Abstract
The paraneoplastic Ma antigen (PNMA) genes are associated with cancer-induced paraneoplastic syndromes that present with neurological symptoms and autoantibody production. How PNMA proteins trigger a severe autoimmune disease is unclear. PNMA genes are predominately expressed in the central nervous system with little known functions but are ectopically expressed in some tumors. Here, we show that PNMA2 is derived from a Ty3 retrotransposon that encodes a protein which forms virus-like capsids released from cells as non-enveloped particles. Recombinant PNMA2 capsids injected into mice induce a robust autoimmune reaction with significant generation of autoantibodies that preferentially bind external “spike” PNMA2 capsid epitopes, while capsid-assembly-defective PNMA2 protein is not immunogenic. PNMA2 autoantibodies present in cerebrospinal fluid of patients with anti-Ma2 paraneoplastic neurologic disease show similar preferential binding to PNMA2 “spike” capsid epitopes. These observations suggest that PNMA2 capsids released from tumors trigger an autoimmune response that underlies Ma2 paraneoplastic neurological syndrome.
Introduction
The PNMA family of genes are predominately expressed in the brain (1–4), but little is known about their biological function. PNMA1-2 were first identified as genes encoding proteins that are targets of neuronal autoantibodies in blood and cerebrospinal fluid (CSF) from patients with certain paraneoplastic neurological syndromes (1–3). Patients with elevated levels of PNMA1 (Ma1) or PNMA2 (Ma2) autoantibodies experience limbic/brainstem encephalitis and cerebellar degeneration (5). These patients often present with solid peripheral tumors (1, 6, 7) that are a potential source of PNMA protein (8–11) and antibodies to PNMA proteins can be diagnostic of specific cancers (12). Prognosis in this paraneoplastic syndrome is poor (7). It has remained enigmatic why these proteins elicit such a strong autoimmune response, but given the severity of disease, it is imperative to gain an understanding of the pathophysiology to better target immunomodulatory treatments.
The PNMA family of genes is predicted to have Gag-homology domains (13, 14). Recent studies identified Gag-derived proteins co-opted from long terminal repeat (LTR) retrotransposons that mediate a new and unexpected virus-like intercellular communication pathway (13–17). These genes include the neuronal Arc genes that are critical for synaptic plasticity, memory, and cognition (15, 16, 18, 19). The Gag domain contained in LTR retrotransposons and retroviruses is necessary and sufficient to produce viral-like particles that package their mRNA during replication. The Arc gene, which was co-opted from a Ty3/mdg4 (formerly known as Ty3/gypsy) retrotransposon in the common ancestor of tetrapod vertebrates, encodes a protein that has retained virus-like biology. Arc assembles into virus-like capsids that are released from neurons in membrane-enveloped extracellular vesicles (EVs) that transfer RNA and protein cell-to-cell (16). The Drosophila Arc (dArc) homologs also assemble capsids from a Gag domain that are released in EVs (16, 20), but originated independently from a distinct lineage of Ty3/mdg4 retrotransposons (18, 21). The Gag-like gene PEG10, which was derived from yet another lineage of Ty3/mdg4 retrotransposons during mammalian evolution, has also retained ancestral virus-like properties such as RNA binding, capsid formation, and release in EVs (14, 22–25). Together these studies suggest that a diverse set of co-opted Gag genes have preserved biochemical properties of ancestral retroelements (26), but their physiological functions and role in human diseases are poorly characterized.
In this study, we investigated whether putative virus-like properties of PNMA proteins trigger an autoimmune reaction that underlies paraneoplastic neurological disorders. Concentrating on the PNMA2 gene, we find that PNMA2 is normally exclusively and highly expressed in neurons. PNMA2 protein self-assembles into virus-like capsids that are released from cells as non-enveloped capsids. Based on these results, we posited that PNMA2 capsids released outside the central nervous system (CNS) may cause autoantibody generation. Consistent with this, mice injected with PNMA2 capsids produce strong autoantibody generation without adjuvant. Based on atomic resolution structures of PNMA2 capsids, we designed PNMA2 mutants that are unable to form capsids. Injection of capsid mutant protein does not elicit autoantibody production. Finally, we find that PNMA2 autoantibodies from paraneoplastic patients preferentially bind to exterior surface “spike” epitopes of PNMA2 capsids. Taken together, our results show that PNMA2 is a Gag-like protein that forms endogenous virus-like capsids that are released without a membrane, which can trigger a robust immune response when ectopically expressed outside the CNS. These properties may underlie the neurological deficits associated with Ma2 paraneoplastic disease.
Results
PNMA2 evolved from a Ty3/mdg4 retrotransposon coopted in the ancestor of placental mammals.
To trace the evolutionary origins of PNMA2, we conducted phylogenomic analysis. The PNMA2 gene is conserved at an orthologous genomic position across all major lineages of placental mammals but absent from the genomes of marsupials and non-mammalian species (Figure 1a). Thus, PNMA2 was coopted in the common ancestor of placental mammals ~100 million years ago. The PNMA2 gene is composed of three exons. The first two are short noncoding exons interrupted by long introns, while the third exon encodes the open reading frame corresponding to the Gag-derived portion of an ancient MamGyp-int element of the Ty3/mdg4 superfamily (16, 21). The predicted PNMA2 promoter region is located only ~150 bp away from its nearest neighboring gene, DPYSL2, which is arranged in the opposite orientation relative to PNMA2 (Figure 1a). This arrangement suggests that PNMA2 and DPYSL2 share a bidirectional promoter. Because the promoter region and the DPYSL2 gene are more deeply conserved across vertebrate species than PNMA2 (see sequence conservation track in Figure 1a), it is likely that the cooption of PNMA2 was facilitated by the capture of a bidirectional promoter from a pre-existing neighboring gene, DPYSL2.
Figure 1. Ty3/mdg4 retrotransposon-derived PNMA2 encodes proteins self-assembling into virus-like capsids.
a. Structure and evolution of PNMA2. The figure is derived from the Vertebrate Multiz Alignment and Conservation track at the UCSC Genome Browser (hg38) and shows the phylogenetic relationship (left) and level of sequence conservation (right) for a subset of vertebrate genomes across the mRNA sequence and shared promoter region of PNMA2 (red) and DPYSL2 (green) gene models. There are three transposable elements annotated by RepeatMasker within the PNMA2 mRNA: a Mam-Gyp-int element (purple box) that gave rise to the Gag-like coding sequence, and two Alu elements (grey boxes) embedded within the 3’ UTR. The arrows depict the predicted transcription start sites for PNMA2 and DPYSL2. The conservation track shows that PNMA2 mRNA sequence is deeply conserved across placental mammals (except for the two Alu elements which are primate-specific insertions), but not other vertebrates, while DPYSL2 and promoter region are conserved more deeply across vertebrate evolution pointing at their earlier origins.
b. Single-cell expression of PNMA2 mRNA in the human cortex (right panel) and mouse cortex and hippocampus (left panel).
c. PNMA2 single molecule fluorescent in situ hybridization probe (RNAscope) and negative control probe were used to detect PNMA2 mRNA in wild-type mouse (2 months old) hippocampal slice. Scale bar: 400μm.
d. Representative negative-stained EM images of purified recombinant mouse PNMA2 (mPNMA2) capsids and human PNMA2 (hPNMA2) capsids. Scale bar: 50nm.
e. Surface representation of mPNMA2 as resolved from cryo-EM, viewed down the two-fold axis. The spike densities are not resolved.
f. Atomic model of the T=1 mPNMA2 capsid. CANTD is depicted in purple and the CACTD in cyan.
g. External view of the isolated five-fold capsomere.
h. One of 60 mPNMA2 monomers required to form the T=1 capsids.
PNMA2 is expressed in mammalian neurons
To gain insight into PNMA2 expression and localization, we first surveyed bulk RNA-seq data from the GTEx project (27), which reveals that PNMA2 is highly and almost exclusively expressed in human brain tissue samples (gtexportal.org/home/gene/ENSG00000240694) (Supplementary Figure S1). Next, we analyzed single-cell RNA-sequencing datasets to examine PNMA2 expression in human motor cortex, mouse cerebral cortex, and mouse hippocampus (28–31). This analysis reveals that PNMA2 mRNA is expressed in excitatory and inhibitory neurons, with very low expression in glia and oligodendrocyte lineages (Fig 1b). To confirm the RNA-seq data, we used fluorescent in situ hybridization (RNAScope) and found that PNMA2 mRNA is highly transcribed in mouse brain (Figure 1c) and primary cultured hippocampal neurons (Supplementary Figure S2). To determine whether PNMA2 expression pattern is conserved across primates, we analyzed single-cell RNA-seq data from brain samples representing 33 anatomical regions from humans (N=132), chimpanzees (N=96), and macaques (N=96) ) (32). PNMA2 expression is remarkably conserved in grey matter areas of all three species, suggesting that the expression of PNMA2 in the brain is under strong purifying selection (Supplemental Figure S3).
PNMA2 proteins self-assemble into virus-like capsids
Purified recombinant mammalian and Drosophila Arc proteins spontaneously form virus-like capsids (15, 16). PNMA 3, 5, and 6a also form virus-like particles (14). To determine if PNMA2 also forms capsids, we purified recombinant PNMA2 in an E. coli expression system and imaged the samples by negative stain EM. Purified mouse PNMA2 (mPNMA2) and human PNMA2 (hPNMA2) protein spontaneously form ordered virus-like capsids (Figure 1d). PNMA2 capsids were highly stable, even when treated with high concentrations of detergent (Supplemental Figure S4). Using single-particle cryo-EM we reconstructed density maps of the entire icosahedral mPNMA2 capsid as well as of local regions of the capsid surface after symmetry relaxation at a resolution of 3.2 Å (Figure 1e, Supplemental Figure S5–6). mPNMA2 forms icosahedral capsids with an outer diameter of ~200 Å and a triangulation number T = 1, composed of 12 pentameric capsomeres and 60 individual mPNMA2 molecules (Figure 1f). The protein shell of the capsid is roughly 30 Å thick. Based on the obtained maps we built an atomic model of the mPNMA2 capsid (Figure 1f; Supplemental Figure S7; Supplementary Movie 1; Supplementary Table 1). The capsid shell spans residues 157–336 which fold into a nine-helix bilobar CA domain structure, highly similar to Arc and Ty3 capsids (18, 33) (Supplemental Movie 2). Both the CANTD and CACTD of mPNMA2 are compact domains with a hydrophobic core and hydrophilic surface. The 28 residues C-terminal to CACTD are located on the inside of the capsid shell, whereas the 156 N-terminal residues protrude from the centre of the five-fold capsomers to form a disordered spike (Figure 1e; Supplemental Figure S7). The protein-protein packing within the mPNMA2 capsid is highly similar to the Ty3 retrotransposon capsid and the mature HIV-1 capsid, confirming the conservation of Gag capsid-assembly (Supplemental Figure S8). However, unlike dArc1, mPNMA2 does not contain any conserved nucleic acid binding motifs in the capsid interior part of the protein. The electrostatic potential of the interior capsid shell reveals only one exposed positively charged patch located at the twofold symmetry axis and this interface is likely transiently shielded by a highly negatively charged motif in the extended C-terminus of the protein (Supplemental Figure S9). These observations suggest that specific interactions with oligonucleotides are unlikely.
PNMA2 is released from cells as non-enveloped capsids.
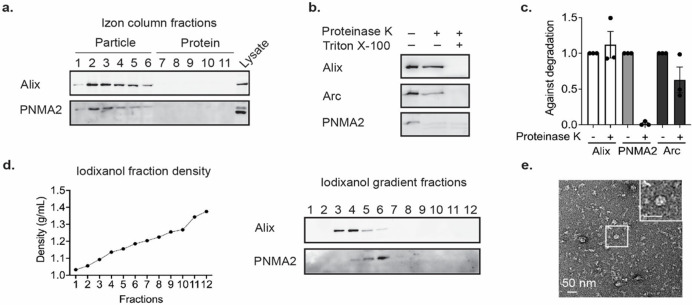
Retrovirus capsids are released from cells as membrane-enveloped particles, similar to EVs. Arc protein is also released in EVs (16). To determine whether mPNMA2 is released from cells, we harvested media from primary cultured cortical neurons and fractionated the media using size-exclusion chromatography. mPNMA2 protein was isolated in early fractions that are also enriched with canonical EV proteins such as the ESCRT protein ALIX (Figure 2a). To determine whether released mPNMA2 protein is enveloped by a membrane, we performed a Proteinase K protection assay. Surprisingly, mPNMA2 protein was highly degraded by Proteinase K even without detergent present, unlike EV proteins, suggesting a lack of membrane protection (Figure 2b and c). These results contrasted with endogenous Arc protein release, which showed significant protection from Proteinase K (Figure 2b and c). To further determine whether mPNMA2 is released in EV fractions, we used iodixanol gradient ultracentrifugation and found that endogenous PNMA2 protein is enriched in a different fraction (fraction 6) to neuronal EV proteins (fraction 3), indicating released mPNMA2 differs in density from proteins found in canonical EV fractions (Figure 2d). Capsid structures were observed in iodixanol fraction 6, as determined by negative-staining EM (Figure 2e). To measure enough capsids to determine average size, myc-mPNMA2 was transfected HEK293T cells (Supplemental Figure S10). Capsids isolated from iodixanol fraction 6 and imaged on EM grids showed an average size similar to capsids from purified recombinant PNMA2 protein (Supplemental Figure S10e).
Figure 2. Endogenous mPNMA2 is released by neurons as non-enveloped capsids.
a. Media was collected from primary cultured cortical neurons (DIV15-16) after 24h incubation and fractionated using size exclusion chromatography (SEC). Fractions were run on a gel and blotted for mPNMA2 protein and ALIX, a canonical EV marker. mPNMA2 protein is released in early fractions that contain EV proteins.
b. The early fractions (1–4) from SEC were pooled and blotted for Arc, mPNMA2, and ALIX. Fractions were incubated with Proteinase K (200μg/mL) with or without detergent present (1% Triton-X) for 10mins. Representative Western blots show that mPNMA2 protein was sensitive to Proteinase K degradation without detergent present.
c. Quantification of Western blot in b (n=3 experiments). Error bars indicate mean ± s.e.m.
d. The early fractions (1–4) from SEC were fractionated using ultracentrifugation. An iodixanol gradient was used to separate proteins by density and size. mPNMA2 protein was enriched in fraction 6, while ALIX was enriched in fractions 3 and 4.
e. Representative negative-stained EM image of non-enveloped mPNMA2 capsids isolated from iodixanol gradient fraction 6 in d.
Patients with PNMA2-related paraneoplastic syndrome often present with small cell lung cancer (6). To test whether tumor cells release hPNMA2, we used a human small cell lung cancer cell line (NCI-H378) that expresses a high level of hPNMA2 (34). We grew cells, harvested media after 24 hours, and performed size exclusion chromatography. hPNMA2 was released and present in early fractions, like ALIX (Supplemental Figure S11a). Similar to neurons, the released hPNMA2 was not protected from Proteinase K degradation (Supplemental Figure S11b) indicating release as non-enveloped capsids.
To determine whether mPNMA2 release requires assembled capsids, we designed point mutations predicted to disrupt the five-fold axis (Y162A – Y/A) or CANTD-CACTD capsid interface (L270Q/L325Q – L/Q) between PNMA2 subunits (Supplemental Figure S7b and e) as mutations in the five-fold axis or CANTD-CACTD interface disrupt retrovirus capsid assembly (35, 36). As predicted, purified mPNMA2 Y/A and mPNMA2 L/Q proteins were unable to form capsids as analyzed by size exclusion chromatography, negative-stain EM, and mass photometry (Supplemental Figure S12 a–c). We overexpressed mPNMA2, mPNMA2 L/Q or mPNMA2 Y/A in HEK293T cells and measured protein release. The release of mPNMA2 L/Q and Y/A were significantly reduced compared with WT protein (Supplemental Figure S12 d–e). Together, these data show that mPNMA2 is released from cells as non-enveloped capsids.
PNMA2 capsids induce autoantibody production.
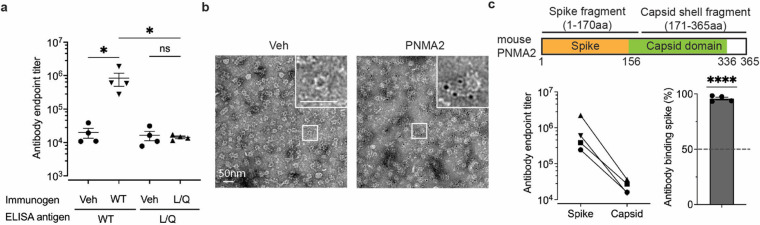
We next determined whether PNMA2’s virus-like properties are responsible for eliciting autoantibody production. Since PNMA2 appears to be exclusively expressed in neurons of the CNS (Figure 1b, Supplemental Figure 1), we hypothesize that aberrant expression of PNMA2 in tumor cells (8–12, 37) and release of capsids outside of the CNS may be immunogenic. Virus capsids induce a stronger immune response than soluble proteins due to the capsid’s influence on antigen transport, adaptive immune response, and cross-presentation (38–41). Thus, the ability of PNMA2 protein to form capsids that are released without membranes may explain its immunogenicity. To examine the immunogenicity of mPNMA2 capsids, we injected mice with 5μg purified recombinant mPNMA2 capsids without adjuvant and collected blood 3 weeks after injection. mPNMA2 capsid-injected mice produced high titers of antibodies that bind to mPNMA2 capsids, as assessed by ELISA (Figure 3a) and by immunogold EM (Figure 3b). The production of antibodies in mPNMA2 injected mice was also confirmed by Western blot (Supplemental Figure S13a). However, mice injected with 5μg of purified mPNMA2 L/Q capsid-mutant protein did not produce mPNMA2 L/Q-specific antibodies (Figure 3a, Supplemental Figure S13a). A second injection of 5μg mPNMA2 L/Q 3 weeks after the first injection, which may have primed the immune system, also did not induce autoantibody production further indicating a lack of immunogenicity (Supplemental Figure S13b). Capsid-induced antibodies showed similar endpoint titers against capsids and L/Q mutant protein when used as ELISA antigens, indicating that the lack of antibody signal in mPNMA2 L/Q injected mice is not due to a lack of exposed epitopes (Supplemental Figure S13c).
Figure 3. mPNMA2 capsids injected into mice induce PNMA2 autoantibody production.
a. Mice were injected intraperitoneally with vehicle (n=4), 5μg purified mPNMA2 capsids (n=4) or 5μg mPNMA2 L/Q protein (n=4) and blood sera collected 3 weeks after injections. Sera were analyzed for antibody production using ELISA, using 2μg/mL mPNMA2 capsids or mPNMA2 L/Q protein as the antigen coated on the plates. mPNMA2 capsid-injected mice produced robust PNMA2 autoantibodies, whereas vehicle and mPNMA2 L/Q injections did not elicit autoantibody production. (*One-way ANOVA with post-hoc pairwise comparisons by Tukey’s test, P=0.0146. Vehicle vs. mPNMA2 WT (using mPNMA2 capsids as antigen): P=0.0308; Vehicle vs. mPNMA2 L/Q (using mPNMA2 L/Q as antigen): P>0.9999); mPNMA2 WT vs. mPNMA2 L/Q: P=0.0296.).
b. Representative negative-stained EM images of purified mPNMA2 capsids immunogold labelled with mouse PNMA2 autoantibodies in serum collected from PNMA2 capsid-injected mice.
c. Purified mPNMA2 spike fragments and capsid shell fragments (see schematic) were used as ELISA antigens to map the epitopes of PNMA2 autoantibodies from PNMA2 capsid-injected mice. The schematic shows PNMA2 protein regions located on the spike and the capsid domains of capsids, as determined by the cryo-EM structure. PNMA2 autoantibodies preferentially bind to the spike fragments. (****One sample t-test, P<0.0001, null hypothesis of 50% binding). Error bars indicate mean ± s.e.m. WT: wild type; L/Q: L270QL325Q; Veh: vehicle.
The structure of mPNMA2 capsids shows that the exposed “spikes” may be more immunogenic than the capsid body since these residues are more likely to be exposed to adaptive immune cells (42, 43). To test this, we purified the N-terminal region of mPNMA2 capsids (the “spike” aa 1–170) and the main capsid shell (aa 171–356) (Figure 3c). Autoantibodies from capsid-injected mice preferentially bind to the N-terminal “spike” fragment (Figure 3c).
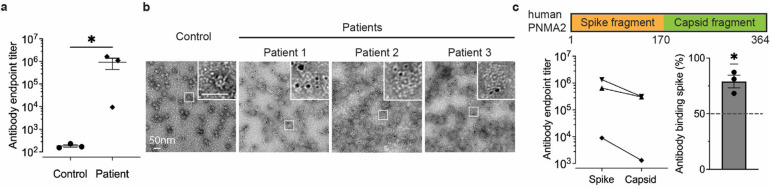
To determine whether PNMA2 autoantibodies from human paraneoplastic patients bind hPNMA2 capsids, we obtained CSF samples from patients associated with paraneoplastic syndromes that have PNMA2 antibodies (see Table 1 for patient information) and control CSF samples (obtained from the Antineuronal Antibodies in Autoimmune Neurological Disease biorepository, University of Utah – IRB #00001919) that did not have PNMA2 autoantibodies (see Table 2). Using an ELISA coated with recombinant hPNMA2 capsids, we found that patient CSF contains high levels of PNMA2 autoantibodies capable of binding capsids (Figure 4a). PNMA2 specific antibodies in patient CSF also robustly labelled hPNMA2 capsids, as determined by immunogold EM (Figure 4b). Patient CSF PNMA2 autoantibodies preferentially bind to N-terminal “spike” epitopes predicted to be on the exterior surface of hPNMA2 capsids (Figure 4c), similar to autoantibodies produced in capsid-injected mice. These data strongly support the hypothesis that PNMA2 protein elicits an autoantibody response due to the high immunogenicity of PNMA2 virus-like capsids.
Table 1 -.
Patient sample information
| Patient Sample 1, PNMA2 positive in CSF |
| 27-year-old previously healthy male presented with a 2-month history of lethargy and decreased libido. Neurological examination demonstrated normal cognition, with mild dysarthria, bilateral ptosis, slowing of upward saccades, and decreased arm swing. Screening blood work confirmed low testosterone, follicular stimulating hormone, and luteinizing hormone. Brain MRI disclosed an enhancing mass in the tectal region with dilation of the ventricles. Cerebrospinal fluid analyses (CSF) showed pleocytosis (13 cells per mm3; reference <6 cells per mm3) with lymphocytic predominance, elevated protein (58 mg/dL; reference 15–45 mg/dL), and normal glucose. There were no unique oligoclonal bands. CSF infectious studies were negative including herpes simplex virus polymerase chain reaction. CSF cytology and cytometry were negative for malignant cells. No disease-associated autoantibodies were detected on serum or CSF screening (Mayo Clinical Laboratories, “autoimmune encephalitis panel”, and anti-aquaporin4 antibodies). Stereotactic-guided tectal biopsy was performed, with pathology confirming a lymphoplasmacytic inflammatory process. Over the ensuing two months, the patient complained of increasing fatigue / lethargy, with double vision attributed to vertical gaze restriction. Repeat neuroimaging showed mild interval improvement in the tectal mass, with normal sized ventricles, with emergence of bilateral T2-FLAIR signal within the temporal lobes. Atypical paraneoplastic causes were considered, and body imaging was pursued. Body CT demonstrated numerous subcentimeter nonspecific nodes in the abdomen and pelvis. Testicular ultrasound confirmed bilateral testicular microlithiasis. A diagnosis of ma1/ma2 autoimmune encephalitis was considered and confirmed through testing of CSF and serum via Athena Diagnostics. Bilateral orchiectomy was performed given concern of disease-associated tumor. Histopathology was notable for dystrophic calcifications and fibrosis, suggestive of a burnt-out germ cell tumor. High-dose intravenous steroids were provided, with a corresponding brief improvement in alertness and temporal lobe T2-hyperintensities. Long-term follow-up was complicated by treatment-refractory temporal lobe seizures, necessitating escalation of immunotherapy, including monthly intravenous cyclophosphamide. Severe fatigue, lethargy, double vision, and vertical gaze palsy persisted. |
| Patient Sample 2, PNMA1 (Ma1) and PNMA2 (Ma2) positive in CSF |
| 54-year-old female with history of renal cell carcinoma status post-resection presented with a 6-month history of progressive diplopia, gaze paresis, abnormal gait with falls, increasing confusion, and new-onset seizures. On exam, the right eye was deviated downward, adduction was impaired in the left eye, and gait was wide-based and apractic. Brain MRI was markedly abnormal, with symmetric subcortical T2-hyperintensities involving the medial temporal lobes, parietooccipital cortices and medial thalami, concerning for posterior reversible encephalopathy syndrome versus neuroinflammation. CSF analyses disclosed 5 white blood cells (per mm3), normal glucose, and protein, with 9 CSF-specific bands. Noting the past history of renal cancer, body imaging was performed leading to detection of mediastinal and bilateral hilar lymphadenopathy, with bilateral pulmonary nodules and masses and a left adrenal mass. Lymph nodes were FDG-avid on follow-up PET scan. Mediastinal biopsy confirmed metastatic renal carcinoma. Paraneoplastic panel (Mayo Clinic Laboratories) including Ma1/Ma2 testing (Athena Diagnostics) identified Ma1/Ma2 antibodies in blood and CSF. High-dose IV methylprednisone and plasmapheresis (5 exchanges) were provided for treatment of Ma1/Ma2 paraneoplastic encephalitis without improvement in symptoms. The patient developed a diencephalic syndrome with ophthalmoparesis (near plegia) with “sunsetting eyes”, progressive encephalopathy, and cortical blindness. Planned treatment of her underlying malignancy was limited by the development of bilateral pulmonary emboli requiring systemic anticoagulation, and a general decline in physical health. The patient died of complications of her illness. |
| Patient Sample 3, PNMA1 (Ma1) and PNMA2 (Ma2) positive in CSF at > 1:32 |
| 66-year-old male with a history of chronic tobacco use presented with two months of worsening balance, diplopia, and confusion. He had no prior medical conditions. Neurological exam revealed short-term memory deficits, a poor fund of knowledge, ataxia, opsoclonus, restricted upgaze, restriction of right eye abduction, and a right extensor toe response. Brain MRI revealed an encephalitis involving the brainstem, mesial temporal lobes, and basal ganglia. Cerebrospinal fluid (CSF) analysis showed a pleocytosis of 75 nucleated cells per mm3 (reference < 6 nucleated cells per mm3) with a lymphocytic predominance, elevated protein of 75 mg/dL (reference 15–45 mg/dL), and normal glucose. Five unique oligoclonal bands were present. PNMA1 (Ma1) and PNMA (Ma2) were positive (titer > 1:32, Athena Diagnostics). Low positive Anti-NMDA-R antibody was detectable only on a neat, undiluted cell-based assay (Mayo Clinical Laboratories), and serum studies demonstrated low positive anti-GAD65 antibodies at a titer present in up to 8% of the normal population and not generally consistent with symptoms or associated neurologic disease (0.12 nmol/L, reference ≤0.02), with the rest of the serum autoimmune encephalopathy evaluation otherwise negative (Mayo Clinical Laboratories). CSF infectious studies were negative including herpes simplex virus polymerase chain reaction. CSF cytology and cytometry were negative for malignant cells. Malignancy screening included a negative testicular ultrasound and a computed tomography (CT) of the chest that demonstrated extensive pulmonary fibrosis, several prominent mediastinal lymph nodes, and multiple segmental pulmonary emboli. A body positron emission tomography CT demonstrated FDG uptake in a right hilar lymph node in addition to a subpleural, sub-centimeter pulmonary nodule in the left upper lobe, but a progressive deterioration of his respiratory status prevented full investigation. |
Table 2 -.
Control CSF patient information
| Control sample 1: 60-year-old male with 2 years of progressive spasticity (bilateral lower extremities, progressed to left upper extremity). Neurologic exam notable initially for spasticity and upper motor neuron signs, but no lower motor neuron signs. Initial EMG with no abnormal spontaneous activity and MRI brain, C-, T- spine unremarkable. CK mildly elevated at 304. CSF with 2 WBC (ref 0–5), prot 52 (ref 14–45), glc 61 (ref 50–80), OCB neg, <0.0 IgG synthesis rate. Mayo autoimmune encephalopathy evaluation of serum and CSF with no informative autoantibodies detected. Glycine receptor antibody also negative in serum (not tested in CSF). Wash University neuromuscular antibody panel on serum also unremarkable. Ultimately progressed, and EMG findings with upper and lower motor neuron findings; diagnosed with amyotrophic lateral sclerosis. |
| Control sample 2: 54-year-old male followed for postherpetic neuralgia. CAF with WBC 0, prot 38, glc 65, OCB 0, <0.0 IgG synthesis rate. CSF culture negative, HSV1/2 Ab screen IgG <0.34, HSV Type 1 Ab IgG 0.0, HSV 1 and or 2 Abs IgM 0.19, VZV IgG <10, VZV IgM 0.02, VZV PCR not detected, CMV IgM <8, HSV 6 Ab IgM by IFA <1:20. Note- all these results are within normal limits. No send out antibody testing done because no concern for immune-mediated process. |
| Control sample 3: 58-year-old female with diffuse paresthesia and pain, diagnosed with small fiber neuropathy, pernicious anemia and positive GAD65 antibody (without neurologic manifestations). CSF WBC 5, prot 37, glc 53, OCB negative (note: matched bands in serum and CSF), IgG synthesis rate <0.0, Mayo Paraneoplastic Autoantibody Evaluation from CSF positive for GAD65 Ab assay 0.04 nmol/L (ref 0.02), negative for AGNA1, Amphiphysin Ab, ANNA1, ANNA2, ANNA3, CRMP5 IgG, PCA1, PCA2, PCA-Tr. Mayo Autoimmune dysautonomia eval (serum) with AChR Ganglionic Neuronal Ab 0.03 nmol/L (ref <0.02), GAD65 Ab 20.1 nmol/L (ref <0.02), negative for ANNA1, Striational Ab, N-type Calcium Channel, Ach Receptor binding Ab, Neuronal VGKC Ab, P/Q Type Calcium channel. Intrinsic factor blocking antibody positive. GAD65 Ab at ARUP >250 IU/mL (ref 0–5) |
Figure 4. PNMA2 autoantibodies in CSF from human patients with paraneoplastic neurological syndrome preferentially bind to hPNMA2 capsids.
a. An ELISA, using purified hPNMA2 capsids as the antigen, was used to quantify PNMA2 autoantibodies in CSF from patients diagnosed with or without PNMA2-related paraneoplastic neurological syndrome.(n=3 for each group, *Mann-Whitney test, P=0.05).
b. Representative negative-stained EM images of hPNMA2 capsids labeled with immunogold using patient CSF from control or paraneoplastic patients.
c. ELISAs using hPNMA2 spike and capsid shell fragments as antigens show patient CSF PNMA2 autoantibodies preferentially bind to the spike fragment (*One sample t-test, P=0.0356, null hypothesis of 50% binding). Error bars indicate mean ± s.e.m.
Discussion
The discovery that Arc proteins form virus-like capsids intimated that other coopted Gag-derived proteins may also retain the ability to form capsids and that this property may have been repurposed repeatedly during evolution to function in intercellular communication (14–16). Other predicted Gag-like genes, including other PNMA genes, have also been shown to form virus-like particles (14). In this study we found that PNMA2 protein, which is highly expressed in mammalian cortex and hippocampus, forms small virus-like capsids. This indicates that there is a repertoire of mammalian endogenous virus-like capsids that have been evolutionarily re-purposed from retrotransposons (26).
In contrast to Arc release in EVs, we found that PNMA2 is released from cells as non-enveloped capsids. These capsids may be released via an unknown secretory pathway that could occur through non-canonical autophagy or lysosomal pathways (44, 45). Some viruses, such as adenoviruses and adeno-associated viruses (AAVs), are also released without membranes from cells. However, the cell biology and mechanisms of non-enveloped capsid release remain obscure. Since AAVs are used in current gene therapy applications and endogenous capsids may also be harnessed for gene delivery (14), understanding how non-enveloped capsids are released may facilitate gene therapy applications. However, our data caution that ectopic capsid expression may activate the immune system. The structural information we uncovered on PNMA2 capsids may provide a resource to design capsids that mitigate an autoimmune response.
Patients with PNMA2 autoantibodies experience neurological symptoms and signs, including cognitive impairment (5). Since normal expression of these genes is restricted to the brain, we posit that an antibody response may be elicited when tumors release high amounts of PNMA2 capsids outside the CNS. Whether autoantibody production correlates with overall PNMA2 levels or amount of release from tumors remains to be determined. The precise role of immune mechanisms in paraneoplastic neurological syndromes remain poorly characterized. However, prior brain pathologic analysis (autopsy or biopsy) demonstrating perivascular lymphocytic cuffing and interstitial infiltrates of lymphocytes with variable gliosis and neuronal degeneration (7). The inflammatory infiltrates were reported to be comprised mainly of T-cells, with smaller amounts of B-cells, macrophages and microglial activation, with a minority of samples also demonstrating plasma cells in the lymphocytic infiltrates (7). However, we find that PNMA2 autoantibodies in patient CSF can bind PNMA2 capsids, suggesting that these antibodies may also interfere with the putative intercellular signaling function of PNMA2. Capsids injected into mice induce a robust generation of autoantibodies that preferentially bind to external “spike” epitopes on capsids. However, mice injected with capsid disrupting mutant mPNMA2 protein showed no antibody response, highlighting the specific immunogenicity of capsids. Patient CSF PNMA2 autoantibodies also preferentially bind to external “spike” epitopes on PNMA2 capsids. We conclude that high levels of PNMA2 capsids found outside the CNS may induce a robust autoimmune response that may trigger a paraneoplastic neurological syndrome.
While PNMA2 is primarily and highly expressed in the mammalian brain, with conserved expression pattern in primates, the biological function of PNMA2 remains unknown. We have shown that PNMA2 protein forms virus-like capsids that are released from neurons and we speculate that these virus-like properties are important for PNMA2 function in the brain. Future studies will determine the function of PNMA2 capsids and whether other PNMA proteins form virus-like capsids that are released as non-enveloped particles. Our findings may help determine therapies for paraneoplastic neurological syndromes and more generally help reveal mechanisms of capsid-induced immune activation.
Methods
Plasmids
The coding sequence (CDS) of mPNMA2, hPNMA2, myc-mPNMA2 1-170aa, myc-mPNMA2 171-365aa, myc-hPNMA2 1-170aa, and myc-hPNMA2 171-364aa, mPNMA2 L270QL325Q were synthesized by Integrated DNA Technologies (Coralville, IA). These fragments were digested by BamHI and XhoI restriction enzymes (New England BioLabs, Ipswich, MA), and ligated into pGEX-6p1 vector for protein expression and purification. The mPNMA2 CDS was amplified by PCR, digested by SalI and NotI, and cloned into the PRK5-myc vector. Similarly, pLVX-myc-mPNMA2, pLVX-mPNMA2, pLVX-mPNMA2 L270Q/L325Q, and pLVX-mPNMA2 Y162A were constructed by PCR amplification of mPNMA2 WT and L270QL325Q CDS, BamHI and XhoI digestion, and ligation into pLVX backbone between BamHI and XhoI sites. All ligation products were transformed into NEB Stable Competent E. coli (High Efficiency) (New England Biolabs, Ipswich, MA). Individual colonies were inoculated to isolate plasmids and screen for correct constructs by sequencing.
Protein purification
Bacterial protein expression plasmids were transformed into E. coli Rosetta2 strain (Novagen, Gibbstown, NJ) and the protein was expressed in ZY autoinduction media. E. coli were grown to 0.6–0.8 of OD600 at 37°C 150rpm, then switched to 19°C for 16–20h shaking. Afterwards, bacteria were pelleted down at 5000g for 15min at 4°C, resuspended in lysis buffer (500mM NaCl, 50mM Tris, 5% glycerol, 1mM DTT, complete Protease Inhibitor Cocktail, pH 8.0). The resuspended bacteria were frozen by liquid nitrogen and then thawed. The thawed lysates were sonicated and centrifuged at 21,000g for 45mins at 4°C. The supernatant was filtered through 0.45μm filters and incubated with Sepharose 4B beads (Cytiva, Marlborough, MA) overnight at 4°C. GST-tagged proteins were eluted from Sepharose beads by incubating with L-reduced glutathione (20mM, pH 8.0) and exchanged to cleavage buffer (50mM Tris, 150mM NaCl, 1mM DTT, 1mM EDTA, pH 7.2) using a Vivaspin column (Cytiva, Marlborough, MA). The cleavage of GST was performed by PreScission Protease (Cytiva, Marlborough, MA) overnight at 4°C. The cleaved PNMA2 protein was run through a HiLoad 16/600 Superdex 200 pg (Cytiva, Marlborough, MA) size-exclusion column in the capsid assembly buffer (500mM phosphate, 50mM Tris, 0.5mM EDTA, pH 7.5) to further purify PNMA2 and facilitate capsid formation. For protein purified for mice immunization, Triton X-114 was used to wash GST-tagged protein bound to Sepharose beads to remove endotoxin.
Negative-staining electron microscopy
Copper 200-mesh grids coated with formvar and carbon (Electron Microscopy Sciences, Hatfield, PA) were discharged for 30s in a vacuum chamber. Grids were incubated with 3.5μL purified protein (0.5mg/mL) for 45s and excess sample was wicked away. Grids were then immediately washed twice with 30uL water for 5 s and once with 15μL 1% uranyl acetate (UA) for 5s. Excess UA was wicked away and the grids were stained with 15μL 1% UA for 30s. Excess UA was wicked away and grids were air-dried. The grids were imaged using a FEI Tecnai T12 Transmission Electron Microscope operated at 120kV.
Cryo-electron microscopy
4 μl of mPNMA2 capsids at a concentration of ~1 mg/ml were applied to glow-discharged QUANTIFOIL 2/2 Au continuous carbon grids. The sample was blotted and plunge-frozen in liquid ethane using a Vitrobot Mark IV. Cryo-EM images were acquired on a 300 keV FEI Titan Krios microscope with X-FEG emitter equipped with a Falcon III detector operated in counting mode. mPNMA2 datasets were collected using EPU (ThermoFisher) at a nominal magnification of 96000x with a calibrated pixel sizes of 0.832 Å. Micrographs were recorded as movies divided into 75 frames with a final accumulated dose of 40 e/Å2. Defocus values ranged from −0.5 to −2.5 μm.
Image processing
mPMNA2 image processing and reconstructions are summarized in Supplemental Figure S5 and S6 and Supplemental Table S1. Acquired movies were aligned using 5×5 patches, averaged and dose-weighted using RELION4 (46). Contrast transfer function (CTF) parameters were estimated using CTFFIND4 (47). Particles were automatically picked using a retrained topaz model (48). Boxsize used for extraction was 512×512 pixels. Extracted particles were subjected to several rounds of 2D classification to remove false picks and junk particles. 2D classes showed views characteristic for icosahedral symmetry, and icosahedral symmetry (I) was applied for initial model generation and throughout subsequent 3D classification and refinement. Finally, we performed per-particle CTF estimation, Bayesian polishing and Ewald sphere correction. The effective resolutions of the cryo-EM density maps were estimated by Fourier shell correlation (FSC = 0.143) according to the definition of Rosenthal and Henderson.
To further improve the resolution, we performed symmetry expansion as implemented in RELION4, to calculate the positions and orientations for each of the asymmetric units centered either at the five, three or two-fold symmetric axes. We extracted individual capsomeres using box sizes of 220×220 pixels. We performed 3D classification without alignment to remove bad views and to select classes for subsequent refinement. For all maps, local resolutions were calculated using ResMap (49) and maps were locally sharpened using Phenix (50).
Model building
For model building we used Alphafold2 (51) to generate an initial model. After initial rough fitting using chimera (52) , we went through several rounds of manual and auto-refinement in coot, Phenix, and ISOLDE (53, 54). Model statistics are shown in Supplemental Table S1.
Mass photometry
Mass photometry was performed on a Refeyn TwoMP mass photometer. Protein stocks were brought to room temperature and diluted with stock buffer (500mM Na2HPO4, 50mM Tris, 10% glycerol, pH 7.5) to a concentration of 100nM. A 20 l droplet of each sample dilution was placed on top of a clean chambered-coverslip mounted on the oil objective of the instrument. Following autofocus stabilization, a one-minute movie was recorded using the Aquire-MP (Refeyn) software. A protein standard containing equimolar amounts of four proteins (conalbumin, aldolase, ferritin and thyroglobulin) with a molecular mass ranging from 75 to 669 kDa was also measured and used for calibration. Movies were converted to mass using a contrast-to-mass calibration and analyzed using the Discover-MP software (Refeyn).
Primary cortical/hippocampal neuronal culture
Primary neuron cultures were prepared from male and female E18 WT mouse cortex and hippocampus as previously described (55). Tissue was dissociated in DNase (0.01%; Sigma-Aldrich, St. Louis, MO) and papain (0.067%; Worthington Biochemicals, Lakewood, NJ), and then triturated with a fire-polished glass pipette to obtain a single-cell suspension. Cells were pelleted at 500xg for 4 min, the supernatant removed, and cells resuspended and counted with a TC-20 cell counter (Bio-Rad, Hercules, CA). Neurons were plated on glass coverslips (Carolina Biological Supply, Burlington, NC) coated with poly-l-lysine (0.2 mg/mL: Sigma-Aldrich, St. Louis, MO) in 12-well plates (Greiner Bio-One, Monroe, NC) at 90,000 cells/mL, or in 10-cm plastic dishes at 800,000 cells/mL. Neurons were initially plated in Neurobasal media containing 5% serum, 2% GlutaMAX, 2% B-27, and 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA) in a 37°C incubator with 5% CO2. On DIV4, neurons were fed via half media exchange with astrocyte-conditioned BrainPhys™ Neuronal Medium (Stemcell Technologies, Vancouver, Canada) containing 1% serum (Thermo Fisher Scientific, Waltham, MA), 0.25% L-Glutamine (Thermo Fisher Scientific, Waltham, MA), 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA), 2% SM1 (Stemcell Technologies, Vancouver, Canada), and 5 μM cytosine β-d-arabinofuranoside (AraC) (Sigma-Aldrich, St. Louis, MO). Half media exchange of astrocyte-conditioned media was conducted every three days thereafter.
HEK 293T cell culture
HEK 293T cells (#CRL-11268) were purchased from ATCC. Cells were cultured at 37°C with 5% CO2 in media including DMEM (Thermo Fisher Scientific, Waltham, MA), 5% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA) and 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA). Cell cultures were passaged at 70% confluency.
NCI-H378 cell culture
NCI-H378 cells (#CRL-5808) were purchased from ATCC. Cells were cultured at 37°C with 5% CO2 in media including RPMI-1640 (ATCC, Manassas, VA), 5% fetal bovine serum (ATCC, Manassas, VA), and 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA).
HEK 293T cells transfection
10 cm dish transfection: 9μg of plasmids and 25μL 1μg/μL Polyethylenimine (Polysciences, Warrington, PA) were mixed in 1mL Opti-MEM (Thermo Fisher Scientific, Waltham, MA) and incubated at room temperature for 20 minutes. This mixture was added to 60% confluent cell culture in transfection media (DMEM+10% FBS) and incubated overnight.
LDH cytotoxicity assay
The amount of LDH in the cell lysates and media was measured using CyQUANT LDH Cytotoxicity Assay kit (Thermo Fisher Scientific, Waltham, MA). The percentage of cell death was calculated by the total amount of LDH in the media divided by that in the cell lysates.
Lentivirus production and transduction
HEK293 cells in a 10 cm dish were transfected with pLVX-VSVG, pLVX-psPAX 2, and pLVX-GFP or pLVX-myc-mPNMA2 overnight. Transfection media was changed to regular culture media (DMEM+10% FBS+ 1% penicillin/streptomycin). The media was collected 24 hours later and centrifuged at 1,000g for 10min. 20mM HEPES was added to the supernatant and centrifuged at 5,000g overnight. Lentivirus particles were pelleted and resuspended in 400 μL PBS. Cultured neurons in a 10 cm dish were transduced with 200 μL lentivirus particles at DIV13 for 2 days.
Mice immunization
10–12 week-old mice (C57BL/6) were used for immunization. Mice were injected intraperitoneally with vehicle (capsid assembly buffer), 5μg of mPNMA2 capsids, or 5μg of mPNMA2 L270QL325Q capsid mutant on day 0. The immunized proteins were not mixed with any adjuvant. Blood was collected on day 21 via the tail vein. A second injection of 5μg of mPNMA2 L270QL325Q capsid mutant and vehicle were performed on day 21 and blood was collected on day 42. Blood was coagulated at 4°C overnight and serum was harvested by centrifuging twice at 2,000g for 10min to collect the supernatant. Endotoxin was removed from the purified protein by Triton X-114 wash. Preparations were checked for the presence of capsids via negative stain EM, prior to injection.
Extracellular vesicle and capsid purification from culture media
For HEK 293T cells, full media change was performed 12 hours after transfection and then collected 24 hours later. For neuronal culture, full media change was performed at DIV15 (2 days after lentivirus transduction) and then collected 24 hours later. Collected media was centrifuged at 2,000g for 10min and then 20,000g for 20min. The supernatant of the collected media was concentrated into 0.5mL using Vivaspin columns. Concentrated media was loaded onto IZON mini-size exclusion columns (IZON Sciences, Christchurch, New Zealand). The first 3mL run through the column was collected as the void fraction. Then, every 0.5mL was collected for a total of 11 fractions, labeled as fraction 1 to 11.
Proteinase K protection assay
Cultured media was processed using IZON columns as above and treated under three different conditions: 1. 20μL sample + 7μL H2O. 2. 20μL sample + 6μL H2O + 1μL proteinase K (200μg/mL) (New England BioLabs, Ipswich, MA). 3. 20μL particle sample + 3μL H2O + 3μL 10% Triton X-100 + 1μL proteinase K (200μg/mL). Media was incubated at room temperature for 10 min. Then, 3μL of 10mM PMSF was added and incubated for 10min at room temperature. Finally, samples were mixed with laemmli buffer (40% glycerol, 250 mM Tris, 4% SDS, 50 mM DTT, pH 6.8) and boiled at 95°C for 5 min.
Western blot
Samples were mixed with 4x laemlli buffer and incubated at 95°C for 5 mins. Proteins were loaded and separated by 10% or 12% SDS-PAGE gel, followed by wet transfer to a nitrocellulose membrane (GE Healthcare, Chicago, IL). Total protein was stained and destained by Pierce Reversible Protein Stain Kit (Thermo Fisher Scientific, Waltham, MA). The membrane was blocked by 5% milk in TBS for 1h at room temperature. Primary antibodies were diluted in 1% milk in TBS and incubated with the membrane at 4°C overnight. Primary antibodies include: anti-myc (05–724, Sigma-Aldrich, St. Louis, MO; 1:1000), anti-PNMA2 (16445-1-AP, Proteintech, Rosemont, Illinois; 1:1000), anti-Alix (customized antibody from Wesley Sundquist’s lab; 1:500), mice sera (1:1000). The membrane was washed by TBS and then incubated with secondary antibodies (anti-mouse IgG, anti-rat IgG, anti-human IgG, Jackson Laboratory, Bar Harbor, ME; 1:5000) diluted by 1% milk in TBS for 1h at room temperature. Afterwards, the membrane was washed with TBS for three times. Bound antibodies were detected by Clarity™ Western ECL Substrate (Bio-Rad, Hercules, CA) and imaged using an Amersham ImageQuant™ 800 Western blot imaging systems (Cytiva, Marlborough, MA). Images were analyzed and quantified using ImageJ.
RNA-Scope
Brian tissues obtained from 2-month-old mice and primary hippocampal neuronal cultures (DIV15) were used for RNAscope. RNAscope multiplex fluorescent v2 assay kit and the probe were purchased from Advanced Cell Diagnostics (Hayward, CA). The probe was tagged to opal 570 dye from Akoya Biosciences (Marlborough, MA).
Confocal microscope imaging
Coverslips were imaged using a 10x objective for brain tissues and 60x oil objective for cultured neurons on a Nikon FV1000 confocal microscope (Tokyo, Japan). The images were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD).
Iodixanol gradient ultracentrifugation
An Iodixanol gradient was made with 2mL 15% (1M NaCl), 3mL 30%, 3mL 40% and 3mL 50% OptiPrep™ iodixanol (Alere Technologies AS, Oslo, Norway). 500μL of the sample was added to the top of the gradient and centrifuged at 280,000 rpm for 48 hours. After ultracentrifugation, sequential 1mL collections were made from the top of the gradient and labeled as fractions 1 to 12.
Immunogold EM
Copper 200-mesh grids coated with formvar and carbon (Electron Microscopy Sciences, Hatfield, PA) were discharged for 30s in a vacuum chamber. PNMA2 purified protein (0.1mg/mL, 3.5μL) was loaded onto the grid and incubated for 45s. Grids were washed twice with 30uL diH2O for 30s and blocked with 30uL 1% BSA for 30mins. Grids were then incubated with sera/CSF/primary antibody in 30uL 1% BSA for 1h at room temperature and then washed three times with 30uL 1% BSA for 30s. Grids were then incubated with 6 nm immunogold conjugated secondary antibody, including Goat-anti-Human IgG (H&L) and Goat-anti-Mouse IgG (H&L) (Electron Microscopy Sciences, Hatfield, PA) (1:20 dilution in 1% BSA, 30uL) for 1h at room temperature. Afterwards, grids were washed three times with 30uL PBS for 30s and fixed with 30uL 2% glutaraldehyde for 5mins. Finally, grids were washed with 30uL diH20 for three times and stained with 30uL 1% uranyl acid for 1 min. The grids were imaged using a FEI Tecnai T12 Transmission Electron Microscope operated at 120kV.
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISAs were used to determine the titer and binding affinity of antibodies in human CSF and mice sera to different antigens. Maxisorp plates (Thermo Fisher Scientific, Waltham, MA) were coated overnight with 2μg/mL of purified PNMA2 capsids or PNMA 2 L270QL325Q protein in assembly buffer at room temperature. PNMA2 fragments were coated in PBS at 4°C. After coating, plates were washed with wash buffer (Thermo Fisher Scientific, Waltham, MA) 5 times and then blocked with 5% Bovine Serum Albumin (BSA; Sigma-Aldrich, St. Louis, MO) in PBS for 2 hours at 37°C. CSF and sera were diluted serially in 1% BSA and incubated with the plates for 2 hours at 37°C. After washing, plates were incubated with anti-mouse IgG or anti-human IgG (1:5000 in 1% BSA; Jackson Laboratory, Bar Harbor, ME) for 1 hour at 37°C. Afterwards, plates were washed and incubated with 3,3’,5,5’-Tetramethylbenzidine (TMB) substrate (Thermo Fisher Scientific, Waltham, MA) for 30 mins at 37°C. The enzyme reaction was stopped by adding the stop buffer (Thermo Fisher Scientific, Waltham, MA) and the absorbance was measured at 450nm.
Quantification and statistical analysis
Data were analyzed using GraphPad Prism. Mann-Whitney test, paired t-test, or two-tailed t-test were used for the analysis. Not significant (ns) p>0.05; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
RNA-seq data analysis
We obtained the processed single-cell expression matrix (counts) from Allen’s brain map (https://portal.brain-map.org) representing the human cortex and mouse cortex-hippocampus. We then used Seurat (v3.1.1) (https://github.com/satija.lab/seurat) within the R environment (v3.6.0) for processing the dataset. We kept the cells with minimum and maximum of 1,000 and 5,000 genes expressed (≥1 count), respectively. Moreover, cells with more than 5% of counts on mitochondrial genes were filtered out. The data normalization was achieved by scaling it with a factor of 10,000, followed by natural-log transformation. Cell type assignment was performed based on the annotations provided by the original publication, albeit we grouped the clusters into broader lineages of excitatory, inhibitory, Oligodendrocyte precursor cells (OPC), Oligodendrocytes and Glial cells (Astrocytes and Microglia). All the given annotations were further confirmed by their respective markers. The expression levels of each gene in a cluster correspond to the average log2 expression level scaled to the number of unique molecular identification (UMI) values captured in single cells. Finally, the sample replicates for each gene were aggregated per cell type, and their expression was calculated as trimmed means. For the analysis of Human, Chimpanzees and Macaque, we downloaded the human brain transcriptome from 33 major regions representing four adult healthy human individuals, three chimpanzees, and three rhesus macaques (GSE 127898). Trimmed mean of M (TMM) values normalized transcripts per million (TPM) values were imported in R, and variable features were identified when the ratio of variance and mean expression values were greater than one.
Supplementary Material
Acknowledgments
We thank Wesley Sundquist and John McCullough for helpful discussions and experimental support. We thank Tammy Smith for helping provide human CSF samples. We thank Jenifer Einstein and Michael Hantak for generating primary cultured neurons, and all members of the Shepherd lab for technical help and support.
Funding
S.E. – The Novo Nordisk Foundation (NNF17OC0030788). C.F. – NIH R35 GM122550. JAGB – Medical Research Council; Max Planck Institute. J.D.S – Chan-Zuckerberg Initiative Ben Barres Early Acceleration Award; NIH NINDS (R01 NS115716).
Footnotes
Competing interests
C.F. is a consultant for Tessera Therapeutics, Inc. and HAYA Therapeutics, Inc. J.D.S is a co-founder of VNV, LLC and a consultant for Aera Therapeutics, Inc.
Code availability
All statistical and plotting details, including the statistical tests, used, and precision measures for RNA-seq data analysis can be found on the GitHub repository at https://github.com/Manu-1512/MamGyp-int-rides-PNMA2
References
- 1.Rosenfeld M. R., Eichen J. G., Wade D. F., Posner J. B., Dalmau J., Molecular and clinical diversity in paraneoplastic immunity to Ma proteins. Annals of Neurology: Official Journal of the American Neurological Association and the Child Neurology Society 50, 339–348 (2001). [PubMed] [Google Scholar]
- 2.Dalmau J. et al. , Ma1, a novel neuron-and testis-specific protein, is recognized by the serum of patients with paraneoplastic neurological disorders. Brain 122, 27–39 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Voltz R. et al. , A Serologic Marker of Paraneoplastic Limbic and Brain-Stem Encephalitis in Patients with Testicular Cancer. New England Journal of Medicine 340, 1788–1795 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Takaji M., Komatsu Y., Watakabe A., Hashikawa T., Yamamori T., Paraneoplastic antigen-like 5 gene (PNMA5) is preferentially expressed in the association areas in a primate specific manner. Cerebral Cortex 19, 2865–2879 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schüller M., Jenne D., Voltz R., The human PNMA family: novel neuronal proteins implicated in paraneoplastic neurological disease. Journal of neuroimmunology 169, 172–176 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann L. et al. , Anti-Ma and anti-Ta associated paraneoplastic neurological syndromes: 22 newly diagnosed patients and review of previous cases. Journal of Neurology, Neurosurgery & Psychiatry 79, 767–773 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Dalmau J. et al. , Clinical analysis of anti-Ma2-associated encephalitis. Brain 127, 1831–1844 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Kidd M., Modlin I. M., Drozdov I., Gene network-based analysis identifies two potential subtypes of small intestinal neuroendocrine tumors. BMC genomics 15, 1–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Da Gama Duarte J. et al. , Identification of Tumor Antigens in Ovarian Cancers Using Local and Circulating Tumor-Specific Antibodies. International journal of molecular sciences 22, 11220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung M. J. et al. , A differential gene expression profiles by cDNA microarrays in endometrioid endometrial carcinoma: a preliminary study. Korean Journal of Gynecologic Oncology 18, 219–226 (2007). [Google Scholar]
- 11.Chapman M. H. et al. , Whole genome RNA expression profiling of endoscopic biliary brushings provides data suitable for biomarker discovery in cholangiocarcinoma. Journal of hepatology 56, 877–885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui T. et al. , Paraneoplastic antigen Ma2 autoantibodies as specific blood biomarkers for detection of early recurrence of small intestine neuroendocrine tumors. PLoS One 5, e16010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campillos M., Doerks T., Shah P. K., Bork P., Computational characterization of multiple Gag-like human proteins. Trends in Genetics 22, 585–589 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Segel M. et al. , Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 373, 882–889 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashley J. et al. , Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 172, 262–274. e211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastuzyn E. D. et al. , The neuronal gene arc encodes a repurposed retrotransposon gag protein that mediates intercellular RNA transfer. Cell 172, 275–288. e218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch C., Tristem M., A co-opted gypsy-type LTR-retrotransposon is conserved in the genomes of humans, sheep, mice, and rats. Current biology 13, 1518–1523 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Erlendsson S. et al. , Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins. Nature neuroscience 23, 172–175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepherd J. D., Bear M. F., New views of Arc, a master regulator of synaptic plasticity. Nature neuroscience 14, 279–284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abrusán G., Szilágyi A., Zhang Y., Papp B., Turning gold into ‘junk’: transposable elements utilize central proteins of cellular networks. Nucleic acids research 41, 3190–3200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cottee M. A., Letham S. C., Young G. R., Stoye J. P., Taylor I. A., Structure of Drosophila melanogaster ARC1 reveals a repurposed molecule with characteristics of retroviral Gag. Science advances 6, eaay6354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abed M. et al. , The Gag protein PEG10 binds to RNA and regulates trophoblast stem cell lineage specification. PLoS One 14, e0214110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandya N. J. et al. , Secreted retrovirus-like GAG-domain-containing protein PEG10 is regulated by UBE3A and is involved in Angelman syndrome pathophysiology. Cell Reports Medicine 2, 100360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zurowska K., Alam A., Ganser-Pornillos B. K., Pornillos O., Structural evidence that MOAP1 and PEG10 are derived from retrovirus/retrotransposon Gag proteins. Proteins: Structure, Function, and Bioinformatics, (2021). [Google Scholar]
- 25.Ono R. et al. , A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Hantak M. P., Einstein J., Kearns R. B., Shepherd J. D., Intercellular communication in the nervous system goes viral. Trends in Neurosciences 44, 248–259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonsdale J. et al. , The genotype-tissue expression (GTEx) project. Nature genetics 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A. I. f. B. Science. (Allen Hum. Brain Atlas In, 2010). [Google Scholar]
- 29.A. I. f. B. Science, Allen Brain Atlas: Mouse Brain. (2017).
- 30.Hodge R. D. et al. , Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeisel A. et al. , Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Khrameeva E. et al. , Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. Genome research 30, 776–789 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dodonova S. O., Prinz S., Bilanchone V., Sandmeyer S., Briggs J. A., Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses. Proceedings of the National Academy of Sciences 116, 10048–10057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tlemsani C. et al. , SCLC-CellMiner: a resource for small cell lung cancer cell line genomics and pharmacology based on genomic signatures. Cell reports 33, 108296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yufenyuy E. L., Aiken C., The NTD-CTD intersubunit interface plays a critical role in assembly and stabilization of the HIV-1 capsid. Retrovirology 10, 1–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bleker S., Sonntag F., J. r. A. Kleinschmidt, Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. Journal of virology 79, 2528–2540 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang G., Cho M., Wang X., OncoDB: an interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Research 50, D1334–D1339 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachmann M. F., Jennings G. T., Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nature Reviews Immunology 10, 787–796 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Gause K. T. et al. , Immunological principles guiding the rational design of particles for vaccine delivery. ACS nano 11, 54–68 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Bachmann M. F. et al. , The influence of antigen organization on B cell responsiveness. Science 262, 1448–1451 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Jegerlehner A. et al. , Regulation of IgG antibody responses by epitope density and CD21-mediated costimulation. European journal of immunology 32, 3305–3314 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Murin C. D., Wilson I. A., Ward A. B., Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nature microbiology 4, 734–747 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arias C. F., DuBois R. M., The astrovirus capsid: a review. Viruses 9, 15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponpuak M. et al. , Secretory autophagy. Current opinion in cell biology 35, 106–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blott E. J., Griffiths G. M., Secretory lysosomes. Nature reviews Molecular cell biology 3, 122–131 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Kimanius D., Dong L., Sharov G., Nakane T., Scheres S. H., New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochemical Journal 478, 4169–4185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of structural biology 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bepler T., Kelley K., Noble A. J., Berger B., Topaz-Denoise: general deep denoising models for cryoEM and cryoET. Nature communications 11, 5208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nature methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liebschner D. et al. , Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallographica Section D: Structural Biology 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jumper J. et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen E. F. et al. , UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Emsley P., Cowtan K., Coot: model-building tools for molecular graphics. Acta crystallographica section D: biological crystallography 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Croll T. I., ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallographica Section D: Structural Biology 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shepherd J. D. et al. , Arc/Arg3. 1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52, 475–484 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.