Abstract
The aryl hydrocarbon receptor (AHR), a transcription factor best known for mediating toxic responses of environmental pollutants, also integrates metabolic signals to promote anti-inflammatory responses, intestinal homeostasis, and maintain barrier integrity. AHR regulates its target genes through direct DNA-binding to aryl hydrocarbon response elements (AHREs) but also through tethering to other transcription factors in a DNA-binding independent manner. However, it is not known if AHR’s anti-inflammatory role in the gut requires its ability to bind to AHREs. To test this, we determined the sensitivity of Ahrdbd/dbd mice, a genetically modified mouse line that express an AHR protein incapable of binding to AHREs, to dextran sulfate sodium (DSS)-induced colitis. Ahrdbd/dbd mice exhibited more severe symptoms of intestinal inflammation than Ahr+/+ mice. None of the Ahrdbd/dbd mice survived after the 5-day DSS followed by 7-day washout period. By day 6, the Ahrdbd/dbd mice had severe body weight loss, shortening of the colon, higher disease index scores, enlarged spleens, and increased expression of several inflammation genes, including interleukin 1b (Il-1b), Il-6, Il-17, C-x-c motif chemokine ligand 1 (Cxcl1), Cxcl2, Prostaglandin-endoperoxide synthase (Ptgs2), and lipocalin-2. Our findings show that AHR’s DNA-binding domain and ability to bind to AHREs are required to reduce inflammation, maintain a healthy intestinal environment, and protect against DSS-induced colitis.
Keywords: aryl hydrocarbon receptor, DNA-binding domain, dextran sulfate sodium, gut immunity, microbiota, AHR
The aryl hydrocarbon receptor (AHR) is a ligand-dependent transcription factor and member of the basic helix-loop-helix (bHLH)-per-ARNT-sim (PAS) family (Gu et al., 2000; Hankinson, 1995). AHR is best known for its role in 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) toxicity but has emerged as an important factor that transmits environmental and endogenous signals to dampen inflammation and regulate immune cell homeostasis (Stockinger et al., 2014; 2021). Several endogenous and dietary ligands or activators of AHR have been identified, including the tryptophan metabolites kynurenine and 6-formylindolo(3,2-b)carbazole (FICZ), as well as dietary ligands such as indole-3-carbinol and one of its acid condensation products, 3,3′-diindolylmethane (Avilla et al., 2020; DeGroot et al., 2015; Denison and Nagy, 2003; Seok et al., 2018). In the canonical AHR pathway, ligand binding to AHR causes its translocation to the nucleus where it heterodimerizes with AHR nuclear translocator (ARNT). The AHR-ARNT complex binds to aryl hydrocarbon response elements (AHREs; xenobiotic or dioxin response elements) in the regulatory regions of its target genes, including the drug-metabolizing enzymes cytochrome P450 1A1 (CYP1A1) and CYP1B1, cytokines, growth factors, and cell cycle regulators (Hankinson, 2005).
AHR is associated with several inflammatory and immune disorders, including inflammatory bowel disease (IBD), allergic responses, cardiovascular disease, multiple sclerosis, and rheumatoid arthritis (Hui and Dai, 2020; Sartor, 2006; Wheeler et al., 2017; Yi et al., 2018). Because of this, there is considerable interest in targeting AHR to constrain inflammation (Cannon et al., 2021; Pernomian et al., 2020). For example, tapinarof, an AHR agonist, has recently been approved for the treatment of plaque psoriasis and atopic dermatitis (Keam, 2022).
AHR is associated with ulcerative colitis and Crohn’s disease, which are the 2 major forms of IBD. Immune cells isolated from patients suffering from Crohn’s disease have reduced levels of AHR (Monteleone et al., 2011). Treatment with the natural AHR ligand, Indigo Naturalis, for 8 weeks resulted in effective clinical responses in patients with ulcerative colitis (Naganuma et al., 2018). In mouse models of intestinal inflammation, AHR activation significantly improves, while its loss or the reduction of its endogenous ligand levels exacerbates dextran sulfate sodium (DSS)-induced colitis and bacterial-induced mucosal inflammation (Lamas et al., 2016; Li et al., 2011; Monteleone et al., 2011; Schiering et al., 2017). In addition, commensal microbial products such as indole-3-pyruvic acid, urolithin A, short-chain fatty acids, and dihydroxyquinoline may regulate intestinal inflammation in an AHR-dependent manner (Pernomian et al., 2020).
Previous studies have reported that AHR regulates some cellular pathways, and the expression of inflammatory genes, through tethering to other transcription factors in an AHRE-independent manner (Beischlag et al., 2008; Dere et al., 2011). This has led to several proposed models in which AHR regulates cellular events through noncanonical signaling processes that are independent of nuclear translocation, AHRE binding or ARNT dimerization (Grosskopf et al., 2021; Puga et al., 1997; Wright et al., 2017; Zhu et al., 2018). Genome-wide AHR ChIP-chip and Chip-sequencing analyses revealed that approximately 50% of AHR-enriched regions do not contain an AHRE (Dere et al., 2011; Lo and Matthews, 2012). These results further support the notion that tethering to other transcription factors is an important function of AHR regulation in addition to its direct binding to AHREs. Genetically modified mice that express a mutant AHR that is incapable of binding to AHREs, termed Ahrdbd/dbd mice, are resistant to TCDD toxicity and exhibit the same ductus venous development abnormalities that have been reported for Ahr−/− mice (Bunger et al., 2008). These findings confirm the importance of AHR-AHRE binding in developmental and toxicological AHR signaling. However, it is yet to be determined if AHR’s anti-inflammatory role in the gut is dependent on its ability to bind to AHREs.
Here, we used a DSS-induced colitis model to investigate the effect of loss of AHR’s DNA-binding activity on DSS-induced intestinal inflammation. We observed that DSS-exposed Ahrdbd/dbd mice exhibited increased severity of disease symptoms. Our findings provide the first evidence that DNA-binding deficient AHR increases intestinal inflammation in the DSS model of colitis. These data further support the importance of the canonical AHR pathway in the biological and toxicological signaling of AHR.
Materials and methods
Chemicals
DSS salt reagent grade (MP Biomedicals) was dissolved to a concentration of 2% in autoclaved drinking water. TCDD was purchased from Accustandard (New Haven, Connecticut). Dimethyl sulfoxide (DMSO) and all other chemicals and biological reagents were purchased from Merck (Frankfurt, Germany) unless stated otherwise.
Animals, TCDD, and DSS treatment
For all studies, 8-week-old male Ahr+/+ and Ahrdbd/dbd mice were used, which were a generous gift from Christopher Bradfield, McArdle Laboratory for Cancer Research University of Wisconsin, Madison, Wisconsin. The characterization and generation of the Ahrdbd/dbd mice have been described previously (Bunger et al., 2008). The Ahr+/dbd heterozygotes were bred together to give Ahrdbd/dbd and Ahr+/+ littermates that were used for experiments. Tissues for genotyping were clipped from tails at 2 weeks of age. DNA extraction and PCR reactions were performed using the REDExtract-N-Amp tissue PCR kit using forward 5′-CTGAGGGGACGTTTTAATG-3′ and reverse 5’-AACATTTGCACTCATGGATAG-3′ primers and following the manufacturer’s recommendations. The PCR amplicon was digested with BamHI, because a BamHI recognition sequence was introduced in the Ahrdbd/dbd sequence. The digestion reaction was then separated on a 2% agarose gel. For the TCDD treatment studies, male Ahr+/+ and Ahrdbd/dbd littermates were intraperitoneally injected with 100 µg/kg TCDD or an equivalent volume of DMSO vehicle control. After 2, 6, or 24 h, animals were sacrificed by cervical dislocation. Liver tissues were flash frozen in liquid nitrogen immediately after collection and later stored at −80°C. For the DSS studies, male Ahr+/+ and Ahrdbd/dbd mice were housed singly and given either normal drinking water or water containing 2% (w/v) DSS for 5 days to induce IBD (Mizoguchi, 2012). The DSS-containing water was replaced with regular water after 6 days and the mice were monitored until day 12. Water and food consumption were measured daily. Body weight and signs of IBD were monitored daily, including diarrhea, weight loss, dehydration, hematochezia, weakness, and rectal prolapse. The animals received supportive care if needed, which included, but was not limited to a heating pad, mash and/or other fluid supplementation. At the end of the experiment animals were euthanized, and liver, intestines, and colon were removed for gene expression, histological, and biochemical analyses. Feces were collected immediately before the start of the experiment and used to evaluate changes in the microbiome by a real-time qPCR-based method, as we and others have described (Hutin et al., 2022; Lamas et al., 2016; Yang et al., 2015). All animals were bred and cared for at the University of Toronto. Care and treatment of animals followed the guidelines set by the Canadian Council on Animal Care and were approved by the University of Toronto Animal Care Committee.
Ahr gene cloning and sequencing
The liver RNA of one Ahrdbd/dbd mouse and one Ahr+/+ mouse was extracted using Aurum total RNA mini kit, and cDNA was synthesized using Applied Biosystems high-capacity cDNA reverse transcription kit according to the manufacturer’s instructions. An Ahr gene-specific reverse primer with an XhoI restriction site (5′-AATTCTCGAGCTACAGGAATCCACCAGG TGTGATATC-3′) was used instead of the 10× RT random primers. Ahr cDNA was then PCR amplified using Platinum Pfx DNA polymerase using the forward primer containing a NheI site: 5′-AATTGCTAGCGCCACCATGAGCAGCGGCGCCAACATCAC-3′, and the Ahr gene-specific reverse primer. The Ahr PCR products and the pcDNA3.1 plasmids were then cloned into the NheI and XhoI sites of pcDNA3.1. The complete Ahr mRNA sequence from Ahr+/+ and Ahrdbd/dbd was determined by the Centre for Applied Genomics (TCAG) operated by the Hospital for Sick Children (SickKids) (Toronto, Ontario, Canada).
Luciferase reporter gene assay
COS-1 cells were plated on 12-well dishes at a density of 1.00 × 105 to 1.25 × 105 cells with 1 ml media per well. The following day, each well was transfected with 2 µl Lipofectamine 2000 and 1 µg DNA consisting of 300 ng pGudLuc 4.1, 100 ng pCH110, and 50 ng pEGFP, 400 ng of pcDNA3.1-Ahrwt or pcDNA3.1-Ahrdbd. pGudLuc 4.1 is an AHR-driven reporter construct containing a luciferase gene downstream of the Cyp1a1 promoter. After 6 h, every well was treated with either DMSO or 100 nM TCDD. The next morning, the cells were lysed, and the luciferase activity was measured using ONE-Glo luciferase assay and normalized to β-galactosidase activity. To confirm the expression of AHRwt and AHRdbd in COS-1 cells after transfection, COS-1 cells were again plated on 12-well dishes at a density of 1.00 × 105 to 1.25 × 105 cells with 1 ml medium per well. On each plate, 4 wells were transfected with each of pcDNA3.1, pcDNA3.1-Ahrdbd, and pcDNA3.1-Ahrwt. The cells were washed with PBS once, and then scraped and suspended in 250 ml RIPA buffer as described below.
RNA extraction, cDNA synthesis, and RT-qPCR
The Aurum total RNA mini kit was used for RNA extraction (BioRad, Hercules, California). For the liver tissue, 20–40 mg liver tissue was homogenized in 700 µl lysis solution. For colon tissue, fecal matter was pushed out, and the tissue was flushed with PBS. The samples were divided in 3, flash frozen in liquid nitrogen and then stored at −80°C. The distal end of the colon was homogenized in 500 µl TRizol and after extraction with 100 µl chloroform and centrifugation, the upper layer was transferred to a new tube containing 300 µl of 70% ethanol. The RNA was added to Aurum RNA purification columns and the RNA was purified according to the manufacturer’s instructions. One milligram of total RNA was reverse transcribed with High-capacity cDNA reverse transcription kit using standard conditions (Applied Biosystems). For real-time qPCR, 1 µl of cDNA was amplified with 5 µl KAPA SYBR fast qPCR master mix (2×) universal, 0.1 µl forward primer at 10 µM, 0.1 µl reverse primer at 10 µM, and 3.8 µl water. The cycling conditions were 95°C for 3 min, followed by 45 cycles of 95°C for 10 s and 60°C for 20 s. Data analysis was performed using the ΔΔCt method. TATA-box binding protein (Tbp) was used as the reference gene for normalization. The primer sequences used for qPCR are provided in Supplementary Table 1.
Western blots
For AHR protein detection in cell culture experiments, cells were lysed in RIPA buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 µg/ml leupeptin). For detection of hepatic AHR, frozen liver tissue (50–80 mg) was homogenized in 400 µl RIPA buffer. The homogenate was sonicated with Bioruptor on the low setting at 4°C for 5 min, 30 s on, and 30 s off. The samples were then centrifuged at 20 000 × g for 10 min at 4°C. Twenty micrograms of protein were separated by SDS-PAGE and transferred to a PVDF membrane. Membranes were blocked for 2 h at room temperature (RT) in 5% nonfat milk dissolved in Tris-buffered saline (TBS)-0.1% Tween20. Membranes were then incubated overnight at 4°C with anti-AHR (1:10 000; Enzo Life Sciences SA210; lot no. 04011942) followed by incubation with anti-rabbit IgG antibody; 1:6250; 1 h incubation RT. PVDF membranes were stripped and incubated with anti-β-actin antibody 1:4000 (Sigma-Aldrich; A-2228) followed by incubation with anti-mouse IgG antibody for 1 h at RT. After membranes were washed with TBST, SuperSignal West Dura (ThermoScientific) was used for detection. The pixel density of protein bands was analyzed with the ImageJ software (imagej.nih.gov/ij). The corresponding βACTIN band intensity normalized AHR signal.
DNA extraction from feces
DNA was extracted from frozen stool samples using QIAamp DNA Stool Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol with the following modifications. Feces were thawed in lysis buffer at 37°C until fully solubilized. To increase the yield of DNA, 600–900 µl of supernatant was passed through the filter column. Differences in the levels of bacteria at the phylum level were determined from fecal DNA by qPCR using PCR primers described previously (Yang et al., 2015). Each reaction consisted of 5 µl KAPA SYBR green, 1 µl DNA with a concentration of 5 ng/μl, 0.1 µl each of forward and reverse primer with a concentration of 10 nM, and 3.8 µl molecular grade H2O to a final volume of 10 μl.
Statistical analysis
Statistical analysis for significance (p < 0.05) was determined using GraphPad Prism 8.0. Comparison of 2 groups were made with an unpaired, 2-tailed Student’s t-test, whereas comparisons of multiple groups were made with analysis of variance (ANOVA) followed by a Tukey’s test. A 2-way ANOVA followed by Tukey’s test was performed when analyzing data with more than one factor.
Results
Mutation of AHR DNA-binding domain prevents TCDD-induced increases of hepatic CYP1A1 and CYP1B1 levels
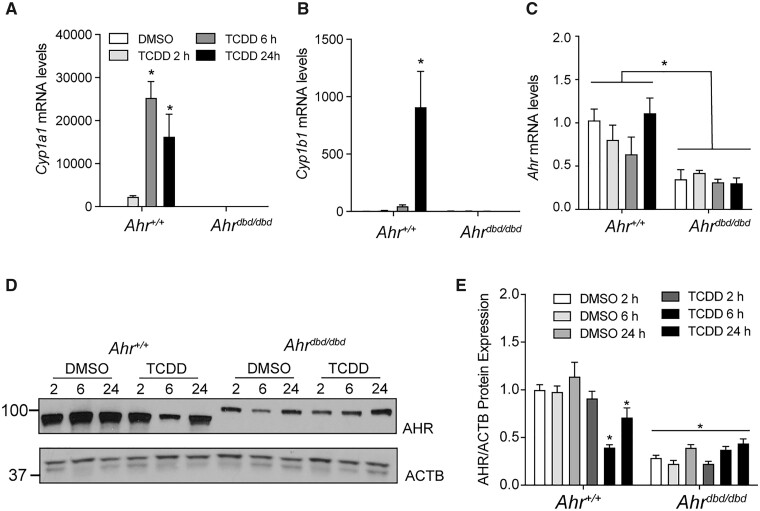
To confirm that mutation of the AHR DBD prevents TCDD-dependent induction of the Cyp1a1 and Cyp1b1 in vivo, Ahr+/+ and Ahrdbd/dbd mice were injected intraperitoneally with 100 µg/kg TCDD. After 2, 4, 6, and 24 h, hepatic RNA was isolated and analyzed for Cyp1a1, Cyp1b1, and Ahr expression levels. TCDD-induced Cyp1a1 and Cyp1b1 mRNA levels in wild-type mice with peaks at 6 and 24 h, respectively (Figs. 1A and 1B). In contrast, extracts from Ahrdbd/dbd mice showed very low levels of Cyp1a1 and Cyp1b1 that were unaffected by TCDD treatment. Ahr mRNA levels were unaltered by TCDD treatment in Ahr+/+ and Ahrdbd/dbd mice but were significantly lower in Ahrdbd/dbd than in Ahr+/+ mice (Figure 1C). Western blotting was then used to quantify AHR protein levels in Ahrdbd/dbd compared with Ahr+/+ mice (Figure 1D). Similar to the mRNA results, AHR protein levels were significantly lower in DMSO-treated Ahrdbd/dbd mice than in Ahr+/+ mice at all time points and were not altered after TCDD treatment (Figure 1E). In contrast, TCDD treatment reduced AHR protein expression in wild-type mice at 6 and 24 h, which most likely reflects ligand-induced proteolytic degradation of AHR (Pollenz, 1996). Collectively, these data support previous findings and show that AHRdbd lacks the ability to activate gene transcription from AHRE elements in vivo (Bunger et al., 2008). The western blots also revealed that the AHR proteins from Ahr+/+ and Ahrdbd/dbd were different in size, which is due to the different Ahr alleles in the different genotypes.
Figure 1.
Characterization of hepatic TCDD-induced gene expression in Ahrdbd/dbd mice and AHRdbd protein levels. Hepatic (A) Cyp1a1, (B) Cyp1b1, and (C) Ahr mRNA levels in Ahr+/+ and Ahrdbd/dbd after treatment with 100 µg/kg TCDD at the times indicated. D, AHRwt and AHRdbd hepatic protein levels and (E) quantification after treatment with DMSO or 100 µg/kg TCDD at the indicated times. Data represent mean ± SEM (N = 3). For A-C, *p ≤ .05 2-way ANOVA compared with DMSO 2 h. For E, *p ≤ .01 2-way ANOVA compared with DMSO 2 h. Abbreviations: DMSO, dimethyl sulfoxide; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Gene sequencing of AHR transcript from AHRwt and AHRdbd
Ahrdbd/dbd mice were originally generated by mutating the DNA-binding domain of the Ahr gene from 129/SvJ embryonic stem cells prior to injection into C57BL/6J blastocysts. Correctly mutated mice were backcrossed for 3 generations to C57BL/6J mice (Bunger et al., 2008), and then for another 10 generations, to C57BL/6J mice (Sarill et al., 2015). Although the Ahrdbd/dbd mice are congenic C57BL/6J, the Ahrdbd gene originated from the 129/SvJ strain that expresses the Ahrd allele, which is similar to that described for Ahrfl/fl mice (Wilson et al., 2021). To confirm this, we cloned and sequenced the gene products from Ahr+/+ and Ahrdbd/dbd mice. The DNA sequences from Ahr+/+ and Ahrdbd/dbd were aligned with sequences of Ahr in C57BL/6J and 129/SvJ that are available on GenBank as AF405563.1 and AF325111.1, respectively. The cloned Ahr+/+ was an exact match with the Ahrb1 from the C57BL/6J strain, whereas the Ahrdbd was identical with Ahrd from 129/SvJ, except for the ATC to GGG changes at nucleic acids 73 to 75, and an insertion of GGATCC between nucleic acids 117 and 118. As expected, these 2 artificial mutations in Ahrdbd result in an I25G substitution as well as a GS insertion after amino acid 39, which are responsible for producing a nonfunctional DNA-binding domain (Bunger et al., 2008). All other differences between the cloned Ahr+/+ and the cloned Ahrdbd were due to differences between Ahrb1 from C57BL/6J and Ahrd from 129/SvJ. Compared with the AHR in 129/SvJ mice, the AHR in C57BL/6J mice had: M324I, V375A, P471L, N533S, M589L, and R806X (Supplementary Figure 1). V375A has been shown to be responsible for the enhanced ligand affinity of AHR in C57BL/6, whereas all other changes have no or very limited impact at the functional level (Chang et al., 1993; Poland et al., 1994). The R806X difference results in an AHR protein with only 805 amino acids in C57BL/6J, compared with the 848 amino acids in 129/SvJ.
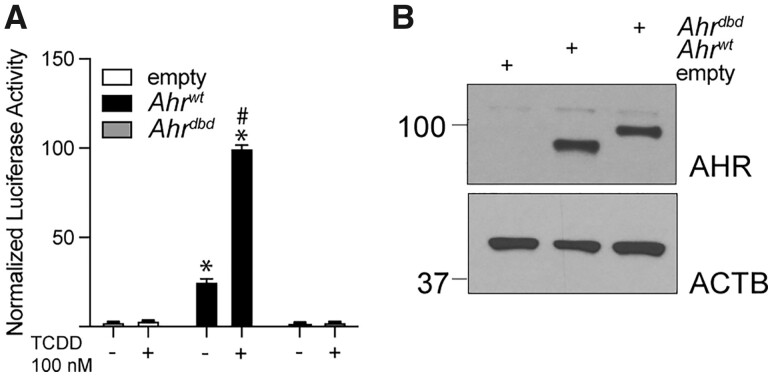
To determine the ability of AHRs from wild-type and Ahrdbd/dbd mice to regulate CYP1A1 reporter gene, COS-1 cells were transfected with pGudLuc 4.1, and pcDNA3.1-Ahrwt, or pcDNA3.1-Ahrdbd plasmids. Compared with empty pcDNA3.1, transfection with Ahrwt alone increased luciferase activity, and this was further increased after TCDD treatment. No significant differences in luciferase activity were observed in the presence of Ahrdbd transfected cells compared with empty pcDNA3.1. TCDD did not affect luciferase activity in Ahrdbd transfected cells (Figure 2A). Western blots were used to confirm that the AHRwt and AHRdbd proteins were equally expressed after transfection (Figure 2B). In summary, the AHRdbd has a nonfunctional DNA-binding domain and fails to regulate TCDD-dependent AHR canonical signaling compared with AHRwt.
Figure 2.
Cloning and characterization of Ahr from Ahr+/+ and Ahrdbd/dbd mice. A, Relative Cyp1a1-regulated luciferase activity of COS-1 cells transfected with empty pcDNA3.1, pcDNA3.1-Ahrwt, and pcDNA3.1-Ahrdbd plasmids and treated with DMSO or 100 nM TCDD. B, Western blot of AHRwt and AHRdbd protein after transfection of COS-1 cells. Data represent mean ± SEM (N = 3). *p < .05 2-way ANOVA compared with DMSO treated empty plasmid. #p < .05 2-way ANOVA compared with transfection matched DMSO treated sample. Abbreviations: DMSO, dimethyl sulfoxide; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Increased sensitivity of Ahrdbd/Dbd mice to DSS-induced colitis
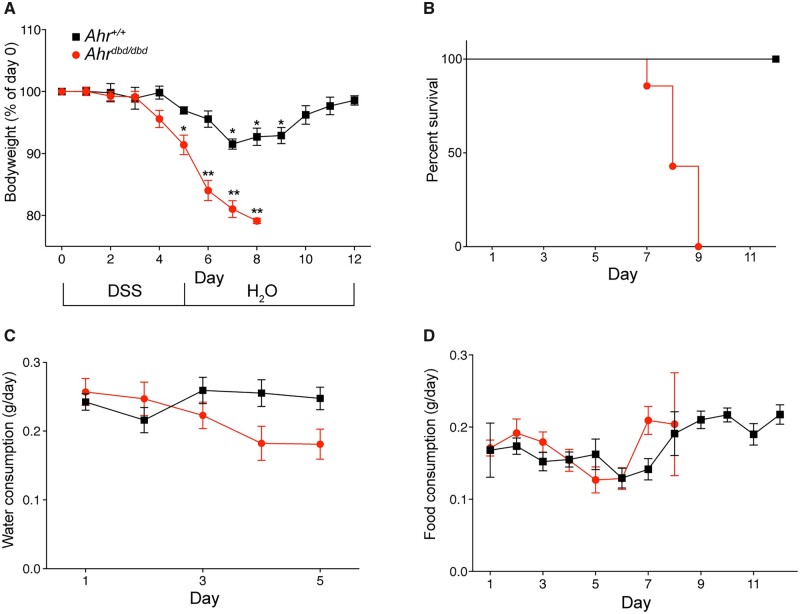
AHR integrates metabolic signals to promote anti-inflammatory responses, intestinal homeostasis, and maintain barrier integrity. Whether the anti-inflammatory actions and overall intestinal protection effects of AHR require its ability to bind to AHREs is not known. To this end, we determined the sensitivity of Ahrdbd/dbd mice to DSS-induced colitis. Mice were exposed to 2% DSS in their drinking water starting on day 0 and continuing to day 5, at which time the DSS-containing water was replaced with normal water. The mice were then monitored for an additional 7 days until day 12. DSS exposure caused a significant body weight loss in wild-type mice starting on day 7 and continuing through day 9, before recovering by day 10 (Figure 3A). All wild-type animals recovered from the DSS treatment. By contrast, DSS exposure caused a significant body weight loss in Ahrdbd/dbd mice by day 5 that continued to day 8. None of the DSS-exposed Ahrdbd/dbd mice survived the treatment as all animals lost greater than 20% of their body weight and were humanely euthanized. No differences in water or food intake between DSS exposed wild-type and Ahrdbd/dbd mice were observed (Figs. 3B and 3C), suggesting that the body weight loss differences between the genotypes were not due to different exposures to DSS.
Figure 3.
Ahrdbd/dbd mice have increased sensitivity to DSS-induced colitis. A, Mice were given 2% DSS in the drinking water for 5 days, before switching to water for an additional 7 days. Controls were given water only. Body weight was measured daily. B, Survival curve of DSS-treated mice. (C, Water and (D) food consumption at the days indicated. *p < .01, **p < .001 2-way ANOVA compared with day 1 for each genotype. Abbreviation: DSS, dextran sulfate sodium.
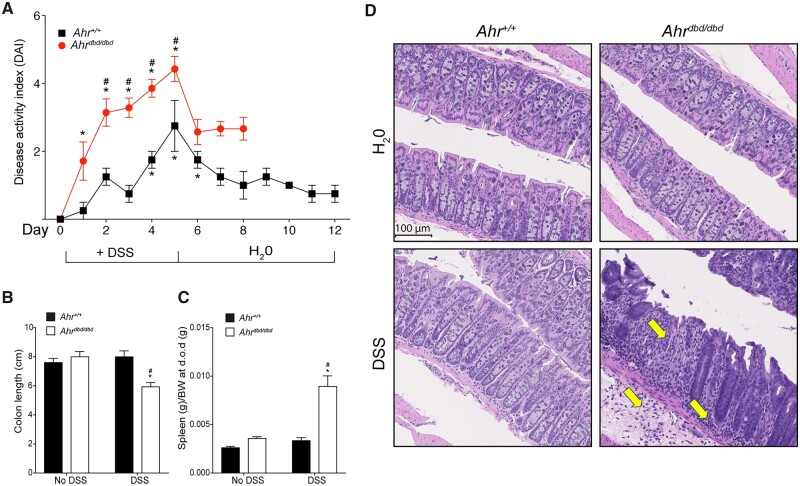
We next assessed the severity of inflammation using a daily disease activity index (DAI) assessment (Figure 4A) as described previously (Hutin et al., 2022; Kim et al., 2012). DSS exposed Ahrdbd/dbd mice displayed a significantly greater DAI than wild-type mice. Colon lengths were reduced in Ahrdbd/dbd mice but not in wild-type mice after DSS exposure (Figure 4B). DSS-exposed Ahrdbd/dbd mice had increased spleen weight compared with DSS exposed Ahr+/+ mice and Ahrdbd/dbd mice given water only (Figure 4C). Histological analysis of colon sections from Ahrdbd/dbd mice exposed to DSS for 7–9 days displayed prototypical features of colitis, including regions of major crypt loss and mucosal inflammation with neutrophilic infiltration (Figure 4D). In contrast, day 12 Ahr+/+ mice displayed normal colon mucosal morphology with little evidence for crypt damage and immune cell infiltration. Vehicle-treated mice from both genotypes displayed normal colon mucosal morphology.
Figure 4.
Increased sensitivity of Ahrdbd/dbd mice to DSS-induced colitis. A, Disease index score (DAI) was assessed daily in all groups. Parameters measured were blood in stool, diarrhea, rectal bleeding, and rectal prolapse. *p < .05 2-way ANOVA compared with genotype-matched and no DSS treated mice. #p < .05 2-way ANOVA compared with day-matched and DSS treated mice. B, Colon length and (C) spleen wet weight corrected for body weight were assessed upon dissection. *p < .05 2-way ANOVA compared with genotype-matched and no DSS-treated mice. #p < .05 2-way ANOVA compared with genotype-matched and DSS-treated mice (D) H&E stain of colon tissue in healthy control mice and mice treated with 2% DSS. Ahrdbd/dbd mice exhibit extensive tissue damage and immune cell infiltration compared with Ahr+/+ mice. The arrows indicate infiltration of inflammatory cells and crypt loss. Abbreviation: DSS, dextran sulfate sodium.
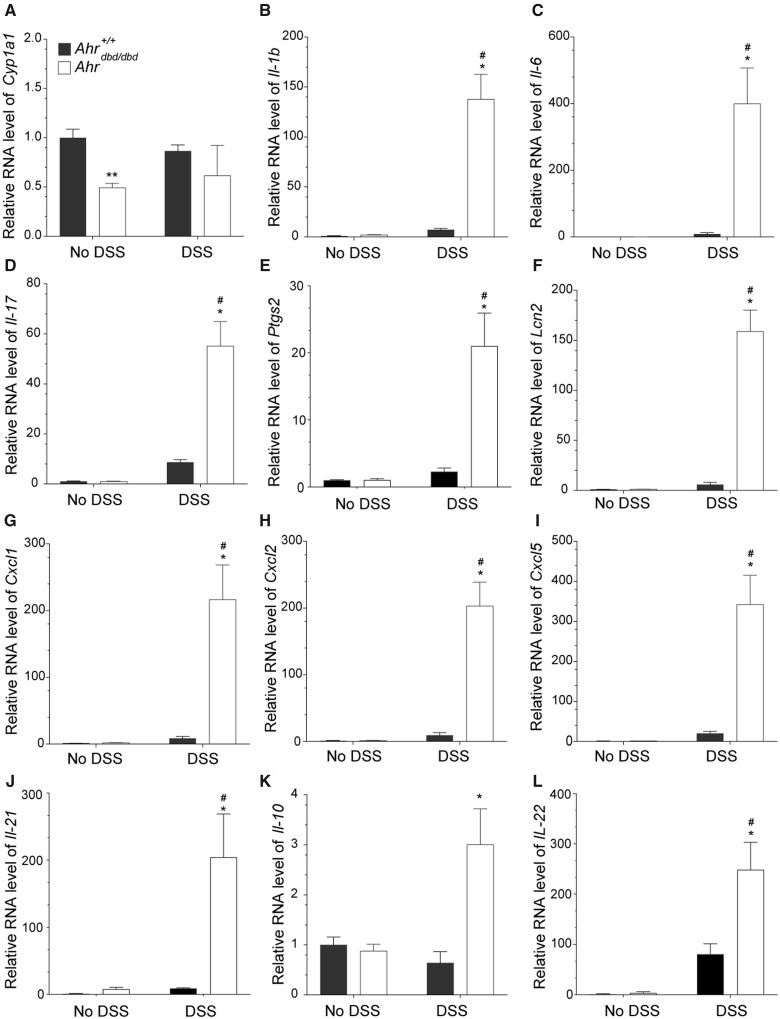
Increased expression of proinflammatory genes in DSS-treated Ahrdbd/Dbd compared with Ahr+/+ mice
We then examined the mRNA levels of Cyp1a1, several cytokines, chemokines, prostaglandin synthase 2 (Ptgs2), and lipocalin-2 (Lcn2), a potential biomarker for IBD. Cyp1a1 mRNA levels in water control Ahrdbd/dbd mice were lower than wild-type mice, but this difference was lost after DSS treatment (Figure 5A). The levels of all cytokines and chemokines examined, as well as Ptgs2 and Lcn2, were significantly increased in colon tissue from DSS-treated Ahrdbd/dbd compared with wild-type mice (Figs. 5B–L). These data demonstrate that the increased sensitivity to DSS-induced colitis in mice expressing a DNA-binding deficient AHR is due to hyperinflammation.
Figure 5.
Increased expression of proinflammatory genes in DSS-exposed Ahrdbd/dbd compared with wild-type mice. Intestinal mRNA expression levels of (A) Cyp1a1, (B) Il-1b, (C) Il-6, (D) Il-17, (E) Ptgs2, (F) Lcn2 (G) Cxcl1, (H) Cxcl2, (I) Cxcl5, (J) Il-21, (K) Il-10, and (L) Il-22 in colon tissue isolated from Ahr+/+ and Ahrdbd/dbd mice that were not exposed to DSS (No DSS) or exposed to 2% DSS in their drinking water. The relative mRNA levels of the indicated genes were determined with RT-qPCR. **p < .05 Student’s t-test compared with genotype and treatment-matched Ahr+/+ mice. *p < .05 2-way ANOVA compared with genotype-matched and DSS-treated animals. #p < .05 2-way ANOVA compared with treatment-matched Ahr+/+ mice. Abbreviation: DSS, dextran sulfate sodium.
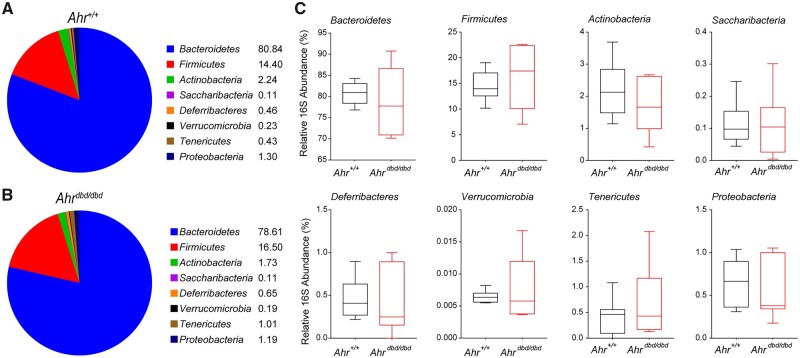
Microbial composition is similar in Ahr+/+ and Ahrdbd/Dbd mice
We next analyzed the microbial composition at the phylum level using a qPCR-based approach to determine if the Ahrdbd/dbd mice exhibit a dysbiosis that predisposes them to intestinal inflammation. For these studies, we analyzed feces from water control animals for both genotypes (Figs. 6A and 6B). We did not observe any significant differences in microbial composition between genotypes (Figure 6C).
Figure 6.
Microbiota at the phylum level were measured before the start of experiment to assess differences between genotypes. A, Average relative amounts of bacterial phyla in healthy Ahr+/+ mice. B, Average relative amounts of bacterial phyla in healthy Ahrdbd/dbd mice. Detailed view of average amounts in (C) Bacteroidetes, Firmicutes, Actinobacteria, Saccharibacteria, Deferribacteres, Verrucomicrobia, Tenericutes, and Proteobacteria.
Discussion
AHR is a key regulator of intestinal homeostasis and barrier function by promoting an anti-inflammatory intestinal environment (Busbee et al., 2013; Cho and Kelsall, 2014; Stockinger et al., 2014). Loss of AHR expression or reduced endogenous AHR ligand levels in mice increases their susceptibility to DSS-induced colitis or Citrobacter rodentium infection by dampening inflammatory responses (Stockinger et al., 2021). However, whether canonical AHR signaling and AHR-AHRE binding is required for these outcomes is not known. We show here that mice harboring a DNA-binding deficient AHR, which is incapable of binding to AHREs, exhibit a similar sensitivity to DSS-induced colitis as Ahr−/− mice. Our findings reveal the importance of canonical AHR signaling, and specifically AHR-AHRE binding in protecting against intestinal inflammation and DSS-induced colitis.
Canonical AHR signaling is fundamental for the biological and toxicological effects of AHR ligands (Bunger et al., 2003, 2008), including immune regulation as many cytokines have AHREs in their upstream regulatory regions (DiNatale et al., 2010). Interactions between AHR and musculoaponeurotic fibrosarcoma (cMAF) are important for the regulation of Il-10 and Il-21 in the development of T cell subset, Tr1 cells (Apetoh et al., 2010). Moreover, lipopolysaccharide-induced increases in Il-10 are impaired in macrophages isolated from Ahr−/− mice (Zhu et al., 2018). IL-10 protects against intestinal inflammation by preventing pro-inflammatory responses and maintaining mucosal homeostasis. This is supported by the findings that Il-10−/− and IL-10 receptor b null (Ilbrb−/−) mice develop spontaneous enterocolitis (Kuhn et al., 1993; Spencer et al., 1998), and that polymorphisms in IL-10R and IL-10 are associated with UC in early childhood (Engelhardt and Grimbacher, 2014). IL-21 is produced by T cells and natural killer T cells and effects a wide range of immune and nonimmune cells. Treatment with DSS increases intestinal IL-21 levels, whereas DSS-treated Il-21ko mice have reduced gut pathology, lower immune cell infiltration, and reduced IL-17 production compared with wild-type mice (Fina et al., 2008). Neutralization of IL-21 ameliorates clinical and pathological findings primarily in experimental T cell-driven colitis. However, IL-21 may play a protective role in the DSS-induced colitis model through its induction of IL-22, a member of the IL-10 family that promotes tissue regeneration and repair (Wei et al., 2020). Thus, it is possible that the increased Il-10 and Il-21 mRNA levels observed in DSS-exposed Ahrdbd/dbd compared Ahr+/+ mice may be due to the increased inflammation in the DNA-binding deficient AHR mice, and not necessarily due to AHRE-independent regulation of these cytokines by AHR.
AHR plays an essential role in the development and maintenance of intestinal RORγt+ type 3 innate lymphoid cells (ILC3s), which are important for gut immunity through their production of IL-22. IL-22 plays a crucial role in the early phase of host defense against C. rodentium infection and barrier integrity (Dumoutier et al., 2000; Sugimoto et al., 2008; Zheng et al., 2008). DSS exposed Il-22−/− mice exhibit increased inflammation, more severe weight loss, and impaired recovery compared with wild-type mice (Zindl et al., 2013). Moreover, exposure of Ahr−/− or CYP1A1 transgenic mice (endogenous AHR ligand deficient) to C. rodentium results in reduced barrier integrity and lower IL-22 levels, increased inflammation and mortality (Qiu et al., 2012; Schiering et al., 2017). AHR-dependent increases in IL-22 levels are also lost in colon explant cultures from Ahr−/− or CYP1A1 transgenic mice (Schiering et al., 2017), whereas CD4+ T cells isolated from Ahrb-1 mice have higher IL-21-dependent increases in IL-22 compared with Ahrd mice (Yeste et al., 2014). Taken together, these studies support a role for AHR in the regulation of IL-22. Although we did not determine the number of intestinal ILC3s in Ahrdbd/dbd mice, we observed that Il-22 levels were increased in DSS-exposed Ahrdbd/dbd compared with Ahr+/+ mice. This suggests that AHR might regulate Il-22 in an AHRE-independent manner. Many different types of immune cells also produce IL-22 including αβ and γδ T cells, and NKT cells (Parks et al., 2015; Sonnenberg et al., 2011a,b). However, in models of colitis and skin inflammation several cytokines and chemokines are increased in Ahr−/− compared with Ahr+/+ mice (Di Meglio et al., 2014; Wang et al., 2018). Thus, we cannot exclude that the increased Il-22 levels observed in DSS-exposed Ahrdbd/dbd mice might be due to other immune cells independently of AHR, as has been reported in skin inflammation models (Di Meglio et al., 2014).
Ahrdbd/dbd mice express the Ahrd allele with a I25G mutation and GS insertion after amino acid 39 which abolishes AHRE binding (Bunger et al., 2008). C57BL/6 mice harboring Ahrd allele fed purified rodent chow (AIN-93G) and exposed to 3.5% DSS were reported to exhibit similar weight loss and DAI compared with correspondingly fed and DSS exposed C57BL/6 Ahrb-1 mice (Hubbard et al., 2017). DSS-induced weight loss was delayed, and DAI reduced in C57BL/6 Ahrb-1 mice fed a diet rich in AHR ligands (broccoli diet). C57BL/6 Ahrd broccoli-fed mice exhibit some improvement in weight loss but no change in DAI, suggesting that AHR ligand rich diets are not as effective at protecting against DSS-induced colitis in mice that harbor the Ahrd allele. In another study, exposure of C57BL/6 Ahr+/+ (Ahrb-1) to 3% DSS resulted in rapid weight loss in standard chow fed mice followed by full recovery after DSS withdrawal (Li et al., 2011). In contrast, similarly exposed Ahr−/− mice experienced accelerated weight loss and extreme colon shortening with 13 out of 16 mice losing over 20% of body weight by 1 day after DSS withdrawal (Li et al., 2011). The sensitivity of Ahrdbd/dbd mice to DSS-induced colitis we observed in the present study is very similar with that reported for Ahr−/− mice fed standard chow (Li et al., 2011). Taken together, this suggests that the increased sensitivity of Ahrdbd/dbd mice to DSS-induced colitis is due to the expression of DNA-binding deficient AHR rather than reduced ligand affinity of AHRdbd compared with AHRwt protein.
Genome-wide profiling of AHR-binding sites revealed that the majority of AHR bound regions do not contain AHREs (Dere et al., 2011; Lo and Matthews, 2012). AHR regulates several cellular pathways through interactions with important regulatory proteins and transcription factors. AHR interacts with retinoblastoma protein (pRB) resulting in G1 cell-cycle arrest (Puga et al., 2002). Ligand-activated AHR also regulates inflammation through nongenomic pathways. TCDD- and polycyclic aromatic hydrocarbon (PAH)-treatment results in activation of MAPK, and Ca2+-induced cPLA2 and PTGS2 activation generating inflammatory prostaglandins (Puga et al., 1997). AHR modulates SRC-dependent STAT3 phosphorylation and the subsequent production of the anti-inflammatory cytokine IL-10 (Zhu et al., 2018). In addition, AHR inhibits the PI3K-AKT signaling by increasing the ubiquitination of RAC1 which results in a mitigated inflammatory response in macrophages (Grosskopf et al., 2021). Although there are no changes in the nuclear localization sequence or nuclear export sequence between AHRwt and AHRdbd, the AHRdbd protein has been reported to be located exclusively in the nucleus (Bunger et al., 2008). Therefore, in addition to the inability of AHR to bind to AHREs, cytosolic AHR signaling is also impaired in Ahrdbd/dbd mice. We cannot exclude that cytosolic AHR signaling may contribute to the anti-inflammatory and protective role of AHR in the gut. To test this, we could perform similar DSS exposure studies in Ahrnls/nls mice that harbor a mutation in the nuclear localization sequence of AHR (Bunger et al., 2003). These mice are resistant to TCDD-induced toxicity and show similar development abnormalities as Ahr−/− mice, but their barrier integrity and sensitivity to colitis have not been studied. Like Ahrdbd/dbd mice, Ahrnls/nls mice were generated using embryonic stem cells derived from 129Sv mice and harbor the Ahrd allele. The original Ahr+/nls mice were backcrossed onto C57BL/6-Ahrd mice to generate appropriate controls (Bunger et al., 2003). This strategy prevented the potential confounding results that may arise when comparing mice expressing Ahrb-1 with Ahrnls/nls (Ahrd). A similar backcrossing strategy onto C57BL/6-Ahrd mice could be done for the Ahrdbd/dbd mice. Alternatively, target gene editing strategies could be used to convert the Ahrdbd/dbd (Ahrd) to Ahrdbd/dbd (Ahrb-1) as have been described for Ahr conditional null (Ahrfx) mice (Wilson et al., 2021). Regardless, both approaches would provide appropriate Ahr allele control animals and eliminate one of the weaknesses of using the congenic C57BL/6 Ahrdbd/dbd strain in the present study.
Dysbiosis to a less diverse and less beneficial composition is commonly observed in IBD. Patients with IBD present with increased Proteobacteria, whereas Firmicutes are often reduced in amount and diversity (Frank et al., 2007). AHR expression and its activation help shape the composition of the microbiota, whereas several microbial produced metabolites act as AHR ligands or activators (Stockinger et al., 2021). In contrast to reports of dysbiosis between mice expressing Ahrd compared with those expressing Ahrb-1 and between Ahr−/− and Ahr+/+ mice (Hubbard et al., 2017; Yang et al., 2020), we did not observe any significant differences in microbiota composition at the phylum level between Ahrdbd/dbd and Ahr+/+ mice. It is possible that if metagenomic analyses of the microbiomes were done, we might have detected differences in microbial communities between Ahrdbd/dbd and Ahr+/+ mice.
In summary, our findings show that AHR-AHRE interactions are required to reduce inflammation, maintain a healthy intestinal environment, and protect against DSS-induced colitis. These findings further support targeting AHR for the therapeutic treatment of inflammatory diseases, including IBD.
Supplementary Material
Contributor Information
Karoline Alvik, Department of Nutrition, Institute of Basic Medical Sciences, University of Oslo, Oslo, Norway.
Peng Shao, Department of Pharmacology and Toxicology, University of Toronto, Toronto M5S1A8, Canada.
David Hutin, Department of Pharmacology and Toxicology, University of Toronto, Toronto M5S1A8, Canada.
Carolyn Baglole, Department of Medicine, McGill University, Montreal H4A3J1, Canada; Department of Pathology, McGill University, Montreal H4A3J1, Canada; Department of Pharmacology and Therapeutics, McGill University, Montreal H3G1Y6, Canada.
Denis M Grant, Department of Pharmacology and Toxicology, University of Toronto, Toronto M5S1A8, Canada.
Jason Matthews, Department of Nutrition, Institute of Basic Medical Sciences, University of Oslo, Oslo, Norway; Department of Pharmacology and Toxicology, University of Toronto, Toronto M5S1A8, Canada.
Supplementary data
Supplementary data are available at Toxicological Sciences online.
Acknowledgments
The authors thank Solveig Pettersen for her technical assistance.
Funding
Canadian Institutes of Health Research (CIHR) (MOP-494265 and MOP-125919 to D.M.G. and J.M.), the Throne Holst Foundation, and the University of Oslo (to J.M.).
Declaration of conflicting interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Apetoh L., Quintana F. J., Pot C., Joller N., Xiao S., Kumar D., Burns E. J., Sherr D. H., Weiner H. L., Kuchroo V. K. (2010). The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 11, 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilla M. N., Malecki K. M. C., Hahn M. E., Wilson R. H., Bradfield C. A. (2020). The Ah receptor: Adaptive metabolism, ligand diversity, and the xenokine model. Chem. Res. Toxicol. 33, 860–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag T. V., Luis Morales J., Hollingshead B. D., Perdew G. H. (2008). The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 18, 207–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger M. K., Glover E., Moran S. M., Walisser J. A., Lahvis G. P., Hsu E. L., Bradfield C. A. (2008). Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor. Toxicol. Sci. 106, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger M. K., Moran S. M., Glover E., Thomae T. L., Lahvis G. P., Lin B. C., Bradfield C. A. (2003). Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J. Biol. Chem. 278, 17767–17774. [DOI] [PubMed] [Google Scholar]
- Busbee P. B., Rouse M., Nagarkatti M., Nagarkatti P. S. (2013). Use of natural AhR ligands as potential therapeutic modalities against inflammatory disorders. Nutr. Rev. 71, 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon A. S., Nagarkatti P. S., Nagarkatti M. (2021). Targeting AhR as a novel therapeutic modality against inflammatory diseases. Int. J. Mol. Sci. 23, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Smith D. R., Prasad V. S., Sidman C. L., Nebert D. W., Puga A. (1993). Ten nucleotide differences, five of which cause amino acid changes, are associated with the Ah receptor locus polymorphism of C57BL/6 and DBA/2 mice. Pharmacogenetics 3, 312–321. [DOI] [PubMed] [Google Scholar]
- Cho H., Kelsall B. L. (2014). The role of type I interferons in intestinal infection, homeostasis, and inflammation. Immunol. Rev. 260, 145–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGroot D. E., Franks D. G., Higa T., Tanaka J., Hahn M. E., Denison M. S. (2015). Naturally occurring marine brominated indoles are aryl hydrocarbon receptor ligands/agonists. Chem. Res. Toxicol. 28, 1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. S., Nagy S. R. (2003). Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43, 309–334. [DOI] [PubMed] [Google Scholar]
- Dere E., Lo R., Celius T., Matthews J., Zacharewski T. R. (2011). Integration of genome-wide computation DRE search, AhR chip-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics 12, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meglio P., Duarte J. H., Ahlfors H., Owens N. D., Li Y., Villanova F., Tosi I., Hirota K., Nestle F. O., Mrowietz U., et al. (2014). Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity 40, 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNatale B. C., Schroeder J. C., Francey L. J., Kusnadi A., Perdew G. H. (2010). Mechanistic insights into the events that lead to synergistic induction of interleukin 6 transcription upon activation of the aryl hydrocarbon receptor and inflammatory signaling. J. Biol. Chem. 285, 24388–24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoutier L., Louahed J., Renauld J. C. (2000). Cloning and characterization of IL-10-related T cell-derived inducible factor (IL-TIF), a novel cytokine structurally related to IL-10 and inducible by IL-9. J. Immunol. 164, 1814–1819. [DOI] [PubMed] [Google Scholar]
- Engelhardt K. R., Grimbacher B. (2014). IL-10 in humans: Lessons from the gut, IL-10/IL-10 receptor deficiencies, and IL-10 polymorphisms. Curr. Top. Microbiol. Immunol. 380, 1–18. [DOI] [PubMed] [Google Scholar]
- Fina D., Sarra M., Fantini M. C., Rizzo A., Caruso R., Caprioli F., Stolfi C., Cardolini I., Dottori M., Boirivant M., et al. (2008). Regulation of gut inflammation and th17 cell response by interleukin-21. Gastroenterology 134, 1038–1048. [DOI] [PubMed] [Google Scholar]
- Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskopf H., Walter K., Karkossa I., von Bergen M., Schubert K. (2021). Non-genomic AhR-signaling modulates the immune response in endotoxin-activated macrophages after activation by the environmental stressor BaP. Front. Immunol. 12, 620270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y. Z., Hogenesch J. B., Bradfield C. A. (2000). The PAS superfamily: Sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40, 519–561. [DOI] [PubMed] [Google Scholar]
- Hankinson O. (1995). The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35, 307–340. [DOI] [PubMed] [Google Scholar]
- Hankinson O. (2005). Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch. Biochem. Biophys. 433, 379–386. [DOI] [PubMed] [Google Scholar]
- Hubbard T. D., Murray I. A., Nichols R. G., Cassel K., Podolsky M., Kuzu G., Tian Y., Smith P., Kennett M. J., Patterson A. D., et al. (2017). Dietary broccoli impacts microbial community structure and attenuates chemically induced colitis in mice in an Ah receptor dependent manner. J. Funct. Foods 37, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui W., Dai Y. (2020). Therapeutic potential of aryl hydrocarbon receptor ligands derived from natural products in rheumatoid arthritis. Basic Clin. Pharmacol. Toxicol. 126, 469–474. [DOI] [PubMed] [Google Scholar]
- Hutin D., Hagen K. A., Shao P., Sugamori K., Grant D. M., Matthews J. (2022). Reduced colonic mucosal injury in 2,3,7,8-tetrachlorodibenzo-p-dioxin poly ADP-ribose polymerase (TIPARP/PARP7)-deficient mice. Int. J. Mol. Sci. 23, 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam S. J. (2022). Tapinarof cream 1%: First approval. Drugs 82, 1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J., Shajib M. S., Manocha M. M., Khan W. I. (2012). Investigating intestinal inflammation in DSS-induced model of IBD. J. Vis. Exp. 60, 3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R., Lohler J., Rennick D., Rajewsky K., Muller W. (1993). Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274. [DOI] [PubMed] [Google Scholar]
- Lamas B., Richard M. L., Leducq V., Pham H. P., Michel M. L., Da Costa G., Bridonneau C., Jegou S., Hoffmann T. W., Natividad J. M., et al. (2016). CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Innocentin S., Withers D. R., Roberts N. A., Gallagher A. R., Grigorieva E. F., Wilhelm C., Veldhoen M. (2011). Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147, 629–640. [DOI] [PubMed] [Google Scholar]
- Lo R., Matthews J. (2012). High-resolution genome-wide mapping of AhR and ARNT binding sites by chip-seq. Toxicol. Sci. 130, 349–361. [DOI] [PubMed] [Google Scholar]
- Mizoguchi A. (2012). Animal models of inflammatory bowel disease. Prog. Mol. Biol. Transl. Sci. 105, 263–320. [DOI] [PubMed] [Google Scholar]
- Monteleone I., Rizzo A., Sarra M., Sica G., Sileri P., Biancone L., MacDonald T. T., Pallone F., Monteleone G. (2011). Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 141, 237–248, 248.e1. [DOI] [PubMed] [Google Scholar]
- Naganuma M., Sugimoto S., Mitsuyama K., Kobayashi T., Yoshimura N., Ohi H., Tanaka S., Andoh A., Ohmiya N., Saigusa K., et al. (2018). Efficacy of indigo naturalis in a multicenter randomized controlled trial of patients with ulcerative colitis. Gastroenterology 154, 935–947. [DOI] [PubMed] [Google Scholar]
- Parks O. B., Pociask D. A., Hodzic Z., Kolls J. K., Good M. (2015). Interleukin-22 signaling in the regulation of intestinal health and disease. Front. Cell Dev. Biol. 3, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernomian L., Duarte-Silva M., de Barros Cardoso C. R. (2020). The aryl hydrocarbon receptor (AhR) as a potential target for the control of intestinal inflammation: Insights from an immune and bacteria sensor receptor. Clin. Rev. Allergy Immunol. 59, 382–390. [DOI] [PubMed] [Google Scholar]
- Poland A., Palen D., Glover E. (1994). Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol. Pharmacol. 46, 915–921. [PubMed] [Google Scholar]
- Pollenz R. S. (1996). The aryl-hydrocarbon receptor, but not the aryl-hydrocarbon receptor nuclear translocator protein, is rapidly depleted in hepatic and nonhepatic culture cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol. Pharmacol. 49, 391–398. [PubMed] [Google Scholar]
- Puga A., Hoffer A., Zhou S., Bohm J. M., Leikauf G. D., Shertzer H. G. (1997). Sustained increase in intracellular free calcium and activation of cyclooxygenase-2 expression in mouse hepatoma cells treated with dioxin. Biochem. Pharmacol. 54, 1287–1296. [DOI] [PubMed] [Google Scholar]
- Puga A., Xia Y., Elferink C. (2002). Role of the aryl hydrocarbon receptor in cell cycle regulation. Chem. Biol. Interact. 141, 117–130. [DOI] [PubMed] [Google Scholar]
- Qiu J., Heller J. J., Guo X., Chen Z. M., Fish K., Fu Y. X., Zhou L. (2012). The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarill M., Zago M., Sheridan J. A., Nair P., Matthews J., Gomez A., Roussel L., Rousseau S., Hamid Q., Eidelman D. H., et al. (2015). The aryl hydrocarbon receptor suppresses cigarette-smoke-induced oxidative stress in association with dioxin response element (DRE)-independent regulation of sulfiredoxin 1. Free Radic. Biol. Med. 89, 342–357. [DOI] [PubMed] [Google Scholar]
- Sartor R. B. (2006). Mechanisms of disease: Pathogenesis of Crohn's disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol. 3, 390–407. [DOI] [PubMed] [Google Scholar]
- Schiering C., Wincent E., Metidji A., Iseppon A., Li Y., Potocnik A. J., Omenetti S., Henderson C. J., Wolf C. R., Nebert D. W., et al. (2017). Feedback control of AhR signalling regulates intestinal immunity. Nature 542, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok S. H., Ma Z. X., Feltenberger J. B., Chen H., Chen H., Scarlett C., Lin Z., Satyshur K. A., Cortopassi M., Jefcoate C. R., et al. (2018). Trace derivatives of kynurenine potently activate the aryl hydrocarbon receptor (AHR). J. Biol. Chem. 293, 1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg G. F., Fouser L. A., Artis D. (2011a). Border patrol: Regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390. [DOI] [PubMed] [Google Scholar]
- Sonnenberg G. F., Monticelli L. A., Elloso M. M., Fouser L. A., Artis D. (2011b). Cd4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S. D., Di Marco F., Hooley J., Pitts-Meek S., Bauer M., Ryan A. M., Sordat B., Gibbs V. C., Aguet M. (1998). The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J. Exp. Med. 187, 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B., Di Meglio P., Gialitakis M., Duarte J. H. (2014). The aryl hydrocarbon receptor: Multitasking in the immune system. Annu. Rev. Immunol. 32, 403–432. [DOI] [PubMed] [Google Scholar]
- Stockinger B., Shah K., Wincent E. (2021). AHR in the intestinal microenvironment: Safeguarding barrier function. Nat. Rev. Gastroenterol. Hepatol. 18, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Ogawa A., Mizoguchi E., Shimomura Y., Andoh A., Bhan A. K., Blumberg R. S., Xavier R. J., Mizoguchi A. (2008). IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 118, 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Yang K., Han B., Sheng B., Yin J., Pu A., Li L., Sun L., Yu M., Qiu Y., et al. (2018). Aryl hydrocarbon receptor inhibits inflammation in DSS-induced colitis via the MK2/p-MK2/TTP pathway. Int. J. Mol. Med. 41, 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H. X., Wang B., Li B. (2020). IL-10 and IL-22 in mucosal immunity: Driving protection and pathology. Front. Immunol. 11, 1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M. A., Rothhammer V., Quintana F. J. (2017). Control of immune-mediated pathology via the aryl hydrocarbon receptor. J. Biol. Chem. 292, 12383–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. H., Carney P. R., Glover E., Parrott J. C., Rojas B. L., Moran S. M., Yee J. S., Nukaya M., Goetz N. A., Rubinstein C. D., et al. (2021). Generation of an allelic series at the ahr locus using an edited recombinant approach. Toxicol. Sci. 180, 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. J., De Castro K. P., Joshi A. D., Elferink C. J. (2017). Canonical and non-canonical aryl hydrocarbon receptor signaling pathways. Curr. Opin. Toxicol. 2, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., DeLuca J. A. A., Menon R., Garcia-Vilarato E., Callaway E., Landrock K. K., Lee K., Safe S. H., Chapkin R. S., Allred C. D., et al. (2020). Effect of diet and intestinal AhR expression on fecal microbiome and metabolomic profiles. Microb. Cell Fact. 19, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. W., Chen M. K., Yang B. Y., Huang X. J., Zhang X. R., He L. Q., Zhang J., Hua Z. C. (2015). Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl. Environ. Microbiol. 81, 6749–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeste A., Mascanfroni I. D., Nadeau M., Burns E. J., Tukpah A. M., Santiago A., Wu C., Patel B., Kumar D., Quintana F. J. (2014). IL-21 induces IL-22 production in CD4+ T cells. Nat. Commun. 5, 3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T., Wang J., Zhu K., Tang Y., Huang S., Shui X., Ding Y., Chen C., Lei W. (2018). Aryl hydrocarbon receptor: A new player of pathogenesis and therapy in cardiovascular diseases. Biomed Res. Int. 2018, 6058784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Valdez P. A., Danilenko D. M., Hu Y., Sa S. M., Gong Q., Abbas A. R., Modrusan Z., Ghilardi N., de Sauvage F. J., et al. (2008). Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289. [DOI] [PubMed] [Google Scholar]
- Zhu J., Luo L., Tian L., Yin S., Ma X., Cheng S., Tang W., Yu J., Ma W., Zhou X., et al. (2018). Aryl hydrocarbon receptor promotes IL-10 expression in inflammatory macrophages through Src-STAT3 signaling pathway. Front. Immunol. 9, 2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindl C. L., Lai J. F., Lee Y. K., Maynard C. L., Harbour S. N., Ouyang W., Chaplin D. D., Weaver C. T. (2013). IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc. Natl. Acad. Sci. U.S.A. 110, 12768–12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.