Abstract
Systemic lupus erythematous (SLE) is a complex chronic autoimmune disease with difficult early treatment and accurate diagnosis. Circulating exosomes containing proteins, lipids and nucleic acids can be ideal diagnostic biomarkers and disease management strategies for SLE. Our aim was to examine the unique expression profiles of circulating exosomal miRNAs and proteins in patients with SLE patients.
Using RNA-sequencing and proteomic approaches, we compared the expression patterns of exosomal miRNAs and proteins in the plasma of SLE patients and healthy subjects, and discussed the underlying signaling network of circulating exosomes. We also summarize common molecules (miRNAs and proteins) and pathways shared by our plasma exosomes, as well as previously reported data (PBMC, T cells, B cells and plasma).
We identified groups of differentially expressed exosomal miRNAs and proteins in the plasma of SLE patients and healthy controls. We obtained consensus molecules (39 miRNAs, 14 proteins) and 21 signaling pathways that are common in our current study and previous reports.
Common molecules (miRNAs and proteins) and pathways shared by our plasma exosomes data and other circulating components data reported previously indicate their potential application in the clinical treatment and diagnosis of SLE disease.
Keywords: Systemic lupus erythematosus, Plasma, Exosomes, miRNAs, Proteins
1. Introduction
Systemic lupus erythematous (SLE) is a clinically heterogeneous disease worldwide, with a global prevalence of approximately 20–150 cases per 100,000 people [1]. Although the survival rates and longevity of SLE patients have improved in recent decades, SLE patients still suffer from both high medical costs of disease management [2]. Therefore, more research is needed to investigate better SLE therapies.
Exosomes are tiny vesicles with a diameter of 50–100 nm. Circulating exosomes may be ideal non-invasive diagnostic biomarkers and represent an excellent disease management strategy for SLE disease management, as they can deliver information to activate target cells or tissues in remote locations [3,4].
Exosomes are rich in proteins, deoxyribonucleic acid and ribonucleic acid. MicroRNAs (miRNAs) are noncoding RNAs of approximately 19–25 nucleotides. Exosomal miRNAs can act as intercellular messengers [5], and their unique expression plays a crucial role in the pathology of SLE. For example, miRNAs (miR-29c [6], miR-26a [7] and miR-146a [8]) in urinary exosomes exhibited distinct differential expression profiles in SLE patients, which correlate significantly with disease activity [4]. Furthermore, one study found that circulating exosomal miR-21 and miR-155 potentially serve as diagnostic biomarkers for SLE and LN [5]. Protein may be a useful diagnostic auxiliary marker for SLE. Several specific and highly sensitive protein biomarkers are currently available for the diagnosis of SLE [9]. Some protein biomarkers summarized in the previous article, such as autoantibodies (ANA, anti-dsDNA), chemokines (CXCL12), complements (C1q, C3, C4), cytokines (IL-16, IL-2), CRP, VCAM-1 and urinary biomarkers (sVCAM-1, S100) [9]. However, most of the existing SLE protein marker studies have focused on peripheral blood mononuclear cells (PBMC), whole plasma (containing both exosomal and non-exosomal proteins) or urine [[9], [10], [11]]. We know very little about plasma exosome-specific proteins and more efforts are needed to fill this knowledge gap.
Therefore, from the above introduction, we understand that previous reports on circulating miRNAs and proteins in SLE have mainly focused on monocytes (different types of PBMCs), whole serum/plasma [10]. There is limited information on exosomal miRNAs, proteins and related signaling pathways in plasma.
In this project, we obtained the unique expression profiles and underlying regulatory networks of both exosomal miRNAs and proteins in the plasma of SLE patients. Our findings can help us understand the role of exosomes in SLE pathology and suggest possible applications of circulating exosomes in the clinical treatment and diagnosis of SLE disease.
2. Methods and materials
Data deposition: The raw data for small RNA sequencing and ribosomal RNA-depleted sequencing in our study have been uploaded to the Genome Sequence Archive (GSA) (Accession number: HRA000985) (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA000985). The protein data was uploaded to the supplemental data (Table S1).
2.1. Blood sample collection
Blood samples were collected from 20 healthy controls (H_M) and 10 SLE patients (SL_M) in Shenzhen People's Hospital (Guangdong, China). Recruitment criteria: SL_M patients were diagnosed with SLE disease, and H_M patients were not diagnosed with SLE or other immune diseases or treated with immunosuppressants. The SLE classification criteria and the characteristics of all patients are presented in Table 1. The study was approved by the ethics committee of Shenzhen People's Hospital (Medical ethical number: LL-KY-2019589), and the experiments were undertaken and carried out according to the guidelines of the Declaration of Helsinki of 1975 (as revised in 2013). In addition, informed consent letters were signed by all entrants. Plasma used for exosome isolation were separated by blood centrifugation.
Table 1.
Clinical data of participants.
| Characteristics | SLE(n = 10) | H_M(n = 20) |
|---|---|---|
| Age | 41.2 ± 15.23 | 37.80 ± 11.0 |
| SLEDAI score, median (range) | 8(5–20) | NA |
| proteinuria(g/24h), μ±σ | 1.248 ± 1.42 | NA |
| ESR (mm/h), μ±σ | 44 ± 23.18 | NA |
| CRP(mg/l), μ±σ | 14.2 ± 19.3 | NA |
| C3(g/l), μ±σ | 0.73 ± 0.394 | NA |
| C4(g/l), μ±σ | 0.16 ± 0.144 | NA |
| Anti-dsDNA, +/n | 7(10) | NA |
| Anti-SmD1, +/n | 3(10) | NA |
| Anti-RNP, +/n | 2(10) | NA |
| Anti-SSA/Ro, +/n | 5(10) | NA |
| Sex, Female/Male | 5/5 | 10/10 |
SLE: systemic lupus erythematosus; H_M: healthy control; SLEDAI: SLE disease activity index; ESR: erythrocyte sedimentation rate; CRP: C reactive protein. C3: complement 3; C4: complement 4. NA, not applicable. μ: Arithmetic mean; σ: Standard Deviation.
σ = , = Value of each data point, = Mean, n = Number of data points.
2.2. Exosome isolation and purification by differential centrifugation
Rapid thaw the freezing sample (plasma) at 37 °C. Transfer the samples to a new centrifuge tube, and then centrifuge at 2000×g, 4 °C for 30 min. Carefully transfer the supernatant to a new centrifuge tube, and centrifuge again at 10,000×g, 4 °C for 45 min to remove the larger vesicles. The supernatant was filtered through a 0.45 μm filter, and the filtrate was collected. The filtered liquid was transferred to a new centrifuge tube, centrifugation at 4 °C, 100,000×g for 70 min. After the supernatant was removed, the pellet was re-suspended with 10 mL of pre-cooled 1 × PBS, and then centrifuged again at 4 °C, 100,000×g for 70 min. Then removed the supernatant, and the pellet was re-suspended with 100 μL pre-cooled 1 × PBS. The purified exosomes can be used in further experiments or stored at −80 °C.
2.3. Transmission electron microscopy (TEM) [3]
Approximately 15 μl exosomes samples were dried onto a copper grid with a lacy carbon film. The grid was negatively stained with 15 μl of 2% uranyl acetate for 1 min at room temperature and imaged with a G2 spirit TWIN FEI (Tecnai, Hillsboro, USA) with an accelerating voltage set to 120 kV. Images were taken with CCD camera.
2.4. Nanoparticle tracking analysis (NTA) particle size and concentration measurements
Size distribution and concentration of exosomes were analyzed by a Nanosight LM10 HS-BF instrument (Nanosight Ltd, UK). Laser: 405-nm 65-mW; Camera: EMCCD Andor Luca camera. Camera settings (Shutter: 850, Gain: 450, Lower Threshold: 910, Higher Threshold: 11180, 60 s). The exosomes (10 mg) were diluted with 1 mL particle-free PBS and sonicated for 1 min to distribute the exosomes evenly. Measurements repeats:15–25, collected particles >5000.
2.5. Immunological confirmation for exosomes by Western blots
PBMC and plasma-derived exosomes were lysed in RIPA reagent containing protease inhibitor cocktail (P8340; Sigma-Aldrich) and the protein concentrations were determined using the BCA method. Total protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride membrane (Millipore). After blocking in 5% skim milk, the membrane was incubated overnight with primary antibodies (diluted to 1:1000). Primary antibodies include Flotillin 1 (A3023), CD63(ABclonal, A19023), CD81 (ABclonal, A4863) and Calnexin (ABclonal, A4846). Followed by 3 times’ washing with TBST, membranes were incubated for 2 h with secondary antibodies. Then, protein bands were visualized by enhanced chemiluminescence (ECL) and photos taken by the LI-COR Odyssey Imagers system.
2.6. Exosomal RNA isolation and qualification
The Norgen's Exosomal RNA Isolation Kit (58000, NORGEN) was used to extract exosome RNA. The quantity and quality of RNA samples were validated by Agilent 2100 bioanalyzer. Quality control: RNA concentration (1–100pg/uL), No peaks of 18S and 28S, and only a small peak after 25 nt. We used all the available plasma sample volumes in the experiments and then normalized the sample with 0.01μg of RNA (the lowest RNA amount in our study).
2.7. Small-RNA library construction and sequencing
Exosomal small-RNA library construction followed the protocol of the TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA), and then the library was sequenced by an Illumina HiSeq 2500 with 50 base single-end reads.
2.8. Bioinformatics analysis of miRNAs
The program ACGT101-miR (LC Sciences, Houston, Texas, USA) was applied to filter non-miRNA sequences in repeats and RNA families (rRNA, tRNA, snRNA, snoRNA). Then, the remaining sequences were mapped to human precursors in miRBase 22.0 by BLAST to identify both novel and known miRNAs. For detailed information, please refer to the “Supplemental Material-Bioinformatics analysis of miRNAs”. To predict the targeted genes of the most abundant miRNAs, two computational target prediction algorithms (TargetScan 5.0 and Miranda 3.3a) were used to identify miRNA binding sites. Finally, the data predicted by both algorithms were combined, and the overlaps were calculated. The GO terms and KEGG pathways of these most abundant miRNAs and miRNA targets were also annotated.
2.9. Proteomic analysis of exosome
Samples were eluted in lysis buffer and further processed by the procedures in a previous report [12,13], and finally, supernatant aliquots were obtained. Protein was digested with Trypsin Gold (Promega). Then, peptides were dried and reconstituted in 0.5 M TEAB and processed according to the manufacturer's introduction. The peptides were subjected to nanoelectrospray ionization. Intact peptides were detected in the orbitrap (Q Exactive Orbitrap Mass Spectrometers, Thermofisher Scientific). The details of the protocol were described in a previous study [12].
2.10. Bioinformatics analysis for proteomics
The bioinformatics analysis tool (Proteome Discoverer™ Software 2.2) and Protein Database (ZJH-Homo_sapiens.GRCh38.pep.all.fasta) were utilized for protein identification. Each confident protein contained at least one unique peptide. The median peptide ratio in Mascot was used for normalization of the quantitative protein ratios.
2.11. Functional enrichment analysis
The Gene Oncology (GO) (http://www.geneontology.org), Cluster of Orthologous Groups of proteins (COG) (http://archive-dtd.ncbi.nlm.nih.gov/COG/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (http://www. genome.jp/kegg) were utilized to predict the underlying biological functions and pathways of the candidate target genes. miRNA- and protein-related genes were input into the database for annotation, visualization and integrated discovery (DAVID) for GO and KEGG pathway analysis.
2.12. Prediction of miRNA-protein targeting pairs
We predicted the potential regulation between the highly differentially expressed proteins and miRNAs by two computational target prediction algorithms (TargetScan 5.0 and Miranda 3.3a), and got some miRNA-protein pairs.
2.13. Verification of identified molecules in the exosome in plasma
The miRNAs identified by sequencing were verified by Quantitative real-time polymerase chain reaction (qRT-PCR). Total RNA was extracted from the exosome. The specific primers for the qRT-PCR are listed in this manuscript (Table 4), and qRT-PCR was carried out by SYBR PrimeScript miRNA RT-PCR Kit (TianGen Biotech, Beijing, China). U6 was used as internal controls. Each gene has three independent biological replicates. The comparative 2-ΔΔCt analyzed method was used in this study. For Targeted protein quantification: in an independent test cohort, according to the manufacturer's instructions, the absolute concentrations of the selected protein was quantified by human enzyme-linked immunosorbent assay (ELISA) kits. Briefly, 50 μL plasma exsomes lysis from each subject was added to the antibody pre-coated plates and incubated at 37 °C for 1 h. After that, 100 μL of HRP-conjugated detecting antibodies were then added and incubated for another 1 h. Then, the plates were washed 3 times, and the HRP substrates were added for 15 min. After the reaction was stopped, the OD value at 450 nm was measured. At the same time, by detecting the OD value of a series of dilutions with known protein concentration, the standard curve of each protein was generated. (Antibodis: IGHG1: PA5-75428, Invitrogen; C1QB: PA5-102737, Invitrogen; Antibodis: LGALS3BP: PA5-51411, Invitrogen; C4: PA5-119047, Invitrogen).
Table 4.
The primers for PCR amplification.
| NO. | Target | Sequence | TM |
|---|---|---|---|
| 1 | hsa-miR-25-5p_R-1 | F: AACAAGAGGCGGAGACTTGG R: GTCGTATCCAGTGCAGGGT |
60 |
| 2 | hsa-miR-873-5p_L+1R-3 | F: AACAAGTGCAGGAACTTGTGAG R: GTCGTATCCAGTGCAGGGT |
60 |
| 3 | hsa-miR-340-5p | F: AACACGCTTATAAAGCAATGAGACT R: GTCGTATCCAGTGCAGGGT |
60 |
| 4 | hsa-miR-142-3p_L-2R-1 | F: AACACGCTAGTGTTTCCTACTTTAT R: GTCGTATCCAGTGCAGGGT |
60 |
| 5 | hsa-miR-181c-5p_R-1 | F: AACAAGAACATTCAACCTGTCGG R:GTCGTATCCAGTGCAGGGT |
60 |
| 6 | hsa-miR-324-3p_L-3R+1 | F:AACAAGACTGCCCCAGGTG R:GTCGTATCCAGTGCAGGGT |
60 |
Note: F, Forward Primer; R, Reverse Primer; TM, Annealing Temperature.
2.14. Statistical analysis
Before further analysing of data, all variables went through normality and equal tests. Normal distribution data was analyzed by Unpaired Student's t-test, while non-normal distribution data was committed to nonparametric tests. Data were analyzed by GraphPad Prism (Version 6, CA) (mean ± SEM, Standard Error of the Mean). Data with P < 0.05 was considered to be statistical difference.
3. Results
Ten SLE patients (SL_M) and Twenty healthy people (H_M) without autoimmune disease were recruited. The SLE diagnostic criteria in our study was based on the ACR classification criteria for SLE, and the characteristics of the enrolled people are presented in Table 1.
3.1. Characterization of exosomes and expression outline of both miRNAs and proteins in exosome
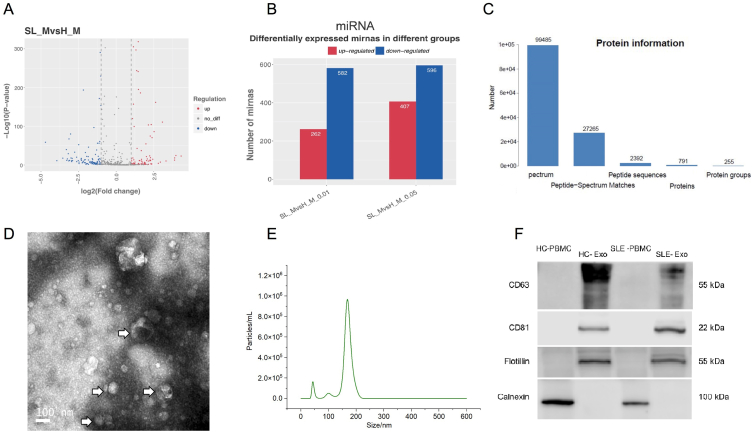
We isolated plasma exosomes from SLE patients and healthy. Exosomes were detected by Transmission electron microscopy (TEM) and Nanoparticle tracking analysis (NTA) and the size of exosomes was ranging from 30 to 200 nm (NTA data from a representative sample, Fig. 1D–E). Western blot analysis of the expression of proteins markers in exosomes, and data showed that proteins including CD63, CD81 and flotillin were positive in exosome samples, while calnexin was negative (Fig. 1F).
Fig. 1.
Characterization of exosomes and the expression outline of exosomal miRNAs and proteins. A. The volcano plots depicted differential expression of microRNAs between H_M and SL_M in plasma. The X-axis of the volcano plots represents the fold changes in the differential expression levels of miRNAs in different samples, while the Y-axis represents the statistical significance of miRNA expression. Dots in red represent the upregulated significantly differentially expressed genes, blue dots represent downregulated significantly differentially expressed genes, and the grey dots represent the nonsignificant differentially expressed genes. B. Bar chart presenting the statistical data for the number of differentially expressed miRNAs with different p-values (p < 0.01; 0.01=<p < 0.05). C. Protein profile results in the protein spectrum of exosomes. D. Transmission electron microscopy(TEM) scans purified plasma. The exosomes were 30–200 nm in size (arrows) and had a lipid layer (insert). E. Nanoparticle tracking analysis (NTA) to quantify the nanosize and concentration of exosomes. F. Western blot analysis of the expression of proteins enriched in exosomes and PBMC cells, including CD63, CD81 and flotillin. As the control, calnexin was not detected in exosomes (For original blotting data please refer to Fig. S1). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Our data showed the distribution of differentially expressed miRNAs in SLE (Fig. 1A), including 407 upregulated and 596 downregulated microRNAs (Fig. 1B). The upregulated miRNAs included hsa-miR-9-3p, hsa-miR-25-5p_R-1, hsa-miR-142-3p_L-2R-1, hsa-miR-181c-5p_R-1, hsa-miR-324-3p_L-3R+1 and so forth. The downregulated miRNAs included hsa-miR-340-5p, hsa-miR-873-5p_L+1R-3, et cetera. For proteomics, 791 proteins were detected (Figs. 1C), 97 of which contain significantly different expression levels (Table S1). There were 84 upregulated and differentially expressed proteins, such as C-reactive protein (CRP), LGALS3BP, IGHG1, C1QB, C4BPA, etc. 13 proteins, including IGHV3OR15-7, HPR and HYDIN, were downregulated significantly in SLE.
3.2. Expression profile of exosomal miRNAs in the plasma and functional analysis of miRNA-targeted genes
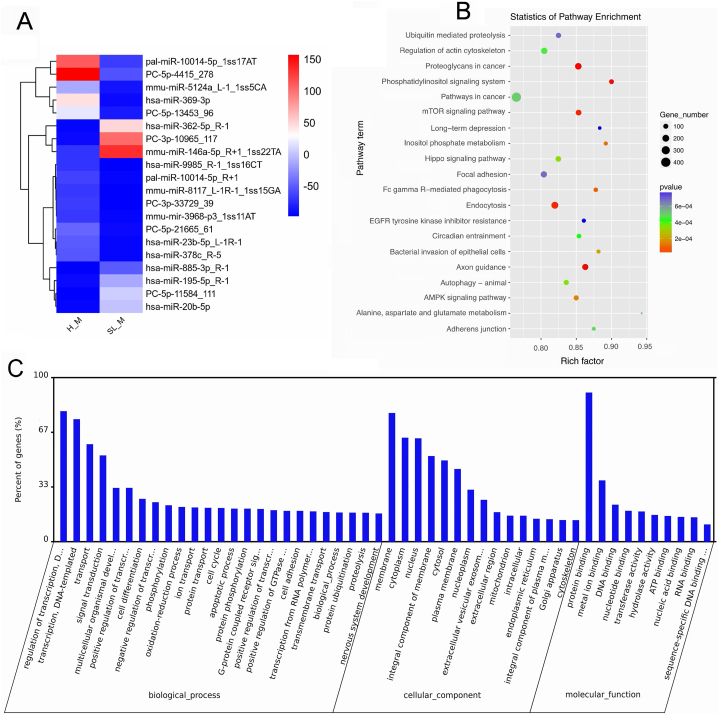
The characteristics of exosomal miRNAs with differential expression swere analyzed, and present partially in heatmap (Fig. 2A). The detailed data are shown in the supplemental data (Table S2).
Fig. 2.
The profile of miRNA expression levels in exosomes and the GO and KEGG enrichment analysis of miRNA-targeted genes. A. Top differentially expressed miRNAs, p-value <0.01. B. KEGG enrichment analysis of exosomal miRNA-targeted genes. C. GO enrichment analysis of exosomal miRNA-targeted genes. The GO terms of miRNA-targeted genes were sorted by the data selection criteria: p-value <0.05 and |log2-fold changes| ≥1.
KEGG enrichment was used to analysis the targeted genes of miRNAs (Fig. 2B). Our data revealed some top significant pathways, including the mTOR signalling pathway and Hippo signalling pathway (Table 2). Targeted genes of differentially expressed miRNAs were analyzed by GO analysis. Top GO items in this study included Transport, Signal transduction, Protein binding, Apoptotic process, Membrane, and Cell differentiation (Fig. 2C).
Table 2.
Common elements between exosomes and mononuclear cells.
| Common elements | “Exosome” and "PBMCs" | “Exosome” and "T cells" | “Exosome” and "B cells" “Exosome” and "DCs" |
“Exosome” and "Macrophages" | |||
|---|---|---|---|---|---|---|---|
| miRNAs | miR-9 [14,15] | miR-410 [15] | miR-3130 [14] | miR-224 [16] | miR-25 [17] | miR-324 [19] | miR-129 [6] |
| miR25 [[14], [15], [16]] | miR-431 [15] | miR-3200 [14] | miR-873 [17] | miR-324 [18] | miR-340 [19] | miR-142 [6] | |
| miR-31 [14,15] | miR-433 [14] | miR-5683 [14] | miR-362 [18] | miR-362 [19] | miR-340 [6] | ||
| miR-134 [14,15] | miR-493 [15] | miR-92b [14] | miR-579 [18] | miR-27b [19] | miR-200a [6] | ||
| miR-142 [[17], [18], [19]] | miR-638 [14,16] | miR-196a [[14], [15], [16]] | miR-181c [18] | miR-148b [6] | |||
| miR-324 [14,15] | miR-766 [[14], [16]] | miR-148b [15,16] | |||||
| miR-330 [14] | miR-769 [[14], [16]] | miR-378a [14] | |||||
| miR-337 [15] | miR-939 [[14], [16]] | miR-23a [14] | |||||
| miR-363 [14] | |||||||
| miR-382 [15] | |||||||
| miR-409 [[17], [18], [19]] | |||||||
| proteins (genes) | C1Q[13] | SERPINA1 [13] | LGALS3BP [7] C3 [8] | IGHG1 [7] C4 [8] | C1Q [20] C3 [20] | ||
| C5[13] | C3 [13] | ||||||
| CRP[13] | APOA4 [11,13] | ||||||
| JCHAIN[11, 13] | C4 [13] | ||||||
| KEGG pathways | mTOR signalling pathway [21] | Estrogen signalling pathway [21,24] | Complement and coagulation cascades [21,26,27] | Phagosome [28] | Chagas disease [6,26] | ||
| Adherens junction [22] | Fc gamma R-mediated phagocytosis [21,24] | Fc gamma R-mediated phagocytosis [21,26,27] | Systemic lupus erythematosus [28] | Complement and coagulation cascades [6,26] | |||
| AMPK signaling pathway [[21], [23]] | Focal adhesion [21,24] | FcγRIIb signalling [21] | Tuberculosis [28] | Endocytosis [26] | |||
| Chagas disease [[21], [23]] | Metabolic pathways [21,24] | Long term depression [21,26,27] | Lysosome [26] | ||||
| Cholesterol metabolism [[21], [23]] | mTOR signaling pathway [25] | NF-kappa B signaling pathway [21,26,27] | Pertussis [6,26] | ||||
| Complement and coagulation cascades [[21], [23]] | NF-kappa B signalling pathway [21] | Pertussis [21,26,27] | Staphylococcus aureus infection [6,26] | ||||
| EGFR tyrosine kinase inhibitor resistance [[21], [23]] | Pathways in cancer [21,24] | PI3K-Akt signaling pathway [24] | Systemic lupus erythematosus [6,26] | ||||
| Fc gamma R-mediated phagocytosis [22] | PI3K-Akt signaling pathway [21,24] | Primary immunodeficiency [21,26,27] | |||||
| HIF-1 signaling pathway [22] | Systemic lupus erythematosus [21,24] | Systemic lupus erythematosus [21,26,27] | |||||
| Malaria [[21], [23]] | Wnt signaling pathway [21,24] | ||||||
| NF-kappa B signaling pathway [22] | |||||||
| Pathways in cancer [[21], [23]] | |||||||
| Pertussis [[21], [23]] | |||||||
| Phagosome [[21], [23]] | |||||||
| PI3K-Akt signaling pathway [23] | |||||||
| Primary immunodeficiency [[21], [23]] | |||||||
| Staphylococcus aureus infection [[21], [23]] | |||||||
| Systemic lupus erythematosus [[21], [23]] | |||||||
| Toll-like receptor signaling pathway [22] | |||||||
| Transcriptional misregulation in cancer [[21], [23]] | |||||||
| Tuberculosis [[21], [23]] | |||||||
3.3. Functional analysis of exosomal proteins in plasma
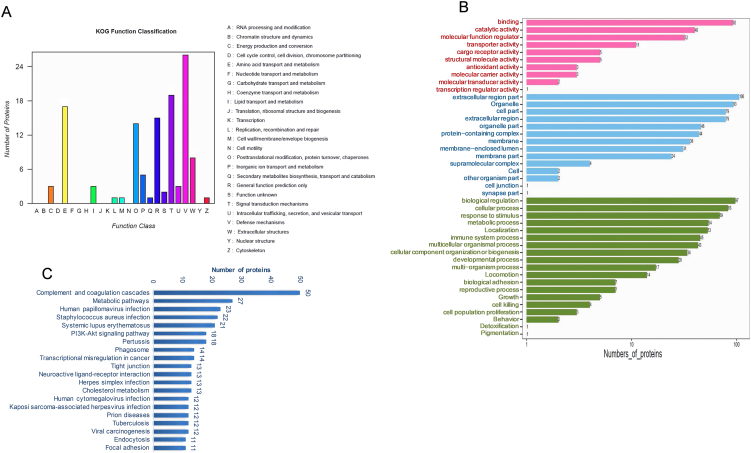
Three databases (COG, GO and KEGG) were used for functional analysis in our study. Our study revealed that KOG (Animal COG database) in SLE has 15 functional categories (Fig. 3A), including [T] signal transduction mechanisms (19 proteins), [V] defence mechanisms (26 proteins), [E] amino acid transport and metabolism (17 proteins) and other functional items. The GO analysis of differentially expressed proteins (DEPs) are shown in Fig. 3 (Fig. 3B). The molecular function events of differentially expressed proteins included Binding, Transporter activity, and Molecular carrier activity. Cellular components of DEPs included extracellular region part and extracellular region, among others. Biological processes of DEPs included biological regulation, cellular processes and immune system processes. KEGG enrichment analysis data of proteins are shown in Fig. 3 (Fig. 3C). KEGG pathways of DEPs included complement and coagulation cascades, systemic lupus erythematosus, the PI3K-Akt signalling pathway and others.
Fig. 3.
GO and KEGG enrichment analyses of proteins in exosomes. A. Exosomal proteins were analyzed by proteins database of orthologous groups' cluster (COG). The horizontal axis indicates the names of COG items, and the vertical axis displays the number of proteins. B. GO enrichment analysis of proteins in exosomes. C. KEGG enrichment analysis of proteins in exosomes. The GO terms and KEGG were sorted by the data selection criteria: p-value <0.05 and |log2-fold changes| ≥1.
3.4. Common miRNAs/proteins/pathways between our plasma exosomal data and other previously reported circulating data
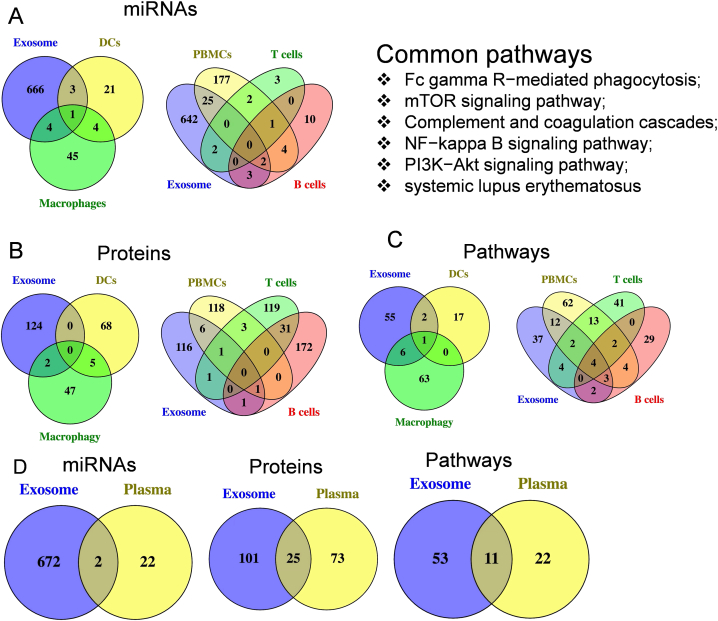
A-C: Common elements between exosomes (our study) and mononuclear cells (PBMCs, DCs, macrophages, T cells and B cells) in peripheral blood (previous reports). A. Common miRNAs; B. Common proteins; C. Common pathways.
D: Common elements (miRNAs, proteins, pathways) between exosomes (our study) and whole plasma.
Our study showed that some common elements (miRNAs/proteins/pathways) were shared by our exosomal data and other circulation components data reported previously (PBMCs, DCs, macrophages, T cells, B cells and the peripheral blood plasma) (Fig. 4A–D). For example, some miRNAs reported previously from non-exosomes contain similar expression profiles to our exosomal data: 4 in dendritic cells (DCs), 5 in macrophages, 2 in T cells, 5 in B cells and 27 in PBMCs. The numbers of common proteins shared by our plasma exosomal data and the earlier research data is as follows: none in DCs, 2 in macrophages, 2 in T cells, 2 in B cells and 8 in PBMCs. The numbers of common signalling pathways shared by our plasma exosomal data and the earlier research data is as follows: 3 in DCs, 7 in macrophages, 10 in T cells, 9 in B cells and 21 in PBMCs. We also summarized the common miRNAs/proteins/pathways shared by the exosomes in plasma and the whole plasma (Fig. 4D). The details for the common miRNAs/proteins/pathways between exosomes in plasma and other blood components are shown in the table (Table 2, Table 3).
Fig. 4.
Common miRNAs/proteins/pathways between plasma exosomes and other blood components.
Table 3.
Common elements between exosomes in plasma and whole plasma.
| Common elements | “Exosome” and “Plasma " |
|---|---|
| miRNAs [15] | miR-142, miR-200a |
| proteins (genes) [10,27,29] | A1BG, A2M, ALB, APOB, APOC3, C1Q, C3, C4, CFH, CP, CRP, FGA, FGB, FGG, HBA1, HBB, HP, HPX, IGHG4, IGHV4-4, ORM1, ORM2, SERPINA1, SERPING1, TF. |
| KEGG pathways [10,27,29] | African trypanosomiasis, Chagas disease, Complement and coagulation cascades, HIF-1 signaling pathway, Malaria, Pertussis, PI3K-Akt signaling pathway, PPAR signaling pathway, Prion diseases, Staphylococcus aureus infection, Systemic lupus erythematosus. |
3.4.1. Verification of data
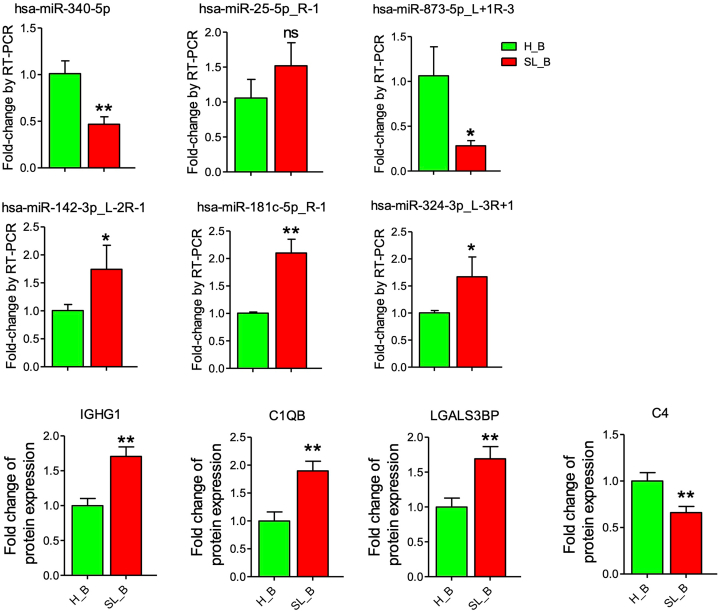
Some common molecules shared by our plasma exosome data and other data reported previously (PBMCs, T cells, B cells and Plasma) have been verified. Compared to health subjects, some exosomal molecules in SLE were up regulated in our sequencing data, such as hsa-miR-25-5p_R-1, hsa-miR-142-3p_L-2R-1, hsa-miR-181c-5p_R-1, hsa-miR-324-3p_L-3R+1, IGHG1, C1QB and LGALS3BP. However, some were down regulated, such as hsa-miR-873-5p_L+1R-3, hsa-miR-340-5p and C4. The above common molecules were confirmed by RT-PCR or ELISA (Fig. 5).
Fig. 5.
Technical validation of sequencing data. Six miRNAs and four proteins were analyzed by qRT-PCR and ELISA. The data are represented in the way of means ± SEM. *p < 0.05 and **p < 0 01 mean the data was significant difference between SLE patient and healthy subject, while ns means no significance with p≥0.05.
4. Discussion
Previous reports have shown that circulating exosomes are critical to the pathogenesis of SLE by inducing the expression of some inflammatory genes [3,4]. However, earlier studies of both circulating miRNAs and proteins in SLE have mainly focused on monocytes, whole serum/plasma/urine [10]. The information about the roles of miRNAs/proteins in plasma exosome in SLE patients is far from enough, and more research is needed. In this project, we obtained the unique expression profiles of plasma exosome miRNAs and proteins in SLE patients, which filled the information gaps of plasma exosomes of SLE patients.
In our study, we found differential expression of 1003 miRNAs and 97 proteins. 39 common miRNAs were recognized in study, such as miRNA-181b was reported to be involved in B cell activation in SLE [17], and miR-224 [15] was overexpressed in T cells. miR-324, miR-340, miR-362 and miR-27b downregulated in circulating dendritic cells (DCs) in SLE patients [15]. Previous studies demonstrated that miR-129 [6], miR-142, and miR-148b [6,17] were overexpressed in circulating macrophages of SLE patients, which was consistent to our plasma exosomal data. One study reported that serum exosomal miR-155 was upregulated in SLE patients [5], while our data showed that the expression of plasma exosomal miR-155 was higher but not significant in SLE. One possible reason for the difference is that plasma contains fibrinogen but serum does not, which leads to the loss of some exosomes in serum.
There were 14 common proteins (genes) shared by our current exosomal data and previously reported data. The proteins C1Q and C4, as inhibitors of the complemental classical pathway, show expression defects in SLE patients [8]. In our study, C1Qb and C4BPA expression was increased in plasma exosomes in SLE patient. Studies showed that the expression level of IGHG 1 gene in B cells of active SLE patients is three times higher than that of healthy subjects [7], which is consistent to our plasma exosomal data.
For the functional analysis, approximately 60 pathways closely related to exosomal miRNAs/proteins were summarized (Table S3), 21 of which were shared with previous studies. The 6 highest frequency common pathways shared by our exosomal data and previously reported data were Fc gamma R-mediated phagocytosis, mTOR signalling pathway, NF-kappa B signalling pathway, PI3K-Akt signalling pathway, systemic lupus erythematosus, and complement and coagulation cascades (Fig. 4C).
In summary, batches of differentially expressed plasma exosomal miRNAs and proteins in SLE patients were identified. Furthermore, miRNAs and proteins shared by our current exosome study and previous research (whole plasma, serum, PBMCs, T cells, B cells, DCs or macrophages) were identified. A total of 39 miRNAs, 14 proteins (genes) and 21 pathways were summarized as common elements.
miRNA is one of the epigenetic regulator of protein expression. We got some potential miRNA-protein targeting pairs, for example, our data showed plasma exosomal miRNA (PC-5p-335271_5) down-regulated while its predictive negative regulated target (predicted by TargetScan and miRanda), IGHG1 up-regulated in SLE. There are many others similar potential miRNA-protein targeting pairs, such as miRNA(PC-5p-322305_6)-protein(LGALS3BP), and miRNA(PC-5p-297304_6)-protein(C1QB) (Table S4). However, it should be noted that the regulatory factors of proteins in plasma exosomes are highly complex. Therefore, the predictive miRNA-protein targeting pairs obtained in our research can only be treated as speculative reference, rather than representing the real regulatory relationship. We need more work to further prove whether the predictive miRNA-protein targeting pairs is true or not, which is the limitation of our research.
Common molecules (miRNAs and proteins) and pathways between our plasma exosome data and previously reported data indicate their potential applications in clinical treatment and diagnosis of SLE disease. In the future, further studies may focus on the specific function of these common molecules and how these common molecules communicate with the target cells or tissues in SLE.
Author contribution statement
Wencong Song: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Chunhong Li: Conceived and designed the experiments; Analyzed and interpreted the data.
Jie Qiu: Analyzed and interpreted the data; Wrote the paper.
Jiyou Dong: Analyzed and interpreted the data; Wrote the paper; Final approval of the version submitted.
Dongzhou Liu: Contributed reagents, materials, analysis tools or data; Analyzed and interpreted the data.
Yong Dai: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Analyzed and interpreted the data.
Funding statement
Wencong Song was supported by Young Scientists Fund [32000641].
Dr yong dai was supported by Shenzhen Key Medical Discipline Construction Fund [SZXK011].
Dongzhou Liu was supported by National Natural Science Foundation of China [81971464], the Key Research and Development Program of Guangdong Province [No. 2019B020229001], the Science and Technology Plan of Shenzhen [JCYJ20190807145815129], Sanming Project of Medicine in Shenzhen [SYJY201704, SYJY201705].
Data availability statement
Data associated with this study has been deposited at The raw data for small RNA sequencing and ribosomal RNA-depleted sequencing in our study have been uploaded to the Genome Sequence Archive (GSA) (Accession number: HRA000985) (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA000985).
Declaration of interest’s statement
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13345.
Contributor Information
Jiyou Dong, Email: xtwdjy@126.com.
Dongzhou Liu, Email: liu_dz2001@sina.com.
Yong Dai, Email: daiyong22@aliyun.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Tsokos G.C. Systemic lupus erythematosus. N. Engl. J. Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Li T., Carls G.S., Panopalis P., Wang S., Gibson T.B., Goetzel R.Z. Long-term medical costs and resource utilization in systemic lupus erythematosus and lupus nephritis: a five-year analysis of a large medicaid population. Arthritis Rheum. 2009;61(6):755–763. doi: 10.1002/art.24545. [DOI] [PubMed] [Google Scholar]
- 3.Lee J.Y., Park J.K., Lee E.Y., Lee E.B., Song Y.W. Circulating exosomes from patients with systemic lupus erythematosus induce an proinflammatory immune response. Arthritis Res. Ther. 2016;18(1):264. doi: 10.1186/s13075-016-1159-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu T., Wang Y., Jin H., Li L. The role of exosome in autoimmune connective tissue disease. Ann. Med. 2019;51(2):101–108. doi: 10.1080/07853890.2019.1592215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W., Liu S., Chen Y., Weng R., Zhang K., He X., He C. Circulating exosomal microRNAs as biomarkers of systemic lupus erythematosus. Clinics. 2020;75:e1528. doi: 10.6061/clinics/2020/e1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao P., Dong C., Yue Y., Xiong S. Dynamic expression of microRNAs in M2b polarized macrophages associated with systemic lupus erythematosus. Gene. 2014;547(2):300–309. doi: 10.1016/j.gene.2014.06.065. [DOI] [PubMed] [Google Scholar]
- 7.Becker A.M., Dao K.H., Han B.K., Kornu R., Lakhanpal S., Mobley A.B., Li Q.Z., Lian Y., Wu T., Reimold A.M., Olsen N.J., Karp D.R., Chowdhury F.Z., Farrar J.D., Satterthwaite A.B., Mohan C., Lipsky P.E., Wakeland E.K., Davis L.S. SLE peripheral blood B cell, T cell and myeloid cell transcriptomes display unique profiles and each subset contributes to the interferon signature. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leffler J., Bengtsson A.A., Blom A.M. The complement system in systemic lupus erythematosus: an update. Ann. Rheum. Dis. 2014;73(9):1601–1606. doi: 10.1136/annrheumdis-2014-205287. [DOI] [PubMed] [Google Scholar]
- 9.Qi S., Chen Q., Xu D., Xie N., Dai Y. Clinical application of protein biomarkers in lupus erythematosus and lupus nephritis. Lupus. 2018;27(10):1582–1590. doi: 10.1177/0961203318773643. [DOI] [PubMed] [Google Scholar]
- 10.Madda R., Lin S.C., Sun W.H., Huang S.L. Differential expressions of plasma proteins in systemic lupus erythematosus patients identified by proteomic analysis. J. Microbiol. Immun. Inf. Wei mian yu gan ran za zhi. 2019;52(5):816–826. doi: 10.1016/j.jmii.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Dai Y., Hu C., Huang Y., Huang H., Liu J., Lv T. A proteomic study of peripheral blood mononuclear cells in systemic lupus erythematosus. Lupus. 2008;17(9):799–804. doi: 10.1177/0961203308089444. [DOI] [PubMed] [Google Scholar]
- 12.Liang Y., Xie S.-B., Wu C.-H., Hu Y., Zhang Q., Li S., Fan Y.-G., Leng R.-X., Pan H.-F., Xiong H.-B. Coagulation cascade and complement system in systemic lupus erythematosus. Oncotarget. 2018;9(19) doi: 10.18632/oncotarget.23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Y., Xie S.B., Wu C.H., Hu Y., Zhang Q., Li S., Fan Y.G., Leng R.X., Pan H.F., Xiong H.B., Ye D.Q. Coagulation cascade and complement system in systemic lupus erythematosus. Oncotarget. 2018;9(19):14862–14881. doi: 10.18632/oncotarget.23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song W., Tang D., Chen D., Zheng F., Huang S., Xu Y., Yu H., He J., Hong X., Yin L., Liu D., Dai W., Dai Y. Advances in applying of multi-omics approaches in the research of systemic lupus erythematosus. Int. Rev. Immunol. 2020;39(4):163–173. doi: 10.1080/08830185.2020.1736058. [DOI] [PubMed] [Google Scholar]
- 15.Husakova M. MicroRNAs in the key events of systemic lupus erythematosus pathogenesis, Biomedical papers of the Medical Faculty of the University Palacky. Olomouc, Czechoslovakia. 2016;160(3):327–342. doi: 10.5507/bp.2016.004. [DOI] [PubMed] [Google Scholar]
- 16.Yan S., Yim L.Y., Lu L., Lau C.S., Chan V.S. MicroRNA regulation in systemic lupus erythematosus pathogenesis. Immune network. 2014;Imm. Net.(3):138–148. doi: 10.4110/in.2014.14.3.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S., Wang D., Liu Y. Extracellular RNA in systemic lupus erythematosus. ExRNA. 2019;1(1):33. [Google Scholar]
- 18.Duroux-Richard I., Cuenca J., Ponsolles C., Piñeiro A.B., Gonzalez F., Roubert C., Areny R., Chea R., Pefaur J., Pers Y.M., Figueroa F.E., Jorgensen C., Khoury M., Apparailly F. MicroRNA profiling of B cell subsets from systemic lupus erythematosus patients reveals promising novel biomarkers. Int. J. Mol. Sci. 2015;16(8):16953–16965. doi: 10.3390/ijms160816953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Hoogen L.L., Rossato M., Lopes A.P., Pandit A., Bekker C.P.J., Fritsch-Stork R.D.E., van Roon J.A.G., Radstake T. microRNA downregulation in plasmacytoid dendritic cells in interferon-positive systemic lupus erythematosus and antiphospholipid syndrome. Rheumatology. 2018;57(9):1669–1674. doi: 10.1093/rheumatology/key159. [DOI] [PubMed] [Google Scholar]
- 20.Ma C., Xia Y., Yang Q., Zhao Y. The contribution of macrophages to systemic lupus erythematosus. Clin. Immun.(Orlando, Fla. 2019;207:1–9. doi: 10.1016/j.clim.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Peng S.L. Altered T and B lymphocyte signaling pathways in lupus. Autoimmun. Rev. 2009;8(3):179–183. doi: 10.1016/j.autrev.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 22.Sandling J.K., Pucholt P. Molecular pathways in patients with systemic lupus erythematosus revealed by gene-centred DNA sequencing. 2021;80(1):109–117. doi: 10.1136/annrheumdis-2020-218636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maleknia S., Salehi Z., Rezaei Tabar V., Sharifi-Zarchi A., Kavousi K. An integrative Bayesian network approach to highlight key drivers in. systemic lupus erythematosus. 2020;22(1):156. doi: 10.1186/s13075-020-02239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zharkova O., Celhar T., Cravens P.D., Satterthwaite A.B., Fairhurst A.M., Davis L.S. Pathways leading to an immunological disease: systemic lupus erythematosus. Rheumatology. 2017;56(suppl_1):i55–i66. doi: 10.1093/rheumatology/kew427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katsuyama T., Tsokos G.C., Moulton V.R. Aberrant T cell signaling and subsets in systemic lupus erythematosus. Front. Immunol. 2018;9:1088. doi: 10.3389/fimmu.2018.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labonte A.C., Kegerreis B., Geraci N.S., Bachali P., Madamanchi S., Robl R., Catalina M.D., Lipsky P.E., Grammer A.C. Identification of alterations in macrophage activation associated with disease activity in systemic lupus erythematosus. PLoS One. 2018;13(12) doi: 10.1371/journal.pone.0208132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madda R., Lin S.C., Sun W.H., Huang S.L. Plasma proteomic analysis of systemic lupus erythematosus patients using liquid chromatography/tandem mass spectrometry with label-free quantification. PeerJ. 2018;6 doi: 10.7717/peerj.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashton M.P., Eugster A., Dietz S., Loebel D., Lindner A., Kuehn D., Taranko A.E., Heschel B., Gavrisan A., Ziegler A.G., Aringer M., Bonifacio E. Association of dendritic cell signatures with autoimmune inflammation revealed by single-cell profiling. Arthritis Rheumatol. 2019;71(5):817–828. doi: 10.1002/art.40793. [DOI] [PubMed] [Google Scholar]
- 29.Chen C., Geng L., Xu X., Kong W., Hou Y., Yao G., Feng X., Zhang H., Liang J. Comparative proteomics analysis of plasma protein in patients with neuropsychiatric systemic lupus erythematosus. Ann. Transl. Med. 2020;8(9):579. doi: 10.21037/atm.2020.04.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at The raw data for small RNA sequencing and ribosomal RNA-depleted sequencing in our study have been uploaded to the Genome Sequence Archive (GSA) (Accession number: HRA000985) (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA000985).