Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV) has become widely dispersed worldwide since it was first reported in 1994, but the seroprevalence of KSHV varies geographically. KSHV is relatively ubiquitous in Mediterranean areas and the Xinjiang Uygur Autonomous Region, China. The origin of KSHV has long been puzzling. In the present study, we collected and analysed 154 KSHV ORF-K1 sequences obtained from samples originating from Xinjiang, Italy, Greece, Iran and southern Siberia using Bayesian evolutionary analysis in BEAST to test the hypothesis that KSHV was introduced into Xinjiang via the ancient Silk Road. According to the phylogenetic analysis, 72 sequences were subtype A and 82 subtype C, with C2 (n = 56) being the predominant subtype. The times to the most recent common ancestors (tMRCAs) of KSHV were 29,872 years (95% highest probability density [HPD], 26,851–32,760 years) for all analysed sequences and 2037 years (95% HPD, 1843–2229 years) for Xinjiang sequences in particular. The tMRCA of Xinjiang KSHV was exactly matched with the time period of the ancient Silk Road approximately two thousand years ago. This route began in Chang’an, the capital of the Han dynasty of China, and crossed Central Asia, ending in the Roman Empire. The evolution rate of KSHV was slow, with 3.44 × 10−6 substitutions per site per year (95% HPD, 2.26 × 10−6 to 4.71 × 10−6), although 11 codons were discovered to be under positive selection pressure. The geographic distances from Italy to Iran and Xinjiang are more than 4000 and 7000 kilometres, respectively, but no explicit relationship between genetic distance and geographic distance was detected.
Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV, also known as human herpesvirus 8), is the aetiological agent of Kaposi’s sarcoma (KS), primary effusion lymphoma, and a form of multicentric Castleman’s disease [8, 35]. A myriad of previous studies have documented that the seroprevalence of KSHV is very high in Africa and the Mediterranean region, where KSHV seroprevalence ranges from 20% to 80% in adult populations [12, 20, 36], whereas its prevalence among the general population in North America and Northern Europe remains low [5, 19, 32]. The prevalence of KSHV in Asia differs among geographical regions, and although the epidemiological features of KSHV in Asia have not been extensively explored, it has been proposed that KSHV is widespread in the Xinjiang autonomous region [44], which is located in northwest China and is surrounded by Mongolia to the east, Russia to the north and Kazakhstan to the west. The region consists of many ethnicities, predominantly Han and Uygur. Previous studies have shown that the prevalence of KSHV in Xinjiang is as high as 40% among different populations [6, 14], which is much higher than that in other regions in China. Xinjiang served as an important staging post for the ancient Silk Road thousands of years ago. The goods produced in the Han dynasty and Rome were traded frequently via the ancient Silk Road. The ancient Roman Empire, now Italy, is an endemic area of KS, suggesting that KSHV was likely introduced into Xinjiang, China, from Rome or the Middle East via the Silk Road. Moreover, previous reports have shown that the KS in Xinjiang can be classified as classical KS and shares common characteristics with classical KS cases occurring in the Mediterranean region [2, 24]. This may indicate potential associations between KS in Xinjiang and the Mediterranean region. Phylogenetic analysis also revealed that KSHV isolates in Xinjiang belong to subtypes A and C, which are ubiquitous in Eurasia [28, 43].
The KSHV genome is composed of double-stranded linear DNA that is 165–170 kb long [34]. To the best of our knowledge, large blocks of KSHV sequence are highly conserved and do not exhibit much variation among isolates. However, some genomic regions in KSHV display remarkable variability, such as ORF-K1, which is located in the left terminal repeat region of the KSHV genome. Previous studies have revealed that ORF-K1 is a transmembrane protein, with amino acid sequence divergence levels between geographically disparate isolates ranging from 30% to 60% [4, 9, 45]. KSHV is grouped into six main genotypes (A to F) that show high divergence in the ORF-K1 gene [45]. Therefore, ORF-K1 is regarded as a marker of strain diversity and is used to track the epidemiological spread of the virus.
Based on these observations, we hypothesized that the ancestor of KSHV in Xinjiang, China, may come from Eurasian areas situated on the ancient Silk Road. Therefore, a phylogenetic and Bayesian evolutionary analysis based on sequences of the ORF-K1 gene was conducted to verify our hypothesis and to better understand the molecular epidemiological features of KSHV in China.
Materials and methods
Sequence collection
A comprehensive search of the KSHV ORF-K1 gene was conducted in GenBank (http://www.ncbi.nlm.nih.gov). A total of 261 sequences were obtained with basic information, such as accession number, genotype and collection date, if provided. The number of sequences obtained from China, Italy, Greece, Iran and southern Siberia was 53, 92, 9, 89 and 18, respectively. Of these, 154 sequences with explicit isolation dates, including 53 from China, 24 from Italy, 9 from Greece, 50 from Iran and 18 from southern Siberia, were included in further analyses. All sequences from China collected between 1997 and 2009 were from Xinjiang.
Sequence alignment and phylogenetic analysis
A total of 154 sequences (Appendix Table 1) with unequivocal isolation dates were aligned using MEGA6 [37] to determine their subtype. The collection date for each sequence was obtained either from GenBank annotations or from the description of the target sequence in the corresponding research article. The resulting alignment was imported into PAUP* for neighbour-joining analysis [42] as follows: 1,000 bootstrap replicates, randomly broken site, and general time-reversible distance measure with a gamma distribution shape parameter of 0.5. The output tree was a bootstrap 50% majority rule consensus tree. Moreover, the sequence diversity was estimated via the Tamura-Nei model in MEGA6.
Bayesian MCMC molecular dating analysis
The recombinant alignments were identified with SplitsTree [23]. A GTR (general time reversible) + I (proportion of invariant sites) + Γ4 (gamma-distributed rate variation with four rate categories) nucleotide substitution model was the best-fitting model based on ModelTest3.7 [31]. The evolutionary rates of strains were calculated using the Bayesian substitution model implemented in BEASTv1.8.3 [16]. Additionally, the times of the most recent common ancestors (tMRCAs) of strains from each country were also estimated using BEAST. Both strict and relaxed (uncorrelated exponential and uncorrelated lognormal) molecular clocks [15] were applied for the analyses. A Bayesian skyline coalescent tree prior was used to estimate the effective population size. The MCMC chain was run for 100 million steps, with parameter values sampled at every 5000 steps. The resulting log files were imported into the program TRACERv1.6.0 to ascertain the convergence of the MCMC chain, which suggested that an effective sample size (ESS) of more than 200 for all parameters was reached. The uncertainty of all parameter estimates was assessed based on 95% HPD (highest probability density) intervals. Both the comparisons of different models and the reconstruction of the Bayesian skyline plot were implemented in TRACER. A lognormal relaxed uncorrelated clock with an exponential growth demographic population was the best-fit model by Bayesian factor analysis. The maximum clade credibility (MCC) tree with the initial 10% of trees removed as the burn-in was generated using Tree Annotator 1.8.3, which is available in the BEAST package. Subsequently, the MCC tree was visualized in FigTree v1.4.2. Finally, we constructed the migration pathways of KSHV among the different locations on the ancient Silk Road on the basis of the tMRCA of KSHV in each country.
Geographic distance versus genetic distance
The pairwise genetic distance of each sequence included in this analysis and the genetic distance between and within each group were estimated using MEGA6, with application of the Tamura-Nei model. The resulting distance matrix combined with the geographic distance obtained from ArcGIS (version 10.1) was then imported into R software (version 3.2.4, http://www.r-project.org/) to assess the correlation between the genetic distance and the geographic distance. A sample collected in Italy (GenBank accession number FJ884611.1) was set as the reference, which has zero genetic and geographic distance from itself.
Selection pressure analysis
To identify codons under positive selection, relative rates of synonymous substitutions per site (dS) and nonsynonymous substitutions per site (dN) were calculated using the HyPhy package in Datamonkey [29, 30] (http://www.datamonkey.org). Two datasets were used for this analysis, one consisting of coding sequences with 115 codons originating from China and a second consisting of all sequences with 71 codons after alignment. The HKY85 model of nucleotide substitution and a neighbour-joining tree were used for the analysis. The ω ratios (dN/dS) were calculated via three different likelihood approaches, including single-likelihood ancestor counting (SLAC), fixed effects likelihood (FEL), and random effects likelihood (REL). Sites showing evidence of positive selection by at least two of the methods listed above with high statistical significance (p < 0.1 or Bayes factor > 50) were considered to be under positive selection.
Results
Phylogenetic analysis
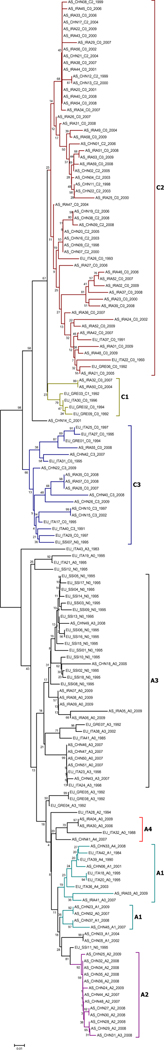
A total of 154 strain sequences obtained from 1983 to 2009 were used for reconstruction of the neighbour-joining tree (Fig. 1), maximum-likelihood tree, and maximum-parsimony tree (Appendix Figures 1 and 2). Of these, 72 were of subtype A and 82 were of subtype C, with the most common KSHV genotype being C2 (n = 56). Strains from Xinjiang were explicitly subdivided into six different genotypes: C2 (n = 18), A2 (n = 12), A3 (n = 8), A1 (n = 7), C3 (n = 6) and A4 (n = 2). Similarly, the predominant genotype in Iran was C2 (n = 34), followed by A3 (n = 5), C3 (n = 7), A4 (n = 2) and A1 (n = 2). The genotype distribution in Italy and Greece was A3 (n = 8), C3 (n = 7), C1 (n = 4), C2 (n = 4), A4 (n = 4) and six uncertain subtypes. Most strains collected from southern Siberia were A3 (n = 15), followed by C3 (n = 2) and A2 (n = 1). The overall mean sequence divergence of KSHV was 10.4% across different areas, and the diversity of the KSHV sequence of Xinjiang was 8.3%.
Fig. 1.
Neighbour-joining tree of KSHV sequences with the following parameters: 1,000 bootstrap replicates, randomly broken site, and general time-reversible distance measure with a gamma distribution shape parameter of 0.5. The length of the alignment was 212 bp, corresponding to nt 159–370 of the KSHV genome
Bayesian MCMC analysis
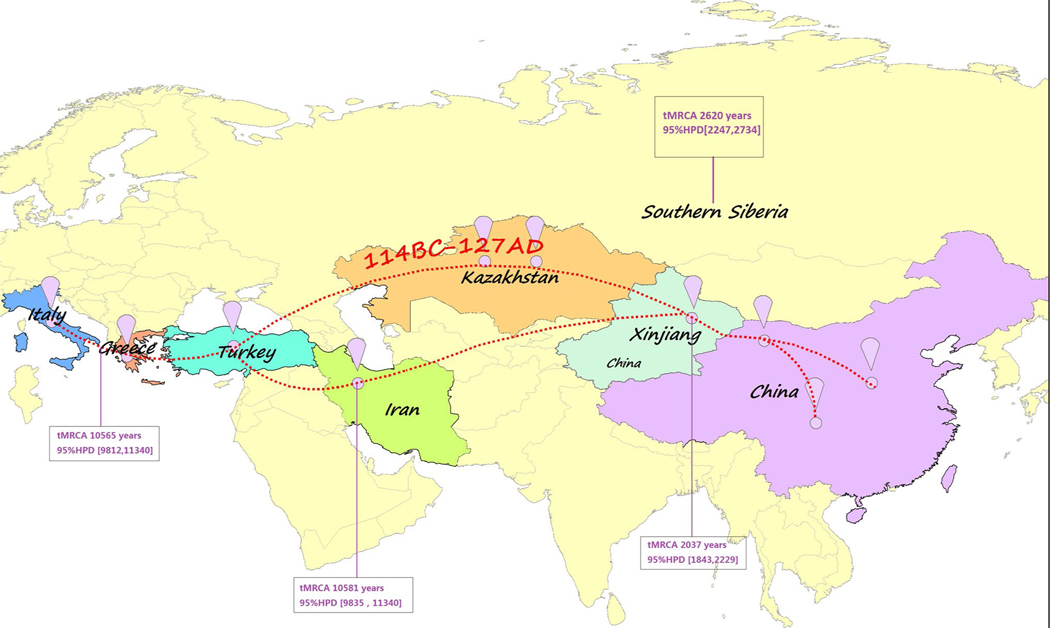
No recombinants were detected using the Phi-test (P = 1.0). The root of the tree was calculated to be 29,872 years old (95% HPD, 26,851–32,760 years), suggesting that the tMRCA of KSHV in Italy, Greece, Iran, China and southern Siberia was approximately 30,000 years ago. Subsequently, we separately assessed the tMRCA of KSHV from each country. The sequences from Greece were analysed together with sequences from Italy because only nine strains from Greece were available. The tMRCA of Italian and Greek sequences was determined to be 10,565 years (95% HPD, 9,812–11,340 years), which was similar to the tMRCA of Iranian sequences (10,581 years, with 95% HPD, 9,835–11,340 years). The tMRCA of sequences from southern Siberia was 2,620 years (95% HPD, 2,247–2,734 years). Moreover, the tMRCA of KSHV in Xinjiang was 2,037 years (95% HPD 1,843–2,229 years), indicating that KSHV in China is much younger than KSHV in Iran and the Mediterranean but similar to that in southern Siberia (Table 1). We then constructed the potential KSHV transmission routes across the five areas based on the MCC trees (Fig. 2), which are shown in Appendix Figures 3-6. The overall mean nucleotide substitution rate was 3.44 × 10−6 substitutions per site per year (95% HPD, 2.26×10−6 to 4.71×10−6).
Table 1.
The statistics generated from Bayesian MCMC analysis
| Summary statistics | TreeModel.tree Height | China | Italy & Greece | Iran | Southern Siberia |
|---|---|---|---|---|---|
| Mean (tMRCA) | 29872 | 2037 | 10565 | 10581 | 2620 |
| Stderr of mean | 51.48 | 0.53 | 8.87 | 3.39 | 0.76 |
| Stdev | 5324.74 | 98.67 | 405.42 | 384.39 | 125.23 |
| Median (tMRCA) | 32651.40 | 2037.53 | 10569.12 | 10574.64 | 2620.46 |
| Geometric mean | 32118.27 | 2035.21 | 10557.75 | 10574.09 | 2614.87 |
| 95% HPD | [26851, 32760] | [1843, 2229] | [9812, 11340] | [9835, 11340] | [2247, 2734] |
| ESS | 19608.87 | 4171.44 | 2086.75 | 12856.79 | 1248.44 |
tMRCA, time to the the most recent common ancestor; Stderr of mean, standard error of mean tMRCA; Stdev, standard deviation; HPD, highest posterior density; ESS, effective sample size
Fig. 2.
The suspected transmission pathway of KSHV along the Silk Road. The maximum clade credibility tree was manually plotted onto the geographic space showing the most parsimonious route of dispersal for KSHV. The red dotted line shows the main route of the Silk Road 2000 years ago, and “114 BC-127 AD” represents the time period of the ancient Silk Road
Genetic distance versus geographic distance
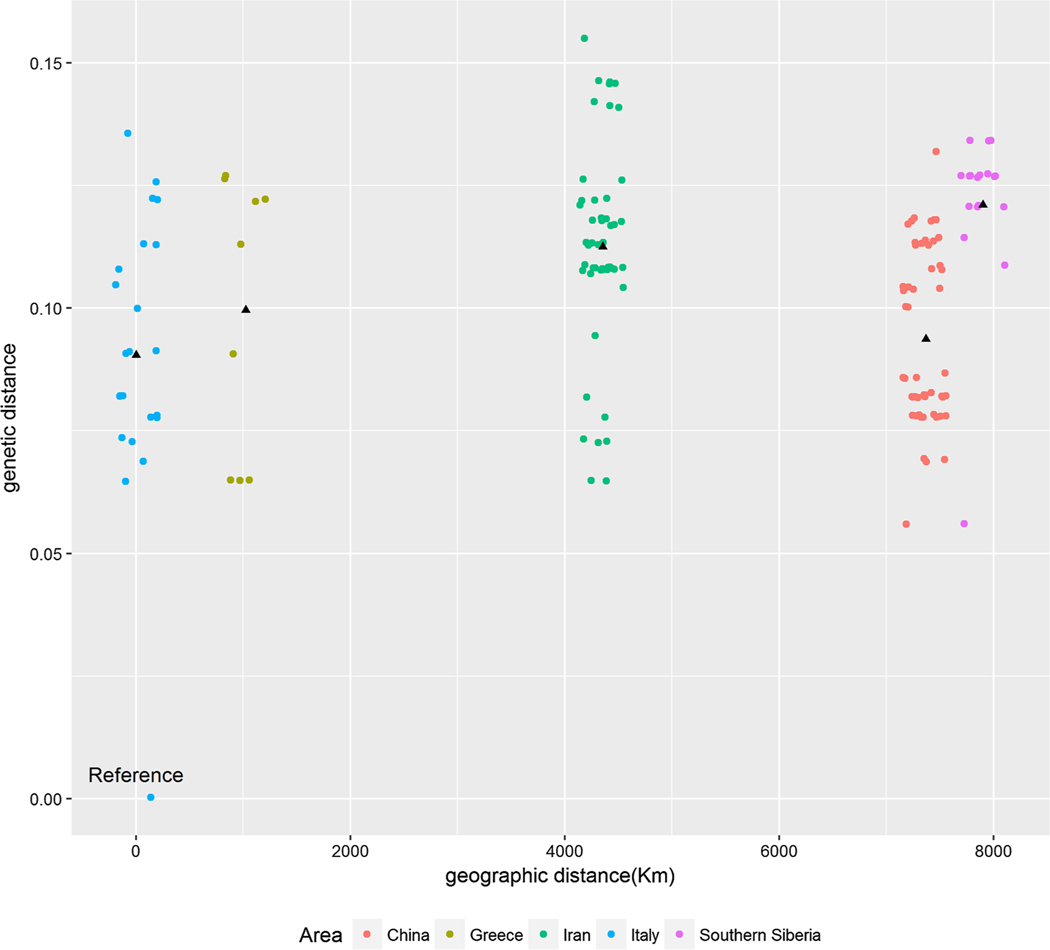
The overall mean genetic distance among all KSHV sequences included in the analysis was 0.104, and the mean genetic distance varied across groups (China, 0.094; Iran, 0.074; Iran and Greece, 0.097; southern Siberia, 0.036) (Appendix Table 2). Plotting the genetic versus geographic distance from a pre-set reference to all residual sequences did not show any explicit trends (Fig. 3). Concordantly, no significant difference between KSHV strains collected from different countries and the reference sequences was detected for the genetic distance (p = 0.452).
Fig. 3.
Plot of genetic distance versus geographic distance of KSHV sequences. The black triangle indicates the average genetic distance of KSHV within each group. Samples in each area were jittered to avoid overlap
Reconstruction of the Bayesian skyline plot
A Bayesian skyline plot (BSP) for the KSHV sequence was constructed (Appendix Figures 7-9). The plot shows changes in the median estimate of relative genetic diversity (Ne τ) of the virus with time, where Ne τ is the product of the effective population size (Ne) and generation time (τ). The plot also displays the 95% HPD interval that represents both the phylogenetic and the coalescent uncertainty. The population size of Chinese KSHV remained relatively stable until 1000 AD, after which it experienced a fluctuation within a narrow range from 1000 AD to 1900 AD. Then, the population size experienced a sharp increase after approximately 1900 AD, peaking at 2000 AD. Interestingly, a similar pattern of effective population size was found in KSHV sequences from the other three countries, although no sharp increases were observed over time.
Selection pressure analysis
The mean ω ratio was 2.54 in the codons of all KSHV sequences included in this analysis, which is highly similar to the mean ω ratios derived from the codons of Chinese sequences (ω = 2.62). Nearly 17% of the codons (12/71) were under positive pressure, as determined by our pre-set criterion when all sequences were included for analysis. Nine of those 12 codons were also under positive selection pressure when sequences not from China were excluded (Appendix Tables 3 and 4). No codon was under negative selection pressure according to the SLAC and REL methods; however, six codons were under negative selection pressure based on the FEL method (data not shown).
Discussion
In this study, we analysed KSHV ORF-K1 sequences from samples originating from China, Iran, Italy, Greece and southern Siberia [7], important staging posts located on the ancient Silk Road two thousand years ago. No recombination was detected in any of the sequences included in the present study, although recombination of KSHV ORF-K1 has been described previously [17]. We found that the tMRCA of KSHV sequences was approximately 30,000 years, with a slower mutation rate than that reported in a prior study conducted by Cordiali-Fei et al. [10]. The following reasons may explain this discrepancy: 1) We included 154 strains collected from five different areas, whereas Cordiali-Fei included 27 strains collected from Italy. 2) The length and location of the alignments were different in our study and the study by Cordiali-Fei. 3) The tMRCA of strains in our study were different from those in their study. Nevertheless, our results indicate that KSHV in Iran and near the Mediterranean Sea, both near Africa, was transmitted via hominin migration from Africa [45]. This finding is consistent with results from ancient DNA analysis showing that the earliest discoveries of distinctively modern populations in both Europe and most parts of Asia can be dated no earlier than 40,000–45,000 years ago [41].
Modern human origin and migration studies have revealed that early migrations out of Africa into the Levant (eastern Mediterranean) and Arabian peninsula occurred approximately 120,000 to 90,000 years ago, but further dispersal of humans halfway around the world did not begin until approximately 60,000 years ago [13]. This out-of-Africa migration was pulsed, with waves of dispersal eastward to South Asia, Indonesia and Australia by 50,000 years ago, migration westward to Europe by 45,000 years ago [13], migration into North Asia by 20,000 years ago, and to the Americas by 15,000 years ago [21]. For China, the earliest modern humans arrived in southern China at least as early as ~80,000 years ago [26], suggesting that modern humans did not appear in Central Asia until long after they left Africa. Therefore, we propose that the first large-scale population migration between Europe and Central Asia occurred along the ancient Silk Road.
Many epidemiological studies have demonstrated that Xinjiang, Iran and Italy, particularly Sicily, are endemic areas for classical KS [22, 25, 27, 40]. In addition, northern Israel and Greece, which were also part of the Silk Road, are high-KSHV-prevalence areas, and classical KS is also endemic there [11, 39]. Furthermore, it is now widely accepted that the ancient Silk Road, for centuries an ancient network of trade routes, was central to cultural interactions across regions of the Asian continent, connecting the East and West from China to the Mediterranean Sea [18]. The route appeared approximately 2000 years ago (114 BC-127 AD, according to Chinese history), and merchants from the Han dynasty (207 BC-220 AD) and the Roman Empire (27 BC-395 AD) crisscrossed Xinjiang, a land of deserts and mountains at the heart of the Silk Road, avoiding the Taklamakan Desert.
The results of BEAST MCMC analysis showed that the tMRCA of KSHV in both Italy and Iran was approximately 10,000 years, whereas the tMRCA of Xinjiang was 2037 years ago (95% HPD, 1843–2229 years), with subtype A originating 1200 years ago and subtype C originating 1100 years ago. These results partially support our hypothesis that KSHV was transmitted to Xinjiang via the migration of people along the Silk Road, according to the temporal sequence.
Phylogenetic analysis revealed that KSHV in these four countries was either subtype A or subtype C, which is consistent with previous studies [38, 43]. Furthermore, the genetic distances of KSHV strains within different countries were close to the mean genetic distance of all KSHV strains in the study. Moreover, no significant trend of increasing genetic distance with increasing geographic distance was detected. These results demonstrate the low evolutionary rate of the KSHV sequence and suggest the presence of an orthologue of KSHV in these four countries. Additionally, selection pressure analysis demonstrated that KSHV subtypes A and C were under pervasive positive selection pressure, which was the principal evolutionary force underlying the genetic differentiation of populations and may explain the relatively large diversity of ORF-K1. Importantly, we found nine sites under positive selection pressure among the Chinese sequences, and these nine codons were also under positive selection pressure in the analysis of all sequences. This finding strongly suggests that KSHV in China and the other three countries may share similar mutation patterns.
Moreover, the Bayesian skyline plot displayed a marked rise in the effective population size of KSHV over time, peaking near the year 2000. Because HIV is the most important risk factor for KSHV infection [33], we postulate that this increase may be attributable to the global outbreak of HIV in the last decades of the 20th century [1, 3]. The subsequent ESS decline may be attributable to HIV prevention efforts.
Some limitations should be noted here. First, more prior distribution should be applied to calibrate the molecular clock to identify discrepancies and for corroboration. Second, a longer alignment is much better for robust phylogenetic results, but the alignment in our study was relatively short.
In conclusion, this study is the first to show, using Bayesian evolutionary analysis, that KSHV in Xinjiang, China, was introduced via the ancient Silk Road and demonstrates for the first time that the transmission route of KSHV has a strong relationship to modern human migration. However, to better understand the global dispersal of KSHV and the molecular features of each subtype, more extensive studies will be required.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00705-017-3467-1) contains supplementary material, which is available to authorized users.
References
- 1.Centers for Disease Control (1982) Current trends update on acquired immune deficiency syndrome (AIDS)—United States. MMWR 31:277–279 [PubMed] [Google Scholar]
- 2.Antman K, Chang Y (2000) Kaposi’s sarcoma. N Engl J Med 342:1027–1038 [DOI] [PubMed] [Google Scholar]
- 3.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L (1983) Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science (New York, NY) 220:868–871 [DOI] [PubMed] [Google Scholar]
- 4.Biggar RJ, Whitby D, Marshall V, Linhares AC, Black F (2000) Human herpesvirus 8 in Brazilian Amerindians: a hyperendemic population with a new subtype. J Infect Dis 181:1562–1568 [DOI] [PubMed] [Google Scholar]
- 5.Blauvelt A, Sei S, Cook PM, Schulz TF, Jeang KT (1997) Human herpesvirus 8 infection occurs following adolescence in the United States. J Infect Dis 176:771–774 [DOI] [PubMed] [Google Scholar]
- 6.Cao Y, Minhas V, Tan X, Huang J, Wang B, Zhu M, Gao Y, Zhao T, Yang L, Wood C (2014) High prevalence of early childhood infection by Kaposi’s sarcoma-associated herpesvirus in a minority population in China. Clin Microbiol Infect: Off Publ Eur Soc Clin Microbiol Infect Dis 20:475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassar O, Bassot S, Plancoulaine S, Quintana-Murci L, Harmant C, Gurtsevitch V, Senyuta NB, Yakovleva LS, de The G, Gessain A (2010) Human herpesvirus 8, Southern Siberia. Emerg Infect Dis 16:580–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM (1995) Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332:1186–1191 [DOI] [PubMed] [Google Scholar]
- 9.Cook PM, Whitby D, Calabro ML, Luppi M, Kakoola DN, Hjalgrim H, Ariyoshi K, Ensoli B, Davison AJ, Schulz TF (1999) Variability and evolution of Kaposi’s sarcoma-associated herpesvirus in Europe and Africa. International Collaborative Group. AIDS (London, England) 13:1165–1176 [DOI] [PubMed] [Google Scholar]
- 10.Cordiali-Fei P, Trento E, Giovanetti M, Lo Presti A, Latini A, Giuliani M, D’Agosto G, Bordignon V, Cella E, Farchi F, Ferraro C, Lesnoni La Parola I, Cota C, Sperduti I, Vento A, Cristaudo A, Ciccozzi M, Ensoli F (2015) Analysis of the ORF-K1 hypervariable regions reveal distinct HHV-8 clustering in Kaposi’s sarcoma and non-Kaposi’s cases. J Exp Clin Cancer Res: CR 34:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidovici B, Karakis I, Bourboulia D, Ariad S, Zong J, Benharroch D, Dupin N, Weiss R, Hayward G, Sarov B, Boshoff C (2001) Seroepidemiology and molecular epidemiology of Kaposi’s sarcoma-associated herpesvirus among Jewish population groups in Israel. J Natl Cancer Inst 93:194–202 [DOI] [PubMed] [Google Scholar]
- 12.Dedicoat M, Newton R (2003) Review of the distribution of Kaposi’s sarcoma-associated herpesvirus (KSHV) in Africa in relation to the incidence of Kaposi’s sarcoma. Br J Cancer 88:1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.deMenocal PB, Stringer C (2016) Human migration: climate and the peopling of the world. Nature 538:49–50 [DOI] [PubMed] [Google Scholar]
- 14.Dilnur P, Katano H, Wang ZH, Osakabe Y, Kudo M, Sata T, Ebihara Y (2001) Classic type of Kaposi’s sarcoma and human herpesvirus 8 infection in Xinjiang, China. Pathol Int 51:845–852 [DOI] [PubMed] [Google Scholar]
- 15.Drummond AJ, Ho SY, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duprez R, Hbid O, Afonso P, Quach H, Belloul L, Fajali N, Ismaili N, Benomar H, Hassane Tahri E, Huerre M, Quintana-Murci L, Gessain A (2006) Molecular epidemiology of the HHV-8 K1 gene from Moroccan patients with Kaposi’s sarcoma. Virology 353:121–132 [DOI] [PubMed] [Google Scholar]
- 18.Elisseeff V (2001) The Silk Road: highways of culture and commerce. UNESCO Publishing/Berghahn Books, New York [Google Scholar]
- 19.Engels EA, Atkinson JO, Graubard BI, McQuillan GM, Gamache C, Mbisa G, Cohn S, Whitby D, Goedert JJ (2007) Risk factors for human herpesvirus 8 infection among adults in the United States and evidence for sexual transmission. J Infect Dis 196:199–207 [DOI] [PubMed] [Google Scholar]
- 20.Gao SJ, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo CR, Saah A, Phair J, Detels R, Chang Y, Moore PS (1996) KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi’s sarcoma. Nat Med 2:925–928 [DOI] [PubMed] [Google Scholar]
- 21.Goebel T, Waters MR, O’Rourke DH (2008) The late Pleistocene dispersal of modern humans in the Americas. Science (New York, NY) 319:1497–1502 [DOI] [PubMed] [Google Scholar]
- 22.Goedert JJ, Martin MP, Vitale F, Lauria C, Whitby D, Qi Y, Gao X, Carrington M (2016) Risk of classic Kaposi sarcoma with combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci: a population-based case–control study. J Infect Dis 213:432–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267 [DOI] [PubMed] [Google Scholar]
- 24.Iscovich J, Boffetta P, Franceschi S, Azizi E, Sarid R (2000) Classic kaposi sarcoma: epidemiology and risk factors. Cancer 88:500–517 [PubMed] [Google Scholar]
- 25.Jalilvand S, Shoja Z, Mokhtari-Azad T, Nategh R, Gharehbaghian A (2011) Seroprevalence of human herpesvirus 8 (HHV-8) and incidence of Kaposi’s sarcoma in Iran. Infect Agents Cancer 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Martinon-Torres M, Cai YJ, Xing S, Tong HW, Pei SW, Sier MJ, Wu XH, Edwards RL, Cheng H, Li YY, Yang XX, de Castro JM, Wu XJ (2015) The earliest unequivocally modern humans in southern China. Nature 526:696–699 [DOI] [PubMed] [Google Scholar]
- 27.Mancuso R, Biffi R, Valli M, Bellinvia M, Tourlaki A, Ferrucci S, Brambilla L, Delbue S, Ferrante P, Tinelli C, Clerici M (2008) HHV8 a subtype is associated with rapidly evolving classic Kaposi’s sarcoma. J Med Virol 80:2153–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouyang X, Zeng Y, Fu B, Wang X, Chen W, Fang Y, Luo M, Wang L (2014) Genotypic analysis of Kaposi’s sarcoma-associated herpesvirus from patients with Kaposi’s sarcoma in Xinjiang, China. Viruses 6:4800–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pond SL, Frost SD (2005) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics (Oxford, England) 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 30.Pond SL, Frost SD, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics (Oxford, England) 21:676–679 [DOI] [PubMed] [Google Scholar]
- 31.Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics (Oxford, England) 14:817–818 [DOI] [PubMed] [Google Scholar]
- 32.Regamey N, Cathomas G, Schwager M, Wernli M, Harr T, Erb P (1998) High human herpesvirus 8 seroprevalence in the homosexual population in Switzerland. J Clin Microbiol 36:1784–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohner E, Wyss N, Heg Z, Faralli Z, Mbulaiteye SM, Novak U, Zwahlen M, Egger M, Bohlius J (2016) HIV and human herpesvirus 8 co-infection across the globe: systematic review and meta-analysis. Int J Cancer 138:45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS (1996) Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci USA 93:14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L et al. (1995) Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 86:1276–1280 [PubMed] [Google Scholar]
- 36.Stolka K, Ndom P, Hemingway-Foday J, Iriondo-Perez J, Miley W, Labo N, Stella J, Abassora M, Woelk G, Ryder R, Whitby D, Smith JS (2014) Risk factors for Kaposi’s sarcoma among HIV-positive individuals in a case control study in Cameroon. Cancer Epidemiol 38:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 38.Tornesello ML, Biryahwaho B, Downing R, Hatzakis A, Alessi E, Cusini M, Ruocco V, Katongole-Mbidde E, Loquercio G, Buonaguro L, Buonaguro FM (2010) Human herpesvirus type 8 variants circulating in Europe, Africa and North America in classic, endemic and epidemic Kaposi’s sarcoma lesions during pre-AIDS and AIDS era. Virology 398:280–289 [DOI] [PubMed] [Google Scholar]
- 39.Touloumi G, Kaklamanis L, Potouridou I, Katsika-Hatziolou E, Stratigos J, Mueller N, Hatzakis A (1997) The epidemiologic profile of Kaposi’s sarcoma in Greece prior to and during the AIDS era. Int J Cancer 70:538–541 [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Wang H, He B, Hui Y, Lv G, Li L, Wen H (2012) Virological and molecular characterization of Kaposi’s sarcoma-associated herpesvirus strains from Xinjiang, China. Eur J Clin Microbiol Infect Dis 31:53–59 [DOI] [PubMed] [Google Scholar]
- 41.White TD, Asfaw B, DeGusta D, Gilbert H, Richards GD, Suwa G, Howell FC (2003) Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature 423:742–747 [DOI] [PubMed] [Google Scholar]
- 42.Wu X, Wan XF, Wu G, Xu D, Lin G (2006) Phylogenetic analysis using complete signature information of whole genomes and clustered Neighbour-Joining method. Int J Bioinform Res Appl 2:219–248 [DOI] [PubMed] [Google Scholar]
- 43.Zhang D, Pu X, Wu W, Jin Y, Juhear M, Wu X (2008) Genotypic analysis on the ORF-K1 gene of human herpesvirus 8 from patients with Kaposi’s sarcoma in Xinjiang, China. J Genet Genomics = Yi chuan xue bao 35:657–663 [DOI] [PubMed] [Google Scholar]
- 44.Zhang T, Shao X, Chen Y, Zhang T, Minhas V, Wood C, He N (2012) Human herpesvirus 8 seroprevalence, China. Emerg Infect Dis 18:150–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zong JC, Ciufo DM, Alcendor DJ, Wan X, Nicholas J, Browning PJ, Rady PL, Tyring SK, Orenstein JM, Rabkin CS, Su IJ, Powell KF, Croxson M, Foreman KE, Nickoloff BJ, Alkan S, Hayward GS (1999) High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol 73:4156–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.