Abstract
The Fusarium oxysporum species complex (FOSC) includes both plant and human pathogens that cause devastating plant vascular wilt diseases and threaten public health. Each F. oxysporum genome comprises core chromosomes (CCs) for housekeeping functions and accessory chromosomes (ACs) that contribute to host-specific adaptation. This study inspected global transcription factor profiles (TFomes) and their potential roles in coordinating CCs and ACs functions to accomplish host-specific pathogenicity. Remarkably, we found a clear positive correlation between the sizes of TFome and proteome of an organism, and FOSC TFomes are larger due to the acquisition of ACs. Among a total of 48 classified TF families, 14 families involved in transcription/translation regulations and cell cycle controls are highly conserved. Among 30 FOSC expanded families, Zn2-C6 and Znf_C2H2 are most significantly expanded to 671 and 167 genes per family, including well-characterized homologs of Ftf1 (Zn2-C6) and PacC (Znf_C2H2) involved in host-specific interactions. Manual curation of characterized TFs increased the TFome repertoires by 3%, including a disordered protein Ren1. Expression profiles revealed a steady expression of conserved TF families and specific activation of AC TFs. Functional characterization of these TFs could enhance our understanding of transcriptional regulation involved in FOSC cross-kingdom interactions, disentangle species-specific adaptation, and identify targets to combat diverse diseases caused by this group of fungal pathogens.
Keywords: Fusarium oxysporum species complex, transcription factors, TFome, accessory chromosome, conservation, expansion
INTRODUCTION
The fungal species complex of Fusarium oxysporum (FOSC) has been used as a model to study cross-kingdom fungal pathogenesis. Members within FOSC can cause devastating fusarium wilt diseases among economically important crops (Ma et al. 2013; Ma 2014; Michielse and Rep 2009; Ploetz 2015; Edel-Hermann and Lecomte 2019; Pegg et al. 2019; Yang et al. 2020; Dean et al. 2012; Rahman et al. 2021; Viljoen et al. 2020; Halpern et al. 2018) and is listed among the top five most important plant pathogens that have a direct impact on the global economy and food security (Dean et al. 2012). With strong host specificity, plant pathogenic F. oxysporum strains are further grouped as formae speciales (Armstrong and Armstrong 1981). For instance, tomato pathogens are named F. oxysporum f.sp. lycopersici, cotton pathogens F. oxysporum f.sp. vasinfectum (Halpern et al. 2018), and banana pathogen F. oxysporum f.sp. cubense (Viljoen et al. 2020). Recently, members within FOSC have also been reported to be responsible for fusariosis, the top emerging opportunistic mycosis (Ma et al. 2013; Yang et al. 2020), and fusarium keratitis, one of the major causes of cornea infections in the developing world and the leading cause of blindness among fungal keratitis patients (Kredics et al. 2015; Hassan et al. 2016).
Comparative genomics studies on this cross-kingdom pathogen revealed that the FOSC genomes, both human and plant pathogens, are compartmented into two components: the core chromosomes (CCs) and accessory chromosomes (ACs). While CCs are conserved and vertically inherited to execute essential housekeeping functions, horizontally transmitted ACs are lineage- or strain-specific and related to fungal adaptation and pathogenicity, conferred by virulent factors such as SIX (Secreted in Xylem) proteins (Ma et al. 2013; Yang et al. 2020; Rep et al. 2004; Yu et al. 2023). ACs and CCs must coordinate their gene expression to coexist within the same genome.
A few characterized transcription factors (TFs) coordinate the crosstalk between CCs and ACs, two compartments. One intriguing example is the cross-regulation among F. oxysporum transcription factors Sge1 (SIX Gene Expression 1), Ftfs, and effector genes. Sge1 is a highly conserved, CC-encoding TF. By name definition, Sge1 regulates the expression of SIX proteins (Michielse et al. 2009; van der Does et al. 2016). AC-encoding Ftf1 proteins (Ftf1 and its AC homologs) and a CC-encoding Ftf2 (Ftf1 CC homolog) are reported in the reference genome of F. oxysporum f.sp. lycopersici Fol4287 (van der Does et al. 2016). Constitutive expression of either Ftf1 genes or Ftf2 induced the expression of effector genes (van der Does et al. 2016). Furthermore, It was documented that DNA binding sites of Sge1 and Ftf1 are enriched among the cis-regulatory elements of in planta transcriptionally up-regulated genes (van der Does et al. 2016). Another CCs and ACs cross-talking example is the alkaline pH-responsive transcription factor PacC/Rim1p reported in F. oxysporum clinical strains (Zhang et al. 2020). In addition to the full-length PacC ortholog (PacC_O), located on a CC, the clinical isolate NRRL32931 genome encodes three truncated PacC homologs, named PacC_a, PacC_b, and PacC_c in ACs (Zhang et al. 2020).
To thoroughly understand the coordination of the crosstalk between genome compartments and their contribution to the cross-kingdom fungal pathogenesis, this study compared the repertoire of TFs (i.e., TFome) among 15 F. oxysporum and 15 other ascomycete fungal genomes, which was organized into 48 families based on the InterPro classification of proteins. Remarkably, we discovered a strong positive correlation (y = 0.07264x - 190.9, r2= 0.9361) between the number of genes (x) and TFome size (y) of an organism. Primarily due to the acquisition of ACs, we observed increased TFome sizes among FOSC genomes. Fourteen out of 48 families involved in transcription/translation regulations and cell cycle controls are highly conserved. Thirty, accounting for ¾ of all families, are expanded in various degrees among FOSC genomes. Unique TF expansions driven by ACs include members of Zn2-C6 fungal-type (Zn2-C6) and Zinc Finger C2H2 (Znf_C2H2) families. This comparative study first highlights the conserved regulatory mechanisms of F. oxysporum, which are essential for variability and plant colonization. With the foundation established by functional conservation, this study further emphasizes potential modifications of existing regulatory pathways by acquiring additional TFs. In combination with existing expression data, this study may provide clues to the fine-tuning of networks in the environmental adaptation of this group of diverse organisms to engage in complex cross-kingdom interactions with different hosts.
MATERIALS AND METHODS
Generation of fungal TFomes
The annotation pipeline is briefly summarized in Figure S1A–B. The fungal proteomes of 30 strains were downloaded from the JGI MycoCosm portal (Grigoriev et al. 2014). Protein annotation was performed using InterProScan/5.38-76.0 (https://www.ebi.ac.uk/interpro/search/sequence/) (Jones et al. 2014). Annotations of proteins putatively serve as TFs were filtered out using a table containing InterPro terms related to transcriptional regulatory functions summarized by literature (Park et al. 2008; Shelest 2017), with further addition by manual curation (Table S1). Orthologous analysis to probe orthologs of functionally validated TFs (Table S3–4 and Table 3) in Fusarium was done with OrthoFinder 2.5.4 (https://github.com/davidemms/OrthoFinder) (Emms and Kelly 2019).
Table 3.
Ortholog copy number and expansion index (EIf) of characterized and expanded TFs in F. oxysporum
| TF | Reported species | References | Family | Overlap | Average_Fo | Average_non-Fo | EIf |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Ftf1/Ftf2 | F. oxysporum | (Niño-Sánchez et al. 2016) | Zn2-C6 | Yes | 4.80 | 1.11 | 2.75 |
| Ebr1/Ebr2 | F. oxysporum | (Jonkers et al. 2014) | Zn2-C6 | Yes | 5.27 | 1.56 | 2.45 |
| Znf1 | M. oryzae | (Yue et al. 2016) | Zn2-C6 | Yes | 6.47 | 2.78 | 1.98 |
| Ctf2 | F. oxysporum | (Bravo-Ruiz et al. 2013) | Zn2-C6 | Yes | 2.93 | 1.33 | 1.69 |
| Fow2 | F. oxysporum | (Imazaki et al. 2007) | Zn2-C6 | Yes | 2.07 | 1.00 | 1.53 |
| Dep6 | A. brassicicola | (Wight et al. 2009) | Zn2-C6 | Yes | 0.93 | 0.67 | 1.16 |
| Pf2 | A. brassicicola | (Jones et al. 2019) | Zn2-C6 | Yes | 1.20 | 1.00 | 1.10 |
| Art1 | F. verticilioides | (Oh et al. 2016) | Zn2-C6 | Yes | 1.00 | 0.89 | 1.06 |
| Clta1 | C. lindemuthianum | (Dufresne et al. 2000) | Zn2-C6 | Yes | 1.07 | 1.00 | 1.03 |
| Fhs1 | F. graminearum | (Son et al. 2016a) | Zn2-C6 | Yes | 1.07 | 1.00 | 1.03 |
| Cos1 | M. oryzae | (Li et al. 2013) | Znf_C2H2 | Yes | 1.80 | 0.00 | 2.80 |
| PacC | F. oxysporum | (Caracuel et al. 2003) | Znf_C2H2 | Yes | 2.13 | 1.00 | 1.57 |
| Fug1 | F. verticillioides | (Ridenour and Bluhm 2017) | AreA_GATA | No | 7.27 | 1.33 | 3.54 |
| Ren1 | F. oxysporum | (Ohara et al. 2004) | disordered | No | 3.00 | 1.00 | 2.00 |
| Tri10 | F. graminearum | (Jiang et al. 2016) | Fun_TF | No | 1.13 | 0.33 | 1.60 |
| Ltf1 | B. cinerea | (Schumacher et al. 2014) | Znf_GATA | Yes | 4.00 | 2.44 | 1.45 |
| Ndt80 | U. maydis | (Doyle et al. 2016) | NDT80 | Yes | 1.73 | 1.11 | 1.29 |
| Hap3p | F. verticillioides | (Ridenour and Bluhm 2014) | CBFA_NFYB | Yes | 1.33 | 1.00 | 1.17 |
| Sod1 | F. oxysporum | (Wang et al. 2021) | SOD_Cu_Zn | No | 1.47 | 1.22 | 1.11 |
| Prf1 | F. oxysporum | (Mendoza-Mendoza et al. 2009) | HMG_box | Yes | 1.07 | 1.00 | 1.03 |
RNA-seq analysis
The RNA-seq datasets were previously described (Guo et al. 2021; Redkar et al. 2022) and deposited by those authors to the NCBI Short Read Archive with accession number GSE87352 and to the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-10597, respectively. For data reprocessing, reads were mapped to reference genomes of Arabidopsis [annotation version Araport11 (Cheng et al. 2017)], Fo5176 (Fokkens et al. 2021), Fo47 (Wang et al. 2020) and Fol4287 (Ma et al. 2010) using HISAT2 version 2.0.5 (Kim et al. 2019). Mapped reads were used to quantify the transcriptome by StringTie version 1.3.4 (Pertea et al. 2015), at which step TPM (transcript per million) normalization was applied. Normalized read counts were first averaged per condition and then transformed by log2 (normalized read count + 1) and Z-scaled, then visualized in pheatmap (version 1.0.12).
Genome partition
The genome partition results for chromosome-level assemblies were retrieved from previous reports for Fol4287 (Ma et al. 2010), FoII5 (Zhang 2019), Fo5176 (Fokkens et al. 2021), and Fo47 (Wang et al. 2020). Fo47 has a clear genome partition with 11 core chromosomes and one accessory chromosome, therefore serving as the reference for the genome partition of other F. oxysporum genomes. mummer/3.22 was applied to align scaffolds of genome assemblies against 11 core chromosomes of the reference genome Fo47 using default parameters. The scaffolds aligned to the core chromosomes of Fo47 with a coverage larger than 5% were annotated as core scaffolds. The rest of the scaffolds were partitioned as accessory scaffolds. Genes residing on core and accessory scaffolds were annotated as core and accessory genes, respectively.
Phylogenetics analysis
Protein sequences were aligned via MAFFT/7.313 (Katoh and Standley 2013). Then the iqtree/1.6.3 (Minh et al. 2020; Nguyen et al. 2015) was run on the sequence alignment to generate the phylogeny (by maximum likelihood method and bootstrapped using 1000 replicates) (Hoang et al. 2018) and was then visualized via the Interactive Tree of Life (Letunic and Bork 2021), producing the phylogram. OrthoFinder 2.5.4 (Emms and Kelly 2019) was used for orthogroup determination. To build a species phylogram, randomly selected 500 conserved proteins (single-copy orthologs) were aligned first. Then the alignment was concatenated, and phylogeny was determined and visualized using the above methods.
RESULTS
1. FOSC TFome expansion resulted from the acquisition of ACs
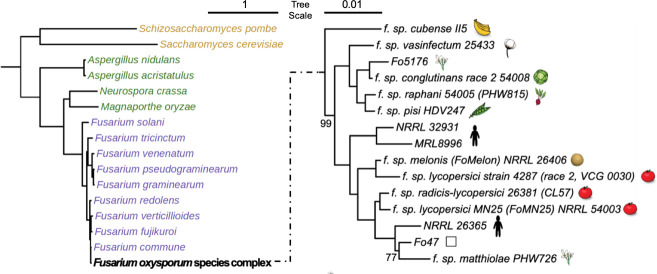
We compared 30 ascomycete fungal genomes (Figure 1 and Table 1), including 15 strains within the FOSC, nine sister species close to F. oxysporum, two yeast genomes (Saccharomyces cerevisiae and Schizosaccharomyces pombe), four other filamentous fungal species (Neurospora crassa, Aspergillus nidulans, Aspergillus acristatulus, and Magnaporthe oryzae). To maintain consistency, the protein sequences for all these genomes were retrieved from the MycoCosm portal (Grigoriev et al. 2014).
Figure 1. Phylogeny of fungal genomes included in this study.
Both left and right phylograms were constructed by concatenated alignment of randomly selected 500 single-copy orthologous proteins, followed by the maximum likelihood method with 1000 bootstraps. Left shows a phylogram of FOSC (represented by the reference genome Fol4287) together with the other 15 ascomycetes. The right shows a phylogram of members within FOSC, rooted by F. verticillioides (not shown). Only bootstrap values not equal to 100 are shown.
Table 1.
Fungal genomes used in this study
|
|
||||||
|---|---|---|---|---|---|---|
| Fungal species or strains | MycoCosm identifier | Genome Size (MB) | No. of genes | TFome Size | Host | Reference |
|
| ||||||
| Saccharomyces cerevisiae | Sacce1 | 12.07 | 6575 | 284 | (Goffeau et al. 1996) | |
| Schizosaccharomyces pombe | Schpo1 | 12.61 | 5134 | 228 | (Wood et al. 2002) | |
| Aspergillus nidulans | Aspnid1 | 30.48 | 10680 | 635 | (Galagan et al. 2005) | |
| Aspergillus acristatulus | Aspacri1 | 32.59 | 11221 | 666 | (Vesth et al. 2018) | |
| Neurospora crassa | Neucr2 | 41.04 | 9730 | 447 | (Galagan et al. 2003) | |
| Magnaporthe oryzae | Magor1 | 40.49 | 12673 | 520 | Rice | (Dean et al. 2005) |
| Fusarium solani | Fusso1 | 52.93 | 17656 | 1137 | broad hosts | (Mesny et al. 2021) |
| F. pseudograminearum | Fusps1 | 36.33 | 12395 | 627 | Wheat | (Gardiner et al. 2012) |
| F. graminearum | Fusgr1 | 36.45 | 13321 | 608 | Wheat | (Cuomo et al. 2007) |
| F. venenatum | Fusven1 | 37.45 | 12845 | 802 | (Mesny et al. 2021) | |
| F. tricinctum | Fustr1 | 43.69 | 14106 | 925 | Broad hosts | (Mesny et al. 2021) |
| F. verticillioides | Fusve2 | 41.78 | 15869 | 917 | Corn | (Ma et al. 2010) |
| F. fujikuroi | Fusfu1 | 43.83 | 14813 | 901 | Broad hosts | (Wiemann et al. 2013) |
| F. redolens | Fusre1 | 52.56 | 17051 | 1098 | Broad hosts | (Mesny et al. 2021) |
| F. commune | Fusco1 | 48.37 | 15731 | 1012 | Broad hosts | (Mesny et al. 2021) |
| F. oxysporum f.sp. cubense (II5) | FoxII5 | 49.43 | 16048 | 1047 | Banana | (Zhang 2019) |
| F. oxysporum f. sp. radicis-lycopersici (CL57) | Fusoxrad1 | 49.36 | 18238 | 1151 | Tomato | (DeIulio et al. 2018) |
| F. oxysporum Fo47 (Fo47) | FusoxFo47_2 | 50.36 | 16207 | 1082 | (Wang et al. 2020) | |
| F. oxysporum f. sp. lycopersici (MN25) | Fusoxlyc1 | 48.64 | 17931 | 1119 | Tomato | (DeIulio et al. 2018) |
| F. oxysporum NRRL26365 (NRRL26365) | Fox26365_1 | 48.46 | 16047 | 1036 | Human | (Yang 2020) |
| F. oxysporum f. sp. melonis (FoMelon) | Fusoxmel1 | 54.03 | 19661 | 1219 | Melon | (Ma et al. 2014) |
| F. oxysporum f. sp. lycopersici (Fol4287) | Fusox2 | 61.36 | 20925 | 1292 | Tomato | (Ma et al. 2010) |
| F. oxysporum NRRL32931 (NRRL32931) | Fusox32931 | 47.91 | 17280 | 1072 | Human | (Zhang et al. 2020) |
| F. oxysporum MRL8996 (MRL8996) | FoxMRL8996 | 50.07 | 16631 | 1057 | Human | (Zhang et al. 2020) |
| F. oxysporum f. sp. matthiolae (PHW726) | FoxPHW726_1 | 57.22 | 17996 | 1157 | Brassica | (Yu et al. 2020) |
| F. oxysporum f. sp. vasinfectum (FoCotton) | Fusoxvas1 | 52.91 | 19143 | 1189 | Cotton | (DeIulio et al. 2018) |
| F. oxysporum f. sp. pisi (HDV247) | Fusoxpis1 | 55.19 | 19623 | 1229 | Pea | (Williams et al. 2016) |
| F. oxysporum f. sp. raphani (PHW815) | Fusoxrap1 | 53.5 | 19306 | 1132 | Brassica | (DeIulio et al. 2018) |
| F. oxysporum f. sp. conglutinans (PHW808) | Fusoxcon1 | 53.58 | 19854 | 1142 | Brassica | (DeIulio et al. 2018) |
| F. oxysporum Fo5176 (Fo5176) | FoxFo5176 | 67.98 | 19130 | 1236 | Arabidopsis | (Fokkens et al. 2021) |
To have a comprehensive TFome annotation, we started with reported InterProScan (IPR) terms associated with fungal transcriptional regulation (Park et al. 2008; Shelest 2017) and curated a mapping with updated IPR classification (interproscan version: 5.38-76.0) (Blum et al. 2021). In addition, we searched the IPR classification of protein families and obtained all other terms related to the transcriptional regulation activity. This resulted in 234 TF-related IPR terms (Table S1). Since most of the terms are initially defined in the mammalian systems, it was not surprising to see that overall, our fungal genomes are only associated with 71 IPR terms out of the total 234 TF-related IPR terms (Table S1, Materials and Methods, and Figure S1A–B for annotation pipeline). After filtering out 13 and 10 terms for redundancy (two terms describing the identical domain) and minimal presentation (< 4 among the 30 genomes), respectively, this comparative TFome study focused on the rest 48 IPR terms, which represented a total of 27967 TFs (Table S1–S2). Notably, 12 out of 48 terms were not reported to be affiliated with fungal transcriptional regulation by either Park et al. 2008 or Shelest 2017 (Table S1), adding values to our manual IPR term search.
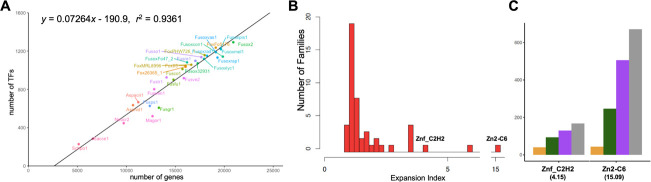
Comparing the total number of genes in a genome (x) and the total number of TFs within that genome (y), we observed a strong positive correlation (y = 0.07264x - 190.9, r2= 0.9361) (Figure 2A). Among all genomes included in this study, FOSC TFomes are the largest, with an average of 1144 TFs per genome (Figure 2A, Table 1). After partitioning each FOSC genome into core and accessory regions (see Materials and Methods for details), we observed a positive correlation between the number of TFs encoded in the accessory chromosomal region of each strain (defined as accessory TFs hereafter) with the size of accessory genomes (Mb) (y = 17.239x + 3.553) (Figure S2), suggesting that accessory chromosomes contribute directly to the expanded TFome.
Figure 2. TFome conservation and variation among ascomycete fungi: baseline description.
(A) There is a positive correlation between the number of genes and TFome size of an organism. JGI fungal genome identifiers were used as labels. (B) Histogram illustrates the distribution of expansion indexes among different families. (C) Average number of TFs of two most drastically expanded families (Znf_C2H2 and Zn2-C6) within each genome set. Genome Set 1 (G1) includes two yeast genomes (S. cerevisiae and S. pombe). Genome Set 2 (G2) includes four filamentous fungal species (N. crassa, A. nidulans, A. acristatulus, and M. oryzae). Genome Set 3 (G3) includes nine sister species close to F. oxysporum. Genome Set 4 (G4) includes 15 FOSC genomes.
To understand genome regulation among FOSC, we developed an expansion index score using two yeast lineages as the baseline (EIy):
Based on this index value, we classified TF families into three major groups (Table 2, Table S1). Group 1 contains 14 TF families with an expansion score of 1, indicating high conservation. Group 2 includes four families with an index score of less than 1, reflecting some level of gene family contraction. Group 3 contains 30 families with an expansion index greater than 1, indicating gene expansion.
Table 2.
Expansion Index (EIy) of 48 TF families
| IPR | Term | EIy |
|---|---|---|
|
| ||
| Group 1 | ||
| IPR000814 | TBP | 1 |
| IPR003228 | TFIID_TAF12 | 1 |
| IPR004595 | TFIIH_C1-like | 1 |
| IPR006809 | TAFII28 | 1 |
| IPR042225 | Ncb2 | 1 |
| IPR008570 | Vps25 | 1 |
| IPR008895 | Vps72/YL1 | 1 |
| IPR007196 | CNOT1 | 1 |
| IPR005612 | CBF | 1 |
| IPR001289 | NFYA | 1 |
| IPR018004 | APSES-type HTH | 1 |
| IPR003150 | RFX | 1 |
| IPR033896 | MADS_MEF2-like | 1 |
| IPR018501 | DDT | 1 |
| Group 2 | ||
| IPR006856 | MATalpha_HMGbox | 0.8 |
| IPR039515 | NOT4 | 0.9 |
| IPR033897 | MADS_SRF-like | 0.95 |
| IPR000232 | HSF | 0.98 |
| Group 3 | ||
| IPR003163 | Tscrpt_reg_HTH_APSES-type | 1.04 |
| IPR001766 | Fork head | 1.05 |
| IPR011016 | Znf_RING-CH | 1.11 |
| IPR001965 | Znf_PHD | 1.11 |
| IPR009071 | HMG_box | 1.12 |
| IPR004181 | Znf_MIZ | 1.24 |
| IPR001606 | ARID | 1.25 |
| IPR000679 | Znf_GATA | 1.3 |
| IPR001005 | SANT/Myb | 1.32 |
| IPR000818 | TEA/ATTS | 1.33 |
| IPR003120 | Ste12 | 1.33 |
| IPR003958 | CBFA_NFYB | 1.35 |
| IPR001083 | Cu_fist | 1.37 |
| IPR000967 | Znf_NFX1 | 1.4 |
| IPR006565 | Bromodomain | 1.52 |
| IPR001387 | Cro/C1-type_HTH | 1.6 |
| IPR001841 | Znf_RING | 1.64 |
| IPR000571 | Znf_CCCH | 1.74 |
| IPR001878 | Znf_CCHC | 1.83 |
| IPR010666 | Znf_GRF | 2 |
| IPR018060 | HTH_AraC* | 2 |
| IPR001356 | Homeobox | 2.28 |
| IPR007604 | CP2* | 2.73 |
| IPR007396 | PAI2 | 3.42 |
| IPR024061 | NDT80 | 3.47 |
| IPR011598 | bHLH | 3.48 |
| IPR007889 | HTH_Psq* | 3.53 |
| IPR013087 | Znf_C2H2 | 4.15 |
| IPR004827 | bZIP | 5.8 |
| IPR001138 | Zn2-C6 | 15.09 |
Asterisk indicates the families without a presence in yeasts
2. Conserved TF families that are primarily associated with general/global transcription factors
About 30% of the TF families, fourteen, are associated with strong orthologous conservation in all genomes we included in this study (Figure 2B; Table 2; Table S1). Because most of these conserved TF families are single-copied TF families, these 30% conserved TF families only account for less than 2% of the total TFomes. Based on a detailed study on S. cerevisiae and other model organisms, these TF families are involved in transcription/translation regulation and cell cycle controls.
2.1. Transcription/Translation regulation
Either TF families are related to transcription initiation and elongation, including TATA box-binding protein (TBP), TBP-associated factors (TAFs), and RNA polymerase II elongation regulator Vps25. CCAAT-Binding Factors (CBFs) are related to ribosomal biogenesis. These families overall play conserved roles in general transcriptional and translational regulation across Ascomycota.
Transcription initiation TBP is one of the most conserved TF families, TBP binds directly to the TATA box to define the transcription start and initiate transcription facilitated by all three RNA polymerases. The function of TBP is so conserved as the yeast homolog can complement TBP mutations in humans (Yamaguchi et al. 2001; Roberts and Winston 1996).
Transcription positive/negative regulators, including TAF12 and TAFII28, are parts of the transcription factor TFIID complex. Interacting with TBP, TAFs form the TFIID complex and positively participate in the assembly of the transcription preinitiation complex (Green 2000). Similarly, TFIIH works synergistically with TFIID to promote the transcription (Fribourg et al. 2000). In contrast, Negative cofactor 2 (Ncb2) inhibits the preinitiation complex assembly (Goppelt et al. 1996). Other factors include the CNOT1, a global regulator involved in transcription initiation and RNA degradation (Chalabi Hagkarim and Grand 2020), and Vps72/YL1 that contributes to transcriptional regulation through chromatin remodeling as reported in the yeast (Liang et al. 2016; Latrick et al. 2016).
Transcription elongation: Vps25 is a subunit of the ESCRT-II complex, which binds to RNA polymerase II elongation factor to exert transcriptional control in mammalian systems (Kamura et al. 2001).
Translational regulation: CCAAT box is a common cis-acting element found in the promoter and enhancer regions of genes in the eukaryotes (Vuorio et al. 1990; Becker et al. 1991). CBFs are necessary for the 60S ribosomal subunit biogenesis and therefore involved in the translational control (Milkereit et al. 2001; Fromont-Racine et al. 2003; Edskes et al. 1998). This family, including Noc3, Noc4, and Mak21 in S. cerevisiae, has three members in each genome, and a clear single-copy orthologous relationship can be observed for each member (Figure S3A).
2.2. Cell cycle control
Five TF families are related to cell cycle control, including cell cycle progression, DNA repair, and machinery/cell integrity maintenance.
APSES-type HTH represents a family of fungal TFs involved in cell-cycle control and is crucial to the development (Xin et al. 2020). Every genome maintains four copies of genes encoding APSES-type HTH (Figure S3B), and they form single-copy orthologs in all genomes except yeasts. Genes in Clade 1, including StuA homologs, are targets of the cyclic AMP (cAMP)-dependent protein kinase A (PKA) signal transduction pathway and were reported to be involved in dimorphic switch (Pan and Heitman 2000; Gimeno and Fink 1994), fungal spore development and the production of secondary metabolites (Lysøe et al. 2011). Genes in Clade 2 and Clade 3 include S. cerevisiae Swi4 and Swi6, which form a protein complex and regulate genes essential during cell cycle progression from G1 to S phase (Koch et al. 1993), as well as meiosis (Son et al. 2016b). Genes in Clade 4 include homologs of S. pombe Bqt4 that connect telomeres to the nuclear envelope (Chikashige et al. 2009). Since this family of TFs is highly conserved across ascomycetes, similar functions can be proposed in F. oxysporum.
DTT, represented by the S. cerevisiae homolog Itc1, is a subunit of ATP-dependent Isw2p-Itc1p chromatin remodeling complex and is required for repression of early meiotic gene expression during the mitotic growth (Sugiyama and Nikawa 2001).
RFX represents a family of fungal TFs involved in DNA repair. Each strain encodes two orthologous copies, except F. venenatum encodes two copies within the RFX1 clade (Figure S3C). A major transcriptional repressor of DNA-damage-regulated genes in S. cerevisiae, Rfx1, is involved in DNA damage repair and replication checkpoint pathways (Lubelsky et al. 2005). In F. graminearum, Rfx1 is essential for maintaining the genome integrity (Min et al. 2014). The other copy, Rsc9 in S. cerevisiae, is a chromatin structure-remodeling complex RSC involved in transcription regulation and nucleosome positioning (Cairns et al. 1996; Hsu et al. 2003).
NFYA can bind to the CCAAT box. All strains maintain one copy of this family. The yeast homolog Hap2 induces the expression of mitochondrial electron transport genes (Olesen et al. 1991). F. verticillioides NFYA Hap2 is essential for fungal growth and the virulence on maize stalks (Ridenour and Bluhm 2014). The conservativeness suggests the functional importance of these TFs across Ascomycota, possibly linked to cellular machinery control (e.g. mitochondrial electron transport chain).
MADS MEF2-like family includes S. cerevisiae Rlm1, a component of the protein kinase C-mediated MAP kinase pathway involved in maintaining cell integrity (Jung et al. 2002). Rlm1 has a paralog from the whole genome duplication in S. cerevisiae, and all filamentous fungi encode one copy. In F. verticilioides, Mef2 plays a vital role in the sexual development (Ortiz and Shim 2013).
3. Minimal gene family contractions in FOSC partially caused by whole genome duplication in yeast
Four TF families, MATalpha_HMGbox, NOT4, MADS_SRF-like, and HSF (Heat Shock Factor), have an expansion score of less than 1, reflecting some level of gene family contraction among members of FOSC compared to the two yeast genomes (Figure S4).
MATalpha_HMGbox is a TF family that includes S. cerevisiae mating type protein alpha 1, a transcription activator that activates mating-type alpha-specific genes (Martin et al. 2010). All F. oxysporum Mat1-1 type strains contain this TF, but Mat1-2 strains do not. The contraction reflects the heterothallic mating strategy, even though sexual reproduction has not been observed in FOSC (Arie et al. 2000).
NOT4 is a component of the multifunctional CCR4-NOT complex, a global transcriptional repressor of the RNA polymerase II transcription (Albert et al. 2002). This TF family remains a single copy in most genomes but is lost in some filamentous fungal genomes, including A. nidulans, F. redolens, F. oxysporum strains NRRL26365, MRL8666, and PHW726. It remains to be discovered why this gene is lost in some of these strains.
The contractions of the other two TF families, MADS SRF-like and HSF, are primarily caused by the whole genome duplication in yeast. In both cases, some degree of expansion was found in FOSC compared to other filamentous fungi (Figure S4).
MADS SRF-like is important for microconidium production and virulence in host plants, as reported in M. oryzae (Ding et al. 2020), and is essential for transcriptional regulation of growth-factor-inducible genes (Messenguy and Dubois 2003). The average copy number of phytopathogenic FOSC strains is 2.73, and the Fo5176 genome has the highest copy number of 6, while most other genomes only contain a single copy (Table S1).
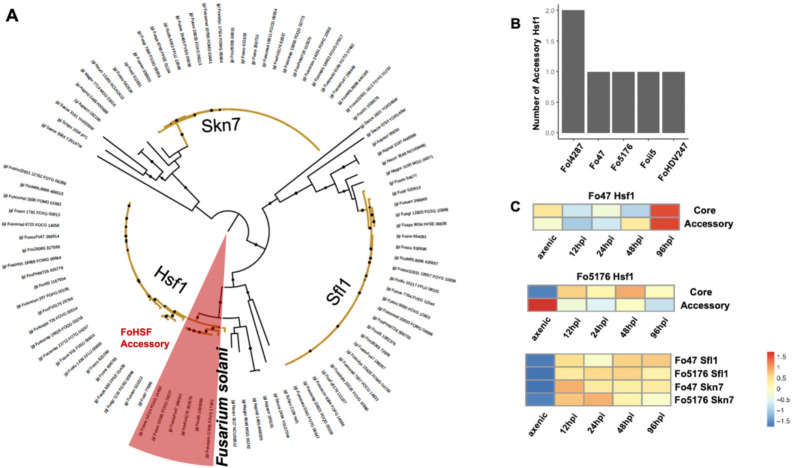
HSF is a family of transcription factors that activate the production of many heat shock proteins that prevent or mitigate protein misfolding under abiotic/biotic stresses (Feder and Hofmann 1999). All non-FOSC filamentous fungi have three copies, while members of FOSC show expansion (e.g., Fo47: 4, Fol4287: 5, II5: 4, HDV274: 4, and Fo5176: 4) (Figure 3A–B). Interestingly, all expanded HSFs are phylogenetically close to Hsf1, which cluster together with the Hsf1 paralog of Fusarium solani, suggesting their horizontal transfer origin (Figure 3A). We then examined the Hsf1 expression during the plant colonization (Guo et al. 2021). We found that the core copies of Hsf1 of both strains Fo47 and Fo5176 were up-regulated during plant colonization. In contrast, the Hsf1 accessory copies of these two strains were under opposite regulations, with Fo47 one being up-regulated and Fo5176 one being down-regulated post infection (Figure 3C), suggesting distinct regulatory adaptations after expansion. Here we noted that transcriptome data could be powerful in understanding the functional importance of TFs (see Section 6 for systematic analysis). In filamentous fungi, there are experimental reports for the other two clades. Sfl1 is essential for vegetative growth, conidiation, sexual reproduction, and pathogenesis, as shown in M. oryzae (Li et al. 2011); Skn7 is a regulator of the oxidative stress response and is essential for pathogenicity in F. graminearum (Jiang et al. 2015). Not surprisingly, both genes of Fo5176 and Fo47 were upregulated during plant colonization (Figure 3C).
Figure 3. Evolutional trajectory of heat shock factors (HSFs) suggesting genome expansion and adaptation.
(A) Phylograms of HSFs were constructed by maximum likelihood method with 1000 bootstraps. Branches of Fusarium HSFs were colored in yellow. Accessory HSFs of FOSC are shared in red. (B) Number of accessory HSFs in some FOSC genomes. (C) Expression of HSF genes during plant colonization (hpi indicates hours post inoculation), compared to axenic growth. Transcriptome data was previously described in Guo et al. 2021. See Materials and Methods for details of data reprocessing and visualization.
4. Significant TFome expansion in FOSC driven by a small number of exceedingly expanded TF families
4.1. Gain-of-function among filamentous ascomycete fungi
Three TF families, CP2 (EIy = 2.73), HTH_AraC (EIy = 2), and HTH_Psq (EIy = 3.53), are absent in both yeast genomes, suggesting a gain of function among filamentous ascomycete fungi (Table S1). CP2 has been studied in animal and fungal kingdoms with a function related to differentiation and development (Paré et al. 2012). Both HTH_AraC and HTH_Psq belong to helix-turn-helix (HTH) superfamily. First reported in bacteria, HTH_AraC is a positive regulator associated with the arabinose operon regulatory protein AraC (Schleif 2010; Gallegos et al. 1993; Bustos and Schleif 1993). HTH_Psq, as part of the eukaryotic Pipsqueak protein family, reported in vertebrates, insects, nematodes, and fungi, regulates the cell death (Siegmund and Lehmann 2002). Most FOSC genomes have a single copy of HTH_AraC, while the count of proteins containing the HTH_Psq ranges from 0 to 9 in the FOSC and ranges from 0 to 3 in other Fusarium relatives. Since the HTH_Psq domain also exists in transposases (Siegmund and Lehmann 2002), and ACs in FOSC are transposon-rich, it remains to be studied whether proteins containing the Psq domain are bona fide TFs.
4.2. Seven exceedingly expanded TF families
Among others, seven TF families have expansion indexes greater than 2 (Table 2 and Figure 2B). Because of their drastic expansion, these seven families overall account for more than 75% of the total TFome. These families include Zn2-C6 (EIy = 15.09), bZIP (EIy = 5.80), and Znf_C2H2 (EIy = 4.15), Homeobox (EIy = 2.28), PAI2 (EIy = 3.42), NDT80 (EIy = 3.47), and bHLH (EIy = 3.48). All seven families show gradual expansion, reflected by the average copy number increment (FOSC > non-FOSC Fusarium > non-Fusarium filamentous fungi > yeasts, table S1). Furthermore, Zn2-C6 (44 in yeasts versus 671 in FOSC) and Znf_C2H2 (40 in yeasts versus 167 in FOSC) have the most drastic number increment along the evolutionary trajectory (Figure 2C and Table S1). Based on both high expansion index and large number increment, we considered Zn2-C6 and Znf_C2H2 as the most significantly expanded families.
The large copy number makes it hard to interpret functions from the protein domain annotation. Here we describe a couple of TFs reported in F. oxysporum and other systems and will introduce orthologous analysis to further survey the functionally validated TFs in the later section.
Zn2-C6, a fungal family TF (MacPherson et al. 2006), has the most significant expansion, reaching over 600 members among FOSC genomes and accounting for more than half of the total TFome. This group of TFs can form a homodimer and bind to the specific palindromic DNA sequence through direct contact with the major groove of the double-stranded DNA molecules (MacPherson et al. 2006). The versatility of this group of TFs can be achieved by domain shuffling and by changing the nucleotide binding specificity. In addition to the well-documented Ftf1 (Niño-Sánchez et al. 2016; van der Does et al. 2016; Ramos et al. 2007; Zuriegat et al. 2021; Zhao et al. 2020), five additional TFs within this family have been characterized in F. oxysporum, including Ctf1 (Rocha et al. 2008), Ctf2 (Rocha et al. 2008), Fow2 (Imazaki et al. 2007), XlnR (Calero-Nieto et al. 2007) and Ebr1 (Jonkers et al. 2014). They are involved in the development, metabolism, stress response, and pathogenicity.
Znf_C2H2 is the most common DNA-binding motif found in the eukaryotic transcription factors (Fedotova et al. 2017). Five F. oxysprum TFs have been reported: Czf1 (Yun et al. 2019), Con7-1 (Ruiz-Roldán et al. 2015), PacC (Caracuel et al. 2003; Zhang et al. 2020), ZafA (López-Berges 2020) and St12 (Asunción García-Sánchez et al. 2010; Rispail and Di Pietro 2009). Particularly, PacC was linked to the pathogenicity of both plant and human host (Zhang et al. 2020; Caracuel et al. 2003).
Other five families include bZIP, Homeobox, PAI2, Ndt80 and bHLH. bZIP domain contains a region for sequence-specific DNA binding followed by a leucine zipper region required for dimerization (Bader and Vogt 2006). Three F. oxysporum bZIP TFs have been reported, including Atf1 (Li et al. 2013), Hapx (López-Berges et al. 2012), and MeaB (López-Berges et al. 2010), all of which are important for fungal pathogenicity. Homeobox is a DNA binding motif with a helix-turn-helix structure. In S. pombe, Phx1 is a transcriptional coactivator that plays a role in yeast fission. In M. oryzae, Hox plays roles in the conidiation and appressorium development (Kim et al. 2009). PAI2 is involved in the negative regulation of protease synthesis and sporulation of the Bacillus subtilis (Honjo et al. 1990). Ndt80 is essential for completing meiosis in S. cerevisiae (Pierce et al. 2003; Tsuchiya et al. 2014) and Ustilago maydis (Doyle et al. 2016). It also promotes the expression of sporulation genes that are essential for the fulfillment of meiotic chromosome segregation (Hepworth et al. 1998). bHLH proteins form a large superfamily of transcriptional regulators found in almost all eukaryotes and function in critical developmental processes (Jones 2004). F. graminearum Gra2 is involved in the biosynthesis of phytotoxin gramillin (Bahadoor et al. 2018). P. digitatum encoding SreA is required for anti-fungal resistance and full virulence in citrus fruits (Liu et al. 2015).
4.3. other families
Other 20 TF families (expanded but with EIy <= 2) account for 20% of the TFome; on average, each of these 20 families contains 9.6 copies in each genome examined (Table S1). These TFs are involved in chromatin remodeling and pheromone response, among other functions.
Four TF families are functionally linked to chromatin remodeling, including Bromodomain (EIy = 1.52), CBFA_NFYB (EIy = 1.35), Znf_RING-CH (EIy = 1.11), and ARID (EIy = 1.25). Bromodomain containing Spt7 is a crucial part of the SAGA complex in yeast. The SAGA complex is required to transcribe many genes in the genome. The bromodomain that is part of this subunit can recognize acetylated lysines of histones and eventually lead it to a more chromatin unwinding (Donczew et al. 2020). CBFA_NFYB is found in the proteins (e.g., S. cerevisiae Dls1) that regulate RNA polymerase II transcription through controlling chromatin accessibility (e.g., telomeric silencing) (Iida and Araki 2004). Znf_RING-CH has a functional connection to chromatin modification (e.g., S. cerevisiae Rkr1) (Braun et al. 2007). ARID is a 100 amino acid motif found in many eukaryotic TFs (Iwahara 2002). S. cerevisiae Swi1 plays a role in chromatin remodeling and is required to transcribe a diverse set of genes, including HO and Ty retrotransposons (Breeden and Nasmyth 1987; Hirschhorn et al. 1992).
Ste12 is a family of TFs that regulate fungal development and pathogenicity (Rispail and Di Pietro 2010). These TFs are found only in the fungal kingdom. Ste12 binds to the DNA sequence that mediates pheromone response. It is involved in haploid mating and pseudohyphae formation in the diploid (Gancedo 2001). F. oxysporum Ste12 controls invasive growth and virulence downstream of the Fmk1-mediated MAPK cascade (Rispail and Di Pietro 2009). Except for S. pombe (missing one), every genome encodes one copy.
Among others, Znf_NFX1 domain is found in the NK-X1, a repressor of the human disease-associated gene HLA-DRA (Song et al. 1994). HMG_box (high mobility group box) in S. cerevisiae, Spp41, is involved in negative expression regulation of spliceosome components (Maddock et al. 1994); Nhp6a is required for the fidelity of some tRNA genes (Braglia et al. 2007); Ixr1 is a transcriptional repressor that regulates hypoxic genes (Vizoso-Vázquez et al. 2012). One example of Znf_GATA is Fep1, a transcription factor that represses the expression of particular iron transporter genes under a high iron concentration (Kim et al. 2016). S. cerevisiae Mbf1, belonging to Cro/C1-type HTH, is a transcriptional coactivator (Takemaru et al. 1997).
5. Orthologous survey of TF families that were manually curated
To further understand expanded TFs and their impacts on transcriptional regulation, we curated a list of 102 TFs reported in literature focusing on F. oxysporum, F. graminearum, and other phytopathogenic fungi (Table S3 and examples as described in the previous section). Compared to this list of curated TFs using Orthofinder, we define 80 orthologous groups among Fusarium genomes (Table S4). 62 out of the 80 orthogroups have been identified using the above IPR-annotated pipeline, which enables the dissection of vastly expanded and high copy number TF families such as Zn2-C6 and Znf_C2H2, which are further mapped to 27 orthologous groups, including 17 in Zn2C6, 9 in Znf_C2H2, and 1 containing both Znf_C2H2 and Zn2-C6 domains (Table S4).
This effort also results in additional annotation to 18 TF families (Table S4), accounting for 32 genes per genome (3% of average Fusarium TFome size). These newly annotated TFs include homologs of those without domain annotation, e.g., disordered proteins F. oxysporum Ren1 (Ohara et al. 2004) and M. oryzae Som1 (Yan et al. 2011), and homologs of those with noncanonical TF domains such as Ankyrin_rpt and WD40_repeat.
We then directly compared F. oxysporum with its Fusarium relatives to calculate the expansion index as follows:
The EIf ranged, with the highest score being 3.54 (Fug, AreA_GATA) and the lowest being 0.5 (Fox1, Fork_head) (Table S4). Among these 80 orthogroups, 36 groups show high conservation (EIf = 1) as they are single-copy orthologs across Fusarium, among which ten were functionally validated in F. oxysporum (Table S4). 24 groups have gene contraction in F. oxysporum (EIf < 1). A total of 20 groups are expanded in F. oxysporum (EIf > 1, Table 3, Table S4), including five groups Fug1 (AreA_GATA, EIf = 3.54), Cos1 (Znf_C2H2, EIf = 2.8), Ftf1/Ftf2 (Zn2-C6, EIf = 2.7), Ebr1/Ebr2 (Zn2-C6, EIf = 2.5) and Ren1 (disordered, EIf = 2), with an EIf value equal or greater than 2. We also identified PacC (EIf = 1.57) as the second most expanded group within the highly expanded Znf_C2H2 family. We will further discuss these six groups (highlighted in bold, Table 3)
Both Ftf1/Ftf2 and Ebr1/Ebr2 belong to the Zn2-C6 family and contribute directly to the fungal virulence (Michielse et al. 2009; van der Does et al. 2016; Ramos et al. 2007). Deletion of accessory copy Ftf1 reduced the pathogenicity of F. oxysporum f. sp. phaseoli (Ramos et al. 2007), highlighting the direct functional involvement of AC TF in virulence. In Fol, deletion of either Ftf1 (AC encoding) or Ftf2 (CC encoding) reduced the virulence towards the host (de Vega-Bartol et al. 2011; Niño-Sánchez et al. 2016). Constitutive expression of either Ftf1 or Ftf2 induced the expression of effector genes (van der Does et al. 2016). The core copy Ftf2 is conserved among all Fusarium species, and the AC copy Ftf1 is only found in F. oxysporum and Fusarium redolens (Figure 4). Ebr1 and paralogues are responsible for virulence and general metabolism. In F. oxysporum, Ebr1 is found as multiple homologs, whereas in F. graminearum, it is seen as a single copy (Jonkers et al. 2014). In F. oxysporum, three paralogous copies, Ebr2, Ebr3, and Ebr4, are encoded in ACs and regulated by core copy Ebr1. The importance of the core paralog has been shown by the reduced pathogenicity and growth defects when it was knocked out (Jonkers et al. 2014). It is worth noting that the Ebr2 coding sequence driven by an Ebr1 promoter was able to rescue the Ebr1 knockout mutation, indicating some functional redundancy of this family.
Figure 4. Unique expansion of some TFs, driven by ACs, may provide clues to host-specific adaptation.
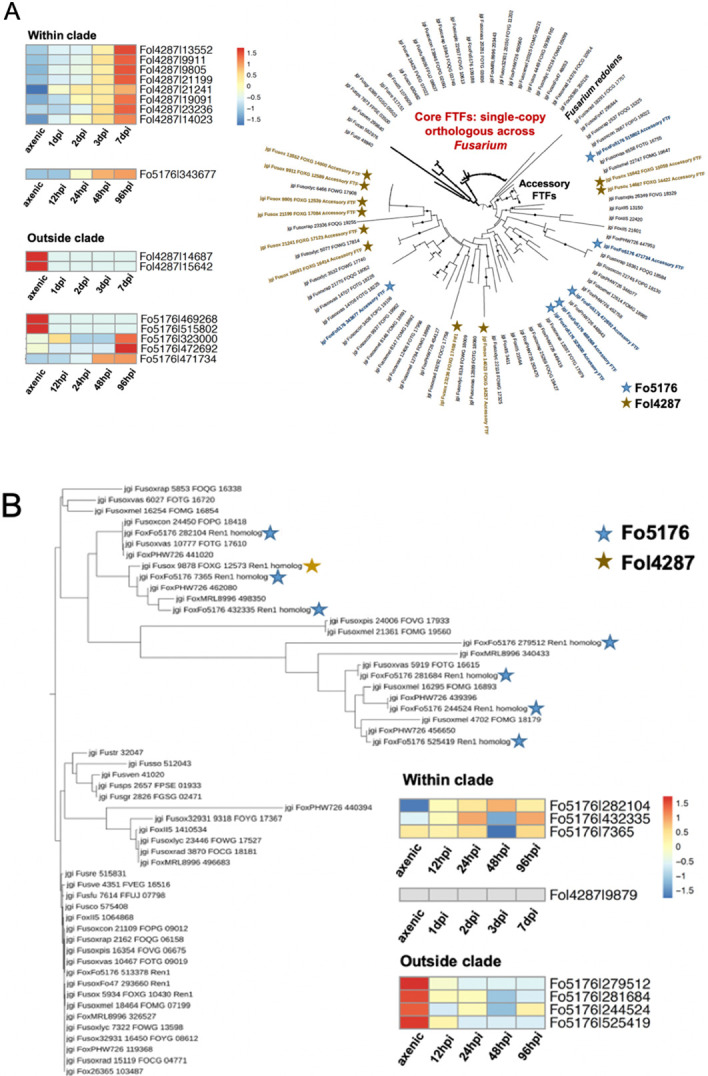
RNA-seq data were previously described (Guo et al., 2021; Redkar et al., 2021). (A) Ftf1, the TF involved in the tomato pathogenicity is most significantly expanded (10 copies of accessory FTFs) in the tomato pathogen Fol4287 genome and the expression of eight out of 10 were induced during plant colonization. (B) Ren1 is most significantly expanded (seven copies of accessory RENs) in the Arabidopsis pathogen Fo5176 genome, and two of them were induced during plant colonization.
Both Cos1 and PacC belong to the Znf_C2H2 family. Mutation to M. oryzae Cos1 resulted in developmental failure of the conidiophores (Li et al. 2013). Furthermore, mutation to Cos1 aggravated the plant infection of leaf blades and sheaths, indicating a negative role in the pathogenicity (Zhou et al. 2009). PacC is an important pH-responsive TF in F. oxysporum (Caracuel et al. 2003; Zhang et al. 2020). PacC homologs are expanded in clinical strains (average accessory copy number 3.7) of FOSC, compared to non-clinical strains (average accessory copy number 0.5), while all Fusarium relatives’ genomes examined only contain a single copy of core PacC. Our previous study revealed that in F. oxysporum clinical strains, the expression of one expanded PacC gene on ACs was induced and the protein localized in the nucleus at mammalian physiological pH (7.4), indicating a potential role in host adaptation (Zhang et al. 2020). Interestingly, the induction of AC-encoding PacC genes was CC-encoding PacC gene-dependent, as the induction disappeared in the CC-encoding PacC knockout mutant, further supporting a cross-talking between core and accessory TFs (Yang 2020). Similar to EBR1, the expression of AC PacC genes is much lower than that of the CC PacC gene, and knockouts of one AC PacC gene affected a small subset of genes compared with the CC PacC knockout, which has a broader effect on cellular processes (Yang 2020).
Fug1 has a role in pathogenicity (maize kernel colonization) and fumonisin biosynthesis in F. verticillioides (Ridenour and Bluhm 2017). In addition, the deletion of Fug1 increased sensitivity to the antimicrobial compound 2-benzoxazolinone and to hydrogen peroxide, which indicates that Fug1 plays a role in mitigating stresses associated with the host defense (Ridenour and Bluhm 2017). Neither core copies nor accessory copies of these two genes were experimentally examined in FOSC. Ren1 is a disordered protein with no IPR functional domain. The expansion score EIf = 2 suggests a unique expansion among FOSC. However, the only reported study on its function is in F. oxysporum f. sp. melonis regulating the development of the conidiation (Ohara et al. 2004).
6. Transcriptome analysis to probe the essential TFs during host colonization
To understand the functional importance of FOSC TFs, we take advantage of two recently reported transcriptomics datasets (Redkar et al. 2022; Guo et al. 2021), including pathogenic interactions (Fo5176 infecting Arabidopsis and Fol4287 infecting tomato) and endophytic interactions (Fo47 colonizing Arabidopsis) (Supplemental Dataset).
We first asked what proportion of genes was expressed in conserved and expanded categories (Table S5). We found that almost all genes (58 out of 60) within the conserved category (Group 1) were consistently expressed (TPM > 1 across all conditions), supporting their general roles in controlling life processes. Within the expanded category (Group 3), the proportion of genes being consistently expressed ranges from 41% to 59% for core TFs, and ranges from 5% to 16% for accessory TFs. With a less strict filter (TPM > 1 at minimum 1 condition), we found that all genes within the conserved category were expressed. Within the expanded category, the proportion of genes being expressed accounts for 93% of core TFs across all strains and ranges from 49% to 67% for accessory TFs. When we compared genes being consistently expressed versus genes being expressed at a minimum one condition, the more dramatic number increase for the expanded category (especially when we only consider the accessory TFs) highlighted that the expanded category, especially the accessory TFs, are more likely to be conditional expressed, further supporting their role in niche adaptation.
With the goal of examining the expression and probing important core and accessory TFs, we aimed to develop filtering parameters. Since most validated TFs were reported in the reference Fol4287 strain, we first reviewed for the reported TFs, both core, and accessory, the expression pattern during Fol4287 infecting tomatoes (Table S6). Out of 27 TFs encoded on the core genome, 18 show up-regulation (defined by at least three out of four in planta conditions show up-regulation compared to the axenic growth) during plant colonization from 1 day post-inoculation (dpi) to 7 dpi, consistent with their reported roles in pathogenicity. The accessory Ftf1 has been exclusively demonstrated to play essential functions in fungal pathogenicity (Niño-Sánchez et al. 2016). Eight of 10 accessory Ftfs were upregulated using the same criteria during plant colonization. Our results illustrate the power of using transcriptome data to probe the functionally important players during plant colonization/infection.
We further developed strict criteria to filter important TFs from TFome (Figure S5), by which half (nine) of previously described upregulated core TFs meet the ‘core’ criteria, and all eight up-regulated accessory Ftfs meet the ‘accessory’ criteria (Table S6). We then apply such measures to the transcriptome of all the TFomes of Fol4287, Fo5176, and Fo47 to probe two types of TFs: 1) The conserved core TFs related to plant colonization; 2) Expanded accessory TFs related to host-specific pathogenicity.
Fol4287, Fo5176, and Fo47 upregulated 95, 62, and 44 core TFs during plant colonization. Among them, ten copies are highly conserved (Table S7), as they are single-copy orthologs across all 15 F. oxysporum strains. Two out of ten were previously reported, Fow2 and Sfl1. Fow2, Zn2C6 TF, is required for full virulence but not hyphal growth and conidiation in F. oxysporum f. sp. melonis (Imazaki et al. 2007). The downstream targets of Fow2 remain unknown in F. oxysporum, thus meriting further analysis. Sfl1 has been described in the previous section and is essential for vegetative growth, conidiation, sexual reproduction, and pathogenesis, as shown in M. oryzae (Li et al. 2011). The functions of FoSfl1 remain to be validated.
Fol4287, Fo5176, and Fo47 upregulated 29, 34, and 9 accessory TFs. Ftf1 and Ren1 are particularly interesting (Figure 4 and Table S8). Though Ftfs have been shown to play an essential role in pathogenicity in Fol, whether this pathway is restricted to the same strain remains a question. Compared to Fol4287 which contains ten accessory Ftfs and eight were upregulated during plant colonization, Fo5176 includes six copies of accessory Ftfs, but only one copy was upregulated. Interestingly, eight upregulated Fol4287 and one upregulated Fo5176 Ftfs are clustered together (Figure 4). The unique expansion with regulatory adaptation (i.e., fine-tuned expression regulation) seems to be restricted to Fol4287 but not another pathogenic strain, Fo5176, when they infect the hosts. Among Fo5176 expanded TFs, we identified Ren1. Compared to Fol4287 which encodes only one accessory Ren1 that was not upregulated during plant colonization, Fo5176 encodes seven accessory copies, among which two were upregulated (Figure 4). Though functional validation is needed, the strain-specific expansion followed by fine-tuned expression regulation when infecting host species exists and likely contributes to the host-specific pathogenicity.
DISCUSSION
For a soilborne pathogen with strong host specificity like FOSC, the adjustment of growth and cell cycle control in response to environmental cues is likely essential for survival. At the same time, expanded families likely contribute to the enhanced functions related to niche adaptation. TFs transmit external and internal signals and regulate complex cellular signaling responses to the sensed stimuli. Transmitted through the soil and vascular wounds of plants causing vascular wilt (Gordon 2017), F. oxysporum must adapt to stresses encountered both outside and inside its host. Therefore, it is not surprising to see that genomes of FOSC have larger TFome than other fungi included in the study. The expansion of TFs among FOSC resulted in a positive correlation between the total number of proteins and the size of the fungal TFome, which was also observed before (Shelest 2017).
A total of 14 TF families that control the global transcriptional event, such as TBP, are highly conserved within the ascomycete fungal lineages. Conserved regulatory mechanisms revealed through this study suggest that the plant colonization process could be a common process among FOSC strains regardless of their host-specific pathogenesis. The notion was also supported by recent studies that highlighted the ability of FOSC as a root colonizer facilitated by the conserved genomics components (Martínez-Soto et al. 2022; Redkar et al. 2022).
In contrast to these stable TFs, 30 families are expanded in various degrees and most significant expansions occurred in Zn2-C6 and Znf_C2H2 TF families among FOSC genomes. The number of Zn2-C6 TFs increases significantly (with the highest expansion score) and makes up most of the TFs (56.7%) found within the FOSC TFome. For example, Ftf1, a TF belonging to the Zn2-C6 and involved in the tomato pathogenicity, is most significantly expanded (10 copies of accessory Ftfs) in the tomato pathogen Fol4287 genome, and the expression of eight out of 10 was induced during plant colonization. The continuous expansion suggests the functional importance of these understudied TFs, further supported by the genetic studies (Niño-Sánchez et al. 2016; van der Does et al. 2016; Ramos et al. 2007; Zuriegat et al. 2021; Zhao et al. 2020) and their induction during host invasion revealed by our RNA-seq data.
Unique expansion of some TFs, driven by ACs, may provide a clue to host-specific interactions. Acquiring additional TFs will modify existing regulatory pathways. No question, this will require the fine-tuning of existing networks for this group of organisms to successfully adapt to different hosts under diverse environments. A previous survey of kinome (the complete set of protein kinases encoded in an organism’s genome) among FOSC and other Ascomycetes revealed a positive correlation between the size of the kinome and the size of the genome (DeIulio et al. 2018), exactly the same as we reported here for TFomes. As kinases and TFs are key regulators that modulate all important signaling pathways and are essential for the proper functions of almost all molecular and cellular processes. Strong correlations among kinome and TFome suggest an ordered, instead of chaotic, recruitment and establishment of ACs among FOSC genomes.
This realization further emphasizes the importance of additional functional studies. Reverse genetics is a powerful tool in defining the functional importance of a TF. For example, TF Ren1, a disordered protein, was identified by genetic and molecular characterization (Ohara et al. 2004). This TF is most significantly expanded (seven copies of accessory Rens) in the Arabidopsis pathogen Fo5176 genome, and two of them were induced during plant colonization. Experiments such as chromatin immunoprecipitation sequencing (CHIP-Seq) and DNA affinity purification sequencing (DAP-seq) to profile the cis-regulatory elements globally are high throughput approaches to define specific binding sites (cis-regulatory elements) of TFs. DAP-seq was used successfully to profile the Cistrome for the entire TFome of the bacterial organism (Baumgart et al. 2021), holding the promise for a better understanding of transcriptional regulation in the fungal model F. oxysporum. TFs can function individually or with other proteins in a complex, and can act as an activator that promotes transcription or a repressor that blocks the recruitment of RNA polymerase. Therefore, defining specific functions of these identified binding sites through DAP-seq can be difficult. Gene regulatory networks based on gene co-expression and other phenotypic and multi-omics data as reported in Fusarium (Guo et al. 2016, 2020) can add more resolution to these complex regulatory processes. However, the ultimate understanding of the regulatory roles of each TF will come from careful molecular and biochemical characterization.
A systematic understanding of transcriptional regulation is essential to get the fine-tuned footprint of the gene regulatory network. Our study not only offered a comprehensive look at the regulation from the evolutionary perspective, but also provided an easily implemented computational pipeline to compare TFs and other functional groups in fungi. A better understanding of their functions would not only inform Fusarium biology but also could be extrapolated to other filamentous fungi and complex basidiomycetes.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Massachusetts Green High-Performance Computing Center for providing high-performance computing capacity
FUNDING
This project was supported by the Natural Science Foundation (IOS-165241), the National Institutes of Health (R01EY030150), and the USDA National Institute of Food and Agriculture (MAS00496). The work (proposal:10.46936/10.25585/60001360) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231. H.Y. is also supported by Lotta M. Crabtree Fellowship and Constantine J. Gilgut Fellowship. S.M.C. is also supported by the Vaadia-BARD Postdoctoral Fellowship.
Footnotes
CONFLICTS OF INTERESTS
The authors declare no conflict of interest. The sponsors had no role in the design, execution, interpretation, or writing of the study.
REFERENCES
- Armstrong G.M., and Armstrong J.K. (1981). Formae speciales and races of Fusarium oxysporum causing wilt disease. In Fusarium: Disease, Biology, and Taxonomy, Nelson P.E., Toussoun T.A., and Cook R.J., eds (University Park, PA: Pennsylvania State University Press; ), pp. 391–399. [Google Scholar]
- Albert T. K., Hanzawa H., Legtenberg Y. I. A., de Ruwe M. J., van den Heuvel F. A. J., Collart M. A., Boelens R., and Timmers H., Th M. 2002. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. The EMBO Journal. 21:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arie T., Kaneko I., Yoshida T., Noguchi M., Nomura Y., and Yamaguchi I. 2000. Mating-type genes from asexual phytopathogenic ascomycetes Fusarium oxysporum and Alternaria alternata. Mol Plant Microbe Interact. 13:1330–1339 [DOI] [PubMed] [Google Scholar]
- Asunción García-Sánchez M., Martín-Rodrigues N., Ramos B., de Vega-Bartol J. J., Perlin M. H., and Díaz-Mínguez J. M. 2010. fost12, the Fusarium oxysporum homolog of the transcription factor Ste12, is upregulated during plant infection and required for virulence. Fungal Genetics and Biology. 47:216–225 [DOI] [PubMed] [Google Scholar]
- Bader A. G., and Vogt P. K. 2006. Leucine Zipper Transcription Factors: bZIP Proteins. Pages 964–967 in: Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine, Springer, Berlin, Heidelberg. [Google Scholar]
- Bahadoor A., Brauer E. K., Bosnich W., Schneiderman D., Johnston A., Aubin Y., Blackwell B., Melanson J. E., and Harris L. J. 2018. Gramillin A and B: Cyclic Lipopeptides Identified as the Nonribosomal Biosynthetic Products of Fusarium graminearum. J. Am. Chem. Soc. 140:16783–16791 [DOI] [PubMed] [Google Scholar]
- Baumgart L. A., Lee J. E., Salamov A., Dilworth D. J., Na H., Mingay M., Blow M. J., Zhang Y., Yoshinaga Y., Daum C. G., and O’Malley R. C. 2021. Persistence and plasticity in bacterial gene regulation. Nat Methods. 18:1499–1505 [DOI] [PubMed] [Google Scholar]
- Becker D. M., Fikes J. D., and Guarente L. 1991. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc. Natl. Acad. Sci. U.S.A. 88:1968–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M., Chang H.-Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A., Nuka G., Paysan-Lafosse T., Qureshi M., Raj S., Richardson L., Salazar G. A., Williams L., Bork P., Bridge A., Gough J., Haft D. H., Letunic I., Marchler-Bauer A., Mi H., Natale D. A., Necci M., Orengo C. A., Pandurangan A. P., Rivoire C., Sigrist C. J. A., Sillitoe I., Thanki N., Thomas P. D., Tosatto S. C. E., Wu C. H., Bateman A., and Finn R. D. 2021. The InterPro protein families and domains database: 20 years on. Nucleic Acids Research. 49:D344–D354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braglia P., Dugas S. L., Donze D., and Dieci G. 2007. Requirement of Nhp6 Proteins for Transcription of a Subset of tRNA Genes and Heterochromatin Barrier Function in Saccharomyces cerevisiae. Mol Cell Biol. 27:1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M. A., Costa P. J., Crisucci E. M., and Arndt K. M. 2007. Identification of Rkr1, a Nuclear RING Domain Protein with Functional Connections to Chromatin Modification in Saccharomyces cerevisiae. Mol Cell Biol. 27:2800–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Ruiz G., Ruiz-Roldán C., and Roncero M. I. G. 2013. Lipolytic system of the tomato pathogen Fusarium oxysporum f. sp. lycopersici. Mol Plant Microbe Interact. 26:1054–1067 [DOI] [PubMed] [Google Scholar]
- Breeden L., and Nasmyth K. 1987. Cell cycle control of the yeast HO gene: Cis- and Trans-acting regulators. Cell. 48:389–397 [DOI] [PubMed] [Google Scholar]
- Bustos S. A., and Schleif R. F. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. U.S.A. 90:5638–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B. R., Lorch Y., Li Y., Zhang M., Lacomis L., Erdjument-Bromage H., Tempst P., Du J., Laurent B., and Kornberg R. D. 1996. RSC, an Essential, Abundant Chromatin-Remodeling Complex. Cell. 87:1249–1260 [DOI] [PubMed] [Google Scholar]
- Calero-Nieto F., Di Pietro A., Roncero M. I. G., and Hera C. 2007. Role of the Transcriptional Activator XlnR of Fusarium oxysporum in Regulation of Xylanase Genes and Virulence. MPMI. 20:977–985 [DOI] [PubMed] [Google Scholar]
- Caracuel Z., Roncero M. I. G., Espeso E. A., González-Verdejo C. I., García-Maceira F. I., and Di Pietro A. 2003. The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum: PacC controls virulence in Fusarium. Molecular Microbiology. 48:765–779 [DOI] [PubMed] [Google Scholar]
- Chalabi Hagkarim N., and Grand R. J. 2020. The Regulatory Properties of the Ccr4–Not Complex. Cells. 9:2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-Y., Krishnakumar V., Chan A. P., Thibaud-Nissen F., Schobel S., and Town C. D. 2017. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89:789–804 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Yamane M., Okamasa K., Tsutsumi C., Kojidani T., Sato M., Haraguchi T., and Hiraoka Y. 2009. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. Journal of Cell Biology. 187:413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo C. A., Guldener U., Xu J.-R., Trail F., Turgeon B. G., Di Pietro A., Walton J. D., Ma L.-J., Baker S. E., Rep M., Adam G., Antoniw J., Baldwin T., Calvo S., Chang Y.-L., DeCaprio D., Gale L. R., Gnerre S., Goswami R. S., Hammond-Kosack K., Harris L. J., Hilburn K., Kennell J. C., Kroken S., Magnuson J. K., Mannhaupt G., Mauceli E., Mewes H.-W., Mitterbauer R., Muehlbauer G., Munsterkotter M., Nelson D., O’Donnell K., Ouellet T., Qi W., Quesneville H., Roncero M. I. G., Seong K.-Y., Tetko I. V., Urban M., Waalwijk C., Ward T. J., Yao J., Birren B. W., and Kistler H. C. 2007. The Fusarium graminearum Genome Reveals a Link Between Localized Polymorphism and Pathogen Specialization. Science. 317:1400–1402 [DOI] [PubMed] [Google Scholar]
- Dean R. A., Talbot N. J., Ebbole D. J., Farman M. L., Mitchell T. K., Orbach M. J., Thon M., Kulkarni R., Xu J.-R., Pan H., Read N. D., Lee Y.-H., Carbone I., Brown D., Oh Y. Y., Donofrio N., Jeong J. S., Soanes D. M., Djonovic S., Kolomiets E., Rehmeyer C., Li W., Harding M., Kim S., Lebrun M.-H., Bohnert H., Coughlan S., Butler J., Calvo S., Ma L.-J., Nicol R., Purcell S., Nusbaum C., Galagan J. E., and Birren B. W. 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 434:980–986 [DOI] [PubMed] [Google Scholar]
- Dean R., Van Kan J. A. L., Pretorius Z. A., Hammond-Kosack K. E., Di Pietro A., Spanu P. D., Rudd J. J., Dickman M., Kahmann R., Ellis J., and Foster G. D. 2012. The Top 10 fungal pathogens in molecular plant pathology: Top 10 fungal pathogens. Molecular Plant Pathology. 13:414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeIulio G. A., Guo L., Zhang Y., Goldberg J. M., Kistler H. C., and Ma L.-J. 2018. Kinome Expansion in the Fusarium oxysporum Species Complex Driven by Accessory Chromosomes Mitchell A.P., ed. mSphere. 3:e00231–18, /msphere/3/3/mSphere231-18.atom [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Xu T., Zhu W., Li L., and Fu Q. 2020. A MADS-box transcription factor FoRlm1 regulates aerial hyphal growth, oxidative stress, cell wall biosynthesis and virulence in Fusarium oxysporum f. sp. cubense. Fungal Biology. 124:183–193 [DOI] [PubMed] [Google Scholar]
- van der Does H. C., Fokkens L., Yang A., Schmidt S. M., Langereis L., Lukasiewicz J. M., Hughes T. R., and Rep M. 2016. Transcription Factors Encoded on Core and Accessory Chromosomes of Fusarium oxysporum Induce Expression of Effector Genes Stukenbrock E.H., ed. PLoS Genet. 12:e1006401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donczew R., Warfield L., Pacheco D., Erijman A., and Hahn S. 2020. Two roles for the yeast transcription coactivator SAGA and a set of genes redundantly regulated by TFIID and SAGA. eLife. 9:e50109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle C. E., Kitty Cheung H. Y., Spence K. L., and Saville B. J. 2016. Unh1, an Ustilago maydis Ndt80-like protein, controls completion of tumor maturation, teliospore development, and meiosis. Fungal Genetics and Biology. 94:54–68 [DOI] [PubMed] [Google Scholar]
- Dufresne M., Perfect S., Pellier A.-L., Bailey J. A., and Langin T. 2000. A GAL4-like Protein Is Involved in the Switch between Biotrophic and Necrotrophic Phases of the Infection Process of Colletotrichum lindemuthianum on Common Bean. Plant Cell. 12:1579–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel-Hermann V., and Lecomte C. 2019. Current Status of Fusarium oxysporum Formae Speciales and Races. Phytopathology. 109:512–530 [DOI] [PubMed] [Google Scholar]
- Edskes H. K., Ohtake Y., and Wickner R. B. 1998. Mak21p of Saccharomyces cerevisiae, a Homolog of Human CAATT-binding Protein, Is Essential for 60 S Ribosomal Subunit Biogenesis. Journal of Biological Chemistry. 273:28912–28920 [DOI] [PubMed] [Google Scholar]
- Emms D. M., and Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder M. E., and Hofmann G. E. 1999. HEAT-SHOCK PROTEINS, MOLECULAR CHAPERONES, AND THE STRESS RESPONSE: Evolutionary and Ecological Physiology. Annu. Rev. Physiol. 61:243–282 [DOI] [PubMed] [Google Scholar]
- Fedotova A. A., Bonchuk A. N., Mogila V. A., and Georgiev P. G. 2017. C2H2 Zinc Finger Proteins: The Largest but Poorly Explored Family of Higher Eukaryotic Transcription Factors. Acta Naturae. 9:47–58 [PMC free article] [PubMed] [Google Scholar]
- Fokkens L., Guo L., Dora S., Wang B., Ye K., Sánchez-Rodríguez C., and Croll D. 2021. A Chromosome-Scale Genome Assembly for the Fusarium oxysporum Strain Fo5176 To Establish a Model Arabidopsis-Fungal Pathosystem. :7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S., Kellenberger E., Rogniaux H., Poterszman A., Van Dorsselaer A., Thierry J.-C., Egly J.-M., Moras D., and Kieffer B. 2000. Structural Characterization of the Cysteine-rich Domain of TFIIH p44 Subunit. Journal of Biological Chemistry. 275:31963–31971 [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., and Fasiolo F. 2003. Ribosome assembly in eukaryotes. Gene. 313:17–42 [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D., Jaffe D., FitzHugh W., Ma L.-J., Smirnov S., Purcell S., Rehman B., Elkins T., Engels R., Wang S., Nielsen C. B., Butler J., Endrizzi M., Qui D., Ianakiev P., Bell-Pedersen D., Nelson M. A., Werner-Washburne M., Selitrennikoff C. P., Kinsey J. A., Braun E. L., Zelter A., Schulte U., Kothe G. O., Jedd G., Mewes W., Staben C., Marcotte E., Greenberg D., Roy A., Foley K., Naylor J., Stange-Thomann N., Barrett R., Gnerre S., Kamal M., Kamvysselis M., Mauceli E., Bielke C., Rudd S., Frishman D., Krystofova S., Rasmussen C., Metzenberg R. L., Perkins D. D., Kroken S., Cogoni C., Macino G., Catcheside D., Li W., Pratt R. J., Osmani S. A., DeSouza C. P. C., Glass L., Orbach M. J., Berglund J. A., Voelker R., Yarden O., Plamann M., Seiler S., Dunlap J., Radford A., Aramayo R., Natvig D. O., Alex L. A., Mannhaupt G., Ebbole D. J., Freitag M., Paulsen I., Sachs M. S., Lander E. S., Nusbaum C., and Birren B. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 422:859–868 [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Cuomo C., Ma L.-J., Wortman J. R., Batzoglou S., Lee S.-I., Baştürkmen M., Spevak C. C., Clutterbuck J., Kapitonov V., Jurka J., Scazzocchio C., Farman M., Butler J., Purcell S., Harris S., Braus G. H., Draht O., Busch S., D’Enfert C., Bouchier C., Goldman G. H., Bell-Pedersen D., Griffiths-Jones S., Doonan J. H., Yu J., Vienken K., Pain A., Freitag M., Selker E. U., Archer D. B., Peñalva M. Á., Oakley B. R., Momany M., Tanaka T., Kumagai T., Asai K., Machida M., Nierman W. C., Denning D. W., Caddick M., Hynes M., Paoletti M., Fischer R., Miller B., Dyer P., Sachs M. S., Osmani S. A., and Birren B. W. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- Gallegos M.-T., Michán C., and Ramos J. L. 1993. The XylS/AraC family of regulators. Nucl Acids Res. 21:807–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiology Reviews. 25:107–123 [DOI] [PubMed] [Google Scholar]
- Gardiner D. M., McDonald M. C., Covarelli L., Solomon P. S., Rusu A. G., Marshall M., Kazan K., Chakraborty S., McDonald B. A., and Manners J. M. 2012. Comparative Pathogenomics Reveals Horizontally Acquired Novel Virulence Genes in Fungi Infecting Cereal Hosts. PLOS Pathogens. 8:e1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C. J., and Fink G. R. 1994. Induction of Pseudohyphal Growth by Overexpression of PHD1, a Saccharomyces cerevisiae Gene Related to Transcriptional Regulators of Fungal Developmentt. MOL. CELL. BIOL. 14:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., Galibert F., Hoheisel J. D., Jacq C., Johnston M., Louis E. J., Mewes H. W., Murakami Y., Philippsen P., Tettelin H., and Oliver S. G. 1996. Life with 6000 Genes. Science. 274:546–567 [DOI] [PubMed] [Google Scholar]
- Goppelt A., Stelzer G., Lottspeich F., and Meisterernst M. 1996. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. The EMBO Journal. 15:3105–3116 [PMC free article] [PubMed] [Google Scholar]
- Gordon T. R. 2017. Fusarium oxysporum and the Fusarium Wilt Syndrome. Annu. Rev. Phytopathol. 55:23–39 [DOI] [PubMed] [Google Scholar]
- Green M. R. 2000. TBP-associated factors (TAF II s): multiple, selective transcriptional mediators in common complexes. Trends in Biochemical Sciences. 25:59–63 [DOI] [PubMed] [Google Scholar]
- Grigoriev I. V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., Riley R., Salamov A., Zhao X., Korzeniewski F., Smirnova T., Nordberg H., Dubchak I., and Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucl. Acids Res. 42:D699–D704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ji M., and Ye K. 2020. Dynamic network inference and association computation discover gene modules regulating virulence, mycotoxin and sexual reproduction in Fusarium graminearum. BMC Genomics. 21:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yu H., Wang B., Vescio K., DeIulio G. A., Yang H., Berg A., Zhang L., Edel-Hermann V., Steinberg C., Kistler H. C., and Ma L.-J. 2021. Metatranscriptomic Comparison of Endophytic and Pathogenic Fusarium – Arabidopsis Interactions Reveals Plant Transcriptional Plasticity. MPMI. 34:1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Zhao G., Xu J., Kistler H. C., Gao L., and Ma L. 2016. Compartmentalized gene regulatory network of the pathogenic fungus Fusarium graminearum. New Phytol. 211:527–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern H. C., Bell A. A., Wagner T. A., Liu J., Nichols R. L., Olvey J., Woodward J. E., Sanogo S., Jones C. A., Chan C. T., and Brewer M. T. 2018. First Report of Fusarium Wilt of Cotton Caused by Fusarium oxysporum f. sp. vasinfectum Race 4 in Texas, U.S.A. Plant Disease. 102:446–446 [Google Scholar]
- Hassan A. S., Al-Hatmi A. M. S., Shobana C. S., van Diepeningen A. D., Kredics L., Vágvölgyi C., Homa M., Meis J. F., de Hoog G. S., Narendran V., Manikandan P., and IHFK Working Group. 2016. Antifungal Susceptibility and Phylogeny of Opportunistic Members of the Genus Fusarium Causing Human Keratomycosis in South India. Med. Myco. 54:287–294 [DOI] [PubMed] [Google Scholar]
- Hepworth S. R., Friesen H., and Segall J. 1998. NDT80 and the Meiotic Recombination Checkpoint Regulate Expression of Middle Sporulation-Specific Genes in Saccharomyces cerevisiae. Mol Cell Biol. 18:5750–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J. N., Brown S. A., Clark C. D., and Winston F. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes & Development. 6:2288–2298 [DOI] [PubMed] [Google Scholar]
- Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., and Vinh L. S. 2018. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular Biology and Evolution. 35:518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo M., Nakayama A., Fukazawa K., Kawamura K., Ando K., Hori M., and Furutani Y. 1990. A novel Bacillus subtilis gene involved in negative control of sporulation and degradative-enzyme production. Journal of Bacteriology. 172:1783–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J., Huang J., Meluh P. B., and Laurent B. C. 2003. The Yeast RSC Chromatin-Remodeling Complex Is Required for Kinetochore Function in Chromosome Segregation. Mol Cell Biol. 23:3202–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T., and Araki H. 2004. Noncompetitive Counteractions of DNA Polymerase ε and ISW2/yCHRAC for Epigenetic Inheritance of Telomere Position Effect in Saccharomyces cerevisiae. Mol Cell Biol. 24:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imazaki I., Kurahashi M., Iida Y., and Tsuge T. 2007. Fow2, a Zn(II)2Cys6-type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum. Mol Microbiol. 63 [DOI] [PubMed] [Google Scholar]
- Iwahara J. 2002. The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA. The EMBO Journal. 21:1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Zhang C., Wu C., Sun P., Hou R., Liu H., Wang C., and Xu J.-R. 2016. TRI6 and TRI10 play different roles in the regulation of deoxynivalenol (DON) production by cAMP signalling in Fusarium graminearum. Environmental Microbiology. 18:3689–3701 [DOI] [PubMed] [Google Scholar]
- Jiang C., Zhang S., Zhang Q., Tao Y., Wang C., and Xu J.-R. 2015. FgSKN 7 and FgATF 1 have overlapping functions in ascosporogenesis, pathogenesis and stress responses in F usarium graminearum: Overlapping functions between FgSKN7 and FgATF1. Environ Microbiol. 17:1245–1260 [DOI] [PubMed] [Google Scholar]
- Jones D. A. B., John E., Rybak K., Phan H. T. T., Singh K. B., Lin S.-Y., Solomon P. S., Oliver R. P., and Tan K.-C. 2019. A specific fungal transcription factor controls effector gene expression and orchestrates the establishment of the necrotrophic pathogen lifestyle on wheat. Sci Rep. 9:15884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A. F., Sangrador-Vegas A., Scheremetjew M., Yong S.-Y., Lopez R., and Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics. 30:1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. 2004. An overview of the basic helix-loop-helix proteins. Genome Biology. :6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers W., Xayamongkhon H., Haas M., Olivain C., van der Does H. C., Broz K., Rep M., Alabouvette C., Steinberg C., and Kistler H. C. 2014. EBR1 genomic expansion and its role in virulence of F usarium species: EBR1 in virulence of Fusarium species. Environ Microbiol. 16:1982–2003 [DOI] [PubMed] [Google Scholar]
- Jung U. S., Sobering A. K., Romeo M. J., and Levin D. E. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase: Reporters for cell wall integrity signalling. Molecular Microbiology. 46:781–789 [DOI] [PubMed] [Google Scholar]
- Kamura T., Burian D., Khalili H., Schmidt S. L., Sato S., Liu W. J., Conrad M. N., Conaway R. C., Conaway J. W., and Shilatifard A. 2001. Cloning and characterization of ELL-associated proteins EAP45 and EAP20. a role for yeast EAP-like proteins in regulation of gene expression by glucose. J Biol Chem. 276:16528–16533 [DOI] [PubMed] [Google Scholar]
- Katoh K., and Standley D. M. 2013. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution. 30:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J. M., Park C., Bennett C., and Salzberg S. L. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37:907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-J., Lee K.-L., Kim K.-D., and Roe J.-H. 2016. The iron uptake repressor Fep1 in the fission yeast binds Fe-S cluster through conserved cysteines. Biochemical and Biophysical Research Communications. 478:187–192 [DOI] [PubMed] [Google Scholar]
- Kim S., Park S.-Y., Kim K. S., Rho H.-S., Chi M.-H., Choi J., Park J., Kong S., Park J., Goh J., and Lee Y.-H. 2009. Homeobox Transcription Factors Are Required for Conidiation and Appressorium Development in the Rice Blast Fungus Magnaporthe oryzae Copenhaver G.P., ed. PLoS Genet. 5:e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Moll T., Neuberg M., Ahorn H., and Nasmyth K. 1993. A Role for the Transcription Factors Mbp1 and Swi4 in Progression from G1 to S Phase. Science. 261:1551–1557 [DOI] [PubMed] [Google Scholar]
- Kredics L., Narendran V., Shobana C. S., Vágvölgyi C., Manikandan P., and Indo-Hungarian Fungal Keratitis Working Group. 2015. Filamentous fungal infections of the cornea: a global overview of epidemiology and drug sensitivity. Mycoses. 58:243–260 [DOI] [PubMed] [Google Scholar]
- Latrick C. M., Marek M., Ouararhni K., Papin C., Stoll I., Ignatyeva M., Obri A., Ennifar E., Dimitrov S., Romier C., and Hamiche A. 2016. Molecular basis and specificity of H2A.Z–H2B recognition and deposition by the histone chaperone YL1. Nat Struct Mol Biol. 23:309–316 [DOI] [PubMed] [Google Scholar]
- Letunic I., and Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research. 49:W293–W296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zhou X., Kong L., Wang Y., Zhang H., Zhu H., Mitchell T. K., Dean R. A., and Xu J.-R. 2011. MoSfl1 Is Important for Virulence and Heat Tolerance in Magnaporthe oryzae Idnurm A., ed. PLoS ONE. 6:e19951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Han X., Liu Z., and He C. 2013. The function and properties of the transcriptional regulator COS1 in Magnaporthe oryzae. Fungal Biology. 117:239–249 [DOI] [PubMed] [Google Scholar]
- Liang X., Shan S., Pan L., Zhao J., Ranjan A., Wang F., Zhang Z., Huang Y., Feng H., Wei D., Huang L., Liu X., Zhong Q., Lou J., Li G., Wu C., and Zhou Z. 2016. Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1. Nat Struct Mol Biol. 23:317–323 [DOI] [PubMed] [Google Scholar]
- Liu J., Yuan Y., Wu Z., Li N., Chen Y., Qin T., Geng H., Xiong L., and Liu D. 2015. A Novel Sterol Regulatory Element-Binding Protein Gene (sreA) Identified in Penicillium digitatum Is Required for Prochloraz Resistance, Full Virulence and erg11 (cyp51) Regulation Cramer R.A., ed. PLoS ONE. 10:e0117115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Berges M. S. 2020. ZafA-mediated regulation of zinc homeostasis is required for virulence in the plant pathogen Fusarium oxysporum. Molecular Plant Pathology. 21:244–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Berges M. S., Capilla J., Turrà D., Schafferer L., Matthijs S., Jöchl C., Cornelis P., Guarro J., Haas H., and Di Pietro A. 2012. HapX-Mediated Iron Homeostasis Is Essential for Rhizosphere Competence and Virulence of the Soilborne Pathogen Fusarium oxysporum. The Plant Cell. 24:3805–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Berges M. S., Rispail N., Prados-Rosales R. C., and Di Pietro A. 2010. A Nitrogen Response Pathway Regulates Virulence Functions in Fusarium oxysporum via the Protein Kinase TOR and the bZIP Protein MeaB. Plant Cell. 22:2459–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y., Reuven N., and Shaul Y. 2005. Autorepression of Rfx1 Gene Expression: Functional Conservation from Yeast to Humans in Response to DNA Replication Arrest. Mol Cell Biol. 25:10665–10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysøe E., Pasquali M., Breakspear A., and Kistler H. C. 2011. The Transcription Factor FgStuAp Influences Spore Development, Pathogenicity, and Secondary Metabolism in Fusarium graminearum. MPMI. 24:54–67 [DOI] [PubMed] [Google Scholar]
- Ma L.-J. 2014. Horizontal chromosome transfer and rational strategies to manage Fusarium vascular wilt diseases: Translating genomics for Fusarium vascular wilts. Molecular Plant Pathology. 15:763–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.-J., van der Does H. C., Borkovich K. A., Coleman J. J., Daboussi M.-J., Di Pietro A., Dufresne M., Freitag M., Grabherr M., Henrissat B., Houterman P. M., Kang S., Shim W.-B., Woloshuk C., Xie X., Xu J.-R., Antoniw J., Baker S. E., Bluhm B. H., Breakspear A., Brown D. W., Butchko R. A. E., Chapman S., Coulson R., Coutinho P. M., Danchin E. G. J., Diener A., Gale L. R., Gardiner D. M., Goff S., Hammond-Kosack K. E., Hilburn K., Hua-Van A., Jonkers W., Kazan K., Kodira C. D., Koehrsen M., Kumar L., Lee Y.-H., Li L., Manners J. M., Miranda-Saavedra D., Mukherjee M., Park G., Park J., Park S.-Y., Proctor R. H., Regev A., Ruiz-Roldan M. C., Sain D., Sakthikumar S., Sykes S., Schwartz D. C., Turgeon B. G., Wapinski I., Yoder O., Young S., Zeng Q., Zhou S., Galagan J., Cuomo C. A., Kistler H. C., and Rep M. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 464:367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.-J., Geiser D. M., Proctor R. H., Rooney A. P., O’Donnell K., Trail F., Gardiner D. M., Manners J. M., and Kazan K. 2013. Fusarium Pathogenomics. Annu. Rev. Microbiol. 67:399–416 [DOI] [PubMed] [Google Scholar]
- Ma L.-J., Shea T., Young S., Zeng Q., and Kistler H. C. 2014. Genome Sequence of Fusarium oxysporum f. sp. melonis Strain NRRL 26406, a Fungus Causing Wilt Disease on Melon. Genome Announcements. 2:e00730–14, 2/4/e00730-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson S., Larochelle M., and Turcotte B. 2006. A Fungal Family of Transcriptional Regulators: the Zinc Cluster Proteins. Microbiol Mol Biol Rev. 70:583–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock J. R., Weidenhammer E. M., Adams C. C., Lunz R. L., and Woolford J. L. 1994. Extragenic suppressors of Saccharomyces cerevisiae prp4 mutations identify a negative regulator of PRP genes. Genetics. 136:833–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T., Lu S.-W., Tilbeurgh H. van, Ripoll D. R., Dixelius C., Turgeon B. G., and Debuchy R. 2010. Tracing the Origin of the Fungal α1 Domain Places Its Ancestor in the HMG-Box Superfamily: Implication for Fungal Mating-Type Evolution. PLOS ONE. 5:e15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Soto D., Yu H., Allen K. S., and Ma L.-J. 2022. Differential colonization of the plant vasculature between endophytic versus pathogenic Fusarium oxysporum strains. MPMI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Mendoza A., Eskova A., Weise C., Czajkowski R., and Kahmann R. 2009. Hap2 regulates the pheromone response transcription factor prf1 in Ustilago maydis. Molecular Microbiology. 72:683–698 [DOI] [PubMed] [Google Scholar]
- Mercier A., Watt S., Bähler J., and Labbé S. 2008. Key Function for the CCAAT-Binding Factor Php4 To Regulate Gene Expression in Response to Iron Deficiency in Fission Yeast. Eukaryot Cell. 7:493–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesny F., Miyauchi S., Thiergart T., Pickel B., Atanasova L., Karlsson M., Hüttel B., Barry K. W., Haridas S., Chen C., Bauer D., Andreopoulos W., Pangilinan J., LaButti K., Riley R., Lipzen A., Clum A., Drula E., Henrissat B., Kohler A., Grigoriev I. V., Martin F. M., and Hacquard S. 2021. Genetic determinants of endophytism in the Arabidopsis root mycobiome. Nat Commun. 12:7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., and Dubois E. 2003. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 316:1–21 [DOI] [PubMed] [Google Scholar]
- Michielse C. B., and Rep M. 2009. Pathogen profile update: Fusarium oxysporum. Molecular Plant Pathology. 10:311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse C. B., van Wijk R., Reijnen L., Manders E. M. M., Boas S., Olivain C., Alabouvette C., and Rep M. 2009. The Nuclear Protein Sge1 of Fusarium oxysporum Is Required for Parasitic Growth Howlett B.J., ed. PLoS Pathog. 5:e1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P., Gadal O., Podtelejnikov A., Trumtel S., Gas N., Petfalski E., Tollervey D., Mann M., Hurt E., and Tschochner H. 2001. Maturation and Intranuclear Transport of Pre-Ribosomes Requires Noc Proteins. Cell. 105:499–509 [DOI] [PubMed] [Google Scholar]
- Min K., Son H., Lim J. Y., Choi G. J., Kim J.-C., Harris S. D., and Lee Y.-W. 2014. Transcription Factor RFX1 Is Crucial for Maintenance of Genome Integrity in Fusarium graminearum. Eukaryot Cell. 13:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., von Haeseler A., and Lanfear R. 2020. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular Biology and Evolution. 37:1530–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.-T., Schmidt H. A., von Haeseler A., and Minh B. Q. 2015. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution. 32:268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niño-Sánchez J., Casado-Del Castillo V., Tello V., De Vega-Bartol J. J., Ramos B., Sukno S. A., and Díaz Mínguez J. M. 2016. The FTF gene family regulates virulence and expression of SIX effectors in Fusarium oxysporum. Molecular Plant Pathology. 17:1124–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M., Son H., Choi G. J., Lee C., Kim J.-C., Kim H., and Lee Y.-W. 2016. Transcription factor ART1 mediates starch hydrolysis and mycotoxin production in Fusarium graminearum and F. verticillioides. Molecular Plant Pathology. 17:755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T., Inoue I., Namiki F., Kunoh H., and Tsuge T. 2004. REN1 Is Required for Development of Microconidia and Macroconidia, but Not of Chlamydospores, in the Plant Pathogenic Fungus Fusarium oxysporum. Genetics. 166:113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen J. T., Fikes J. D., and Guarente L. 1991. The Schizosaccharomyces pombe homolog of Saccharomyces cerevisiae HAP2 reveals selective and stringent conservation of the small essential core protein domain. 11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C. S., and Shim W.-B. 2013. The role of MADS-box transcription factors in secondary metabolism and sexual development in the maize pathogen Fusarium verticillioides. Microbiology. 159:2259–2268 [DOI] [PubMed] [Google Scholar]
- Pan X., and Heitman J. 2000. Sok2 Regulates Yeast Pseudohyphal Differentiation via a Transcription Factor Cascade That Regulates Cell-Cell Adhesion. Mol Cell Biol. 20:8364–8372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré A., Kim M., Juarez M. T., Brody S., and McGinnis W. 2012. The Functions of Grainy Head-Like Proteins in Animals and Fungi and the Evolution of Apical Extracellular Barriers Stajich J.E., ed. PLoS ONE. 7:e36254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Park J., Jang S., Kim S., Kong S., Choi J., Ahn K., Kim J., Lee S., Kim S., Park B., Jung K., Kim S., Kang S., and Lee Y.-H. 2008. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics. 24:1024–1025 [DOI] [PubMed] [Google Scholar]
- Pegg K. G., Coates L. M., O’Neill W. T., and Turner D. W. 2019. The Epidemiology of Fusarium Wilt of Banana. Front. Plant Sci. 10:1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., and Salzberg S. L. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33:290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce M., Benjamin K. R., Montano S. P., Georgiadis M. M., Winter E., and Vershon A. K. 2003. Sum1 and Ndt80 Proteins Compete for Binding to Middle Sporulation Element Sequences That Control Meiotic Gene Expression. Mol Cell Biol. 23:4814–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploetz R. C. 2015. Fusarium Wilt of Banana. Phytopathology. 105:1512–1521 [DOI] [PubMed] [Google Scholar]
- Rahman M. Z., Ahmad K., Siddiqui Y., Saad N., Hun T. G., Mohd Hata E., Rashed O., Hossain M. I., and Kutawa A. B. 2021. First Report of Fusarium wilt disease on Watermelon Caused by Fusarium oxysporum f. sp. niveum (FON) in Malaysia. Plant Dis. [Google Scholar]
- Ramos B., Alves-Santos F. M., García-Sánchez M. A., Martín-Rodrigues N., Eslava A. P., and Díaz-Mínguez J. M. 2007. The gene coding for a new transcription factor (ftf1) of Fusarium oxysporum is only expressed during infection of common bean. Fungal Genetics and Biology. 44:864–876 [DOI] [PubMed] [Google Scholar]
- Redkar A., Sabale M., Schudoma C., Zechmann B., Gupta Y. K., López-Berges M. S., Venturini G., Gimenez-Ibanez S., Turrà D., Solano R., and Di Pietro A. 2022. Conserved secreted effectors contribute to endophytic growth and multihost plant compatibility in a vascular wilt fungus. The Plant Cell. :koac174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Van Der Does H. C., Meijer M., Van Wijk R., Houterman P. M., Dekker H. L., De Koster C. G., and Cornelissen B. J. C. 2004. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato: Cysteine-rich protein required for I-3-mediated resistance. Molecular Microbiology. 53:1373–1383 [DOI] [PubMed] [Google Scholar]
- Ridenour J. B., and Bluhm B. H. 2014. The HAP complex in Fusarium verticillioides is a key regulator of growth, morphogenesis, secondary metabolism, and pathogenesis. Fungal Genetics and Biology. 69:52–64 [DOI] [PubMed] [Google Scholar]
- Ridenour J. B., and Bluhm B. H. 2017. The novel fungal-specific gene FUG1 has a role in pathogenicity and fumonisin biosynthesis in Fusarium verticillioides. Molecular Plant Pathology. 18:513–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispail N., and Di Pietro A. 2009. Fusarium oxysporum Ste12 Controls Invasive Growth and Virulence Downstream of the Fmk1 MAPK Cascade. MPMI. 22:830–839 [DOI] [PubMed] [Google Scholar]
- Rispail N., and Di Pietro A. 2010. The homeodomain transcription factor Ste12: Connecting fungal MAPK signaling to plant pathogenicity. Communicative & Integrative Biology. 3:327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. M., and Winston F. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 16:3206–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha A. L. M., Di Pietro A., Ruiz-Roldán C., and Roncero M. I. G. 2008. Ctf1, a transcriptional activator of cutinase and lipase genes in Fusarium oxysporum is dispensable for virulence. Molecular Plant Pathology. 9:293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Roldán C., Pareja-Jaime Y., González-Reyes J. A., and Roncero G., M. I. 2015. The Transcription Factor Con7-1 Is a Master Regulator of Morphogenesis and Virulence in Fusarium oxysporum. MPMI. 28:55–68 [DOI] [PubMed] [Google Scholar]
- Schleif R. 2010. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol Rev. 34:779–796 [DOI] [PubMed] [Google Scholar]
- Schumacher J., Simon A., Cohrs K. C., Viaud M., and Tudzynski P. 2014. The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen Botrytis cinerea. PLOS Genetics. 10:e1004040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelest E. 2017. Transcription Factors in Fungi: TFome Dynamics, Three Major Families, and Dual-Specificity TFs. Front. Genet. 8:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund T., and Lehmann M. 2002. The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev Genes Evol. 212:152–157 [DOI] [PubMed] [Google Scholar]
- Son H., Fu M., Lee Y., Lim J. Y., Min K., Kim J.-C., Choi G. J., and Lee Y.-W. 2016a. A novel transcription factor gene FHS1 is involved in the DNA damage response in Fusarium graminearum. Sci Rep. 6:21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M., Lee Y., and Kim K.-H. 2016b. The Transcription Cofactor Swi6 of the Fusarium graminearum Is Involved in Fusarium Graminearum Virus 1 Infection-Induced Phenotypic Alterations. The Plant Pathology Journal. 32:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Krishna S., Thanos D., Strominger J. L., and Ono S. J. 1994. A novel cysteine-rich sequence-specific DNA-binding protein interacts with the conserved X-box motif of the human major histocompatibility complex class II genes via a repeated Cys-His domain and functions as a transcriptional repressor. J Exp Med. 180:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M., and Nikawa J.-I. 2001. The Saccharomyces cerevisiae Isw2p-Itc1p Complex Represses INO1 Expression and Maintains Cell Morphology. J Bacteriol. 183:4985–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K., Li F.-Q., Ueda H., and Hirose S. 1997. Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc. Natl. Acad. Sci. U.S.A. 94:7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya D., Yang Y., and Lacefield S. 2014. Positive Feedback of NDT80 Expression Ensures Irreversible Meiotic Commitment in Budding Yeast Lichten M., ed. PLoS Genet. 10:e1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vega-Bartol J. J., Martín-Dominguez R., Ramos B., García-Sánchez M.-A., and Díaz-Mínguez J. M. 2011. New Virulence Groups in Fusarium oxysporum f. sp. phaseoli: The Expression of the Gene Coding for the Transcription Factor ftf1 Correlates with Virulence. Phytopathology. 101:470–479 [DOI] [PubMed] [Google Scholar]
- Vesth T. C., Nybo J. L., Theobald S., Frisvad J. C., Larsen T. O., Nielsen K. F., Hoof J. B., Brandl J., Salamov A., Riley R., Gladden J. M., Phatale P., Nielsen M. T., Lyhne E. K., Kogle M. E., Strasser K., McDonnell E., Barry K., Clum A., Chen C., LaButti K., Haridas S., Nolan M., Sandor L., Kuo A., Lipzen A., Hainaut M., Drula E., Tsang A., Magnuson J. K., Henrissat B., Wiebenga A., Simmons B. A., Mäkelä M. R., de Vries R. P., Grigoriev I. V., Mortensen U. H., Baker S. E., and Andersen M. R. 2018. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat Genet. 50:1688–1695 [DOI] [PubMed] [Google Scholar]
- Viljoen A., Mostert D., Chiconela T., Beukes I., Fraser C., Dwyer J., Murray H., Amisse J., Matabuana E. L., Tazan G., Amugoli O. M., Mondjana A., Vaz A., Pretorius A., Bothma S., Rose L. J., Beed F., Dusunceli F., Chao C.-P., and Molina A. B. 2020. Occurrence and spread of the banana fungus Fusarium oxysporum f. sp. cubense TR4 in Mozambique. South African Journal of Science. 116:1–11 [Google Scholar]
- Vizoso-Vázquez Á., Lamas-Maceiras M., Becerra M., González-Siso M. I., Rodríguez-Belmonte E., and Cerdán M. E. 2012. Ixr1p and the control of the Saccharomyces cerevisiae hypoxic response. Appl Microbiol Biotechnol. 94:173–184 [DOI] [PubMed] [Google Scholar]
- Vuorio T., Maity S. N., and de Crombrugghe B. 1990. Purification and molecular cloning of the “A” chain of a rat heteromeric CCAAT-binding protein. Sequence identity with the yeast HAP3 transcription factor. Journal of Biological Chemistry. 265:22480–22486 [PubMed] [Google Scholar]
- Wang B., Yu H., Jia Y., Dong Q., Steinberg C., Alabouvette C., Edel-Hermann V., Kistler H. C., Ye K., Ma L.-J., and Guo L. 2020. Chromosome-Scale Genome Assembly of Fusarium oxysporum Strain Fo47, a Fungal Endophyte and Biocontrol Agent. MPMI. :MPMI-05-20-0116 [DOI] [PubMed] [Google Scholar]
- Wang Q., Pokhrel A., and Coleman J. J. 2021. The Extracellular Superoxide Dismutase Sod5 From Fusarium oxysporum Is Localized in Response to External Stimuli and Contributes to Fungal Pathogenicity. Front Plant Sci. 12:608861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P., Sieber C. M. K., Bargen K. W. von, Studt L., Niehaus E.-M., Espino J. J., Huß K., Michielse C. B., Albermann S., Wagner D., Bergner S. V., Connolly L. R., Fischer A., Reuter G., Kleigrewe K., Bald T., Wingfield B. D., Ophir R., Freeman S., Hippler M., Smith K. M., Brown D. W., Proctor R. H., Münsterkötter M., Freitag M., Humpf H.-U., Güldener U., and Tudzynski B. 2013. Deciphering the Cryptic Genome: Genome-wide Analyses of the Rice Pathogen Fusarium fujikuroi Reveal Complex Regulation of Secondary Metabolism and Novel Metabolites. PLOS Pathogens. 9:e1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight W. D., Kim K.-H., Lawrence C. B., and Walton J. D. 2009. Biosynthesis and role in virulence of the histone deacetylase inhibitor depudecin from Alternaria brassicicola. Mol Plant Microbe Interact. 22:1258–1267 [DOI] [PubMed] [Google Scholar]
- Williams A. H., Sharma M., Thatcher L. F., Azam S., Hane J. K., Sperschneider J., Kidd B. N., Anderson J. P., Ghosh R., Garg G., Lichtenzveig J., Kistler H. C., Shea T., Young S., Buck S.-A. G., Kamphuis L. G., Saxena R., Pande S., Ma L.-J., Varshney R. K., and Singh K. B. 2016. Comparative genomics and prediction of conditionally dispensable sequences in legume–infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genomics. 17:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V., Gwilliam R., Rajandream M.-A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., Basham D., Bowman S., Brooks K., Brown D., Brown S., Chillingworth T., Churcher C., Collins M., Connor R., Cronin A., Davis P., Feltwell T., Fraser A., Gentles S., Goble A., Hamlin N., Harris D., Hidalgo J., Hodgson G., Holroyd S., Hornsby T., Howarth S., Huckle E. J., Hunt S., Jagels K., James K., Jones L., Jones M., Leather S., McDonald S., McLean J., Mooney P., Moule S., Mungall K., Murphy L., Niblett D., Odell C., Oliver K., O’Neil S., Pearson D., Quail M. A., Rabbinowitsch E., Rutherford K., Rutter S., Saunders D., Seeger K., Sharp S., Skelton J., Simmonds M., Squares R., Squares S., Stevens K., Taylor K., Taylor R. G., Tivey A., Walsh S., Warren T., Whitehead S., Woodward J., Volckaert G., Aert R., Robben J., Grymonprez B., Weltjens I., Vanstreels E., Rieger M., Schäfer M., Müller-Auer S., Gabel C., Fuchs M., Fritzc C., Holzer E., Moestl D., Hilbert H., Borzym K., Langer I., Beck A., Lehrach H., Reinhardt R., Pohl T. M., Eger P., Zimmermann W., Wedler H., Wambutt R., Purnelle B., Goffeau A., Cadieu E., Dréano S., Gloux S., Lelaure V., Mottier S., Galibert F., Aves S. J., Xiang Z., Hunt C., Moore K., Hurst S. M., Lucas M., Rochet M., Gaillardin C., Tallada V. A., Garzon A., Thode G., Daga R. R., Cruzado L., Jimenez J., Sánchez M., del Rey F., Benito J., Domínguez A., Revuelta J. L., Moreno S., Armstrong J., Forsburg S. L., Cerrutti L., Lowe T., McCombie W. R., Paulsen I., Potashkin J., Shpakovski G. V., Ussery D., Barrell B. G., and Nurse P. 2002. The genome sequence of Schizosaccharomyces pombe. Nature. 415:871–880 [DOI] [PubMed] [Google Scholar]
- Xin C., Zhang J., Nian S., Wang G., Wang Z., Song Z., and Ren G. 2020. Analogous and Diverse Functions of APSES-Type Transcription Factors in the Morphogenesis of the Entomopathogenic Fungus Metarhizium rileyi Zhou N.-Y., ed. Appl Environ Microbiol. 86:e02928–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Narita T., Inukai N., Wada T., and Handa H. 2001. SPT Genes: Key Players in the Regulation of Transcription, Chromatin Structure and Other Cellular Processes. Journal of Biochemistry. 129:185–191 [DOI] [PubMed] [Google Scholar]
- Yan X., Li Y., Yue X., Wang C., Que Y., Kong D., Ma Z., Talbot N. J., and Wang Z. 2011. Two Novel Transcriptional Regulators Are Essential for Infection-related Morphogenesis and Pathogenicity of the Rice Blast Fungus Magnaporthe oryzae. PLOS Pathogens. 7:e1002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. 2020. ACCESSORY GENES CONTRIBUTE TO REWIRING THE TRANSCRIPTIONAL NETWORK IN FUSARIUM OXYSPORUM. Doctoral Dissertations. [Google Scholar]
- Yang H., Yu H., and Ma L.-J. 2020. Accessory Chromosomes in Fusarium oxysporum. Phytopathology®. :PHYTO-03-20-006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Ayhan D. H., Diener A. C., and Ma L.-J. 2020. Genome Sequence of Fusarium oxysporum f. sp. matthiolae, a Brassicaceae Pathogen. MPMI. 33:569–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Ayhan D. H., Martínez-Soto D., Cochavi S. M., and Ma L.-J. 2023. Accessory Chromosomes of the Fusarium oxysporum Species Complex and Their Contribution to Host Niche Adaptation. Pages 371–388 in: Plant Relationships, The Mycota. Scott B. and Mesarich C., eds. Springer International Publishing, Cham. [Google Scholar]
- Yue X., Que Y., Xu L., Deng S., Peng Y., Talbot N. J., and Wang Z. 2016. ZNF1 Encodes a Putative C2H2 Zinc-Finger Protein Essential for Appressorium Differentiation by the Rice Blast Fungus Magnaporthe oryzae. Mol Plant Microbe Interact. 29:22–35 [DOI] [PubMed] [Google Scholar]
- Yun Y., Zhou X., Yang S., Wen Y., You H., Zheng Y., Norvienyeku J., Shim W.-B., and Wang Z. 2019. Fusarium oxysporum f. sp. lycopersici C2H2 transcription factor FolCzf1 is required for conidiation, fusaric acid production, and early host infection. Curr Genet. 65:773–783 [DOI] [PubMed] [Google Scholar]
- ZHANG Y. 2019. EVOLUTION OF THE PATHOGENIC FUSARIUM OXYSPORUM THROUGH THE LENS OF COMPARATIVE GENOMICS. Doctoral Dissertations. [Google Scholar]
- Zhang Y., Yang H., Turra D., Zhou S., Ayhan D. H., DeIulio G. A., Guo L., Broz K., Wiederhold N., Coleman J. J., Donnell K. O., Youngster I., McAdam A. J., Savinov S., Shea T., Young S., Zeng Q., Rep M., Pearlman E., Schwartz D. C., Di Pietro A., Kistler H. C., and Ma L.-J. 2020. The genome of opportunistic fungal pathogen Fusarium oxysporum carries a unique set of lineage-specific chromosomes. Commun Biol. 3:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., An B., Guo Y., Hou X., Luo H., He C., and Wang Q. 2020. Label free proteomics and systematic analysis of secretome reveals effector candidates regulated by SGE1 and FTF1 in the plant pathogen Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genomics. 21:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Li G., Lin C., and He C. 2009. Conidiophore Stalk-less1 Encodes a Putative Zinc-Finger Protein Involved in the Early Stage of Conidiation and Mycelial Infection in Magnaporthe oryzae. MPMI. 22:402–410 [DOI] [PubMed] [Google Scholar]
- Zuriegat Q., Zheng Y., Liu H., Wang Z., and Yun Y. 2021. Current progress on pathogenicity-related transcription factors in Fusarium oxysporum. Mol Plant Pathol. 22:882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.