Abstract
Methylmercury (MeHg) is a potent neurotoxin that bioaccumulates along food chains. The conversion of MeHg from mercury (Hg) is mediated by a variety of anaerobic microorganisms carrying hgcAB genes. Mangrove sediments are potential hotspots of microbial Hg methylation; however, the microorganisms responsible for Hg methylation are poorly understood. Here, we conducted metagenomic and metatranscriptomic analyses to investigate the diversity and distribution of putative microbial Hg-methylators in mangrove ecosystems. The highest hgcA abundance and expression occurred in surface sediments in Shenzhen, where the highest MeHg concentration was also observed. We reconstructed 157 metagenome-assembled genomes (MAGs) carrying hgcA and identified several putative novel Hg-methylators, including one Asgard archaea (Lokiarchaeota). Further analysis of MAGs revealed that Deltaproteobacteria, Euryarchaeota, Bacteroidetes, Chloroflexi, and Lokiarchaeota were the most abundant and active Hg-methylating groups, implying their crucial role in MeHg production. By screening publicly available MAGs, 104 additional Asgard MAGs carrying hgcA genes were identified from a wide range of coast, marine, permafrost, and lake sediments. Protein homology modelling predicts that Lokiarchaeota HgcAB proteins contained the highly conserved amino acid sequences and folding structures required for Hg methylation. Phylogenetic tree revealed that hgcA genes from Asgard clustered with fused hgcAB genes, indicating a transitional stage of Asgard hgcA genes. Our findings thus suggest that Asgard archaea are potential novel Hg-methylating microorganisms and play an important role in hgcA evolution.
Subject terms: Microbial ecology, Biogeography
Introduction
Due to natural processes and anthropogenic effects, mercury (Hg) is widespread in terrestrial and aquatic ecosystems such as sediments, rice paddy soils, and the water column [1]. Inorganic Hg in the environment can be converted to potent neurotoxic methylmercury (MeHg) by a variety of anaerobic microorganisms [2, 3]. The synthesized MeHg can subsequently bioaccumulate and biomagnify in food chains, posing a risk to wildlife and to human health [4]. Therefore, it is critical to investigate MeHg production and Hg-methylating microorganisms in aquatic environments.
The hgcAB gene cluster provides a molecular marker for identifying Hg-methylating microorganisms [5]. The hgcA gene encodes a homolog of corrinoid iron-sulfur proteins, and the hgcB gene is generally present next to hgcA by encoding an iron-sulfur cluster protein in various microorganisms including bacteria and archaea [6]. Previous structure model of the HgcB suggested that conserved cysteines in HgcB are involved in shuttling HgII, methymercury, or both [7]. The gene pair hgcAB presumably functions in methyltransferase and corrinoid reduction, endowing microorganisms with the ability to produce MeHg [5]. All experimentally confirmed Hg-methylators contain hgcA homologues, which predominately affiliate to sulfate-reducing bacteria and iron-reducing bacteria belonging to the class δ-Proteobacteria within the phylum Proteobacteria, fermentative bacteria within the phylum Firmicutes (Clostridia), and methanogenic archaea within the phylum Euryarchaeota [8, 9]. More diverse hgcA carriers, including Chloroflexi, Spirochaetes [10], Nitrospina [11], Planctomycetes, Verrucomicrobia [12], Acidobacteria, Actinobacteria, Aminicenantes, Elusimicrobia, and Nitrospirae [6, 13–15] have been discovered in other microbial phyla using culture-independent approaches. It is believed that microorganisms carrying hgcAB gene sequences are phylogenetically diverse and prevalent in a wide range of habitats [16, 17].
Asgard is a recently discovered archaeal superphylum that contains more than 16 distinct phyla, such as Loki-, Thor-, Odin-, Heimdall-, Hel-, Gerd-, Sif-, Baldr-, Hermod-, Borr-, Hod-, Kari-, Wukong-, Sigyn-, Freyr- and Njordarchaeota [18–25]. They are widely distributed in anoxic environments such as sediments, cold seeps, hot springs, and soils [26–28]. Previous surveys have revealed high concentrations of heavy metals (e.g., Cu, Pb, Hg, As) in certain anoxic wetlands, lake, marine, and paddy sediments [29–31]. These findings imply that Asgard archaea can adapt to sediment conditions with high concentrations of heavy metals, suggesting that they are involved in heavy metal metabolism [32]. However, whether Asgard archaea are able to methylate Hg remains unknown.
Mangroves, as hotspots of microbial Hg methylation, are ideal niches in which to search for novel putative Hg-methylators [33]. Mangrove sediments receive Hg inputs of anthropogenic origin as a substantial source for methylation [34–36]. In addition, abiotic factors such as relatively warm temperatures and low oxygen and high carbon contents in mangrove sediments could accelerate MeHg production by facilitating the activity of microbial methylators [29, 37, 38]. In this study, we measured the concentrations of total mercury (THg) and MeHg and applied metagenomic and metatranscriptomic analyses to investigate Hg-methylating microorganisms in the sediments of mangrove ecosystems across southeastern China (Fig. S1). We provide evidence of the potential novel Hg-methylators and their key roles in hgcA evolution.
Results and discussion
Hg and MeHg biogeochemistry in mangroves
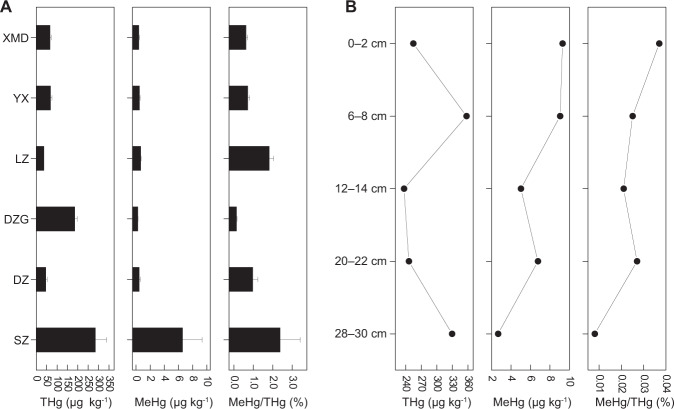
Total Hg (THg) and MeHg concentrations in mangrove sediments are listed in Fig. 1 and Table S1. The concentrations of THg ranged from 36 to 357 µg kg−1 dry sediment, and MeHg ranged from 0.25 to 9.3 µg kg−1 dry sediment. Similar MeHg concentrations have been reported in other sediments [29]. Of all the sampling sites, Shenzhen (SZ) had the highest concentrations of THg and MeHg, which might be due to intensive anthropogenic emissions, such as wastewater discharge, that increase the Hg accumulation in mangrove sediments in Shenzhen [33]. In addition, the ratio of MeHg to THg decreased from 3.65% in surface (0–2 cm) sediments to 0.81% in bottom (28–30 cm) sediments in Shenzhen mangroves, suggesting a stronger activity of Hg-methylating microorganisms in the top sediments.
Fig. 1. Mercury profiles in mangrove sediments.
A Concentrations of total Hg (THg), MeHg and MeHg as a percentage of THg at different sites. B Concentrations of total Hg (THg), MeHg and MeHg as a percentage of THg at different depths in Shenzhen.
Identification of putative mercury-methylating microorganisms in mangrove sediments
By hidden Markov model (HMM) search, we identified a final set of 1087 hgcA genes from 10 mangrove metagenomic assemblies (Table S2). Among these, 706 hgcA gene sequences had downstream hgcB genes, suggesting that these are likely functional Hg-methylation genes. HgcA sequences that binned to MAGs were assigned taxonomic classifications according to GTDB taxonomy of MAGs phylogenies. Those HgcA sequences that not binned to MAGs were searched against reference HgcA and HgcAB sequences database published recently [39] using BLASTp (Table S3). The 1087 identified hgcA sequences were clustered into 28 phyla (Table S2). According to the phylogeny, HgcA sequences from the same taxonomy were not clustered into one clade (Fig. S2), which might be due to horizontal gene transfer (HGT) [40, 41]. The hgcA gene abundances varied among different sites and were highest in Shenzhen (Fig. S3), suggesting that Hg-methylators were most abundant at this site. Moreover, the transcripts of hgcA genes were highest at 6–8 cm in Shenzhen, suggesting that Hg-methylators were more active in subsurface sediment layers. We further found that some environmental factors (pH, Cl−, and SO42− concentrations) were significantly correlated to MeHg/THg ratio (Fig. S4). It is widely accepted that TOC and pH affect Hg bioavailability and methylators’ activities [42–44]. Indeed, previous studies had suggested that lower pH could increase Hg mobility and provide more available Hg for microbial Hg methylation [45]. We also observed a significantly positive correlation between the RPKM (reads per kilobase of transcript per million mapped reads) values of hgcA genes and MeHg/THg ratio (p < 0.01). Further correlation analysis showed that the most abundant mercury methylators such as Deltaproteobacteria, Chloroflexi, Planctomycetes, Spirochaetes, Nitrospirae, Euryarchaeota were significantly correlated with MeHg/THg ratio (Fig. S5), suggesting all these microorganisms play important roles in mercury methylation. Overall, the most abundant hgcA genes were observed in subsurface sediments of Shenzhen, where they coincided with the MeHg peaks. Since large amounts of terrestrial and riverine Hg reach and accumulate in coastal environments, the top layers of the mangrove sediments supply substrates for Hg-methylators [36, 46].
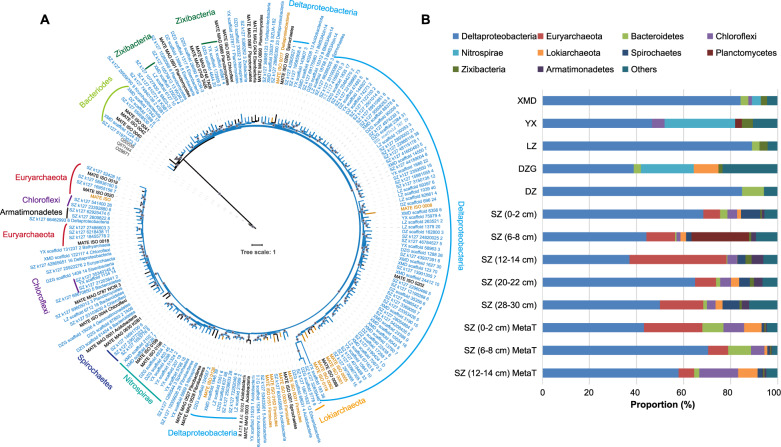
A total of 1183 MAGs with completeness >50% and contamination <10% were recovered from 10 mangrove metagenomics datasets after dereplication. Among these, 157 MAGs belonging to 20 groups at phylum level (Acidobacteria, Armatimonadetes, Bacteroidetes, Bathyarchaeota, Chloroflexi, Edwardsbacteria, Eisenbacteria, Euryarchaeota, Fibrobacteres, Gemmatimonadetes, Krumholzibacteriota, KSB1, Lokiarchaeota, Nitrospirae, Planctomycetes, Proteobacteria, Spirochaetes, TA06, WOR-3, Zixibacteria) have hgcA genes (Fig. 2A). Genome size, completeness, and contamination of the 157 MAGs are summarized in Table S4. Among these, 139 MAGs had an hgcB sequence paired with hgcA. Well-known methylators Deltaproteobacteria and Euryarchaeota accounted for 81 and 7 MAGs, respectively, representing more than one-half of all identified putative methylators. Putative methylators belonging to the Acidobacteria, Bacteroidetes, Chloroflexi, Eisenbacteria, Fibrobacteres, KSB1, Nitrospirae, Planctomycetes, Spirochaetes, TA06, and WOR-3 have previously been identified from isolate genomes or reconstructed MAGs [6]. We also detected hgcA in phyla that, to our knowledge, have not previously been shown to carry the genes. These phyla include two bacterial phyla (Armatimonadetes and Gemmatimonadetes), two candidate archaeal phyla (Bathyarchaeota and Lokiarchaeota), and three candidate bacterial phyla (Edwardsbacteria, Krumholzibacteriota, and Zixibacteria). The most abundant putative Hg-methylator was Deltaproteobacteria, followed by Euryarchaeota, Bacteroidetes, Chloroflexi, Nitrospirae, and Lokiarchaeota (Fig. 2B). Deltaproteobacteria were abundant and active in all the samples, showing that they are predominant and widespread Hg-methylators across all mangrove sediments in Southeastern China. Previous studies have also shown that Deltaproteobacteria are the most abundant Hg-methylators in coastal sediments [10, 14]. In the present study, Desulfatiglandales, Desulfobacterales, Desulfobulbales, Desulfuromonadales, and Syntrophobacterales were the most frequent and abundant five orders in Deltaproteobacteria (Table S4). The relative abundances of Euryarchaeota hgcA-carriers were highest in the Shenzhen (SZ) site. Within Chloroflexi, hgcA-carriers were all in the Anaerolineae class, while experimental confirmation of Hg-methylation capacity in this phylum is still needed. Nitrospirae hgcA-carriers were more abundant in Yunxiao (YX), which is characterized by high nitrate concentration and nitrite oxidation rates [47]. The relative abundances of Lokiarchaeota hgcA-carriers account for 10 and 8% of the total hgcA-containing MAGs reads in Dongzhaigang and Shenzhen, respectively (Fig. 2). Our findings suggest a broad diversity of putative Hg-methylators.
Fig. 2. Summary of 157 Metagenome-Assembled Genomes (MAGs) recovered with hgcA from mangrove sediments.
A Maximum-likelihood phylogenetic tree of HgcA amino acid sequences (1000 bootstraps; values >90% are shown by gray dots at the nodes). The 157 HgcA sequences from corresponded MAGs (Table S4) obtained in this study are highlighted in blue. HgcA sequences retrieved from public databases are shown in black. Experimentally confirmed HgcA are shown in brown. HgcA paralogues from non-methylating microorganisms were used as outgroups and are shown in gray. B Relative abundances of hgcA-carrying MAGs affiliated to different taxonomic groups in different mangrove sediment samples. Different taxonomic groups are represented by different colors. MetaT represent metatranscriptomic.
The potential for Hg methylation by Asgard archaea
In this study, we identified hgcA gene sequence in Lokiarchaeota MAGs (DZG_bin1.240 and SZ_1_bins.287) (Table S4). To investigate the Hg methylation potentials of Asgard MAGs, we searched publicly available Asgard MAGs and obtained 104 Asgard MAGs that contain hgcA gene (Table S5). The 104 hgcA-carrying Asgard MAGs comprised 8 phyla: Borr-, Kari-, Hod-, Heimdall-, Loki-, Hel-, Hermod-, and Thorarchaeota, respectively. Asgard HgcA sequences contained the conserved NVWCAA motif (or is NVWCGA in one genome of Lokiarchaeota) in the cap helix (Fig. S6). Asgard HgcB sequences contained two tandem CX2CX2CX3C motifs (Fig. S6). To eliminate the possibility of contamination, all genes identified on the contigs containing hgcA genes in Asgard MAGs were blast against 201,003 Asgard amino acid sequences downloaded from NCBI as references (Table S6). All the identity of blast results were above 20% and 3318 of 3743 sequences were above 50%, suggesting these genes on the contigs likely belong to Asgard archaea.
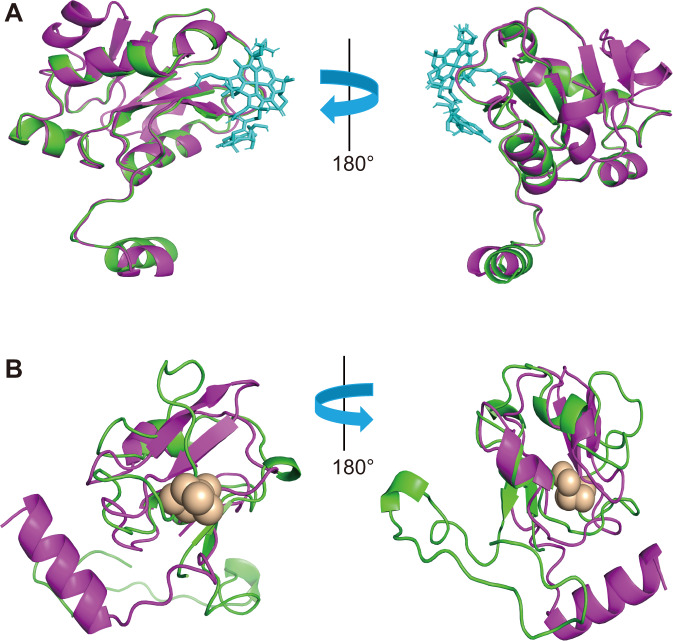
Asgard archaea have never been implicated as putative methylators. The complete genome of the only one co-cultured representative of Lokiarchaeota archaea, Candidatus Prometheoarchaeum syntrophicum strain MK-D1 [48], showed that it contained hgcAB genes. However, it has not been experimentally tested for Hg-methylation capacity. Based on its HgcAB protein sequence and structure, we propose Lokiarchaeota as a potential mercury methylator. Homology models were constructed to predict the three-dimensional structure of Lokiarchaeota HgcA and HgcB amino acid sequences (Fig. 3A and B). Lokiarchaeota HgcA protein structure consisted of a corrinoid binding domain at the N-terminus and five transmembrane spanning helices at the C-terminus. The ligand cobalamin facilitates the transfer of methyl groups to inorganic Hg [49]. This structure is similar to the previously reported HgcA from the experimentally validated Hg-methylating strain Desulfovibrio desulfuricans ND132 [5, 50]. Modelling of the Lokiarchaeota HgcB protein demonstrated two 4Fe-4S motifs at the N-terminal and the Cys tail at the C-terminal, with the function of transferring electrons and binding the Hg(II) substrate, respectively. Overall, our findings strongly suggest that Asgard archaea represent novel potential Hg-methylators. The extension of Hg methylators’ phylogeny into Asgard also broadly expands the niches associated with MeHg production. Asgard archaea carrying hgcA have been retrieved from a range of anoxic environments, including marine/coastal, lake environments, hydrothermal vent, cold seep, and permafrost sediments (Table S5), suggesting MeHg production by Asgard archaea across a wide range of ecological environments might be ignored previously.
Fig. 3. Structure comparison between HgcA and HgcB from Lokiarchaeota and Desulfovibrio desulfuricans ND132.
A Superposition of Lokiarcheaota-HgcA (green) and Desulfovibrio desulfuricans ND132-HgcA (purple) functional domain homology models. The functional domain is bound to cobalamin shown cyan-blue stick; B Superposition of Lokiarchaeota-HgcB (green) and Desulfovibrio desulfuricans ND132-HgcB (purple) homology models. Model of full-length Lokiarchaeota-HgcB in complex with two [4Fe-4S] clusters; Interactions between the protein and iron are shown by brown ball.
Hg-methylation process includes reduction of the corrinoid cofactor to form Co(I), methylation of Co(I) to form CH3-Co(III), and methyl transfer to Hg(II) to form CH3Hg(II). The presence of hgcAB in Asgard archaea might be insufficient for Hg methylation, and additional genes functioning in uptake and transport of Hg(II) and MeHg export might be needed for effective MeHg production [50]. No experimental assays have been conducted for mercury methylation of Asgard archaea. Therefore, experimental confirmation of Hg-methylation capacity in Asgard archaea needs to be provided by future research.
Additional metabolic features of hgcA-carrying Asgard MAGs
The functional potentials of 104 hgcA-carrying Asgard MAGs were inferred by reference to eggNOGmapper annotations (Fig. S7, Table S7). Asgard lineages mostly inhabited anoxic marine, coastal, and lake sediments. However, some of them contained genes encoding for terminal oxidases, which allowed them to accommodate the lower levels of oxygen in the benthic habitats of shallow layers. Genome analysis reveals that Hod- and Kariarchaeota contain genes encoding for respiratory complexes cytochrome c oxidase (coxBCP). Hodarchaeota also contain genes encoding for cytochrome bd ubiquinol oxidase (cydAB). The presence of cox and cyd genes indicates that Hod- and Kariarchaeota might represent facultative aerobes under fluctuating oxygen conditions. Furthermore, genes encoding for flagellar proteins (flg, fli, and mot) were identified in Hermod-, Hod-, and Thorarchaeota, enabling them to reposition optimally to suitable O2 levels.
Asgard archaea appear to have diverse metabolic capabilities in C, N, S cycles (Fig. S7, Table S7). The presence of an almost-complete archaeal Wood-Ljungdahl (WL) pathway and carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in Borr-, Hela-, Hermoda-, Loki-, and Thorarchaeota suggests that they can fix CO2 or, in a reverse function, produce it [51]. HgcA and the CODH have structural and functional similarities, suggesting associations between the two pathways [2]. Genes encoding for trimethylamine-specific methyltransferase (MttB) were found in Hermod-, Hod-, Loki-, and Thorarchaeota, suggesting they have potential for utilization of trimethylamine. Helarchaeota from marine and coastal sediments have acetyl-coenzyme M reductase (acr) coding genes, suggesting they have the potential for short-chain hydrocarbon oxidation [19, 23]. Kariarchaeota MAGs contain genes encoding nitrate reductase (Nar). Hel-, Hod-, and Thorarchaeota MAGs contain genes encoding nitrite reductase (Nir), suggesting nitrite can be a nitrogen source for them. We also detected the sulfate reduction–related genes sat and cys within most Asgard archaea. Above all, Hg methylators in Asgard are also involved in diverse metabolic functions and growth strategies.
Although MeHg is much more toxic than Hg(II), MeHg can be exported from the cell [52]. Similarly, As(V) is reduced to the more toxic As(III) to export from the cell. Hg methylation and As reduction are both detoxification mechanisms deployed against environmental stress. In our study, most methylators in Asgard also contained genes encoding for arsenate reductase (Ars), Cu transport detoxification protein (CopA), and Fe2+ transport system protein (FeoAB) (Table S7), protecting them against other toxin compounds. Interestingly, we selected several Lokiarchaeota MAGs containing hgcAB and ars regions and found ars were in the neighborhood of hgcAB within the same scaffolds (Fig. S8, Table S8). Gene clusters hgcAB were flanked by arsR, encoding an arsenical resistance operon repressor; arsB, encoding an arsenical pump membrane protein; arsM, an arsenite S-adenosylmethionine methyltransferase; arsC, encoding a thioredoxin- or glutaredoxin-dependent arsenite reductase; and arsA, encoding arsenite-activated ATPase. The potential for metal resistance and transformation may allow Asgard to adapt to metal-containing environments.
The diversity of archaeal hgcA carriers
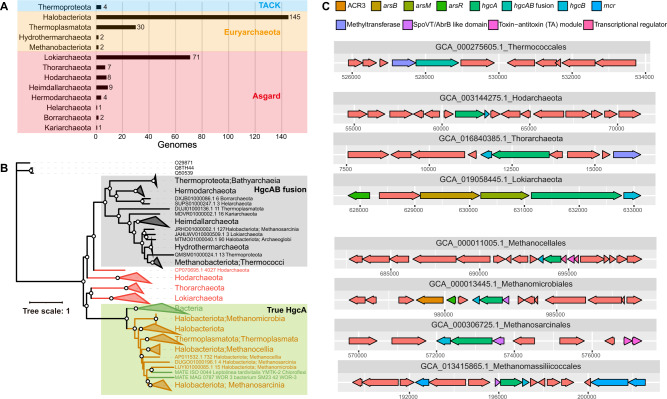
To investigate the diversity of hgcAB genes in archaea, we searched publicly available archaeal isolate genomes and MAGs and obtained 287 archaeal potential Hg methylators (Table S9). The 287 archaeal hgcA-carriers belongs to three superphyla: TACK, Euryarchaeota, and Asgard (Fig. 4A). HgcA-encoding genes were found less frequently in TACK. HgcA-encoding genes were found in Bathyarchaeia and Thermoproteia in TACK. The occurrence of hgcAB among Euryarchaeota was confined to seven classes: Archaeoglobi, Methanocellia, Methanomicrobia, Methanosarcinia, Hydrothermarchaeia, Thermococci, and Thermoplasmata. Methanocellia with hgcAB are widespread in rice paddies [53], and Methanomassiliicoccus luminyensis with hgcAB was isolated from human feces [54], suggesting a potential human health risk. Some hgcA-carriers in Methanomicrobia, Methanosarcinia, and Methanomassiliicoccales were obtained from permafrost soils, which also need further attention concerning elevated Hg deposition in the Arctic ecosystem [55]. This study remarkably expands the diversity and distribution of known hgcA-carriers in the domain Archaea.
Fig. 4. Diversity and phylogenetic analyses of archaeal HgcA amino acid sequences.
A Numbers of putative methylating genomes belonging to each taxa. B Maximum-likelihood phylogenetic tree of HgcA amino acid sequences (1000 bootstraps; values >90% are shown by white dots at the nodes). Gray-shaded clades represent the fused HgcAB sequences. Green-shaded clades represent the true HgcA sequences. HgcA sequences belonging to Euryarchaeota and Asgard are represented by orange and red, respectively. C Gene clusters of contigs containing hgcA in various archaeal taxa. Colors correspond to predicted functions of each gene. HgcAB sequences were identified based on HMMs, while all other genes were based on prodigal predictions.
A phylogenetic tree was constructed of HgcA amino acid sequences from 287 archaeal hgcA-carrying MAGs and 3 HgcA paralogues from non-methylating microorganisms (Fig. 4B). According to the phylogeny, some hgcA and hgcB sequences are fused into a single open reading frame in 30 hgcA-carrying genomes (Table S9), as shown in the gray-shaded clades in Fig. 4B. Among these, only two (Methanococcoides methylutens and Pyrococcus furiosus) were cultured organisms, which were experimentally validated to be unable to produce MeHg, indicating that the fused HgcAB may not retain the function of Hg methylation [2, 56]. HgcA from five Asgard phyla (Borrarchaeota, Kariarchaeota, Heimdallarchaeota, Helarchaeota, and Hermodarchaeota) were fused with HgcB sequences. Most hgcAB fusion carriers were obtained from extreme environments, including a hydrothermal vent and a saline lake (Table S9). Some HgcA amino acid sequences from three Asgard phyla (Hod-, Loki-, and Thorarchaeota) were in a transitional stage between fused HgcAB and true HgcA, which have implications for the underlying evolutionary mechanisms of the methylation pathway (Fig. 4B). Experimental confirmation of Hg-methylation capacity in Asgard archaea could be launched to fill this gap. Since HgcA and carbon monoxide dehydrogenase/acetyl-CoA synthase gamma subunit (CFeSP) had the paralogous relationship, we constructed phylogenetic tree of CFeSP (Fig. S9). We found CFeSP in Euryarchaeota appear in all clades, suggesting CFeSP might originate from Euryarchaeota, which is consistent with previous propose [2]. Microorganisms with syntrophic interactions acquired hgcAB through HGT.
We provided the blast hits of 291 HgcA amino acid sequences to the 32 experimentally confirmed HgcA sequences (Table S10). Corresponded to the phylogenetic tree, HgcAB fusion clade sequences had a lower identity (<47%) and lower align length (<266 bp). Asgard HgcA clade sequences had a lower identity (<47%) and higher align length (>268 bp). Euryarchaeota and bacterial true HgcA clade sequences had a higher identity (>50%) and higher align length (>266 bp). Moreover, contig synteny (Fig. 4C) was combined with a phylogenetic tree (Fig. 4B). The contig synteny varied among the three clades (Table S11). HgcAB fusion-containing contigs contained methyltransferase. Asgard HgcA-containing contigs contained methyltransferase and Ars-related proteins. Euryarchaeota true HgcA-containing contigs contained Ars-related proteins, SpoVT/AbrB like domain and toxin-antitoxin (TA) module. The variation of gene clusters have an implication for evolution of Hg methylation function.
In summary, MeHg concentrations and hgcA gene abundances were reported, and both were highest in Shenzhen mangrove sediments. A total of 157 hgcA-carrying MAGs from mangrove sediments and 104 additional Asgard hgcA-carrying MAGs from publicly available databases were obtained. According to the relative abundance and expression of these Hg-methylators, Deltaproteobacteria, Euryarchaeota, Bacteroidetes, Chloroflexi were the most abundant and transcriptionally active Hg-methylating groups, suggesting that they could contribute most to the MeHg production in mangrove sediments. Multiple lines of evidence support the conclusion that Asgard are previously unrecognized microbial Hg-methylators. Further studies should experimentally confirm Hg-methylation function in Lokiarchaeota enrichments.
Materials and methods
Sampling and chemical analysis
A total of 83 sediment samples were collected from six mangrove nature reserves: Ximendao National Marine Reserve in Zhejiang Province (XMD), Yunxiao Zhangjiangkou National Nature Reserve in Fujian Province (YX), Shenzhen Futian National Nature Reserve in Guangdong Province (SZ), Leizhou Nature Reserve in Guangdong Province (LZ), Dongzhaigang National Nature Reserve in Hainan Province (DZG), and Danzhou Xinyinggang Nature Reserve in Hainan Province (DZ) (Fig. S1, Table S1). Sediment samples in XMD, YX, LZ, DZG, and DZ were collected from October to December 2017 and stored at −40 °C. In SZ, samples from 0–2, 6–8, 12–14, 20–22, and 28–30 cm of a 32 cm sediment core were collected in April 2017 and stored at −40 °C. The physiochemical properties of these sediments were distinct from one another, as reported previously [57, 58]. Total Hg (THg) and methylmercury (MeHg) contents in the sediments were measured as previously [29]. Among all the samples, 10 representative sediment samples (bold in Table S1) were chosen for metagenomics sequencing. Three subsurface samples in XMD, YX, LZ, DZG, and DZ were combined as one sample, respectively.
Nucleic acid extraction, metagenome/metatranscriptome sequencing and assembly
Genomic DNA was extracted from 5 g of wet sediment samples using DNeasy PowerSoil kit (Qiagen, Germany) according to the manufacturer’s protocol. Shotgun metagenomic sequencing (2 × 150 bp) of the above 10 samples was performed using HiSeq 2000 (Illumina, USA) at Novogene Bioinformatics Technology Co., Ltd. (Tianjin, China). About 110 Gbp of sequence data were generated for each sample. For each sample, raw reads of metagenomic datasets (2 × 150 bp paired-end) were dereplicated and trimmed to remove replicated and low-quality reads using Sickle (https://github.com/najoshi/sickle) with the default settings. The trimmed reads were de novo assembled into longer scaffolds using IDBA-UD (v1.1.1) with the following parameters: -mink 65, -maxk 145, and -step 10 [59]. For five sediment samples in SZ, the trimmed reads were co-assembled using MEGAHIT v1.2.8 [60]. The assembled scaffolds longer than 500 bp were translated by Prodigal (v.2.6.3) using the “-p meta” parameters [61]. RNA extraction and metatranscriptomic analysis from the 0–2, 6–8, and 12–14 cm layers in SZ has been described in a previous study [62].
Metagenomic binning and annotation
Assembled scaffolds were binned using MetaBAT (v.2.12.1) [63], then aggregated using DAS Tool [64]. The completeness, contamination, and strain heterogeneity of recovered MAGs were evaluated by using CheckM (v1.0.11) [65]. Only those bins which were of medium to high quality (i.e., ≥50% complete, <10% contaminated) were analyzed further. The MAGs obtained from six sites were dereplicated using dRep (v2.5.4) [66] at 97% cutoff. After dereplication, we retrieved 125 bins in XMD, 96 bins in YX, 89 bins in LZ, 100 bins in DZG, 68 bins in DZ, and 705 bins in SZ. The dereplicated bins were selected for further analysis. The taxonomy of each bin was estimated using GTDB-Tk (v1.5.0) [67]. The MAGs were translated by Prodigal using the “-p meta” parameters and annotated using the KEGG Automatic Annotation Server (www.genome.jp/tools/kaas/) [68] and eggNOG-mapper (http://eggnog-mapper.embl.de/) [69].
Identification of hgcAB
The hgcAB gene identification in shotgun metagenomics datasets using HMM is a reliable method for determining Hg-methylator abundance and diversity [13]. A custom HMM of the HgcA sequence was built with hmmbuild from the hmmer softerware [70] using HgcA amino acid sequences with and without experimental validation from Hg-MATE database [39] as reference. Putative hgcA sequences were searched from each assembly using hmmsearch (v3.1b2) [71] with score >131.8 and E value ≤ 1e−05 as the cutoff. Each hit was then manually confirmed by validating the presence of conserved sequence domains (cap helix domain (N(V/I)WCA(A/G) and at least four transmembrane domains) [5]. The transmembrane domains were predicted by TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). Sequences annotated as hgcA were extracted, resulting in 1087 hgcA sequences (Table S2). The genes immediately downstream of each hgcA gene were extracted, and sequences that contained the conserved CX2CX2CX3C motif were scored as hgcB genes. HgcA sequences that binned to MAGs were assigned taxonomic classifications according to GTDB taxonomy of MAGs phylogenies. Those HgcA sequences that not binned to MAGs were searched against reference HgcA and HgcAB sequences database published recently [39] using BLASTp (Table S3).
Phylogenetic analysis
For HgcA phylogenies, putative HgcA amino acid sequences and reference HgcA sequences [39] were aligned using MUSCLE (v3.8.31) [72] and trimmed using trimAL with option “-gt 0.95” [73]. The HgcA maximum likelihood (ML) tree was constructed with IQ-TREE (v1.3.10) [74] under the LG + F + G4 protein model chosen according to BIC [75] with option “-bb 1000”.
We also investigated the phylogenetic relationship between HgcA and the gamma subunit of the acetyl-CoA synthase complex (CFeSP) involved in the WL pathway. The MAGs were translated by Prodigal using the “-p meta” parameters and annotated using the KEGG Automatic Annotation Server (www.genome.jp/tools/kaas/). CFeSP corresponds to KO number K00197. The CFeSP ML tree was conducted with IQ-TREE (v1.3.10) under the LG + F + G4 protein model chosen according to BIC with option “-bb 1000”. All trees were visualized by using iTOL [76].
Protein homology modeling
The amino acid sequences of HgcA and HgcB was extracted from the ‘Candidatus Prometheoarchaeum syntrophicum’ strain MK-D1 and Desulfovibrio desulfuricans ND132 genomes. Computational homology modeling was performed to identify novel Asgard Hg-methylators. The three-dimensional structures of the putative HgcA and HgcB sequences were built using AlphaFold [77]. The cobalamin and [4Fe-4S] clusters were docked to the HgcA and HgcB using AutoDock VinA [78], respectively. All of the structure were visualized, and exported as images using PyMOL (http://www.pymol.org).
Relative abundance and expression of hgcA-carrying MAGs and hgcA genes
The 157 hgcA-carrying MAGs and 1087 hgcA gene sequences were used to recruit reads from metagenomics and metatranscriptomic datasets to calculate relative abundance and expression, respectively. The abundance/expression of dereplicated MAGs was estimated by mapping the metagenomic/metatranscriptomic reads, respectively, to the contigs of the MAGs using Bowtie2 (v2.2.8) [79]. BEDTools was applied to calculate coverage values for the contigs [80]. The relative abundance of each hgcA-carrying MAG is defined as the number of reads mapped to individual hgcA-carrying MAG divided by the total number of metagenomic/metatranscriptomic reads mapped to all hgcA-carrying MAGs in every sample.
The abundance and transcription of hgcA genes were determined by mapping metagenomic/metatranscriptomic reads to annotated hgcA gene sequences using Burrows-Wheeler Aligner (BWA, v0.7.5a-r405) with default settings [81]. The read coverage in genes was calculated using BEDTools. The mapped reads were then normalized to the length of the gene and the number of mapped reads using the “Fragments per kilobase per million” or “FPKM” metric. For hgcA, the relative abundance of each group was calculated by dividing the FPKM value of the individual group by the sum of FPKM values of all groups.
Explore the diversity of archaeal hgcA carriers
To investigate the diversity of hgcAB genes in archaea, we downloaded publicly available archaeal metagenome-assembled genomes (MAGs) from the GenBank database in December 2021. Using the criteria described above, we identified 287 hgcA-carrying archaeal MAGs, including 104 hgcA-carrying Asgard, 4 TACK, and 179 Euryarchaeota genomes, from this data set. The taxonomy of archaeal MAGs was estimated using GTDB-Tk (v1.5.0) [67].
Supplementary information
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (grant no. 92251306, 42007217, 32225003, 31970105, 92051102, and 32070108), the Shenzhen Science and Technology Program (grant no. JCYJ20200109105010363 and KCXFZ20201221173404012), and the Innovation Team Project of Universities in Guangdong Province (No. 2020KCXTD023).
Author contributions
C-JZ, Y-RL and ML conceived this study. C-JZ, YL, JP and MC performed sampling, DNA extraction and the data analyses. GC and ZL performed protein structure computation. X-QZ performed the laboratory measurement of physiochemical properties and Hg concentrations. C-JZ wrote, and all authors edited and approved the paper.
Data availability
The genome bins generated and analyzed during the current study are available in the National Omics Data Encyclopedia (NODE, https://www.biosino.org/node/) database and can be viewed under Project OEP003752. The HgcAB amino acid sequences, and their alignment and phylogenetic tree have also been uploaded to the NODE database under Project OEP003752. The metagenome and metatranscriptome raw data generated during the current study can be viewed in NODE under Project OEP001892 and OEP000712.
Code availability
The sources of the code and programs used for analyses are mentioned in the Methods, and are also available at GitHub (https://github.com/cui-jing/Mangrove-hgcAB).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cui-Jing Zhang, Yu-Rong Liu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01360-w.
References
- 1.Hsu-Kim H, Kucharzyk KH, Zhang T, Deshusses MA. Mechanisms regulating mercury bioavailability for methylating microorganisms in the aquatic environment: A critical review. Environ Sci Technol. 2013;47:2441–56. doi: 10.1021/es304370g. [DOI] [PubMed] [Google Scholar]
- 2.Podar M, Gilmour CC, Brandt CC, Soren A, Brown SD, Crable BR, et al. Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci Adv. 2015;1:e1500675. doi: 10.1126/sciadv.1500675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu YR, Johs A, Bi L, Lu X, Hu HW, Sun D, et al. Unraveling microbial communities associated with methylmercury production in paddy soils. Environ Sci Technol. 2018;52:13110–8. doi: 10.1021/acs.est.8b03052. [DOI] [PubMed] [Google Scholar]
- 4.Lee C-S, Fisher NS. Bioaccumulation of methylmercury in a marine copepod. Environ Toxicol Chem. 2017;36:1287–93. doi: 10.1002/etc.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parks JM, Johs A, Podar M, Bridou R, Hurt RAJ, Smith SD, et al. The genetic basis for bacterial mercury methylation. Science. 2013;339:1332–5. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 6.McDaniel EA, Peterson BD, Stevens SLR, Tran PQ, Anantharaman K, McMahon KD. Expanded phylogenetic diversity and metabolic flexibility of mercury-methylating microorganisms. mSystems. 2020;5:e00299–20. doi: 10.1128/mSystems.00299-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper CJ, Zheng K, Rush KW, Johs A, Sanders BC, Pavlopoulos GA, et al. Structure determination of the HgcAB complex using metagenome sequence data: Insights into microbial mercury methylation. Commun Biol. 2020;3:320. doi: 10.1038/s42003-020-1047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerin EJ, Gilmour CC, Roden E, Suzuki MT, Coates JD, Mason RP. Mercury methylation by dissimilatory iron-reducing bacteria. Appl Environ Microbiol. 2006;72:7919–21. doi: 10.1128/AEM.01602-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilmour CC, Podar M, Bullock AL, Graham AM, Brown SD, Somenahally AC, et al. Mercury methylation by novel microorganisms from new environments. Environ Sci Technol. 2013;47:11810–20. doi: 10.1021/es403075t. [DOI] [PubMed] [Google Scholar]
- 10.Capo E, Bravo AG, Soerensen AL, Bertilsson S, Pinhassi J, Feng C, et al. Deltaproteobacteria and Spirochaetes-like bacteria are abundant putative mercury methylators in oxygen-deficient water and marine particles in the Baltic Sea. Front Microbiol. 2020;11:574080. doi: 10.3389/fmicb.2020.574080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gionfriddo CM, Tate MT, Wick RR, Schultz MB, Zemla A, Thelen MP, et al. Microbial mercury methylation in Antarctic sea ice. Nat Microbiol. 2016;1:16127. doi: 10.1038/nmicrobiol.2016.127. [DOI] [PubMed] [Google Scholar]
- 12.Jones DS, Walker GM, Johnson NW, Mitchell CPJ, Coleman Wasik JK, Bailey JV. Molecular evidence for novel mercury methylating microorganisms in sulfate-impacted lakes. ISME J. 2019;13:1659–75. doi: 10.1038/s41396-019-0376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen GA, Gionfriddo CM, King AJ, Moberly JG, Miller CL, Somenahally AC, et al. Determining the reliability of measuring mercury cycling gene abundance with correlations with mercury and methylmercury concentrations. Environ Sci Technol. 2019;53:8649–63. doi: 10.1021/acs.est.8b06389. [DOI] [PubMed] [Google Scholar]
- 14.Villar E, Cabrol L, Heimburger-Boavida LE. Widespread microbial mercury methylation genes in the global ocean. Environ Microbiol Rep. 2020;12:277–87. doi: 10.1111/1758-2229.12829. [DOI] [PubMed] [Google Scholar]
- 15.Lin H, Ascher DB, Myung Y, Lamborg CH, Hallam SJ, Gionfriddo CM, et al. Mercury methylation by metabolically versatile and cosmopolitan marine bacteria. ISME J. 2021;15:1810–25. doi: 10.1038/s41396-020-00889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King JK, Kostka JE, Frischer ME, Saunders FM, Jahnke RA. A quantitative relationship that demonstrates mercury methylation rates in marine sediments are based on the community composition and activity of sulfate-reducing bacteria. Environ Sci Technol. 2001;35:2491–6. doi: 10.1021/es001813q. [DOI] [PubMed] [Google Scholar]
- 17.Regnell O, Watras CJ. Microbial mercury methylation in aquatic environments: A critical review of published field and laboratory studies. Environ Sci Technol. 2019;53:4–19. doi: 10.1021/acs.est.8b02709. [DOI] [PubMed] [Google Scholar]
- 18.Xie R, Wang Y, Huang D, Hou J, Li L, Hu H, et al. Expanding Asgard members in the domain of Archaea sheds new light on the origin of eukaryotes. Sci China Life Sci. 2022;65:818–29. doi: 10.1007/s11427-021-1969-6. [DOI] [PubMed] [Google Scholar]
- 19.Seitz KW, Dombrowski N, Eme L, Spang A, Lombard J, Sieber JR, et al. Asgard archaea capable of anaerobic hydrocarbon cycling. Nat Commun. 2019;10:1822. doi: 10.1038/s41467-019-09364-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Backstrom D, Juzokaite L, Vancaester E, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–8. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Makarova KS, Huang W-C, Wolf YI, Nikolskaya AN, Zhang X, et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature. 2021;593:553–7. doi: 10.1038/s41586-021-03494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang JW, Dong HP, Hou LJ, Liu Y, Ou YF, Zheng YL, et al. Newly discovered Asgard archaea Hermodarchaeota potentially degrade alkanes and aromatics via alkyl/benzyl-succinate synthase and benzoyl-CoA pathway. ISME J. 2021;15:1826–43. doi: 10.1038/s41396-020-00890-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai M, Liu Y, Yin X, Zhou Z, Friedrich MW, Richter-Heitmann T, et al. Diverse Asgard archaea including the novel phylum Gerdarchaeota participate in organic matter degradation. Sci China Life Sci. 2020;63:886–97. doi: 10.1007/s11427-020-1679-1. [DOI] [PubMed] [Google Scholar]
- 24.Baker BJ, De Anda V, Seitz KW, Dombrowski N, Santoro AE, Lloyd KG. Diversity, ecology and evolution of Archaea. Nat Microbiol. 2020;5:887–900. doi: 10.1038/s41564-020-0715-z. [DOI] [PubMed] [Google Scholar]
- 25.Farag Ibrahim F, Zhao R, Biddle Jennifer F, Atomi H. “Sifarchaeota,” a novel Asgard phylum from Costa Rican sediment capable of polysaccharide degradation and anaerobic methylotrophy. Appl Environ Micro. 2021;87:e02584–20. doi: 10.1128/AEM.02584-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adam PS, Borrel G, Brochier-Armanet C, Gribaldo S. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J. 2017;11:2407–25. doi: 10.1038/ismej.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai M, Richter-Heitmann T, Yin X, Huang W-C, Yang Y, Zhang C, et al. Ecological features and global distribution of Asgard archaea. Sci Total Environ. 2021;758:143581. doi: 10.1016/j.scitotenv.2020.143581. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C-J, Chen Y-L, Sun Y-H, Pan J, Cai M-W, Li M. Diversity, metabolism and cultivation of archaea in mangrove ecosystems. Mar Life Sci Tech. 2020;3:252–62. doi: 10.1007/s42995-020-00081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai SS, Yang Z, Tong Y, Chen L, Liu SY, Pan R, et al. Global distribution and environmental drivers of methylmercury production in sediments. J Hazard Mater. 2021;407:124700. doi: 10.1016/j.jhazmat.2020.124700. [DOI] [PubMed] [Google Scholar]
- 30.Tang WL, Liu YR, Guan WY, Zhong H, Qu XM, Zhang T. Understanding mercury methylation in the changing environment: Recent advances in assessing microbial methylators and mercury bioavailability. Sci Total Environ. 2020;714:136827. doi: 10.1016/j.scitotenv.2020.136827. [DOI] [PubMed] [Google Scholar]
- 31.Tsui MTK, Finlay JC, Balogh SJ, Nollet YH. In situ production of methylmercury within a stream channel in northern California. Environ Sci Technol. 2010;44:6998–7004. doi: 10.1021/es101374y. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Zhou Z, Pan J, Baker BJ, Gu JD, Li M. Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota. ISME J. 2018;12:1021–31. doi: 10.1038/s41396-018-0060-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei P, Zhong H, Duan D, Pan K. A review on mercury biogeochemistry in mangrove sediments: Hotspots of methylmercury production? Sci Total Environ. 2019;680:140–50. doi: 10.1016/j.scitotenv.2019.04.451. [DOI] [PubMed] [Google Scholar]
- 34.Beckers F, Rinklebe J. Cycling of mercury in the environment: Sources, fate, and human health implications: A review. Crit Rev Env Sci Tec. 2017;47:693–794. doi: 10.1080/10643389.2017.1326277. [DOI] [Google Scholar]
- 35.de Oliveira DC, Correia RR, Marinho CC, Guimaraes JR. Mercury methylation in sediments of a Brazilian mangrove under different vegetation covers and salinities. Chemosphere. 2015;127:214–21. doi: 10.1016/j.chemosphere.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Li R, Xu H, Chai M, Qiu GY. Distribution and accumulation of mercury and copper in mangrove sediments in Shenzhen, the world’s most rapid urbanized city. Environ Moni Assess. 2016;188:87. doi: 10.1007/s10661-016-5103-z. [DOI] [PubMed] [Google Scholar]
- 37.O’Connor D, Hou D, Ok YS, Mulder J, Duan L, Wu Q, et al. Mercury speciation, transformation, and transportation in soils, atmospheric flux, and implications for risk management: A critical review. Environ Int. 2019;126:747–61. doi: 10.1016/j.envint.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 38.Obrist D, Kirk JL, Zhang L, Sunderland EM, Jiskra M, Selin NE. A review of global environmental mercury processes in response to human and natural perturbations: Changes of emissions, climate, and land use. Ambio. 2018;47:116–40. doi: 10.1007/s13280-017-1004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capo E, Peterson BD, Kim M, Jones DS, Acinas SG, Amyot M, et al. A consensus protocol for the recovery of mercury methylation genes from metagenomes. Mol Ecol Resour. 2022; 10.1111/1755-0998.13687. [DOI] [PMC free article] [PubMed]
- 40.Gionfriddo CM, Wymore AM, Jones DS, Wilpiszeski RL, Lynes MM, Christensen GA, et al. An improved hgcAB primer set and direct high-throughput sequencing expand Hg-methylator diversity in nature. Front Microbiol. 2020;11:541554. doi: 10.3389/fmicb.2020.541554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu R-Q, Barkay T. Chapter two - microbial mercury transformations: Molecules, functions and organisms. Adv Appl Microbiol. 2022;118:31–90. doi: 10.1016/bs.aambs.2022.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Chételat J, Richardson MC, MacMillan GA, Amyot M, Poulain AJ. Ratio of methylmercury to dissolved organic carbon in water explains methylmercury bioaccumulation across a latitudinal gradient from north-temperate to arctic lakes. Environ Sci Technol. 2018;52:79–88. doi: 10.1021/acs.est.7b04180. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y-R, Dong J-X, Han L-L, Zheng Y-M, He J-Z. Influence of rice straw amendment on mercury methylation and nitrification in paddy soils. Environ Pollut. 2016;209:53–9. doi: 10.1016/j.envpol.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 44.Moreau JW, Gionfriddo CM, Krabbenhoft DP, Ogorek JM, DeWild JF, Aiken GR, et al. The effect of natural organic matter on mercury methylation by Desulfobulbus propionicus 1pr3. Front Microbiol. 2015;6:1389. doi: 10.3389/fmicb.2015.01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C-F, Ju Y-R, Chen C-W, Dong C-D. The distribution of methylmercury in estuary and harbor sediments. Sci Total Environ. 2019;691:55–63. doi: 10.1016/j.scitotenv.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Bravo AG, Bouchet S, Guédron S, Amouroux D, Dominik J, Zopfi J. High methylmercury production under ferruginous conditions in sediments impacted by sewage treatment plant discharges. Water Res. 2015;80:245–55. doi: 10.1016/j.watres.2015.04.039. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Su J, Zheng T, Yang X. Insights into the role of plant on ammonia-oxidizing bacteria and archaea in the mangrove ecosystem. J Soil Sediment. 2015;15:1212–23. doi: 10.1007/s11368-015-1074-x. [DOI] [Google Scholar]
- 48.Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, et al. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature. 2020;577:519–25. doi: 10.1038/s41586-019-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Riccardi D, Beste A, Smith JC, Parks JM. Mercury methylation by HgcA: Theory supports carbanion transfer to Hg(II) Inorg Chem. 2014;53:772–7. doi: 10.1021/ic401992y. [DOI] [PubMed] [Google Scholar]
- 50.Smith Steven D, Bridou R, Johs A, Parks Jerry M, Elias Dwayne A, Hurt Richard A, et al. Site-directed mutagenesis of HgcA and HgcB reveals amino acid residues important for mercury methylation. Appl Environ Micro. 2015;81:3205–17. doi: 10.1128/AEM.00217-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sousa FL, Neukirchen S, Allen JF, Lane N, Martin WF. Lokiarchaeon is hydrogen dependent. Nat Microbiol. 2016;1:16034. doi: 10.1038/nmicrobiol.2016.34. [DOI] [PubMed] [Google Scholar]
- 52.Schaefer JK, Rocks SS, Zheng W, Liang L, Gu B, Morel FMM. Active transport, substrate specificity, and methylation of Hg(II) in anaerobic bacteria. Proc Natl Acad Sci USA. 2011;108:8714. doi: 10.1073/pnas.1105781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakai S, Imachi H, Hanada S, Ohashi A, Harada H, Kamagata Y. Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage ‘Rice Cluster I’, and proposal of the new archaeal order Methanocellales ord. nov. Int J Syst Evol Microbiol. 2008;58:929–36. doi: 10.1099/ijs.0.65571-0. [DOI] [PubMed] [Google Scholar]
- 54.Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol. 2012;62:1902–7. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- 55.Dietz R, Sonne C, Basu N, Braune B, O’Hara T, Letcher RJ, et al. What are the toxicological effects of mercury in arctic biota? Sci Total Environ. 2013;443:775–90. doi: 10.1016/j.scitotenv.2012.11.046. [DOI] [PubMed] [Google Scholar]
- 56.Gilmour Cynthia C, Bullock Allyson L, McBurney A, Podar M, Elias Dwayne A, Lovley Derek R. Robust mercury methylation across diverse methanogenic archaea. mBio. 2018;9:e02403–17.. doi: 10.1128/mBio.02403-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan J, Chen Y, Wang Y, Zhou Z, Li M. Vertical distribution of Bathyarchaeotal communities in mangrove wetlands suggests distinct niche preference of Bathyarchaeota subgroup 6. Micro Ecol. 2019;77:417–28. doi: 10.1007/s00248-018-1309-7. [DOI] [PubMed] [Google Scholar]
- 58.Zhang C-J, Pan J, Duan C-H, Wang Y-M, Liu Y, Sun J, et al. Prokaryotic diversity in mangrove sediments across southeastern China fundamentally differs from that in other biomes. mSystems. 2019;4:e00442–19. doi: 10.1128/mSystems.00442-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 60.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 61.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang C-J, Pan J, Liu Y, Duan C-H, Li M. Genomic and transcriptomic insights into methanogenesis potential of novel methanogens from mangrove sediments. Microbiome. 2020;8:94. doi: 10.1186/s40168-020-00876-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3:836–43. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olm MR, Brown CT, Brooks B, Banfield JF. dRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–8. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–31. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 69.Huerta-Cepas J, Forslund K, Szklarczyk D, Jensen LJ, von Mering C, Bork P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–22. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. Plos ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinforma (Oxf, Engl) 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome bins generated and analyzed during the current study are available in the National Omics Data Encyclopedia (NODE, https://www.biosino.org/node/) database and can be viewed under Project OEP003752. The HgcAB amino acid sequences, and their alignment and phylogenetic tree have also been uploaded to the NODE database under Project OEP003752. The metagenome and metatranscriptome raw data generated during the current study can be viewed in NODE under Project OEP001892 and OEP000712.
The sources of the code and programs used for analyses are mentioned in the Methods, and are also available at GitHub (https://github.com/cui-jing/Mangrove-hgcAB).