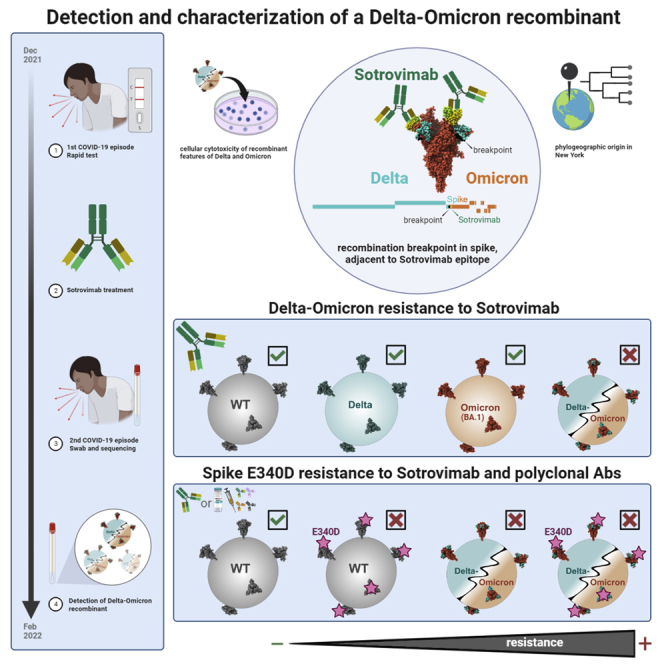
Summary
The emergence of recombinant viruses is a threat to public health, as recombination may integrate variant-specific features that together result in escape from treatment or immunity. The selective advantages of recombinant SARS-CoV-2 isolates over their parental lineages remain unknown. We identified a Delta-Omicron (AY.45-BA.1) recombinant in an immunosuppressed transplant recipient treated with monoclonal antibody Sotrovimab. The single recombination breakpoint is located in the spike N-terminal domain adjacent to the Sotrovimab binding site. While Delta and BA.1 are sensitive to Sotrovimab neutralization, the Delta-Omicron recombinant is highly resistant. To our knowledge, this is the first described instance of recombination between circulating SARS-CoV-2 variants as a functional mechanism of resistance to treatment and immune escape.
Subject areas: biological sciences, immunology, immunity
Graphical abstract
Highlights
-
•
Delta-Omicron recombinant in an immunosuppressed individual treated with Sotrovimab
-
•
Delta and Omicron (BA.1) are sensitive to Sotrovimab, but recombinant is resistant
-
•
Spike E340D mutation confers resistance to Sotrovimab and vaccine sera
-
•
Delta-Omicron recombinant is resistant to Sotrovimab irrespective of E340D
Biological sciences; Immunology; Immunity.
Introduction
In individuals infected with more than one viral variant, template switching during viral RNA synthesis can lead to RNA recombination, resulting in mosaic viruses.1 As known from influenza and HIV, recombination can accelerate viral adaptive evolution.2,3 Recombination events have been recorded in seasonal coronaviruses and MERS-CoV, and are discussed as cause for the initial zoonotic spillover of SARS-CoV-2.4 Within the first year of the SARS-CoV-2 pandemic, there was little evidence of recombination, similar to the short-lived SARS-CoV-1 epidemic.5 Since then, high case numbers and the co-circulation of variants have increased the likelihood of coinfections6,7 and the opportunity for inter-variant recombination.6,8,9,10,11,12,13 Recombination may result in viruses that have increased transmissibility or evade the host immune response or antiviral treatment. However, causal links between recombination and functional selective advantages have not yet been described for SARS-CoV-2.
Omicron has caused an ongoing global wave beginning in November 2021, which overlapped with Delta for several weeks.14 In New York City (NYC), Omicron arrived when the only circulating variant was Delta and its sublineages.15 Delta-Omicron recombinants were identified in Europe and the U.S.6,8,12 In these recombinants, genome break points frequently occurred in the N-terminal domain (NTD) of spike, resulting in hybrid genomes where non-structural ORF1a/b genes are from Delta and C-terminal spike regions, including the receptor-binding domain (RBD) from Omicron. Data on the functional consequences of recombination are scarce. Two studies using pseudotyped virus neutralization or recombinant spike infectious reporter assays suggested that a Delta-Omicron recombinant was comparable to BA.1 with regards to resistance to convalescent or vaccine-induced antibodies.16,17 The sensitivity to monoclonal antibodies (mAbs) was not tested, though mAbs is key in the treatment of immunocompromised patients, who are a prominent source for the development of new mutations or variants.18,19,20,21 Overall, there is a lack of data on which underlying conditions and treatment pressures favor the selection of recombinant SARS-CoV-2 over their parental lineages. Here, we report a Delta-Omicron recombinant, detected in an immunosuppressed, unvaccinated COVID-19 patient who was treated with Sotrovimab, a leading therapeutic monoclonal antibody (mAb) against the BA.1 Omicron lineage. We show that, whereas Delta and BA.1 are sensitive, the recombinant virus is resistant to neutralization by Sotrovimab. To our knowledge, this is the first report of a recombinant SARS-CoV-2 spike conferring escape from a therapeutic mAb regimen.
Results
An immunosuppressed transplant recipient with two COVID-19 episodes, treated with Sotrovimab
A male in his late twenties had end-stage renal disease due to hypertensive nephrosclerosis and received a deceased donor kidney transplant in June 2021. He was unvaccinated against COVID-19. His immunosuppressive regimen consisted of thymoglobulin induction at transplant and tacrolimus, prednisone, and mycophenolate mofetil as maintenance (Table 1). First COVID-19 episode: In late December 2021, he developed cough and fever and tested positive for COVID-19 by rapid antigen test on the same day. No PCR test was done. He received an infusion of Sotrovimab 500 mg two days after the onset of symptoms which resolved the next day, and he did not require hospitalization at this time. Second COVID-19 episode: In the middle of February 2022, he presented with a fever of up to 102°F, chills, body aches, and severe fatigue for one day, prompting hospitalization. He had no cough, shortness of breath, or chest pain. A chest X-ray showed left-sided basilar opacities. A CT scan of the chest showed patchy airspace opacities with consolidation in the left lower lobe consistent with pneumonia. A respiratory viral pathogen panel multiplex PCR on a nasopharyngeal swab specimen was positive for SARS-CoV-2 (Ct 26.4). He received empiric antibiotics for a presumed bacterial superinfection for seven days, and his immunosuppressive regimen was modified by halving the dose of mycophenolate. Tacrolimus and prednisone were continued at previous doses. The patient became afebrile two days into hospitalization and was discharged two days later. He did not receive antiviral medications during or after hospitalization. No further RT-PCR test was done and no hypoxia was observed (Table 1).
Table 1.
Demographic and clinical features of the patient infected with the Delta-Omicron recombinant SARS-CoV-2 virus
| Demographics | |
| Age range | 25–29 |
| Sex | Male |
| Clinical features | |
| COVID-19 vaccination status | Unvaccinated |
| Immune status | Immunosuppressed |
| Comorbidities | Hypertension, |
| End-stage renal disease (ESRD) due to hypertensive nephrosclerosis | |
| Deceased donor kidney transplant recipient in 2021 | |
| Immunosuppression at baseline | Induction (at transplant): Thymoglobulin |
| Maintenance immunosuppression | Tacrolimus 2 mg q AM/3 mg q PM, prednisone 5 mg daily, mycophenolate mofetil 1000 mg q12h |
| COVID-19 clinical course | |
| 1) December 2021 | COVID-19 positive by rapid antigen test (no PCR done) |
| Cough and fever | |
| Not hospitalized | |
| Received Sotrovimab 500 mg infusion once | |
| Symptoms resolved the next day | |
| 2) February 2022 | Fever (up to 102°F), chills, body aches, and fatigue for 4 days |
| Hospitalized | |
| No cough, shortness of breath, hypoxia, or chest pain. Not on supplemental oxygen | |
| Chest X-ray: right basilar opacities | |
| Chest CT: left lower lobe pneumonia | |
| Tests for potential pathogens | Respiratory viral pathogen panel multiplex PCR positive for SARS-CoV-2 (Ct 26.4) |
| Negative for blood cultures, Legionella and Streptococcus pneumoniae urinary antigens, serum cryptococcal antigen, beta-D- glucan, and Aspergillus galactomannan. | |
| Sputum culture was not performed due to lack of a good quality specimen. | |
| Treatment | No antivirals given |
| Antibiotics for 7 days (2 days Piperacillin-tazobactam + vancomycin, then 2 days piperacillin-tazobactam + azithromycin, then 3 days Levofloxacin) | |
| Immunosuppression reduced by halving mycophenolate dose, and maintaining the doses of tacrolimus and prednisone | |
| Outcome | Afebrile after 2 days of antibiotic treatment |
| Discharged after 4 days | |
Identification of a recombinant virus composed of Delta AY.45 and Omicron BA.1 sequences
Genomic analysis of the SARS-CoV-2-positive nasopharyngeal swab specimen from the patient’s second COVID-19 episode in late February 2022 revealed an unusual SARS-CoV-2 sequence, based on its outlier Omicron BA.1 placement in a global phylogenetic tree and its high number of mutations (Figures 1A and 1B). No lineage was assigned by Pangolin,22 and further inspection of each individual mutation and comparison with the mutations from different lineages revealed that the entire ORF1ab genomic region and 5′region of the spike gene up to position 22,035 contained Delta-specific mutations most similar to Delta sublineage AY.45 and lacked Omicron-specific mutations. The remainder of the genome, specifically after position 22,193 contained Omicron BA.1-specific mutations and had no Delta-specific mutations, indicative of a Delta-Omicron recombinant with a single breakpoint in spike NTD (Figure 1A). Phylogenetic analyses of the separate subregions confirmed the relatedness of the 5′ and -3′ segments with Delta (AY.45) and Omicron (BA.1) clades, respectively (Figure 1B). Nine multi-method sequencing runs on nasopharyngeal swab and cultured virus specimens, including xGen amplicon sequencing, metagenomics, Ion AmpliSeq Insight, and in vitro virus growth with ARTIC sequencing confirmed the identity of a Delta-Omicron recombinant (Table S1). The sequence was deposited in GISAID: EPI_ISL_10792641, flagged as a recombinant.
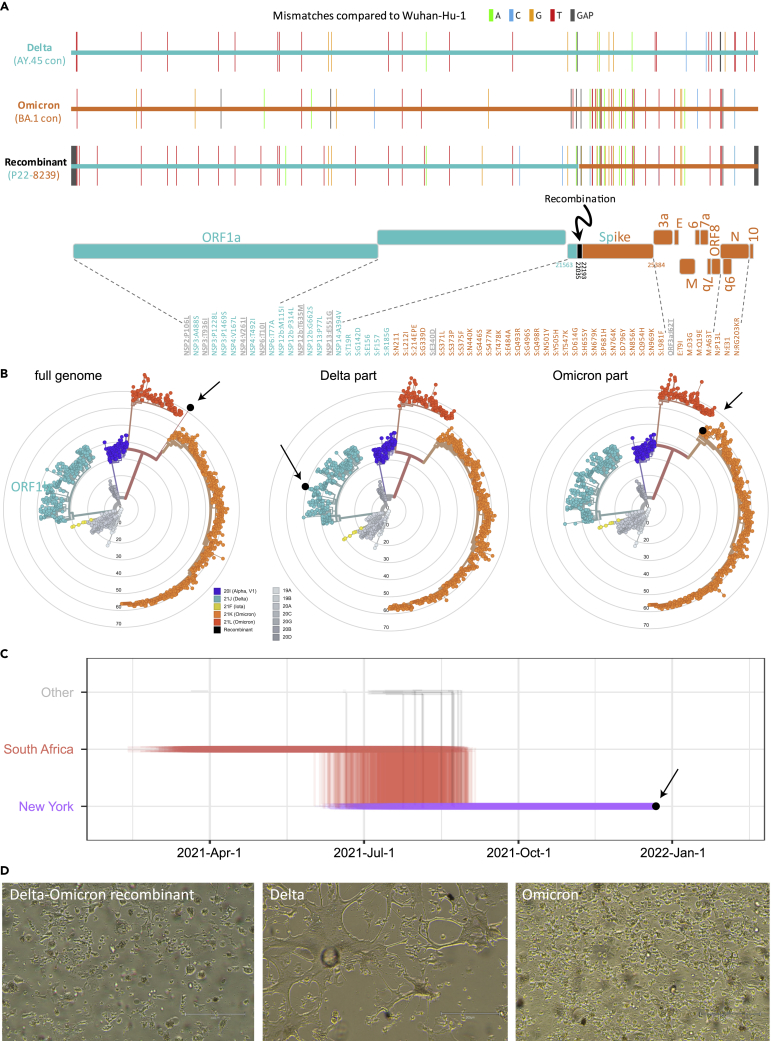
Figure 1.
Identification of a Delta (AY.45)-Omicron (BA.1) recombinant virus with traces to New York and cytopathic effects
(A) Full genome mutations of the Delta-Omicron recombinant in comparison with Delta AY.45 and Omicron BA.1 consensus (con) sequences. In the schematic of the Delta and Omicron portions, all non-synonymous mutations are listed and shown in teal (Delta), orange (Omicron), or gray (not Delta or Omicron).
(B) Maximum likelihood phylogenies of 1557 global SARS-CoV-2 sequences based on a New York, New Jersey, and Connecticut-focused subsampling. The recombinant is shown in black. The phylogenetic analysis was done using the recombinant’s full genome (left) and Delta or Omicron parts only (middle and right). Concentrical rings indicate the number of mutations compared to Wuhan-Hu-1 (root).
(C) Markov jump trajectory plot depicting the ancestral location history of the AY.45 segment of the recombinant sequence. Each individual trajectory corresponds to the Markov jumps in a single tree from the posterior distribution of 1,800 trees (Figure S2B). Horizontal lines represent the time maintained at an ancestral location and vertical lines represent a Markov jump between two locations. The first SARS-CoV-2 positive test of the patient is indicated in black.
(D) Cytopathic effects observed in various lineages of SARS-CoV-2 on VeroE6/TMPRSS2 cells. Images were taken with an EVOS M5000 inverted microscope (ThermoFisher Scientific, Waltham, MA); 10X magnification. (see also Figures S1 and S2 and Tables S1 and S2).
Phylogeographic origin of the Delta AY.45 infection lies in New York
While phylogenetic inference was not reliably feasible for BA.1 due to the large number of infections in conjunction with a low number of mutations, a maximum clade credibility (phylogenetic) tree using >1000 Delta AY.45 genomes and the Delta piece of the recombinant (Figure S2) revealed a monophyletic clade consisting entirely of United States AY.45 genomes including the recombinant (100% posterior support) as a descendant of a South African ancestral AY.45 backbone. The Markov jump trajectory plot (Figure 1C) supports an ancestral origin in South Africa and points to an introduction from South Africa into New York State between June and July 2021. The lineage associated with the recombinant genome was circulating in New York for >100 days before the positive COVID-19 test in December 2021, suggesting that infection with AY.45 occurred in New York. The phylogeographic reconstruction further shows that all US infections within this AY.45 clade originated from New York.
Cytopathic effect of the recombinant is characteristic of Delta and Omicron
Cytopathic effects (CPE) of the in vitro-grown recombinant virus were first observed at 48 hpi on VeroE6/TMPRSS2 cells, with minor cell rounding in small areas of the monolayer, progressing to widespread cell rounding, detachment, debris, and cell death by 96 hpi (Figure S1), consistent with Omicron. However, affected cells appeared larger and more prominent in the CPE produced by the recombinant virus, and cell detachment was more advanced. As with Omicron, syncytia formation was not apparent in the recombinant virus cultures. Overall, the recombinant virus-induced CPE was more similar to those seen with Omicron than Delta (Figure 1D). By 96 hpi (Ct 13.5), viral production had increased more than 5 log10 from the 24h timepoint in the culture supernatant (Table S2).
The recombinant breakpoint is adjacent to the quaternary Sotrovimab epitope in spike
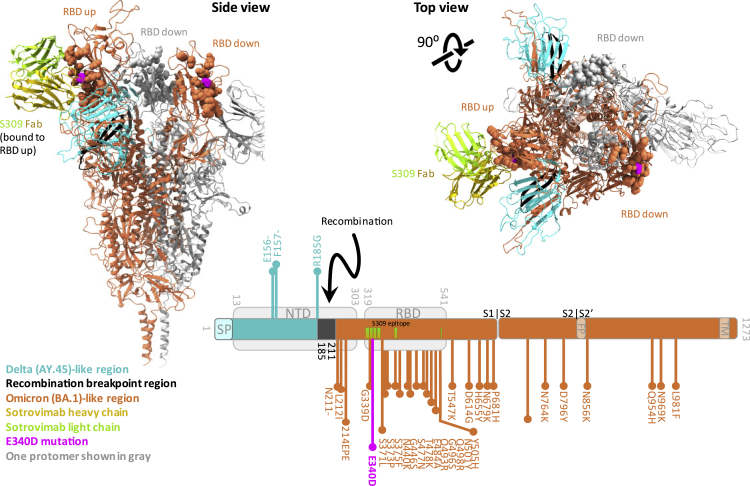
Since recombination occurred in the carboxy-terminal portion of spike NTD (bp 22035-22193) (Figure 1A), most of the NTD is composed of Delta sequence, while RBD and the C-terminal regions are Omicron (Figure 2). During the first symptomatic COVID-19 episode in December 2021, the patient was treated with Sotrovimab, a class 3 anti-spike RBD mAb with broad antiviral activity against SARS-CoV-2, including Delta and BA.1.23,24,25,26 The spike binding epitope for Sotrovimab involves the N-terminus of RBD, and, in a 3D structural model, the Fab moiety engages the space between RBD and the neighboring NTD (Figure 2). The recombinant virus sequence harbors one atypical spike mutation (E340D) not related to Delta or Omicron and rarely found in global SARS-CoV-2 sequences, with a frequency of less than 3 × 10−5.14,27 Interestingly, E340 is in the middle of the Sotrovimab-binding epitope and is the primary site of resistance to Sotrovimab.23,28,29 E340D is a conservative replacement without a change in charge, in contrast to the more abundant resistance mutations E340 A/K,23,28 yet we show that E340D can confer resistance (Figures 3, 4, and 5).
Figure 2.
Breakpoint region of the Delta-Omicron recombinant is adjacent to the Sotrovimab (S309) binding epitope in spike
Trimer spike structure of the Delta-Omicron recombinant in the open, one RBD-up conformation with one Sotrovimab Fab molecule bound to the RBD in up-position. Sotrovimab is a slightly refined, engineered version of its precursor S309 that has been used in the structures shown here. The spike portions are shown as ribbons and the Sotrovimab epitopes as spheres. One spike protomer is shown in gray; the other two protomers are colored according to the legend. Lower right: schematic of the location of the Delta (teal) and Omicron (orange) spike portions as well as the recombinant breakpoint region (black) including the mutations related to Delta (teal, facing up) or Omicron (orange, facing down). The mutation E340D, which is not related to Delta or Omicron, is highlighted in pink. The recombinant spike structure is a homology model of the recombinant spike based on pdb: 7TO4 (1 RBD-up Omicron spike trimer). The S309 (Sotrovimab) molecule was added by structural overlay of a S309-bound spike co-structure in 1 RBD-up position (pdb: 7TM0). FP: fusion peptide, NTD: N-terminal domain, RBD: receptor-binding domain, SP: signal peptide, TM: transmembrane domain.
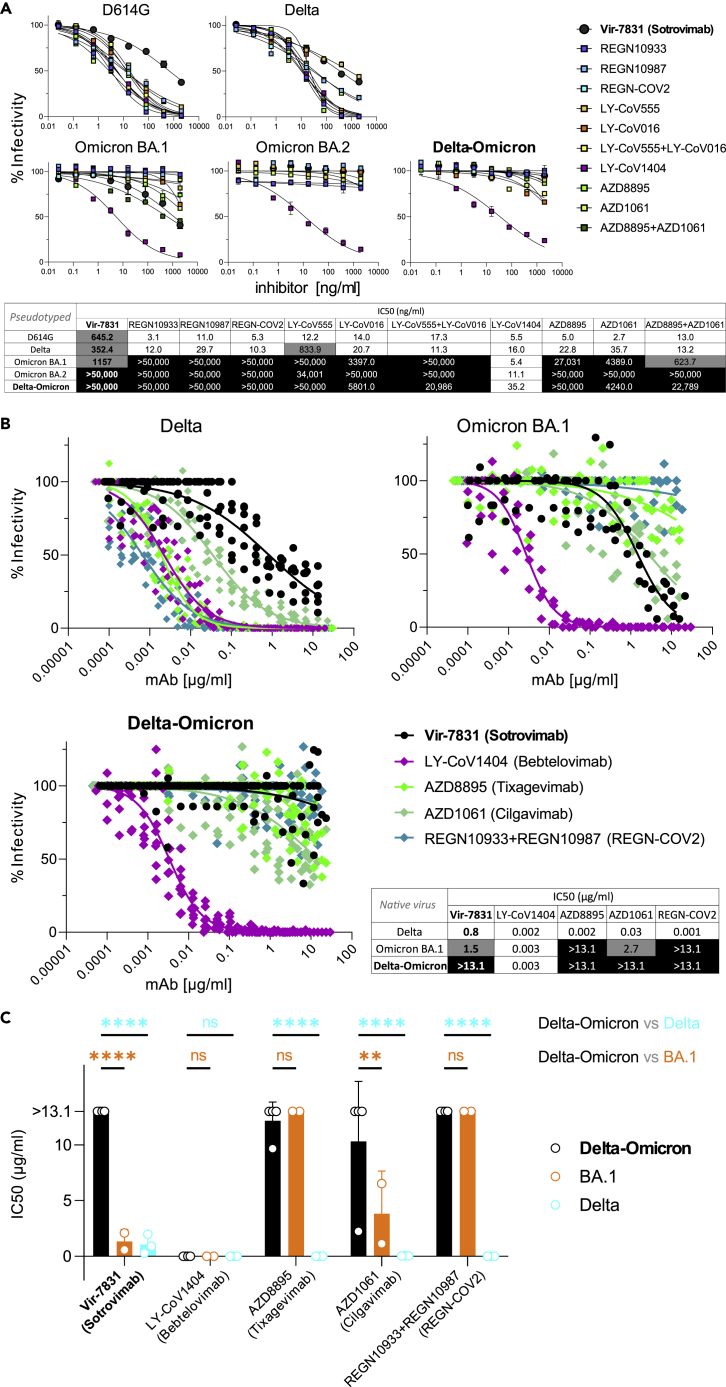
Figure 3.
Delta and Omicron (BA.1) are sensitive to Sotrovimab, but Delta-Omicron is resistant
(A) Neutralization of viruses with D614G, Delta, Omicron, and Delta-Omicron spike proteins by monoclonal antibodies (mAbs) Regeneron REGN10933 (Casirivamab), REGN10987 (Imdevimab), the REGN-COV-2 cocktail (Casirivimab + Imdevimab), LY-CoV555 (Bamlanivimab), LY-CoV016 (Etesevimab), the LY-CoV555+LY-CoV016 combination, LY-CoV1404 (Bebtelovimab), AZD8895 (Tixagevimab), AZD1061 (Cilgavimab), the AZD8895 + AZD1061 combination (Evusheld), and VIR-7831 (Sotrovimab). The heatmap table shows the half-maximal inhibitory concentrations (IC50) of the therapeutic mAbs calculated using the data from the antibody neutralization curves shown above. High resistance (IC50 > 3000 ng/mL) is highlighted in black, moderate resistance (50 ng/mL < IC50 < 3000 ng/mL) in gray, and no/low resistance (IC50 < 50 ng/mL) shown in white background.
(B) Neutralization of infectious native Delta-Omicron virus in comparison with Delta and BA.1 by a selection of five mAbs used in A, including Vir-7831 (Sotrovimab). Inhibition was determined in plaque reduction neutralization tests using serially diluted mAb doses. The graphs display all data points (staggered) from two to four biological replicates with technical duplicates (Delta-Omicron: four biological replicates). Neutralization curves are shown as non-linear regression fits for each mAb. In the IC50 heatmap table, high resistance (IC50 > 10 μg/mL) is highlighted in black, moderate resistance (1 μg/mL < IC50 < 10 μg/mL) in gray, and no/low resistance (IC50 < 1 μg/mL) shown in white background.
(C) Statistical comparison of IC50 values from all biological replicates in B (two-way ANOVA with Tukey’s multiple comparison test, ∗∗p < 0.005, ∗∗∗∗p < 0.0001, ns: not significant). Data in A and C are shown as mean with SD. (see also Table S3).
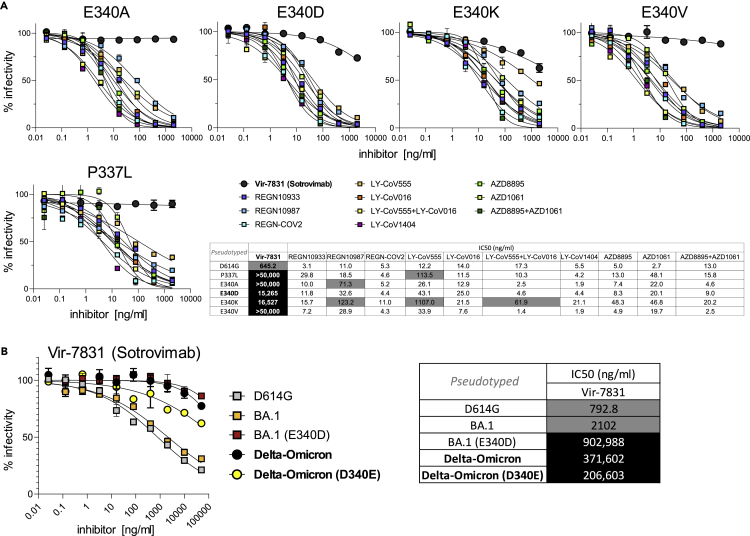
Figure 4.
Delta-Omicron is resistant to Sotrovimab, irrespective of the E340D mutation in RBD
(A) Neutralization of D614G (backbone) and P337L, E340A, E340D, E340K, and E340V point-mutated spike protein-pseudotyped viruses by the same set of mAbs as in Figure 3A. Experiments in Figures 3A and 4A were conducted simultaneously with D614G virus as a reference.
(B) Neutralization of D614G (E340), BA.1 (E340), E340D point-mutated BA.1, Delta-Omicron (D340), and D340E point-mutated Delta-Omicron spike protein-pseudotyped viruses by VIR-7831 (Sotrovimab). The tables show the half-maximal inhibitory concentrations (IC50) of the therapeutic mAbs calculated using the data from the antibody neutralization curves shown above and to the left, respectively. High resistance (IC50 > 3000 ng/mL) is highlighted in black, moderate resistance (50 ng/mL < IC50 < 3000 ng/mL) in gray, and no/low resistance (IC50 < 50 ng/mL) shown in white background. Data points in A and B are shown as mean with SD. (see also Table S3).
Figure 5.
Resistance of the Delta-Omicron recombinant and P337L or E340X-mutated viruses to vaccine-elicited polyclonal antibodies
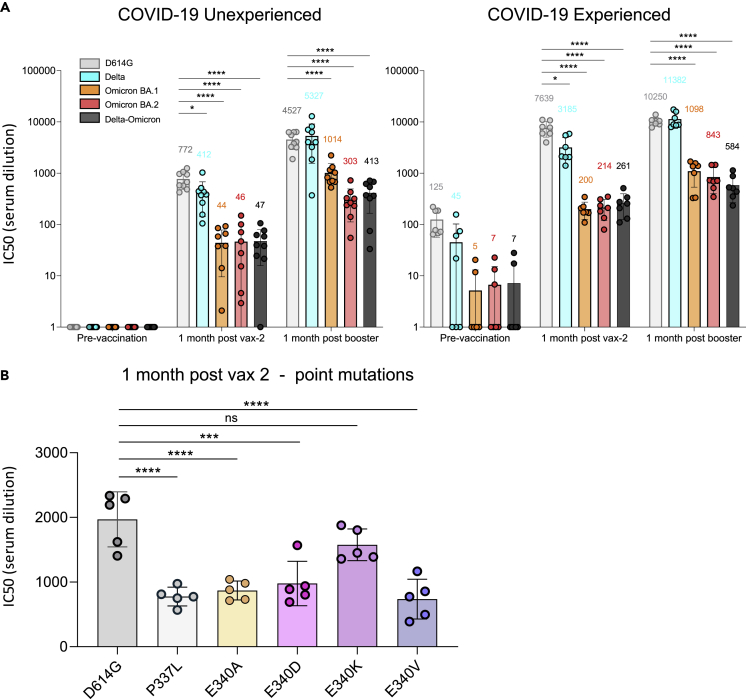
(A) Neutralizing antibody titers of participants with or without previous history of SARS-CoV-2 infection and/or vaccination were measured using pseudotyped viruses with D614G, Delta, Omicron, and Delta-Omicron spike. Sera were collected from study participants pre-vaccination, 1-month post-second vaccination with Pfizer BNT162b2, and 1-month post-boost (third vaccination). Study participants were without previous SARS-CoV-2 infection (unexperienced) (left) (n = 9) or previously infected (experienced) (right) (n = 7). COVID-19 history was determined by symptoms and a PCR-positive test or serology. Equivalent amounts of D614G, Delta, Omicron, and Delta-Omicron spike protein-pseudotyped viruses were mixed with a 2-fold serial dilution of donor serum and then applied to ACE2.293T cells. Luciferase activity was measured two days post-infection. Each serum dilution was measured in triplicate and the experiment was done twice with similar results. Half-maximal inhibitory concentrations (IC50) are shown for one representative experiment. Mean values for each group are shown above the bar.
(B) Neutralizing antibody titers of sera from COVID-19 unexperienced individuals (n = 5) collected 1 month after second vaccination against D614G (as a backbone) and P337L, E340A, E340D, E340K, and E340V point mutated spike protein-pseudotyped viruses. Statistical significance was calculated by two-sided testing. (one-way ANOVA with Tukey’s multiple comparison test, ∗p ≤ 0.05, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, ns: not significant). Bars are shown as mean with SD. (see also Tables S3 and S4).
Delta-Omicron recombinant exhibits enhanced Sotrovimab resistance compared to Delta and BA.1
Delta viruses are sensitive to most therapeutic anti-SARS-CoV-2 antibodies, including all 11 mAbs and cocktails we tested, whereas BA.1 is resistant to most of them (8/11; Figure 3A). Among the few mAbs that retained activity against BA.1 are LY-CoV1404 and the cocktail AZD8895 + AZD1061 (Evusheld) as well as the class 3 anti-RBD mAb VIR-7831 (Sotrovimab), the latter binding RBD outside the ACE2 binding site, in contrast to the more frequent class 1 and 2 mAbs.26 Notably, while Delta and Omicron BA.1 are sensitive to Sotrovimab, the Delta-Omicron recombinant was resistant, in assays using pseudotyped (>43× increase in IC50 over BA.1) and infectious virus (>9× increase in IC50 over BA.1) (Figures 3A–3C, Table S3). The Delta-Omicron recombinant was resistant to 10/11 tested mAbs, comparable to Omicron BA.2, and only the cocktail LY-CoV1404 retained activity (Figure 3A). In addition to the differential resistance to Sotrovimab, the recombinant exhibited enhanced resistance to AZD8895 + AZD1061 (Evusheld) in pseudotyped virus assays, and also AZD1061 (Cilgavimab) alone in infectious virus assays when compared to BA.1. Both Sotrovimab and Cilgavimab belong to class III nAbs with overlapping RBD binding epitopes.30 Because this particular Delta-Omicron recombinant carries an unusual E340D mutation in the Sotrovimab epitope at a site that has been linked with Sotrovimab resistance (E340 A/K),28 we compared the impact of E340D on Sotrovimab neutralization with other mutations that were recorded upon Sotrovimab treatment/resistance (Figure 4A). While E340 A/V and P337L conferred complete Sotrovimab resistance in D614G pseudotyped virus (IC50 > 50,000 ng/mL), the impact of E340D impact was lower (IC50 = 15,265 ng/mL) but substantial (24× IC50 increase) and comparable to E340K (26× IC50 increase). All five mutations interfered with Sotrovimab neutralization exclusively but not with any of the other 10 mAbs tested (Figure 4A).
To deconvolute the specific contributions of E340D and the recombination event itself in conferring Sotrovimab resistance, we performed site-directed mutagenesis in the recombinant and BA.1 pseudotyped viruses (Figure 4B). E340D in the BA.1 background conferred Sotrovimab resistance, as shown before for D614G. To test the effect of Delta-Omicron recombination in the absence of E340D, we reverted 340D to 340E in the chimeric spike. Notably, the D340E revertant pseudotyped virus retained resistance to Sotrovimab (IC50 > 50,000 ng/mL) with a minor increase in sensitivity compared to the original (D340) version (1.8× change from 371,602 to 206,603 ng/mL). Thus, we conclude that the recombination of Delta and Omicron spike was sufficient to confer resistance to Sotrovimab and that E340D marginally enhanced the resistance phenotype (Figure 4B).
Resistance of Delta-Omicron virus to polyclonal antibody responses and clinical impact of E340X and P337L mutations
The Delta-Omicron recombinant was resistant to naturally and vaccine-induced anti-SARS-CoV-2 neutralizing antibodies (nAbs) to a level comparable with that of BA.1 or BA.2 (Figure 5A, Table S4). The neutralization sensitivity of the recombinant remained low (IC50 < 600 serum dilution) even in COVID-19-experienced individuals 1 month after the vaccine boost, compared to IC50s > 10,000 with Delta or D614G viruses. In boosted individuals, the recombinant had 2.5× (COVID-19-naïve) and 1.9× (COVID-19-experienced) lower IC50 values compared to BA.1, suggesting a slightly enhanced resistant phenotype. Immune responses induced 1-month post-second vaccine dose were significantly reduced against the D614G virus carrying E340 A/V/D or P337L but not E340K (Figure 5B). This finding suggests that Sotrovimab-like nAbs are present in vaccinee sera and that Sotrovimab resistance mutations have an impact on vaccine-induced polyclonal nAb responses.
Discussion
Drivers for recombinant variant selection have remained obscure, and functionally distinctive features of SARS-CoV-2 recombinants are poorly understood. Here, we show for the first time that recombination between two variants can functionally contribute to neutralization resistance (Figure 3). These results highlight recombination as a mechanism for SARS-CoV-2 immune escape irrespective of specific resistance mutations. In contrast to other recombinants reported so far,6,8,12,31 we describe a SARS-CoV-2 recombinant detected in an immunosuppressed transplant recipient after mAb treatment (Figures 1A and 1B). Immunocompromised patients are an important source of new variants due to prolonged viral replication/evolution, particularly under selective pressure by mAb treatment.18,19,20,21 The identified Delta-Omicron recombinant was resistant to Sotrovimab, as well as other mAbs and vaccine-elicited antibodies (Figures 3, 4, and 5). In line with the latter finding, a recent study suggested a more vaccine-resistant phenotype of a Delta-Omicron pseudotyped virus compared with D614G and BA.316 but therapeutic mAbs were not tested. Another report using a spike reporter assay indicated that Delta-Omicron recombinants have similar resistance to natural infection or COVID-19 vaccine-induced nAbs as BA.1, but less pronounced as the BA.4/5 subvariants.17
Our neutralization data further showed that both the recombination event in spike NTD and the RBD mutation E340D added up to a combined resistance phenotype (Figure 4). It is plausible that the Sotrovimab treatment selected for the recombinant and/or its E340D mutation, and recombination and E340D co-evolved reinforcing each other. High-resolution structures will be needed to determine whether recombination altered the conformation/accessibility of the Sotrovimab epitope, thus rendering the virus resistant to Sotrovimab. Our titration experiments suggest that E340D confers partial Sotrovimab escape comparable to E340K when tested against the D614G background virus. E340D, located in the center of the Sotrovimab binding epitope in RBD, is extremely rare and neither related to a specific lineage nor community spread and might thus be an indicator of exerted Sotrovimab immune pressure. Besides mAbs (Sotrovimab), E340D confers resistance to vaccine or infection-induced polyclonal nAbs (Figures 4A and 5B) and thus needs to be considered an immune escape mutation.
Since Delta had disappeared from circulation by February 2022 in the NYC metro area, we suspect that the patient became co-infected with Delta and Omicron in December 2021 during his first COVID-19 episode, when Delta and Omicron were at ∼10% and 90% prevalence in New York, respectively. Alternatively, since the patient was tested by rapid antigen test only during the first COVID-19 episode when there was no RNA available for sequencing, it is possible that the participant became infected with the recombinant from an undefined source. Phylogenetically related BA.1 and AY.45 sequences (Figure 1C) were highly prevalent in New York at this time, which renders a regional Delta and Omicron co-infection and recombination event more likely than the introduction of an unknown recombinant from an undefined source. Delta-Omicron recombinants remained rare without rampant community transmissions, possibly because of their selective advantage preferentially against mAbs compared to polyclonal antibodies (Figures 3 and 5A). Such recombinants might thus be more common and alarming in high-risk/immunocompromised patients treated with therapeutic mAbs. Therefore, genomic surveillance of immunocompromised individuals remains a priority. This case stresses the importance of functional testing of emerging variants to identify new mechanisms and pathways of immune escape that might involve functional resistance based on recombination.
Limitations of the study
No nasopharyngeal swab was available upon the primary SARS-CoV-2 infection of the patient, detected by rapid test in December 2021, and thus a detailed reconstruction of the recombination even was not possible. Since the recombinant was the only virus detected in the February 2022 specimen in the absence of parental Delta or Omicron lineages, we can only speculate whether the recombinant was generated after Delta and Omicron co-infection in December 2021 with a subsequent escape from and selection against Sotrovimab, possibly supported by other selection/replication advantages. Further studies to examine the replicative and competitive fitness between the recombinant and its parental Delta and Omicron lineages could elucidate some of these speculations. Although other Delta-Omicron recombinants have been reported, they remained on a low level, and the current recombinant has not been detected in other patients.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Casirivimab | Regeneron Pharmaceuticals | N/A |
| Imdevimab | Regeneron Pharmaceuticals | N/A |
| Casirivimab + Imdevimab cocktail | Regeneron Pharmaceuticals | N/A |
| Bamlanivimab | Discarded vials, NYU Langone Health | N/A |
| Etesevimab | Discarded vials, NYU Langone Health | N/A |
| Bamlanivimab + Etesevimab cocktail | Discarded vials, NYU Langone Health | N/A |
| Bebtelovimab | Discarded vials, NYU Langone Health | N/A |
| Tixagevimab | Discarded vials, NYU Langone Health | N/A |
| Cilgavimab | Discarded vials, NYU Langone Health | N/A |
| Tixagevimab + Cilgavimab cocktail | Discarded vials, NYU Langone Health | N/A |
| Sotrovimab | Discarded vials, NYU Langone Health | N/A |
| Bacterial and virus strains | ||
| Infectious SARS-CoV-2 Delta-Omicron | This study | N/A |
| Infectious SARS-CoV-2 Delta | This study | N/A |
| Infectious SARS-CoV-2 BA.1 | This study | N/A |
| SARS-CoV-2 Δ19 SD614G pseudotyped reporter virus | Tada et al.32 | N/A |
| SARS-CoV-2 Δ19 S Delta pseudotyped reporter virus | Tada et al.33 | N/A |
| SARS-CoV-2 Δ19 S BA.1 pseudotyped reporter virus | Tada et al.34 | N/A |
| SARS-CoV-2 Δ19 S BA.2 pseudotyped reporter virus | Zhou et al.35 | N/A |
| SARS-CoV-2 Δ19 S Delta-Omicron pseudotyped reporter virus | This paper | N/A |
| Biological samples | ||
| Sera from individuals vaccinated with BNT162b2. See Table S4 | NYU Vaccine Center with written consent under Institutional Review Board approval | Deidentified, IRB 20-00595 and IRB 18-02037 |
| Chemicals, peptides, and recombinant proteins | ||
| Polyethyleneimine | Polysciences | Cat# 23966-1 |
| Luciferase substrate | NanoLight Technology | Cat# 325-50 |
| Critical commercial assays | ||
| GIBCO ExpiFectamine Transfection Kits | Thermo Fisher Scientific | Cat# A29130 |
| Nano-Glo® Luciferase Assay System | Promega | Cat# N1120 |
| Deposited data | ||
| Full genome sequence of SARS-CoV-2 Delta-Omicron recombinant virus | GISAID | EPI_ISL_10792641 |
| xGen and shotgun deep sequencing data (raw fastq files; NYU). See Table S1 | NCBI SRA | PRJNA915368 |
| AmpliSeq, harvest and 24h–96h in vitro culture deep sequencing data (raw fastq files; NY State DOH). See Table S1 | NCBI SRA | PRJNA915368 |
| Experimental models: Cell lines | ||
| VeroE6/TMPRSS2 cells | JCRB Cell Bank | JCRB1819 |
| 293T cells | ATTC | CRL-3216™ |
| ACE2.293T cells | Tada et al.32 | N/A |
| Oligonucleotides | ||
| Primers. See Table S3 | This study | IDT |
| Recombinant DNA | ||
| HIV-1 Gag/Pol expression vector pMDL | Tada et al.32 | N/A |
| HIV-1 Rev expression vector pRSV.Rev | Tada et al.32 | N/A |
| Lentiviral reporter vector pLenti.GFP-NLuc | Tada et al.32 | N/A |
| Synthesized cDNA fragment, SARS-CoV-2 Delta-Omicron recombinant spike. See Table S3 | This study | GenScript |
| SARS-CoV-2 Δ19 S D614G spike plasmid | Tada et al.32 | N/A |
| SARS-CoV-2 Δ19 S D614G spike P337L plasmid | This study | N/A |
| SARS-CoV-2 Δ19 S D614G spike E340A plasmid | This study | N/A |
| SARS-CoV-2 Δ19 S D614G spike E340D plasmid | This study | N/A |
| SARS-CoV-2 Δ19 S D614G spike E340K plasmid | This study | N/A |
| SARS-CoV-2 Δ19 S D614G spike E340V plasmid | This study | N/A |
| SARS-CoV-2 Δ19 S Delta spike plasmid | Tada et al.33 | N/A |
| SARS-CoV-2 Δ19 S BA.1 spike plasmid | Tada et al.34 | N/A |
| SARS-CoV-2 Δ19 S BA.2 spike plasmid | Zhou et al.35 | N/A |
| SARS-CoV-2 Δ19 S Delta-Omicron spike plasmid | This study | N/A |
| SARS-CoV-2 Δ19 S BA.1 spike E340D plasmid | This study | N/A |
| SARS-CoV-2 Δ19 S Delta-Omicron spike D340E plasmid | This study | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.0e | GraphPad Prism Software, Inc. | N/A |
| IQ-TREE v2.2.0 | IQ-TREE | N/A |
| BEAST v1.10.5 | BEAST | N/A |
| Nextstrain CLI v6.0.0 | Nextstrain | N/A |
| Nextclade v1.14 | Nextclade | N/A |
| Swiss-Model | Swiss-Model | N/A |
| UCSF ChimeraX 1.3 | UCSF Chimera | N/A |
| BioRender | BioRender | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Ralf Duerr (Ralf.Duerr@nyulangone.org).
Materials availability
-
•
The in vitro-propagated infectious Delta-Omicron virus can be made available on request after completion of a Materials Transfer Agreement and proof of BSL3 certification from Kirsten St. George (kirsten.st.george@health.ny.gov).
-
•
Pseudotyped virus plasmids encoding the recombinant spike or spike mutants can be made available on request after completion of a Materials Transfer Agreement from Nathaniel R. Landau (Nathaniel.Landau@nyulangone.org).
Experimental model and subject details
Study approval
This study was approved by the NYULH Institutional Review Board, protocol numbers i21-00493, i21-00561, i20-00595, and i18-02037 (NYU Langone Vaccine Center). Virus was sequenced and isolated from one study subject, a male in his late twenties.
Cells and media for infectious virus culture
VeroE6/TMPRSS2 cells, a modified VeroE6 cell line expressing the transmembrane serine protease, TMPRSS2,36 were used for virus isolation, obtained from the Japanese Collection of Research Bioresources (JCRB Cell Bank) cell number JCRB1819, through Sekisui Xenotech, LLC (Kansas City, KS), agreement # A2000230. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with sodium bicarbonate and 10% fetal bovine serum (FBS, all from Millipore Sigma, St. Louis, MO), as well as 1mg/1 mL geneticin G418 (Gibco). VeroE6/TMPRSS2 cells were seeded three days prior to infection, to reach 85–90% confluency. Infected monolayers were checked daily for cytopathic effect (CPE), and 110 μL of supernatant was removed at 24, 48, 72, and 96 hours post-infection (hpi) for further analysis. At 96 hpi, cells and supernatant were harvested together.
Virus isolation and culture
VeroE6/TMPRSS2 cells were seeded in T25 flasks with 5 mL cell suspension (1.5 × 105 cells/mL), three days prior to infection, to reach 85–90% confluency. Before inoculation, 300 μL of specimen were combined with 300 μL penicillin/streptomycin (pen/strep, 10 units/mL Millipore Sigma, St. Louis, MO), 150 μL of nystatin (1 mg/mL; Millipore Sigma), and 250 μL of gelatin-Tris-Hanks (GTH). The cell monolayer was infected with the processed specimen, adsorbed 1.5 h at 37°C with 5% CO2, then overlaid with 5 mL viral growth medium as described above, except FBS was reduced to 2%. In accordance with NY State DOH safety policy, the SARS-CoV-2 inoculations were performed at biosafety level 2 (BSL-2) but all subsequent incubations, culture samplings and harvesting were performed at biosafety level 3 (BSL-3). Infected monolayers were checked daily for cytopathic effect (CPE), and 110 μL of supernatant were removed at 24, 48, 72, and 96 hours post-infection (hpi) and immediately placed in NucliSENS lysis buffer for further analysis. At 96 hpi, cells and supernatant were harvested together, and 110uL of the harvest was also lysed for further analysis. All samples were extracted and tested by real-time RT-PCR to confirm viral growth.
Plasmids and SARS-CoV-2 spike lentiviral pseudotypes
The SARS-CoV-2 Delta, BA.1, and BA.2-spike were previously generated.33,34,35 The Delta-like N-terminal domain of the SARS-CoV-2 Delta-Omicron recombinant spike was chemically synthesized as a short fragment (Genscript Biotech Corporation, Piscataway, New Jersey, USA). The N-terminal fragment was amplified with a forward primer containing a Kpn-I site and a reverse primer containing an inner Omicron BA.1 sequence. This fragment was fused with another fragment which encodes RBD and C-terminal parts of Omicron BA.1 spike including an Xho-I site by overlapping PCR. 19 amino acids at the spike C-terminus, a reported endoplasmic reticulum retention sequence, were deleted (Δ19).37 The full Delta-Omicron recombinant spike was cloned into the Kpn-I and Xho-I sites of pcDNA6 (Invitrogen). Point mutations were introduced by overlap extension PCR. All spike expression vectors encode a termination codon that deletes the carboxy-terminal 19 amino acids (Δ19).32 HIV-1 Gag/Pol expression vector pMDL and HIV-1 Rev expression vector pRSV.Rev have been previously described.32
SARS-CoV-2 variant spike pseudotyped lentivirus stocks were produced by cotransfection of 293T cells with pMDL, pLenti.GFP-NLuc, pcCoV2.S-Δ19 (or variants thereof) and pRSV.Rev as previously described.32 Real-time PCR reverse transcriptase activity was used to measure the lentivirus stocks and normalize them.38
Human sera and monoclonal antibodies
Sera from individuals vaccinated at NYULH with BNT162b2 were collected through the NYU Vaccine Center with written consent under Institutional review Board (IRB) approval and were deidentified. Casirivimab, Imdevimab, and the cocktail Casirivimab + Imdevimab were provided by Regeneron Pharmaceuticals. Bamlanivimab, Etesevimab, the cocktail Bamlanivimab + Etesevimab, Bebtelovimab, Tixagevimab, Cilgavimab, the cocktail Tixagevimab + Cilgavimab, and Sotrovimab were obtained from discarded vials.
Cells for pseudotyped virus neutralization
293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (P/S) at 37°C in 5% CO2. Stable ACE2.293T cell line were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (P/S) and 1 μg/mL puromycin, as previously described.32
Method details
SARS-CoV-2 sequencing and bioinformatic analysis
SARS-CoV-2 full-genome sequencing was performed in two independent laboratories using four different approaches to confirm the identity of the recombinant and to exclude artifacts (e.g. by mixed reads) as reason for the detection of recombinant sequences. The methods included xGen Amplicon39 and metagenomics sequencing (NYU), as well as AmpliSeq Insight and ARTIC Amplicon sequencing (NY State DOH).
IDT xGen amplicon and metagenomics approach (NYU)
Genomic surveillance was carried out as described previously,39 using an amplicon-based library prep method. The web-based Nextclade v1.1440 and Auspice v2.36 phylogenomic visualization features of Nextstrain14 were used to examine viral genome clade assignment and mutation calling. For metagenomics, a shotgun library from total RNA extracted from the nasal swab was generated using the Illumina Stranded Total RNA prep Ligation with Ribo-Zero Plus. Sequencing was performed on an Illumina Nova Seq 6000, 150 PE, dual index.
Sequencing reads were demultiplexed using the Illumina bcl2fastq2 Conversion Software v2.20 and adapters and low-quality bases were trimmed with Trimmomatic v0.36.41 BWA v0.7.1742 was utilized for mapping reads to the SARS-CoV-2 reference genome (NC_045512.2, wuhCor1) and duplicate reads were removed using Sambamba v0.6.8.43 GATK v3.8 DepthOfCoverage and HaplotypeCaller tools44 were used to determine on-target viral coverage and call mutations.
Ion AmpliSeq Insight and ARTIC amplicon-based approach (NY state DOH)
Total nucleic acid was extracted from the primary clinical specimen and samples of cultured isolate using the bioMerieux NucliSens easyMAG platform (bioMerieux Inc, Durham, NC), using 110 μL of sample. Real-time RT-PCR was performed on the original primary specimen, as well as the cultured isolate, using the New York SARS-CoV-2 Real-Time Reverse Transcriptase (RT)-PCR Diagnostic Panel, according to the protocol indicated in the Instructions for Use.45 Library preparation was performed on RNA from primary specimen with an Ion Chef and sequencing with an Ion S5 XL, using the Ion AmpliSeq SARS-CoV-2 Insight Research Assay (ThermoFisher Scientific, Waltham, MA, USA). The assay was performed with 27 cycles and a 4-min extension time. Library preparation was performed on RNA from the cultured isolates using a modified ARTIC protocol with ARTIC V4 primers, as previously described.46 Libraries were sequenced on an Illumina NextSeq instrument.
Consensus genomes were generated as previously described46 with the ncov2019-artic-nf pipeline (https://github.com/connor-lab/ncov2019-artic-nf). Mutations were identified with Nextclade (https://clades.nextstrain.org/) and lineage assignment was performed with Pangolin v3.1.19 (pangoLearn 2022-01-22)22 for both consensus genomes, which identified the first part of the recombinant genome as AY.45, i.e., a sublineage of the Delta variant of concern (VOC), and the second part as Omicron BA.1. Additionally, we used Usher (https://genome.ucsc.edu/cgi-bin/hgPhyloPlace) to place consensus genomes in a global SARS-CoV-2 phylogenetic tree to identify closest relatives. Lineage composition of the sequencing libraries was determined by Freyja (https://github.com/andersen-lab/Freyja) to evaluate the possibility of a mixed infection.
Combined multi-method SARS-CoV-2 sequencing approach
The full-genome SARS-CoV-2 sequence was initially determined by xGen amplicon sequencing of the nasopharyngeal swab. The original swab was then re-extracted, re-sequenced, and processed in the same manner, which revealed the same 5′ Delta and 3′ Omicron-specific mutations with a breakpoint between 22,035-22,193 bp in the near full-length high-quality sequence (Table S1). No mixed bases were observed, suggesting that this was not a co-infection or technical artifact. For further confirmation, we prepared and sequenced libraries from the same sample using a ribodepletion shotgun metagenomics approach. GATK variant detection revealed a 100% concordance between mutations called with both the amplicon and independent shotgun metagenomics approach. The metagenomics approach yielded a single additional Omicron-specific change at the beginning of the Omicron-like portion of the genome, i.e., a 9 bp insertion at position 22,204 (spike 214EPE). The specimen was also separately extracted and sequenced by AmpliSeq Insight at a second site, NY State Department of Health (DOH) at Wadsworth, further confirming the sequencing results. Additionally, the virus was grown (NY State DOH) in VeroE6/TMPRSS2 cells and the progeny sequenced using another amplicon-based method, ARTIC V4, producing a fourth confirmation of the recombinant identity. Delta and Omicron-derived mutations were supported by 99–100% of the reads, consistent with the presence of a single recombinant. ARTIC sequencing confirmed the spike 214EPE insertion. Table S1 summarizes all mutations detected by nine multi-method sequencing runs on different specimens.
Mutation analysis
Highlighter analyses were performed on MAFFT-aligned SARS-CoV-2 full-genome sequences using the Highlighter tool provided by the Los Alamos HIV sequence database47 against Wuhan-Hu-1 as master. Delta AY.45 and Omicron BA.1 consensus sequences were used for comparison. They were generated with the EMBOSS cons tool (50% consensus threshold)48 using complete, high-coverage AY.45 and BA.1 sequences from North-America sampled between June and December 2021 and downloaded from GISAID.49
Phylogenetic analysis
Phylogenetic analyses were done using the Nextstrain CLI package WSL on Windows.14 As input, we used the full genome sequence of the recombinant virus (hCoV-19/USA/NY-NYULH6045/2022) and a North-America-focused global dataset until the end of February 2022 (n = 2259), complemented with a global Delta AY.45 dataset of complete, high-coverage sequences from November 2021 until the end of February 2022 (n = 335), all of which were downloaded (FASTA files and metadata) from GISAID.49 The trees were constructed using a focal-contextual subsampling, focusing on sequences from the Tri-state area of New York, New Jersey, and Connecticut (including the recombinant) and complemented with a maximum of 1500 contextual sequences outside the Tri-state area and prioritized by proximity to the focal dataset (total of n = 1557 sequences). Default parameters were used including the masking of the first 100 and last 50 bp of the SARS-CoV-2 alignment and creating a maximum-likelihood IQ time-calibrated tree, rooted to Wuhan-Hu-1.14 The phylogenetic analyses of the Delta and Omicron subregions of the recombinant were done by masking the complementary regions (Omicron or Delta genomic regions, respectively) and skipping the minimum length criterion.
Phylogeographic analysis
We performed Bayesian phylogeographic analysis to determine the plausible geographic origins of AY.45 and BA.1 infections. We combined the AY.45 part of the recombinant with all available high-quality (with high coverage, low coverage excluded and with complete sampling dates) AY.45 genomes on GISAID49 until December 31st, 2021. We used Nextalign v1.11.040 to align all genomes to the reference and trimmed the resulting alignment. Next, we estimated an unrooted maximum-likelihood phylogeny using IQ-TREE v2.2.050 with automated model selection, which determined the GTR + F + R3 model as yielding the highest fit to the data. We performed 1000 bootstrap replicates using the ultrafast bootstrap approximation (UFBOOT) in IQ-TREE v2.2.0. Given the high bootstrap support obtained for the clade that contains the recombinant (bootstrap ≥ 92%), we performed a subsequent discrete phylogeographic analysis51 with Bayesian stochastic search variable selection (BSSVS) using BEAST v1.10.552 in combination with BEAGLE v3.253 on a large subclade of the consensus phylogeny that consists of 1122 AY.45 genomes, by employing an empirical tree distribution with default priors. In order to attain proper statistical mixing, we grouped USA states that only reported a single AY.45 genome into a “USA other” location state and removed all other locations with only a single AY.45 genome. We inspected convergence and mixing aspects of all relevant parameters using Tracer 1.754 to ensure that their associated effective sample size (ESS) values were all >200. After having discarded 10% of sampled posterior trees as burn-in, we constructed a maximum clade credibility (MCC) tree using TreeAnnotator 1.10.5.52 Given the large number of available BA.1 genomes until December 31st, 2021, the analysis pipeline as described for AY.45 could not be performed. We hence used the BA.1 part of the recombinant genome and employed BLAST to consider only the closest sequences to the BA.1 recombinant segment. We first downloaded and aligned all BA.1 genomes collected up to December 31st, 2021 (n = 138,350) from GISAID and removed identical sequences originating from the same country or state in the case of sequences originating from the US. We used the resulting alignment of 105,176 sequences to create a local BLAST database, against which we queried the BA.1 part of the recombinant genome with a megablast search in BLAST + v2.12.0.55 We allowed for a minimum word size of 100 and a maximum number of 10 high scoring segment pairs per target sequence in order to obtain as many non-overlapping local alignments. After manually checking that segments do not overlap, we aggregated all hits on the same target sequence and sorted by highest combined bitscore. A total of 4,706 sequences originating from 79 different countries were tied with the highest combined bitscore. We subsequently proceeded to align these highest scoring BA.1 genomes and obtain an unrooted maximum-likelihood phylogeny as described for the AY.45 dataset, with IQ-TREE v2.2.0 this time selecting the GTR + F + R2 substitution model as the optimal model choice. However, the bootstrap analysis yielded an almost entirely unresolved phylogeny in the form of a large multifurcation (data not shown). As a result, no subsequent phylogeographic analysis was performed.
Structural analysis
Molecular graphics and analyses were performed with UCSF ChimeraX 1.3.56 Homology models of the recombinant spike were generated with the protein structure homology-modelling server SWISS-MODEL.57 The template structure was chosen based on coverage, sequence similarity, Global Model Quality Estimation (GMQE) and Quaternary Structure Quality Estimation (QSQE) scores, oligo state, and open/closed state (PDB: 7TO4, 1 RBD-up Omicron spike trimer). The Fab moiety of S309 (Sotrovimab) was added to the recombinant spike trimer model by structural overlay of an S309-bound spike co-structure in the open (1 RBD-up) position (PDB: 7TM0) using MatchMaker with pairing of the RBDs in up position.
Infectious virus neutralization assay
Wadsworth Center plaque reduction neutralization tests (PRNTs) were performed by mixing 100 μL of low passage, sequence-confirmed SARS-CoV-2 virus isolates containing approximately 100–180 plaque forming units with 100 μL of 2-fold serially diluted monoclonal antibody preparations or test sera and incubating at 37°C in 5% CO2 for one hour. Confluent Vero E6 with TMPRSS2 cells (JCRB1819, Sekusi XeonTech) seeded in 6 well plates were inoculated with 100 μL of the virus: antibody mixtures and adsorption proceeded for one hour at 37°C in 5% CO2. A 0.6% agar overlay prepared in maintenance medium (Dulbecco’s Modified Eagle Medium, 2% heat-inactivated fetal bovine serum, 100 μg/mL Penicillin G, 100 U/mL Streptomycin, 1 mg/mL Geneticin) was added after adsorption and the assay was incubated at 37°C in 5% CO2. A second agar overlay containing 0.2% Neutral red was added two days post infection. The number of plaques in each well was recorded after two additional days of incubation. All procedures were conducted in biosafety level 3 (BSL-3) laboratory conditions. Viruses used include BA.1: hCoV-19/USA/NY-Wadsworth-21103366-01/2021; Delta: hCoV-19/USA/NY-Wadsworth-2200014356-01/2021; Delta-Omicron. IC50 values were calculated by determining the percent neutralization (relative to virus only controls) for technical duplicates from two to four biological replicates of each well and using non-linear regression ([inhibitor] vs. normalized response with variable slope, GraphPad Prism).
Infectious virus plaque assay
Quantification of infectious virus during culture of the SARS-CoV-2 recombinant was performed using a solid double overlay plaque assay method. Confluent 6-well plates, seeded with VeroE6/TMPRSS2 cells, were inoculated with 100 μl of serially 10-fold diluted virus supernatant or harvest, in duplicate. After a one-hour adsorption at 37°C in 5% CO2, a 3 mL overlay containing maintenance medium (DMEM +10% FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, 1 mg/mL G418) and 0.6% Oxoid agar was added to each well. Plates were incubated for 48 h at 37°C in 5% CO2, after which a second agar overlay was added, containing maintenance medium +2% FBS, 0.6% agar, and 0.01% neutral red. Plates were incubated an additional 24 h, and the number of plaques in each well was recorded on days three, four and seven post-infection. Infectious titers were calculated, averaged, and expressed as log10 PFU/mL.
Pseudotyped virus neutralization assay
To measure neutralizing titer, sera or mAbs were serially diluted (2-fold for sera and 5-fold for mAbs) and incubated for 30 min at room temperature with pseudotyped virus. The mixture was added on 1 × 104 of ACE2.293T cells (MOI of 0.2). After 1 day of infection, medium was removed and infectivity was measured with Nano-Glo luciferase substrate (Nanolight). Luminescence was read in an Envision 2103 microplate luminometer (PerkinElmer). D614G virus (original Wuhan virus carrying the D614G spike mutation that was selected early during the pandemic) was used as reference.
Quantification and statistical analysis
Infectious virus neutralization assay
Replicate experiments were performed in technical duplicates and analyzed using GraphPad Prism 8. Antibodies that did not allow the virus to reach <50% infectivity at the highest concentration tested are listed as IC50 > 13.1 μg/mL. IC50 values were calculated by determining the percent neutralization (relative to virus only controls) for technical duplicates from two to four biological replicates of each well and using non-linear regression ([inhibitor] vs. normalized response with variable slope, GraphPad Prism). Statistical comparisons of IC50 values determined in neutralization experiments with multiple biological replicates were compared using two-way ANOVA with Tukey’s multiple comparison test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Data are shown as the mean ± standard deviation.
Pseudotyped virus neutralization assay
All experiments were performed in technical duplicates and data were analyzed using GraphPad Prism 8. Antibodies that did not allow the virus to reach <70% infectivity at the highest concentration tested are listed as IC50 > 50,000 ng/mL; for antibodies that reached 50–70% infectivity at the highest concentration tested, the IC50 was extrapolated.
Statistical significance of differences between groups were determined by one-way ANOVA followed by two-tailed, unpaired t-tests for direct comparisons (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Data are shown as the mean ± standard deviation.
Illustration
The graphical abstract was created with BioRender (BioRender.com) and UCSF ChimeraX 1.3.
Acknowledgments
We thank Drs. Joan Cangiarella and Dafna Bar-Sagi for institutional funding and support of SARS-CoV-2 genomic surveillance at NYULH. We thank Drs Marie I. Samanovic-Golden and Mark J. Mulligan from the NYU Langone Vaccine Center and the Department of Medicine for the clinical samples and related information. We also thank the staff of the Wadsworth Center Applied Genomics Technology Core for performing ARTIC sequencing, and Ashley Fisher for proofreading of the article. We are grateful to the authors and submitting laboratories who deposited data in GISAID, in particular to those whose sequences we used to create the phylogenetic tree. We thank Cornelius Roemer from the Biozentrum, University of Basel, and Nextclade for the initial assessment of our sequence.
Funding. This work was supported by: the National Institutes of Health P30CA016087 (AH). National Institutes of Health Clinical and Translational Science award 3UL1TR001445-06A1S1 (MBO). National Institutes of Health R01DA046100, R01AI122390, and R01AI120898 (NRL). Vilcek/Goldfarb Fellowship Endowment Fund (TT). Centers for Disease Control and Prevention Cooperative Agreement Number NU50CK000516 (KSG). Research Foundation Flanders (“Fonds voor Wetenschappelijk Onderzoek - Vlaanderen”) G0E1420N (GB and SLH). Research Foundation Flanders (“Fonds voor Wetenschappelijk Onderzoek - Vlaanderen”) G098321N, V420922N (GB). Internal Funds KU Leuven, Grant C14/18/094 (GB).
Author contributions
Conceptualization: RD, PJP, KSG, and AH. Methodology: RD, HZ, TT, DD, CM, PZ, GW, JP, SBG, RG, JM, SB, EL-N, SLH, GB, PJP, and AH. Visualization: RD, HZ, TT, and GB. Funding acquisition: MBO, NRL, KSG, and AH. Supervision: MD, KMcD, NRL, KSG, and AH. Writing – original draft: RD and AH. Writing – review & editing: RD, HZ, TT, DD, CM, PZ, GW, JP, SBG, RG, JM, SB, EL-N, SLH, GB, MD, MBO, PJP, KMcD, NRL, KSG, and AH.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: February 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106075.
Contributor Information
Ralf Duerr, Email: ralf.duerr@nyulangone.org.
Adriana Heguy, Email: adriana.heguy@nyulangone.org.
Supplemental information
Data and code availability
-
•
The sequence of the Delta-Omicron recombinant virus was deposited in GISAID: EPI_ISL_10792641.
-
•
The raw fastq files of xGen and shotgun deep sequencing (NYU, see Table S1), and AmpliSeq and ARTIC deep sequencing (NY State DOH, see Table S1) were deposited in NCBI SRA: PRJNA915368.
-
•
All other data are available in the main text or supplemental information, or can be made available from the lead contact upon request.
-
•
This paper does not report original code.
References
- 1.Simon-Loriere E., Holmes E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bean W.J., Jr., Cox N.J., Kendal A.P. Recombination of human influenza A viruses in nature. Nature. 1980;284:638–640. doi: 10.1038/284638a0. [DOI] [PubMed] [Google Scholar]
- 3.Moradigaravand D., Kouyos R., Hinkley T., Haddad M., Petropoulos C.J., Engelstädter J., Bonhoeffer S. Recombination accelerates adaptation on a large-scale empirical fitness landscape in HIV-1. PLoS Genet. 2014;10:e1004439. doi: 10.1371/journal.pgen.1004439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Z., Meng K., Meng G. Genomic recombination events may reveal the evolution of coronavirus and the origin of SARS-CoV-2. Sci. Rep. 2020;10:21617. doi: 10.1038/s41598-020-78703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollett S., Conte M.A., Sanborn M., Jarman R.G., Lidl G.M., Modjarrad K., Maljkovic Berry I. A comparative recombination analysis of human coronaviruses and implications for the SARS-CoV-2 pandemic. Sci. Rep. 2021;11:17365. doi: 10.1038/s41598-021-96626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolze A., Basler T., White S., Dei Rossi A., Wyman D., Dai H., Roychoudhury P., Greninger A.L., Hayashibara K., Beatty M., et al. Evidence for SARS-CoV-2 Delta and Omicron co-infections and recombination. medrXiv. 2022;3:848–859.e4. doi: 10.1101/2022.03.09.22272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rockett R.J., Draper J., Gall M., Sim E.M., Arnott A., Agius J.E., Johnson-Mackinnon J., Fong W., Martinez E., Drew A.P., et al. Co-infection with SARS-CoV-2 Omicron and Delta variants revealed by genomic surveillance. Nat. Commun. 2022;13:2745. doi: 10.1038/s41467-022-30518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacek K.A., Rambo-Martin B.L., Batra D., Zheng X.-y., Sakaguchi H., Peacock T., Keller M., Wilson M.M., Sheth M., Davis M.L., et al. Identification of a novel SARS-CoV-2 delta-omicron recombinant virus in the United States. bioRxiv. 2022 doi: 10.1101/2022.03.19.484981. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson B., Boni M.F., Bull M.J., Colleran A., Colquhoun R.M., Darby A.C., Haldenby S., Hill V., Lucaci A., McCrone J.T., et al. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell. 2021;184:5179–5188.e8. doi: 10.1016/j.cell.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekizuka T., Saito M., Itokawa K., Sasaki N., Tanaka R., Eto S., Someno R., Ogamino A., Yokota E., Saito T., Kuroda M. Recombination between SARS-CoV-2 Omicron BA.1 and BA.2 variants identified in a traveller from Nepal at the airport quarantine facility in Japan. J. Trav. Med. 2022;29:taac051. doi: 10.1093/jtm/taac051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu H., Ng D.Y.M., Liu G.Y.Z., Cheng S.S.M., Krishnan P., Chang L.D.J., Cheuk S.S.Y., Hui M.M.Y., Lam T.T.Y., Peiris M., Poon L.L.M. Recombinant BA.1/BA.2 SARS-CoV-2 virus in arriving travelers, Hong Kong, february 2022. Emerg. Infect. Dis. 2022;28:1276–1278. doi: 10.3201/eid2806.220523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colson P., Fournier P.E., Delerce J., Million M., Bedotto M., Houhamdi L., Yahi N., Bayette J., Levasseur A., Fantini J., et al. Culture and identification of a "Deltamicron" SARS-CoV-2 in a three cases cluster in southern France. J. Med. Virol. 2022;94:3739–3749. doi: 10.1002/jmv.27789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Zhou H.Y., Li J.Y., Cheng Y.X., Zhang S., Aliyari S., Wu A., Cheng G. Potential inter-variant and intra-variant recombination of Delta and Omicron variants. J. Med. Virol. 2022;94:4830–4838. doi: 10.1002/jmv.27939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duerr R., Dimartino D., Marier C., Zappile P., Levine S., François F., Iturrate E., Wang G., Dittmann M., Lighter J., et al. Clinical and genomic signatures of rising SARS-CoV-2 Delta breakthrough infections in New York. medRxiv. 2021 doi: 10.1101/2021.12.07.21267431. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans J.P., Qu P., Zeng C., Zheng Y.-M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., Oltz E.M., et al. Neutralization of the SARS-CoV-2 deltacron and BA.3 variants. N. Engl. J. Med. 2022;386:2340–2342. doi: 10.1056/NEJMc2205019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurhade C., Zou J., Xia H., Liu M., Yang Q., Cutler M., Cooper D., Muik A., Sahin U., Jansen K.U., et al. Neutralization of Omicron sublineages and Deltacron SARS-CoV-2 by 3 doses of BNT162b2 vaccine or BA.1 infection. bioRxiv. 2022 doi: 10.1101/2022.06.05.494889. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.-H., Boucau J., Bowman K., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weigang S., Fuchs J., Zimmer G., Schnepf D., Kern L., Beer J., Luxenburger H., Ankerhold J., Falcone V., Kemming J., et al. Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants. Nat. Commun. 2021;12:6405. doi: 10.1038/s41467-021-26602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nussenblatt V., Roder A.E., Das S., de Wit E., Youn J.H., Banakis S., Mushegian A., Mederos C., Wang W., Chung M., et al. Yearlong COVID-19 infection reveals within-host evolution of SARS-CoV-2 in a patient with B-cell depletion. J. Infect. Dis. 2022;225:1118–1123. doi: 10.1093/infdis/jiab622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Toole Á., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rockett R., Basile K., Maddocks S., Fong W., Agius J.E., Johnson-Mackinnon J., Arnott A., Chandra S., Gall M., Draper J., et al. Resistance mutations in SARS-CoV-2 Delta variant after Sotrovimab use. N. Engl. J. Med. 2022;386:1477–1479. doi: 10.1056/NEJMc2120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta A., Gonzalez-Rojas Y., Juarez E., Crespo Casal M., Moya J., Falci D.R., Sarkis E., Solis J., Zheng H., Scott N., et al. Early treatment for covid-19 with SARS-CoV-2 neutralizing antibody Sotrovimab. N. Engl. J. Med. 2021;385:1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- 25.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CoV-GLUE . 2022. CoV-GLUE - amino acid replacements.http://cov-glue.cvr.gla.ac.uk/#/replacement [Google Scholar]
- 28.Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.-J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birnie E., Biemond J.J., Appelman B., de Bree G.J., Jonges M., Welkers M.R.A., Wiersinga W.J. Development of resistance-associated mutations after Sotrovimab administration in high-risk individuals infected with the SARS-CoV-2 Omicron variant. JAMA. 2022;328:1104–1107. doi: 10.1001/jama.2022.13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almagro J.C., Mellado-Sánchez G., Pedraza-Escalona M., Pérez-Tapia S.M. Evolution of anti-SARS-CoV-2 therapeutic antibodies. Int. J. Mol. Sci. 2022;23:9763. doi: 10.3390/ijms23179763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Y. 2022. Possible English Recombinant Descended from Omicron and Delta (UKHSA "signal under monitoring") #422.https://github.com/cov-lineages/pango-designation/issues/422 [Google Scholar]
- 32.Tada T., Fan C., Chen J.S., Kaur R., Stapleford K.A., Gristick H., Dcosta B.M., Wilen C.B., Nimigean C.M., Landau N.R. An ACE2 microbody containing a single immunoglobulin fc domain is a potent inhibitor of SARS-CoV-2. Cell Rep. 2020;33:108528. doi: 10.1016/j.celrep.2020.108528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Mulligan M.J., Landau N.R. Partial resistance of SARS-CoV-2 Delta variants to vaccine-elicited antibodies and convalescent sera. iScience. 2021;24:103341. doi: 10.1016/j.isci.2021.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Chivukula V., Herati R.S., Hubbard S.R., Mulligan M.J., Landau N.R. Increased resistance of SARS-CoV-2 Omicron variant to neutralization by vaccine-elicited and therapeutic antibodies. EBioMedicine. 2022;78:103944. doi: 10.1016/j.ebiom.2022.103944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou H., Dcosta B.M., Landau N.R., Tada T. Resistance of SARS-CoV-2 Omicron BA.1 and BA.2 variants to vaccine-elicited sera and therapeutic monoclonal antibodies. Viruses. 2022;14:1334. doi: 10.3390/v14061334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giroglou T., Cinatl J., Rabenau H., Drosten C., Schwalbe H., Doerr H.W., von Laer D. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J. Virol. 2004;78:9007–9015. doi: 10.1128/JVI.78.17.9007-9015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vermeire J., Naessens E., Vanderstraeten H., Landi A., Iannucci V., Van Nuffel A., Taghon T., Pizzato M., Verhasselt B. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One. 2012;7:e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duerr R., Dimartino D., Marier C., Zappile P., Wang G., Lighter J., Elbel B., Troxel A.B., Heguy A. Dominance of Alpha and Iota variants in SARS-CoV-2 vaccine breakthrough infections in New York City. J. Clin. Invest. 2021;131:e152702. doi: 10.1172/JCI152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. Zenodo; 2021. Nextclade: Clade Assignment, Mutation Calling and Quality Control for Viral Genomes (1.4.5) [Google Scholar]
- 41.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wadsworth Center New York State Department of Health . 2020. New York SARS-CoV-2 Real-Time Reverse Transcriptase (RT)-PCR Diagnostic Panel, Instructions for Use.https://www.fda.gov/media/135847/download [Google Scholar]
- 46.Plitnick J., Griesemer S., Lasek-Nesselquist E., Singh N., Lamson D.M., St George K. Whole-genome sequencing of SARS-CoV-2: assessment of the Ion torrent AmpliSeq panel and comparison with the Illumina MiSeq ARTIC protocol. J. Clin. Microbiol. 2021;59:e0064921. doi: 10.1128/JCM.00649-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Los Alamos National Laboratory tools . 2022. Highlighter Tool.http://www.hiv.lanl.gov/ [Google Scholar]
- 48.explorer; E. 2022. EMBOSS Cons.https://www.bioinformatics.nl/cgi-bin/emboss/cons [Google Scholar]
- 49.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.Es.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemey P., Rambaut A., Drummond A.J., Suchard M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ayres D.L., Cummings M.P., Baele G., Darling A.E., Lewis P.O., Swofford D.L., Huelsenbeck J.P., Lemey P., Rambaut A., Suchard M.A. Beagle 3: improved performance, scaling, and usability for a high-performance computing library for statistical phylogenetics. Syst. Biol. 2019;68:1052–1061. doi: 10.1093/sysbio/syz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morgulis A., Coulouris G., Raytselis Y., Madden T.L., Agarwala R., Schäffer A.A. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:296–303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The sequence of the Delta-Omicron recombinant virus was deposited in GISAID: EPI_ISL_10792641.
-
•
The raw fastq files of xGen and shotgun deep sequencing (NYU, see Table S1), and AmpliSeq and ARTIC deep sequencing (NY State DOH, see Table S1) were deposited in NCBI SRA: PRJNA915368.
-
•
All other data are available in the main text or supplemental information, or can be made available from the lead contact upon request.
-
•
This paper does not report original code.