Abstract
Objective
Skeletal muscle oxidative capacity is central to physical activity, exercise capacity and whole-body metabolism. The three estrogen-related receptors (ERRs) are regulators of oxidative metabolism in many cell types, yet their roles in skeletal muscle remain unclear. The main aim of this study was to compare the relative contributions of ERRs to oxidative capacity in glycolytic and oxidative muscle, and to determine defects associated with loss of skeletal muscle ERR function.
Methods
We assessed ERR expression, generated mice lacking one or two ERRs specifically in skeletal muscle and compared the effects of ERR loss on the transcriptomes of EDL (predominantly glycolytic) and soleus (oxidative) muscles. We also determined the consequences of the loss of ERRs for exercise capacity and energy metabolism in mice with the most severe loss of ERR activity.
Results
ERRs were induced in human skeletal muscle in response to an exercise bout. Mice lacking both ERRα and ERRγ (ERRα/γ dmKO) had the broadest and most dramatic disruption in skeletal muscle gene expression. The most affected pathway was “mitochondrial function”, in particular Oxphos and TCA cycle genes, and transcriptional defects were more pronounced in the glycolytic EDL than the oxidative soleus. Mice lacking ERRβ and ERRγ, the two isoforms expressed highly in oxidative muscles, also exhibited defects in lipid and branch chain amino acid metabolism genes, specifically in the soleus. The pronounced disruption of oxidative metabolism in ERRα/γ dmKO mice led to pale muscles, decreased oxidative capacity, histochemical patterns reminiscent of minicore myopathies, and severe exercise intolerance, with the dmKO mice unable to switch to lipid utilization upon running. ERRα/γ dmKO mice showed no defects in whole-body glucose and energy homeostasis.
Conclusions
Our findings define gene expression programs in skeletal muscle that depend on different combinations of ERRs, and establish a central role for ERRs in skeletal muscle oxidative metabolism and exercise capacity. Our data reveal a high degree of functional redundancy among muscle ERR isoforms for the protection of oxidative capacity, and show that ERR isoform-specific phenotypes are driven in part, but not exclusively, by their relative levels in different muscles.
Keywords: Skeletal muscle, Exercise capacity, Exercise intolerance, Mitochondrial oxidative metabolism, Estrogen-related receptor (ERR), Multi-minicore myopathy
HIGHLIGHTS
-
•
All three ERRs are induced by an endurance exercise bout in human skeletal muscle.
-
•
ERRs act redundantly to each other to drive oxidative metabolism genes in muscle.
-
•
ERRs regulate genes of BCAA and lipid catabolism in skeletal muscle.
-
•
Skeletal muscle ERRs are essential for exercise capacity.
-
•
Mice lacking muscle ERRs show no apparent defects in glucose or energy metabolism.
ABBREVIATIONS
- BCAA
branched chain amino acid
- BF
biceps femoris
- COX
cytochrome c oxidase
- DBD
DNA binding domain
- EDL
extensor digitorum longus
- ERR
estrogen-related receptor
- FAO
fatty acid oxidation
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GC
gastrocnemius
- KO
knock-out
- LBD
ligand binding domain
- OxPhos
oxidative phosphorylation
- PPAR
peroxisome proliferator-activated receptor
- PGC-1
peroxisome proliferator-activated receptor γ coactivator 1
- SDH
succinate dehydrogenase
- SRC
steroid receptor coactivator
- MHC
myosin heavy chain
- TCA
tricarboxylic acid
- TEM
transmission electron microscopy
1. Introduction
Physical fitness and high exercise capacity are among the best predictors of lifespan [1,2]. Moreover, consistent engagement in physical activity improves metabolic health, enhances insulin sensitivity and improves lipid parameters [3,4]. Conversely, poor physical fitness and inactivity are risk factors for type 2 diabetes, obesity, and cardiovascular disease [5,6]. While the benefits of physical activity and exercise are well recognized, the molecular determinants of exercise capacity are not fully elucidated.
The estrogen-related receptors (ERRα, ERRβ and ERRγ) are orphan members of the nuclear receptor family and target multiple facets of mitochondrial biogenesis and oxidative metabolism, i.e. pathways important for physical activity and exercise, in different cell systems [[7], [8], [9], [10], [11], [12]]. The three ERRs share a high degree of amino acid sequence similarity in their DNA binding and ligand binding domains (DBD and LBD, respectively) (Suppl. Fig. 1). Consistent with the high sequence conservation in the DBD (98% similarity between ERRα and the other two ERRs; 100% between ERRβ and ERRγ), all three ERRs recognize the same DNA sequence motif, and ERRα and ERRγ show similar genomic occupancies [8,13]. The LBDs of ERRs (78–81% similarity between ERRα and the other two ERRs; 88% between ERRβ and ERRγ) share some functional similarities, (e.g., constitutively active conformations in the absence of a ligand), but also have isoform-specific properties (e.g., ERRβ and ERRγ interact with coactivators of both the p160 SRC and PGC-1 families, while ERRα is restricted to PGC-1 family members) [[14], [15], [16], [17], [18], [19], [20]]. Outside the DBD and LBD, ERRα has further diverged from the other two ERRs, having distinct N-terminal and hinge domains, while ERRβ and ERRγ retain high similarity to each other in these two regions (73% and 80%, respectively) (Suppl. Fig.1). Fitting the high level of similarity between ERRs, all three ERRs can enhance the expression of oxidative phosphorylation (OxPhos) genes and promote oxidative metabolism [7,11,[21], [22], [23]]. However, the degree to which ERRs are physiologically important for oxidative metabolism in skeletal muscle is still unclear.
The strongest support for a role of ERRs in the regulation of skeletal muscle mitochondrial function comes from transgenic overexpression mouse models. Muscle-specific overexpression of ERRγ or ERRα enhances oxidative metabolism and leads to fiber-type switching, toward type I or IIa oxidative fibers, respectively [[24], [25], [26]]. ERRγ transgenic mice also show higher exercise capacity [25]. These studies, coupled to others showing that human ERRα is induced in response to an acute exercise bout [27,28] and that mouse ERRγ levels increase with training [24], i.e., signals known to promote muscle oxidative metabolism [29,30], have implicated ERRs as exercise-responsive activators of oxidative metabolism programs.
Surprisingly, loss of function approaches have so far provided little support for a physiological role of endogenous skeletal muscle ERRs in oxidative metabolism. Mice lacking ERRγ globally, i.e., in all tissues, die shortly after birth [7]; ERRγ heterozygote mice are viable and have mildly impaired exercise performance but show only modest and limited decreases in a few muscle oxidative metabolism genes [24]. Mice lacking ERRα globally are hypoactive and have impaired exercise capacity, but similarly show only a modest reduction in skeletal muscle oxidative metabolism gene expression [31]. Notably, the degree to which impaired exercise capacity in these mouse models can be attributed to a role of ERRs in skeletal muscle as opposed to other cell types, (e.g., neurons innervating the muscles or CNS nuclei controlling locomotor activity), has not been addressed. Muscle-specific ERRα KO mice show just mild or transient defects in the recovery of mitochondrial energetic capacity during regeneration in response to injury, but no differences in oxidative metabolism genes in non-injured muscles [32]. Mice with a double muscle-specific KO of ERRβ and ERRγ were reported to have decreased expression of type I fiber-specific genes in gastrocnemius, with no reference to changes in oxidative metabolism genes [33]. Thus, it remains to be established whether skeletal muscle ERRs have essential roles for the regulation of oxidative metabolism in vivo. Presently, it cannot be excluded that endogenous ERRs are minor contributors to the regulation of muscle energy metabolism. The ability of ERRs to activate mitochondrial biogenesis and oxidative function overlaps with that of other transcription factors that activate diverse aspects of mitochondrial biogenesis and oxidative capacity [e.g., PPARs, NRF1, GABP, KLFs and YY1 [34]], and these other factors may be the major drivers of mitochondrial oxidative metabolism in skeletal muscle in vivo. Alternatively, ERRs may have essential functions for the regulation of oxidative metabolism in muscle but act in a redundant manner to each other, as seen in adipocytes and cardiomyocytes [21,23,35]. A critical role of ERRs in muscle oxidative metabolism may thus be masked in single KO models. If this is the case, the question is which of the three ERRs are important for skeletal muscle function, and if any or all ERRs are similarly important in glycolytic and oxidative muscles.
To address the roles and significance of skeletal muscle ERRs for muscle mitochondria and muscle function, we generated mice lacking combinations of ERRs specifically in skeletal muscle and compared the impact of ERR loss on the transcriptomes of EDL and soleus, i.e., muscles rich in glycolytic or oxidative fibers, respectively. We then focused on the ERR combination that led to a robust defect in total ERR levels and function and addressed the importance of ERRs for mitochondrial oxidative metabolism, exercise capacity, and glucose and energy homeostasis.
2. Materials and methods
2.1. Human muscle biopsies and acute endurance exercise
Skeletal muscle samples were from the belly of the vastus lateralis muscle of male subjects that performed single leg cycling at 65% single leg VO2 peak until exhaustion and have been described previously [28].
2.2. Mice
Mice with floxed ERR alleles were backcrossed to the C57BL/6JBom background (Taconic Biosciences) for at least 10 generations and have been described previously [32,33,35]. To generate mice with muscle-specific deletions of ERRs, ERR floxed mice were crossed to mice expressing CRE recombinase under the control of the human α-skeletal actin (HSA) promoter [36]. ERR floxed littermates that did not carry the CRE transgene were used as controls and are referred to as WT. Mice were born and housed at room temperature (∼22 °C), fed a standard chow breeder diet, and euthanized between noon and 4 pm, after a 5–6 h fast, unless specified differently in procedures below. Tissues were rapidly excised, weighed, snap frozen in liquid nitrogen and stored at −80 °C. All animal procedures were approved by the Institutional Animal Care and Use Committees of the Scripps Research Institute and Johns Hopkins University. Male and female cohorts of mice were used in separate, sex- and age-matched experiments; sex, age and numbers of mice are specified in the figure legends. Full assessments of whole-body metabolism by indirect calorimetry, body composition, glucose- and insulin-tolerance tests, exercise capacity on a treadmill, and grip strength, as well as partial assessments of histology (H&E and enzymatic assays), mRNA and protein levels were done in both male and female mice. We did not find an effect of sex on the phenotypic changes seen in ERRα/γ dmKO mice. A male only group was tested for voluntary wheel running; and a female only group was assessed by indirect calorimetry while running in an enclosed metabolic treadmill.
2.3. RNA extraction and quantitative PCR
Total RNA was isolated from whole muscles using TRIzol Reagent (LifeTechnologies), following the manufacturer's protocol. cDNA was synthesized using SuperScript II Reverse Transcriptase, and qPCR performed using gene specific primers (see Supplementary Table for primers), the HotStart-IT SYBR green qPCR master mix (Affymetrix) and the QuantStudio 6 Flex Real Time PCR system (Applied Biosystems). Relative mRNA levels were determined after normalization to cyclophilin (Ppia, all ERR mKO comparisons) or 36B4 (Rpl0, used in data of Figure 1) mRNA as reference gene.
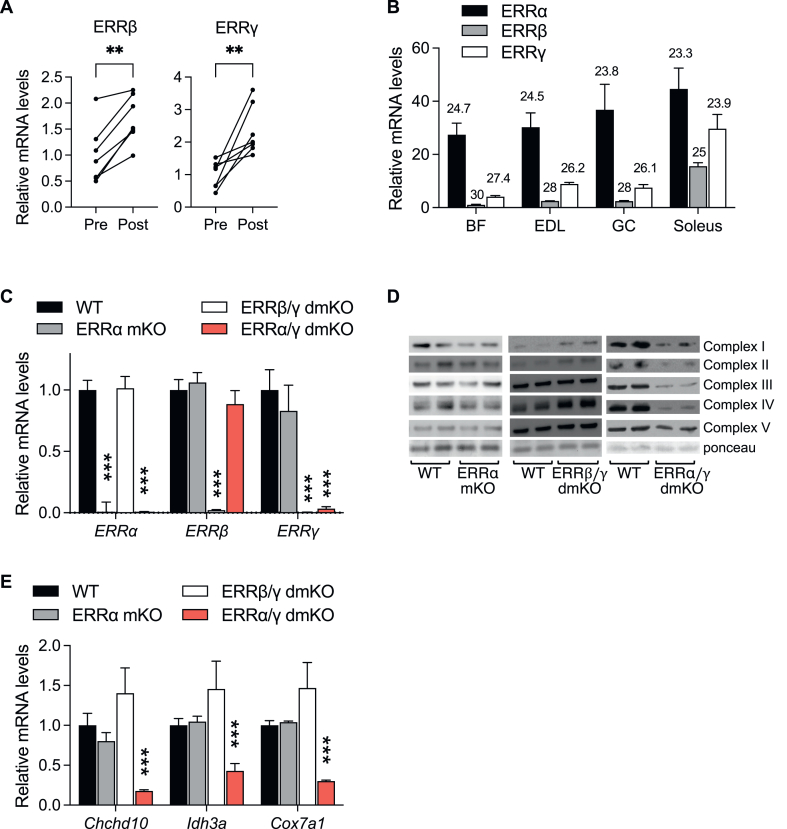
Figure 1.
Skeletal muscle expression of ERRs, and loss of ERR and ERR targets in skeletal muscle - specific ERR KO mouse models. (A) ERRβ and ERRγ mRNA levels in muscle biopsies taken from human male subjects (n = 7) before (Pre) or 3 h after (Post) an acute cycling bout (60 min at ∼70% of their VO2 peak) were quantified by RT-qPCR as described [28]. (B) ERR mRNA levels in biceps femoris (BF), extensor digitorum longus (EDL), gastrocnemius (GC) and soleus of 8-week-old male mice (n = 3) were determined by RT-qPCR and expressed as mean ± SD of ERR mRNA copy number per 1,000 copies of 36B4 mRNA. Numbers above bars represent Ct (threshold cycle) values for each ERR isoform in the different muscles. (C) Relative ERR mRNA levels in GC of WT or littermate mice with skeletal muscle-specific deletions of ERRα (ERRα mKO), ERRβ and ERRγ (ERRβ/γ dmKO), or ERRα and ERRγ (ERRα/γ/dmKO), normalized to levels of 36B4 mRNA, and expressed relative to the levels of each gene in WT muscle. (D) Protein levels of OxPhos complexes I–V and ponceau stain (for loading control) in lysates of GC muscles from ERRα mKO, ERRβ/γ dmKO, ERRα/γ/dmKO and control WT littermates. (E) Relative mRNA levels of ERR-regulated genes Chchd10, Idh3a and Cox7a1, acting in oxidative phosphorylation (OxPhos) and/or the tricarboxylic acid (TCA) cycle. ∗∗, p < 0.01; ∗∗∗, p < 0.001 for differences in mRNA levels (post vs. pre-exercise in A; ERR KO vs. WT littermates in C&E).
2.4. RNA sequencing
For RNA sequencing, EDL and soleus total RNA was isolated using Trizol (Invitrogen) and the RNeasy mini kit (Qiagen). RNA libraries were prepared using 400 ng of RNA amplified via 10 PCR cycles and the NEBNext UltraII Directional RNA Library Prep Kit (NEB) for Illumina, following the manufacturer's instructions. Approximately 16 million 75bp single-reads were generated for each sample by the NextSeq500 Analyzer (Illumina) at the Scripps Research Institute (TSRI) DNA Sequencing Facility and exported as 36 single-end FASTQ sequence files for processing and analysis at the JHMI Single Cell & Transcriptomics Core. The files were imported into the CLCGenomicsServer 9.1.1 platform (QIAGEN Bioinformatics) and their reads aligned to the NCBI April '19 mouse transcriptome, GRCm38.p6, of 45,802 transcripts. Gene expression levels were calculated as fragments per kilobase of exon per million reads mapped (FPKM), and transcript identifiers derived from these FPKM files' “Name” column were updated to current HGNC/NCBI nomenclature. More than 80% of the reads for each library were mapped uniquely to the mouse genome reference mm10. Data were imported into the Partek Genomics Suite 6.6 (Partek Inc St. Louis MO, USA) for statistical analysis. Values were log2 converted and quantile normalized in order to minimize technical inter-sample variation, then compared between biological classes using the appropriate two-tailed one-way ANOVA test to determine differential expression as fold change and its statistical significance as p-value for each transcript. Annotated transcripts that demonstrated adequately high FPKM expression levels were used to assess differential expression between the experimental biological class groups, and filtered to include only transcripts for which the average number of reads was >40 reads (in at least one group), to minimize noise. Differentially expressed genes (log2FC>±0.4 and p-value <0.05) were subjected to Ingenuity Pathway Analysis (QIAGEN, Bioinformatics) to decipher major biological pathways associated with these genes. Heat maps were generated using Displayr. Unsupervised PCA analysis was performed on the normalized expression values of all transcripts and of all samples to evaluate the samples' class similarities and groupings in an unbiased manner. The analysis was computed using a correlation dispersion matrix and normalized eigenvector scaling for the first three principal components.
2.5. Western blot analysis
Whole tissues were homogenized as previously described [35,37]. Protein lysates were resolved by SDS-PAGE and transferred to nitrocellulose membrane (Hybond C Extra; Amersham Biosciences). Blots were probed using the anti-Rt/Ms Total OxPhos Complex Kit (Invitrogen), anti-ERRα (ab76228, Abcam) or anti-ERRγ antibodies [35]. Ponceau staining was used to control for loading.
2.6. Histology, immunohistochemistry, fiber type composition analysis
Skeletal muscle samples were frozen in liquid nitrogen-cooled isopentane. Cross-sections (10 μm thick) were cut from the mid-belly of the gastrocnemius and soleus on a cryostat at −18 °C (Leica CM3050S). For hematoxylin and eosin (H&E) staining, sections were fixed with 4% PFA and stained with Harry's Hematoxylin and Eosin Y. Succinate dehydrogenase (SDH), cytochrome c oxidase (COX), and immunostaining for laminin and myosin heavy chain (MHC) isoforms were performed as described [37]. Fiber type percentiles are based on counting 2 sections (and a total of 700–950 fibers) per mouse. Laminin-stained cross-sections were used to determine muscle fiber size distribution, by measuring the minimal “Feret's diameter” with ImageJ (NIH, Bethesda, MD, USA).
2.7. Transmission electron microscopy (TEM)
Samples were fixed in 3.5% glutaraldehyde, 3 mM MgCl2 in 0.1 M sodium cacodylate buffer, pH 7.2 for overnight at 4 °C. After buffer rinses, samples were postfixed in 2% osmium tetroxide in 0.1 M sodium cacodylate for at least one hour (no more than two) on ice in the dark. Samples were then rinsed in 0.1 M Maleate buffer (pH 6.2), followed by uranyl acetate in 0.1 M Maleate (0.22 μm filtered, 1hr, dark), dehydrated in a graded series of ethanol and embedded in Epon (PolySci) resin. Samples were polymerized at 60 °C overnight. Thin longitudinal sections, 60–90 nm, were cut with a diamond knife on a Leica UCT ultramicrotome and picked up with 2 × 1 mm Formvar copper slot grids. Grids were stained with 2% uranyl acetate (aq.) followed by lead citrate and observed with a Hitachi 7600 TEM at 80 kV. Images were captured with an AMT CCD XR80 (8 megapixel camera - side mount AMT XR80 – high-resolution high-speed camera). For quantification, the perimeters of mitochondria on EM images were outlined in a blinded to genotype manner, and digital files were processed using ImageJ (NIH, Bethesda, MD, USA) to determine area of each “mitochondrial unit” and % of total area. Data shown are the means of 27 images (at 15,000 magnification) of soleus from 2 WT mice, and 15 images of soleus from 2 ERRα/γ dmKO mice.
2.8. Mitochondrial respiration in frozen muscle homogenates
Frozen muscles were homogenized in MAS buffer (70 mM sucrose, 220 mM mannitol, 5 mM KH2PO4, 5 mM MgCl2, 1 mM EGTA, 2 mM HEPES pH 7.4), and 7 μg of muscle homogenates were loaded into a Seahorse XF96 microplate in 150 ul of MAS, as described [38]. Oxygen consumption rates (OCR) were measured in the presence of 10 mM cytochrome c, using a Seahorse XF96 analyzer, at basal and after injection of substrate or inhibitor (final concentration), as follows: port A, NADH (1 mM) or succinate with rotenone (5 mM and 2 μM respectively); port B, rotenone with antimycin A (2 μM and 4 μM); port C, TMPD in ascorbic acid (0.5 mM and 1 mM); and port D: azide (50 mM). Complex I was defined as OCRNADH – OCRrotenone+antimycin; Complex II as OCRSuccinate/rotenone – OCRrotenone/antimycin, and Complex IV as OCRTMPD/ascorbate – OCRazide. For each soleus muscle tested, complex activity was determined as the average of 4–6 technical replicates in the Seahorse XF96 microplate.
2.9. In vivo assessment of muscle strength
Grip strength was determined using an automated grip strength meter (Columbus Instruments, Columbus, OH). Each mouse was tested 5 times, with several minutes rest in between each trial, and forces were recorded in Newtons (N). The average force of the 5 trials per mouse, as well as the maximum force out of the 5 trials, were used for analyses.
2.10. Treadmill exercise and glucose/lactate measurements
Mice were habituated to a motorized treadmill (Exer3/6M, Columbus Instruments) for three days prior to testing (10 min sessions) and given at least 1 day break between habituation and testing. Exercise tolerance was determined by a stepwise progressive test, during which the mice initially ran at a speed of 7.5 m/min and a 5% incline for 5 min. The speed of the treadmill was then increased incrementally until mice were unable to avoid an electrical shock grid at the back of the treadmill, at which time the electrical stimulus was turned off and running times and distances were recorded. Lactate and glucose were determined pre- and post-exercise using a Lactate Plus lactate meter (Nova Biomedical) and a glucometer (Accu-chek Aviva Plus), respectively. Habituation and exercise tests were performed during the day, between 10 am and 4 pm.
2.11. Indirect calorimetry during exercise
Oxygen consumption rates (VO2) and respiratory exchange ratios (RER) were measured with a one-lane enclosed metabolic treadmill for mice coupled to the Oxymax Equal Flow indirect calorimeter (Columbus Instruments). The Oxymax was calibrated before each test, and inflow and outflow gases were sampled every 10 s. Mice were habituated to the treadmill for three consecutive days prior to testing, with at least 1 day break between habituation and testing, which were done during the day (10 am–3 pm). The treadmill was set to 10° incline. The exercise protocol consisted of the following 3 steps: 1) 0 m/min for 2 min; 2) 7 m/min for 1 min; 3) step up of the speed by 1 m/min every minute, until the mouse was exhausted (stayed on the shock grid continuously for 5 s). Mice were euthanized and muscles were collected immediately after the exercise test.
2.12. Muscle glycogen content
Glycogen was measured using a protocol adapted from the NIH-funded Mouse Metabolic Phenotyping Centers (https://mmpc.org/shared/protocols.aspx). Frozen gastrocnemius muscle was pulverized in liquid nitrogen and homogenized in 9 volumes of 0.6 N perchloric acid. Samples were clarified by centrifuging at 10,000 g for 1 min. Supernatant (30 μl) was transferred to each of 2 reaction tubes, along with 15 μl 1 M potassium bicarbonate and 75 μl 0.4 M sodium acetate buffer (pH 4.8), with or without 2.5 mg/ml amyloglucosidase. Reactions were incubated for 2 h at 38 °C, and then neutralized with NaOH and centrifuged for 1 min at 10,000 g. Glucose was measured in 10 μl of supernatants, using the Amplex Red glucose assay kit (Thermo Fisher Scientific) and according to the manufacturer's protocol. For each muscle sample, the amount of glucose in tubes without amyloglucosidase (measuring free glucose) was subtracted from the amount of glucose in tubes with amyloglucosidase (measuring free glucose + glucose from glycogen) to yield the amount of glucose from glycogen. This value was then normalized to tissue weight.
2.13. Running wheels
Mice were singly housed in Coulbourn Instruments (cat No. ACT-551-MS-SS) cages containing a running wheel attached to an activity monitor, in standard 12h light-12h dark cycle, and with ad libitum access to food and water. Voluntary running-wheel data were collected and analyzed using Actimetrics' Clocklab Analysis Software.
2.14. Metabolic and body composition measurements
Whole body metabolism was determined in individually housed mice using the 24-cage Comprehensive Lab Animal Monitoring System indirect calorimeter for mice (CLAMS, Columbus Instruments). Mice were acclimated to the cages for at least 3 days before measurements. Rates of oxygen consumption (VO2) and carbon dioxide production (VCO2), and physical activity were measured continuously for 72 h, at room temperature (∼22 °C). Body composition data for lean mass and fat mass were obtained using an EchoMRI-100 at the Johns Hopkins University Phenotyping service core.
2.15. Glucose and insulin tolerance tests
Intraperitoneal Glucose Tolerance Tests (GTT) were performed in the morning, after an overnight fasting. Blood glucose concentrations were determined with a glucometer (Accu-chek, Aviva Plus) at 20, 40, 60, 90 and 120 min following glucose injection (1 g/kg). Intraperitoneal Insulin Tolerance Tests (ITT) were performed between noon and 2 pm, after 5 h fasting. Blood glucose concentrations were determined with a glucometer (Accu-chek Aviva Plus) at 20, 40, 60, 90 and 120 min following insulin injection (0.70 U/kg).
2.16. Statistical analyses
Data shown are mean ± SEM, unless specified otherwise, and were analyzed using a two-tailed Student's t-test for single variables, with the level of significance set to P < 0.05, or a two-way ANOVA test (GraphPad Software) for multiple variables, followed by a Tukey's multiple comparisons test, with the adjusted p value set to P < 0.05.
2.17. Data availability
GSE214876RNA-seq data generated in this work have been deposited into the Gene Expression Omnibus (GEO) database (GSE214876).
3. Results
3.1. ERR expression and generation of animal models
To provide a link between exercise and ERR expression, we asked how ERRβ and ERRγ mRNA levels change in response to an exercise bout. ERRα has already been shown to be increased in human skeletal muscle after exercise [27,28]. Similarly, levels of mRNA for both ERRβ and ERRγ were increased at 3 h post an acute endurance exercise bout in human subjects, showing that the expression of all three ERRs is regulated by physical activity (Figure 1A and Refs. [27,28]).
Next, to gain insight into where ERRs are expressed, we determined the relative mRNA levels of the three isoforms in different mouse skeletal muscles. ERRα was the most abundant isoform of the three ERRs in all muscles tested, with its expression being only mildly higher in muscles rich in type I (slow) and type IIa oxidative fibers (e.g., soleus), compared to muscles with mixed oxidative/glycolytic fibers [e.g. gastrocnemius (GC)] and muscles rich in type IIb glycolytic fibers [e.g. biceps femoris (BF) and extensor digitorum longus (EDL)] (Figure 1B). ERRβ and ERRγ were significantly enriched in oxidative muscles, as reported earlier for ERRγ [24,25], but were still detectable in all muscles (Figure 1B). Collectively, total ERR mRNA content was higher in oxidative-than glycolytic-fiber rich muscles. ERRα was the predominant ERR in glycolytic muscles (84% of total ERR in BF) and ∼50% of total ERR in soleus. In agreement with our findings, recent single-nucleus RNA-seq analyses of mouse skeletal muscles show that the expression of all three ERRs is lowest in myonuclei of glycolytic fibers type IIb, and highest in myonuclei of oxidative fibers types I and IIa [39].
Based on the relative ERR expression levels, as well as the degree of sequence/function similarity between the three isoforms, we went on to generate three lines of mice lacking different combinations of ERRs specifically in skeletal muscle. The first mouse line, (ERRα mKO, for muscle KO), lacked just ERRα, the isoform that is more abundant, broadly expressed, and relying on PGC-1 coactivators for its transcriptional activity. The second line, (ERRβ/γ dmKO, for double muscle KO) lacked ERRβ and ERRγ, the two isoforms that are preferentially expressed in oxidative muscles, share the highest similarity to each other across all protein domains (Suppl. Fig. 1), and can interact with a wide range of coactivators, including members of the PGC-1 and p160 SRC families. The third mouse line, ERRα/γ dmKO, lacked ERRα and ERRγ, the two most abundant muscle ERRs, in an effort to generate a severe loss of total ERR (based on Figure 1B, ∼94% and ∼83% loss of total ERR in EDL and soleus muscles, respectively). All single and double ERR muscle KOs were born and weaned at the expected Mendelian ratio and were indistinguishable from control littermates by in-cage appearance and behavior. Loss of the targeted ERR isoforms was efficient and did not lead to significant changes in the levels of other non-targeted ERRs (Figure 1C).
For a first assessment of whether ERRs may act redundantly to each other in skeletal muscle, we next determined the abundance of known ERR targets in the gastrocnemius (GC). OxPhos protein levels were mildly affected in the ERRα mKO and ERRβ/γ dmKO (down and up, respectively), but dramatically decreased in the ERRα/γ dmKO (Figure 1D). In a similar pattern, RT-qPCR for three ERR-regulated genes involved in mitochondrial oxidative metabolism showed that the double deletion of ERRα and ERRγ led to significant decreases in the expression of these genes, whereas deleting ERRα alone or ERRβ and ERRγ together had no significant effect (Figure 1E), consistent with ERRα and ERRγ having overlapping and redundant roles in muscle oxidative pathways.
3.2. ERRs act collectively to drive expression of skeletal muscle mitochondrial oxidative metabolism programs
To gain an unbiased understanding of the relative contributions of the different ERRs to skeletal muscle expression, we next used RNA sequencing to compare the global transcriptional changes caused by the tissue-specific loss of ERRα, ERRβ/γ and ERRα/γ. Because of the differential expression of ERRs in glycolytic and oxidative muscles, we assessed the transcriptomes of two muscles: EDL, as representative of glycolytic muscles; and soleus, as an oxidative fiber-rich muscle. Unsupervised Principal Component Analysis (PCA) of normalized expression values of all transcripts and samples was used to visualize in an unbiased manner the similarities and groupings of the different datasets. All datasets segregated according to muscle group identity (EDL vs. soleus) (Figure 2A). Both EDL and soleus of ERRα/γ dmKO mice diverged significantly from their control muscles, as well as from other ERR KO sets, but did not shift from EDL space to soleus space (or vice versa). In addition, ERRβ/γ dmKO soleus separated from the control WT sets, though the variability and spread in this group was large and did not allow for a clear picture of divergence. ERRα mKO EDL, ERRα mKO soleus, and ERRβ/γ dmKO EDL datasets grouped close to control datasets (Figure 2A). A comparison of the number of genes that were differentially expressed (DE, using a log2 fold change ± 0.4 and p < 0.05) in ERR KO vs. control littermate samples, further demonstrated that deletion of ERRα/γ had the greatest impact, with 1962 and 1603 DE genes in EDL and soleus, respectively, far more than the number of DE genes in muscles lacking ERRα only or ERRβ/ERRγ (Figure 2B). Deletion of ERRα alone had a greater impact in EDL (680 DE genes) than soleus (433 DE genes), whereas the loss of ERRβ/ERRγ was more impactful in soleus (684 DE genes) than in EDL (305 DE genes), consistent with the relative levels of ERRs in these muscles.
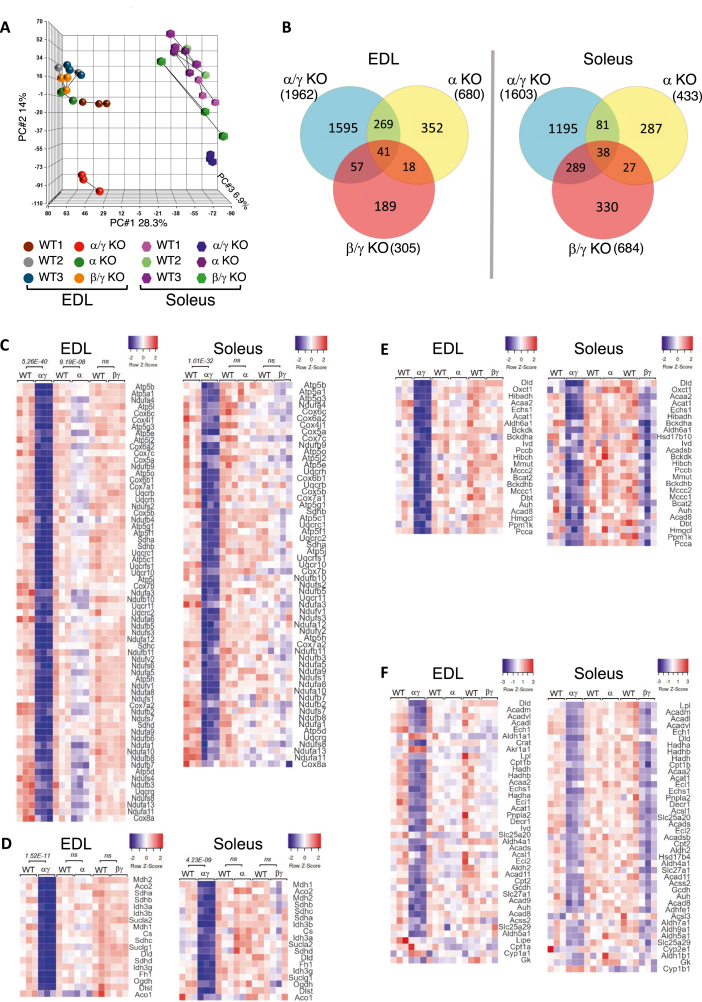
Figure 2.
ERRs act collectively to drive expression of skeletal muscle mitochondrial oxidative metabolism programs. A) Unsupervised PCA, performed on normalized values of all transcripts of all samples, and showing the first three principal components. B) Venn diagrams of differentially expressed genes (DE, log2 FC ± 0.4; p < 0.05) in EDL and soleus muscles of ERRα mKO, ERRβ/γ dmKO, and ERRα/γ/dmKO mice, compared to their respective control littermates. (C–F) Heatmaps showing relative expression (red, high; blue, low) of genes that were differentially expressed (log2 FC ± 0.4; p < 0.05) in at least one of the three ERR KO mouse model, compared to their control littermates. Heat maps are organized by metabolic pathway: (C) OxPhos; (D) TCA cycle, (E) BCAA and (F) Lipid catabolism genes; note that a few genes may be shown in more than one pathway. RNA-seq data are from 12-month-old male mice (n = 3 per group).
Pathway analyses of the DE genes in all comparisons identified “Mitochondrial Dysfunction” and “Oxidative Phosphorylation” (OxPhos) as the top pathways under the control of ERRs in both EDL and soleus (Table 1, Table 2). Heat maps of the DE OxPhos genes (Figure 2C) highlighted: i) the large number of genes affected by the loss of ERRs; ii) the high degree of redundancy between ERRs in both EDL and soleus, with most genes only significantly decreased upon deletion of both ERRα and ERRγ, while moderately (or not) affected in ERRα mKO and ERRβ/ERRγ dmKO muscles; and iii) the differential dependence of OxPhos genes on distinct ERR isoforms in EDL and soleus, with ERRα having a more prominent and statistically significant role in EDL, and ERRβ/γ affecting expression of OxPhos genes in soleus more than in EDL (Figure 2C). A very similar pattern is seen in the heatmap of TCA cycle genes, where only the combined loss of ERRα and ERRγ led to a significant and pronounced downregulation of the pathway (Table 1, Table 2 and Figure 2D).
Table 1.
Top Canonical Pathways differentially expressed in the EDL of ERRα/γ dmKO mice and their Rank and P-value in all ERR KO lines.
| Canonical pathways | ERRα/γ dmKO |
ERRα mKO |
ERRβ/γ dmKO |
|||
|---|---|---|---|---|---|---|
| Rank | P-Value | Rank | P-Value | Rank | P-Value | |
| Oxidative phosphorylation | #1 | 5.4E-40 | #1 | 9.2E-08 | #289 | 1 |
| Mitochondrial dysfunction | #2 | 7.1E-40 | #2 | 2.9E-07 | #209 | 1 |
| Sirtuin Signaling Pathway | #3 | 7.4E-14 | #26 | 2.3E-02 | #176 | 0.5 |
| EIF2 Signaling | #4 | 2.2E-12 | #394 | 1 | NA | – |
| TCA Cycle | #5 | 1.5E-11 | #421 | 1 | NA | – |
| Valine Degradation | #6 | 4.3E-08 | NA | – | #120 | 0.3 |
| Leucine Degradation | #7 | 4.8E-06 | NA | – | NA | – |
| Isoleucine Degradation | #8 | 5.1E-05 | NA | – | #23 | 4.3E-02 |
NA, non-applicable; these pathways were not identified as regulated in ERRα mKO or ERRβ/γ dmKO EDL muscles. For ERRα/γ dmKO EDL, only pathways with p < 1 × 10E-04 are shown; for ERRα mKO EDL, only the two pathways shown here had P-Value <1 × 10E-04; For ERR β/γ dmKO EDL, only one pathway was decreased with p < 1 × 10E-4 (Agranulocyte Adhesion and Diapedesis, P = 5.9E-05).
Table 2.
Top Canonical Pathways differentially expressed in the soleus of ERRα/γ dmKO mice and their Rank and P-value in all ERR KO lines.
| Canonical pathways | ERRα/γ dmKO |
ERRα mKO |
ERRβ/γ dmKO |
|||
|---|---|---|---|---|---|---|
| Rank | P-Value | Rank | P-Value | Rank | P-Value | |
| Mitochondrial dysfunction | #1 | 2.4E-33 | #354 | 1 | #242 | 1 |
| Oxidative phosphorylation | #2 | 1.0E-32 | #404 | 1 | #398 | 1 |
| Sirtuin Signaling Pathway | #3 | 5.8E-12 | #312 | 0.5 | #447 | 1 |
| Valine Degradation | #4 | 3.4E-09 | NA | – | #2 | 8.3E-08 |
| TCA Cycle | #5 | 4.2E-09 | NA | – | #162 | 0.4 |
| Fatty Acid Oxidation | #6 | 4.9E-06 | NA | – | #1 | 4.6E-08 |
| Isoleucine Degradation | #7 | 9.5E-06 | NA | – | #5 | 3.6E-06 |
| Glycolysis | #8 | 5.1E-05 | #124 | 0.1 | #122 | 0.2 |
| Acetyl-CoA Biosynthesis | #9 | 5.8E-05 | NA | – | #134 | 0.3 |
NA, non-applicable; these pathways were not identified as regulated in ERRα mKO or ERRβ/γ dmKO soleus. For ERRα/γ dmKO soleus, only pathways with p < 1 × 10E-04 are shown. For ERRα mKO soleus, only one pathway was decreased with p < 1 × 10E-4 (TR/RXR Activation, P = 6.0E-05). Pathways significantly deregulated in ERR β/γ dmKO soleus are shown in Table 3.
Besides impacting “Mitochondrial Dysfunction”, “OxPhos”, and “TCA cycle”, the loss of ERRs led to downregulation of genes of branched chain amino acid (BCAA) degradation and fatty acid oxidation FAO (Table 1, Table 2, Table 3). Heat maps of BCAA and a broader class of lipid catabolism genes that were differentially expressed in ERR mKO muscles, showed that in EDL these pathways were significantly affected only in the ERRα/γ dmKO mice (Figure 2E&F). In soleus, genes of BCAA and lipid catabolism were significantly decreased in both ERRβ/γ dmKO and ERRα/γ dmKO mice (Table 3 and Figure 2E&F), suggesting an increased reliance of BCAA/lipid pathways (compared to OxPhos and TCA cycle) on ERRβ and/or ERRγ.
Table 3.
Top Canonical Pathways differentially expressed in the soleus of ERRβ/γ dmKO mice and their Rank and P-value in all ERR KO lines.
| Canonical pathways | ERRα/γ dmKO |
ERRα mKO |
ERRβ/γ dmKO |
|||
|---|---|---|---|---|---|---|
| Rank | P-Value | Rank | P-Value | Rank | P-Value | |
| Fatty Acid β-oxidation | #6 | 4.9E-06 | NA | – | #1 | 4.6E-08 |
| Valine Degradation | #4 | 3.4E-09 | NA | – | #2 | 8.3E-08 |
| Glutaryl-CoA Degradation | #15 | 4.1E-04 | NA | – | #3 | 7.8E-07 |
| Tryptophan Degradation | #11 | 1.0E-04 | #293 | 0.5 | #4 | 1.5E-06 |
| Isoleucine Degradation | #7 | 9.5E-06 | NA | – | #5 | 3.6E-06 |
| Ethanol Degradation II | #30 | 1.2E-02 | NA | – | #6 | 7.4E-06 |
| Ethanol Degradation IV | #44 | 2.5E-02 | NA | – | #7 | 3.4E-05 |
| Ketolysis | #21 | 2.2E-03 | NA | – | #8 | 3.9E-05 |
| Oxidat. Ethanol Degrad. III | #48 | 3.8E-02 | NA | – | #9 | 9.5E-05 |
NA, non-applicable; these pathways were not identified as regulated in ERRα mKO soleus. For ERRβ/γ dmKO EDL, only pathways with p < 1 × 10E-4 are shown.
Other notably impacted pathways included the upregulation of “EiF2 signaling” in the EDL of ERRα/γ dmKO mice, a change caused by a broad upregulation of mostly ribosomal genes (Table 1 and Suppl. Fig. 2). A subset of these genes was also mildly increased in the soleus of ERRα/γ dmKO. We validated by RT-qPCR the increase in EDL for a few genes in the pathway (Suppl. Fig. 2). The physiological significance of the changes in this pathway is unclear at this stage; an interesting possibility is that an upregulation of the translational machinery may endow the ERRα/γ dmKO with higher translational capacity, thereby compensating for the decreased levels of mRNAs vital for skeletal muscle bioenergetics. Finally, we noted “Glycolysis” among the canonical pathways upregulated in ERRα/γ dmKO mice, specifically in the soleus (Table 2 and Suppl. Fig. 2).
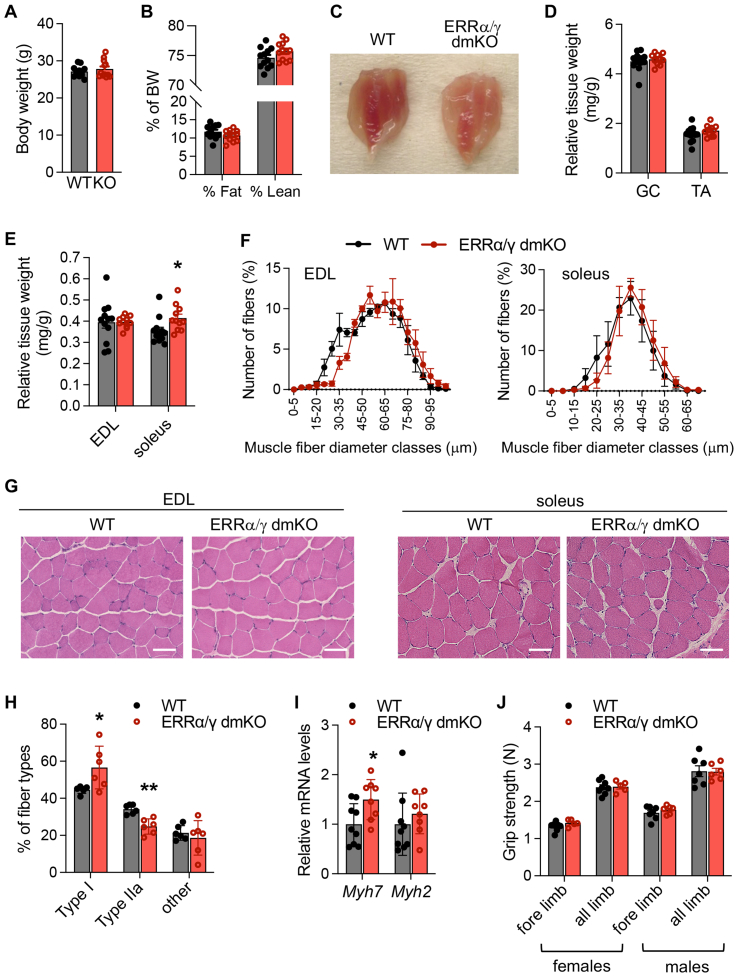
3.3. Mice lacking skeletal muscle ERRα and ERRγ have paler muscles but no differences in body weight, muscle mass, or muscle strength
To understand the functional significance of ERRs and ERR-regulated pathways in skeletal muscle, we next focused on the model with the most severe loss of ERR activity, i.e., mice lacking both ERRα and ERRγ (Suppl. Fig 3A for ERR protein levels). ERRα/γ dmKO male and female mice had similar body weights (BW) and BW composition as their control littermates and were indistinguishable from controls in their physical appearance and in-cage spontaneous physical activity (Figure 3A,B & data not shown). Upon dissection, we noted that ERRα/γ dmKO muscles had a distinctly paler color, compared to muscles of WT mice (Figure 3C). Consistent with the similar BW and BW composition, the mass of most tissues and muscles were similar in ERRα/γ dmKO and control mice (Figure 3D&E and Suppl. Fig. 3B), except for the soleus, which consistently had a mildly elevated mass in dmKO mice (Figure 3E). Quantitation of muscle fiber size showed no significant differences, though we noted a decreased frequency of small size fibers in the EDL (Figure 3F). H&E staining showed regular muscle fiber morphology (Figure 3G). Fiber typing by immunofluorescence showed a small shift from type IIa to type I fibers in soleus (Suppl. Fig. 3C and Figure 3H); consistent with this finding, there was a small but statistically significant increase in Myh7 (type I MHC) mRNA levels in muscles from ERRα/γ dmKO compared to controls (Figure 3I). The physiological relevance of this is unclear, as the type I contractile program did not appear to be significantly upregulated in the transcriptome data of ERRα/γ dmKO soleus. All findings suggested relatively normal muscle structure and size, so we next assessed grip strength, as an indicator of innervation and strength. No differences were observed between ERRα/γ dmKO and WT littermates, in average or maximal grip strength measurements (Figure 3J and Suppl. Fig. 3D), consistent with the absence of gross defects in skeletal muscle morphology.
Figure 3.
Mice lacking skeletal muscle ERRα and ERRγ have similar BW, muscle mass and muscle strength as control littermates. A) Body weight (BW) and B) Body composition, determined by EchoMRI, of ERRα/γ dmKO (red bars) and control (gray bars) littermates (n = 12 males/group, 6-month-old). C) Representative images of hind limb muscles of WT and ERRα/γ/dmKO littermates. D,E) Weights of the indicated skeletal muscles, expressed relative to body weight (n = 12/group, 6-month-old). F) Distribution of fiber sizes, measured as minimal Feret's diameter (μm) and expressed as % of total fibers, in EDL and soleus of ERRα/γ dmKO compared to control littermates (n = 3 mice/group, 6-month-old, with at least 1400 individual fibers quantified per muscle and genotype). G) Representative H&E staining of cross sections of EDL and soleus muscles of ERRα/γ dmKO and control littermates (scale bar: 50 μm) (n = 3/group, 6-month-old). H) Percentage of fiber types, quantified in soleus of ERRα/γ dmKO and control littermates by immunofluorescence for type I and type IIa (n = 3/group, 6-month-old); “other” fibers were negative for type I/IIa or their staining was ambiguous and not clearly assigned to type I or IIa. I) Relative mRNA levels of Myh7 (type I MHC) and Myh2 (type IIa MHC) in soleus of ERRα/γ dmKO compared to control littermates (9 WT and 8 KO males, 6-month-old). J) Average fore limb and all-four limb grip strength of ERRα/γ dmKO and control littermates, expressed as force (N, Newtons). Data are from 10-week-old mice (7 WT and 7 KO males; 8 WT and 5 KO females). Data in all panels are the mean ± SEM. ∗, ∗∗, ∗∗∗, p < 0.05, 0.01, 0.001.
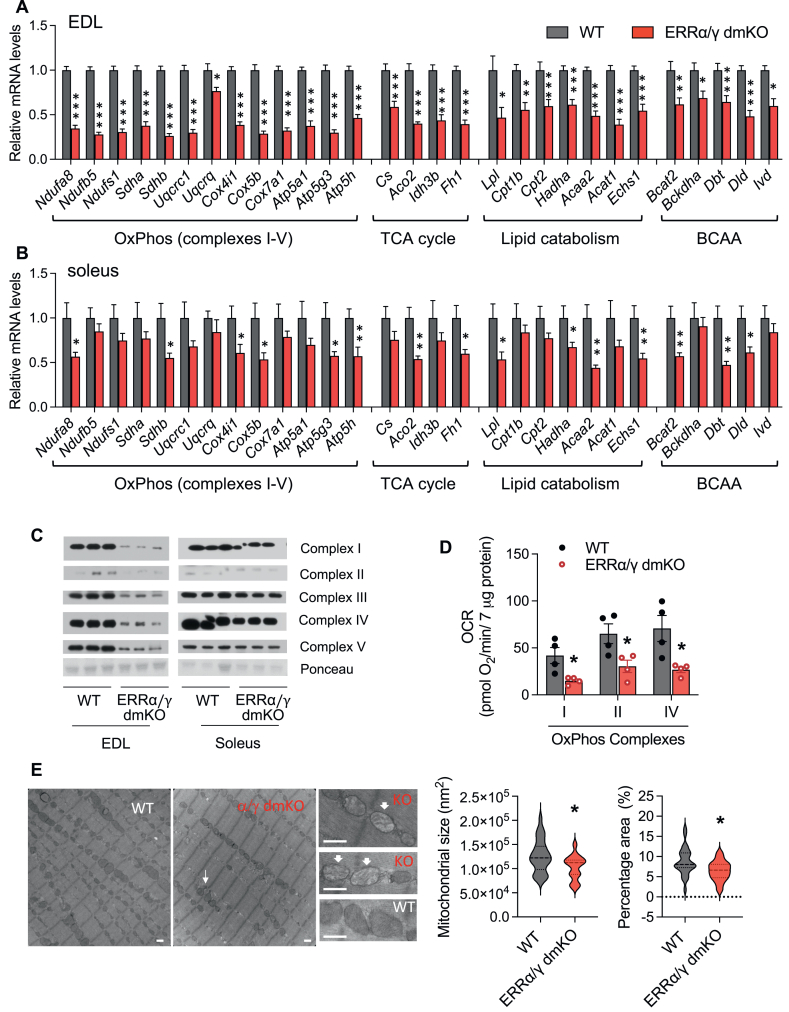
3.4. ERRα/γ dmKO muscles have profound defects in mitochondrial function, and pathology reminiscent of multi-minicore myopathies
The most profoundly dysregulated pathways in ERRα/γ dmKO muscles are those associated with oxidative metabolism (Figure 2). To validate and better understand how these pathways are affected, we assessed facets of oxidative metabolism at different levels. First, RT-qPCR confirmed the concerted downregulation of genes of all five OxPhos complexes, TCA cycle, lipid and BCAA catabolism pathways in EDL and soleus (Figure 4A–B). The expression of these genes was more highly dependent on ERRα/ERRγ in EDL than in soleus, as seen in the RNA-seq data (Figure 2). In addition to the oxidative metabolism genes, we observed a decrease in the levels of PPARα, a regulator of FAO genes, but not Klf15 or PPARδ (regulators of BCAA genes and other oxidative metabolism genes, respectively) (Suppl. Fig. 4A). Consistent with the OxPhos complex mRNA levels, western blot analyses showed a severe decrease in OxPhos complex proteins in the EDL of ERRα/γ dmKO mice, compared to WT controls (Figure 4C). Decreases in OxPhos proteins were mild in soleus, compared to EDL (Figure 4A–C); thus, we next assessed OxPhos complex activity in soleus homogenates, using respirometry. Complex I, II and IV activities were greatly reduced (Figure 4D), suggesting that even though decreases in RNA and protein levels were relatively modest, the wide range of genes affected in ERRα/γ dmKO soleus strongly compromised oxidative capacity. Finally, to visualize mitochondrial structure in ERRα/γ dmKO muscle, we used transmission electron microscopy (TEM) to image sections of soleus, the muscle that was relatively less affected by the deletion of ERRα/γ, so that we could readily assess changes in mitochondrial size or structure. Representative TEM images are shown in Figure 4E; quantitation showed a decrease in the size of intermyofibrillar mitochondria, and a decrease in total mitochondrial area. In addition, we observed mitochondria with abnormal density and cristae morphology, as shown by arrows in the small panels of Figure 4E, indicative of dysfunctional mitochondria.
Figure 4.
ERRα/γ dmKO muscles have decreased expression of oxidative metabolism genes and decreased respiratory capacity. A–B) Relative mRNA levels of representative genes of the different downregulated metabolic pathways, determined by RT-qPCR, in A) EDL and B) soleus (n = 9 WT and 8 KO males, 6-month-old). C) Representative western blot for OxPhos complex I–V proteins in lysates of EDL and soleus (n = 3 males/group, 6-month-old). D) OxPhos complex I, II and IV activities in soleus homogenates, measured by respirometry (n = 4 males/group, 6-month-old). E) TEM images and quantification of average mitochondrial size and mitochondrial percentage area in soleus sections of ERRα/γ dmKO and control WT littermates (n = 2 females/group, 6-month-old, with 15–21 images quantified/group). Arrows point to mitochondria with abnormal density and cristae morphology, indicative of dysfunctional mitochondria (scale bar: 500 nm).
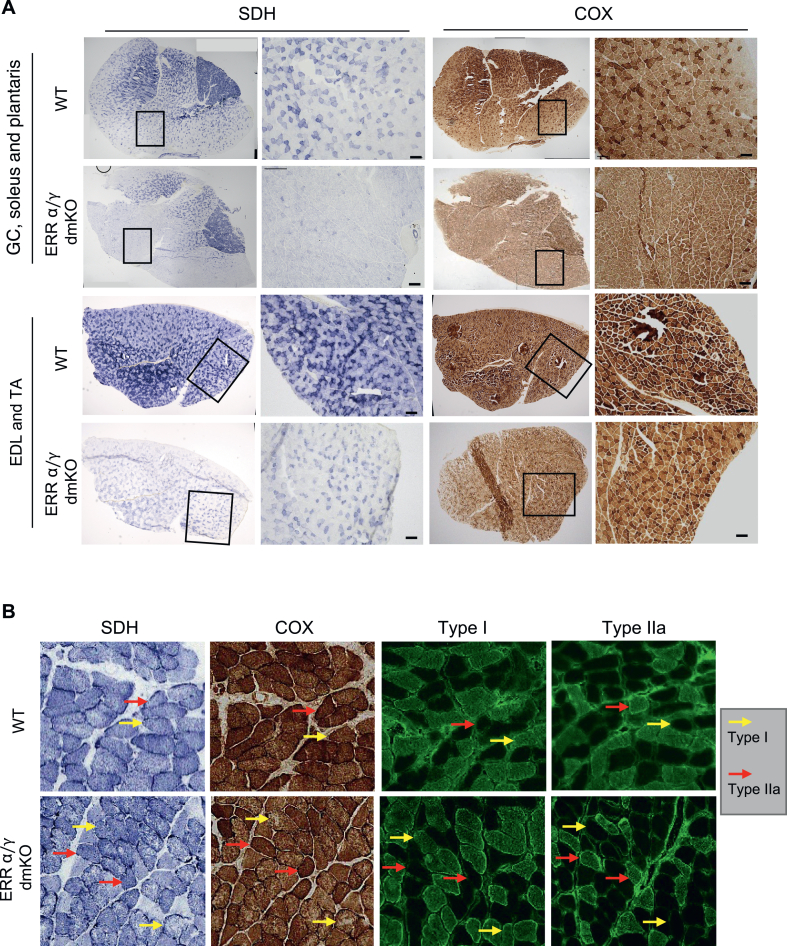
To gain spatial resolution of the oxidative capacity decreases in glycolytic and oxidative parts of skeletal muscles, hindlimb muscle sections were assayed for succinate dehydrogenase (SDH, complex II) and cytochrome c oxidase (COX, complex IV) activity. Decreased SDH and COX staining was seen throughout all muscles, and was particularly pronounced in the mixed fiber regions, white GC, EDL and TA (Figure 5A and Suppl. Fig 4B&C). While SDH and COX activities were retained to some degree in the highly oxidative soleus, higher magnification images of this area revealed abnormal and spotty mitochondrial distribution, with areas devoid of SDH/COX activity (Figure 5B and Suppl. Fig 4D, yellow arrows). These SDH/COX-deficient areas resemble histochemistry seen in muscle biopsies of individuals with multi-minicore myopathies [40,41], and were observed preferentially in type I, not IIa fibers (Figure 5C). About 50% of minicore myopathies are caused by genetic defects in Ryr1 or Senp1 [40,41]. We did not observe changes in Ryr1 or Senp1 expression in the dmKO mice (data not shown).
Figure 5.
SDH and COX staining shows oxidative defects in multiple ERRα/γ dmKO muscles, and pathology reminiscent of minicore myopathies in soleus. A) SDH and COX staining of GC, soleus and plantaris (top two rows) and TA and EDL (bottom two rows) of ERRα/γ dmKO and control WT littermates. Whole muscle sections are composites of multiple images; scale bar in magnified images is 100 μm). B) Consecutives sections of soleus, stained for SDH, COX, type I and type IIa MHC, highlight pathological features in ERRα/γ dmKO muscles, compared to control WT littermates. Yellow and red arrows point to examples of type I and type IIa fibers, respectively. Yellow arrows in sections of ERRα/γ dmKO soleus indicate examples of fibers with areas devoid of SDH, and in particular COX activity. Additional examples are in Suppl. Fig. 3D. All images are representative of sections from 3 mice/group, 6-month-old.
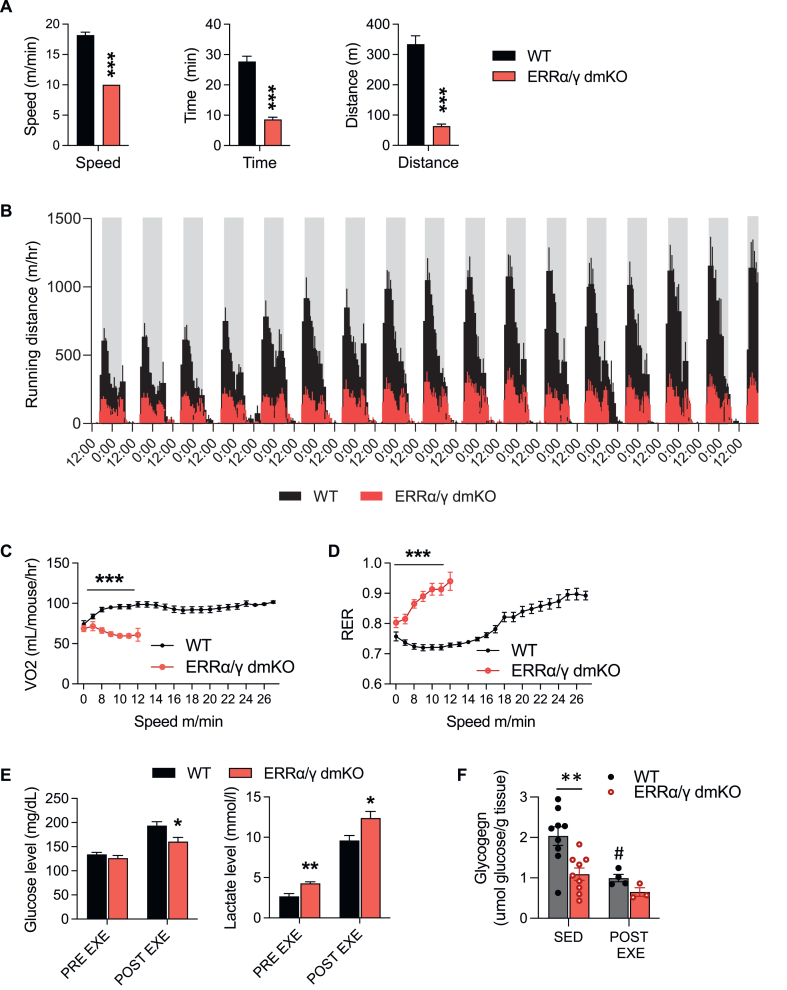
3.5. ERRα/γ dmKO mice show severe exercise intolerance
To determine the physiological consequences of loss of ERRs for skeletal muscle function in the context of exercise, we next tested the ability of ERR KO mice to run on a treadmill, using a high intensity running protocol that assesses maximal running speed. In this type of running paradigm, mice warm up at speeds of 7.5 or 10 m/min, after which the belt speed is increased stepwise to determine how fast the mice can run. Male and female ERRα/γ dmKO had severely impaired running capacity, with 24 out of 25 mice unable to exceed a speed of 10 m/min (Figure 6A and Suppl. Fig. 5A). By comparison, the single ERRα mKO mice on the same running protocol showed only a mild reduction in their maximal speed (p = 0.07), with significant but smaller (compared to ERRα/γ dmKO mice) magnitude decreases in time and distance run (Suppl. Fig. 5B). ERRβ/γ dmKO mice showed no defects in their maximal speed or performance in this high intensity protocol (Suppl. Fig. 5C), even though they show earlier fatigue when running on a low intensity endurance protocol [33].
Figure 6.
ERRα/γ dmKO mice show severe exercise intolerance. A) Maximum speed and time and distance run by ERRα/γ dmKO and control littermates on a treadmill (7 WT and 5 KO females, 4.5-month-old). B) Running distance for ERRα/γ dmKO and control littermates on voluntary running wheels (7 WT and 6 KO males, 6-month-old). C) Oxygen consumption (VO2) and D) RER during a high intensity exercise bout on an enclosed treadmill (7 WT and 6 KO females, 7-month-old). E) Lactate and glucose levels pre- and post-treadmill exercise bout (7 WT and 5 KO females, 4.5-month-old). F) Glycogen content of GC muscle of 7-month-old female mice that were either sedentary (SED, n = 9 per group) or immediately after the acute exercise bout of Figure 6C (Post-Exe, 4 WT and 3 KO mice). Data are the mean ± SEM. ∗, ∗∗, ∗∗∗, p < 0.05, 0.01, 0.001 (effect of genotype); #, p < 0.05 (effect of exercise).
Because the treadmill can be stressful for mice, particularly if they cannot run, we also assessed the ability of ERRα/γ dmKO mice to engage in voluntary wheel running. When placed in cages with wheels, ERRα/γ dmKO run significantly less than their control littermates (Figure 6B and Suppl. Fig. 5D), despite spending a comparable amount of time using the wheels [80% of time that control mice spent on wheels (Suppl. Fig. 5D)].
To gain an understanding of the metabolism of ERRα/γ dmKO mice when they engage in running, we placed the mice in a closed chamber treadmill, where rates of oxygen consumption (VO2), CO2 production (VCO2) and respiratory exchange ratio (RER) could be determined. When WT mice started running, they increased their oxygen consumption (VO2, Figure 6C) and utilization of lipids as energy substrate (seen as a decrease in RER) (Figure 6D). As the speed increased, WT mice progressively switched from lipid to glucose utilization (seen as an increase in RER) (Figure 6D). ERRα/γ dmKO mice, however, were unable to increase their VO2 and immediately relied on using glucose (increased RER) upon running, demonstrating an inability to utilize lipids as energy source (Figure 6C&D). Consistent with metabolic inflexibility and over-reliance on glucose, ERRα/γ dmKO had lower blood glucose and higher lactate levels than control littermates at the end of a run, even though they had run considerably less (Figure 6E). To gain a better insight into glucose utilization, we measured muscle glycogen content. The muscles of ERRα/γ dmKO mice had lower glycogen content, compared to WT muscles, even when the mice were sedentary (Figure 6F). At the end of a running bout, glycogen levels decreased in both WT and ERRα/γ dmKO mice; the decrease did not reach statistical significance in KO mice, where the number of available muscle samples was small (Figure 6F). The decreased muscle glycogen stores in sedentary ERRα/γ dmKO mice, together with the higher basal blood lactate levels (Figure 6E), are consistent with an increased reliance on muscle glycolysis at all times, not just during exercise. The decrease in glycogen content also suggests that the ERRα/γ dmKO mice start running with a deficit in available nutrient stores, which likely contributes to their impaired running capacity.
Finally, to examine other possible sources for the fast fatigue seen in ERRα/γ dmKO mice, we determined the extent to which leaky expression of the HSA-CRE transgene in the heart could affect cardiac ERR expression. ERRα and ERRβ mRNA levels were similar in WT and ERRα/γ dmKO hearts; ERRγ mRNA levels were decreased by 25% (Suppl. Fig. 5E). Even though the decrease in ERRγ was statistically significant, it seems unlikely that this decrease can impact cardiac function as the deletion of ERRγ alone specifically in the heart does not affect the expression of cardiac oxidative metabolism genes or cardiac function [23].
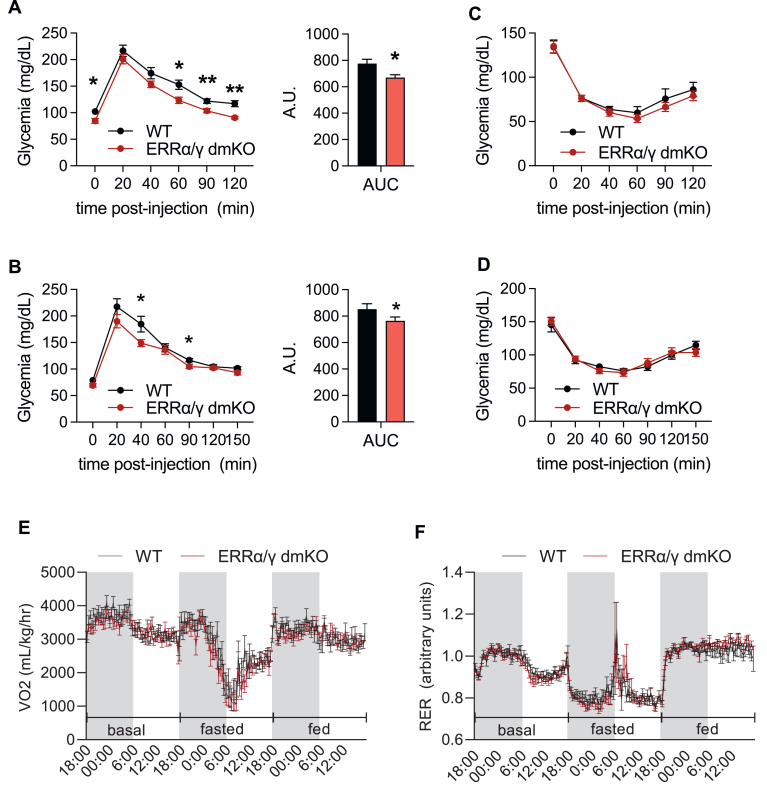
3.6. ERRα/γ dmKO mice show no defects in whole-body glucose homeostasis or energy metabolism
Decreased mitochondrial function and exercise capacity are associated with defects in glucose homeostasis [42,43]. Thus, we next determined the ability of mice to dispose of glucose and respond to insulin. In a glucose tolerance test (GTT), ERRα/γ dmKO male and female mice showed mildly improved glucose homeostasis, with glucose levels dropping faster (Figure 7A&B). In an insulin tolerance test (ITT), ERRα/γ dmKO and control littermates were indistinguishable from control mice (Figure 7C&D), suggesting they have no defects in whole-body glucose homeostasis, at least in chow-fed mice.
Figure 7.
ERRα/γ dmKO mice show no defects in whole-body glucose homeostasis or energy metabolism. Intraperitoneal glucose tolerance test and the associated area under the curve in A) female (n = 12/group, 5-month-old) and B) male (n = 13 WT and 11 KO, 6-month-old) ERRα/γ dmKO and WT littermate mice. Intraperitoneal insulin tolerance in C) female and D) male (n = 11–13/group, 5- to 6-month-old) ERRα/γ dmKO and WT littermate mice. E) VO2 and F) RER of ERRα/γ dmKO and control WT littermates under basal, fed and fasted conditions (n = 12 females/group, 5-month-old). Similar findings, i.e., no changes in VO2 and RER, were seen in a group of 5-month-old ERRα/γ dmKO and WT male mice. Data are the mean ± SEM. ∗, p < 0.05; ∗∗, 0.01.
Finally, to determine if the decreased oxidative capacity could be linked to changes in whole body energy metabolism, we measured the metabolic rate of mice using indirect calorimetry. ERRα/γ dmKO and control littermates showed no differences in VO2, RER, or physical activity in the metabolic cages (Figure 7E&F and Suppl. Fig. 6), suggesting that decreased skeletal muscle oxidative capacity of ERRα/γ dmKO mice is not an important determinant for whole-body energy metabolism in sedentary mice.
4. Discussion
ERRs are recognized as regulators of mitochondrial and oxidative function gene expression in several different cell types and tissues [44]. Yet, there is little in vivo evidence so far for a physiologically significant role of endogenous skeletal muscle ERRs in muscle oxidative capacity. In this study we show that collective loss of the two most abundant muscle ERRs (ERRα and ERRγ) results in a dramatic loss of oxidative capacity, histochemical pathology reminiscent to that of minicore myopathies, severe exercise intolerance, and an inability to utilize lipids as fuel to support exercise. Our findings highlight a central and thus far unappreciated role of ERRs in skeletal muscle oxidative function. We also show that ERRβ and ERRγ levels in human skeletal muscle are acutely induced by an endurance exercise bout, similar to what has been reported for ERRα [27], [28], suggesting that all three ERRs are part of a transcriptional network that enables exercise-driven enhancements of oxidative capacity.
A strength of our study is that it dissects the roles of skeletal muscle ERRs using tissue-specific KO models. Previous studies have determined exercise capacity in whole body KO mice of ERRα or ERRγ [24,31]. However, ERRs, and in particular ERRγ, are highly expressed in the nervous system, including motor neurons [45], and it is possible that defects in muscle innervation (or in other cell types outside muscle and neurons) contribute to the phenotypes seen in whole body ERRα or ERRγ mice. In support of this notion, the impaired exercise capacity in the single ERRα mKO mice (Suppl. Fig. 5) is considerably milder than that observed in whole body ERRα KO mice [ [31] and our unpublished studies]. It will be interesting to test in the future the extent to which neuronal and skeletal muscle deletions of a given ERR synergistically compromise muscle function and exercise capacity.
Comparisons of the EDL and soleus transcriptomes of the ERR mKO models give insights into how the ERR network functions in skeletal muscle. First, they highlight a remarkable degree of redundancy, with the OxPhos and TCA cycle pathways severely decreased only when the two most abundant ERRs are deleted (double ERRα/γ dmKO). The redundancy between ERRs likely endows myotubes with a robust system to protect oxidative capacity, where perturbations in a single ERR can be largely buffered by the presence of another ERR family member. Second, the transcriptomes point to a certain degree of role partitioning that is consistent with their relative expression levels, with ERRα bearing more of the collective ERR role in EDL; and ERRβ/ERRγ doing the same in soleus. Third, although both ERRα and ERRγ are more highly expressed in the oxidative soleus than the glycolytic EDL, the combined ablation of ERRα and ERRγ leads, somewhat surprisingly, to a more dramatic loss of OxPhos and TCA gene expression in EDL than in soleus. The lower dependency of these pathways on ERRα/ERRγ in soleus, compared to EDL, may be due to the presence of ERRβ, which is still expressed in soleus and possibly sufficient to drive some expression of OxPhos and TCA genes. In addition, there may be other soleus-enriched transcription factors that drive oxidative metabolism gene expression, rendering these pathways less dependent on ERRs in this muscle. Overall, our findings underscore that an understanding of the physiological roles of transcription factors must take into account not only their absolute levels of expression in different cell types, but also their relative expression to other factors that converge on the same pathways.
In addition to OxPhos and TCA pathways, which have been associated with ERR function before, the transcriptome data highlight the importance of skeletal muscle ERRs for two other metabolic programs: BCAA and lipid catabolism. In agreement with a recent report showing that ERRα activates BCAA catabolism genes in human primary myotubes, our loss-of-function studies in vivo in mice reveal a major role for ERRs in the regulation of most genes of BCAA catabolism, in both EDL and soleus. Similarly, muscles of ERRα/γ dmKO mice show a broad and significant decrease in genes important for lipolysis (Pnpla2/Atgl, Lpl), the mitochondrial carnitine cycle (Cpt1b, Cpt2), and mitochondrial FAO (including genes coding for Mcad, Vlcad, Scad, and subunits of the Mitochondrial Trifunctional Protein). Some of the FAO targets may be indirectly regulated by ERRs, as we noted a decrease in levels of PPARα, a transcriptional regulator of FAO genes. Lipid and BCAA oxidation provide fuel to sustain exercise, and the decreased expression of these pathways is likely to contribute to the exercise intolerance of the ERRα/γ dmKO mice.
The pathways of BCAA and lipid oxidation are clearly co-regulated by ERRα and ERRγ in EDL, but they are also largely dependent on ERRβ and ERRγ in soleus (Table 1 and Figure 2). This finding suggests that while ERRα can activate genes of these pathways, soleus ERRα alone is insufficient to maintain their expression. The inability of ERRα by itself to sustain BCAA and lipid oxidation genes in soleus contrasts its efficient support of OxPhos and TCA cycle gene expression in the same muscle (Figure 2). A possible explanation for this differential action is that ERRα and ERRβ/γ have, to some degree, isoform-specific roles at different targets and pathways.
The mechanism(s) by which ERRβ/ERRγ may target genes differentially from ERRα are not clear. The three ERRs have almost identical DBDs and recognize the same DNA sequence motif [8,13]; thus, any isoform-specific roles likely rely on functions encoded by other protein domains. The NTD and hinge regions of ERRβ and ERRγ share a high degree of similarity and differ from the corresponding domains of ERRα (Suppl. Fig. 1), suggesting they could carry some isoform-specific functions. Interestingly, we do not know much about the roles of these domains in ERR transcriptional function. Finally, although the LBD is conserved in all three ERRs, it is structurally and functionally distinct in ERRα vs. ERRβ/ERRγ [16,17,46,47]. Notably, the LBDs of ERRβ and ERRγ are transcriptionally active in cellular contexts where the LBD of ERRα is not [15], consistent with ERRβ and ERRγ interacting with a wide range of coactivators. Thus, more than one ERRβ/ERRγ domain may enable distinct partnering of ERRβ/ERRγ with factors that ERRα does not recognize and allowing ERRβ/ERRγ to activate BCAA and lipid metabolism genes. Clearly more studies are required to elucidate the degree to which ERRβ/γ have isoform-specific roles, and the mechanisms by which they do so.
Histochemistry of skeletal muscle sections of ERRα/γ dmKO for SDH (OxPhos complex II) and COX (OxPhos complex IV) validated the decreases in enzymatic activities, consistent with the reductions seen at the RNA and protein levels. The SDH/COX staining also provided spatial resolution to the mitochondrial dysfunction and revealed distinct muscle-specific patterns in how respiratory activity is lost. In EDL, the characteristic checkerboard pattern of fibers with high and low oxidative metabolism is largely maintained, but all fibers have lower complex II/IV activities. In GC, the checkerboard pattern is diminished, particularly in the superficial parts of the muscle; SDH/COX activity is retained in the deeper regions of the muscle. In the ERRα/γ dmKO soleus, closer examination reveals areas devoid of SDH and COX activities, reminiscent of the pathology seen in multi-minicore diseases [40,41]. These “cores” are found specifically in type I fibers, which seem more affected than type IIa fibers. Altogether, the SDH/COX histochemistry highlights a high level of complexity in how respiratory complex activity is reduced across different muscle groups and fibers in the absence of ERRα/ERRγ. The findings suggest that the reliance of oxidative metabolism genes on ERRs depends not just on muscle group and fiber type but possibly also other factors, e.g., possibly recruitment of muscles during daily physical activity. The heterogeneity in the effects of ERRα/ERRγ loss on OxPhos activity also suggests that further understanding of the transcriptional mechanisms that regulate oxidative metabolism in muscle will require approaches with single fiber resolution, rather than the bulk approach we have taken in this study.
The most striking defect in ERRα/γ dmKO mice is their intolerance to exercise. The KO mice stop running at 10 m/min, a speed typically used by mice for warm-up on the treadmill. This exercise defect is comparable to that of old (>15 months of age) mdx mice, a model of muscular dystrophy [48]. ERRα/γ dmKO mice also show dramatic decreases in voluntary wheel running; as a comparison, mice lacking both skeletal muscle PGC-1α and PGC-1β, i.e., factors widely considered as the master regulators of mitochondrial function, have only a mild, statistically non-significant decrease in running distance on voluntary wheels [49]. The intolerance of ERRα/γ dmKO mice to exercise is particularly remarkable when put into the context of the multiple other aspects of skeletal muscle that are unaffected or only minorly affected in the dmKO mice: no large changes in muscle morphology, mass or fiber type composition, similar grip strength, and no changes in spontaneous physical activity under normal housing conditions. The ERRα/γ dmKO mouse makes a striking case for the importance of oxidative metabolism for even light exercise.
Not surprisingly, ERRα/γ dmKO mice have no defects in whole-body glucose homeostasis, as the dmKO muscles are likely to rely predominantly on glucose uptake and glycolysis. This high dependence on glucose may explain the somewhat improved glucose tolerance observed in a glucose challenge, and the mild increase in glycolysis genes seen in soleus. The ERRα/γ dmKO mice also show no differences in whole-body energy expenditure under the normal housing conditions used in calorimetry measurements. It will be interesting to determine in the future the metabolic consequences of loss of skeletal muscle ERRs in an enriched environment, where mice are more physically active, and in particular to address whether ERRα/γ dmKO can benefit from such enriched environments to a similar extent as WT mice [50,51].
5. Conclusions
ERRα, ERRβ and ERRγ carry a collective and essential role in skeletal muscle oxidative metabolism, driving the expression of genes important for OxPhos, TCA, BCAA and lipid catabolism. Deletion of two of the highest expressed isoforms, ERRα and ERRγ, causes a severe intolerance to exercise, in otherwise metabolically healthy mice. Our findings support a role of ERRs as exercise-induced activators of oxidative metabolism programs.
Author contributions
J.W, E.E, B.H., A.P.R and A.K. designed and performed experiments and analyzed data; Y.C., A.W, performed experiments and analyzed data. S.C. developed methods and helped with experiments; S-C.O. analyzed data. J.W., E.E. and A.K. wrote and all authors reviewed and edited the manuscript.
Acknowledgements
We thank ex-lab members Josep A. Villena (VHIR Vall d’Hebron Research Institute, Spain) and Paulo Garda (University of Campinas, Brazil) for sample collections; Qinchuan Wang and Susan Aja (Johns Hopkins University School of Medicine) for advice on closed chamber treadmill and calorimetry studies, Dylan Sarver and Nanami Senoo (Johns Hopkins University School of Medicine) for help with respirometry assays; Connie Talbot (Johns Hopkins University School of Medicine) for help with RNA-seq analyses; Ulrich Mueller for comments on the manuscript. The work was supported by NIH grant DK105126 to A.K.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101670.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
RNA-seq data generated in this work have been deposited into the Gene Expression Omnibus (GEO) database (GSE214876).
References
- 1.Kokkinos P. Physical activity, health benefits, and mortality risk. ISRN Cardiol. 2012;2012:718789. doi: 10.5402/2012/718789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kokkinos P., Myers J., Faselis C., Panagiotakos D.B., Doumas M., Pittaras A., et al. Exercise capacity and mortality in older men: a 20-year follow-up study. Circulation. 2010;122:790–797. doi: 10.1161/CIRCULATIONAHA.110.938852. [DOI] [PubMed] [Google Scholar]
- 3.Boule N.G., Weisnagel S.J., Lakka T.A., Tremblay A., Bergman R.N., Rankinen T., et al. Effects of exercise training on glucose homeostasis: the HERITAGE Family Study. Diabetes Care. 2005;28:108–114. doi: 10.2337/diacare.28.1.108. [DOI] [PubMed] [Google Scholar]
- 4.Myers J., Kokkinos P., Nyelin E. Physical activity, cardiorespiratory fitness, and the metabolic syndrome. Nutrients. 2019;11 doi: 10.3390/nu11071652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikus C.R., Oberlin D.J., Libla J.L., Taylor A.M., Booth F.W., Thyfault J.P. Lowering physical activity impairs glycemic control in healthy volunteers. Med Sci Sports Exerc. 2012;44:225–231. doi: 10.1249/MSS.0b013e31822ac0c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thyfault J.P., Booth F.W. Lack of regular physical exercise or too much inactivity. Curr Opin Clin Nutr Metab Care. 2011;14:374–378. doi: 10.1097/MCO.0b013e3283468e69. [DOI] [PubMed] [Google Scholar]
- 7.Alaynick W.A., Kondo R.P., Xie W., He W., Dufour C.R., Downes M., et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Dufour C.R., Wilson B.J., Huss J.M., Kelly D.P., Alaynick W.A., Downes M., et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Huss J.M., Torra I.P., Staels B., Giguere V., Kelly D.P. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mootha V.K., Handschin C., Arlow D., Xie X., St Pierre J., Sihag S., et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schreiber S.N., Emter R., Hock M.B., Knutti D., Cardenas J., Podvinec M., et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villena J.A., Hock M.B., Chang W.Y., Barcas J.E., Giguere V., Kralli A. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc Natl Acad Sci U S A. 2007;104:1418–1423. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 14.Gaillard S., Dwyer M.A., McDonnell D.P. Definition of the molecular basis for estrogen receptor-related receptor-alpha-cofactor interactions. Mol Endocrinol. 2007;21:62–76. doi: 10.1210/me.2006-0179. [DOI] [PubMed] [Google Scholar]
- 15.Gantner M.L., Hazen B.C., Conkright J., Kralli A. GADD45gamma regulates the thermogenic capacity of brown adipose tissue. Proc Natl Acad Sci U S A. 2014;111:11870–11875. doi: 10.1073/pnas.1406638111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greschik H., Wurtz J.M., Sanglier S., Bourguet W., van Dorsselaer A., Moras D., et al. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol Cell. 2002;9:303–313. doi: 10.1016/s1097-2765(02)00444-6. [DOI] [PubMed] [Google Scholar]
- 17.Kallen J., Schlaeppi J.M., Bitsch F., Filipuzzi I., Schilb A., Riou V., et al. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (ERRalpha): crystal structure of ERRalpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. J Biol Chem. 2004;279:49330–49337. doi: 10.1074/jbc.M407999200. [DOI] [PubMed] [Google Scholar]
- 18.Kamei Y., Ohizumi H., Fujitani Y., Nemoto T., Tanaka T., Takahashi N., et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci U S A. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Percharde M., Lavial F., Ng J.H., Kumar V., Tomaz R.A., Martin N., et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 2012;26:2286–2298. doi: 10.1101/gad.195545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreiber S.N., Knutti D., Brogli K., Uhlmann T., Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 21.Gantner M.L., Hazen B.C., Eury E., Brown E.L., Kralli A. Complementary roles of estrogen-related receptors in Brown adipocyte thermogenic function. Endocrinology. 2016;157:4770–4781. doi: 10.1210/en.2016-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sone M., Morone N., Nakamura T., Tanaka A., Okita K., Woltjen K., et al. Hybrid cellular metabolism coordinated by Zic3 and Esrrb synergistically enhances induction of naive pluripotency. Cell Metab. 2017;25:1103–1117 e1106. doi: 10.1016/j.cmet.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Wang T., McDonald C., Petrenko N.B., Leblanc M., Wang T., Giguere V., et al. Estrogen-related receptor alpha (ERRalpha) and ERRgamma are essential coordinators of cardiac metabolism and function. Mol Cell Biol. 2015;35:1281–1298. doi: 10.1128/MCB.01156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rangwala S.M., Wang X., Calvo J.A., Lindsley L., Zhang Y., Deyneko G., et al. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. J Biol Chem. 2010;285:22619–22629. doi: 10.1074/jbc.M110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narkar V.A., Fan W., Downes M., Yu R.T., Jonker J.W., Alaynick W.A., et al. Exercise and PGC-1alpha-independent synchronization of type I muscle metabolism and vasculature by ERRgamma. Cell Metab. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sopariwala D.H., Likhite N., Pei G., Haroon F., Lin L., Yadav V., et al. Estrogen-related receptor alpha is involved in angiogenesis and skeletal muscle revascularization in hindlimb ischemia. Faseb J. 2021;35 doi: 10.1096/fj.202001794RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CCartoni R., Leger B., Hock M.B., Praz M., Crettenand A., Pich S., et al. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. J Physiol. 2005;567:349–358. doi: 10.1113/jphysiol.2005.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace M.A., Hock M.B., Hazen B.C., Kralli A., Snow R.J., Russell A.P. Striated muscle activator of Rho signalling (STARS) is a PGC-1alpha/oestrogen-related receptor-alpha target gene and is upregulated in human skeletal muscle after endurance exercise. J Physiol. 2011;589:2027–2039. doi: 10.1113/jphysiol.2011.205468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holloszy J.O., Coyle E.F. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol. 1984;56:831–838. doi: 10.1152/jappl.1984.56.4.831. [DOI] [PubMed] [Google Scholar]
- 30.Joseph A.M., Pilegaard H., Litvintsev A., Leick L., Hood D.A. Control of gene expression and mitochondrial biogenesis in the muscular adaptation to endurance exercise. Essays Biochem. 2006;42:13–29. doi: 10.1042/bse0420013. [DOI] [PubMed] [Google Scholar]
- 31.Perry M.C., Dufour C.R., Tam I.S., B'Chir W., Giguere V. Estrogen-related receptor-alpha coordinates transcriptional programs essential for exercise tolerance and muscle fitness. Mol Endocrinol. 2014;28:2060–2071. doi: 10.1210/me.2014-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaBarge S., McDonald M., Smith-Powell L., Auwerx J., Huss J.M. Estrogen-related receptor-alpha (ERRalpha) deficiency in skeletal muscle impairs regeneration in response to injury. Faseb J. 2014;28:1082–1097. doi: 10.1096/fj.13-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan Z., Rumsey J., Hazen B.C., Lai L., Leone T.C., Vega R.B., et al. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J Clin Invest. 2013;123:2564–2575. doi: 10.1172/JCI67652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gan Z., Fu T., Kelly D.P., Vega R.B. Skeletal muscle mitochondrial remodeling in exercise and diseases. Cell Res. 2018;28:969–980. doi: 10.1038/s41422-018-0078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown E.L., Hazen B.C., Eury E., Wattez J.S., Gantner M.L., Albert V., et al. Estrogen-related receptors mediate the adaptive response of Brown adipose tissue to adrenergic stimulation. iScience. 2018;2:221–237. doi: 10.1016/j.isci.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwander M., Leu M., Stumm M., Dorchies O.M., Ruegg U.T., Schittny J., et al. Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev Cell. 2003;4:673–685. doi: 10.1016/s1534-5807(03)00118-7. [DOI] [PubMed] [Google Scholar]
- 37.Cho Y., Hazen B.C., Gandra P.G., Ward S.R., Schenk S., Russell A.P., et al. Perm1 enhances mitochondrial biogenesis, oxidative capacity, and fatigue resistance in adult skeletal muscle. Faseb J. 2016;30:674–687. doi: 10.1096/fj.15-276360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acin-Perez R., Benador I.Y., Petcherski A., Veliova M., Benavides G.A., Lagarrigue S., et al. A novel approach to measure mitochondrial respiration in frozen biological samples. EMBO J. 2020;39 doi: 10.15252/embj.2019104073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrany M.J., Swoboda C.O., Sun C., Chetal K., Chen X., Weirauch M.T., et al. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat Commun. 2020;11:6374. doi: 10.1038/s41467-020-20063-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fusto A., Moyle L.A., Gilbert P.M., Pegoraro E. Cored in the act: the use of models to understand core myopathies. Dis Model Mech. 2019;12 doi: 10.1242/dmm.041368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogasawara M., Nishino I. A review of core myopathy: central core disease, multiminicore disease, dusty core disease, and core-rod myopathy. Neuromuscul Disord. 2021;31:968–977. doi: 10.1016/j.nmd.2021.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Shulman G.I. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N Engl J Med. 2014;371:1131–1141. doi: 10.1056/NEJMra1011035. [DOI] [PubMed] [Google Scholar]
- 43.Stephenson E.J., Hawley J.A. Mitochondrial function in metabolic health: a genetic and environmental tug of war. Biochim Biophys Acta. 2014;1840:1285–1294. doi: 10.1016/j.bbagen.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Eichner L.J., Giguere V. Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11:544–552. doi: 10.1016/j.mito.2011.03.121. [DOI] [PubMed] [Google Scholar]
- 45.Yu G., Zilundu P.L.M., Liu L., Zhong K., Tang Y., Ling Z., et al. ERRgamma is downregulated in injured motor neuron subpopulations following brachial plexus root avulsion. Exp Ther Med. 2020;19:205–213. doi: 10.3892/etm.2019.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kallen J., Lattmann R., Beerli R., Blechschmidt A., Blommers M.J., Geiser M., et al. Crystal structure of human estrogen-related receptor alpha in complex with a synthetic inverse agonist reveals its novel molecular mechanism. J Biol Chem. 2007;282:23231–23239. doi: 10.1074/jbc.M703337200. [DOI] [PubMed] [Google Scholar]
- 47.Wang L., Zuercher W.J., Consler T.G., Lambert M.H., Miller A.B., Orband-Miller L.A., et al. X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation. J Biol Chem. 2006;281:37773–37781. doi: 10.1074/jbc.M608410200. [DOI] [PubMed] [Google Scholar]
- 48.Aartsma-Rus A., van Putten M. Assessing functional performance in the mdx mouse model. J Vis Exp. 2014;85:51303. doi: 10.3791/51303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowe G.C., Patten I.S., Zsengeller Z.K., El-Khoury R., Okutsu M., Bampoh S., et al. Disconnecting mitochondrial content from respiratory chain capacity in PGC-1-deficient skeletal muscle. Cell Rep. 2013;3:1449–1456. doi: 10.1016/j.celrep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao L., Choi E.Y., Liu X., Martin A., Wang C., Xu X., et al. White to brown fat phenotypic switch induced by genetic and environmental activation of a hypothalamic-adipocyte axis. Cell Metab. 2011;14:324–338. doi: 10.1016/j.cmet.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slater A.M., Cao L. A protocol for housing mice in an enriched environment. J Vis Exp. 2015 doi: 10.3791/52874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GSE214876RNA-seq data generated in this work have been deposited into the Gene Expression Omnibus (GEO) database (GSE214876).
RNA-seq data generated in this work have been deposited into the Gene Expression Omnibus (GEO) database (GSE214876).