Abstract
Circular RNA (circRNAs) is a covalently closed circular non-coding RNA formed by reverse back-splicing from precursor messenger RNA. It is found widely in eukaryotic cells and can be released to the surrounding environment and captured by other cell types. This, circRNAs serve as connections between different cell types for the mediation of multiple signaling pathways. CircRNAs reshape the tumor microenvironment (TME), a key factor involved in all stages of cancer development, by regulating epithelial-stromal transformation, tumor vascularization, immune cell function, and inflammatory responses. Immune cells are the most abundant cellular TME components, and they have profound toxicity to cancer cells. This review summarizes circRNA regulation of immune cells, including T cells, natural killer cells, and macrophages; highlights the impact of circRNAs on tumor progression, treatment, and prognosis; and indicates new targets for tumor immunotherapy.
Subject terms: Cancer microenvironment, Immunotherapy
Facts
The expression of circRNAs is frequently dysregulated in human cancers.
CircRNAs play different roles during tumorigenesis and cancer progression.
CircRNAs regulate T cells, NK cells, and macrophages to reshape the tumor microenvironment.
CircRNA regulation of the tumor microenvironment provides potential therapeutic opportunities for cancer treatment.
Questions
Why circRNAs have multiple functions in the same or different human cancers. What are the underlining molecular determinants of this specificity?
Does dose-dependent targeting of circRNAs work in mouse models, at least in three-dimensional tumor organoid models?
Is circRNA targeting applicable in clinical trials?
Can we design prophylactic or therapeutic anti-cancer approaches based on genetic of polymorphisms of circRNAs?
Introduction
Circular RNAs (circRNA) is a closed circular molecule that is resistant to exonucleases, and is thus stable and widespread in animals and plants. CircRNA was discovered in 1976 when the Sanger team studied virus-like RNAs [1]. In 1991, Nigro et al. [2] accidentally discovered a normal novel RNA product. Due to its low expression and the limitations of detection technology, circRNA was originally considered to be an aberrant product of RNA splicing. Recently, with advances in high-throughput sequencing technology, increasing numbers of circRNAs have been characterized and their roles and mechanisms have become active areas of investigation [3–5].
The immune system maintains homeostasis through immunomodulation, surveillance, and the prevention of pathogen invasion. The immune response coordinates a variety of immune cells and has antiviral, antibacterial, and antitumor functions. With rapid developments in oncology, immunology, molecular biology, and related disciplines, immunotherapies such as immune checkpoint inhibitors, tumor vaccines, and adoptive cell therapy have revolutionized cancer treatment. However, therapeutic responses, especially those of solid tumors, have been unsatisfactory in clinical trials and clinical applications. Recent studies have demonstrated that circRNAs are involved in cancer development [6–8] and immune responses [9–12]. In this review, we discuss the roles of circRNAs in the regulation of immune cells, immune-related molecules, and tumor immunity. We anticipate that this summary of current knowledge will facilitate the development of strategies to target circRNAs in the immune microenvironments of human cancers.
Biogenesis and function of circRNAs
CircRNA is a class of non-coding RNA generated from precursor messenger RNA (mRNA). Most circRNAs originate from exons in gene coding regions; others originate from 3′–untranslated regions (UTRs), 5'-UTRs, introns, intergenic regions, and antisense RNA [13, 14]. CircRNAs can be divided into four categories based on their sequence origin: (1) exonic circular RNAs (EciRNAs) derived from exons of the parent gene; (2) lasso-type or circular intronic RNAs (ciRNAs) derived from introns; (3) exonic–intronic circular RNAs (EIciRNAs) derived from both exons and introns; and (4) other circRNAs, including those derived from antisense strand transcripts (antisense circRNAs) and those derived from intergenic sequences or other unannotated genomic sequences (intergenic circRNAs) [15]. About 80% of circRNAs are EciRNAs localized mainly to the cytoplasm, whereas ciRNAs and EIciRNAs are often localized to the nucleus. CircRNAs are relatively evolutionarily conserved in different species. Jeck et al. [16] used the genome-wide RNase R enrichment method to detect >25,000 circRNAs in fibroblasts. Wang et al. [17] observed circRNA expression in fungi, plants, and prokaryotes, reflecting a high degree of conservation and widespread distribution among species. The expression of the same circRNA varies greatly under diseased and non-diseased conditions, among tissues, and during different time periods. The half-life of circRNAs exceeds that of the associated linear mRNA, as the covalent closed-loop structure lacks 5′ and 3′ends, which makes circRNAs more resistant to the exonuclease RNase R [18].
CircRNAs have four main biological functions (Fig. 1). (1) As they contain a large number of micro-RNA (miRNA) binding sites, they serve as molecular sponges and compete for miRNA binding to target mRNAs, thereby upregulating the expression of target genes [19–22]. (2) They participate in regulatory protein binding. Various RNA-binding proteins (RBPs) play crucial roles in RNA splicing, RNA stabilization, and mRNA translation. They bind to RNA and facilitate its processing and translation. CircRNAs interact with RBPs to form an RNA-protein complex, affecting RBP-mediated gene expression [23–25]. (3) They participate in protein encoding, as some circRNAs can be translated into peptides by ribosomes [26, 27]. (4) They regulate gene transcription, promoting parental gene expression by interacting with U1 small ribonucleoprotein or enhancing RNA polymerase activity [28, 29].
Fig. 1. Biological functions of circRNAs.
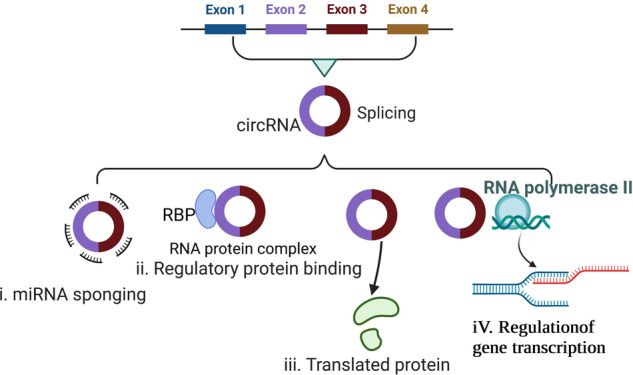
(i) Serving as molecular sponges for miRNA. (ii) Regulating protein binding. (iii) Encoding protein. (iv) Regulating gene transcription. RBP RNA-binding proteins.
circRNAs regulate T cells
T cells play an important role in the antitumor immune response [30]. T-cell activation is initiated through interaction with antigenic ligands, which are short peptide fragments bound to major histocompatibility complex class I (MHC I) and class II (MHC II) molecules. CD4+ T cells recognize mainly exogenous antigens presented by MHC II molecules, whereas CD8+ T cells recognize mainly endogenous antigens presented by MHC I molecules [31, 32]. Endogenous tumor antigens are processed intracellularly into antigenic peptides, and CD8+ T cells are activated upon recognizing antigenic peptide–MHC I complexes on tumor cell surfaces; these activated cells kill tumor cells by secreting perforin and granzymes, tumor necrosis factor, and lymphatic toxins [33]. They also kill tumor cells directly through apoptotic signals by interacting with human factor–related apoptosis and its ligand [34, 35]. Soluble antigens secreted by tumor cells are presented to CD4+ T cells by antigen-presenting cells, activating the CD4+ T cells. Primed CD4+ T cells activate B and CD8+ T cells to kill tumor cells [36, 37].
Mounting evidence indicates that tumor cells secrete exosomes into the circulation, which deliver certain intracellular components, such as circRNAs, into the tumor microenvironment (TME), reshaping it [38–40]. Tumor cell-derived circRNAs have recently been reported to play a vital and direct role in tumor immune escape (Table 1). Mechanically, circRNAs enhance the interaction between the immunosuppressive molecule programmed death receptor 1 (PD-1) and its ligand (PD-L1) by upregulating PD-1 expression in T cells, suppressing T-cell activation and cytokine secretion. Exosomes derived from different tumor cells deliver various circRNAs to T cells to inhibit their killing ability via PD-1 upregulation. Those derived from ovarian cancer cells were found to deliver circ-0001068 into T cells, increasing PD-1 expression via miR-28-5p sponging and thereby causing T-cell exhaustion [41]. In lung adenocarcinoma, circRNA-002178 was found to enter CD8+ T cells via exosomes and upregulate PD-1 expression by absorbing miR-34a [42]. circRNA can also upregulate the expression of the immune checkpoint molecules PD-L1 and CD73 on tumor cell surfaces via miRNA sponging, which helps tumor cells to escape recognition and death by T cells. Multiple studies have shown that circRNAs regulate PD-L1 expression via the circRNA–miRNA–mRNA axis, for instance, the circRNA of vimentin, CDR1-AS, hsa_circ_0003288, hsa_circ_0000190, hsa_circ_0046523, circ-CPA4, hsa-circRNA-002178, circ_0000284, circ_001678, circ-HSP90A, and circIGF2BP3 (Table 1) [43–54]. Mechanistically, they upregulate PD-L1 expression by sponging miRNAs in tumor cells, which induces T-cell apoptosis and immune escape (Fig. 2). A recent study showed that circ_0136666 induces regulatory T (Treg) cell activation by increasing PD-L1 expression through miR-497, leading to the immune escape of colorectal cancer (CRC) cells. Preclinical studies have shown that the upregulation of CD73, believed to be a novel immune checkpoint molecule, promotes tumor growth and disease progression by TME remodeling [55]. The inhibition of CD73 may promote the activity of T cells and other immune cells, enhancing antitumor immune surveillance via the adenosine pathway [56]. Xu and colleagues found that the expression of circHMGCS1-016 was upregulated in intrahepatic cholangiocarcinoma tissue, and that this upregulation correlated with poor survival; CD73 and GAL-8 were also upregulated in this tissue. Mechanistically, circHMGCS1-016 induced CD73 and GAL-8 expression by sequestering miR-1236-3p [57].
Table 1.
circRNAs regulate T cells.
| circRNA | T cells | Targets/pathways | regulation of immune responses | Cancer | References |
|---|---|---|---|---|---|
| circ-0001068 | T cell | circ-0001068 induce PD1 expression and T cell exhaustion by sponging miR-28-5p | prompt T cell exhaustion | ovarian cancer | [41] |
| CircRNA-002178 | CD8+T cell | CircRNA-002178/miR-28-5p /PD-L1 in CD8+T cell induce T-cell exhaustion | induce T-cell exhaustion | lung adenocarcinoma | [42] |
| hsa_circ_0136666 | Treg cell | hsa_circ_0136666 /miR-497/ PD-L1 b inducing the activation of Treg cells and leading to the immune escape | inducing the activation of Treg cell | colorectal cancer | [43] |
| circ-VIM | CD8+T cell | circ-VIM / miR-124/PD-L1 in CD8+T cell induces immunity escape | damage the viability and cytotoxicity of CD8+ T cells | esophageal cancer | [44] |
| hsa_circ_0000190 | T cell | hsa_circ_0000190 /PD-L1 mRNA/soluble PD-L1 (sPD-L1) interfering with T-cell activation induces immunity escape | interfering with T-cell activation, | Non-Small-Cell Lung Cancer | [47] |
| Hsa_circ_0046523 | CD4+T cell CD8+T cell Treg cell | circ_0046523/miR-148a-3p/PD-L1 axis mediates immunosuppressive microenvironment | decrease the proportion of CD4 + and CD8 + T cells, and increase the proportion of Tregs; promoted the apoptosis and exhaustion of CD8 + T cell, inhibited CD8 + T cell function | pancreatic cancer | [48] |
| circ_001678 | CD8+T cell | circ_001678 /miR-326/ZEB1 /PD-1/PD-L1/axis inducing immune escape | promote CD8+ T cell apoptosis; decrease the percentage of CD8+ T cells | non-small cell lung cancer | [51] |
| Circ-HSP90A | CD8+ T cell | circ-HSP90A promoted CD8 + T cells apoptosis via upregulating PD-L1 expression | induce CD8 + T cell apoptosis | non-small cell lung cancer | [52] |
| hsa_circ_0079587 (circIGF2BP3) | CD8+T cell | hsa_circ_0079587/PXP3/PD-L1 axis inducing immune escape | inducing the inactivity and exhaustion of T cells | non-small-cell lung cancer | [53] |
| CircKRT1 | CD8+ T cell | circKRT1/miR-495-3p/PDL1 axis induces immune evasion | weaken CD8 + T cell cytotoxicity and induce CD8 + T cell apoptosis | oral squamous cell carcinoma | [54] |
| circHMGCS1–016 | CD4+T cell CD8+T cell | circHMGCS1–016/miR-1236-3p/CD73 and GAL-8 axis induces immunosuppression | Number of T cell decrease and function of T cell damaged | intrahepatic cholangiocarcinoma | [57] |
| circ_002172 | CD8+T cell | circ_002172/miR-296-5p/CXCL12 axis inducing immune escape | inhibits cytotoxic T lymphocytes (CTL) infiltration | breast cancer | [58] |
| Circ_0008287 | CD8 + T cell | circ_0008287/miR-548c-3p/CLIC1 axis | damage the function of CD8+ T cell and induce apoptosis | gastric cancer | [64] |
| circTRPS1 | CD8+ T cell | circTRPS1/miR141-3p/GLS1 axis CD8+ T cell inducing exhaustion | CD8+ T cell exhaustion | bladder cancer | [65] |
| has_circ_0069313 | Treg cell | has_circ_0069313 /miR-325-3p/Foxp3 axis in Treg cell induces immunity escape | promotes Treg function | oral squamous cell carcinoma | [69] |
| circGSE1 | Treg cell CD8+ T | circGSE1/miR-324-5p/TGFBR1/ Smad3 axis inducing Tregs expansion | Tregs suppress the function of CD8+ T cell | hepatocellular carcinoma | [70] |
Fig. 2. Circular RNAs modulate T-cell function.
TGF-β transforming growth factor-β. TGF-βR I transforming growth factor beta receptor I. IFN-γ interferon γ. Treg regulatory T cells. PD1 programmed cell death. PDL1 programmed cell death ligand 1.
CircRNAs modulate antitumor T-cell activity through various mechanisms. In addition to regulating immune checkpoint molecules, circ_002172 inhibited cytotoxic T cell (CTL) infiltration and promoted breast cancer development by upregulating C-X-C motif chemokine ligand 12 (CXCL12) expression via miR-296-5p [58]. CXCL12 upregulation promotes tumor growth and leads to the recruitment of immunosuppressive cells to prevent CTL infiltration of tumors [59]. The dysfunctional expression of chloride intracellular channel 1 (CLIC1), a member of the chloride channel protein family, is related closely to tumor invasion, metastasis, treatment resistance, and prognosis [60]. High CLIC1 levels have been found in a variety of malignant tumors, including cervical cancer, breast cancer, hepatocellular carcinoma, gastric cancer, gallbladder carcinoma, and CRC [61–63]. Li et al. [64] found that circ_0008287 promotes the immune escape of gastric cancer cells by impairing miR-548c-3p–dependent CLIC1 inhibition. CLIC1 depletion in these cells suppresses CD8+ T-cell apoptosis, thereby increasing interferon (IFN)-γ secretion. CircRNAs have been demonstrated to suppress CD8+ T-cell activity by regulating tumor cell metabolism. Glutamine metabolism is partly responsible for the detoxification of reactive oxygen species, which profoundly influences the TME. circTRPS1 was recently reported to promote the malignant phenotype of bladder cancer (BCa) and CD8+ T-cell exhaustion therein. Mechanistically, it regulates redox equilibrium by altering glutaminase 1–mediated glutamine metabolism [65]. circRNAs have also been found to affect glutamate, glucose, and lipid metabolism, among others [66, 67].
Treg cells are subsets of T lymphocytes that mediate immune suppression through inhibitory cytokine secretion and in many other ways; they thus play important roles in the TME [68]. Mounting evidence indicates that circRNAs derived from tumor cells can induce immune escape by regulating Treg cell function. For example, circ_0069313 induces immune escape via the miR-325-3p–Foxp3 axis in Treg cells. Consistently, CD8 effector T cells were less infiltrative in oral squamous cell carcinoma (OSCC) tissues with high circ_0069313 expression. Treatment with OSCC cell–derived exosomes increased circ_0069313 and PD-L1 expression in Treg cells. Circ_0069313 depletion inhibited PD-L1 and CD25 expression in OSCC cells, whereas its ectopic expression increased the expression of CD25, but not PD-L1 [69]. These findings indicate that circ_0069313 modulates Treg cell activity. A recent study showed that circGSE1 not only promotes Treg cell function, but also expands the Treg cell population by regulating the miR-324-5p–transforming growth factor (TGF)-β receptor 1–Smad3 axis in hepatocellular carcinoma (HCC). The Treg: CD8+ T cell ratio is increased when T cells are cultured with HCC-derived exosomes [70]. In addition, circ_0136666 was shown to activate Treg cells by targeting the miR-497–Akt–mammalian target of rapamycin signaling pathway, to reduce the forkhead box P3 (FOXP3)+:CD8+ T cell ratio, and to increase the FOXP3+:CD4+ and FOXP3+: CD25+ T cell ratios [43].
These findings indicate that circRNAs inhibit antitumor effector T cells and promote Treg-cell expansion and activity via miRNA sponging (Fig. 2). As we described as above, the regulation of circRNAs on T cells and tumor cells is the activation of PD1 / PDL1 signaling pathway. It also indicated that circRNAs are promising potential targets in cancer immunotherapy [71]. However, the binding of PD1 and PDL1 not only reduces the viability and proliferation ability of T cells, but also affects the treatment of immune checkpoint inhibitors in tumors. In addition, the tumor microenvironment of different cancers of different patients are heterogeneous, the roles of circRNAs in cancer immunotherapy is also complicated. Given the important roles of circRNAs in the regulation of T-cell functions, thorough assessment of whether circRNAs regulate the TME in vivo and the translation of these findings into applicable clinical practice would be of interest.
CircRNAs regulate natural killer cells
Natural killer (NK) cells are composed mainly of T-cell receptor–, B lymphocyte antigen receptor–, CD56+, CD16+ lymphoid cells that spontaneously kill tumor cells. Their function depends mainly on the balance between their surface activating receptor natural killer group 2 member D (NKG2D) and the inhibitory killer immunoglobulin-like receptor (KIR). The binding of KIR on NK cells to MHC molecules on a tumor cell inhibits the killing function, whereas activation of the NKG2D ligand on NK cells promotes this function [72].
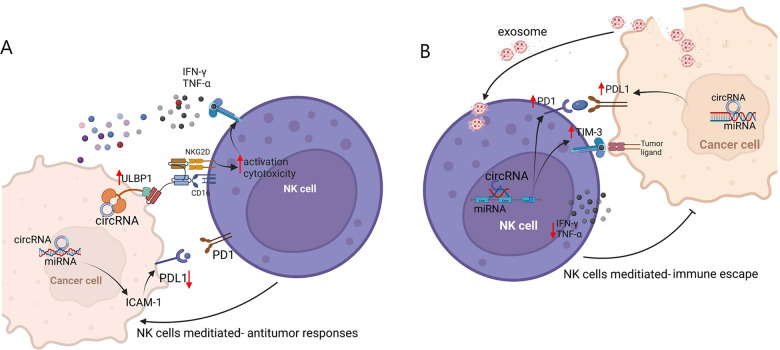
NK cells kill tumor cells and mediate cytotoxicity mainly by secreting perforin and granzymes. They also express death receptors that mediate the apoptosis of target cells. Increased CD16 and chemokine receptor-3 expression allows NK cells to accumulate and kill cells expressing the chemokine ligand. NK cell dysfunction has been reported to play crucial roles in tumorigenesis and cancer progression [73]. Emerging evidence indicates that circRNAs induce NK cell disability and exhaustion in the TME (Table 2). Various circRNAs, including circUHRF1, circARSP91, circ_0007456, and circ_0048674, play different roles in NK cell regulation via signaling pathways, contributing to the development of cancers including HCC, renal cell carcinoma, and pancreatic cancer (Table 2). In human HCC tissue, high expression levels of circUHRF1 (circ0048677), which originates from ubiquitin-like containing PHD and RING finger domains 1 (UHRF1), are associated with poor clinical prognosis and NK cell dysfunction. Mechanistically, circUHRF1 inhibits NK cell–derived IFN-γ and tumor necrosis factor (TNF)-α secretion and decreases the proportion and tumor infiltration of NK cells by regulating the miR-449c-5p–TIM-3 axis. TIM-3 upregulation induces NK cell exhaustion and promotes HCC progression [74]. Circ_0048674, which also originates from UHRF1, facilitates HCC progression and NK cell exhaustion through a different mechanism; it serves as an miR-223-3p sponge to alter PD-L1 expression. Circ_0048674 knockdown inhibits tumor cell proliferation, migration, and apoptosis and impairs NK cell function [75]. Whether these circRNAs work cooperatively or competitively to support tumor immune evasion remains largely unknown.
Table 2.
circRNAs regulate NK cells.
| circRNA | Targets/pathways | regulation of immune responses | Cancer | References |
|---|---|---|---|---|
| circUHRF1 | KLF16/circUHRF1/ sponging miR-449c-5p/Tim-3 | suppress NK cell secretion of IFN-γ and TNF-α | hepatocellular carcinoma | [74] |
| Hsa_circ_0048674 | Hsa_circ_0048674 /miR-223-3p/PDL1 | inhibit the functions and promotes NK cells exhaustion | hepatocellular carcinoma | [75] |
| CircARSP91 | CircARSP91/ULBP1 in HCC | enhances the cytotoxicity of NK cells | hepatocellular carcinoma | [76] |
| Hsa_circ_0007456 | Hsa_circ_0007456/miR-6852-3p/ICAM-1 axis | regulated the susceptibility of HCC to NK cells | hepatocellular carcinoma | [77] |
| circ_0000977 | circ_0000977/miR-153 axis modulates HIF1A-mediated immune escape | inhibit the killing effect of NK cells on pancreatic cancer tumor cells | pancreatic carcinoma | [80] |
| circZKSCAN1 | KLF2-induced circZKSCAN1/miR-1294/PIM1 axis | attenuate NK cell-mediated cytotoxicity | renal cell carcinoma | [81] |
| circFOXO3 | circFOXO3/miR-29a-3p and miR-122-5p | attenuate NK Cell-Mediated Cytotoxicity | renal cell carcinoma | [85] |
| circ_0001005 | AR/circ_0001005/PD-L1 axis in impacting NK cell antitumor efficacy | damage the function of NK cell antitumor efficacy | urinary bladder cancer | [87] |
In contrast to the role of circRNAs in suppressing NK cells described as above, mounting evidence has indicated that some circRNAs enhance the cytotoxicity of NK cells and inhibit the malignancy of HCC. CircARSP91 was reported to coordinate with tumor suppressors to exert anti-HCC effects. Specifically, it boosts the expression of UL16 binding protein 1, which binds to the NKG2D ligand on NK cell surfaces, mediating NK cell activation and cytotoxicity [76]. Similarly, circ_0007456 influences HCC susceptibility to NK cells by enhancing intercellular cell adhesion molecule-1 (ICAM-1) expression through miR-6852-3p sponging [77]. ICAM-1, also called CD54, is a member of the immunoglobulin superfamily of adhesion molecules that plays a crucial role in adhesion reaction mediation. Recent studies show that ICAM-1 on tumor exosome surfaces mediates the adhesion of the exosomes to CD8+ T cells, which is prerequisite for PDL1–mediated immunosuppressive effects [78, 79]. Accordingly, ICAM-1 may function as an essential checkpoint or potential therapeutic target downstream of circ_0007456 in the setting of HCC.
CircRNAs have also been reported to regulate the NK cell immune response against many other cancer types. For example, circ_0000977 was reported to be upregulated in pancreatic cancer cells under hypoxia and to induce tumor immune escape via the miR-153–hypoxia inducible factor 1α–a disintegrin and metalloprotease (ADAM) 10 axis. ADAM10 upregulation prompted membrane major histocompatibility complex class I chain-related gene A (MICA) shedding from pancreatic ductal adenocarcinoma cell surfaces and conversion to soluble MICA to degrade NKG2D on NK cells. This reduction of NKG2D expression resulted in NK cell hyporesponsiveness, and thus the inactivation of innate and adaptive immune responses and escape from immune surveillance [80]. CircZKSCAN1 is generally upregulated in clear cell renal cell carcinoma (ccRCC), and its downregulation significantly enhanced NK cell–mediated toxicity to RCC cells. It was found to modulate proviral integration site for Moloney murine leukemia virus-1 (PIM1) expression to inhibit NK cell–mediated toxicity to ccRCC cells via miR-1294 sponging [81]. However, the exact molecular mechanism underlying the role of PIM1 in NK cells remains unknown. Kruppel-like factor (KLF) is a transcription factor with a zinc finger structure that participates in the regulation of gene transcription, which is related to cell proliferation and differentiation and tumorigenesis [82, 83]. KLF2 inhibits early-stage NK cell proliferation and maintains a static late-stage NK cell state [84]. In addition, KLF16 has been shown to have an important role in suppressing NK cell–mediated cytotoxicity. It transcriptionally activates circFOXO3, which sponges miR-29a-3p and miR-122-5p to aggravate NK cell toxicity to ccRCC cells [85]. The androgen receptor, an oncogene, is associated closely with invasion and drug resistance in different cancers, including prostate cancer (PCa) and BCa [86]. It has been reported to upregulate circ_0001005 expression, attenuating NK cell killing efficacy by affecting PD-L1 expression via miR-200a-3p sequestration in BCa [87]. In addition, circRHOT1 upregulation has been found in BCa and is associated with the attenuation of NK cell–mediated toxicity to BCa cells. ZNF652, a member of the largest family of transcription factors that plays roles in the proliferation, invasion, and metastasis of many cancer types, induces circRHOT1 expression [88–90]. Although many circRNAs serve as miRNA sponges, some regulate the antitumor toxicity of NK cells through distinct mechanisms. circARSP91 was recently reported to bind directly to tumor suppressors to exert anti-HCC effects [91]. Thus, circRNAs can act as pleiotropic TME modulators by regulating tumor and NK cells (Fig. 3).
Fig. 3. Circular RNAs have bidirectional modulatory effect on NK cells.
A Circular RNAs promoting NK-mediated antitumor responses. B Circular RNAs suppressing NK cell activity to trigger tumor immune escape. ULBP1 human UL 16-binding protein 1, ICAM-1 intercellular cell adhesion molecule-1, NKG2D natural killer group 2 member D, TIM-3 T cell immunoglobulin and mucin domain-containing protein 3.
CircRNAs regulate macrophages
Macrophages are major lymphocytes that infiltrate solid tumors. Those infiltrating tumor tissues or distributed in the TME are called tumor-associated macrophages (TAMs) and have a central role in initiating the innate immune response, which leads to activation of the adaptive response in the later phase. Macrophages present two polarized states: classical (M1) and alternative (M2) activation, which occur through distinct pathways during mature differentiation. M1 TAMs are induced by IFN-γ, granulocyte-macrophage colony-stimulating factor (CSF), TNF-α, and many other cytokines and are able to kill tumor cells, whereas M2 TAMs are activated mainly by interleukin (IL)-4, IL-13, TGF-β, macrophage CSF, and other cytokines to promote tumor progression by activating a type 2 helper T cell–type immune response [92]. In addition to causing immunosuppression, M2 TAMs promote tumor growth and metastasis through many other mechanisms, such as tumor invasion, leakage to blood vessels, and angiogenesis promotion [93]. Most tumors do not have M1 macrophages without specific antigens and other factors. Thus, most TAMs have the M2, which promotes tumor occurrence, development, and metastasis, although they have the potential to repolarize to M1 macrophages [94].
Some circRNAs have been reported to regulate macrophage polarization in many cancers. CircTMEM181 prompts T-cell exhaustion by sponging miR-4883p to upregulate CD39 expression in macrophages, indicating that it affects mainly the macrophages in the immune microenvironment, rather than HCC cells [95]. Consistently, elevated circTMEM181 expression is correlated with anti–PD-1 treatment resistance and poor prognosis in patients with HCC. As another example, circ_0110102 upregulates C–C motif chemokine ligand (CCL) 2 expression by inhibiting miR-580-5p in HCC. CCL2 then activates the cyclooxygenase-2/prostaglandin E2 pathway in macrophages via FoxO1 in a p38 mitogen-activated protein kinase–dependent manner [96]. Tumor cells can recruit macrophages into tumor tissue through the secretion of many chemokines; tumor cells then secrete various cytokines, metabolites, and exosomes to alter and polarize the function of TAMs. Circ_0003410 was shown to promote HCC cell proliferation and migration via miR-139-3p sponging and thus the upregulation of CCL5 expression, which recruits M2 macrophages to enhance HCC deterioration in vitro and in vivo [97]. Similarly, circ_0074854 was shown to inhibit HCC tumorigenesis, mainly through the suppression of M2 macrophage polarization in vitro and in vivo [98]. However, the mechanism underlying this polarization regulation needs to be investigated further. CircASAP1 was found to regulate the expression of CSF-1, which controls the macrophage production, differentiation, and function [99], through the miR326/ miR-532-5p–CSF-1 signaling pathway, resulting in CD68+ TAM infiltration and HCC growth and metastasis [91].
Many reports describe circRNA regulation of macrophages in the microenvironments of tumors other than HCC, such as esophageal squamous cell carcinoma (ESCC), non-small cell lung cancer (NSCLC), and PCa. Different circRNAs affect TAMs through different pathways or regulators. For example, circRNA TCFL5 promotes esophageal cancer progression by regulating M2 macrophage polarization via the miR-543–formin-like protein 2 axis [100]. Similarly, circ-0048117 upregulates toll-like receptor 4 expression via miR-140 sponging to promote M2 macrophage polarization, prompting ESCC invasion and metastasis [101]. CircPLCE1 and Circ_0006990 have been demonstrated to motivate TAM M2 polarization in the TME through the miR-485-5p–actin-γ1 and miR-132-3p–mucin 13 cell surface associated axes, respectively, in CRC [102, 103]. A recent study showed that the flavonoid quercetin significantly reversed the promotion of M2-TAMS on proliferation of CRC cell by downregulating circ_0006990 [103]. CircSHKBP1 and circFARSA promote NSCLC migration and invasion by inducing M2 macrophage polarization and impairing CD8+ T cell function in vitro and in vivo [104, 105]. Similarly, Gao et al have demonstrated that exosomal circZNF451 could induce M2 polarization of macrophages and exhaustion of cytotoxic CD8 + T cells to reshape the TME via the FXR1- ELF4-IRF4 axis. More importantly, they will limit the sensitivity of anti-PD1 treatment in vitro and in vivo (or in C57BL/6 J mice) [106]. Myeloid-derived suppressor cells (MDSCs) are bone marrow–derived immature cells that suppress T cells and are activated and mobilized under pathological conditions, such as cancer. Their main functions are to promote tumor development and tumor-related TAM transformation [107, 108]. In lung cancer, circPTK2 and circHIPK3 play important roles in monocytic MDSC differentiation into CD163+ M2 macrophages [109].
Cytokines and chemokines play crucial roles in M2 macrophage polarization. CircSMARCC1 has been shown to regulate CCL20 expression by suppressing miR-1322 activity, thereby mediating M2 macrophage polarization and infiltration, in PCa [110]. In breast cancer cells, circWWC3 upregulates IL-4 expression and secretion to induce M2 macrophage polarization [111] and T-cell inactivation, leading to immune escape [112]. In addition, CircITGB6, circsafb2, and circNEIL3 promote M2 macrophage polarization in ovarian cancer, kidney cancer, and gliomas, respectively [113–115].
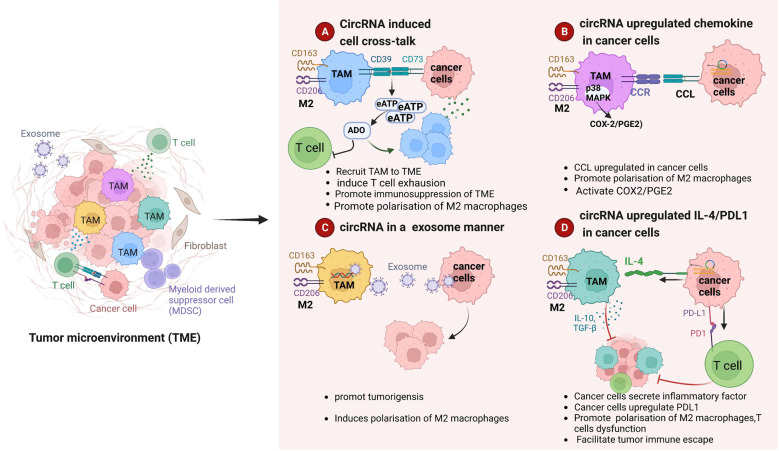
TAMs form the most abundant immune cell population in the TME. CircRNAs in tumor cells can regulate macrophage polarization through multiple pathways in the TME (Table 3, Fig. 4): (1) they induce crosstalk between tumor cells and macrophages (Fig. 4A), (2) they promote chemokine secretion from tumor cells (Fig. 4B), (3) tumor cell-derived circRNAs in exosomes enter macrophages to play a regulatory role (Fig. 4C), and (4) they promote tumor-cell expression of cytokines such as IL-4 and PD-L1 (Fig. 4D). In-depth investigation of the mechanisms underlying these roles and preclinical studies are urgently needed.
Table 3.
circRNAs regulate .macrophages.
| circRNA | Targets/pathways | regulation of immune responses | Cancer | References |
|---|---|---|---|---|
| CircASAP1 | CircASAP1/ miR326/miR-532-5p-CSF-1/ | promotes TAM infiltration | hepatocellular carcinoma | [91] |
| hsa_circ_0001663 (circTMEM181) | circTMEM181/ sponging with miR-488-3/ inducingCD39 expression in macrophages | induces macrophage M2-like polarization | hepatocellular carcinoma | [95] |
| hsa_circ_0110102 | circ_0110102/miR-580-5p/PPARα/CCL2 | inhibits the pro-inflammatory cytokine release from macrophages | hepatocellular carcinoma | [96] |
| hsa_circ_0003410 | hsa_circ_0003410/miR-139-3p/CCL5 axis/increasing the ratio of M2/M1 macrophages | increases the ratio of M2/M1 macrophages | hepatocellular carcinoma | [97] |
| hsa_circ_0074854 | hsa_circ_00074854 / macrophage M2 polarization/immune escape | promotes Macrophage M2 Polarization | hepatocellular carcinoma | [98] |
| circ TCFL5 | circRNA TCFL5 /miR-543-FMNL2 axis / modulating M2 macrophage polarization | modulates M2 macrophage polarization | esophageal squamous cell carcinoma | [100] |
| circ0048117 | hypoxic/exosomal-circ0048117/miR-140 / TLR4/promoting M2 polarization | promoting M2 polarization | esophageal squamous cell carcinoma | [101] |
| hsa_circ_0006990 | circ_0006990/miR-132-3p/MUC13 axis/promotes the polarization of M2 macrophages | promotes the polarization of M2 macrophages | colorectal cancer | [103] |
| CircPLCE1 | Circ PLCE1/miR485-5p/ACTG1 axis/ modulates TAM M2 polarization | modulates TAM M2 polarization | colorectal cancer | [102] |
| circFARSA | exosomal-circFARSA /PTEN/PI3K/AKT /mediates M2 macrophage polarization | promotes M2 polarization | non-small cell lung cancer | [105] |
| circSHKBP1 | circSHKBP1/promoting M2 polarization and macrophage recruitment. | promotes M2 polarization and macrophage recruitment | non-small cell lung cancer | [104] |
| circHIPK3 | circHIPK3/PTK2/infiltration of M2 polarization | lung cancer | [109] | |
| CircSMARCC1 | CircSMARCC1/miR-1322/CCL20/CCR6/inducing TAMs infltration and M2 polarization | induces M2 macrophage polarization | prostate cancer | [110] |
| circWWC3 | circWWC3/ IL-4, PD-L1/ M2-like TAM polarization | modulates M2 macrophage polarization | breast cancer | [112] |
| circITGB6 | circITGB6 / FGF9/ induces polarization of M2 macrophages | induces polarization of TAMs toward M2 phenotype | Ovarian cancer | [113] |
| circSAFB2 | circSAFB2 /miR-620/JAK1/ STAT3 axis /inducing polarization of M2 macrophages | induces polarization of M2 macrophages | renal cell carcinoma | [114] |
| circNEIL3 | exosomes-EWSR1 / circNEIL3/ stabilizing IGF2BP3/macrophage immunosuppressive polarization | Macrophage immunosuppressive polarization | glioma | [115] |
Fig. 4. Circular RNAs (circRNAs) regulate macrophage polarization to promote tumor progression.
A CircRNAs mediate crosstalk between tumor cells and macrophages, induce M2 macrophage polarization, and impair T cell function, resulting in the formation of an immunosuppressive tumor microenvironment (TME). B CircRNAs upregulate chemokine expression in tumor cells and induce M2 macrophage polarization. C Tumor cells release exosomes containing circRNAs into macrophages to enhance M2 macrophage polarization. D CircRNAs promote the secretion of inflammatory factors and immunosuppressive molecules in tumor cells to recruit and induce M2 macrophage polarization and disable T cells.
CircRNAs regulate neutrophils, myeloid-derived suppressor cells, and cancer-associated fibroblast
The neutrophils are also an important component in the TME, participating in different stages of tumor development and progression such as tumorigenesis, proliferation and metastasis. Neutrophils could play dual roles as a pro-tumor(N2) or tumor suppressor (N1) in the tumor microenvironment due to heterogeneous phenotypes and functional diversity. Recently, mounting evidence show that circular RNA affects tumor development by regulating the function of neutrophils. In bladder cancer, circDHTKD1 recruited and activated neutrophils by inducing CXCL5 expression, and then neutrophils participated in lymphangiogenesis by secreting VEGF-C, facilitating lymphatic metastasis of bladder cancer cells [116]. But in CRC, circPACRGL mainly promoted differentiation of N1 to N2 neutrophils by sponging miR-142-3p/miR-506-3p, N2 neutrophils increased the expression of transforming growth factor-β1 (TGF-β1), which promoted CRC cell proliferation, migration and invasion [117]. Although the underlying mechanism is not very clear, but the diversity and plasticity of neutrophils maybe act as a potential and promising immunotherapy target in clinical treatment.
Myeloid-derived suppressor cells (MDSC) are also another key player in TME. In addition to the immunosuppressive effect, MDSC can also exert tumor-promoting effects by promoting angiogenesis, invasion and metastasis. More details about non-coding RNAs including circRNAs modulate MDSCs in TME have been summarized elsewhere [118].
Cancer-associated fibroblasts (CAFs), also named as tumor-associated fibroblast, are a key factor in tumor microenvironment. It plays important role in tumor growth and metastasis due to diverse functions, such as interactions with cancer cells and crosstalk with infiltrating leukocytes and so on. In pancreatic cancer, Hu et al found that circFARP1 upregulated the expression and secretion of LIF via CAV1 in CAFs to induce chemoresistance [119]. In addition, in other cancers, circRNAs derived from CAFs also can promote tumor progression. For example, circEIF3K from CAF promotes CRC progression via miR-214/PD-L1 axis [120]. Exosomal circSLC7A6 from CAF promote tumorigenesis of CRC by regulating CXCR5 [121]. CAF-derived CXCL11 modulates HCC migration and metastasis through the circUBAP2/miR-4756/IFIT1/3 axis [122]. All evidence suggesting an oncogenic role of CAFs in tumorigenesis and indicating CAFs or circRNAs can considered as potential target in immunotherapy.
Conclusion and perspectives
For several decades following the discovery of RNA viruses in 1976 and eukaryotes in 1979 [1, 123], circRNAs were considered to be splicing errors. With the rapid development of RNA sequencing technologies and bioinformatics, numerous circRNAs have been identified and their roles in various diseases, especially cancer, have been investigated extensively. As reviewed here, circRNAs form a multifaceted class of regulators that play multiple roles in tumorigenesis, tumor progression and metastasis, and treatment resistance. They can act as miRNA sponges or interact with RBPs. Different circRNAs may regulate the same downstream gene expression by sequestering different miRNAs; for instance, circ_0046523 induces PD-L1 expression via miR-148a-3p sponging in pancreatic cancer [48], whereas circKRT1 induces PD-L1 expression via miR-495-3p sponging in OSCC [54]. However, our current knowledge of circRNA functions has been obtained mainly from cell-based studies. The examination of whether circRNA loss regulates tumor immune responses and cancer development in genetic mouse models would be of great interest. Despite much progress in past decades, the establishment of circRNA nomenclature rules remains a pressing issue. The production of different circRNAs from the same gene due to alternative splicing can cause confusion. Additionally, the molecular mechanisms underlying this process and the different roles of these circRNAs need to be elucidated.
The TME is a complex integrated system containing tumor cells, tumor-infiltrated immune cells, blood vessels, extracellular matrix, and signaling molecules. Accumulating evidence has revealed that circRNAs play crucial roles in TME regulation, such as tumor immune evasion, metastasis, and metabolism. However, the precise physiological and pathological roles of circRNAs in the TME and related underlying mechanisms remain largely unclear. In this review, we have described the roles of circRNAs in the TME, especially in TME-related immune cells such as T cells, NK cells, and macrophages. CircRNA in the TME can upregulate the expression of the immune checkpoint molecules PD-L1, PD-1, and CD73 on tumor cell surfaces via miRNA sponging, helping tumor cells to escape recognition and death by T cells [51–53, 57]. Stromal cells, such as cancer-associated fibroblasts, endothelial cells, and pericytes, are important TME components, and much more research is warranted to explore their potential regulation. Although TME reprogramming is considered to be a potentially effective strategy for tumor eradication and the improvement of tumor immunotherapy efficacy, there is still a long way to go before we can conquer cancer. For example, does dose targeting of circRNAs work in mouse models, at least in three-dimensional tumor organoid models? Is circRNA targeting applicable in clinical trials? Thorough investigations of circRNAs using animal models would help to accelerate the translation of basic research into clinical practice. We believe that an improved understanding of circRNA functions and mechanisms related to tumorigenesis and immunotherapy would certainly contribute to the development of new therapeutic strategies for cancer.
Acknowledgements
We thank all of the members from the Han and Zhou laboratory for their critical comments and helpful suggestions. We apologize for being unable to cite many important papers in this field due to space limitations. We thank Medjaden Inc. for scientific editing of this manuscript.
Author contributions
Conceptualization, W.R., T.H. and L.G.; writing—original draft preparation, T.H., L.G., and X.Z.; writing—review and editing, L.G, Q.H., B.G., and M.W.; visualization, L.G. and Q.H.; supervision, W.R., T.H., and X.Z.; funding acquisition, T.H., L.G., Q.H., and X.Z. All authors have read the final version of the manuscript and agreed to its publication.
Funding
This study was supported in part by the Key Science and Technology Program of Henan Province (no. 222102310098 to L.G.), the National Natural Science Foundation of China (nos. 82002731 and 82172891 to T.H., no. 82173022 to Q.H., no. 82273098 to X.Z., no. 82002731 to the Article Publishing Charge), the Henan National Science Fund for Excellent Young Scholars (no. 212300410067 to T.H.), the Doctoral Foundation of Xinxiang Medical University (no. XYBSKYZZ202001 to T.H.), the Key Research and Development Program of Henan province (No. 221111310100 to W.R.).
Competing interests
The authors declare no competing interests.
Footnotes
Edited by Professor Hans-Uwe Simon
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors have contributed equally: Liping Guan, Qian Hao.
Contributor Information
Xiang Zhou, Email: xiangzhou@fudan.edu.cn.
Tao Han, Email: doubleyingtao@gmail.com.
Wenjie Ren, Email: wjren1966@163.com.
References
- 1.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–6. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell. 1991;64:607–13. doi: 10.1016/0092-8674(91)90244-S. [DOI] [PubMed] [Google Scholar]
- 3.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–8. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 5.Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 6.Han J, Zhao G, Ma X, Dong Q, Zhang H, Wang Y, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503:2429–35. doi: 10.1016/j.bbrc.2018.06.172. [DOI] [PubMed] [Google Scholar]
- 7.Yang X, Ye T, Liu H, Lv P, Duan C, Wu X, et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol Cancer. 2021;20:4. doi: 10.1186/s12943-020-01300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li YF, Zhang J, Yu L. Circular RNAs regulate cancer onset and progression via Wnt/β-Catenin signaling pathway. Yonsei Med J. 2019;60:1117–28. doi: 10.3349/ymj.2019.60.12.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan S, Wang S, Huang T, Wang J, Yuan X. circRNAs: Insight into their role in tumor-associated macrophages. Front Oncol. 2021;11:780744. doi: 10.3389/fonc.2021.780744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz-Munoz MD, Turner M. Uncovering the role of RNA-binding proteins in gene expression in the immune system. Front Immunol. 2018;9:1094. doi: 10.3389/fimmu.2018.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Cheng Y, Wu F, Wu L, Cao H, Wang Q, et al. The emerging landscape of circular RNAs in immunity: breakthroughs and challenges. Biomark Res. 2020;8:25. doi: 10.1186/s40364-020-00204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taheri M, Barth DA, Kargl J, Rezaei O, Ghafouri-Fard S, Pichler M. Emerging role of non-coding RNAs in regulation of T-lymphocyte function. Front Immunol. 2021;12:756042. doi: 10.3389/fimmu.2021.756042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 14.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–42. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 16.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shuwen H, Qing Z, Yan Z, Xi Y. Competitive endogenous RNA in colorectal cancer: a systematic review. Gene. 2018;645:157–62. doi: 10.1016/j.gene.2017.12.036. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S. The characteristics of circRNA as competing endogenous RNA in pathogenesis of acute myeloid leukemia. BMC Cancer. 2021;21:277. doi: 10.1186/s12885-021-08029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin Y, Yu LL, Zhang B, Liu CF, Chen Y. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res. 2018;51:e7811. doi: 10.1590/1414-431x20187811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Corrigendum: Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2017;24:194. doi: 10.1038/nsmb0217-194a. [DOI] [PubMed] [Google Scholar]
- 24.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 2019;30:157–173. doi: 10.1016/j.cmet.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang J, et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer. 2020;19:71. doi: 10.1186/s12943-020-01179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Liu Y, Liu Z, Lin C, Meng F, Xu L, et al. CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Mol Cancer. 2021;20:114. doi: 10.1186/s12943-021-01412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Liu G, Wu Y, Ma J, Wu H, Xie Z, et al. Correction to: CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of beta-catenin/LEF1 complex via effects on chromatin remodeling. Mol Cancer. 2020;19:75. doi: 10.1186/s12943-020-01193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12:269–81. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forman J. T cells, the MHC, and function. Immunol Rev. 1984;81:203–19. doi: 10.1111/j.1600-065X.1984.tb01111.x. [DOI] [PubMed] [Google Scholar]
- 32.Petrova G, Ferrante A, Gorski J. Cross-reactivity of T cells and its role in the immune system. Crit Rev Immunol. 2012;32:349–72. doi: 10.1615/CritRevImmunol.v32.i4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pittet MJ, Speiser DE, Valmori D, Rimoldi D, Lienard D, Lejeune F, et al. Ex vivo analysis of tumor antigen specific CD8+ T cell responses using MHC/peptide tetramers in cancer patients. Int Immunopharmacol. 2001;1:1235–47. doi: 10.1016/S1567-5769(01)00048-0. [DOI] [PubMed] [Google Scholar]
- 34.Villa-Morales M, Fernandez-Piqueras J. Targeting the Fas/FasL signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:85–101. doi: 10.1517/14728222.2011.628937. [DOI] [PubMed] [Google Scholar]
- 35.Wajant H. CD95L/FasL and TRAIL in tumour surveillance and cancer therapy. Cancer Treat Res. 2006;130:141–65. doi: 10.1007/0-387-26283-0_7. [DOI] [PubMed] [Google Scholar]
- 36.Law LW, Appella E, Henriksen O, Rogers M. Some biologic and biochemical properties of soluble tumor antigens. Ann N. Y Acad Sci. 1976;276:11–25. doi: 10.1111/j.1749-6632.1976.tb41633.x. [DOI] [PubMed] [Google Scholar]
- 37.Zusman I. Soluble tumor-associated antigens in cancer detection, prevention and therapy. Med Sci Monit. 2004;10:RA317–24. [PubMed] [Google Scholar]
- 38.Kok VC, Yu CC. Cancer-derived exosomes: their role in cancer biology and biomarker development. Int J Nanomed. 2020;15:8019–36. doi: 10.2147/IJN.S272378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He L, Zhu W, Chen Q, Yuan Y, Wang Y, Wang J, et al. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics. 2019;9:8206–20. doi: 10.7150/thno.37455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–94. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Yao Y, Jin M. Circ-0001068 is a novel biomarker for ovarian cancer and inducer of PD1 expression in T cells. Aging (Albany NY) 2020;12:19095–106. doi: 10.18632/aging.103706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu YJ, Zhao JM, Gao C, Ni XF, Wang W, Hu WW, et al. Hsa_circ_0136666 activates Treg-mediated immune escape of colorectal cancer via miR-497/PD-L1 pathway. Cell Signal. 2021;86:110095. doi: 10.1016/j.cellsig.2021.110095. [DOI] [PubMed] [Google Scholar]
- 44.Gao C, Xu YJ, Qi L, Bao YF, Zhang L, Zheng L. CircRNA VIM silence synergizes with sevoflurane to inhibit immune escape and multiple oncogenic activities of esophageal cancer by simultaneously regulating miR-124/PD-L1 axis. Cell Biol Toxicol. 2022;38:825–45. doi: 10.1007/s10565-021-09613-0. [DOI] [PubMed] [Google Scholar]
- 45.Zou Y, Zheng S, Deng X, Yang A, Xie X, Tang H, et al. The role of circular RNA CDR1as/ciRS-7 in regulating tumor microenvironment: a pan-cancer analysis. Biomolecules. 2019;9:429. doi: 10.3390/biom9090429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu G, Zhang P, Liang H, Xu Y, Shen J, Wang W, et al. Circular RNA hsa_circ_0003288 induces EMT and invasion by regulating hsa_circ_0003288/miR-145/PD-L1 axis in hepatocellular carcinoma. Cancer Cell Int. 2021;21:212. doi: 10.1186/s12935-021-01902-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo YH, Yang YP, Chien CS, Yarmishyn AA, Adekunle Ishola A, Chien Y, et al. Circular RNA hsa_circ_0000190 facilitates the tumorigenesis and immune evasion by upregulating the expression of soluble PD-L1 in non-small-cell lung cancer. Int J Mol Sci. 2021;23:64. doi: 10.3390/ijms23010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu X, Sun G, Tu S, Fang K, Xiong Y, Tu Y, et al. Hsa_circ_0046523 mediates an immunosuppressive tumor microenvironment by regulating MiR-148a-3p/PD-L1 axis in pancreatic cancer. Front Oncol. 2022;12:877376. doi: 10.3389/fonc.2022.877376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC) J Exp Clin Cancer Res. 2020;39:149. doi: 10.1186/s13046-020-01648-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Li L, Zhang Q, Lian K. Circular RNA circ_0000284 plays an oncogenic role in the progression of non-small cell lung cancer through the miR-377-3p-mediated PD-L1 promotion. Cancer Cell Int. 2020;20:247. doi: 10.1186/s12935-020-01310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian Q, Wu T, Zhang X, Xu K, Yin X, Wang X, et al. Immunomodulatory functions of the circ_001678/miRNA-326/ZEB1 axis in non-small cell lung cancer via the regulation of PD-1/PD-L1 pathway. Hum Mol Genet. 2022;31:4094–106. doi: 10.1093/hmg/ddac155. [DOI] [PubMed] [Google Scholar]
- 52.Lei J, Zhu J, Hui B, Jia C, Yan X, Jiang T, et al. Circ-HSP90A expedites cell growth, stemness, and immune evasion in non-small cell lung cancer by regulating STAT3 signaling and PD-1/PD-L1 checkpoint. Cancer Immunol Immunother. 2023;72:101–24. doi: 10.1007/s00262-022-03235-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021;20:105. doi: 10.1186/s12943-021-01398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Chen W, Wang Y, Qin M, Ji Y. CircKRT1 drives tumor progression and immune evasion in oral squamous cell carcinoma by sponging miR‐495‐3p to regulate PDL1 expression. Cell Biol Int. 2021;45:1423–35. doi: 10.1002/cbin.11581. [DOI] [PubMed] [Google Scholar]
- 55.Ghalamfarsa G, Kazemi MH, Raoofi Mohseni S, Masjedi A, Hojjat-Farsangi M, Azizi G, et al. CD73 as a potential opportunity for cancer immunotherapy. Expert Opin Ther Targets. 2019;23:127–42. doi: 10.1080/14728222.2019.1559829. [DOI] [PubMed] [Google Scholar]
- 56.Allard D, Chrobak P, Allard B, Messaoudi N, Stagg J. Targeting the CD73-adenosine axis in immuno-oncology. Immunol Lett. 2019;205:31–39. doi: 10.1016/j.imlet.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 57.Xu YP, Dong ZN, Wang SW, Zheng YM, Zhang C, Zhou YQ, et al. circHMGCS1-016 reshapes immune environment by sponging miR-1236-3p to regulate CD73 and GAL-8 expression in intrahepatic cholangiocarcinoma. J Exp Clin Cancer Res. 2021;40:290. doi: 10.1186/s13046-021-02095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li P, Ren X, Zheng Y, Sun J, Ye G. Tumor promoting effect of circ_002172 associates with induced immune escape in breast cancer via the miR-296-5p/CXCL12 axis. Int Immunopharmacol. 2022;106:108530. doi: 10.1016/j.intimp.2022.108530. [DOI] [PubMed] [Google Scholar]
- 59.Garg B, Giri B, Modi S, Sethi V, Castro I, Umland O, et al. NFkappaB in pancreatic Stellate cells reduces infiltration of tumors by cytotoxic T cells and killing of cancer cells, via up-regulation of CXCL12. Gastroenterology. 2018;155:880–891. doi: 10.1053/j.gastro.2018.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen CD, Wang CS, Huang YH, Chien KY, Liang Y, Chen WJ, et al. Overexpression of CLIC1 in human gastric carcinoma and its clinicopathological significance. Proteomics. 2007;7:155–67. doi: 10.1002/pmic.200600663. [DOI] [PubMed] [Google Scholar]
- 61.Xia J, Wang Q, Ju F, Luo X, Wang F, Zhou Y, et al. Chloride intracellular channel 1 is a potential biomarker for breast cancer. Breast Cancer. 2022;14:247–58. doi: 10.2147/BCTT.S367519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yasuda Y, Nagano T, Jimbo N, Kiriu T, Suraya R, Hazama D, et al. Chloride intracellular channel 1 expression is associated with poor prognosis of lung adenocarcinoma. Anticancer Res. 2022;42:271–7. doi: 10.21873/anticanres.15482. [DOI] [PubMed] [Google Scholar]
- 63.Zhang S, Wang XM, Yin ZY, Zhao WX, Zhou JY, Zhao BX, et al. Chloride intracellular channel 1 is overexpression in hepatic tumor and correlates with a poor prognosis. APMIS. 2013;121:1047–53. doi: 10.1111/apm.12093. [DOI] [PubMed] [Google Scholar]
- 64.Li B, Liang L, Chen Y, Liu J, Wang Z, Mao Y, et al. Circ_0008287 promotes immune escape of gastric cancer cells through impairing microRNA-548c-3p-dependent inhibition of CLIC1. Int Immunopharmacol. 2022;111:108918. doi: 10.1016/j.intimp.2022.108918. [DOI] [PubMed] [Google Scholar]
- 65.Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y, et al. Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Mol Ther. 2022;30:1054–70. doi: 10.1016/j.ymthe.2022.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao X, Lu C, Chu W, Zhang B, Zhen Q, Wang R, et al. MicroRNA-124 suppresses proliferation and glycolysis in non-small cell lung cancer cells by targeting AKT-GLUT1/HKII. Tumour Biol. 2017;39:1010428317706215. doi: 10.1177/1010428317706215. [DOI] [PubMed] [Google Scholar]
- 67.Guo XY, Chen JN, Sun F, Wang YQ, Pan Q, Fan JG. circRNA_0046367 prevents hepatoxicity of lipid peroxidation: an inhibitory role against hepatic steatosis. Oxid Med Cell Longev. 2017;2017:3960197. doi: 10.1155/2017/3960197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka A, Sakaguchi S. Targeting Treg cells in cancer immunotherapy. Eur J Immunol. 2019;49:1140–6. doi: 10.1002/eji.201847659. [DOI] [PubMed] [Google Scholar]
- 69.Chen Y, Li Z, Liang J, Liu J, Hao J, Wan Q, et al. CircRNA has_circ_0069313 induced OSCC immunity escape by miR-325-3p-Foxp3 axes in both OSCC cells and Treg cells. Aging (Albany NY) 2022;14:4376–89. doi: 10.18632/aging.204068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang M, Huang X, Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022;113:1968–83. doi: 10.1111/cas.15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang Z, Jiang C, Li S. The potential regulatory roles of circular RNAs in tumor immunology and immunotherapy. Front Immunol. 2021;11:617583. doi: 10.3389/fimmu.2020.617583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu H, Wang S, Xin J, Wang J, Yao C, Zhang Z. Role of NKG2D and its ligands in cancer immunotherapy. Am J Cancer Res. 2019;9:2064–78. [PMC free article] [PubMed] [Google Scholar]
- 73.Guillerey C. NK Cells in the Tumor Microenvironment. Adv Exp Med Biol. 2020;1273:69–90. doi: 10.1007/978-3-030-49270-0_4. [DOI] [PubMed] [Google Scholar]
- 74.Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li S, Chen Z, Zhou R, Wang S, Wang W, Liu D, et al. Hsa_circ_0048674 facilitates hepatocellular carcinoma progression and natural killer cell exhaustion depending on the regulation of miR-223-3p/PDL1. Histol Histopathol. 2022;37:1185–1199. doi: 10.14670/HH-18-440. [DOI] [PubMed] [Google Scholar]
- 76.Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett. 2019;17:388–97. doi: 10.3892/ol.2018.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH, Wang YG, et al. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 2021;12:94. doi: 10.1038/s41419-020-03334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang W, Zhong W, Wang B, Yang J, Yang J, Yu Z, et al. ICAM-1-mediated adhesion is a prerequisite for exosome-induced T cell suppression. Dev Cell. 2022;57:329–343. doi: 10.1016/j.devcel.2022.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dieterich LC. Mechanisms of extracellular vesicle-mediated immune evasion in melanoma. Front Immunol. 2022;13:1002551. doi: 10.3389/fimmu.2022.1002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ou ZL, Luo Z, Wei W, Liang S, Gao TL, Lu YB. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: role of circ_0000977/miR-153 axis. RNA Biol. 2019;16:1592–603. doi: 10.1080/15476286.2019.1649585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li M, Zhang M, Chen M, Xiao J, Mu X, Peng J. KLF2-induced circZKSCAN1 potentiates the tumorigenic properties of clear cell renal cell carcinoma by targeting the miR-1294/PIM1 axis. Cell Cycle. 2022;21:1376–90. doi: 10.1080/15384101.2022.2051293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luchtenborg AM, Metzger P, Cosenza Contreras M, Oria V, Biniossek ML, Lindner F, et al. Kruppel-like factor 7 influences translation and pathways involved in ribosomal biogenesis in breast cancer. Breast Cancer Res. 2022;24:65. doi: 10.1186/s13058-022-01562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiu DH, Chen Y, Liu L, Yang HS, Liu GF. Tumor-suppressive role of Kruppel-like factor 4 (KLF-4) in colorectal cancer. Genet Mol Res. 2017;16. 10.4238/gmr16019272. [DOI] [PubMed]
- 84.Xiao S, Jin-Xiang Y, Long T, Xiu-Rong L, Hong G, Jie-Cheng Y, et al. Kruppel-like factor 2 disturb non-small cell lung cancer energy metabolism by inhibited glutamine consumption. J Pharm Pharm. 2020;72:843–51. doi: 10.1111/jphp.13252. [DOI] [PubMed] [Google Scholar]
- 85.Yang F, Chen Y, Luo L, Nong S, Li T. circFOXO3 induced by KLF16 modulates clear cell renal cell carcinoma growth and natural killer cell cytotoxic activity through sponging miR-29a-3p and miR-122-5p. Dis Markers. 2022;2022:6062236. doi: 10.1155/2022/6062236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martinez-Rojo E, Berumen LC, Garcia-Alcocer G, Escobar-Cabrera J. The role of androgens and androgen receptor in human bladder cancer. Biomolecules. 2021;11:594. doi: 10.3390/biom11040594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Q, You B, Meng J, Huang CP, Dong G, Wang R, et al. Targeting the androgen receptor to enhance NK cell killing efficacy in bladder cancer by modulating ADAR2/circ_0001005/PD-L1 signaling. Cancer Gene Ther. 2022;29:1988–2000. doi: 10.1038/s41417-022-00506-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ke H, Zhang J, Wang F, Xiong Y. ZNF652-Induced circRHOT1 promotes SMAD5 expression to modulate tumorigenic properties and nature killer cell-mediated Ttoxicity in bladder cancer via targeting miR-3666. J Immunol Res. 2021;2021:7608178. doi: 10.1155/2021/7608178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li X, Han M, Zhang H, Liu F, Pan Y, Zhu J, et al. Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma. Biomark Res. 2022;10:2. doi: 10.1186/s40364-021-00345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heyliger SO, Soliman KFA, Saulsbury MD, Reams RR. The identification of zinc-finger protein 433 as a possible prognostic biomarker for clear-cell renal cell carcinoma. Biomolecules. 2021;11:1193. doi: 10.3390/biom11081193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 2020;72:906–22. doi: 10.1002/hep.31068. [DOI] [PubMed] [Google Scholar]
- 92.Han S, Wang W, Wang S, Yang T, Zhang G, Wang D, et al. Tumor microenvironment remodeling and tumor therapy based on M2-like tumor associated macrophage-targeting nano-complexes. Theranostics. 2021;11:2892–916. doi: 10.7150/thno.50928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Niedobitek G, Barros MH, Dreyer JH, Hauck F, Al-Sheikhyaqoob D. Tumor-associated macrophages: function and differentiation. Pathologe. 2015;36:477–84. doi: 10.1007/s00292-015-0054-7. [DOI] [PubMed] [Google Scholar]
- 94.Wu K, Lin K, Li X, Yuan X, Xu P, Ni P, et al. Redefining tumor-associated macrophage subpopulations and functions in the tumor microenvironment. Front Immunol. 2020;11:1731. doi: 10.3389/fimmu.2020.01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu JC, Zhang PF, Huang XY, Guo XJ, Gao C, Zeng HY, et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J Hematol Oncol. 2021;14:200. doi: 10.1186/s13045-021-01207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang X, Sheng W, Xu T, Xu J, Gao R, Zhang Z. CircRNA hsa_circ_0110102 inhibited macrophage activation and hepatocellular carcinoma progression via miR-580-5p/PPARalpha/CCL2 pathway. Aging (Albany NY) 2021;13:11969–87. doi: 10.18632/aging.202900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cao P, Ma B, Sun D, Zhang W, Qiu J, Qin L, et al. hsa_circ_0003410 promotes hepatocellular carcinoma progression by increasing the ratio of M2/M1 macrophages through the miR-139-3p/CCL5 axis. Cancer Sci. 2022;113:634–47. doi: 10.1111/cas.15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int J Nanomed. 2021;16:2803–18. doi: 10.2147/IJN.S284560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hume DA, MacDonald KP. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood. 2012;119:1810–20. doi: 10.1182/blood-2011-09-379214. [DOI] [PubMed] [Google Scholar]
- 100.Lin C, Xi Y, Yu H, Wang Z, Chen X, Shen W. circRNA TCFL5 promote esophageal cancer progression by modulating M2 macrophage polarization via the miR-543-FMNL2 axis. J Oncol. 2022;2022:5075615. doi: 10.1155/2022/5075615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu Q, Wang X, Zhu J, Fei X, Chen H, Li C. Hypoxic tumor-derived exosomal Circ0048117 facilitates M2 macrophage polarization acting as miR-140 sponge in esophageal squamous cell carcinoma. Onco Targets Ther. 2020;13:11883–97. doi: 10.2147/OTT.S284192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yi B, Dai K, Yan Z, Yin Z. Circular RNA PLCE1 promotes epithelial mesenchymal transformation, glycolysis in colorectal cancer and M2 polarization of tumor-associated macrophages. Bioengineered. 2022;13:6243–56. doi: 10.1080/21655979.2021.2003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen B, Wu L, Tang X, Wang T, Wang S, Yu H, et al. Quercetin inhibits tumorigenesis of colorectal cancer through downregulation of hsa_circ_0006990. Front Pharm. 2022;13:874696. doi: 10.3389/fphar.2022.874696. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Chen W, Tang D, Lin J, Huang X, Lin S, Shen G, et al. Exosomal circSHKBP1 participates in non-small cell lung cancer progression through PKM2-mediated glycolysis. Mol Ther Oncolytics. 2022;24:470–85. doi: 10.1016/j.omto.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen T, Liu Y, Li C, Xu C, Ding C, Chen J, et al. Tumor-derived exosomal circFARSA mediates M2 macrophage polarization via the PTEN/PI3K/AKT pathway to promote non-small cell lung cancer metastasis. Cancer Treat Res Commun. 2021;28:100412. doi: 10.1016/j.ctarc.2021.100412. [DOI] [PubMed] [Google Scholar]
- 106.Gao J, Ao YQ, Zhang LX, Deng J, Wang S, Wang HK, et al. Exosomal circZNF451 restrains anti-PD1 treatment in lung adenocarcinoma via polarizing macrophages by complexing with TRIM56 and FXR1. J Exp Clin Cancer Res. 2022;41:295. doi: 10.1186/s13046-022-02505-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hegde S, Leader AM, Merad M. MDSC: Markers, development, states, and unaddressed complexity. Immunity. 2021;54:875–84. doi: 10.1016/j.immuni.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bruger AM, Dorhoi A, Esendagli G, Barczyk-Kahlert K, van der Bruggen P, Lipoldova M, et al. How to measure the immunosuppressive activity of MDSC: assays, problems and potential solutions. Cancer Immunol Immunother. 2019;68:631–44. doi: 10.1007/s00262-018-2170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Katopodi T, Petanidis S, Domvri K, Zarogoulidis P, Anestakis D, Charalampidis C, et al. Kras-driven intratumoral heterogeneity triggers infiltration of M2 polarized macrophages via the circHIPK3/PTK2 immunosuppressive circuit. Sci Rep. 2021;11:15455. doi: 10.1038/s41598-021-94671-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xie T, Fu DJ, Li ZM, Lv DJ, Song XL, Yu YZ, et al. CircSMARCC1 facilitates tumor progression by disrupting the crosstalk between prostate cancer cells and tumor-associated macrophages via miR-1322/CCL20/CCR6 signaling. Mol Cancer. 2022;21:173. doi: 10.1186/s12943-022-01630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gunassekaran GR, Poongkavithai Vadevoo SM, Baek MC, Lee B. M1 macrophage exosomes engineered to foster M1 polarization and target the IL-4 receptor inhibit tumor growth by reprogramming tumor-associated macrophages into M1-like macrophages. Biomaterials. 2021;278:121137. doi: 10.1016/j.biomaterials.2021.121137. [DOI] [PubMed] [Google Scholar]
- 112.Zheng Y, Ren S, Zhang Y, Liu S, Meng L, Liu F, et al. Circular RNA circWWC3 augments breast cancer progression through promoting M2 macrophage polarization and tumor immune escape via regulating the expression and secretion of IL-4. Cancer Cell Int. 2022;22:264. doi: 10.1186/s12935-022-02686-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li H, Luo F, Jiang X, Zhang W, Xiang T, Pan Q, et al. CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype. J Immunother Cancer. 2022;10:e004029. doi: 10.1136/jitc-2021-004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang X, Wang J, Guan J, Zheng Z, Hao J, Sheng Z, et al. Exosomal Circsafb2 reshaping tumor environment to promote renal cell carcinoma progression by mediating M2 macrophage polarization. Front Oncol. 2022;12:808888. doi: 10.3389/fonc.2022.808888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q, et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer. 2022;21:16. doi: 10.1186/s12943-021-01485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu Q, Yin H, Deng Y, Chen W, Diao W, Ding M, et al. circDHTKD1 promotes lymphatic metastasis of bladder cancer by upregulating CXCL5. Cell Death Disco. 2022;8:243. doi: 10.1038/s41420-022-01037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu X, Zhao S, Sui H, Liu H, Yao M, Su Y, et al. MicroRNAs/LncRNAs modulate MDSCs in tumor microenvironment. Front Oncol. 2022;12:772351. doi: 10.3389/fonc.2022.772351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hu C, Xia R, Zhang X, Li T, Ye Y, Li G, et al. CircFARP1 enables cancer-associated fibroblasts to promote gemcitabine resistance in pancreatic cancer via the LIF/STAT3 axis. Mol Cancer. 2022;21:24. doi: 10.1186/s12943-022-01501-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang K, Zhang J, Bao C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer. 2021;21:933. doi: 10.1186/s12885-021-08669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gu C, Lu H, Qian Z. Matrine reduces the secretion of exosomal circSLC7A6 from cancer-associated fibroblast to inhibit tumorigenesis of colorectal cancer by regulating CXCR5. Biochem Biophys Res Commun. 2020;527:638–45. doi: 10.1016/j.bbrc.2020.04.142. [DOI] [PubMed] [Google Scholar]
- 122.Liu G, Sun J, Yang ZF, Zhou C, Zhou PY, Guan RY, et al. Cancer-associated fibroblast-derived CXCL11 modulates hepatocellular carcinoma cell migration and tumor metastasis through the circUBAP2/miR-4756/IFIT1/3 axis. Cell Death Dis. 2021;12:260. doi: 10.1038/s41419-021-03545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–40. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]