Abstract
Invasive fungal infections caused by non-albicans Candida species are increasingly reported. Recent advances in diagnostic and molecular tools enabled better identification and detection of emerging pathogenic yeasts. The Candida haemulonii species complex accommodates several rare and recently described pathogenic species, C. duobushaemulonii, C. pseudohaemulonii, C. vulturna, and the most notorious example is the outbreak-causing multi-drug resistant member C. auris. Here, we describe a new clinically relevant yeast isolated from geographically distinct regions, representing the proposed novel species C. khanbhai, a member of the C. haemulonii species complex. Moreover, several members of the C. haemulonii species complex were observed to be invalidly described, including the clinically relevant species C. auris and C. vulturna. Hence, the opportunity was taken to correct this here, formally validating the names of C. auris, C. chanthaburiensis, C. konsanensis, C. metrosideri, C. ohialehuae, and C. vulturna.
Keywords: Candida auris, Candida vulturna, Candida haemulonii species complex, emerging pathogen, antifungal resistant, misidentification
Introduction
The Candida genus (Ascomycota, Saccharomycotina, Saccharomycetes, Saccharomycetales) contains some of the most well-known human fungal pathogens. Although Candida albicans remains the primary cause of candidiasis, infections caused by non-albicans Candida species are increasingly reported.1 Globalization, climate change, and a steadily growing population of immunocompromised patients over the last few decades paved the way for rare and new species to emerge as fungal pathogens.2–4 Recent advances in diagnostic and molecular tools enabled better detection and identification of these emerging fungi. Therefore, the list of reported species causing human infection is rapidly expanding.
Several of these emerging pathogens group within the Candida haemulonii species complex. Members of this complex belong to the family of the Metschnikowiaceae and are closely related to the genus Clavispora.5 Lately, the C. haemulonii species complex has drawn much attention because of its multi-drug resistant nature and the rapid worldwide spread of one of its members, Candida auris.6 Next to C. auris other members also cause nosocomial infections, including C. haemulonii, Candida duobushaemulonii, Candida pseudohaemulonii, and Candida vulturna.5,7 Some of these species have been isolated from ecological sources such as fish (C. haemulonii), flowers (C. vulturna), or insects (C. duobushaemulonii).5,8 However, most reported strains have a human origin. For example, C. pseudohaemulonii and C. auris were described from clinical cases since no environmental strains were available.9,10 Only very recent sampling efforts linked C. auris to the marine ecosystem, but the ecological niche of C. pseudohaemulonii remains to be explored.11,12 In the present study, we describe a new yeast isolated from geographically distinct clinical samples, representing a proposed novel species, Candida khanbhai, related to members of the C. haemulonii species complex. Additionally, it was observed that several members of the C. haemulonii species complex were invalidly described. Hence, the opportunity was taken to correct this here, formally validating the names of C. auris, Candida chanthaburiensis, Candida konsanensis, Candida metrosideri, Candida ohialehuae, and C. vulturna.
Case descriptions
Two clinical strains were obtained from two patients (Table 1). Case 1: The strain (Kw2195/19 = CBS 16213T) was collected in 2019 from a 73-year-old male patient during a suspected C. auris outbreak involving two patients in the Mubarak Al-Kabeer Hospital, Jabriya, Kuwait. Besides reinforcing infection control measures and isolation precautions, all patients in the same medical ward were screened for C. auris nasal carriage. The patient's nasal swab was subcultured onto Sabouraud dextrose agar, which grew cream-colored colonies. Identification by VITEK MS (bioMérieux, Marcy-l’Étoile, France) could not reliably identify the strain. Instead, VITEK 2 (bioMérieux) was used. The result was indicative of C. duobushaemulonii (99% score). Of note, the patient did not receive antibacterial or antifungal agents in the last 3 months and was not infected or colonized with Candida species throughout his stay in the hospital. Given the discrepant identification results the strain was sent to the Westerdijk Fungal Biodiversity Institute for further analyses.
Table 1.
Isolation sources and NCBI GenBank accession numbers of the yeast strains characterized in this study.
Case 2. The strain (UZ687/14 = CBS 16555) originated from the blood culture of a 55-year-old male patient admitted in July 2014 to the Tengku Ampuan Afzan Hospital, Pahang, Malaysia, due to having generalized body weakness, lethargy, nausea, and slight fever of 37.8°C. Blood cultures taken on day one and day four were negative. On day nine, the patient developed a high-grade fever of 39.5°C. Chest X-ray showed pulmonary infiltration in the bilateral lower zones. The patient was treated for new-onset hospital-acquired pneumonia. Intravenous piperacillin/tazobactam (562 mg/6 h) was given for 7 days. The blood culture taken on day nine grew yeast-like colonies that were identified biochemically by API 20C (bioMérieux) as Clavispora lusitaniae (81% score) after 72 h of incubation. A dosage of 400 mg/day intravenous fluconazole was started at day 12. Nevertheless, the patient's clinical condition deteriorated further, and he succumbed to the infection two days later. The strain was sent to the Westerdijk Fungal Biodiversity Institute for further analyses.
Materials and Methods
DNA sequencing and phylogenetic analysis
After receiving both strains at the mycological reference center (Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands) they were subcultured onto 2% glucose, 0.5% yeast extract, 1% peptone, and 1.5% agar (GYPA) plates. After 48 h of incubation at 35°C, the strains were further processed for molecular characterization and culture collection deposition. DNA was extracted using a manual standardized cetyltrimethylammonium bromide-based method, as previously described.13 Similarly, this was done for a reference set of type-strains, representing all members of the C. haemulonii species complex: C. auris (CBS 10913T, clade II; AR0383, clade III; AR0385, clade IV; AR0387, clade I; AR1097, clade V), C. chanthaburiensis (CBS 10926T), C. duobushaemulonii (CBS 7798T), C. heveicola (CBS 10701T), C. haemulonii (CBS 5149T), C. konsanensis (CBS 12666T), C. pseudohaemulonii (CBS 10004T), C. metrosideri (CBS 16091T), C. ohialehuae (CBS 16092T), C. ruelliae (CBS 10815T), C. vulturna (CBS 14366T), and C. lusitaniae (CBS 6936T). Type strains were obtained from the CBS culture collection of the Westerdijk Fungal Biodiversity Institute, while most of the C. auris reference strains came from the CDC Isolate Bank (US Centers for Disease Control & Prevention, Atlanta, GA, USA).
Amplified fragment length polymorphism (AFLP) fingerprint analysis was performed as previously described, except that the fragment analysis was performed using the ABI3700xL Genetic Analyzer platform (Applied Biosystems, Palo Alto, CA, USA).14 Raw fingerprint data were analyzed in Bionumerics v7.6 (Applied Math, St. Martens-Latem, Belgium) and a dendrogram was created using the Pearson correlation similarity coefficient and unweighted pair group method with arithmetic mean (UPGMA) cluster analysis algorithms.
Amplification of the D1/D2 region of the large subunit (LSU) ribosomal RNA gene and the internal transcribed spacer ITS1-5.8S-ITS2 regions was done using the NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) plus NL4 (5′-GGTCCGTGTTTCAAGACGG-3′), and ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) plus ITS4 (5′ TCCTCCGCTTATTGATATGC 3′) primer pairs, respectively.15 Sequencing was performed with the same primers using BigDye chemistry v3.1 and sequences were generated onto the ABI3700xL Genetic Analyzer platform (Applied Biosystems). Raw data bi-directional sequences were manually checked and corrected using the SeqMan module of the Lasergene v17 software package (DNASTAR, Madison, WI, USA).
The aligned ITS (455 bp) and LSU (535 bp) datasets were concatenated in SequenceMatrix v1.8.16 The concatenated sequences were aligned using MAFFT v7.409 with FFT-NS-i option.17 The best model of substitution was established by ModelFinder resulting in TNe + G4 according to Bayesian Information Criterion (BIC).18 Phylogenetic trees were estimated using maximum likelihood with IQ-tree v1.6.12 calculating likelihood ratio test, Bayes inference, and ultrafast bootstrap values running 1000 bootstraps showing the values in this order on the nodes of the tree.19
Phenotypic/physiological characterization
The colony morphology of both strains was determined using plate cultures grown on GYPA for 7 days at 25°C or up to 1 month at room temperature. Microscopic pictures were taken of cells grown in YPD broth (1% yeast extract, 2% bacto peptone, 2% glucose) at 200 rpm, 25°C for 24 h, using an Axioskop 2 plus (Carl Zeiss, Jena, Germany) microscope fitted with a Nikon DS-Ri2 microscope camera (Nikon Instruments, Melville, NY, USA). Additionally, the mating compatibility of both the strains was tested by mixing them onto GYPA and Malt-Extract Agar (MEA; Oxoid, Basingstoke, U.K.) plates and incubated at 25°C for up to 12 weeks. Given the close phylogenetic relatedness of the studied strains to C. auris and C. haemulonii (see results), the colony phenotype was compared with the type strains of C. haemulonii (CBS 5149T) and C. auris (CBS 10913T) on the new CHROMagar Candida Plus (CCP) (CHROMagar, Paris, France), specifically designed to differentiate C. auris from other pathogenic yeasts. Finally, (mis)identification of the strains by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI–TOF MS) was tested using a Bruker MALDI Biotyper system (database v11). MALDI–TOF MS main spectra (MSP) of both strains were generated using the manufacturers’ instructions. Physiological characterization of both strains was done using standard procedures previously described.20 Fermentation and assimilation of carbohydrates were performed in liquid media at 25°C up to 21 days. Assimilation of nitrogen compounds was assessed with the auxanographic method.
Antifungal susceptibility testing
Due to the clinical origin of both strains and their relatedness to the multidrug resistant members of the C. haemulonii complex, their in vitro antifungal susceptibility was determined by the broth dilution method using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) protocol.21 Antifungals were obtained from Sigma–Aldrich (St. Louis, MI, USA). Included in this test were: amphotericin B (AMB), anidulafungin (ANI), micafungin (MCF), 5-flucytosine (5FC), and various azoles including fluconazole (FLU), itraconazole (ITR), voriconazole (VOR), posaconazole (POS), and isavuconazole (ISA). The concentrations tested ranged between 0.03 and 16 μg/ml for all drugs, except fluconazole (0.125–64 μg/ml). MIC values were determined after 24 h and 48 h.
Results
Identification and phylogeny
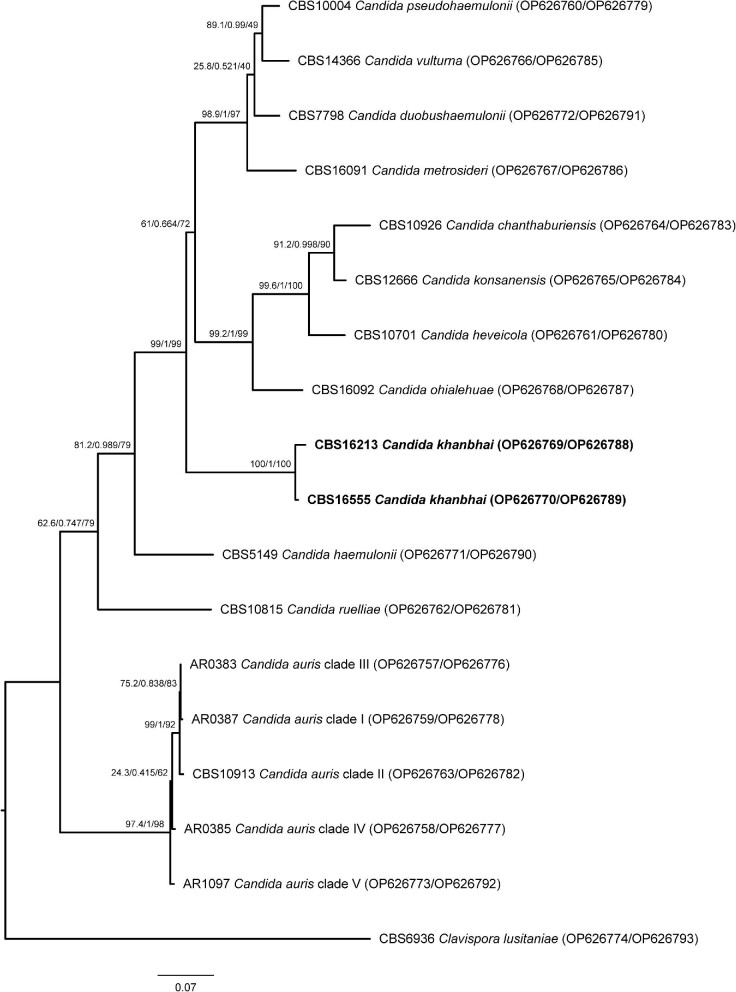
Molecular based identification of CBS 16213T and CBS 16555 by ITS sequencing resulted in low-score NCBI Nucleotide database BLAST hits with a closest hit of 86% similarity to C. duobushaemulonii. Similarly, the D1/D2 sequence of both these strains had a similarity of 92% with C. heveicola using the NCBI Nucleotide database (Table 1). By AFLP analysis CBS 16213T and CBS 16555 shared an 84% fingerprint similarity but had only 35% similarity compared with the fingerprints of C. heveicola and C. chanthaburiensis type strains and 24% similarity with C. konsanensis (Fig. 1). Phylogenetic analysis of the concatenated ITS and D1/D2 sequence data showed that CBS 16213T and CBS 16555 clustered tightly together and form a lineage that is basal to a group of eight C. haemulonii species complex members, of which three (C. duobushaemulonii, C. pseudohaemulonii, and C. vulturna) are clinically relevant (Fig. 2).
Figure 1.
AFLP fingerprint analysis of the C. haemulonii species complex. An AFLP fingerprint analysis was initially performed to investigate if both C. khanbhai strains CBS 16213 and CBS 16555 (in bold) had similar fragment patterns as any of the other described species within the C. haemulonii species complex. C. khanbhai strains CBS 16213 and CBS 16555 had an 84% similarity, but had only 35% similarity compared with the AFLP fingerprints of C. heveicola and C. chanthaburiensis type strains and 24% similarity with C. konsanensis. Less than 20% similarity was observed with any of the other C. haemulonii species complex members.
Figure 2.
Phylogenetic analysis of the C. haemulonii species complex. Sequenced based analysis was done by using the aligned and concatenated internal transcribed spacer (ITS1-5.8S-ITS2) region and the D1/D2 region of the large subunit (LSU) of the ribosomal DNA for the type strains representing the members of the C. haemulonii species complex plus the here described C. khanbhai strains CBS 16213T and CBS 16555 (in bold). Clavispora lusitaniae (CBS 6936) was used as outgroup. The best model of substitution was observed to be TNe + G4 according to Bayesian Information Criterion (BIC) (18). Phylogenetic trees were estimated using maximum likelihood with IQ-tree v1.6.12 (19). The likelihood ratio test, Bayes inference, and ultrafast bootstrap values (1000 bootstraps) are in this order placed on the nodes of the tree.
Phenotypic characterization
Both clinical strains CBS 16213T and CBS 16555 did not show any morphological differences. Pictures of the type strain (CBS 16213T) are given in Figure 3A–C. On GYPA plates, colonies are off-white colored, glossy, soft, butyrous, and with an entire margin and do not change morphology from 7 days up to 1 month of incubation (Fig. 3A). After 24 h in YPD broth, cells were ovoid, ellipsoidal to spherical (1.5–3.5 × 2.5–4.5 μm in diameter) (Fig. 3B). After 7 days of growth on GYPA plates, cells are shaped similarly as in liquid culture, but the formation of a sub-population of enlarged cells that are up to 6 μm in diameter can be observed (Fig. 3C). Cells occur singly or in pairs and occasionally form short chains of up to three cells. Reproduction took place by monopolar budding (apical or sub-apical). (Pseudo)hyphae were not present on GYPA or MEA after up to 12 weeks of incubation. In addition, ascospore formation was not observed in pure cultures or when mixing both strains on GYPA and MEA plates at 25°C for up to 12 weeks. On CHROMagar Candida Plus, colonies of both strains appeared pale cream to lavender with a distinctive blue halo surrounding the colony (Fig. 3D3–4). This was the exact same phenotype as the type strain of C. auris (CBS 10913T, Fig. 3D2). Identification by MALDI–TOF MS did not result in any matched patterns (score values of 0.00–1.69). The closest hits were C. pseudohaemulonii (score value 1.56) for CBS 16213T and Clostridium cadaveris (score value 1.40) for CBS 16555. To this end, new MALDI-TOF MS main spectra (MSPs) were created for both strains, which can be added to the repository to allow future reliable identification by Bruker Daltonics MALDI–TOF MS. Finally, physiological characteristics were determined and are summarized in Table 2. Compared with close phylogenetic relatives, unique characteristics seem to be the delayed but positive fermentation of maltose, negative growth on 50%–60% glucose, tolerance to 0.1% cycloheximide and growth up to 42°C for both strains.
Figure 3.

Morphology of strain CBS 16213T. (A) Colony morphology on GYPA after incubation for 7 days. (B) Yeast cells from an overnight culture in YPD. (C) Yeast cells from a 7-day old culture on GYPA showing a sub-population of larger cells (arrows). (D) Colonies grown on CHROMagar Candida Plus: (1) C. haemulonii CBS 5149T, (2) C. auris CBS 10913T, (3) C. khanbhai CBS 16213T, (4) C. khanbhai CBS 16555. Bars, 1 cm for (A), 10 μm for (B, C).
Table 2.
Complete phenotypic characteristics of C. khanbhai strains CBS 16213T and CBS 16555, and their close phylogenetic relatives.
| Reference strains | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| C. khanbhai | |||||||||
| Characteristic | C. vulturna CBS 14366 | C. duobus-haemulonii CBS 7798 | C. pseudohaemulonii CBS 10004 | C. haemulonii CBS 5149 | C. konsanensis CBS 12666 | C. heveicola CBS 10701 | C. auris CBS 10913 | CBS 16213T | CBS 16555 |
| Fermentation | |||||||||
| Galactose | − | ND | ND | − | − | ND | − | − | − |
| Sucrose | + | + | ND | + | +/w | + | w | +, s | +, s |
| Maltose | − | ND | ND | − | − | w/d | − | +/d | +/d |
| Raffinose | + | + | ND | − | − | ND | − | w/d | +/d |
| Trehalose | + | ND | ND | w/d | ND | ND | w | + | + |
| Assimilation | |||||||||
| Inulin | + | +/− | − | − | w | − | w | + | + |
| Raffinose | + | + | + | + | − | + | + | + | + |
| Melibiose | − | −/v | − | − | − | − | − | −/w | − |
| Galactose | + | + | + | + | + | + | − | + | + |
| Melezitose | + | + | + | + | + | + | + | + | + |
| Methyl-α-d-glucoside | w/d/+ | −/w/d | v | − | + | w | − | + | + |
| Soluble starch | + | + | + | + | + | − | + | + | + |
| Cellobiose | −/+ | − | − | − | ND | − | − | w | + |
| Salicin | −/w/d | − | − | − | − | + | − | − | − |
| l-Sorbose | − | + | d | − | + | + | − | + | + |
| l-Rhamnose | + | + | ND | + | + | − | − | w/d | + |
| d-Xylose | −/w/d | d | d | d | W | D | − | + | + |
| l-Arabinose | + | −/w | v | − | D | − | − | + | + |
| d-Arabinose | −,w | d | − | − | W | ND | − | w | s |
| d-Ribose | − | −/w | + | D | D | − | − | + | + |
| Methanol | − | w/d | − | w/d | − | − | − | − | − |
| Ethanol | − | w/d | d | d | D | + | − | − | − |
| meso-Erythritol | − | −/d | − | − | − | − | − | w/d | + |
| Ribitol | + | + | + | d | + | + | w | + | + |
| Xylitol | + | + | + | w/d | − | ND | ND | + | + |
| Galactitol | ND | ND | + | ND | + | + | + | + | + |
| myo-Inositol | − | −/w | − | − | − | − | − | − | − |
| Gluconolactone | + | + | + | + | + | ND | ND | + | + |
| 2-Keto-d-gluconate | ND | ND | + | ND | + | ND | − | + | + |
| dl-Lactate | − | −/w/d | ND | − | − | − | − | − | −/w |
| Succinate | + | + | + | + | + | + | − | + | + |
| Citrate | + | + | + | + | + | + | + | − | − |
| d-Gluconate | + | + | + | + | + | ND | − | + | + |
| d-Glucosamine | + | + | + | − | + | + | − | + | + |
| Nitrate | w | − | − | − | − | − | − | − | − |
| Nitrite | w | − | − | − | − | − | − | − | − |
| Ethylamine | − | + | + | + | + | + | − | + | + |
| Cadaverine | − | + | + | + | + | + | + | + | + |
| d-Tryptophan | + | ND | ND | − | ND | ND | ND | − | − |
| Pepton | ND | ND | ND | ND | ND | ND | ND | + | + |
| Growth on YM agar at: | |||||||||
| 4°C | ND | ND | ND | ND | ND | ND | ND | w | w |
| 10°C | ND | ND | ND | ND | ND | ND | ND | s | + |
| 30°C | + | + | + | + | + | + | + | + | + |
| 37°C | + | + | + | + | + | + | + | + | + |
| 40°C | ND | − | − | − | − | − | + | + | + |
| 42°C | ND | − | − | − | − | − | w/s | w/s | w/s |
| Other phenotypic tests: | |||||||||
| Tolerance to NaCl (10% w/v) | ND | + | ND | + | + | ND | + | + | + |
| Osmotolerance (50% glucose, w/w) | + | + | ND | + | + | ND | + | − | − |
| Osmotolerance (60% glucose, w/w) | + | + | ND | + | w | ND | ND | − | − |
| Tolerance to cycloheximide 0.01% (w/v) | − | + | ND | + | + | ND | − | + | + |
| Tolerance to cycloheximide 0.1% (w/v) | ND | ND | ND | ND | − | ND | − | + | + |
| Starch production | ND | ND | ND | ND | ND | − | ND | − | − |
| DBB reaction | ND | ND | ND | ND | ND | − | ND | − | − |
| Hydrolysis of urea | ND | ND | − | ND | − | − | − | − | − |
Data for the reference strains originates from references5,8−10,24 and the CBS collection database (https://wi.knaw.nl/page/Collection). All strains were positive for the fermentation of glucose, and assimilation of d-glucose, sucrose, trehalose, maltose, glycerol, d-mannitol, d-glucitol, and l-lysine, and negative for fermentation of lactose. All strains grow at a temperature of 25 °C. Scoring system according to Kurtzman et al. (20): +, positive; −, negative; d, delayed positive (latent); s, slowly positive; w, weakly positive; v, variable; ND, no data available.
Antifungal susceptibilities
The in vitro antifungal susceptibility of both strains was tested for AMB, 5FC, multiple azoles and two echinocandins according to EUCAST standards. Table 3 shows the MIC results. Both strains had elevated MICs for AMB (2–4 μg/ml). Increased MICs were also observed for most of the azoles, especially after 48 h. Notably, CBS 16213T is sensitive to 5FC (0.03–0.125 μg/ml), whereas CBS 16555 has a strongly increased MIC value for 5FC (4–16 μg/ml). Both strains were sensitive to ANI and MCF. However, again moderately elevated MICs for ANI (0.5–1 μg/ml) and MCF (0.25–0.5 μg/ml) were observed for CBS 16555 compared with CBS 16213T.
Table 3.
Antifungal susceptibility profiles of strains CBS 16213T and CBS 16555 characterized in this study
| MIC (μg/ml)a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | AMB | 5FC | ANI | MCF | FLU | ISA | ITR | POS | VOR | |
| CBS16213T | 24h | 2 | 0.03 | 0.125 | 0.06 | 16 | 4 | 4 | 0.5 | 0.125 |
| 48h | 4 | 0.125 | 0.5 | 0.125 | 32 | 4 | 8 | >16 | 2 | |
| CBS16555 | 24h | 2 | 4 | 0.5 | 0.25 | >64 | 0.125 | 1 | 2 | 0.5 |
| 48h | 4 | 16 | 1 | 0.5 | >64 | 8 | 16 | 4 | 16 | |
aMinimal inhibitory concentration (MIC) to: Amphotericin B (AMB), 5-flucytosine (5FC), anidulafungin (ANI), micafungin (MCF), fluconazole (FLU), isavuconazole (ISA), itraconazole (ITR), posaconazole (POS), and voriconazole (VOR).
Discussion
The two new clinical yeast strains in this study came from geographically distinct places. They were isolated from patients in Kuwait (CBS 16213T) and Malaysia (CBS 16555). Nevertheless, AFLP fingerprint analysis and phylogenetic analysis showed that both strains cluster together and represent a distinct novel species with a close relationship to members of the C. haemulonii complex (Figs. 1 and 2). Although the clinical record of the holotype (CBS 16213) only indicates colonization of the patient, the second strain (CBS 16555) came from a blood culture. The patient infected with strain CBS 16555 passed away despite the use of intravenous fluconazole, suggesting a serious case of resistant candidemia caused by the new species introduced as C. khanbhai below. Antifungal susceptibility testing indeed showed reduced sensitivity to multiple antifungal agents (Table 3). When the tentative MIC breakpoints given for C. auris by the CDC are used as references, both strains are resistant to AMB, and at least CBS 16555 is resistant to FLU.22 In addition, both strains also showed increased MICs for the triazoles tested. A steep increase in MIC after 48 h of incubation for the triazoles indicated a trailing effect (Table 3). The trailing effect is defined as reduced but persistent visible growth of Candida species at azole concentrations above the MIC.23 Since both C. khanbhai strains did not show poor growth after 24 h it is recommended to read the MIC results after 24 h according to the EUCAST protocol, to avoid overestimation of the MIC.21 However, to evaluate the clinical impact of trailing it could be useful to check the MIC of future clinical isolates also after 48 h. The observed multi-resistance pattern is the same as described for other closely related members of the C. haemulonii species complex.5 This resistance together with the ability to infect humans make correct and rapid identification of C. khanbhai essential. Notably, standard biochemical identification systems such as MALDI–TOF MS, VITEK 2, and API 20C AUX misidentified this new species for C. pseudohaemulonii, C. duobushaemulonii, and C. lusitaniae, respectively. Hence, databases should first be updated to enable proper identification of this new pathogenic yeast species. The new CHROMagar Candida plus (CCP) is seen as a cost-effective and easy to use alternative to differentiate C. auris from its close relatives and more common Candida pathogens.23 However, both C. khanbhai strains have the exact same phenotype as C. auris on this medium (Fig. 1D). Therefore, future infections by C. khanbhai are likely to be misidentified as C. auris when using CCP. Although C. khanbhai and C. auris do share the same phenotype on chromogenic agar and the important trait of growth at 42°C, they have significantly different assimilation patters, which could be used to develop future differentiation assays (Table 2).
Taken together with the description of C. khanbhai we add a relevant clinical Candida species to the growing list of emerging fungal pathogens. The isolation from clinical sources at geographically distinct locations together with a worrisome multidrug resistance pattern and misidentification by commonly used laboratory methods shows many similarities with the story of C. auris.6 Therefore, monitoring and correct identification of this species are essential.
Description of Candida khanbhai A.W. de Jong and F. Hagen sp. nov.
Candida khanbhai (khan.bhai, ‘khan’ referring to the surname of Prof. Dr. Ziauddin Khan who was a medical mycologist, and ‘bhai’ referring to the Hindi word for ‘friend’ and ‘brother’). MycoBank number: MB 846114. Holotype: CBS 16213 preserved in a metabolically inactive state at the CBS culture collection hosted at the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. Ex-type cultures: CBS 16213, Kw2195/19, 2MG-A0803-17.
On GYPA after 7 days of incubation at 25°C the colonies are off-white colored, glossy, soft, butyrous, and with an entire margin (Fig. 3A). In YPD broth after 1 day of incubation cells are ovoid, ellipsoidal to spherical (1.5–3.5 × 2.5–4.5 μm in diameter), occur singly or in pairs and reproduce by monopolar budding (apical or sub-apical) (Fig. 3B,C). On GYPA and MEA (pseudo)hyphae are not formed. Sporulation is not observed in pure cultures or when mixing both strains on GYPA and MEA at 25°C for up to 12 weeks. A summary of the fermentation tests, assimilation tests, and other growth characteristics of strains CBS 16213T and CBS 16555 is provided in Table 2. MIC of the main classes of antifungal drugs are given in Table 3. The holotype CBS 16213 was isolated in 2019 from a nasal swab of a 73-year-old patient hospitalized in Mubarak Al-Kabeer Hospital (Jabriya, Kuwait) and is permanently preserved in a metabolically inactive state in the CBS yeast collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands.
In addition to the description of C. khanbhai it was observed that several members of the C. haemulonii species complex were invalidly described. Hence, the opportunity was taken to correct this here, formally validating the names:
Candida auris Satoh and Makimura ex F. Hagen sp. nov. MB 846663. For a detailed description see Satoh et al., Microbiol. Immunol. 53 (1): 43 (2009) (reference 9). Holotype: CBS 10913 preserved in a metabolically inactive state. Ex-type cultures: JCM 15448; CBS 10913; DSM 21092. [originally described as: Candida auris Satoh and Makimura, Microbiol. Immunol. 53 (1): 43 (2009), nom. inval., Art. 40.7 (Shenzhen)].
Candida vulturna Sipiczki and Tap ex F. Hagen sp. nov. MB 846664. For a detailed description see Sipiczki and Tap, Int. J. Syst. Evol. Microbiol. 66 (10): 4014 (2016) (reference 8). Holotype: CBS 14366 preserved in a metabolically inactive state. Ex-type cultures: CBS 14366; 11-1170; NCAIM-Y02177; CCY 094-001-001. [originally described as: Candida vulturna Sipiczki and Tap, Int. J. Syst. Evol. Microbiol. 66: 4014 (2016), nom. inval., Arts Art. 36.1(b) and 40.7 (Shenzhen)].
Candida metrosideri Klaps, C. Vega, C.M. Herrera, Junker, B. Lievens and Álvarez-Pérez ex F. Hagen sp. nov. MB 846665. For a detailed description see Klaps et al., PLoS One 15(10): e0240093 (2020) (reference 26). Holotype: CBS 16091 preserved in a metabolically inactive state. Ex-type cultures: CBS 16091; JK22; MUCL 57821. [originally described as: Candida metrosideri Klaps, C. Vega, C.M. Herrera, Junker, B. Lievens, Álvarez-Pérez, PLoS One 15 (10): e0240093 (2020), nom. inval., Art. 36.1(a) (Shenzhen)].
Candida ohialehuae Klaps, C. Vega, C.M. Herrera, Junker, B. Lievens, Álvarez-Pérez ex F. Hagen sp. nov. MB 846666. For a detailed description see Klaps et al., PLoS One 15(10): e0240093 (2020) (reference 26). Holotype: CBS 16092 preserved in a metabolically inactive state. Ex-type cultures: CBS 16092; JK58.2; MUCL 57822. [originally described as: Candida ohialehuae Klaps, C. Vega, C.M. Herrera, Junker, B. Lievens, Álvarez-Pérez, PLoS One 15 (10): e0240093 (2020), nom. inval., Art. 36.1(a) (Shenzhen)].
Candida chanthaburiensis Limtong and Yongman. ex F. Hagen sp. nov. MB 846667. For a detailed description see Limtong and Yongmanitchai, Antonie van Leeuwenhoek 98: 383 (2010) (reference 27). Holotype: CBS 10926 preserved in a metabolic inactive state. Ex-type cultures: CBS 10926; EM33; NBRC 102176. [originally described as: Candida chanthaburiensis Limtong and Yongman., Antonie van Leeuwenhoek 98 (3): 383 (2010), nom. inval., Art. 40.7 (Shenzhen)]
Candida konsanensis Sarawan, Mahakhan, Jindam., K. Vichitph., S. Vichitph, Sawaengk. ex F. Hagen sp. nov. MB 846668. For a detailed description, see Sarawan et al., World J Microbiol Biotechnol. 29: 1483 (2013) (reference 25). Holotype: CBS 12666 preserved in a metabolic inactive state. Ex-type cultures: CBS 12666; KKU-FW10; BCC 52588; NBRC 109082. [originally described as: Candida konsanensis Sarawan, Mahakhan, Jindam., K. Vichitph., S. Vichitph. & Sawaengk., World J. Microbiol. Biotechnol. 29: 1483 (2013), nom. inval., Arts 40.7, F.5.1 (Shenzhen)]
Although C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii were validly described, these names were found to be linguistically incorrect. Several orthographic variants were introduced, with the ‘haemulonii’ part of these epithets also written as ‘haemuloni’ and ‘haemulonis’. We were aware of a discussion between taxonomists—with expertise in Greek and Latin language—which has resulted in a more drastic name change as they proposed to change the epithets of C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii into C. haemuli, C. duobushaemuli, and C. pseudohaemuli (see https://www.mycobank.org/). However, we argue that this and previously proposed minor name corrections are all purely linguistic driven and do not serve the clinical and diagnostic community. Hence, we advocate to keep the naming of C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii as they were originally described up to four decades ago.5,10,28 Moreover, these species epithets are familiar for clinicians, diagnostic laboratory staff, and epidemiologists since then.24,29–33
Acknowledgements
We thank Konstanze Bensch for her valuable input related to the taxonomic status of the here described novel species and formalizing the invalidly described species. The Centers for Disease Control & Prevention (Atlanta, GA, USA) is acknowledged for providing majority of the Candida auris reference strains via the CDC Isolate Bank.
Contributor Information
Auke W de Jong, Department of Medical Mycology, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; Institute for Biodiversity and Ecosystem Dynamics (IBED), University of Amsterdam, Amsterdam, The Netherlands.
Khaled Al-Obaid, Department of Microbiology, Mubarak Al-Kabir Hospital, Jabriya, Kuwait.
Ratna Mohd Tap, Mycology Section, Bacteriology Unit, Infectious Disease Research Centre, Institute for Medical Research, National Institutes of Health, Selangor, Malaysia.
Bert Gerrits van den Ende, Department of Medical Mycology, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands.
Marizeth Groenewald, Department of Medical Mycology, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands.
Leena Joseph, Department of Microbiology, Faculty of Medicine, Kuwait University, Jabriya, Kuwait.
Suhail Ahmad, Department of Microbiology, Faculty of Medicine, Kuwait University, Jabriya, Kuwait.
Ferry Hagen, Department of Medical Mycology, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; Institute for Biodiversity and Ecosystem Dynamics (IBED), University of Amsterdam, Amsterdam, The Netherlands; Department of Medical Microbiology, University Medical Center Utrecht, Utrecht, The Netherlands.
Conflict of interest
The authors declared no conflict of interest.
Data availability
Yeast strains used in this study have been deposited in the CBS culture collection (hosted at the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands), or are available via the CDC Isolate Bank (hosted at the Centers for Disease Control & Prevention, Atlanta, GA, USA). The generated sequences have been deposited in the NCBI GenBank repository. The strains and sequence data accession numbers are indicated throughout the manuscript, all accession numbers are provided in Figure 2. The MALDI TOF MS main spectra (MSP) of CBS 16213 and CBS 16555 have been added to the CDC MicrobeNet online identification tool.
(CRediT)
A.W.J.: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing—original draft, writing—review and editing. K.A.: conceptualization, resources, validation, writing—review and editing. R.M.T.: conceptualization, resources, validation, writing—review and editing. B.G.E.: conceptualization, data curation, investigation, methodology, software, validation, writing—review and editing. M.G.: conceptualization, data curation, investigation, methodology, resources, validation, writing—review and editing. L.J.: conceptualization, resources, validation, writing—review and editing. S.A.: conceptualization, resources, validation, writing—review and editing. F.H.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing—original draft, writing—review and editing.
References
- 1. Stavrou AA, Lackner M, Lass-Flörl C, Boekhout T.. The changing spectrum of Saccharomycotina yeasts causing candidemia: phylogeny mirrors antifungal susceptibility patterns for azole drugs and amphothericin B. FEMS Yeast Res. 2019; 19: foz037. [DOI] [PubMed] [Google Scholar]
- 2. Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ.. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 2018; 360: 739–742. [DOI] [PubMed] [Google Scholar]
- 3. Garcia-Solache MA, Casadevall A.. Global warming will bring new fungal diseases for mammals. mBio. 2010; 1: e00061–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geddes-McAlister J, Shapiro RS.. New pathogens, new tricks: emerging, drug-resistant fungal pathogens and future prospects for antifungal therapeutics: drug-resistant fungal pathogens. Ann N Y Acad Sci. 2019; 1435: 57–78. [DOI] [PubMed] [Google Scholar]
- 5. Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo Aet al. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: Three multiresistant human pathogenic yeasts. J Clin Microbiol. 2012; 50: 3641–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Jong AW, Hagen F.. Attack, defend and persist: How the fungal pathogen Candida auris was able to emerge globally in healthcare environments. Mycopathologia. 2019; 184: 353–365. [DOI] [PubMed] [Google Scholar]
- 7. Gade L, Muñoz JF, Sheth Met al. Understanding the emergence of multidrug-resistant Candida: using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front Genet. 2020; 11: 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sipiczki M, Tap RM.. Candida vulturna pro tempore sp. nov., a dimorphic yeast species related to the Candida haemulonis species complex isolated from flowers and clinical sample. Int J Syst Evol Microbiol. 2016; 66: 4009–4015. [DOI] [PubMed] [Google Scholar]
- 9. Satoh K, Makimura K, Hasumi Yet al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009; 53: 41–44. [DOI] [PubMed] [Google Scholar]
- 10. Sugita T, Takashima M, Poonwan N, Mekha N.. Candida pseudohaemulonii sp. nov., an amphotericin B-and azole-resistant yeast species, isolated from the blood of a patient from Thailand. Microbiol Immunol. 2006; 50: 469–473. [DOI] [PubMed] [Google Scholar]
- 11. Arora P, Singh P, Wang Yet al. Environmental isolation of Candida auris from the coastal wetlands of Andaman Islands, India. mBio. 2021; 12: e03181–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Escandón P. Novel environmental niches for Candida auris: Isolation from a coastal habitat in Colombia. J Fungi. 2022; 8: 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Hoog GS, van den Ende AG.. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses. 1998; 41: 183–189. [DOI] [PubMed] [Google Scholar]
- 14. Prakash A, Sharma C, Singh Aet al. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect. 2016; 22: 277.e1–277.e9. [DOI] [PubMed] [Google Scholar]
- 15. Leaw SN, Chang HC, Sun HFet al. Identification of medically important yeast species by sequence analysis of the Internal transcribed spacer regions. J Clin Microbiol. 2006; 44: 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vaidya G, Lohman DJ, Meier R.. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011; 27: 171–180. [DOI] [PubMed] [Google Scholar]
- 17. Katoh K. MAFFT: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002; 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 2017; 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018; 35: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kurtzman CP, Fell JW, Boekhout T, Robert V.. Methods for isolation, phenotypic characterization and maintenance of yeasts. The Yeasts Elsevier 87–110. 2011. doi:10.1016/B978-0-444-52149-1.00007-0
- 21. Arendrup MC, Meletiadis J, Mouton JWet al. EUCAST definitive document E.DEF 7.3.2. Published online2020; 2020: 21. [Google Scholar]
- 22. Centers for Disease Control and Prevention . Antifungal susceptibility testing and interpretation. https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html. Published 2018. Accessed November 2022.
- 23. Marcos-Zambrano LJ, Escribano P, Sánchez-Carrillo C, Bouza E, Guinea J.. Scope and frequency of fluconazole trailing assessed using EUCAST in invasive Candida spp. isolates. Med Mycol. 2016; 54: 733–739. [DOI] [PubMed] [Google Scholar]
- 24. de Jong AW, Dieleman C, Carbia M, Mohd Tap R, Hagen F.. Performance of two novel chromogenic media for the identification of multidrug-resistant Candida auris compared with other commercially available formulations. J Clin Microbiol. 2021; 59: e03220–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarawan S, Mahakhan P, Jindamorakot Set al. Candida konsanensis sp. nov., a new yeast species isolated from Jasminum adenophyllum in Thailand with potentially carboxymethyl cellulase-producing capability. World J Microbiol Biotechnol. 2013; 29: 1481–1486. [DOI] [PubMed] [Google Scholar]
- 26. Klaps J, de Vega C, Herrera CMet al. Candida metrosideri pro tempore sp. nov. and Candida ohialehuae pro tempore sp. nov., two antifungal-resistant yeasts associated with Metrosideros polymorpha flowers in Hawaii. PLOS ONE. 2020; 15: e0240093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Limtong S, Yongmanitchai W.. Candida chanthaburiensis sp. nov., Candida kungkrabaensis sp. nov., and Candida suratensis sp. nov., three novel yeast species from decaying plant materials submerged in water of mangrove forests. Antonie Van Leeuwenhoek. 2010; 98: 379–388. [DOI] [PubMed] [Google Scholar]
- 28. Yarrow D, Meyer SA.. Proposal for amendment of the diagnosis of the genus Candida Berkhout nom. cons. Int J System Bacteriol. 1978; 28: 611–615. [Google Scholar]
- 29. Gade L, Muñoz JF, Sheth Met al. Understanding the emergence of multidrug-resistant Candida: Using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front Genet. 2020; 11: 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar A, Prakash A, Singh Aet al. Candida haemulonii species complex: an emerging species in India and its genetic diversity assessed with multilocus sequence and amplified fragment-length polymorphism analyses. Emerg Microbes Infect. 2016; 5: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramos R, Caceres DH, Perez Met al. Red Nacional de Vigilancia Epidemiologica en Microbiologia Clinica; Vallabhaneni S.. Emerging multidrug-resistant Candida duobushaemulonii infections in Panama hospitals: Importance of laboratory surveillance and accurate identification. J Clin Microbiol. 2018; 56: e00371–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramos LS, Figueiredo-Carvalho MH, Barbedo LSet al. Candida haemulonii complex: species identification and antifungal susceptibility profiles of clinical isolates from Brazil. J Antimicrob Chemother. 2015; 70: 111–115. [DOI] [PubMed] [Google Scholar]
- 33. Shin JH, Kim MN, Jang SJet al. Detection of amphotericin B resistance in Candida haemulonii and closely related species by use of the Etest, Vitek-2 yeast susceptibility system, and CLSI and EUCAST broth microdilution methods. J Clin Microbiol. 2012; 50: 1852–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Yeast strains used in this study have been deposited in the CBS culture collection (hosted at the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands), or are available via the CDC Isolate Bank (hosted at the Centers for Disease Control & Prevention, Atlanta, GA, USA). The generated sequences have been deposited in the NCBI GenBank repository. The strains and sequence data accession numbers are indicated throughout the manuscript, all accession numbers are provided in Figure 2. The MALDI TOF MS main spectra (MSP) of CBS 16213 and CBS 16555 have been added to the CDC MicrobeNet online identification tool.