Abstract
Objectives
To assess the duration of viable virus shedding and polymerase chain reaction (PCR) positivity of the SARS-CoV-2 Omicron variant in the upper respiratory tract.
Methods
We systematically searched PubMed, Cochrane, and Web of Science for original articles reporting the duration of viable virus shedding and PCR positivity of the SARS-CoV-2 Omicron variant in the upper respiratory tract from November 11, 2021 to December 11, 2022. This meta-analysis was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines and was registered with PROSPERO (CRD42022357349). We used the DerSimonian-Laird random-effects meta-analyses to obtain the pooled value and the 95% confidence intervals.
Results
We included 29 studies and 230,227 patients. The pooled duration of viable virus shedding of the SARS-CoV-2 Omicron variant in the upper respiratory tract was 5.16 days (95% CI: 4.18-6.14), and the average duration of PCR positivity was 10.82 days (95% CI: 10.23-11.42). The duration of viable virus shedding and PCR positivity of the SARS-CoV-2 Omicron variant in symptomatic patients was slightly higher than that in asymptomatic patients, but the difference was not significant (P >0.05).
Conclusion
The current study improves our understanding of the status of the literature on the duration of viable virus shedding and PCR positivity of Omicron in the upper respiratory tract. Our findings have implications for pandemic control strategies and infection control measures.
Keywords: Viable virus shedding, PCR positivity, SARS-CoV-2 Omicron variant, Upper respiratory tract, Meta
Introduction
SARS-CoV-2 is a new coronavirus responsible for the COVID-19 pandemic. Over 546 and 6.33 million confirmed COVID-19 cases and deaths, respectively, had been reported to the World Health Organization as of July 4, 2022 [1]. Since the beginning of the pandemic, SARS-CoV-2 has continually evolved and mutated, producing variants with different transmissibility and virulence levels. Currently, the Omicron variant is the dominant strain of SARS-CoV-2. The first case of Omicron infection was reported to the World Health Organization by South Africa on November 24, 2021. Since then, the variant has spread rapidly to many countries and regions worldwide [2]. Omicron appears to have a shorter incubation period and series interval but stronger infectivity and immune evasion than Delta and other strains [3,4], which complicates the prevention and control of Omicron.
The duration of viral shedding is a key determinant of disease transmission. This indicates the duration of infectiousness, which is a crucial parameter essential for the effective control and modeling of diseases [5]. Reverse transcription-polymerase chain reaction (PCR) is the gold standard for the diagnosis and screening of COVID-19. However, reverse transcription PCR results may be persistently positive without necessarily indicating the presence of viable virus (e.g., infectious or replication-competent virus) and viral transmissibility [6]. In their systematic review and meta-analysis, Cevik et al. [5] indicated that the mean duration of SARS-CoV-2 PCR positivity was 17.0 days (95% confidence interval [CI]: 15.5-18.6; studies, 43; individuals, 3229) in the upper respiratory tract; they further revealed that no study reported a live virus beyond day 9 of illness.
The epidemiological characteristics of Omicron are different from those of the original strain and other variants of SARS-CoV-2. Few studies have focused on the duration of viable virus shedding and PCR positivity of the Omicron variant. Studies have reported inconsistent findings regarding the viral load dynamics and viral shedding duration. Therefore, we reviewed the literature available since the emergence of Omicron, assessed the duration of viable virus shedding and PCR positivity of this variant in the upper respiratory tract, and compared the duration of viable virus shedding and PCR positivity of the Omicron variant in different populations.
Methods
Search strategy
This meta-analysis was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guideline and was registered with PROSPERO (CRD42022357349). The literature (November 11, 2021 to December 11, 2022) was searched for relevant studies. Articles published before November 11, 2021 were excluded because the first confirmed case of the SARS-CoV-2 Omicron variant was reported on November 11, 2021. PubMed, Cochrane, and Web of Science were searched using the following keywords: “SARS-CoV-2” OR “2019-nCoV” OR “COVID-19” AND “virus” OR “viral shedding” OR “rna” OR “ribonucleic” (Supplementary Table S1). No restriction on language or publication status was imposed provided that an English abstract was available. Furthermore, we manually screened the references of the included original studies to obtain additional studies. The initial searches were performed by six investigators (Yu Wu, Zirui Guo, Jie Yuan, Yaping Wang, Guiying Cao, and Peng Gao).
Outcome measures and study selection
The following outcome variables were assessed: conversion time to negative viral culture and conversion time to negative PCR results. For both outcomes, we regarded the earliest date of symptom onset or first positive PCR test as the index date of observation. The first day after the last positive PCR or positive culture was regarded as the end date. The duration of viable virus shedding and PCR positivity was estimated in terms of the conversion time to negative viral culture and conversion time to negative PCR, respectively.
Studies on the Omicron infection that reported viral load kinetics, viral shedding duration, or viable virus shedding duration were included in the current study. We excluded review studies, animal studies, environmental sampling studies, studies lacking clear data on virus shedding and PCR positivity duration, and modeling studies with no original data.
The search results were screened in two stages. First, the titles and abstracts of the entries were screened, and only relevant articles were retained. Next, the articles were read in detail. The studies were selected for meta-analysis if they reported relevant parameters and CIs or sufficient information to facilitate the calculation of these values.
From the selected studies, the following data were extracted: the name of the first author, area of study, time period for data collection, characteristics of the study population, type of strain, duration of viable virus shedding or PCR positivity, and 95% CI values. Some studies reported only the median and interquartile range values; for such studies, we calculated the corresponding mean and SD values through an appropriate approximation to ensure consistency in synthesizing data for the meta-analysis [7].
Quality assessment
Two authors (Yu Wu, Zirui Guo) independently appraised the quality of the included studies. To assess the quality of observational studies, we used a scale modified from the Newcastle-Ottawa scale [8] by McAloon et al. [9] (Supplementary Table S2). This scale comprises two parts, with a total score of 5 stars. The first part is “external validity”, with a maximum score of 1 star. The second part is “internal validity”, which includes the “exposure window”, with a maximum score of 2 stars, and the “outcomes”, with a maximum score of 2 stars. By combing the scores of the included studies on each part, we divided the studies into three categories (≤1 star: weak; 2-3 stars: moderate; ≥4 stars: strong). Two investigators independently evaluated the studies; the results were compared, and the differences in ratings were resolved through discussion until a consensus was achieved.
Meta-analysis
A meta-analysis of continuous outcomes was performed. We analyzed the data obtained during the incubation period. After extracting all essential data into Excel (Microsoft Corporation, Redmond, WA, USA), Stata (version 14.1) was used for the meta-analysis. A random-effects (DerSimonian and Laird method) meta-analysis was performed. The pooled average estimates with 95% CIs were presented using forest plots. To determine the extent of variation across studies, we performed a heterogeneity test using the Higgins method, which was quantified in terms of I2 statistics.
Results
Search results
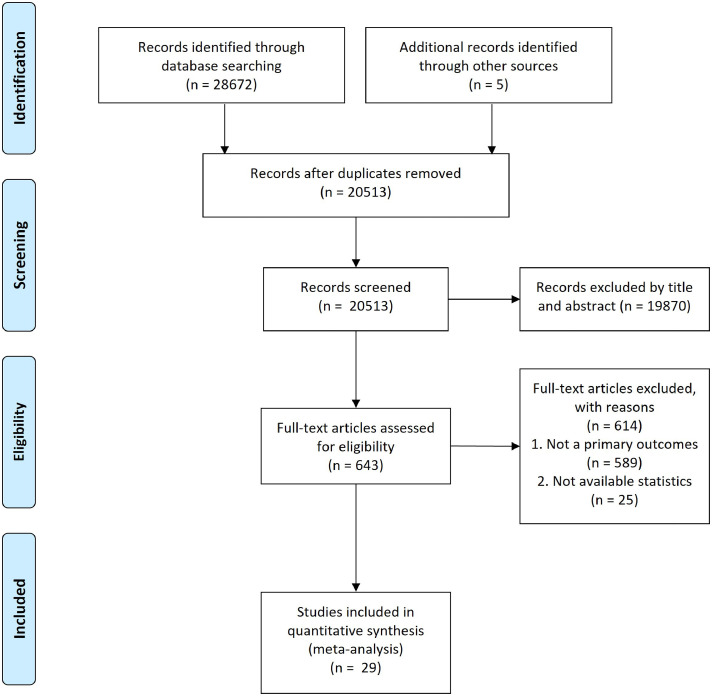
We identified 28,672 studies by searching the databases and the reference lists of relevant articles. Of these studies, 643 were subjected to full-text review. Finally, 29 studies (230,227 patients) containing information regarding the duration of viable virus shedding and PCR positivity of the Omicron variant were analyzed (Figure 1 ). Table 1 summarizes the characteristics of the studies included in the current systematic review and meta-analysis.
Figure 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses study flow diagram for search up to May 28, 2022.
Table 1.
Characteristics of the studies included in the systematic review and meta-analysis.
| Study | Area of study | Time period for data | Type of study | Detection method | Sample size |
|---|---|---|---|---|---|
| Boucau et al. [10] | US | 2021.07-2022.01 | Cohort study | Viral culture & RT-PCR | 19 |
| Bouton et al. [11] | US | 2021.11-2022.04 | Cohort study | Viral culture | 85 |
| Chen et al. [12] | US | 2022.03.20-2022.05.10 | Cohort study | RT-PCR | 847 |
| Hay et al. [37] | US | 2021.07.05-2022.01.10 | Case series | RT-PCR | 97 |
| Hua et al. [13] | China | 2022.07.11-2022.07.26 | Cohort study | RT-PCR | 225 |
| Jang et al. [14] | Korea | 2021.12 | Case series | Viral culture | 9 |
| Jung et al. [15] | Korea | 2022.03.14-2022.04.03 | Cohort study | Viral culture & RT-PCR | 32 |
| Kang et al. [16] | Korea | 2021.11-2022.05 | Cohort study | Viral culture & RT-PCR | 34 |
| Keske et al. [17] | Turkey | 2022.01.08-2022.02.17 | Case series | Viral culture & RT-PCR | 55 |
| Kim et al. [18] | Korea | 2022.01-2022.03 | Cohort study | Viral culture | 37 |
| Kojima et al. [19] | US | 2021.12 | Case series | RT-PCR | 734 |
| Lin et al. [38] | US | 2022.01 | Cohort study | RT-PCR | 3 |
| Lu et al. [20] | China | 2022.04-2022.05 | Cohort study | RT-PCR | 1377 |
| Luna-Muschi et al. [21] | Brazil | 2022.01.11-2022.01.24 | Case series | Viral culture & RT-PCR | 30 |
| Ma et al. [22] | China | By 2022.06.16 | Cohort study | RT-PCR | 11 |
| Mack et al. [23] | US | 2021.12.14-2021.12.19 | Case series | RT-PCR | 173 |
| Okumura et al. [24] | Japan | 2021.11-2021.12 | Case series | RT-PCR | 11 |
| Pei et al. [25] | China | 2022.04.05-2022.05.08 | Cohort study | RT-PCR | 25168 |
| Saade et al. [26] | France | 2021.11-2022.02 | Case series | Viral culture | 44 |
| Shen et al. [27] | China | 2022.03.08-2022.03.24 | Randomized controlled trial | RT-PCR | 76 |
| Takahashi et al. [28] | Japan | 2021.11.29-2021.12.18 | Case series | Viral culture | 10 |
| Tassetto et al. [29] | US | 2021.07-2022.03 | Cohort study | Viral culture | 39 |
| Tillmann et al. [30] | Germany | - | Cohort study | RT-PCR | 20 |
| Wang et al. [31] | China | By 2022.03.31 | Case series | RT-PCR | 376 |
| Xu et al. [32] | China | 2022.04.16-2022.05.05 | Cohort study | RT-PCR | 458 |
| Yin et al. [33] | China | 2022.03.26-2022.05.20 | Case series | RT-PCR | 199590 |
| Yu et al. [34] | China | 2022.04.05-2022.04.29 | Cohort study | RT-PCR | 331 |
| Zeng et al. [35] | China | 2022.01.08-2022.01.29 | Cohort study | RT-PCR | 380 |
| Zhong et al. [36] | China | 2022.04.24-2022.05.28 | Non-randomized clinical trial | RT-PCR | 36 |
RT-PCR, reverse transcription-polymerase chain reaction; US, United States.
Among the 29 studies, 12, 11, and six exhibited strong, moderate, and weak power, respectively (Supplementary Table S3). A total of 27 were published articles [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], and two were preprints [37,38]. All 29 studies reported the duration of either viable virus shedding (11 studies [10,11,[14], [15], [16], [17], [18],21,26,28,29]) or PCR positivity (23 studies [10,12,13,[15], [16], [17],[19], [20], [21], [22], [23], [24], [25],27,[30], [31], [32], [33], [34], [35], [36], [37], [38]]) of the Omicron variant in the upper respiratory tract. Of the included studies, seven were conducted in the United States, [10,11,19,23,29,37,38], 12 in China [12,13,20,22,25,27,[31], [32], [33], [34], [35], [36]], two in Japan [24,28], and three in Korea [14,16,18]. A total of 11 studies were case series [14,17,19,21,23,24,26,28,31,33,37], 16 were cohort studies [[10], [11], [12], [13],15,16,18,20,22,25,29,30,32,34,35,38], one was a randomized controlled trial [27], and one was a nonrandomized clinical trial [36].
Duration of viable virus shedding
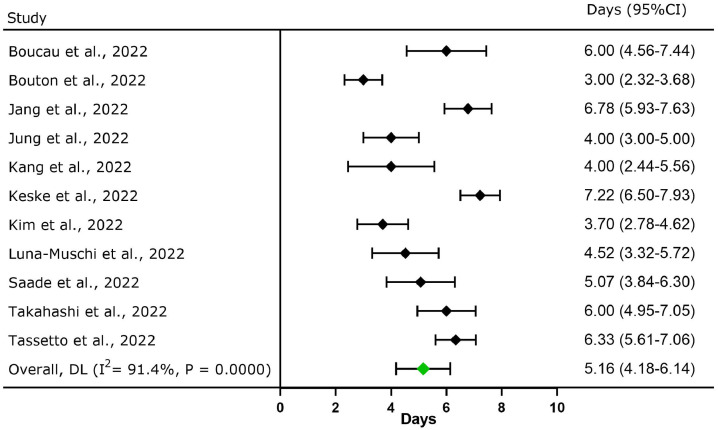
A total of 11 studies (n = 384) reported the duration of viable virus shedding of Omicron; the pooled viable virus shedding duration was 5.16 days (95% CI: 4.18-6.14; Figure 2 ). The maximum duration of viable virus shedding was 15 days in the upper respiratory tract [11].
Figure 2.
Forest plot for the meta-analysis of viable virus shedding duration of the SARS-CoV-2 Omicron variant in upper respiratory tract. CI, confidence interval; DL, DerSimonian and Laird method.
Boucau et al. [10] revealed that >50% of all patients carried replication-competent, culturable virus on day 5 and 25% of all patients had culturable virus on day 8. Takahashi et al. [28] detected an infectious virus in 10 patients with Omicron infection, and the highest proportion (41.7%) of virus isolates was detected in samples collected 2-5 days after diagnosis. Jang et al. [14] reported that the rate of Omicron culture positivity was 22-100% in unvaccinated patients within 0-8 days after symptom onset and the rate of viral culture positivity on day 5 was the highest (100%). The preliminary data obtained from the National Institute of Infectious Diseases [39], which conducts disease surveillance in Japan, indicated that 41.2% of all patients with Omicron infection had a culturable virus 3-6 days after diagnosis, and no infectious virus was detected in the respiratory samples collected 10 days after diagnosis or symptom onset. Keske et al. [17] revealed that in 19% of all patients, the duration of shedding was longer than that of symptoms. On day 5, four patients reported the absence of symptoms; although, the viral culture was positive.
Duration of PCR positivity
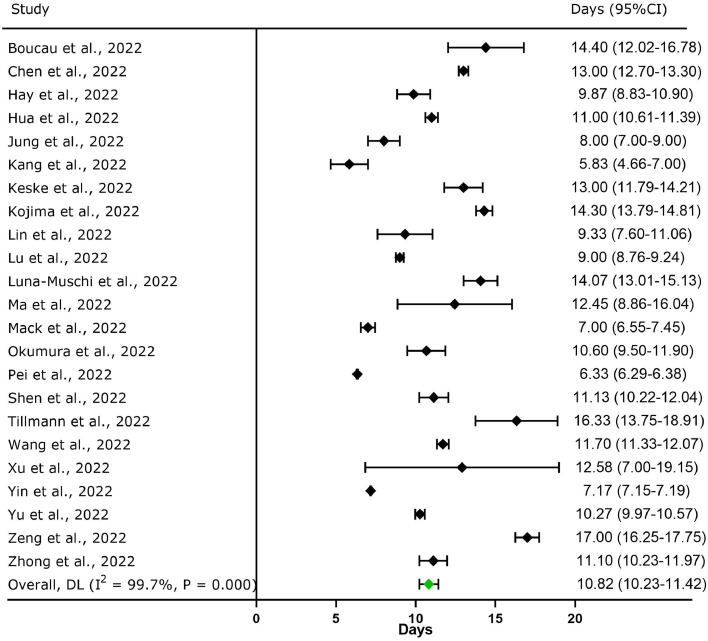
A total of 23 studies (n = 230,013) reported the duration of PCR positivity of Omicron; the pooled PCR positivity duration was 10.82 days (95% CI: 10.23-11.42; Figure 3 ). The maximum duration of PCR positivity was 23 days in the upper respiratory tract [10].
Figure 3.
Forest plot for the meta-analysis of PCR positivity duration of the SARS-CoV-2 Omicron variant in upper respiratory tract. CI, confidence interval; DL, DerSimonian and Laird method.
Boucau et al. [10] simultaneously reported the duration of viable virus shedding and PCR positivity of Omicron in the upper respiratory tract. The mean duration of PCR positivity was 14.40 days (95% CI: 12.02-16.78), which was higher than that of viable virus shedding (6.00 days; 95% CI: 4.56-7.44).
Takahashi et al. [28] reported that the highest amount of Omicron RNA was detected 2-5 days after diagnosis or symptom onset and then decreased gradually until it was markedly reduced 10 days after diagnosis or symptom onset. The lowest quantitation cycle (Cq) value (i.e., viral RNA levels) was 18.7-30. The period with the lowest Cq value was that from 1 day before symptoms onset to 5 days after it. At least three patients were infectious during the incubation period.
Subgroup analysis
Duration of viable virus shedding and PCR positivity of Omicron in symptomatic and asymptomatic patients
Two studies reported the duration of PCR positivity in symptomatic and asymptomatic patients [27,31]. Shen et al. [27] indicated that the mean duration of PCR positivity was slightly higher in symptomatic patients than in asymptomatic patients; however, the difference was nonsignificant (P >0.05). By contrast, Wang et al. [31] revealed no difference between symptomatic and asymptomatic patients; both were 11.70 days (P = 0.064, Table 2 ).
Table 2.
Duration of viable virus shedding and PCR positivity of the SARS-CoV-2 Omicron variant in patients with different characteristics.
| Study | Outcome variable | Characteristics of patients | Sample Size | Mean duration (95% confidence interval) |
|---|---|---|---|---|
| Shen et al [27] | Duration of PCR positivity | Symptomatic | 39 | 12.25 (10.99-13.51) |
| Asymptomatic | 37 | 9.95 (8.69-11.20) | ||
| Wang et al [31] | Duration of PCR positivity | Symptomatic | 257 | 11.70 (11.26-12.14) |
| Asymptomatic | 119 | 11.70(11.00-12.40) | ||
| Takahashi et al [28] | Duration of viable virus shedding | Symptomatic | 8 | 6.25(4.98-7.52) |
| Asymptomatic | 2 | 5.00(5.00-5.00) | ||
| Bouton et al [11] | Duration of viable virus shedding | Fully vaccinated by two doses | 44 | 3.00(2.09-3.91) |
| Vaccinated with a third booster dose | 41 | 2.67(1.96-3.37) | ||
| Shen et al [27] | Duration of PCR positivity | Unvaccinated | 12 | 13.05 (11.34-14.76) |
| Fully vaccinated by two doses | 36 | 10.46 (9.07-11.85) | ||
| Vaccinated with a third booster dose | 27 | 11.13 (9.48-12.79) | ||
| Zeng et al. [35] | Duration of PCR positivity | Full inactivated vaccination | 355 | 17.00 (16.23-17.77) |
| Full recombinant vaccination | 14 | 21.53 (17.87-25.20) | ||
| Partial vaccination | 11 | 16.67 (8.65-24.69) | ||
| Ma et al. [22] | Duration of PCR positivity | Unvaccinated | 3 | 11.05 (5.19-16.90) |
| Vaccinated | 11 | 11.67 (7.16-16.18) | ||
| Hua et al. [13] | Duration of PCR positivity | Unvaccinated | 22 | 10.67 (9.01-12.32) |
| Fully vaccinated | 64 | 10.33 (9.78-10.89) | ||
| Booster vaccination | 139 | 11.00 (10.50-11.50) | ||
| Chen et al. [12] | Duration of PCR positivity | Fully vaccinated or booster | 339 | 10.00 (9.52-10.48) |
| Not fully vaccinated | 268 | 14.33 (13.71-14.96) |
PCR, polymerase chain reaction.
Only Takahashi et al. [28] reported the duration of viable virus shedding in symptomatic and asymptomatic patients. They detected the infectious virus in 10 patients (symptomatic, eight; asymptomatic, two) and found that the duration of viable virus shedding was similar between symptomatic and asymptomatic patients (6.25 vs 5.00 days, respectively; P >0.05). Two symptomatic patients shed the live virus before symptom onset (i.e., incubation period).
Duration of viable virus shedding and PCR positivity of the Omicron in patients with different vaccination statuses
Six studies compared the duration of viable virus shedding and PCR positivity of Omicron across patients with different vaccination statuses [[11], [12], [13],22,27,35]. Bouton et al. [11] revealed that the mean duration of viable virus shedding was 3.00 days (95% CI: 2.09-3.91) for fully vaccinated (two doses) patients and 2.67 days (95% CI: 1.96-3.37) for booster-vaccinated (third dose) patients.
Chen et al. [12] indicated that the full or booster vaccination shortened the duration of PCR positivity (adjusted hazard ratio = 1.40, P = 0.001). Shen et al. [27] reported that the mean duration of PCR positivity in unvaccinated, fully vaccinated, and booster-vaccinated patients were 13.05, 10.46, and 11.13 days, respectively; however, the difference was nonsignificant (P >0.05). As shown in Table 2, the findings reported by Hua et al. [13] were similar to those reported by Shen et al. [27].
Duration of viable virus shedding and PCR positivity of SARS-CoV-2 in patients with Omicron infection and Delta infection
Boucau et al. [10] compared the duration of viable virus shedding and PCR positivity between patients with the Omicron infection and those with the Delta infection. They found no difference between the two groups in the duration of viable virus shedding (both 6.00 days) or PCR positivity (Omicron: 12.00; Delta: 12.67 days; P >0.05).
Kang et al. [16] revealed that Omicron exhibited a shorter duration of PCR positivity (genomic and subgenomic RNA) and viable virus shedding than Delta (P <0.01, Table 3 ). Hay et al. [37] reported that the mean duration of PCR positivity was 9.87 days for Omicron (95% CI: 8.83-10.9) compared with 10.9 days (95% CI: 9.41-12.4) for Delta (Table 3). The peak viral RNA load (cycle threshold [Ct] value) was lower for Omicron than for Delta (Omicron: Ct, 23.3; 95% CI: 22.4-24.3; Delta: Ct, 20.5; 95% CI: 19.2-21.8).
Table 3.
Duration of viable virus shedding and PCR positivity of the SARS-CoV-2 Omicron variant in patients with Omicron variant and Delta variant.
| Study | Outcome variable | Variant | Sample size | Mean duration (95%CI) |
|---|---|---|---|---|
| Boucau et al. [10] | Duration of viable virus shedding | Omicron | 19 | 6.00 (5.10-6.90) |
| Delta | 37 | 6.00 (5.68-6.32) | ||
| Duration of PCR positivity | Omicron | 19 | 12.00 (10.20-13.80) | |
| Delta | 37 | 12.67 (11.70-13.63) | ||
| Bouton et al. [11] | Duration of viable virus shedding | Omicron | 75 | 3.00 (2.32-3.68) |
| Delta | 16 | 3.67 (1.28-6.06) | ||
| Hay et al. [37] | Duration of PCR positivity | Omicron | 107 | 9.87 (8.83-10.90) |
| Delta | 97 | 10.90 (9.41-12.40) | ||
| Kang et al. [16] | Duration of viable virus shedding | Omicron | 34 | 4.00 (2.44-5.56) |
| Delta | 48 | 8.17 (6.44-9.90) | ||
| Duration of PCR positivity | Omicron | 34 | 9.00 (8.04-9.96) | |
| Delta | 48 | 20.33 (18.39-22.28) | ||
| Hua et al. [13] | Duration of PCR positivity | Omicron | 225 | 11.00 (10.61-11.39) |
| Delta | 326 | 16.50 (15.61-17.39) |
PCR, polymerase chain reaction.
Discussion
We reviewed the viral dynamics of Omicron, including the duration of viable virus shedding and PCR positivity. The mean duration of viable virus shedding and PCR positivity were 5.16 (95% CI: 4.18-6.14) and 10.82 (95% CI: 10.23-11.42) days, respectively.
Few studies have been conducted on the viable virus shedding of the Omicron variant. Because of the difficulty in obtaining information on the duration of viable virus shedding, most studies have assessed the duration of PCR positivity (i.e., the time of viral RNA shedding). Cevik et al. [5] reviewed and meta-analyzed 43 studies (3229 individuals) published until June 2020. They stated that the mean duration of SARS-CoV-2 RNA shedding was 17.0 days (95% CI: 15.5-18.6) in the upper respiratory tract. Fontana et al. [40] reviewed 28 studies published through September 8, 2020; the overall pooled median duration of RNA shedding from respiratory sources was 18.4 days (95% CI: 15.5-21.3; I2 = 98.87%; P <0.01). Viable virus was isolated (culture) between -6 and 20 days from symptom onset. The aforementioned studies focused mainly on the original strain of SARS-CoV-2, and the duration of PCR positivity was higher in these studies than in the current study. Omicron has a shorter incubation period [41] and a higher transmission rate [42] than the previously prevalent SARS-CoV-2 variants. Because of the shortened duration of viable virus shedding of Omicron, recent public health guidelines in several countries have recommended withdrawing isolation-related restrictions for asymptomatic patients who test positive for COVID-19 and shortening the quarantine time for the patients’ close contacts.
In this study, the duration of PCR positivity of Omicron in the upper respiratory tract was considerably longer than that of viable virus shedding. Although Omicron may exhibit a long duration of PCR positivity (approximately 23 days), most studies reported no live virus isolation beyond day 10 after symptom onset. Cevik et al. [5], however, indicated that patients with SARS-CoV-2 infection have prolonged RNA shedding for approximately 83 days; yet, no live virus was isolated beyond day 9 after symptom onset, despite the persistently high loads of viral RNA. Fontana et al. [40] found that the duration of RNA shedding of SARS-CoV-2 exceeded that of viable virus shedding (45 vs 13 days, respectively). These findings indicated that, in clinical practice, repeat testing might not be indicated to deem patients no longer infectious. Prevention and control strategies should be adjusted according to the duration of viable virus shedding.
Patients with SARS-CoV-2 infection are infectious during the incubation period. In their study (February 24, 2020 to April 2, 2021), Jones et al. [43] examined 415,935 patients with COVID-19 in and around Berlin, Germany to evaluate the viral load over the disease course. The viral load peaked 1-3 days before symptom onset, indicating that the virus shedding in patients with high infectivity could begin a few days before the onset of symptoms. Takahashi et al. [28] demonstrated that at least three of 18 patients were infectious during the incubation period. Studies on viable virus shedding and viral dynamics in asymptomatic and symptomatic patients with Omicron infection are scarce, thus necessitating further studies.
In the current study, the duration of viable virus shedding and PCR positivity was slightly higher in symptomatic patients than in asymptomatic patients; although, the difference was nonsignificant (P >0.05). Studies have indicated that viral loads are similar between asymptomatic and symptomatic patients with SARS-CoV-2 infection, and viral load is the predominant predictive factor for virus viability in asymptomatic carriers [5,6]. Nevertheless, most studies have reported faster viral clearance in asymptomatic patients than in symptomatic patients [44]. This finding is consistent with the viral kinetics observed for other respiratory viruses, such as influenza and Middle East respiratory syndrome-related coronavirus, in which patients with asymptomatic infection have a shorter duration of viral shedding than those with symptomatic infection [5]. In the study conducted by Yan et al. [44], the mean duration of viral shedding in symptomatic patients was 19.7 days (95% CI: 17.2-22.7), which was significantly longer than that in asymptomatic infections (10.9 days; 95% CI: 8.3-14.3; P <0.05). One reason for this is that viral clearance is faster in asymptomatic patients than in symptomatic patients [45,46]. Another reason may be that the duration of viral shedding in asymptomatic patients was calculated from the first positive PCR result and was dependent primarily on close contact tracking. Viral shedding in these individuals might have begun before the first positive PCR result and may have been missed because of the absence of clinical symptoms.
Although we found no difference among unvaccinated, fully vaccinated, and booster-vaccinated patients in terms of the duration of viable virus shedding, complete vaccination with inactivated vaccines has been reported to promote early viral clearance. The magnitude of protection against prolonged viral shedding may be correlated with vaccine-induced antiviral immunity [47].
To the best of our knowledge, this study, a meta-analysis of the duration of viable virus shedding and PCR positivity of Omicron in the upper respiratory tract, is the first to compare the aforementioned parameters among symptomatic and asymptomatic patients, vaccinated and unvaccinated patients, and patients with Omicron infection and those with Delta infection. Our findings may facilitate public health-related policy making for the prevention and control of COVID-19.
Our study has some limitations. First, almost all patients assessed in the included studies received various treatments, which might have modified the shedding dynamics. Second, many studies provided no information on the daily virus culture and PCR test results of the patients, which may have affected the measurement accuracy of the duration of viable virus shedding and PCR positivity. Finally, few studies have evaluated the duration of viable virus shedding of Omicron. The duration of viable virus shedding was estimated in a limited number of patients within a short period, which limits the generalizability of our findings. Furthermore, most studies reported the shedding duration in terms of median and interquartile range values, but the meta-analysis necessitated their conversion to mean and SD values. The validity of this conversion was based on the assumption that the data corresponding to the duration of viral shedding exhibited a normal distribution, which might not have been true for some studies.
Conclusion
The current study improves our understanding of the status of the literature on the duration of viable virus shedding and PCR positivity of Omicron in the upper respiratory tract. Our findings have implications for pandemic control strategies and infection control measures. The mean durations of viable virus shedding and PCR positivity were 5.16 and 10.82 days, respectively. Although Omicron RNA shedding from respiratory samples can be prolonged, the duration of viable virus is relatively short. The current study may provide important insights for the policy makers engaged in making public health-related policies.
Funding
This study was supported by the grant from the National Key Research and Development Project of China (2021ZD0114104, 2021ZD0114105, 2021ZD0114101) and the National Natural Science Foundation of China (71934002, 72122001).
Ethical approval
This study did not involve individual patient information; so, there was no requirement for written informed consent.
Data sharing
No additional data are available.
Access to Data statement
Min Liu and Yu Wu had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
CRediT authorship contribution statement
Yu Wu: Conceptualization, Writing – original draft, Data curation, Formal analysis. Zirui Guo: Data curation, Formal analysis. Jie Yuan: Data curation, Formal analysis. Guiying Cao: Data curation, Formal analysis. Yaping Wang: Data curation, Formal analysis. Peng Gao: Data curation, Formal analysis. Jue Liu: Conceptualization, Writing – original draft, Formal analysis. Min Liu: Conceptualization, Writing – original draft, Formal analysis.
Declaration of competing interest
The authors have no competing interests to declare.
Acknowledgments
This manuscript was edited by Wallace Academic Editing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2023.02.011.
Appendix. Supplementary materials
References
- 1.World Health Organization. Coronavirus disease (COVID-19) pandemic, https://www.who.int/emergencies/diseases/novel-coronavirus-2019; 2022 [accessed 04 July 2022].
- 2.World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern, https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern); 2021 [accessed 26 November 2021].
- 3.Wu Y, Liu J, Liu M, Liang W. Epidemiologic features and containment of SARS-CoV-2 omicron variant. Chin Gen Pract. 2022;25:14–19. https://kns.cnki.net/kcms/detail/13.1222.R.20211215.0857.002.html [Google Scholar]
- 4.Chen J, Wang R, Gilby NB, Wei GW. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022;62:412–422. doi: 10.1021/acs.jcim.1c01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murata T, Sakurai A, Suzuki M, et al. Shedding of viable virus in asymptomatic SARS-CoV-2 carriers. mSphere. 2021;6:e00019–e00021. doi: 10.1128/mSphere.00019-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells G, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Department of Epidemiology and Commuunity Medicine, University of Ottawa; 2000. [Google Scholar]; https://www.ohri.ca//programs/clinical_epidemiology/oxford.asp [accessed 09 August 2021].
- 9.McAloon C, Collins Á, Hunt K, et al. Incubation period of COVID-19: a rapid systematic review and meta-analysis of observational research. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-039652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucau J, Marino C, Regan J, et al. Duration of shedding of culturable virus in SARS-CoV-2 omicron (BA.1) infection. N Engl J Med. 2022;387:275–277. doi: 10.1056/NEJMc2202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouton TC, Atarere J, Turcinovic J, et al. Viral dynamics of Omicron and Delta SARS-CoV-2 variants with implications for timing of release from isolation: a longitudinal cohort study. Clin Infect Dis. 2022 doi: 10.1093/cid/ciac510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Wang H, Ai J, et al. Identification of CKD, bedridden history and cancer as higher-risk comorbidities and their impact on prognosis of hospitalized Omicron patients: a multi-centre cohort study. Emerg Microbes Infect. 2022;11:2501–2509. doi: 10.1080/22221751.2022.2122581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua Q, Zheng D, Yu B, et al. Effectiveness of inactivated COVID-19 vaccines against COVID-19 caused by the SARS-CoV-2 Delta and omicron variants: a retrospective cohort study. Vaccines (Basel) 2022;10:1753. doi: 10.3390/vaccines10101753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang YR, Jeong-Min K, Rhee JE, et al. Clinical features and duration of viral shedding individuals with SARS-CoV-2 omicron variant infection. Open Forum Infect Dis. 2022;9 doi: 10.1093/ofid/ofac237. ofac237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung J, Kang SW, Lee S, et al. Risk of transmission of COVID-19 from healthcare workers returning to work after a 5-day isolation, and kinetics of shedding of viable SARS-CoV-2 variant B.1.1.529 (Omicron) J Hosp Infect. 2023;131:228–233. doi: 10.1016/j.jhin.2022.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang SW, Kim JY, Park H, et al. Comparison of secondary attack rate and viable virus shedding between patients with SARS-CoV-2 Delta and omicron variants: a prospective cohort study. J Med Virol. 2023;95:e28369. doi: 10.1002/jmv.28369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keske Ş, Güney-Esken G, Vatansever C, et al. Duration of infectious shedding of SARS-CoV-2 Omicron variant and its relation with symptoms. Clin Microbiol Infect. 2023;29:221–224. doi: 10.1016/j.cmi.2022.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H, Yang JS, Ko JH, et al. Can nirmatrelvir/ritonavir treatment shorten the duration of COVID-19 isolation? Front Med (Lausanne) 2022;9 doi: 10.3389/fmed.2022.988559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima N, Roshani A, Klausner JD. Duration of COVID-19 PCR positivity for Omicron vs earlier variants. J Clin Virol Plus. 2022;2 doi: 10.1016/j.jcvp.2022.100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu G, Zhang Y, Zhang H, et al. Geriatric risk and protective factors for serious COVID-19 outcomes among older adults in Shanghai Omicron wave. Emerg Microbes Infect. 2022;11:2045–2054. doi: 10.1080/22221751.2022.2109517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luna-Muschi A, Noguera SV, Borges IC, et al. Characterization of severe acute respiratory syndrome coronavirus 2 omicron variant shedding and predictors of viral culture positivity on vaccinated healthcare workers with mild coronavirus disease 2019. J Infect Dis. 2022;226:1726–1730. doi: 10.1093/infdis/jiac391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma E, Ai J, Zhang Y, et al. Omicron infections profile and vaccination status among 1881 liver transplant recipients: a multi-centre retrospective cohort. Emerg Microbes Infect. 2022;11:2636–2644. doi: 10.1080/22221751.2022.2136535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack CD, Wasserman EB, Killerby ME, et al. Results from a test-to-release from isolation strategy among fully vaccinated National Football League players and staff members with COVID-19 - United States, December 14–19, 2021. MMWR Morb Mortal Wkly Rep. 2022;71:299–305. doi: 10.15585/mmwr.mm7108a4. [DOI] [PubMed] [Google Scholar]
- 24.Okumura N, Tsuzuki S, Saito S, et al. The first eleven cases of SARS-CoV-2 Omicron variant infection in Japan: a focus on viral dynamics. Glob Health Med. 2022;4:133–136. doi: 10.35772/ghm.2021.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ying-Hao P, Yuan-Yuan G, Hai-Dong Z, Qiu-Hua C, Xue-Ran G, Hai-Qi Z, Hua J. Clinical characteristics and analysis of risk factors for disease progression of patients with SARS-CoV-2 Omicron variant infection: a retrospective study of 25207 cases in a Fangcang hospital. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.1009894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saade C, Brengel-Pesce K, Gaymard A, et al. Dynamics of viral shedding during ancestral or Omicron BA.1 SARS-CoV-2 infection and enhancement of pre-existing immunity during breakthrough infections. Emerg Microbes Infect. 2022;11:2423–2432. doi: 10.1080/22221751.2022.2122578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen Y, Ai J, Lin N, et al. An open, prospective cohort study of VV116 in Chinese participants infected with SARS-CoV-2 omicron variants. Emerg Microbes Infect. 2022;11:1518–1523. doi: 10.1080/22221751.2022.2078230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi K, Ishikane M, Ujiie M, et al. Duration of infectious virus shedding by SARS-CoV-2 omicron variant-infected vaccinees. Emerg Infect Dis. 2022;28:998–1001. doi: 10.3201/eid2805.220197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tassetto M, Garcia-Knight M, Anglin K, et al. Detection of higher cycle threshold values in culturable SARS-CoV-2 omicron BA.1 sublineage compared with pre-Omicron variant specimens – San Francisco Bay Area, California, July 2021-March 2022. MMWR Morb Mortal Wkly Rep. 2022;71:1151–1154. doi: 10.15585/mmwr.mm7136a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tillmann FP, Figiel L, Ricken J, et al. Effect of third and fourth mRNA-based booster vaccinations on SARS-CoV-2 neutralizing antibody titer formation, risk factors for non-response, and outcome after SARS-CoV-2 omicron breakthrough infections in patients on chronic hemodialysis: a prospective multicenter cohort study. J Clin Med. 2022;11:3187. doi: 10.3390/jcm11113187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Chang H, Tian H, et al. Epidemiological and clinical features of SARS-CoV-2 Infection in children during the outbreak of Omicron Variant in Shanghai, March 7-31, 2022. Influenza Other Respir Viruses. 2022;16:1059–1065. doi: 10.1111/irv.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu N, Pan J, Sun L, et al. Interferon α-2b spray shortened viral shedding time of SARS-CoV-2 Omicron variant: an open prospective cohort study. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.967716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin Y, Lin J, Yuan S, et al. The relationship between early isolation and the duration of viral shedding of mild and asymptomatic infection with SARS-CoV-2 Omicron BA.2 variant. J Infect. 2022;85:e184–e186. doi: 10.1016/j.jinf.2022.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu SY, Xie JR, Luo JJ, et al. Liver test abnormalities in asymptomatic and mild COVID-19 patients and their association with viral shedding time. World J Hepatol. 2022;14:1953–1963. doi: 10.4254/wjh.v14.i11.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng QL, Lv YJ, Liu XJ, et al. Clinical characteristics of omicron SARS-CoV-2 variant infection after Non-mRNA-Based vaccination in China. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.901826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong W, Jiang X, Yang X, et al. The efficacy of Paxlovid in elderly patients infected with SARS-CoV-2 omicron variants: results of a non-randomized clinical trial. Front Med (Lausanne) 2022;9 doi: 10.3389/fmed.2022.980002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay JA, Kissler SM, Fauver JR, et al. Viral dynamics and duration of PCR positivity of the SARS-CoV-2 Omicron variant. medRxiv. 2022:22269257. doi: 10.1101/2022.01.13.22269257. [DOI] [Google Scholar]
- 38.Lin J, Frediani JK, Damhorst GL, et al. Where is omicron? Comparison of SARS-CoV-2 RT-PCR and antigen test sensitivity at commonly sampled anatomic sites over the course of disease. Medrxiv. 2022 https://www.medrxiv.org/content/10.1101/2022.02.08.22270685v1 09 February. [accessed 09 February 2022] [Google Scholar]
- 39.National Institute of Infectious Diseases disease control and prevention center. Active epidemiological investigation on SARS-CoV-2 infection caused by Omicron variant (Pango lineage B.1.1.529) in Japan: preliminary report on infectious period, https://www.niid.go.jp/niid/en/2019-ncov-e/10884-covid19-66-en.html; 2022 [accessed 05 January 2022].
- 40.Fontana LM, Villamagna AH, Sikka MK, McGregor JC. Understanding viral shedding of severe acute respiratory coronavirus virus 2 (SARS-CoV-2): review of current literature. Infect Control Hosp Epidemiol. 2021;42:659–668. doi: 10.1017/ice.2020.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Kang L, Guo Z, Liu J, Liu M, Liang W. Incubation period of COVID-19 caused by unique SARS-CoV-2 strains: a systematic review and meta-analysis. JAMA Netw Open. 2022;5 doi: 10.1001/jamanetworkopen.2022.28008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Rocklöv J. The effective reproductive number of the Omicron variant of SARS-CoV-2 is several times relative to Delta. J Travel Med. 2022;29 doi: 10.1093/jtm/taac037. taac037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones TC, Biele G, Mühlemann B, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021;373:eabi5273. doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan D, Zhang X, Chen C, et al. Characteristics of viral shedding time in SARS-CoV-2 infections: a systematic review and meta-analysis. Front Public Health. 2021;9 doi: 10.3389/fpubh.2021.652842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Y, Li Y, Deng W, et al. Symptomatic infection is associated with prolonged duration of viral shedding in mild coronavirus disease 2019: a retrospective study of 110 children in wuhan. Pediatr Infect Dis J. 2020;39:e95–e99. doi: 10.1097/INF.0000000000002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao T, Wang Y, Yuan J, et al. Early viral clearance and antibody kinetics of COVID-19 among asymptomatic carriers. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.595773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Z, Yin Y, Wang K, et al. Impact of inactivated COVID-19 vaccines on viral shedding in B.1.617.2 (Delta) variant-infected patients. Sci China Life Sci. 2022;65:2556–2559. doi: 10.1007/s11427-021-2115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.