PURPOSE
To explore the novel diagnostic value of epigenetic imprinting biomarkers in thyroid nodules.
PATIENTS AND METHODS
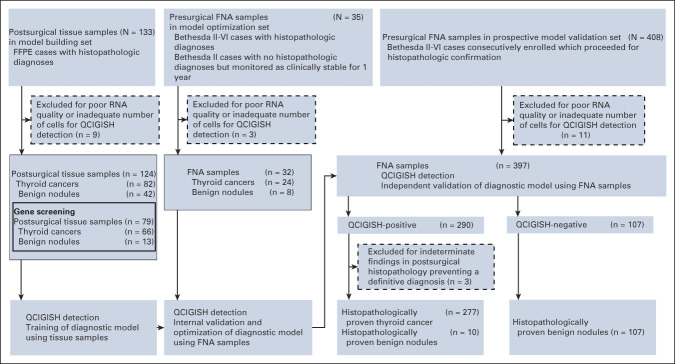
A total of 550 patients with fine-needle aspiration (FNA)–evaluated and histopathologically confirmed thyroid nodules were consecutively recruited from eight medical centers. Quantitative chromogenic imprinted gene in situ hybridization (QCIGISH) was used to assess the allelic expression of imprinted genes SNRPN and HM13, on the basis of which a diagnostic grading model for thyroid nodules was developed. The model was retrospectively trained on 124 postsurgical thyroid samples, optimized on 32 presurgical FNA samples, and prospectively validated on 394 presurgical FNA samples. Blinded central review–based cytopathologic and histopathologic diagnoses were used as the reference standard.
RESULTS
For thyroid malignancy, the QCIGISH test achieved an overall diagnostic sensitivity of 100% (277/277), a specificity of 91.5% (107/117; 95% CI, 86.4 to 96.5), a positive predictive value (PPV) of 96.5% (95% CI, 94.4 to 98.6), and a negative predictive value (NPV) of 100% in the prospective validation, with a diagnostic accuracy of 97.5% (384/394; 95% CI, 95.9 to 99.0). QCIGISH demonstrated a PPV of 97.8% (95% CI, 94.7 to 100) and NPV of 100%, with a diagnostic accuracy of 98.2% (111/113; 95% CI, 95.8 to 100), for indeterminate Bethesda III-V thyroid nodules. QCIGISH demonstrated a PPV of 96.6% (95% CI, 91.9 to 100) and a NPV of 100%, with a diagnostic accuracy of 97.5% (79/81; 95% CI, 94.2 to 100), for Bethesda III-IV. For Bethesda VI, QCIGISH showed a 100% (184/184) accuracy.
CONCLUSION
This imprinting biomarker-based test can effectively distinguish malignant from benign thyroid nodules. The high PPV and NPV make the test both an excellent rule-in and rule-out diagnostic tool. With such a diagnostic performance and its technical simplicity, this novel thyroid molecular test is clinically widely applicable.
INTRODUCTION
Thyroid nodules are common with a prevalence of about 70% on thyroid ultrasonography, and about 5% of them are malignant.1,2 Accurate assessment to distinguish malignant from benign thyroid nodules is critical for their appropriate clinical management. Although ultrasound imaging combined with fine-needle aspiration (FNA) biopsy is the diagnostic mainstay for thyroid nodules, about 20%-30% are diagnostically indeterminate with this approach.3,4 This diagnostic dilemma often causes confusion on how to treat a thyroid nodule clinically.5
CONTEXT
Key Objective
To explore the diagnostic value of imprinted genes for thyroid nodules, as they have emerged to be cancer-relevant, implying their potential to be a novel category of effective diagnostic biomarkers for thyroid nodules.
Knowledge Generated
Imprinted gene biomarkers are demonstrated here to have a high diagnostic value for thyroid nodules, which can effectively distinguish malignant from benign thyroid nodules; the high diagnostic sensitivity and specificity make the test both an excellent rule-in and rule-out test across all Bethesda cytologic categories of thyroid nodules, including diagnostically most challenging indeterminate thyroid nodules.
Relevance (M.L. Gillison)
A novel assay for imprinted gene biomarkers has considerable potential for clinical diagnosis of malignant thyroid nodules.*
*Relevance section written by JCO Associate Editor Maura L. Gillison, MD, PhD.
The American College of Radiology Thyroid Imaging, Reporting, and Data System (ACR TI-RADS) category classification6 and Bethesda cytology classification7 are widely used to estimate the malignancy risk of thyroid nodules. The presurgical diagnosis can be challenging, particularly in indeterminate cytologic categories, including Bethesda III, IV, and V—atypia of undetermined significance or follicular lesion of undetermined significance, follicular neoplasm or suspicious for follicular neoplasm, and suspicious for malignancy, respectively. There are several thyroid diagnostic biomarker systems used variably around the world. These include mostly genetic alterations, gene expression, DNA methylation, and microRNAs, with each being associated with certain limitations.8-12 A more effective biomarker-based diagnostic approach is needed for thyroid nodules.
Genomic imprinting is an epigenetic regulatory mechanism in mammalian embryo development and tumorigenesis.13,14 In normal somatic cells, paternal and maternal alleles of an imprinted gene are differentially methylated in an allele-specific manner, resulting in the silencing of one allele and activation of the other. In cancers, the normally silenced allele is often aberrantly activated in certain imprinted genes, resulting in the expressions of both alleles.15 This phenomenon is termed loss of imprinting (LOI), which is associated with various cancers.15 A nascent RNA in situ hybridization (ISH) method, targeting the short-lived introns to label and visualize transcription sites, has been widely used to study the transcriptional regulations of both imprinted and nonimprinted genes.16-21 We have previously adopted this approach to developing a sensitive and specific objective quantification of imprinting alterations through measuring the biallelic expression (BAE), multiallelic expression (MAE), and total expression (TE) of a panel of imprinted genes, which we termed quantitative chromogenic imprinted gene in situ hybridization (QCIGISH).22 Using this method, we previously identified three imprinted genes with diagnostic potential for cancer—guanine nucleotide-binding protein, alpha-stimulating complex locus (GNAS), growth factor receptor–bound protein (GRB10), and small nuclear ribonucleoprotein polypeptide N (SNRPN).22 Here, we used the QCIGISH technique to investigate the diagnostic value of the expression status of these three imprinted genes and a new imprinted gene minor histocompatibility antigen H13 (HM13) on presurgical thyroid FNA specimens of thyroid nodules and matched histopathologic tissues. The combination of SNRPN and HM13 was found to be particularly efficient for developing an accurate diagnostic grading model for thyroid nodules, which we investigated for its diagnostic value in various Bethesda categories of thyroid nodules.
PATIENTS AND METHODS
Study Design and Participants
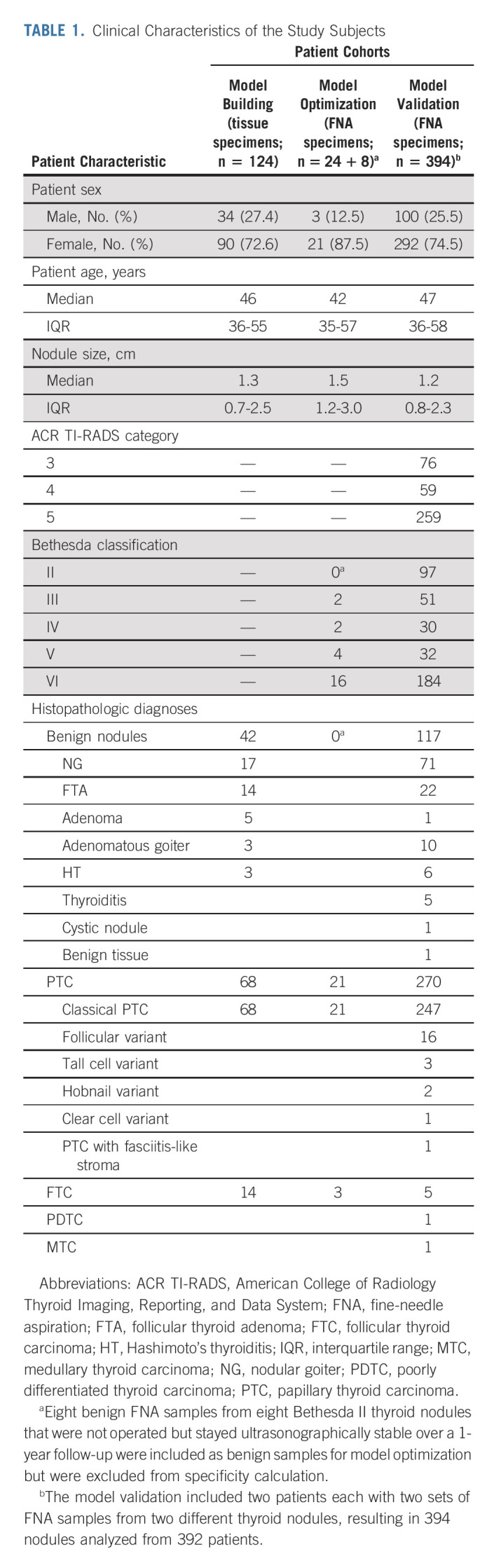
Patients with ultrasound-detected thyroid nodules recommended to have FNA evaluations were recruited from eight medical centers, including Shanghai Tenth People's Hospital of Tongji University School of Medicine, Jiangyuan Hospital affiliated to Jiangsu Institute of Nuclear Medicine, Taizhou People's Hospital, Taizhou Third People's Hospital, Shengjing Hospital of China Medical University, Nanjing First Hospital, Cancer Hospital of the University of Chinese Academy of Sciences, and Henan Cancer Hospital. Patients were divided into three groups for different test purposes as illustrated in Figure 1. Thyroid ultrasound examination and fine-needle aspiration biopsy were performed as described in the Data Supplement (online only). Clinical characteristics of the subjects are shown in Table 1. We used 124 formalin-fixed paraffin-embedded (FFPE) postsurgical thyroid specimens for initial diagnostic model building (Fig 1). These consisted of 42 benign nodules and 82 malignant nodules (68 papillary thyroid carcinoma [PTC] and 14 follicular thyroid carcinoma [FTC]). We used 32 presurgical thyroid FNA specimens for model optimization, including eight benign nodules with Bethesda II cytologic diagnosis and 24 malignant nodules (21 PTC and three FTC, all pathologically confirmed; Fig 1). A total of 408 cases of thyroid nodules (ACR TI-RADS category 3-5; Data Supplement) were consecutively recruited for prospective model validation of the QCIGISH test (Fig 1); they all had postsurgical histopathology diagnoses. Among these, 11 cases of poor-quality FNA specimen and three cases with indeterminate postsurgical histopathology were excluded. The operated benign (Bethesda II) nodules were so treated for clinical symptoms or patient's preference per standard clinical practice guidelines.23 A total of 394 FNA samples that had postsurgical histopathology were finally included for the prospective validation study. These included histopathologically confirmed 117 benign nodules and 277 malignant nodules (270 PTC, five FTC, one poorly differentiated thyroid cancer, and one medullary thyroid carcinoma). The QCIGISH testing on FNA specimen was performed in a blinded manner: persons conducting the QCIGISH test were blinded to the clinical, cytologic, and histopathologic diagnoses. The FNA samples from each patient were subjected to simultaneous Bethesda cytology classification (Data Supplement) and QCIGISH testing with the specimens analyzed and blindly scored according to the diagnostic QCIGISH grading criteria.
FIG 1.
Study design and workflow diagram. FFPE, formalin-fixed paraffin-embedded; FNA, fine-needle aspiration; QCIGISH, quantitative chromogenic imprinted gene in situ hybridization.
TABLE 1.
Clinical Characteristics of the Study Subjects
All cytopathologic and histopathologic diagnoses were from a central review by an independent committee of three experienced thyroid pathologists who were blinded to the QCIGISH results (Data Supplement). This study was approved by the ethics committees of Shanghai Tenth People's Hospital, Nanjing First Hospital, and Cancer Hospital of the University of Chinese Academy of Sciences (approval number SHSY-IEC-4.1/19-6/01), Jiangyuan Hospital (approval number YL201811), Taizhou People's Hospital and Taizhou Third People's Hospital (approval number TZ20190520), Shengjing Hospital (approval number 2020PS377K), and Henan Cancer Hospital (approval number 2019122504). All participants were age older than 18 years and provided informed consents. This study was registered in the Chinese Clinical Trial Registry (registration number: ChiCTR1900025265).
Sample Preparation and QCIGISH Detection
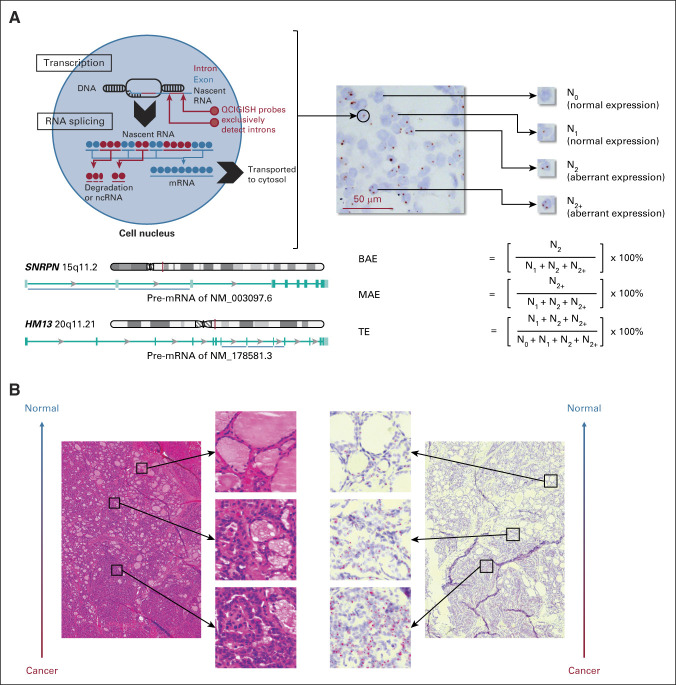
For model building, surgical tissues were prepared using a previously described procedure.22 For model optimization and blinded prospective validation, a thyroid FNA specimen was divided into two parts for simultaneous cytopathology evaluation and blinded QCIGISH testing (Data Supplement). The FNA specimens for QCIGISH testing were fixed in 10% formalin neutral buffer immediately after sampling and were mechanically separated before being mounted on positively charged slides. Each FNA sample was mounted in a well of 10-mm2 area with hydrophobic barrier and dried overnight at 60°C. For ISH, the sample slides were pretreated following the RNAscope sample preparation procedures.24 ISH was performed as described previously22 using probes targeting the noncoding intronic regions of nascent RNAs from GNAS, GRB10, SNRPN, and HM13, as detailed in Figure 2A and the Data Supplement. The detected gene-expressing site appeared as a distinct red or brown dot under common bright-field microscope (Fig 2A). For surgical tissue samples, four representative 400× high-power fields were selected for nuclei counting. For FNA specimens, scanned microscopic images with at least 1,200 nuclei for each sample were randomly selected for nuclei counting. The number of nuclei with various gene expression signals were manually counted for FFPE tissue sections and automatically counted using an image recognition software program with manual verification for FNA specimens. BAE, MAE, and TE were determined as previously described (Fig 2A).22
FIG 2.
The principle of QCIGISH—visualization, quantification, and pathologic confirmation of the allelic expression status of imprinted genes. (A) Conceptual framework of QCIGISH. Shown is a QCIGISH-stained tissue section of a case of PTC. Blue components in the image are cell nuclei stained using hematoxylin. The red vertical lines on the chromosome map indicate the gene loci. The blue horizontal lines under the intron/exon map of pre-mRNAs indicate the targeted introns of the in situ hybridization probes. (B) Pathologic confirmation of the QCIGISH results using simultaneous hematoxylin and eosin staining examination of a case of PTC. The low-magnification image was captured at a particular tumor region showing both malignant (lower subregion) and benign (upper subregion) morphologic characteristics. BAE, biallelic expression; MAE, multiallelic expression; ncRNA, noncoding RNA; PTC, papillary thyroid carcinoma; QCIGISH, quantitative chromogenic imprinted gene in situ hybridization; TE, total expression.
Gene Screening Study, Diagnostic Grading Model Building, and Model Optimization
We identified two imprinted genes—SNRPN and HM13 as the most efficient thyroid cancer biomarkers from a gene screening study of four candidate imprinted genes (GNAS, GRB10, SNRPN, and HM13; Data Supplement); this two-gene combination achieved optimally high sensitivity with minimal compromise in specificity. QCIGISH test of the two genes was applied to the model training set of 124 surgical thyroid tissue samples, with the development of a five-grade thyroid cancer prediction model (Data Supplement). We showed a correlation between the allelic expression signal numbers of these genes and the morphologic malignancy level by comparing the QCIGISH and hematoxylin and eosin staining on serial tissue sections of the tumor (Fig 2B). Taking grades 0 and I as negative predictions and grades II, III, and IV as positive predictions, this model demonstrated an optimism-corrected 93.9% sensitivity (95% CI, 93.6 to 94.1) and 86.3% specificity (95% CI, 86.0 to 86.6). We independently applied this QCIGISH model established on FFPE to 32 FNA specimens for optimization in presurgical diagnosis (Data Supplement) and achieved an optimism-corrected sensitivity of 88.6% (95% CI, 88.2 to 89.0) and an optimism-corrected false-positive rate of 1.9% (95% CI, 1.3 to 2.4; Data Supplement).
Statistical Analysis
Continuous variables were reported as medians with interquartile ranges, while frequencies and proportions were reported for categorical variables. For the imprinted gene panel from the model building set, a robust rank-order nonparametric test was used for the comparison of the benign and malignant groups.25 Area under the curve was used to evaluate and compare the discrimination performance of the imprinted gene panel for the gene screening, model building, and optimization sets.26 All computed area under the curve values were generated using the ROCR package in R.27 Hypothesis testing was done in a two-sided manner, with computed P < .05 considered to be significant. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), diagnostic accuracy, and 95% CIs were calculated using standard methods. All statistical analyses and visualizations were conducted using R software (version 3.5.0). Sample size calculation is described in the Data Supplement.
RESULTS
Test of the Clinical Applicability of QCIGISH in a Blinded Prospective Validation Cohort
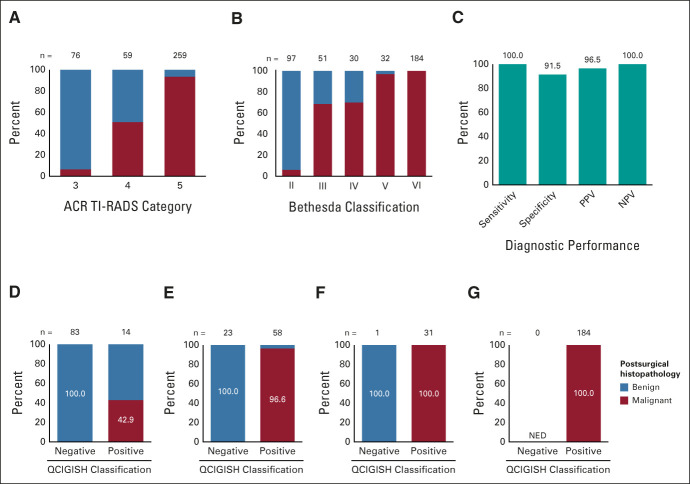
We tested the clinical applicability of the QCIGISH model in an independent blinded prospective validation set of 394 thyroid nodules (Data Supplement). An increasing histopathologic malignancy rate was observed from ACR TI-RADS categories 3-5 on ultrasonography (Fig 3A). The histopathologic malignancy rate was also increased from Bethesda II to Bethesda VI thyroid nodules, as can be expected (Fig 3B). Using histopathologic diagnoses as the diagnostic reference standard, the final QCIGISH grading model on FNA specimen testing achieved an overall diagnostic sensitivity of 100% (277/277), a specificity of 91.5% (107/117; 95% CI, 86.4 to 96.5), a PPV of 96.5% (95% CI, 94.4 to 98.6), and a NPV of 100% for malignancy in the validation set of 394 nodules (Table 2, Fig 3C). The corresponding overall diagnostic accuracy was 97.5% (384/394; 95% CI, 95.9 to 99.0).
FIG 3.
Performance of QCIGISH test in the blinded prospective validation cohort. (A) Distribution of histopathologically benign and malignant cases in different ultrasound imaging categories. (B) Distribution of histopathologically benign and malignant cases in different Bethesda cytopathology categories. (C) Sensitivity, specificity, PPV, and NPV of the QCIGISH test in the blinded prospective validation study. Distribution of histopathologically benign and malignant cases in QCIGISH-negative and QCIGISH-positive groups for (D) Bethesda II, (E) Bethesda III and IV, (F) Bethesda V, and (G) Bethesda IV categories. Histopathologic benign and malignant diagnoses were confirmed by postsurgical thyroid examination in all cases. ACR TI-RADS, American College of Radiology Thyroid Imaging, Reporting, and Data System; NED, no evidence of disease; NPV, negative predictive value; PPV, positive predictive value; QCIGISH, quantitative chromogenic imprinted gene in situ hybridization.
TABLE 2.
Performance of Quantitative Chromogenic Imprinted Gene In Situ Hybridization Test on Fine-Needle Aspiration Specimens in Different Bethesda Categories of Thyroid Nodules in the Prospective Model Validation
The Diagnostic Values of QCIGISH in Different Bethesda Cytologic Categories
The diagnostic performance of the QCIGISH test in different Bethesda cytologic categories is summarized in Table 2. For Bethesda II thyroid nodules, 100% (83/83) of QCIGISH-negative cases were histopathologically benign, while 42.9% (6/14) of QCIGISH-positive cases were histopathologically proven to be malignant (Table 2, Fig 3D). For Bethesda III and IV thyroid nodules, 100% (23/23) of QCIGISH-negative cases on FNA specimen testing were histopathologically proven to be benign, while 96.6% (56/58) of QCIGISH-positive cases on FNA specimen testing were histopathologically proven to be malignant, demonstrating a PPV of 96.6% (95% CI, 91.9 to 100) and a NPV of 100%, with a diagnostic accuracy of 97.5% (79/81; 95% CI, 94.2 to 100) for diagnosing the malignancy of thyroid nodules (Table 2, Fig 3E). For Bethesda V thyroid nodules, the only case classified as QCIGISH-negative was histopathologically proven to be benign, and 100% (31/31) of QCIGISH-positive cases on FNA specimens proved to be histopathologically malignant (Table 2, Fig 3F). For Bethesda VI thyroid nodules, 100% (184/184) of QCIGISH-positive cases on FNA specimens proved to be histopathologically malignant (Table 2, Fig 3G). When cytologically indeterminate Bethesda III, IV, and V were combined, 92.3% (24/26; 95% CI, 82.1 to 100) of the FNA cases that were histopathologically proven to be benign were negative on QCIGISH test, and 100% (87/87) of the FNA cases that were histopathologically proven to be malignant were positive on QCIGISH test, demonstrating a PPV of 97.8% (95% CI, 94.7 to 100) and a NPV of 100% for diagnosing the malignancy of thyroid nodules (Table 2). The overall diagnostic accuracy for combined Bethesda III, IV, and V cases was 98.2% (111/113; 95% CI, 95.8 to 100).
In two cases, presurgical QCIGISH test on two nodules in the same patient distinguished the malignant from the benign one, which were histopathologically confirmed. In addition, 52 Bethesda II cases of thyroid nodules not surgically operated were all negative on QCGISH testing.
As a case illustration of the excellent diagnostic performance of QCIGISH test on indeterminate Bethesda thyroid nodules, the Data Supplement shows histopathologically confirmed one benign and one malignant thyroid nodule, which were both ultrasonographically ACR TI-RADS category 5 and cytologically Bethesda III and were indeterminate nodules. QCIGISH test was able to presurgically distinguish the benign from the malignant case as histopathologically confirmed.
DISCUSSION
Although combined ultrasonographic and cytologic evaluation with FNA is currently the diagnostic mainstay for thyroid nodules, it can be challenging, particularly in the case of indeterminate cytology. This is true even with several currently used molecular diagnostic systems, as they each have limitations. In this study, we tested the value of the imprinted gene-based QCIGISH in diagnosing thyroid nodules and demonstrated an excellent diagnostic performance, including a high PPV of 97.8% and a NPV of 100% in cytologically indeterminate Bethesda III-V thyroid nodules. For Bethesda III and IV thyroid nodules, which are most challenging diagnostically, QCIGISH demonstrated a PPV of 96.6% and a NPV of 100%. The high NPV of QCIGISH can effectively help rule out malignancy of thyroid nodules, while its high PPV makes it also an effective rule-in test.
Aberrant expressing status of an imprinted gene often occurs at an early stage of carcinogenesis. An efficient and practical detection method to quantify the imprinting changes to reliably assess malignancy has been lacking. We have recently developed the novel QCIGISH method targeting noncoding intronic nascent RNAs to directly visualize transcription loci of imprinted genes in cell nuclei.22 The BAE observed with this method most likely represents LOI, with activation of both the paternal and maternal alleles, but it could also be from the copy-number variation (CNV) of the normally activated allele.28 Similarly, MAE could represent activation of the normally imprinted allele plus its CNV or just CNV. Our previous studies have demonstrated that BAE, MAE, and TE indeed represented the combined LOI and CNV of imprinted genes in various cancers.29,30 Regardless of the mechanism, an increase in the expression signals of the imprinted gene suggests malignancy. We have previously identified three common imprinted genes GNAS, GRB10, and SNRPN showing aberrant expression in 10 cancer types.22 In this study, we investigated the diagnostic value of these genes and additionally also a novel imprinted gene HM13 in thyroid nodules. Through model building and optimization, we demonstrated that the combination of SNRPN and HM13 had the most efficient diagnostic value that could effectively improve the presurgical diagnosis of thyroid nodules. Following the model optimization using FNA specimens, the prospective validation study with the grading model demonstrated a high diagnostic performance across all thyroid nodules, including Bethesda III-V nodules, which account for about 30% of thyroid nodules, representing a considerable diagnostic challenge in clinical thyroidology.3,4 There is well-known diagnostic variability in thyroid cytopathology, especially in indeterminate categories.31 To be consistent with this, there were several histopathologically confirmed malignant cases of cytologically Bethesda II thyroid nodules in this study. The QCIGISH can now effectively help mitigate this challenge as its diagnostic accuracy is remarkably high across all thyroid nodules regardless of the cytologic categories.
In this study, the malignancy rate in the Bethesda II cases was higher than reported.7 This is likely because many of these surgically treated Bethesda II thyroid nodules were clinically symptomatic and might thus have an intrinsically increased malignancy risk. The malignancy rates of Bethesda III and IV thyroid nodules in this study were also relatively high. This was likely because of the use of high-risk ultrasonographic characteristics or BRAF mutation to guide treatment of Bethesda III and IV nodules toward surgery in the hospitals participating in this study. Regardless, QCIGISH demonstrated a robust diagnostic performance across all Bethesda categories. Interestingly, among the three cases of QCIGISH-positive indeterminate histopathology, one case showed BRAF V600E mutation (Data Supplement), the second showed double immunohistochemistry staining for CK19 and Hbme-1 (Data Supplement), and the third showed double immunohistochemistry staining for CK19 and Galectin-3 (Data Supplement). These molecular markers are known to suggest carcinogenesis or early-stage cancer.32-34 As epigenetic alterations occur even before malignant morphologic changes, such apparently false QCIGISH-positive cases might actually have malignant potential and warrant careful clinical follow-up.
The QCIGISH test demonstrated a diagnostic sensitivity and NPV of both 100% in all Bethesda categories of the large cohort of thyroid nodules in this study. As such, this test can effectively help avoid unnecessary thyroid surgeries for benign nodules that are negative on presurgical QCIGISH test but are otherwise cytologically indeterminate. It is remarkable that the QCIGISH test showed such a high diagnostic performance using only two imprinted genes, making it an economically favorable test. Moreover, QCIGISH is based on ISH, which offers an easy and inexpensive yet accurate and robust diagnostic test. The technical simplicity of this test makes it widely applicable practically, which is different than some other molecular tests that are often too complex or too specialized technically to be widely applicable.8,10,35,36
The malignant cases of thyroid nodules consisted of mainly classical PTC and some FTC in this study, with some cases of nonclassical PTC. There was only one case of medullary thyroid carcinoma. Consistent with their rarity in the Asian population,37,38 Hürthle cell adenoma and carcinoma and noninvasive follicular thyroid neoplasm with papillary-like nuclear features were not included in this study. Also, the oncogenic functionality, if any, of imprinted genes SNRPN and HM13 in the thyroid gland is not well known, making it only speculative to understand their functional relevance with respect to their high diagnostic performance for thyroid malignancy. These are the limitations of this study.
In summary, to our knowledge, this study for the first time demonstrates that the imprinted gene-based QCIGISH test has a robust diagnostic performance for thyroid nodules. Its high NPV makes this test highly effective in ruling out malignancy, while its high PPV makes it also an excellent rule-in test, which will be particularly helpful in assisting the evaluation of cytologically indeterminate thyroid nodules. As such, this novel thyroid molecular diagnostic test will likely have a significant clinical impact.
ACKNOWLEDGMENT
The authors thank Rulong Shen at Ohio State University Wexner Medical Center, Wenbin Huang at Nanjing First Hospital in Nanjing, and Hongyu Yu at Changzheng Hospital in Shanghai for their role as experienced expert pathologists on the independent central committee to provide blinded cytopathologic and histopathologic review of all the thyroid specimens used in this study. The authors thank Han Si and Weiwei Shi at Lisen Imprinting Diagnostics Inc for their assistance in the performance of laboratory experiments and data collection. The authors also thank Chen Yuan at Lisen Imprinting Diagnostics Inc, for his coordination of the collaboration between the different medical centers and Lisen Imprinting Diagnostics Inc.
Ning Zhou
Employment: Lisen Imprinting Diagnostics
Xing Li
Employment: Lisen Imprinting Diagnostics
John P. Pineda
Employment: Lisen Imprinting Diagnostics
Xiaonan Wang
Employment: Lisen Imprinting Diagnostics Inc
Tong Cheng
Employment: Lisen Imprinting Diagnostics
Mingzhao Xing
Consulting or Advisory Role: Lisen Imprinting Diagnostics Inc
No other potential conflicts of interest were reported.
DISCLAIMER
The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
SUPPORT
Supported by the National Natural Science Foundation of China (grant Nos. 81772849 and 82171942), Innovation Capacity Development Plan of Jiangsu Province (grant No. BM2018023), Science and Technology Commission of Shanghai Municipality (grant Nos. 19DZ2251100, 19441903200, and 20ZR1443400), Shanghai Municipal Health Commission (grant Nos. 2019LJ21 and SHSLCZDZK03502), and Taizhou Science and Technology Support (Social Development) Project (grant No. TS201825). The study was partially sponsored by Lisen Imprinting Diagnostics Inc.
H.X., Y.Z., H.W., and N.Z. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Huixiong Xu, Yifeng Zhang, Hongxun Wu, Ning Zhou, Xing Li, Tong Cheng, Mingzhao Xing
Administrative support: Yifeng Zhang, Hongxun Wu, Qian Li
Provision of study materials or patients: Hongxun Wu, Yun Zhu, Jianwu Qin
Collection and assembly of data: Yifeng Zhang, Hongxun Wu, Yun Zhu, Huijun Fu, Ming Ying, Shufang Yang, Jiandong Bao, Lulu Yang, Bingjie Zhang, Lehang Guo, Liping Sun, Feng Lu, Hanxiang Wang, Ying Huang, Tiantong Zhu, Xiaonan Wang, Qing Wei, Chunjun Sheng, Shen Qu, Zhongwei Lv, Dong Xu, Qian Li, Yongling Dong
Data analysis and interpretation: Hongxun Wu, Ning Zhou, Xing Li, John P. Pineda, Lulu Yang, Feng Lu, Qian Li, Jianwu Qin, Tong Cheng, Mingzhao Xing
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
High Diagnostic Accuracy of Epigenetic Imprinting Biomarkers in Thyroid Nodules
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Ning Zhou
Employment: Lisen Imprinting Diagnostics
Xing Li
Employment: Lisen Imprinting Diagnostics
John P. Pineda
Employment: Lisen Imprinting Diagnostics
Xiaonan Wang
Employment: Lisen Imprinting Diagnostics Inc
Tong Cheng
Employment: Lisen Imprinting Diagnostics
Mingzhao Xing
Consulting or Advisory Role: Lisen Imprinting Diagnostics Inc
No other potential conflicts of interest were reported.
REFERENCES
- 1.Guth S, Theune U, Aberle J, et al. : Very high prevalence of thyroid nodules detected by high frequency (13 MHz) ultrasound examination. Eur J Clin Invest 39:699-706, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Yeung MJ, Serpell JW: Management of the solitary thyroid nodule. Oncologist 13:105-112, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Wang CC, Friedman L, Kennedy GC, et al. : A large multicenter correlation study of thyroid nodule cytopathology and histopathology. Thyroid 21:243-251, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongiovanni M, Spitale A, Faquin WC, et al. : The Bethesda system for reporting thyroid cytopathology: A meta-analysis. Acta Cytol 56:333-339, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Haugen BR, Alexander EK, Bible KC, et al. : 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid 26:1-133, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tessler FN, Middleton WD, Grant EG, et al. : ACR thyroid imaging, reporting and data system (TI-RADS): White paper of the ACR TI-RADS committee. J Am Coll Radiol 14:587-595, 2017 [DOI] [PubMed] [Google Scholar]
- 7.Cibas ES, Ali SZ: The 2017 Bethesda system for reporting thyroid cytopathology. Thyroid 27:1341-1346, 2017 [DOI] [PubMed] [Google Scholar]
- 8.Nikiforova MN, Mercurio S, Wald AI, et al. : Analytical performance of the ThyroSeq v3 genomic classifier for cancer diagnosis in thyroid nodules. Cancer 124:1682-1690, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel KN, Angell TE, Babiarz J, et al. : Performance of a genomic sequencing classifier for the preoperative diagnosis of cytologically indeterminate thyroid nodules. JAMA Surg 153:817-824, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos MTD, Buzolin AL, Gama RR, et al. : Molecular classification of thyroid nodules with indeterminate cytology: Development and validation of a highly sensitive and specific new miRNA-based classifier test using fine-needle aspiration smear slides. Thyroid 28:1618-1626, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarkesh M, Zadeh-Vakili A, Azizi F, et al. : Altered epigenetic mechanisms in thyroid cancer subtypes. Mol Diagn Ther 22:41-56, 2018 [DOI] [PubMed] [Google Scholar]
- 12.Ahmed AA, Essa MEA: Potential of epigenetic events in human thyroid cancer. Cancer Genet 239:13-21, 2019 [DOI] [PubMed] [Google Scholar]
- 13.Barlow DP, Bartolomei MS: Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 6:a018382, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murrell A: Genomic imprinting and cancer: From primordial germ cells to somatic cells. ScientificWorldJournal 6:1888-1910, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jelinic P, Shaw P: Loss of imprinting and cancer. J Pathol 211:261-268, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Wissink EM, Vihervaara A, Tippens ND, et al. : Nascent RNA analyses: Tracking transcription and its regulation. Nat Rev Genet 20:705-723, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown JM, Buckle VJ: Detection of nascent RNA transcripts by fluorescence in situ hybridization. Methods Mol Biol 659:33-50, 2010 [DOI] [PubMed] [Google Scholar]
- 18.Misteli T, Spector DL: RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell 3:697-705, 1999 [DOI] [PubMed] [Google Scholar]
- 19.Borensztein M: Investigating the inner cell mass of the mouse blastocyst by combined immunofluorescence staining and RNA fluorescence in situ hybridization. Methods Mol Biol 2214:157-173, 2021 [DOI] [PubMed] [Google Scholar]
- 20.Jouvenot Y, Poirier F, Jami J, et al. : Biallelic transcription of Igf2 and H19 in individual cells suggests a post-transcriptional contribution to genomic imprinting. Curr Biol 9:1199-1202, 1999 [DOI] [PubMed] [Google Scholar]
- 21.Kohda A, Taguchi H, Okumura K: Visualization of biallelic expression of the imprinted SNRPN gene induced by inhibitors of DNA methylation and histone deacetylation. Biosci Biotechnol Biochem 65:1236-1239, 2001 [DOI] [PubMed] [Google Scholar]
- 22.Shen R, Cheng T, Xu C, et al. : Novel visualized quantitative epigenetic imprinted gene biomarkers diagnose the malignancy of ten cancer types. Clin Epigenetics 12:71, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel KN, Yip L, Lubitz CC, et al. : The American Association of Endocrine Surgeons guidelines for the definitive surgical management of thyroid disease in adults. Ann Surg 271:e21-e93, 2020 [DOI] [PubMed] [Google Scholar]
- 24.Wang F, Flanagan J, Su N, et al. : RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14:22-29, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fligner MA, Policello GE: Robust rank procedures for the Behrens-Fisher problem. J Am Stat Assoc 76:162-168, 1981 [Google Scholar]
- 26.DeLong ER, DeLong DM, Clarke-Pearson DL: Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 44:837-845, 1988 [PubMed] [Google Scholar]
- 27.Chan BKC: Applied statistics for human genetics using R. Adv Exp Med Biol 1082:123-144, 2018 [DOI] [PubMed] [Google Scholar]
- 28.Martin-Trujillo A, Vidal E, Monteagudo-Sanchez A, et al. : Copy number rather than epigenetic alterations are the major dictator of imprinted methylation in tumors. Nat Commun 8:467, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen R, Wu HX, Cheng T, et al. : Epigenetic panel of imprinted genes can enhance the accuracy of FNA cytopathology for thyroid cancer. Thyroid 29:A-44, 2019 [Google Scholar]
- 30.Yung R, Cheng T, Li X, et al. : P1.09-12 In-situ hybridization visual scoring of epigenetic imprinting genes improves early diagnosis and grading of lung cancers. J Thorac Oncol 14:S500-S501, 2019 [Google Scholar]
- 31.Cibas ES, Baloch ZW, Fellegara G, et al. : A prospective assessment defining the limitations of thyroid nodule pathologic evaluation. Ann Intern Med 159:325-332, 2013 [DOI] [PubMed] [Google Scholar]
- 32.Xing M: BRAF mutation in thyroid cancer. Endocr Relat Cancer 12:245-262, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Yu P, Xiong Y, et al. : Significance of CK19, TPO, and HBME-1 expression for diagnosis of papillary thyroid carcinoma. Int J Clin Exp Med 8:4369-4374, 2015 [PMC free article] [PubMed] [Google Scholar]
- 34.Bartolazzi A, Orlandi F, Saggiorato E, et al. : Galectin-3-expression analysis in the surgical selection of follicular thyroid nodules with indeterminate fine-needle aspiration cytology: A prospective multicentre study. Lancet Oncol 9:543-549, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Steward DL, Carty SE, Sippel RS, et al. : Performance of a multigene genomic classifier in thyroid nodules with indeterminate cytology: A prospective blinded multicenter study. JAMA Oncol 5:204-212, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrell RM, Eyerly-Webb SA, Golding AC, et al. : Statistical comparison of afirma gsc and afirma gec outcomes in a community endocrine surgical practice: Early findings. Endocr Pract 25:161-164, 2019 [DOI] [PubMed] [Google Scholar]
- 37.Agarwal S, Bychkov A, Jung CK, et al. : The prevalence and surgical outcomes of Hurthle cell lesions in FNAs of the thyroid: A multi-institutional study in 6 Asian countries. Cancer Cytopathol 127:181-191, 2019 [DOI] [PubMed] [Google Scholar]
- 38.Bychkov A, Jung CK, Liu Z, et al. : Noninvasive follicular thyroid neoplasm with papillary-like nuclear features in asian practice: Perspectives for surgical pathology and cytopathology. Endocr Pathol 29:276-288, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]