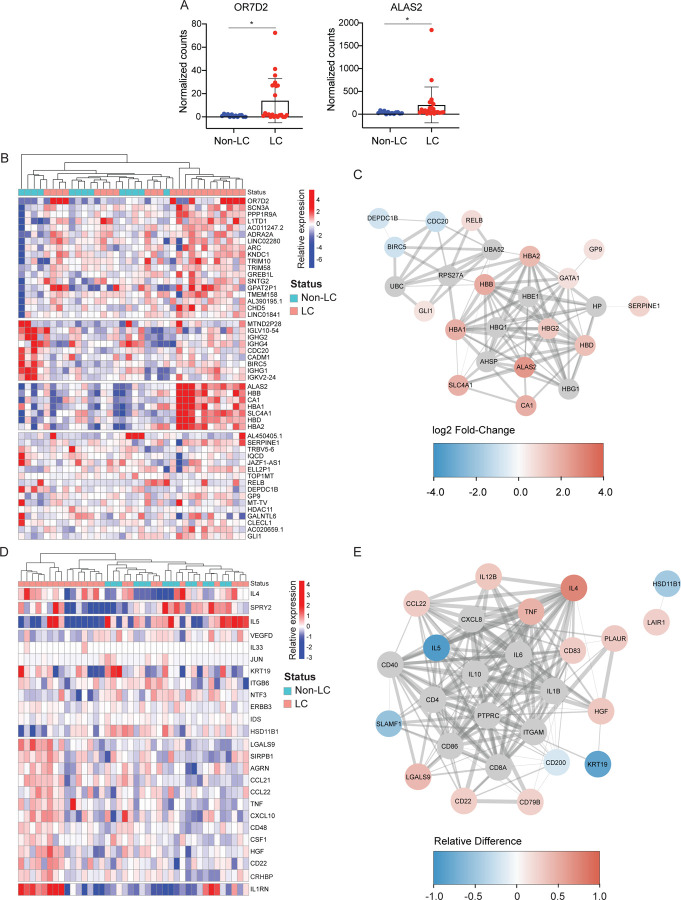
To determine whether the differences between LC and non-LC individuals extended beyond T cells and the humoral response, we examined the transcriptome of the PBMCs and the proteome of the sera included in the participant specimens. Assessing for total changes in gene expression in PBMCs by RNAseq, we found only two genes that remained significantly differentially expressed after multiple comparison adjustments: OR7D2 (Olfactory Receptor 7D2) and ALAS2 (5’-Aminolevulinate Synthase 2), both of which were over-expressed in the individuals with LC (Fig. 7A). OR7D2 encodes a G-protein-coupled receptor that responds to odorant molecules in the nose, while ALAS2 encodes a protein that catalyzes the first step in heme synthesis, defects in which can lead to anemia. A number of other genes were upregulated in individuals with LC, although not significantly so after conservative adjustments for multiple comparisons; these included a module of genes that regulate heme synthesis and carbon dioxide transport (ALAS3, HBB, CA1, HA, HBD, HBA2) (Fig. 7B). By contrast, a module consisting of immunoglobin kappa, lambda, and heavy chain genes, along with BIRC5, which plays an important role in T cell survival and function 37, were more highly expressed in non-LC individuals (Fig. 7B). Gene ontology analysis revealed that genes from both of these modules were highly networked together (Fig. 7C), strongly suggesting that these genes are indeed linked to LC.
Fig. 7. Global differential gene and gene product expression in participants with and without LC.
A. Relative gene expression levels of top two significantly differentially expressed genes (DEGs) in LC vs. non-LC individuals. OR7D2 corresponds to Olfactory Receptor family 7D2 (log2 fold-change=3.63), and ALAS2 to 5’Aminolevulinate Synthase 2 (log2 fold-change=2.58). *p < 0.05 (Wald test, with Benjamini-Hochberg correction). B. Clustered heatmap of the top 50 DEGs in PBMCs in LC compared to non-LC individuals. Genes are grouped into k-clusters based on similarity. Note four modules of gene expression, with the second corresponding to immunoglobulin and T cell genes (under-expressed in LC), and the third corresponding to heme synthesis and carbon dioxide transport (over-expressed in LC). C. Network mapping of related DEGs. Each node corresponds to a gene, and colors of nodes indicate the extent of change as indicated in the heatmap scale bar, with red corresponding to upregulation in individuals with LC, and blue corresponding to downregulation in individuals with LC. Edges depict the functional relevance between pairs of genes, where the thickness of the edge corresponds to the confidence of the evidence. The highly networked nature of the indicated genes supports their association with LC. D. Clustered heatmap of the top 25 differentially expressed plasma proteins from Olink Proximity Extension Assay with markers grouped into k-clusters based on similarity. Note a dominant module of inflammatory-related genes including LGALS9, CCL21, CCL22, TNF, CXCL10, and CD48. E. Network mapping of related differentially expressed proteins as detected by Olink. Graph representations are as described in panel C. Note the simultaneous over-expression of IL4 and CCL22 (in red) with under-expression of IL5 (in blue), all three proteins of which are involved in Th2 immune responses.
Olink analysis also revealed global changes associated with LC, including a module consisting of elevated levels of proteins associated with inflammation (LGALS9, CCL21, CCL22, TNF, CXCL10, CD48) (Fig. 7D). Proteins associated with immune regulation (IL1RN, CD22) were also elevated in individuals with LC (Fig. 7D). Interestingly, although IL4 and IL5 are both canonical cytokines for Th2 responses, these two cytokines exhibited very different expression patterns (Fig. 7C), and individuals with LC overall exhibited elevated levels of IL4 yet lower levels of IL5 (Fig. 7E). CCL22, a ligand for the Th2 marker CCR4, was expressed at elevated levels in individuals with LC (Fig. 7E). Together, these results suggest an elevated yet mis-coordinated Th2 response (elevated IL4 and CCL22 but diminished IL5) in individuals with LC. As for the RNAseq data, networking of genes from the inflammatory and immunoregulatory modules, as well as from the Th2 markers IL4, IL5, and CCL22, suggests a biologically-relevant plausible association of all of these genes with LC. Overall, our findings suggest that LC is associated with unique, and likely complex, global immune dysregulation.
Discussion
Using multiple “omics” analytical approaches on specimens from individuals exhibiting consistent long COVID trajectories, we demonstrate that individuals with LC exhibit perturbations in both total and SARS-CoV-2-specific T cells, which manifests at a global level as mis-coordination between the two main arms of adaptive immunity and overall changes in gene expression. In this analysis, we took care to limit several confounders that often constrain studies of LC. First, we carefully selected a cohort of individuals who consistently met the case definition for LC over an 8-month period and compared them with individuals who, when measured in the same way, using the same study instruments at the same timepoints, consistently demonstrated complete recovery. Second, to avoid surveillance bias, all assays were applied on samples from the same timepoint (8 months post-COVID), and we chose this relatively late timepoint so that we would not be confounded by individuals only exhibiting shorter-term LC (e.g., which resolve spontaneously after 4–6 months). Third, we restricted our analysis to only those individuals who prior to the time of sampling had not yet received a SARS-CoV-2 vaccine and who had not had a known or suspected SARS-CoV-2 re-infection, as either could markedly affect our SARS-CoV-2-specific antibody and T cell measurements.
Our CyTOF data revealed profound changes in classical subset distribution among total CD4+ T cells in individuals with LC, specifically a significantly higher proportion of CD4+ Tcm, Tfh, and Treg cells. Elevated frequencies of Tcm in LC have been reported previously 28, although another group reported the opposite observation that Tcm frequencies were decreased in LC 19. The reason for these discrepancies is not clear, but may reflect the composition of the clinical cohorts studied: while our study and the other one which also reported elevated Tcm cells examined only non-vaccinated individuals, the study where Tcm were decreased included some individuals who received a SARS-CoV-2 vaccine prior to sampling. Higher frequencies of Tfh and Treg cells in individuals with LC have, to our knowledge, not been previously reported. Interestingly, however, a prior study reported that elevated frequencies of activated Treg cells during acute SARS-CoV-2 infection predicted development of LC two to three months later 12, which together with our findings is consistent with Tregs being involved in both LC initiation and maintenance.
Elevated frequencies of Tcm, Tfh, and Treg in individuals with LC indicate an ongoing immune response persisting at 8 months post-infection. This immune response, however, may not necessarily be directed against SARS-CoV-2, and could potentially be directed against other viruses (e.g., reactivated EBV or other herpes viruses) or auto-antigens 12. Indeed, we did not find significantly higher magnitude of the SARS-CoV-2-specific T cell response as determined by intracellular cytokine staining in individuals with LC, consistent with prior observations reported from the activation-induced marker (AIM) assay 28. We also did not find individuals with LC to harbor more polyfunctional SARS-CoV-2-specific T cells, and in fact polyfunctionality trended lower in both the CD4 and CD8 compartments. At the same time, other studies have reported higher 38 or lower 29 SARS-CoV-2-specific T cell responses in the context of LC. Discrepancies may stem from differences in the LC cohorts analyzed, and in the assays used to quantitate T cell responses (including the SARS-CoV-2 proteins examined, and the approaches used to identify responding cells). We note that our approach was comprehensive in that we monitored expression levels of 10 different effectors, and settled on a subset of five of these (IFNγ, TNFα, IL2, MIP1β, IL6) using strict criteria, to define SARS-CoV-2-specific T cells.
One aspect highly consistent between all studies to date is the ability to detect SARS-CoV-2-specific T cells in both LC and non-LC individuals, months after infection. This could simply be attributed to the long-term persistence of memory T cells elicited by SARS-CoV-2, but may also indicate the persistence of a long-lived tissue viral reservoir. Indeed, we found that in LC relative to non-LC individuals, SARS-CoV-2-specific CD8+ T cells, but not total CD8+ T cells, more frequently expressed the exhaustion markers PD1 and CTLA4, which is consistent with ongoing stimulation with viral antigens. Also in support of a potential persistent reservoir is our observation of higher SARS-CoV-2 antibody levels in LC as compared to non-LC individuals, which has also been previously seen with Spike-specific IgG levels 28. Interestingly, our data revealed that the individuals with the highest frequencies of exhausted (PD1+CTLA4+) SARS-CoV-2-specific CD8+ T cells were not those with the highest SARS-CoV-2 antibody levels, suggesting that there may be multiple endotypes of LC being driven by persisting virus. Consistent with this possibility, a recent RNAseq study identified two types of LC: one being driven by high expression of Ig-related genes, and the other being associated with low levels of Ig-related genes 39. Based on these observations as well as case reports of improvement in LC symptoms following antiviral treatment 40–42, nirmatrelvir-ritonavir treatment as an antiviral strategy to clear this putative LC-associated SARS-CoV-2 reservoir is underway (NCT05576662, NCT05595369, NCT05668091). Future studies could evaluate other antivirals or monoclonal antibodies, and might consider incorporating checkpoint inhibition in conjunction with antivirals to reinvigorate T cells’ ability to help eliminate residual viremia.
One intriguing aspect of LC that emerged from our study is sex-dimorphism in T cell phenotypes. This perhaps is not so surprising given the different trajectories of COVID-19 between males and females 43 and the observation that LC is more common in females 5,44. Our data revealed that, among females, a subset of activated and cytotoxic T cells was more elevated in LC than in non-LC individuals; intriguingly, the opposite pattern was observed in males. The presence of cytotoxic T cells has been associated with gastrointestinal LC symptoms 12 and it will be of interest in future studies to establish whether biological sex impacts LC-associated cytotoxic T cell function. Intriguingly, biological sex was recently shown to manifest in the context of differential responses to influenza vaccines after COVID-19 convalescence 45, although individuals with LC were not examined therein.
While SARS-CoV-2-specific CD8+ T cells from individuals with LC showed signs of exhaustion, their CD4+ counterparts preferentially expressed the tissue-homing receptors CXCR4, CXCR5, and CCR6. Of note, these receptors can all direct immune cells to the lung, and CXCR4 is of particular interest as its expression on bystander T cells has been associated with severe/fatal COVID-19 32. More recently, elevated expression of CXCR4 was also observed on pulmonary neutrophils from severe COVID-19 cases, suggesting it as a potential target for constraining ARDS induced by SARS-CoV-2 infection 46. As we found elevated expression of CXCR4 not only on SARS-CoV-2-specific but also total CD4+ T cells in the context of LC, targeting of this receptor as well as other chemokine receptors may be useful to limit immune cell infiltration into the lung, which may persist in an elevated state of inflammation in individuals with LC.
Another intriguing observation we made about the SARS-CoV-2-specific CD4+ T cells from individuals with LC is their production of IL6 in response to spike peptide stimulation. Although this was observed in only a small minority of individuals with LC, it suggests that a highly inflammatory response directed against the virus, persisting for at least 8 months post-infection, could be a driver of the sequelae. Interestingly, elevated IL6 levels have been associated with pulmonary LC 38, and IL6 production induced by broad-spectrum mitogen PMA was found to be elevated in individuals with LC 19. These data together bolster the notion of targeting IL6 as a potential LC therapeutic strategy.
Most striking from our study was the finding that while fully recovered individuals exhibited coordinated humoral and cellular immune responses to SARS-CoV-2, this coordination was lost in the LC group. This finding is consistent with observations that about half of individuals with LC with no detectable SARS-CoV-2 antibodies have detectable SARS-CoV-2-specific T cell responses 47. That improper crosstalk between T and B cells may be involved in the etiology of LC is also supported by our RNAseq data, which showed that a cluster of genes including both immunoglobulin synthesis and T cell function were co-upregulated in those without LC, but not in individuals with LC. The downregulation of immunoglobulin-related genes in the context of LC has previously been reported and shown to be independent of spike antibody levels 39, which is in line with our finding higher levels of spike antibodies in our individuals with LC. How the humoral response becomes divorced from the cellular response is unclear, and could potentially involve a mis-alignment between IL4 and IL5 production by Th2 cells which emerged from our Olink analysis. Potential upstream initiators leading to the mis-coordination include a long-lived SARS-CoV-2 reservoir, reactivation of viral co-infections, or autoimmune responses.
Finally, our datasets taken together point to not only a dysregulated but also a highly pro-inflammatory signature in LC, consistent with prior data suggesting elevated and persistent inflammation in LC 22,29,48–50. Of particular interest was the elevation of the SGALS9 gene product in LC. LGALS9 encodes for Galectin 9, which has previously been shown to be upregulated during acute COVID-19 and may be a contributing factor in cytokine release and subsequent disease severity 51–53. The high inflammatory state observed in our individuals with LC may be in part driven by immune dysregulation, which could initiate from improper cross-talk between T and B cells as discussed above, or potentially faulty regulatory mechanisms as supported by our observation that the individuals with LC with the highest frequencies of exhausted SARS-CoV-2-specific CD8+ T cells were those that had the lowest frequencies SARS-CoV-2-specific CD4+ Treg cells. Non-immune mechanisms may also be at play, as supported by our findings that genes involved in olfactory sensing and heme synthesis were also upregulated in those with LC. The findings of increased heme synthesis were interesting in light of the fact that higher expression of genes involved in heme biosynthesis are observed during acute COVID-19 54,55, and that SARS-CoV-2 can bind hemoglobin and dysregulate heme metabolism 56–58. It is also possible that increased heme synthesis may reflect fibrin amyloid microclot formation that has been observed in individuals with LC 10,59. These microclots appear to be resistant to fibrinolizes and may trap potential circulating biomarkers of the coagulopathy 60,61. As a result, heme synthesis may play a useful role in determining the extent of microclot formation. Further studies of iron metabolism and red blood cell function, and their relationships to coagulopathy in the setting of LC, are warranted.
Our study has several limitations. The analysis cohort included only 43 participants, but this small sample was mitigated by our strict definitions of LC and complete recovery as detailed above. The parent cohort is a convenience sample and certainly not representative of all individuals with a history of SARS-CoV-2 infection, although it did reflect the characteristics of the pandemic in our geographic region. A second limitation was our focus on blood specimens, when the source of immune dysregulation, including SARS-CoV-2 persistence, likely originates from tissues. The infrastructure supporting LIINC has the ability for non-invasive tissue sampling via gut biopsies and fine needle aspirates 62,63, and future studies will take advantage of these capabilities to better understand the tissue-based mechanisms underlying the immune dysregulation that manifests in LC.
Overall, we found using multiple analytical approaches in a carefully selected cohort of individuals with consistent post-COVID symptom trajectories that LC is associated with dysregulation between humoral and cellular immunity. While LC exhibits both clinical and biological complexity, this work contributes to a growing understanding of the potential pathophysiological contributors and suggests several mechanisms warranting further exploration and/or disruption in future therapeutic trials.
Methods
Study participants
Participants were volunteers in the University of California, San Francisco (UCSF)-based Long-term Impact of Infection with Novel Coronavirus (LIINC) cohort (www.liincstudy.org; NCT04362150). Details of cohort recruitment, enrollment, and measurement procedures have been described in detail previously 35. Briefly, LIINC is a prospective observational study enrolling individuals with prior nucleic acid-confirmed SARS-CoV-2 infection in the San Francisco Bay Area, regardless of the presence or absence of post-acute symptoms. At each study visit, participants underwent an interviewer-administered assessment of 32 physical symptoms that were newly developed or had worsened since COVID-19 diagnosis, as well as assessment of mental health and quality of life. Pre-existing and unchanged symptoms were not considered to be attributable to COVID-19. In addition, detailed data regarding medical history, COVID-19 history, SARS-CoV-2 vaccination, and SARS-CoV-2 reinfection were collected. Two participants enrolled in LIINC had biospecimens collected previously via the UCSF COVID-19 Host Immune Response Pathogenesis (CHIRP) study, which utilizes identical procedures for ascertainment of clinical history as the LIINC study 33.
Because of challenges in the measurement of LC as outlined in prior work from the LIINC cohort, including within-participant symptom variability as well as the fact that some individuals with LC demonstrate symptomatic improvement and resolution of symptoms over time 35, we selected for this analysis participants who consistently met a case definition for LC based on the presence or absence of at least one symptom attributable to COVID-19 for the 8-month period following SARS-CoV-2 infection. The LC group (n=27) included individuals who consistently reported at least 1 COVID-19 attributed symptom during the entire study period, while the non-LC group (n=16) included individuals who consistently reported no COVID-19 attributed symptoms during the entire study period. Because of the potential effects of SARS-CoV-2 vaccination on clinical symptoms of LC as well as the immunologic measurements conducted in this study, we restricted the participants to those who provided a post-COVID blood sample prior to having ever received a SARS-CoV-2 vaccination. Blood samples were collected between September 16, 2020, and April 6, 2021.
Biospecimen Collection
At each visit, whole blood was collected in EDTA tubes followed by density gradient separation and isolation of PBMCs and plasma, as described previously 29. Serum was obtained concomitantly from serum-separator tubes. Serum and plasma were stored at −8°C and PBMCs were cryopreserved and stored in liquid nitrogen.
Antibody Assays
Antibody responses against SARS-CoV-2 spike RBD were measured on sera using the Pylon COVID-19 total antibody assay (ET Health). The assay’s lower limit of detection was 10 relative fluorescence units (RFUs).
SARS-CoV-2 peptide pool
Peptides used for T cell stimulation comprised a mix of overlapping 15-mers spanning the entire SARS-CoV-2 spike protein (PM-WCPV-S-1, purchased from JPT), and peptides corresponding to CD8+ T cell epitopes identified by T-scan 64 which were synthesized in-house (Table S3). The final concentration of 15-mer peptides was 300 nM and the final concentration of T-scan peptides was 450 nM.
Sample preparation for CyTOF
Sample preparation was performed similarly to methods previously described 30–33 with some modifications. Briefly, PBMCs were isolated from fresh blood draws from the LIINC participants and cryopreserved. Upon revival, cells were rested overnight and then divided equally into two aliquots. To the first aliquot, we added 3 µg/ml brefeldin A (BFA, to enable intracellular cytokine detection), the co-stimulation agonists anti-CD28 (2 µg/ml, BD Biosciences) and anti-CD49d (1 µg/ml, BD Biosciences), and the SARS-CoV-2 peptide pool prepared as described above. To the second aliquot, we added only 1% DMSO (Sigma) and 3 µg/ml BFA. Cells from both treatments were incubated at 37°C for 6 h. Thereafter, cells were treated with cisplatin (Sigma) as a live/dead distinguisher and then fixed in paraformaldehyde (PFA, Electron Microscopy Science) using methods similar to those recently implemented 30–33. Briefly, 6 million cells were resuspended in 4 ml of PBS (Rockland) containing 2 mM EDTA (Corning) and 25 µM cisplatin (Sigma), and incubated for 60 seconds. Cells were then washed twice in 1 ml CyFACS (containing 0.1% Bovine Serum Albumin (Sigma) and 0.1% Sodium Azide (Sigma) in PBS) and fixed for 10 mins at room temperature in 1.2 ml 2% PFA diluted in CyFACS. Cells were then washed twice with 1 ml CyFACS, resuspended in 100 µl 10% DMSO (Sigma) diluted in CyFACS, and frozen at −80℃ until CyTOF staining. PBMCs from a healthy donor were also subjected to the same cisplatin/fixation protocol and then aliquoted, and served as bridge samples for batch correction.
CyTOF staining was performed similar to methods recently described 30–33. Cisplatin-treated and PFA-fixed specimens were barcoded using Cell-ID™ 20-Plex Pd Barcoding Kit (Standard BioTools) according to manufacturer’s instructions. After barcoding, cells from up to 20 different samples were then combined into a single sample, at a concentration of 6 million cells per sample in 100 µl. The cells were then blocked at 4℃ for 15 mins in 200 µl CyFACS buffer containing 3 µl rat serum (Invitrogen), 3 µl mouse serum (Invitrogen), and 0.6 µl human serum (Sigma). After two washes with CyFACS, cells were subjected to surface antibody staining by resuspending the cells in 100 µl of freshly-prepared cell surface antibody mix (Table S2). Staining was allowed to proceed for 45 mins at 4℃. After 3 washes with CyFACS, cells were fixed overnight in 100 µl 2% PFA diluted in PBS. The next day, cells were washed with CyFACS and permeabilized by resuspension in 200 µl Foxp3 fix/perm buffer (eBioscience), and incubated at 4℃ for 30 mins. The cells were pelleted and then blocked at 4℃ for 15 mins by addition of a pre-mixed solution of 15 µl mouse serum, 15 µl rat serum, and 70 µl permeabilization Buffer (eBioscience). The cells were then washed in 800 µl permeabilization buffer (eBioscience), and incubated at 4℃ for 45 mins in 100 µl freshly-prepared intracellular staining antibody mix (Table S2). After another two washes with CyFACS, cells were stained with 250 nM Intercalator-IR (Standard BioTools) at room temperature for 20 mins. Finally, after two additional washes with CyFACS, the cells were then fixed with 1 ml 2% PFA diluted in CyFACS.
CyTOF data acquisition
The PFA-fixed samples were washed twice with CAS buffer (Standard BioTools) and then spiked with 10% (v/v) EQ™ Four Element Calibration Beads (Standard BioTools) diluted in CAS buffer, before loading onto a Helios CyTOF instrument (UCSF Parnassus Flow Core). A running speed of 200 to 400 events per second was maintained during sample collection, and the loading voltage was controlled between 4 and 5 to minimize clogging. Data were normalized to EQ beads by CyTOF software provided by Standard BioTools to batch-correct for instrument sensitivity during sample collection. Data matrices were exported as flow cytometry standard (fcs) files for data analyses as described below.
T cell data analyses
Data preprocessing.
EQ bead-normalized CyTOF datasets were concatenated, de-barcoded, and normalized using CyTOF software provided by Standard BioTools (version 6.7) according to manufacturer’s instructions. Cells were then analyzed by FlowJo (version 10.8.1, BD Biosciences). Intact (Ir191+Ir193+), live (Pt195-), singlet events were identified as described in Fig. 2A. Those events were then gated on T cells (CD3+) followed by sub-gating on CD4+ T cells and CD8+ T cells (Fig. 2A).
Identification of SARS-CoV-2-specific T cells.
For identification and definition of SARS-CoV-2-specific T cells, we compared unstimulated specimens to their peptide-stimulated counterparts. Effector cytokines (IFNγ, TNFα, IL2, IL4, IL6, IL17, MIP1β), cytolytic effectors (Granzyme B and perforin) and LAMP1 were assessed for the ability to identify antigen-specific T cells. The following criteria were established to identify effector molecules appropriate for identifying SARS-CoV-2-specific T cells: 1) counts of positive cells in unstimulated sample (not receiving peptide) was less than 5 events, or the frequency of positive cells was lower than 0.1%; 2) counts of positive cells in the peptide-stimulated sample was not less than 5, or the frequency was higher than 0.1%; 3) differences in frequencies of positive cells between unstimulated and peptide-stimulated samples cells was not less than 1%, 4) the fold-change in frequencies of positive cells between unstimulated and peptide-stimulated samples cells was greater than 10; and 5) the aforementioned 4 criteria could identify SARS-CoV-2-specific T cells among >50% of participants. Effectors which fulfilled all five criteria for CD4+ T cells were IFNγ, TNFα, and IL2, and those which fulfilled all five criteria for CD8+ T cells were IFNγ, TNFα, and MIP1β. For a sub-analysis to identify responding cells that may only exist in a small subset of individuals, we removed criteria #5 and reduced the positive cell counts to number 3 within criteria #1 and #2. This approach allowed us to determine that SARS-CoV-2-specific CD4+ T cells producing IL6 were exclusively detected from LC (Fig. S3D). SARS-CoV-2-specific T cells, once identified, were analyzed by Boolean gating 65 and exported for further analyses.
SPICE.
SPICE analyses were performed similar to previously described methods 66,67. Briefly, CD4+ and CD8+ T cells were subjected to manual gating based on expression of cytokines used to define SARS-CoV-2-specific T cells (IFNγ, TNFα, IL2, and MIP1β, see above) using operations of Boolean logic. The dataset matrix generated from Boolean gating was then inputted into SPICE software (version 6.1) for polyfunctional analysis. The parameters for running the dataset were: iterations for permutation test = 10,000, and highlight values = 0.05. The parameters for the query structure were set as follows: values = frequency of single cytokine positive cells in total CD4+/CD8+ T cells (generated directly from FlowJo); category = IFNγ, TNFα, IL2, and MIP1β; overlay = patient type (LC vs. Non-LC); group = all other variables in the data matrix (including sex, PID, cell type, hospitalization status, and batch). All other parameters for SPICE analyses were kept as default.
T cell subsetting.
Manual gating was performed using R (version 4.1.3). Briefly, arcsinh-transformed data corresponding to total or SARS-CoV-2-specific T cells were plotted as 2D plots using the CytoExploreR package. Statistical data were then exported for further analyses. Visualization of datasets by tSNE were performed using R (version 4.1.3), using methods similar to those previously described 30–33. Briefly, CytoExploreR and tidyr packages were used to load the data. tSNE was performed using Rtsne and RColorBrewer packages on arcsinh-transformed markers. Total CD4+/CD8+ T cells were downsampled to n = 8000 (maximal cell number for individual samples) before tSNE analysis. The parameters for tSNE were set as follows: iteration = 1000, perplexity = 30, and theta= 0.5.
T cell clustering analysis.
FCS files corresponding to total and SARS-CoV-2 specific CD4+ and CD8+ T cells were imported in R for data transformation. Packages of flowcore, expss, class, and openxlsx were loaded in R for training FCS files. Arcsinh-transformed data were then exported as csv files for clustering analyses. Biological (LC status, gender, hospitalization status) and technical (batch/run of processing) variables were visualized using the DimPlot function in the Seurat package 68. As batch effects associated with the processing run were evident, batch correction was performed across the 6 batches using the harmony 69 batch correction function RunHarmony, applied to the marker levels in cells. The optimal clustering resolution parameters were determined using Random Forests 70 and a silhouette score-based assessment of clustering validity and subject-wise cross-validation. This procedure is described in greater detail in George et al. 62. A generalized linear mixed model (GLMM) (implemented in the lme4 71 package in R with family argument set to the binomial probability distribution) was used to estimate the association between cluster membership and LC status and the gender of the participant, with the participant modeled as a random effect. For each given participant, cluster membership of cells is encoded as a pair of numbers representing the number of cells in the cluster and the number of cells not in the cluster. Clusters having fewer than 3 cells were discarded. The gender-specific log odds ratio of cluster membership association with LC status was estimated using the emmeans 72 R package using the GLMM model fit.
RNAseq
RNAseq was performed on total PBMCs from n=36 individuals in our cohort using the AllPrep kit as per manufacturer’s instructions (Qiagen). RNA libraries, next generation Illumina sequencing, quality control analysis, trimming, and alignment to the human genome (hg19) were performed by Genewiz (Azenta Inc.). Briefly, following oligo dT enrichment, fragmentation and random priming, cDNA syntheses was completed. End repair, 5’ phosphorylation and dA-tailing were performed, followed by adaptor ligation, PCR enrichment, and sequencing on an Illumina HiSeq platform using PE150 (paired-end sequencing, 150 bp for reads 1 and reads 2). Raw reads (480 Gb in total) were trimmed using Trimmomatic (version 0.36) to remove adapter sequences and poor-quality reads. Trimmed reads were then mapped to Homo sapiens GRCh37 using star aligner (version 2.5.2b). Log2 fold-changes were calculated between those with or without any LC symptoms. P values were adjusted controlling for false discovery rates. We used DESeq2’s Wald test for normalizing raw reads counts. Genes with an adjusted p-value < 0.05 and absolute log2 fold-change > 1 were considered as significantly differentially expressed genes (DEGs). Clustered heatmaps of DEG were constructed with groups of genes (rows) defined using the k-means algorithm to cluster genes into k clusters based on their similarity. K = 4 was determined using the HOPACH (Hierarchical Ordered Partitioning and Collapsing Hybrid) algorithm, which recursively partitions a hierarchical tree while ordering and collapsing clusters at each level to identify the level of the tree with maximally homogeneous clusters.
Olink
We performed the Olink EXPLORE 384 inflammation Protein Extension Assay (PEA) from plasma from n=40 individuals in our cohort to characterize 384 unique plasma proteins associated with inflammation and immune signaling. PEA involves dual-recognition of two matched antibodies labelled with unique DNA oligonucleotides that simultaneously bind to specific target proteins. The simultaneous antibody binding leads to hybridization of unique DNA oligonucleotides that serve as templates for polymerase-dependent extension (DNA barcoding) followed by PCR amplification and NovaSeq (Illumina) DNA sequencing as published 73–77. A similar analysis pipeline was applied to the protein biomarkers as described for the gene expression data as above.
Data visualization for RNAseq and Olink
To generate heatmaps, the R package HOPACH 78 was used to find the best cluster number. Gene expression values were log-transformed and centered using the average expression value for each gene. Genes were clustered by running the Kmeans algorithm using the best cluster number K found, and the results were plotted using the pheatmap package 79. For gene network analyses, the STRING interaction database was used to reconstruct gene networks using stringApp 80 for Cytoscape 81. For the network, the top 50 genes or 25 proteins with the lowest p-values were selected from the RNAseq data and Olink data, respectively. Then genes were subjected to stringApp with an interaction score cutoff = 0.5, and the number of maximum additional indirect interactors cutoff = 10. The analysis integrated STRING data with our gene inputs, resulting in a network of 24 nodes and 100 edges in for the RNAseq data, and a network of 26 nodes and 165 edges for the Olink data. In each network, a node corresponds to a gene, an edge represents the functional relevance between a pair of genes, with the thickness of each edge reflecting the confidence level. Node color indicates the degree of log2 fold-change and the difference between protein expression values for the RNAseq and Olink data, respectively.
Supplementary Material
Fig. S1. Clinical characteristics of LC and non-LC individuals analyzed in this study. A. Gender distribution of individuals from the LIINC cohort analyzed in this study. Overall, 55.8% of the participants were female; the non-LC group comprised 43.75% females and the LC group 62.96% females. B. The proportion of LC and non-LC individuals that were hospitalized at the time of acute COVID-19 infection. Overall, 20.9% of the participants were hospitalized: 12.5% among the non-LC individuals, and 25.9% among the individuals with LC. C. The number of sequelae symptoms significantly increased with time among the individuals with LC. Number of sequelae symptoms at four (M4) and eight (M8) months are shown. *p<0.05 (student’s t-test).
Fig. S2. Cytokine and effector molecule expression on T cells in the absence or presence of SARS-CoV-2 peptides. A. CD4+ T cells from a representative donor, in the absence or presence of SARS-CoV-2 peptides. The red box highlights the three cytokines used to define SARS-CoV-2-specific CD4+ T cells. B. CD8+ T cells from a representative donor, in the absence or presence of SARS-CoV-2 peptides. The red boxes highlight the three cytokines used to define SARS-CoV-2-specific CD8+ T cells.
Fig. S3. SARS-CoV-2-specific T cell responses defined individually as those producing IFNγ, IL2, TNFα, and MIP1β do not differ between LC and non-LC individuals. A. Shown are the proportions of SARS-CoV-2-specific CD4+ T cells as defined by cells producing IFNγ, IL2, or TNFα in response to SARS-CoV-2 peptide stimulation. n.s.: non-significant as determined by student’s t-test. B. Shown are the proportions of SARS-CoV-2-specific CD8+ T cells as defined by cells producing IFNγ, MIP1β, or TNFα in response to SARS-CoV-2 peptide stimulation. n.s.: non-significant as determined by student’s t-test. C. IL6-producing CD4+ T cells are observed after SARS-CoV-2 peptide stimulation in some donors. Shown are cells from a representative individual with LC. D. IL6+ SARS-CoV-2-specific CD4+ T cells are exclusively observed from participants with LC. *p<0.05 (Welch’s t-test).
Fig. S4. Gating strategy to define classical T cell subsets. Shown are gating strategies to define the indicated subsets of CD4+ (A) and CD8+ (B) T cells.
Fig. S5. Subset distribution of total and SARS-CoV-2-specific CD8+ T cells among LC and Non-LC individuals. A. Tem frequencies trended higher among total CD8+ T cells from LC as compared to non-LC individuals (student’s t-test). B. Temra frequencies trended lower among SARS-CoV-2-specific CD8+ T cells from LC as compared to non-LC individuals (student’s t-test).
Fig. S6. Sex-dimorphic CD4+ T cell cluster distribution in individuals with LC. A. Cluster distribution among baseline CD4+ T cells as depicted by UMAP. B. Female individuals with LC harbor significantly lower frequencies of cluster A1 cells relative to female non-LC individuals, and male individuals with LC harbor significantly lower frequencies of cluster A4 cells relative to male non-LC individuals. *p<0.05 (GLMM fit - see methods). C. Relative to total CD4+ T cells, cluster A1 is characterized by expression of naïve cell markers (CD45RAhighCD45ROlowCD27high), and low expression of inflammatory tissue homing receptors (CD29lowCXCR4low) but high expression lymph node homing receptors (CD62LhighCCR7high). The activation markers HLA-DR and OX40 were also lowly expressed in cluster A1 cells. D. Relative to total CD4+ T cells, cluster A4 is characterized by expression of terminally differentiated effector memory T cell markers (CD45RAlowCD45ROhighCD27lowCCR7lowCD57high), and high expression of homing receptors for inflamed tissues (CD29highCXCR4highCCR5high) but low expression of lymph node homing receptors (CD62LlowCCR7low). The cytolysis markers perforin, granzyme, and LAMP1 were expressed at elevated levels in cluster A4 cells. ****p<0.0001 (student’s t-test). Shown are data of concatenated data depicted as histograms, or violin plots showing distribution of cells, among the indicated cluster as compared to total CD4+ T cells.
Fig. S7. Sex-dimorphic differential CD8+ T cell cluster distribution in individuals with LC. A. Cluster distribution among baseline CD8+ T cells as depicted by UMAP. B. Female individuals with LC harbor significantly lower frequencies of cluster B1 cells and significantly higher frequencies of cluster B2 cells, relative to female non-LC individuals. *p<0.05 (GLMM fit - see methods). C. Relative to total CD8+ T cells, cluster B1 is characterized by expression of naïve cell markers (CD45RAhighCD45ROlowCD27high), and low expression of tissue homing receptors (CD29lowCXCR4low) but high expression lymph node homing receptors (CD62LhighCCR7high). The activation markers HLA-DR and OX40 were also lowly expressed in cluster B1 cells. D. Relative to total CD8+ T cells, cluster B2 is characterized by expression of terminally differentiated effector memory T cell markers (CD45RAlowCD45ROhighCD27lowCCR7lowCD57high), and high expression of homing receptors for inflamed tissues (CD29highCXCR4highCCR5high) but low expression of lymph node homing receptors (CD62LlowCCR7low). The cytolysis markers perforin, granzyme, and LAMP1 were expressed at elevated levels in cluster B2 cells. Shown are data of concatenated data depicted as histograms, or violin plots showing distribution of cells, among the indicated cluster as compared to total CD8+ T cells.
ACKNOWLEDGEMENTS
This work was supported by the Van Auken Private Foundation, David Henke, and Pamela and Edward Taft; philanthropic funds donated to Gladstone Institutes by The Roddenberry Foundation and individual donors devoted to COVID-19 research; the Program for Breakthrough Biomedical Research, which is partly funded by the Sandler Foundation; and Awards #2164 and #2208 from Fast Grants, a part of Emergent Ventures at the Mercatus Center, George Mason University. We acknowledge the NIH DRC Center Grant P30 DK063720 and the S10 1S10OD018040–01 for use of the CyTOF instrument. This study was also funded by the Ministerium für Wissenschaft, Forschung und Kunst, Baden Württemberg, Germany (KNKC.031) and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 316249678—SFB 1279. The parent cohort (LIINC) is supported by NIH/NIAID 3R01AI141003–03S1 and NIH/NIAID R01AI158013. We thank Stanley Tamaki, Vinh Nguyen, Pricsilla Sanchez and Claudia Bispo for CyTOF assistance at the Parnassus Flow Core, Merve Karacan and Nico Preising for technical assistance in peptide synthesis, John Carroll for assistance on graphics, Françoise Chanut for editorial assistance, and Robin Givens for administrative assistance.
We are grateful to the study participants and their medical providers. We acknowledge current and former LIINC clinical study team members Tamara Abualhsan, Andrea Alvarez, Mireya Arreguin, Melissa Buitrago, Monika Deswal, Nicole DelCastillo, Emily Fehrman, Halle Grebe, Heather Hartig, Yanel Hernandez, Marian Kerbleski, Raushun Kirtikar, James Lombardo, Michael Luna, Sadie Munter, Lynn Ngo, Enrique Martinez Ortiz, Antonio Rodriguez, Justin Romero, Dylan Ryder, Ruth Diaz Sanchez, Matthew So, Celina Song, Viva Tai, Alex Tang, Cassandra Thanh, Fatima Ticas, Leonel Torres, Brandon Tran, Deepshika Varma, and Meghann Williams; and LIINC laboratory team members Amanda Buck, Tyler-Marie Deveau, Joanna Donatelli, Jill Hakim, Nikita Iyer, Owen Janson, Brian LaFranchi, Christopher Nixon, Isaac Thomas, and Keirstinne Turcios. We thank Jessica Chen, Aidan Donovan, and Carrie Forman for assistance with data entry and review. We thank the UCSF AIDS Specimen Bank for processing specimens and maintaining the LIINC biospecimen repository. We are grateful to Elnaz Eilkhani and Monika Deswal for regulatory support. We are also grateful for the contributions of additional current and former LIINC leadership team members: Matthew Durstenfeld, Priscilla Hsue, Bryan Greenhouse, Isabelle Rodriguez-Barraquer, and Rachel Rutishauser.
Footnotes
CONFLICTS OF INTERESTS
MJP reports consulting fees from Gilead Sciences and AstraZeneca, outside the submitted work. SGD reports grants and/or personal fees from Gilead Sciences, Merck & Co., Viiv, AbbVie, Eli Lilly, ByroLogyx, and Enochian Biosciences, outside the submitted work. TJH receives grant support from Merck and consults for Roche. All other authors report no potential conflicts.
Statistical tests
Unless otherwise indicated, permutation tests, student’s t-tests, and Welch’s t test were used for statistical analyses. *p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0001, and n.s. non-significant. Graphs were plotted by GraphPad Prism (version 9.4.1).
Informed consent
All participants provided written informed consent and study protocols were approved by the UCSF Institutional Review Board.
Data availability
The raw CyTOF datasets for this study corresponding to total and SARS-CoV-2-specific CD4+ and CD8+ T cells are publicly accessible through the following link: https://datadryad.org/stash/share/TE_QuY0JX23V2n2CIMO2PgsR6afIp6GGusdQ5nXVGnk. The raw RNAseq data from this study are deposited in the GEO (Gene Expression Omnibus) database: GSE224615. All other raw datasets from this study are available from the corresponding authors upon request.
REFERENCES
- 1.Nalbandian A. et al. Post-acute COVID-19 syndrome. Nat. Med., doi: 10.1038/s41591-021-01283-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peluso M. J. & Deeks S. G. Early clues regarding the pathogenesis of long-COVID. Trends Immunol. 43, 268–270, doi: 10.1016/j.it.2022.02.008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis H. E., McCorkell L., Vogel J. M. & Topol E. J. Long COVID: major findings, mechanisms and recommendations. Nature Reviews Microbiology, 1–14 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azzolini E. et al. Association Between BNT162b2 Vaccination and Long COVID After Infections Not Requiring Hospitalization in Health Care Workers. JAMA, doi: 10.1001/jama.2022.11691 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayoubkhani D. et al. Trajectory of long covid symptoms after covid-19 vaccination: community based cohort study. BMJ 377, e069676, doi: 10.1136/bmj-2021-069676 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Aly Z., Bowe B. & Xie Y. Long COVID after breakthrough SARS-CoV-2 infection. Nat. Med., 1–7, doi: 10.1038/s41591-022-01840-0 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Y., Choi T. & Al-Aly Z. Nirmatrelvir and the Risk of Post-Acute Sequelae of COVID-19. doi: 10.1101/2022.11.03.22281783. [DOI] [Google Scholar]
- 8.Wynberg E. et al. The effect of SARS-CoV-2 vaccination on post-acute sequelae of COVID-19 (PASC): A prospective cohort study. Vaccine 40, 4424–4431, doi: 10.1016/j.vaccine.2022.05.090 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambrosino P. et al. Clinical assessment of endothelial function in convalescent COVID-19 patients: a meta-analysis with meta-regressions. Ann. Med. 54, 3234–3249, doi: 10.1080/07853890.2022.2136403 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pretorius E. et al. Prevalence of symptoms, comorbidities, fibrin amyloid microclots and platelet pathology in individuals with Long COVID/Post-Acute Sequelae of COVID-19 (PASC). Cardiovasc. Diabetol. 21, 148, doi: 10.1186/s12933-022-01579-5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeßle J. et al. Persistent symptoms in adult patients one year after COVID-19: a prospective cohort study. Clin. Infect. Dis., doi: 10.1093/cid/ciab611 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Y. et al. Multiple Early Factors Anticipate Post-Acute COVID-19 Sequelae. Cell 0, doi: 10.1016/j.cell.2022.01.014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Son K. et al. Circulating anti-nuclear autoantibodies in COVID-19 survivors predict long-COVID symptoms. Eur. Respir. J., doi: 10.1183/13993003.00970-2022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold J. E., Okyay R. A., Licht W. E. & Hurley D. J. Investigation of long COVID prevalence and its relationship to Epstein-Barr virus reactivation. Pathogens 10, 763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peluso M. J. et al. Impact of pre-existing chronic viral infection and reactivation on the development of long COVID. The Journal of Clinical Investigation (2022). [Google Scholar]
- 16.Peluso M. J. et al. SARS-CoV-2 and Mitochondrial Proteins in Neural-Derived Exosomes of COVID-19. Ann. Neurol., doi: 10.1002/ana.26350 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swank Z. et al. Persistent circulating SARS-CoV-2 spike is associated with post-acute COVID-19 sequelae. Clin. Infect. Dis., doi: 10.1093/cid/ciac722 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natarajan A. et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med (N Y), doi: 10.1016/j.medj.2022.04.001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein J. et al. Distinguishing features of Long COVID identified through immune profiling. medRxiv, doi: 10.1101/2022.08.09.22278592 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phetsouphanh C. et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol., 1–7, doi: 10.1038/s41590-021-01113-x (2022). [DOI] [PubMed] [Google Scholar]
- 21.Peluso M. J. et al. Markers of Immune Activation and Inflammation in Individuals With Postacute Sequelae of Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J. Infect. Dis., doi: 10.1093/infdis/jiab490 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giron L. B. et al. Markers of fungal translocation are elevated during post-acute sequelae of SARS-CoV-2 and induce NF-κB signaling. JCI Insight 7, doi: 10.1172/jci.insight.164813 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vijayakumar B. et al. Immuno-proteomic profiling reveals aberrant immune cell regulation in the airways of individuals with ongoing post-COVD-19 respiratory disease. Immunity 0, doi: 10.1016/j.immuni.2022.01.017 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Littlefield K. M. et al. SARS-CoV-2-specific T cells associate with inflammation and reduced lung function in pulmonary post-acute sequalae of SARS-CoV-2. PLoS Pathog. 18, e1010359, doi: 10.1371/journal.ppat.1010359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultheiß C. et al. The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep Med 3, 100663, doi: 10.1016/j.xcrm.2022.100663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peluso M. J., Thomas I. J., Munter S. E., Deeks S. G. & Henrich T. J. Lack of Antinuclear Antibodies in Convalescent Coronavirus Disease 2019 Patients With Persistent Symptoms. Clinical Infectious Diseases 74, 2083–2084 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodansky A et al. Autoantigen profiling reveals a shared post-COVID signature in fully recovered and Long COVID patients. medRxiv (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Files J. K. et al. Duration of post-COVID-19 symptoms is associated with sustained SARS-CoV-2-specific immune responses. JCI Insight 6, doi: 10.1172/jci.insight.151544 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peluso M. J. et al. Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms. Cell Rep 36, 109518, doi: 10.1016/j.celrep.2021.109518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma T. et al. Protracted yet Coordinated Differentiation of Long-Lived SARS-CoV-2-Specific CD8(+) T Cells during Convalescence. J Immunol 207, 1344–1356, doi: 10.4049/jimmunol.2100465 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neidleman J. et al. SARS-CoV-2-Specific T Cells Exhibit Phenotypic Features of Helper Function, Lack of Terminal Differentiation, and High Proliferation Potential. Cell Rep Med 1, 100081, doi: 10.1016/j.xcrm.2020.100081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neidleman J. et al. Distinctive features of SARS-CoV-2-specific T cells predict recovery from severe COVID-19. Cell Rep 36, 109414, doi: 10.1016/j.celrep.2021.109414 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neidleman J. et al. mRNA vaccine-induced T cells respond identically to SARS-CoV-2 variants of concern but differ in longevity and homing properties depending on prior infection status. Elife 10, doi: 10.7554/eLife.72619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suryawanshi R. K. et al. Limited cross-variant immunity from SARS-CoV-2 Omicron without vaccination. Nature 607, 351–355, doi: 10.1038/s41586-022-04865-0 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peluso M. J. et al. Persistence, Magnitude, and Patterns of Postacute Symptoms and Quality of Life Following Onset of SARS-CoV-2 Infection: Cohort Description and Approaches for Measurement. Open Forum Infect Dis 9, ofab640, doi: 10.1093/ofid/ofab640 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rydyznski Moderbacher C. et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 183, 996–1012 e1019, doi: 10.1016/j.cell.2020.09.038 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson K. M. et al. Survivin co-ordinates formation of follicular T-cells acting in synergy with Bcl-6. Oncotarget 6, 20043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Littlefield K. M. et al. SARS-CoV-2-specific T cells associate with inflammation and reduced lung function in pulmonary post-acute sequalae of SARS-CoV-2. PLoS pathogens 18, e1010359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson R. C. et al. Molecular states during acute COVID-19 reveal distinct etiologies of long-term sequelae. Nature Medicine, 1–11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peluso M. J. et al. Effect of oral nirmatrelvir on long COVID Symptoms: 4 cases and rationale for systematic studies. Pathogens and Immunity 7, 95 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geng L. N., Bonilla H. F., Shafer R. W., Miglis M. G. & Yang P. C. Case report of breakthrough long COVID and the use of nirmatrelvir-ritonavir. (2022). [Google Scholar]
- 42.Visvabharathy L., Orban Z. S. & Koralnik I. J. Treatment of Long COVID with nirmatrelvir/ritonavir and tocilizumab in a patient with rheumatoid arthritis and SARS-CoV-2 antigen persistence: a case report. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi T. et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 588, 315–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sudre C. H. et al. Attributes and predictors of long COVID. Nature medicine 27, 626–631 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sparks R. et al. Influenza vaccination reveals sex dimorphic imprints of prior mild COVID-19. Nature, 1–3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddins D. J. et al. Transcriptional reprogramming of infiltrating neutrophils drives lung disease in severe COVID-19 despite low viral load. Blood Advances (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krishna B. A. et al. Evidence of previous SARS-CoV-2 infection in seronegative patients with long COVID. EBioMedicine 81, 104129 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peluso M. J. et al. Markers of immune activation and inflammation in individuals with postacute sequelae of severe acute respiratory syndrome coronavirus 2 infection. The Journal of infectious diseases 224, 1839–1848 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durstenfeld M. S. et al. Role of antibodies, inflammatory markers, and echocardiographic findings in post-acute cardiopulmonary symptoms after SARS-CoV-2 infection. MedRxiv (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peluso M. J. et al. Plasma markers of neurologic injury and inflammation in people with self-reported neurologic postacute sequelae of SARS-CoV-2 infection. Neurology-Neuroimmunology Neuroinflammation 9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel H. et al. Proteomic blood profiling in mild, severe and critical COVID-19 patients. Scientific reports 11, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bozorgmehr N. et al. Galectin-9, a player in cytokine release syndrome and a surrogate diagnostic biomarker in SARS-CoV-2 infection. MBio 12, e00384–00321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du L. et al. Human Galectin-9 Potently Enhances SARS-CoV-2 Replication and Inflammation in Airway Epithelial Cells (preprint). (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gardinassi L. G., Souza C. O., Sales-Campos H. & Fonseca S. G. Immune and metabolic signatures of COVID-19 revealed by transcriptomics data reuse. Frontiers in immunology 11, 1636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su W.-L. et al. Desaturation and heme elevation during COVID-19 infection: A potential prognostic factor of heme oxygenase-1. Journal of Microbiology, Immunology and Infection 54, 113–116 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hopp M. T., Rathod D. C. & Imhof D. Host and viral proteins involved in SARS-CoV-2 infection differentially bind heme. Protein Science 31, e4451 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kronstein-Wiedemann R. et al. SARS-CoV-2 Infects red blood cell progenitors and dysregulates hemoglobin and iron metabolism. Stem Cell Reviews and Reports, 1–13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lechuga G. C. et al. SARS-CoV-2 proteins bind to hemoglobin and its metabolites. International Journal of Molecular Sciences 22, 9035 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grobbelaar L. M. et al. in Seminars in thrombosis and hemostasis. (Thieme Medical Publishers, Inc.). [Google Scholar]
- 60.Kruger A. et al. Proteomics of fibrin amyloid microclots in long COVID/post-acute sequelae of COVID-19 (PASC) shows many entrapped pro-inflammatory molecules that may also contribute to a failed fibrinolytic system. Cardiovascular Diabetology 21, 1–23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kell D. B., Laubscher G. J. & Pretorius E. A central role for amyloid fibrin microclots in long COVID/PASC: origins and therapeutic implications. Biochemical Journal 479, 537–559 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.George A. F. et al. Deep Phenotypic Analysis of Blood and Lymphoid T and NK Cells From HIV+ Controllers and ART-Suppressed Individuals. Front Immunol 13, 803417, doi: 10.3389/fimmu.2022.803417 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neidleman J. et al. Phenotypic analysis of the unstimulated in vivo HIV CD4 T cell reservoir. Elife 9, e60933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferretti A. P. et al. Unbiased Screens Show CD8(+) T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity 53, 1095–1107 e1093, doi: 10.1016/j.immuni.2020.10.006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steiner S. et al. SARS-CoV-2 T Cell Response in Severe and Fatal COVID-19 in Primary Antibody Deficiency Patients Without Specific Humoral Immunity. Front Immunol 13, 840126, doi: 10.3389/fimmu.2022.840126 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Littlefield K. M. et al. SARS-CoV-2-specific T cells associate with inflammation and reduced lung function in pulmonary post-acute sequalae of SARS-CoV-2. PLoS Pathog 18, e1010359, doi: 10.1371/journal.ppat.1010359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roederer M., Nozzi J. L. & Nason M. C. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 79, 167–174, doi: 10.1002/cyto.a.21015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hao Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529, doi: 10.1016/j.cell.2021.04.048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korsunsky I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296, doi: 10.1038/s41592-019-0619-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Breiman L. Random forests. Machine learning 45, 5–32 (2001). [Google Scholar]
- 71.Bates D., Mächler M., Bolker B. & Walker S. Fitting linear mixed-effects models using lme4. arXiv preprint arXiv:1406.5823 (2014). [Google Scholar]
- 72.Lenth R., Singmann H., Love J., Buerkner P. & Herve M. Emmeans: Estimated marginal means, aka least-squares means. R package version 1, 3 (2018). [Google Scholar]
- 73.Assarsson E. et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PloS one 9, e95192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bäckryd E., Tanum L., Lind A.-L., Larsson A. & Gordh T. Evidence of both systemic inflammation and neuroinflammation in fibromyalgia patients, as assessed by a multiplex protein panel applied to the cerebrospinal fluid and to plasma. Journal of pain research 10, 515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Björkesten J. et al. Stability of proteins in dried blood spot biobanks. Molecular & Cellular Proteomics 16, 1286–1296 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wigren M. et al. Decreased levels of stem cell factor in subjects with incident coronary events. Journal of internal medicine 279, 180–191 (2016). [DOI] [PubMed] [Google Scholar]
- 77.Loskog A. et al. Immunostimulatory AdCD40L gene therapy combined with low-dose cyclophosphamide in metastatic melanoma patients. British journal of cancer 114, 872–880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vvan der Laan M. & Pollard K. Hybrid clustering of gene expression data with visualization and the bootstrap. Journal of Statistical Planning and Inference 117, 275–303 (2003). [Google Scholar]
- 79.Kolde R. & Kolde M. R. Package ‘pheatmap’. R package 1 (2018). [Google Scholar]
- 80.Doncheva N. T., Morris J. H., Gorodkin J. & Jensen L. J. Cytoscape StringApp: network analysis and visualization of proteomics data. Journal of proteome research 18, 623–632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Clinical characteristics of LC and non-LC individuals analyzed in this study. A. Gender distribution of individuals from the LIINC cohort analyzed in this study. Overall, 55.8% of the participants were female; the non-LC group comprised 43.75% females and the LC group 62.96% females. B. The proportion of LC and non-LC individuals that were hospitalized at the time of acute COVID-19 infection. Overall, 20.9% of the participants were hospitalized: 12.5% among the non-LC individuals, and 25.9% among the individuals with LC. C. The number of sequelae symptoms significantly increased with time among the individuals with LC. Number of sequelae symptoms at four (M4) and eight (M8) months are shown. *p<0.05 (student’s t-test).
Fig. S2. Cytokine and effector molecule expression on T cells in the absence or presence of SARS-CoV-2 peptides. A. CD4+ T cells from a representative donor, in the absence or presence of SARS-CoV-2 peptides. The red box highlights the three cytokines used to define SARS-CoV-2-specific CD4+ T cells. B. CD8+ T cells from a representative donor, in the absence or presence of SARS-CoV-2 peptides. The red boxes highlight the three cytokines used to define SARS-CoV-2-specific CD8+ T cells.
Fig. S3. SARS-CoV-2-specific T cell responses defined individually as those producing IFNγ, IL2, TNFα, and MIP1β do not differ between LC and non-LC individuals. A. Shown are the proportions of SARS-CoV-2-specific CD4+ T cells as defined by cells producing IFNγ, IL2, or TNFα in response to SARS-CoV-2 peptide stimulation. n.s.: non-significant as determined by student’s t-test. B. Shown are the proportions of SARS-CoV-2-specific CD8+ T cells as defined by cells producing IFNγ, MIP1β, or TNFα in response to SARS-CoV-2 peptide stimulation. n.s.: non-significant as determined by student’s t-test. C. IL6-producing CD4+ T cells are observed after SARS-CoV-2 peptide stimulation in some donors. Shown are cells from a representative individual with LC. D. IL6+ SARS-CoV-2-specific CD4+ T cells are exclusively observed from participants with LC. *p<0.05 (Welch’s t-test).
Fig. S4. Gating strategy to define classical T cell subsets. Shown are gating strategies to define the indicated subsets of CD4+ (A) and CD8+ (B) T cells.
Fig. S5. Subset distribution of total and SARS-CoV-2-specific CD8+ T cells among LC and Non-LC individuals. A. Tem frequencies trended higher among total CD8+ T cells from LC as compared to non-LC individuals (student’s t-test). B. Temra frequencies trended lower among SARS-CoV-2-specific CD8+ T cells from LC as compared to non-LC individuals (student’s t-test).
Fig. S6. Sex-dimorphic CD4+ T cell cluster distribution in individuals with LC. A. Cluster distribution among baseline CD4+ T cells as depicted by UMAP. B. Female individuals with LC harbor significantly lower frequencies of cluster A1 cells relative to female non-LC individuals, and male individuals with LC harbor significantly lower frequencies of cluster A4 cells relative to male non-LC individuals. *p<0.05 (GLMM fit - see methods). C. Relative to total CD4+ T cells, cluster A1 is characterized by expression of naïve cell markers (CD45RAhighCD45ROlowCD27high), and low expression of inflammatory tissue homing receptors (CD29lowCXCR4low) but high expression lymph node homing receptors (CD62LhighCCR7high). The activation markers HLA-DR and OX40 were also lowly expressed in cluster A1 cells. D. Relative to total CD4+ T cells, cluster A4 is characterized by expression of terminally differentiated effector memory T cell markers (CD45RAlowCD45ROhighCD27lowCCR7lowCD57high), and high expression of homing receptors for inflamed tissues (CD29highCXCR4highCCR5high) but low expression of lymph node homing receptors (CD62LlowCCR7low). The cytolysis markers perforin, granzyme, and LAMP1 were expressed at elevated levels in cluster A4 cells. ****p<0.0001 (student’s t-test). Shown are data of concatenated data depicted as histograms, or violin plots showing distribution of cells, among the indicated cluster as compared to total CD4+ T cells.
Fig. S7. Sex-dimorphic differential CD8+ T cell cluster distribution in individuals with LC. A. Cluster distribution among baseline CD8+ T cells as depicted by UMAP. B. Female individuals with LC harbor significantly lower frequencies of cluster B1 cells and significantly higher frequencies of cluster B2 cells, relative to female non-LC individuals. *p<0.05 (GLMM fit - see methods). C. Relative to total CD8+ T cells, cluster B1 is characterized by expression of naïve cell markers (CD45RAhighCD45ROlowCD27high), and low expression of tissue homing receptors (CD29lowCXCR4low) but high expression lymph node homing receptors (CD62LhighCCR7high). The activation markers HLA-DR and OX40 were also lowly expressed in cluster B1 cells. D. Relative to total CD8+ T cells, cluster B2 is characterized by expression of terminally differentiated effector memory T cell markers (CD45RAlowCD45ROhighCD27lowCCR7lowCD57high), and high expression of homing receptors for inflamed tissues (CD29highCXCR4highCCR5high) but low expression of lymph node homing receptors (CD62LlowCCR7low). The cytolysis markers perforin, granzyme, and LAMP1 were expressed at elevated levels in cluster B2 cells. Shown are data of concatenated data depicted as histograms, or violin plots showing distribution of cells, among the indicated cluster as compared to total CD8+ T cells.
Data Availability Statement
The raw CyTOF datasets for this study corresponding to total and SARS-CoV-2-specific CD4+ and CD8+ T cells are publicly accessible through the following link: https://datadryad.org/stash/share/TE_QuY0JX23V2n2CIMO2PgsR6afIp6GGusdQ5nXVGnk. The raw RNAseq data from this study are deposited in the GEO (Gene Expression Omnibus) database: GSE224615. All other raw datasets from this study are available from the corresponding authors upon request.