Graphical abstract
Keywords: Microbiome assembly, Co-occurrence, Trade-off, Tea plant, Phyllosphere, Homeostasis
Highlights
-
•
The critical mechanisms underlying the microbiome assembly in the tea plants are revealed.
-
•
There is a trade-off between stochastic and deterministic processes in microbiome assembly in the tea plants.
-
•
Assembly processes were dominated by deterministic processes in bulk and rhizosphere soils.
-
•
The stochastic processes in roots and leaves was critically driven by amino acids for environmental selection.
-
•
The core taxa assembled in phyllosphere could attenuate the virulence of a prevalent foliar pathogen.
Abstract
Introduction
Assembly and co-occurrence of the host co-evolved microbiota are essential ecological and evolutionary processes, which is not only crucial for managing individual plant fitness but also ecological function. However, understanding of the microbiome assembly and co-occurrence in higher plants is not well understood. The tea plant was shown to contribute the forest fitness due to the microbiome assembled in the phyllosphere; the landscape of microbiome assembly in the tea plants and its potential implication on phyllosphere homestasis still remains untangled.
Objectives
This study aimed to deciphering of the microbiome networks of the tea plants at a continental scale. It would provide fundamental insights into the factors driving the microbiome assembly, with an extended focus on the resilience towards the potential pathogen in the phyllosphere.
Methods
We collected 225 samples from 45 locations spanning approximately 2000-km tea growing regions across China. By integration of high-throughput sequencing data, physicochemical properties profiling and bioinformatics analyses, we investigated continental scale microbiome assembly and co-occurrence in the tea plants. Synthetic assemblages, interaction assay and RT-qPCR were further implemented to analyze the microbial interaction indexed in phyllosphere.
Results
A trade-off between stochastic and deterministic processes in microbiomes community assembly was highlighted. Assembly processes were dominated by deterministic processes in bulk and rhizosphere soils, and followed by stochastic processes in roots and leaves with amino acids as critical drivers for environmental selection. Sphingobacteria and Proteobacteria ascended from soils to leaves to sustain a core leaf taxa. The core taxa formed a close association with a prevalent foliar pathogen in the co-occurrence network and significantly attenuated the expression of a set of essential virulence genes in pathogen.
Conclusion
Our study unveils the mechanism underpinning microbiome assembly in the tea plants, and a potential implication of the microbiome-mediated resilience framework on the phyllosphere homeostasis.
Introduction
Plant microbiomes are transmitted through seed dispersal or recruited from a plant’s surrounding environment [1]. Root microbiomes enter the host plant through the cracks formed in the lateral root junctions or through wounds caused by microbe or nematode phytopathogens and quickly spread to the endorhizosphere [2]. Entry of endophytic bacteria in plant roots also occurs via root hairs and the spaces between epidermal cells [2]. Utilizing a dynamic infection process, endophytic bacteria such as Rhizobia have been found to ascendingly migrate from roots to leaves in rice plants, where they transiently grow to large local populations [3]. Root and rhizosphere microbiomes benefit host plants by enhancing nutritional acquisition and improving resistance to pathogenic infections [4]. The synonymous aerial compartments are more frequently exposed to high fluctuations in temperature, humidity, UV radiation and anthropogenic practice, and the resident microbiome likely co-evovle with their host in adaption to various stress [5]. Understanding of assembly of the co-evovled microbiome in plant is crucial for managing both individual plant fitness and ecological function [6]. However, how microbiome assembly spatially and temporally is yet not well understood.
Tea plant (Camellia sinensis), one of the oldest widely-distributed plants on earth, has been considered to gain strong disease-resilience during long-term domestication [7], [8]. The resilience towards bacterial disease is conventionally attributed to the abundant and unique flavors derived from secondary metabolites involved in phyllosphere defense such as flavonoids, theanine and purine alkaloids [9], [10]. More recently, the production of these aforementioned secondary metabolites has been associated with the co-evolved microbiomes [11]. Endophytic and rhizosphere bacteria promote the production of secondary metabolites in C. sinensis through plant growth hormone production, phosphate solubilization, nutrient acquisition and N2 fixation [12]. Characteristic secondary metabolites, or their precursors, can also be produced by endophytic bacteria associated with C. sinensis. The co-evolution in terms of this metabolite-microbiome synergism forementioned might explain the disease-resilience to some extent [11]. It is eatablished that co-occurrence of the microbiome assembled shapes the co-evolution under scenario of biogeography and consequent plasticity towards disease in mammals [13], [14], but remains so far largely elusive in higher plants. Deciphering of such co-occurrence pattern of microbiome assembled in the disease-resilient model plant would provide fundamental insights into the phyllosphere homeostasis-shaped disease resilience in Kingdom Planta.
Here, we investigated the continental scale microbiome assembly and co-occurrence in disease-resilient model plant by including 225 samples collected from 45 locations spanning 2000-km tea growing regions across China. Integration of high-throughput data, plant primary and secondary metabolites, soil physicochemical properties, and heavy metals profiling, microbiome assembly driven by the stochastic and deterministic processes throughout the below-to-above ground compartments was characterized. Subsequent analysis of co-occurrence pattern showed that a core microbiome sustained in the phyllosphere underpinning homeostasis in the term of the resilience against disease. Our study outlines the continental scale of microbiome networks underlying the resilience framework against disease.
Results
Microbial signatures of tea plants from soil to leaves
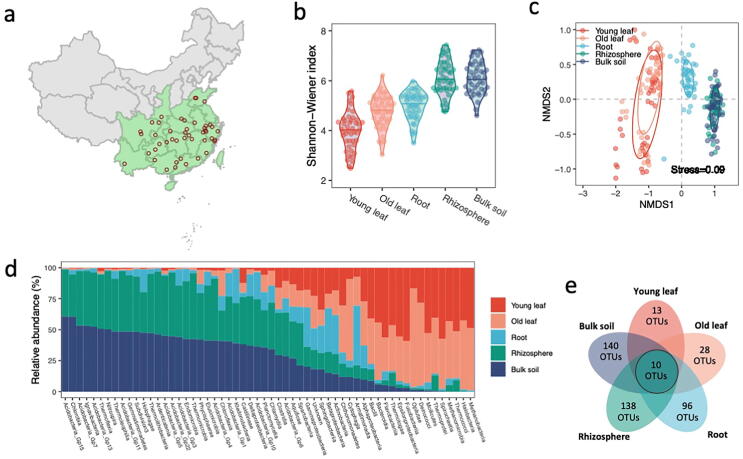
A total of 225 samples from 45 tea plantations in prominent tea-producing regions of China (Fig. 1a, Table S1) were collected to explore the microbiomes of bulk soil, rhizosphere soil (hereafter ‘rhizosphere’), roots, young leaves and old leaves. These 45 locations included four soil types, two climate types, and contrasting altitude (20–1600 m) and pH (4.1–7.4). We sequenced the 16S rRNA gene amplicon for each compartment of all samples (approximately 79.8 million high-quality sequence tags). An average of 0.27 million tags were generated for each sample after removal of sequences associated with chloroplast and mitochondrial DNA.
Fig. 1.
Compositions and core microbiota of microbiomes in different compartments of the tea plants. (a) Location of sampling sites in China. (b) microbiome α-diversity (Shannon-Wiener index) (n = 45). (c) microbiome β-diversity (weighted UniFrac distance) (n = 45). (d) Microbial taxa compartment relative abundance. (e) Unique and shared core microbiota.
Microbial α-diversity (Shannon-Wiener) decreased with compartment distance from soil (ANOVA, DF = 4, F = 75.3, P < 0.001) (Fig. 1b). β-diversity (UniFrac distance) also revealed that the community composition differentiated between soils (bulk and rhizosphere), roots and leaves (young and old) (ANOSIM, R = 0.76, P = 0.001) (Fig. 1c). Core taxa were defined as the OTUs present in all samples. The number of core operational taxonomic units (OTUs) decreased sequentially from bulk soils to rhizosphere, root, old leaves, and young leaves (Fig. 1d). Bulk soil, rhizosphere, roots, young leaves and old leaves contained 140, 138, 96, 28 and 13 core OTUs, respectively (Fig. 1e). There were a further 10 generalist OTUs present in all compartments.
Enrichment and depletion of tea plants microbiomes from soils to leaves
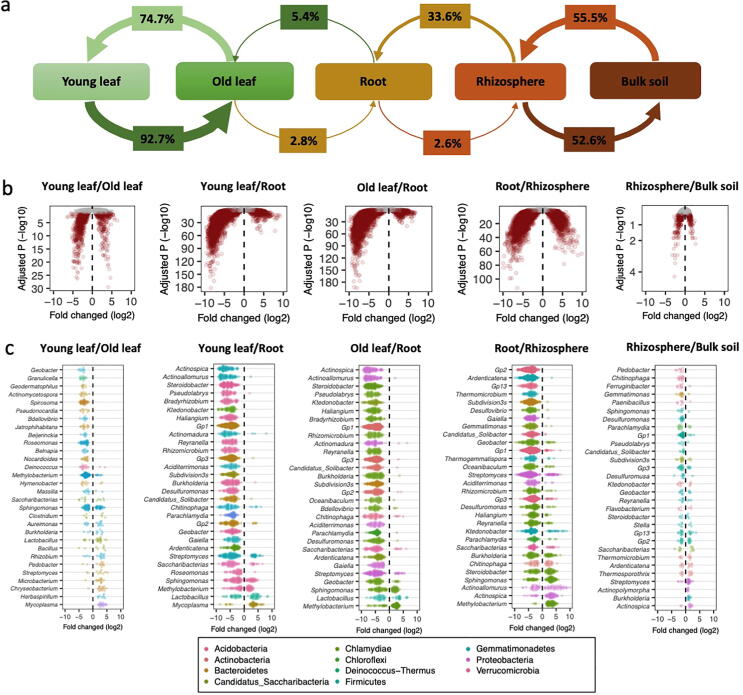
SourceTracker analysis revealed substantial similarities within the two leaf compartments and within the two soil compartments, and also indicated that there were rare exchanges between roots and leaves, therefore indicating partitioning between above and belowground microbiomes (Fig. 2a). This transfer bottleneck was observed as a 34% similarity between the rhizosphere and roots but only a 5% similarity between roots and old leaves. When genera were compared sequentially across the compartments, each ascension in a compartment (e.g. from soil to rhizosphere or old leaf to young leaf) lead to substantially more depleted OTUs than enriched OTUs (Fig. 2b). Depletion ratios (depletion/enrichment) ranged from 0.5 in rhizosphere/bulk soil to 7.5 for both old leaf/root and root/rhizosphere. Sphingomonas, Methylobacterium and Burkholderia were frequently associated with these changes (Fig. 2c). These results show strong filtering effects from rhizosphere samples to roots and from roots to leaves.
Fig. 2.
Microbiome associations in different compartments of the tea plants. (a) Source proportions for sequential compartments; arrow direction indicates potential transfer of microbiomes and arrow weight varies with proportion. (b) Enrichment (positive) and depletion (negative) of OTUs between ascending compartments; red points indicate significantly enriched or depleted OTUs (Wald test, P < 0.05). (c) Genera of enriched and depleted OTUs between ascending compartments. Colors of points indicate phylum classification.
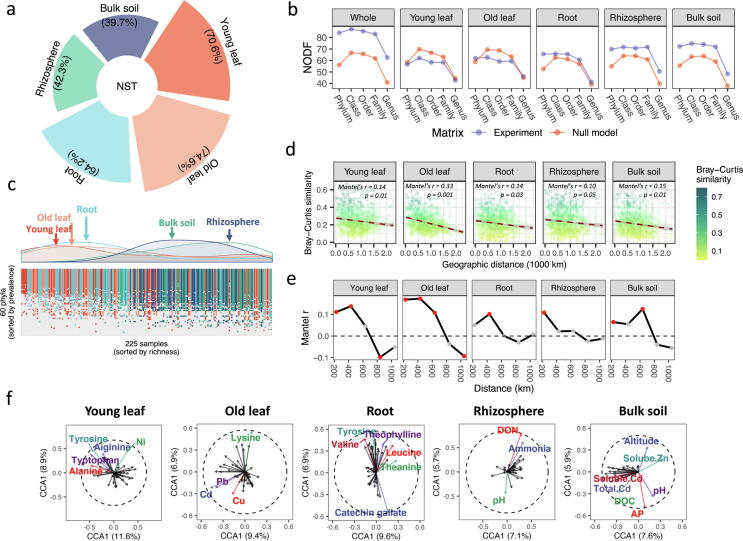
Community assembly mechanisms of tea plants microbiomes from soils to leaves
Community assembly normalized stochasticity ratio (NST) increased from 40% in bulk soil to more than 70% in leaves (Fig. 3a). Nestedness was significant for rhizosphere (t-test, P < 0.05) and bulk soil microbiomes (t-test, P < 0.05) but non-significant for root and leaf microbiomes (t-tests, P > 0.05) at all ranks from genus to phylum (Fig. 3b). Microbiomes in soils, roots and leaves were nested as subsets of rhizosphere microbiomes and showed distinct patterns in species richness (Fig. 3c). When considered using Bray-Cutis similarity and after controlling the impact of the environmental matrix using Partial Mantel tests, all compartments were negatively correlated with geographic distance (Fig. 3d). This response was particularly strong in old leaves; Mantel correlograms show that only this compartment linearly changed with geographic distance (Fig. 3e). The environmental selection effect of physicochemical properties in rhizosphere and bulk soils, amino acids and catechin in roots and leaves, and heavy metals on microbial communities was assessed using constrained corresponding analysis (Fig. 3f). The microbial community structures in plant tissues were closely associated with amino acids in young leaves, old leaves and roots; microbial community structures in old leaves were additionally closely associated with metals. Bulk soil microbial community structures were closely associated with environmental variables such as altitude, soil pH, phosphorus, carbon and metals, while rhizosphere microbial communities were closely associated with soil pH and nitrogen variables.
Fig. 3.
The contribution of stochasticity, dispersal limitation and environmental selection to microbiome community assembly processes in different compartments of the tea plants. (a) Normalized stochastically ratio (NST). (b) Nestedness (NODF) at five taxon ranks. (c) Nestedness at phylum level. (d) The relationship between community similarity and geographic distance. Mantel’s r and p values indicate the results of partial Mantel tests while controlling with the environmental matrix. (e) Mantel correlograms. Red points indicate significant Mantel’s r (P < 0.05). (f) Constrained corresponding analysis (CCA) of compartment microbiomes; text labels indicate significant environmental drivers (permutation test for CCA, P < 0.05).
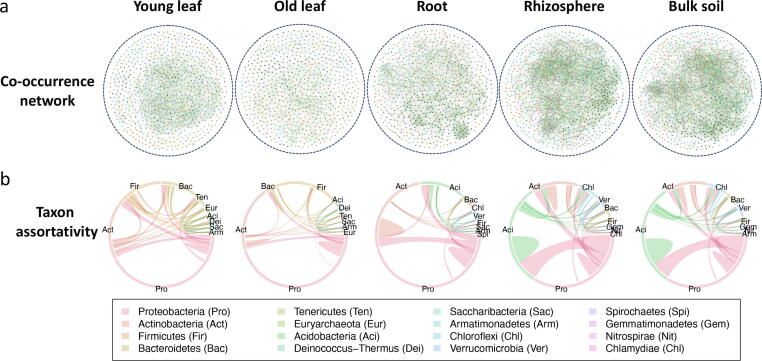
Microbial co-occurrence networks of tea plants microbiomes from soils to leaves
Potential interaction patterns within microbial communities were compared by inferring co-occurrence networks using the abundance matrix of the 1000 most abundant OTUs. The number of links, representing the intensity of potential microbial interactions, was the largest in bulk and rhizosphere soils, followed by roots and young leaves, and was the smallest in old leaves (Fig. 4a). The network diameter, representing the longest path in a network, was the longest in young and shortest in old leaves; transitivity, representing network modularity, was highest in roots and lowest in young leaves (Table S2). The co-occurrence networks also displayed different taxon assortativity (Fig. 4b). Overrepresented links include Proteobacteria to Firmicutes in young leaves, Proteobacteria to Bacteroidetes in old leaves, Proteobacteria to Actinobacteria and Proteobacteria to Acidobacteria in roots, rhizosphere and bulk soils (Fig. 4b). Intra-Acidobacteria links were substantially overrepresented in rhizosphere and bulk soils.
Fig. 4.
Links and taxon assortativity in microbiome co-occurrence network of different compartments of the tea plants. (a) co-occurrence networks in different compartments; (b) Taxa associations in different compartments.
Coordination of phyllosphere homeostasis by non-pathogenic core taxa
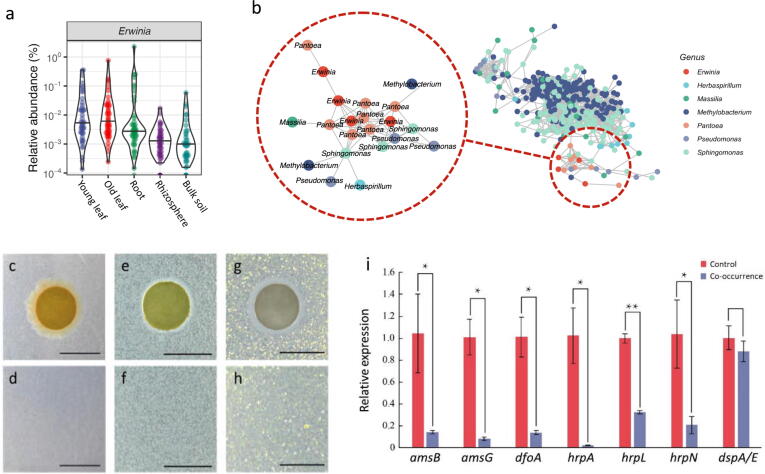
To reveal the underlying phyllosphere homeostasis of tea plant bacterial disease-resilience hinted at by the co-occurrence patterns, we carried deepening analyses of the co-occurrence patterns among phyllosphere core taxa and foliar pathogens. Results showed that phytopathogenic Erwinia spp. were a major and prevalent presence in all tea plant eco-niches, with the highest relative abundance occurring in the phyllosphere (Fig. 5a). Erwinia formed tight clusters of links with the non-pathogenic core taxa, consisting of six genera (Herbaspirillum, Massilia, Methylobacterium, Pantoea, Pseudomonas and Sphingomonas) in the phyllosphere (Fig. 5b). Erwinia amylovora and the core taxa members that co-occurred with it were simultaneously retrieved, identified and subjected to interaction assays; synthetic assemblages (SynAss) were constructed from the six isolates that occurred at the highest frequency in isolated genera belonging to each core taxon. Growth of E. amylovora was not affected by SynAss (Fig. 5c & d). In contrast, the growth of SynAss was slightly promoted in response to E. amylovora (Fig. 5e & f). A dual culture interaction assay further showed that Sphingomonas exclusion from the core taxa stimulated E. amylovora growth (Fig. 5g & h). Even though E. amylovora growth was unaffected, virulence gene expression required for colonization and infection of host plant leaves were drastically depressed by 2 to 46 fold by the co-occurrence SynAss (Fig. 5i); removal of Sphingomonas from the SynAss reduced the effectiveness of SynAss as a virulence-moderator.
Fig. 5.
Interaction between bacterial phytopathogen and nonpathogenic core taxa underlying the phyllosphere homeostasis. (a) Relative abundance of major foliar pathogens present in microbiome of the tea plants; (b) Co-occurrence association between the pathogen Erwinia and non-pathogenic core taxa including Herbaspirillum, Massilia, Methylobacterium, Pantoea, Pseudomonas and Sphingomonas in the phyllosphere; (c-h) Co-culture assays for the interaction between E. amylovora and SynAss consisting of Herbaspirillum, Massilia, Methylobacterium, Pantoea, Pseudomonas and Sphingomonas isolates. SynAss was loaded on the paper disc and co-cultured on the E. amylovora-impregnated media plate (c); E. amylovora-impregnated media plate without treatment of SynAss (d); E. amylovora was loaded on the paper disc and co-cultured on the SynAss-impregnated media plate (e); the SynAss-impregnated media plate without treatment of E. amylovora (f); E. amylovora was loaded on the paper disc and co-cultured on the Sphingomonas-impregnated media plate (g); the Sphingomonas-impregnated media plate without treatment of E. amylovora (h). Essential virulence genes expression in E. amylovora (i). Control: mono-culture of E. amylovora; Co-culture: E. amylovora co-cultured with SynAss that was derived from the six non-pathogenic core taxa. Scale bars, 0.6 cm; * P<0.05, ** P<0.01.
Discussion
The core microbiomes of C. sinensis are known to be vitally important for manipulating phytopathogenic bacteria but the underlying mechanisms of this process is largely unknown. Here we collected C. sinensis samples across China to identity the core microbiomes of C. sinensis and examined their impact on co-cultivating phytopathogenic E. amylovora. The key finding of this research is that phyllosphere C. sinensis core microbiomes enabled a resilience to phytopathogenic bacteria by manipulating phytopathogen virulence (Fig. S1). This finding allows us to engineer microbiomes to manage the resilience of higher plants against foliar bacterial diseases.
The NST values suggest that microbial community assembly processes were dominated by deterministic processes in bulk soil and rhizosphere samples, and by stochastic processes in root and leaf samples; this result is supported by the microbiome nestedness patterns and may be due to decreased environmental selection strength from leaf to root to soil. Belowground microbes face environmental selection pressures of oxygen content, nutrient supply and space limitation while phyllosphere microbes are more randomly distributed on leaves through atmospheric deposition, seed-associated dispersion and animal sources [15]. The increasing community similarity from leaf to root and soil in our study supports this inference as it indicates an increasing environmental selection strength [16]. It is generally accepted that stochasticity dominates the early stages of community establishment while deterministic processes become progressively important over time [17]. Thus, the decreased contribution of stochastic processes from leaf to root and soil may also result from time since community establishment.
Deterministic processes mainly consist of dispersal limitation, environmental selection and biological interactions [18]. The importance of dispersal limitation and environmental selection as ecological processes shaping tea microbiomes can be observed in the significant decline of microbial similarity with geographic distance. The associations between microbial communities and geographic distance after controlling for the impact of the environmental matrix suggest a significant impact of dispersal limitation on all compartment microbiomes. This impact may be due to the interactions of drift and dispersal limitation among leaves, rhizosphere and soil. As the movement of a single taxon from air or animal to leaves is a random event, leaf microbial composition may largely depend on stochastic migration processes. However, dispersal limitation is also important in maintaining phyllosphere community structure as microbes are host-associated; therefore the successful establishment of a newly arrived taxon is more limited than new taxa in soils [19]. In addition, drift processes are more important when α-diversity and microbial community abundances are low [20]. Accordingly, the low Shannon-Wiener index for leaf microbiomes suggests that both drift and dispersal limitation are important ecological processes for shaping the microbial assembly of leaf microbiomes; thus, both drift and dispersal limitation strengthen the geographic community similarity decay in old leaves. However, this drift effect may be weaker in new leaves and is masked by the strong selection pressures in soil [21].
The discrepancy in driving factors for environmental selection from soils to leaves suggests that divergent environmental selection processes are active in the compartments. It is not surprising to find that soil pH, the best predictor of soil bacterial and archaeal community composition across global [22], regional [23] and local scales [24], is an important driver for microbiomes in both the rhizosphere and bulk soil. In addition to soil pH, soil organic carbon quality and quantity and nitrogen and phosphorus availability also have notable influences on soil microbial community structure [25]. The results here indicate that microbial communities were closely associated with dissolved organic carbon and available phosphorus in bulk soils and dissolved organic nitrogen (DON) in rhizosphere. Organic carbon substrates and phosphorus compounds exhibited enormous ranges and are persistent abiotic stressors affecting microbial survival and growth in soils [25]. However, carbon and phosphorus limitation in the rhizosphere can be alleviated by root exudates and soil pH neutralization [26]. Decreased nitrogen availability in the rhizosphere also induces microbial DON stress [2]. The significant impact of Cd on microbiomes in bulk soil relative to rhizosphere samples may be due to the alleviation of metal toxicity by increased rhizosphere soil pH [27].
The associations between amino acids and endophytic bacteria in roots and leaves suggest that plant tissue amino acids play a vital role in endophytic bacterial community assembly. Endophytic bacteria produce alkaloids as secondary metabolites; these compounds have diverse potentials as anti-fungal and -viral agents [10]. Most alkaloids are derived from amines produced by the decarboxylation of amino acids [10], such as tyrosine in roots and young leaves, and lysine in old leaves. There was an association between endophytic bacteria and metals such as Cd, Pb and Cu in old leaves, and metals such as Ni in young leaves. Endophytic bacteria can potentially bioaccumulate metals from contaminated mediums as shown by LK11 reprograming its amino acids and proteomic expressions to maintain steady growth during Cd stress [28]. Phytohormone producing endophytic bacteria could therefore be an ideal approach to increase the phytoextraction potential of metal contamination bioremediation plants. The association between Ni and young leaves is unsurprising as it is a urease activator and is required for young tissue growth [29].
The topological properties of microbial co-occurrence networks indicate different microbial interaction contributions for compartment microbial community assembly. The decreasing gradient in linking intensity from soils to leaves suggests that potential microbial interactions in soils are more complex than those in plant tissues. Although the linking intensity was higher for young leaves than old leaves, the longer network diameter for young leaves suggests that the interaction is still inefficient and that young leaf microbial communities may not be capable of reaching a stable status. Co-occurrence network modules indicate potential niche numbers in corresponding microbial communities [30]. The low modularity for young leaves indicates sparse niches in this compartment while the high modularity for root indicates the opposite. The high number of negative associations in plant tissues suggests a high prevalence of adverse interactions as negative associations in microbial communities represent potential antagonistic relationships [31].
Core microbiota are the consequence of long-term environmental selection in specific environments. The core microbiota in leaves of C. sinensis, including Bacilli, Sphingobacteriia and α-Proteobacteria, have been widely identified as endophytic bacteria in pine leaves [32], maple leaves [33] and several plant species found in Tallgrass Prairie communities [34], suggesting that these microbial taxa are adapted to leaf tissue environments. Genomic analyses of plant-associated α-Proteobacteria has shown that they possess a remarkable number of regulators, sugar transporters, metabolic enzymes and nodulation genes [35]. Although the pattern of depletion was not consistent between ascending compartments, Methylobacterium (α-Proteobacteria) and Sphingomonas (Sphingobacteriia) were consistently enriched in each compartment from soils to leaves. This enrichment effect also explains the formation of core microbiota in C. sinensis leaves; overrepresented taxon-associations in each microbial co-occurrence network were mainly associated with the core microbiota in corresponding compartments, indicating an essential role of core microbiota in community assembly.
Understanding of core microbiota function is essential for both individual plant health and agroecosystem health [36]; however, phenotype-based “top-down” strategies using guided observation of microbiome function is still rare, especially in the phyllosphere [37]. Tea is a resilient plant species towards bacterial disease, which was conventionally attributed to the presence of abundant secondary metabolites in its leaves. However, unfolding evidence indicates that a core microbiota-mediated action in higher organisms is an enhanced immune system [38], [39].
Phyllosphere homeostasis has been reported to be governed by genetic networks and coordinated by core microbiota to resist pathogen infection [40]. It was remarkable that our samples indicated a prevalence of Erwinia spp. in the tea phyllosphere that was present without causing any disease symptoms. During E. amylovora infection, DspA/E is a Hrp (Type III Secretion) pathway dependent pathogenicity factor responsible for initial infection [41], dfoA induces electrolyte leakage of host cell for symptom development, and amS gene clusters control exopolysaccharides production to trigger biofilm formation as well as subsequent movement through host xylem vessels [42], [43]. Interestingly, we observed that these genes essential for successful establishment of E. amylovora in host plants were down-regulated by the phyllosphere core microbiota (Fig. 5i), despite E. amylovora maintaining normal survival and growth rates in the presence of the tea phyllospheric core microbiota (Fig. 5g & h). The virulence system of E. amylovora was uniquely repressed through non-antagonistic action, suggesting that core microbiota likely targeted signaling networks.
Similar to the other phytopathogens, virulence gene expression is coordinated by a complex regulatory signaling network involving intracellular two component signal transduction systems, cyclic di-GMP, and upstream intercellular signaling quorum sensing (QS) in E. amylovora [44], [45]. In our study, the action of QS was largely dependent on relevant micro-environmental conditions (Fig. 5b). N-acyl-homoserine lactone (AHL)-QS is highly conserved in E. amylovora to enable successful infection and abolishment of extracellularly accumulated AHLs by extrinsic acyl-homoserine lactonase may lead to defective pathogenicity [46]. Interestingly, we found that AHLs-QS was absent in Sphingomonas spp., and the AHL-degrading gene homolog (N-acyl-homoserine lactonase) was conversly prevalent and conserved in Sphingomonas spp. genomes (Fig. S2). This homolog possibly enabled Sphingomonas to be a key member of the core microbiome for sustaining phyllosphere homeostasis by causing the inactivation of AHL-QS dependent signaling in foliar pathogens such as E. amylovora.
Conclusions and perspectives
While this study provides the continental scale microbiome networks in C. sinensis from soils to leaves, our understanding of these systems is still in its infancy. Our study comprehensively deciphered the driving mechanisms for community assembly and co-occurrence, and provided experimental evidence for the co-occurrence pattern that underlies phyllosphere homeostasis for disease resilience. Collectively, these findings provide novel insight into the co-evolution of higher plants and phyllosphere microbiome, which is potentially exploitable for managing upward prevalence of phytopathogenic bacteria.
Materials and Methods
Sample collection
To capture biogeographical differences in C. sinensis microbiomes, we collected samples from 45 locations spanning all 15 tea growing provinces in China (Fig. 1a). The samples were collected from diverse C. sinensis varieties, soil types and climate types (Table S1); soil characteristics are listed in Fig. S3-4. Five compartments (bulk soil, rhizosphere, roots, young leaves and old leaves) were collected at each plantation using the following protocol. At least 15 healthy C. sinensis plants (10 m of separation) were selected from each tea plantation for sample collection. For each plant, 200 g young leaves and 200 g old leaves were clipped, the top 5 cm of soil was removed and fine roots (approximately 1 mm diameter) from a depth of 5–20 cm were collected. The roots were removed from the soil with a shovel and then gently shaken to remove the soil not tightly attached to the roots. Soil tightly attached to the roots was termed rhizosphere soils. Soil from the same 5–20 cm depth without any roots near the selected trees was termed bulk soil. All subsamples for each compartment were mixed throughout to create a representative sample for each plantation. Samples were transported to the laboratory within 3 days and stored at −20 °C until DNA extraction.
DNA extraction and sequencing
Rhizosphere and bulk soil DNA was extracted from each sample using an MP FastDNA soil extraction kit (MP Laboratories Inc. Carlsbad, CA, USA). Root and leaf DNA was extracted from each sample using a MinkaGene Plant DNA Kit (mChip BioTech, Guangzhou, China). Extracting protocols followed the manufacturer’s instructions with some modifications. DNA quality and quantity were determined by using a NanoDrop device (Thermo Scientific, Wilmington, DE). The 16S rDNA amplicon library preparation and sequencing were performed according to the manufacturer’s protocol at Novogene, China. For the amplicon library preparation, amplification of 16S rDNA V5-V7 region fragments was performed using primers 799F and 1193R [47]. After quality control and quantification and normalization of the DNA libraries, 250-bp paired-end reads were generated using an Illumina NovaSeq 6000 system (Illumina inc, San Diego, US) according to the manufacturer’s instructions.
Amplicon data analysis
Microbial community composition was determined by sequencing 16S rDNA amplicons. The high-quality paired-end reads of the 16S V5-V7 region were merged using USEARCH v11 [48]. OTUs were obtained using the UPARSE pipeline based on the merged sequences [48]. OTU taxonomy was generated using representative sequences of each OTU and aligned against the RDP II databases. OTUs and merged sequences that were defined as originating from unknown, chloroplast, mitochondria or plant sources were removed.
Diversity and ordination
α-diversity was calculated for each sample using the Shannon-Wiener index based on the normalized OTU abundance table using the binomial method [49]. Community dissimilarity analysis between samples was performed using Principal Coordinate Analysis (PCoA) with unweighted UniFrac distances.
Core microbiota identification
The core microbiota for C. sinensis was defined as cohort of OTUs present in all 225 samples. Unique core microbiota for each compartment was defined as the OTUs present in all 45 samples of the corresponding compartment and also absent in all other compartments.
Source tracker analysis
The potential source of microbial compositions among various compartments was estimated using SourceTracker [50].
Enrichment and depletion from soils to leaves
OTU enrichment and depletion were calculated using the DESeq function of the R package DESeq2 based on the negative binomial distribution algorithm [49].
Normalized stochastic ratio estimation
Normalized stochasticity ratio (NST) of communities in samples and null model was calculated with tNST function of the R package NST [16]. The null model calculation was repeated 10 times.
Nestedness estimation
Community nestedness at various taxonomic ranks was calculated with nestednodf function in vegan package. The corresponding nestedness using a null model was calculated based on a matrix generated with the permatfull function in vegan package.
Dispersal limitation
Community similarity geographic decay was tested by estimating the relationship between community similarity and geographic distance. Dispersal limitation was tested using correlations between community similarity and geographic distance under a partial correlation condition for environmental properties using partial mantel statistic with mantel.partial function in vegan package. The spatial correlation of communities along geographic distance was determined with Mantel correlogram using mantel. correlog function in vegan package.
Environmental selection
Amino acid and catechin concentrations in roots, young leaves and old leaves, the physicochemical properties of rhizosphere and bulk soils, and the concentrations of 8 heavy metals were measured in all samples (Figs. S3-S7). The impact of these environmental factors on microbial communities for corresponding compartments was estimated using constrained corresponding analysis with cca function in vegan.
Microbial co-occurrence network
Microbial co-occurrence networks for each compartment were constructed based on a spearman correlation matrix for the OTUs presenting in each compartment. In order to avoid taxon number biases, each community dataset was trimmed to the dominant 400 taxa. p-values were adjusted for multiple tests using the Benjamini and Hochberg FDR controlling procedure with the R package multtest [51]. Direct correlation dependencies were distinguished using the network enhancement method [48] and thresholds values for Spearman cutoffs were determined by Random matrix theory (RMT) method [52]). Network properties were calculated with the igraph package for R [52].
Construction of synthetic assemblages
Tea leaf samples were ground with 10 mL of sterilized distilled water premixed with quartz sand in a sterilized mortar. The resulting suspension was used for a serial dilution and as an inoculant. Agar plates using Modified Winogradsky and Luria-Bertani (LB) media were employed as the culture medium for bacteria. Plates were incubated for 5–10 days at 25 °C, and culturable and distinguishable bacterial colonies were purified and identified using universal PCR primers for amplification of 16S rRNA gene sequence [53]. Consequently, a foliar pathogen (Erwinia amylovora T11), and several non-pathogenic isolates belonging to Herbaspirillum, Massilia, Methylobacterium, Pantoea, Pseudomonas and Sphingomonas were identified, from which the six most abundant strains of each genera was selected for construction of the synthetic assemblages (SynAss) [11], [54]. These strains were: Herbaspirillum sp. T32, Massilia sp. T02, Methylobacterium sp. T27, Pantoea sp. T05, Pseudomonas sp. T29 and Sphingomonas sp. T41. Each strain was cultured in the dark at 25 °C overnight, and re-suspended in sterile PBS buffer after removal of the media. According the OD value-calibrated density, the six suspension was mixed evenly to generate SynAss with a final density of each strain of 106 CFU/mL.
Experimental evidence for the co-occurrence pattern in phyllosphere
To investigate growth performance during the interaction between E. amylovora T11 and the SynAss, we carried out a confrontation assay using a paper disc-media system as previously reported. E. amylovora T11 or SynAss (or each strain) was impregnated in the agar media or charged on the paper disc at equivalent density (106 CFU/mL). In the same agar plate, the region without the charged paper disc was observed as the control. To analyze virulence gene expression in E. amylovora T11, E. amylovora T11 was inoculated with SynAss at an equivalent density of 103 CFU/mL in LB after which the cultures were shaken overnight and then subjected to total RNA isolation. The construction of a cDNA library was conducted as previously reported [55] and subsequent quantitative PCR were performed using a ChamQ Universal SYBR qPCR Master Mix Kit (Vazyme, Nanjing, China) with specific primer pairs (Table S3) in a thermal cycler real time dice (ABI 7500, USA). Holding stage was 300 s at 95 °C; cycling stage was 10 s at 95 °C, 30 s at 58 °C, repeat for 40 cycles; melt curve stage was 15 s at 95 °C, 60 s at 60 °C, +0.3 °C, 15 s at 95 °C. Each gene was calculated and expressed as fold change in comparison to the housekeeping gene rlsA for each treatment according to the 2-△△CT method [56].
Statistics
All the statistics were performed using R 3.6.0 [57]. Significant differences in α-diversity across compartments were determined using one-way analysis of variance (ANOVA) and Tukey’s ‘Honest Significant Difference’ (HSD). Significant differences in community dissimilarity were determined using analysis of similarities (ANOSIM). Significant differences in NST between communities in samples and the null model were determined using one-tailed one-sample t-tests.
Data and material availability
The sequence data are deposited in the National Genomics Data Center (bigd.big.ac.cn) with accession number PRJCA002571. The R code for all analysis is freely accessible at https://www.github.org/microbma/teamicrobiome.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Ping Xu: Conceptualization, Funding acquisition, Investigation, Writing – original draft. Erinne Stirling: Writing – original draft. Hengtong Xie: Investigation, Formal analysis. Wenbing Li: Conceptualization, Formal analysis. Xiaofei Lv: Data curation, Formal analysis. Haruna Matsumoto: Investigation, Formal analysis. Haiyan Cheng: Investigation, Formal analysis. Anan Xu: Investigation, Formal analysis. Wanyi Lai: Investigation, Formal analysis. Yuefei Wang: Investigation, Formal analysis. Zuntao Zheng: Investigation, Formal analysis. Mengcen Wang: Conceptualization, Investigation, Writing – original draft. Xingmei Liu: Investigation, Formal analysis. Bin Ma: Conceptualization, Investigation, Formal analysis, Writing – original draft. Jianming Xu: Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (42090060) and the Zhejiang Provincial Natural Science Foundation of China (LD19D060001).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.04.002.
Contributor Information
Mengcen Wang, Email: wmctz@zju.edu.cn.
Bin Ma, Email: bma@zju.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Agler M.T., Ruhe J., Kroll S., Morhenn C., Kim S.-T., Weigel D., et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14(1):e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Santi Ferrara F.I., Oliveira Z.M., Gonzales H.H.S., Floh E.I.S., Barbosa H.R. Endophytic and rhizospheric enterobacteria isolated from sugar cane have different potentials for producing plant growth-promoting substances. Plant Soil. 2012;353:409–417. [Google Scholar]
- 3.Chi F., Shen S.-H., Cheng H.-P., Jing Y.-X., Yanni Y.G., Dazzo F.B. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol. 2005;71(11):7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch P.R., Mauchline T.H. Who’s who in the plant root microbiome? Nat Biotechnol. 2012;30:961–962. doi: 10.1038/nbt.2387. [DOI] [PubMed] [Google Scholar]
- 5.Wang M., Cernava T. Overhauling the assessment of agrochemical-driven interferences with microbial communities for improved global ecosystem integrity. Environ Sci Ecotechnol. 2020;4:100061. doi: 10.1016/j.ese.2020.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan J., Kerstetter J.E., Turcotte M.M. Eco-evolutionary interaction between microbiome presence and rapid biofilm evolution determines plant host fitness. Nat Ecol Evol. 2021;5(5):670–676. doi: 10.1038/s41559-021-01406-2. [DOI] [PubMed] [Google Scholar]
- 7.Xia E.-H., Zhang H.-B., Sheng J., Li K., Zhang Q.-J., Kim C., et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 2017;10(6):866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Wang X., Feng H.u., Chang Y., Ma C., Wang L., Hao X., et al. Population sequencing enhances understanding of tea plant evolution. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-18228-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei C., Yang H., Wang S., Zhao J., Liu C., Gao L., et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci U S A. 2018;115(18) doi: 10.1073/pnas.1719622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C.-F., Zhu Y., Yu Y., Zhao Q.-Y., Wang S.-J., Wang X.-C., et al. Global transcriptome and gene regulation network for secondary metabolite biosynthesis of tea plant (Camellia sinensis) BMC Genomics. 2015;16(1) doi: 10.1186/s12864-015-1773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu P., Fan X., Mao Y., Cheng H., Xu A., Lai W., et al. Temporal metabolite responsiveness of microbiota in the tea plant phyllosphere promotes continuous suppression of fungal pathogens. J Adv Res. 2022;39:49–60. doi: 10.1016/j.jare.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh M., Kumar A., Singh R., Pandey K.D. Endophytic bacteria: a new source of bioactive compounds. 3 Biotech. 2017;7(5) doi: 10.1007/s13205-017-0942-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaldson G., Lee S., Mazmanian S. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarkar A., Harty S., Johnson K.-A., Moeller A.H., Archie E.A., Schell L.D., et al. Microbial transmission in animal social networks and the social microbiome. Nat Ecol Evol. 2020;4(8):1020–1035. doi: 10.1038/s41559-020-1220-8. [DOI] [PubMed] [Google Scholar]
- 15.Whipps J.M., Hand P., Pink D., Bending G.D. Phyllosphere microbiology with special reference to diversity and plant genotype. J Appl Microbiol. 2008;105(6):1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x. [DOI] [PubMed] [Google Scholar]
- 16.Ning D., Deng Y., Tiedje J.M., Zhou J. A general framework for quantitatively assessing ecological stochasticity. Proc Natl Acad Sci U S A. 2019;116(34):16892–16898. doi: 10.1073/pnas.1904623116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dini-Andreote F., Stegen J.C., van Elsas J.D., Salles J.F. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci U S A. 2015;112(11):E1326–E1332. doi: 10.1073/pnas.1414261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonze D., Lahti L., Raes J., Faust K. Multi-stability and the origin of microbial community types. ISME J. 2017;11(10):2159–2166. doi: 10.1038/ismej.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vorholt J.A. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10(12):828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 20.Chase J.M., Myers J.A. Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans Roy Soc Lond B Biol Sci. 2011;366(1576):2351–2363. doi: 10.1098/rstb.2011.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vellend M. Conceptual synthesis in community ecology. Q Rev Biol. 2010;85(2):183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 22.Fierer N., Strickland M.S., Liptzin D., Bradford M.A., Cleveland C.C. Global patterns in belowground communities. Ecol Lett. 2009;12(11):1238–1249. doi: 10.1111/j.1461-0248.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths R.I., Thomson B.C., James P., Bell T., Bailey M., Whiteley A.S. The bacterial biogeography of British soils. Environ Microbiol. 2011;13(6):1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x. [DOI] [PubMed] [Google Scholar]
- 24.Ma B., Lv X., Cai Y., Chang S.X., Dyck M.F. Liming does not counteract the influence of long-term fertilization on soil bacterial community structure and its co-occurrence pattern. Soil Biol Biochem. 2018;123:45–53. [Google Scholar]
- 25.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Na Rev Microbiol. 2017;15(10):579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 26.Ahkami A.H., Allen White R., Handakumbura P.P., Jansson C. Rhizosphere engineering: enhancing sustainable plant ecosystem productivity. Rhizosphere. 2017;3:233–243. [Google Scholar]
- 27.Blossfeld STEPHAN, Gansert DIRK. A novel non-invasive optical method for quantitative visualization of pH dynamics in the rhizosphere of plants. Plant Cell Environ. 2007;30(2):176–186. doi: 10.1111/j.1365-3040.2006.01616.x. [DOI] [PubMed] [Google Scholar]
- 28.Khan A.L., Ullah I., Hussain J., Kang S.-M., Al-Harrasi A., Al-Rawahi A., et al. Regulations of essential amino acids and proteomics of bacterial endophytes Sphingomonas sp. Lk11 during cadmium uptake. Environ Toxicol. 2016;31(7):887–896. doi: 10.1002/tox.22100. [DOI] [PubMed] [Google Scholar]
- 29.Fabiano C.C., Tezotto T., Favarin J.L., Polacco J.C., Mazzafera P. Essentiality of nickel in plants: A role in plant stresses. Front Plant Sci. 2015;6:754. doi: 10.3389/fpls.2015.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Röttjers L., Faust K. From hairballs to hypotheses-biological insights from microbial networks. FEMS Microbiol Rev. 2018;42(6):761–780. doi: 10.1093/femsre/fuy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faust K., Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10(8):538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 32.Proença D.N., Francisco R., Kublik S., Schöler A., Vestergaard G., Schloter M., et al. The Microbiome of endophytic, wood colonizing bacteria from pine trees as affected by pine wilt disease. Sci Rep. 2017;7:4205. doi: 10.1038/s41598-017-04141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wemheuer F., Wemheuer B., Daniel R., Vidal S. Deciphering bacterial and fungal endophyte communities in leaves of two maple trees with green islands. Sci Rep. 2019;9:14183. doi: 10.1038/s41598-019-50540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding T., Melcher U. Influences of plant species, season and location on leaf endophytic bacterial communities of non-cultivated plants. PLoS ONE. 2016;11(3) doi: 10.1371/journal.pone.0150895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pini F., Galardini M., Bazzicalupo M., Mengoni A. Plant-bacteria association and symbiosis: are there common genomic traits in Alphaproteobacteria? Genes. 2011;2(4):1017–1032. doi: 10.3390/genes2041017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toju H., Peay K.G., Yamamichi M., Narisawa K., Hiruma K., Naito K., et al. Core microbiomes for sustainable agroecosystems. Nat Plants. 2018;4(5):247–257. doi: 10.1038/s41477-018-0139-4. [DOI] [PubMed] [Google Scholar]
- 37.Liu H., Brettell L.E., Singh B. Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 2020;25(9):841–844. doi: 10.1016/j.tplants.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto H., Fan X., Wang Y., Kusstatscher P., Duan J., Wu S., et al. Bacterial seed endophyte shapes disease resistance in rice. Nat Plants. 2021;7(1):60–72. doi: 10.1038/s41477-020-00826-5. [DOI] [PubMed] [Google Scholar]
- 39.Xie H., Feng X., Wang M., Wang Y., Kumar Awasthi M., Xu P. Implications of endophytic microbiota in Camellia sinensis: a review on current understanding and future insights. Bioengineered. 2020;11(1):1001–1015. doi: 10.1080/21655979.2020.1816788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen T., Nomura K., Wang X., Sohrabi R., Xu J., Yao L., et al. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature. 2020;580(7805):653–657. doi: 10.1038/s41586-020-2185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogdanove A.J., Bauer D.W., Beer S.V. Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. J Bacteriol. 1998;180(8):2244–2247. doi: 10.1128/jb.180.8.2244-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salomone-Stagni M., Barthoa J.D., Polsinellia I., Bellini D., Walsh M.A., Demitri N., et al. A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora. J Struct Biol. 2018;202(3):236–249. doi: 10.1016/j.jsb.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Koczan J.M., McGrath M.J., Zhao Y., Sundin G.W. Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology. 2009;99(11):1237–1244. doi: 10.1094/PHYTO-99-11-1237. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto H., Qian Y., Fan X., Chen S., Nie Y., Qiao K., et al. Reprogramming of phytopathogen transcriptome by a non-bactericidal pesticide residue alleviates its virulence in rice. Fund Res. 2022;2(2):198–207. doi: 10.1016/j.fmre.2021.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piqué N., Miñana-Galbis D., Merino S., Tomás J.M. Virulence Factors of Erwinia amylovora: A Review. Int J Mol Sci. 2015;16(6):12836–12854. doi: 10.3390/ijms160612836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molina Lázaro, Rezzonico F., Défago Geneviève, Duffy B. Autoinduction in Erwinia amylovora: evidence of an acyl-homoserine lactone signal in the fire blight pathogen. J Bacteriol. 2005;187(9):3206–3213. doi: 10.1128/JB.187.9.3206-3213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horton M.W., Bodenhausen N., Beilsmith K., Meng D., Muegge B.D., Subramanian S., et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat Commun. 2014;5(1) doi: 10.1038/ncomms6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 49.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knights D., Kuczynski J., Charlson E.S., Zaneveld J., Mozer M.C., Collman R.G., et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8(9):761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollard K.S., Dudoit S., van der Laan M.J. In: Bioinformatics and computational biology solutions using R and Bioconductor. Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Springer; New York(NY): 2005. Multiple testing procedures: R multtest package and applications to genomics; pp. 249–271. [Google Scholar]
- 52.Csardi G, Nepusz T. 2006. The igraph software package for complex network research. Int J Complex Sys. 1695.
- 53.Liu X., Fan X., Matsumoto H., Nie Y., Sha Z., Yi K., et al. Biotoxin tropolone contamination associated with nationwide occurrence of pathogen Burkholderia plantarii in agricultural environments in China. Environ Sci Technol. 2018;52(9):5105–5114. doi: 10.1021/acs.est.7b05915. [DOI] [PubMed] [Google Scholar]
- 54.Fan X., Fu Y., Nie Y., Matsumoto H., Wang Y., Hu T., et al. Keystone taxa-mediated bacteriome response shapes the resilience of the paddy ecosystem to fungicide triadimefon contamination. J Hazard Mater. 2021;417:126061. doi: 10.1016/j.jhazmat.2021.126061. [DOI] [PubMed] [Google Scholar]
- 55.Wang M., Hashimoto M., Hashidoko Y. Repression of tropolone production and induction of a Burkholderia plantarii pseudo-biofilm by carot-4-en-9,10-diol, a cell-to-cell signaling disrupter produced by Trichoderma virens. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0078024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan X., Matsumoto H., Wang Y., Hu Y., Liu Y., Fang H., et al. Microenvironmental interplay predominated by beneficial Aspergillus abates fungal pathogen incidence in paddy environment. Environ Sci Technol. 2019;53(22):13042–13052. doi: 10.1021/acs.est.9b04616. [DOI] [PubMed] [Google Scholar]
- 57.R Core Team. R: A language and environment for statistical computing. Vienna(Austria): R Foundation for Statistical Computing; 2019. Available from: https://www.R-project.org/. Accessed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.