CONSPECTUS:
DNA G-quadruplex secondary structures formed in guanine-rich human telomeres and oncogene promoters are functionally important and have emerged as a promising new class of cancer-specific drug targets. These globular intramolecular structures are stabilized by K+ or Na+ and form readily under physiological solution conditions. Moreover, G-quadruplexes are epigenetic features and can alter chromatin structure and function together with interactive proteins. Here, we discuss our efforts over the last two decades to understand the structures and functions of DNA G-quadruplexes formed in key oncogene promoters and human telomeres and their interactions with small molecules. Using high-field NMR spectroscopy, we determined the high-resolution structures of physiologically relevant telomeric G-quadruplexes in K+ solution with a major form (hybrid-2) and a minor form (hybrid-1), as well as a two-tetrad intermediate. The intrinsic structural polymorphism of telomeric DNA may be important for the biology of human telomeres, and we proposed a model for the interconversion. More recently, we have worked on G-quadruplexes of MYC, BCL2, PDGFR-β, VEGF, and k-RAS oncogene promoters. We determined the structure of the major G-quadruplex formed in the MYC promoter, a prototype for parallel G-quadruplexes. It is the first example of the parallel-stranded G3NG3 structure motif with a 1-nt loop, which is prevalent in promoter sequences and likely evolutionarily selected to initiate folding. Remarkably, the parallel MYC promoter G-quadruplexes are highly stable. Additionally, we determined the molecular structures of G-quadruplexes formed in human BCL2, VEGF, and PDGFR-β promoters, each adopting a unique structure. For example, the BCL2 promoter contains distinct interchangeable G-quadruplexes in two adjacent regions, suggesting precise regulation by different proteins. The PDGFR-β promoter adopts unique “broken-strand” and vacancy G-quadruplexes, which can be recognized by cellular guanine metabolites for a potential regulatory role.
Structural information on G-quadruplexes in complex with small-molecules is critical for understanding specific recognition and structure-based rational drug design. Our studies show that many G-quadruplexes contain unique structural features such as capping and loop structures, allowing specific recognition by drugs and protein. This represents a paradigm shift in understanding DNA as a drug target: Rather than a uniform, nonselective binding site in duplex DNA, the G-quadruplex is being pursued as a new class of selectively targetable drug receptors. We focus on targeting the biologically relevant MYC promoter G-quadruplex (MycG4) with small molecules and have determined its first and additional drug complex structures. Very recently, we have discovered clinically tested indenoisoquinolines as strong MycG4 binders and potent MYC inhibitors. We have also discovered drugs targeting the unique dGMP-bound-vG4 formed in the PDGFR-β promoter. Moreover, we determined the complex structures of the first small molecules that specifically recognize the physiologically relevant human telomeric G-quadruplexes. Unlike the previously recognized dogma that the optimal G-quadruplex ligands are large aromatic or cyclic compounds, our results suggest that smaller asymmetric compounds with appropriate functional groups are better choices to specifically bind G-quadruplexes. This body of work lays a strong foundation for future work aimed at understanding the cellular functions of G-quadruplexes and G-quadruplex-targeted drug design.
Graphical Abstract

INTRODUCTION
The DNA G-quadruplex has become one of the most exciting nucleic acid secondary structures. They are noncanonical structures formed in guanine-rich sequences that are functionally important, most prominently in human telomeres and key oncogene promoter regions. G-quadruplexes are amenable to small molecule targeting and have emerged as a promising new class of cancer-specific molecular targets for drug development.5–9
Our laboratory has been fascinated by G-quadruplex structures and functions and started this exciting journey in 2002 to study the biologically relevant intramolecular G-quadruplexes formed in human telomeres and oncogene promoters.10–24 In 2005 and 2006, we described the potassium solution structures of the G-quadruplexes formed in the c-Myc oncogene promoter and in human telomeres.10,11 Since then, we have described the potassium solution structures and functions of G-quadruplexes formed in several other oncogene promoters, such as BCL2, VEGF, and PDGFR-β.12–19 Toward G-quadruplex-targeted small molecules, we have characterized the recognition of biologically relevant human telomeric G-quadruplexes and promoter G-quadruplexes, particularly c-Myc.1,3,4,25–32 One of our primary approaches, high-field nuclear magnetic resonance (NMR) spectroscopy, represents a major method for the determination of high-resolution G-quadruplex structures under physiologically relevant solution conditions.
DNA G-quadruplexes are four-stranded secondary structures stabilized by cellular monovalent cations such as K+ or Na+. Most biologically relevant G-quadruplexes are intramolecular (monomeric) structures with a three-tetrad core (Figure 1). Intramolecular G-quadruplexes are specifically determined by nucleotide sequences and can readily form in solution under physiological conditions. Unlike duplex DNA, one important feature of DNA G-quadruplexes is their globular structure. Intramolecular G-quadruplexes exhibit great structural diversity such as folding topology, loop conformations, and capping structures (Figure 1).
Figure 1.
G-quadruplex secondary structures. (A) The G-quadruplex core consists of stacked guanine-tetrads (G-tetrads), with four guanine bases connected with cyclic Hoogsteen hydrogen-bonding in a square planar platform. G-quadruplexes are stabilized by monovalent cations, specifically K+ or Na+, which coordinate the O6 atoms of tetrad guanines. (B) Based on G-strand directionality, a G-quadruplex can be parallel with all four G-strands in the same direction, hybrid or mixed with adjacent strands being both co-directional (parallel) and anti-directional (antiparallel), or antiparallel with all adjacent G-strands anti-directional to each other. Intramolecular G-quadruplexes contain different types of loops, such as propeller for connecting parallel strands, lateral for connecting adjacent antiparallel strands, and diagonal for connecting antiparallel strands across the G-tetrad core. (C) The tetrad guanines can adopt anti or syn glycosidic conformation, same for guanines from parallel strands, different for antiparallel strands.
In this Account, we survey the structural and functional studies in our lab related to the biologically relevant DNA G-quadruplexes formed in human telomeres and oncogene promoters as well as their interactions with small molecules and proteins. We also present critical insights and conceptual advances that were pivotal to our understanding of G-quadruplex structures, functions, and drug targeting.
G-QUADRUPLEXES IN HUMAN TELOMERES
Physiologically Relevant Human Telomeric G-Quadruplexes
We started this exciting journey driven by our curiosity about the G-quadruplexes formed in human telomeres under physiologically relevant K+ solution conditions. Human telomeres consist of d(TTAGGG) tandem repeats terminating in a single-stranded 3′-overhang.33–35 Telomeric G-quadruplexes were the first biologically relevant G-quadruplexes found in the human genome. Telomeric G-quadruplexes are considered attractive anticancer drug targets because their formation renders the telomeres inadequate for extension by the cancer-specific telomerase and alternative lengthening of telomere (ALT) pathways.36–39
A 22-nt human telomeric DNA sequence was reported to form a basket-type three-tetrad G-quadruplex in Na+ solution in 1993.40 In 2002, the same sequence was reported to form a parallel-stranded G-quadruplex in crystalline form in the presence of K+.41 After we saw the crystal structure at the 2002 AACR annual meeting in San Francisco, we were curious about the solution structure in the presence of K+ and decided to conduct an NMR structural study. To our surprise, this 22-nt human telomeric sequence forms neither the parallel nor the basket structure in K+ solution. Instead, the human telomeric DNA forms a mixture of two three-tetrad hybrid-type G-quadruplexes in K+-solution, that is, a major form (hybrid-2) (Figure 2A) and a minor form (hybrid-1) (Figure 2B), and we determined their high-resolution NMR structures.11,20,21 Both hybrid-type structures contain three G-strands co-directional (parallel) and one anti-directional (antiparallel) G-strand; however, the two structures differ in strand and loop arrangements as well as capping structures. Moreover, we determined the high-resolution NMR structure of a two-tetrad basket-type G-quadruplex in a truncated telomeric sequence without 5′-flanking in K+ solution (Figure 2C).22 Notably, the two-tetrad structure can be seen as a three-tetrad quadruplex with a G-vacancy (vG4) (Figure 2 bottom).
Figure 2.
G-quadruplexes formed in human telomeres in K+ solution. The folding (top), NMR structure (middle), and schematics with capping (bottom) of (A) the major form hybrid-2, (B) the minor form hybrid-1, and (C) the two-tetrad basket-form (or three-tetrad vG4). Both hybrid structures contain three G-strands in co-directional (parallel) and one in anti-directional (antiparallel) orientation, but in different order. The hybrid-2 structure has sequential lateral–lateral–propeller loops, whereas the hybrid-1 has propeller–lateral–lateral loops. Each hybrid-type forms specific capping structures: 3′ T8:A9:T25 triad for hybrid-2, and 5′ adenine-triple (A3, A9, A21) and 3′ T14:A25 base-pair for hybrid-1.
Structure Polymorphism and Interconversion of Human Telomeric G-Quadruplexes
The DNA sequence d(TTAGGG)n is highly conserved in higher eukaryote telomeres. While lacking sequence diversity, we demonstrated the structural polymorphism of the human telomeric DNA sequence in physiologically relevant K+ solution, with a major form (hybrid-2) and a minor form (hybrid-1), and a two-tetrad intermediate (Figure 2).11,20–22 Our physiologically relevant solution structures together with other forms suggest that structure polymorphism is an intrinsic property of the human telomeric DNA sequence.40–42 The three-guanine runs generate three-tetrad G-quadruplexes, while the 3-nt asymmetric TTA adenine-containing loop can form lateral, diagonal, and propeller loops at different positions and provides a basis for distinct loop and capping structures (Figure 2). Interestingly, while the coexistence of two hybrid G-quadruplexes indicates a small energy difference, the interconversion between the structures is kinetically slow, suggesting a high-energy intermediate.21 Based on the folding structures and insights that the 3-nt propeller loop is more dynamic,10,14 we proposed the first folding model with a strand-reorientation mechanism for interconversion of hybrid and basket forms of human telomeric G-quadruplexes assuming the 3-nt propeller loop unfolds first and folds last (Figure 3A).11,22,42 Notably, the hybrid-type structures allow effective packing of G-quadruplex multimers in human telomeres (Figure 3B);11,21 however, the presence of a less-favored 3nt-loop in the parallel motif appears to contribute to the structure dynamics and polymorphism. The hybrid-type G-quadruplexes have been shown by others to form in cells,43,44 and the packed multimer conformation has been studied by Chaires and Trent.45 Evolution possibly selected the specific telomeric sequence for its intrinsic structural polymorphism allowing for unique protein recognition and biological function.
Figure 3.
(A) A model for interconversion of different human telomeric G-quadruplexes. Both hybrid-type telomeric G-quadruplexes likely interconvert through a G-triplex or a three-vacancy G-quadruplex formed after unfolding of the G-strand connected with the propeller loop. The three G-strands in the G-triplex are dynamic and can switch Hoogsteen hydrogen-bond partners. The refolding process likely involves strand insertion of the detached G-strand, in which our determined two-tetrad structure may represent an intermediate step between the interconversion of the two possible G-triplexes and hybrid G-quadruplexes, as well as with the basket-type form. (B) With the 5′- and 3′-ends pointing to the opposite directions, hybrid-type structures allow effective packing of G-quadruplex multimers in human telomeres.
HUMAN ONCOGENE PROMOTER G-QUADRUPLEXES
Since the seminal study of the MYC promoter G-quadruplex by Laurence Hurley in 2002,46 DNA G-quadruplexes have been found to form in the proximal promoters of a number of key oncogenes and act as transcriptional regulators. Both negative and positive regulatory roles have been proposed, and the underlying complexity has yet to be fully understood. Unlike telomeric DNA, the promoter G-rich sequences are significantly more diverse. They often contain more than four guanine runs, separated by one or more non-guanine bases, and can form multiple G-quadruplexes. Moreover, G-quadruplex formation in gene promoters needs to compete with the formation of B-DNA. Transcription-induced negative supercoiling has been proposed as the driving force;47 however, it is still under debate.48,49 Over the last 20 years, we have worked on G-quadruplexes formed in the oncogene promoters of MYC, BCL2, PDGFR-β, VEGF, and k-RAS.10,12–19,23,24,50
The MYC Oncogene Promoter
The Major G-Quadruplex Formed in the MYC Oncogene Promoter Is a Parallel-Stranded Structure with the G3NG3 Motif.
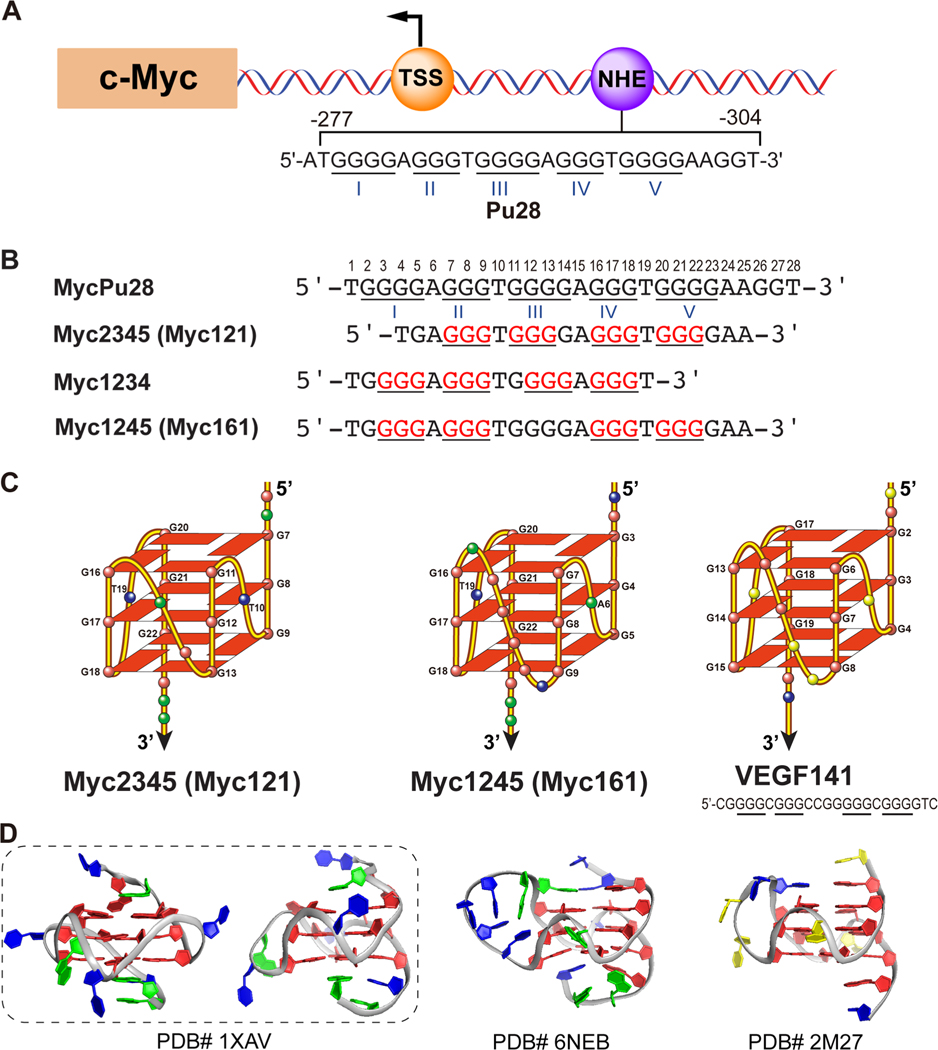
In 2002, Laurence Hurley discovered that the G-quadruplex formed in the MYC proximal promoter nuclease hypersensitive element (NHE) III1 functions as a transcriptional silencer.46 The 28-nt G-rich strand of the MYC NHE III1 (Pu28) consists of five consecutive runs of guanine, with three G4 runs and two G3 runs each separated by a single non-guanine nucleotide, and can form multiple G-quadruplexes (Figure 4A).46,51 The major G-quadruplex formed in K+ solution involves the 3′ four consecutive G runs, II, III, IV, and V, as shown using dimethyl sulfate (DMS) footprinting in conjunction with mutational analysis,46 and adopts parallel structures.52,53 DMS footprinting reveals the residues involved in the G-tetrads because they are protected from methylation by DMS and subsequent cleavage. We determined the high-resolution NMR structure of the major G-quadruplex formed in the MYC promoter in K+ solution, which is a parallel-stranded three-tetrad G-quadruplex with a 1:2:1 loop-length arrangement (Myc2345 or Myc121, Figure 4B–D).10 This was the first promoter G-quadruplex structure and the first example of the three-tetrad G3NG3 parallel-stranded structure motif with a 1-nt loop. Remarkably, we found that the 1-nt loop within the parallel G3NG3 motif is highly favored because of the right-handed twist of the DNA strands, and that the major MYC G-quadruplex, Myc121, is highly stable with a Tm over 85 °C in 100 mM K+ solution.10,54 Since then, the parallel G3NG3 motif has been found to be a robust structural motif and highly prevalent in promoter G-quadruplexes.55
Figure 4.
(A) Human c-Myc gene promoter. (B) MYC promoter G-quadruplex forming sequences. (C, D) Folding (C) and NMR structures (D) of parallel promoter G-quadruplexes. (left) Myc121, the major MYC promoter G-quadruplex. Its 2-nt middle loop is entirely restricted to the groove, and the capping structures are formed solely by the flanking segments. (middle) Myc161, the minor MYC promoter G-quadruplex; (right) VEGF promoter G-quadruplex and sequence. The middle-loops of Myc161 and VEGF stretch over the 5′-tetrad to form a unique capping structure with the sequence-specific 5′-flanking segment.
All Other MYC Promoter G-Quadruplexes Adopt Parallel Structures in K+ Solution.
The MYC promoter NHE III1 can form a minor species involving G runs I, II, IV, and V with a longer central loop (Figure 4B) as shown by DMS footprinting data.46 Recently, we found that the nucleolin protein preferably binds this G-quadruplex, and we determined its NMR structure in K+ solution, which is a parallel-stranded structure with 1:6:1 loop-length arrangement (Myc1245 or Myc161, Figure 4C,D).24 Interestingly, the 6-nt middle loop of Myc161 stretches over the 5′-tetrad to form a unique capping structure with the 5′-flanking segment.
An additional G-quadruplex was found to form with the G runs I, II, III, and IV (Myc1234, Figure 4) in a supercoiled plasmid system,56 and we determined its NMR K+ solution structure.23 Myc1234 also adopts a parallel structure with 1:2:1 folding like Myc2345 but different loop sequences. However, both Myc1234 and Myc1245 are notably less stable than Myc2345, indicating a connection between the stabilities of parallel G-quadruplexes and loop length and composition.23,54
Remarkably, the MYC promoter sequence is unique in that all its G-quadruplexes adopt parallel structures with two 1-nt loop parallel-stranded G3NG3 motifs, which are thermodynamically and kinetically highly favorable. In fact, we found ourselves very fortunate that the MYC promoter sequence, the first G-quadruplex-forming gene promoter we studied, appears to be one of the simplest but most robust G-quadruplex-forming sequences. The robustness of the parallel G-quadruplexes formed in the MYC promoter is likely associated with the prevalence of MYC promoter G-quadruplex observed in cancer cells.57
BCL2 Promoter Contains Two Adjacent G-Rich Regions That Form Interchangeable G-Quadruplexes
G-quadruplex-forming promoters can be more complex with more than one G-quadruplex-forming regions, which may form multiple interchangeable G-quadruplexes. During our collaboration with Laurence Hurley in 2006, we found a 39-nt G-rich region in the BCL2 proximal promoter with six G runs separated by one or more bases (Bcl2Pu39, Figure 5A) that is involved in transcriptional regulation.58 We identified a stable G-quadruplex formed by the middle four consecutive G runs (II–V) and determined its NMR structure in K+ solution (Bcl2-Mid; Figure 5B).12,14 Bcl2-Mid quadruplex adopts a three-tetrad hybrid-2 structure (Figure 5C, right). Bcl2-Mid’s 1-nt third propeller loop makes it significantly more stable than the hybrid-2 telomeric G-quadruplex, indicating the preference for the 1-nt loop in the parallel motif (Figure 6, bottom).
Figure 5.
(A) Human BCL2 gene promoter. (B) BCL2 promoter G-quadruplex forming sequences for P1G4 and Pu39 regions. (C) (left) Two equilibrating parallel G-quadruplexes formed in Bcl2-P1G4 with a unique hairpin structure. (right) Two distinct interchangeable three-tetrad G-quadruplexes formed in Bcl2-Pu39. Bcl2-Mid adopts a hybrid-2 structure, while Bcl2-1245 adopts a parallel structure. Loop arrangements are shown in parentheses.
Figure 6.
Comparison of G-quadruplex-forming sequences in human gene promoters and telomeres. Sequences denoted with an * have been studied by other groups.65–68
In further studies, we found that the full length Bcl2–Pu39 sequence forms a more stable G-quadruplex involving the 5′ nonconsecutive G runs, that are, I, II, IV, and V (Bcl2–1245; Figure 5B) in K+ solution using a combination of DMS footprinting and NMR.15 The determined Bcl2–1245 structure is a three-tetrad parallel G-quadruplex with two 1-nt G3NG3 motifs and a long 13-nt middle loop. This study reveals that having two 1-nt loops can lead to a stable three-tetrad parallel G-quadruplex with a long middle loop (13-nt). This is remarkable because 7-nt had been considered the maximum loop length in a stable G-quadruplex and was used in the available quadruplex-predicting software at the time.59–61
To our surprise, a functional study of the human BCL2 promoter activity in 2014 led us to discover an additional 28-nt G-rich region immediately upstream of the P1 promoter, which functions as a transcription repressor (Bcl2-P1G4, Figure 5A,B).16 P1G4 is located 13-nt downstream of Pu39 and appears to play a more dominant role in transcriptional repression. G-quadruplexes from P1G4 and Pu39 were shown to form independently within the extended BCL2 P1 promoter region. Using NMR spectroscopy, we determined that in K+ solution the P1G4 sequence adopts an equilibrium of two three-tetrad parallel G-quadruplexes with a unique hairpin structure adopted by the long middle-loop (Figure 5C, left). This unique loop structure of P1G4 may provide a specific recognition site for small molecules or proteins. Notably, both P1G4 parallel G-quadruplexes contain two or three 1-nt G3NG3 motifs.
Intriguingly, the presence of two G-quadruplex forming sequences, P1G4 and Pu39, in adjacent regions of the BCL2 promoter that can each adopt two different conformations suggests a potential mechanism for precise regulation of gene transcription by protein interactions.
VEGF Gene Promoter and Parallel G-Quadruplexes
Since the discovery of the MYC and BCL2 promoter G-quadruplexes, the list of G-quadruplexes found in oncogene promoters has been growing. For example, a G-quadruplex formed in the proximal promoter region of the vascular endothelial growth factor (VEGF) gene is involved in transcription regulation.62,63 We determined the NMR structure of the major VEGF promoter G-quadruplex in K+ solution, which is a parallel-stranded structure with a 1:4:1 loop-length arrangement (Figure 4C,D right).13 This major VEGF promoter G-quadruplex again contains the 1-nt G3NG3 motif in its first and third loop and a 4-nt middle loop and forms a unique capping structure.
Together with the parallel G-quadruplexes formed in the MYC and the BCL2 promoters, our studies have elucidated that parallel-stranded G-quadruplexes with three tetrads are common in the human promoter sequences. Most of these parallel structures contain a 1-nt first and third loop, and a middle loop of variable length (Figure 6). Notably, a three-tetrad parallel G-quadruplex with two 1-nt loop motifs can contain a long middle loop as shown by our Bcl2–1245 and Bcl2-P1G4 structures with a 13-nt and 12-nt middle loop, respectively. The robust G3NG3 parallel-structure motif is highly prevalent in promoter G-quadruplexes and may be evolutionarily selected as a stable foundation and to initiate folding for the parallel G-quadruplexes.64 Although parallel G-quadruplexes with short loops have a similar basic structure, they often form unique capping and loop structures determined by the specific central loop and flanking sequences. As parallel structures are common to the promoter and RNA G-quadruplexes, our studies suggest that the variations in parallel G-quadruplexes give rise to different structure features, allowing specific recognition by proteins or small molecules.
PDGFR-β Gene Promoter Forms Broken-Strand Parallel G-Quadruplexes
Another variation in parallel structures comes from the broken-strand and vacancy G-quadruplexes. In 2010, Laurence Hurley’s group found a highly GC-rich nuclease hypersensitive element (NHE) in the proximal promoter of the human platelet derived growth factor receptor beta (PDGFR-β), which forms G-quadruplexes that are transcriptional repressors.69 The 39-nt G-rich strand contains seven guanine runs, with one G7 run, one G4 run, three G3 runs, and two G2runs, and can form multiple G-quadruplexes. Among these, the most stable G-quadruplex is formed by G runs II–V (PD-5Mid, Figure 7). Using NMR in combination with DMS footprinting and mutational analysis, we determined that PD-5Mid adopts a novel “broken-strand” three-tetrad parallel G-quadruplex in K+ solution (Figure 7).19 Intriguingly, this novel “broken-strand” parallel structure maximizes the number of 1-nt loops (three).
Figure 7.
(A) Human PDGFR-β gene promoter. (B) PDGFR-β promoter G-quadruplex forming sequences. (C) (top) PD-5Mid forms a novel broken-strand G-quadruplex. Instead of using four G runs of three or more consecutive guanines, PD-5Mid uses the G2 run III to form a “broken” G-strand consisting of guanines from different G runs (left). Two equilibrating vG4 (middle), which can be filled-in and stabilized by endogenous guanine metabolites, and the NMR structure of the dGMP complex (right). (bottom) PD-3End forms two equilibrating broken-strand G-quadruplexes. The G2 run VII moves up or down within the G-core to accommodate an inserted guanine from the 3′ or 5′ nonadjacent flanking regions into the external tetrad.
The less stable PDGFR-β NHE 3′-end G-quadruplex sequence (G runs III–VII) can be selectively targeted by the small molecule GSA1129 for gene downregulation.70 Interestingly, we found using NMR spectroscopy and DMS footprinting that the 3′-end G-quadruplex sequence forms two coexisting end-insertion (or broken-strand) intramolecular structures in K+ solution (PD-3End, Figure 7C, bottom left).17 The G run VII shifts up or down to accommodate an inserted distal guanine within the 3′- or 5′-tetrad.
Notably, the novel “broken-strand” structures formed in the PDGFR promoter appear to be a variation of parallel-stranded G-quadruplexes with a 1-nt loop parallel-stranded G3NG2 or G2NG3 motif, a variant of the G3NG3 motif (Figure 6). The novel broken-strand structures expand our understanding of sequence requirements for stable quadruplexes. Thus, parallel G-quadruplexes can exist in various forms, such as broken-strands, end-insertions, or with an additional hairpin motif.
The Broken-Strand PDGFR-β Promoter G-Quadruplexes Are Able to Form vG4s That Can Be Filled-In by Guanine Metabolites
It is intriguing that the broken-strand structures appear to be a general feature of the PDGFR-β promoter G-quadruplexes. Broken-strand G-quadruplexes have the potential to form vacancy G-quadruplexes (vG4) if the intramolecularly filled-in guanine becomes more dynamic and vacates its G-tetrad. We were curious whether the PDGFR-β broken-strand structures could form vG4s and whether these could be filled-in with external guanine or guanine derivatives. Amazingly, using NMR and other biophysical and biochemical methods, we found that the PD-5Mid sequence can form two coexisting 3′- and 5′-end vG4s that can be readily filled-in and stabilized by guanine metabolites, such as dGMP, GMP, and cGMP, as well as guanine-derivative drugs such as acyclovir and ganciclovir (Figure 7C).18 Interestingly, all guanine metabolites and drugs prefer the 5′-end fill-in, except for cGMP, which can equally bind at both ends. We determined the NMR structure of the PDGFR-β dGMP-fill-in vG4 in K+ solution, which shows specific vG4 recognition by guanine metabolites involving Hoogsteen hydrogen-bonding and sugar′phosphate backbone interactions. This was the first example of a guanine-metabolite-fill-in vG4 formed by a human gene promoter sequence, suggesting potential regulatory effects of cellular endogenous guanine metabolites on gene transcription through interactions with the unique vG4s, a new role of quadruplexes similar to RNA riboswitches.
TARGETING DNA G-QUADRUPLEXES WITH SMALL MOLECULES
DNA G-quadruplexes formed in promoter regions of key oncogenes and human telomeres have become a promising new class of cancer-specific molecular targets for drug development.7,36,37,71 Our high-resolution structures of G-quadruplexes in complex with small molecules reveal structural features that are specifically recognizable by ligands and provide a structural basis for rational drug design. Importantly, our studies show that G-quadruplexes can contain unique capping and loop structures determined by their specific sequence.
Small Molecule Recognition of Physiologically Relevant G-Quadruplexes in Human Telomeres
Recognition of the Hybrid-2 Human Telomeric G-Quadruplex.
Human telomeric G-quadruplex is an attractive anticancer drug target and has been intensively pursued because small molecules stabilizing telomeric G-quadruplex can stall cancer-specific telomerase/ALT pathways and lead to apoptosis of cancer cells. 36–39 Although we determined the human telomeric G-quadruplex structures in K+ solution in 2006 and 2007, it was not until 2015 when we discovered in collaboration with Yong Shao the first small molecule that binds specifically to the human telomeric DNA in K+ solution, the medicinal natural isoquinoline alkaloid epiberberine (EPI).72
We determined the NMR solution structure of EPI in complex with the hybrid-2 structure formed by the wild-type 26-nt telomeric DNA (wtTel26) in K+ solution (Figure 8A).1 EPI specifically binds the 5′-end of the hybrid-2 telomeric G-quadruplex and induces extensive rearrangement of the flanking segment and the second TTA loop to form a well-defined, four-layer binding pocket (Figure 8A). EPI covers two guanines and recruits the 5′-flanking A3 to form a hydrogen-bonded “intercalative quasi-triad” ligand–base plane stacking over the 5′-external-tetrad. The positively charged N7 atom of EPI is positioned above the central pore of the 5′-tetrad. The ligand–base plane is capped by a T2:T13:A15 triad layer and another T1:T14 base-pair layer. Surprisingly, other structurally similar berberine alkaloids, such as berberine, palmatine, or coptisine (Figure 8A), are unable to bind the hybrid-2 structure well. Our determined structure reveals that the closed methylenedioxy ring E of EPI provides an appropriate hydrogen-bond acceptor for the recruited adenine, while the open methoxy moieties at ring A avoid a clash with the DNA backbone, demonstrating the critical insights that are only provided by high-resolution atomic structures.
Figure 8.
(A) NMR structure of EPI in complex with the hybrid-2 human telomeric G-quadruplex showing a well-defined, extensive four-layer binding pocket at the 5′-end. EPI shows minimal binding at the 3′-end, which forms the same TAT triad as in the unbound hybrid-2 structure. (B) EPI binding converts other human telomeric G-quadruplex forms to a hybrid-2 structure in solution. (C) NMR structure of the 1:1 complex of Pt-tripod and hybrid-1 human telomeric G-quadruplex. In the complex, Pt-tripod recruits the 5′-flanking A21 from the third lateral loop to form a ligand–base plane covering the external-tetrad, with the aromatic moieties of arms 1 and 2 each stacking over a tetrad guanine. The ligand–base plane is covered with the H-bonded triad A9, A3:T20, replacing the A3:A9:A21 A-triple from the unbound form.
The EPI binding pocket is specific to the hybrid-2 folding and human telomeric sequence. Remarkably, the preference of EPI binding for the hybrid-2 form is so strong that it converts other human telomeric G-quadruplex forms to this conformation, such as hybrid-1 and basket-type structures, as well as the unfolded form (Figure 8B). EPI is the only known compound that can convert other telomeric G-quadruplexes to the hybrid-2 structure and thus presents a valuable chemical probe to investigate the protein binding and biology of human telomeric G-quadruplexes, whose intrinsic polymorphism often presents a challenge.
Recognition of the Hybrid-1 Human Telomeric G-quadruplex.
In collaboration with Zong-wan Mao, we discovered the first-in-class Pt(II)-based Pt-tripod compound that specifically recognizes the hybrid-1 human telomeric G-quadruplex (Figure 8C).27 The Pt-tripod contains a nonplanar central tertiary amine with three symmetric arms each containing two aromatic rings and a terminal positively charged platinum complex. At 1:1 ratio, Pt-tripod binds to the 5′-end of the hybrid-1 structure to form a monomeric complex; at higher ratio, a second Pt-tripod binds the 3′-end and induces the formation of a 4:2 dimeric complex. We determined the NMR structures of both complexes in K+ solution (Figure 8C). Pt-tripod binding induces a complete conformational rearrangement at both the 5′- and 3′-end. A unique feature of the selective G-quadruplex recognition by Pt-tripod is anchoring the three arms into three G-quadruplex grooves to enable specific hydrogen-bonding, electrostatic, and π-π stacking interactions with the DNA bases and sugar-backbone.
Targeting the MYC Promoter G-Quadruplex for Drug Discovery
MYC is one of the most commonly deregulated genes in human cancers. It is the most widely validated “driver” oncogene in the context of hematologic malignancies, and overexpressed in most solid cancers.73 However, the MYC protein is often considered undruggable due to difficulties associated with targeting transcription factors and its disordered structure,74,75 whereas interesting work is being carried out on MYC protein interactors.76 Importantly, compounds that bind and stabilize the MYC promoter G-quadruplex can lower MYC levels and induce cancer cell death.3,46,77–79 Therefore, the MYC promoter G-quadruplex represents a promising target for MYC inhibition and cancer therapeutics. After we determined the structure of the major MYC promoter G-quadruplex in 2005, we have been intensively working on understanding its small molecule recognition.
First Ligand Complex Structure with Quindoline.
However, finding small molecules that bind the MYC G-quadruplex with sufficient affinity and specificity for structure determination has not been easy. We found those previously recognized optimal G-quadruplex-interactive compounds with large symmetric cyclic rings, such as telomestatin80 and TMPyP4,81 do not bind G-quadruplexes specifically. In 2008, we found that the quindoline reported by Zhi-shu Huang binds MycG4 specifically to generate good-quality NMR spectra.78 We determined the high-resolution NMR solution structure of the 2:1 complex of quindoline and the major MYC promoter G-quadruplex Myc121 (Figure 9), the first drug complex structure of a biologically relevant promoter G-quadruplex.25 Quindoline binds at both ends with large conformational rearrangements of the flanking regions to form specific binding pockets. It specifically recruits the immediate flanking residue to form an intercalated “quasi-triad” ligand–base plane that covers the external-tetrad. Absolute binding free energy calculations show that the flanking residues at the two binding sites undergo significant reorganization as the ligand unbinds.31
Figure 9.
(A) NMR structure of the 2:1 complex of quindoline and the major MYC promoter G-quadruplex in two views. Quindoline binds at both the 3′- and 5′-end and recruits the immediate flanking residue to anchor its position and form an intercalated “ligand–base quasi-triad” plane that covers the external-tetrad, with additional flanking residues covering the ligand–base plane. (B) Comparison of the binding recognition of four different compounds of the major MYC promoter G-quadruplex with the wild-type and mutant 3′-flanking GAA or TAA.
High Affinity Binding of BMVC.
BMVC (3,6-bis(1-methyl-4-vinylpyridinium) carbazole diiodide) is the first fluorescent probe to detect telomeric G-quadruplexes in human cells developed by Ta-Chau Chang.82–84 However, we found that BMVC binds the parallel-stranded MYC promoter G-quadruplex with higher specificity and affinity than the human telomeric G-quadruplexes and determined the high-resolution NMR solution structures of the 1:1 and 2:1 BMVC-Myc121 complexes (Figure 9B).26 The high-affinity binding of BMVC with Myc121 leads to an unusual slow-exchange rate on the NMR time scale. At 1:1 ratio, BMVC first binds the 5′-end of Myc121; at higher ratio, BMVC also binds the 3′-end to form a 2:1 complex. In both complexes, BMVC recruits the immediate flanking DNA base to lock its position and form a “quasi-triad” ligand–base plane stacking over the external G-tetrad.
Interestingly, while BMVC binds the more stacking-accessible 5′-end with higher affinity, it shows a greater sequence selectivity at the weaker-bound 3′-end. Remarkably, in each complex BMVC adjusts the conformation of its flexible arms to a contracted form to match the external G-tetrad for an optimal stacking interaction. This is the first structural example showing the importance of ligand conformational adjustment in G-quadruplex recognition, which is likely responsible for G-quadruplex-specific strong fluorescence enhancement of BMVC.
Drug-like PEQ as a Specific MycG4 Binder.
In another fruitful collaboration with Laurence Hurley, we screened the National Cancer Institute (NCI) chemical diversity library set using a combination of FRET assay, drug similarity search, and 1D 1H NMR titration to discover drug-like molecules that can specifically recognize the MYC promoter G-quadruplex. We found NSC85697, a phenyl-ethenyl-quinoline (PEQ), specifically binds the major MYC promoter G-quadruplex Myc121. PEQ has low molecular weight and is less rigid without a large aromatic moiety. These drug-like properties make it a promising lead compound for future optimization. We determined the high-resolution NMR solution structures of the 2:1 complex of PEQ with Myc121 with the wild-type and mutant 3′-flanking GAA or TAA, respectively (Figure 9B).28 Like quindoline, PEQ binds both ends of the parallel-stranded Myc121 with conformational rearrangements of the flanking regions to form specific binding pockets using a base-recruiting recognition mechanism. Most previous studies of Myc121 used a G23-to-T mutation to avoid conformational polymorphism. Notably, a comparison of the two complex structures shows a different binding interface of the recruited 3′-flanking wild-type G23 compared to the mutated T23, which will help design Myc121-selective drugs.
Rational Drug Design Targeting MycG4 and Parallel G-Quadruplexes
Parallel G-quadruplexes are prevalent in promoter and RNA G-quadruplexes. The major MYC promoter G-quadruplex Myc121 is the model system for parallel G-quadruplexes with short loops, whose ligand binding pockets only involve the 5′- and 3′-flanking sequences. Our ligand complex structures demonstrate the importance of the shape of the ligand and the two flanking sequences in determining the binding specificity of Myc121.25,26,28,29 Base recruitment is likely to be the key step for ligand recognition. Intriguingly, our systematic analysis of the four available NMR structures with Myc121 revealed that the flanking base is recruited in a conserved residue-specific way (Figure 9B).28 This finding suggests that MycG4 and more generally parallel G-quadruplexes with short loops can be selectively targeted based on different flanking sequences. Moreover, the ligand-DNA interface bears the potential to improve sequence selectivity by optimization of complementary hydrogen-bond interactions between the ligand and the recruited base. These insights underline the important benefit of experimentally determined structures for rational design and in situ screening of drugs targeting MycG4 and parallel G-quadruplexes.
Notably, in all complex structures, the 5′-flanking residue is more dynamic and adopts different conformations in the bound and unbound forms. On the other hand, the recruited 3′-flanking residue appears to be less dynamic upon ligand binding and adopts similar conformations in the bound and unbound state. Combined with the lower stacking accessibility of the 3′-tetrad, this finding indicates a more prominent binding selectivity for the 3′-binding, as observed in binding of BMVC to MycG4 and parallel G-quadruplexes.26,32
G4 DNA Microarray for Drug Development and Binding Selectivity.
In collaboration with Charles Vinson at NCI, we designed a custom G4 DNA microarray with over 15 000 potential G-quadruplex sequences to provide a large-scale and high-throughput platform for assessing the G-quadruplex-binding selectivities of proteins, antibodies, and small molecules.85 Remarkably, the large-scale G4 microarray study revealed that BMVC preferentially binds the parallel MycG4, with a clear sequence selectivity for the 3′-flanking thymine of MycG4 and parallel G-quadruplexes (Figure 10A),32 consistent with the NMR study.26 Custom DNA microarrays will be especially useful for G-quadruplex-targeted drug development as a platform to broadly and unbiasedly assess ligand binding selectivity.
Figure 10.
(A) Large-scale G4 microarray data (left) shows that BMVC preferentially binds the parallel MycG4, with a sequence selectivity for the 3′-flanking thymine of MycG4 and parallel G-quadruplexes. (B) Clinically tested indenoisoquinolines are strong MycG4 binders and potent MYC inhibitors. (C) Synergistic effect of MYC inhibition by transcriptional silencing and topoisomerase I inhibition for anticancer activity of indenoisoquinolines.
Clinical Indenoisoquinolines Strongly Bind and Stabilize the MYC Promoter G-Quadruplex and Downregulate MYC.
Very recently, in collaboration with Mark Cushman at Purdue, we have discovered that clinically tested indenoisoquinolines with excellent drug-like properties are strong MycG4 binders and potent MYC inhibitors.3 Indenoisoquinolines were developed as topoisomerase I inhibitors, with three compounds in phase I–II clinical trials (Figure 10B).
However, some indenoisoquinolines lack strong topoisomerase I inhibition despite potent anticancer activity, suggesting a separate mechanism of action. Using a combination of biophysical, biochemical, and cellular assays, we demonstrated that many anticancer indenoisoquinolines strongly bind and stabilize the MycG4 in vitro and potently lower MYC mRNA and protein levels in cancer cells. Intriguingly, we found a synergistic effect of MYC and topoisomerase I inhibition for anticancer activity, suggesting that dual targeting of MYC and topoisomerase I could be a novel strategy for cancer therapeutics (Figure 10C).
Molecular Recognition of vG4s Formed in the PDGFR-B Promoter
The PDGFR-β gene promoter forms unique vG4s that can be filled-in and stabilized by endogenous guanine metabolites. An interesting question is whether the novel PDGFR-β promoter vG4 in complex with endogenous guanine metabolites can be recognized by small molecules. We found that berberine, a clinically important natural product with anticancer activity, binds and stabilizes the unique PDGFR-β dGMP-fill-in vG4. We characterized the binding of berberine to the vG4–dGMP complex and determined the high-resolution NMR structure of this ternary complex in K+ solution (Figure 11).4 The NMR structure provides insights and structural basis for designing small molecules targeting the unique vG4s in the PDGFR-β promoter, particularly using the G-vacancy site as an anchor point to design guanine conjugates with G-quadruplex ligands.
Figure 11.
NMR structure of the ternary complex of PDGFR-β vG4–dGMP and berberine. Berberine binding at the 5′-end significantly stabilizes the fill-in dGMP in the 5′-tetrad of the vG4–dGMP complex. Each berberine recruits the flanking adenine at 5′- or 3′-end to form a ligand–base plane.
Insights into Small Molecule Recognition of G-Quadruplexes
Our determined NMR solution structures of ligand–G-quadruplex complexes provide structural details and insights into targeting MycG4 and parallel G-quadruplexes with short loops, as well as nonparallel hybrid-type G-quadruplexes in human telomeres. In parallel G-quadruplexes with short loops, the ligand binding pockets solely involve the 5′- and 3′- flanking segments. In contrast, loop regions of hybrid-type structures play a pivotal role in binding pocket formation. The multilayer binding pockets of both hybrid-type telomeric G-quadruplexes involve the external tetrad, flanking, and loop residues and are markedly more extensive than those observed with the parallel G-quadruplexes lacking the lateral loops. This indicates the importance of interactions with flanking segments and various loops to achieve specific recognition of the nonparallel G-quadruplexes.
Unlike the previously proposed concept that the best G-quadruplex-interactive compounds are large symmetric cyclic or aromatic molecules that maximize stacking interactions with the external G-tetrads, our structural studies indicate that a smaller asymmetric pharmacophore with appropriate functional groups is more likely to bind in a specific manner to the biologically relevant intramolecular G-quadruplexes. The base-recruitment is observed in all our ligand complexes in which the ligand–base plane stacks over the external G-tetrad. Only an asymmetric compound with a smaller pharmacophore, which does not completely cover the tetrad, can effectively recruit the DNA flanking base to achieve a specific binding orientation. Our ligand complex structures show that the optimal small-molecule recognition of G-quadruplexes utilizes stacking, H-bonds, and electrostatic interactions. It may involve first ligand–base recognition by the shape of the pharmacophore and the specific flanking residue, and second the groove/loop recognition by functional groups of the compound, followed by interactions with DNA capping structures.
Our complex structures show that a specific type of electrostatic interaction is important for ligand recognition of G-quadruplexes. A central positive charge in the aromatic pharmacophore of the ligand is often positioned above the electron-rich central pore of the G-quadruplex, resembling the coordinated cations within the G-tetrad core, as observed for quindoline, EPI, Pt-tripod, PEQ, and berberine (Figures 8, 9, and 11). In addition, positive charges located at the pharmacophore edges or on side chains often have favorable electrostatic interactions with the negatively charged DNA sugar-phosphate backbone, as seen for quindoline, BMVC, and Pt-tripod (Figures 8, 9, and 11).
Although there are common features of G-quadruplex ligand recognition, there are clear differences in binding interactions as well as binding stoichiometry. Notably, these important differences related to ligand interactions can only be monitored in solution and are amenable to NMR studies. For example, the 5′-binding in the quindoline–MycG4 complex is driven by stacking interactions and is [K+]-independent, whereas the 3′-binding is stabilized by H-bond interactions and is [K+]-dependent such that a well-defined conformation can only be achieved at low salt concentration.25 Such a subtle difference in stacking and electronic interactions is only observable in solution. For BMVC, the 5′-end binding of MycG4 is much stronger than the 3′-end binding; however, a clear sequence selectivity is observed in the 3′-end binding as shown by the NMR study in K+ solution.26 In the case of EPI, it not only specifically binds the hybrid-2 wtTel26 but also is able to convert the hybrid-1 and the basket G-quadruplexes to the hybrid-2 structure in solution.1 The Pt-tripod binds the 5′-end of hybrid-1 TelG4 at 1:1 ratio but also binds to the 3′-end at higher equivalences and induces dimer formation in K+ solution.27 Moreover, we have very recently shown by NMR that the solution binding mode and stoichiometry of berberine to a parallel-stranded G-quadruplex are substantially different from the crystal structure: berberine binds as monomer instead of dimer with a reversed orientation.29 Therefore, it is crucial to validate drug–DNA interactions under physiologically relevant solution conditions for drug development efforts.
CONCLUDING REMARKS
With our growing understanding, G-quadruplexes have developed from a laboratory curiosity to biologically important regulatory motifs and attractive molecular targets for cancer therapeutics. Our long-term goal is to understand structure–function relationships of biologically relevant DNA G-quadruplexes formed in human telomeres and oncogene promoters and rationally develop G-quadruplex-targeted anticancer drugs. Our studies show that biologically relevant intramolecular G-quadruplexes exhibit great structural diversity based on the specific DNA sequences and form readily under physiological conditions stabilized by cellular monovalent cations. While several principles of G-quadruplex folding have been recognized, G-quadruplex conformations are difficult to predict and require experimental structure determination. For small molecule recognition of G-quadruplexes, the molecular-level information can only be obtained from detailed structural studies. Our recent systematic evaluation of four commonly used docking programs (AutoDock Vina, DOCK 6, Glide, and RxDock) showed clear differences in performance, with the docking success rate of the best performing DOCK6 still notably low compared to protein targets.30 This result suggests that it is challenging to correctly predict small-molecule binding modes with G-quadruplex by current docking programs. Beyond G-quadruplexes formed natively, cellular oxidative damage can induce structural changes in genomic DNA G-quadruplexes.86 We very recently demonstrated for the BLM gene promoter that G-oxidation induces a vacancy G-quadruplex, which subsequently can be filled-in and stabilized by guanine metabolites, suggesting that oxidative damage and cellular metabolites may work together through a G-quadruplex-based epigenetic mechanism for gene regulation.87 In addition to G-quadruplex, the complementary C-rich strands in gene promoters can form imotif DNA secondary structures, such as those we characterized in the promoters of MYC and BCL2.56,88,89
Furthermore, we are working on discovering and understanding proteins that interact with the MYC promoter G-quadruplex to regulate gene transcription (Figure 12). The MYC promoter G-quadruplex is extremely stable and requires a resolving protein/helicase to generate the transcriptionally active single-stranded or duplex forms of DNA. We have very recently discovered that the DDX5 helicase proficiently and actively unfolds MycG4 and activates MYC gene transcription in a MycG4-dependent manner in human cancer cells.2 DDX5 is one of the founding members of the DEAD-box RNA helicase family, and our study is the first description of a DEAD-box helicase activity on a DNA substrate. Importantly, the DDX5-mediated MycG4-unfolding and MYC activation can be targeted by small molecules and thus represents a novel molecular target for MYC silencing. On the other hand, the nucleolin protein specifically binds and stabilizes the MYC promoter G-quadruplex and functions as a transcription repressor.90 We are studying the molecular-level interactions of these proteins with the MYC promoter G-quadruplex to exploit their cellular functions and the potential for small molecule drug targeting.
Figure 12.
Proposed model for the MYC promoter G-quadruplex and the involvement of DDX5, nucleolin, topoisomerase I, and a G-quadruplex ligand in modulating transcriptional activation and silencing. The G-quadruplex formed in the NHE III1 under transcription-induced negative supercoiling is a silencer element. Topoisomerase I relaxes negative supercoiling back to the duplex form. DDX5 helicase unfolds the G-quadruplex to the transcriptionally active form, whereas binding of nucleolin stabilizes the G-quadruplex in its silencer form. Stabilization of the G-quadruplex by a small-molecule prevents DDX5-mediated MycG4 unfolding and MYC activation, and inhibition of topoisomerase I by a small-molecule preserves the G-quadruplex and transcriptional silencing.
We anticipate that the body of work detailed in this Account lays a strong foundation and provides a critical structural basis for future work aimed at understanding the cellular functions of G-quadruplexes and G-quadruplex-targeted rational drug design. We expect continued growth in the G-quadruplex field and a plethora of exciting developments, such as protein interactions of G-quadruplexes at structural and cellular levels, chromatin interactions and epigenetic roles, and newly discovered liquid–liquid phase separation. We anticipate that decades of drug development and ever-improving understanding of small molecule recognition will result in more drugs entering preclinical and clinical studies and ultimately offering new hope for cancer patients.
KEY REFERENCES.
Lin, C.; Wu, G.; Wang, K.; Onel, B.; Sakai, S.; Shao, Y.; Yang, D. Molecular Recognition of the Hybrid-2 Human Telomeric G-Quadruplex by Epiberberine: Insights into Conversion of Telomeric G-Quadruplex Structures. Angew. Chem., Int. Ed. 2018, 57, 10888–10893.1 High-resolution NMR solution structure that shows the specific recognition of the physiologically relevant hybrid-2 human telomeric G-quadruplex by the small molecule epiberberine. We show for the first time that a compound can convert other telomeric G-quadruplexes to hybrid-2 structure.
Wu, G.; Xing, Z.; Tran, E. J.; Yang, D. DDX5 Helicase Resolves G-Quadruplex and Is Involved in MYC Gene Transcriptional Activation. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 20453–20461.2 We discovered that DDX5 helicase actively resolves the highly stable MYC promoter G-quadruplex to activate MYC transcription, a missing link in MycG4 transcriptional control. This is the first description of a DEAD-box RNA helicase activity on DNA.
Wang, K.-B.; Elsayed, M. S. A.; Wu, G.; Deng, N.; Cushman, M.; Yang, D. Indenoisoquinoline Topoisomerase Inhibitors Strongly Bind and Stabilize the MYC Promoter G-Quadruplex and Downregulate MYC. J. Am. Chem. Soc. 2019, 141, 11059–11070.3 We discovered that clinical indenoisoquinolines strongly bind the MYC promoter G-quadruplex and are potent MYC inhibitors. We further established dual actions of MYC and topoisomerase I inhibition as an effective strategy for cancer therapeutics.
Wang, K.-B.; Dickerhoff, J.; Yang, D. Solution Structure of Ternary Complex of Berberine Bound to a DGMP-FillIn Vacancy G-Quadruplex Formed in the PDGFR-β Promoter. J. Am. Chem. Soc. 2021, 143, 16549–16555.4 We previously discovered that the PDGFR-β gene promoter forms broken-strand and vacancy G-quadruplexes that can be recognized by endogenous guanine metabolites. This report describes the first ternary complex structure of a biologically relevant vacancy G-quadruplex in the human oncogene promoter bound with guanine metabolite and a small molecule.
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health R01CA177585, U01CA240346, R01CA153821, R01GM083117, R01CA122952, S10 RR016659, and K01CA83886 (D.Y.), P30CA023168 (Purdue Center for Cancer Research), Grants-in-aid for the Promotion of Joint International Research by Japan Society for the Promotion of Science 15KK0179 (S.S.), and the Deutsche Forschungsge-meinschaft Projektnummer 427347592 (J.D.).
Biographies
Luying Chen received her bachelor degree in engineering from the South China University of Technology and is a Ph.D. student in Professor Danzhou Yang’s laboratory at Purdue University. Her research interests include DNA G-quadruplexes and protein interactions.
Jonathan Dickerhoff obtained his Ph.D. in 2017 from the University of Greifswald. He then joined the laboratory of Professor Danzhou Yang at Purdue University as postdoctoral researcher. His research focuses on the structural characterization of G-quadruplexes and drug complexes.
Saburo Sakai is a senior research scientist at Japan Agency for MarineEarth Science and Technology. He started collaborative work on G-quadruplexes with Danzhou Yang in 2012. He received his Ph.D. from Hiroshima University, Japan, in 2000. His research focuses on the isotope biogeochemistry and laser spectroscopy.
Danzhou Yang is a Distinguished Professor and Martha and Fred Borch Chair of Cancer Therapeutics at Purdue University. Previously, she was a professor at the University of Arizona (2000–2016). She received her Ph.D. in Biophysics from the University of Illinois at Urbana–Champaign in 1996. Her research focuses on the structures and functions of DNA G-quadruplexes and their interactions with small molecules and proteins.
Footnotes
Notes
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.accounts.2c00337
Contributor Information
Luying Chen, Department of Medicinal Chemistry and Molecular Pharmacology, College of Pharmacy, Purdue University, West Lafayette, Indiana 47907, United States.
Jonathan Dickerhoff, Department of Medicinal Chemistry and Molecular Pharmacology, College of Pharmacy, Purdue University, West Lafayette, Indiana 47907, United States.
Saburo Sakai, Department of Medicinal Chemistry and Molecular Pharmacology, College of Pharmacy, Purdue University, West Lafayette, Indiana 47907, United States; Biogeochemistry Research Center, Japan Agency for Marine-Earth Science and Technology, Yokosuka-city, Kanagawa 237-0061, Japan.
Danzhou Yang, Department of Medicinal Chemistry and Molecular Pharmacology, College of Pharmacy, Purdue University, West Lafayette, Indiana 47907, United States; Purdue Center for Cancer Research, Purdue University, West Lafayette, Indiana 47907, United States; Department of Chemistry, Purdue University, West Lafayette, Indiana 47907, United States; Purdue Institute for Drug Discovery, Purdue University, West Lafayette, Indiana 47907, United States.
REFERENCES
- (1).Lin C; Wu G; Wang K; Onel B; Sakai S; Shao Y; Yang D Molecular Recognition of the Hybrid-2 Human Telomeric G-quadruplex by Epiberberine: Insights into Conversion of Telomeric G-Quadruplex Structures. Angew. Chem., Int. Ed 2018, 57, 10888–10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wu G; Xing Z; Tran EJ; Yang D DDX5 Helicase Resolves G-Quadruplex and Is Involved in MYC Gene Transcriptional Activation. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 20453–20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wang K-B; Elsayed MSA; Wu G; Deng N; Cushman M; Yang D Indenoisoquinoline Topoisomerase Inhibitors Strongly Bind and Stabilize the MYC Promoter G-Quadruplex and Downregulate MYC. J. Am. Chem. Soc 2019, 141, 11059–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wang K-B; Dickerhoff J; Yang D Solution Structure of Ternary Complex of Berberine Bound to a DGMP–Fill-In Vacancy G-Quadruplex Formed in the PDGFR-β Promoter. J. Am. Chem. Soc 2021, 143, 16549–16555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Carvalho J; Mergny J-L; Salgado GF; Queiroz JA; Cruz C G-Quadruplex, Friend or Foe: The Role of the G-Quartet in Anticancer Strategies. Trends Mol. Med 2020, 26, 848–861. [DOI] [PubMed] [Google Scholar]
- (6).Spiegel J; Adhikari S; Balasubramanian S The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yang D; Okamoto K Structural Insights into G-Quadruplexes: Towards New Anticancer Drugs. Future Med. Chem 2010, 2 (4), 619–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Neidle S Quadruplex Nucleic Acids as Targets for Anticancer Therapeutics. Nat. Rev. Chem 2017, 1, 0041. [Google Scholar]
- (9).Brooks TA; Hurley LH The Role of Supercoiling in Transcriptional Control of MYC and Its Importance in Molecular Therapeutics. Nat. Rev. Cancer 2009, 9, 849–861. [DOI] [PubMed] [Google Scholar]
- (10).Ambrus A; Chen D; Dai J; Jones RA; Yang D Solution Structure of the Biologically Relevant G-Quadruplex Element in the Human c-MYC Promoter. Implications for G-Quadruplex Stabilization. Biochemistry 2005, 44, 2048–2058. [DOI] [PubMed] [Google Scholar]
- (11).Ambrus A; Chen D; Dai J; Bialis T; Jones RA; Yang D Human Telomeric Sequence Forms a Hybrid-Type Intramolecular G-Quadruplex Structure with Mixed Parallel/Antiparallel Strands in Potassium Solution. Nucleic Acids Res. 2006, 34, 2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Dai J; Chen D; Jones RA; Hurley LH; Yang D NMR Solution Structure of the Major G-Quadruplex Structure Formed in the Human BCL2 Promoter Region. Nucleic Acids Res. 2006, 34, 5133–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Agrawal P; Hatzakis E; Guo K; Carver M; Yang D Solution Structure of the Major G-Quadruplex Formed in the Human VEGF Promoter in K+: Insights into Loop Interactions of the Parallel G-Quadruplexes. Nucleic Acids Res. 2013, 41, 10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dai J; Dexheimer TS; Chen D; Carver M; Ambrus A; Jones RA; Yang D An Intramolecular G-Quadruplex Structure with Mixed Parallel/Antiparallel G-Strands Formed in the Human BCL-2 Promoter Region in Solution. J. Am. Chem. Soc 2006, 128, 1096–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Agrawal P; Lin C; Mathad RI; Carver M; Yang D The Major G-Quadruplex Formed in the Human BCL-2 Proximal Promoter Adopts a Parallel Structure with a 13-Nt Loop in K+ Solution. J. Am. Chem. Soc 2014, 136, 1750–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Onel B; Carver M; Wu G; Timonina D; Kalarn S; Larriva M; Yang D A New G-Quadruplex with Hairpin Loop Immediately Upstream of the Human BCL2 P1 Promoter Modulates Transcription. J. Am. Chem. Soc 2016, 138, 2563–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Onel B; Carver M; Agrawal P; Hurley LH; Yang D The 3′-End Region of the Human PDGFR-β Core Promoter Nuclease Hypersensitive Element Forms a Mixture of Two Unique End-Insertion G-Quadruplexes. Biochim. Biophys. Acta BBA - Gen. Subj 2018, 1862, 846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wang K-B; Dickerhoff J; Wu G; Yang D PDGFR-β Promoter Forms a Vacancy G-Quadruplex That Can Be Filled in by DGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs. J. Am. Chem. Soc 2020, 142, 5204–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chen Y; Agrawal P; Brown RV; Hatzakis E; Hurley L; Yang D The Major G-Quadruplex Formed in the Human Platelet-Derived Growth Factor Receptor β Promoter Adopts a Novel Broken-Strand Structure in K+ Solution. J. Am. Chem. Soc 2012, 134, 13220–13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Dai J; Punchihewa C; Ambrus A; Chen D; Jones RA; Yang D Structure of the Intramolecular Human Telomeric G-Quadruplex in Potassium Solution: A Novel Adenine Triple Formation. Nucleic Acids Res. 2007, 35, 2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Dai J; Carver M; Punchihewa C; Jones RA; Yang D Structure of the Hybrid-2 Type Intramolecular Human Telomeric G-Quadruplex in K+ Solution: Insights into Structure Polymorphism of the Human Telomeric Sequence. Nucleic Acids Res. 2007, 35, 4927–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhang Z; Dai J; Veliath E; Jones RA; Yang D Structure of a Two-G-Tetrad Intramolecular G-Quadruplex Formed by a Variant Human Telomeric Sequence in K+ Solution: Insights into the Interconversion of Human Telomeric G-Quadruplex Structures. Nucleic Acids Res. 2010, 38, 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Mathad RI; Hatzakis E; Dai J; Yang D C-MYC Promoter G-quadruplex Formed at the 5′-End of NHE III1 Element: Insights into Biological Relevance and Parallel-Stranded G-Quadruplex Stability. Nucleic Acids Res. 2011, 39, 9023–9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Dickerhoff J; Onel B; Chen L; Chen Y; Yang D Solution Structure of a MYC Promoter G-Quadruplex with 1:6:1 Loop Length. ACS Omega 2019, 4, 2533–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dai J; Carver M; Hurley LH; Yang D Solution Structure of a 2:1 Quindoline-c-MYC G-Quadruplex: Insights into G-Quadruplex-Interactive Small Molecule Drug Design. J. Am. Chem. Soc 2011, 133, 17673–17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Liu W; Lin C; Wu G; Dai J; Chang T-C; Yang D Structures of 1:1 and 2:1 Complexes of BMVC and MYC Promoter G-Quadruplex Reveal a Mechanism of Ligand Conformation Adjustment for G4-Recognition. Nucleic Acids Res. 2019, 47, 11931–11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Liu W; Zhong Y-F; Liu L-Y; Shen C-T; Zeng W; Wang F; Yang D; Mao Z-W Solution Structures of Multiple G-Quadruplex Complexes Induced by a Platinum(II)-Based Tripod Reveal Dynamic Binding. Nat. Commun 2018, 9, 3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Dickerhoff J; Dai J; Yang D Structural Recognition of the MYC Promoter G-Quadruplex by a Quinoline Derivative: Insights into Molecular Targeting of Parallel G-Quadruplexes. Nucleic Acids Res. 2021, 49, 5905–5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Dickerhoff J; Brundridge N; McLuckey SA; Yang D Berberine Molecular Recognition of the Parallel MYC G-Quadruplex in Solution. J. Med. Chem 2021, 64, 16205–16212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Dickerhoff J; Warnecke KR; Wang K; Deng N; Yang D Evaluating Molecular Docking Software for Small Molecule Binding to G-Quadruplex DNA. Int. J. Mol. Sci 2021, 22, 10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Deng N; Wickstrom L; Cieplak P; Lin C; Yang D Resolving the Ligand-Binding Specificity in c-MYC G-Quadruplex DNA: Absolute Binding Free Energy Calculations and SPR Experiment. J. Phys. Chem. B 2017, 121, 10484–10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wu G; Tillo D; Ray S; Chang T-C; Schneekloth JS; Vinson C; Yang D Custom G4 Microarrays Reveal Selective G-Quadruplex Recognition of Small Molecule BMVC: A Large-Scale Assessment of Ligand Binding Selectivity. Molecules 2020, 25, 3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Moyzis RK; Buckingham JM; Cram LS; Dani M; Deaven LL; Jones MD; Meyne J; Ratliff RL; Wu JR A Highly Conserved Repetitive DNA Sequence, (TTAGGG)n, Present at the Telomeres of Human Chromosomes. Proc. Natl. Acad. Sci. U. S. A 1988, 85, 6622–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wright WE; Tesmer VM; Huffman KE; Levene SD; Shay JW Normal Human Chromosomes Have Long G-Rich Telomeric Overhangs at One End. Genes Dev. 1997, 11, 2801–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sfeir AJ; Chai W; Shay JW; Wright WE Telomere-End Processing the Terminal Nucleotides of Human Chromosomes. Mol. Cell 2005, 18, 131–138. [DOI] [PubMed] [Google Scholar]
- (36).Sun D; Thompson B; Cathers BE; Salazar M; Kerwin SM; Trent JO; Jenkins TC; Neidle S; Hurley LH Inhibition of Human Telomerase by a G-Quadruplex-Interactive Compound. J. Med. Chem 1997, 40, 2113–2116. [DOI] [PubMed] [Google Scholar]
- (37).Neidle S; Parkinson G Telomere Maintenance as a Target for Anticancer Drug Discovery. Nat. Rev. Drug Discovery 2002, 1, 383–393. [DOI] [PubMed] [Google Scholar]
- (38).Riou JF; Guittat L; Mailliet P; Laoui A; Renou E; Petitgenet O; Megnin-Chanet F; Helene C; Mergny JL Cell Senescence and Telomere Shortening Induced by a New Series of Specific G-Quadruplex DNA Ligands. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 2672–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Shay JW; Reddel RR; Wright WE Cancer and TelomeresAn ALTernative to Telomerase. Science 2012, 336, 1388–1390. [DOI] [PubMed] [Google Scholar]
- (40).Wang Y; Patel DJ Solution Structure of the Human Telomeric Repeat d[AG3(T2AG3)3] G-Tetraplex. Structure 1993, 1, 263–282. [DOI] [PubMed] [Google Scholar]
- (41).Parkinson GN; Lee MPH; Neidle S Crystal Structure of Parallel Quadruplexes from Human Telomeric DNA. Nature 2002, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- (42).Dai J; Carver M; Yang D Polymorphism of Human Telomeric Quadruplex Structures. Biochimie 2008, 90, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Manna S; Sarkar D; Srivatsan SG A Dual-App Nucleoside Probe Provides Structural Insights into the Human Telomeric Overhang in Live Cells. J. Am. Chem. Soc 2018, 140, 12622–12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hänsel R; Löhr F; Foldynová-Trantírková S; Bamberg E; Trantírek L; Dötsch V The Parallel G-Quadruplex Structure of Vertebrate Telomeric Repeat Sequences Is Not the Preferred Folding Topology under Physiological Conditions. Nucleic Acids Res. 2011, 39, 5768–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Monsen RC; Chakravarthy S; Dean WL; Chaires JB; Trent JO The Solution Structures of Higher-Order Human Telomere G-Quadruplex Multimers. Nucleic Acids Res. 2021, 49, 1749–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Siddiqui-Jain A; Grand CL; Bearss DJ; Hurley LH Direct Evidence for a G-Quadruplex in a Promoter Region and Its Targeting with a Small Molecule to Repress c-MYC Transcription. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zheng K; He Y; Liu H; Li X; Hao Y; Tan Z Superhelicity Constrains a Localized and R-Loop-Dependent Formation of G-Quadruplexes at the Upstream Region of Transcription. ACS Chem. Biol 2017, 12, 2609–2618. [DOI] [PubMed] [Google Scholar]
- (48).Sekibo DAT; Fox KR The Effects of DNA Supercoiling on G-Quadruplex Formation. Nucleic Acids Res. 2017, 45, 12069–12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Spiegel J; Cuesta SM; Adhikari S; Hänsel-Hertsch R; Tannahill D; Balasubramanian S G-Quadruplexes Are Transcription Factor Binding Hubs in Human Chromatin. Genome Biol. 2021, 22, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kaiser CE; Van Ert NA; Agrawal P; Chawla R; Yang DZ; Hurley LH Insight into the Complexity of the I-Motif and G-Quadruplex DNA Structures Formed in the KRAS Promoter and Subsequent Drug Induced Gene Repression. J. Am. Chem. Soc 2017, 139, 8522–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Simonsson T; Pecinka P; Kubista M DNA Tetraplex Formation in the Control Region of C-Myc. Nucleic Acids Res. 1998, 26, 1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Seenisamy J; Rezler EM; Powell TJ; Tye D; Gokhale V; Joshi CS; Siddiqui-Jain A; Hurley LH The Dynamic Character of the G-Quadruplex Element in the c-MYC Promoter and Modification by TMPyP4. J. Am. Chem. Soc 2004, 126, 8702–8709. [DOI] [PubMed] [Google Scholar]
- (53).Phan AT; Modi YS; Patel DJ Propeller-Type Parallel-Stranded G-Quadruplexes in the Human c-Myc Promoter. J. Am. Chem. Soc 2004, 126, 8710–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hatzakis E; Okamoto K; Yang D Thermodynamic Stability and Folding Kinetics of the Major G-Quadruplex and Its Loop Isomers Formed in the Nuclease Hypersensitive Element in the Human c-Myc Promoter: Effect of Loops and Flanking Segments on the Stability of Parallel-Stranded Intramolecular G-Quadruplexes. Biochemistry 2010, 49, 9152–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Chen Y; Yang D Sequence, Stability, and Structure of G-Quadruplexes and Their Interactions with Drugs. Curr. Protoc. Nucleic Acid Chem 2012, 50, 17.5.1–17.5.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sun D; Hurley LH The Importance of Negative Superhelicity in Inducing the Formation of G-Quadruplex and i-Motif Structures in the c-Myc Promoter: Implications for Drug Targeting and Control of Gene Expression. J. Med. Chem 2009, 52, 2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Hänsel-Hertsch R; Beraldi D; Lensing SV; Marsico G; Zyner K; Parry A; Di Antonio M; Pike J; Kimura H; Narita M; Tannahill D; Balasubramanian S G-Quadruplex Structures Mark Human Regulatory Chromatin. Nat. Genet 2016, 48, 1267–1272. [DOI] [PubMed] [Google Scholar]
- (58).Dexheimer TS; Sun D; Hurley LH Deconvoluting the Structural and Drug-Recognition Complexity of the G-Quadruplex-Forming Region Upstream of the Bcl-2 P1 Promoter. J. Am. Chem. Soc 2006, 128, 5404–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kikin O; D’Antonio L; Bagga PS QGRS Mapper: A Web-Based Server for Predicting G-Quadruplexes in Nucleotide Sequences. Nucleic Acids Res. 2006, 34, W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Huppert JL; Balasubramanian S Prevalence of Quadruplexes in the Human Genome. Nucleic Acids Res. 2005, 33, 2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Todd AK; Johnston M; Neidle S Highly Prevalent Putative Quadruplex Sequence Motifs in Human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Sun D; Guo K; Rusche JJ; Hurley LH Facilitation of a Structural Transition in the Polypurine/Polypyrimidine Tract within the Proximal Promoter Region of the Human VEGF Gene by the Presence of Potassium and G-Quadruplex-Interactive Agents. Nucleic Acids Res. 2005, 33, 6070–6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Guo K; Gokhale V; Hurley LH; Sun D Intramolecularly Folded G-Quadruplex and i-Motif Structures in the Proximal Promoter of the Vascular Endothelial Growth Factor Gene. Nucleic Acids Res. 2008, 36, 4598–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Onel B; Lin C; Yang DZ DNA G-Quadruplex and Its Potential as Anticancer Drug Target. Sci. China Chem 2014, 57, 1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).De Armond R; Wood S; Sun D; Hurley LH; Ebbinghaus SW Evidence for the Presence of a Guanine Quadruplex Forming Region within a Polypurine Tract of the Hypoxia Inducible Factor 1α Promoter. Biochemistry 2005, 44, 16341–16350. [DOI] [PubMed] [Google Scholar]
- (66).Fernando H; Reszka AP; Huppert J; Ladame S; Rankin S; Venkitaraman AR; Neidle S; Balasubramanian S A Conserved Quadruplex Motif Located in a Transcription Activation Site of the Human C-Kit Oncogene. Biochemistry 2006, 45, 7854–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Guo K; Pourpak A; Beetz-Rogers K; Gokhale V; Sun D; Hurley LH Formation of Pseudosymmetrical G-Quadruplex and i-Motif Structures in the Proximal Promoter Region of the RET Oncogene. J. Am. Chem. Soc 2007, 129, 10220–10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Palumbo SL; Ebbinghaus SW; Hurley LH Formation of a Unique End-to-End Stacked Pair of G-Quadruplexes in the HTERT Core Promoter with Implications for Inhibition of Telomerase by G-Quadruplex-Interactive Ligands. J. Am. Chem. Soc 2009, 131, 10878–10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Qin Y; Fortin JS; Tye D; Gleason-Guzman M; Brooks TA; Hurley LH Molecular Cloning of the Human Platelet-Derived Growth Factor Receptor Beta (PDGFR-Beta) Promoter and Drug Targeting of the G-Quadruplex-Forming Region to Repress PDGFR-Beta Expression. Biochemistry 2010, 49, 4208–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Brown RV; Wang T; Chappeta VR; Wu G; Onel B; Chawla R; Quijada H; Camp SM; Chiang ET; Lassiter QR; Lee C; Phanse S; Turnidge MA; Zhao P; Garcia JGN; Gokhale V; Yang D; Hurley LH The Consequences of Overlapping G-Quadruplexes and i-Motifs in the Platelet-Derived Growth Factor Receptor β Core Promoter Nuclease Hypersensitive Element Can Explain the Unexpected Effects of Mutations and Provide Opportunities for Selective Targeting of Both Structures by Small Molecules To Downregulate Gene Expression. J. Am. Chem. Soc 2017, 139, 7456–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Balasubramanian S; Hurley LH; Neidle S Targeting G-Quadruplexes in Gene Promoters: A Novel Anticancer Strategy? Nat. Rev. Drug Discovery 2011, 10, 261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Zhang L; Liu H; Shao Y; Lin C; Jia H; Chen G; Yang D; Wang Y Selective Lighting Up of Epiberberine Alkaloid Fluorescence by Fluorophore-Switching Aptamer and Stoichiometric Targeting of Human Telomeric DNA G-Quadruplex Multimer. Anal. Chem 2015, 87, 730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Vita M; Henriksson M The Myc Oncoprotein as a Therapeutic Target for Human Cancer. Semin Cancer Biol. 2006, 16, 318–330. [DOI] [PubMed] [Google Scholar]
- (74).Horiuchi D; Anderton B; Goga A Taking on Challenging Targets: Making MYC Druggable. Am. Soc. Clin. Oncol. Educ. Book 2014, 34, e497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Chen H; Liu H; Qing G Targeting Oncogenic Myc as a Strategy for Cancer Treatment. Signal Transduct. Target. Ther 2018, 3, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Lourenco C; Resetca D; Redel C; Lin P; MacDonald AS; Ciaccio R; Kenney TMG; Wei Y; Andrews DW; Sunnerhagen M; Arrowsmith CH; Raught B; Penn LZ MYC Protein Interactors in Gene Transcription and Cancer. Nat. Rev. Cancer 2021, 21, 579–591. [DOI] [PubMed] [Google Scholar]
- (77).Brown RV; Danford FL; Gokhale V; Hurley LH; Brooks TA Demonstration That Drug-Targeted Down-Regulation of MYC in Non-Hodgkins Lymphoma Is Directly Mediated through the Promoter G-Quadruplex. J. Biol. Chem 2011, 286, 41018–41027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Ou T-M; Lu Y-J; Zhang C; Huang Z-S; Wang X-D; Tan J-H; Chen Y; Ma D-L; Wong K-Y; Tang JC-O; Chan AS-C; Gu L-Q Stabilization of G-Quadruplex DNA and Down-Regulation of Oncogene c-Myc by Quindoline Derivatives. J. Med. Chem 2007, 50, 1465–1474. [DOI] [PubMed] [Google Scholar]
- (79).Kang HJ; Park HJ Novel Molecular Mechanism for Actinomycin D Activity as an Oncogenic Promoter G-Quadruplex Binder. Biochemistry 2009, 48, 7392–7398. [DOI] [PubMed] [Google Scholar]
- (80).Shin-ya K; Wierzba K; Matsuo K; Ohtani T; Yamada Y; Furihata K; Hayakawa Y; Seto H Telomestatin, a Novel Telomerase Inhibitor from Streptomyces Anulatus. J. Am. Chem. Soc 2001, 123, 1262–1263. [DOI] [PubMed] [Google Scholar]
- (81).Han FXG; Wheelhouse RT; Hurley LH Interactions of TMPyP4 and TMPyP2 with Quadruplex DNA. Structural Basis for the Differential Effects on Telomerase Inhibition. J. Am. Chem. Soc 1999, 121, 3561–3570. [Google Scholar]
- (82).Chang CC; Wu JY; Chien CW; Wu WS; Liu H; Kang CC; Yu LJ; Chang TC A Fluorescent Carbazole Derivative: High Sensitivity for Quadruplex DNA. Anal. Chem 2003, 75, 6177–6183. [DOI] [PubMed] [Google Scholar]
- (83).Chang CC; Kuo IC; Ling IF; Chen CT; Chen HC; Lou PJ; Lin JJ; Chang TC Detection of Quadruplex DNA Structures in Human Telomeres by a Fluorescent Carbazole Derivative. Anal. Chem 2004, 76, 4490–4494. [DOI] [PubMed] [Google Scholar]
- (84).Chang CC; Chu JF; Kao FJ; Chiu YC; Lou PJ; Chen HC; Chang TC Verification of Antiparallel G-Quadruplex Structure in Human Telomeres by Using Two-Photon Excitation Fluorescence Lifetime Imaging Microscopy of the 3,6-Bis(1-Methyl-4-Vinylpyridinium)Carbazole Diiodide Molecule. Anal. Chem 2006, 78, 2810–2815. [DOI] [PubMed] [Google Scholar]
- (85).Ray S; Tillo D; Boer RE; Assad N; Barshai M; Wu G; Orenstein Y; Yang D; Schneekloth JS; Vinson C Custom DNA Microarrays Reveal Diverse Binding Preferences of Proteins and Small Molecules to Thousands of G-Quadruplexes. ACS Chem. Biol 2020, 15, 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Fleming AM; Burrows CJ Interplay of Guanine Oxidation and G-Quadruplex Folding in Gene Promoters. J. Am. Chem. Soc 2020, 142, 1115–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Wang K-B; Liu Y; Li Y; Dickerhoff J; Li J; Yang M-H; Yang D; Kong L-Y Oxidative Damage Induces a Vacancy G-Quadruplex That Binds Guanine Metabolites: Solution Structure of a CGMP Fill-in Vacancy G-Quadruplex in the Oxidized BLM Gene Promoter. J. Am. Chem. Soc 2022, 144, 6361–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Kendrick S; Kang HJ; Alam MP; Madathil MM; Agrawal P; Gokhale V; Yang DZ; Hecht SM; Hurley LH The Dynamic Character of the BCL2 Promoter I-Motif Provides a Mechanism for Modulation of Gene Expression by Compounds That Bind Selectively to the Alternative DNA Hairpin Structure. J. Am. Chem. Soc 2014, 136, 4161–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Dai JX; Hatzakis E; Hurley LH; Yang DZ I-Motif Structures Formed in the Human c-MYC Promoter Are Highly Dynamic-Insights into Sequence Redundancy and I-Motif Stability. PLoS One 2010, 5, e11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).González V; Guo K; Hurley L; Sun D Identification and Characterization of Nucleolin as a C-Myc G-Quadruplex-Binding Protein. J. Biol. Chem 2009, 284, 23622–23635. [DOI] [PMC free article] [PubMed] [Google Scholar]