Abstract
Importance
Undetected biological heterogeneity adversely impacts trials in Alzheimer’s disease because rate of cognitive decline — and perhaps response to treatment — differs in subgroups. Recent results show that data-driven approaches can unravel the heterogeneity of Alzheimer’s disease progression. The resulting stratification is yet to be leveraged in clinical trials.
Objective
Investigate whether image-based data-driven disease progression modelling could identify baseline biological heterogeneity in a clinical trial, and whether these subgroups have prognostic or predictive value.
Design
Screening data from the Anti-Amyloid Treatment in Asymptomatic Alzheimer Disease (A4) Study collected between April 2014 and December 2017, and longitudinal data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) observational study downloaded in February 2022 were used.
Setting
The A4 Study is an interventional trial involving 67 sites in the US, Canada, Australia, and Japan. ADNI is a multi-center observational study in North America.
Participants
Cognitively unimpaired amyloid-positive participants with a 3-Tesla T1-weighted MRI scan. Amyloid positivity was determined using florbetapir PET imaging (in A4) and CSF Aβ(1-42) (in ADNI).
Main Outcomes and Measures
Regional volumes estimated from MRI scans were used as input to the Subtype and Stage Inference (SuStaIn) algorithm. Outcomes included cognitive test scores and SUVr values from florbetapir and flortaucipir PET.
Results
We included 1,240 Aβ+ participants (and 407 Aβ− controls) from the A4 Study, and 731 A4-eligible ADNI participants. SuStaIn identified three neurodegeneration subtypes — Typical, Cortical, Subcortical — comprising 523 (42%) individuals. The remainder are designated subtype zero (insufficient atrophy). Baseline PACC scores (A4 primary outcome) were significantly worse in the Cortical subtype (median = −1.27, IQR=[−3.34,0.83]) relative to both subtype zero (median=−0.013, IQR=[−1.85,1.67], P<.0001) and the Subcortical subtype (median=0.03, IQR=[−1.78,1.61], P=.0006). In ADNI, over a four-year period (comparable to A4), greater cognitive decline in the mPACC was observed in both the Typical (−0.23/yr; 95% CI, [−0.41,−0.05]; P=.01) and Cortical (−0.24/yr; [−0.42,−0.06]; P=.009) subtypes, as well as the CDR-SB (Typical: +0.09/yr, [0.06,0.12], P<.0001; and Cortical: +0.07/yr, [0.04,0.10], P<.0001).
Conclusions and Relevance
In a large secondary prevention trial, our image-based model detected neurodegenerative heterogeneity predictive of cognitive heterogeneity. We argue that such a model is a valuable tool to be considered in future trial design to control for previously undetected variance.
Introduction
There is increasing evidence of biological heterogeneity to accompany the observed clinical heterogeneity in Alzheimer’s disease (AD), highlighting a range of potential disease subtypes and disease trajectories1-6. This heterogeneity reflects the inherent complexity of the disease and may have contributed to the high failure rate of clinical trials7, where study cohorts likely contain individuals potentially belonging not only to different subtypes, but also being at different stages of biological severity (even within a clinical/biomarker-based group), resulting in heterogeneous rates of progression independent of treatment effect8,9. This compounds the already difficult task of identifying treatment effects in AD clinical trials, where neuropsychological test scores or single biomarkers (e.g. amyloid PET SUVr) are used as outcomes10,11. Nearly half of currently ongoing Phase 3 AD trials (for disease-modifying therapies) did not use biomarkers as inclusion criteria, relying instead on neuropsychological tests10. Although some composite neuropsychological tests are sensitive to relatively early, subtle changes in cognitive function12, underlying pathology in the preclinical phase cannot be fully characterized without biomarkers12,13.
Recognizing this limitation in trial designs, biomarkers are increasingly used in screening protocols10,11,14,15, yet still incur high failure rates likely due to the limited temporal resolution of a single biomarker, and the presence of undetected subtypes having differing cognitive trajectories/prognoses4,16. Data-driven methods such as disease progression modelling (DPM) have previously characterized heterogeneity (both in variation of severity and the presence of subtypes) typically using image-based input features5,17. The ability to utilize high-dimensional medical imaging data enables deeper comparisons between individuals beyond univariate cut-offs, providing utility in assessing heterogeneity across a cohort. In this study, we use DPM to identify undetected MRI-based heterogeneity at screening in the Anti-Amyloid Treatment in Asymptomatic AD (A4) Study18, analyze these subtypes for differences in trial outcome measures (at baseline), and investigate differences across subtypes in longitudinal outcomes in the Alzheimer’s Disease Neuroimaging Initiative (ADNI)19 observational study to characterize the potential impact such heterogeneity may have on the A4 interventional trial outcomes.
Methods
Our primary aim is to test whether biological heterogeneity is predictive of cognitive decline in the carefully selected A4 Study clinical trial cohort. We train a subtyping DPM on imaging (MRI) data, then compare cognitive outcomes across subtypes cross-sectionally (in A4) and longitudinally (in ADNI).
Participants
Data comes from cognitively unimpaired participants in the A4 Study interventional trial (NCT02008357) and the ADNI observational study (eMethods 1 in supplementary material). The A4 Study is a prevention trial investigating whether solanezumab reduces cognitive decline in an Aβ+ preclinical cohort14,20, with the primary outcome being the change from baseline of the Preclinical Alzheimer Cognitive Composite (PACC)21. Secondary outcomes include changes from baseline in the Clinical Dementia Rating Sum of Boxes (CDR-SB) score and the Cognitive Function Index (CFI) total score (combining participant and study partner scores)22. ADNI is an observational study investigating biomarker and clinical changes in individuals diagnosed with, or at-risk of, AD.
The A4 Study selected individuals using the following criteria14: Mini-Mental State Examination (MMSE) score of 25-30; Clinical Dementia Rating (CDR) global score of 0; Logical Memory Delayed Recall (LMDR-IIa) score of 6-18; and, amyloid positivity (Aβ+). Amyloid positivity was defined following florbetapir PET imaging, using a mean cortical standardized uptake value ratio (SUVr) with a whole cerebellar reference region of ≥1.15 (or ≥1.10 on a positive visual read with a 2-reader consensus). To maximize data availability in ADNI, CSF Aβ(1-42) was instead used to determine amyloid positivity using a cut-off of 977pg/mL23. Cognitively unimpaired individuals in ADNI were selected using the same inclusion criteria as the A4 Study. ADNI data was downloaded from the LONI Image & Data Archive on 27th February 2022.
Control subjects for our study were Aβ− and APOE ε4 non-carriers from the A4 Study. These controls (N=407) were used for covariate adjustment and as a normative reference when analyzing both A4 and ADNI cohorts (see below).
Data Pre-Processing
All available 3-Tesla (3T) T1-weighted MRI were obtained for both the A4 and ADNI cohorts. Images from ADNI were selected having passed overall quality control. All images were processed using FreeSurfer v7.1.124 within a publicly available containerized pipeline (https://github.com/e-dads/freesurfer), with cortical and subcortical volumes combined into bilateral averages in 13 regions of interest25. Original image acquisition protocols for both studies can be found in their respective documentation.
Prior to input into our DPM, FreeSurfer volumes were adjusted for healthy/normal linear trends (in controls) in age, years of education, sex, and intracranial volume, estimated using a general linear model26. The covariate-adjusted volumes were transformed into z-scores relative to controls.
The amyloid (florbetapir) and tau (flortaucipir) PET images acquired in the A4 Study were analyzed to investigate differences across subtypes. For this analysis, we used a newly-developed multi-platform software AmyPET (https://github.com/AMYPAD/AmyPET), extending NiftyPET27. AmyPET enables robust quantification of PET scans while using SPM12 for core MR-PET image registration28, estimating amyloid/tau load with high quantitative accuracy and precision from raw count PET data to accurately account for common head motion. Protocol details for amyloid and tau PET acquisition are provided in eMethods 2 (supplementary material).
Disease Progression Modelling
To characterize biological heterogeneity in the A4 Study cohort, we used the Subtype and Stage Inference (SuStaIn) algorithm5 as our DPM. This approach combines clustering with event-based modelling17,29 to reconstruct one (or more) sequences of disease progression from cross-sectional data, simultaneously disentangling pathological and temporal heterogeneity to identify both disease subtype(s) and severity (i.e. stage) across a cohort. SuStaIn has been applied to identify image-based subtypes and spreading patterns in AD5,16,30, other forms of dementia such as frontotemporal dementia31, and other neurodegenerative diseases such as multiple sclerosis32.
Disease “events” (transitions of a regional volume to abnormality) are defined by reaching progressive z-scores of 1 and 2 (relative to controls), from which SuStaIn determines the event sequence that maximizes the data likelihood. SuStaIn then iteratively determines multiple such sequences, referred to here as disease subtypes, that maximize the likelihood for subsets of the data from one subtype up to a pre-defined maximum (set as Nmax = 4), with the optimal number of subtypes selected using cross-validation (described below). Markov chain Monte Carlo (MCMC) sampling is used to estimate uncertainty in the sequence(s).
SuStaIn was trained using the 1,240 Aβ+ participants with complete MRI data from the A4 Study. This produced a model for each number of subtypes investigated. To select a single model, we performed 10-fold cross-validation and calculated the cross-validation information criterion (CVIC)13, which balances minimizing model complexity (i.e. fewer subtypes) and maximizing how well the model fits the data. The model with the minimal CVIC is selected, and then used to assign both a maximum-likelihood subtype and stage to each participant (for both A4 and ADNI cohorts)5,17.
Statistical Analyses
The original A4 Study analysis14 found significant differences between the Aβ+ and Aβ− groups at baseline for both the PACC and CFI scores, with the Aβ+ group performing worse on both scores. We performed a similar analysis stratified by DPM subtype assignment, using Pearson‘s X2 (adjusted for multiple comparisons using Holm-Bonferroni correction). This analysis was repeated for the ADNI subset to compare differences between the subtypes, and between the A4 and ADNI cohorts to identify any systematic differences in demographic and cognitive variables common to both studies.
Associations between DPM subtypes and cognitive outcomes are assessed for (pairwise) differences using a two-tailed Mann-Whitney U test (with Holm-Bonferroni correction). This was performed cross-sectionally in A4 using the PACC and CFI (total) scores to potentially identify undetected baseline cognitive heterogeneity revealed by the DPM.
We then analyzed longitudinal cognitive trajectories in ADNI for two similar trial outcomes available in ADNI: a modified version of the PACC (mPACC), which uses the (log-transformed) Trail-Making Test B to replace the unavailable Digit Symbol Substitution test33, and the CDR-SB. The impact of subtype assignment and resulting differences in cognitive decline was analyzed using a linear mixed effects model, including age at baseline, time since baseline (continuous), and subtype-by-time interactions as fixed effects, as well as allowing for participant-specific random intercepts34. To mirror the A4 Study, we restricted this analysis using data obtained over a similar period of observation (4 years from baseline).
Results
A4 Study MRI-based Disease Progression Subtypes
A total of 1,240 Aβ+ participants with complete MRI data were used as input into SuStaIn, following covariate adjustment and z-scoring using the 407 Aβ− screen-failures. Cross-validation indicated that the 3-subtype model best fit the data. Table 1 shows demographic and cognitive score variables for the Aβ− control group (for reference), the whole Aβ+ group, and the Aβ+ subtype groups which we designate as Typical, Cortical, and Subcortical based on the observed patterns of atrophy (Figure 1). There were no significant differences in demographic variables between the subtypes nor in the CDR-SB or MMSE, but the PACC, CFI (total), and Digit Symbol Substitution test scores were significantly different across the subtypes.
Table 1 –
Characteristics of the A4 Study cohort, separated between Aβ− (control) and Aβ+ (SuStaIn input) groups, further separated by subtype assignment. There were no significant differences among subtypes in demographic variables, the CDR-SB and the MMSE according to either an ANOVA or Pearson’s X2 test (following Holm-Bonferroni adjustment for multiple comparisons). The PACC, CFI (total), and Digital Symbol Substitution test scores showed significant (P < .001) differences between subtypes. Abbreviations: SUVr – Standardized Uptake Value ratio; PACC – Preclinical Alzheimer Cognitive Composite; CFI – Cognitive Function Index; MMSE – Mini-Mental State Examination; CDR-SB – Clinical Dementia Rating Sum of Boxes
| Characteristic | Aβ− (Control group) |
Aβ+ (Whole group) |
Subtype Zero (sub- threshold) |
Typical Subtype |
Cortical Subtype |
Subcortical Subtype |
Adjusted P-value (across subtypes) |
|---|---|---|---|---|---|---|---|
| No. individuals | 407 | 1240 | 717 | 170 | 179 | 174 | |
| Age, yrs, mean (SD) | 70.8 (4.4) | 72.0 (4.9) | 71.9 (4.6) | 72.4 (5.3) | 72.6 (5.2) | 71.6 (4.9) | .58a |
| Female, (%) | 258 (63.4) | 730 (59.0) | 445 (62.1) | 98 (57.6) | 103 (57.5) | 84 (48.3) | .08 b |
| Education, yrs, mean (SD) | 16.7 (2.5) | 16.6 (2.8) | 16.6 (2.8) | 16.0 (2.8) | 16.6 (2.8) | 16.8 (2.9) | .18 a |
| Family history of dementia (%) | 249 (61.2) | 924 (74.5) | 525 (73.2) | 130 (76.5) | 136 (76.0) | 133 (76.4) | .58 b |
| APOE ε4 alleles (%) | |||||||
| 0 | 407 (100.0) | 506 (40.8) | 297 (41.4) | 62 (36.5) | 73 (40.8) | 74 (42.5) | .68 b |
| 1 | 0 (0.0) | 621 (50.1) | 364 (50.8) | 92 (54.1) | 82 (45.8) | 83 (47.7) | |
| 2 | 0 (0.0) | 101 (8.2) | 49 (6.8) | 12 (7.1) | 23 (12.9) | 17 (9.8) | |
| Missing | 0 (0.0) | 12 (1.0) | 7 (1.0) | 4 (2.4) | 1 (0.6) | 0 (0.00) | |
| PET SUVr, mean (SD) | 0.99 (0.07) | 1.33 (0.18) | 1.32 (0.17) | 1.35 (0.19) | 1.35 (0.19) | 1.33 (0.18) | .58 a |
| PACC score, mean (SD) | 0.22 (2.38) | −0.42 (2.69) | −0.15 (2.57) | −0.89 (2.72) | −1.30 (3.04) | −0.13 (2.49) | < .001 a |
| CDR-SB score, mean (SD) | 0.05 (0.16) | 0.06 (0.17) | 0.05 (0.15) | 0.08 (0.19) | 0.06 (0.16) | 0.07 (0.22) | .50 a |
| MMSE score, mean (SD) | 28.93 (1.13) | 28.74 (1.28) | 28.80 (1.23) | 28.55 (1.38) | 28.65 (1.41) | 28.78 (1.21) | .50 a |
| Digit Symbol, mean (SD) | 44.21 (8.85) | 42.59 (8.93) | 43.52 (8.91) | 41.08 (9.19) | 40.55 (8.82) | 42.29 (8.39) | < .001 a |
| CFI total score, mean (SD) | 2.92 (3.07) | 3.82 (3.49) | 3.43 (3.10) | 4.29 (3.83) | 4.81 (4.17) | 3.93 (3.68) | < .001 a |
ANOVA
Pearson’s X2
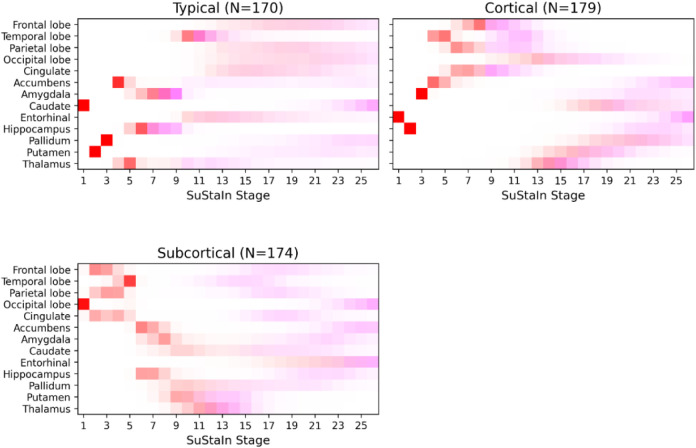
Figure 1 –
Positional variance diagram showing the disease progression sequences of the final 3-subtype disease progression model trained on the A4 Aβ+ cohort. The red squares indicate a z-score 1 event (abnormality), and magenta squares indicate a z-score 2 event (atrophy). Color intensity is proportional to positional confidence/certainty of the model.
Figure 1 shows a positional variance diagram illustrating the MCMC-sampled posterior distributions for each subtype in the final 3-subtype SuStaIn model. The vertical axis of brain volumes is ordered from cortical to subcortical regions, top-to-bottom. Z-score events along the horizontal axis are color-coded, with z=1 indicating subtle abnormality (in red), and z=2 indicating atrophy (in magenta). Subtypes were named according to the earliest atrophy events (z=2, magenta). In Figure 1, the Typical subtype shows early atrophy in the hippocampus, amygdala, and temporal lobe, alongside early yet subtle abnormality in the caudate, putamen, and pallidum. The Cortical subtype shows early atrophy in cortical regions (including the cingulate gyrus) and subtle abnormality in the entorhinal cortex, hippocampus, and amygdala. The Subcortical subtype shows early atrophy in the putamen and thalamus, albeit with high uncertainty (possibly indicating slow atrophy), alongside subtle abnormality in cortical lobes.
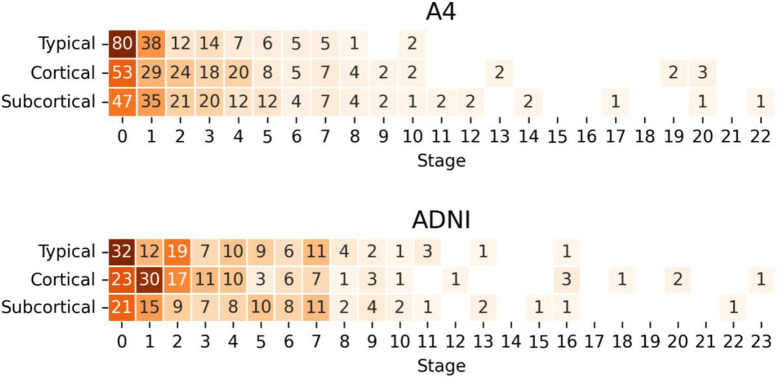
Figure 2 shows the distribution of stage assignments stratified by subtype in the A4 cohort (upper panel) and ADNI cohort (lower panel). Across A4, 523 (42.2%) individuals were assigned to a subtype: 170 (32.5%) Typical, 179 (34.2%) Cortical, and 174 (33.3%) Subcortical. The remaining 717 (57.8%) individuals were not assigned a subtype due to all regional volumes scoring z<1. We refer to these as subtype zero. The Typical, Cortical, and Subcortical subtypes have median stage assignments of 2 (IQR=1–3), 3 (IQR=1–5), and 3 (IQR=1–5) respectively, consistent with having subtle abnormality reflective of their cognitively unimpaired status.
Figure 2 –
Heatmap and counts of stage assignments per subtype for the A4 Study cohort (upper panel) and ADNI cohort (lower panel).
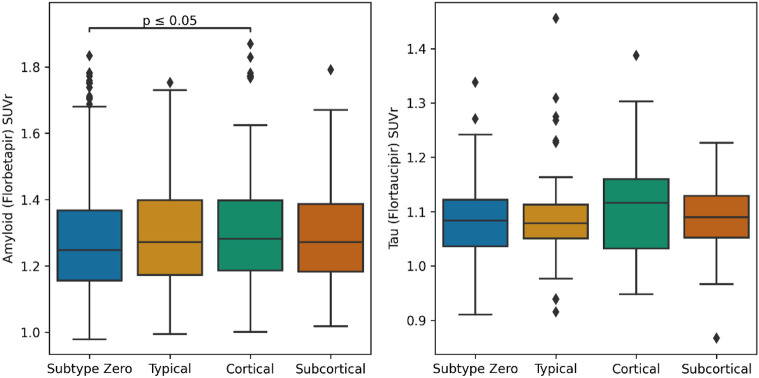
Mean cortical SUVr used in the A4 Study screening did not show any significant differences across subtypes (Table 1). In Figure 3, our image processing using AmyPET (see Methods) shows SUVr (with a whole cerebellar reference region) values for amyloid (N=1234) and tau (N=392) PET imaging stratified by subtype. Following multiple comparison correction, there was a significant difference between subtype zero and the Cortical subtype in amyloid SUVr (P=.04), though the Cortical subtype also had a notably higher tau burden (median=1.12; IQR=1.03–1.16) relative to subtype zero (median=1.08; IQR=1.04–1.12), the Typical subtype (median=1.08; IQR=1.05–1.11), and the Subcortical subtype (median=1.09; IQR=1.05–1.13).
Figure 3 –
Boxplots showing SUVr values (stratified by model-based subtype) for amyloid (left) and tau (right) PET scans, each using the whole cerebellum as a reference region. Lines are added to show pairwise significant differences (two-tailed Mann-Whitney U test, adjusted for multiple comparisons). Abbreviations: SUVr – standardized uptake value ratio
Biological Heterogeneity Predicts Cognitive Heterogeneity
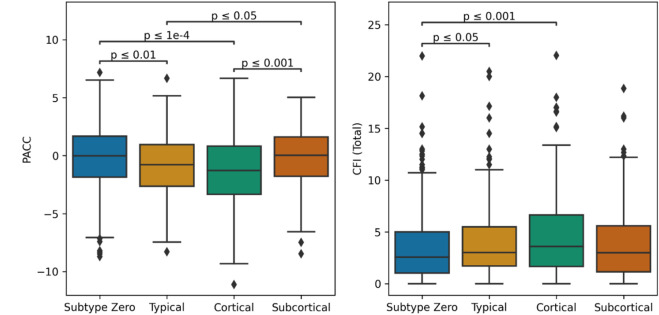
Error! Reference source not found. shows baseline PACC and CFI (total) scores in the A4 cohort stratified by subtype assignment. Broadly, baseline cognitive test scores were better in subtype zero and the Subcortical subtype. For the PACC score, there were significant differences between subtype zero (median = −0.013; IQR = −1.85 – 1.67) and both the Typical subtype (median = −0.78; IQR = −2.65 – 0.96; P = .004) and the Cortical subtype (median = −1.27; IQR = −3.34 – 0.83; P < .0001), but not for the Subcortical subtype (median = 0.03; IQR = −1.78 – 1.61; P = .87). Among the atrophy subtypes, PACC scores in both the Typical and Cortical subtypes were significantly different from those in the Subcortical subtype (P = .017 and P = .0006 respectively). For the CFI (total), there were also significant differences between subtype zero (median = 2.57; IQR = 1.04 – 5.00) and the Typical subtype (median = 3.02; IQR = 1.71 – 5.50; P = .04) and the Cortical subtype (median = 3.61; IQR = 1.67 – 6.65; P = .0003), but not the Subcortical subtype (median = 3.00, IQR = 1.14 – 5.58, P = .69). Among the atrophy subtypes, CFI did not differ statistically.
Simulating A4 Study Subtype Cognitive Decline using ADNI
Error! Reference source not found. (lower panel) shows the distribution of subtype and staging for the ADNI subset selected using A4 inclusion criteria. A total of 731 individuals (726 CN, 5 MCI) from ADNI matched the criteria and had a baseline 3T scan. There were significant differences across in the subtypes in baseline age, sex, and amount of education, but not in APOE ε4 carriers or cognitive variables (eTable 1). Notably, each subtype had a much higher percentage of the female sex compared to subtype zero. The ADNI subset significantly differed from A4 in age and number of APOE ε4 alleles (but not sex or education) and had small but significant differences in the baseline MMSE and CDR-SB scores (eTable 2). Using the A4-trained SuStaIn model, these individuals were assigned a stage and subtype as follows: 390 (53.4%) were unclassifiable (subtype zero), 118 (16.1%) were Typical, 120 (16.4%) Cortical, and 103 (14.1%) Subcortical. The Typical, Cortical, and Subcortical subtypes had median stage assignments of 3 (IQR = 1 – 6), 3 (IQR = 2 – 5), and 4 (IQR = 2 – 7) respectively, similar to the A4 cohort.
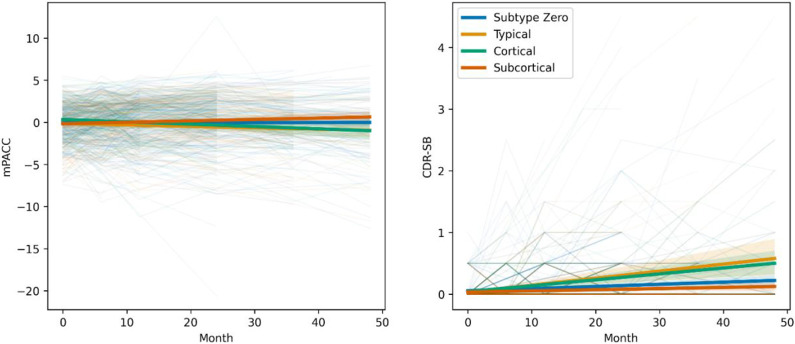
Error! Reference source not found. shows cognitive decline measured by longitudinal scores over 4 years on mPACC (left) and CDR-SB (right) in the ADNI cohort, stratified by baseline subtype. Most subtypes showed minimal cognitive decline. For the mPACC, both the Typical (−0.23/yr; 95% CI, −0.41 to −0.05; P = .01) and Cortical (−0.24/yr; 95% CI, −0.42 to −0.06; P = .009) subtypes were associated with greater cognitive decline relative to subtype zero. For the CDR-SB, both the Typical (+0.09/yr; 95% CI, 0.06 to 0.12; P < .0001) and Cortical (+0.07/yr; 95% CI, 0.04 to 0.10; P < .0001) subtypes were associated with greater cognitive decline relative to subtype zero. Higher age at baseline was significantly associated with faster cognitive decline over the period for both CDR-SB (P = .04) and mPACC (P < .0001).
Discussion
The A4 Study represents one of the largest studies of preclinical Alzheimer’s disease, providing an opportunity to leverage data-driven methods for analysis and potential applications, such as prediction of future decline and subgroup discovery. Using such a data-driven disease progression model on baseline MRI, the Subtype and Stage Inference (SuStaIn) algorithm uncovered previously undetected neurodegenerative heterogeneity in the A4 Study, despite enriching for amyloid (PET) positivity and extensive cognitive screening. Our DPM suggested three image-based subtypes in 523/1240 (42%) of participants, characterized as Typical, Cortical, and Subcortical, which share similarities with subtypes previously observed in cognitively impaired cohorts5,16. Our subtypes showed no significant differences in demographic, biomarker, or genetic variables which might otherwise be used to identify such differences.
The A4 cohort had 42% of individuals with sufficient neurodegeneration to be assigned to one of the three subtypes, which were approximately equal in size. Individuals within each subtype were spread across model stages (i.e. the extent of neurodegeneration), though the majority exhibited abnormality/atrophy in only a few regions, as expected by their preclinical status. This neurodegenerative heterogeneity was not captured by an amyloid and/or tau PET signal in the neocortex — we did not find significant differences between subtypes in the SUVr used for trial screening, and minimal differences across subtypes. Median amyloid and tau burden was greater in those assigned to a non-zero subtype, and numerically greatest in the Cortical subtype (not statistically significant for tau). This supports the notion that data-driven multivariate methods such as SuStaIn are uniquely useful for identifying (statistically significant) clinical heterogeneity in the absence of symptoms, above and beyond traditional screening tools.
Our image-based subtypes predicted cognitive heterogeneity, both cross-sectionally and longitudinally. Differences in baseline cognitive performance and longitudinal cognitive decline were both associated with DPM subtypes. In particular, the Cortical subtype displayed both poorer PACC scores at screening in A4 and more severe cognitive decline on mPACC and CDR-SB in ADNI (alongside the Typical subtype), suggesting that the screening protocol did not sufficiently capture (the variability in) underlying neurodegeneration. The ability to identify biological heterogeneity and predict cognitive heterogeneity supports the notion that image-based DPM can be used to inform clinical trial design, whether through screening (which could increase screen failures and initial costs), stratification, or covariate adjustment short of direct stratification35. Another potential design innovation could use image-based DPM to match subtypes to one of several targeted therapies in an umbrella design.
Limitations
Our forecasts of cognitive decline were produced by combining MRI-based neurodegenerative heterogeneity in the A4 cohort with longitudinal data from an A4-like ADNI subset. These cohorts are not perfectly matched, despite using the A4 inclusion criteria to select participants from ADNI. We found small but statistically significant differences in the number of APOE ε4 alleles (more non-carriers in ADNI), age, baseline MMSE (0.34 points higher) and CDR-SB (0.02 points lower), indicating that the ADNI subset is at lower risk of AD decline than A4. This suggests that our longitudinal predictions may be underestimating effect sizes of decline (favoring better cognition), but only by testing directly on longitudinal data from A4 (when released) will our predictions be realized.
While longitudinal data is available in ADNI, only 160 (of 731) individuals had cognitive scores available after 4 years. This limits the sample size (particularly after stratification) and thus analysis, potentially reducing the translatability of our observations into the prospective outcomes of the A4 Study.
Conclusions
The A4 Study represents a significant effort and investment into curating a preclinical cohort where anti-amyloid therapy may be more effective at slowing cognitive decline. Despite the study’s strict eligibility criteria and extensive screening, our disease progression model identified considerable neurodegenerative heterogeneity in 42% of participants, characterized by three MRI-based subtypes of neurodegeneration that can also be found in more advanced AD cohorts, associated with differences in cognition at baseline. Longitudinal analysis of an A4-matched subset of ADNI revealed significant differences in cognitive decline across the observed subtypes. These findings have potential ramifications for preclinical (and symptomatic) trials, where heterogeneity may obscure treatment effects. The use of data-driven disease progression modelling in trial design may increase statistical power of trials by accounting for the potential confounding effects of such heterogeneity on trial outcomes.
Supplementary Material
Figure 4 –
Boxplots of PACC (left) and CFI Total (right) scores in the A4 Study cohort, stratified by model-based subtype. Lines are added to show pairwise significant differences (two-tailed Mann-Whitney U test, adjusted for multiple comparisons). Abbreviations: PACC – Preclinical Alzheimer Cognitive Composite; CFI – Cognitive Function Index.
Figure 5 –
Analysis of A4 Study outcomes available in ADNI (CDR-SB and mPACC) over a period of four years, with individuals stratified by their baseline subtype assignment using the final disease progression model. Abbreviations: mPACC – Modified Preclinical Alzheimer Cognitive Composite; CDR-SB – Clinical Dementia Rating Sum of Boxes.
Key Points.
Question
Can MRI-based computational subtypes of preclinical neurodegeneration predict cognitive outcomes?
Findings
In this cross-sectional analysis of magnetic resonance imaging (MRI) data at screening (pre-randomization) in the preclinical Anti-Amyloid Treatment in Asymptomatic Alzheimer disease (A4) Study, we detected considerable neurodegenerative heterogeneity using data-driven disease progression modelling. The MRI-based computational subtypes identified by Subtype and Stage Inference (SuStaIn) differed in baseline cognitive test scores (A4) and in longitudinal cognitive decline (ADNI), with sufficient heterogeneity to potentially obscure treatment effect in A4 trial outcomes.
Meaning
Data-driven disease progression modelling of screening MRI scans can predict heterogeneity in cognitive performance/decline and potentially reduce heterogeneity in future clinical trials.
Acknowledgements
NPO and CS acknowledge funding from a UKRI Future Leaders Fellowship (MR/S03546X/1). NPO, CS, FB and DCA acknowledge funding from the Early Detection of Alzheimer's Disease Subtypes project (E-DADS; EU JPND 2019; MR/T046422/1). NPO, FB, and DCA acknowledge funding from the National Institute for Health Research University College London Hospitals Biomedical Research Centre. DCA also acknowledges support from Wellcome Trust award 221915/Z/20/Z. PJM is supported by EU-EFPIA Innovative Medicines Initiative 2 Joint Undertaking (IMI 2 JU) Amyloid Imaging to Prevent Alzheimer’s Disease (AMYPAD, grant agreement number: 115952). DMC is supported by the UK Dementia Research Institute which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK (ARUK-PG2017-1946), the UCL/UCLH NIHR Biomedical Research Centre, and the UKRI Innovation Scholars: Data Science Training in Health and Bioscience (MR/V03863X/1). MCD acknowledges funding from Eli Lilly and Company, Alzheimer’s Association, and NIH (R01AG063689 and U24AG057437).
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012).
The A4 Study is a secondary prevention trial in preclinical Alzheimer's disease, aiming to slow cognitive decline associated with brain amyloid accumulation in clinically normal older individuals. The A4 Study is funded by a public-private-philanthropic partnership, including funding from the National Institutes of Health-National Institute on Aging, Eli Lilly and Company, Alzheimer's Association, Accelerating Medicines Partnership, GHR Foundation, an anonymous foundation and additional private donors, with in-kind support from Avid and Cogstate. The companion observational Longitudinal Evaluation of Amyloid Risk and Neurodegeneration (LEARN) Study is funded by the Alzheimer's Association and GHR Foundation. The A4 and LEARN Studies are led by Dr. Reisa Sperling at Brigham and Women's Hospital, Harvard Medical School and Dr. Paul Aisen at the Alzheimer's Therapeutic Research Institute (ATRI), University of Southern California. The A4 and LEARN Studies are coordinated by ATRI at the University of Southern California, and the data are made available through the Laboratory for Neuro Imaging at the University of Southern California. The participants screening for the A4 Study provided permission to share their de-identified data in order to advance the quest to find a successful treatment for Alzheimer's disease. We would like to acknowledge the dedication of all the participants, the site personnel, and all of the partnership team members who continue to make the A4 and LEARN Studies possible. The complete A4 Study Team list is available on: a4study.org/a4-study-team.
References
- 1.Risacher SL, Anderson WH, Charil A, et al. Alzheimer disease brain atrophy subtypes are associated with cognition and rate of decline. Neurology. 2017;89(21):2176–2186. doi: 10.1212/WNL.0000000000004670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitwell JL, Dickson DW, Murray ME, et al. Neuroimaging correlates of pathologically defined subtypes of Alzheimer’s disease: A case-control study. Lancet Neurol. 2012;11(10):868–877. doi: 10.1016/S1474-4422(12)70200-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray ME, Graff-Radford NR, Ross OA, Petersen RC, Duara R, Dickson DW. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: A retrospective study. Lancet Neurol. 2011;10(9):785–796. doi: 10.1016/S1474-4422(11)70156-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira D, Nordberg A, Westman E. Biological subtypes of Alzheimer disease. Neurology. 2020;94(10):436–448. doi: 10.1212/WNL.0000000000009058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young AL, Marinescu R v., Oxtoby NP, et al. Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with Subtype and Stage Inference. Nat Commun. 2018;9(1). doi: 10.1038/s41467-018-05892-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong A, Toledo JB, Honnorat N, et al. Heterogeneity of neuroanatomical patterns in prodromal Alzheimer’s disease: links to cognition, progression and biomarkers. Brain. 2017;140(3):735–747. doi: 10.1093/BRAIN/AWW319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummings JL, Morstorf T, Zhong K. Alzheimer’s disease drug-development pipeline: Few candidates, frequent failures. Alzheimers Res Ther. 2014;6(4):1–7.doi: 10.1186/ALZRT269/TABLES/3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jutten RJ, Sikkes SAM, van der Flier WM, Scheltens P, Visser PJ, Tijms BM. Finding Treatment Effects in Alzheimer Trials in the Face of Disease Progression Heterogeneity. Neurology. 2021;96(22):e2673–e2684. doi: 10.1212/WNL.0000000000012022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oxtoby NP, Shand C, Cash DM, Alexander DC, Barkhof F. Targeted Screening for Alzheimer’s Disease Clinical Trials Using Data-Driven Disease Progression Models. Front Artif Intell. 2022;5. doi: 10.3389/frai.2022.660581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings J, Lee G, Nahed P, et al. Alzheimer’s disease drug development pipeline: 2022. Alzheimer’s & Dementia: Translational Research & Clinical Interventions. 2022;8(1):e12295. doi: 10.1002/TRC2.12295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen K, Langbaum JBS, Fleisher AS, et al. Twelve-month metabolic declines in probable Alzheimer’s disease and amnestic mild cognitive impairment assessed using an empirically pre-defined statistical region-of-interest: Findings from the Alzheimer’s Disease Neuroimaging Initiative. Neuroimage. 2010;51(2):654–664. doi: 10.1016/J.NEUROIMAGE.2010.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & Dementia. 2011;7(3):280–292. doi: 10.1016/J.JALZ.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jack CR, Therneau TM, Weigand SD, et al. Prevalence of Biologically vs Clinically Defined Alzheimer Spectrum Entities Using the National Institute on Aging-Alzheimer’s Association Research Framework. JAMA Neurol. 2019;76(10):1174–1183. doi: 10.1001/jamaneurol.2019.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sperling RA, Donohue MC, Raman R, et al. Association of Factors with Elevated Amyloid Burden in Clinically Normal Older Individuals. JAMA Neurol. 2020;77(6):735–745. doi: 10.1001/jamaneurol.2020.0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langford O, Raman R, Sperling RA, et al. Predicting Amyloid Burden to Accelerate Recruitment of Secondary Prevention Clinical Trials. Journal of Prevention of Alzheimer’s Disease. 2020;7(4):213–218. doi: 10.14283/jpad.2020.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogel JW, Young AL, Oxtoby NP, et al. Four distinct trajectories of tau deposition identified in Alzheimer’s disease. Nature Medicine 2021 27:5. 2021;27(5):871–881. doi: 10.1038/s41591-021-01309-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young AL, Oxtoby NP, Daga P, et al. A data-driven model of biomarker changes in sporadic Alzheimer’s disease. Brain. 2014;137(9):2564–2577. doi: 10.1093/BRAIN/AWU176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sperling RA, Rentz DM, Johnson KA, et al. The A4 Study: Stopping AD Before Symptoms Begin? Sci Transl Med. 2014;6(228). doi: 10.1126/SCITRANSLMED.3007941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI). Neurology. 2010;74(3):201–209. doi: 10.1212/WNL.0B013E3181CB3E25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doody RS, Thomas RG, Farlow M, et al. Phase 3 Trials of Solanezumab for Mild-to-Moderate Alzheimer’s Disease A BS TR AC T. n engl j med. 2014;370:311–332. doi: 10.1056/NEJMoa1312889 [DOI] [PubMed] [Google Scholar]
- 21.Mormino EC, Papp K v., Rentz DM, et al. Early and late change on the preclinical Alzheimer’s cognitive composite in clinically normal older individuals with elevated amyloid β. Alzheimer’s & Dementia. 2017;13(9):1004–1012. doi: 10.1016/J.JALZ.2017.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amariglio RE, Donohue MC, Marshall GA, et al. Tracking Early Decline in Cognitive Function in Older Individuals at Risk for Alzheimer Disease Dementia: The Alzheimer’s Disease Cooperative Study Cognitive Function Instrument. JAMA Neurol. 2015;72(4):446–454. doi: 10.1001/JAMANEUROL.2014.3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansson O, Seibyl J, Stomrud E, et al. CSF biomarkers of Alzheimer’s disease concord with amyloid-β PET and predict clinical progression: A study of fully automated immunoassays in BioFINDER and ADNI cohorts. Alzheimer’s & Dementia. 2018;14(11):1470–1481. doi: 10.1016/J.JALZ.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/J.NEUROIMAGE.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein A, Tourville J. 101 labeled brain images and a consistent human cortical labeling protocol. Front Neurosci. 2012;0(DEC):171. doi: 10.3389/FNINS.2012.00171/ABSTRACT [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voevodskaya O, Simmons A, Nordenskjöld R, et al. The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer’s disease. Front Aging Neurosci. 2014;6(OCT). doi: 10.3389/FNAGI.2014.00264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markiewicz PJ, Ehrhardt MJ, Erlandsson K, et al. NiftyPET: a High-throughput Software Platform for High Quantitative Accuracy and Precision PET Imaging and Analysis. Neuroinformatics. 2018;16(1):95–115. doi: 10.1007/s12021-017-9352-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markiewicz PJ, Matthews JC, Ashburner J, et al. Uncertainty analysis of MR-PET image registration for precision neuro-PET imaging. Neuroimage. 2021;232:117821. doi: 10.1016/J.NEUROIMAGE.2021.117821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fonteijn HM, Modat M, Clarkson MJ, et al. An event-based model for disease progression and its application in familial Alzheimer’s disease and Huntington’s disease. Neuroimage. 2012;60(3):1880–1889. doi: 10.1016/J.NEUROIMAGE.2012.01.062 [DOI] [PubMed] [Google Scholar]
- 30.Collij LE, Salvadó G, Wottschel V, et al. Spatial-Temporal Patterns of β-Amyloid Accumulation. Neurology. 2022;98(17):e1692–e1703. doi: 10.1212/WNL.0000000000200148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young AL, Bocchetta M, Russell LL, et al. Characterizing the Clinical Features and Atrophy Patterns of MAPT-Related Frontotemporal Dementia With Disease Progression Modeling. Neurology. 2021;97(9):e941–e952. doi: 10.1212/WNL.0000000000012410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eshaghi A, Young AL, Wijeratne PA, et al. Identifying multiple sclerosis subtypes using unsupervised machine learning and MRI data. Nature Communications 2021 12:1. 2021;12(1):1–12. doi: 10.1038/s41467-021-22265-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donohue MC, Sperling RA, Petersen R, Sun CK, Weiner M, Aisen PS. Association Between Elevated Brain Amyloid and Subsequent Cognitive Decline Among Cognitively Normal Persons. JAMA. 2017;317(22):2305–2316. doi: 10.1001/JAMA.2017.6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Dyck CH, Nygaard HB, Chen K, et al. Effect of AZD0530 on Cerebral Metabolic Decline in Alzheimer Disease: A Randomized Clinical Trial. JAMA Neurol. 2019;76(10):1219–1229. doi: 10.1001/jamaneurol.2019.2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frost C, Mulick A, Scahill RI, et al. Design optimization for clinical trials in early-stage manifest Huntington’s disease. Movement Disorders. 2017;32(11):1610–1619. doi: 10.1002/MDS.27122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.