Summary
Identifying the molecular mechanisms that promote optimal immune responses to coronavirus disease 2019 (COVID-19) vaccination is critical for future rational vaccine design. Here, we longitudinally profile innate and adaptive immune responses in 102 adults after the first, second, and third doses of mRNA or adenovirus-vectored COVID-19 vaccines. Using a multi-omics approach, we identify key differences in the immune responses induced by ChAdOx1-S and BNT162b2 that correlate with antigen-specific antibody and T cell responses or vaccine reactogenicity. Unexpectedly, we observe that vaccination with ChAdOx1-S, but not BNT162b2, induces an adenoviral vector-specific memory response after the first dose, which correlates with the expression of proteins with roles in thrombosis with potential implications for thrombosis with thrombocytopenia syndrome (TTS), a rare but serious adverse event linked to adenovirus-vectored vaccines. The COVID-19 Vaccine Immune Responses Study thus represents a major resource that can be used to understand the immunogenicity and reactogenicity of these COVID-19 vaccines.
Keywords: SARS-CoV-2, COVID-19 vaccine, RNA-seq, T cell, antibody responses, vaccination, immunophenotyping, proteomics, lipidomics, cytokines
Graphical abstract
Highlights
-
•
Multi-omics profiling of responses in 102 adults after COVID-19 vaccination
-
•
Baseline and innate responses correlate with vaccine immunogenicity/reactogenicity
-
•
ChAdOx1-S, but not BNT162b2, induces an adenoviral memory response after the first dose
-
•
ChAdOx1-S memory response correlates with expression of pro-thrombotic proteins
Ryan et al. use a multi-omics approach to longitudinally profile innate and adaptive immune responses in blood collected from 102 adults at baseline and post-vaccination with the ChAdOx1-S, BNT162b2, or mRNA-1273 vaccines. The study reveals key differences in immune responses to adenovirus-vectored compared with mRNA COVID-19 vaccines.
Introduction
The BNT162b2 (Pfizer-BioNTech), ChAdOx1-S (Oxford-AstraZeneca), and mRNA-1273 (Moderna) vaccines are highly effective at preventing severe coronavirus disease 2019 (COVID-19),1,2,3 although protection against infection wanes within 4–6 months after the second dose and two doses provide decreased protection against infection by more recent variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).4,5 A third dose of mRNA vaccine has been shown to induce superior immunogenicity and effectiveness.4,6,7 While the adaptive immune responses induced by these vaccines are increasingly well understood,8,9,10,11 recent studies are also shedding light on how immune responses induced in the hours and days after vaccination relate to subsequent immunogenicity.12,13,14 Despite these efforts there remain significant gaps in our knowledge of the immune processes underpinning the differences in immunogenicity between vaccines.
These vaccines are also associated with relatively high levels of reactogenicity ranging from self-resolving symptoms (e.g., pain at the injection site, headache, fever, chills, and myalgia) to rare, serious adverse events (AEs) such myocarditis,15,16,17 or thrombosis with thrombocytopenia syndrome (TTS; also known as vaccine-induced immune thrombotic thrombocytopenia).18,19,20,21 To date, no studies have systematically compared innate and early adaptive immune responses with vaccination with BNT162b2 and ChAdOx1-S at a systems level in the same cohort, nor has it been investigated how immune responses induced immediately after the third dose compares with those after the first and second doses. Here, we report the results of the COVID-19 Vaccine Immune Responses Study (COVIRS), a systems vaccinology study in which we have longitudinally profiled early and late immune responses after the first, second, and third doses of vaccine in 102 healthy adults who received the BNT162b2 (mRNA) or ChAdOx1-S (adenovirus vectored) vaccines as their first vaccinations in the absence of community transmission of SARS-CoV-2. A subset of participants were also assessed after a third dose of either the BNT162b2 or mRNA-1273 vaccines.
Results
We recruited 146 adults living in Adelaide, South Australia, in 2021 to COVIRS. The first and second doses of vaccine were administered in the absence of SARS-CoV-2 community transmission because of strict quarantine and border control measures in place in South Australia. Forty-four participants withdrew before sample collection. The remaining participants received two doses of either the BNT162b2 (n = 86) or ChAdOx1-S (n = 16) vaccines for their first and second doses. Samples were also collected from a subset of participants who received a third dose of either BNT162b2 (n = 38) or mRNA-1273 (n = 14) (Figure 1A). Participants were 70% female with a mean age of 39 ± 11 years at enrollment. There was no significant difference between those who received two doses of the BNT162b2 compared with those who received two doses of ChAdOx1-S in terms of their age, gender, body mass index, comorbidities (e.g., hypertension, asthma), self-reported alcohol consumption or cigarette smoking (Figure 1B, Table S1). Participants completed a detailed survey (Data S1) to report any AEs 1 week after each vaccine dose (>95% completion rate).
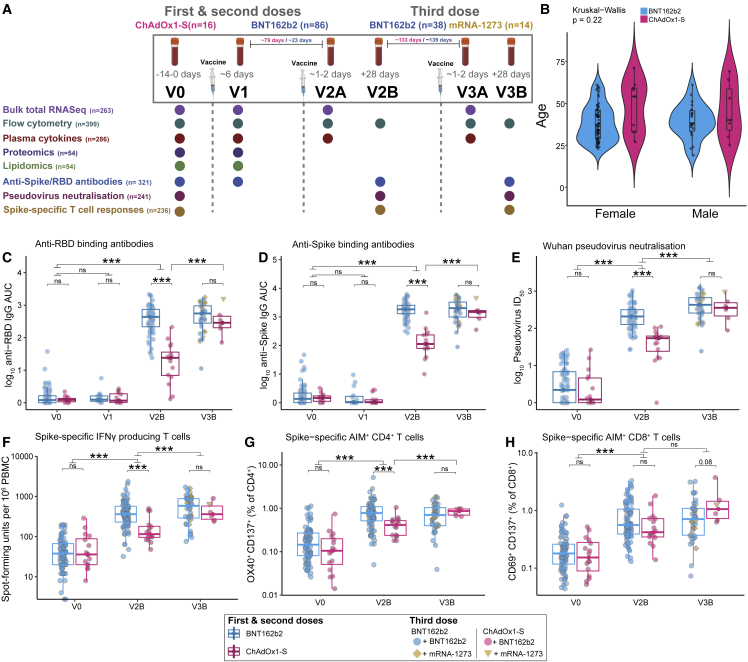
Figure 1.
Antigen-specific antibody and T cell responses in adults vaccinated with the BNT162b2, mRNA-1273, or ChAdOx1-S vaccines
(A) Sample collection timepoints and assays performed. Assay sample numbers represent the total number of samples analyzed. ChAdOx1-S participants n = 16, BNT162b2 participants n = 86.
(B) Age and gender of participants.
(C–H) (C) Anti-RBD and (D) anti-Spike IgG titers, (E) Wuhan-Hu-1 pseudovirus NAb titers (ID50), (F) Spike-specific IFN-γ spot-forming units (SFU), (G) Spike-specific AIM+ CD4+ T cells, (H) Spike-specific AIM+ CD8+ T cells, before and after vaccination with BNT162b2 or ChAdOx1-S (first and second doses) and a third dose of either BNT162b2 or mRNA-1273. Data in (B–H) are represented as violin (B) or boxplots (C–H), with the box denoting the 25th and 75th percentiles, the whiskers the 5th and 95th percentiles; the middle bar is the median. Statistical significance was assessed in (B) using a Kruskal-Wallis test, in (C–H) using Wilcoxon rank-sum tests. ∗p < 0.05, ∗∗∗p < 0.001, ns = not significant.
Three doses of COVID-19 vaccines induce high levels of protective antibodies and Spike-specific T cells in healthy, SARS-CoV-2 naive adults irrespective of the first vaccine
Anti-Spike and anti-RBD total IgG titers and Wuhan-Hu-1 pseudovirus neutralizing antibody (NAb) titers were measured pre-vaccination (V0), and approximately 28 days after the second (V2B) and third (V3B) doses. Anti-Spike/RBD IgG titers were also measured in a subset of participants (n = 32) approximately 6 days after the first dose (V1). Anti-Spike/RBD IgG titers at V2B and V3B were positively correlated with each other and with NAb titers (Figures S1A–S1F). All participants had robust anti-RBD (Figure 1C) and anti-Spike (Figure 1D) IgG responses, with at least a 35-fold increase in anti-Spike IgG titers at V2B. Similar increases were observed in NAb titers (Figure 1E). We observed a significant but relatively weak negative correlation between age and anti-RBD/Spike and NAb titers at V2B (Figures S1G–S1I), consistent with previous reports of age-dependent vaccine responses.22 No correlation with age was detected after the third dose (Figures S1J–S1L). Participants who received the ChAdOx1-S vaccine for their first and second vaccinations had lower anti-Spike/RBD and NAb titers at V2B compared with those who received BNT162b2 (Figures 1C–1E). After a third dose of either BTN162b2 or mRNA-1273, however, antibody titers were comparable in participants who received either ChAdOx1-S or BTN162b2 initially (Figures 1C–1E). NAb titers were also measured against Omicron (B.1.1.529) pseudovirus at V3B and were significantly lower compared with neutralization of Wuhan-Hu-1 pseudovirus (Figure S1M). Omicron NAb titers were strongly correlated with both Wuhan-Hu-1 NAb and anti-RBD IgG titers (Figures S1N and S1O).
Spike-specific T cell responses were assessed at V0, V2B and V3B by activation-induced marker (AIM) and ELISpot assays after stimulation of peripheral blood mononuclear cells (PBMCs) with a peptide pool covering the full length of the Spike protein. AIM+ CD4+ and CD8+ T cell responses were correlated with each other, and with the ELISpot data (Figures S2A–S2F). A small population of Spike-reactive T cells was also present at baseline, which may be cross-reactive T cells derived from prior exposure to other endemic coronaviruses and/or environmental antigens.23,24,25 There was a positive correlation (p = 0.07) between the proportion of AIM+CD4+ T cells before vaccination and anti-RBD/Spike and NAb titers after the second dose (data not shown). Vaccination with either BNT162b2 or ChAdOx1-S induced significantly increased antigen-specific T cell responses, relative to baseline, as measured by IFN-γ secretion in ELISpot assays (Figure 1F). T cell responses in ChAdOx1-S-vaccinated participants were significantly lower at V2B relative to BNT162b2-vaccinated participants; however, responses were not different after the third dose of BTN162b2 or mRNA-1273 (Figure 1F). AIM assays demonstrated a similar decreased frequency of AIM+CD4+ T cells after ChAdOx1-S at V2B, which recovered after a third vaccination (Figure 1G). There was no difference in AIM+CD8+ T cells in participants vaccinated with ChAdOx1-S or BNT162b2 at either V2B or V3B (Figure 1H). The majority of AIM+CD4+ T cells had an effector memory (Tem) surface phenotype (Figure S2G), whereas AIM+CD8+ T cells predominantly exhibited a Tem/TemRA surface phenotype (Figure S2H). The proportions of AIM+CD4+ or CD8+ T cells were not correlated with anti-RBD/Spike/NAb titers at V2B in participants vaccinated with BNT162b2. In contrast, after ChAdOx1-S, AIM+CD4+ T cells were positively correlated with Wuhan-Hu-1 pseudovirus NAb titers (Figure S2I), and the proportion of TemRA AIM+CD4+ T cells was positively correlated with anti-Spike IgG titers (Figure S2J). After the third dose of either BNT162b2 or mRNA-1273, CD8+ Tem cells were positively correlated with anti-Spike/RBD and Wuhan-Hu-1 pseudovirus NAb titers (Figure S2K). The proportion of CD8+ TemRA cells, in contrast, was negatively correlated with anti-RBD and Wuhan-Hu-1 pseudovirus NAb titers at V3B.
Vaccination with the ChAdOx1-S, but not BNT162b2, vaccine induces a memory-like cTfh and plasmablast response after the first dose
We performed total RNA sequencing to assess transcriptome-wide changes in whole blood at V0, V1, and approximately 1–2 days after the second (V2A) and third (V3A) doses, generating more than 12 billion reads across 263 samples (Figure S3A, Table S2). Consistent with a previous study reporting only a transient increase in the expression of type I interferon (IFN-I) inducible genes 1–2 days after the first dose of BNT162b2,12 no differentially expressed genes (DEGs) were identified at V1 (mean of 6 days after the first dose) in BNT162b2-vaccinated participants (Figure 2A). Gene Set Enrichment Analysis (GSEA) did, however, detect an enrichment for IFN-α-inducible genes (Table S2), and we observed a correlation between days after vaccination and the expression of the gene, IFI27 (Figure S3B), indicating that this IFN-I signature is still detectable in whole blood from these participants up to approximately 6 days after vaccination.
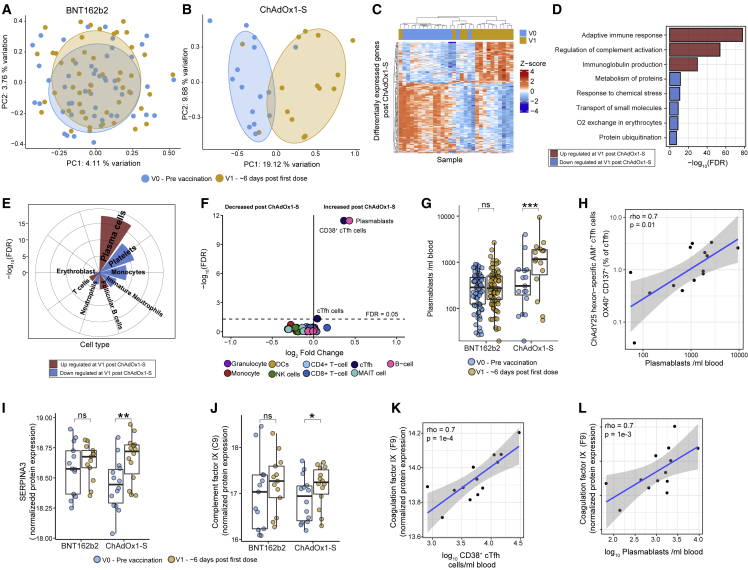
Figure 2.
Multi-omics assessment of responses to a single dose of BNT162b2 or ChAdOx1-S at approximately 6 days after vaccination (V1)
(A and B) Multidimensional scaling analysis (MDS) of whole-blood gene expression profiles (RNA-seq) at V0 and V1 after (A) BNT162b2 (n = 66) or (B) ChAdOx1-S (n = 15).
(C) Heamap of DEGs at V1 compared with V0 in participants vaccinated with ChAdOx1-S.
(D and E) Selected reactome pathways and Gene Ontology (GO) terms and (E) cell types, enriched among DEGs at V1 in participants vaccinated with ChAdOx1-S.
(F) Volcano plot of immune cell populations after one dose of ChAdOx1-S.
(G) The number of plasmablasts at V1 in participants vaccinated with ChAdOx1-S (n = 16) or BNT162b2 (n = 77).
(H) Correlation between ChAdY25 hexon-specific AIM+ cTfh cells and the number of plasmablasts at V1 in participants vaccinated with ChAdOx1-S.
(I and J) Normalized expression of selected proteins identified as differentially abundant in plasma at V1 compared with V0 in after ChAdOx1-S (n = 14) or BNT162b2 (n = 13).
(K and L) Correlation (Spearman) between coagulation factor IX protein expression in plasma at V1 and (K) CD38+ cTfh cells and (L) plasmablasts at V1 in participants vaccinated with ChAdOx1-S. Data in (G), (I), and (J) are represented as boxplots (see Figure 1). Statistical significance was assessed in (D) and (E) using a hypergeometric test, in (F–H) using a generalized linear model, and in (I, J) with limma. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = not significant, FDR = false discovery rate.
In contrast, 424 DEGs were found at approximately 6 days after vaccination with ChAdOx1-S (Figures 2B and 2C, Table S2). Up-regulated genes were enriched for roles in the adaptive immune response, complement activation, and immunoglobulin production (Figure 2D) and were strongly enriched for genes expressed in plasma cells (Figure 2E), particularly immunoglobulin genes (Figure S3C). In contrast, down-regulated genes were enriched for genes expressed in platelets and monocytes. To further investigate these signatures, flow cytometry analysis was used to characterize changes in 54 different immune cell sub-populations (Table S3, Methods S1 and S2). Consistent with our RNA sequencing (RNA-seq) data, we did not detect any changes in immune cell populations at V1 after BNT162b2 vaccination (Table S3). In contrast, vaccination with ChAdOx1-S had a striking effect on plasmablasts (CD19+/dimCD20-/dimCD38++CD27++)26 and circulating T follicular helper cells (cTfh; CD3+CD4+CXCR5+PD-1+), in particular CD38+ cTfh cells (recently activated), which were increased after the first dose of ChAdOx1-S (Figures 2F and 2G, S3D and S3E). Four ChAdOx1-S participants did not have increased plasmablasts or CD38+ cTfh cells (Figures 2G, S2E); however, these individuals had samples collected at 5 days after vaccination (Figure S3F), suggesting that this plasmablast/cTfh response is only evident on or after day 6 after vaccination. Programmed cell death 1 (PD-1) expression has been correlated with the activation of cTfh cells27 and consistent with this, we observed increased PD-1 expression (mean fluorescent intensity) on cTfh cells after the first dose of ChAdOx1-S, but not BNT162b2 (Figure S3G).
The induction of plasmablasts and cTfh cells after the first dose of ChAdOx1-S is analogous to what is observed after seasonal influenza vaccination, where pre-existing immunity to influenza from previous infection or vaccination results in an increase in CD38+ cTfh cells and plasmablasts in the blood at approximately 7 days after vaccination.27,28 The magnitude of the plasmablast and cTfh response after influenza vaccination correlates with subsequent serum antibody titers28; however, we did not detect a correlation between the magnitude of the plasmablast or CD38+ cTfh cell response induced after the first dose of the ChAdOx1-S vaccine and antibody titers at V2B (Figure S3H), nor did we detect anti-Spike/RBD binding antibodies at V1 (Figures 1C and 1D). Taken together, these data suggest a memory response to a component of the ChAdOx1-S vaccine or a complex involving the vaccine.29 To determine if this was a recall response directed against the ChAdOx1 adenoviral vector, we stimulated V0 and V1 PBMCs with a peptide pool derived from the Chimpanzee Adenovirus type Y25 (ChAdY25) hexon protein (the adenoviral vector in the ChAdOx1-S vaccine) and assessed immune responses to stimulation by ELISpot and AIM assays. IFN-γ-secreting cells and AIM+CD4+ cTfh cells were significantly increased at V1 relative to V0 samples after ChAdY25 hexon peptide stimulation in ChAdOx1-S, but not BNT162b2, vaccinated participants (Figures S3I and S3J). In contrast, stimulation of V0 and V1 samples with a Spike-derived peptide pool did not lead to an increase in IFN-γ-secreting cells or AIM+CD4+ cTfh cells in any of the vaccinated participants (Figures S3K and S3L). Most significantly, the frequency of ChAdY25 hexon-specific AIM+CD4+ cTfh cells observed at V1 in ChAdOx1-S vaccinated participants was strongly positively correlated with the number of circulating plasmablasts induced in these participants after the first dose of ChAdOx1-S (Figure 2H). Taken together, these data suggest that there is pre-existing cross-reactive cellular immunity to the ChAdOx1 vector that is induced after the first dose of ChAdOx1-S and is correlated with the induction of circulating plasmablasts.
The expression of complement and coagulation-related proteins is correlated with the memory-like cTfh and plasmablast response induced after the first dose of ChAdOx1-S
To further characterize the molecular changes after the first dose of ChAdOx1-S, we performed untargeted proteomics on plasma samples (n = 54) collected from these participants and a matched cohort of BNT162b2 vaccinated participants. Vaccination with ChAdOx1-S led to changes in the expression of nine proteins in plasma at V1 relative to V0 (Table S4), whereas no changes were detected following BNT162b2 at V1 relative to V0. After ChAdOx1-S, for example, there was increased expression of SERPINA3, an acute phase protein (Figure 2I), complement component 9 (Figure 2J), and APOC2 (Figure S3M), a cofactor activating lipoprotein lipase. The expression of other proteins including the glycoprotein HRG (Figure S3N, Table S4), were decreased following vaccination with ChAdOx1-S. Pathway enrichment analysis revealed complement activation related pathways were enriched among up-regulated proteins (Table S4). The expression of multiple coagulation and complement-related proteins including coagulation factors IX and XII and protein C, as well as complement component C3, C8A, C8B, and CFH, were strongly correlated with the number of CD38+ cTfh cells and plasmablasts induced after the first dose of ChAdOx1-S (Figures 2K and 2L, Table S4). Taken together these data suggest that the magnitude of the memory response induced after the first dose of the ChAdOx1-S vaccine is correlated with the expression of proteins involved in thrombosis.
Given these data, it is important to consider that vaccination with ChAdOx1-S has, in rare instances, been associated with TTS after the first dose, a condition in which antibodies are generated that recognize platelet factor 4 (PF4), leading to activation of the coagulation cascade.20 We speculated that the plasmablasts induced after the first dose of ChAdOx1-S could produce anti-PF4 antibodies; however, we measured anti-PF4 IgG titers in serum collected from participants at V0 and at V1 after vaccination with ChAdOx1-S (n = 14) and from an age- and gender-matched subset of BNT162b2 vaccinated participants (n = 13), and there was no statistically significant difference detected (Figure S3O). An important caveat is that none of the participants in our study developed TTS and further research is needed to assess whether similar responses, perhaps of greater magnitude, occur in patients who go on to develop TTS.
We also performed untargeted lipidomics on the same set of 54 plasma samples. Consistent with our proteomics and RNA-seq data, we found no changes in plasma lipid levels at V1 after BNT162b2; however, nine lipid species were decreased after the first dose of ChAdOx1-S (Figures S3P–S3R and Table S4). Lipids decreased after ChAdOx1-S were predominantly phosphatidylcholines, an abundant cellular phospholipid required for the generation of antibody-secreting cells.30 Sixty-eight lipid species were correlated with the number of CD38+ cTfh cells and/or plasmablasts induced at V1 after ChAdOx1-S, including multiple negative correlations between the levels of phosphatidylcholine species and the number of plasmablasts (Figure S3S, Table S4), consistent with increased energy requirements of these cells.30
Similar changes to the whole blood transcriptome are induced approximately 1–2 days after the second and third doses, irrespective of vaccine type
Next, we assessed transcriptome-wide changes in whole blood at V2A and V3A. Surprisingly, multidimensional scaling analysis revealed that a second dose of either BNT162b2 or ChAdOx1-S induced remarkably similar changes to the transcriptome at V2A (Figure 3A). Furthermore, gene expression changes induced after a third dose of BNT162b2 or mRNA-1273 were indistinguishable from each other or from those changes induced after the second dose of BNT162b2 or ChAdOx1-S. Compared with pre-vaccination baseline, after BNT162b2, there were 611 and 1,015 DEGs (with a fold-change of at least 1.5-fold) at V2A and V3A, respectively (Figure 3B, Table S2) including genes encoding transcription factors, such as STAT1 and chemokines, such as CXCL10 (Figures 3C and 3D). Similar numbers of DEGs were observed following vaccination with ChAdOx1-S at V2A (533 DEGs) or mRNA-1273 (1,549 DEGs) at V3A (Table S2). A direct statistical comparison of gene expression responses after the second (BNT162b2 vs. ChAdOx1-S at V2A) or a third dose (BNT162b2 vs. mRNA-1273 at V3A) did not identify any DEGs (FDR < 0.05), nor did comparing V2A with V3A, indicating that changes in gene expression induced at both timepoints were very similar, irrespective of the vaccine type. Genes up-regulated at either V2A or V3A were strongly enriched for roles in IFN and cytokine signaling, antigen presentation, and the complement cascade (Figure 3E, Table S2). The robust IFN signature observed after the second and third doses is consistent with a previous study of responses to the BNT162b2 vaccine, which also reported a robust IFN-I signature after first and second doses of BNT162b2.12 Experiments in Ifnar−/− mice (IFN-α/β receptor deficient) provide strong support for the functional important of IFN signaling in the generation of optimal vaccine-specific T and B cell responses.31
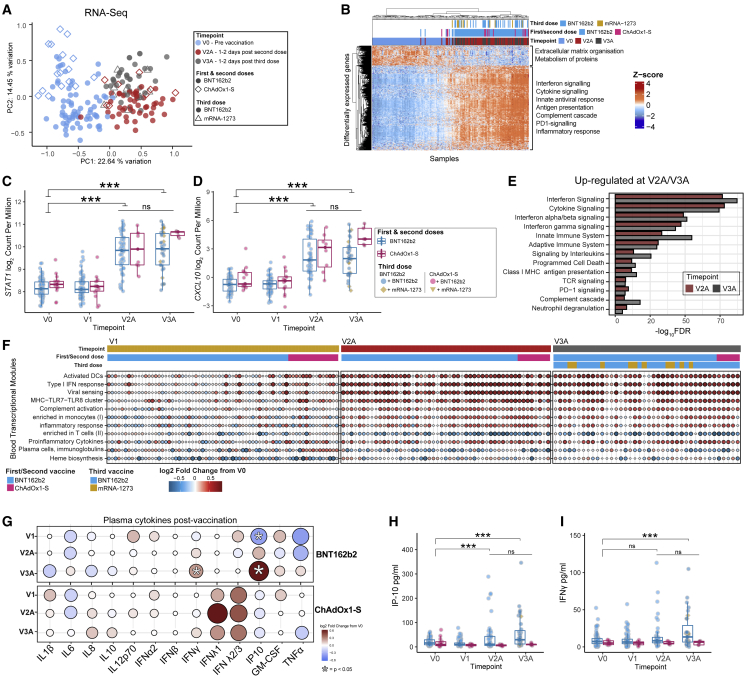
Figure 3.
Gene expression and cytokine responses approximately 1–2 days after the second and third doses of vaccine
(A) MDS of whole blood gene expression data at V0, V2A (BNT162b2 n = 46, ChAdOx1-S n = 8), and V3A (BNT162b2 n = 32, mRNA-1273 n = 10).
(B–D) (B) Heatmap of DEGs at V2A. Normalized gene expression of (C) STAT1 and (D) CXCL10 after vaccination.
(E) Selected reactome pathways and Gene Ontology terms enriched among DEGs at V2A/3A.
(F) Selected BTMs that were differentially expressed at V1, V2A, or V3A after vaccination.
(G) Fold-change in plasma cytokine levels at V1, V2A, and V3A after vaccination. The concentration of (H) IP-10 and (I) IFN-γ in plasma before and after vaccination. Data in (C, D) and (H, I) are represented as boxplots (see Figure 1). Statistical significance was assessed in (C, D) using edgeR, in (E) using a hypergeometric test, and in (G–I) with a generalized linear model. ∗p < 0.05, ∗∗∗p < 0.001, ns = not significant.
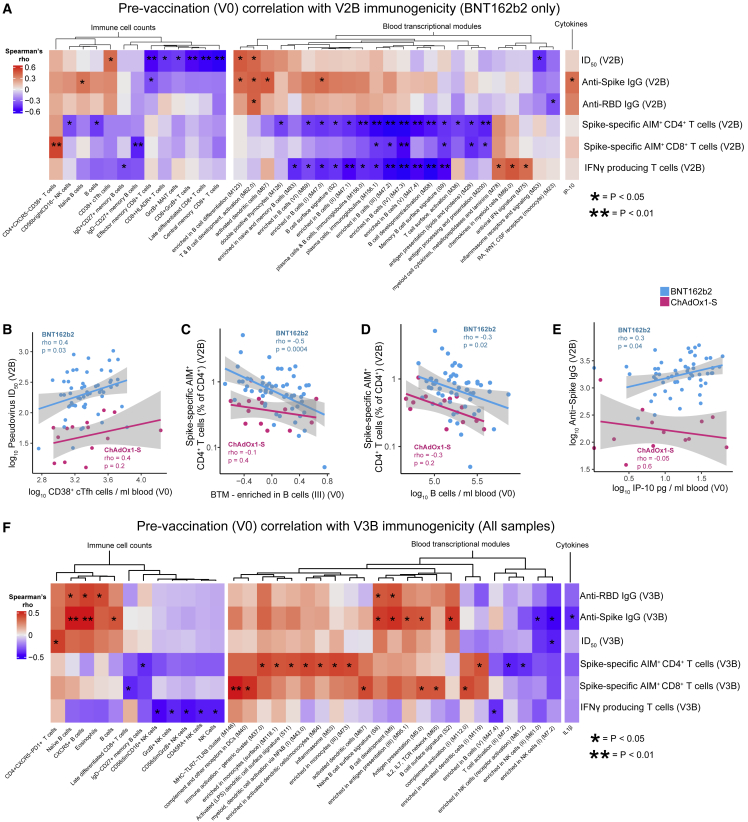
We next assessed transcriptional changes using gene set variation analysis (GSVA).32 We used this approach to calculate an activity score for a set of more than 200 pre-defined blood transcriptional modules (BTMs),31 reflecting changes in the expression of these BTMs in each participant at each timepoint after vaccination (Figure 3F, Table S2). At V1, this approach detected the increase in plasma cell gene expression after ChAdOx1-S reported above, but also decreased activity of BTMs related to heme biosynthesis and platelet activation. Consistent with our pathway analysis, there was an increase in the activity of BTMs related to antigen presentation, IFN-I response, and inflammation at V2A and V3A, irrespective of vaccine (Figure 3F, Table S2). No BTMs were identified as differentially active when comparing responses at V2A with those at V3A, further emphasizing the similarity in transcriptional responses after the second and third doses. Next, we assessed whether the activity of any BTMs at V2A or V3A were correlated with antigen-specific antibody or T cell responses at V2B or V3B (including anti-Spike/RBD/NAb titers, AIM+ CD4+/CD8+ cells, or the number of IFN-γ-producing T cells). Several interesting and biologically plausible correlations were detected at p < 0.05 (Figures S4A–S4H); however, given the extremely large number of features measured across our multi-omics analyses, the majority of these correlations were not statistically significant after correction for multiple testing. Correlations detected included a positive correlation between a T and B cell activation-related BTM (M62) and Wuhan-Hu-1 pseudovirus NAb titers at V2B after BNT162b2 (Figure S4B) and a positive correlation between an IFN-related BTM (M75) and anti-Spike IgG titers (Figure S4C), consistent with the important role of IFN signaling in optimal responses to this vaccine. Additionally, the signaling in T cells (II) BTM (M35.1) was positively correlated with the proportion of AIM+CD4+ T cells at V2B (Figure S4D). Interestingly, those BTMs that correlated with ChAdOx1-S immunogenicity were different from those identified for BNT162b2 and included several negative correlations between antigen presentation-related BTMs and the proportion of AIM+CD4+ T cells at V2B (Figure S4A, Table S5). We also assessed correlations at the level of individuals genes, identifying hundreds of correlations before correction for multiple testing (Table S5). For example, CD40 expression in blood at V2A was positively correlated with anti-RBD titers at V2B (Figure S4F). Similarly, the expression of RNF115 (Figure S4G), part of the RIG-I signaling pathway induced upon viral infection, was positively correlated with the number of IFN-γ-producing T cells at V2B. A further 15 BTMs at V3A were correlated with antibody titers or T cell responses at V3B (Figure S4H, Table S5). Notably, there were several positive correlations between the activity of antigen presentation, monocyte, and dendritic cell (DC)-related BTMs at V3A and the proportion of AIM+CD8+ T cells or IFN-γ-producing T cells at V3B, whereas plasma cell and memory B cell-related modules were negatively correlated. Only one BTM (M58, B cell development/activation) was positively correlated with anti-RBD, anti-Spike and NAb titers.
To further examine which transcriptional signatures were associated with robust antibody responses, we divided participants into high (top quartile of neutralizing antibody titers at V3B) versus low (bottom quartile of neutralizing antibody titers at V3B) responders after the third dose. GSEA revealed that low responders had increased expression of neutrophil and inflammation-related BTMs at V3A, while high responders were enriched for B cell, plasma cell, and antigen presentation modules (Table S2), further emphasizing the relationship between the induction of innate immune responses immediately after vaccination and subsequent immunogenicity.
We next used a multiplex immunoassay (LEGENDplex Human Anti-Virus Response 13-plex Panel, BioLegend) to quantify 13 cytokines/chemokines in plasma at V0, V1, V2A, and V3A (Figures 3G–3I, S5). We did not detect a difference in cytokine levels at V1 in participants vaccinated with either BNT162b2 or ChAdOx1-S. Consistent with previous reports14,33 and our gene expression data, plasma IP-10 levels were elevated in BNT162b2 recipients approximately 1–2 days after the second and third doses (Figure 3K). IFN-γ levels were also significantly increased after the third dose (Figure 3L). Although no cytokines or chemokines were significantly elevated after ChAdOx1-S (Figures 3G–3I), this could be due to the smaller sample size compared with the BNT162b2 group.
Lymphopenia and activation of immune cells approximately 1–2 days after the second and third doses of vaccine
As reported above, we performed flow cytometry analysis to characterize changes in 54 immune cell sub-populations at V0 and all timepoints after vaccination. BNT162b2 induced a mild lymphopenia at V2A and V3A (Figures 4A, 4B, S6A, Table S3), which included a decrease in CD3+ T cells (Figure 4C), multiple natural killer (NK) cell subsets (Figures 4A, 4B, and 4D), both conventional DCs (cDC) and plasmacytoid DCs (pDC), multiple subsets of CD4+ and CD8+ T cells, cTfh cells, and memory B cell subsets (Figures 4A and 4B, Table S3). Intermediate monocytes were increased at V2A and V3A after BNT162b2 (Figure 4E). No populations were detected as significantly altered at any time point after ChAdOx1-S vaccination compared with V0; however, as can be observed (Figure 4B), the trend was similar to BNT162b2 and the lack of statistical significance is likely due to the smaller sample size of ChAdOx1-S participants. When directly statistically comparing immune cell populations after ChAdOx1-S and BNT162b2, aside from the differences at V1 reported above, only classical monocytes were significantly different and were increased at V2A after ChAdOx1-S (Figure 4F). No differences in immune cell populations were detected at V3A or V3B when comparing between the different vaccine groups.
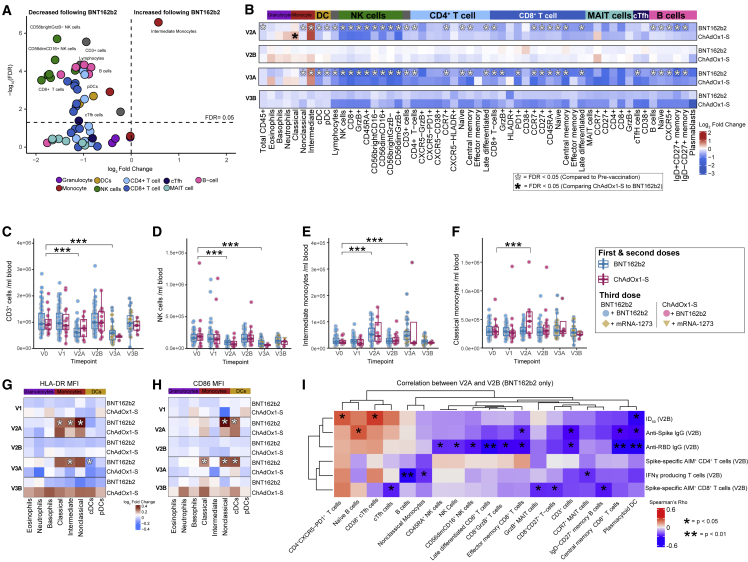
Figure 4.
Flow cytometry analysis of major immune cell populations in peripheral blood approximately 1–2 days after the second and third doses of vaccine
(A) Volcano plot showing changes in immune cell populations in BNT162b2 vaccinated participants (n = 44) at V2A relative to V0.
(B) Heatmap showing the fold-change (log2 transformed) in immune cell populations in BNT162b2 or ChAdOx1-S vaccinated participants at V2A, V2B, V3A, and V3B relative to V0.
(C–H) (C) CD3+ cells, (D) NK cells, (E) intermediate monocytes, and (F) classical monocytes (counts/mL) in peripheral blood pre- and post-vaccination. Heatmap showing fold-change in the expression of myeloid cell activation markers (G) HLA-DR and (H) CD86 at V1-V3B relative to V0.
(I) Heatmap of correlations between immune cell populations at V2A and antigen-specific antibody and T cell responses at V2B after BNT162b2. Only populations with at least one significant correlation (p < 0.05) shown. Statistical significance was assessed in (A–H) with a generalized linear model. Spearman correlations shown in (I). ∗p < 0.05, ∗∗∗p < 0.001.
We also examined myeloid activation markers and found that, after two doses of BNT162b2, the expression of HLA-DR was increased on all monocyte subsets assessed (Figure 4G). ChAdOx1-S recipients showed a similar trend, but again these differences did not attain statistical significance. HLA-DR expression was similarly increased on monocyte subsets at V3A after BNT162b2, although these differences were only statistically significant for intermediate monocytes. Expression of CD86 was increased on nonclassical monocytes and cDCs at V2A and V3A, and on classical monocytes at V3A, in participants vaccinated with BNT162b2 (Figure 4H).
The frequencies of multiple immune cell populations at V2A were correlated (p < 0.05) with antigen-specific antibody or T cell responses at V2B (Figures 4I and S6B). For example, the number of pDCs was negatively correlated with antibody and T cell responses at V2B, while CD38+ cTfh cells and PD-1 expressing CD4+ T cells were positively correlated with NAb titers (Figure 4I). The correlations observed after ChAdOx1-S were different from those observed after BTN162b2 (Figure S6B). For example, multiple CD8+ T cell and innate immune cell subsets were negatively correlated with anti-RBD titers at V2B, while multiple NK cell subsets were positively correlated with the proportion of AIM+CD8+ T cells at V2B (Figure S6B). After BNT162b2, the number of CD38+ cTfh cells at V2A was positively correlated with Wuhan-Hu-1 pseudovirus Nabs at V2B (Figure S6C). In some cases, opposing correlations were observed. For example, CD38+ cTfh cells at V2A were negatively correlated with Spike-specific IFN-γ-secreting cells at V2B after ChAdOx1-S, but were positively correlated after BNT162b2 (Figure S6D). Similarly, the frequency of NK cells at V2A were positively correlated with AIM+CD8+ T cells at V2B after ChAdOx1-S, but negatively correlated after BNT162b2 (Figure S6E). No immune cell populations at V3A were correlated with antibody responses at V3B; however, 10 immune cell populations were negatively correlated with T cell responses (Table S5), including negative correlations between naive CD4+ T cells at V3A and the number of Spike-specific IFN-γ-secreting cells at V3B, and between CD4+ MAIT cells at V3A and the proportion of AIM+CD8+ T cells at V3B (Table S5).
Pre-vaccination immune status is correlated with vaccine immunogenicity
As previous studies suggest that the pre-vaccination immune state is predictive of antigen-specific responses to other vaccines,34 we sought to assess whether this is also true for COVID-19 mRNA or adenovirus vectored vaccines. We found multiple immune cell populations, BTMs, and cytokine and chemokine levels before vaccination were correlated (p < 0.05) with antigen-specific antibody or T cell responses at V2B after BNT162b2 (Figure 5A) or ChAdOx1-S (Figure S6B). For example, the number of CD38+ cTfh cells before vaccination was positively correlated with NAb titers at V2B after vaccination with BNT162b2 (Figure 5B). Unexpectedly, we found that both the transcriptional activity of B cell-related BTMs before vaccination and number of B cells in blood, as assessed by flow cytometry, were negatively correlated with antigen-specific T cell responses at V2A after vaccination with either BNT162b2 or ChAdOx1-S (Figures 5A, 5C, and 5D, S6F, Table S5). We also identified that the pre-vaccination concentration of IP-10 was positively correlated with anti-Spike IgG titers after BNT162b2 (Figure 5E). Correlations between BTMs, immune cell populations, and cytokine concentrations with antibody and T cell responses induced after the third dose (either BNT162b2 or mRNA-1273) were also identified (Figure 5F, Table S5). For example, the number of naive B cells before vaccination was correlated with anti-Spike titers at V3B, whereas BTM M146 (MHC-TLR7-TLR8 cluster) was positively correlated with the proportion of AIM+CD8+ T cells at V3B (Figure 5F).
Figure 5.
Pre-vaccination immune status is correlated with antigen-specific antibody and T cell responses after the second or third doses of vaccine
(A) Heatmap showing correlations between BTM activity score, immune cell counts, or cytokine concentration at V0, and antigen-specific antibody and T cell responses at V2B (n = 86).
(B) Correlation between CD38+ cTfh cells at V0 and Wuhan-Hu-1 ID50 at V2B.
(C) Correlation between activity of BTM 47.2 (enriched in B cells III) at V0 and Spike-specific AIM+ CD4+ T cells at V2B.
(D) Correlation between B cells (counts/mL) at V0 and Spike-specific AIM+ CD4+ T cells at V2B.
(E) Correlation between IP-10 in plasma at V0 and anti-Spike IgG titers at V2B.
(F) Heatmap of correlations between BTM activity, immune cell counts, or cytokine concentration at V0 and antigen-specific responses at V3B.
(A and F) Only BTMs, immune cell populations, or cytokines with at least one statistically significant correlation (p < 0.05) are shown. Spearman correlations shown in (A–F). ∗p < 0.05, ∗∗p < 0.01.
Reactogenicity after vaccination is associated with higher T cell responses, but not antibody responses
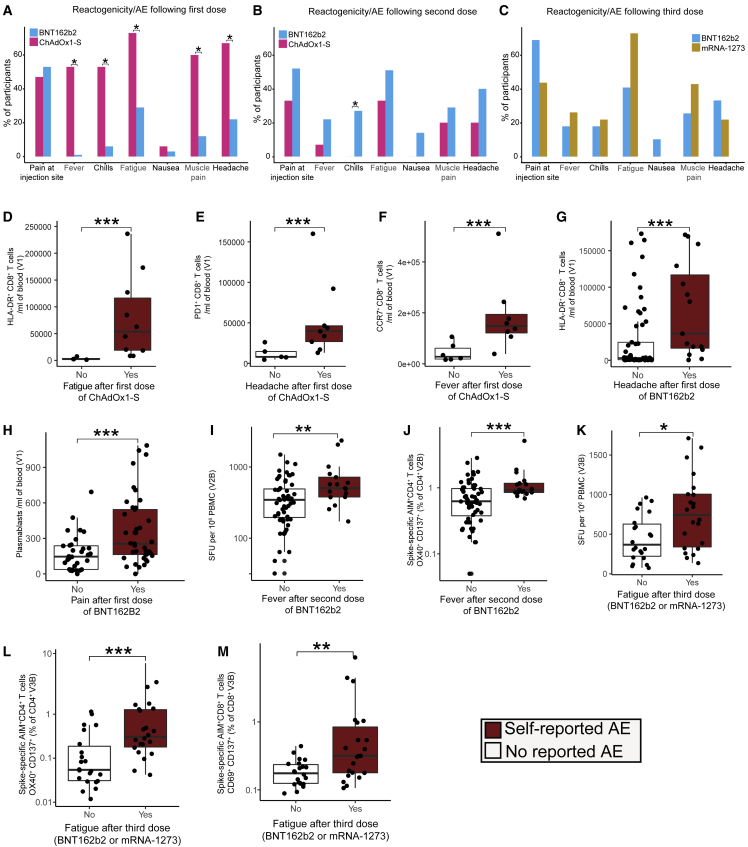
A survey recording symptoms of vaccine reactogenicity was sent to participants 1 week after each vaccination (>95% completion rate, Table S6, Data S1). No participants reported serious AEs (e.g., TTS). After the first dose, participants who received ChAdOx1-S reported higher rates of fatigue, fever, chills, muscle pain, and headache compared with those vaccinated with BNT162b2 (Figure 6A). In contrast, after the second dose of vaccine, higher rates of chills were detected in those vaccinated with BNT162b2 (Figure 6B). There was no significant difference in reported AEs between those vaccinated with BNT162b2 or mRNA-1273 for their third dose (Figure 6C). These patterns of AEs were consistent with prior reports.35 In contrast with prior reports,14 we did not detect associations between reported AEs and antigen-specific anti-Spike/RBD or NAb titers induced by either vaccine at any timepoint (data not shown). We also did not detect a relationship between reactogenicity and the pre-vaccination transcriptome, cytokine levels or circulating immune cell populations. However, higher frequencies of HLA-DR+CD8+ T cells, PD-1+CD8+ T cells, and CCR7+CD8+ T cells were observed in those who experienced fatigue, headache, or fever, respectively, after the first dose of ChAdOx1-S (Figures 6D–6F). HLA-DR+CD8+ T cells were also increased in those who experienced a headache after the first dose of BNT162b2 (Figure 6G). The frequency of plasmablasts at V1 was also associated with pain at the injection site and headache after the first dose of BNT162b2 (Figures 6H, S6G) but was not correlated with subsequent antibody or T cell responses. Participants reporting chills at V2A after vaccination with BNT162b2 had lower levels of IL8, IP-10, and tumor necrosis factor-α (Figures S6H–S6J). Interestingly, those who reported fever after the second dose of BNT162b2 had increased numbers of Spike-specific IFN-γ-secreting cells at V2B (Figure 6I) and an increased proportion of Spike-specific AIM+CD4+ T cells (Figure 6J). After the third dose (BNT162b2 or mRNA-1273), we observed that fatigue was correlated with an increased proportion of Spike-specific IFN-γ-secreting cells and AIM+CD4+ and AIM+CD8+ T cells (Figures 6K-6M). These data suggest a relationship between reactogenicity and increased T cell responses after vaccination with COVID-19 mRNA vaccines.
Figure 6.
Reactogenicity following the BNT162b2, mRNA-1273 or ChAdOx1-S vaccines
(A–F) The proportion of participants who reported AE after the (A) first, (B) second dose of BNT162b2 (n = 86) or ChAdOx1-S (n = 16) or (C) third dose of BNT162b2 (n = 38) or mRNA-1273 (n = 14). Activated CD8+ T cells subsets after the first dose of ChAdOx1-S in participants who reported (D) fatigue (E) headache or (F) fever.
(G) HLA-DR+ CD8+ T cells in participants who reported headache after the first dose of BNT162b2.
(H) Plasmablasts in participants who reported pain at the injection site after the first dose of BNT162b2.
(I) Spike-specific IFN-γ SFU at V2B in participants who reported fever after a second dose of BNT162b2.
(J) AIM+ CD4+ T cells at V2B in participants who reported fever after a second dose of BTN162b2.
(K) Spike-specific IFN-γ SFU at V3B in participants who reported fatigue after a third dose (BNT162b2/mRNA-1273).
(L) AIM+ CD4+ T cells at V3B in participants who reported fatigue after a third dose of BNT162b2 or mRNA-1273.
(M) The proportion of Spike-specific AIM+ CD8+ T cells at V3B in participants who reported fatigue after a third dose of BNT162b2 or mRNA-1273. Data in (D–M) are represented as boxplots (see Figure 1). Statistical significance was assessed in (A–C) using Fisher’s exact tests, and in (D–M) using Wilcoxon rank-sum tests. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
Vaccination is a cornerstone of the global strategy to control the spread of SARS-CoV-2 with tens of millions of lives saved to date.36 While randomized controlled trials have demonstrated the efficacy of both mRNA and adenoviral vectored vaccines against severe COVID-19,2,3,37,38 such trials typically do not include systems-level assessments of the immune response to vaccination, limiting our knowledge of the precise mechanisms that determine optimal immune responses to different COVID-19 vaccine types. Here, we present a longitudinal, systems-level analysis of the early (days) and late (weeks to months) immune responses induced in a cohort of 102 SARS-CoV-2-naive, healthy adults, vaccinated with either mRNA (BNT162b2, mRNA-1273) or adenoviral vectored (ChAdOx1-S) vaccines. We comprehensively profiled immune responses in blood before vaccination, and at 1–7 days after the first, second, and third doses, using a multi-omics approach that included transcriptomics, proteomics, lipidomics, cytokine profiling, and multi-parameter flow cytometry, and compared these responses with antigen-specific antibody and T cell responses induced approximately 28 days after the second and third doses.
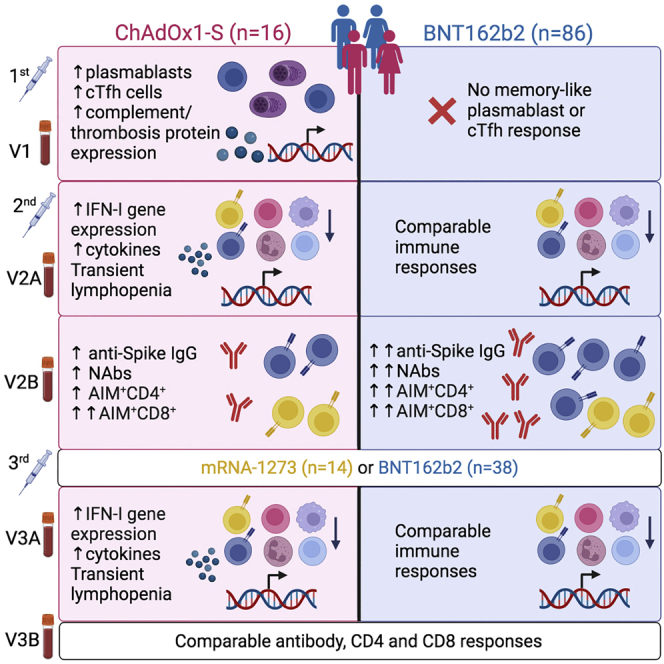
After the first dose of ChAdOx1-S, we detected significant differences in the blood transcriptome that were not observed in BNT162b2 vaccinated participants, which suggested that the induction of a plasma cell response within 6 days of the first dose of ChAdOx1-S. Multiparametric flow cytometry confirmed this, revealing a significant increase in both plasmablasts and recently activated cTfh cells in ChAdOx1-S vaccinated individuals 6–7 days after the first dose. This response is remarkably similar to the anamnestic responses observed after seasonal influenza vaccination28,39 and suggested pre-existing immunity to a component of the ChAdOx1-S vaccine or vector. Pre-existing immunity in the form of ChAdOx1-neutralizing antibodies is thought to be uncommon,40,41 but there has been little investigation into ChAdOx1 cross-reactive T cell memory. ChAdOx1-S vaccination has recently been shown to expand a pre-existing CD4+ memory T cell pool that also responds to the human adenovirus 5 hexon protein, which shares approximately 80% sequence homology with the ChAdOx1 hexon protein,42 strongly suggesting that cross-reactive T cell epitopes are shared between human adenoviruses and the ChAdOx1 viral vector. We demonstrate that a proportion of the cTfh cells induced by ChAdOx1-S vaccination at V1 are ChAdOx1 hexon specific and are strongly and positively correlated with the number of circulating plasmablasts at this time point. We also observe a concomitant increase in the number of ChAdOx1-specific IFN-γ-secreting cells. Importantly, we do not see any increase in the number of Spike-reactive T cells at this time point, suggesting that this is likely expansion of pre-existing ChAdOx1-specific memory cells. Together, these data strongly support the existence of pre-existing cellular immunity to the ChAdOx1 adenoviral vector, likely a form of cross-reactive memory derived from prior exposure to human adenoviruses, which may have implications for future adenovirus-vectored vaccines and therapies.
Interestingly, our proteomics analyses revealed that there was a strong positive correlation between the abundance of multiple coagulation and complement proteins in plasma and the magnitude of the cTfh and plasmablast response induced 6–7 days after the first dose of ChAdOx1-S, suggesting a link between the induction of this memory-like response and the expression of proteins involved in thrombosis. Adenoviral vectored vaccines have been associated with rare cases of TTS, driven in part by the production of auto-antibodies against PF4, with clinical symptoms manifesting from 7 days after vaccination.18,19,20 How these PF4 auto-antibodies manifest in those rare cases of TTS is unclear, but it has been demonstrated that several adenovirus vaccine vector candidates, including ChAdOx1, bind to PF4 in vitro.29,43 If the same phenomenon occurs in vivo, PF4-specific B cells could internalize and present PF4-ChAdOx1 complexes to ChAdOx1 cross-reactive memory T cells, leading to intermolecular epitope spreading and the production of PF4 auto-antibodies,44,45 consistent with a proposed mechanism for TTS.29 Although we were unable to detect a statistically significant increase in PF4 auto-antibodies in ChAdOx1-S vaccinated participants, none of whom developed TTS, further investigation is warranted to assess the antigen specificity of the plasmablasts that appear after the first dose of adenovirus-vectored vaccines and the consequences this may have for vaccine-induced AEs.
Interestingly, given the differences observed after the first dose, similar transcriptional, cytokine/chemokine, and immune cell population changes were induced approximately 1–2 days after the second or third dose, irrespective of vaccine type. Several hundred genes changed in response to vaccination, and through correlation analyses we were able to link the expression of specific genes (e.g., CD40) and BTMs, induced approximately 1–2 days after vaccination, to the magnitude of subsequent antigen-specific antibody and T cell responses assessed approximately 28 days later. A mild lymphopenia was induced approximately 1–2 days after the second and third vaccine doses, which was observed irrespective of vaccine administered. This lymphopenia was primarily driven by decreased numbers of T, B, and NK cells, likely due to migration from the peripheral blood to lymphoid organs. The magnitude of the mild lymphopenia was also correlated with antigen-specific responses approximately 28 days later. Interestingly, the only cell populations detected as increased in this period after vaccination were classical (CD14+CD16−) and intermediate (CD14+CD16+) monocytes, which were increased by ChAdOx1-S or BNT162b2 vaccination, respectively. The increase in classical monocytes is particularly noteworthy; they were negatively correlated with anti-RBD antibody responses induced by ChAdOx1-S. In other contexts, monocyte recruitment has been shown to be capable of disrupting formation of germinal centers.46,47 We also identified a positive correlation between the number of CD38+ cTfh cells at V2A and pseudovirus NAb titers at V2B in BNT162b2-vaccinated, but not ChAdOx1-S-vaccinated, participants. Additionally, the frequency of specific immune cell populations and the transcriptional activation of BTMs before vaccination were correlated with subsequent vaccine-specific B and T cell responses. Irrespective of vaccine, an overt B cell signature before vaccination was associated with lower vaccine-specific T cell responses at V2B, while more abundant cytotoxic CD8+ T cell subsets before BNT162b2 vaccination was associated with lower NAb titers at V2B. These data suggest that pre-vaccine immune signatures can be used to predict the magnitude of subsequent vaccine-specific immune responses.
Consistent with prior reports,11,48,49 the proportion of AIM+CD4+ T cells was lower in participants who received ChAdOx1-S, relative to BNT162b2-vaccinated participants. However, after a third dose of a mRNA vaccine, AIM+CD4+ T cells in ChAdOx1-S-vaccinated participants recovered to levels comparable with triple mRNA vaccinated participants.48,50 In contrast, there was no difference in the number of Spike-specific CD8+ T cells at any timepoint after vaccination between ChAdOx1-S- and BNT162b2-vaccinated participants, demonstrating a specific deficit in the generation of CD4+ T cell responses after ChAdOx1-S vaccination. The proportion of AIM+CD4+ T cells at V2B in ChAdOx1-S-vaccinated participants was also positively correlated with NAb titers, which were decreased in ChAdOx1-S-vaccinated participants relative to BNT162b2 participants at V2B, but recovered after a heterologous mRNA vaccination. These data suggest that a possible underlying cause for the lower antibody titers after ChAdOx1-S vaccination may be suboptimal CD4+ T cell responses, which provide less help to promote humoral responses. Additionally, the number of IFN-γ-secreting cells at V2B in ChAdOx1-S-vaccinated participants was negatively correlated with the number of cTfh cells as determined by flow cytometry. As Th1 and Tfh cells represent mutually exclusive T cell fates,51,52 ChAdOx1-S vaccination may drive suboptimal CD4+ T cell expansion that is Th1 biased at the expense of Tfh differentiation, resulting in poorer antibody responses as compared with the BNT162b2 vaccine. Further experiments in animal models are needed to prove causality.
Whether vaccine reactogenicity translates into enhanced immunogenicity is currently not well understood, with conflicting reports.53,54 Here, reactogenicity after the first dose of ChAdOx1-S was associated with an increased activation of CD8+ T cells and increased numbers of circulating plasmablasts in the blood of BNT162b2 vaccinated individuals. Interestingly, we did not detect any significant associations between reactogenicity and anti-Spike/RBD or NAb titers. However, fever and fatigue were associated with more robust AIM+ T cell responses after the second and third doses of vaccine, respectively. We also identified a significant association between reactogenicity and CD8+ T cell activation that, in turn, was negatively correlated with anti-Spike IgG titers. Together these data suggest that reactogenicity is associated with enhanced vaccine-specific T cell responses, but not vaccine-specific antibody titers.
In conclusion, COVIRS represents a significant resource for understanding the early innate and adaptive immune responses to mRNA and adenovirus vectored COVID-19 vaccines and how these responses relate to immunogenicity and reactogenicity. The generation of high titers of Spike-specific binding and neutralizing antibodies is currently the best correlate of protection after vaccination against SARS-CoV-2, although the contribution of vaccine-induced T cells in protection from SARS-CoV-2 infection is becoming increasingly apparent.55,56 Identifying the factors that promote the optimal generation of vaccine-specific T and B cell responses is a key priority and our study provides a wealth of data to inform future rational vaccine design.
Limitations of the study
Our study has several limitations, including an imbalance of in the number of participants receiving BNT162b2 compared with the ChAdOx1-S and mRNA-1273 vaccines. This was unavoidable; BNT162b2 was the recommended vaccine for the majority of participants in our cohort and vaccine administration was not under our control. The cost and complexity of the multi-omics analyses performed also limits the number of participants that can be included in such systems vaccinology studies, although we note that our sample size is at the upper end of comparable studies in the field. The large number of different parameters assessed in our multi-omics analyses is also a challenge for correlation analyses that account for multiple testing. For this reason, we report correlations that were statistically significant without correction for multiple testing, accepting this approach as a limitation of the large number of parameters versus the sample size. Analyses were performed on peripheral blood; analysis of immune responses in draining lymph nodes or the mucosa would be an exciting addition in future studies. We focused on the use of pre-defined BTMs in much of our transcriptomic analysis to enable comparisons to other recently published systems vaccinology studies.57,58 However, one should be cognizant of potential differences in the composition of BTMs in whole blood compared with PBMC used in others.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-Human IgG (H + L) Secondary Antibody, HRP | Invitrogen | Cat#31410; RRID:AB_228269 |
| CD3 FITC | BD Biosciences | Cat#555339; RRID:AB_395745 |
| CD3 BUV395 | BD Biosciences | Cat#564117; RRID:AB_2738603 |
| CD4 M-T477 BV480 | BD Biosciences | Cat#746671; RRID:AB_2743943 |
| CD4 BV605 | BD Biosciences | Cat#562658; RRID:AB_2744420 |
| CD8 BV650 | BD Biosciences | Cat#563821; RRID:AB_2744462 |
| CD8 G42-8 BV711 | BD Biosciences | Cat#743068; RRID:AB_2741262 |
| CD11c PE | BD Biosciences | Cat#555392; RRID:AB_395793 |
| CD14 Alexa Fluor 647 | BD Biosciences | Cat#562690; RRID:AB_2737724 |
| CD16 BV421 | BD Biosciences | Cat#562874; RRID:AB_2716865 |
| CD19 BV786 | BD Biosciences | Cat#563325; RRID:AB_2744314 |
| CD19 PE | BD Biosciences | Cat#555413; RRID:AB_395813 |
| CD20 BV786 | Biolegend | Cat#302355; RRID:AB_2566315 |
| CD27 BV711 | BD Biosciences | Cat#563167; RRID:AB_2738042 |
| CD38 BV510 | BD Biosciences | Cat#563251; RRID:AB_2738097 |
| CD45 BV395 | BD Biosciences | Cat#563792; RRID:AB_2869519 |
| CD45RA FITC | Biolegend | Cat# 304106; RRID:AB_314410 |
| CD56 BUV563 | BD Biosciences | Cat#612928; RRID:AB_2870213 |
| CD69 FN50 PE-CF594 | BD Biosciences | Cat# 562617; RRID:AB_2737680 |
| CD86 PerCP-Cy5.5 | BD Biosciences | Cat#561129; RRID:AB_10562395 |
| CD123 BV711 | BD Biosciences | Cat#563161; RRID:AB_2738038 |
| CD134 (OX40) Ber-ACT35 PE-Cy7 | Biolegend | Cat#350012; RRID:AB_10901161 |
| CD137 (4-1BB) 4B4-1 APC | Biolegend | Cat#309810; RRID:AB_830672 |
| CD185(CXCR5) PE-Vio770 | Miltenyi Biotec | Cat#130-117-508; RRID:AB_2733204 |
| CD197(CCR7) Alexa Fluor 647 | BD Biosciences | Cat#560816; RRID:AB_2033948 |
| CD279 (PD1) BUV737 | BD Biosciences | Cat#612791; RRID:AB_2870118 |
| CCR7 (CD197) 2-L1-A BV421 | BD Biosciences | Cat# 566743; RRID:AB_2869843 |
| BD Horizon PE-CFS594 mouse anti-human Granzyme B | BD Biosciences | Cat#562462; RRID:AB_2737618 |
| Fixable Viability Stain 780 | BD Biosciences | Cat#565388; RRID:AB_2869673 |
| HLA-DR APC-H7 | BD Biosciences | Cat#561358; RRID:AB_10611876 |
| IFN-γ 1D1K purified | MabTech | Cat#3420-3-250; RRID:AB_907283 |
| IFN-γ 7-B6-1 biotin | MabTech | Cat#3420-6-250; RRID:AB_907273 |
| Siglec-8 PE-dazzle594 | Biolegend | Cat#347109; RRID:AB_2629717 |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 ectodomain (isolate WHU1, residues1-1208) with HexaPro mutations | Hsieh et al. 202059 (Provided by Adam Wheatley) | N/A |
| SARS-Cov-2 receptor-binding domain (RBD) with C-terminal His-tag (residues 319–541) | Amanat et al. 202060 (Provided by Florian Krammer) | N/A |
| SARS-CoV-2 Spike Glycoprotein-crude | GenScript | Cat#RP30020 |
| Comassie Blue | Thermo Fisher Scientific | Cat#20278 |
| 1-Step™ Ultra TMB Substrate | Thermo Fisher Scientific | Cat#34028 |
| ELISA/ELISpot buffer | Thermo Fisher Scientific | Cat# 00-4202-56 |
| 2M sulfuric acid | Merck | Cat#1.60313 |
| Murine leukemia virus (MLV) gag/pol and luciferase vectors | Keck et al., 2009, Bartosch et al., 2003,61,62 (Provided by Prof. Francois-Loic Cosset) | N/A |
| DMSO | Sigma | Cat#D8418 |
| Fetal Calf Serum, Australian origin | CellSera | Cat# F31904 |
| RNase free, PCR grade water | Fisher Biotec | Cat#UPW-100 |
| RPMI-1640 Medium | Sigma | Cat#R8758 |
| RNAlater™ Stabilization Solution | Thermo Fisher | Cat#AM7021 |
| Dulbecco’s PBS | Merck | Cat#D8537 |
| BSA | AusGeneX | Cat#PBSA-250G |
| EDTA | Merck | Cat#E8008 |
| Ethanol | Merck | Cat#E7023 |
| FACS-Lyse 10X Concentrate | BD Biosciences | Cat#349202 |
| Sodium acetate solution | Merck | Cat#71196 |
| Tris (2-carboxyethyl) phosphine hydrochloride | Merck | Cat#C4705 |
| 2-chloracetamide | Merck | Cat#C0267 |
| PureCube NHS Activated MagBeads | Cube Biotech | Cat#50405 |
| Acetonitrile | Merck | Cat#271004 |
| Lysyl Endopeptidase®, Mass Spectrometry Grade (Lys-C) | Fujifilm Wako Pure Chemical Corporation | Cat#121-05063 |
| SOLu-trypsin | Merck | Cat#EMS0004 |
| AEC Substrate Set | BD Biosciences | Cat#551951 |
| Streptavidin-HRP | BD Biosciences | Cat#554066; RRID:AB_2868972 |
| Human Tru-stain FC Block | Biolegend | Cat#422301 |
| Human MR1 6-FP tetramer BV421 | NIH tetramer facility | MR1 |
| Formic Acid | Merck | Cat# 100264 |
| ELISpot plates | Merck | Cat# MSIPS4W10 |
| Plasmid encoding SARS-CoV-2 Spike | Hoffmann et al., 202063 | N/A |
| Critical commercial assays | ||
| MagMAX Viral/Pathogen Nucleic Acid Isolation Kit | Thermo Fisher Scientific | Cat#A42352 |
| Superscript III one step RT-PCR system | Thermo Fisher Scientific | Cat#12574018 |
| Mammalian Calphos transfection kit | Takara Bio | Cat#631312 |
| Polybrene Infection/Transfection Reagent | Sigma-Aldrich | Cat#TR-1003 |
| Bright-Glo™ Luciferase Assay System | Promega | Cat#E2610 |
| Brilliant Violet Stain Buffer | BD Biosciences | Cat#566349 |
| Lymphoprep | Stem Cell Technologies | Cat#07851 |
| Foxp3/Transcription Factor Fixation/Permeabilization Concentrate and Diluent | eBioscience | Cat#00-5521-00 |
| Perm/Wash Buffer | BD Biosciences | Cat#554723 |
| Liquid counting beads | BD Biosciences | Cat#335925 |
| RiboPure™ RNA Purification kit, Blood RNA kit | Thermo Fisher Scientific | Cat#AM1928 |
| Human Anti-Heparin/Platelet Factor 4 Antibody (Anti-HPF4) ELISA Kit | Abbexa | Cat#abx150683 |
| LEGENDplex™ Human Anti-Virus Response Panel (13-plex) with V-bottom Plate | BioLegend | Cat#740390 |
| Nugen Universal Plus Total RNA-Seq library kit with Anydeplete | Tecan | Cat# 0520-A01 |
| DNBSEQ-G400RS High-throughput Sequencing Set (FCL PE150)∗ | MGI | Cat# 1000016952 |
| Deposited data | ||
| Supporting data | BitBucket | https://bitbucket.org/lynnlab/covirs |
| Raw Sequence data | Gene expression omnibus | GSE199750 |
| Proteomics data | PRIDE | PXD036608 |
| Additional supplementary data | Mendelay data | https://doi.org/10.17632/cd3fxgssc5.1 |
| Experimental models: Cell lines | ||
| Expi293 cells | Thermo Fisher Scientific | A14635 |
| CD81KO 293T cells | Kalemera et al.64 (provided by Dr. Joe Grove) | N/A |
| 293T-ACE2 over-expressed cells | Crawford et al.65 (A/Prof. Jesse Bloom) | N/A |
| Software and algorithms | ||
| MultiQC | Ewels et al.66 | v 1.10.1 |
| Trimmomatic | Bolger et al.67 | v 0.38 |
| HISAT2 | Kim et al.68 | v 2.1.0 |
| FeatureCounts | Liao et al.69 | v 1.5.0-p2 |
| Ensembl | Howe et al.70 | v 101 |
| R | R Foundation for Statistical Computing | v 4.2 |
| EdgeR | Robinson et al.71 | v 3.38 |
| SVASeq | Leek72 | v 3.44 |
| Hmisc | Harrell Jr. and Harrell Jr.73 | v4.7 |
| Prism Graphpad | GraphPad Software, Inc. | v 8 and 9 |
| ggplot2 | Wickham74 | v3.3.6 |
| limma | Ritchie et al.75 | v3.51 |
| Msigdbr | Dolgalev76 | v7.5.1 |
| Spectre | Ashhurst et al.77 | v1.0 |
| GSVA | Hänzelmann et al.32 | v1.45.2 |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact, Prof. David Lynn (david.lynn@sahmri.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Participant recruitment
This study was performed in accordance with the ethical principles consistent with the latest version of the Declaration of Helsinki (version Fortaleza 2013), Good Clinical Practice (GCP) and according to the Australian National Health and Medical Research Council Guidelines for Research published in the National Statement on the Ethical Conduct in Human Research (2007; updated 2018). 146 healthy adult participants were recruited in Adelaide, South Australia between April 8th and November 1st, 2021 under protocols approved by the CALHN Human Research Ethics Committee, Adelaide, Australia (Approval No. 14778) or by the Royal Children’s Hospital, Melbourne, Australia, Human Research Ethics Committee (Approval No. 62586). Strict international and interstate border control measures to prevent community transmission of SARS-CoV2 in South Australia were in place until late November 2021. Participant inclusion criteria were being healthy, aged 18+ years, voluntary informed consent, and the ability to attend study follow-up visits. Exclusion criteria included any previous positive SARS-CoV-2 test prior to vaccination, a COVID-19 vaccine administered prior to baseline blood sample collection, or any other vaccines received 14 days prior to a COVID-19 vaccine.
Participants received two doses of either the BNT162b2 (n = 86) or ChAdOx1-S (n = 16) vaccines for their first/second vaccinations. Participants were asked to provide a blood sample before their first COVID-19 vaccine and then ∼6 days after the first dose (V1), and ∼1–2 days (V2A) and ∼28 days after the second dose (V2B). In a subset of participants, blood samples were collected ∼1–2 days (V3A) and ∼28 days (V3B) after a third dose of BNT162b2 (n = 38) or mRNA-1273 (Moderna, n = 14) vaccine. Venous blood (∼29 mL/individual) was collected in multiple blood tubes including serum separator (CAT serum separator clot activator) tubes processed for serum; sodium citrate tubes processed for plasma; sodium heparin tubes for peripheral blood mononuclear cell (PBMC) isolation; and lithium heparin tubes for whole blood preservation in RNAlater solution (Thermo Fisher Scientific, Waltham, USA) or FACSlyse buffer (BD Biosciences, Franklin Lakes, USA) used for RNA extraction and flow cytometry analysis, respectively. Blood tubes were processed within 3 h of collection. A survey recording any symptoms of vaccine reactogenicity was sent to participants for completion one week after each vaccination (Data S1). The third survey also captured any known COVID-19 positive events among participants when community transmission began to occur in South Australia in December 2021, following the lifting of border restrictions.
Method details
SARS-CoV-2 protein purification and ELISA
Recombinant SARS-CoV-2 spike protein and receptor-binding domain (RBD) were produced for ELISA. Prefusion SARS-CoV-2 ectodomain (isolate WHU1, residues1-1208) with HexaPro mutations59 (kindly provided by Adam Wheatley) and SARS-Cov-2 receptor-binding domain (RBD) with C-terminal His-tag60 (residues 319–541; kindly provided by Florian Krammer) were over-expressed in Expi293 cells and purified by Ni-NTA affinity and size-exclusion chromatography. Recombinant proteins were analyzed via a standard SDS-PAGE gel to check protein integrity. Gels were stained with Comassie Blue (Invitrogen) for 2 h and de-stained in distilled water overnight. For ELISA, MaxiSorp 96-well plates were coated overnight at 4°C with 5 μg/mL of recombinant RBD or Spike proteins. After blocking with 5% w/v skim milk in 0.05% Tween 20/PBS (PBST) at room temperature (RT), serially diluted (heat inactivated) sera were added and incubated for 2 h at RT. Plates were washed 4 times with 0.05% PBST and secondary antibodies added. Secondary antibodies were diluted in 5% skim milk in PBST as follows: Goat anti-Human IgG (H + L) Secondary Antibody HRP (1:30,000; Invitrogen) and incubated for 1 h at RT. Plates were developed with 1-Step™ Ultra TMB Substrate (Thermo Fisher) and stopped with 2M sulfuric acid. OD readings were read at 450 nm on a Synergy HTX Multi-Mode Microplate Reader. AUC calculation was performed using Prism (GraphPad, CA, USA), where the X axis is half log10 of sera dilution against OD450 on the Y axis.
SARS-CoV-2 pseudovirus neutralization assay
SARS-CoV-2 neutralization assays were performed as previously described.78 SARS-CoV-2 pseudovirus were generated by co-transfecting expression plasmids containing SARS-CoV-2 Spike (kindly provided by Dr Markus Hoffmann63), or the Omicron variant B.1.1.529 (cat no. 179907, Addgene, MA, USA)79 and the murine leukemia virus (MLV) gag/pol and luciferase vectors (kindly provided by Prof. Francois-Loic Cosset61,62), in CD81KO 293T cells (kindly provided by Dr Joe Grove64), using the mammalian Calphos transfection kit (Takara Bio, Shiga, Japan). Pseudovirus culture supernatants were harvested 48 h post transfection and concentrated 10-fold using 100,000 MWCO Vivaspin centrifugal concentrators (Sartorius, Göttingen, Germany) by centrifugation at 2000 x g and stored at −80°C. For neutralization assays, pseudovirus was diluted in media to be 1000- to 5000-fold more infectious than negative background (based on pseudovirus lacking SARS-CoV-2 Spike). Diluted pseudovirus were incubated for 1 h with heat inactivated (56°C for 30 min) participant serum, followed by the addition of polybrene at a final concentration of 4 μg/mL (Merck, St. Louis, USA), prior to addition to 293T-ACE2 over-expressed cells (kindly provided by A/Prof Jesse Bloom65). 293T-ACE2 cells were seeded 24 h earlier at 1.5 × 104 cells per well in 96-well, white flat-bottom plates (Merck). Cells were spinoculated at 800 x g for 2 h and incubated for 2 h at 37°C, prior to media change. After 72 h, the cells were lysed with a lysis buffer (Promega, Madison, USA) and Bright Glo reagent (Promega) was added at a 1:1 ratio. Luminescence (RLU) was measured using CLARIOstar microplate reader (BMG Labtech, Ortenberg, Germany). Neutralization assays were performed in triplicates. Percentage neutralization of pseudovirus was calculated as (1 – RLUtreatment/RLUno treatment) × 100. The 50% inhibitory concentration (ID50) titer was calculated using a non-linear regression model in Prism (GraphPad, CA, USA).
PBMC isolation
Sodium-heparin anticoagulated whole blood (8–12 mL) was diluted in 12 mL of endotoxin-free sterile PBS, layered onto 12 mL Lymphoprep (Stem Cell Technologies, Vancouver, Canada) and spun at 800 x g for 30 min, RT, with no brake. PBMC were isolated from the resultant interphase layer via pipetting, washed twice in PBS, pelleted at 600 x g for 5 min at RT, and resuspended in 1.5 mL freezing medium (10% DMSO, 90% FCS). PBMC (500 μL) were aliquoted into cryovials, placed in a Mr. Frosty™ Freezing Container (Nalgene, Rochester, USA) and stored at −80°C overnight. Frozen PBMC cryovials were transferred to liquid nitrogen for long-term storage within 1–2 days.
Flow cytometry analysis of immune cell populations in whole blood
Sodium-heparin anticoagulated whole blood (200 μL) was incubated with 1700 μL FACS™ Lysing Solution (BD Biosciences) for 10 min at RT, mixing 3 times. Samples were centrifuged at 600 x g for 5 min, the supernatant aspirated and the cells resuspended in 400 μL FACSLyse. 200 μL aliquots were snap frozen at −80°C and transferred to liquid nitrogen within 1 week for long term storage. For analysis, samples were rapidly thawed at 37°C in batches, plated into a 96 well U-bottom plate on ice and spun at 600 x g for 5 min. Samples were then washed twice in 240 μL FACS buffer (PBS, 0.1% BSA, 2mM EDTA), and split into two different panels. Cells were incubated with 3 μL TruStain FcX (Biolegend, San Diego, USA) in 15 μL Brilliant Violet Stain buffer (BD Biosciences) on ice for 15 min and then stained with of one of two master-mixes of antibodies (a 13-color pan-leukocyte panel or a 17-color lymphocyte panel) in 30 μL final volume for 30 min on ice. The stained cells were washed with 200 μL FACS buffer, centrifuged 600 x g for 5 min and the pan-leukocyte panel was resuspended in FACS buffer for analysis on a flow cytometer, while the lymphocyte panel was resuspended in 100 μL 1x Permeabilization Buffer (Foxp3/Transcription Factor Staining Buffer Set, eBioscience, San Diego, USA) and incubated on ice for 10 min. The lymphocyte panel was spun at 600 x g for 5 min and stained with anti-Granzyme B (BD Biosciences) in 30 μL 1 x Perm buffer (BD Biosciences) on ice for 30 min. Samples were then washed once with 200 μL Perm buffer and once with 200 μL FACS buffer, centrifuging for 600 x g for 5 min between washes. Cells were then resuspended in FACS buffer and run immediately on a BD FACS Symphony flow cytometer (BD Biosciences). Liquid counting beads (BD Biosciences) were added to each sample to enumerate cells.
Flow cytometry data acquisition & analysis
To control for batch effects, the BD FACS symphony lasers were calibrated with dye conjugated standards (Cytometer Set &Track beads) run every day. All voltages of photomultiplier tubes (PMT)s were adjusted to negative unstained control baseline typically log scale 102. Antibodies were titrated for optimal signal over background prior to use. Compensation was set with beads matched to each panel antibody combination using spectral compensation using FlowJo Software v18 (FlowJo, Ashland, USA). Additionally, all samples across timepoints from each individual participant were analyzed in a single batch. An SSC-A/FSC-A plot was used to determine sample size and complexity. Lymphocytes, monocyte, and granulocyte gates were based on physical parameters. Events were gated for FCS-A as well as SSC-A linearity, and restricted FSC-H values for doublet discrimination. Populations of cells were expressed as a proportion of parent gated cells, proportion of the highest order lineage gate namely: Lymphocytes, B cells, T cells or CD45+ cells, or as absolute cells per mL of blood. Gating strategy for both panels are shown in (Methods S1 and S2). Flow cytometry data were imported into R v4.2 for further statistical analyses. Changes in cell counts or activation was assessed using a mixed effect linear model implemented in R v4.2 (glm function) controlling for individual and processing batch. The Benjamini and Hochberg method was employed to correct for multiple comparisons. Statistical significance was determined as FDR <0.05. Cell density plots of cTfhs and Plasmablasts were generated using the Spectre v1.0.077 package. Florescence data was transformed using the arcsinh normalization with a cofactor of 500 prior to plotting. Other visualizations were generated using ggplot2 v3.3.674 packages.
Activation induced marker (AIM) assay
Cryopreserved PBMCs were thawed and 106 live cells were plated out in 96 well U-bottom plates. Spike protein peptide pool (GenScript, New Jersey, USA), or Chimpanzee Adenovirus Y25 hexon protein peptide pool (JPT Peptide Technologies, Berlin, Germany) was added to each well at 1 μg/mL and cells incubated for 20–24 h at 37°C. For surface staining of AIM markers, cells were incubated in 1 μg/mL human Fc block (BD Biosciences) and fixable viability solution 780 (BD Biosciences) in PBS for 15 min and washed in PBS. An antibody cocktail containing antibodies against CD3 (clone UCHT1, BD Biosciences), CD4 (clone M-T477, BD Biosciences), CD8 (G42-8, BD Biosciences), CXCR5 (J252D4, BD Biosciences), CD38 (HIT2, BD Biosciences), PD-1 (EHI2.1, BD Biosciences), CD69 (clone FN50, BD Biosciences), CD137 (clone 4B4-1, Biolegend), OX40 (clone Ber-ACT35, Biolegend), CCR7 (clone 2-L1-A, BD Biosciences), and CD45RA (clone HI100, BD Biosciences)were added directly to cells and incubated for a further 30 min at 4°C. Following surface staining, cells were washed twice in PBS. All samples were acquired on a BD FACS Symphony and analyzed using FlowJO software v18 (FlowJo, Ashland, USA). The gating strategy for AIM+ T cells is shown in (Methods S3). Data were imported into R v4.2 and visualized with the ggplot2 v3.3.6 package.
ELISpot assay
ELISpot plates (MERCK-Millipore, Burlington, Vermont, USA) were coated with human IFNγ capture antibody (1D1K, Mabtech, Stockholm, Sweden; 5 μg/mL) overnight at 4°C. Plates were blocked with 1x ELISA/ELISpot blocking buffer (Thermo Fisher Scientific) for at least 1 h at RT then 2 × 105 thawed PBMCs were seeded per well and stimulated for 20-24h with pools of SARS-CoV-2 peptides (GenScript, 1 μg/mL), or ChAdY25 hexon peptides (JPT peptides 1 μg/mL). After stimulation, plates were washed 3x in PBS-T and incubated for 2 h with human biotinylated IFNγ detection antibody (7-B6-1, Mabtech; 1:500). Plates were then washed 3x with PBS-T followed by a 1 h incubation with streptavidin-HRP (BD biosciences, 1:1000) and developed with 3-Amino-9-ethylcarbazole (AEC) substrate (BD Biosciences). Spot forming units (SFU) were quantified with ImmunoSpot software (Cellular Technology Limited, Ohio, USA). Results were expressed as IFNγ-SFU/106 PBMCs. Data were imported into R v4.2 and visualized with ggplot2 v3.3.6.
Human anti-PF4 ELISA
For anti-PF4 ELISAs, a commercial kit (Abbexa, Cambridge, UK) was used as per manufacturer’s instructions. Human serum samples were diluted 1:50 in PBS. Plates were read on a Synergy HTX Multi-mode plate reader (Biotek) at 450nm. Data were imported into R v4.2 and visualized with ggplot2 v3.3.6.
Whole blood RNA extraction and library preparation
RNA extraction and genomic DNA elimination was carried out using the RiboPure™ RNA Purification kit for blood (Invitrogen) as per the manufacturer’s instructions. Final elution into 50 μL RNase-free water. A further RNA precipitation reaction was carried out. RNA was resuspended 2.5x in 100% ethanol and 10% sodium acetate (Merck) and spun at 12,000 x g for 15 min at 4°C. Samples were washed in 75% ethanol. Pellets were air dried and resuspended in 29 μL RNase free water and total RNA yield was determined by analysis of samples using a TapeStation (Agilent, Santa Clara, USA) and Qubit (ThermoFisher Scientific, Waltham, USA). Total RNA was converted to strand-specific Illumina compatible sequencing libraries using the Nugen Universal Plus Total RNASeq library kit from Tecan (Mannedorf, Switzerland) as per the manufacturer’s instructions (MO1523 v2) using 12 cycles of PCR amplification for the final libraries. An Anydeplete probe mix, targeting both human ribosomal and adult globin transcripts (HBA1, HBA2, HBB, HBD), was used to deplete these transcripts. The final library pool was converted to an MGI compatible circularized template using the MGIEasy Universal Library Conversion Kit (MGI, Shenzhen, China) before sequencing of the library pool (2 × 150 bp paired-end reads) was performed on an MGI DNBSEQ-G400.
Proteomics
Plasma samples were processed using the USP3 protocol as outlined previously.80 2 μL plasma samples were reduced/alkylated in 1% SDS lysis buffer using 10 mM Tris (2-Carboxyethyl) phosphine (TCEP, Merck) and 40 mM 2-chloracetamide (2-CAA, Merck) for 10 min at 95°C. Sample volumes containing 25 μg protein were transferred to a 0.5 mL low-bind deep-well plate (Eppendorf, Hamburg, Germany) and were incubated with 20 μL of magnetic PureCube Carboxy agarose beads (Cube Biotech, Monheim, Germany) (pre-washed three times in MilliQ water) and acetonitrile (Merck, 70% v/v) for 20 min at RT using the ThermoMixer C (Eppendorf) shaking at 400 rpm. Samples were placed on a magnetic rack, supernatant removed, and three 200 μL washes performed: twice with 70% ethanol, and one with neat acetonitrile (ACN). After washing, all traces of solvent were evaporated using a CentriVap (Labconco, Kansas City, USA), and digestion buffer (100 mM Tris-HCl, pH 8) was added to the beads containing Lys-C (Wako, Chuo-Ku, Japan) and SOLu-Trypsin (Merck) each at a 1:50 enzyme to protein ratio, and digestion performed for 1 h at 37°C with shaking at 400 rpm. Digests were desalted using in-house C18 stage tips containing 2 plugs as previously described,81 vacuum centrifuged to dryness, and peptides reconstituted in 75 μL 0.1% formic acid (Merck) and 2% ACN (Merck) in preparation for LC-MS/MS. Mass spectrometry analyses were performed on an M-class (Waters, Milford, USA) coupled to a timsTOF Pro (Bruker, Billerica, USA) equipped with a CaptiveSpray source. Peptides (1 μL) were separated by reverse-phase chromatography on a C18 fused silica column (inner diameter 75 μm, OD 360 μm × 15 cm length, 1.6 μm C18 beads) packed into an emitter tip (IonOpticks, Middle Camberwell, Australia), using a nano-flow HPLC (M-class, Waters). The HPLC was coupled to a timsTOF Pro (Bruker) equipped with a CaptiveSpray source. Peptides were loaded directly onto the column at a constant flow rate of 400 nL/min with buffer A (99.9% Milli-Q water, 0.1% FA) and eluted with a 90-min linear gradient from 2 to 34% buffer B (99.9% ACN, 0.1% FA). The timsTOF Pro (Bruker) was operated in diaPASEF mode using Compass Hystar 5.1. The settings on the TIMS analyzer were as follows: Lock Duty Cycle to 100% with equal accumulation and ramp times of 100 ms, and 1/K0 Start 0.6 V s/cm2 End 1.6 V s/cm2, Capillary Voltage 1400V, Dry Gas 3 L/min, Dry Temp 180°C. Methods were set up using the instrument firmware (timsTOF control 2.0.18.0) for data-independent isolation of multiple precursor windows within a single TIMS scan. The method included two windows in each diaPASEF scan, with window placement overlapping the diagonal scan line for doubly and triply charged peptides in the m/z – ion mobility plane across 16 × 25 m/z precursor isolation windows (resulting in 32 windows) defined from m/z 400 to 1,200, with 1 Da overlap, with CID collision energy ramped stepwise from 20 eV at 0.8 V s/cm2 to 59 eV at 1.3 V s/cm2. DIA-NN 1.882 was used for searching of timsTOF diaPASEF.d files. The option for library-free search was enabled, and data were searched against reviewed sequences from the human Uniprot Reference Proteome (downloaded May 2021). The search was set to trypsin specificity, peptide length of 7–30 residues, cysteine carbidomethylation as a fixed modification, and the maximum number of missed cleavages at 2. Additionally, n-terminal acetylation and oxidation of methionine were set as variable modifications with the maximum number of variable modifications set to 1. In addition, the maximum mass accuracy was set to 10 ppm for both MS1 and MS2 spectra and the quantification strategy to robust LC (high precision). Match between runs (MBR) was enabled, and all other settings left as default. For reporting precursor and protein numbers, outputs were filtered at precursor q-value <1% and PG protein q-value <1%, and the PG.MaxLFQ was used to obtain the normalized quantity for protein groups based on proteotypic peptides (i.e. unique proteins).
Lipidomics
Lipid extraction was performed as previously described.83 10 μL of plasma was mixed with 100 μL of butanol:methanol (1:1) with 10 mM ammonium formate which contained a mixture of internal standards. Samples were then vortexed and set in a sonicator bath for 1 h at RT. Samples were then centrifuged (14,000 x g, 10 min, 20°C) before transferring into the sample vials for analysis. Analysis of plasma extracts was performed on an Agilent 6495C QQQ mass spectrometer with an Agilent 1290 series HPLC system and a ZORBAX eclipse plus C18 column (2.1 × 100 mm 1.8 mm, Agilent) with the thermostat set at 45°C. Mass spectrometry analysis was performed with dynamic scheduled multiple reaction monitoring (dMRM). Chromatogram integration was performed using Agilent MassHunter v10.0 and quantification of lipid species was determined by comparison to the relevant internal standard.
Multiplex cytokine analysis
A flow cytometric bead multiplex assay (LEGENDplex™ Human Anti-Virus Response 13-plex Panel, Biolegend) was used to quantify 13 cytokines (IL-1β, IL-6, IL-8, IL-10, IL-12p70, IFN-α, IFN-β, IFN-λ1, IFN-λ2/3, IFN-γ, TNF-α, IP-10/CXCL10 & GM-CSF) in sodium citrate plasma samples collected at V0, V1, V2A and V3A, according to the manufacturer’s instructions. Samples were thawed at RT and centrifuged (2000 x g, 5 min, 4°C) prior to 1:1 dilution with Assay Buffer (Biolegend). 9.5 μL of diluted sample was mixed with 9.5 μL Assay Buffer and 9.5 μL premixed capture beads in a V-bottom plate and incubated protected from light (200 x g, 2 h, RT). Samples were washed (200 μL LEGENDplex™ wash buffer) and incubated for a further 1 h (200 x g RT) with 9.5 μL premixed detection antibody before the addition of 9.5 μL Streptavidin-PE and another 30 min incubation. Samples were washed (150 μL wash buffer) before resuspension in 150 μL washing buffer for acquisition. Samples were immediately acquired on a BD LSR Fortessa X-20 (BD Biosciences) with High Throughput Sampler. Bead populations were identified based on forward/side scatter and the degree of APC-H7 fluorescence. PE fluorescence (geometric mean fluorescence intensity) was determined for each sample and cytokine concentrations were interpolated from a standard curve (sigmoidal 4-parameter logistic curve) using GraphPad Prism 9 software. Differential analysis was performed using a mixed effect linear model implemented in R (glm function). Data were visualized with the ggplot2 v3.3.6 package.
Quantification and statistical analysis
Whole blood RNASeq analysis
Sequence read quality was assessed using FastQC version 0.11.9 and summarized with MultiQC version 1.10.166 prior to quality control with Trimmomatic version 0.3867 with a window size of 4 nucleotides and an average quality score of 30. Following this, reads which were <50 nucleotides after trimming were discarded. Reads that passed all quality control steps were then aligned to the human genome (GRCh38 assembly) using HISAT2 version 2.1.0.68 The gene count matrix was generated with FeatureCounts version 1.5.0-p269 with Ensembl version 101 annotation.70 The count matrix was then imported into R 4.2 for further analysis and visualization in ggplot2 v3.3.6. Counts were normalized using the trimmed mean of M values (TMM) method in EdgeR version 3.3871 and represented as counts per million (cpm). Prior to multidimensional scaling analysis svaseq v3.44 was applied to remove batch effects and other unwanted sources of variation in the data.72 Differential gene expression analysis was performed using the glmLRT function in EdgeR adjusting for gender and batch (run) in the model. Genes with <3 cpm in at least 15 samples were excluded from the differential expression analysis. The Benjamini and Hochberg method was employed to correct for multiple comparisons. Statistical significance was determined as FDR <0.05. Pathway and Gene Ontology (GO) overrepresentation analysis was carried out in R using a hypergeometric test. Cell type enrichment analysis was carried out in R using the using the camera function in the EdgeR library with cell type gene sets from the human cell atlas bone marrow dataset in the Molecular Signatures database collection C8 (R package msigdbr v7.5.1).76 Blood Transcriptional Module (BTM) analysis was carried out using a pre-defined set of modules defined by Li et al. as an alternative to pathway-based analyses.31 Gene Set Variation Analysis (GSVA)32 was used to calculate a per sample activity score for each of the modules (excluding unannotated modules labeled ‘TBA’) using the R Bioconductor package GSVA v1.44.1. limma v3.51.075 was used to identify modules that were differentially active at at least one timepoint. See data and code availability statement for access to the R code.
Proteomics
Data processing and analysis were performed using R v4.2. Only proteins that were quantified in at least 50% of replicates in at least one condition were kept. The data were normalized using variance stabilizing method implemented in limma v3.51.0 (Figures S7A-S7G). Differential analysis was performed using limma v3.51.0. The Benjamini and Hochberg method was employed to correct for multiple comparisons. Statistical significance was determined as FDR <0.1. See data and code availability statement for access to the R code.
Lipidomics
The data were normalized using variance stabilizing method implemented in limma v3.51.0 (Figures S8A-S8D). Differential analysis was performed using a mixed effect linear model implemented in R (glm function). The Benjamini and Hochberg method was employed to correct for multiple comparisons. Statistical significance was determined as FDR <0.1. Data were visualized with ggplot2 v3.3.6. See data and code availability statement for access to the R code.
Multi-omics correlation analysis
Spearman correlation analysis was performed using the Hmisc v4.7-0 package in R v4.2.73 Statistical significance was defined as p < 0.05. In order to avoid spurious correlations driven by differences in antibody titers or T cell responses induced following the ChAdOx1-S and BNT162b2 vaccines, the correlation analyses were performed for each vaccine group separately for antibody tires at V2B. At V3B we did not detect significant differences in antibody titers between the different vaccine groups, so all samples were used for the correlation analysis.
Acknowledgments
The authors thank all of the study participants. This work was funded by BioPlatforms Australia, the Flinders Foundation, and EMBL Australia Group Leader funding awarded to D.J.L. The proteomics component of this study was funded by AstraZeneca. This work was also supported in part by the MRFF (APP2015305) and The Hospital Research Foundation Group. We thank SA Pathology for help with sample collection and the South Australian Genomics Center (SAGC) for help with RNA sequencing. The SAGC is supported by the National Collaborative Research Infrastructure Strategy (NCRIS) via BioPlatforms Australia and by the SAGC partner institutes. We thank the Garvan Institute for providing the sequin controls for RNA sequencing. The MR1 tetramer technology was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. Flow cytometry analysis was performed at the ACRF Cellular Imaging and Cytometry Core Facility in SAHMRI. We thank Prof. Peter Meikle and Natalie Mellett for their assistance with generating the lipidomics data.
Author contributions
The study was conceived and designed by D.J.L. F.J.R. performed the multi-omics analyses under the supervision of D.J.L. with input from S.J.B., T.S.N., C.M., and N.E.S.; S.J.B. and N.E.S. generated the flow cytometry data with help from S.C.B.; N.E.S. generated the cytokine data. T.S.N. conducted the T cell ELISpot and AIM assays. J.J., G.L.E., Y.C.T., and M.A.L managed and processed the clinical samples. C.M., V.I., J.Z., and P.M. generated the proteomics data. H.S., H.T., and J.D.M. performed the lipidomics analysis. M.G.M., A.E.L.Y., and B.G.-B. generated the binding antibody data. A.A., A.D., and R.B. generated the NAb data. Sample collection protocols were adapted from protocols originally developed by N.L.M. and N.C. Participant surveys were developed by N.L.M. and N.C.; R.B. and S.E.B. were responsible for participant coordination. The manuscript was written by F.J.R., T.S.N., N.E.S, S.J.B., and D.J.L., with contributions and approval from all the authors.
Declaration of interests
V.I., P.M., and C.M. received funding from AstraZeneca to undertake the proteomics component of this study. J.Z. is an employee of, and holds or may hold stock, in AstraZeneca.
Published: February 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.100971.
Supplemental information
Data and code availability
-
•
RNASeq data have been deposited at GEO and are publicly available as of the date of publication. Proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Supplementary data have been deposited at Mendeley. The DOI is listed in the key resources table. All the multi-omics datasets (Proteomic, lipidomic, serology, flow cytometry, multiplex immunoassay) are available via the Lynn Laboratory BitBucket (https://bitbucket.org/lynnlab/covirs).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Falsey A.R., Sobieszczyk M.E., Hirsch I., Sproule S., Robb M.L., Corey L., Neuzil K.M., Hahn W., Hunt J., Mulligan M.J., et al. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 vaccine. N. Engl. J. Med. 2021;385:2348–2360. doi: 10.1056/NEJMoa2105290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lustig Y., Gonen T., Meltzer L., Gilboa M., Indenbaum V., Cohen C., Amit S., Jaber H., Doolman R., Asraf K., et al. Superior immunogenicity and effectiveness of the third compared to the second BNT162b2 vaccine dose. Nat. Immunol. 2022;23:940–946. doi: 10.1038/s41590-022-01212-3. [DOI] [PubMed] [Google Scholar]
- 5.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O'Connell A.M., et al. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng H.F., Ackerson B.K., Luo Y., Sy L.S., Talarico C.A., Tian Y., Bruxvoort K.J., Tubert J.E., Florea A., Ku J.H., et al. Effectiveness of mRNA-1273 against SARS-CoV-2 Omicron and delta variants. Nat. Med. 2022;28:1063–1071. doi: 10.1038/s41591-022-01753-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon S.K., Hegmann K.T., Thiese M.S., Burgess J.L., Ellingson K., Lutrick K., Olsho L.E.W., Edwards L.J., Sokol B., Caban-Martinez A.J., et al. Protection with a third dose of mRNA vaccine against SARS-CoV-2 variants in frontline workers. N. Engl. J. Med. 2022;386:1855–1857. doi: 10.1056/NEJMc2201821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GeurtsvanKessel C.H., Geers D., Schmitz K.S., Mykytyn A.Z., Lamers M.M., Bogers S., Scherbeijn S., Gommers L., Sablerolles R.S.G., Nieuwkoop N.N., et al. Divergent SARS-CoV-2 Omicron-reactive T and B cell responses in COVID-19 vaccine recipients. Sci. Immunol. 2022;7:eabo2202. doi: 10.1126/sciimmunol.abo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Mateus J., Coelho C.H., Dan J.M., Moderbacher C.R., Gálvez R.I., Cortes F.H., Grifoni A., Tarke A., Chang J., et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell. 2022;185:2434–2451.e17. doi: 10.1016/j.cell.2022.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provine N.M., Amini A., Garner L.C., Spencer A.J., Dold C., Hutchings C., Silva Reyes L., FitzPatrick M.E.B., Chinnakannan S., Oguti B., et al. MAIT cell activation augments adenovirus vector vaccine immunogenicity. Science. 2021;371:521–526. doi: 10.1126/science.aax8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takano T., Morikawa M., Adachi Y., Kabasawa K., Sax N., Moriyama S., Sun L., Isogawa M., Nishiyama A., Onodera T., et al. Distinct immune cell dynamics correlate with the immunogenicity and reactogenicity of SARS-CoV-2 mRNA vaccine. Cell Rep. Med. 2022;3:100631. doi: 10.1016/j.xcrm.2022.100631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patone M., Mei X.W., Handunnetthi L., Dixon S., Zaccardi F., Shankar-Hari M., Watkinson P., Khunti K., Harnden A., Coupland C.A.C., et al. Risks of myocarditis, pericarditis, and cardiac arrhythmias associated with COVID-19 vaccination or SARS-CoV-2 infection. Nat. Med. 2022;28:410–422. doi: 10.1038/s41591-021-01630-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mevorach D., Anis E., Cedar N., Bromberg M., Haas E.J., Nadir E., Olsha-Castell S., Arad D., Hasin T., Levi N., et al. Myocarditis after BNT162b2 mRNA vaccine against Covid-19 in Israel. N. Engl. J. Med. 2021;385:2140–2149. doi: 10.1056/NEJMoa2109730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barda N., Dagan N., Ben-Shlomo Y., Kepten E., Waxman J., Ohana R., Hernán M.A., Lipsitch M., Kohane I., Netzer D., et al. Safety of the BNT162b2 mRNA Covid-19 vaccine in a nationwide setting. N. Engl. J. Med. 2021;385:1078–1090. doi: 10.1056/NEJMoa2110475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greinacher A., Thiele T., Warkentin T.E., Weisser K., Kyrle P.A., Eichinger S. Thrombotic thrombocytopenia after ChAdOx1 nCov-19 vaccination. N. Engl. J. Med. 2021;384:2092–2101. doi: 10.1056/NEJMoa2104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz N.H., Sørvoll I.H., Michelsen A.E., Munthe L.A., Lund-Johansen F., Ahlen M.T., Wiedmann M., Aamodt A.H., Skattør T.H., Tjønnfjord G.E., Holme P.A. Thrombosis and thrombocytopenia after ChAdOx1 nCoV-19 vaccination. N. Engl. J. Med. 2021;384:2124–2130. doi: 10.1056/NEJMoa2104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scully M., Singh D., Lown R., Poles A., Solomon T., Levi M., Goldblatt D., Kotoucek P., Thomas W., Lester W. Pathologic antibodies to platelet factor 4 after ChAdOx1 nCoV-19 vaccination. N. Engl. J. Med. 2021;384:2202–2211. doi: 10.1056/NEJMoa2105385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simpson C.R., Shi T., Vasileiou E., Katikireddi S.V., Kerr S., Moore E., McCowan C., Agrawal U., Shah S.A., Ritchie L.D., et al. First-dose ChAdOx1 and BNT162b2 COVID-19 vaccines and thrombocytopenic, thromboembolic and hemorrhagic events in Scotland. Nat. Med. 2021;27:1290–1297. doi: 10.1038/s41591-021-01408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collier D.A., Ferreira I.A.T.M., Kotagiri P., Datir R.P., Lim E.Y., Touizer E., Meng B., Abdullahi A., CITIID-NIHR BioResource COVID-19 Collaboration. Elmer A., Kingston N., et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature. 2021;596:417–422. doi: 10.1038/s41586-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartolo L., Afroz S., Pan Y.G., Xu R., Williams L., Lin C.F., Tanes C., Bittinger K., Friedman E.S., Gimotty P.A., et al. SARS-CoV-2-specific T cells in unexposed adults display broad trafficking potential and cross-react with commensal antigens. Sci. Immunol. 2022;7:eabn3127. doi: 10.1126/sciimmunol.abn3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mateus J., Dan J.M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. Cross-reactive CD4(+) T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374:eabh1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanz I., Wei C., Jenks S.A., Cashman K.S., Tipton C., Woodruff M.C., Hom J., Lee F.E.H. Challenges and opportunities for consistent classification of human B cell and plasma cell populations. Front. Immunol. 2019;10:2458. doi: 10.3389/fimmu.2019.02458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herati R.S., Muselman A., Vella L., Bengsch B., Parkhouse K., Del Alcazar D., Kotzin J., Doyle S.A., Tebas P., Hensley S.E., et al. Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells. Sci. Immunol. 2017;2:eaag2152. doi: 10.1126/sciimmunol.aag2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentebibel S.E., Lopez S., Obermoser G., Schmitt N., Mueller C., Harrod C., Flano E., Mejias A., Albrecht R.A., Blankenship D., et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci. Transl. Med. 2013;5:176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker A.T., Boyd R.J., Sarkar D., Teijeira-Crespo A., Chan C.K., Bates E., Waraich K., Vant J., Wilson E., Truong C.D., et al. ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome. Sci. Adv. 2021;7:eabl8213. doi: 10.1126/sciadv.abl8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brewer J.W., Solodushko V., Aragon I., Barrington R.A. Phosphatidylcholine as a metabolic cue for determining B cell fate and function. Cell. Immunol. 2016;310:78–88. doi: 10.1016/j.cellimm.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S., Rouphael N., Duraisingham S., Romero-Steiner S., Presnell S., Davis C., Schmidt D.S., Johnson S.E., Milton A., Rajam G., et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergamaschi C., Terpos E., Rosati M., Angel M., Bear J., Stellas D., Karaliota S., Apostolakou F., Bagratuni T., Patseas D., et al. Systemic IL-15, IFN-gamma, and IP-10/CXCL10 signature associated with effective immune response to SARS-CoV-2 in BNT162b2 mRNA vaccine recipients. Cell Rep. 2021;36:109504. doi: 10.1016/j.celrep.2021.109504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang J.S., Schwartzberg P.L., Kotliarov Y., Biancotto A., Xie Z., Germain R.N., Wang E., Olnes M.J., Narayanan M., Golding H., et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Therapeutic Goods Administration A.D.o.H. 2022. COVID-19 vaccine adverse events.https://www.health.gov.au/initiatives-and-programs/covid-19-vaccines/advice-for-providers/clinical-guidance/adverse-events [Google Scholar]
- 36.Watson O.J., Barnsley G., Toor J., Hogan A.B., Winskill P., Ghani A.C. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect. Dis. 2022;22:1293–1302. doi: 10.1016/S1473-3099(22)00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koutsakos M., Wheatley A.K., Loh L., Clemens E.B., Sant S., Nüssing S., Fox A., Chung A.W., Laurie K.L., Hurt A.C., et al. Circulating TFH cells, serological memory, and tissue compartmentalization shape human influenza-specific B cell immunity. Sci. Transl. Med. 2018;10:eaan8405. doi: 10.1126/scitranslmed.aan8405. [DOI] [PubMed] [Google Scholar]
- 40.Dicks M.D.J., Spencer A.J., Edwards N.J., Wadell G., Bojang K., Gilbert S.C., Hill A.V.S., Cottingham M.G. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One. 2012;7:e40385. doi: 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramasamy M.N., Minassian A.M., Ewer K.J., Flaxman A.L., Folegatti P.M., Owens D.R., Voysey M., Aley P.K., Angus B., Babbage G., et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2021;396:1979–1993. doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saggau C., Martini G.R., Rosati E., Meise S., Messner B., Kamps A.-K., Bekel N., Gigla J., Rose R., Voß M., et al. The pre-exposure SARS-CoV-2-specific T cell repertoire determines the quality of the immune response to vaccination. Immunity. 2022;55:1924–1939.e5. doi: 10.1016/j.immuni.2022.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michalik S., Siegerist F., Palankar R., Franzke K., Schindler M., Reder A., Seifert U., Cammann C., Wesche J., Steil L., et al. Comparative analysis of ChAdOx1 nCoV-19 and Ad26.COV2.S SARS-CoV-2 vector vaccines. Haematologica. 2022;107:947–957. doi: 10.3324/haematol.2021.280154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanderlugt C.L., Miller S.D. Epitope spreading in immune-mediated diseases: implications for immunotherapy. Nat. Rev. Immunol. 2002;2:85–95. doi: 10.1038/nri724. [DOI] [PubMed] [Google Scholar]
- 45.Cornaby C., Gibbons L., Mayhew V., Sloan C.S., Welling A., Poole B.D. B cell epitope spreading: mechanisms and contribution to autoimmune diseases. Immunol. Lett. 2015;163:56–68. doi: 10.1016/j.imlet.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Biram A., Liu J., Hezroni H., Davidzohn N., Schmiedel D., Khatib-Massalha E., Haddad M., Grenov A., Lebon S., Salame T.M., et al. Bacterial infection disrupts established germinal center reactions through monocyte recruitment and impaired metabolic adaptation. Immunity. 2022;55:442–458.e8. doi: 10.1016/j.immuni.2022.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Sammicheli S., Kuka M., Di Lucia P., de Oya N.J., De Giovanni M., Fioravanti J., Cristofani C., Maganuco C.G., Fallet B., Ganzer L., et al. Inflammatory monocytes hinder antiviral B cell responses. Sci. Immunol. 2016;1:eaah6789. doi: 10.1126/sciimmunol.aah6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khoo N.K.H., Lim J.M.E., Gill U.S., de Alwis R., Tan N., Toh J.Z.N., Abbott J.E., Usai C., Ooi E.E., Low J.G.H., et al. Differential immunogenicity of homologous versus heterologous boost in Ad26.COV2.S vaccine recipients. Med. 2022;3:104–118.e4. doi: 10.1016/j.medj.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bánki Z., Mateus J., Rössler A., Schäfer H., Bante D., Riepler L., Grifoni A., Sette A., Simon V., Falkensammer B., et al. Heterologous ChAdOx1/BNT162b2 vaccination induces stronger immune response than homologous ChAdOx1 vaccination: the pragmatic, multi-center, three-arm, partially randomized HEVACC trial. EBioMedicine. 2022;80:104073. doi: 10.1016/j.ebiom.2022.104073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lönnberg T., Svensson V., James K.R., Fernandez-Ruiz D., Sebina I., Montandon R., Soon M.S.F., Fogg L.G., Nair A.S., Liligeto U., et al. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci. Immunol. 2017;2:eaal2192. doi: 10.1126/sciimmunol.aal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakayamada S., Kanno Y., Takahashi H., Jankovic D., Lu K.T., Johnson T.A., Sun H.W., Vahedi G., Hakim O., Handon R., et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., Yang F., Wirz O.F., Solis D., Hoh R.A., et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185:1025–1040.e14. doi: 10.1016/j.cell.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S.W., Moon J.Y., Lee S.K., Lee H., Moon S., Chung S.J., Yeo Y., Park T.S., Park D.W., Kim T.H., et al. Anti-SARS-CoV-2 spike protein RBD antibody levels after receiving a second dose of ChAdOx1 nCov-19 (AZD1222) vaccine in healthcare workers: lack of association with age, sex, obesity, and adverse reactions. Front. Immunol. 2021;12:779212. doi: 10.3389/fimmu.2021.779212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J., Yu J., McMahan K., Jacob-Dolan C., He X., Giffin V., Wu C., Sciacca M., Powers O., Nampanya F., et al. CD8 T cells contribute to vaccine protection against SARS-CoV-2 in macaques. Sci. Immunol. 2022;7:eabq7647. doi: 10.1126/sciimmunol.abq7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagan T., Gerritsen B., Tomalin L.E., Fourati S., Mulè M.P., Chawla D.G., Rychkov D., Henrich E., Miller H.E.R., Diray-Arce J., et al. Transcriptional atlas of the human immune response to 13 vaccines reveals a common predictor of vaccine-induced antibody responses. Nat. Immunol. 2022;23:1788–1798. doi: 10.1038/s41590-022-01328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fourati S., Tomalin L.E., Mulè M.P., Chawla D.G., Gerritsen B., Rychkov D., Henrich E., Miller H.E.R., Hagan T., Diray-Arce J., et al. Pan-vaccine analysis reveals innate immune endotypes predictive of antibody responses to vaccination. Nat. Immunol. 2022;23:1777–1787. doi: 10.1038/s41590-022-01329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1101/2020.05.30.125484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1101/2020.03.17.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keck Z.Y., Li S.H., Xia J., von Hahn T., Balfe P., McKeating J.A., Witteveldt J., Patel A.H., Alter H., Rice C.M., Foung S.K.H. Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J. Virol. 2009;83:6149–6160. doi: 10.1128/JVI.00248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartosch B., Bukh J., Meunier J.C., Granier C., Engle R.E., Blackwelder W.C., Emerson S.U., Cosset F.L., Purcell R.H. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA. 2003;100:14199–14204. doi: 10.1073/pnas.2335981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalemera M.D., Cappella-Pujol J., Chumbe A., Underwood A., Bull R.A., Schinkel J., Sliepen K., Grove J. Optimised cell systems for the investigation of hepatitis C virus E1E2 glycoproteins. bioRxiv. 2020 doi: 10.1101/2020.06.18.159442. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 70.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leek J.T. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014;42:e161. doi: 10.1093/nar/gku864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrell F.E., Jr., Harrell M.F.E., Jr. 2019. Package ‘hmisc’. CRAN2018 2019; pp. 235–236. [Google Scholar]
- 74.Wickham H. Springer-Verlag; 2016. Package ‘ggplot2’: Elegant Graphics for Data Analysis. 978–970. [Google Scholar]
- 75.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dolgalev I. 2020. msigdbr: MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format. R Package Version 7. [Google Scholar]
- 77.Ashhurst T.M., Marsh-Wakefield F., Putri G.H., Spiteri A.G., Shinko D., Read M.N., Smith A.L., King N.J.C. Integration, exploration, and analysis of high-dimensional single-cell cytometry data using Spectre. Cytometry A. 2022;101:237–253. doi: 10.1002/cyto.a.24350. [DOI] [PubMed] [Google Scholar]
- 78.Abayasingam A., Balachandran H., Agapiou D., Hammoud M., Rodrigo C., Keoshkerian E., Li H., Brasher N.A., Christ D., Rouet R., et al. Long-term persistence of RBD(+) memory B cells encoding neutralizing antibodies in SARS-CoV-2 infection. Cell Rep. Med. 2021;2:100228. doi: 10.1016/j.xcrm.2021.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dagley L.F., Infusini G., Larsen R.H., Sandow J.J., Webb A.I. Universal solid-phase protein preparation (USP3) for bottom-up and top-down proteomics. J. Proteome Res. 2019;18:2915–2924. doi: 10.1021/acs.jproteome.9b00217. [DOI] [PubMed] [Google Scholar]
- 81.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 82.Demichev V., Yu F., Teo G.C., Szyrwiel L., Rosenberger G.A., Decker J., Kaspar-Schoenefeld S., Lilley K.S., Mülleder M., Nesvizhskii A.I., Ralser M. High sensitivity dia-PASEF proteomics with DIA-NN and FragPipe. bioRxiv. 2021 doi: 10.1101/2021.03.08.434385. Preprint at. [DOI] [Google Scholar]
- 83.Huynh K., Barlow C.K., Jayawardana K.S., Weir J.M., Mellett N.A., Cinel M., Magliano D.J., Shaw J.E., Drew B.G., Meikle P.J. High-throughput plasma lipidomics: detailed mapping of the associations with cardiometabolic risk factors. Cell Chem. Biol. 2019;26:71–84.e4. doi: 10.1016/j.chembiol.2018.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNASeq data have been deposited at GEO and are publicly available as of the date of publication. Proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Supplementary data have been deposited at Mendeley. The DOI is listed in the key resources table. All the multi-omics datasets (Proteomic, lipidomic, serology, flow cytometry, multiplex immunoassay) are available via the Lynn Laboratory BitBucket (https://bitbucket.org/lynnlab/covirs).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.