Abstract
The rapid divergence of repetitive sequences makes them desirable markers for phylogenetic studies of closely related groups, provided that a high level of sequence homogeneity has been maintained within species. Intraspecific polymorphisms are found in an increasing number of studies now, and this highlights the need to determine why these occur. In this study we examined intraindividual variation present in the first ribosomal internal transcribed spacer (ITS1) from a group of cryptic mosquito species. Individuals of the Anopheles punctulatus group contained multiple ITS1 length variants that ranged from 1.2 to 8.0 kb. Nucleotide and copy number variation for several homologous internal repeats is common, yet the intraspecific sequence divergence of cloned PCR isolates is comparable to that of other mosquito species (~0.2–1.5%). Most of the length variation is comprised of a 5′-ITS1 repeat that was identified as a duplication of a conserved ITS2 region. Secondary structure conservation for this repeat is pronounced and several repeat types that are highly homogenized have formed. Significant interspecific divergence indicates a high rate of evolutionary change for this spacer. A maximum likelihood tree constructed here was congruent with previous phylogenetic hypotheses and suggests that concerted evolution is also accompanied by interpopulation divergence. The lack of interindividual differences and the presence of homogenized internal repeats suggest that a high rate of turnover has reduced the overall level of variation. However, the intraindividual variation also appears to be maintained by the absence of a single turnover rate and the complex dynamics of ongoing recombination within the spacer.
Keywords: Concerted evolution, DNA turnover, rDNA, ITS1, Anopheles punctulatus
Introduction
The process of gene duplication is considered a fundamental driving force in evolution because the generation of additional gene copies leads to a relaxation of selective constraint and the possibility for the evolution of new functions (Ohno 1970). The evolution of these repeated sequences forms a range of homologous relationships which requires an awareness of this and their effect on the phylogenetic congruence of data sets (Fitch 2000). For example, gene duplication and speciation events do not always co-occur because paralogous sequences reflect the phylogeny of the gene family and orthologous sequences reflect the duplication of taxa via speciation. Paralogous sequences evolve in the absence of recombinant interactions, via a continual process of birth-by-duplication and death-by-mutation (Nei and Rooney 2005). However, many repeated sequences evolve in a nonindependent fashion, by ongoing intra- and interlocus recombination within a species that homogenizes paralogous sequences into what are called plerologues. This accumulation of species-specific differences is termed concerted evolution (Graur and Li 2000). Here the distinction between orthology and paralogy is blurred by concerted evolution and depending on the level of homogenization, whole families of repeated sequences may appear to evolve like single-copy genes and so reflect speciation events (Sanderson and Doyle 1992).
Homogenization occurs through a variety of non-Mendelian recombination processes that transfer DNA either reciprocally or nonreciprocally within and between chromosomes (Dover 2000). The continual turnover of DNA involved in these exchanges results in the gain or loss of repeated sequences during the lifetime of an individual and includes unequal crossing-over, gene conversion, replication slippage, duplicative transposition, and retroposition (see Graur and Li 2000). The level of homogenization is maintained in a balance between the processes that generate variation and those that eliminate it. This level is expected to decrease with increasing mutation rate, population size, number of repeats, and number of nonhomologous loci, as well as when interchromosomal exchanges are rare (Ohta and Dover 1983). DNA turnover can also generate variation and plays a similar, though not equivalent, role to mutation (Kimura and Ohta 1979). Recombinant exchanges occurring at the length of a repeat or greater may lead to its homogenization, but its rate of turnover in the genome also has to exceed the mutation rate and the turnover of other repeats present within or overlapping it. This is an important consideration because several recombinant processes can operate simultaneously on the same stretch of DNA.
More attention is usually given to differences in the rates of turnover that occur within and between chromosomes (i.e., intra- and interlocus recombination) (e.g., Gonzalez and Sylvester 2001), as this forms the core of a debate over the relative importance of population-level versus genomic processes for concerted evolution. Low rates of recombinant exchanges between chromosomes can lead to the formation of independent chromosomal lineages within a population that can be observed by differences between individuals. For example, physical constraints to X–Y chromosome pairing in Drosophila melanogaster lead to incomplete homogenization of the ITS1, in which a 24-bp deletion is restricted to males (Schlötterer and Tautz 1994). The presence of interindividual differences enables natural selection and random genetic drift to affect the frequency of variant repeats. However, without restrictions to interchromosomal exchange, the coupling of DNA turnover to meiotic recombination (i.e., sex) can reduce interindividual differences and spread variation throughout a population in a genomic-level process called molecular drive (Dover 2000).
The ribosomal DNA (rDNA) multigene family, with its contrasting regions of phylogenetically conserved rRNA genes and divergent spacers, has long provided insights into the underlying processes of concerted evolution. Of these, the first internal transcribed spacer (ITS1) diverges rapidly between closely related species, which has enabled it to be utilized for phylogenetic studies at or below the species level (e.g., Fabry et al. 1999; Rodriguez-Perez et al. 2006). Despite some success at investigating population relationships, intraindividual variation is common for this spacer, and unless a high level of sequence homogeneity has been maintained within species, it can obscure the resolution of phylogenetic trees (Miller et al. 1996; Famà et al. 2000). This variation is often due to internal repetition, in the form of microsatellites (Harris and Crandall 2000), or longer repeated sequences (Paskewitz et al. 1993; van Herwerden et al. 1999). The increasing incidence at which such intraspecific polymorphisms are found highlights the need to determine the effect these have on the level of homogenization and the subsequent evolution of these repeated sequences within a species.
Members of the Anopheles (Cellia) punctulatus group are major vectors of malaria in the southwest Pacific region which contain morphologically identical mosquito species that require the use of rDNA for their identification (Beebe and Cooper 2002). Intraindividual length variation has been demonstrated in the ITS1 of An. farauti and includes the presence of population-specific lengths (Beebe et al. 2000b). Here we surveyed the intraindividual variation within this species group to determine its association with internal repetition and its effect on the concerted evolution of this spacer. Length variation was examined using cloned PCR isolates and Southern blot hybridization. The species relationships are well supported by two concordant phylogenetic hypotheses, using the mitochondrial COII gene (Foley et al. 1998) and 18S rDNA (Beebe et al. 2000a). This enabled us to use the phylogeny to interpret the molecular evolution of the internal repeats. The effect these have on the level of homogenization was determined by an analysis of the spacer sequences and their repeat types. A phylogeny was estimated using the ITS1 for comparison to the previous hypothesis. The possibility that some restriction to interchromosomal exchange was also generating variation was explored. We suggest that intraspecific variation is maintained because multiple units of turnover are generated from the internal repeats with differential rates of recombination.
Materials and Methods
Mosquito specimens used here came from field collections and five colonies maintained by the Australian Army Malaria Institute. Included are 10 of the 12 known species of the Anopheles punctulatus group and an outgroup species, An. annulipes sensu lato (Table 1). The An. farauti colony was founded by a single gravid female collected from the island of Rabaul in 1968. Colonies from other species were established from the pooled egg batches of several females. The An. hinesorum and An. farauti 3 colonies were established in 1987 and 1983, respectively, from collections from the Innisfail area, northern Queensland, Australia. The An. punctulatus and An. species near punctulatus colonies were established in 1993 and 1994, respectively. The former species was collected from Buksak village, Madang Province, Papua New Guinea (PNG), and the latter from the Komo area, Southern Highlands Province, PNG. Note that there have been some recent name changes within the An. farauti complex (Schmidt et al. 2001, 2003).
Table 1.
Anopheles species used in this study with length and G+C variation of cloned ITS sequences
| Species | Geographic origin | No. of individuals | No. of clones | ITS1 length (bp) | ITS1 G+C% | ITS1 mean p-distance | ITS2 length (bp) | ITS2 G+C% |
|---|---|---|---|---|---|---|---|---|
| An. farauti | Rabaul (PNG) | 12 | 22 | 841–1138 | 54–58 | 0.0076 | 554* | 69 |
| Northern Territory (AUS) | 1 | 3 | 1204–1491 | 58–59.8 | 0.0017 | 549 | 68.9 | |
| Central Province (PNG) | 1 | 4 | 1167–1194 | 57.2–57.3 | 0.0034 | 562* | 68.5 | |
| An. irenicus | Guadalcanal (Sol Is) | 3 | 9 | 1197–1584 | 57.6–60 | 0.0045 | 560* | 69.3 |
| An. farauti 6 | Irian Jaya (Indo) | 2 | 3 | 1169–1175 | 55.9–56 | 0.0046 | 549 | 66.5 |
| An. hinesorum | Queensland (Aus) | 4 | 10 | 1162–1486 | 55.6–57.8 | 0.0068 | 555 | 66.3 |
| An. torresiensis | Queensland (Aus) | 5 | 8 | 1183–1200 | 62.4–63.2 | 0.0058 | 533 | 71.3 |
| An. koliensis | Madang Province (PNG) | 2 | 3 | 1467–1469 | 61.2 | 0.0143 | 591 | 67.9 |
| Central Province (PNG) | 1 | 2 | 1465–1480 | 61.1 | 0.0055 | – | – | |
| An. punctulatus | Madang Province (PNG) | 3 | 7 | 1590–1799 | 63–64.1 | 0.0059 | 551 | 67.3 |
| An. sp. nr punctulatus | Southern Highlands (PNG) | 3 | 7 | 1706–2006 | 60.5–61.3 | 0.0038 | 557 | 64.8 |
| An. clowi | Irian Jaya (Indo) | 1 | 3 | 548–731 | 54.4–55.1 | 0.0030 | 579* | 64.9 |
| An. annulipes sp. l | Karumba (Aus) | 1 | 3 | 1370 | 53.1–53.2 | 0.0024 | 415* | 65.3 |
Note: Aus Australia, PNG Papua New Guinea, Sol Is Solomon Islands, Indo Indonesia
* ITS2 sequences isolated in this study; all others from Beebe et al. (1999)
DNA Extraction, PCR Amplification, Cloning, and Sequencing
Genomic DNA was extracted using a CTAB-based method (Reineke et al. 1998). Each sample of DNA was resuspended in 30 μl of TE buffer (10 mM Tris-HCl, pH 8, 0.1 mM EDTA) containing 250 ng of Rnase (Roche). Specimens were identified using the PCR-RFLP method of Beebe and Saul (1995), which discriminates 11 of the 12 cryptic species of the An. punctulatus group. This method was modified using primers that amplify a slightly longer ITS2 PCR product: primers F-5.8S (5′-ACCGACACGTTGAACGCATATGGC-3′), which amplifies 73 bp of the 5.8S gene, and R-ITS/28S (5′-CCGTTTCGCTCGCAGCTACTCAGG-3′), which amplifies 108 bp of the 28S gene.
The whole ITS was isolated by PCR amplification, using primers R-ITS/28S and F-ITS/18S (5′-GATTCCACCGCGAGTTGCAGTTGG-3′), which amplifies 112 bp of the 18S gene. PCRs were performed with ~10 ng of DNA, each primer at 500 nM, 5% DMSO, each dNTP at 50 μM, 1 unit of Pfu DNA polymerase (MBI Fermentas), 1× PCR buffer supplied (pH 8.8), and 2 mM MgSO4. Thermocycling conditions were one cycle at 95°C for 3 min, followed by 20–25 cycles at 95°C for 30 s, 69°C for 30 s, and 75°C for 3.5 min, then one cycle at 75°C for 3 min. PCR products were electrophoresed on 1% agarose gel, and the bands excised, then purified using a gel extraction kit (QIAGEN). The ITS1 was amplified under the same conditions using primers F-ITS/18S and R-5.8S (5′-CGGTCTTCATCGATCCATGAGC-3′), which amplifies 45 bp of the 5.8S gene.
An aliquot of each PCR gel extract was phosphorylated with 1 unit of T4 Polynucleotide Kinase, 1× ligase buffer, and 5% PEG 4000 (all MBI Fermentas). These were incubated at 37°C for 20 min, then denatured at 65°C for 20 min. A 50-ng aliquot of dephosphorylated SmaI-cut pGEM-3Zf(+) (Promega) was added during the last 5 min and gradually cooled to 4°C. Two units of T4 DNA ligase (MBI Fermentas), 1× ligase buffer, and 5% PEG 4000 were added and the samples incubated at 4°C for 16 h. Ligation reactions were denatured at 70°C for 15 min. Standard techniques for transformation and identification of recombinant cells (e.g., Sambrook and Russell 2001) were carried out using CaCl2-competent Eschericha coli cells of the DH5α strain. Cells were recovered for no longer than 40 min after transformation. Insert-containing plasmids were purified using the GeneElute miniprep kit (Sigma-Aldrich).
Both forward and reverse DNA strands were sequenced using the BigDye terminator cycle sequencing kit v2.0 and an ABI 377 sequencer (AppliedBiosystems). The PCR primers F-ITS/18S, F-5.8S, R-5.8S, and R-ITS/28S were used, in addition to the sequencing primers for each species (Table 2). Cycle sequencing conditions were one cycle at 95°C for 3 min, followed by 28 cycles of 95°C for 15 s and 60°C for 3.5 min. Longer reads for DNA sequencing (1000–1100 bp) were performed according to an AppliedBiosystems user bulletin (http://www.appliedbiosystems.com), for clones with numerous long internal repeats.
Table 2.
Sequencing primers for the ITS1 of the An. punctulatus group and an outgroup taxon
| Primer | Oligonucleotide sequence | Species |
|---|---|---|
| R-ITS1precore | 5′-CCT GTC GGA TTG TGA CAT GTT C-3′ | An. farauti |
| F-Anf-central | 5′-TCC TAC CAC CAA CAG TCG TAG-3′ | An. farauti, An. hinesorum, An. farauti 6, & An. irenicus |
| R-Anf-central | 5′-GCG TGC TAC GAC TGT TGG TG-3′ | – |
| R-HinfI/ITS2 | 5′-CGG GAG CGG GCA ACG GAT TG-3′ | – |
| F-Anfarauti3 | 5′-GAC GCG CCG CCC AAC CAA GGT G-3′ | An. torresiensis |
| R-Anfarauti3 | 5′-GCT TGA CCT TTT ACA CCT TGG-3′ | – |
| F-Ankoliensis | 5′-CGT GAG CCA CGT ACG GGA TCG CTC-3′ | An. koliensis |
| R-Ankoliensis | 5′-GAG CGA TCC CGT ACG TGG CTC ACG-3′ | – |
| F-Anpunct1 | 5′-AGT CAT ACG GGG AAA AGC GAC C-3′ | An. punctulatus |
| R-Anpunct1 | 5′-GGT CGC TTT TCC CCG TAT GAC TC-3′ | – |
| R-Anp1retro | 5′-GTA CGT GGC TAA CGC CTT ATC AGA C-3′ | – |
| F-Anpunct2 | 5′-CGA GTG AGA GTT GAA CAA GTC AC-3′ | An. sp. nr punctulatus |
| R-Anpunct2 | 5′-GTG ACT TGT TCA ACT CTC ACT CG-3′ | – |
| R-Anp2retro | 5′-CTA CAG TGC GCA ACA TCT AAC GACC-3′ | – |
| F-ITS1/Anclowi | 5′-CGT AGA GGT GGC CGA ACT ACC GC-3′ | An. clowi |
| R-ITS1/Anclowi | 5′-CCG GAC AGT TGG ACA CCA TGT GC-3′ | – |
| R-ITS2/Anclowi | 5′-AAG GCC CGC GTG GCA TGT TGT AG-3′ | An. clowi & An. annulipes sp. l |
| F-ITS1/Anannul | 5′-CGT ACA CGT GAG GGC AAG TCT GG-3′ | An. annulipes sp. 1 |
| R-ITS1/Anannul | 5′-CGT CCA GAA CAG TTA GCG CAC TG-3′ | – |
Southern Probe Labeling, Genomic DNA Digests, and Southern Hybridization
An 18S probe was generated from an An. farauti individual (Rabaul) for Southern hybridization. The primers For-18Sprobe (5′-TGTACACACCGCCCGTCGCTAC-3′) and Rev-18Sprobe (5′-TAATGATCCTTCCGCAGGTTCAC-3′) were used to amplify the 3′-terminal 179 bp of the 18S rDNA. The primer For-18Sprobe includes a SspBI restriction site (underlined) present in the genomic DNA, 22 bp downstream from an EcoR47III site subsequently used for genomic digests. Approximately 5 ng of DNA was used in a PCR containing the above primers at 500 nM each, 10 mM Tris-HCl, pH 8.9, 50 mM KCl, 1.5 mM MgCl2, 5% DMSO, each dNTP at 100 μM, and 0.5 unit of Taq DNA polymerase. Thermocycling conditions were one cycle at 95°C for 3 min, followed by 25 cycles at 95°C for 30 s, 62°C for 30 s, and 72°C for 30 s, then one cycle at 72°C for 3 min. A PCR product of the expected size was cloned as described above, except the 3′-overhangs introduced by Taq polymerase were removed by the addition of 0.5 unit of Klenow (MBI Fermentas) and 200 μM of each dNTP during phosphorylation. A recombinant clone was selected containing the insert in the orientation, allowing its release from the vector by BamHI and SspBI digestion (Roche). The sequence of this 18S probe template was determined from an undigested aliquot using the M13F and M13R universal vector primers.
An 18S probe was 32P-labeled for a high specific activity using imbalanced nucleotides (Mertz and Rashtchian 1994). Approximately 5 ng of DNA was used in a PCR reaction containing 500 mM each of primers For-18Sprobe and Rev-18Sprobe, 40 mM KCl, 10 mM Tris-HCl, pH 8.9, 1.5 mM MgCl2, 6.25 μM each dATP, dGTP, and dTTP, 1 μM α32P-labeled dCTP (6 μl of 3000 μCi; PerkinElmer), 5% DMSO, and 1.5 units of Taq DNA polymerase. Thermocycling conditions were one cycle at 95°C for 3 min, followed by 45 cycles at 95°C for 30 s, 62°C for 30 s, and 72°C for 1 min 10 s, then one cycle at 72°C for 3 min. An internal ITS1 probe (215 bp) was made from an ITS clone using the same method and individual with primers F-ITS1postcore (5′-AAGCAGCAAGTAGAGCATCCC-3′) and R-Chi (5′-GCAACGGCGTTGTAAAGCG-3′). Another probe was made from GeneRuler DNA ladder mix (MBI Fermentas). This was 3′-end-labeled with 2 μg of DNA, 1.3 μM α32P-labeled dCTP, 1.5 mM MgCl2, 5 units of Taq DNA polymerase, 1 × PCR buffer supplied (Sigma-Aldrich) and incubated for 5 h at 72°C. Unincorporated labeled nucleotides were removed using G-50 Sephadex columns (Amersham Biosciences).
Southern hybridization was performed to separate ITS1 length variants in An. farauti, An. hinesorum, An. torresiensis, An. farauti 6, An. irenicus, An. koliensis, and An. sp. nr punctulatus. Between 100 and 300 ng of genomic DNA was double-digested overnight with 2 units each of EcoR47III and PstI in 1 × bufferH (Roche). Anopheles sp. nr punctulatus was digested with EcoR47III and BlnI because of internal ITS1 PstI sites. The ITS1 PCR products are 97 bp smaller than the EcoR47III and PstI hybridized fragments, as the primers used lie within these sites. Digests were electrophoresed at 25 V for 15 h in 1.2% superfine-resolution (SFR) agarose gel (Amresco). The DNA was transferred to a Hybond N+ nylon membrane (Amersham Biosciences) and fixed by baking for 2 h at 80°C, according to Sambrook and Russell (2001). The membrane was prehybridized for 2 h at 62°C in standard hybridization buffer (5 × SSC, 0.1% N-lauroylsarcosine, 0.02% SDS, 1% nonfat milk powder, 0.45 μM filtered). Hybridization was carried out for 18 h at 62°C after the addition of denatured probe. The membrane was washed twice for 5 min at room temperature in 2 × SSC and 0.1% SDS, then twice for 15 min at 65°C in 0.5 × SSC and 0.1% SDS. The plastic-wrapped membrane was exposed overnight to a phosphor screen (Molecular Dynamics) and hybridizations were detected using the Storm 860 laser scanning system.
Sequence Analysis
Secondary structure predictions were generated using the mfold server (www.bioinfo.rpi.edu/applications/mfold/old/rna) with the default setting of 5% suboptimality (Zuker 2003), and structure drawings were produced using RnaViz 2 (De Rijk et al. 2003). Predicted structures were chosen on the basis of comparative sequence analysis within the species group and were used to identify putatively homologous regions. Multiple sequence alignments were aligned visually because of internal repetition. An unambiguous ITS1 sequence alignment was possible only for four species of the An. farauti complex: An. farauti, An. irenicus, An. hinesorum, and An. farauti 6. BLAST searches (Tatusova and Madden 1999) were also used to identify sequence similarities. The stringency of matches was reduced using low gap opening and extension penalties to allow for expected length differences. Sequence alignment restrictions caused by internal repetition meant that the uncorrected distance measure (p-distance) was used for comparison of pairwise nucleotide differences among species, with gaps excluded. Substitutional models were assessed using MODELTEST (Posada and Crandall 1998). A maximum likelihood (ML) 50% majority-rule consensus tree was produced from 500 bootstrap replications. Heuristic searches with random stepwise addition were employed using PAUP* v4.0 (Swofford 2002).
Results
Sequence Variation in the An. punctulatus Group
The rDNA ITS1 contains multiple length variants within each of the individuals and species sampled, which indicates the presence of internal repeats. The ITS1 ranges between 900 and 3000 bp in PCR amplification products (Fig. 1). The possibility that some intraindividual length variants were concatamers of a main PCR product or heteroduplexes was explored by comparison to a Southern blot of seven species (Fig. 2). This was the largest species sample able to be cleaved by the same pair of restriction enzymes. Some PCR products do appear to be heteroduplexes, for example, the intermediate band among the three smallest bands in lane 4 of the gel (Fig. 1). It appears unlikely that PCR concatamers were formed; indeed many larger lengths are seen in the Southern blot (>2300 bp) that were not isolated by PCR. This is likely to have resulted from an inherent PCR and cloning bias for shorter lengths.
Fig. 1.
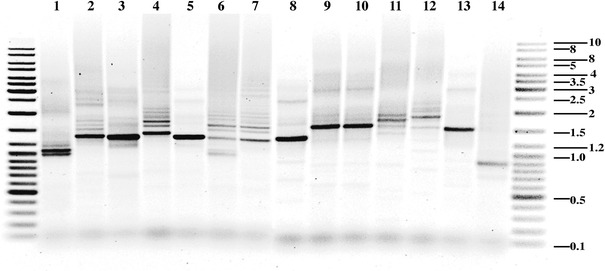
PCR products of the ITS1 from primers F-ITS/18S and R-5.8S resolved on 1.2% agarose gel. Samples are shown in the following order: An. farauti (lanes 1–3; Rabaul, Northern Territory and Central Province), An. irenicus (lane 4), An. farauti 6 (lane 5), An. hinesorum (lanes 6–7; Central Province and QLD), An. torresiensis (lane 8), An. koliensis (lanes 9–10; Central and Madang Provinces), An. punctulatus (lane 11), An. sp. nr punctulatus (lane 12), An. annulipes s.l. (lane 13), and An. clowi (lane 14). Fragment sizes of the flanking DNA ladders are in kilobases
Fig. 2.
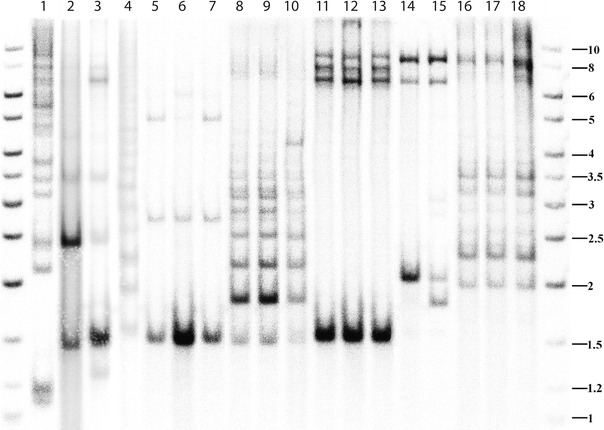
Southern blot of genomic DNA digested with EcoR47III and PstI, probed with a 32P-labeled 18S rDNA fragment. Samples are shown from An. farauti (lanes 1–3; Rabaul, Northern Territory, and Central Province), An. irenicus (lane 4), An. farauti 6 (lanes 5–7), An. hinesorum (lanes 8 and 9, QLD; and lane 10, Central Province), An. torresiensis (lanes 11–13), An. koliensis (lanes 14 and 15, Central and Madang Provinces), plus EcoR47III and BlnI digested DNA from An. sp. nr punctulatus (lanes 16–18). Fragment sizes of the flanking DNA ladders are in kilobases
Each band in the blot contains a single hybridized copy of the 18S probe (Fig. 2), therefore differences for band intensity within lanes are expected to reflect the relative abundance of each ITS1 length. This type of intraindividual variation can be seen, but was not quantified here. The number of bands and their relative abundance appeared similar within population samples, though interpopulation differences were observed for An. farauti, An. hinesorum, and An. koliensis. ITS1 bands with a maximal length of between 6.5 and 8.0 kb were observed in all species and are clearly seen in An. torresiensis and An. koliensis. An apparent ladder-like arrangement of ITS1 sizes in An. irenicus, An. hinesorum, and An. sp. nr punctulatus indicates the presence of a ~300-bp repeat. The internal ITS1 probe confirmed the presence of this spacer, as the same fragments were hybridized with this probe. Multiple ITS1 lengths were also observed over a range of genomic DNA and restriction enzyme concentrations, thereby ruling out the possibility of partial digestion.
The ITS1 was sequenced from a total of 84 clones and 39 individuals, with one to five clones sampled from each in order to isolate length variants (Table 1; GenBank accession numbers EF042693–EF042776). No variable sites were found in the partial sequence of the 18S gene. The whole 5.8S gene (161 bp) was sequenced from most clones except for An. sp. nr punctulatus and An. farauti 6. The remaining species had an identical 5.8S, except for An. annulipes sp. l, An. clowi, and an intraspecific variant found in the four clones from a single inland sample of An. farauti (Central Province, PNG). No variation was observed for the PstI site in this gene. The ITS1 contained a high level of sequence divergence between species and was usually accompanied by significant intraindividual length variation formed by several long internal repeats (Table 1). Sequenced ITS1 lengths ranged from 548 bp in An. clowi to 2006 bp in An. sp. nr punctulatus. These lengths were also observed in the Southern blot, though in most cases only the shortest one or two bands were isolated by cloning. The ITS1 is less G+C-rich than the ITS2, which varies within individuals primarily because of copy number variation of a G+C-rich repeat at the 5′-end.
Mean p-distance was estimated from several species as an indicator of the level of homogenization at the nucleotide level, which could be compared to other mosquito species (Table 1). Nucleotide and length variation have been maintained in all species, including those maintained in colonies. Pairwise substitutional differences were calculated by multiplying mean p-distance and average ITS1 length per species for those with three or more individuals: An. farauti (Rabaul) = 7.3, An. hinesorum = 7.9, An. torresiensis = 6.9, An. punctulatus = 9.9, An. sp. nr punctulatus = 6.9, and An. irenicus = 5.7. Anopheles punctulatus contains the most substitutional variation, though the only noncolony sample, An. irenicus (a species restricted to the island of Guadalcanal), appears to be more homogenized at this level.
Internal Repeats and Secondary Structure
Internal repeats were identified by comparison of aligned sequences, BLAST analysis, and secondary structural analysis. The most inclusive ITS1 sequence alignment possible among the species contained four species from the An. farauti complex: An. farauti, An. hinesorum, An. farauti 6, and An. irenicus (supplementary alignment). Interspecific sequence divergence also limited construction of a secondary structure model, as was also found for the ITS2 (Beebe et al. 1999). The 5′-end usually has multiple tandem copies of a G+C-rich repeat (63.5–73%), 109–188 bp that folds into a conserved stem structure (Fig. 3). This internal repeat, hereafter called the 5′-subrepeat, constitutes most of the sequenced length variation but is noticeably absent from the basal taxa, An. clowi and An. annulipes sp. l, and from most clones from the An. farauti Rabaul colony. The rest of the spacer is 811–966 bp long and forms a Y-shaped RNA structure in the An. punctulatus group (except An. sp. nr punctulatus), characterized by two long central stems (Fig. 3). This ‘core’ region has a lower G+C content (52–61.2%) and several smaller repeats are found at the apex of its two central stems and the terminal 3′-end (Fig. 4). The first central stem was the only shared ITS1 region BLAST analysis identified between the An. punctulatus group and An. annulipes sp. l: 219 bp in the latter (Fig. 4). No significant similarity was found between any species and An. clowi.
Fig. 3.
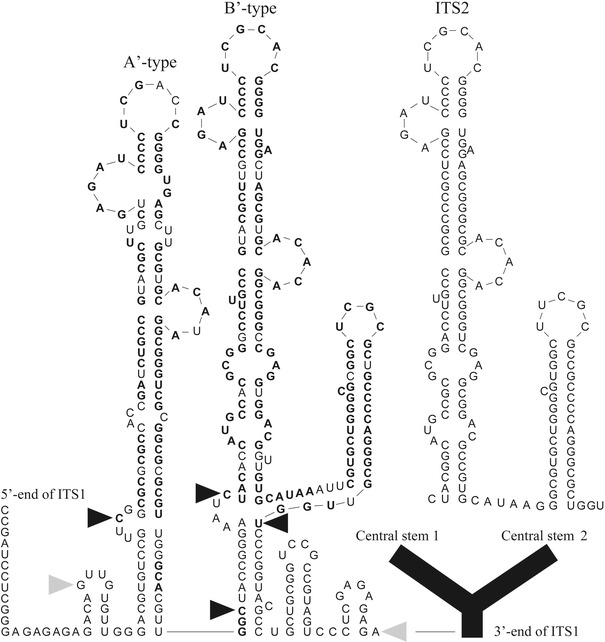
Secondary structure prediction of a 5′-subrepeat from An. irenicus showing the dual A- and B-type repeats and their ITS2 paralogue (inset) (GenBank accession no. EF042725). Identical nucleotides between the ITS structures are shown in boldface for both ITS1 repeats. Black arrows show the sequence match boundaries to the ITS2 structure. Gray arrows denote the beginning and end of the dual repeat. The downstream ITS1 sequence, or ‘core’ region, is drawn in black as a generalized Y-shaped structure with two long central stems
Fig. 4.
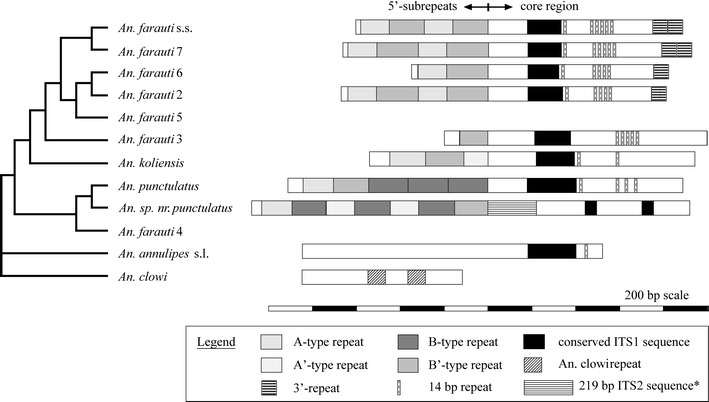
Scaled representations of the ITS1 from members of the An. punctulatus group (using the longest clone) alongside the most parsimonious tree of the 18S rDNA (taken from Beebe and Cooper 2002). The 5′-subrepeat and core region are shown for each species (except the two basal taxa). Different shading patterns illustrate the internal repeats and a conserved core region. *Additional part of the translocated ITS2 sequence
A surprising BLAST result was the identification a single paralogous copy (142–151 bp) of the 5′-subrepeat in the ITS2 of all species, including the basal taxa. A similar G+C content has been maintained (67.5–75.7%), but there is greater sequence divergence and length variation in the ITS1 copies. These were not readily alignable among species, unlike their more conserved ITS2 paralogues. Despite this divergence, the central region of each subrepeat matches the boundary of a conserved RNA stem structure it shares with the ITS2 (Fig. 3). A BLAST search to previously published sequences also resulted in a match to the ITS2 from two other Cellia species, An. dirus and An. baimaii. These results indicate that its original position within the rDNA array was likely to be the ITS2, though no reverse transcriptase gene or terminal repeats were identified to indicate recent translocation. Only in An. sp. nr punctulatus does similarity between the two spacers extend beyond the 5′-subrepeats to include an additional 219 bp: a 370-bp region in all (Fig. 4). This ITS1 sequence matches a continuous part of the ITS2 found in all species.
Most clones contained two divergent 5′-subrepeat types, termed the A type and B type (Figs. 3, 4), that represent paralogous copies. The B type and ITS2 paralogues are more similar in sequence and secondary structure, while the A type is characterized by the deletion of a short stem (Fig. 3). Further differentiation of types occurs in three species, but these are termed A′ and B′ for convenience only. Anopheles torresiensis is exceptional, having just a single shortened B-type copy (126–129 bp). The 5′-subrepeats vary in copy number by multiples of one (126–155 bp) or two (288–324 bp), with alternating A and B types (Fig. 4; Table 3). This dual A+B repeat structure was seen as ~300-bp length variants in the Southern blot (Fig. 2). There is some evidence from the blot for single-repeat length variants of ~150 bp in An. hinesorum and An. sp. nr punctulatus, but these appear to be rare and were not isolated by cloning.
Table 3.
Length variation observed among cloned 5′-subrepeat types from eight species within the An. punctulatus group
| Species | No. of subrepeats | Overall lengtha | Repeat type A | Repeat type A′ | Repeat type B | Repeat type B′ |
|---|---|---|---|---|---|---|
| An. farauti | 0, 2, or 4 | 288–297 | 135–139 | 152–158 | ||
| An. irenicus | 2 or 4 | 320–324 | 135–137 | 185–189 | ||
| An. farauti 6 | 2 | 328–332 | 135 | 193 | ||
| An. hinesorum | 2 or 4 | 321–323 | 135 | 186–188 | ||
| An. torresiensis | 1 | 126–129 | ||||
| An. koliensis | 3 | 448–451 | 159 | 109 | 180–181 | |
| An. punctulatus | 4 or 5 | 175–176 | 144 | 155 | 175–176 | |
| An. sp. nr punctulatus | 5, 6, or 7 | 126–294 | 129–131 | 126–127 | 151 | 168 |
aOverall length variation of tandemly paired repeat types (base pairs)
Despite interspecific divergence and the formation of paralogous 5′-subrepeats, plerologous copies were present within and among ITS1 clones. Some plerologous copies differed by a few point mutations, yet most were completely homogenized within species. The pattern of internal repeat types among the An. punctulatus clones was 4 × A+B+B′+B′ and 3 × A+B+B′+B′+B′, of which 12 of 17 B′-type sequences were identical. In An. sp. nr punctulatus there were 4 × A+B′+A′+B′+B, 2 × A+B′+A′+B′+A′+B′+B and 1 × A+B′+A′+A′+B′+B clones, of which 15 of 16 B′-type and 7 of 10 A′-type sequences were identical. The ITS1 within the An. farauti complex usually contained equal amounts of two subrepeat types. There were 4 × A+B and 6 × A+B+A+B among the An. hinesorum clones, of which all but 1 of 16 A-type sequences were identical. All the B-type sequences were identical except for a variable region at the 3′-end of the first B-type copy in A+B+A+B clones. A similar variable region was also found in the A+B+A+B clones from the Northern Territory An. farauti and An. irenicus. This is likely to be a recombination breakage point since it is present at the same junction between pairs of A+B subrepeats. In An. irenicus there were seven A+B and two A+B+A+B clones. Eight of the 11 A types were identical, though the B types were more variable. Homogenization of these repeats in An. farauti occurred within the populations. Among the six A+B clones from Rabaul, five of the A types and all B types were identical, as were all four A+B clones from the Central Province.
The core region contains three additional internal repeats. A 23-bp perfect tandem repeat is present at the apex of the first central stem and is found only in the An. farauti individual from the Central Province. A repeat of varying length with a conserved 14-bp sequence (5′-GGTGTCCAARGGTC-3′) occurs in all species except An. sp. nr punctulatus and An. clowi (Fig. 4). The An. farauti complex usually has a dispersed copy and a tandem array of four to six copies at the apex of the second central stem. Just the single element occurs in An. annulipes sp. l. Another tandem repeat (65–67 bp) was generated by a duplication of the 3′-end of the core region in An. farauti and An. irenicus. The ITS1 of An. clowi has an 84-bp-long repeat, present as either a single copy or two dispersed repeats, again, near the apex of a long central stem. Numerous microsatellites (2–3 bp) are also common among the species, in particular, the dinucleotides CAn, GTn, and GAn and the trinucleotides GGTn and GCTn.
Sequence Heterogeneity in the An. farauti Complex
An alignment of the ITS1 was constructed from four species of the An. farauti complex: An. farauti, An. irenicus, An. hinesorum, and An. farauti 6. Identical and ‘duplicate’ sequences, i.e., those differing by one or more dinucleotide repeats, were removed from the alignment given the redundancy of indels under a likelihood model. In all, there were three identical pairs of sequences (all An. farauti Rabaul) and five duplicate pairs (An. farauti = 2, An. irenicus = 2, An. hinesorum = 1). Sixteen of the 22 An. farauti clones had no 5′-subrepeat and were excluded from these alignments, but were used to analyze interindividual variation in the core region. The alignment contained 29 unique sequences from 35 clones and 86% constant characters among 1380 sites, gaps excluded. Sequences were also edited to remove the large gap introduced by a second copy of the 5′-subrepeat. The 3′-copy was removed for interspecific comparisons, an arbitrary choice that did not significantly affect our analyses.
A phylogenetic estimate of the ITS1 was constructed for comparison to the previously hypothesized relationships within the An. farauti complex (Foley et al. 1998; Beebe et al. 2000a). The DNA substitutional model chosen by MODELTEST (Posada and Crandall 1998) was the general time-reversible (GTR) model (Rodríguez et al. 1990) with a proportion of sites treated as potentially variable that are drawn from a gamma distribution (Г) and the remainder considered invariant (I). The proportion of invariant nucleotides was 0.51 and the shape parameter of the gamma-distribution α was 0.86. Base frequency estimates from this model were A = 23.3%, C = 25.8%, G = 30.2%, and T = 20.7%. One An. hinesorum sequence was arbitrarily chosen to root the trees in order to resolve any relationship between the An. farauti population samples and its inferred sister species, An. irenicus (Foley et al. 1998; Beebe et al. 2000a). The well-supported clades in the consensus tree clearly demonstrate species-specific divergence of the ITS1 at the nucleotide level that is concordant with the previously estimated relationships for these species (Fig. 5). The bootstrap proportions also indicate the presence of interpopulation differences within An. farauti.
Fig. 5.
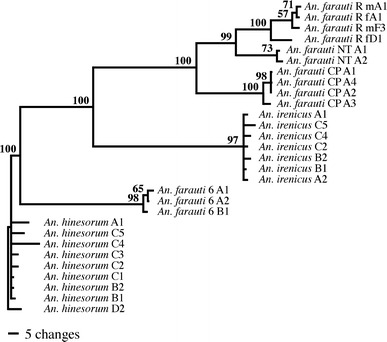
Phylogram showing phylogenetic relationships of the ITS1 (50% majority-rule consensus tree from 500 bootstrap replicates using ML analysis) from a sample of the An. farauti complex. Bootstrap proportions are shown above each node of the phylogram and branch lengths are proportional to the number of changes (see scale bar). Individuals are denoted ‘A’ to ‘F’ after species name, and clones are numbered 1 to 5. The An. farauti populations are denoted R = Rabaul (wherein m = male and f = female), NT = Northern Territory, and CP = Central Province
An analysis of interindividual variation was carried out by sequence comparisons between both sexes from the An. farauti Rabaul colony. We aimed to detect any significant restrictions to interchromosomal exchanges that might explain the presence of intraindividual variation. The 5′-subrepeat is relatively uncommon in this colony, therefore only the core region was analyzed. A total of 22 clones were compared from six males and six females (11 from each sex; 1–4 clones per individual). The average pairwise difference between males and females was very similar (mean p-distance = 0.0069 and 0.0074, respectively). There was no copy number variation for the 21-bp repeats, though a total of four repeat types were found in positions 2 and 3 within the tandem array. These formed two common repeat pairings in 20 clones, while two others were unique. Both sexes also shared three variant types of a 67-bp repeat, copy number variation for this and several di-nucleotide repeats, and a total of 17 substitutions (Table 4). Although there appear to be five singletons, four of these actually represent the unique positioning of a 21-bp (Table 4; nucleotides 1001 and 1006) and a 67-bp (Table 4; nucleotides 1361 and 1373) repeat type. Just two putative sex-specific variants were observed: an ACn1–2 dinucleotide in two males (Table 4; nucleotides 586–587) and a transition in two identical clones from the same male (nucleotide 692). Neither variant was observed in any other species.
Table 4.
Variable sites present in the ITS1 core of An. farauti Rabaul males (m) and females (f)
| Nucleotide position | 3 | 4 | 44 | 4 | 5 | 5 | 5 | 55 | 6 | 6 | 6 | 7 | 8 | 9 | 1 | 1 | 1 | 1 | 11 | 11 | 1 | 1 | 1 | 1 | 1 | 1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 1 | 22 | 6 | 4 | 4 | 4 | 88 | 3 | 8 | 9 | 4 | 2 | 8 | 0 | 0 | 0 | 1 | 11 | 11 | 2 | 2 | 2 | 3 | 3 | 3 | ||
| 4 | 4 | 89 | 6 | 4 | 6 | 8 | 67 | 8 | 3 | 2 | 3 | 4 | 0 | 0 | 0 | 0 | 2 | 88 | 88 | 3 | 3 | 9 | 0 | 6 | 7 | ||
| 1 | 2 | 6 | 6 | 56 | 78 | 5 | 6 | 4 | 6 | 1 | 3 | ||||||||||||||||
| Sex | Clone | ||||||||||||||||||||||||||
| m | A1 | T | G | − | A | T | C | G | − | T | T | G | G | T | T | T | A | A | C | GT | GT | G | G | T | C | − | − |
| m | A2 | . | . | − | . | . | . | . | − | . | . | . | . | . | . | . | . | . | . | .. | .. | . | . | . | . | − | − |
| m | B1 | . | . | − | . | . | G | A | − | . | . | A | . | A | A | . | . | . | A | .. | .. | A | . | . | A | C | A |
| m | C1 | . | . | CA | . | . | . | . | − | C | . | A | . | . | . | . | . | . | . | .. | − | . | A | C | A | C | A |
| m | D1 | . | . | − | . | . | . | . | AC | . | A | A | A | . | A | A | G | T | . | − | − | . | A | C | A | T | C |
| m | D2 | C | . | − | . | . | . | . | − | . | . | A | . | A | . | . | . | . | . | − | − | . | . | . | A | − | − |
| m | E1 | . | . | − | . | . | . | . | − | . | . | A | . | A | . | . | . | . | . | .. | .. | . | . | . | A | − | − |
| m | F1 | . | . | − | . | . | . | . | AC | . | . | A | . | A | . | . | . | . | . | .. | − | . | A | C | A | C | A |
| m | F2 | . | A | CA | T | A | G | A | − | . | A | A | . | . | . | . | . | . | . | .. | − | . | A | C | A | C | A |
| m | F3 | . | . | − | . | . | . | . | − | . | . | A | . | A | . | . | . | . | . | .. | .. | . | . | . | A | − | − |
| m | F4 | . | . | − | . | . | . | . | − | . | . | A | . | A | A | . | . | . | . | .. | .. | . | . | . | A | − | − |
| f | A1 | . | . | − | . | . | . | . | − | . | . | A | . | . | . | . | . | . | . | .. | .. | . | . | . | . | − | − |
| f | A2 | . | . | − | . | . | . | . | − | . | A | A | . | . | . | . | . | . | . | .. | − | . | A | C | A | − | − |
| f | B1 | . | . | − | . | . | . | . | − | . | . | A | . | A | . | . | . | . | . | .. | .. | . | . | . | A | − | − |
| f | B2 | . | A | CA | T | . | . | . | − | C | . | A | . | . | . | . | . | . | . | .. | − | . | A | C | A | C | A |
| f | C1 | . | A | CA | T | A | G | A | − | . | A | A | A | . | A | . | . | . | . | .. | − | . | A | C | A | − | − |
| f | C2 | . | A | CA | T | A | G | A | − | . | A | A | A | A | A | . | . | . | . | .. | − | . | A | C | A | − | − |
| f | D1 | . | . | − | . | . | . | . | − | . | . | A | . | . | . | . | G | . | . | .. | − | . | . | . | A | − | − |
| f | E1 | . | . | CA | . | . | . | . | − | C | . | A | . | A | A | . | . | . | . | − | − | . | A | . | A | C | A |
| f | E2 | . | . | − | . | . | G | A | − | . | . | A | . | A | A | . | . | . | A | .. | .. | A | . | . | A | C | A |
| f | F1 | . | . | − | . | . | . | . | − | . | . | A | . | A | A | . | . | . | . | − | − | A | . | . | A | − | − |
| f | F2 | . | . | − | . | . | . | . | − | . | . | A | . | A | A | . | . | . | . | .. | .. | . | . | . | A | − | − |
| ** | * | + | + | + | + | + | + | + | |||||||||||||||||||
Note: Nucleotide and gap (−) positions are shown above each column, with nucleotides identical to the top sequence encoded by a dot. Putative sex-specific variants (*) and variants found in a repeat (+) are shown. Individuals are coded A to F for each sex, followed by the clone number. Nucleotide positions correspond to the ITS1 alignment
Discussion
Intraindividual Variation
Multiple ITS1 length variants were observed within individuals and appear to be characteristic for these members of the An. punctulatus group. Shorter lengths were preferentially amplified during PCR compared to those seen in the Southern blot (Figs. 1, 2). Previous studies have shown that PCR tends to underestimate of the degree of intraindividual length variation present (e.g., Mateos and Markow 2005). Longer ITS1 lengths of 6.5–8.0 kb were detected in the seven species examined by Southern analysis, though most were between 1.2 and 4.0 kb. These ITS1 lengths are large considering they are generally <1 kb, although within the same subgenus, Cellia, the spacer ranges between 1.25 and 5.5 kb in An. merus and An. melas (Paskewitz et al. 1993). ITS1 lengths of up to 2.5 kb also occur in ladybird beetles (von der Schulenburg et al. 2001) and can exceed 3 kb in pinyon pines (Gernandt et al. 2001). The wide range of observed lengths attests to the repeated recombination of internal repeats, which has led to some interpopulation differences, as seen in An. farauti, An. hinesorum, and An. koliensis. Most of this length variation may be accounted for by a long internal repeat, the 5′-subrepeat, as was seen in the ITS1 of the An. gambiae complex (Paskewitz et al. 1993).
The partial 18S sequence and complete 5.8S rDNA were homogenized, as expected, and the only instance of intraspecific variation in these gene regions was from the An. farauti Central Province individual. No variation was found in the 5.8S PstI restriction site that might have accounted for some of the length variants in the Southern blot. Length variation among the PCR products also makes it unlikely that alternative restriction sites were present in the conserved rDNA. Preliminary analyses using a range of DNA and restriction enzyme concentrations also excluded the possibility of partial digestion. In comparison, the ITS1 was only partially homogenized and the sequenced lengths ranged from 548 bp in An. clowi to 2006 bp in An. sp. nr punctulatus. However, the level of intraspecific sequence divergence at the nucleotide level (~0.2–1.5%) is still well within the range measured in other mosquitoes (Paskewitz et al. 1993; Miller et al. 1996; Fairley et al. 2005), which indicates that internal repetition itself has not significantly reduced the level of homogenization. The presence of pseudogenic copies can also contribute significantly to the level of divergence (e.g., Harpke and Peterson 2006; Li and Wilkerson 2007). It is unlikely that these were isolated here, given that the only instance of variation in the rRNA genes was restricted to one individual and since most G+C variation resulted from copy number differences of the 5′-subrepeat.
ITS1 Internal Repeats
A high level of interspecific divergence limited ITS1 sequence alignments to within the An. farauti complex and restricted RNA structural predictions to the 5′-subrepeat. The dual repeats of the 5′-subrepeat were predicted to fold into two long stems, and the core into a Y-shaped structure (Fig. 3). Internal repetition appears to be characteristic for the evolution of the ITS1 in several invertebrate taxa. Long repeats occur at the 5′-end in a closely related group of mosquitoes (Paskewitz et al. 1993), in trematodes (van Herwerden et al. 1999), and also within the middle of the spacer in ladybird beetles (von der Schulenburg et al. 2001). The 5′-ITS1 repeat in the An. gambiae complex is also ~250 bp long and forms a long stem-loop structure (Paskewitz et al. 1993), although no similarity was found to the An. punctulatus group. Similar long stem-loop secondary structures have also been observed in the tandemly arrayed subrepeats present in the ITS1 of pinyon pines (Gernandt et al. 2001).
The 5′-subrepeat was only detected in the ITS1 of the An. punctulatus group. However, BLAST analysis identified a single paralogous copy in the ITS2 from all species, including An. dirus and An. baimaii from Southeast Asia. The central region of each subrepeat matches the boundary of a conserved RNA stem structure shared with the ITS2, and both have a similar G+C content (Fig. 3). In An. sp. nr punctulatus this region of similarity between the spacers extends a further 219 bp downstream, which corresponds to a conserved ITS2 region in these species (Fig. 4). These results indicate that the original chromosome position of this duplicated sequence was within the ITS2. It is possible that the 5′-subrepeat might be a remnant of an ITS2 sequence reverse transcribed from a pre-rRNA template and reintegrated upstream into the ITS1 in the stem species of the An. punctulatus group. RNA molecules are known to frequently serve as templates for what are called retrosequences, which, unlike retrotransposons, contain neither terminal repeats nor a reverse transcriptase gene (Brosius 2003). Two conditions would have favored this: the abundance of rRNA template and the fact that in all arthropods reverse transcriptase is expressed by two site-specific retrotransposons, the R1 and R2 elements, located just downstream in the 28S rDNA (Jakubczak et al. 1991).
The 5′-subrepeat region of the spacer usually consists of two structurally different paralogous repeat types which vary in copy number in either series or alternate pairs (Fig. 3; Table 3). In six of the species analyzed, two paralogous copies form into a dual subrepeat 288–324 bp long, which may be repeated up to 20 times. The copy number variation of the sequenced clones was also observed in the Southern blot. Clustering of the same repeat type occurred to a lesser degree (2 or 3 × B′ type and 2 × A′ type in An. punctulatus and An. sp. nr punctulatus, respectively), though it indicates that recombinant exchanges include either the single or the dual repeat. Long ITS1 repeats in ladybird beetles and pinyon pines have also formed into paralogous repeat types within a tandem array (a.k.a. positional homologues), which may also promote recombination (von der Schulenburg et al. 2001; Gernandt et al. 2001). It is significant that a high level of homogenization was observed within species for each repeat type and within the An. farauti populations. Although this is contrary to the expectation of a reduced level of homogenization with increased copy number (Ohta and Dover 1983), it may reflect differences in the turnover rate of the 5′-subrepeat relative to the rest of the spacer.
Sequence Heterogeneity in the An. farauti Complex
The ITS1 phylogeny illustrates the presence of both species-specific and population-specific variation (Fig. 5). A sister relationship between An. farauti and An. irenicus had previously been inferred in two other studies (Foley et al. 1998; Beebe et al. 2000a) and was also shown by the ITS1. There was high bootstrap support for the grouping of each clone with its species and population source, which clearly indicates the occurrence of concerted evolution at the nucleotide level. However, a sister relationship between Rabaul and Northern Territory An. farauti was unexpected, given the closer proximity of Rabaul to the Central Province of PNG. There were also numerous differences in the ITS1 and a substitution in the 5.8S gene, which leaves the possibility that the Central Province specimen represents yet another cryptic species. Fortunately, the resolution of these groups means that this spacer might be applied to such a question. Intraindividual variation in the ITS1 of An. aquasalis proved too problematic to generate a population phylogeny (Fairley et al. 2005). Most of the length variation consists of two to four internal repeats 75–88 bp in length that are flanked by a series of GAn di-nucleotide repeats, which were also observed flanking the 5′-subrepeats in the An. punctulatus group.
The presence of the rDNA on the heterologous X and Y chromosomes in Anopheles (Marchi and Pili 1994) enabled an analysis of interindividual differences through a comparison of variation between the sexes. The An. gambiae complex is an exception to this because the rDNA is X-Y-linked in some species but only X-linked in others (Collins et al. 1989). Although intraindividual ITS1 length variation occurs in this complex, it is only seen in the X-Y-linked species (Paskewitz et al. 1993). The absence of significant interindividual differences in An. farauti strongly suggests that recombination within and between the two rDNA loci is likely to occur and that intraindividual variation is not a by-product of restrictions to X-Y exchanges. Similarly, intraindividual variation in the An. albitarsis complex was not associated with differences between males and females (Li and Wilkerson 2007). Two sex-specific variants were observed in the Rabaul males, one isolated from the same individual and the other a CAn dinucleotide repeat. These are likely to be new variants and, as such, are expected to have a restricted chromosomal distribution (Dover 1989).
We have demonstrated the widespread presence of intraindividual ITS1 variation in members of the An. punctulatus group. This spacer is characterized by internal repeats which indicate the occurrence of recombination over a number of lengths, or what are termed units of turnover. Intraindividual variation in a monofunctional multigene family like rDNA is sometimes explained as inefficient homogenization at the level of either the chromosome or a set of chromosomes (e.g., von der Schulenburg et al. 2001). However, there are cases where this is not accompanied by interindividual differences, as shown in this study, black flies (Tang et al. 1994), and freshwater crayfish (Harris and Crandall 2000). This reinforces the point that homogenization is not an instantaneous process. Although the ITS1 does not consist of a single unit of turnover within the rDNA locus, the intragenomic heterogeneity that prevails in this species group has not impeded the concerted evolution of the spacer as a whole. Indeed, different regions within a single rDNA repeat are known to experience different rates of turnover in the genome (e.g., Gonzalez and Sylvester 2001; Averbeck and Eickbush 2005). What is not generally appreciated is how the presence of multiple units of turnover can generate additional variation. We consider this to have been a significant contributing factor to the observed level of heterogeneity and caution others to consider the complexity of the process of DNA turnover rather than simply invoke the argument of the apparent inefficiencies of homogenization.
Electronic Supplementary Material
This file is unfortunately not in the Publisher's archive anymore: (NEX 44 kb)
Acknowledgments
We thank M. Dowton and D. Ayre for useful suggestions and technical assistance. This research was supported by a graduate student grant from the University of Wollongong to J.E.B., and the National Health and Medical Research Council of Australia 117102 to N.W.B., and was published with the approval of the Director General of Defence Health Services.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Averbeck KT, Eickbush TH. Monitoring the mode and tempo of concerted evolution in the Drosophila melanogaster rDNA locus. Genetics. 2005;171:1837–1846. doi: 10.1534/genetics.105.047670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe NW, Cooper RD. Distribution and evolution of the Anopheles punctulatus group (Diptera: Culicidae) in Australia and Papua New Guinea. Int J Parasitol. 2002;32:563–574. doi: 10.1016/S0020-7519(01)00359-9. [DOI] [PubMed] [Google Scholar]
- Beebe NW, Saul A. Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction-restriction fragment length polymorphism analysis. Am J Trop Med Hyg. 1995;53:478–481. doi: 10.4269/ajtmh.1995.53.478. [DOI] [PubMed] [Google Scholar]
- Beebe NW, Ellis JT, Cooper RD, Saul A. DNA sequence analysis of the ribosomal DNA ITS2 region for the Anopheles punctulatus group of mosquitoes. Insect Mol Biol. 1999;8:381–390. doi: 10.1046/j.1365-2583.1999.83127.x. [DOI] [PubMed] [Google Scholar]
- Beebe NW, Cooper RD, Morrison DA, Ellis JT. A phylogenetic study of the Anopheles punctulatus group of malaria vectors comparing rDNA sequence alignments derived from the mitochondrial and nuclear small ribosomal subunits. Mol Phylogenet Evol. 2000;17:430–436. doi: 10.1006/mpev.2000.0853. [DOI] [PubMed] [Google Scholar]
- Beebe NW, Cooper RD, Foley DH, Ellis JT. Populations of the southwest Pacific malaria vector Anopheles farauti s.s. revealed by ribosomal DNA transcribed spacer polymorphisms. Heredity. 2000;84:244–253. doi: 10.1046/j.1365-2540.2000.00665.x. [DOI] [PubMed] [Google Scholar]
- Brosius J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica. 2003;118:99–116. doi: 10.1023/A:1024141306559. [DOI] [PubMed] [Google Scholar]
- Collins FH, Paskewitz SM, Finnerty V. Ribosomal RNA genes of the Anopheles gambiae species complex. Adv Dis Vector Res. 1989;6:2–28. [Google Scholar]
- De Rijk P, Wuyts J, De Wachter R. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- Dover GA. Linkage disequilibrium and molecular drive in the rDNA gene family. Genetics. 1989;122:249–252. doi: 10.1093/genetics/122.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover GA. How genomic and developmental dynamics affect evolutionary processes. Bio Essays. 2000;22:1153–1159. doi: 10.1002/1521-1878(200012)22:12<1153::AID-BIES13>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Dover GA. Molecular drive. Trends Genet. 2002;18:587–589. doi: 10.1016/S0168-9525(02)02789-0. [DOI] [PubMed] [Google Scholar]
- Fabry S, Köhler A, Coleman AW. Intraspecies analysis: comparison of ITS sequence data and gene intron sequence data with breeding data for a worldwide collection of Gonium pectorale. J Mol Evol. 1999;48:94–101. doi: 10.1007/PL00006449. [DOI] [PubMed] [Google Scholar]
- Fairley TL, Kilpatrick CW, Conn JE. Intragenomic heterogeneity of internal transcribed spacer rDNA in neotropical malaria vector Anopheles aquasalis (Diptera: Culicidae) J Med Entomol. 2005;42:795–800. doi: 10.1093/jmedent/42.5.795. [DOI] [PubMed] [Google Scholar]
- Famà P, Olsen JL, Stam WT, Procaccini G. High levels of intra- and inter-individual polymorphism in the rDNA ITS1 of Caulerpa racemosa (Chlorophyta) Eur J Phycol. 2000;35:349–356. doi: 10.1080/09670260010001735951. [DOI] [Google Scholar]
- Fitch WM. Homology: a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/S0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Foley DH, Bryan JH, Yeates D, Saul A. Evolution and systematics of Anopheles: insights from a molecular phylogeny of Australian mosquitoes. Mol Phylogenet Evol. 1998;9:262–275. doi: 10.1006/mpev.1997.0457. [DOI] [PubMed] [Google Scholar]
- Gernandt DS, Liston A, Piñero D. Variation in the nrDNA ITS of Pinus subsection Cembroides: implications for molecular systematic studies of pine species complexes. Mol Phylogenet Evol. 2001;21:449–467. doi: 10.1006/mpev.2001.1026. [DOI] [PubMed] [Google Scholar]
- Gonzalez IL, Sylvester JE. Human rDNA: evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics. 2001;73:255–263. doi: 10.1006/geno.2001.6540. [DOI] [PubMed] [Google Scholar]
- Graur D, Li W-H. Fundamentals of molecular evolution. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- Harpke D, Peterson A. Non-concerted ITS evolution in Mammillaria (Cactaceae) Mol Phylogenet Evol. 2006;41:579–593. doi: 10.1016/j.ympev.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Harris DJ, Crandall KA. Intragenomic variation within ITS1 and ITS2 of freshwater crayfishes (Decapoda: Cambaridae): implications for phylogenetic and microsatellite studies. Mol Biol Evol. 2000;17:284–291. doi: 10.1093/oxfordjournals.molbev.a026308. [DOI] [PubMed] [Google Scholar]
- Jakubczak J, Burke WD, Eickbush TH. Retrotransposable elements R1 and R2 interrupt the rDNA genes of most insects. Proc Natl Acad Sci USA. 1991;88:3295–3299. doi: 10.1073/pnas.88.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ohta T. Population genetics of multigene family with special reference to decrease of genetic correlation with distance between gene members on a chromosome. Proc Natl Acad Sci USA. 1979;76:4001–4005. doi: 10.1073/pnas.76.8.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wilkerson RC. Intragenomic rDNA ITS2 variation in the neotropical Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae) J Hered. 2007;98:51–59. doi: 10.1093/jhered/esl037. [DOI] [PubMed] [Google Scholar]
- Marchi A, Pili E. Ribosomal RNA genes in mosquitoes: localization by fluorescence in situ hybridization (FISH) Heredity. 1994;72:599–605. doi: 10.1038/hdy.1994.83. [DOI] [PubMed] [Google Scholar]
- Mateos M, Markow TA. Ribosomal intergenic spacer (IGS) length variation across the Drosophilinae (Diptera: Drosophilidae) BMC Evol Biol. 2005;5:46–59. doi: 10.1186/1471-2148-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz LM, Rashtchian A. Nucleotide imbalance and polymerase chain reaction: effects on DNA amplification and synthesis of high specific activity radiolabeled DNA probes. Anal Biochem. 1994;221:160–165. doi: 10.1006/abio.1994.1392. [DOI] [PubMed] [Google Scholar]
- Miller BR, Crabtree MB, Savage HM. Phylogeny of fourteen Culex mosquito species, including the Culex pipiens complex, inferred from the internal transcribed spacers of ribosomal DNA. Insect Mol Biol. 1996;5:93–107. doi: 10.1111/j.1365-2583.1996.tb00044.x. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin: Springer; 1970. [Google Scholar]
- Ohta T, Dover GA. Population genetics of multigene families that are dispersed into two or more chromosomes. Proc Natl Acad Sci USA. 1983;80:4079–4083. doi: 10.1073/pnas.80.13.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paskewitz SM, Wesson DM, Collins FH. The internal transcribed spacers of ribosomal DNA in five members of the Anopheles gambiae species complex. Insect Mol Biol. 1993;2:247–257. doi: 10.1111/j.1365-2583.1994.tb00144.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Reineke A, Karlovsky P, Zebitz CPW. Preparation and purification of DNA from insects for AFLP analysis. Insect Mol Biol. 1998;7:95–99. doi: 10.1046/j.1365-2583.1998.71048.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez F, Oliver JF, Marín A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142:485–501. doi: 10.1016/S0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Pérez MA, Núñez-González CA, Lizarazo-Ortega C, Sánchez-Valera A, Wooten MC, Unnasch TR. Analysis of genetic variation in ribosomal DNA internal transcribed spacer and the NADH dehydrogenase subunit 4 mitochondrial genes of the onchocerciasis vector Similium ochraceum. J Med Entomol. 2006;43:701–706. doi: 10.1093/jmedent/43.4.701. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sanderson MJ, Doyle JJ. Reconstruction of organismal and gene phylogenies from data on multigene families: concerted evolution, homoplasy, and confidence. Syst Biol. 1992;41:4–17. doi: 10.1093/sysbio/41.1.4. [DOI] [Google Scholar]
- Schlötterer C, Tautz D. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr Biol. 1994;4:777–783. doi: 10.1016/S0960-9822(00)00175-5. [DOI] [PubMed] [Google Scholar]
- Schmidt ER, Foley DH, Hartel GF, Williams GM, Bryan JH. Descriptions of the Anopheles (Cellia) farauti complex of sibling species (Diptera: Culicidae) in Australia. Bull Entomol Res. 2001;91:389–410. doi: 10.1079/BER2001113. [DOI] [PubMed] [Google Scholar]
- Schmidt ER, Foley DH, Bugoro H, Bryan JH. A morphological study of the Anopheles punctulatus group (Diptera: Culicidae) in the Solomon Islands, with a description of Anopheles (Cellia) irenicus Schmidt. sp.n. Bull Entomol Res. 2003;93:515–526. doi: 10.1079/BER2003267. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Tang J, Toè L, Black C, Unnasch TR. Intra-specific heterogeneity of the rDNA internal transcribed spacer in the Simulium damnosum (Diptera: Simuliidae) complex. Mol Biol Evol. 1994;13:244–252. doi: 10.1093/oxfordjournals.molbev.a025561. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. Blast 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- van Herwerden L, Blair D, Agatsuma T. Intra- and interindividual variation in ITS1 of Paragonimus westermani (Trematoda: Digenea) and related species: implications for phylogenetic studies. Mol Phylogenet Evol. 1999;12:67–73. doi: 10.1006/mpev.1998.0572. [DOI] [PubMed] [Google Scholar]
- von der Schulenburg JHG, Hancock JM, Pagnamenta A, Sloggett JJ, Majerus MEN, et al. Extreme length and length variation in the first ribosomal internal transcribed spacer of ladybird beetles (Coleoptera: Coccinellidae) Mol Biol Evol. 2001;18:648–660. doi: 10.1093/oxfordjournals.molbev.a003845. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]


