Abstract
Chromobox (CBX) proteins are important epigenetic regulatory proteins and are widely involved in biological processes, such as embryonic development, the maintenance of stem cell characteristics and the regulation of cell proliferation and apoptosis. Disorder and dysfunction of CBXs in cancer usually lead to the blockade or ectoptic activation of developmental pathways, promoting the occurrence, development and progression of cancer. In the present review, the characteristics and functions of CBXs were first introduced. Subsequently, the expression of CBXs in cancers and the relationship between CBXs and clinical characteristics (mainly cancer grade, stage, metastasis and relapse) and prognosis were discussed. Finally, it was described how CBXs regulate cell proliferation and self-renewal, apoptosis and the acquisition of malignant phenotypes, such as invasion, migration and chemoresistance, through mechanisms involving epigenetic modification, nuclear translocation, noncoding RNA interactions, transcriptional regulation, posttranslational modifications, protein-protein interactions, signal transduction and metabolic reprogramming. The study also focused on cancer therapies targeting CBXs. The present review provides new insight and a comprehensive basis for follow-up research on CBXs and cancer.
Keywords: chromobox, cancer, chromatin modification, biomarker, cancer therapy
1. Introduction
In 1978, Lewis (1) discovered the polycomb group (PcG) protein in Drosophila melanogaster, which controls recognition, differentiation and the somatotype in these flies. Subsequently, the PcG protein was identified in all metazoans and indicated to exhibit a high degree of evolutionary conservation (2); this protein regulates a variety of biological processes during embryonic development, such as cell fate and lineage determination, cell memory, stem cell function and tissue homeostasis (3-5). In 1986, the heterochromatin protein 1 (HP1) protein was discovered in Drosophila melanogaster. HP1 is a nonhistone chromosome protein that mediates gene silencing through heterochromatin formation and structural maintenance (6,7). Chromobox (CBX) proteins are important members of the PcG protein family and HP1 protein family. Forming a class of epigenetic regulators, CBXs are extensively involved in various biological processes, including embryonic development, the maintenance of stem cell characteristics, the regulation of cell proliferation and apoptosis (8-10). Increasing evidence demonstrates that CBXs are involved in the regulation of tumor biological processes, such as the cell cycle (11), chemotherapy sensitivity (12), radiotherapy sensitivity (13), tumor cell stemness (14) and tumor metabolism (15), and have a key role in tumor occurrence and development. Identifying the mechanisms by which CBXs regulate tumors may provide promising novel targets for anticancer strategies. To date, numerous chromatin regulatory factors have been identified as targets for anticancer therapy (16). Furthermore, small-molecule inhibitors of these targets have been developed and entered the clinical evaluation stage. Of note, the histone deacetylase (HDAC) inhibitors, such as vorinostat and romidepsin (17), have been approved by the Food and Drug Administration for clinical use. However, they erase/write epigenetic marks throughout the genome, thereby activating/inhibiting a series of genes with carcinogenic and antitumor functions (18,19). Therefore, treatment with nonspecific HDAC inhibitors may lead to unintended consequences. It is thought that targeted readers may specifically activate an antiproliferation pathway or inhibit a tumor growth pathway. Small-molecule regulators of histone readers, such as bromodomain inhibitors, are under investigation in clinical trials, highlighting their great potential as treatments (20,21). As histone readers, CBXs are expected to become a new target in tumor therapy. Therefore, a comprehensive and in-depth understanding of the function of CBXs and information on the recent research status of its role in tumors have important guiding significance for researchers.
Although several recent articles have reviewed the roles of HP1 family proteins (22) and PcG family proteins (23-25) related to CBXs in cancer, none of these articles have summarized or analyzed the biological functions of all of the members of the CBX family (CBX1-8) in cancers. The present review is a comprehensive and systematic review focusing on the biological functions, regulatory mechanisms and prognostic and therapeutic value of all the members of the CBX family in cancers. It is expected that the present review will provide new insight and a comprehensive basis for follow-up research on CBXs and cancer.
2. Constituent members of CBXs
To date, eight genes encoding CBXs have been identified in the mammalian genome: CBX1 (chromosomal location: 17q21.32), CBX2 (chromosomal location: 17q25.3), CBX3 (chromosomal location: 7p15.2), CBX4 (chromosomal location: 17q25.3), CBX5 (chromosomal location: 12q13.13), CBX6 (chromosomal location: 22q13.1), CBX7 (chromosomal location: 22q13.1) and CBX8 (chromosomal location: 17q25.3). The CBXs encoded by these genes have similar chemical structures. Of note, the N-terminal region carries a chromatin-binding domain (chromodomain, CHD). Furthermore, according to the similarity of their C-terminal domains, CBXs may be further classified into CBXs in the PcG family and CBXs in the HP1 family.
3. Characteristics and functions of PcG family CBXs
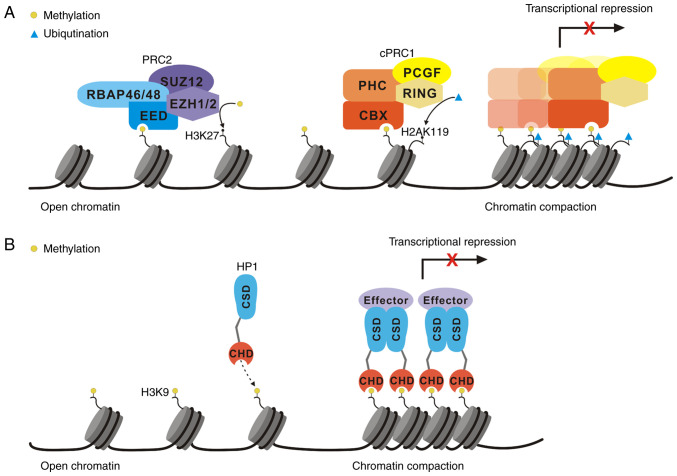
CBXs of the PcG family include CBX2, CBX4, CBX6, CBX7 and CBX8. In addition to the highly conserved CHD at their N-terminal region, all of these CBXs carry a conserved polycomb repressor box in their C-terminus. In addition, adjacent to the CHD, all vertebrate CBXs carry a DNA-binding motif and an AT-hook motif (in CBX2) or an AT-hook-like motif (in the other CBXs) (Fig. 1). AT-hook motifs are basic amino acid clusters that recognize AT-enriched sequences in DNA and are necessary for histone-independent DNA binding (26). The AT-hook motif in CBX2 may direct this CBX to chromatin, which indicates that CBX2 may bind DNA independent of histone H3 lysine K27 trimethylation (H3K27me3) (27). The AT-hooklike motif also binds DNA (28).
Figure 1.
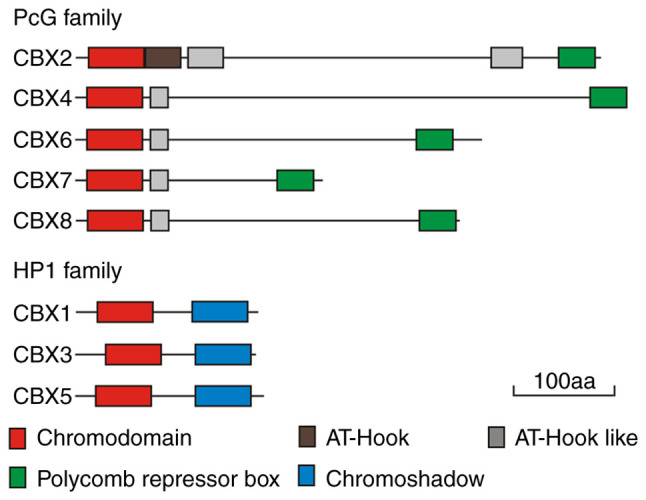
Conserved regions of CBX proteins. CBX, chromobox; aa, amino acid; PcG, polycomb group; HP1, heterochromatin protein 1.
PcG proteins mainly suppress the expression of target genes at the transcriptional level by forming multisubunit complexes called polycomb repression complexes (PRCs) and by modifying histones (29). PRCs comprise two main protein complexes: PRC1 and PRC2. In mammals, PRC1 may be further subclassified into two main complexes, namely, canonical PRC1 (cPRC1) and noncanonical PRC1 (ncPRC1). In contrast to ncPRC1, cPRC1, which carries unique CBXs and polyhomeotic-like protein (PHC), is thought to mainly mediate chromatin contraction (30). In addition to CBXs, the three other core proteins of cPRC1 are PHC, i.e. PHC1/2/3, really interesting new gene protein 1 (RING1), i.e. RING1A/1B, and polycomb group ring finger protein (PCGF), i.e. PCGF2/4 (31). cPRC1 is able to recognize the H3K27me3 mark through the CHD in CBXs (32-34) (Fig. 2A). In addition, the combination of the polycomb repressor box domain and RING1 protein binds CBXs to form cPRC1. Mammalian CBXs are able to recognize both the histone H3 lysine K9 trimethylation (H3K9me3) and H3K27me3 modifications, but the affinity is not identical (32). The chromatin domains of CBX2 and CBX7 have affinity for both H3K9me3 and H3K27me3, CBX4 has a stronger affinity for H3K9me3 and CBX6 has a weak affinity for both modifications (35). In addition, serine 42 in the CHD of CBX2 is the key residue for casein kinase (CK)2 phosphorylation. Unmodified CBX2 preferentially binds H3K9me3, but the phosphorylation of serine 42 induces a shift in the preference of CBX2 from H3K9me3 to H3K27me3 (36). The plasticity of this function contributes to the dynamic regulation of target genes and connects the extracellular environment with changes in chromatin availability. In contrast to cPRC1, ncPRC1 carries RING1 and YY1-binding protein (RYBP)/YY1-associated factor 2 (YAF2), not CBXs and PHC (37). The ncPRC1 complex carrying RYBP/YAF2 has a higher enzymatic activity against histone H2A lysine K119 (H2AK119) (38,39). All PRC1 complexes deposit a ubiquitin group at H2AK119 that is executed via its E3 ubiquitin ligase RING1A/1B, which forms a heterodimer with one of the six PCGFs (PCGF1-6) (38,40).
Figure 2.
Transcriptional repression by (A) PRCs and (B) HP1. PRC, polycomb repression complex; HP1, heterochromatin protein 1; CBX, chromobox; PHC, polyhomeotic-like protein; RING, really interesting new gene protein; PCGF, polycomb group ring finger protein; EZH1/2, enhancer of zeste homolog protein 1/2; EED, embryonic ectoderm development protein; SUZ12, suppressor of zeste protein 12; RBAP46/48, retinoblastoma binding-associated protein 46/48; H3K27, histone H3 lysine K27; CHD, chromodomain; CSD, chromoshadow domain.
Another major PRC is PRC2. PRC2 is also highly conserved among species and is composed of the enhancer of zeste homolog protein (EZH)1/2, embryonic ectoderm development protein (EED), suppressor of zeste 12 protein (SUZ12) and retinoblastoma binding-associated protein 46/48 (RBAP 46/48) (37). As the catalytic subunit of the complex, EZH1/2 catalyzes the H3K27me3, but it needs to be activated by other factors. EED and SUZ12 are essential for histone methyltransferase (HMT) activity. SUZ12 is critical for regulating HMT activity and EED regulates the substrate specificity of the EZH1/2 complex to mediate specific HMT activity against histone H3 lysine K27 or histone H1 lysine K26 (41,42) (Fig. 2A).
Although both PRC1 and PRC2 are involved in the posttranslational modifications (PTMs) of histones, the difference in their targets leads to different biological functions. PcG proteins recruit PRC1 mainly through H3K27me3 induced by PRC2, which leads to the monoubiquitination of H2AK119 (H2AK119ub1) and ultimately inhibits target gene transcription. Furthermore, H2AK119ub1 placed by PRC1 recruits PRC2 (39) (Fig. 2A). Therefore, once directed to chromatin, PcG amplifies its own activity. Further in-depth research revealed the presence of additional PRC recruitment mechanisms. First, the PRC complex may target DNA by interacting with noncoding RNA (ncRNA). For instance, CBXs in the PRC1 complex interact with X inactive specific transcript RNA to target inactive X chromosomes (43) or interact with the ncRNA antisense RNA (AS) in the INK4 locus, ANRIL, to target and inactivate INK4A sites (44). Evidence has indicated that PRC1 is recruited to CpG islands by lysine demethylase 2B and causes the ubiquitination of H2AK119, after which the recruitment of PRC2 leads to an increase in the abundance of H3K27me3, which binds PRC1 through interactions with CBXs (39). In summary, the recognition of H3K27me3 by CBXs is the main mechanism by which transcriptionally repressed polycomb complexes are recruited and proliferate. The recognition of H3K27me3 by CBXs is considered to be the key to PRC1 localization. In fact, genome-wide studies have indicated that PRC1 is clearly located in H3K27me3 marked domains (45). Traditionally, PcG complexes suppress target genes at the transcriptional level mainly through histone modification. However, in recent years, increasing evidence has suggested that PcG complexes are able to activate transcription and modify nonhistone substrates to participate in a variety of biological processes, such as the cell cycle and tumor development (46). One mechanism is recruitment through transcription factors (TFs). The cPRC1 complex colocalizes with runt-related transcription factor (RUNX)1 and core-binding factor subunit β through direct interaction with PCGF4 (47). Therefore, the active sites recruiting PRC1 may interact with TFs and remain bound during the transcriptional activation of target genes. Another mechanism is mediated through interactions with ncRNA and PTMs. The methylation state of CBX4 determines the specific ncRNA it binds, thereby determining whether coactivators or coinhibitors are recruited. Furthermore, the significance of the regulation of PRC1 subunits by PTMs has been demonstrated via CK2-mediated RING1B phosphorylation, which inhibits PRC1 activity, thereby promoting gene activation (48).
4. Characteristics and functions of HP1 family CBXs
In mammals, the HP1 family is composed of three different but highly conserved nonhistone homologs: CBX1/HP1β, CBX3/HP1γ and CBX5/HP1α (49,50). The CHDs of HP1 and PcG share >60% amino acid sequence identity (51). The CHD of HP1 is critical for the association of HP1 with chromatin through the specific CHD interaction with histone H3 lysine K9 dimethylation (H3K9me2)/H3K9me3, and the higher level of H3K9me2/3, the stronger binding affinity to the CHD (52,53). A second unique conserved domain called the chromoshadow domain (CSD) is located in the carboxyl terminal region of the HP1 protein (54) (Fig. 1). The overall structure of the CSD is similar to that of the CHD, but these domains exhibit different functions. The CSD acts mainly as a dimer domain and HP1 proteins easily form homodimers and heterodimers through their CSDs (52,55,56). The most notable function of HP1 is the formation heterochromatin, the compact form of chromatin. In the chromatin structure, the formation of heterochromatin results in transcriptional inactivation or silencing of genes. One model of heterochromatin formation is based on the methylation-induced binding of HP1, the histone H3 lysine K9 (H3K9) and cyclic recruitment. All three HP1 proteins recognize and bind to the H3K9me2 and H3K9me3 marks, which then recruit the H3K9 methyltransferase suppressor of variegation 3-9 homolog 1 (SUV39H1) to methylate adjacent H3K9 residues. This process creates new binding sites for additional HP1 proteins, forming a positive feedback loop that causes the formation of inhibitory H3K9me3 marks along specific chromosome regions (53). The diffusion of H3K9me3 marks is accompanied by the recruitment of numerous proteins, inducing chromatin contraction and gene silencing by embedding genes to make them impossible to transcribe (53,57-59) (Fig. 2B). This model also extends to DNA methylation, as both HP1 and SUV39H1 recruit DNA methyltransferases. Of note, in certain cases, histone H3K9 methylation precedes DNA methylation (60,61), supporting the view that these molecules are involved in a recruitment cycle during gene silencing.
5. Expression of CBXs in cancers and the relationship between CBXs and clinical characteristics and prognosis
Immunohistochemistry, western blot and quantitative PCR analyses of clinical tumor samples have confirmed abnormal changes in CBX expression in numerous cancer types, and these changes have been closely associated with the malignant phenotype of tumors and cancer prognosis. Furthermore, with the recent rapid development of bioinformatics, analyses based on public gene sequencing databases, such as The Cancer Genome Atlas (62), Gene Expression Omnibus (63), GEPIA (64), cBioPortal (65) and the Human Protein Atlas (66), have provided strong evidence that CBXs may be used as biomarkers for cancer prognosis. To date, CBXs have been proven to be differentially expressed in 17 cancers, are closely related to clinical characteristics and may be used as biomarkers for cancer prognosis (Table I).
Table I.
Correlations between the expression of CBXs, clinical characteristics and cancer prognosis.
| Cancer type | Expression
|
Clinical characteristics
|
Prognosis
|
Research methods | (Refs.) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ↑ | ↓ | Tumorsize ↑ | Grade ↑ | Stage↑ | Metastasis ↑ | Relapse ↑ | Others | OS ↓ | Others | |||
| Glioma | CBX2/3/5/8 | CBX6/7 | CBX2/3/8↑ | CBX2/3/8↑ | CBX3/8↑ CBX6↓ | BI | (67) | |||||
| Glioma | CBX2/3/5/8 | CBX6/7 | CBX2/6/7↑: IDH mutation↑ | CBX7↓ CBX8↑ | BI | (68) | ||||||
| Glioma | CBX3 | CBX3↑ | CBX3↑ | CBX3↑ | CBX3↑ | CBX3↑: RFS↓ | BI; WB; qPCR | (69) | ||||
| TSCC | CBX3 | CBX3↑ | CBX3↑ | CBX3↑ | IHC; WB; qPCR | (70) | ||||||
| HNSCC | CBX1/2/3/4/5/6/8 | CBX7 | CBX3↑ | CBX3↑ | CBX3↑ | CBX3↑: vascular invasion↑ | CBX3↑ CBX5↓ | BI; qPCR | (71) | |||
| BC | CBX2 | CBX2↑ | CBX2↑ | CBX2↑: DFS↓ | BI | (72) | ||||||
| BC | CBX2 | CBX2↑ | CBX2↑: PFS↓ | BI; IHC | (73) | |||||||
| BC | CBX2 | CBX2↑ | CBX2↑ | CBX2↑ | CBX2↑: HER-2 positive state↑ | CBX2↑ | IHC | (74) | ||||
| BC | CBX2 | CBX2↑ | BI | (75) | ||||||||
| BC | CBX4 | CBX4↑ | CBX4↑ | CBX4↑ | IHC; WB; qPCR | (76) | ||||||
| BC | CBX1/2/3/5/8 | CBX6/7 | CBX2↑ CBX4/6/7↓ | CBX1/7/8↑: PPS↑ CBX1/2/3/5↑ CBX6/7↓: DMFS↓ | BI; IHC | (77) | ||||||
| BC | CBX2/3/4/7/8 | CBX1/2/3↑ CBX4/5/6/7↓: RFS↓ | BI | (78) | ||||||||
| BC | CBX2 | CBX7 | CBX2↑ CBX7↓ | BI | (15) | |||||||
| BC | CBX8 | CBX8↑ | BI | (79) | ||||||||
| BC | CBX2/3/4/8 | CBX6/7 | CBX3↑ | CBX3/5↑ CBX4↓ | CBX3↑: DFS↓ | BI | (80) | |||||
| NSCLC | LUAD: CBX1/2/3/4/5/8↑ LUSC: CBX1/2/3/4/5↑ | CBX6/7 (LUAD); CBX7/8 (LUSC) | CBX3↑ | CBX1/3 (LUAD)↑ | CBX1/3 (LUAD)↑: DSS↓ DFS↓ PFS↓ | BI | (81) | |||||
| NSCLC | CBX3 | CBX3↑: EGFR mutation↑ | BI; IHC | (82) | ||||||||
| NSCLC | CBX7 | CBX7 (LUAD)↓ | BI | (83) | ||||||||
| LUAD | CBX1/2/3/5 | CBX7 | CBX3/5↑ CBX7↓ | CBX3/5↑ CBX7↓ | CBX1/3/5↑ CBX7/8↓ | BI | (84) | |||||
| LUAD | CBX7 | CBX7↓ | BI; IHC | (125) | ||||||||
| LUAD | CBX4 | CBX4↑ | BI; IHC | (85) | ||||||||
| EC | CBX1/2/3/4/5/8 | CBX1↑ | CBX3/4/8↑ CBX7↓ | CBX1↑: DFS↓ | BI | (86) | ||||||
| EC | CBX1/3/8 | CBX7 | CBX3/4↑ CBX7↓ | CBX1↑: PFS↓ CBX4/5↑: DFS↓ | BI | (87) | ||||||
| EC | CBX3/4/5/8 | CBX7 | CBX3/4/7/8↑ | BI | (88) | |||||||
| EC | CBX2 | CBX2 | ↑ | CBX2↑: DSS↓ | IHC; qPCR | (89) | ||||||
| EC | CBX8 | CBX8↑ | IHC; WB; qPCR | (90) | ||||||||
| GC | CBX3 | CBX3↑ | BI; IHC | (91) | ||||||||
| GC | CBX4 | CBX4↑ | BI | (92) | ||||||||
| GC | CBX1/2/3/4/5 | CBX7 | CBX3/8↑: OS↑ | BI | (93) | |||||||
| GC | CBX1/2/3/4/5/6 | CBX7 | CBX1/3/4/5/6↑ CBX7↓ | BI | (94) | |||||||
| GC | CBX/1/2/3/4/5/8 | CBX7 | CBX4/5/6/7/8↑ | CBX4/5/6/7/8↑: PFS↓ PPS↓ CBX3↑: PFS↑OS↑ | BI | (95) | ||||||
| GC | CBX/1/2/3/4/5/8 | CBX7 | CBX1/5/6/8↑ CBX7↓ | CBX1/5/6/8↑ CBX7↓: PFS↓ | BI | (96) | ||||||
| GC | CBX1/2/3/4/5/8 | CBX7 | CBX1/5/6/8↑ CBX7↓ | CBX1/5/6/8↑ CBX7↓: PFS↓ | BI | (97) | ||||||
| GC | CBX2/3 | CBX6/7 | CBX3↓ CBX4/5/6/7/8↑ | BI | (98) | |||||||
| HCC | CBX1/2/3/4/5/6/7/8 | CBX1/2/3/6/8↑ CBX7↓ | CBX1↑: DFS↓ | BI | (114) | |||||||
| HCC | CBX1 | CBX1↑ | CBX1↑ | CBX1↑: vascular invasion↑ | BI; IHC; WB; qPCR | (115) | ||||||
| HCC | CBX3 | CBX3↑ | CBX3↑ | BI; IHC; WB; qPCR | (116) | |||||||
| HCC | CBX4 | CBX4↑ | CBX4↑ | CBX4↑ | CBX4↑ | CBX4↑ | CBX4↑: RFS↓ | IHC | (11) | |||
| HCC | CBX6 | CBX6↑ | CBX6↑: RFS↓ | IHC; WB; qPCR | (117) | |||||||
| HCC | CBX6 | CBX6↑ | CBX6↑ | IHC | (118) | |||||||
| HCC | CBX7 | CBX7↓ | IHC | (119) | ||||||||
| HCC | CBX8 | CBX8↑ | IHC; WB; qPCR | (120) | ||||||||
| HCC | CBX8 | CBX8↑ | IHC; WB; qPCR | (121) | ||||||||
| HCC | CBX1/3/4/5/6/7/8 | CBX3↑ | CBX1/2/3/6/8↑ | BI | (122) | |||||||
| PAAD | CBX1/3/5/8 | CBX1/5/6/7↑ | CBX3↑ CBX2/6/7/8↓ | BI | (99) | |||||||
| PAAD | CBX7 | CBX7↓ | IHC | (109) | ||||||||
| PAAD | CBX8 | CBX8↑ | qPCR | (100) | ||||||||
| CRC | CBX3 | CBX3↑ | CBX3↑ | CBX3↑ | CBX3↑ | IHC; WB | (101) | |||||
| CRC | CBX8 | CBX8↑ | IHC | (102) | ||||||||
| CRC | CBX1/2/3/4/5/8 | CBX6/7 | CBX3↑ | CBX5/6↑ | CBX3↑: DFS↓ | BI | (103) | |||||
| CRC | CBX1/2/3/4/5/8 | CBX6/7 | CBX2↑ | CBX2↑: DFS↓ | BI | (104) | ||||||
| ccRCC | CBX3/4 | CBX1/5/6/7 | CBX3/4/8↑ CBX1/5/6/7↓ | BI | (110) | |||||||
| UBC | CBX8 | CBX8↑ | CBX8↑ | CBX8↑: RFS↓ | IHC; WB | (105) | ||||||
| OC | CBX1/2/3 | CBX1/3↑: chemoresistance↑ | CBX1/2/3↑ | CBX1/2/3↑: PFS↓ | BI | (123) | ||||||
| OC | CBX3/8 | CBX1/6/7 | CBX2/4/5/8↑ | CBX2/4/5/8↑: chemoresistance↑ | CBX1/2/3↑ | CBX1/2/3↑: PFS↓ | BI | (111) | ||||
| OC | CBX2 | CBX2↑ | BI | (124) | ||||||||
| CCA | CBX7 | CBX7↓ | CBX7↓ | CBX7↓ | CBX7↓: vascular invasion↑ | CBX7↓ | IHC | (112) | ||||
| Sarcoma | CBX1/3/4/5/6/8 | CBX1/3/5↑ CBX7↓ | CBX2↑ CBX6/7↓: RFS↓ | BI | (106) | |||||||
| Osteosarcoma | CBX3 | CBX3↑ | CBX3↑ | CBX3↑ | CBX3↑ | CBX3↑: DFS↓ | BI | (107) | ||||
| Osteosarcoma | CBX4 | CBX4↑ | qPCR | (108) | ||||||||
| SKCM | CBX2/3/5/6 | CBX7/8 | CBX5↑ CBX7↓ | BI | (113) | |||||||
CBX, chromobox; ↑, upregulation; ↓, downregulation; TSCC, tongue squamous cell carcinoma; HNSCC, head and neck squamous cell carcinoma; BC, breast cancer; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; EC, esophageal cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; CRC, colorectal cancer; ccRCC, clear cell renal cell carcinoma; UBC, urinary bladder cancer; OC, ovarian cancer; CCA, cervical carcinoma; SKCM, skin cutaneous melanoma; IDH, isocitrate dehydrogenase; HER-2, human epidermal growth factor receptor-2; EGFR, epidermal growth factor receptor; OS, overall survival; RFS, relapse-free survival; DFS, disease-free survival; PFS, progression-free survival; PPS, post-progression survival; DMFS, distant metastasis-free survival; DSS, disease-specific survival; BI, bioinformatics; WB, western blot; qPCR, quantitative PCR; IHC, immunohistochemistry.
Expression of CBXs in cancers
Compared with normal tissues or paracancerous tissues, the expression of all or some of the CBX1/2/3/4/5/8 members is upregulated in most cancer tissues, including glioma (67-69), tongue squamous cell carcinoma (TSCC) (70), head and neck squamous cell carcinoma (HNSCC) (71), breast cancer (BC) (15,72-80), non-small cell lung cancer (NSCLC) (81-85), esophageal cancer (EC) (86-90), gastric cancer (GC) (91-98), pancreatic adenocarcinoma (PAAD) (99,100), colorectal cancer (CRC) (101-104), urinary bladder cancer (UBC) (105), sarcoma (106) and osteosarcoma (107,108). As a tumor suppressor, CBX7 is expressed at low levels in most tumors, such as glioma, HNSCC (71), EC (87,88), GC (93-98), PAAD (109), CRC (103,104), clear cell renal cell carcinoma (ccRCC) (110), ovarian cancer (OC) (111), cervical carcinoma (CCA) (112) and skin cutaneous melanoma (SKCM) (113), while all CBX members, including CBX7 in hepatocellular carcinoma (HCC) (11,114-122), have been proven to be tumor-promoting factors. The expression of CBX6 is different in different cancer types, with low expression in glioma (67,68), BC (77,80), CRC (103,104), ccRCC (110) and OC (111), but high expression in HNSCC (71), sarcoma (106) and SKCM (113). The results of studies on the expression of CBX in the same type of cancer are not completely consistent. For instance, the results of Hu et al (111) indicated that the expression of CBX1 was low in OC, while Xu et al (123) reported that CBX1 was highly expressed in OC. The expression of CBX6 in GC was high in one study (94) and low in another (98). In addition, three studies have shown low expression of CBX7 in BC (15,77,80), while one study showed high expression of CBX7 in BC (78). This may be due to sample heterogeneity, different data sources or research methods; therefore, larger sample sizes, multiple analytic methods and multicenter research designs are required to obtain more credible results. The details of CBX expression are presented in Table I.
CBXs are related to the clinical characteristics of patients with cancer
As indicated in Table I, the expression of CBX3 is positively correlated with the tumor grade and/or stage of most cancer types, such as glioma and TSCC (70), lung adenocarcinoma (LUAD) (84), GC (91), HCC (116), CRC (101) and osteosarcoma (107), while the expression of CBX7 is negatively correlated with the tumor stage and grade of LUAD (84) and CCA (112). In addition, higher expression of CBX1/2/4/5/6/8 was associated with a higher tumor stage and grade in certain cancer types (11,67,69,74,76,80,84,86,90, 99,104,108,111,118). Furthermore, higher expression of CBX3 was associated with a larger size of glioma (69) and osteosarcoma (107). Similarly, the expression of CBX2 in BC (74) and CBX1/4 in HCC was proven to be positively correlated with the tumor size (11,115). High expression of CBX3 in TSCC (70), HNSCC (71), NSCLC (81), CRC (101) and osteosarcoma (107) was reported to be associated with more robust metastatic characteristics, indicating that CBX3 has an important role in tumor metastasis. High expression of CBX2/3/8 in glioma (67,69), CBX2 in EC (89) and CBX1 in HCC (115) was indicated to lead to a high probability of tumor recurrence. In terms of other cancer characteristics, CBX2/6/7 is closely related to the isocitrate dehydrogenase mutation in glioma (68), CBX2 is related to a human epidermal growth factor receptor (EGFR)2-positive status of BC (74) and CBX3 is related to EGFR mutation in NSCLC (82). CBX1, CBX3 and CBX7 are also associated with vascular invasion in HCC (115), HNSCC (71) and CCA (112), respectively. The expression of CBX1/2/3/4/5/8 in OC was reported to be related to an increase in chemoresistance (111,123).
CBXs are cancer prognostic biomarkers
The differential expression of CBX family members is closely related to the overall survival (OS), relapse-free survival, disease-free survival, progression-free survival, disease-specific survival, post-progression survival and distant metastasis-free survival of patients with cancer, and it has great potential as a prognostic marker of cancer (Table I). High expression of CBX1 is associated with shorter OS of patients with LUAD (84), GC (94), HCC (114), OC (123) and sarcoma (106). Furthermore, increased expression of CBX2 in BC (15,72-75,77), HCC (114,122) and OC (111,123,124) indicates poor prognosis. CBX3 is a poor prognostic factor in as many as 13 tumor types (67,69-71,80,81,84,86-88,101,106,110,111,114,116,122,123). CBX4 is of prognostic value in BC (76,80), LUAD (85), EC (86-88), GC (92,94,95,98), HCC (11) and ccRCC (110). In addition, patients with BC (80), LUAD (84), GC (94-98), CRC (103), sarcoma (106), SKCM (113) with upregulated CBX5 and HNSCC (71) and ccRCC (110) with downregulated CBX5 have poor prognosis. CBX6 may be either a poor prognostic factor or a favorable prognostic factor, depending on the type of cancer (67,77,94,96-99,103,110,117,122). CBX7 acts as a tumor suppressor in glioma (68), BC (15,77), LUAD (83,84,125), HCC (114,119), PAAD (99,109), ccRCC (110), CCA (112), sarcoma (106) and SKCM (113). When the expression of CBX7 is low, the OS of patients is shorter, but its relationship to survival in EC (86-88) and GC (94-98) is controversial. High expression of CBX8 in patients with glioma (67), BC (79), HCC (120,121), CRC (102), ccRCC (110) and UBC (105) lead to unfavorable prognosis, but patients with LUAD (84) with low CBX8 expression exhibit a shorter OS. In diffuse large B-cell lymphoma (DLBCL), CBX1/2/3/5/6/8 are expressed at high levels and CBX7 at low levels, but no significant correlation has been identified between CBX1-8 expression and prognosis, indicating that these CBXs may not be used as prognostic markers in patients with DLBCL (126).
Compared with their expression in normal tissues or paracancerous tissues, CBXs may be upregulated or downregulated in different types of cancers and differences in their expression levels are closely related to clinical characteristics, such as tumor size, clinical grade and stage, metastasis, relapse, vascular invasion, gene mutation, chemoresistance and prognosis. In general, CBX1/2/3/4/5/6/8 are tumor-promoting factors in most cancers, while CBX7 is a tumor-suppressing factor in almost all cancers. In addition to the abnormal expression of CBX genes, single nucleotide polymorphisms (SNPs) of CBX genes are closely related to cancer and may be prognostic biomarkers for cancer. The SNPs CBX4 rs2289728 and CBX7 rs139394 confer protection against HCC. These two SNPs inhibit the expression of CBX4 and CBX7, reducing the risk of HCC (127). The survival rate of patients with HCC with the homozygous CBX4 SNP AA (rs77447679-AA) is significantly decreased (128). Considerable evidence indicates that CBXs exhibit broad clinical application prospects as markers for cancer diagnosis and prognosis.
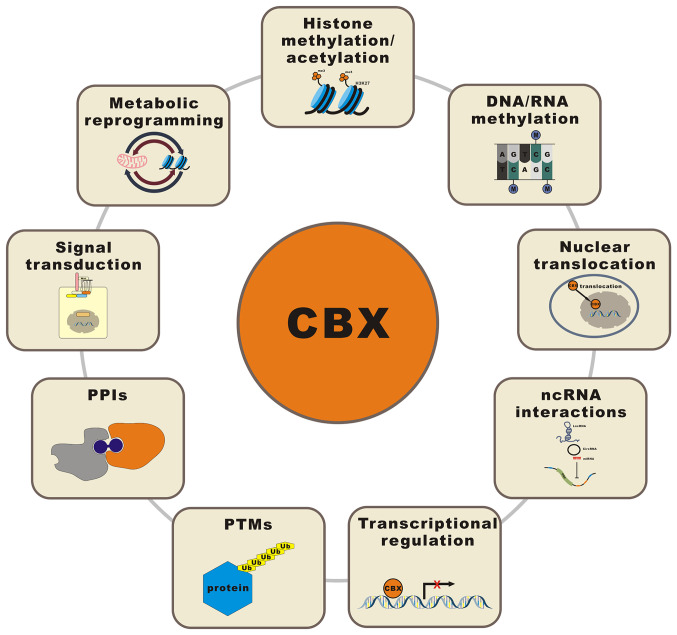
6. CBXs regulate biological tumor processes through epigenetic modification, nuclear translocation, ncRNA interactions, transcriptional regulation, PTMs, protein-protein interactions (PPIs), signal transduction and metabolic reprogramming
The mechanisms underlying the involvement of CBXs in regulating the occurrence and development of cancer are complex and multifaceted. As epigenetic regulators, CBXs classically regulate chromatin status and the expression of target genes via epigenetic modification, such as histone methylation/acetylation and DNA/RNA methylation. CBXs also promote/inhibit a variety of biological processes in tumors, including cell proliferation, migration, invasion and drug resistance; the cell cycle; and tumor cell stemness, through novel regulatory mechanisms, such as nuclear translocation, ncRNA interactions, transcriptional regulation, PTMs, PPIs, signal transduction and metabolic reprogramming. Fig. 3 illustrates the various regulatory mechanisms of CBXs in cancer.
Figure 3.
Schematic representation of various regulatory mechanisms of CBXs in cancer. CBX, chromobox; PPI, protein-protein interaction; Ub, ubiquitin; PTM, posttranslational modification; ncRNA, noncoding RNA; lncRNA, long noncoding RNA; miRNA, microRNA; circRNA, circular RNA; ORF, open reading frame; H3K27me3, histone H3 lysine K27 trimethylation.
Histone methylation/acetylation
As outlined in Table II, CBXs regulate malignant phenotype changes in tumors through histone modification. The depletion of CBX3 in prostate carcinoma (PCa) cells inhibits their proliferation, induces apoptosis and inhibits tumorigenicity. Mechanistically, c-Myc may upregulate CBX3 by directly binding the E-box element in the first intron of the CBX3 gene, and upregulated CBX3 in turn inhibits the expression of miR-451a by enhancing H3K9 methylation at the promoter region (129). CBX4 and histone H3 lysine K4 trimethylation (H3K4me3) coordinate and combine with the cell division cycle (CDC)20 promoter region to promote CDC20 expression and significantly enhance and maintain GC cell proliferation, migration and metastasis in vivo (92). Dicer is upregulated in cholangiocellular carcinoma (CCCA) and its nuclear form may interact with CBX5. The nuclear Dicer/CBX5 complex appears to promote H3K9me3 and DNA methylation of the secreted frizzled-related protein 1 (SFRP1) promoter and promote the proliferation and invasion of CCCA cells by inhibiting SFRP1 (130). CBX5-knockout in neuroendocrine prostate cancer (NEPC) cells inhibits proliferation and induces apoptotic death, resulting in tumor growth arrest. Mechanistically, CBX5 reduces the expression of androgen receptor (AR) and repressor element-1 (RE1)-mediated silencing of TF by enriching H3K9me3 on the respective gene promoter (131). CBX8 positively regulates Notch signal transduction to promote breast cell tumorigenesis by maintaining the level of H3K4me3 at the promoter of Notch network genes (79). Forced overexpression of CBX8 induced the epithelial-mesenchymal transition (EMT), invasive cell migration and stem cell-like characteristics, all of which are related to increased tumor growth and metastasis, in mice. Mechanistically, CBX8 regulates the H3K27me3 modification at the promoter of the bone morphogenetic protein (BMP)4 gene, which is related to active BMP4 transcription and therefore to the activation of mothers against decapentaplegic homolog (SMAD) and mitogen-activated protein kinase (MAPK) (120).
Table II.
CBXs regulating malignant phenotype changes in tumors through histone modification.
| Cancer type | Malignant phenotype changes | Regulation mechanism | (Refs.) |
|---|---|---|---|
| PCa | Proliferation; tumorigenesis; apoptosis | CBX3 inhibits the expression of miR-451a by increasing H3K9 methylation at the promoter regions | (129) |
| GC | Proliferation; migration; metastases; stemness | CBX4 coordinates with H3K4me3 to bind the CDC20 promoter region and thus promote CDC20 expression | (92) |
| CCCA | Proliferation; invasion | The Dicer/CBX5 complex inhibits SFRP1 by promoting the H3K9me3 modification and DNA methylation of the SFRP1 promoter | (130) |
| NEPC | Proliferation; cell cycle; apoptosis | CBX5 reduces the expression of AR and RE1-mediated silencing of TF by enriching H3K9me3 at the gene promoter | (131) |
| BC | Growth; stemness | CBX8 positively regulates Notch network genes by maintaining the level of H3K4me3 at the promoter | (79) |
| HCC | Growth; migration; invasion; stemness; EMT | CBX8 actives BMP4 transcription by regulating H3K27me3 at the promoter of BMP4 | (120) |
| CRC | Migration; invasion; metastases | CBX4 inhibits RUNX2 expression by recruiting HDAC3 to the RUNX2 promoter | (132) |
| ccRCC | Proliferation; growth; migration | CBX4 interacts with HDAC1 to inhibit the expression of KLF6 | (133) |
| Glioma | Proliferation; cell cycle; migration; invasion | CBX7 silences cyclin E1 by binding to its promoter and recruiting HDAC2 | (134) |
| TCA | Malignant progression | CBX7 upregulates the expression of E-cadherin by interacting with HDAC2 to increase the acetylation of histone H3 and H4 at the E-cadherin promoter | (135) |
| OC | Growth; migration | CBX8 and SET bind to the promoter of SUSD2 to establish H2AK119ub1 and block the acetylation of histone H3, resulting in transcriptional inhibition of SUSD2 | (136) |
CBX, chromobox; PCa, prostate carcinoma; GC, gastric cancer; CCCA, cholangiocellular carcinoma; NEPC, neuroendocrine prostate cancer; BC, breast cancer; HCC, hepatocellular carcinoma; CRC, colorectal cancer; ccRCC, clear cell renal cell carcinoma; TCA, thyroid carcinoma; OC, ovarian cancer; EMT, epithelial to mesenchymal transition; H3K9, histone H3 lysine K9; H3K4me3, histone H3 lysine K4 trimethylation; CDC20, cell division cycle 20; SFRP1, secreted frizzled-related protein 1; AR, androgen receptor; RE1, repressor element-1; TF, transcription factor; BMP4, bone morphogenetic protein 4; RUNX2, runt-related transcription factor 2; HDAC, histone deacetylase; KLF6, Kruppel-like factor 6; SET, SE translocation protein; SUSD2, sushi domain containing 2; H2AK119ub1, monoubiquitination of histone H2A lysine 119.
Evidence suggests that CBX4 has an inhibitory role in CRC; it inhibits CRC metastasis by HDAC3 to the RUNX2 promoter to inhibit RUNX2 expression (132). CBX4 promotes the proliferation and migration of ccRCC cells by interacting with HDAC1 to transcriptionally inhibit the expression of Kruppel-like factor 6 (133). By binding the cyclin E1 promoter and recruiting HDAC2, CBX7 silences cyclin E1 and causes glioma cell cycle arrest in the G0/G1 phase (134). CBX7 increases the acetylation of histone H3 and H4 at the E-cadherin promoter by interacting with HDAC2 and upregulates the expression of E-cadherin, explaining the correlation between the loss of CBX7 expression and the highly malignant phenotype of thyroid carcinoma (135). CBX8 is upregulated in OC. Overexpression and knockdown experiments have indicated that CBX8 promotes the growth and migration of OC cells in vitro. Mechanistically, CBX8 and SE translocation protein (SET) bind the promoter of sushi domain containing 2 (SUSD2) to establish H2AK119ub1 and block the acetylation of histone H3, resulting in the transcriptional inhibition of SUSD2 (136).
DNA/RNA methylation
CBX1 promotes the proliferation of hepatoma cells and CBX1 knockdown regulates the level of methionine adenosyltransferase 2A, leading to a decrease in the totals level of S-adenosylmethionine and methylated DNA and inhibiting the proliferation of hepatoma cells (137). HMT G9a cooperates with CBX5 and DNA methyltransferase (DNMT)1 to regulate epigenetic gene expression through H3K9me2 and DNA methylation, activates the Wnt/β-catenin signaling pathway and promotes the growth of NSCLC in vitro and in vivo (138). In HCC, Toll-like receptor 4 enhances the interaction between CBX5 and DNMT3B and inhibits the attachment and extension of RNA polymerase II in the promoter region of telomere repeat-containing RNA (TERRA) with telomere duplication, thereby inhibiting the transcription of TERRA (139). The downregulation of CBX5 in CCCA may reduce H3K9me3 enrichment and the DNA methylation rate of the SFRP1 promoter, thus restoring the expression of SFRP1 and inhibiting CCCA cell proliferation (140). When m6A methylation is increased, CBX8 interacts with lysine methyltransferase 2B and RNA polymerase II to promote leucine-rich repeat-containing G-protein coupled receptor 5 expression, which helps to increase the stemness of colon cancer (CC) and reduce the chemical sensitivity of CC (12). Details are provided in Table III.
Table III.
CBXs regulating malignant phenotype changes in tumors through DNA/RNA methylation.
| Cancer type | Malignant phenotype changes | Regulation mechanism | (Refs.) |
|---|---|---|---|
| HCC | Proliferation | CBX1 knockdown regulates the level of MAT2A, leading to a decrease in the total level of SAM and methylated DNA | (137) |
| NSCLC | Proliferation | HMT G9a cooperates with CBX5 and DNMT1 to regulate epigenetic gene expression through H3K9me2 and DNA methylation | (138) |
| HCC | Stemness | TLR4 enhances the interaction between CBX5 and DNMT3B, resulting in transcriptional inhibition of TERRA | (139) |
| CCCA | Proliferation | Downregulation of CBX5 reduces H3K9me3 enrichment and the DNA methylation rate of the SFRP1 promoter, thus restoring the expression of SFRP1 | (140) |
| CC | Stemness; chemosensitivity | When m6A methylation is increased, CBX8 interacts with KMT2B and RNA polymerase II to promote LGR5 expression | (12) |
CBX, chromobox; HCC, hepatocellular carcinoma; NSCLC, non-small cell lung cancer; CCCA, cholangiocellular carcinoma; CC, colon cancer; MAT2A, methionine adenosyltransferase 2A; SAM, S-adenosylmethionine; HMT, histone methyltransferase; DNMT, DNA methyltransferase; H3K9me2, histone H3 lysine K9 dimethylation; TERRA, telomere repeat-containing RNA; SFRP1, secreted frizzled-related protein 1; KMT2B, lysine methyltransferase 2B; LGR5, leucine-rich repeat-containing G-protein coupled receptor 5.
Nuclear translocation
Yi et al (141) suggested that the subcellular localization of CBX3, not its expression, is closely related to the progression of CCA. Details are presented in Table IV. The nuclear output of high-risk human papillomavirus-mediated CBX3 reduces the stability of p53 in the progression of CCA through ubiquitin-conjugating enzyme E2 L3 (UBE2L3)-mediated polyubiquitination of p53. Exosomal circ_0006790 derived from bone marrow mesenchymal stem cells promotes the nuclear translocation of CBX7 and recruits DNA methyltransferase to its promoter region to increase the DNA methylation of S100A11; thus, inhibiting S100A11 transcription may downregulate S100A11 in pancreatic ductal adenocarcinoma (PDAC) cells and inhibit PDAC growth, metastasis and immune escape (142). In osteosarcoma, metabolic glutamate receptor 4 may interact with CBX4 to limit its nuclear localization and affect the transcriptional activity of hypoxia-inducible factor (HIF)-1α, which affects cell proliferation, migration and invasion (143).
Table IV.
CBXs regulating malignant phenotype changes in tumors through nuclear translocation.
| Cancer type | Malignant phenotype changes | Regulation mechanism | (Refs.) |
|---|---|---|---|
| CCA | Proliferation | The nuclear output of CBX3 reduces the stability of p53 through UBE2L3-mediated polyubiquitination of p53 | (141) |
| PAAD | Growth; migration; invasion; immune escape | Exosomal circ_0006790 promotes the nuclear translocation of CBX7 and recruits DNA methyltransferase to its promoter region to increase the DNA methylation of S100A11 | (142) |
| Osteosarcoma | Proliferation; migration; invasion | GRM4 may interact with CBX4 to limit its nuclear localization and affect the transcriptional activity of HIF-1α | (143) |
CBX, chromobox; CCA, cervical carcinoma; PAAD, pancreatic adenocarcinoma; UBE2L3, ubiquitin-conjugating enzyme E2 L3; GRM4, metabolic glutamate receptor 4; HIF-1α, hypoxia-inducible factor-1α.
ncRNA interactions
CBXs may interact with ncRNA [long ncRNA (lncRNA), microRNA (miRNA) or circular RNA (circRNA)] to regulate target genes or to be regulated as target genes, participating in the occurrence and development of tumors. Details are presented in Table V.
Table V.
CBXs regulating malignant phenotype changes in tumors through ncRNA interactions.
| Cancer type | Malignant phenotype changes | ncRNA interactions | (Refs.) |
|---|---|---|---|
| PAAD | Proliferation; migration; invasion | lncRNA PCAT6/miR-185-5p/CBX2 | (144) |
| UBC | Proliferation | lncRNA CASC9/miR-497-5p/CBX2 | (145) |
| NEPC | Proliferation; metastases | LINC00261/miR-8485/CBX2/FOXA2 | (146) |
| OC | Proliferation; apoptosis; migration; invasion | miR-342-5p/CBX2/Wnt/β-catenin | (147) |
| Osteosarcoma | Proliferation; growth; invasion | miRNA let-7a/CBX2 | (148) |
| OC | Proliferation; apoptosis; migration; invasion; chemosensitivity | circ_0061140/miR-136/CBX2 | (149) |
| Glioma | Proliferation; migration; invasion; self-renewal | lncRNA RP11-279C4.1/miR-1273g-3p/CBX3 | (150) |
| HCC | Proliferation; apoptosis; migration; invasion | lncRNA KCNQ1OT1/miR-29a-3p/CBX3 | (151) |
| HCC | Proliferation; migration; invasion | LINC01006/miR-433-3/CBX3 | (152) |
| COAD | Proliferation; migration; invasion | lncRNA SNHG17/miR-375/CBX3 | (153) |
| DLBCL | Proliferation; apoptosis | LINC00857/miR-370-3p/CBX3 | (154) |
| Glioma | Proliferation; cell cycle | LINC00998/CBX3/c-Met/AKT/mTOR | (155) |
| HCC | Growth; migration; invasion | miR-139/CBX3 | (156) |
| CRC | Proliferation; growth | miR-30a/CBX3/CDKN1A | (157) |
| Glioma | Growth; apoptosis; migration; invasion | circ_EZH2/miR-1265/CBX3 | (158) |
| NSCLC | Proliferation | lncRNA SNHG5/miR-181c-5p/CBX4 | (159) |
| GC | Proliferation | LINC00265/miR-144-3p/CBX4 | (160) |
| CCA | Proliferation; migration; invasion | lncRNA FOXP4-AS1/miR-136-5p/CBX4 | (161) |
| PCa | Proliferation; migration; invasion | lncRNA RAMS11/CBX4/TOP2α | (162) |
| BC | Proliferation | miR-129-5p/CBX4 | (163) |
| BC | Proliferation; migration; invasion | miR-515-5p/CBX4 | (164) |
| GC | Proliferation; apoptosis; invasion | miR-507/CBX4/Wnt/β-catenin/HIF-1α | (165) |
| HCC | Proliferation; metastases; angiogenesis; self-renewal | miR-6838-5p/CBX4/ERK | (166) |
| CCA | Proliferation; cell cycle | miR-497-5p/CBX4 | (167) |
| ACP | Osteoblast differentiation and calcium deposition | BMP2/miR-181b/CBX4/HDAC3/RUNX2 | (168) |
| LC | Proliferation; migration; invasion | circ_PVT1/miR-21-5p/CBX4/Wnt4/β-catenin | (170) |
| BC | Proliferation; migration | CBX4/miR-137/Notch1 | (76) |
| HCC | Proliferation; stemness | miR-424/CBX4/YAP1 | (169) |
| BC | Proliferation; migration; invasion | circ_0008039/miR-515-5p/CBX4 | (171) |
| Glioma | Proliferation; apoptosis | LINC02381/CEBPβ/CBX5 | (172) |
| BC | Proliferation; migration | lncRNA SNHG11/miR-2355-5p/CBX5 | (173) |
| HCC | Proliferation | miR-675/CBX5/EGR1/H19/PKM2 | (174) |
| HCC | Stemness | miR-675/PKM2/CBX5/c-Myc | (175) |
| RCC | Proliferation; migration | lncRNA LOXL1-AS1/miR-589-5p/CBX5 | (176) |
| RCC | Proliferation; migration; invasion | circ_0037866/miR-384/CBX5 | (177) |
| HCC | Proliferation; migration; invasion | lncRNA MIR100HG/miR-146b-5p/CBX6 | (178) |
| LUAD | Proliferation; migration | lnc RNA SNHG7/miR-181/CBX7/Wnt/β-catenin | (179) |
| NSCLC | Proliferation; migration; invasion | miRNA-19/CBX7 | (180) |
| UBC | Invasion | miR-9/CBX7 | (181) |
| PCa | Proliferation; migration; invasion | miR-375/CBX7 | (182) |
| OC | Proliferation; migration; invasion; EMT | miR-18a/CBX7/ERK/MAPK | (183) |
| BC | Proliferation; cell cycle | HMGA1/CBX7/miR-181b | (184) |
| PCa | Proliferation; cell cycle; apoptosis | circ_GOLPH3/CBX7 | (185) |
| HCC | Growth; migration | CBX8/EGR1/miR-365-3p/AKT/β-catenin | (121) |
| CC | Proliferation; apoptosis; migration; invasion | CBX8/miR-378a-3p/PDIA4/caspase 3/caspase 7 | (186) |
| DLBCL | Proliferation; apoptosis | miR-429/CBX8 | (187) |
| BC | Proliferation; migration; invasion | circ_0005230/miR-618/CBX8 | (188) |
| CCA | Proliferation; migration; invasion | circ_8924/miR-518d-5p/519-5p/CBX8 | (189) |
CBX, chromobox; PAAD, pancreatic adenocarcinoma; UBC, urinary bladder cancer; NEPC, neuroendocrine prostate cancer; OC, ovarian cancer; HCC, hepatocellular carcinoma; COAD, colon adenocarcinoma; DLBCL, diffuse large B-cell lymphoma; CRC, colorectal cancer; NSCLC, non-small cell lung cancer; GC, gastric cancer; CCA, cervical carcinoma; PCa, prostate carcinoma; BC, breast cancer; ACP, adamantinomatous craniopharyngioma; LC, laryngocarcinoma; RCC, renal cell carcinoma; LUAD, lung adenocarcinoma; CC, colon cancer; EMT, epithelial to mesenchymal transition; ncRNA, noncoding RNA; lncRNA, long noncoding RNA; miR, microRNA; circ, circular RNA; PCAT6, prostate cancer associated transcript 6; CASC9, cancer susceptibility 9; FOXA2, forkhead box A2; KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; SNHG, small nucleolar RNA host gene; CDKN1A, cyclin dependent kinase inhibitor 1A; EZH2, enhancer of zeste homolog 2; FOXP4-AS1, forkhead box P4 antisense RNA 1; RAMS11, RNA associated with metastasis-11; TOP2α, topoisomerase IIα; HIF-1α, hypoxia-inducible factor-1α; ERK, extracellular signal-regulated kinase; BMP2, bone morphogenetic protein 2; HDAC, histone deacetylase; RUNX2, runt-related transcription factor 2; YAP1, Yes-associated protein 1; CEBPβ, CCAAT/enhancer-binding protein β; EGR1, early growth response 1; PKM2, pyruvate kinase M2; LOXL1-AS1, lysyl oxidase like 1 antisense RNA 1; MAPK, mitogen-activated protein kinase; HMGA1, high mobility group AT-hook 1; GOLPH3, Golgi phosphoprotein 3; PDIA4, protein disulfide-isomerase A4.
The expression of CBX2 is positively regulated by the lncRNA prostate cancer associated transcript 6 (PCAT6) sponging of miR-185-5p in PDAC (144), lncRNA cancer susceptibility 9 (CASC9) sponging of miR-497-5p in UBC (145) and LINC00261 sponging of miR-8485 in NEPC (146), which increases the acquisition of the respective malignant cancer phenotype. Targeting CBX2 with miR-342-5p mediates the inhibition of the Wnt/β-catenin signaling pathway, which significantly reduces the proliferation, invasion, migration and viability of OC cells and promotes their apoptosis (147). The let-7a/CBX2 axis has an important role in the progression of osteosarcoma (148). Circ_0061140 is able to mediate the proliferation, migration, invasion and paclitaxel sensitivity of OC cells by regulating the miR-136/CBX2 axis in vivo (149).
CBX3, a target gene, is regulated by a competing endogenous RNA axis, which includes the lncRNA RP11-279C4.1/miR-1273g-3p/CBX3 axis in glioma (150), the lncRNA KCNQ1 opposite strand/antisense transcript 1/miR-29a-3p/CBX3 (151) and LINC01006/miR-433-3/CBX3 axis (152) in HCC, the lncRNA small nucleolar RNA host gene (SNHG) 17/miR-375/CBX3 axis in colon adenocarcinoma cells (153) and the LINC00857/miR-370-3p/CBX3 axis in DLBCL (154). LINC00998 may stabilize CBX3 to promote H3K9me3 in the c-Met promoter region and further weaken the activation of the c-Met/AKT/mammalian target of rapamycin (mTOR) signaling pathway, which inhibits the proliferation of glioma in vitro and in vivo (155). CBX3 regulated by miR-139 (156) and miR-30a (157) promotes HCC growth, migration and invasion by regulating cell cycle progression and CRC growth, respectively. Overexpression of circ_EZH2 significantly promotes the growth, migration and invasion of glioma cells and inhibits their apoptosis. The carcinogenic function of CBX3 depends on its inhibition of dimethylarginine dimethylaminohydrolase 1 and sponging of miR-1265 (158).
The lncRNA SNHG5/miR-181c-5p axis in NSCLC (159), the LINC00265/miR-144-3p axis in GC (160) and the lncRNA forkhead box P4 AS1/miR-136-5p axis in CCA (161) upregulate the expression of CBX4 and promote cancer progression. The lncRNA RNA associated with metastasis-11 (RAMS11) promotes the growth and metastasis of PCa cells by binding CBX4 and activating the expression of topoisomerase IIα (TOP2α) (162). miR-129-5p (163) and miR-515-5p (164) in BC, miR-507 in GC (165), miR-6838-5p in HCC (166) and miR-497-5p in CCA (167) target CBX4 to regulate the biological characteristics of human cancers. BMP2 increases miR-181b levels to directly target and inhibit CBX4 expression in adamantinomatous craniopharyngioma (ACP), resulting in reduced regulation of CBX4-dependent HDAC3 nuclear translocation, RUNX2 activation/osteoblast differentiation and calcium deposition in ACP (168). CBX4 is upregulated in BC and shows carcinogenic activity mediated through the activation of the miR-137-mediated Notch1 signaling pathway (76). MiR-424 inhibits the nuclear translocation of the Yes-associated protein (YAP)1 that has been induced by CBX4, and CBX4 and inhibits the proliferation and stem cell-like characteristics of HCC cells (169). Circ_PVT1 (170) and circ_0008039 (171) also enhance the expression of CBX4 separately through competitive binding to miR-21-5p and miR-515-5p, respectively, thereby promoting the progression of laryngocarcinoma and BC.
LINC02381 may interact and cooperate with CCAAT/enhancer-binding protein β to bind the CBX5 promoter and transcriptionally activate CBX5 to promote glioma cell proliferation and apoptosis (172). The lncRNA SNHG11/miR-2355-5p/CBX5 axis regulates the proliferation and migration of triple-negative BC cells (173). Overexpression of miR-675 promotes the growth of hepatoma cells in vitro and in vivo. Mechanistically, miR-675 inhibits the expression of CBX5 in human hepatoma cells, leading to a decrease in H3K9me3 and H3K27me3 abundance and triggering the transcription, translation, small ubiquitin-like modifier (SUMOylation) and activation of early growth response 1 (EGR1), which upregulates the lncRNA H19 and induces and activates tumor-specific pyruvate kinase M2 (PKM2) (174). MiR-675 in conjunction with PKM2 triggers the upregulation of c-Myc by increasing the interaction between H3K9me3 and CBX5, which contributes to the malignant progression of liver cancer stem cells (175). Overexpression of CBX5 or inhibition of miR-589-5p in renal cell carcinoma (RCC) reverses the inhibitory effect of silencing lysyl oxidase like 1-AS1 on the proliferation and migration of RCC cells (176). Circ_0037866 may sponge miR-384 to increase the expression of its target, CBX5, thereby promoting the survival, invasion and migration of RCC cells in vitro and in vivo (177).
The expression of the lncRNA miR-100HG and CBX6 was enhanced in HCC cells. Knocking out miR-100HG inhibited the viability, migration and invasion of HCC cells by targeting the miR-146b-5p/CBX6 axis (178).
The lncRNA SNHG7 interacts with miR-181, upregulates CBX7 and inhibits the proliferation and migration of LUAD cells in vitro and in vivo via the Wnt/β-catenin pathway (179). CBX7 has been confirmed to be a functional target of miRNA-19 in NSCLC (180), miR-9 in UBC (181), miR-375 in PCa (182) and miR-18a in OC (183). CBX7, which is negatively regulated by high mobility group AT-hook (HMGA)1, negatively regulates the expression of miR-181b, which leads to BC progression (184). CircRNA Golgi phosphoprotein 3 and its binding protein CBX7 may promote the proliferation of PCa cells and inhibit their apoptosis (185).
CBX8, an oncogene, upregulates EGR1 and miR-365-3p to stimulate the AKT/β-catenin pathway, which promotes the growth and metastasis of HCC (121). CBX8 may be an independent RNA-binding protein (RBP) that regulates the maturation of miRNAs. CBX8 may inhibit the nuclear output of premiR-378a depending on its own nuclear localization and interaction with premiR-378a, thus inhibiting the maturation of miR-378a. MiR-378a-3p inhibits the malignant expression of human CC cells by targeting protein disulfide-isomerase A4, resulting in an increase in caspase-3 and caspase-7 activity (186). MiR-429 targets CBX8 to promote apoptosis in DLBCL (187). The increased expression of circ_0005230 in BC (188) and circ_8924 in CCA (189) promotes CBX8 expression by sponging miR-618 and miR-518d-5p/519-5p, respectively, to regulate cell proliferation, migration and invasion and is associated with poor prognosis.
Transcriptional regulation
All CBXs may upregulate or downregulate the expression of oncogenes or tumor suppressor genes at the transcriptional level (Table VI).
Table VI.
CBXs influencing malignant phenotype changes in tumors through regulation at the transcrip-tional level.
| Cancer type | Malignant phenotype changes | Transcriptionally regulated gene | (Refs.) |
|---|---|---|---|
| CC | Migration | CBX1-MMP2 | (190) |
| PCa | Proliferation; apoptosis | CBX2-AURKA/AURKB/cyclin B1/MKI67/CDK1/CDC25A | (191) |
| LUAD | Proliferation; migration | CBX3-NCOR2/ZBTB7A | (192) |
| CC | Proliferation; cell cycle | CBX3-CDK6/p21 | (193) |
| PAAD | Proliferation; growth; migration; invasion | CBX3-CDK1/PCNA | (194) |
| PAAD | Proliferation; growth; migration; invasion | CBX3-SMURF2 | (195) |
| BC | Migration; invasion | CBX4-cadherin 1 | (196) |
| NSCLC | Proliferation; cell cycle; migration | CBX4-CDK2/cyclin E/PCGF4 | (197) |
| HCC | Proliferation; cell cycle | CBX4-cyclin E2/p16 | (11) |
| Osteosarcoma | Migration; invasion | CK1α-CBX4-GCN5/RUNX2 | (198) |
| BC | Migration; invasion | E2F-CBX5 | (199) |
| AML | Proliferation; apoptosis | RBMXL1-CBX5 | (200) |
| BC | Proliferation; migration; invasion | EZH2-CBX6 | (201) |
| PM | Migration; invasion | CBX6-MMP2 | (202) |
| HCC | Proliferation; migration; invasion | CBX6-Snail/ZEB1 | (118) |
| Glioma | Proliferation; cell cycle | miR-18a-CBX7-CDK2/cyclinA2 | (203) |
| Glioma | Proliferation; migration; invasion | CBX7-DKK1/ZEB1 | (204) |
| blBC | Growth; migration; invasion; metastases | CBX7-TWIST1/EPHA2 | (205) |
| PCa | Proliferation | CBX7-p16INK4A/Rb; CBX7-p14ARF/p53 | (206) |
| UBC | Proliferation; migration; invasion | CBX7-PDE4B | (207) |
| UBC | Proliferation; migration; invasion; stemness | CBX7-AKR1B10 | (14) |
| Glioma, BC, NSCLC | Migration; invasion | CBX8-WNK2/MMP2/RAC1 | (208) |
| ESCC | Proliferation; migration; invasion; metastases | CBX8-Snail | (209) |
| CC | Proliferation | IGF1-CBX8 | (210) |
| CRC | Proliferation; migration; invasion; metastases | CBX8-p53/ITGB4 | (102) |
| Leukemia | Tumorigenesis | CBX8-HOX | (211) |
CBX, chromobox; CC, colon cancer; PCa, prostate carcinoma; LUAD, lung adenocarcinoma; PAAD, pancreatic adenocarcinoma; BC, breast cancer; NSCLC, non-small cell lung cancer; HCC, hepatocellular carcinoma; AML, acute myeloid leukemia; PM, pleural mesothelioma; blBC, basal-like BC; UBC, urinary bladder cancer; ESCC, esophageal squamous cell carcinoma; CRC, colorectal cancer; MMP2, matrix metallopeptidase 2; AURK, aurora kinase; MKI67, marker of proliferation Ki-67; CDK, cyclin dependent kinase; CDC25A, cell division cycle 25A; NCOR2, nuclear receptor corepressor 2; ZBTB7A, zinc finger and BTB domain containing 7A; PCNA, proliferating cell nuclear antigen; SMURF2, SMAD specific E3 ubiquitin protein ligase 2; PCGF4, polycomb group ring finger protein 4; GCN5, general control non-derepressible 5; RUNX2, runt-related transcription factor 2; RBMXL1, RNA binding motif protein X-linked like 1; EZH2, enhancer of zeste homolog 2; ZEB1, zinc finger E-box binding homeobox 1; DKK1, Dickkopf-related protein 1; TWIST1, twist family bHLH transcription factor 1; EPHA2, EPH receptor A2; PDE4B, phosphodiesterase 4B; AKR1B10, aldo-keto reductase family 1 member B10; WNK2, with-no-lysine kinase 2; RAC1, Rac family small GTPase 1; IGF1, insulin like growth factor 1; ITGB4, integrin subunit β4; HOX, homeobox.
In CC, CBX1 inhibits the expression of matrix metallopeptidase (MMP)2 at the transcriptional level and regulates CC cell metastasis (190).
CBX2 depletion decreases cell viability and induces apoptosis in metastatic PCa cell lines. Mechanistically, numerous key regulatory factors, such as aurora kinase (AURK)A, AURKB, cyclin B1, marker of proliferation Ki-67 (MKI67), cyclin dependent kinase (CDK) 1 and CDC25A, are downregulated by CBX2 to control cell proliferation and metastasis (191).
CBX3 promotes cell proliferation by directly suppressing the expression of nuclear receptor corepressor 2 (NCOR2) and zinc finger and BTB domain containing 7A in LUAC (192) and CDK6/p21 in CC (193). CBX3 also mediates tumor promotion by regulating the expression of CDK1 and proliferating cell nuclear antigen (PCNA) in PAAD cells (194), inhibiting the expression of SMAD-specific E3 ubiquitin protein ligase 2 and promoting the activation of the TGF-β signaling pathway (195).
CBX4 regulates telomerase reverse transcriptase-mediated cadherin 1 transcription and promotes the migration and invasion of BC cells (196). In lung cancer, CBX4 knockdown effectively blocks the cell cycle in the G0/G1 phase by inhibiting the expression of CDK2 and cyclin E and reduces the formation of filamentous pseudopodia by inhibiting MMP2, MMP9 and C-X-C motif chemokine receptor 4 (CXCR4). In addition, CBX4 promotes cell proliferation and metastasis by regulating PCGF4 expression (197). Knocking down CBX4 results in the downregulation of PCNA and cyclin E2 and the upregulation of p16, followed by decreased cell proliferation and blocked cell cycle progression (11). CBX4 promotes osteosarcoma metastasis by recruiting general control non-derepressible 5 to the RUNX2 promoter to upregulate RUNX2 at the transcriptional level, and CK1α inhibits osteosarcoma cell migration and invasion by inhibiting CBX4 (198).
CBX5 inhibits BC cell migration and invasion. The E2F transcription factor 5 (E2F5) regulates CBX5 transcription, and E2F5 consumption increases the expression of CBX5 in invasive BC cells (199). The RNA binding motif protein X-linked (RBMX) reverse transcriptional gene product RBMX like 1 (RBMXL1) is an RBP that directly binds mRNA and affects the transcription of the CBX5 locus in acute myeloid leukemia. RBMX/L1 controls leukemic cell survival by regulating chromatin status through its downstream target CBX5 (200).
The expression of CBX6 is negatively regulated by EZH2, which may inhibit cell proliferation and induce G0/G1 cell cycle arrest in BC cells (201). Knocking out CBX6 promotes MMP2 expression and tumor invasion in pleural mesothelioma (202). CBX6 upregulates the expression of Snail and zinc finger E-box binding homeobox 1 (ZEB1) promotes the proliferation, migration and invasion of HCC cells (118).
In glioma, overexpression of exogenous CBX7 induces apoptosis and inhibits cell proliferation, migration and invasion, as it reduces the expression of CDK2 and cyclin A2 (203) and the core EMT factor ZEB1 (204). CBX7 blocks the binding of twist family bHLH transcription factor 1 (TWIST1) to the EPH receptor A2 (EPHA2) promoter, inhibits the expression of EPHA2, and inhibits the growth and metastasis of basal-like BC (205). CBX7 inhibits the expression of p16INK4A and p14ARF in PCa cells and affects their growth (206). CBX7 acts as a tumor suppressor to downregulate the expression of the oncogenes phosphodiesterase 4B (207) and aldo-keto reductase family 1 member B10 (14), promoting the proliferation, migration and invasion of UBC cells at the transcriptional level.
CBX8 is overexpressed in numerous cancers and has been indicated to promote the invasion and migration of glioma, BC and lung cancer in vitro and in vivo. Mechanistically, CBX8 promotes cell invasion and migration by targeting with-no-lysine kinase 2, resulting in increased expression and activity of Rac family small GTPase 1 (RAC1) and MMP2 (208). CBX8 may have contradictory roles in esophageal squamous cell carcinoma (ESCC), promoting cell proliferation and inhibiting metastasis, and this newly reported function of CBX8 depends on its binding to the Snail promoter, thereby inhibiting the transcription of Snail (209). Insulin-like growth factor 1 promotes the proliferation of CC cells by promoting CBX8 expression (210). Knocking down CBX8 inhibits the proliferation of CRC cells in vitro and in vivo, mainly by increasing p53 and its downstream effectors. However, the knockdown of CBX8 enhances the migration, invasion and metastasis of CRC cells in vitro and in vivo, partially by directly upregulating integrin subunit β4, thereby reducing the activity of ras homolog (Rho)A (102). CBX8 is necessary for mixed lineage leukemia (MLL)-AF9-induced transcriptional activation and leukemia development. By contrast, the elimination of CBX8 by a point mutation in MLL-AF9 and the specific elimination of the MLL-AF9-CBX8 interaction abrogates both the upregulation of the homeobox gene and the transformation of MLL-AF9-positive leukemia (211).
PTMs
Protein PTMs, such as phosphorylation, acetylation, SUMOylation and ubiquitination, reveal the great complexity of the proteome. PTMs have important roles in signal transduction, protein stability and conversion, protein-to-protein recognition and interaction, and spatial localization by changing the structure and function of the protein. CBXs activate or inhibit cancer-related signaling pathways through the PTM of key proteins in a pathway (Table VII).
Table VII.
CBXs regulating malignant phenotype changes in tumors through posttranslational modifications.
| Cancer type | Malignant phenotype changes | Regulation mechanism | (Refs.) |
|---|---|---|---|
| HCC | Proliferation; apoptosis | CBX2 knockdown inhibits the expression of WTIP, stimulates the Hippo pathway, and leads to the phosphorylation-induced inactivation of YAP | (212) |
| Glioma | Proliferation; tumorigenesis | CBX3 directly suppresses PARK2 and STUB1 at the transcriptional level to reduce the ubiquitination of EGFR | (213) |
| HCC | Angiogenesis | CBX4 promotes HCC via HIF-1α ubiquitination and VEGF upregulation | (214) |
| BC | EMT | Increased SENP7L decreases the SUMOylation of CBX5 | (215) |
| BC | Chemosensitivity | Ubiquitinated CBX5 is recruited to ncRNA-rich chromatin loci to promote DNA damage and is associated with chemosensitivity in BC mediated via SUMOylated CBX5/ncRNA | (216) |
| EC | Proliferation; radiosensitivity | The inhibition of CBX8 increases the phosphorylation of p21, Wee1 and choline kinase 1 | (90) |
CBX, chromobox; HCC, hepatocellular carcinoma; BC, breast cancer; EC, esophageal cancer; EMT, epithelial to mesenchymal transition; WTIP, Wilms' tumor protein 1-interacting protein; YAP, Yes-associated protein; PARK2, Parkinson disease 2; STUB1, stress induced phosphoprotein 1 homology and U-box containing protein 1; EGFR, epidermal growth factor receptor; HIF-1α, hypoxia-inducible factor-1α; VEGF, vascular endothelial growth factor; SENP7L, SUMO specific peptidase 7 long transcript; SUMOylation, small ubiquitin-like modifier.
CBX2 knockdown in HCC inhibits the proliferation of HCC cells and promotes their apoptosis. The following mechanisms underlie these effects: CBX2 knockdown inhibits the expression of Wilms' tumor protein 1-interacting protein, stimulates the Hippo pathway and leads to the phosphorylation-induced inactivation of YAP (212). CBX3 directly suppresses Parkinson disease 2 and stress-induced phosphoprotein 1 homology and U-box containing protein 1 at the transcriptional level to reduce the ubiquitination of EGFR, significantly promoting the proliferation, invasion and tumorigenesis of glioblastoma multiforme cells in vitro and in vivo (213). CBX4 promotes HCC via HIF-1α ubiquitnation and vascular endothelial growth factor upregulation (214). In BC, SUMO specific peptidase 7 long transcript (SENP7L) has enhanced abundance. Increased SENP7L decreases the SUMOylation rate of CBX5 and promotes abnormal proliferation and the EMT (215). Ubiquitinated CBX5 is recruited to ncRNA-rich chromatin loci to promote DNA damage and is associated with chemosensitivity in BC (216). The inhibition of CBX8 decreases cell proliferation in vitro and in vivo and increases the phosphorylation of p21, Wee1 and choline kinase 1, resulting in CDK inhibition and cell cycle delay (90).
PPIs
Table VIII indicates that CBX regulates tumor progression through PPIs. CBX1 interacts with the TF HMGA2 to activate the Wnt/β-catenin signaling pathway, which promotes cell proliferation and migration in HCC (115). In PCa, CBX1 is an androgen/AR coactivator involved in the proliferation and progression of AR-expressing PCa cells into castration-resistant PCa (217). Mechanistically, CBX1 interacts with AR to enhance the DNA-binding capacity of AR, particularly prostate-specific antigen enhancers and androgen response elements in promoter regions, and to increase the transcription of AR target genes. CBX2 cooperates with EZH2 to downregulate several peroxisome proliferator-activated receptor signaling pathway genes and tumor suppressor genes by cooperating with or binding their promoters, respectively (218). CBX7 binds the E-box to inhibit TWIST1 function and tumorigenicity and reduce the metastatic potential of secondary epithelial ovarian cancer cells (219). CBX8 promotes tumorigenesis and radioresistance of ESCC cells by targeting apoptotic peptidase activating factor 1 (13). CBX8 is upregulated in HCC, interacts with Y-box-binding protein 1 and regulates the cell cycle to promote the proliferation of HCC cells (220). Karyopherin subunit alpha 2 (KPNA2) and CBX8 are highly expressed in UBC. The interaction between KPNA2 and CBX8 promotes the proliferation, migration and invasion of bladder cancer cells by mediating the PR/SET domain 1/c-Fos pathway (221).
Table VIII.
CBXs regulating malignant phenotype changes in tumors through PPIs.
| Cancer type | Malignant phenotype changes | PPIs | (Refs.) |
|---|---|---|---|
| HCC | Proliferation; migration | CBX1-HMGA2 | (115) |
| PCa | Proliferation; malignant; progression | CBX1-AR | (217) |
| NSCLC | Growth; metastases | CBX2-EZH2 | (218) |
| seOC | Growth; metastases | CBX7-E-box | (219) |
| ESCC | Proliferation; apoptosis; radiosensitivity | CBX8-APAF1 | (13) |
| HCC | Proliferation; cell cycle | CBX8-YBX1 | (220) |
| UBC | Proliferation; migration; invasion | CBX8-KPNA2 | (221) |
CBX, chromobox; PPI, protein-protein interaction; HCC, hepatocellular carcinoma; PCa, prostate carcinoma; NSCLC, non-small cell lung cancer; seOC, secondary epithelial ovarian cancer; ESCC, esophageal squamous cell carcinoma; UBC, urinary bladder cancer; HMGA2, high mobility group AT-hook 2; AR, androgen receptor; EZH2, enhancer of zeste homolog 2; APAF1, apoptotic peptidase activating factor 1; YBX1, Y-Box binding protein 1; KPNA2, karyopherin subunit alpha 2.
Signal transduction
Although certain studies have not clarified the tumor regulatory mechanism of CBX at the molecular level, they have made clear that CBX functions through a specific signal transduction pathway (Table IX).
Table IX.
CBXs regulating malignant phenotype changes in tumors through signal transduction.
| Cancer type | Malignant phenotype changes | Correlated signaling pathway | (Refs.) |
|---|---|---|---|
| BC | Proliferation; growth | CBX2/mTORC1/DREAM | (222) |
| GC | Proliferation; migration; invasion | CBX2/YAP/β-catenin | (223) |
| Glioma | Proliferation; tumorigenesis; invasion; stemness | CBX2/PI3K/AKT | (224) |
| BC | Proliferation; growth; invasion | CBX2/PI3K/AKT | (73) |
| TSCC | Proliferation | CBX3/p21 | (70) |
| LUAD | Growth; invasion | CBX3/ARHGAP24/RAC1 | (225) |
| LUAD | Proliferation; invasion | CBX4/Wnt/β-catenin | (85) |
| Osteosarcoma | Growth; cell cycle; apoptosis | CBX4/HIF-1α | (108) |
| HCC | Growth | CBX6/S100A9/NF-κB/MAPK | (117) |
| Glioma | Migration | CBX7/YAP/Tafazzin/CTGF/JNK | (226) |
| BC | Tumorigenesis | CBX7/DKK1/Wnt/β-catenin | (227) |
| PAAD | Proliferation; migration; invasion | CBX7/PTEN/AKT | (228) |
| NSCLC | Proliferation; apoptosis; migration; invasion | CBX7/ERK/MAPK | (83) |
| GC | Stemness | CBX7/p16/AKT/NF-κB/miR-21 | (229) |
| GC | Tumorigenesis; migration; metastases | CBX7/p16INK4A | (230) |
| PAAD | Proliferation | HIF-1α/CBX8/IRS1/PI3K/AKT | (100) |
| UBC | Proliferation; cell cycle | CBX8/p53 | (105) |
CBX, chromobox; BC, breast cancer; GC, gastric cancer; TSCC, tongue squamous cell carcinoma; LUAD, lung adenocarcinoma; HCC, hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; NSCLC, non-small cell lung cancer; UBC, urinary bladder cancer; mTORC1, mechanistic target of rapamycin complex 1; DREAM, dimerisation partner, retinoblastoma link, E2F and MuvB; YAP, Yes-associated protein; PI3K, phosphatidylinositol-3-kinase; AKT, protein kinase B; ARHGAP24, Rho GTPase activating protein 24; RAC1, Rac family small GTPase 1; HIF-1α, hypoxia-inducible factor-1α; NF-κB, noncanonical nuclear factor-kappaB; MAPK, mitogen-activated protein kinase; CTGF, connective tissue growth factor; JNK, c-Jun N-terminal kinase; DKK1, Dickkopf-related protein 1; PTEN, phosphatase and tensin homolog; ERK, extracellular signal-regulated kinase; IRS1, insulin receptor substrate 1.
It has been indicated that CBX2 promotes mTORC1 signal transduction and inhibits the activity of the dimerisation partner, retinoblastoma link, E2F and MuvB-complex to drive the growth of BC cells (222). CBX2 depletion inhibits the proliferation, migration and invasion of GC cells by inactivating the YAP/β-catenin pathway (223). CBX3 induces the proliferation and invasion of glioma (224) and BC cells (73) through the activation of the PI3K/AKT pathway. CBX3 decreases the G1/S phase transition mediated through p21 to promote tumor proliferation and is associated with poor prognosis in TSCC (70). Upregulation of CBX3 promotes smoking-related LUAD progression by activating the RAC1 pathway via the inhibition of Rho GTPase activating protein 24 (225). Wang et al (85) found that the overexpression of CBX4 significantly promoted the proliferation and invasive growth of human and mouse LUAD cells by activating the Wnt/β-catenin pathway. CBX4 may promote tumor growth by activating the HIF-1α signaling pathway in osteosarcoma (108). Zheng et al (117) confirmed that CBX6 significantly promoted the growth of HCC cells both in vitro and in vivo through S100A9 and the noncanonical NF-κB/MAPK pathway. CBX7 inhibits cell proliferation, migration and invasion by inhibiting the YAP/Tafazzin/connective tissue growth factor/JNK pathway in glioma (226), the Wnt/β-catenin pathway in BC (227), the ERK/MAPK pathway in lung squamous cell carcinoma (83), and the phosphatase and tensin homolog/AKT axis in pancreatic cancer (228). CBX7 positively regulates the stem cell-like characteristics of GC cells by inhibiting p16 and activating the AKT/NF-κB/miR-21 pathway (229). CBX7, an oncogene, is involved in the occurrence and development of GC, partially through the p16INK4A regulatory pathway, to mediate tumorigenesis, cell migration and cancer metastasis (230). CBX8 effectively activates PI3K/AKT signaling by upregulating insulin receptor substrate 1, which has been indicated to drive the proliferation of PAAD (100). CBX8 depletion delays the cell cycle progression of UBC cells at the G2 and M phases mediated through the p53 pathway (105).
Metabolic reprogramming
As indicated in Table X, in BC, CBX2/CBX7 and metabolic reprogramming are directly related. Upregulated CBX2 expression leads to enhanced glycolysis, which in turn promotes the proliferation of BC cells, while decreased CBX7 expression leads to increased glycolysis, which in turn promotes the proliferation of BC cells (15). CBX3 promotes cell proliferation and regulates glycolysis by inhibiting fructose-bisphosphatase 1 (FBP1) in pancreatic cancer, and abrogating the CBX3-FBP1 signaling axis may effectively prevent aerobic glycolysis and inhibit cell proliferation (231).
Table X.
CBXs regulating malignant phenotype changes in tumors through metabolic reprogramming.
| Cancer type | Malignant phenotype changes | Regulation mechanism | (Refs.) |
|---|---|---|---|
| BC | Proliferation | CBX2/glycolysis | (15) |
| BC | Proliferation | CBX7/glycolysis | (15) |
| PAAD | Proliferation | CBX3/glycolysis | (231) |
CBX, chromobox; BC, breast cancer; PAAD, pancreatic adenocarcinoma.
7. Cancer therapies targeting CBXs
Studies have indicated that proteins mediate chemotherapy and radiosensitivity in cancer. The HDAC inhibitor vorinostat exerts its anti-leukemic effect by enhancing SUMO-triggered ubiquitin-mediated CBX2 stability (232). CBX3 inhibits UBE2L3, which enhances the stability of the tumor suppressor p53 in CCA cells and makes CCA cells sensitive to cisplatin (233). RAMS11-dependent CBX4 recruitment of transcriptionally activated TOP2α increases the resistance of CRC to topoisomerase inhibitors (234). A chimera composed of the CBX5 protein fused to the estrogen receptor-DNA-binding domain and AR-ligand-binding domain is an effective transcriptional inhibitor and participates in the gene silencing effect associated with long-term 4-hydroxytamoxifen (OHT) therapy, inducing drug resistance to OHT (235). Resveratrol inhibits the proliferation of oral squamous cell carcinoma and induces apoptosis by inhibiting CBX7/AKT and activating the p16 signaling pathway (236). The retention of CBX7 decreases lung cancer cell proliferation (at least partially through the downregulation of phosphorylated ERK and phosphorylated p38) and increases the apoptosis rate after irinotecan and etoposide therapy (at least partially through the downregulation of Bcl-2, phosphorylated AKT and phosphorylated JNK) (237). CBX7 downregulates ETS proto-oncogene 1 to inactivate the tumor necrosis factor signaling pathway, which inhibits the proliferation of ccRCC cells and enhances the sensitivity of ccRCC cells to tyrosine kinase inhibitors (238) With decreases in EZH2 and EED, CBX8 depletion leads to the accumulation of spontaneous DNA damage and increases the sensitivity of tumor cells to radiation or H2O2 exposure (90). CBX8 antagonizes the effect of the sirtuin 1 inhibitor sirtinol on the premature senescence of K562 chronic myeloid leukemia cells through the AKT/Rb/E2F transcription factor 1 pathway (239).
Although small-molecule inhibitors targeting histone-modifying enzymes have been used in the clinic, these treatments nonspecifically erase/write epigenetic marks throughout the entire genome, which may lead to unintended consequences. CBXs, as epigenetic readers, show broad prospects for cancer treatment, and certain small molecule inhibitors targeting CBXs have been found.
The CBX2 chromatin domain-selective probe SW2_152F has good cell permeability, selectively inhibits CBX2-chromatin binding in cells and blocks the neuroendocrine differentiation of PCa cell lines in response to androgen deprivation (240). UNC3866 is a recently reported polypeptide inhibitor of methyl lysine reading function in CBXs, i.e. CBX2/4/6/7/8. UNC3866 inhibits the proliferation of PCa cells (241). Compared with its affinity for CBX2, UNC3866 has a higher affinity for CBX7 (242). When UNC3866 is used to inhibit CBX4, the tumorigenicity and stem cell-like characteristics of stem cells are markedly reduced (169). Milosevich et al (243,244) developed several CBX6/8-biactive CBX inhibitors based on peptide mimics in 2016 and 2021, respectively. These inhibitors were effective against both CBX6 and CBX8 and affected the proliferation of rhabdomyoma tumor cell lines. CBX7i was the first generation of chromatin domain small-molecule inhibitors (245). CBX7i increases DNA damage and apoptosis induced by adriamycin and increases the toxicity of doxorubicin in BC (246). Simhadri et al (247) optimized the scaffolds of trimethyl lysine, a series of effective peptide antagonists, to target CBX7. Ren et al (248) also found two different kinds of small-molecule CBX7 CHD antagonists. Class A MS452 derivatives inhibit the binding of CBX7 CHD/methyl lysine by blocking H3K27me3 binding, while the class B compound MS351 uniquely inhibits the binding of the CBX7 CHD to H3K27me3. Lamb et al (249) reported that the first potent positive allosteric modulator (PAM) peptidomimetic, UNC4976, a PRC1-specific chemical probe with high cellular activity, is an effective inhibitor of CBX7. PAM activation of UNC4976 regulates PRC1, driving it away from the target region of H3K27me3, by antagonizing the specific recruitment of CBX7 to the target gene via H3K27me3 and increasing nonspecific binding to DNA and RNA. Ren et al (245) found a small molecule, MS37452, that inhibits the binding of the CBX7 CHD to H3K27me3 and suppresses the transcription of p16/CDK inhibitor 2A, the target gene of the polycomb inhibitory complex, by abrogating the binding of CBX7 to INK4A/ARF loci in PCa cells. Simhadri et al (250) created a low-molecular-weight inhibitor of CBX7 (33F) via the rational modification of the structure of methyl-reading protein lethal 3 malignant brain tumor-like protein inhibitor 1. Denton et al (251) identified an effective and selective inhibitory peptide, PSL, against CBX7 and CBX8 and confirmed that the acylation of this inhibitory peptide by 5-methylisoxazole-3-carboxylic acid (PSL-81) increased the potency and selectivity of CBX8. Treatment with UNC7040 effectively and selectively removes PRC1-carrying CBX8 from chromatin, abrogates gene silencing and reduces the proliferation of different cancer cell lines (252).
8. Conclusion and prospects
In the present review, the characteristics and functions of CBX proteins were introduced and the expression of CBX1-8 in cancers and the relationship between the expression of CBXs and clinical characteristics (mainly cancer grade, stage, metastasis and relapse) and prognosis were comprehensively discussed. How CBXs regulate cell proliferation and self-renewal, apoptosis and the acquisition of malignant phenotypes such as invasion, migration and chemoresistance through mechanisms involving epigenetic modification, nuclear translocation, noncoding RNA interactions, transcriptional regulation, posttranslational modifications, protein-protein interactions, signal transduction and metabolic reprogramming, were also discussed in-depth.
In summary, CBXs have key roles in the occurrence and development of cancers. Several issues related to the roles of CBXs in cancer remain to be addressed. For instance, different CBXs have different roles in promoting/suppressing cancer. However, it remains elusive which CBX protein is the key driver of all cancers. As noncoding RNAs and signaling pathways regulate the expression of CBXs, targeting these noncoding RNAs and pathways is an alternative approach to control CBX expression. CBXs regulate tumor development through a variety of signaling pathways. It remains elusive whether activators or inhibitors of these pathways work in synergy with CBX inhibitors. Due to the key role of CBXs in carcinogenesis, targeting CBXs may be a cancer treatment method. Numerous compounds have been proven to target several CBXs. The identification of specific inhibitors of CBXs for the individualized treatment of cancer would be an optimal outcome. In addition, the safety and efficacy of these CBX inhibitors in clinical cancer treatment remain to be determined. Therefore, further study of the roles played by CBXs in the occurrence and development of cancers will help us to design new cancer treatment strategies that work by targeting CBXs.
Acknowledgments
Not applicable.
Funding Statement
The present study was supported by grants from the Science and Technology Department of Jilin Province (grant nos. 20200201123JC and 20220402066GH to DY).
Availability of data and materials
Not applicable.
Authors' contributions
Conceptualization, WW, DY and JW; investigation, JW and BY; writing-original draft, JW and BY; writing-review and editing, JW, BY, XZ, SL, XP, CM and SM; supervision, WW; project administration, DY; funding acquisition, DY. All authors have read and agreed to the published version of the manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 2.Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 4.Kerppola TK. Polycomb group complexes-many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 7.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai D, Pethe P. Polycomb repressive complex 1: Regulators of neurogenesis from embryonic to adult stage. J Cell Physiol. 2020;235:4031–4045. doi: 10.1002/jcp.29299. [DOI] [PubMed] [Google Scholar]
- 9.Ma RG, Zhang Y, Sun TT, Cheng B. Epigenetic regulation by polycomb group complexes: Focus on roles of CBX proteins. J Zhejiang Univ Sci B. 2014;15:412–428. doi: 10.1631/jzus.B1400077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camahort R, Cowan CA. Cbx proteins help ESCs walk the line between self-renewal and differentiation. Cell Stem Cell. 2012;10:4–6. doi: 10.1016/j.stem.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Wang B, Tang J, Liao D, Wang G, Zhang M, Sang Y, Cao J, Wu Y, Zhang R, Li S, et al. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S684–692. doi: 10.1245/s10434-013-3171-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Kang M, Zhang B, Meng F, Song J, Kaneko H, Shimamoto F, Tang B. m6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer. 2019;18:185. doi: 10.1186/s12943-019-1116-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Zhang Y, Chen H, Zhu H, Sun X. CBX8 promotes tumorigenesis and confers radioresistance in esophageal squamous cell carcinoma cells through targeting APAF1. Gene. 2019;711:143949. doi: 10.1016/j.gene.2019.143949. [DOI] [PubMed] [Google Scholar]
- 14.Huang Z, Yan Y, Zhu Z, Liu J, He X, Dalangood S, Li M, Tan M, Cai J, Tang P, et al. CBX7 suppresses urinary bladder cancer progression via modulating AKR1B10-ERK signaling. Cell Death Dis. 2021;12:537. doi: 10.1038/s41419-021-03819-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iqbal MA, Siddiqui S, Ur Rehman A, Siddiqui FA, Singh P, Kumar B, Saluja D. Multiomics integrative analysis reveals antagonistic roles of CBX2 and CBX7 in metabolic reprogramming of breast cancer. Mol Oncol. 2021;15:1450–1465. doi: 10.1002/1878-0261.12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai HC, Baylin SB. Cancer epigenetics: Linking basic biology to clinical medicine. Cell Res. 2011;21:502–517. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simhadri C, Daze KD, Douglas SF, Quon TT, Dev A, Gignac MC, Peng F, Heller M, Boulanger MJ, Wulff JE, et al. Chromodomain antagonists that target the polycomb-group methyllysine reader protein chromobox homolog 7 (CBX7) J Med Chem. 2014;57:2874–2883. doi: 10.1021/jm401487x. [DOI] [PubMed] [Google Scholar]
- 18.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eich ML, Athar M, Ferguson JE, III, Varambally S. EZH2-targeted therapies in cancer: Hype or a reality. Cancer Res. 2020;80:5449–5458. doi: 10.1158/0008-5472.CAN-20-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 2016;3:e186–e195. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 21.Shorstova T, Marques M, Su J, Johnston J, Kleinman CL, Hamel N, Huang S, Alaoui-Jamali MA, Foulkes WD, Witcher M. SWI/SNF-compromised cancers are susceptible to bromodomain inhibitors. Cancer Res. 2019;79:2761–2774. doi: 10.1158/0008-5472.CAN-18-1545. [DOI] [PubMed] [Google Scholar]
- 22.Jeon YH, Kim GW, Kim SY, Yi SA, Yoo J, Kim JY, Lee SW, Kwon SH. Heterochromatin protein 1: A Multiplayer in cancer progression. Cancers (Basel) 2022;14:763. doi: 10.3390/cancers14030763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.German B, Ellis L. Polycomb directed cell fate decisions in development and cancer. Epigenomes. 2022;6:28. doi: 10.3390/epigenomes6030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parreno V, Martinez AM, Cavalli G. Mechanisms of Polycomb group protein function in cancer. Cell Res. 2022;32:231–253. doi: 10.1038/s41422-021-00606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong GJ, Xu JL, Qi YR, Yuan ZQ, Zhao W. Critical roles of polycomb repressive complexes in transcription and cancer. Int J Mol Sci. 2022;23:9574. doi: 10.3390/ijms23179574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fonfría-Subirós E, Acosta-Reyes F, Saperas N, Pous J, Subirana JA, Campos JL. Crystal structure of a complex of DNA with one AT-hook of HMGA1. PLoS One. 2012;7:e37120. doi: 10.1371/journal.pone.0037120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tardat M, Albert M, Kunzmann R, Liu Z, Kaustov L, Thierry R, Duan S, Brykczynska U, Arrowsmith CH, Peters AH. Cbx2 targets PRC1 to constitutive heterochromatin in mouse zygotes in a parent-of-origin-dependent manner. Mol Cell. 2015;58:157–171. doi: 10.1016/j.molcel.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Senthilkumar R, Mishra RK. Novel motifs distinguish multiple homologues of Polycomb in vertebrates: Expansion and diversification of the epigenetic toolkit. BMC Genomics. 2009;10:549. doi: 10.1186/1471-2164-10-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine SS, King IF, Kingston RE. Division of labor in polycomb group repression. Trends Biochem Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 31.Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 32.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T, Dent SY. Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nat Rev Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Kingston RE. The CBX family of proteins in transcriptional repression and memory. J Biosci. 2020;45:16. doi: 10.1007/s12038-019-9972-5. [DOI] [PubMed] [Google Scholar]
- 35.Jangal M, Lebeau B, Witcher M. Beyond EZH2: Is the poly-comb protein CBX2 an emerging target for anti-cancer therapy. Expert Opin Ther Targets. 2019;23:565–578. doi: 10.1080/14728222.2019.1627329. [DOI] [PubMed] [Google Scholar]
- 36.Kawaguchi T, Machida S, Kurumizaka H, Tagami H, Nakayama JI. Phosphorylation of CBX2 controls its nucleosome-binding specificity. J Biochem. 2017;162:343–355. doi: 10.1093/jb/mvx040. [DOI] [PubMed] [Google Scholar]
- 37.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen L, Ito S, Cooper S, Kondo K, Koseki Y, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 42.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/S1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 43.Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol. 2004;167:1025–1035. doi: 10.1083/jcb.200409026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan HL, Morey L. Emerging roles for polycomb-group proteins in stem cells and cancer. Trends Biochem Sci. 2019;44:688–700. doi: 10.1016/j.tibs.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, Chouinard CR, Labadorf A, Akie TE, Moran TB, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45:330–343. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature. 2014;516:349–354. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saunders WS, Chue C, Goebl M, Craig C, Clark RF, Powers JA, Eissenberg JC, Elgin SC, Rothfield NF, Earnshaw WC. Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J Cell Sci. 1993;104:573–582. doi: 10.1242/jcs.104.2.573. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R. Heterochromatin formation in mammalian cells: Interaction between histones and HP1 proteins. Mol Cell. 2001;7:729–739. doi: 10.1016/S1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 53.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 54.Aasland R, Stewart AF. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1,HP1. Nucleic Acids Res. 1995;23:3168–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, Broadhurst RW, Ball LJ, Murzina NV, Laue ED. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol. 2000;10:517–525. doi: 10.1016/S0960-9822(00)00467-X. [DOI] [PubMed] [Google Scholar]
- 57.Azzaz AM, Vitalini MW, Thomas AS, Price JP, Blacketer MJ, Cryderman DE, Zirbel LN, Woodcock CL, Elcock AH, Wallrath LL, et al. Human heterochromatin protein 1α promotes nucleosome associations that drive chromatin condensation. J Biol Chem. 2014;289:6850–6861. doi: 10.1074/jbc.M113.512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casale AM, Cappucci U, Fanti L, Piacentini L. Heterochromatin protein 1 (HP1) is intrinsically required for post-transcriptional regulation of Drosophila Germline Stem Cell (GSC) maintenance. Sci Rep. 2019;9:4372. doi: 10.1038/s41598-019-40152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ligresti G, Caporarello N, Meridew JA, Jones DL, Tan Q, Choi KM, Haak AJ, Aravamudhan A, Roden AC, Prakash YS, et al. CBX5/G9a/H3K9me-mediated gene repression is essential to fibroblast activation during lung fibrosis. JCI Insight. 2019;5:e127111. doi: 10.1172/jci.insight.127111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 61.Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 62.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis S, Meltzer PS. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 64.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asplund A, Edqvist PH, Schwenk JM, Pontén F. Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research. Proteomics. 2012;12:2067–2077. doi: 10.1002/pmic.201100504. [DOI] [PubMed] [Google Scholar]
- 67.Li J, Xu Z, Zhou L, Hu K. Expression profile and prognostic values of Chromobox family members in human glioblastoma. Aging (Albany NY) 2022;14:1910–1931. doi: 10.18632/aging.203912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng ZQ, Yuan GQ, Kang NL, Nie QQ, Zhang GG, Wang Z. Chromobox 7/8 serve as independent indicators for glioblastoma via promoting proliferation and invasion of glioma cells. Front Neurol. 2022;13:912039. doi: 10.3389/fneur.2022.912039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao SP, Wang F, Yang M, Wang XY, Jin CL, Ji QK, Li S, Zhao XL. CBX3 promotes glioma U87 cell proliferation and predicts an unfavorable prognosis. J Neurooncol. 2019;145:35–48. doi: 10.1007/s11060-019-03286-w. [DOI] [PubMed] [Google Scholar]
- 70.Zhang H, Chen W, Fu X, Su X, Yang A. CBX3 promotes tumor proliferation by regulating G1/S phase via p21 downregulation and associates with poor prognosis in tongue squamous cell carcinoma. Gene. 2018;654:49–56. doi: 10.1016/j.gene.2018.02.043. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X, Zhou W, Zhang Y, Liu Z. CBX3 is a prognostic biomarker correlated with ATR activation and immune infiltration in head and neck squamous cell carcinoma. Int J Gen Med. 2022;15:1497–1508. doi: 10.2147/IJGM.S344390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo XH, Zhang JY, Jiao DC, Zhu JJ, Ma YZ, Yang Y, Xiao H, Liu ZZ. The expression and significance of chromobox protein homolog 2 in breast cancer. Zhonghua Yi Xue Za Zhi. 2020;100:130–135. doi: 10.3760/cma.j.issn.0376-2491.2020.02.010. In Chinese. [DOI] [PubMed] [Google Scholar]
- 73.Zheng S, Lv P, Su J, Miao K, Xu H, Li M. Overexpression of CBX2 in breast cancer promotes tumor progression through the PI3K/AKT signaling pathway. Am J Transl Res. 2019;11:1668–1682. [PMC free article] [PubMed] [Google Scholar]
- 74.Chen WY, Zhang XY, Liu T, Liu Y, Zhao YS, Pang D. Chromobox homolog 2 protein: A novel biomarker for predicting prognosis and Taxol sensitivity in patients with breast cancer. Oncol Lett. 2017;13:1149–1156. doi: 10.3892/ol.2016.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Piqué DG, Montagna C, Greally JM, Mar JC. A novel approach to modelling transcriptional heterogeneity identifies the oncogene candidate CBX2 in invasive breast carcinoma. Br J Cancer. 2019;120:746–753. doi: 10.1038/s41416-019-0387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeng JS, Zhang ZD, Pei L, Bai ZZ, Yang Y, Yang H, Tian QH. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. Int J Biochem Cell Biol. 2018;95:1–8. doi: 10.1016/j.biocel.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 77.Li X, Gou J, Li H, Yang X. Bioinformatic analysis of the expression and prognostic value of chromobox family proteins in human breast cancer. Sci Rep. 2020;10:17739. doi: 10.1038/s41598-020-74792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang YK, Lin HY, Chen CF, Zeng D. Prognostic values of distinct CBX family members in breast cancer. Oncotarget. 2017;8:92375–92387. doi: 10.18632/oncotarget.21325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chung CY, Sun Z, Mullokandov G, Bosch A, Qadeer ZA, Cihan E, Rapp Z, Parsons R, Aguirre-Ghiso JA, Farias EF, et al. Cbx8 Acts Non-canonically with Wdr5 to promote mammary tumorigenesis. Cell Rep. 2016;16:472–486. doi: 10.1016/j.celrep.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mao G, Zheng Y, Lin S, Ma L, Zhou Z, Zhang S. Bioinformatic analysis of prognostic value, genetic interaction, and immune infiltration of chromobox family proteins in breast cancer. Int J Gen Med. 2021;14:9181–9191. doi: 10.2147/IJGM.S343948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie X, Ning Y, Long J, Wang H, Chen X. Diverse CBX family members as potential prognostic biomarkers in non-small-cell lung cancer. FEBS Open Bio. 2020;10:2206–2215. doi: 10.1002/2211-5463.12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang SC, Lai YC, Chen YC, Wang NK, Wang WS, Lai JI. CBX3/heterochromatin protein 1 gamma is significantly upregulated in patients with non-small cell lung cancer. Asia Pac J Clin Oncol. 2018;14:e283–e288. doi: 10.1111/ajco.12820. [DOI] [PubMed] [Google Scholar]
- 83.Huang J, Zhang W, Lin D, Lian L, Hong W, Xu Z. Chromobox Homologue 7 acts as a tumor suppressor in both lung adenocarcinoma and lung squamous cell carcinoma via inhibiting ERK/MAPK signaling pathway. Evid Based Complement Alternat Med. 2022;2022:4952185. doi: 10.1155/2022/4952185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang C, Chang L, Yao Y, Chao C, Ge Z, Fan C, Yu H, Wang B, Yang J. Role of the CBX molecular family in lung adenocarcinoma tumorigenesis and immune infiltration. Front Genet. 2021;12:771062. doi: 10.3389/fgene.2021.771062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z, Fang Z, Chen G, Liu B, Xu J, Li F, Li F, Liu H, Zhang H, Sun Y, et al. Chromobox 4 facilitates tumorigenesis of lung adenocarcinoma through the Wnt/β-catenin pathway. Neoplasia. 2021;23:222–233. doi: 10.1016/j.neo.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu J, Shen H, Chen X, Ding Y, Wang H, Xu N, Teng L. Expression and prognostic value of chromobox family proteins in esophageal cancer. Genes (Basel) 2022;13:1582. doi: 10.3390/genes13091582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fang X, Wang J, Chen J, Zhuang M, Huang T, Chen Z, Huang Y, Zheng B, Wang X. Identification and validation of chromobox family members as potential prognostic biomarkers and therapeutic targets for human esophageal cancer. Front Genet. 2022;13:851390. doi: 10.3389/fgene.2022.851390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hou J, Yang Y, Gao H, Ouyang T, Liu Q, Ding R, Kan H. Systematic investigation of the clinical significance and prognostic value of the CBXs in esophageal cancer. Medicine (Baltimore) 2022;101:e30888. doi: 10.1097/MD.0000000000030888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ueda S, Kanda M, Sato Y, Baba H, Nakamura S, Sawaki K, Shimizu D, Motoyama S, Fujii T, Kodera Y, et al. Chromobox 2 expression predicts prognosis after curative resection of oesophageal squamous cell carcinoma. Cancer Genomics Proteomics. 2020;17:391–400. doi: 10.21873/cgp.20198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao W, Ou C, Qin J, Xing F, Sun Y, Li Z, Qiu J. CBX8, a novel DNA repair protein, promotes tumorigenesis in human esophageal carcinoma. Int J Clin Exp Pathol. 2014;7:4817–4826. [PMC free article] [PubMed] [Google Scholar]
- 91.Lin H, Lian J, Xia L, Guan G, You J. CBX3 promotes gastric cancer progression and affects factors related to immunotherapeutic responses. Cancer Manag Res. 2020;12:10113–10125. doi: 10.2147/CMAR.S271807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li W, Chen H, Wang Z, Liu J, Lei X, Chen W. Chromobox 4 (CBX4) promotes tumor progression and stemness via activating CDC20 in gastric cancer. J Gastrointest Oncol. 2022;13:1058–1072. doi: 10.21037/jgo-22-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He M, Yue L, Wang H, Yu F, Yu M, Ni P, Zhang K, Chen S, Duan G, Zhang R. Evaluation of the prognostic value of CBXs in gastric cancer patients. Sci Rep. 2021;11:12375. doi: 10.1038/s41598-021-91649-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma T, Ma N, Chen JL, Tang FX, Zong Z, Yu ZM, Chen S, Zhou TC. Expression and prognostic value of Chromobox family members in gastric cancer. J Gastrointest Oncol. 2020;11:983–998. doi: 10.21037/jgo-20-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin K, Zhu J, Hu C, Bu F, Luo C, Zhu X, Zhu Z. Comprehensive analysis of the prognosis for chromobox family in gastric cancer. J Gastrointest Oncol. 2020;11:932–951. doi: 10.21037/jgo-20-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fisher RS, Acevedo C, Arzimanoglou A, Bogacz A, Cross JH, Elger CE, Engel J, Jr, Forsgren L, French JA, Glynn M, et al. ILAE official report: A practical clinical definition of epilepsy. Epilepsia. 2014;55:475–482. doi: 10.1111/epi.12550. [DOI] [PubMed] [Google Scholar]
- 97.Chen ZY, Sun SX, Zhu SX, Bu J. Identification of the roles of chromobox family members in gastric cancer: A study based on multiple datasets. Biomed Res Int. 2020;2020:5306509. doi: 10.1155/2020/5306509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang YJ, Zhao LY, He X, Yao RF, Lu F, Lu BN, Pang ZR. CBXs-related prognostic gene signature correlates with immune microenvironment in gastric cancer. Aging (Albany NY) 2022;14:6227–6254. doi: 10.18632/aging.204214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Q, Fu L, Wu D, Wang J. Prognostic and immune infiltrates for the Chromobox (CBX) protein family in human pancreatic adenocarcinoma. J Gastrointest Oncol. 2021;12:2310–2324. doi: 10.21037/jgo-21-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Teng BW, Zhang KD, Yang YH, Guo ZY, Chen WW, Qiu ZJ. Genome-wide CRISPR-Cas9 screening identifies that hypoxia-inducible factor-1a-induced CBX8 transcription promotes pancreatic cancer progression via IRS1/AKT axis. World J Gastrointest Oncol. 2021;13:1709–1724. doi: 10.4251/wjgo.v13.i11.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang H, Zhao W, Wang J, Zhang Z. Clinicopathological significance of CBX3 in colorectal cancer: An intensive expression study based on formalin-fixed and paraffin-embedded tissues. Pathol Int. 2022;72:107–116. doi: 10.1111/pin.13194. [DOI] [PubMed] [Google Scholar]
- 102.Tang J, Wang G, Zhang M, Li FY, Sang Y, Wang B, Hu K, Wu Y, Luo R, Liao D, et al. Paradoxical role of CBX8 in proliferation and metastasis of colorectal cancer. Oncotarget. 2014;5:10778–10790. doi: 10.18632/oncotarget.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li Q, Pan Y, Cao Z, Zhao S. Comprehensive analysis of prognostic value and immune infiltration of chromobox family members in colorectal cancer. Front Oncol. 2020;10:582667. doi: 10.3389/fonc.2020.582667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou H, Xiong Y, Liu Z, Hou S, Zhou T. Expression and prognostic significance of CBX2 in colorectal cancer: Database mining for CBX family members in malignancies and vitro analyses. Cancer Cell Int. 2021;21:402. doi: 10.1186/s12935-021-02106-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yuan GJ, Chen X, Lu J, Feng ZH, Chen SL, Chen RX, Wei WS, Zhou FJ, Xie D. Chromobox homolog 8 is a predictor of muscle invasive bladder cancer and promotes cell proliferation by repressing the p53 pathway. Cancer Sci. 2017;108:2166–2175. doi: 10.1111/cas.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou J, Chen Z, Zou M, Wan R, Wu T, Luo Y, Wu G, Wang W, Liu T. Prognosis and immune infiltration of chromobox family genes in sarcoma. Front Oncol. 2021;11:657595. doi: 10.3389/fonc.2021.657595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma C, Nie XG, Wang YL, Liu XH, Liang X, Zhou QL, Wu DP. CBX3 predicts an unfavorable prognosis and promotes tumorigenesis in osteosarcoma. Mol Med Rep. 2019;19:4205–4212. doi: 10.3892/mmr.2019.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang J, Cheng D, Zhu B, Zhou S, Ying T, Yang Q. Chromobox Homolog 4 is positively correlated to tumor growth, survival and activation of HIF-1α signaling in human osteosarcoma under normoxic condition. J Cancer. 2016;7:427–435. doi: 10.7150/jca.13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Karamitopoulou E, Pallante P, Zlobec I, Tornillo L, Carafa V, Schaffner T, Borner M, Diamantis I, Esposito F, Brunner T, et al. Loss of the CBX7 protein expression correlates with a more aggressive phenotype in pancreatic cancer. Eur J Cancer. 2010;46:1438–1444. doi: 10.1016/j.ejca.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 110.Zhu Y, Pu Z, Li Z, Lin Y, Li N, Peng F. Comprehensive analysis of the expression and prognosis value of chromobox family members in clear cell renal cell carcinoma. Front Oncol. 2021;11:700528. doi: 10.3389/fonc.2021.700528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hu K, Yao L, Xu Z, Yan Y, Li J. Prognostic value and therapeutic potential of CBX family members in ovarian cancer. Front Cell Dev Biol. 2022;10:832354. doi: 10.3389/fcell.2022.832354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tian P, Zhang C, Ma C, Ding L, Tao N, Ning L, Wang Y, Yong X, Yan Q, Lin X, et al. Decreased chromobox homologue 7 expression is associated with epithelial-mesenchymal transition and poor prognosis in cervical cancer. Open Med (Wars) 2021;16:410–418. doi: 10.1515/med-2021-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li D, Liu Y, Hao S, Chen B, Li A. Mining database for the clinical significance and prognostic value of CBX family in skin cutaneous melanoma. J Clin Lab Anal. 2020;34:e23537. doi: 10.1002/jcla.23537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ning G, Huang YL, Zhen LM, Xu WX, Jiao Q, Yang FJ, Wu LN, Zheng YY, Song J, Wang YS, et al. Transcriptional expressions of Chromobox 1/2/3/6/8 as independent indicators for survivals in hepatocellular carcinoma patients. Aging (Albany NY) 2018;10:3450–3473. doi: 10.18632/aging.101658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang YF, Pan YH, Tian QH, Wu DC, Su SG. CBX1 indicates poor outcomes and exerts oncogenic activity in hepatocellular carcinoma. Transl Oncol. 2018;11:1110–1118. doi: 10.1016/j.tranon.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhong X, Kan A, Zhang W, Zhou J, Zhang H, Chen J, Tang S. CBX3/HP1γ promotes tumor proliferation and predicts poor survival in hepatocellular carcinoma. Aging (Albany NY) 2019;11:5483–5497. doi: 10.18632/aging.102132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zheng H, Jiang WH, Tian T, Tan HS, Chen Y, Qiao GL, Han J, Huang SY, Yang Y, Li S, et al. CBX6 overexpression contributes to tumor progression and is predictive of a poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8:18872–18884. doi: 10.18632/oncotarget.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang J, He H, Jiang Q, Wang Y, Jia S. CBX6 promotes HCC metastasis via transcription factors Snail/Zeb1-mediated EMT mechanism. Onco Targets Ther. 2020;13:12489–12500. doi: 10.2147/OTT.S257363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhu X, Qin M, Li C, Zeng W, Bei C, Tan C, Zhang Y, Shi W, Kong J, Fu Y, et al. Downregulated expression of chromobox homolog 7 in hepatocellular carcinoma. Genet Test Mol Biomarkers. 2019;23:348–352. doi: 10.1089/gtmb.2018.0293. [DOI] [PubMed] [Google Scholar]
- 120.Tang B, Tian Y, Liao Y, Li Z, Yu S, Su H, Zhong F, Yuan G, Wang Y, Yu H, et al. CBX8 exhibits oncogenic properties and serves as a prognostic factor in hepatocellular carcinoma. Cell Death Dis. 2019;10:52. doi: 10.1038/s41419-018-1288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang CZ, Chen SL, Wang CH, He YF, Yang X, Xie D, Yun JP. CBX8 exhibits oncogenic activity via AKT/β-catenin activation in hepatocellular carcinoma. Cancer Res. 2018;78:51–63. doi: 10.1158/0008-5472.CAN-17-0700. [DOI] [PubMed] [Google Scholar]
- 122.Pan C, Luo N, Guo K, Wang W, Li L, Fan N, Tian Y. Members of the chromobox family have prognostic value in hepatocellular carcinoma. Front Genet. 2022;13:887925. doi: 10.3389/fgene.2022.887925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu Y, Pan S, Song Y, Pan C, Chen C, Zhu X. The prognostic value of the chromobox family in human ovarian cancer. J Cancer. 2020;11:5198–5209. doi: 10.7150/jca.44475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wheeler LJ, Watson ZL, Qamar L, Yamamoto TM, Post MD, Berning AA, Spillman MA, Behbakht K, Bitler BG. CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer. Oncogenesis. 2018;7:92. doi: 10.1038/s41389-018-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang Y, Hu Z, Sun H, Yu Q, Yang L, Yin F, Sun Y, Pu L, Zhu X, Li S, et al. CBX7, a potential prognostic biomarker in lung adenocarcinoma. Onco Targets Ther. 2021;14:5477–5492. doi: 10.2147/OTT.S325203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hu CY, Li X, Zeng T, Ye DM, Li YK, Yan HX. Significance of chromobox protein (CBX) expression in diffuse LBCL. Gene. 2022;813:146092. doi: 10.1016/j.gene.2021.146092. [DOI] [PubMed] [Google Scholar]
- 127.Tan C, Bei C, Zhu X, Zhang Y, Qin L, Tan S. Single nucleotide polymorphisms of CBX4 and CBX7 decrease the risk of hepatocellular carcinoma. Biomed Res Int. 2019;2019:6436825. doi: 10.1155/2019/6436825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu XY, Huang MJ, Su QY, Wang XZ, Wang J, Long QQ, Wu XM, Huang XY, Yao JG, Long XD. The predictive potential of genetic single nucleotide polymorphisms in CBX4 for hepatocellular carcinoma survival. Front Biosci (Landmark Ed) 2021;26:1191–1203. doi: 10.52586/5019. [DOI] [PubMed] [Google Scholar]
- 129.Chang C, Liu J, He W, Qu M, Huang X, Deng Y, Shen L, Zhao X, Guo H, Jiang J, et al. A regulatory circuit HP1γ/miR-451a/c-Myc promotes prostate cancer progression. Oncogene. 2018;37:415–426. doi: 10.1038/onc.2017.332. [DOI] [PubMed] [Google Scholar]
- 130.Cheng W, Qi Y, Tian L, Wang B, Huang W, Chen Y. Dicer promotes tumorigenesis by translocating to nucleus to promote SFRP1 promoter methylation in cholangiocarcinoma cells. Cell Death Dis. 2017;8:e2628. doi: 10.1038/cddis.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ci X, Hao J, Dong X, Choi SY, Xue H, Wu R, Qu S, Gout PW, Zhang F, Haegert AM, et al. Heterochromatin protein 1α mediates development and aggressiveness of neuroendocrine prostate cancer. Cancer Res. 2018;78:2691–2704. doi: 10.1158/0008-5472.CAN-17-3677. [DOI] [PubMed] [Google Scholar]
- 132.Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao D, Wang G, Qin G, Xu RH, Kang T. CBX4 suppresses metastasis via recruitment of HDAC3 to the Runx2 promoter in colorectal carcinoma. Cancer Res. 2016;76:7277–7289. doi: 10.1158/0008-5472.CAN-16-2100. [DOI] [PubMed] [Google Scholar]
- 133.Jiang N, Niu G, Pan YH, Pan W, Zhang MF, Zhang CZ, Shen H. CBX4 transcriptionally suppresses KLF6 via interaction with HDAC1 to exert oncogenic activities in clear cell renal cell carcinoma. EBioMedicine. 2020;53:102692. doi: 10.1016/j.ebiom.2020.102692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu T, Wu Y, Hu Q, Zhang J, Nie E, Wu W, Wang X, Wang Y, Liu N. CBX7 is a glioma prognostic marker and induces G1/S arrest via the silencing of CCNE1. Oncotarget. 2017;8:26637–26647. doi: 10.18632/oncotarget.15789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Federico A, Pallante P, Bianco M, Ferraro A, Esposito F, Monti M, Cozzolino M, Keller S, Fedele M, Leone V, et al. Chromobox protein homologue 7 protein, with decreased expression in human carcinomas, positively regulates E-cadherin expression by interacting with the histone deacetylase 2 protein. Cancer Res. 2009;69:7079–7087. doi: 10.1158/0008-5472.CAN-09-1542. [DOI] [PubMed] [Google Scholar]
- 136.Wu Y, Duan Y, Li X, Zhao R, Lan B, Zhang X, Wang X, Chen H, Feng S, Liu Z, et al. CBX8 together with SET facilitates ovarian carcinoma growth and metastasis by suppressing the transcription of SUSD2. Mol Cancer Res. 2022;20:1611–1622. doi: 10.1158/1541-7786.MCR-22-0139. [DOI] [PubMed] [Google Scholar]
- 137.Mancini M, Papon L, Mangé A, Cammas F, Fabbrizio E. HP1s modulate the S-Adenosyl Methionine synthesis pathway in liver cancer cells. Biochem J. 2020;477:1033–1047. doi: 10.1042/BCJ20190621. [DOI] [PubMed] [Google Scholar]
- 138.Zhang K, Wang J, Yang L, Yuan YC, Tong TR, Wu J, Yun X, Bonner M, Pangeni R, Liu Z, et al. Targeting histone methyltransferase G9a inhibits growth and Wnt signaling pathway by epigenetically regulating HP1α and APC2 gene expression in non-small cell lung cancer. Mol Cancer. 2018;17:153. doi: 10.1186/s12943-018-0896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng Q, Xu J, Lin Z, Lu Y, Xin X, Li X, Yang Y, Meng Q, Wang C, Xiong W, et al. Inflammatory factor receptor Toll-like receptor 4 controls telomeres through heterochromatin protein 1 isoforms in liver cancer stem cell. J Cell Mol Med. 2018;22:3246–3258. doi: 10.1111/jcmm.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cheng W, Tian L, Wang B, Qi Y, Huang W, Li H, Chen YJ. Downregulation of HP1α suppresses proliferation of cholangiocarcinoma by restoring SFRP1 expression. Oncotarget. 2016;7:48107–48119. doi: 10.18632/oncotarget.10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yi SA, Lee DH, Kim GW, Ryu HW, Park JW, Lee J, Han J, Park JH, Oh H, Lee J, et al. HPV-mediated nuclear export of HP1γ drives cervical tumorigenesis by downregulation of p53. Cell Death Differ. 2020;27:2537–2551. doi: 10.1038/s41418-020-0520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gao G, Wang L, Li C. Circ_0006790 carried by bone marrow mesenchymal stem cell-derived exosomes regulates S100A11 DNA methylation through binding to CBX7 in pancreatic ductal adenocarcinoma. Am J Cancer Res. 2022;12:1934–1959. [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang Z, Li N, Wei X, Chen B, Zhang Y, Zhao Y, Hu X, Hou S. GRM4 inhibits the proliferation, migration, and invasion of human osteosarcoma cells through interaction with CBX4. Biosci Biotechnol Biochem. 2020;84:279–289. doi: 10.1080/09168451.2019.1673147. [DOI] [PubMed] [Google Scholar]
- 144.Wang W, Li X, Guan C, Hu Z, Zhao Y, Li W, Jiang X. LncRNA PCAT6 promotes the proliferation, migration and invasion of pancreatic ductal adenocarcinoma via regulating miR-185-5p/CBX2 axis. Pathol Res Pract. 2020;216:153074. doi: 10.1016/j.prp.2020.153074. [DOI] [PubMed] [Google Scholar]
- 145.Huo W, Tan D, Chen Q. CASC9 facilitates cell proliferation in bladder cancer by regulating CBX2 expression. Nephron. 2020;144:388–399. doi: 10.1159/000507828. [DOI] [PubMed] [Google Scholar]
- 146.Mather RL, Parolia A, Carson SE, Venalainen E, Roig-Carles D, Jaber M, Chu SC, Alborelli I, Wu R, Lin D, et al. The evolutionarily conserved long non-coding RNA LINC00261 drives neuroendocrine prostate cancer proliferation and metastasis via distinct nuclear and cytoplasmic mechanisms. Mol Oncol. 2021;15:1921–1941. doi: 10.1002/1878-0261.12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dou Y, Chen F, Lu Y, Qiu H, Zhang H. Effects of Wnt/β-catenin signal pathway regulated by miR-342-5p targeting CBX2 on proliferation, metastasis and invasion of ovarian cancer cells. Cancer Manag Res. 2020;12:3783–3794. doi: 10.2147/CMAR.S250208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Han Q, Li C, Cao Y, Bao J, Li K, Song R, Chen X, Li J, Wu X. CBX2 is a functional target of miRNA let-7a and acts as a tumor promoter in osteosarcoma. Cancer Med. 2019;8:3981–3991. doi: 10.1002/cam4.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhu J, Luo JE, Chen Y, Wu Q. Circ_0061140 knockdown inhibits tumorigenesis and improves PTX sensitivity by regulating miR-136/CBX2 axis in ovarian cancer. J Ovarian Res. 2021;14:136. doi: 10.1186/s13048-021-00888-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang F, Zhang L, Luo Y, Zhang Q, Zhang Y, Shao Y, Yuan L. The LncRNA RP11-279C4.1 enhances the malignant behaviour of glioma cells and glioma stem-like cells by regulating the miR-1273g-3p/CBX3 axis. Mol Neurobiol. 2021;58:3362–3373. doi: 10.1007/s12035-021-02337-6. [DOI] [PubMed] [Google Scholar]
- 151.Zhou W, Li H, Shang S, Liu F. lncRNA KCNQ1OT1 reverses the effect of sevoflurane on hepatocellular carcinoma progression via regulating the miR-29a-3p/CBX3 axis. Braz J Med Biol Res. 2021;54:e10213. doi: 10.1590/1414-431x2020e10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Song Y, Wang S, Cheng X. LINC01006 regulates the proliferation, migration and invasion of hepatocellular carcinoma cells through regulating miR-433-3p/CBX3 axis. Ann Hepatol. 2021;25:100343. doi: 10.1016/j.aohep.2021.100343. [DOI] [PubMed] [Google Scholar]
- 153.Liu J, Zhan Y, Wang J, Wang J, Guo J, Kong D. lncRNA-SNHG17 promotes colon adenocarcinoma progression and serves as a sponge for miR-375 to regulate CBX3 expression. Am J Transl Res. 2020;12:5283–5295. [PMC free article] [PubMed] [Google Scholar]
- 154.Huang Y, Lin Y, Song X, Wu D. LINC00857 contributes to proliferation and lymphomagenesis by regulating miR-370-3p/CBX3 axis in diffuse large B-cell lymphoma. Carcinogenesis. 2021;42:733–741. doi: 10.1093/carcin/bgab013. [DOI] [PubMed] [Google Scholar]
- 155.Cai H, Yu Y, Ni X, Li C, Hu Y, Wang J, Chen F, Xi S, Chen Z. LncRNA LINC00998 inhibits the malignant glioma phenotype via the CBX3-mediated c-Met/Akt/mTOR axis. Cell Death Dis. 2020;11:1032. doi: 10.1038/s41419-020-03247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang P, Yang X, Zha Z, Zhu Y, Zhang G, Li G. CBX3 regulated by miR-139 promotes the development of HCC by regulating cell cycle progression. Cell Cycle. 2022;21:1740–1752. doi: 10.1080/15384101.2022.2068329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu M, Huang F, Zhang D, Ju J, Wu XB, Wang Y, Wang Y, Wu Y, Nie M, Li Z, et al. Heterochromatin protein HP1γ promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res. 2015;75:4593–4604. doi: 10.1158/0008-5472.CAN-14-3735. [DOI] [PubMed] [Google Scholar]
- 158.Gao F, Du Y, Zhang Y, Ren D, Xu J, Chen D. Circ-EZH2 knockdown reverses DDAH1 and CBX3-mediated cell growth and invasion in glioma through miR-1265 sponge activity. Gene. 2020;726:144196. doi: 10.1016/j.gene.2019.144196. [DOI] [PubMed] [Google Scholar]
- 159.Kang S, Ou C, Yan A, Zhu K, Xue R, Zhang Y, Lai J. Long Noncoding RNA SNHG5 induces the NF-κB pathway by regulating miR-181c-5p/CBX4 axis to promote the progression of Non-small cell lung cancer. Arch Bronconeumol. 2022;59:10–18. doi: 10.1016/j.arbres.2022.07.001. [DOI] [PubMed] [Google Scholar]
- 160.Yang Z, OuYang X, Zheng L, Dai L, Luo W. Long intergenic noncoding RNA00265 promotes proliferation of gastric cancer via the microRNA-144-3p/Chromobox 4 axis. Bioengineered. 2021;12:1012–1025. doi: 10.1080/21655979.2021.1876320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhao J, Yang T, Li L. LncRNA FOXP4-AS1 is involved in cervical cancer progression via regulating miR-136-5p/CBX4 Axis. Onco Targets Ther. 2020;13:2347–2355. doi: 10.2147/OTT.S241818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zheng Z, Qiu K, Huang W. Long Non-coding RNA (lncRNA) RAMS11 promotes metastatis and cell growth of prostate cancer by CBX4 complex binding to Top2α. Cancer Manag Res. 2021;13:913–923. doi: 10.2147/CMAR.S270144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Meng R, Fang J, Yu Y, Hou LK, Chi JR, Chen AX, Zhao Y, Cao XC. miR-129-5p suppresses breast cancer proliferation by targeting CBX4. Neoplasma. 2018;65:572–578. doi: 10.4149/neo_2018_170814N530. [DOI] [PubMed] [Google Scholar]
- 164.Wen LJ, Wang YS, Tan PY. miR-515-5p inhibits the proliferation, migration and invasion of human breast cancer cells by targeting CBX4. Exp Ther Med. 2021;22:1328. doi: 10.3892/etm.2021.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fang X, Pan A. MiR-507 inhibits the progression of gastric carcinoma via targeting CBX4-mediated activation of Wnt/β-catenin and HIF-1α pathways. Clin Transl Oncol. 2022;24:2021–2028. doi: 10.1007/s12094-022-02862-3. [DOI] [PubMed] [Google Scholar]
- 166.Dou Z, Lu F, Hu J, Wang H, Li B, Li X. MicroRNA-6838-5p suppresses the self-renewal and metastasis of human liver cancer stem cells through downregulating CBX4 expression and inactivating ERK signaling. Biol Chem. 2022;404:29–39. doi: 10.1515/hsz-2022-0156. [DOI] [PubMed] [Google Scholar]
- 167.Chen Y, Du J, Wang Y, Shi H, Jiang Q, Wang Y, Zhang H, Wei Y, Xue W, Pu Z, et al. MicroRNA-497-5p induces cell cycle arrest of cervical cancer cells in S phase by targeting CBX4. Onco Targets Ther. 2019;12:10535–10545. doi: 10.2147/OTT.S210059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yan X, Kang D, Lin Y, Qi S, Jiang C. CBX4-dependent regulation of HDAC3 nuclear translocation reduces Bmp2-induced osteoblastic differentiation and calcification in adamantinomatous craniopharyngioma. Cell Commun Signal. 2022;20:3. doi: 10.1186/s12964-021-00797-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhao W, Ma B, Tian Z, Han H, Tang J, Dong B, An G, Cao B, Wang B. Inhibiting CBX4 efficiently protects hepatocellular carcinoma cells against sorafenib resistance. Br J Cancer. 2021;124:1237–1248. doi: 10.1038/s41416-020-01240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yu F, Lin Y, Ai MM, Tan GJ, Huang JL, Zou ZR. Knockdown of circular RNA hsa_circ_PVT1 inhibited laryngeal cancer progression via preventing wnt4/β-catenin signaling pathway activation. Front Cell Dev Biol. 2021;9:658115. doi: 10.3389/fcell.2021.658115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Huang FJ, Dang JQ, Zhang S, Cheng ZY. Circular RNA hsa_ circ_0008039 promotes proliferation, migration and invasion of breast cancer cells through upregulating CBX4 via sponging miR-515-5p. Eur Rev Med Pharmacol Sci. 2020;24:1887–1898. doi: 10.26355/eurrev_202002_20367. [DOI] [PubMed] [Google Scholar]
- 172.Sun Y, Wang X, Bu X. LINC02381 contributes to cell proliferation and hinders cell apoptosis in glioma by transcriptionally enhancing CBX5. Brain Res Bull. 2021;176:121–129. doi: 10.1016/j.brainresbull.2021.07.009. [DOI] [PubMed] [Google Scholar]
- 173.Yu L, Zhang W, Wang P, Zhang Q, Cong A, Yang X, Sang K. LncRNA SNHG11 aggravates cell proliferation and migration in triple-negative breast cancer via sponging miR-2355-5p and targeting CBX5. Exp Ther Med. 2021;22:892. doi: 10.3892/etm.2021.10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li H, Li J, Jia S, Wu M, An J, Zheng Q, Zhang W, Lu D. miR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget. 2015;6:31958–31984. doi: 10.18632/oncotarget.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yang Y, Meng Q, Wang C, Li X, Lu Y, Xin X, Zheng Q, Lu D. MicroRNA 675 cooperates PKM2 to aggravate progression of human liver cancer stem cells induced from embryonic stem cells. J Mol Med (Berl) 2018;96:1119–1130. doi: 10.1007/s00109-018-1687-9. [DOI] [PubMed] [Google Scholar]
- 176.Wu C, Zhang J. Long non-conding RNA LOXL1-AS1 sponges miR-589-5p to up-regulate CBX5 expression in renal cell carcinoma. Biosci Rep. 2020;40:BSR20200212. doi: 10.1042/BSR20200212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shi X, Song S, Gao Y, Cui Z, Wang W, Liu M. Circ_0037866 contributes to the tumorigenesis of renal cell carcinoma by sequestering miR-384 to elevate chromobox 5 expression. Kidney Blood Press Res. 2022;47:329–340. doi: 10.1159/000522190. [DOI] [PubMed] [Google Scholar]
- 178.Li F, Sun X, Liu Q, Liu X, Zhang J. Long Noncoding RNA MIR100HG knockdown attenuates hepatocellular carcinoma progression by regulating MicroRNA-146b-5p/Chromobox 6. Gastroenterol Res Pract. 2021;2021:6832518. doi: 10.1155/2021/6832518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pei YF, He Y, Hu LZ, Zhou B, Xu HY, Liu XQ. The Crosstalk between lncRNA-SNHG7/miRNA-181/cbx7 modulates malignant character in lung adenocarcinoma. Am J Pathol. 2020;190:1343–1354. doi: 10.1016/j.ajpath.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 180.Peng X, Guan L, Gao B. miRNA-19 promotes non-small-cell lung cancer cell proliferation via inhibiting CBX7 expression. Onco Targets Ther. 2018;11:8865–8874. doi: 10.2147/OTT.S181433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xie D, Shang C, Zhang H, Guo Y, Tong X. Up-regulation of miR-9 target CBX7 to regulate invasion ability of bladder transitional cell carcinoma. Med Sci Monit. 2015;21:225–230. doi: 10.12659/MSM.893232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pickl JM, Tichy D, Kuryshev VY, Tolstov Y, Falkenstein M, Schüler J, Reidenbach D, Hotz-Wagenblatt A, Kristiansen G, Roth W, et al. Ago-RIP-Seq identifies Polycomb repressive complex I member CBX7 as a major target of miR-375 in prostate cancer progression. Oncotarget. 2016;7:59589–59603. doi: 10.18632/oncotarget.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhao Y, Liu XL, Huang JH, Yin AJ, Zhang H. MicroRNA-18a suppresses ovarian carcinoma progression by targeting CBX7 and regulating ERK/MAPK signaling pathway and epithelial-to-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2020;24:5292–5302. doi: 10.26355/eurrev_202005_21311. [DOI] [PubMed] [Google Scholar]
- 184.Mansueto G, Forzati F, Ferraro A, Pallante P, Bianco M, Esposito F, Iaccarino A, Troncone G, Fusco A. Identification of a new pathway for tumor progression: MicroRNA-181b Up-regulation and CBX7 Down-regulation by HMGA1 Protein. Genes Cancer. 2010;1:210–224. doi: 10.1177/1947601910366860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gong L, Tang Y, Jiang L, Tang W, Luo S. Regulation of circGOLPH3 and its binding protein CBX7 on the proliferation and apoptosis of prostate cancer cells. Biosci Rep. 2020;40:BSR20200936. doi: 10.1042/BSR20200936. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 186.Song X, Ning W, Niu J, Zhang G, Liu H, Zhou L. CBX8 acts as an independent RNA-binding protein to regulate the maturation of miR-378a-3p in colon cancer cells. Hum Cell. 2021;34:515–529. doi: 10.1007/s13577-020-00477-w. [DOI] [PubMed] [Google Scholar]
- 187.Liang Y, Yu ZJ, Liu M, Liu HM, Zhang JZ, Xiong T, Tang YY, Huang ZP. hsa-miR-429 targets CBX8 to promote cell apoptosis in diffuse large B-cell lymphoma. Mol Med Rep. 2021;24:857. doi: 10.3892/mmr.2021.12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Xu Y, Yao Y, Leng K, Ji D, Qu L, Liu Y, Cui Y. Increased expression of circular RNA circ_0005230 indicates dismal prognosis in breast cancer and regulates cell proliferation and invasion via miR-618/CBX8 signal pathway. Cell Physiol Biochem. 2018;51:1710–1722. doi: 10.1159/000495675. [DOI] [PubMed] [Google Scholar]
- 189.Liu J, Wang D, Long Z, Liu J, Li W. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p/519-5p family and modulating the expression of CBX8. Cell Physiol Biochem. 2018;48:173–184. doi: 10.1159/000491716. [DOI] [PubMed] [Google Scholar]
- 190.Yi SA, Ryu HW, Lee DH, Han JW, Kwon SH. HP1β suppresses metastasis of human cancer cells by decreasing the expression and activation of MMP2. Int J Oncol. 2014;45:2541–2548. doi: 10.3892/ijo.2014.2646. [DOI] [PubMed] [Google Scholar]
- 191.Clermont PL, Crea F, Chiang YT, Lin D, Zhang A, Wang JZ, Parolia A, Wu R, Xue H, Wang Y, et al. Identification of the epigenetic reader CBX2 as a potential drug target in advanced prostate cancer. Clin Epigenetics. 2016;8:16. doi: 10.1186/s13148-016-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Alam H, Li N, Dhar SS, Wu SJ, Lv J, Chen K, Flores ER, Baseler L, Lee MG. HP1γ promotes lung adenocarcinoma by downregulating the transcription-repressive regulators NCOR2 and ZBTB7A. Cancer Res. 2018;78:3834–3848. doi: 10.1158/0008-5472.CAN-17-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fan Y, Li H, Liang X, Xiang Z. CBX3 promotes colon cancer cell proliferation by CDK6 kinase-independent function during cell cycle. Oncotarget. 2017;8:19934–19946. doi: 10.18632/oncotarget.15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen LY, Cheng CS, Qu C, Wang P, Chen H, Meng ZQ, Chen Z. Overexpression of CBX3 in pancreatic adenocarcinoma promotes cell cycle transition-associated tumor progression. Int J Mol Sci. 2018;19:1768. doi: 10.3390/ijms19061768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang H, Yu H, Ren D, Sun Y, Guo F, Cai H, Zhou C, Zhou Y, Jin X, Wu H. CBX3 regulated By YBX1 promotes smoking-induced pancreatic cancer progression via inhibiting SMURF2 expression. Int J Biol Sci. 2022;18:3484–3497. doi: 10.7150/ijbs.68995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sanyal S, Mondal P, Sen S, Sengupta Bandyopadhyay S, Das C. SUMO E3 ligase CBX4 regulates hTERT-mediated transcription of CDH1 and promotes breast cancer cell migration and invasion. Biochem J. 2020;477:3803–3818. doi: 10.1042/BCJ20200359. [DOI] [PubMed] [Google Scholar]
- 197.Hu C, Zhang Q, Tang Q, Zhou H, Liu W, Huang J, Liu Y, Wang Q, Zhang J, Zhou M, et al. CBX4 promotes the proliferation and metastasis via regulating BMI-1 in lung cancer. J Cell Mol Med. 2020;24:618–631. doi: 10.1111/jcmm.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang X, Qin G, Liang X, Wang W, Wang Z, Liao D, Zhong L, Zhang R, Zeng YX, Wu Y, et al. Targeting the CK1α/CBX4 axis for metastasis in osteosarcoma. Nat Commun. 2020;11:1141. doi: 10.1038/s41467-020-14870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Thomsen R, Christensen DB, Rosborg S, Linnet TE, Blechingberg J, Nielsen AL. Analysis of HP1α regulation in human breast cancer cells. Mol Carcinog. 2011;50:601–613. doi: 10.1002/mc.20755. [DOI] [PubMed] [Google Scholar]
- 200.Prieto C, Nguyen D, Liu Z, Wheat J, Perez A, Gourkanti S, Chou T, Barin E, Velleca A, Rohwetter T, et al. Transcriptional control of CBX5 by the RNA binding proteins RBMX and RBMXL1 maintains chromatin state in myeloid leukemia. Nat Cancer. 2021;2:741–757. doi: 10.1038/s43018-021-00220-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Deng H, Guan X, Gong L, Zeng J, Zhang H, Chen MY, Li G. CBX6 is negatively regulated by EZH2 and plays a potential tumor suppressor role in breast cancer. Sci Rep. 2019;9:197. doi: 10.1038/s41598-018-36560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sakai K, Nishiuchi T, Tange S, Suzuki Y, Yano S, Terashima M, Suzuki T, Matsumoto K. Proteasomal degradation of polycomb-group protein CBX6 confers MMP-2 expression essential for mesothelioma invasion. Sci Rep. 2020;10:16678. doi: 10.1038/s41598-020-72448-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wu W, Zhou X, Yu T, Bao Z, Zhi T, Jiang K, Nie E, Wang Y, Zhang J, You Y. The malignancy of miR-18a in human glioblastoma via directly targeting CBX7. Am J Cancer Res. 2017;7:64–76. [PMC free article] [PubMed] [Google Scholar]
- 204.Bao Z, Xu X, Liu Y, Chao H, Lin C, Li Z, You Y, Liu N, Ji J. CBX7 negatively regulates migration and invasion in glioma via Wnt/β-catenin pathway inactivation. Oncotarget. 2017;8:39048–39063. doi: 10.18632/oncotarget.16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dai T, Liu Y, Cao R, Cao J. CBX7 regulates metastasis of basal-like breast cancer through Twist1/EphA2 pathway. Transl Oncol. 2022;24:101468. doi: 10.1016/j.tranon.2022.101468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bernard D, Martinez-Leal JF, Rizzo S, Martinez D, Hudson D, Visakorpi T, Peters G, Carnero A, Beach D, Gil J. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2005;24:5543–5551. doi: 10.1038/sj.onc.1208735. [DOI] [PubMed] [Google Scholar]
- 207.Huang Z, Liu J, Yang J, Yan Y, Yang C, He X, Huang R, Tan M, Wu D, Yan J, et al. PDE4B Induces Epithelial-to-mesenchymal transition in bladder cancer cells and is transcriptionally suppressed by CBX7. Front Cell Dev Biol. 2021;9:783050. doi: 10.3389/fcell.2021.783050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jia Y, Wang Y, Zhang C, Chen MY. Upregulated CBX8 promotes cancer metastasis via the WNK2/MMP2 pathway. Mol Ther Oncolytics. 2020;19:188–196. doi: 10.1016/j.omto.2020.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wang G, Tang J, Zhan W, Zhang R, Zhang M, Liao D, Wang X, Wu Y, Kang T. CBX8 suppresses tumor metastasis via repressing snail in esophageal squamous cell carcinoma. Theranostics. 2017;7:3478–3488. doi: 10.7150/thno.20717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yang S, Liu W, Li M, Wen J, Zhu M, Xu S. Insulin-like growth Factor-1 modulates polycomb Cbx8 expression and inhibits colon cancer cell apoptosis. Cell Biochem Biophys. 2015;71:1503–1507. doi: 10.1007/s12013-014-0373-y. [DOI] [PubMed] [Google Scholar]
- 211.Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mao J, Tian Y, Wang C, Jiang K, Li R, Yao Y, Zhang R, Sun D, Liang R, Gao Z, et al. CBX2 regulates proliferation and apoptosis via the phosphorylation of YAP in hepatocellular carcinoma. J Cancer. 2019;10:2706–2719. doi: 10.7150/jca.31845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Peng W, Shi S, Zhong J, Liang H, Hou J, Hu X, Wang F, Zhang J, Geng S, Sun X, et al. CBX3 accelerates the malignant progression of glioblastoma multiforme by stabilizing EGFR expression. Oncogene. 2022;41:3051–3063. doi: 10.1038/s41388-022-02296-9. [DOI] [PubMed] [Google Scholar]
- 214.Li J, Xu Y, Long XD, Wang W, Jiao HK, Mei Z, Yin QQ, Ma LN, Zhou AW, Wang LS, et al. Cbx4 governs HIF-1α to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25:118–131. doi: 10.1016/j.ccr.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 215.Bawa-Khalfe T, Lu LS, Zuo Y, Huang C, Dere R, Lin FM, Yeh ET. Differential expression of SUMO-specific protease 7 variants regulates epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2012;109:17466–17471. doi: 10.1073/pnas.1209378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lin FM, Kumar S, Ren J, Karami S, Bahnassy S, Li Y, Zheng X, Wang J, Bawa-Khalfe T. SUMOylation of HP1α supports association with ncRNA to define responsiveness of breast cancer cells to chemotherapy. Oncotarget. 2016;7:30336–30349. doi: 10.18632/oncotarget.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Shiota M, Song Y, Yokomizo A, Tada Y, Kuroiwa K, Eto M, Oda Y, Inokuchi J, Uchiumi T, Fujimoto N, et al. Human heterochromatin protein 1 isoform HP1beta enhances androgen receptor activity and is implicated in prostate cancer growth. Endocr Relat Cancer. 2010;17:455–467. doi: 10.1677/ERC-09-0321. [DOI] [PubMed] [Google Scholar]
- 218.Hu FF, Chen H, Duan Y, Lan B, Liu CJ, Hu H, Dong X, Zhang Q, Cheng YM, Liu M, et al. CBX2 and EZH2 cooperatively promote the growth and metastasis of lung adenocarcinoma. Mol Ther Nucleic Acids. 2022;27:670–684. doi: 10.1016/j.omtn.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li J, Alvero AB, Nuti S, Tedja R, Roberts CM, Pitruzzello M, Li Y, Xiao Q, Zhang S, Gan Y, et al. CBX7 binds the E-box to inhibit TWIST-1 function and inhibit tumorigenicity and metastatic potential. Oncogene. 2020;39:3965–3979. doi: 10.1038/s41388-020-1269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Xiao L, Zhou Z, Li W, Peng J, Sun Q, Zhu H, Song Y, Hou JL, Sun J, Cao HC, et al. Chromobox homolog 8 (CBX8) Interacts with Y-Box binding protein 1 (YBX1) to promote cellular proliferation in hepatocellular carcinoma cells. Aging (Albany NY) 2019;11:7123–7149. doi: 10.18632/aging.102241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zeng F, Luo L, Li D, Guo J, Guo M. KPNA2 interaction with CBX8 contributes to the development and progression of bladder cancer by mediating the PRDM1/c-FOS pathway. J Transl Med. 2021;19:112. doi: 10.1186/s12967-021-02709-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bilton LJ, Warren C, Humphries RM, Kalsi S, Waters E, Francis T, Dobrowinski W, Beltran-Alvarez P, Wade MA. The epigenetic regulatory protein CBX2 promotes mTORC1 signalling and inhibits DREAM complex activity to drive breast cancer cell growth. Cancers (Basel) 2022;14:3491. doi: 10.3390/cancers14143491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zeng M, Li B, Yang L, Guan Q. CBX2 depletion inhibits the proliferation, invasion and migration of gastric cancer cells by inactivating the YAP/β-catenin pathway. Mol Med Rep. 2021;23:137. doi: 10.3892/mmr.2020.11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wang L, Ren B, Zhuang H, Zhong Y, Nan Y. CBX2 induces glioma cell proliferation and invasion through the Akt/PI3K pathway. Technol Cancer Res Treat. 2021;20:15330338211045831. doi: 10.1177/15330338211045831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jin X, Zhang B, Zhang H, Yu H. Smoking-associated upregulation of CBX3 suppresses ARHGAP24 expression to activate Rac1 signaling and promote tumor progression in lung adenocarcinoma. Oncogene. 2022;41:538–549. doi: 10.1038/s41388-021-02114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Nawaz Z, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Cbx7 is epigenetically silenced in glioblastoma and inhibits cell migration by targeting YAP/TAZ-dependent transcription. Sci Rep. 2016;6:27753. doi: 10.1038/srep27753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kim HY, Park JH, Won HY, Lee JY, Kong G. CBX7 inhibits breast tumorigenicity through DKK-1-mediated suppression of the Wnt/β-catenin pathway. FASEB J. 2015;29:300–313. doi: 10.1096/fj.14-253997. [DOI] [PubMed] [Google Scholar]
- 228.Ni S, Wang H, Zhu X, Wan C, Xu J, Lu C, Xiao L, He J, Jiang C, Wang W, et al. CBX7 suppresses cell proliferation, migration, and invasion through the inhibition of PTEN/Akt signaling in pancreatic cancer. Oncotarget. 2017;8:8010–8021. doi: 10.18632/oncotarget.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ni SJ, Zhao LQ, Wang XF, Wu ZH, Hua RX, Wan CH, Zhang JY, Zhang XW, Huang MZ, Gan L, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J Hematol Oncol. 2018;11:17. doi: 10.1186/s13045-018-0562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zhang XW, Zhang L, Qin W, Yao XH, Zheng LZ, Liu X, Li J, Guo WJ. Oncogenic role of the chromobox protein CBX7 in gastric cancer. J Exp Clin Cancer Res. 2010;29:114. doi: 10.1186/1756-9966-29-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chen LY, Cheng CS, Qu C, Wang P, Chen H, Meng ZQ, Chen Z. CBX3 promotes proliferation and regulates glycolysis via suppressing FBP1 in pancreatic cancer. Biochem Biophys Res Commun. 2018;500:691–697. doi: 10.1016/j.bbrc.2018.04.137. [DOI] [PubMed] [Google Scholar]
- 232.Di Costanzo A, Del Gaudio N, Conte L, Dell'Aversana C, Vermeulen M, de Thé H, Migliaccio A, Nebbioso A, Altucci L. The HDAC inhibitor SAHA regulates CBX2 stability via a SUMO-triggered ubiquitin-mediated pathway in leukemia. Oncogene. 2018;37:2559–2572. doi: 10.1038/s41388-018-0143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yi SA, Kim GW, Yoo J, Han JW, Kwon SH. HP1γ sensitizes cervical cancer cells to Cisplatin through the Suppression of UBE2L3. Int J Mol Sci. 2020;21:5976. doi: 10.3390/ijms21175976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Silva-Fisher JM, Dang HX, White NM, Strand MS, Krasnick BA, Rozycki EB, Jeffers G, Grossman JG, Highkin MK, Tang C, et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat Commun. 2020;11:2156. doi: 10.1038/s41467-020-15547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Oliva J, El Messaoudi S, Pellestor F, Fuentes M, Georget V, Balaguer P, Cavaillès V, Vignon F, Badia E. Involvement of HP1alpha protein in irreversible transcriptional inactivation by antiestrogens in breast cancer cells. FEBS Lett. 2005;579:4278–4286. doi: 10.1016/j.febslet.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 236.Chen L, Xia JS, Wu JH, Chen YG, Qiu CJ. Resveratrol inhibits oral squamous cell carcinoma cells proliferation while promoting apoptosis through inhibition of CBX7 protein. Environ Toxicol. 2020;35:1234–1240. doi: 10.1002/tox.22988. [DOI] [PubMed] [Google Scholar]
- 237.Cacciola NA, Sepe R, Forzati F, Federico A, Pellecchia S, Malapelle U, De Stefano A, Rocco D, Fusco A, Pallante P. Restoration of CBX7 expression increases the susceptibility of human lung carcinoma cells to irinotecan treatment. Naunyn Schmiedebergs Arch Pharmacol. 2015;388:1179–1186. doi: 10.1007/s00210-015-1153-y. [DOI] [PubMed] [Google Scholar]
- 238.Liu W, Wang H, Jian C, Li W, Ye K, Ren J, Zhu L, Wang Y, Jin X, Yi L. The RNF26/CBX7 axis modulates the TNF pathway to promote cell proliferation and regulate sensitivity to TKIs in ccRCC. Int J Biol Sci. 2022;18:2132–2145. doi: 10.7150/ijbs.69325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lee SH, Um SJ, Kim EJ. CBX8 antagonizes the effect of Sirtinol on premature senescence through the AKT-RB-E2F1 pathway in K562 leukemia cells. Biochem Biophys Res Commun. 2016;469:884–890. doi: 10.1016/j.bbrc.2015.12.070. [DOI] [PubMed] [Google Scholar]
- 240.Wang S, Alpsoy A, Sood S, Ordonez-Rubiano SC, Dhiman A, Sun Y, Jiao G, Krusemark CJ, Dykhuizen EC. A potent, selective CBX2 Chromodomain ligand and its cellular activity during prostate cancer neuroendocrine differentiation. Chembiochem. 2021;22:2335–2344. doi: 10.1002/cbic.202100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Stuckey JI, Dickson BM, Cheng N, Liu Y, Norris JL, Cholensky SH, Tempel W, Qin S, Huber KG, Sagum C, et al. A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1. Nat Chem Biol. 2016;12:180–187. doi: 10.1038/nchembio.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Liu H, Li Z, Li L. The molecular selectivity of UNC3866 inhibitor for Polycomb CBX7 protein from molecular dynamics simulation. Comput Biol Chem. 2018;74:339–346. doi: 10.1016/j.compbiolchem.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 243.Milosevich N, Gignac MC, McFarlane J, Simhadri C, Horvath S, Daze KD, Croft CS, Dheri A, Quon TT, Douglas SF, et al. Selective inhibition of CBX6: A methyllysine reader protein in the polycomb family. ACS Med Chem Lett. 2016;7:139–144. doi: 10.1021/acsmedchemlett.5b00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Milosevich N, Wilson CR, Brown TM, Alpsoy A, Wang S, Connelly KE, Sinclair K, Ponio FR, Hof R, Dykhuizen EC, et al. Polycomb Paralog Chromodomain inhibitors active against both CBX6 and CBX8*. ChemMedChem. 2021;16:3027–3034. doi: 10.1002/cmdc.202100262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ren C, Morohashi K, Plotnikov AN, Jakoncic J, Smith SG, Li J, Zeng L, Rodriguez Y, Stojanoff V, Walsh M, et al. Small-molecule modulators of methyl-lysine binding for the CBX7 chromodomain. Chem Biol. 2015;22:161–168. doi: 10.1016/j.chembiol.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Connelly KE, Martin EC, Dykhuizen EC. CBX Chromodomain inhibition enhances chemotherapy response in glioblastoma multiforme. Yale J Biol Med. 2016;89:431–440. [PMC free article] [PubMed] [Google Scholar]
- 247.Simhadri C, Gignac MC, Anderson CJ, Milosevich N, Dheri A, Prashar N, Flemmer RT, Dev A, Henderson TG, Douglas SF, et al. Structure-activity relationships of Cbx7 inhibitors, including selectivity studies against other Cbx proteins. ACS Omega. 2016;1:541–551. doi: 10.1021/acsomega.6b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ren C, Smith SG, Yap K, Li S, Li J, Mezei M, Rodriguez Y, Vincek A, Aguilo F, Walsh MJ, et al. Structure-guided discovery of selective antagonists for the chromodomain of polycomb repressive protein CBX7. ACS Med Chem Lett. 2016;7:601–605. doi: 10.1021/acsmedchemlett.6b00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lamb KN, Bsteh D, Dishman SN, Moussa HF, Fan H, Stuckey JI, Norris JL, Cholensky SH, Li D, Wang J, et al. Discovery and characterization of a cellular potent positive allosteric modulator of the polycomb repressive Complex 1 chromodomain, CBX7. Cell Chem Biol. 2019;26:1365–1379.e22. doi: 10.1016/j.chembiol.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Simhadri C, Daze KD, Douglas SF, Milosevich N, Monjas L, Dev A, Brown TM, Hirsch A, Wulff JE, Hof F. Rational adaptation of L3MBTL1 inhibitors to create small-molecule Cbx7 antagonists. ChemMedChem. 2019;14:1444–1456. doi: 10.1002/cmdc.201900021. [DOI] [PubMed] [Google Scholar]
- 251.Denton KE, Wang S, Gignac MC, Milosevich N, Hof F, Dykhuizen EC, Krusemark CJ. Robustness of in vitro selection assays of DNA-encoded peptidomimetic ligands to CBX7 and CBX8. SLAS Discov. 2018;23:417–428. doi: 10.1177/2472555217750871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Suh JL, Bsteh D, Hart B, Si Y, Weaver TM, Pribitzer C, Lau R, Soni S, Ogana H, Rectenwald JM, et al. Reprogramming CBX8-PRC1 function with a positive allosteric modulator. Cell Chem Biol. 2022;29:555–571.e11. doi: 10.1016/j.chembiol.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.