Abstract
High-risk individuals (HRIs) with familial and genetic predisposition to pancreatic ductal adenocarcinoma (PDAC) are eligible for screening. There is no accurate biomarker for detecting early-stage PDAC. We previously demonstrated that a panel of methylated DNA markers (MDMs) accurately detect sporadic PDAC. In this study we compared the distribution of MDMs in DNA extracted from tissue of PDAC cases who carry germline mutations and non-carriers with family history, with control tissue and demonstrate high discrimination like that seen in sporadic PDAC. These results provide scientific rationale for examining plasma MDMs in HRIs with the goal of developing a minimally-invasive early detection test.
Keywords: Pancreatic cancer, DNA methylation, Biomarker
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a lethal disease, and most patients are diagnosed at an advanced stage. Although screening for PDAC is not recommended in the average-risk population, for certain high-risk individuals (HRIs) who are either genetically predisposed or belong to familial PDAC kindreds, such screening is considered best practice [1-3]. Currently, magnetic resonance imaging (MRI) and endoscopic ultrasound (EUS) are preferred screening modalities. To improve clinical decision making, there is a need to develop blood-based biomarkers that can accurately detect PDAC in HRIs. In previous efforts, we have identified and validated methylated DNA markers (MDMs) that distinguish sporadic PDAC from normal pancreas; performance of these MDMs in HRIs has not been previously assessed [4-6]. In this study, we aimed to compare distributions of tissue extracted MDMs from PDAC patients with and without predisposing germline mutations and PDAC family history. We hypothesized that the MDM distributions in both groups will demonstrate high discrimination from control tissue samples, and if established as comparable to sporadic PDAC, would support future testing of this candidate marker panel in blood-based early detection applications in HRIs.
2. Methods
The study was approved by the Mayo Clinic Institutional Review Board (IRB) and research biospecimens were obtained from the Mayo Clinic Biospecimen Resource for Pancreas Research (P50 CA102701). Genetic test results were clinical genetic testing when available, and/or derived from the Mayo Clinic Cancer Risk Estimates Related to Susceptibility Genes (CARRIERS) study research protocol in which variant carrier status of PDAC probands was based on 21 cancer-susceptibility genes [7]. Paraffin-embedded case tissues were obtained from patients in 3 groups: Group 1 (Mutation): PDAC with germline genetic mutations in cancer-susceptibility genes (Table 1); Group 2 (FamHx): family history of PDAC in one or more first-degree relatives (FDR) and mutation-negative; and Group 3 (Sporadic): age- and gender-balanced (relative to the combined distribution of Group 1 and Group 2) mutation-negative and family history-negative sporadic PDAC. Paired distant normal pancreas tissues from case patients when available were used as control. Tissues were macro-dissected, and histology reviewed and confirmed by an expert pathologist (R.P.G). Tissue samples were randomized, and blinded. DNA was purified using the QIAamp DNA FFPE Tissue protocol (Qiagen GmbH, Hilden, Germany) and bisulfite converted with Zymo EZ DNA Methylation reagents (Zymo Research, Irvine, CA). Samples were tested by qPCR to ensure sufficient genomic equivalents and marker selection was based on our previously published results, targeting MDMs that demonstrated discrimination in comparisons of PDAC and non-cancer controls. A panel of 19 candidate MDMs (NDRG4, BMP3, ADCY1, C13orf18, GRIN2D, ELMO1, IGF2BP1, CD1D, ZNF781, FER1L4, RYR2, CLEC11A, AK055957, GH05J042948, HOXA1, PRKCB, SHISA9, NTRK3, TBX15) was assayed from 10 ng (per MDM) using methylation specific PCR and marker levels were normalized to ACTB.
Table 1.
Distribution of germline mutations in PDAC patients harboring a pathogenic germline mutation (Mutation).
| Gene Name | Frequency |
|---|---|
| BRCA2 | 13 |
| ATM | 9 |
| FANCC | 3 |
| CHEK2 | 2 |
| ERCC2 | 2 |
| FANCM | 2 |
| PPM1D | 2 |
| APC | 1 |
| BLM | 1 |
| BRCA1 | 1 |
| EPCAM | 1 |
| ERCC3 | 1 |
| FANCC + PPM1D | 1 |
| MRE11A | 1 |
| MUTYH | 1 |
| NF1 | 1 |
| PALB2 | 1 |
| PRSS1 | 1 |
| RAD50 | 1 |
| RAD51C | 1 |
| RECQL | 1 |
| RINT1 | 1 |
A series of uncorrelated linear combination of MDMs was developed on sporadic case and control tissues using principal component analysis (PCA). By definition, the first linear combination captures the highest proportion of total variance in the data, with subsequent uncorrelated linear combinations adding incremental improvements, and was calculated across all case and control data to create a “PCA score” per patient. Discriminate accuracy of the PCA score was depicted using boxplots and receiver operator characteristic curves (ROC) and summarized using the area under the ROC curve (AUC) with corresponding 95% confidence intervals. A minimum of 15 cases and 15 controls per disposition subtype was targeted to provide 80% power to detect an AUC of 0.75 or higher relative to chance discrimination using a one-sided significance level of 0.05. Based on tissue availability and adequacy of DNA recovery, the final set of analyzable data was not limited to patients with paired cancer and control tissue; results presented are based on unpaired data analyses to maximize power given the estimated relative efficiency of a paired analysis relative to an unpaired analysis was approximately 97.5%.
3. Results
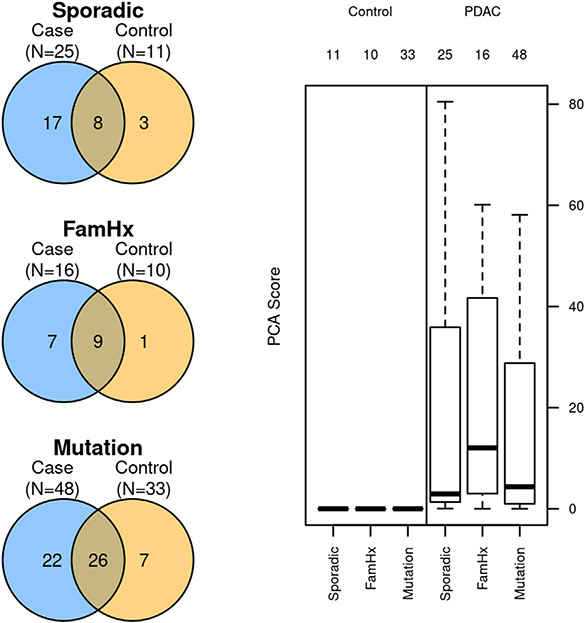
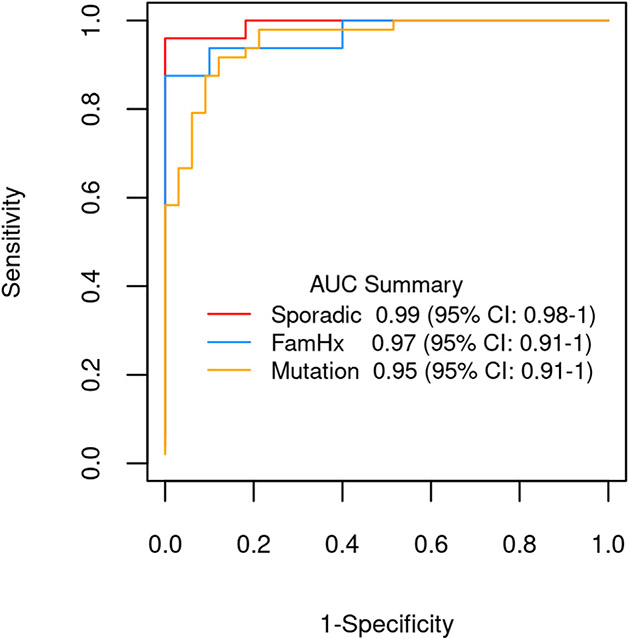
Group 1 (Mutation) (n = 48 case and 33 control tissues; median age 69, 44% male) comprised PDAC patients with known pathogenic germline mutation (BRCA1/BRCA2 (14), other genes (34) Table 1); Group 2 (FamHx) (n = 16 case and 10 control tissues, median age 70, 47% male) included germline mutation negative PDAC patients with family history of PDAC in at least one FDR and Group 3 (Sporadic) (n = 25 case and 11 control tissues, median age 65, 52% male) included PDAC patients negative for both germline mutation and family history. The median AUC for distinguishing case from control tissue was 0.88 (IQR: 0.82—0.92), 0.92 (IQR: 0.86—0.96) and 0.92 (IQR: 0.84—0.96) in groups 1,2 and 3 respectively. The composite PCA score captured 61% of the total variance observed across the 19 MDM panel and was higher in case tissue compared to control (all p < 0.001, Fig. 1) with AUCs ranging from 0.95 (0.91—1) to 0.99 (0.98—1) (Fig. 2). No statistically significant pairwise differences of discrimination were observed between the 3 groups (p > 0.1 for all comparisons). When subjects were grouped based on mutations in ATM, APC, BRCA, EPCAM, PALB2, and PRSS1 (19 controls 27 cases) vs other mutations (14 controls and 21 cases) the AUC of the PCA score was 0.96 (0.92—1) and 0.95 (0.81—1), respectively, and not significantly different (p = 0.75). However, 3 individual MDMs (namely, NDRG4, BMP3, and ADCY1) did show depressed signal in these mutations compared to subjects with other mutations.
Fig. 1.
Left: Case and control tissue distribution in the study groups (Sporadic, Family History and Mutation) with overlapping area indicating paired case and control tissue availability with adequate DNA recovery. Right: Composite principal components analysis (PCA) score comparing sporadic, familial, and mutation-positive pancreatic ductal adenocarcinoma (PDAC) case groups to distant normal pancreatic tissue.
Fig. 2.
Comparative AUCs for discrimination of pancreatic ductal adenocarcinoma (PDAC) from control tissue using the PCA score derived from the methylated DNA marker panel assayed on tissue-extracted DNA from cases and controls from three PDAC groups-sporadic, familial (FamHx) and in those harboring a pathogenic germline mutation (Mutation).
4. Discussion
In this study, we demonstrate that tissue MDM profile in PDAC patients with either germline mutations or family history of PDAC, accurately distinguishes cancer from normal tissue, and the discrimination is comparable to that in sporadic PDAC.
There is expert consensus that HRIs should be considered for PDAC screening [2,8]. Although HRIs are the intended use population for any PDAC screening biomarker test there are several factors that limit discovery and validation of biomarkers specifically in this population. Despite increased lifetime risk of PDAC in HRIs, the number of PDAC events in a prospectively followed HRI cohort is small and the diagnostic signal for any given blood-based biomarker fades in the pre-diagnostic phase [9,10]. Hence, testing diagnostic performance of a pancreatic cancer early detection biomarker in a HRI cohort will require a longitudinal serial sample collection at set time intervals. Although this has been recognized as a critical gap in the early detection paradigm and several efforts are currently underway to assemble these longitudinal serial sampling high-risk cohorts, availability of pre-diagnostic blood samples from HRIs are limited for discovery efforts or feasibility testing for novel biomarker applications [11,12]. This highlights the importance of demonstrating proof-of-concept and feasibility in tissue prior to blood-based evaluation. Thus, our study findings provide scientific rationale and support for exploring MDMs in blood from HRIs in concert with discovery in sporadic PDAC, with the eventual goal of developing a clinically applicable blood-based early detection test.
Declaration of competing interest
Mayo Clinic and Exact Sciences have an intellectual property development agreement. Drs. Majumder & Kisiel and Messrs. Taylor & Mahoney are listed as inventors under this agreement and could share potential future royalties paid to Mayo Clinic. The other authors of this manuscript have no conflicts of interest to declare. The abstract summarizing results from this study was presented at Virtual Digestive Disease Week 2021. Support was provided by the Centene Charitable Foundation (to SM and GMP), CA102701 (to GMP), CA214679 (to JBK), and a sponsored research agreement between Mayo Clinic and Exact Sciences. Additional support from Tissue Registry, Biospecimens Accession & Processing, and Pathology Research Core shared recourses of the Mayo Clinic Cancer Center Support Grant (P30 CA015083).
Abbreviations:
- PDAC
Pancreatic ductal adenocarcinoma
- MDM
Methylated DNA marker
- CA 19-9
Carbohydrate antigen 19-9
References
- [1].Owens DK, Davidson KW, Krist AH, et al. Screening for pancreatic cancer: US preventive services task force reaffirmation recommendation statement. JAMA 2019;322:438–44. [DOI] [PubMed] [Google Scholar]
- [2].Goggins M, Overbeek KA, Brand R, et al. Management of patients with increased risk for familial pancreatic cancer: updated recommendations from the International Cancer of the Pancreas Screening (CAPS) Consortium. Gut 2020;69:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aslanian HR, Lee JH, Canto MI. AGA clinical practice update on pancreas cancer screening in high-risk individuals: expert review. Gastroenterology 2020;159:358–62. [DOI] [PubMed] [Google Scholar]
- [4].Kisiel JB, Raimondo M, Taylor WR, et al. New DNA methylation markers for pancreatic cancer: discovery, tissue validation, and pilot testing in pancreatic juice. Clin Cancer Res 2015;21:4473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Majumder S, Taylor WR, Yab TC, et al. Novel methylated DNA markers discriminate advanced neoplasia in pancreatic cysts: marker discovery, tissue validation, and cyst fluid testing. Am J Gastroenterol 2019;114:1539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Majumder S, Taylor WR, Foote PH, et al. High detection rates of pancreatic cancer across stages by plasma assay of novel methylated DNA markers and CA19-9. Clin Cancer Res 2021;27:2523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hu C, Hart SN, Polley EC, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA 2018;319:2401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sawhney MS, Calderwood AH, Thosani NC, et al. ASGE guideline on screening for pancreatic cancer in individuals with genetic susceptibility: summary and recommendations. Gastrointest Endosc 2022;95:817–26. [DOI] [PubMed] [Google Scholar]
- [9].Canto MI, Almario JA, Schulick RD, et al. Risk of neoplastic progression in individuals at high risk for pancreatic cancer undergoing long-term surveillance. Gastroenterology 2018;155:740–51. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Udgata S, Takenaka N, Bamlet WR, et al. THBS2/CA19-9 detecting pancreatic ductal adenocarcinoma at diagnosis underperforms in prediagnostic detection: implications for biomarker advancement. Cancer Prev Res 2021;14:223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gonda TA, Everett JN, Wallace M, et al. Recommendations for a more organized and effective approach to the early detection of pancreatic cancer from the PRECEDE (pancreatic cancer early detection) consortium. Gastroenterology 2021;161:1751–7. [DOI] [PubMed] [Google Scholar]
- [12].Tanaka H, Tamura K, Abe T, et al. Serum carboxypeptidase activity and genotype-stratified CA19-9 to detect early-stage pancreatic cancer. Clin Gastroenterol Hepatol 2021;S1542-3565:01094–6. 10.1016/j.cgh.2021.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]