Abstract
Understanding transmission routes of SARS-CoV-2 is crucial to establish effective interventions in healthcare institutions. Although the role of surface contamination in SARS-CoV-2 transmission has been controversial, fomites have been proposed as a contributing factor. Longitudinal studies about SARS-CoV-2 surface contamination in hospitals with different infrastructure (presence or absence of negative pressure systems) are needed to improve our understanding of their effectiveness on patient healthcare and to advance our knowledge about the viral spread.
We performed a one-year longitudinal study to evaluate surface contamination with SARS-CoV-2 RNA in reference hospitals. These hospitals have to admit all COVID-19 patients from public health services that require hospitalization. Surfaces samples were molecular tested for SARS-CoV-2 RNA presence considering three factors: the dirtiness by measuring organic material, the circulation of a high transmissibility variant, and the presence or absence of negative pressure systems in hospitalized patients' rooms.
Our results show that: (i) There is no correlation between the amount of organic material dirtiness and SARS-CoV-2 RNA detected on surfaces; (ii) SARS-CoV-2 high transmissible Gamma variant introduction significantly increased surface contamination; (iii) the hospital with negative pressure systems was associated with lower levels of SARS-CoV-2 surface contamination and, iv) most environmental samples recovered from contaminated surfaces were assigned as non-infectious.
This study provides data gathered for one year about the surface contamination with SARS-CoV-2 RNA sampling hospital settings. Our results suggest that spatial dynamics of SARS-CoV-2 RNA contamination varies according with the type of SARS-CoV-2 genetic variant and the presence of negative pressure systems. In addition, we showed that there is no correlation between the amount of organic material dirtiness and the quantity of viral RNA detected in hospital settings. Our findings suggest that SARS CoV-2 RNA surface contamination monitoring might be useful for the understanding of SARS-CoV-2 dissemination with impact on hospital management and public health policies. This is of special relevance for the Latin-American region where ICU rooms with negative pressure are insufficient.
Keywords: SARS-CoV2 RNA, Ct value, Fomites, Organic material dirtiness, ATP measurements, Negative pressure system, Variant of concern
Highlights
-
•
SARS-CoV-2 RNA was detected in a wide range of hospital surfaces.
-
•
There is no correlation between the amount of organic material dirtiness and SARS-CoV-2 RNA detected on surfaces.
-
•
Negative pressure systems were associated with lower levels of SARS-CoV-2 surface contamination in hospitals.
-
•
SARS-CoV-2 high transmissible variant significantly increased surfaces contamination.
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of COVID-19, was first detected in China in December 2019. By December 2022, about 642 million confirmed cases and over 6.5 million deaths were reported with COVID-19 disease as result of the current pandemic (World Health Organization, 2022). SARS-CoV-2 is mainly transmitted by exposure to infectious respiratory fluids, primarily via droplets and aerosols and secondarily by contact with contaminated surfaces [1,2]. Although the role of surface contamination in viral transmission has been controversial [[3], [4], [5]], since few studies that attempted to isolate viable viruses failed, fomites have been proposed as a contributing factor in virus transmission.
In late 2020, it was clear that extremely transmissible and potentially more dangerous variants were arising globally. This was a warning to public health, leading to worldwide efforts to study, these emerging variants, which were named as Variant of Concern (VOCs). In the case of Latin America, the VOC Gamma was discovered in Manaus, Brazil. Genome sequencing analysis revealed point mutations in the viral spike protein associated with increased binding to the human receptor [6]. At that time, we implemented a genomic surveillance algorithm for monitoring the VOC emergence and spread in our region [7]. We found that the Gamma variant was estimated to be introduced in Uruguay multiple times from Brazil between mid-February and early March of 2021 leading to an increase in hospitalizations and deaths for COVID-19 during that year [7].
Later, it was shown that clinical samples from patients infected with the Gamma variant have significantly lower cycle threshold (Ct) values related to a high viral load (approximately ten-fold higher than patients infected with the previous circulating variants of SARS-CoV-2) and high transmissibility capacity [8]. The Ct value is defined as the number of PCR cycles required for the fluorescent signal to cross a given threshold. Thus, Ct levels are inversely proportional to the amount of target nucleic acid (viral genome) in the sample. Moreover, this variant was later associated with increased mortality risk, as well as a higher COVID-19 severity in younger age groups [9].
SARS-CoV-2 surface contamination has been studied in several real-world and laboratory settings [10] and different factors such as temperature, pH, relative humidity, and type of material have been addressed to check viral presence and viability [[11], [12], [13], [14], [15]]. However, the correlation between organic material dirtiness (such as respiratory fluids via droplets and aerosols) and SARS-CoV-2 surface contamination remains to be clarified [16]. Additionally, the COVID-19 pandemic highlighted the need for hospital infrastructure, specifically, negatively pressurized isolation rooms (NPRs) are used in order to avoid airborne microorganisms from spreading to other areas and contaminating other patients, health-care workers as well as sterile equipment [17]. Nevertheless, in low- and middle-income countries a significant proportion of ICUs are unlikely to have NPRs. For example, 37% of ICUs throughout Asian countries do not have NPRs. Since most Latin American countries are classified as middle income, hospitals have insufficient infrastructure and equipment, such as NPRs [[18], [19], [20]]. Therefore, longitudinal studies about SARS-CoV-2 surface contamination in hospitals with different infrastructures are needed to improve our understanding of their effectiveness in patient healthcare and to advance our knowledge about viral spread.
Here, we performed a one-year study to evaluate SARS-CoV-2 RNA surface contamination. We monitored the presence of viral RNA in two COVID-19 reference hospitals considering three factors: the dirtiness by measuring organic material, the introduction of a high transmission variant, and the effect of the presence or absence of negative pressure systems. Thus, we tackled the following questions: (i) Is there any correlation between the dirtiness caused by organic material detected on surfaces and the amount of SARS-CoV-2 RNA contamination in hospital settings? (ii) Do high transmission variants (VOCs) increase RNA contamination on surfaces? And (iii) how does negative pressure impact the prevalence of SARS-CoV-2 RNA on surfaces?
2. Materials and methods
2.1. Sampling collection
Sampling was performed in the two COVID-19 public health reference hospitals in Uruguay where most COVID-19 patients requiring critical care were hospitalized. These centers were: (i) Hospital Español Doctor Juan José Crottogini, referenced as Hospital A, and (ii) Instituto Nacional de Ortopedia y Traumatología, referenced as Hospital B. The role of reference hospitals in Uruguay was to admit every patient from public health services with severe COVID-19 diagnostic regardless of whether they had other comorbidities. There were no differences in cleaning regimen between hospitals. Every room was sanitized with bleach every 6 h in both hospitals. Moreover, the cleaning staff was dressed with personal protective equipment including exclusive work clothes (complete overalls), gloves, N95 mask, eye/face protection, cap and footwear cover. In addition, both hospitals were directed by the same medical board under common policies. Patients were assigned to one or other hospital depending on room availability. Patients were assigned to intermediate or ICU care depending on if they needed invasive mechanical ventilation (ICU) or not (intermediate).
The room layout was similar in both hospitals (intermediate and ICU rooms), except for the presence or absence of negative pressure systems in hospital B. However, there were some individual rooms and others were shared rooms. In Hospital A, 73.3% (22/30) of the beds were in individual rooms, whereas the other 8 beds were in two shared rooms of 4 beds. Similarly, in hospital B, 71.4% (15/21) of the beds were in individual rooms, whereas the other 6 beds were in a shared room. Hospital B has a negative pressure system (20 air exchanges per hour) at each patients' bed, whereas Hospital A does not have it.
This work was conducted from May 2020 to June 2021. Surfaces sampling were carried out using Isohelix mini swabs (Isohelix cat. number MS-02) pre-moistened with Zymo DNA/RNA Shield (Zymo cat. number R1100-250), a stabilization solution that preserves the genetic integrity and expression profiles of samples at ambient temperatures and completely inactivates infectious agents, according to the manufacturer. Swabs were deposited inside tubes pre-filled with 400 μL of Zymo DNA/RNA Shield (Zymo cat. number R1100-250) and stored at room temperature until processing (usually within the week of swab sampling). Samples were collected by swabbing different surfaces in patients' rooms (both intermediate and intensive care rooms) such as bed table, mechanical ventilator, vitals monitor, foot of patient’s bed (foot bed), floor at 1 m of distance from the patient’s bed, floor at 1 m of distance of the foot of patient’s bed, and the room entrance door (Supplementary Fig. S1). Surfaces sampling occurred around the middle of work shifts and before surfaces were cleaned and disinfected. Patient rooms were cleaned at every work shift (every 6 h). In total, we collected 323 swab samples (222 surface samples from hospital A and 101 surface samples from hospital B) that resulted in 969 PCR data without including controls (666 from hospital A and 303 from hospital B) (see summary of swab samples collected in Table 1).
Table 1.
Surface samples collection.
| Gamma introduction | Hospital | Hospital area | Number of swab samples |
|---|---|---|---|
| Before | A | Nursery area | 26 |
| A | Patient's rooms | 114 | |
| After | A | Patient's rooms | 82 |
| B | Patient’s room | 101 | |
| Total | 323 |
2.2. ATP measures
Adenosine triphosphate (ATP) monitoring is utilized as a surrogate marker for hygiene in hospitals [21]. ATP is detected with a biological reaction that produces light in the presence of organic matter, including microbes, feces and other kinds of dirt [22].
ATP bioluminescence measurements were performed using the PocketSwab Plus swabs and the luminometer Novalum (Charm Science Inc). Samples were collected from different surfaces of 20 Intensive Care Unit (ICU) rooms (10 from intermediate care and 10 from intensive care) and four high-touch surfaces from a non-COVID-19 area for one year in hospital A. Sampling sites of non-COVID-19 area were keyboard and PC, table surface and telephone located in the nursing area. Briefly, for each area, the surfaces analyzed for SARS-CoV-2 RNA detection were monitored (see previous point). In all sampling places the same criterion was adopted for the selection of the areas to be monitored, trying to sample, for the same equipment, the most frequently hand-touched sites. Measurements were carried out by 100 cm2 area (10 cm × 10 cm) for 5 s. The results of the ATP measurements were expressed as Relative Light Units (RLU)/cm2. According to the infection committee of Hospital A and previous work [23], the threshold of ATP measurement to consider a dirty surface is 3000 RLUs/100 cm2.
2.3. SARS-CoV-2 detection
RNA extraction was performed with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA extracted were stored at −80 °C until RT-qPCR processing. For each batch of processed samples, at least one extraction control was performed in parallel with nuclease free water. Extracted RNA was used then for the detection of SARS-CoV-2 using a specific RT-qPCR. Briefly, a 20 μL PCR reaction contained 5 μL of 4x TaqMan Fast Virus Master Mix (Thermo Fisher), 8.6 μL of nuclease-free water and 0.75 μL of N1 and N2 combined primer/probe mix each (2019-nCoV CDC EUA Kit - IDT) and 5 μL of RNA. Thermal cycling was run on a Step-One Plus RT-PCR thermal cycler (Applied Biosystems) with the following cycle parameters: 50 °C for 5 min for reverse transcription, inactivation of reverse transcriptase at 95 °C for 20 s and then 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The threshold value was set manually at 0.03 in all assays to determine the threshold cycle (Ct). The Ct value is defined as the number of PCR cycles required to cross a given threshold to detect, in this case, viral nucleic acid. Thus, Ct values are inversely proportional to the amount of target viral genome. Ct values < 40 were considered positive for SARS-CoV-2 [24]. For every qPCR run, a positive control (Plasmid DNA), a non-template control (nuclease-free water) and extraction controls were included. All RT-qPCRs were performed in triplicate. Each sample was considered positive for SARS-CoV-2 when at least two of the three technical replicates showed a Ct value < 40.
2.4. Construction of RNA for quantification standards and analytical sensitivity of the RT-PCR assay
A fragment of 968 bp containing the N1 and N2 targets from SARS-CoV-2, were cloned into pCR™2.1- TOPO® using the TOPO® TA Cloning® Kit (Invitrogen) following manufacturer’s instructions and transformed in NEB® 5-alpha Competent E. coli (High Efficiency) by the heat shock method (42 °C, 30 s). Plasmids were isolated using PureLink Quick Plasmid Miniprep Kit (Invitrogen) and quantified by spectrophotometric analysis (Bio-photometer, Eppendorf). Then, 1 μg of each plasmid was linearized with SpeI and in vitro transcribed with T7 RNA Polymerase (Thermo Fisher) following the manufacturer’s instructions. In vitro transcribed RNA was treated with DNase and purified with TURBO DNA-free™ Kit (Thermo Fisher). RNA purified was checked for size and integrity by gel electrophoresis and quantified by fluorometric analysis (Qubit 2.0, Thermo). The number of copies/μL was calculated as: (NA × C)/MW, where NA is the Avogadro constant expressed in mol−1, C is the concentration expressed in g/μL, and MW is the molecular weight expressed in g/mol.
A stock containing 3 × 1011 copies/μL of in vitro transcribed RNA was used for standard curve and sensitivity determination of the RT-qPCR assays. The calibration curve and analytical sensitivity were determined by 10-fold serial dilutions of RNA in vitro transcribed. Then, 5 μL of the corresponding RNA 10-fold dilution was added to the mix and tested in triplicate. Calibration curves were represented as Ct vs log copy number/reaction. The linear regression curve (y) was determined as y = 3.28x + 34.19 and the coefficient of determination (R2) was 0.9919.
2.5. SARS-CoV-2 genome sequencing and phylogenetic analysis
Sequencing was performed according to the PrimalSeq approach (Grubaugh et al., 2019) on the MinION sequencing platform (Oxford Nanopore Technologies, United Kingdom). Sequencing libraries were prepared based on the PCR-tiling protocol for SARS-CoV-2 (ARTIC Network) using the Ligation Sequencing Kit (SQK-LSK109) and Native Barcoding Expansion (EXP-NBD114) (Oxford Nanopore Technologies, United Kingdom), with minor modifications. Briefly, RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA, USA) and random hexamer primers following the manufacturer’s instructions. Viral genomes were amplified using ARTIC nCoV-2019 V3 primer pools (IDT) and Q5® Hot Start High-Fidelity 2X Master Mix (NEB). Amplified sequences were cleaned up using AMPure XP beads (Beckman Coulter) and repaired using NEBNext Ultra II End repair/dA-tailing Module (NEB). NEBNext Ultra II Ligation Module (NEB) was used for unique barcode ligation. To detect possible barcode cross-contamination between samples, the RT negative control was carried through the barcoding step and a previously sequenced SARS-CoV-2 sample was included as a positive control. Finally, sequencing adapters were ligated using NEBNext Quick Ligation Module (NEB) and a total of ∼20 fmol of pooled samples was loaded into a FLO-FLG001 flow cell and sequenced for 24 h. Subsequently, high accuracy basecalling and demultiplexing was performed using Guppy software v3.6.0. Consensus genomes were generated and variants were called using a modified version of Nextflow [25], a pipeline developed by the COVID-19 Genomics UK consortium [26] which is based on the ARTIC Network bioinformatic protocol (https://artic.network/ncov-2019/ncov2019-bioinformaticssop.html). Minor modifications were made to fit local infrastructure (https://github.com/iferres/ncov2019-artic-nf). Amplicons that were not sequenced or whose depth was less than 20x were not included in the consensus sequences and these positions were represented by “N” stretches. Curation of sequences was performed manually in positions with at least 10x sequencing depth in each position. PANGO lineages were determined using Pangolin v3.1.5 [https://github.com/cov-lineages/pangolin, https://doi.org/10.1038/s41564-020-0770-5] and Nextstrain clade assignment was performed using Nextclade CLI v1.10.3 [https://github.com/nextstrain/nextclade, https://doi.org/10.1093/bioinformatics/bty407]. Time-scaled phylogeny. A maximum likelihood (ML) phylogeny was generated using the consensus sequences from the present study and 804 available SARS-CoV–2 genomes from Uruguay in EpicCoV/GISAID database [(www.gisaid.org, last accessed 2022-06-30] obtained from March 2020 to June 2021 (EPI_SET_221121nk). Sequences were first aligned using Nextalign CLI v1.4.5 with the WIV04 (EPI_ISL_402124) sequence as reference. The ML tree was inferred with IQ-TREE v2.1.4 using the GTR nucleotide substitution model. A time tree was estimated using TreeTime joint-maximum likelihood approach with the oldest reroot option. Time tree visualizations were generated with the ggtree package.
2.6. Statistical analysis
Statistical analyses were performed with SPSS software, version 23 (IBM Corporation). ANOVA was used to compare means. Fisher exact test was used to compare positivity rates (SARS-CoV-2 positive places over all places interrogated). Linear regressions were performed with R Studio. Differences were considered statistically significant when p-value <0.05.
3. Results
3.1. There was no correlation between the amount of organic material dirtiness and SARS-CoV-2 RNA detected on surfaces
Adenosine triphosphate (ATP) bioluminescence assays are used to measure organic material as a proxy for cleanliness in hospitals worldwide [21]. This method does not specifically detect viruses but indicates different forms of biomass, like epithelium from the upper respiratory tract, saliva, and biological material from coughs and sneezes [27]. However, whether the ATP measurements have or not a relationship with the viral RNA concentration on surface contamination remains scarce [22]. To address this, we analyzed organic material contamination using ATP bioluminescence and the presence of SARS-CoV-2 genomes using RT-qPCR. In the case of the analysis of swabs from areas with non-COVID-19 patients, we found that 15.38% (4/26) of the surface swabs displayed RLU/cm2 values above the threshold of ATP whereas the number of surface swabs with Ct < 40 was 7.69% (2/26). However, no swab sample from hospital areas with non-COVID-19 samples tested positive for both ATP and RNA. Regarding the analysis of swabs sampled from patient rooms, we found that 96.5% of the surface swabs (110/114) displayed RLU/cm2 values above the threshold of ATP measurement to consider a surface contaminated with organic material. In contrast, we found that only 43.9% of the surface swabs (50/114) detected SARS-CoV-2 RNA (Ct < 40).
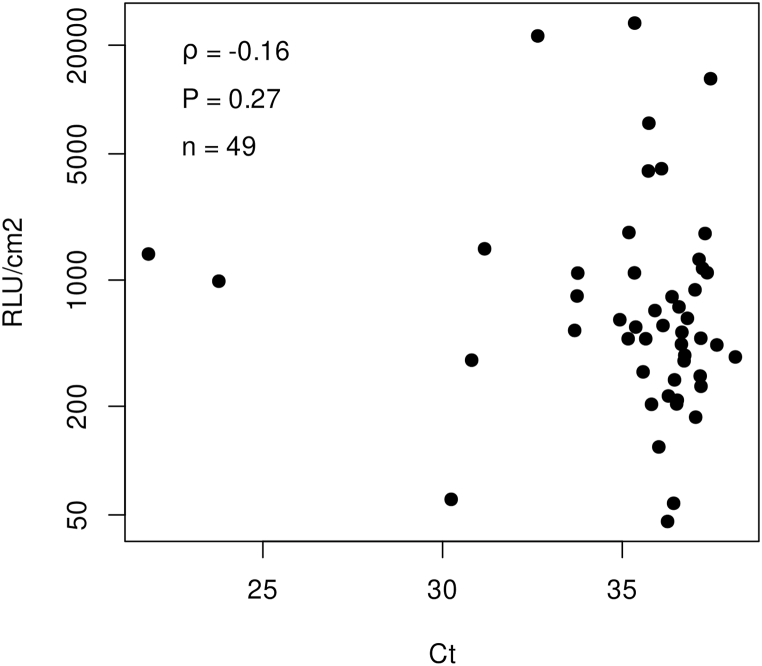
Then, we studied if the positive swabs for both organic material and SARS-RNA contamination were correlated (n = 49). Our study indicates no significant correlation between ATP measurements expressed as RLU/cm2 and the Ct values of SARS-CoV-2 found in ICU rooms (Spearman, r = −0.16, p = 0.27) (Fig. 1).
Fig. 1.
Correlation between ATP relative light units (RLU/cm2) and Ct values obtained from surfaces in hospital settings. The scatter plot includes positive swabs for both organic material (ATP >3000) and SARS-CoV-2 RNA (Ct < 40) (n = 49). The chart shows no linear correlation between Ct values and ATP (Spearman’s rank correlation coefficient ρ = −0.16; p-value = 0.27). The y axis is represented in LOG10 scale.
3.2. SARS-CoV-2 high transmissible Gamma variant introduction significantly increased surface contamination
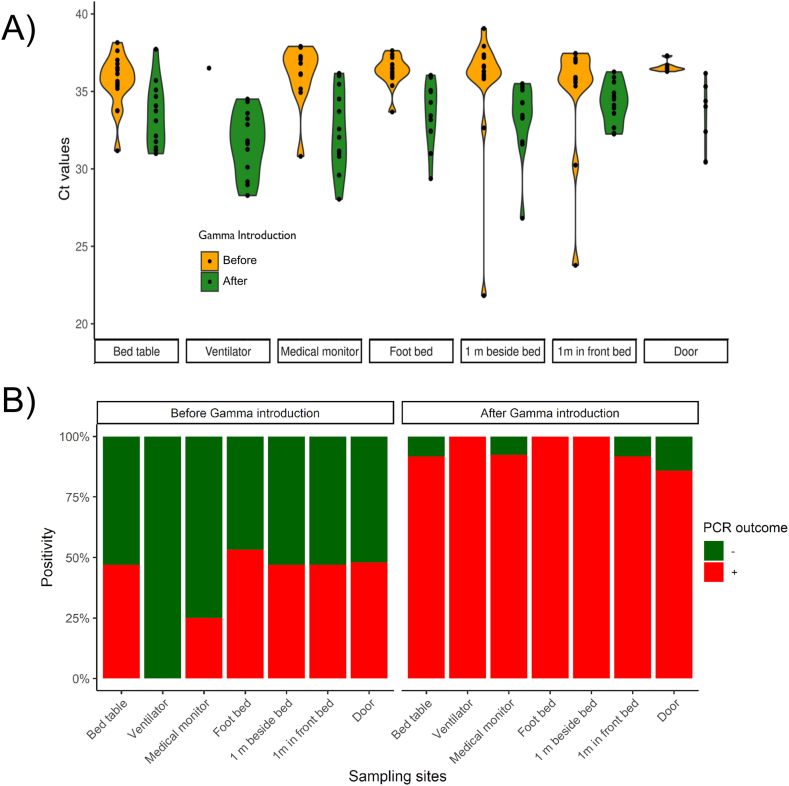
We evaluated SARS-CoV-2 RNA environmental contamination by swabbing seven different places in ICU patients' rooms from COVID-19 reference hospital A. Sampling was done before (n = 114) and after (n = 82) Gamma variant introduction. Environmental surface contamination was slightly lower in intensive care rooms (Ct mean = 35.45 ± 0.16) than those observed in intermediate care rooms (Ct mean = 36.28 ± 0.26) (nested ANOVA, p = 0.006). SARS-CoV-2 Gamma variant introduction produced an increase in surfaces contamination displayed by a decrease of roughly four Ct units on average, from Ct mean = 37.89 ± 0.19 to Ct mean = 33.84 ± 0.22 (p < 0.001) (Fig. 2A). In addition, no significant differences in Ct mean values were observed between sampling sites (p = 0.061), indicating that surface contamination was homogeneous at the different sampling locations.
Fig. 2.
Surface contamination with SARS-CoV-2 RNA observed during one-year follow up study of SARS-CoV-2 RNA contamination in hospital A. Ct values (A) and positivity rates (B) observed before (orange violins) and after (green violins) the circulation of VOC Gamma in Hospital A. Positive and negative PCR outcomes were represented in red and green, respectively. Surfaces sampled are in decreasing order relative to the patient’s bed distance. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The SARS-CoV-2 Gamma variant is highly transmissible due, at least in part, to higher viral loads than previous variants [6]. Therefore, we hypothesized that the Gamma variant introduction would produce an increase in positive tests on surfaces of hospital settings. Overall, our results showed that the SARS-CoV-2 positive sites over all sites tested, defined as the positivity rate, increased from 44.55% (50/114) to 96.88% (78/82). This represented a 2.17-fold increase in the positivity rate after Gamma introduction (Fisher exact test, p < 0.001) (Fig. 2B).
3.3. The hospital with negative pressure systems was associated with lower levels of SARS-CoV-2 RNA surface contamination
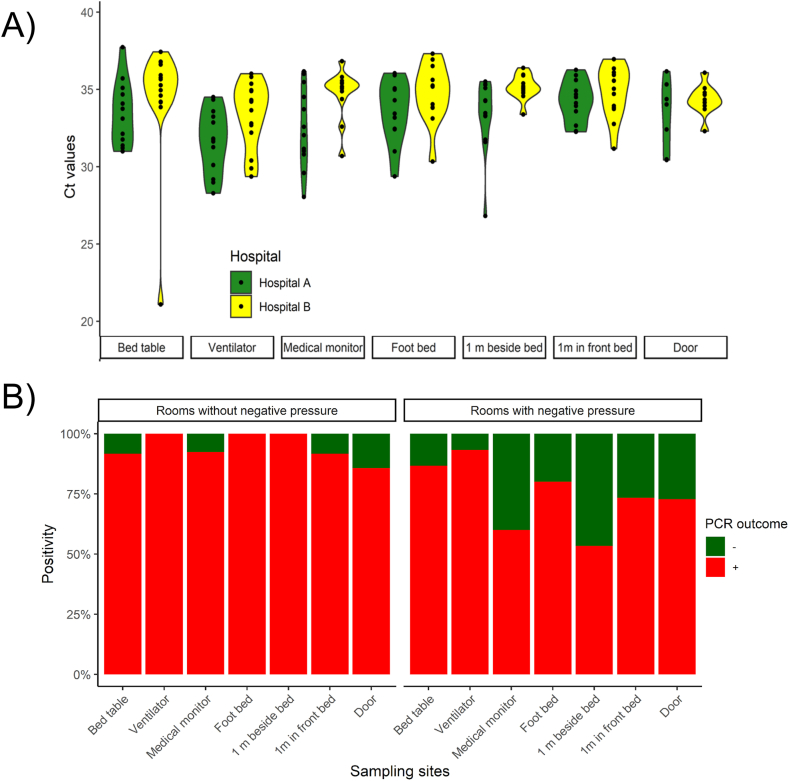
To evaluate the effect of negative pressure on SARS-CoV-2 RNA contamination on surfaces, we compared SARS-CoV-2 RNA detection in samples obtained from hospital A and hospital B after the Gamma variant introduction into the country. Hospital A lacks a negative pressure system whereas Hospital B has rooms submitted to negative pressure. We found that Ct values obtained from surfaces at Hospital A (Ct mean = 33.92 ± 0.24) were lower than those obtained from Hospital B (Ct mean = 35.985 ± 0.175) (nested ANOVA, p < 0.001), showing a higher viral RNA contamination in the hospital without negative pressure systems. Contrary to that observed in Hospital A; in Hospital B, we found significant differences in Ct mean values from different sampling sites (p < 0.001) indicating that Ct values were heterogeneous at different sampling locations (Fig. 3A). A Tukey’s post hoc test showed that the means of the Ct values obtained from the bed table, ventilator, foot bed, 1 m beside the bed, 1 m in the front bed, and the door, form a single homogeneous subgroup (p = 0.149), which differs from those obtained from swabs samples of the medical monitor.
Fig. 3.
Comparison of surface contamination with SARS-CoV-2 RNA among hospital settings. Ct values (A) and positivity rates (B) observed in Hospital A (green violins) and Hospital B (yellow violins) during the circulation of VOC Gamma in Uruguay. Positive and negative PCR outcomes were represented in red and green, respectively. Surfaces sampled are in decreasing order relative to the patient’s bed distance. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In light of these data, we hypothesized that negative pressure could affect the positivity rate for each sampling site should be higher in Hospital A (rooms without negative pressure) compared to Hospital B (rooms under negative pressure). Overall, our findings showed that the positivity rate in Hospital A (96.88%; 78/82) was significantly higher than the observed at Hospital B (75.31%; 76/101) (Fisher exact test, p < 0.001) (Fig. 3B).
Finally, we evaluated the correlation between Ct values obtained from surfaces and the distance to the patient (Table 2). We found that Ct values decrease with the patient proximity in Hospital A, showing a significant positive correlation (Spearman, r = 0.33, p = 0.002). In contrast, no correlation was found at Hospital B (Spearman, r = −0.02, p = 0. 871).
Table 2.
Summary of the surface swabs accessed after Gamma variant introduction.
| Hospital | Sampling sites | Number of positive samples1 | Number of total samples | Rate of positivity (%) | Mean Ct ± SEM | Distance (m) | Distance (ranking) | Spearman coefficient (ρ) | p-value |
|---|---|---|---|---|---|---|---|---|---|
| A | Medical monitor | 12 | 13 | 92.3 | 31.79 ± 0.65 | 1.00 | 1 | ||
| Bed table | 11 | 12 | 91.7 | 33.08 ± 0.51 | 1.12 | 2 | |||
| Ventilator | 13 | 13 | 100.0 | 31.65 ± 0.57 | 1.12 | 2 | |||
| 1 m beside bed | 13 | 13 | 100.0 | 33.33 ± 0.65 | 1.80 | 4 | 0.33 | 0.002 | |
| Foot bed | 12 | 12 | 100.0 | 33.59 ± 0.59 | 2.00 | 5 | |||
| 1 m in front bed | 11 | 12 | 91.7 | 34.25 ± 0.43 | 3.00 | 6 | |||
| Door | 6 | 7 | 85.7 | 32.84 ± 0.85 | 5.00 | 7 | |||
| Total | 78 | 82 | 95.1 | 32.91 ± 0.24 | – | – | |||
| B | Medical monitor | 9 | 15 | 60.0 | 34.47 ± 0.57 | 1.00 | 1 | ||
| Bed table | 13 | 15 | 86.7 | 34.21 ± 1.12 | 1.12 | 2 | |||
| Ventilator | 14 | 15 | 93.3 | 33.27 ± 0.58 | 1.12 | 2 | |||
| 1 m beside bed | 8 | 15 | 53.3 | 35.45 ± 0.20 | 1.80 | 4 | 0.02 | 0.871 | |
| Foot bed | 12 | 15 | 80.0 | 34.76 ± 0.55 | 2.00 | 5 | |||
| 1 m in front bed | 11 | 15 | 73.3 | 34.54 ± 0.51 | 3.00 | 6 | |||
| Door | 8 | 11 | 72.7 | 34.22 ± 0.38 | 5.00 | 7 | |||
| Total | 75 | 101 | 74.3 | 34.34 ± 0.26 | – | – |
Rate of positivity of the sampling sites assayed by RT-qPCR after Gamma introduction and spread in the country.
Distance between patient and sampling sites is expressed as the Euclidean distance (in meters) and as ranking.
Spearman’s rank correlation (ρ) between cycle threshold (Ct) of positive samples and the distance to the patient, per hospital.
Ct value < 40 in at least two of the three RT-qPCR technical replicates.
3.4. SARS-CoV-2 genomes recovered from surfaces were fully sequenced
Recovering complete SARS-CoV-2 genome sequences directly from environmental surface swabs is a difficult task considering their low RNA amount inferred by their showed Ct values. However, we recovered two near-complete genome sequences belonging to two patient rooms (named HosA-008 and HosB-004). The first sequence was recovered from the floor, 1 m beside the bed, before the Gamma variant introduction, with a mean Ct value of 21.82 ± 0.23 and a genome coverage of 407.98X (Accession number EPI_ISL_11782100). The second sequence was recovered after Gamma variant introduction from a room bed table and showed a mean Ct value of 21.10 ± 0.06 and a genome coverage of 69.07X (Accession number EPI_ISL_11783770).
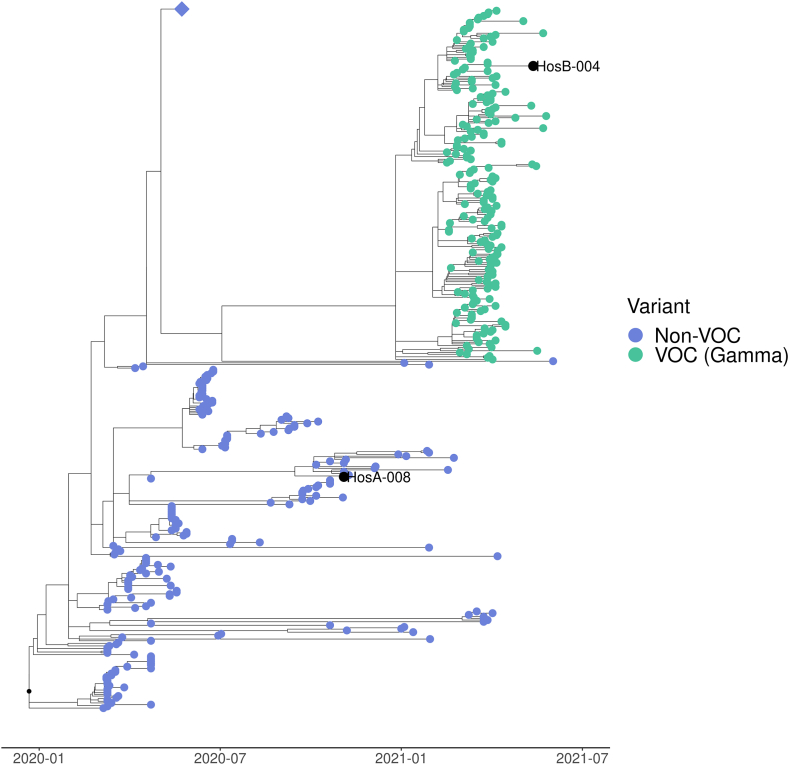
We conducted a phylogenetic analysis using NextStrain (Hadfield et al., 2018) to compare the obtained genomes with others (from Uruguay) available in the database. Sequences recovered from HosA-008A were placed in the “Non-VOC” clade and sequence from HosB-004 was placed in the VOC clade (Gamma) (Fig. 4). These findings are in agreement with the results obtained from national real time genomic surveillance (more detailed phylogenetic tree in Supplementary Fig. S2). We showed previously that strains circulating in the “Non-VOC” clade were dominant in Uruguay until Gamma variant was introduced [7].
Fig. 4.
Time-scaled phylogeny from publicly available Uruguayan sequences from March 2020 to June 2021. Consensus sequences, obtained from surface samples at hospitals A and B (denoted with a black dot), were clustered as non-VOC (HosA-008) and Gamma VOC (HosB-004), respectively. Blue diamond shape represents a collapsed node containing 400 non-VOC sequences from P.6 (n = 315), P.7 (n = 34) and P.2 (n = 51) PANGO lineages. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Most surface swab samples positive for SARS-CoV-2 detection by RT-qPCR are predicted to yield non-infectious virus
Viral RNA detection by RT-qPCR on surfaces does not necessarily indicate virus viability and/or infectivity [28] and indeed we inactivated the infectious agents immediately after swab collection in DNA/RNA Shield solution. Previously it has been shown that it is possible to predict the infectivity of swabs samples in cell culture. Berg and colleagues (2021) found that cytopathic effect (CPE) was observed in Vero cells when samples had a minimum of 4.2 log10 viral genomes copies/mL [29]. On the other hand, another work showed that infectious virus was isolated from samples with at least 3.1 log10 copies/uL [30]. We used a standard curve to infer the viral infectivity based on the Ct values experimentally obtained and the linear regression curve to determine the number of genome copies/unit of volume. Taking this into account, we inferred that in our PCR assay settings, the Ct values corresponding to these copy numbers were between Ct = 27 and Ct = 24, respectively. Since swab samples showed a Ct mean value of 34.03 ± 0.18, we can conclude that most of the swab samples analyzed in this work would not be infectious. There were just two samples, which represent the 0.07% (2/323) of the N used, with Ct values lower than 24 that could be potentially infectious (21.82 ± 0.23 and 21.10 ± 0.06). In addition, these samples corresponded to the two SARS-CoV-2 genomes recovered from surfaces that we were able to fully sequenced.
4. Discussion
Prevention of disease in healthcare workers and successful public health measures for COVID-19 require management based on the understanding of viral spread and modes of transmission. Despite the main mechanism of transmission of SARS-CoV-2 is droplet transmission by close contact between persons [1], there is also evidence that viral transmission is also achieved by aerosols [[3], [4], [5]]. SARS-CoV-2 RNA has been detected in several studies in hospital settings (systematically reviewed in Ref. [31]) suggesting the potential of virus transmission by contact with contaminated surfaces. Importantly, the risk of infections by contact with contaminated surfaces is considered low and difficult to decouple from other transmission mechanisms [32].
Here, we implemented a one-year longitudinal study taking into account factors that have not been fully addressed to assess their impact on SARS-CoV-2 surface contamination in hospitals. These factors were: (i) organic material, which is used as a surrogate for cleanliness; (ii) the introduction of highly transmissible viral variants, and (iii) the presence or absence of negative pressure systems in the patient’s room.
We first tested the feasibility of implementing the organic material measure as a proxy for detecting SARS-CoV-2 RNA contamination in hospitals. Measuring organic material by ATP bioluminescence assays is widely used as a marker for cleanliness in hospitals [21,33]. If there were any correlation, ATP measurement could be used to infer the presence of viral RNA on surfaces during epidemics. At the time of writing this manuscript, one study tried to attempt this goal in hospital settings. However, the authors failed to detect SARS-CoV-2 RNA in any surface sampled [22]. Our results are in agreement with a previous work focused on RNA from bacteriophages [34]. In that study, the ATP measurement was not related to the viral nucleic acid contamination.
We then studied the impact of more transmissible variants on surface contamination in hospital A settings. We analyzed this issue before and after the SARS-CoV-2 Gamma variant was introduced and became prevalent in Uruguay [7]. This Variant of Concern (VOC) displayed a significant increase in transmission capacity [6,8]. Before Gamma variant introduction, the positivity rate determined for Hospital A showed high concordance with results obtained from ICUs in Wuhan, China, in early 2020 (Guo et al., 2020; Wu et al., 2020). Interestingly, after the Gamma variant introduction, we found a significant increase in viral contamination in ICU rooms, both in terms of positivity rate and cycle threshold (Ct) values obtained. Specifically, we observed a 2.17-fold increase in the positivity rate, whereas the Ct decreased on average by 4 units. Importantly, according to the linear regression of our standard curve, this decrease in Ct values indicates a 15-fold increase in the amount of viral RNA detected on surfaces during the circulation of the VOC Gamma. These results are consistent with those obtained from clinical samples in Brazil where Ct values from patients infected with VOC Gamma were around 3 units lower than those observed from patients infected with previous no-VOC variants. This means that the viral load of patients infected with VOC Gamma was ∼10-fold higher than in clinical samples from non-VOC patients [8].
Subsequently, we compared the surfaces contamination results from two hospitals, one of them with negatively pressurized isolation rooms. This was done to test whether we could identify differences of SARS-CoV-2 RNA contamination during the wave of cases produced by VOC Gamma. Isolation rooms are important to prevent the spread of respiratory pathogens to other areas contaminating other patients or healthcare workers [17]. Our results indicate that the amount of viral RNA of contaminated surfaces in hospital A was approximately 4-fold higher than those found in Hospital B. This lower contamination, in terms of amount of viral RNA detected by RT-qPCR in Hospital B, was associated with a 1.3-fold decrease in the positivity rate compared to Hospital A. We have also found a moderate positive correlation between the Ct value from surface samples and patient distance in Hospital A whereas this correlation was lost in Hospital B. Altogether, these results indicate that the hospital with negative pressure systems was associated with lower levels of SARS-CoV-2 RNA surface contamination which is consistent with previous work [35].
We could obtain two full SARS-COV-2 genome sequences from surface samples. The sequences matched with the circulating variants at that time [7].
Finally, our study presents the following limitations. First, as it is an experiment carried out in hospital settings there are different factors with a high degree of heterogeneity. For example, we cannot ignore clinical differences in the patients admitted in both hospitals, (age, seriousness of infection, comorbidities, symptoms and length of time since diagnosis). Secondly, the Gamma variant wave of infections triggers an increase in daily cases and hospitalizations. This fact directly impacted on hospital bed occupancy. It is also important to note that viral RNA detection by RT-qPCR on surfaces does not indicate virus viability and/or infectivity [28]. This particular issue represented a weakness of our work. This issue could be addressed by testing virus viability or infectivity in tissue culture. In addition, we do not have access to a biosafety level 3 facility (BSL3) to experimentally determine this. However, taking into account previous analyses about viral infectivity, we estimated the number of genome copies/mL. Our analysis suggests that 99.93% (321/323) of the samples collected from contaminated surfaces were likely non-infectious. This result is in agreement with previous work [29,30], indicating that despite SARS-CoV-2 fomites transmission is possible, it is significantly low.
Our results highlight the importance of tracking the spatial viral contamination at surfaces may be useful as a surrogate of viral dissemination in medical setting with implications in patient and healthcare worker management.
5. Conclusions
This study provides data gathered for one year about the surface contamination with SARS-CoV-2 RNA in hospital settings. First, despite ATP bioluminescence assays are widely used as marker for cleanliness in hospitals, our results showed that ATP measures are not suitable for evaluate SARS-CoV-2 RNA contamination. Second, our results indicated that SARS-CoV-2 RNA contamination increased after the introduction of more transmissible (Gamma) variants. Third, we provided evidence of lower surface SARS-CoV-2 RNA contamination in a hospital with a negative pressure system. Therefore, this work has valuable implications for hospital management and public health policies. Mainly in low and middle income-countries' hospitals, where negative pressure systems are not widely distributed. Other approaches are needed to distinguish effects from confounder factors in our measurements.
Funding sources
FOCEM (MERCOSUR Structural Convergence Fund), COF 03/11; Fondo de Solidaridad para Proyectos Innovadores, Sociedad Civil, Francofonía y Desarrollo Humano (FSPI), Ambassade de France; Fondo Clemente Estable, Agencia Nacional de Investigación e Innovación (ANII) and Centro Latinoamericano de Biotecnología (CABBIO) and G4 program Institut Pasteur Montevideo.
Acknowledgements
We thanks the Programa para el Desarrollo de las Ciencias básicas (PEDECIBA), and health-workers from Hospital Español Dr. José Crottogini and INOT, Montevideo, Uruguay.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13875.
Contributor Information
Marianoel Pereira-Gómez, Email: mnpereira@pasteur.edu.uy.
Gonzalo Moratorio, Email: moratorio@pasteur.edu.uy.
Abbreviations
- ATP
Adenosine triphosphate
- COVID-19
Coronavirus Disease 2019
- Ct
Cycle threshold
- PCR
Polymerase Chain Reaction
- RT-qPCR
Reverse Transcription Real Time Polymerase Chain Reaction
- RLU
Relative Light Units
- VOC
Variant of Concern
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Sampling sites in individual hospital rooms. Sampling sites are shown and labeled. Figure was Created with BioRender.com.
Time-scaled phylogeny from publicly available Uruguayan sequences from March 2020 to June 2021. Consensus sequences, obtained from surface samples at hospitals A and B (denoted with a black dot), were assigned to the 20B Nextstrain clade (HosA-008) and 20J (Gamma, V3) (HosB-004), respectively. The blue diamond shape represents the collapsed node containing the P.6, P.7 and P.2 lineages.
References
- 1.WHO-Scientific Brief . 2020. Transmission of SARS-CoV-2: implications for infection prevention precautions, 9 July.https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions (accessed July 26, 2021) [Google Scholar]
- 2.Delikhoon M., Guzman M.I., Nabizadeh R., Baghani A.N. Modes of transmission of severe acute respiratory syndrome-coronavirus-2 (Sars-cov-2) and factors influencing on the airborne transmission: a review. Int. J. Environ. Res. Publ. Health. 2021;18:1–18. doi: 10.3390/ijerph18020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/J.ENVINT.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadnytskyi V., Bax C.E., Bax A., Anfinrud P. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc. Natl. Acad. Sci. USA. 2020;117:11875–11877. doi: 10.1073/PNAS.2006874117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R., Li Y., Zhang A.L., Wang Y., Molina M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. USA. 2020;117:14857–14863. doi: 10.1073/PNAS.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D. da S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., de Jesus J.G., Andrade P.S., Coletti T.M., Ferreira G.M., Silva C.A.M., Manuli E.R., Pereira R.H.M., Peixoto P.S., Kraemer M.U.G., Gaburo N., Camilo C. da C., Hoeltgebaum H., Souza W.M., Rocha E.C., de Souza L.M., de Pinho M.C., Araujo L.J.T., Malta F.S.V., de Lima A.B., Silva J. do P., Zauli D.A.G., Ferreira A.C. de S., Schnekenberg R.P., Laydon D.J., Walker P.G.T., Schlüter H.M., dos Santos A.L.P., Vidal M.S., Del Caro V.S., Filho R.M.F., dos Santos H.M., Aguiar R.S., Proença-Modena J.L., Nelson B., Hay J.A., Monod M., Miscouridou X., Coupland H., Sonabend R., Vollmer M., Gandy A., Prete C.A., Nascimento V.H., Suchard M.A., Bowden T.A., Pond S.L.K., Wu C.-H., Ratmann O., Ferguson N.M., Dye C., Loman N.J., Lemey P., Rambaut A., Fraiji N.A., Carvalho M.do P.S.S., Pybus O.G., Flaxman S., Bhatt S., Sabino E.C. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science (80-.) 2021;372:815–821. doi: 10.1126/SCIENCE.ABH2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rego N., Costábile A., Paz M., Salazar C., Perbolianachis P., Spangenberg L., Ferrés I., Arce R., Fajardo A., Arleo M., Possi T., Bellini I., Bilbao L., Reyes N., Bentancor M.N., Lizasoain A., Benítez M.J., Castells M., Victoria M., Maya L., Bortagaray V., Moller A., Bello G., Arantes I., Brandes M., Smircich P., Chappos O., Duquía M., González B., Griffero L., Méndez M., Techera M.P., Zanetti J., Rivera B., Maidana M., Alonso M., Alonso C., Medina J., Albornoz H., Colina R., Noya V., Iraola G., Fernández-Calero T., Moratorio G., Moreno P. Real-time genomic surveillance for SARS-CoV-2 variants of concern. Uruguay, Emerg. Inf. Disp. 2021;27 doi: 10.3201/eid2711.211198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naveca F.G., Nascimento V., de Souza V.C., Corado A. de L., Nascimento F., Silva G., Costa Á., Duarte D., Pessoa K., Mejía M., Brandão M.J., Jesus M., Gonçalves L., da Costa C.F., Sampaio V., Barros D., Silva M., Mattos T., Pontes G., Abdalla L., Santos J.H., Arantes I., Dezordi F.Z., Siqueira M.M., Wallau G.L., Resende P.C., Delatorre E., Gräf T., Bello G. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 2021;277(27):1230–1238. doi: 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- 9.Banho C.A., Sacchetto L., Campos G.R.F., Bittar C., Possebon F.S., Ullmann L.S., Marques B. de C., da Silva G.C.D., Moraes M.M., Parra M.C.P., Negri A.F., Boldrin A.C., Barcelos M.D., dos Santos T.M.I.L., Milhim B.H.G.A., Rocha L.C., Dourado F.S., dos Santos A.L., Ciconi V.B., Patuto C., Versiani A.F., da Silva R.A., de Oliveira Lobl E.E., Hernandes V.M., Zini N., Pacca C.C., Estofolete C.F., Ferreira H.L., Rahal P., Araújo J.P., Cohen J.A., Kerr C.C., Althouse B.M., Vasilakis N., Nogueira M.L. Impact of SARS-CoV-2 Gamma lineage introduction and COVID-19 vaccination on the epidemiological landscape of a Brazilian city. Commun. Med. 2022;2:41. doi: 10.1038/s43856-022-00108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katona P., Kullar R., Zhang K. Bringing transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to the surface: is there a role for fomites? Clin. Infect. Dis. 2022;75:910–916. doi: 10.1093/cid/ciac157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chia P.Y., Coleman K.K., Tan Y.K., Ong S.W.X., Gum M., Lau S.K., Lim X.F., Lim A.S., Sutjipto S., Lee P.H., Son T.T., Young B.E., Milton D.K., Gray G.C., Schuster S., Barkham T., De P.P., Vasoo S., Chan M., Ang B.S.P., Tan B.H., Leo Y.S., Ng O.T., Wong M.S.Y., Marimuthu K., Lye D.C., Lim P.L., Lee C.C., Ling L.M., Lee L., Lee T.H., Wong C.S., Sadarangani S., Lin R.J., Ng D.H.L., Sadasiv M., Yeo T.W., Choy C.Y., Tan G.S.E., Dimatatac F., Santos I.F., Go C.J., Chan Y.K., Tay J.Y., Tan J.Y.L., Pandit N., Ho B.C.H., Mendis S., Chen Y.Y.C., Abdad M.Y., Moses D. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. The Lancet Microbe. 2020;1 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Z., Wang Z., Zhang S., Li X., Li L., Li C., Cui Y., Fu R., Dong Y., Chi X., Zhang M., Liu K., Cao C., Liu B., Zhang K., Gao Y., Lu B., Chen W. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, wuhan, China, 2020. Emerg. Infect. Dis. 2020;26:1586–1591. doi: 10.3201/EID2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santarpia J.L., Rivera D.N., Herrera V.L., Morwitzer M.J., Creager H.M., Santarpia G.W., Crown K.K., Brett-Major D.M., Schnaubelt E.R., Broadhurst M.J., Lawler J.V., Reid S.P., Lowe J.J. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-69286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otter J.A., Zhou J., Price J.R., Reeves L., Zhu N., Randell P., Sriskandan S., Barclay W.S., Holmes A.H. Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London. Clin. Infect. Dis. 2020 doi: 10.1093/CID/CIAA905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marzoli F., Bortolami A., Pezzuto A., Mazzetto E., Piro R., Terregino C., Bonfante F., Belluco S. A systematic review of human coronaviruses survival on environmental surfaces. Sci. Total Environ. 2021;778 doi: 10.1016/j.scitotenv.2021.146191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Benna S. Negative pressure rooms and COVID-19. J. Perioperat. Pract. 2020;31:18–23. doi: 10.1177/1750458920949453. [DOI] [PubMed] [Google Scholar]
- 18.Aguilar-Rodríguez J., Peel L., Stella M., Wagner A., Payne J.L. The architecture of an empirical genotype-phenotype map. Evolution (N. Y). 2018;72:1242–1260. doi: 10.1111/evo.13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwal R., Bhatia R., Soni K.D., Trikha A. Fast tracking intensive care units and operation rooms during the COVID-19 pandemic in resource limited settings. J. Anaesthesiol. Clin. Pharmacol. 2020;36 doi: 10.4103/joacp.JOACP_262_20. https://journals.lww.com/joacp/Fulltext/2020/36001/Fast_tracking_intensive_care_units_and_operation.5.aspx [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West T.E., Schultz M.J., Ahmed H.Y., Shrestha G.S., Papali A. Pragmatic recommendations for tracheostomy, discharge, and rehabilitation measures in hospitalized patients recovering from severe COVID-19 in low- and middle-income countries. Am. J. Trop. Med. Hyg. 2021;104:110–119. doi: 10.4269/ajtmh.20-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alfa M.J., Olson N., Murray B.-L. Adenosine tri-phosphate (ATP)-based cleaning monitoring in health care: how rapidly does environmental ATP deteriorate? J. Hosp. Infect. 2015;90:59–65. doi: 10.1016/j.jhin.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y.M., Kim D.Y., Park K.H., Lee M.S., Kim Y.J. Monitoring environmental contamination caused by SARS-CoV-2 in a healthcare facility by using adenosine triphosphate testing. Am. J. Infect. Control. 2020;48:1280–1281. doi: 10.1016/j.ajic.2020.06.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Arkel A., Willemsen I., Kilsdonk-Bode L., Vlamings-Wagenaars S., Van Oudheusden A., De Waegemaeker P., Leroux-Roels I., Verelst M., Maas E., Van Oosten A., Willemse P., Van Asselen E., Klomp-Berens E., Franssen K., Van Cauwenberg E., Kluytmans J. ATP measurement as an objective method to measure environmental contamination in 9 hospitals in the Dutch/Belgian border area. Antimicrob. Resist. Infect. Control. 2020;9:1–8. doi: 10.1186/S13756-020-00730-9/TABLES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA, J. Am. Med. Assoc. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Tommaso P., Chatzou M., Floden E., Barja P., Palumbo E., Notredame C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017;35:316–319. doi: 10.1038/NBT.3820. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls S.M., Poplawski R., Bull M.J., Underwood A., Chapman M., Abu-Dahab K., Taylor B., Colquhoun R.M., Rowe W.P.M., Jackson B., Hill V., O’Toole Á., Rey S., Southgate J., Amato R., Livett R., Gonçalves S., Harrison E.M., Peacock S.J., Aanensen D.M., Rambaut A., Connor T.R., Loman N.J. CLIMB-COVID: continuous integration supporting decentralised sequencing for SARS-CoV-2 genomic surveillance. Genome Biol. 2021;221(22):1–21. doi: 10.1186/S13059-021-02395-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaughnessy R.J., Cole E.C., Moschandreas D., Haverinen-Shaughnessy U. ATP as a marker for surface contamination of biological origin in schools and as a potential approach to the measurement of cleaning effectiveness. J. Occup. Environ. Hyg. 2013;10:336. doi: 10.1080/15459624.2013.784633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., Doan K., Bastien N., Li Y., Van Caeseele P.G., Poliquin G. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin. Infect. Dis. An Off. Publ. Infect. Dis. Soc. Am. 2020;71:2663–2666. doi: 10.1093/CID/CIAA638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg M.G., Zhen W., Lucic D., Degli-Angeli E.J., Anderson M., Forberg K., Olivo A., Sheikh F., Toolsie D., Greninger A.L., Cloherty G.A., Coombs R.W., Berry G.J. Development of the RealTime SARS-CoV-2 quantitative Laboratory Developed Test and correlation with viral culture as a measure of infectivity. J. Clin. Virol. 2021;143 doi: 10.1016/J.JCV.2021.104945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruce E.A., Mills M.G., Sampoleo R., Perchetti G.A., Huang M.-L., Despres H.W., Schmidt M.M., Roychoudhury P., Shirley D.J., Jerome K.R., Greninger A.L., Botten J.W. Predicting infectivity: comparing four PCR-based assays to detect culturable SARS-CoV-2 in clinical samples. EMBO Mol. Med. 2022;14 doi: 10.15252/EMMM.202115290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onakpoya I.J., Heneghan C.J., Spencer E.A., Brassey J., Plüddemann A., Evans D.H., Conly J.M., Jefferson T. SARS-CoV-2 and the role of fomite transmission: a systematic review. F1000Research. 2021;10:233. doi: 10.12688/f1000research.51590.3. 10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kampf G., Brüggemann Y., Kaba H., Steinmann J., Pfaender S., Scheithauer S., Steinmann E. Potential sources, modes of transmission and effectiveness of prevention measures against SARS-CoV-2. J. Hosp. Infect. 2020;106:678–697. doi: 10.1016/J.JHIN.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanna T., Dallolio L., Raggi A., Mazzetti M., Lorusso G., Zanni A., Farruggia P., Leoni E. ATP bioluminescence assay for evaluating cleaning practices in operating theatres: applicability and limitations. BMC Infect. Dis. 2018;18 doi: 10.1186/S12879-018-3505-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sifuentes L.Y., Fankem S.L.M., Reynolds K., Tamimi A.H., Gerba C.P., Koenig D. Use of ATP readings to predict a successful hygiene intervention in the workplace to reduce the spread of viruses on fomites. Food Environ. Virol. 2017;9:14–19. doi: 10.1007/s12560-016-9256-2. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., Liu X., Xu K., Ho K., Kan H., Fu Q., Lan K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. 5827813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sampling sites in individual hospital rooms. Sampling sites are shown and labeled. Figure was Created with BioRender.com.
Time-scaled phylogeny from publicly available Uruguayan sequences from March 2020 to June 2021. Consensus sequences, obtained from surface samples at hospitals A and B (denoted with a black dot), were assigned to the 20B Nextstrain clade (HosA-008) and 20J (Gamma, V3) (HosB-004), respectively. The blue diamond shape represents the collapsed node containing the P.6, P.7 and P.2 lineages.