Abstract
Metallic nanoparticles are increasingly present in our environment, raising concerns on their interactions with living organisms and potential toxicity. Indeed, metallic nanoparticles release metal ions that can be toxic, bioessential, therapeutically active, or combine several of these features. However, human cell responses to different metallic nanoparticles and ions have rarely been compared so far. We propose here a meta-analysis of the transcriptomic responses of human cells to nanoparticles and ions of various metals (titanium, iron, copper, zinc, silver, cadmium, platinum, gold), in order to identify the commonalities and differences between cell responses to these compounds. This analysis revealed that the chemical properties of metals are more important than their known biological functions (i.e., essential metals, toxicity) in governing the cell transcriptome. Particularly, we evidence that the response to nanoparticles is dominated by the response to the ions they contain, and depend on the nanoparticles’ solubility. The formulation as nanoparticles impacts the cell response at lower intensity than the released ions, by altering genes related to vesicle intracellular transport and the cytoskeleton. Moreover, we put into light that several metals (i.e., copper, zinc, silver, cadmium, and gold) trigger a common cell response governed by metallothioneins, which coexist with singular signatures that are specific to a given element.
Keywords: metallic nanoparticles, transcriptomic response, nanoparticle dissolution, metal ions, metallothioneins
Introduction
Humans are increasingly exposed to nanoparticles (NPs) that originate from various manufactured products, such as pesticides, food, textiles, cosmetics, or paints, that are made by combustion or vehicle emissions, or that are introduced in the body for medical purposes. This growing exposure to NPs calls into question their impact on human health. Among these NPs, metal-based NPs (i.e., metallic, metal oxide, or metal sulfide NPs) represent an important proportion, but their nanotoxicology is still a concern. Metallic pollutants are increasingly considered to have an important impact on human health, although the mechanisms underlying metal toxicity are still widely debated. Indeed, it remains challenging to determine which biological effects on human health could be related to the metal they are composed of, and which are due to their formulation as particles.
Particle matter and nanoparticles undergo multiple transformations and release metal ions at different stages of their life cycle, either before reaching the human cells or after their cellular uptake and processing. However, there is no consensus on how human cells respond to the various forms of metals they are exposed to. Beyond the variety of forms of metals, e.g., their nanoparticle or soluble forms, it is also unclear if human cells can share common response to different metals and how such responses are related to metal toxicity.
Various metal ions are naturally found in the human body with different redox states, concentrations, and locations. Some of them, such as iron, copper, zinc, or manganese are essential to human life, regulating the expression, folding, and activity of vitally important proteins. However, all metals are also toxic, regardless of their essential role, when they exceed a certain concentration. Genetic diseases that lead to metal accumulation (i.e., hypermanganesaemia for manganese, primary hemochromatosis for iron, Wilson and Menkes diseases for copper) highlight the deleterious effect of essential metal ions at high doses when homeostasis mechanisms are disrupted.1,2 In contrast, the so-called heavy metals, i.e., lead, mercury, or cadmium, are known to cause poisoning even at low exposure doses. Several metals, including nonessential and highly toxic ones, are also used in medicine as therapeutic or diagnostic agents, such as platinum anticancer drugs, gold-based drugs against rheumatoid arthritis, or gadolinium contrast agents.
The cellular response to metallic compounds is complex to capture, as it may vary not only with the nature of the metal, but also with its chemical formulation (coordination sphere, counterions, oxidation state, crystallization, size, shape or surface state of NPs...).3,4 This complexity makes it difficult to identify common patterns in the cell response to metallic compounds. However, if many features impact this response, it is likely that some of them outweigh the others and drive most of the cellular response to metallic compounds. This is the goal of the present study, to identify and hierarchize the human cell response to various metals encountered by cells as nanoparticles or ionic forms.
Qualitative meta-analysis could be a valuable tool to identify reproducible patterns between metallic compounds, independently of the exposure conditions and cell type. Using a combination of different studies offers the possibility to overcome batch effects by relying on results produced in different contexts over a wide range of experimental conditions and across different cell lines. Furthermore, the combination of different analyses increases statistical power and gives access to low intensity signals, provided that they are repeated across data sets.
Such a statistical meta-analysis approach has so far been implemented to offer a comprehensive view of NP cytotoxicity,5,6 NP ecotoxicity,7−9 NP delivery to tumors,10,11 or NP protein corona composition,12 relying on a wide variety of indicators including quantitative as well as qualitative factors as raw data. Fewer studies use transcriptomic data as raw materials to unravel the cellular outcome of NPs. Transcriptomics studies measure the expression of several thousands of genes in a single experiment. It is thus an important tool to comprehensively capture the cell status and the affected biological pathways. This approach was used to unravel the impact of titanium dioxide or carbon-based nanomaterials on pulmonary functions,13−15 of silver NPs on epithelial cells,16 of various NPs on plants, fungi or aquatic species,17,18 or to highlight the transcriptional mechanisms of action of engineered nanomaterials.19
We hereby propose a meta-analysis of publicly available transcriptomics data to compare the response of human cells to eight different metals, i.e., cadmium, copper, gold, iron, platinum, silver, titanium, and zinc, either in their ionic or nanoparticulate forms. This list of metal contains essential (i.e., iron, copper, and zinc) and nonessential metals, with different toxicity profiles. We first compared the cell response to the different materials and ions in an unsupervised way with the aim to identify common features of cellular responses without preconceived ideas. It revealed that the cell response is similar for ionic and nanoparticulate forms of the same metal. Furthermore, a general cell response is common for five of the eight metals and driven by metal binding proteins called metallothioneins, regardless of the toxicity or physiological functions of those metals. In addition to this common response, we then identified up-regulated biological functions that are specifically up-regulated by one or a few metals, and that can be linked to the effects of metals as drugs or as poison. Finally, we performed a supervised study to identify commonly up-regulated sets of genes at a lower intensity that would distinguish cell response to the ionic or nanoparticulate form of the metal or to essential versus nonessential metals.
Results and Discussion
Data Treatment and Meta-analysis Pipeline
The NCBI (National Center for Biotechnology Information) database GEO (Gene Expression Omnibus) was screened to identify in vitro data sets that describe human cell response to single exposure to a metallic compound, which could be either metallic NPs or metal ions. This approach thus enabled the comparison of data sets obtained in very different contexts that would not otherwise be brought together. For example, exposure to Pt salts used in chemotherapy or to Au NPs exploited in nanomedicine can be compared to the exposure to TiO2 NPs used as a food additive, with no a priori on their outcome in the cell transcriptome. This naive approach enables the comparison of gene expression across conditions without prior knowledge of the systems of interest and the singular cell responses.
To focus on short-term cell responses, a maximal time lapse of 7 days between the metal exposure and the RNA extraction was considered. The study was also restricted to transcriptomic data generated by DNA microarrays, a standardized method to measure gene expression with a low number of commercial microarrays, standardized protocols, and analysis, allowing reliable comparison of data obtained in different laboratories. Moreover, all data in the GEO database follow the MIAME (Minimum Information About a Microarray Experiment) standard in the microarray submission, allowing an easy access to the experimental features on the data.20
Initially, data were gathered for eight metals: titanium (Ti), iron (Fe), copper (Cu), zinc (Zn), silver (Ag), cadmium (Cd), platinum (Pt), and gold (Au). 51 data sets were identified and analyzed individually, as summarized in Figure 1 (orange dotted square), Supplementary Figure S1, and detailed in the Materials and Methods section. To investigate nonmetallic NPs as control, we chose 5 additional data sets of cells exposed to carbon-based nanomaterials or silicon-based NPs.
Figure 1.
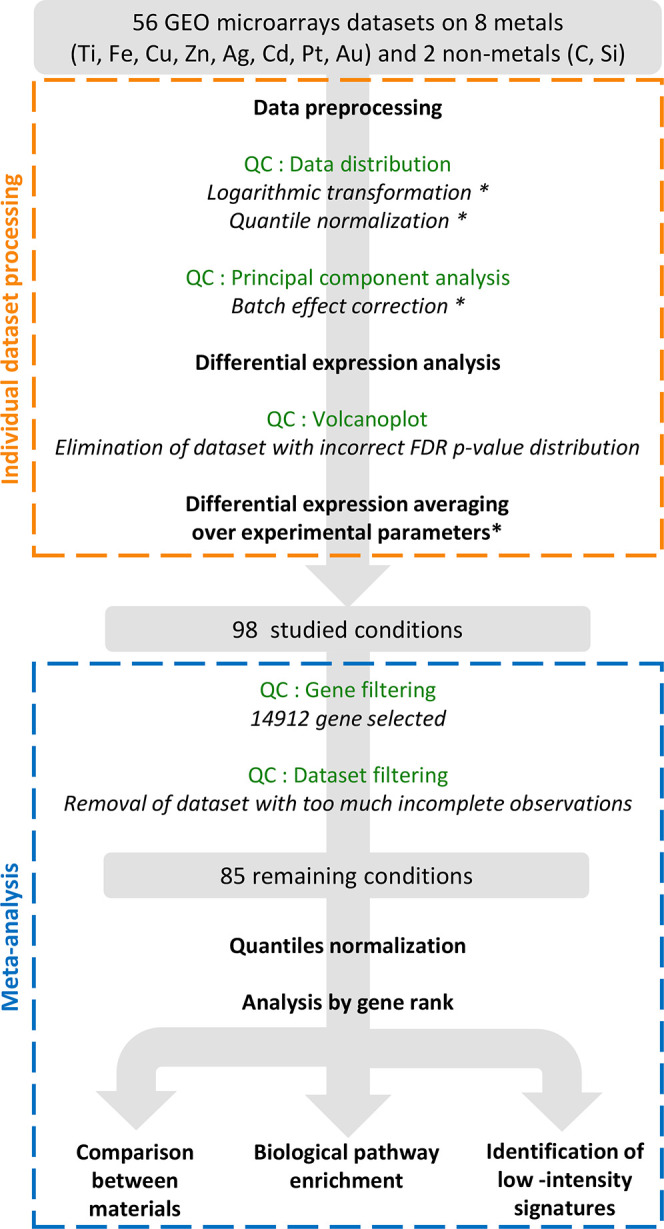
Schematic description of the analysis process. QC stands for Quality Control. Steps followed with * are not systematic and have been applied only if required.
The selected data sets most often compared multiple doses, time points after exposure, cell types, and NPs with different sizes, coatings, and shapes. In order to avoid imbalance between the data sets, all experimental parameters could not be conserved in the final analysis, and gene expressions were averaged on the basis of the correlations between log2 fold change. First, the conditions with different doses and time points were sufficiently correlated to allow averaging gene expression (median Pearson’s correlation coefficients of r = 0.54 ± 0.20 and 0.60 ± 0.25, respectively, Figure S2a–d). More surprisingly, the cell transcriptomic response was only slightly affected by the different sizes, shapes, and coating of NPs constituted of the same material (median Pearson’s correlation coefficient of r = 0.80 ± 0.28, Figure S2e,f). Thus, we could average the responses to NPs for one material or metal.
In contrast, the recipient cell types displayed different responses to NPs made of the same metal or material (median correlation Pearson’s coefficient of r = 0.32 ± 0.29, Figure S2g,h) and thus were treated separately. Indeed, different cell types might elicit different responses: for example, phagocytes such as macrophages can present a different profile than nonphagocytic cells in their response to metallic NPs, because of their ability to generate a high level of oxidative species that might impact and potentially accelerate NP intracellular dissolution. Regarding metal ions, differences could also be expected, for example, in the case of Pt between tumor cells that show resistance or not to Pt based treatment. It is thus not surprising to obtain a lower correlation coefficient when comparing cell types.
From these individual analyses, a general table displaying the expression of 14 912 genes in log2 fold change was generated from the 85 studied conditions (Figure 1, blue dotted square), including eight different metals as NPs or ionic forms, and nonmetallic NPs (Table 1, Supplementary Table S1 and Data File S1).
Table 1. Summary of the Number of Data Set, Cell Type, and Conditions for Each Material and Form.
| metal | Ag |
Au |
Cd | Cu |
Fe |
Pt | Ti | Zn |
C | Si | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| form | ions | NPs | ions | NPs | ions | ions | NPs | ions | NPs | ions | NPs | ions | NPs | NPs | NPs |
| number of GEO data set | 2 | 2 | 1 | 5 | 5 | 3 | 1 | 4 | 1 | 9 | 7 | 4 | 4 | 3 | 2 |
| number of cell line | 2 | 2 | 3a | 5 | 5 | 4 | 1b | 4 | 1c | 18 | 12 | 5d | 7 | 4 | 2 |
| number of conditions | 3 | 3 | 3 | 6 | 5 | 4 | 1 | 4 | 1 | 19 | 13 | 5 | 7 | 8 | 2 |
3 B cell lines.
Bronchial epithelial cells.
Mesenchymal stem cells.
4 B cell lines.
Metal Nanoparticles and Ions Elicit a Common Transcriptomic Signature
We first examined if we could classify metallic stimuli into subgroups according to their impact on cell gene expressions using principal component analysis (PCA). The genes that have the higher contribution to the first principal component were found to be mainly associated with DNA damages and cell death (GADD45B, GADD45A, CDKN1A, PMAIP1). The second principal component was associated with metallothionein (MT) genes, which encode proteins that bind and store metal ions (MT1A, MT1E, MT1G, MT1B, MT2A) (Figure S3 and Table S2).21 No identified function was associated with the genes that contributed the most to the third principal component. Importantly, all materials, either in their ionic or NP form, belong to a single point cloud, with the exception of Pt ions, which generated a separated cluster (Figure S4).
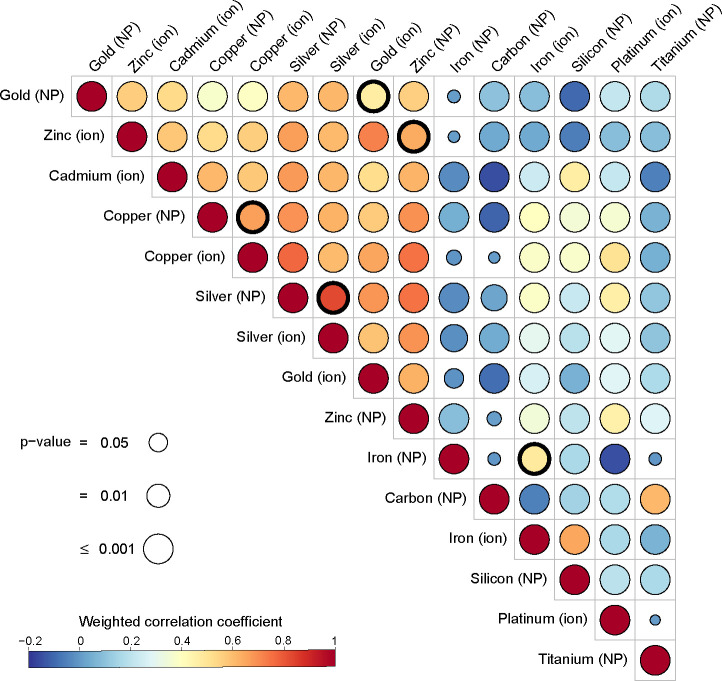
These results show that the log2 fold change is not an effective metric to separate the cell responses to metals. Consequently, we also compared the data sets related to a given material and form (e.g., Au NPs or Pt ions) using the average gene rank, from the highest to the lowest log2 fold change,19 in order to identify the genes that are commonly up regulated. The correlations between the resulting average ranks are displayed in Figure 2. Weighted correlation was implemented in order to give a higher weight to the up-regulated genes, which are considered to be more specific than the down-regulated genes in response to a given stimulus.22P-values were calculated by two different ways fully described in the Materials and Methods, with the one related to weighted linear model being implemented in Figure 2, and the one calculated by bootstrapping being displayed in Table S3 and Figure S5. Correlation coefficients that are described as statistically significant present a p-value under 0.01 using both methods.
Figure 2.
Correlation table between the different materials and forms. Correlation values are calculated from the weighted correlation between average gene ranks. P-values are computed using a Fisher test after modeling with a weighted linear model. Thick circles indicate the correlations between ions and NPs of the same metal.
A first reading of Figure 2 is to analyze to what extent the cell responses to nanoparticulate or ionic forms of the same metal are correlated (thick circles). Remarkably, the correlation coefficient between NPs and ions exceeds 0.6 for Ag, Cu, and Zn (rAg(ion)/Ag(NP) = 0.83, rCu(ion)/Cu(NP) = 0.66, rZn(ion)/Zn(NP) = 0.64) and 0.4 for Fe and Au (rFe(ion)/Fe(NP) = 0.48, rAu(ion)/Au(NP) = 0.47).
This striking result indicates that the cell responses to metal NPs and ions investigated in both form share common up-regulated genes. These similarities can be tentatively explained considering the fate of NPs outside or inside the cells. On the one hand, extracellular dissolution of NPs might take place during cell exposure: several NPs that are insoluble into pure water, such as CuO (copper oxide),23 ZnO (zinc oxide),24 and Ag NPs,25 were found to partially dissolve into the cell culture medium. Indeed, serum proteins can bind metal ions and displace the dissolution equilibrium toward the ionic form.26 On the other hand, intracellular degradation of NPs often take place in the lysosomes, where most NPs converge and accumulate following endocytosis pathway. Endolysosomes are characterized by an oxidative potential, a moderate acidity (minimum of pH 4.5), a high proteolytic activity, and redox regulation pathways, including glutathione and MTs metabolism as well as ferritin regulation and storage. This environment favors the dissolution of several metallic NPs, including iron oxides,27 ZnO,24 CuO,28 Ag,25 and Au29 NPs, and thus intracellular exposure to metallic ions. Lysosomal processing of metal ions has been also described to regulate the metabolism/homeostasis of metals or ensure detoxification through sequestration.30,31
The case of the recently evidenced Au NP degradation within lysosomes is particularly informative: as Au NPs are poorly reactive, their dissolution is observed over weeks to months in cells.29 A longitudinal transcriptomic monitoring during this dissolution process revealed that the cell response to Au NPs was markedly different from that to Au ions at the shortest time point, while both responses converged at the latest time points.32 This finding suggests that the pace of NP degradation within the cells is a key factor to understand the similarities between the cell responses to metallic NPs and their ionic forms.
In addition to these dissolution mechanisms, the commonalities in the responses to ions and NPs can also be explained in light of the recrystallization process of metals inside the cells. As an example, Au ions and NPs have a common intracellular fate as they end up as a single form, called aurosomes, after exposure and internalization by cells.29,32 These intralysosomal structures, which appear quickly after exposure to ionic Au, but within days to months after exposure to Au NPs, are composed of self-assembled crystallized Au nanoclusters. Aurosome formation strikingly illustrates that ions and NPs of the same metal can be processed by cells in the same manner, eliciting similar gene expression. Crystallization processes from ionic species have also been described for other metals, such as Pt, Ag, Zn, and Fe.33,34 It has been demonstrated that intracellular and extracellular medium continuously transform Ag, Cu, Zn, Fe, and Au NPs into their ionic forms, and, conversely, that cells exposed to metal ions can also biomineralize NPs in situ. Thus, we hypothesize that the continuity of intracellular fate can explain that mostly the same genes are involved in the cell response to ionic and nanoparticulate forms of these metals, thus sharing a common metabolism for NPs and ions.
Cu, Zn, Ag, Cd, and Au Induce a Common Cell Response Driven by Metallothioneins
Apart from the correlations between ions and NP forms, we thought to analyze whether the cell response can be similar for different metals. Actually, the second and most evident observation from Figure 2 is that two groups of materials can be distinguished. On the one hand, Cu, Zn, Cd, Ag-based compounds (both ions and NPs) and Au ions present high and statistically significant correlation coefficients between each other (0.52 ≤ r ≤ 0.83). The correlation is less marked when including Au NP, but still important (r ≥ 0.34). This represents a group of metals with a common cellular response, which is clearly distinguished from the cell responses to Fe (both ions and NPs), Pt (ions), Ti (NPs) as well as nonmetallic silicon and carbon-based NPs. The latter group of materials is defined only in contrast to the first group. The transcriptomic response to Pt, Ti, and Fe are much less or even not correlated between each other, which is evidence of the singularity of these three metallic compounds.
To get insight into the similarities within the cell responses to Cu, Zn, Ag, Cd, and Au (disregarding their forms), we investigated the list of the most commonly up-regulated genes for these 5 metals. We then identified the top genes of the list which best separate Cu, Zn, Ag, Cd, and Au treated cells from Ti, Fe, Pt, or nonmetallic NPs treated cells, that show distinct responses. This selection process combines hierarchical clustering and a stepwise approach. Among the 50 most up-regulated genes, the ones that optimized the partition score F1, which was calculated from the clustering precision and recall, were selected.
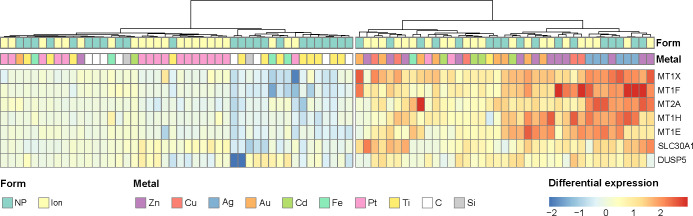
Interestingly, seven genes were enough to optimize the clustering, with a F1 score as high as 90.9%, with an associated p-value of 0.0040 based on bootstrapping (Figure S6). The expression of these seven genes over conditions is displayed in Figure 3. Five of these genes belong to the metallothionein family of proteins (MT1X, MT2A, MT1H, MT1E, MT1F). MTs are small proteins implicated in metal storage and detoxification.21 They contain 30% of cysteine, whose sulfur can strongly interact with metal ions. Among the two last genes, one encodes for the protein SLC30A1. This protein is implicated in the transport of Zn ions, which are known to play a key role in MTs regulation and biosynthesis, as they activate MT transcription factor. Hence, SLC30A1 up-regulation can also be related to MT activation. Lastly, DUSP5 gene is also commonly up-regulated by the five metals sharing a common response. It plays a role in the inhibition of mitogen-activated protein (MAP) kinases, which are implied in cellular division and differentiation. Its role is therefore more general and less metal-specific than the six other genes.
Figure 3.
Heatmap of gene expression for the genes related to Cu, Zn, Ag, Cd, and Au treated cells, across data sets and conditions. Differential expressions are in log2 fold change. Clustering was performed by ascending hierarchical classification using Ward method on squared distances.
Overall, our major finding is that Cu, Zn, Ag, Cd, and Au commonly up-regulate MTs or MT-related genes in exposed cells, in contrast to the other metallic and nonmetallic materials investigated here. This is consistent with the known interactions of these five metals with MT proteins.21 On the contrary, no clear results have been established about the interaction between MTs and Ti ions. The interactions of Pt and Fe with MTs is more complex. It has been observed that Pt ions can interact with MTs in vivo in rats,35 and that Fe(II) can bind metal-free rabbit MTs.36 However, these two studies suggest that Fe and Pt ions could only bind to the free thiol function of MTs, and cannot displace metal ions that are already bound to the protein. As the displacement of Zn ions from MT is necessary to induce MTs biosynthesis,37 Fe and Pt ions could not activate MTs genes expression.
Our results prompt us to draw a first classification of metals: the ones that have a greater affinity than Zn ions for MT thiols, and those which have a lower binding energy that Zn ions. This affinity can be related to several parameters. First, as thiols are known to be a soft base in Pearson acid–base theory, they should interact predominantly with soft acid, namely Cu+, Ag+, Au+, Cd2+, and Pt2+, while Zn2+ and Fe2+ have an intermediate behavior, and Ti4+ is a hard acid. This would explain why Ti ions have not been shown to interact with MTs, while the other have, but also why Zn ions are particularly suitable to regulate MT synthesis, as they have an intermediate behavior which enables other metal ions to displace them from MTs. However, it does not explain why Pt and Fe ions cannot displace Zn from loaded MTs. A second parameter that could influence the ability of metals to bind MTs is the favored configuration of ions within MTs. Metallic ions bind to MTs with a diagonal, trigonal, or tetrahedral coordination,38 which is not favorable for Pt2+ and Fe2+ ions. The similar response to Cu, Zn, Ag, Cd, and Au-based compounds can thus be related to their chemical features, and probably a combination of two factors: their affinity for soft base thiols and their acceptance of MTs binding geometry.
Altogether, the comparison between the metal transcriptomic signatures revealed a common cell response for Cu, Zn, Ag, Cd, and Au that is dominated by MT or MT-related genes up-regulation. In contrast, the three remaining metals, Ti, Fe and Pt, and the nonmetallic carbon and silicon-based NPs do not trigger the up-regulation of these genes, as they probably interact less strongly with MT thiols because of their chemical properties.
Metals Also Induce Singular Transcriptomic Signatures
The latter analysis clearly underlined common mechanisms in the cell response to Cu, Zn, Ag, Cd, and Au. However, paradoxically, these metals have very different impacts on cells or organisms. Indeed, this list contains highly and moderately toxic metals, physiological or essential metals, nonessential metals, and also ions that are widely used as potent medicines. This suggests that beyond a common response mediated by MTs, these compounds might have distinct interactions with cells and proteins.
We thus investigated the most up-regulated genes for all distinct materials. The evolution of the average has been plotted to estimate the impact of the number of conditions, as illustrated for representative cases in Figure S7. To capture the genes that were highly specific to a given metallic form, the 50 genes with the lower gene rank were selected (Data File S2), and associated with several biological functions (Data Files S3–S14), the most represented ones being summarized in Table 2.
Table 2. Summary of the Up-Regulated Biological Functions after Exposure to a Metallic Stimulusa.
| metal | Ag |
Au |
Cd | Cu |
Zn |
Fe |
Ti | Pt | C | Si | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| form | ions | NPs | ions | NPs | ions | ions | NPs | ions | NPs | ions | NPs | NPs | ions | NPs | NPs |
| metal detoxification | + | + | + | + | + | + | + | + | |||||||
| response to oxidative stress | + | + | + | (+) | (+) | ||||||||||
| unfolded protein response | + | + | + | + | |||||||||||
| inflammation | + | + | (+) | ||||||||||||
| apoptosis/cell death | + | + | + | + | + | + | + | + | |||||||
| DNA damage | + | + | |||||||||||||
| cell cycle | + | + | |||||||||||||
These functions have been identified by studying the role of the 50 most commonly up-regulated genes for each condition. The parenthesis indicates functions for which the pathway activation is not certain.
Metal detoxification, which is mostly directed by MT genes, is clearly identified, confirming the above analysis on individual genes. MT up-regulation is also the strongest observed signal for most conditions, including the NP-treated cells. This supports the important finding that the cell response to NPs is widely dominated by the response to metallic ions released after NP dissolution, rather than by the formulation as NPs. An increasing number of studies suggested that most of the observed toxicity of metallic NP might be related to the free metal ions and not to the NP themselves.28,39,40 This effect has been called the Trojan horse effect, as the internalized NPs can release their metallic compounds directly inside the cell, while metal ion uptake is tightly controlled by the cell. Here, we observed that the general response to metallic NPs encompass a primary response to metal ions. Released metal ions thus play the prominent role in the cell response to NPs, both on the transcriptomic and toxicological aspect. This observation further interrogates on the existence and nature of a response to the nanoparticulate form itself at a lower intensity than the response to metal ions, which will be examined in the next section.
Secondary to the MT response, we can notice that Cu, Zn, Ag, Cd, Au-based compounds also partially share other cell responses, such as the response to unfolded protein (heat shock protein genes, DnaJ genes, CRYAB, DDIT3), the response to oxidative stress (glutathione metabolism genes, HMOX1, NQO1, SRXN1, TXNRD1), or to immune stress (TNF, cytokines). Importantly the relationship between metallic ions and oxidative stress has been widely described as a determinant of metal toxicity.41 In contrast fewer studies described the relationship between protein folding and metal stress.42−44 These oxidative, immune or unfolded protein stress could be responsible for the activation of cell death and/or apoptosis genes.
Both Fe ions and Fe-containing NPs do not appear to trigger a clear-cut response in comparison to other metals. This unclear response can either be due to a lack of data sets to describe these conditions, or to a higher variability of the cellular response to Fe compounds between cell types than for other metallic compounds. Similarly, no cellular function is clearly activated by Ti, carbon, or silica NPs. On the contrary, Pt ions appear to induce DNA damage (CDKN1A, GADD45A, GADD45B), dysregulation of the cell cycle (BTG2, BTG3), and apoptosis (PMAIP1, BAK1).
It is interesting to confront the above results on transcriptomic response to the known biological action of these metallic compounds. As an example, Pt ions are commonly used as a potent anticancer drug, which interacts with the DNA and triggers DNA damage.45 This mechanism of action is fully consistent with our meta-analysis, which points out the DNA damage induced specifically by Pt ions. In another medical field, Au ions were used for 50 years as a treatment for rheumatoid arthritis patients.32 Rheumatoid arthritis is a chronic inflammatory disease, and Au ions potentially interfere with the immune system. Here, the meta-analysis reveals the activation of several genes related to inflammation (CCL5, CCL3, TNF). This inflammatory response seems to be mediated by the overexpression of PIR, a protein that can “translate” oxidative stress to immune stress by activating the NFkB pathway.46 This protein, which has been found to be only up-regulated by Au ions in our study, might explain how Au ions can interact with the immune system.
If Pt and Au ions are mostly known for their medical use, other metals, such as Cd ions, are known for their high toxicity. We can notice that Cd ions, contrary to most of the other MT-related ions, do not trigger the overexpression of oxidative stress-related genes, which encode for antioxidant proteins. This is a surprising result, as Cd ions are known to generate free radicals.47 Reactive oxygen species should normally be captured and deactivated by antioxidant proteins to avoid cellular damages. Hence, in the absence of antioxidant protein activation, Cd ions might critically damage the cell, which would explain its toxicity. Overall, the analysis of transcriptomic cellular response highlights some specificity of each metal in line with their known biological outcome. It is worth emphasizing that the metal singular features are secondary to their common signature for the group of Cu, Zn, Ag, Cd, and Au metals.
A Low-Intensity Transcriptomic Signature Is Identified for the NP Form
We highlighted above that the cell response to metal NPs was primarily governed by the response to metal ions. However, we wondered if an intrinsic response to NPs could exist, but that could be hidden by other biological functions that are strongly activated.
To investigate this hypothesis, we performed a supervised study to identify the sets of genes that would distinguish NPs from their ionic forms and that could be hidden by the dominant cellular response. To identify this possible response to the NP form, we averaged, on one part, the ranks for all metallic and nonmetallic NPs, and, on the other part, the genes that are up-regulated by hardly degradable metallic and nonmetallic NPs (i.e., Au, Ti, C).29,40,48 We used the same strategy as described before to cluster the NPs-treated cells from metal ions using hierarchical clustering and optimization of the F1 score.
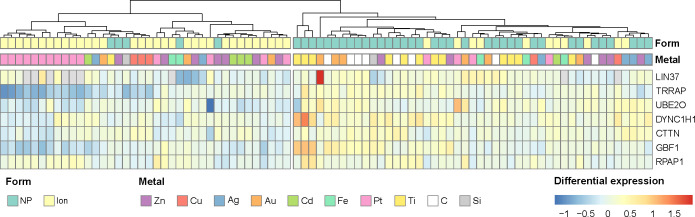
Importantly the clustering obtained with the group of hardly degradable NPs better separates NPs from ions than the clustering taking all NPs into account (maximal F1 score of 81.8% and 66.0%, respectively), with high significance (p-value of 0.0021 after bootstrapping, Figure S8a). This NP-specific clustering relies on seven genes, whose expression across conditions is displayed in Figure 4. These seven genes do not only cluster hardly degradable NPs from the other conditions, but also Cu, Zn, or Ag NPs. This justifies a posteriori that the specific NPs signature could be unveiled from the response to hardly degradable NPs, but also characterizes more degradable NPs.
Figure 4.
Heatmap of NP-related gene expression across data sets and conditions. Differential expressions are in log2 fold change. Clustering was performed by ascending hierarchical classification using the Ward method on squared distances.
Among the NP-induced genes, we observed that several of them are related to intracellular transport and cytoskeleton, such as DYNC1H1, CTTN, and GBF1. As NPs are denser, larger, and stiffer than most of the biological organic compounds, this disturbance can thus be explained by the intracellular trafficking of NPs inside the cells that generate cytoskeleton rearrangement. The relationship between NP internalization and the cytoskeleton has been studied before for a large variety of NPs.49 Here we clearly shed light on a list of related genes that characterize the cell response to NPs across NPs and cell lines. It would be instructive to evaluate how the silencing of these genes would impact NP uptake and intracellular processing.
Two of the NP-induced genes are involved in transcription regulation (TRRAP, UBE2O), and are harder to interpret as they can control many functions. RPAP1 gene encodes for a protein which participates in RNA polymerase II, which could interact with chaperone proteins. Regarding LIN37 protein, its function is still unclear. The roles of these four genes identified in the cell response to NPs are thus not currently elucidated.
Finally, considering the wide variety of biological functions displayed by metals, we interrogated whether the qualification of metals as life-essential (i.e., Cu, Zn, Fe), or highly toxic (i.e., Ag, Cd, Pt) could lead to a segregation of the data sets with an associated list of genes using the same hierarchical approach as reported above for NPs. We found that optimized clustering was associated with F1 scores of respectively 66.7% and 67.7% for bioessential metals and supposedly toxic metals, and respective p-values of 0.020 and 0.022 (Figure S8b,c). These scores are poorly significant and lower than the one distinguishing NPs from metal ions. Moreover, the selected genes could hardly be related to the essential character or the toxicity of the metals (Figure S9). Other metal features could be tested with this approach, such as the concentration of free metal ions compared to the bound fraction, that is to say the metal availability.50 Unfortunately, this notion has so far been studied mostly for essential ions, which limits here our ability to test if some genes are associated with this characteristic.
In conclusion, we designed a hierarchical analysis pipeline enabling us to search for common expression patterns that occur, even at low intensities, behind the dominant response for a subgroup of conditions. Beyond the predominant similarities between NPs and their ionic form for all metals, this approach also shed light on the existence of a general response to the NP form, but not to toxic or essential metals. We evidenced a hierarchy between the activated pathways specifically related to NPs, and observed a clear difference between highly and poorly soluble NPs.
Conclusion
We performed a meta-analysis of publicly available data related to the transcriptomics response of human cells exposed to various metallic or metal oxide NPs (Ti, Fe, Cu, Zn, Ag, and Au NPs), metallic ions (Fe, Cu, Zn, Ag, Cd, Pt, and Au ions), as well as nonmetallic NPs (C and Si NPs). We proposed different stages of analysis with an increasing degree of supervision that enabled us to identify the common and singular signatures in the response to metallic compounds, but also their relative intensities.
The analysis pipeline we propose combines the use of stringent quality controls (redundancy avoidance, p-value distribution check) and a very general formulation that can be extended or adapted to a wide range of databases and biological questions. One of the limitations we encountered is that many steps rely on signal averaging (between the different doses, time points, and NPs types as a first step, between the different cell types as a second step) that might hide singular behaviors.
A first important biological question addressed in this study is the existence of a general cell response to metallic compounds, as a class of chemically defined materials, that would arise from the similarities in transcriptomic profile. Notably, five of the eight considered metals, namely Cu, Zn, Ag, Cd, and Au, emerged as a group that triggered a common and intense transcriptomic response not observed in cells exposed either to nonmetallic carbon and silicon NPs, or to Fe, Ti, and Pt. This signature is driven by the common overexpression of MT or MT-related genes, which are implicated in Zn and Cu homeostasis, heavy metal detoxification, and cellular redox chemistry. One can conclude from this dominant cell response that Cu, Zn, Ag, Cd, and Au-based compounds are primarily processed through a shared MT-related metabolism, regardless of their specific outcome and fate within the cell. Another important finding of the meta-analysis was that Fe, Pt, and Ti, as well as nonmetallic NPs, were excluded from the group of metals whose cellular homeostasis mostly relies on MTs. This can be related to their distinctive chemical properties, which drive metal ions interactions with MTs. Our results are consistent with the assumption that metals can be classified according to their ability to displace Zn ions from MTs and thus induce MTs biosynthesis, owing to the combination of their affinity for soft base thiols and their acceptance of MTs binding geometry.
Apart from the general common impact of metal ions and NPs on the metal binding pathway, the meta-analysis also identified individual responses for each types of metal, such as the activation of oxidative stress, ER stress, or inflammation pathways, that are generally observed at a lower intensity than the MT response. Interestingly, these cellular responses correlate with known singular effects of some metals, such as the biological functions of Au and Pt ions as drugs, which also validates the statistical approach in our study.
Besides the categorization of metals according to their transcriptomic impact, the main original biological question asked through this meta-analysis was if human cells respond similarly to metal ions and metal NPs. This question underpins most of the experimental settings to assess inorganic NP toxicity and the long-lasting debates on the biological impact related to NP structuration with respect to their individual constituents. Our main result is that the response to the various metal NPs included in the meta-analysis is not distinct from that of the corresponding metal ions. PCA analysis as well as gene ranking of the most up-regulated genes did not clearly distinguish metal and ionic forms of the same metal and high correlation coefficients were found when comparing most of the metals in both forms. In particular, metal or metal oxide NPs that are prone to degradation and ion release (i.e., Cu, Zn, Ag) elicit gene transcripts closer to that of their constitutive ions. These important results shed light on the continuum of the cell responses to NPs and ionic forms, which can be explained by common intracellular fate at some points of their journey in the cell.
Our hierarchical analysis of transcriptomic data sets also allowed us to search for low-intensity responses shared by a given set of conditions (e.g., NPs vs ions, highly toxic vs moderately toxic metals, bioessential vs non-bioessential metals). With this supervised strategy, we could interrogate if the dominant similarity between responses to metal ions and NPs could hide a low-intensity signature related to NP specificities. Notably, we shed light on several genes that are more specifically overexpressed after exposure to NPs (metallic or not) and particularly to poorly degradable NPs. This NP-related signature contains cytoskeleton and vesicle traffic-related genes, which can be induced by the intracellular trafficking of NPs involving membrane tension and cytoskeleton rearrangement that are not observed with the molecular form of metals. This signature related to the physical properties of NPs (size, high surface/volume ratio, rigidity, architecture) is important to consider as the primary determinant of cell response which is independent of constitutive elements. Contrary to what we observe regarding the nanoparticulate form, no set of genes could be correlated to the essential character of a metal or to its toxicity with a sufficiently high partition score.
Overall, our important finding that the cell response to metal NPs encompasses the response to metal ions has several consequences. From a toxicology point of view, the impact of metal NPs at the cell level (notwithstanding some differences of biodistribution or bioavailability at the tissue level) could be predicted from the impact of their constitutive ions, which is often better elucidated. This could introduce a paradigm shift in which it is more important to understand and control the biotransformation and life cycle of NPs than their initial chemical properties. Likewise, the therapeutic promises of metal NPs could be envisioned in view of the biological functions and fate of their constitutive ions as well as in situ crystallization process. For example, the common fate of Au NPs and Au ions, both resulting in aurosome intralysosomal formation, encourages one to reconsider gold-based therapeutics and promote the revival of gold salt therapy.32,51
This result can also help to understand the biological action of complex NPs, such as nanoparticulate matter that participates in atmospheric pollution. These particles are hard to describe as they contain multiple chemical elements, with a high variance between particles. Thus, understanding the respective role of the soluble part and the insoluble part is a cornerstone to evaluating their toxicity.52 If the response to the metallic part of the particle could be estimated for its extracellular and intracellular soluble fractions, as shown here for simple NPs, it could be a tremendous improvement in the understanding of the toxicity of such pollutants.53
Materials and Methods
Data Set Selection
Data were extracted from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database. Eight metals were investigated in this database: titanium, iron, copper, zinc, silver, cadmium, platinum, and gold. We selected data acquired in vitro on human cells, after exposure to a single metal and no costimulus. We favored data that provide replicates for each condition. We also selected studies that do not provide replicates, but only if different concentrations or time points were explored after metal stimulation, and if replicates were available for the control condition.
Individual Data Set Extraction and Processing
The data were processed and statistical analysis was performed using the programming software R (version 3.6.0). All analysis pipelines are freely available at https://github.com/AliceBlf/Transcriptomics-meta-analysis-metals.git.
The function getGEO (package GEOquery) was used to extract the raw data using the GEO accession number in order to download both the raw data and its annotations. Data were treated to have only one gene per probe and one probe per gene. Briefly, multigene probes were duplicated to get one value per gene, and redundant genes were treated in order to get the mean values of the different probes.
Then, a value distribution was plotted to determine if the data were expressed as intensity count or log fold change, and were translated to log scale if needed. Data set expressions between conditions were also normalized using the function normalize.quantiles (package preprocessCore). Data sets were then visualized by principal component analysis with the function PCA (package FactoMineR) using as input the genes with the highest variance (720 genes selected). This visualization sometimes helped to correct batch effects, using the function ComBat (package sva).54
Gene differential expressions were then calculated for each metal exposure condition as a comparison to a unique control condition for each cell type using the functions lm.Fit, contrasts.fit, and eBayes (package limma) with default parameter values.55 Volcano plots were performed to control the distribution of differential expression and p-values over the data set.
Correlation of the Different Tested Conditions
For each cell type, metallic compound, dose, and time, gene expression was compared between conditions using Pearson’s correlation. First, the correlation was calculated after one-by-one comparison of the different doses. As the median correlation coefficient for all data sets was higher than 0.5, the different doses were averaged as a single gene expression. Then, the same comparison protocol was applied to the different time points, and then to the different types of NPs. In both cases, the correlation coefficient was found to be sufficiently high to average the gene expression over time and NP types. Finally, the different cell types were compared, but showed different responses from one cell type to another, and were thus treated distinctly.
Data Aggregation and Preprocessing
Once data sets were individually analyzed, they were aggregated into a single 2-dimension matrix of unique genes vs gene expression for each condition. As all gene sets downloaded from the GEO database do not use the same RNA chips, this matrix was not completely filled. To avoid working with genes or data sets that were not sufficiently described, we first eliminated the genes that were not measured in at least two-thirds of the data set, and then the data sets that had missing values in at least two-thirds of the remaining genes. Data sets were then normalized to present a homogeneous range of values using the function normalize.quantiles (preprocessCore package).
Determination of Gene Rank for a Condition
To determine the signature genes for one condition, for each data set, the genes were associated with their rank of expression, the first one being the most up-regulated gene of the data set. Then, for all data sets that describe a given material, the average rank of each gene was calculated using arithmetic mean.
The average rank helps to determine the most commonly up-regulated genes, which have the lower average rank. However, we notice that the amplitude of the average rank was dependent on the number of associated conditions, and could hardly be compared together under their raw form. Thus, all ranks were normalized using the function normalize.quantiles (package preprocessCore) prior to further analysis.
Comparison of Signature between Conditions
To compare average gene ranks, we use weighted correlation function weightedCorr (package wCorr). Basically, correlation was weighted for each gene by the inverse of the product of the square of the average rank:
with wX,Y being the vector of weights applied to the calculate the weighted correlation coefficient between the conditions X and Y, sX the vector of average gene rank for the condition X, and sY the vector of average gene rank for the condition Y.
The significance of the correlation was calculated using two distinct methodologies. First, p-values were calculated using a linear regression between the two lists of ranks using the function lm with the same weights as the weighted correlation. The linear regression calculation generates a Fischer test that estimates the association between the two variables. . Then, lists of ranks were randomly shuffled to calculate 300 correlation coefficients for each pair of two conditions (bootstrapping). The cumulative function was estimated using ecdf function, and the p-value was estimated with this function using the actual correlation coefficient from the nonshuffled vectors.
Selection of the Genes to Predict a Classification
Classical tools to predict a classification (i.e., linear discriminant analysis, logistic regression, random forest regression) could not be implemented here because of the large number of missing values. Hence, the selection of genes was operated using hierarchical clustering and a stepwise-like approach to identify the most important genes.
First step: Classification with a small number of variables. The 5 genes that were most commonly up-regulated were selected and used as input data to generate a tree relying on hierarchical clustering (function hclust, using Ward method on squared distances). The two main clusters were then identified with the function cutree. The adequacy between this clustering and the classification to predict was calculated through a F1 score.
Second step: Addition of new variables. The 45 following genes were successively added at input data to generate new trees, new partitions in two, and an associated F1 score. If one gene increases the F1 score compared to the highest F1 score calculated so far (in the first step or during the second step), it was selected. Else, it was eliminated. This step is similar to the forward selection of a stepwise regression.
Third step: Elimination of selected variables. One by one, the selected genes were removed from the input data, to generate a new tree and F1 score. If the elimination of the gene decreases the F1 score, they were conserved in the final list of genes, else they were eliminated. This step is similar to the backward selection of a stepwise regression.
The final list of genes was used to generate an expression heatmap using pheatmap function (package pheatmap), with the final hierarchical tree.
The significance of the obtained F1 score was evaluated using bootstrapping. Briefly, for an identified set of n genes, n genes were randomly selected in the 50 top genes, and used to generate clustering. This step was repeated 500 times, and the cumulative function of F1 scores distribution was estimated using ecdf function. The p-value was then estimated for the actual F1 score using this distribution.
Calculation of Biological Pathway Enrichment
Enriched biological pathways have been determined from the 50 most up-regulated genes for each metal form or NPs. Data File S2 gathers all the top 50 gene lists. Gene Ontology (GO) database was chosen as a database associating biological function to gene sets.
Significance of the enrichment was calculated using a hyper-geometric test using the phyper function. This test calculated the probability that n genes are present both in the top gene list and a given gene set by chance. It computes the probability to choose n genes belonging to a given gene set by randomly choosing genes among the total number of studied genes, which is here 14 912. The p-values were then corrected by Benjamini–Hochberg method, and the threshold for the resulting false discovery rate (FDR) adjusted p-value was fixed to 0.01. In addition, gene sets that had only three or fewer genes in common with the top 50 gene list were also excluded to limit false positives. The selected gene sets are displayed as Data Files S3–S14, with their associated FDR adjusted p-value and the genes driving the enrichment. For Au NPs, Ti NPs, and Fe NPs, no gene set was selected; thus, no data files are provided for these three types of NPs.
To rationalize biological function enrichment and avoid redundancies, enrichments have been summarized in Table 2 manually. However, this process was supported by several visualizations of the data: for example, as some groups of genes obviously participated to the activation of a group of gene sets, a matrix displaying the occurrence of two genes in the same gene set was established for each type of NPs and ions. These matrices were then visualized using pheatmap function (package pheatmap) with hierarchical clustering, or graph_from_adjacency_matrix function (package igraph). Groups of genes that were responsible for the activation of a group of gene sets were then thoroughly studied to be associated with a small number of large biological functions.
Acknowledgments
We gratefully acknowledge the financial support of the ANR (Agence Nationale de la Recherche) for the projects CarGold-16-CE09-026 and CycLys-18-CE09-0015-01, the French National Research Program for Environmental and Occupational Health of the Anses (Agence nationale de sécurité sanitaire de l’alimentation, de l’environnement et du travail) (2018/1/007) and the European Union’s Horizon 2020 research and innovation program (Grant Agreement 801305). We are grateful to Clotilde Policar and Nathalie Luciani for fruitful discussions, and to Pierre Bost for his technical support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnanoscienceau.2c00035.
Figure S1: Example of a volcano plot that does not match quality standard because of unexpected alignment between points; Figure S2: Distribution of correlation coefficients between log2 fold change gene expression calculated between the different experimental parameters tested in each data set, and representative examples of these correlation coefficients; Figure S3: Principal Component Analysis (PCA) associated graphs and quality controls; Figure S4: Representation of the three first principal components of the principal component analysis performed on the 85 studied conditions and the genes with the highest variance; Figure S5: Distribution of weighted correlation coefficients from randomized data of Figure 2; Figure S6: Distribution of F1 scores obtained for random set of genes from the 50 top genes corresponding to Zn, Ag, Au, Cu, and Cd signatures; Figure S7: Representative average rank plots for three materials/ionic forms; Figure S8: Distribution of F1 scores obtained for random set of genes of the same size of the optimized selection from the 50 top genes corresponding to persistent NPs, essential metals or highly toxic metals; Figure S9: Heatmap of gene expression for the genes related to bioessential or highly toxic metals-treated cells across data sets and conditions; Table S1: GEO references and experimental details of the data sets that have been analyzed; Table S2: Table of the 25 top genes that contribute the most to the first three principal components of the principal component analysis; Table S3: Table of the p-values calculated from randomization method to estimate the significancy of the calculated correlation coefficients displayed in Figure 2 (PDF)
Data File S1: General table of gene expression in log2 fold change for the 85 studied conditions and the 14 912 selected genes; Data File S2: Table of the 50 genes with the lowest average rank for each studied nanoparticulate or ionic form; Data Files S3–S14: Tables of the enriched biological functions for each studied nanoparticulate or ionic form, with the associated FDR adjusted p-value and the gene driving the enrichment (ZIP)
Author Present Address
‡ Laboratoire des BioMolécules (LBM), Département de Chimie, École Normale Supérieure, PSL University, Sorbonne Université, CNRS, 24 rue Lhomond, 75005, Paris, France
The authors declare no competing financial interest.
Supplementary Material
References
- Ferreira C. R.; Gahl W. A. Disorders of metal metabolism. Translational science of rare diseases 2017, 2, 101–139. 10.3233/TRD-170015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umair M.; Alfadhel M. Genetic Disorders Associated with Metal Metabolism. Cells 2019, 8, 1598. 10.3390/cells8121598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova K. S.; Ananikov V. P. Toxicity of metal compounds: knowledge and myths. Organometallics 2017, 36, 4071–4090. 10.1021/acs.organomet.7b00605. [DOI] [Google Scholar]
- Soenen S. J.; Parak W. J.; Rejman J.; Manshian B. (Intra) cellular stability of inorganic nanoparticles: effects on cytotoxicity, particle functionality, and biomedical applications. Chem. Rev. 2015, 115, 2109–2135. 10.1021/cr400714j. [DOI] [PubMed] [Google Scholar]
- Oh E.; Liu R.; Nel A.; Gemill K. B.; Bilal M.; Cohen Y.; Medintz I. L. Meta-analysis of cellular toxicity for cadmium-containing quantum dots. Nature Nanotechnol. 2016, 11, 479. 10.1038/nnano.2015.338. [DOI] [PubMed] [Google Scholar]
- Bilal M.; Oh E.; Liu R.; Breger J. C.; Medintz I. L.; Cohen Y. Bayesian Network Resource for Meta-Analysis: Cellular Toxicity of Quantum Dots. Small 2019, 15, 1900510. 10.1002/smll.201900510. [DOI] [PubMed] [Google Scholar]
- Notter D. A.; Mitrano D. M.; Nowack B. Are nanosized or dissolved metals more toxic in the environment? A meta-analysis. Environmental toxicology and chemistry 2014, 33, 2733–2739. 10.1002/etc.2732. [DOI] [PubMed] [Google Scholar]
- Wang P.; Lombi E.; Menzies N. W.; Zhao F.-J.; Kopittke P. M. Engineered silver nanoparticles in terrestrial environments: a meta-analysis shows that the overall environmental risk is small. Environ. Sci.: Nano 2018, 5, 2531–2544. [Google Scholar]
- Renzi M.; Guerranti C. Ecotoxicity of nanoparticles in aquatic environments: a review based on multivariate statistics of meta-data. J. Environ. Anal. Chem. 2015, 2, 2380–2391. [Google Scholar]
- Cheng Y.-H.; He C.; Riviere J. E.; Monteiro-Riviere N. A.; Lin Z. Meta-Analysis of Nanoparticle Delivery to Tumors Using a Physiologically Based Pharmacokinetic Modeling and Simulation Approach. ACS Nano 2020, 14, 3075–3095. 10.1021/acsnano.9b08142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn Y. J.; Kong T. H.; Choi J. S.; Yun W. S.; Key J.; Seo Y. J. Strategies to enhance efficacy of SPION-labeled stem cell homing by magnetic attraction: a systemic review with meta-analysis. International journal of nanomedicine 2019, 14, 4849. 10.2147/IJN.S204910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Z.; Yuan P.; Yu F.; Peng T.; Zhou Q.; Hu X. Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 10492–10499. 10.1073/pnas.1919755117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikota J.; Williams A.; Yauk C. L.; Wallin H.; Vogel U.; Halappanavar S. Meta-analysis of transcriptomic responses as a means to identify pulmonary disease outcomes for engineered nanomaterials. Part. Fibre Toxicol. 2015, 13, 25. 10.1186/s12989-016-0137-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talikka M.; Belcastro V.; Gubian S.; Martin F.; Peitsch M. C.; Hoeng J. Systems toxicology meta-analysis—From aerosol exposure to nanotoxicology. Current Opinion in Toxicology 2019, 16, 39–48. 10.1016/j.cotox.2019.03.010. [DOI] [Google Scholar]
- Halappanavar S.; Saber A. T.; Decan N.; Jensen K. A.; Wu D.; Jacobsen N. R.; Guo C.; Rogowski J.; Koponen I. K.; Levin M.; et al. Transcriptional profiling identifies physicochemical properties of nanomaterials that are determinants of the in vivo pulmonary response. Environmental and molecular mutagenesis 2015, 56, 245–264. 10.1002/em.21936. [DOI] [PubMed] [Google Scholar]
- Ghojavand S.; Bagheri F.; Tanha H. M. Integrative meta-analysis of publically available microarray datasets of several epithelial cell lines identifies biological processes affected by silver nanoparticles exposure. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 2019, 216, 67–74. 10.1016/j.cbpc.2018.11.003. [DOI] [PubMed] [Google Scholar]
- Burkard M.; Betz A.; Schirmer K.; Zupanic A. Common Gene Expression Patterns in Environmental Model Organisms Exposed to Engineered Nanomaterials: A Meta-Analysis. Environ. Sci. Technol. 2020, 54, 335–344. 10.1021/acs.est.9b05170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruotolo R.; Maestri E.; Pagano L.; Marmiroli M.; White J. C.; Marmiroli N. Plant response to metal-containing engineered nanomaterials: an omics-based perspective. Environ. Sci. Technol. 2018, 52, 2451–2467. 10.1021/acs.est.7b04121. [DOI] [PubMed] [Google Scholar]
- Serra A.; Letunic I.; Fortino V.; Handy R. D.; Fadeel B.; Tagliaferri R.; Greco D. INSIdE NANO: a systems biology framework to contextualize the mechanism-of-action of engineered nanomaterials. Sci. Rep. 2019, 9, 1–10. 10.1038/s41598-018-37411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.; Barrett T. NCBI GEO standards and services for microarray data. Nature biotechnology 2006, 24, 1471–1472. 10.1038/nbt1206-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman M. J. Metallothioneins. Coord. Chem. Rev. 1995, 144, 461–511. 10.1016/0010-8545(95)01173-M. [DOI] [Google Scholar]
- Scott M.; Gunderson C. W.; Mateescu E. M.; Zhang Z.; Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science 2010, 330, 1099–1102. 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- Semisch A.; Ohle J.; Witt B.; Hartwig A. Cytotoxicity and genotoxicity of nano-and microparticulate copper oxide: role of solubility and intracellular bioavailability. Part. Fibre Toxicol. 2014, 11, 1–16. 10.1186/1743-8977-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihai C.; Chrisler W. B.; Xie Y.; Hu D.; Szymanski C. J.; Tolic A.; Klein J. A.; Smith J. N.; Tarasevich B. J.; Orr G. Intracellular accumulation dynamics and fate of zinc ions in alveolar epithelial cells exposed to airborne ZnO nanoparticles at the air–liquid interface. Nanotoxicology 2015, 9, 9–22. 10.3109/17435390.2013.859319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao I.-L.; Hsieh Y.-K.; Wang C.-F.; Chen I.-C.; Huang Y.-J. Trojan-horse mechanism in the cellular uptake of silver nanoparticles verified by direct intra-and extracellular silver speciation analysis. Environ. Sci. Technol. 2015, 49, 3813–3821. 10.1021/es504705p. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Von Dem Bussche A.; Kabadi P. K.; Kane A. B.; Hurt R. H. Biological and environmental transformations of copper-based nanomaterials. ACS Nano 2013, 7, 8715–8727. 10.1021/nn403080y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M.; Luciani N.; Alloyeau D.; Elgrabli D.; Deveaux V.; Pechoux C.; Chat S.; Wang G.; Vats N.; Gendron F.; et al. Long term in vivo biotransformation of iron oxide nanoparticles. Biomaterials 2011, 32, 3988–3999. 10.1016/j.biomaterials.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Strauch B. M.; Hubele W.; Hartwig A. Impact of Endocytosis and Lysosomal Acidification on the Toxicity of Copper Oxide Nano-and Microsized Particles: Uptake and Gene Expression Related to Oxidative Stress and the DNA Damage Response. Nanomaterials 2020, 10, 679. 10.3390/nano10040679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balfourier A.; Luciani N.; Wang G.; Lelong G.; Ersen O.; Khelfa A.; Alloyeau D.; Gazeau F.; Carn F. Unexpected intracellular biodegradation and recrystallization of gold nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 103–113. 10.1073/pnas.1911734116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polishchuk E. V.; Polishchuk R. S. The emerging role of lysosomes in copper homeostasis. Metallomics 2016, 8, 853–862. 10.1039/C6MT00058D. [DOI] [PubMed] [Google Scholar]
- Asano T.; Komatsu M.; Yamaguchi-Iwai Y.; Ishikawa F.; Mizushima N.; Iwai K. Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Molecular and cellular biology 2011, 31, 2040–2052. 10.1128/MCB.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balfourier A.; Kolosnjaj-Tabi J.; Luciani N.; Carn F.; Gazeau F. Gold-based therapy: From past to present. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 22639–22648. 10.1073/pnas.2007285117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman F. U.; Jiang H.; Selke M.; Wang X. Mammalian cells: a unique scaffold for in situ biosynthesis of metallic nanomaterials and biomedical applications. J. Mater. Chem. B 2018, 6, 6501–6514. 10.1039/C8TB01955J. [DOI] [PubMed] [Google Scholar]
- Van de Walle A.; Sangnier A. P.; Abou-Hassan A.; Curcio A.; Hémadi M.; Menguy N.; Lalatonne Y.; Luciani N.; Wilhelm C. Biosynthesis of magnetic nanoparticles from nano-degradation products revealed in human stem cells. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 4044–4053. 10.1073/pnas.1816792116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żelazowski A. J.; Garvey J. S.; Hoeschele J. D. In vivo and in vitro binding of platinum to metallothionein. Archives of biochemistry and biophysics 1984, 229, 246–252. 10.1016/0003-9861(84)90150-4. [DOI] [PubMed] [Google Scholar]
- Good M.; Vasak M. Iron (II)-substituted metallothionein: evidence for the existence of iron-thiolate clusters. Biochemistry 1986, 25, 8353–8356. 10.1021/bi00374a003. [DOI] [PubMed] [Google Scholar]
- Andrews G. K.Zinc Biochemistry, Physiology, and Homeostasis; Springer, 2001; pp 37–51. [Google Scholar]
- Sutherland D. E.; Stillman M. J. The “magic numbers” of metallothionein. Metallomics 2011, 3, 444–463. 10.1039/c0mt00102c. [DOI] [PubMed] [Google Scholar]
- Sabella S.; Carney R. P.; Brunetti V.; Malvindi M. A.; Al-Juffali N.; Vecchio G.; Janes S. M.; Bakr O. M.; Cingolani R.; Stellacci F.; et al. A general mechanism for intracellular toxicity of metal-containing nanoparticles. Nanoscale 2014, 6, 7052–7061. 10.1039/c4nr01234h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffan M.; Rose J.; Wiesner M. R.; Bottero J.-Y. Chemical stability of metallic nanoparticles: a parameter controlling their potential cellular toxicity in vitro. Environ. Pollut. 2009, 157, 1127–1133. 10.1016/j.envpol.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Valko M.; Morris H.; Cronin M. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005, 12, 1161–1208. 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- Tamás M. J.; Sharma S. K.; Ibstedt S.; Jacobson T.; Christen P. Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 2014, 4, 252–267. 10.3390/biom4010252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal S. S.; Botelho H. M.; Gomes C. M. Metal ions as modulators of protein conformation and misfolding in neurodegeneration. Coord. Chem. Rev. 2012, 256, 2253–2270. 10.1016/j.ccr.2012.04.004. [DOI] [Google Scholar]
- Sharma S. K.; Goloubinoff P.; Christen P. Heavy metal ions are potent inhibitors of protein folding. Biochemical and biophysical research communications 2008, 372, 341–345. 10.1016/j.bbrc.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Reed E. Platinum-DNA adduct, nucleotide excision repair and platinum based anti-cancer chemotherapy. Cancer treatment reviews 1998, 24, 331–344. 10.1016/S0305-7372(98)90056-1. [DOI] [PubMed] [Google Scholar]
- Liu F.; Rehmani I.; Esaki S.; Fu R.; Chen L.; de Serrano V.; Liu A. Pirin is an iron-dependent redox regulator of NF-κB. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 9722–9727. 10.1073/pnas.1221743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Qu W.; Kadiiska M. B. Role of oxidative stress in cadmium toxicity and carcinogenesis. Toxicology and applied pharmacology 2009, 238, 209–214. 10.1016/j.taap.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgrabli D.; Dachraoui W.; Menard-Moyon C.; Liu X. J.; Begin D.; Begin-Colin S.; Bianco A.; Gazeau F.; Alloyeau D. Carbon nanotube degradation in macrophages: live nanoscale monitoring and understanding of biological pathway. ACS Nano 2015, 9, 10113–10124. 10.1021/acsnano.5b03708. [DOI] [PubMed] [Google Scholar]
- Ispanixtlahuatl-Meráz O.; Schins R. P.; Chirino Y. I. Cell type specific cytoskeleton disruption induced by engineered nanoparticles. Environ. Sci.: Nano 2018, 5, 228–245. [Google Scholar]
- Robinson N. J.; Glasfeld A. Metalation: nature’s challenge in bioinorganic chemistry. JBIC Journal of Biological Inorganic Chemistry 2020, 25, 543–545. 10.1007/s00775-020-01790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz-Duval A. S.; Konopka C. J.; Moitra P.; Daza E. A.; Srivastava I.; Johnson E. V.; Kampert T. L.; Fayn S.; Haran A.; Dobrucki L. W.; Pan D. Intratumoral generation of photothermal gold nanoparticles through a vectorized biomineralization of ionic gold. Nat. Commun. 2020, 11, 1–18. 10.1038/s41467-020-17595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq B.; Alleman L. Y.; Perdrix E.; Riffault V.; Happillon M.; Strecker A.; Lo-Guidice J.-M.; Garçon G.; Coddeville P. Particulate metal bioaccessibility in physiological fluids and cell culture media: toxicological perspectives. Environmental research 2017, 156, 148–157. 10.1016/j.envres.2017.03.029. [DOI] [PubMed] [Google Scholar]
- Huang Y.-C. T.; Li Z.; Carter J. D.; Soukup J. M.; Schwartz D. A.; Yang I. V. Fine ambient particles induce oxidative stress and metal binding genes in human alveolar macrophages. American journal of respiratory cell and molecular biology 2009, 41, 544–552. 10.1165/rcmb.2008-0064OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T.; Johnson W. E.; Parker H. S.; Jaffe A. E.; Storey J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K.Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer, 2005; pp 397–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.