Abstract
Background
Alzheimer’s disease (AD) is a complex neurodegenerative disorder with β-amyloid pathology as a key underlying process. The relevance of cerebrospinal fluid (CSF) and brain imaging biomarkers is validated in clinical practice for early diagnosis. Yet, their cost and perceived invasiveness are a limitation for large-scale implementation. Based on positive amyloid profiles, blood-based biomarkers should allow to detect people at risk for AD and to monitor patients under therapeutics strategies. Thanks to the recent development of innovative proteomic tools, the sensibility and specificity of blood biomarkers have been considerably improved. However, their diagnosis and prognosis relevance for daily clinical practice is still incomplete.
Methods
The Plasmaboost study included 184 participants from the Montpellier’s hospital NeuroCognition Biobank with AD (n = 73), mild cognitive impairments (MCI) (n = 32), subjective cognitive impairments (SCI) (n = 12), other neurodegenerative diseases (NDD) (n = 31), and other neurological disorders (OND) (n = 36). Dosage of β-amyloid biomarkers was performed on plasma samples using immunoprecipitation-mass spectrometry (IPMS) developed by Shimadzu (IPMS-Shim Aβ42, Aβ40, APP669–711) and Simoa Human Neurology 3-PLEX A assay (Aβ42, Aβ40, t-tau). Links between those biomarkers and demographical and clinical data and CSF AD biomarkers were investigated. Performances of the two technologies to discriminate clinically or biologically based (using the AT(N) framework) diagnosis of AD were compared using receiver operating characteristic (ROC) analyses.
Results
The amyloid IPMS-Shim composite biomarker (combining APP669–711/Aβ42 and Aβ40/Aβ42 ratios) discriminated AD from SCI (AUC: 0.91), OND (0.89), and NDD (0.81). The IPMS-Shim Aβ42/40 ratio also discriminated AD from MCI (0.78). IPMS-Shim biomarkers have similar relevance to discriminate between amyloid-positive and amyloid-negative individuals (0.73 and 0.76 respectively) and A−T−N−/A+T+N+ profiles (0.83 and 0.85). Performances of the Simoa 3-PLEX Aβ42/40 ratio were more modest. Pilot longitudinal analysis on the progression of plasma biomarkers indicates that IPMS-Shim can detect the decrease in plasma Aβ42 that is specific to AD patients.
Conclusions
Our study confirms the potential usefulness of amyloid plasma biomarkers, especially the IPMS-Shim technology, as a screening tool for early AD patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13195-023-01188-8.
Keywords: Alzheimer’s disease, Plasma, Biomarkers, IPMS, Simoa, Diagnosis
Background
Alzheimer’s disease (AD) is a complex, age-related neurodegenerative disorder, whose prevalence is anticipated to triple worldwide by 2050 [1]. With the introduction of molecular biomarkers, AD progressively acquired a biological definition that optimized the traditional clinical symptom-based approach [2]. Briefly, “A”, amyloid-beta (Aβ) plaques with amyloid precursor protein; “T”, neurofibrillary tangles of the hyperphosphorylated tau protein (p-tau); and “(N)”, neurodegeneration, taken together, define the AT(N) system, a biomarker-guided classification scheme categorizing individuals using the core pathophysiological features of the disease.
To date, for clinical routine, the quantification of biomarkers is based on the cerebrospinal fluid (CSF) concentration assessment of the 42-amino acid-long Aβ peptide (Aβ42) and/or the ratio between Aβ42 and the 40-amino acid-long Aβ peptide (Aβ40), hyperphosphorylated tau (p-tau), and total tau (t-tau) proteins [3] and/or neuroimaging techniques such as Aβ-positron emission tomography (PET) [4], tau-PET imaging [5], and structural magnetic resonance imaging (MRI) [6]. However, while highly performant, such tools require expensive imaging equipment, highly trained staff, and, for lumbar puncture, invasive procedures.
To overcome these constraints, blood-based biomarkers have been developed and results are promising, especially concerning the Aβ42/40 ratio; p-tau; neurofilament light chain (NfL), a marker of neuroaxonal injury; or glial fibrillary acidic protein (GFAP), a marker of glial activation [7–9]. Initial conflicting outcomes were later explained by the unavailability of immunoassays sensitive enough or possible misclassification of clinical diagnosis [8]. Recent advances both in mass spectrometry (MS) and immunodetection methods, together with standardization of preanalytical variables, allowed to partly overcome those limitations by improving sensitivity [10, 11].
Eight plasma Aβ42/40 assays were recently compared, in terms of performances, when detecting abnormal cerebral Aβ status (according to CSF Aβ42/40 or Aβ-PET imaging) in early AD patients [12]. Only two of them seem operable on a large scale but involve an arbitration between cost, flow, and performances: ultrasensitive single molecule array (Simoa) technology [13] or immunoprecipitation coupled with MS (IPMS), as developed by the Washington University or Shimadzu (IPMS-Shim) [14, 15].
A study relying on Simoa reported decreased plasma Aβ40 and Aβ42 concentrations and reduced Aβ42/40 in AD patients [16]. Such biomarkers could even discriminate mild cognitive impairment (MCI) from control individuals [16] and were relevant predictive tools of positive amyloid-PET status [17]. MS-based studies found similar results indicating that Aβ42/40 was inversely proportional to brain Aβ burden [15].
However, data are still incomplete in clinical practice and concerning the diagnostic and prognostic accuracy of Aβ plasma biomarkers to discriminate AD from MCI, individuals with subjective cognitive impairments (SCI), other neurodegenerative diseases (NDD), or other neurological disorders (OND). No comparative studies have been done between the most recent and relevant plasma biomarker dosages. In addition, it remains unclear whether plasma biomarkers have a better diagnostic usefulness based on core clinical or biological criteria (Aβ−/Aβ+, AT(N)). Eventually, the temporal changes in plasma amyloid biomarkers remained to be determined and explored using recent ultrasensitive proteomic technologies.
The main objective of our study was to determine, in a cohort of memory clinic patients with differential diagnosis and, for some of them, spread along the AD continuum, the diagnostic and prognosis relevance of the two most operable plasma amyloid biomarkers, ultrasensitive immunoassay and IPMS amyloid biomarker dosages.
Methods
Study participants
Participants were retrospectively selected from the NeuroCognition Biobank of Montpellier’s University Hospital including biological samples (plasma and CSF) from patients recruited in the Resource and Research Memory Center between 2007 and 2016. Patients gave informed and written consent to have their samples stored in an officially registered and ethically approved biological collection (#DC-2008-417) and later used for scientific research.
The diagnosis of AD patients was performed using the National Institute on Aging-Alzheimer’s Association (NIA-AA) criteria by Albert et al. [18] after multidisciplinary collegial meetings evaluating medical history, clinical symptoms, neuropsychological assessments, and neuroimaging (MRI), both prior and after the results of CSF Aβ42, t-tau, and p-tau181 biomarkers.
This allowed us to determine clinical AD diagnosis independently of the results of biomarkers, to dichotomize them into Aβ-positive (Aβ+) and Aβ-negative (Aβ−) participants, and to establish their AT(N) biological profiles [19]. Individuals established as AD patients with additional brain vascular lesions (n = 14) were diagnosed as mixed dementia [20] and combined with AD individuals to constitute the AD group. Twelve of them were amyloid-positive (CSF measurement, see below).
The MCI group was constituted based on Petersen’s criteria with memory or cognitive troubles without loss of autonomy [21]. Among MCI patients, nine were amyloid-positive, 19 negative, and four unknowns. The SCI group was defined as individuals with self-reported experience of worsening or more frequent confusion or memory loss, but without objective impairment in cognitive performance [22]. Details about the diagnosis included in the other neurodegenerative diseases (NDD) and other neurological disorders (OND) groups are available in Table S1.
CSF biomarker measurement and cutoffs
CSF biomarkers were measured using standardized commercial Innotest sandwich ELISA (Fujirebio, n = 152) or Euroimmun ELISA method (n = 2) according to the manufacturer’s instructions. Sandwich ELISA relies on two antibodies for detection, one targeting the first 6 N-terminal amino acids (3D6) and the second the 6 C-terminal amino acids (21F12). The Aβ42/40 cutoff used to distinguish Aβ+ from Aβ− individuals was 0.05, as preconized for clinical setting [23]. For the AT(N) research framework, participants were considered “A+” if CSF Aβ42 was < 500/700 pg/ml depending on the nature of the polypropylene collection tubes or if CSF Aβ42/40 ratio < 0.05/0.1 according to ELISA technic [24, 25]; “T+” if CSF p-tau181 was > 60 pg/ml; and N+ if CSF t-tau was > 400 pg/ml.
Plasma biomarker samples
K2-EDTA blood samples were obtained through venipuncture. After a 10-min centrifugation at 1800×g within 2 h from collection, plasma was divided into 0.5-ml aliquots in 1.5–2-ml polypropylene tubes (Sarstedt, Germany) and stored at −80°C until biochemical assessment.
Ultrasensitive Simoa immunoassay (Quanterix)
Samples were thawed at room temperature and centrifuged at 10,000×g for 10 min. Samples were measured using the commercial Simoa® Human Neurology 3-PLEX A assay (N3PA) (Quanterix). This assay relies on distinct antibodies to capture and to detect amyloid-β species (Aβ42, Aβ40). The capture antibody (6E10) recognizes the N-terminal region of both species (amino acids 4 to 10) while the detection antibodies are specific to the Aβ42 and Aβ40 C-terminal ends to reveal them [26]. Briefly, Aβ42, Aβ40, and t-tau were measured simultaneously in duplicates in 80-μl samples, according to the manufacturer’s instructions.
IPMS-shim
The IPMS-Shim technology was slightly modified from Nakamura et al. [15]. Briefly, Aβ42, Aβ40, and APP669–711 were measured in 250-μl samples using a linear MALDI-TOF mass spectrometer (AXIMA Assurance, Shimadzu) after two consecutive IP steps with Dynabeads M-270 Epoxy used as beads and mouse monoclonal anti-Aβ antibodies to coat the beads. Aβ42 was then expressed relative to APP669–711 (also known as Aβ3–40) and Aβ40, both reflecting basal amyloid-β expression level. The IPMS-Shim composite biomarker was generated by combining the normalized score of APP669–711/Aβ42 and Aβ40/Aβ42 ratios, with Aβ42 as the denominator to obtain a normal distribution, as previously described [15].
Statistical analysis
Data were analyzed using the R statistical software (version 4.0.2) [27]. For each group, quantitative variables were expressed as the median with the interquartile range (IQR, Q1 and Q3). Groups were compared using non-parametric Kruskal–Wallis or Mann–Whitney tests according to the group number. Pairwise comparisons were adjusted with Bonferroni correction. Correlations were assessed using Spearman’s rank correlation coefficient (ρ). The distribution of categorical variables was expressed with percentages and compared using Fisher’s exact test.
For comparisons of biomarker concentrations between diagnosis groups, to avoid the influence of extreme values, outliers were identified using Rosner’s test and discarded (n = 6). For each group, normal distribution was assessed using the Shapiro–Wilk test and homoscedasticity through Levene’s test. The assumption of normality was not obtained in only two groups, IPMS-Shim and Simoa Aβ42/40 for the AD diagnosis. ANCOVA’s assumptions of linearity, homogeneity of variance, non-collinearity of the factors (variance inflation factor <5), non-influential observations, and normality of residuals were evaluated. The impact of diagnosis on plasma amyloid biomarker concentrations was evaluated using ANCOVA controlling for age and APOE ε4 status. Multiple comparisons of the means were achieved using Tukey contrasts with diagnosis as a factor.
Plasma cutoffs were computed using expectation–maximization (EM) algorithms for mixtures of univariate normal distributions [28]. Cutoffs were visually determined at the intersection of two normal distributions.
A receiver operating characteristic (ROC) analysis was used to determine biomarker performances. A predictive formula adjusted for age and APOE ε4 status was built using a logistic regression analysis. The best values for sensitivity (se) and specificity (sp) were computed at an optimal cutoff point. Youden’s index was used to determine this optimal cutoff corresponding to the threshold maximizing the distance to the identity (diagonal) line and giving and equal weight to sensitivity and specificity. The area under the curve (AUC) was compared using the DeLong test. All tests were two-tailed, and significance was set at α = .05.
Results
Demographical, clinical, and CSF biomarker profiles
Characteristics of the 184 participants are summarized in Table 1. As expected, age was different between groups with participants in the OND group being significantly younger. APOE ε4 status also differed with a higher proportion of APOE ε4 carriers observed in AD patients relative to SCI and OND. Median MMSE scores were similar between AD, NDD, and OND groups, but differed, as expected, between AD, MCI, and SCI patients. The CSF biomarker profile supports the diagnosis of AD patients: CSF Aβ42 concentrations and Aβ42/40 ratio were reduced in the AD group relative to all other groups, while p-tau181 and t-tau concentrations were elevated compared to all other diagnoses.
Table 1.
Plasmaboost cohort characteristics (n = 184)
| Characteristic | Median (IQR) | p valuea | ||||||
|---|---|---|---|---|---|---|---|---|
| AD (n = 73) | MCI (n = 32) | SCI (n = 12) | NDD (n = 31) | OND (n = 36) | AD vs. MCI/SCI | AD vs. NDD/OND | NAs | |
| APOE ε4, No. (%) | 24 (33)b | 9 (28) | 0 (0) | 4 (13) | 4 (11) | 0.045 | 0.014 | |
| Female, no. (%) | 36 (49) | 16 (50) | 7 (58) | 10 (32) | 19 (53) | 0.89 | 0.20 | |
| Age, y | 71 (68–76) | 69 (64–77) | 69 (62–72) | 70 (66–77) | 54 (35–67)c | 0.50 | < 0.001 | |
| Education, y | 9 (5–12) | 9 (6–12) | 14 (10–15) | 9 (5–15) | 11 (10–14) | 0.20 | 0.47 | 3 | 1 | 3 | 1 | 26 |
| MMSE, /30 | 24 (20–26)d | 27 (26–29) | 29 (27–30) | 23 (19–28) | 23 (17–27) | < 0.001 | 0.99 | 6 | 1 | 0 | 4 | 26 |
| CSF, pg/ml | ||||||||
| Aβ42 | 578 (486–703)e | 885 (609–1205) | 800 (689–1477) | 738 (590–1049) | 971 (736–1206) | < 0.001 | < 0.001 | 3 | 4 | 4 | 0 | 19 |
| Aβ42/40 | 0.034 (0.027–0.045)e | 0.065 (0.047–0.078) | 0.062 (0.051–0 .072) | 0.083 (0.055–0.096) | 0.086 (0.048–0.097) | < 0.001 | < 0.001 | 33 | 9 | 7 | 12 | 22 |
| p-tau181 | 82 (66–106)e | 45 (34–60) | 41 (39–42) | 33 (26–53) | 33 (26–42) | < 0.001 | < 0.001 | 3 |4 | 4 | 0 | 19 |
| t-tau | 630 (503–840)e | 276 (229–391) | 221 (200–228) | 226 (170–388) | 220 (144–340) | < 0.001 | < 0.001 | 3 | 4 | 4 | 0 | 19 |
aFisher’s exact (APOE ε4 and sex) or Kruskal–Wallis tests
bAD vs. MCI (p = 0.82), SCI (p = 0.017) or NDD (p = 0.052), OND (p = 0.019), Fisher’s exact test
cOND vs. AD (p < 0.001), NDD (p < 0.001), Mann–Whitney U tests with Bonferroni correction
dAD vs. MCI (p < 0.001), SCI (p < 0.001), Mann–Whitney U tests with Bonferroni correction
eAD vs. MCI (p < 0.05), SCI (p < 0.05) or NDD (p < 0.05), OND (p < 0.05), Mann–Whitney U tests with Bonferroni correction
We further explored potential associations between individual amyloid biomarkers and participants’ characteristics (age, education, MMSE, APOE ε4 status, and sex) to identify additional variables that might influence biomarker concentrations and potentially confound the impact of diagnosis (Table S2). The Aβ42/40 ratio, measured with Simoa and IPMS-Shim, decreased with age while the IPMS-Shim composite biomarker significantly increased with age. Same evolutions were observed for the APOE ε4 carriers. None of the other demographic characteristics (sex, education, and MMSE) was linked to plasma biomarker concentrations. At the scale of the cohort considered as a whole, plasma biomarkers are associated with age and APOE ε4 status, both were included in our ANCOVA and GLM models as potential confounding factors.
Plasma amyloid biomarker characteristics
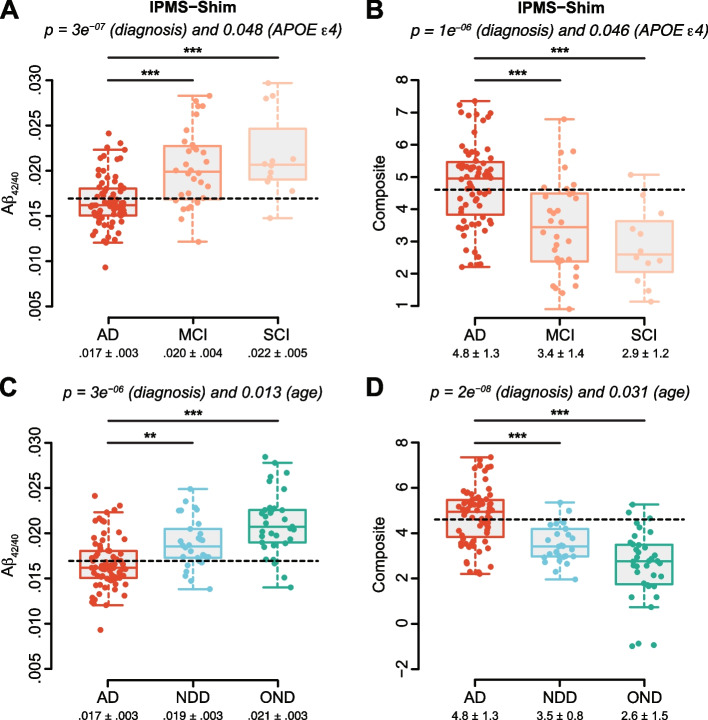
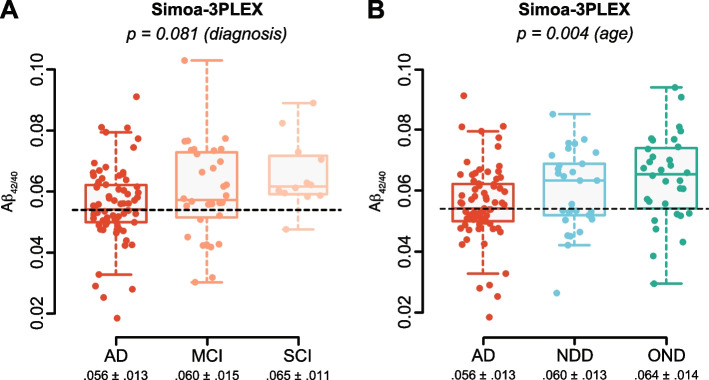
The IPMS-Shim Aβ42/40 ratio was significantly reduced in the AD group relative to both MCI and SCI groups (Fig. 1A) and similarly the IPMS-Shim composite score was increased (Fig. 1B). These two biomarkers were also able to distinguish AD from NDD and OND groups (Fig. 1C, D) with a combined effect of diagnosis and age for IPMS-Shim Aβ42/40 and the composite score. A cutoff to discriminate between AD and MCI/SCI and between AD and NDD/ODN could be established at 0.017 for IPMS-Shim Aβ42/40 ratio and at 4.6 for the composite score. Simoa Aβ42/40 ratio values were not statistically different between AD, MCI, and SCI (Fig. 2A). For Simoa Aβ42/40, only an effect of age reached statistical significance (Fig. 2B). Exclusion of patients with mixed dementia from the AD group did not change the effect of diagnosis. The impact on biomarker concentrations is thus robust and not sensitive to the presence of those samples.
Fig. 1.
Levels of amyloid plasma biomarkers measured by IPMS and according to diagnosis. A Aβ42/40 ratio is decreased in the AD group relative to MCI and SCI with an effect of group and APOE ε4 status. B IPMS-Shim composite score is significantly greater in AD relative to both MCI and SCI groups with an effect of group and APOE ε4 status. C The Aβ42/40 ratio evaluated with IPMS-Shim is decreased in AD relative to NDD and OND groups with an effect of diagnosis and age. D The IPMS-Shim composite score is increased in AD relative to both groups with an effect of diagnosis and age. Plasma cutoffs, 0.017 for IPMS-Shim-Aβ42/40 and 4.6 for IPMS composite score, are represented with dashed lines. Results are boxplot, and mean ± SD are indicated below, n = 12–73. ANCOVA with diagnosis, age, and APOE ε4 status as factors followed by multiple comparisons of means using Tukey contrasts and diagnosis as a factor; ***p < 0.001, **p < 0.01
Fig. 2.
Levels of amyloid plasma biomarkers measured by Simoa-3PLEX and according to diagnosis. A Simoa Aβ42/40 ratio is not significantly different between diagnoses. B The Aβ42/40 ratio is globally different between groups when measured with Simoa but associated with an effect of age only. Plasma cutoff for Simoa-Aβ42/40, 0.054, is represented with dashed lines. Results are boxplot, and mean ± SD are indicated below, n = 12–73. ANCOVA with diagnosis, age, and APOE ε4 status as factors followed by multiple comparisons of means using Tukey contrasts and diagnosis as a factor
Diagnosis relevance
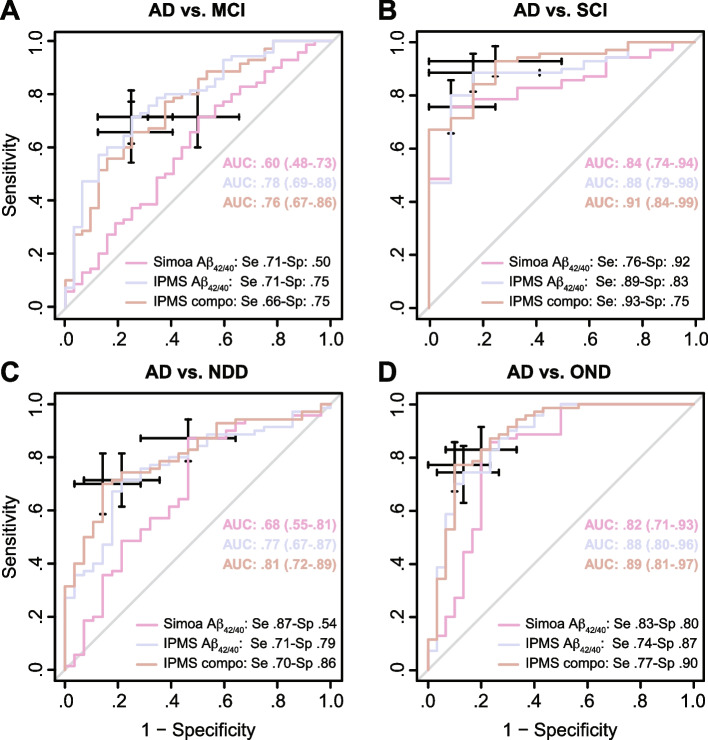
Diagnosis relevance of the biomarkers was computed for each diagnostic class individually, and models were adjusted for age and APOE ε4 status. Only the IPMS-Shim Aβ42/40 ratio and IPMS-Shim composite score were able to differentiate AD from MCI even with multivariate GLM adjustment, but not Simoa Aβ42/40 (Fig. 3A). IPMS-Shim Aβ42/40 had the largest AUC (0.78, 95% CI: 0.69–0.88) relative to the composite biomarker and both were significantly larger than Simoa Aβ42/40 (DeLong’s test; p < 0.01; Fig. 3A).
Fig. 3.
Performance of the plasma amyloid biomarkers to discriminate AD from other clinical diagnoses. A Receiver operating characteristic (ROC) analysis adjusted for age and APOE ε4 status of Simoa Aβ42/40, IPMS-Shim Aβ42/40, and composite biomarkers between AD and MCI groups. B ROC analysis of the three plasma amyloid biomarkers between AD and SCI groups. C ROC analysis of the biomarkers between AD and NDD groups. D ROC analysis of the biomarkers between AD and OND groups. AUC (area under the curve) is presented with 95% confidence interval (CI). The best values for sensitivity (se) and specificity (sp) were computed at an optimal cutoff point determined using Youden’s index
All three plasma biomarkers were able to discriminate AD from SCI in both univariate and multivariate models. While the IPMS-Shim composite score had the largest AUC (0.91, 0.84–0.99), it was not statistically different from the Aβ42/40 biomarkers (Fig. 3B).
To discriminate AD from NDD, the IPMS-Shim composite score had the largest adjusted AUC (0.81, 0.72–0.89) among the three biomarkers (Fig. 3C), even if not significantly different from Simoa Aβ42/40 and IPMS-Shim Aβ42/40. The IPMS-Shim Aβ42/40 ratio and IPMS-Shim composite score were both significant univariate and multivariate predictors of AD.
Simoa, IPMS-Shim Aβ42/40, and IPMS-Shim composite were able to discriminate AD from OND, but only IPMS-Shim Aβ42/40 and IPMS-Shim composite score remained significant after multivariate GLM adjustments (Fig. 3D). Taken together, the IPMS-Shim composite score had the largest adjusted AUC (0.89, 0.81–0.97). Simoa Aβ42/40 AUC was significantly smaller than the IPMS-Shim composite score (Fig. 3D).
Consistency between plasma biomarkers and CSF Aβ42/40, AT(N) profile, and core clinical diagnosis
The CSF Aβ42/40 ratio was available for 103 individuals and the complete AT(N) profile for 112 among the 184 participants of the Plasmaboost cohort (Table S3). We assessed the ability of blood-based biomarkers to discriminate biologically confirmed cases using the dichotomy based on the CSF Aβ42/40 ratio and the AT(N) classification.
Aβ+/Aβ− dichotomy
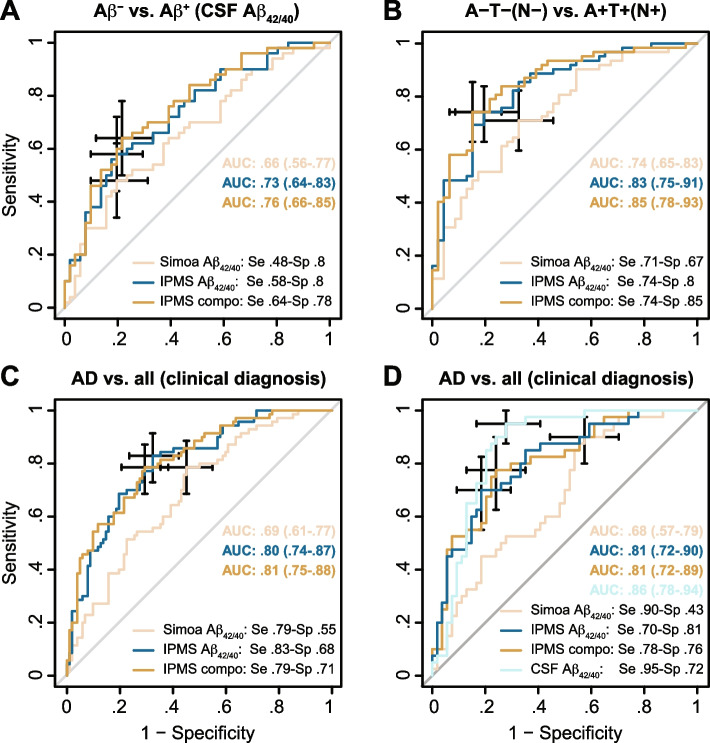
As expected, a greater proportion of AD patients belong to the Aβ+ group; however, the other characteristics were similar when comparing the Aβ− and Aβ+ individuals (Table S3). The three plasma biomarkers were equivalent in discriminating Aβ− from Aβ+ (Fig. 4A). AUC were between 0.66 and 0.76. Simoa Aβ42/40 was a significant predictor of Aβ+ status in a univariate model but the effect soothes to a statistical trend in a multivariate model (p = 0.06); this was not the case for the IPMS-Shim biomarkers which were both significant predictors in each type of model.
Fig. 4.
Performance of the plasma amyloid biomarkers to discriminate amyloid-positive (Aβ+) from amyloid-negative (Aβ−) subjects. A ROC analysis adjusted for age and APOE ε4 status of plasma Simoa Aβ42/40, IPMS-Shim Aβ42/40, and IPMS-Shim composite biomarkers between Aβ+ individuals and Aβ−. B ROC analysis of the amyloid biomarkers between A−T−N− and A+T+N+ participants. C ROC analysis of the three amyloid biomarkers between AD and all other diagnoses pooled together (non-AD). D ROC analysis of amyloid biomarkers between AD and other diagnoses for individuals with available CSF Aβ42/40 ratio. AUC is presented with 95% confidence interval (CI). The best values for sensitivity (se) and specificity (sp) were computed at an optimal cutoff point determined using Youden’s index
AT(N) research framework
As for the Aβ group, there was a greater proportion of AD patients in the A+T+ and A+T+N+ groups (Table S3). Except for age, all other characteristics were similar between A−T− and A+T+ participants; A+T+N+ also had a greater proportion of APOE ε4 carriers (Table S3). All three plasma amyloid biomarkers significantly discriminate A−T− from A+T+ profiles even when models were adjusted for age and APOE ε4 status (Fig. S1). Both IPMS-Shim biomarkers have higher performances than Simoa Aβ42/40. All three biomarkers similarly discriminate A−T−N− from A+T+N+ profiles including after adjustment for age and APOE ε4 status (Fig. 4B). Interestingly, Simoa Aβ42/40 had significantly lower AUC than IPMS-Shim composite but similar to IPMS-Shim Aβ42/40.
To compare the discriminative ability of the plasma biomarkers using a biological or a clinical set-up, we pulled together all non-AD diagnoses (see Table 1 and Fig. 1). For all three plasma biomarkers, AUC ranged from 0.69 to 0.81 to discriminate AD against other diagnoses with the IPMS-Shim scores having significantly higher AUC than Simoa Aβ42/40 (Fig. 4C). Interestingly, Simoa was able to discriminate AD from other types of diagnosis even when adjusted for age and APOE ε4 status.
Eventually, to assess the performances of plasma biomarkers versus CSF, all participants with CSF Aβ42/40 and a diagnosis were compared (Fig. 4D). IPMS-Shim plasma biomarkers and CSF Aβ42/40 showed comparable discriminative power but CSF Aβ42/40 performance remained better than Simoa Aβ42/40.
Correlations between CSF and plasma biomarkers
We compared technological platforms using the Passing-Bablok regression fit [29]. Results indicated that IPMS and Simoa were not equivalent when quantifying Aβ42, Aβ40, and Aβ42/40 ratio, confirming the performances measured with AUC, sensitivity, and sensibility. We further explored correlations between Simoa and IPMS-Shim Aβ42, Aβ40, and Aβ42/40 ratio plasma measurements with CSF Aβ42, Aβ40, and Aβ42/40 ratio results (Table S4). IPMS-Shim Aβ42 and Simoa Aβ42 correlated, as Aβ40 and Aβ42/40 ratios. Only IPMS-Shim Aβ42 was weakly correlated with CSF Aβ42 but not Simoa Aβ42. However, IPMS-Shim Aβ42/40 was significantly correlated with CSF Aβ42/40 as Simoa Aβ42/40. Eventually, none of the plasma Aβ40 was correlated with CSF Aβ40.
Evolution of plasma biomarkers through time
Plasma biomarker levels were measured a second time, after 2 years (± 352 days), in 29 participants (10 AD, 8 MCI, 5 SCI, 3 NDD, and 3 OND). Characteristics were equivalent between AD and non-AD (Table S5). Individuals from the AD group exhibited lower MMSE in relation with the cognitive decline expected in AD. None of the SCI or MCI subjects converted to AD. Among CSF biomarkers, only p-tau181 and t-tau were increased in the AD group; however, results should be taken with caution given the small number of subjects in this exploratory study. Interestingly, both IPMS-Shim plasma biomarkers were significantly altered in the AD group at baseline while this was not the case for Simoa Aβ42/40 ratio.
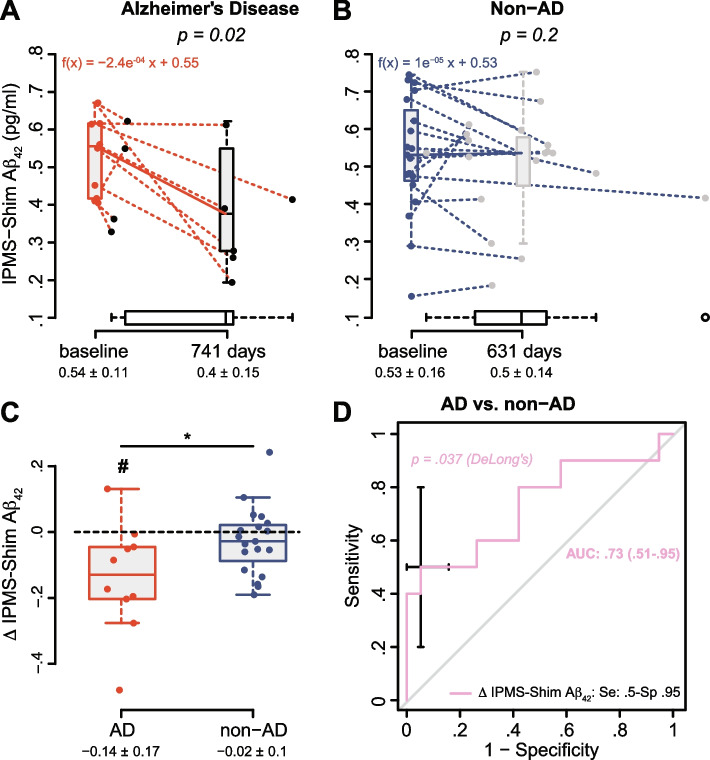
Among all plasma biomarkers tested, only IPMS Aβ40, Aβ42, and Simoa Aβ42/40 exhibited significant changes after ~2 years (Table 2). Interestingly, the decrease in IPMS-Shim Aβ42 was specific to the AD group. We thus further explored IPMS Aβ42 and showed a significant decrease after a follow-up of 741 days on average and with a mean rate of decline of 0.08 pg/ml/year (Fig. 5A). This was not the case for the non-AD group (Fig. 5B). We further plotted the variation in Aβ42 concentrations to show the average change over 2 years that was statistically different from 0 and from the non-AD group (Fig. 5C). We then estimated the ability of the decrease in IPMS-shim Aβ42 to predict AD status (GLM/ROC) and showed an AUC of 0.73 (Fig. 5D) which was statistically different from random and with a specificity of 0.95. Eventually, for the four participants with a change in IPMS-shim Aβ42 below −0.188, we showed that this decrease could be predicted (GLM) by a low MMSE at baseline (p = 0.03).
Table 2.
Baseline and follow-up amyloid biomarker concentrations according to diagnosis and analytical technique
| Biomarker | AD (n = 10) | p valuea | Non-AD (n = 19) | p valuea | ||
|---|---|---|---|---|---|---|
| Baseline | Follow-up | Baseline | Follow-up | |||
| Simoa Aβ40 | 233 (194–283) | 198 (180–274) | 0.94 | 230 (180–254) | 197 (149–224) | 0.18 |
| IPMS-Shim Aβ40 | 36 (28–38) | 23 (18–33) | 0.027 | 29 (24–33) | 27 (22–30) | 0.045 |
| Simoa Aβ42 | 11 (9–13) | 10 (8–13) | 0.94 | 13 (11–15) | 12 (9–13) | 0.71 |
| IPMS-Shim Aβ42 | 0.55 (0.42–0.61) | 0.37 (0.29–0.51) | 0.020 | 0.53 (0.46–0.65) | 0.54 (0.45–0.58) | 0.23 |
| Simoa Aβ42/40 | 0.055 (0.041–0.057) | 0.054 (0.042–0.057) | 0.69 | 0.057 (0.052–0.062) | 0.058 (0.054–0.077) | 0.044 |
| IPMS-Shim Aβ42/40 | 0.016 (0.015–0.018) | 0.016 (0.015–0.017) | 0.56 | 0.018 (0.017–0.021) | 0.02 (0.018–0.023) | 0.17 |
| IPMS-Shim composite | 5.1 (4.7–5.3) | 5.4 (4.5–6.2) | 0.77 | 4 (2.7–4.7) | 3.1 (2.1–3.9) | 0.14 |
Biomarker concentrations are expressed as the median with (IQR, Q1–Q3) in pg/ml
Abbreviations: AD, Alzheimer’s disease; non-AD: 8 MCI, 5 SCI, 3 NDD, and 3 OND; Aβ40, 40-amino acid-long Aβ peptide; Aβ42 42-amino acid-long Aβ peptide
aWilcoxon signed-rank test (for paired data)
Fig. 5.
Evolution of the plasma IPMS-Shim Aβ42 biomarker through time and ability to discriminate AD subjects. A Variation in IPMS-Shim Aβ42 concentrations after a median of 741 (172–783) days in AD subjects. B Evolution in IPMS-Shim Aβ42 levels after a median of 631 (366–773) days in non-AD subjects. Results are boxplots with individual values at baseline and follow-up, and mean ± SD are indicated below with n = 10 and 19. Dotted lines connect the values of the same individual. Horizontal boxplots represent delay distribution. The decline was expressed as a linear function because only two time points were available. Wilcoxon signed-rank test for paired data. C Change (Δ) in IPMS-Shim Aβ42 for the AD and non-AD groups. One-sample Mann–Whitney test, #p < 0.05; Mann–Whitney test, *p < 0.05. D ROC/AUC analysis of the change in IPMS-Shim Aβ42 between AD and non-AD groups. AUC is presented with 95% confidence interval (CI). The best values for sensitivity (se) and specificity (sp) were computed at an optimal cutoff point determined using Youden’s index
Discussion
We validated, using samples obtained in a memory clinic, the diagnostic relevance of the IPMS-Shim composite score to discriminate clinical AD—in the early stages of the disease—from MCI, SCI, OND, and NDD (Fig. 3). IPMS-Shim plasma Aβ42 measurements and Aβ42/40 ratio were weakly but significantly correlated with CSF Aβ42 and Aβ42/40 results (Table S2). In contrast, Simoa 3-PLEX did not achieve IPMS-Shim diagnostic performances and failed to correlate with the core biomarkers, at least for Aβ42.
The positive correlation between plasma IPMS-Shim Aβ42/40 measurements and CSF values was replicated, as previously indicated [14]. Moreover, as described by Janelidze and colleagues, we confirmed the lack of correlation between CSF and plasma using the Simoa 3-PLEX technology [16]. It was later discovered that a substantial non-specific Aβ3–42 signal was measured using this assay due to the region targeted by the capture antibody (amino acid 4 to 10) [30]. Alternatively, quantification in the CSF employs the highly specific sandwich ELISA technique, potentially explaining the lack of correlation with the 3-PLEX and the modest performances of the 3-PLEX assay. Thijsenn et al. recently developed full-length antibodies against Aβ40 and Aβ42 that indeed revealed better sensitivity and specificity than the 3-PLEX [30] and used for the development of a new assay (4-PLEX).
IPMS-Shim-based biomarkers revealed better diagnostic performances in all clinical categories, which could be explained by the high specificity of MS-based technologies, in general [31], and the better performances compared with those of immunoassays [12]. Moreover, MS minimizes the matrix effect observed in the blood [32]. Eventually, multiple pathological conditions (inflammation, renal dysfunction…) alter or at least affect basal amyloid-β expression and might cause inter-individual variations, especially in the plasma. As shown by others, expressing Aβ42 relative to a reference, as APP669–711, improves its discriminative performance [33]. Expressing Aβ42 relative to two references, combined in a composite score, exhibit even higher performances [15].
We were able to reveal a decrease of IPMS-Shim Aβ42 that seemed to be specific to the AD diagnosis (Table S5 and Fig. 5A, B). Even if these results are exploratory and should be confirmed on a larger cohort, this is the first description, to our knowledge, of such evolution of plasma Aβ42 using ultrasensitive methods. An important change (< −0.188 over ~2 years) in plasma Aβ42 concentrations could reveal a useful biomarker to detect AD patients as it is highly specific of the disease (Sp = 0.95). The data available in the literature indicate a drop in plasma Aβ42 in the early phases of the disease in healthy controls transitioning to MCI [34] or MCI to AD [35], consistent with the decrease observed with CSF Aβ42 [36]. Additional studies incorporating multiple time points and using state-of-the-art technologies will be necessary to conclude on the evolution of Aβ42 in the plasma.
Aβ42/40 ratio was further explored since there is growing evidence emphasizing its role as a potentially better diagnostic biomarker than the absolute value of Aβ42, at least in CSF analyses [37]. Plasma Aβ42/40 was reduced in AD patients relative to NDD, OND, MCI, and SCI participants (Fig. 1). We confirmed the results found by other studies that used Simoa [16] and MS [15, 38, 39]. Moreover, when Aβ+, A+T+, or A+T+N+ individuals were investigated, this ratio was reduced using all strategies (Table S3). Taken together, those results emphasize the potential role of low plasma Aβ42/40 concentrations as a robust indicator of both AD clinical diagnosis and biologically confirmed cases.
Plasma p-tau is assumed to be another attractive blood-based candidate biomarker for AD clinical diagnosis. However, p-tau, which is stable in CSF, exhibits a very short half-life (around 10 hours) in blood [40] and may appear later during the progression of the disease [41]. Eventually, t-tau, considered as a biomarker of neuronal injury in the CSF but susceptible to degradation by proteases in the plasma, might be replaced, for an initial blood-based diagnostic, by NfL. NfL is a more promising biomarker, robust in the plasma, whose concentration increases with neurodegeneration, that would allow to identify patients at risk of cognitive decline and to track disease progression [8].
Our study presents some limits. First, the number of individuals in the SCI group and with available CSF Aβ42/40 concentrations was limited. This was also the case for the longitudinal analysis; however, given the specificity in Aβ42 decrease, it appeared worth reporting. Second, a few characteristics (MMSE, education, CSF biomarkers) were not available for the OND group because it did not require the same set of procedures as the other groups in a memory clinic.
One of the strengths of our analysis is that it was conducted on a sample that reflects the population that attends memory clinics in France. None of the highly selective inclusion or exclusion criteria generally used for clinical research was used. Our sample, while heterogeneous, thus mirror the diversity of AD presentations. The most up-to-date and operable proteomic techniques were used for biomarker quantification. Our results confirm that they could be implemented for AD pre-screenings in memory clinics before further expensive or invasive tests and with diagnosis performances similar to CSF measures.
Conclusions
There is no doubt that the real diagnostic potential of plasma biomarkers will be achieved by developing molecular panels combining several of them [42]. Indeed, mounting evidence indicates that AD may present, even at the preclinical stage, a complex molecular signature; this can be deduced from peripheral blood analyses. Hence, using panels of blood biomarkers is supposed to outperform single candidate biomarkers in terms of AD diagnosis and prognosis [40, 43]. Unquestionably, blood (plasma)-based biomarkers are expected to play a crucial role in both AD diagnosis and prognosis, and in the therapeutic practice of the disease, in the upcoming future.
Supplementary Information
Additional file 1: Table S1. Details about the diagnosis in the other neurodegenerative diseases (NDD) and other neurological disorders (OND) groups. Table S2. Correlations between plasma Biomarkers with Simoa and IPMS-Shim and Demographic Features. Table S3. Characteristics of Study Participants According to CSF Aβ42/40 Status and AT or AT(N) Profiles. Table S4. Correlations between Plasma Amyloid Biomarkers and CSF/Plasma Amyloid Biomarkers. Table S5. Baseline Characteristics of the 29 Individuals with follow-up and repeated biomarkers assessment. Fig. S1. Performance of the plasma amyloid biomarkers to discriminate A+T+ from A-T- subjects. ROC analysis of the amyloid biomarkers. AUC is presented with 95% confidence interval (CI).
Acknowledgements
Mass spectrometry experiments were carried out using the facilities of the Montpellier Proteomics Platform (PPM-PPC, BioCampus Montpellier).
Abbreviations
- IQR
Interquartile range (Q1–Q3)
- AD
Alzheimer’s disease
- MCI
Mild cognitive impairment
- SCI
Subjective cognitive impairment
- NDD
Other neurodegenerative diseases
- OND
Other neurological disorders
- NA
Non-available
- y
Years
- APOE ε4
ε4 allele of the gene encoding for the apolipoprotein E
- No.
Number
- MMSE
Mini-Mental State Examination
- CSF
Cerebrospinal fluid
- Aβ42 (also known as Aβ1–42)
42-Amino acid-long Aβ peptide
- Aβ40 (also known as Aβ1–40)
40-Amino acid-long Aβ peptide
- p-tau181
Tau hyperphosphorylated at threonine 181
- t-tau
Total tau
Authors’ contributions
CH, YD, CB, SLe and AG contributed to study concept and design. GB, JK, SN, LT and JV had a major role in the acquisition of data. CH, GB, LAG, SLe and AG analyzed or interpreted the data. CH, GB, SLi, SLe and AG drafted/revised the manuscript for content, including medical writing with assistance of all authors. CH, KB, YD, CB, SLe and AG obtained funding. The authors read and approved the final manuscript.
Funding
This work was funded, for its realization, by a grant from the “Union France Alzheimer.”
Availability of data and materials
Plasmaboost de-identifed data are available to qualified researchers upon approved request to the corresponding author.
Declarations
Ethics approval and consent to participate
Patients signed an informed consent form validated by the ethical committee “CPP Sud Méditerranée IV” and had their samples stored in an officially registered and ethically approved biological collection (#DC-2008-417) by The French Ministry of Health at the ISO 20387 certified biological resource center of the CHU of Montpellier. Research was conducted according to the Declaration of Helsinki.
Consent for publication
NA.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Christophe Hirtz and Germain U. Busto contributed equally as co-first authors.
Sylvain Lehmann and Audrey Gabelle contributed equally to this work as co-senior authors.
References
- 1.Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chételat G, Teunissen CE, et al. Alzheimer’s disease. Lancet Lond Engl. 2021;397:1577-90. [DOI] [PMC free article] [PubMed]
- 2.Jack CR, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement J Alzheimers Assoc. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. 2010;6(3):131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- 4.Chételat G, Arbizu J, Barthel H, Garibotto V, Law I, Morbelli S, et al. Amyloid-PET and 18F-FDG-PET in the diagnostic investigation of Alzheimer’s disease and other dementias. Lancet Neurol. 2020;19(11):951–962. doi: 10.1016/S1474-4422(20)30314-8. [DOI] [PubMed] [Google Scholar]
- 5.Okamura N, Harada R, Furumoto S, Arai H, Yanai K, Kudo Y. Tau PET imaging in Alzheimer’s disease. Curr Neurol Neurosci Rep. 2014;14(11):500. doi: 10.1007/s11910-014-0500-6. [DOI] [PubMed] [Google Scholar]
- 6.Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6(2):67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chong JR, Ashton NJ, Karikari TK, Tanaka T, Schöll M, Zetterberg H, et al. Blood-based high sensitivity measurements of beta-amyloid and phosphorylated tau as biomarkers of Alzheimer’s disease: a focused review on recent advances. J Neurol Neurosurg Psychiatry. 2021;92(11):1231–1241. doi: 10.1136/jnnp-2021-327370. [DOI] [PubMed] [Google Scholar]
- 8.Alawode DOT, Heslegrave AJ, Ashton NJ, Karikari TK, Simrén J, Montoliu-Gaya L, et al. Transitioning from cerebrospinal fluid to blood tests to facilitate diagnosis and disease monitoring in Alzheimer’s disease. J Intern Med. 2021;290(3):583–601. doi: 10.1111/joim.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansson O, Edelmayer RM, Boxer AL, Carrillo MC, Mielke MM, Rabinovici GD, et al. The Alzheimer’s association appropriate use recommendations for blood biomarkers in Alzheimer’s disease. Alzheimers Dement. 2022;18:2669-86. [DOI] [PMC free article] [PubMed]
- 10.O’Bryant SE, Gupta V, Henriksen K, Edwards M, Jeromin A, Lista S, et al. Guidelines for the standardization of preanalytic variables for blood-based biomarker studies in Alzheimer’s disease research. Alzheimers Dement J Alzheimers Assoc. 2015;11(5):549–560. doi: 10.1016/j.jalz.2014.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verberk IMW, Misdorp EO, Koelewijn J, Ball AJ, Blennow K, Dage JL, et al. Characterization of pre-analytical sample handling effects on a panel of Alzheimer’s disease-related blood-based biomarkers: results from the standardization of Alzheimer’s blood biomarkers (SABB) working group. Alzheimers Dement J Alzheimers Assoc. 2022;18(8):1484–1497. doi: 10.1002/alz.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janelidze S, Teunissen CE, Zetterberg H, Allué JA, Sarasa L, Eichenlaub U, et al. Head-to-head comparison of 8 plasma amyloid-β 42/40 assays in Alzheimer disease. JAMA Neurol. 2021; [cited 2021 Oct 9]; Available from: https://jamanetwork.com/journals/jamaneurology/fullarticle/2784411. [DOI] [PMC free article] [PubMed]
- 13.Chang L, Rissin DM, Fournier DR, Piech T, Patel PP, Wilson DH, et al. Single molecule enzyme-linked immunosorbent assays: theoretical considerations. J Immunol Methods. 2012;378(1–2):102–115. doi: 10.1016/j.jim.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ovod V, Ramsey KN, Mawuenyega KG, Bollinger JG, Hicks T, Schneider T, et al. Amyloid β concentrations and stable isotope labeling kinetics of human plasma specific to central nervous system amyloidosis. Alzheimers Dement J Alzheimers Assoc. 2017;13(8):841–849. doi: 10.1016/j.jalz.2017.06.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura A, Kaneko N, Villemagne VL, Kato T, Doecke J, Doré V, et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature. 2018;554(7691):249–254. doi: 10.1038/nature25456. [DOI] [PubMed] [Google Scholar]
- 16.Janelidze S, Stomrud E, Palmqvist S, Zetterberg H, van Westen D, Jeromin A, et al. Plasma β-amyloid in Alzheimer’s disease and vascular disease. Sci Rep. 2016;6:26801. doi: 10.1038/srep26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmqvist S, Janelidze S, Stomrud E, Zetterberg H, Karl J, Zink K, et al. Performance of fully automated plasma assays as screening tests for Alzheimer disease-related β-amyloid status. JAMA Neurol. 2019;76(9):1060–1069. doi: 10.1001/jamaneurol.2019.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement J Alzheimers Assoc. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jack CR, Bennett DA, Blennow K, Carrillo MC, Feldman HH, Frisoni GB, et al. A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology. 2016;87(5):539–547. doi: 10.1212/WNL.0000000000002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zekry D, Hauw JJ, Gold G. Mixed dementia: epidemiology, diagnosis, and treatment. J Am Geriatr Soc. 2002;50(8):1431–1438. doi: 10.1046/j.1532-5415.2002.50367.x. [DOI] [PubMed] [Google Scholar]
- 21.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56(3):303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 22.Jessen F, Amariglio RE, van Boxtel M, Breteler M, Ceccaldi M, Chételat G, et al. A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer’s disease. Alzheimers Dement J Alzheimers Assoc. 2014;10(6):844–852. doi: 10.1016/j.jalz.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumurgier J, Schraen S, Gabelle A, Vercruysse O, Bombois S, Laplanche JL, et al. Cerebrospinal fluid amyloid-β 42/40 ratio in clinical setting of memory centers: a multicentric study. Alzheimers Res Ther. 2015;7(1):30. doi: 10.1186/s13195-015-0114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perret-Liaudet A, Pelpel M, Tholance Y, Dumont B, Vanderstichele H, Zorzi W, et al. Risk of Alzheimer’s disease biological misdiagnosis linked to cerebrospinal collection tubes. J Alzheimers Dis. 2012;31(1):13–20. doi: 10.3233/JAD-2012-120361. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann S, Delaby C, Boursier G, Catteau C, Ginestet N, Tiers L, et al. Relevance of Aβ42/40 ratio for detection of Alzheimer disease pathology in clinical routine: the PLMR scale. Front Aging Neurosci. 2018;10:138. doi: 10.3389/fnagi.2018.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baghallab I, Reyes-Ruiz JM, Abulnaja K, Huwait E, Glabe C. Epitomic characterization of the specificity of the anti-amyloid Aβ monoclonal antibodies 6E10 and 4G8. J Alzheimers Dis JAD. 2018;66(3):1235–1244. doi: 10.3233/JAD-180582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Core Team . R: a language and environment for statistical computing [internet] Vienna: R Foundation for Statistical Computing; 2021. [Google Scholar]
- 28.Benaglia T, Chauveau D, Hunter DR, Young D. mixtools : an R package for analyzing finite mixture models. J Stat Softw. 2009;32(6) [cited 2022 Apr 28]. Available from: http://www.jstatsoft.org/v32/i06/.
- 29.Passing H, Bablok W. A new biometrical procedure for testing the equality of measurements from two different analytical methods. Application of linear regression procedures for method comparison studies in clinical chemistry. Part I. J Clin Chem Clin Biochem. 1983;21(11):709–720. doi: 10.1515/cclm.1983.21.11.709. [DOI] [PubMed] [Google Scholar]
- 30.Thijssen EH, Verberk IMW, Vanbrabant J, Koelewijn A, Heijst H, Scheltens P, et al. Highly specific and ultrasensitive plasma test detects Abeta(1–42) and Abeta(1–40) in Alzheimer’s disease. Sci Rep. 2021;11(1):9736. doi: 10.1038/s41598-021-89004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neubert H, Shuford CM, Olah TV, Garofolo F, Schultz GA, Jones BR, et al. Protein biomarker quantification by immunoaffinity liquid chromatography-tandem mass spectrometry: current state and future vision. Clin Chem. 2020;66(2):282–301. doi: 10.1093/clinchem/hvz022. [DOI] [PubMed] [Google Scholar]
- 32.Oeckl P, Otto M. A review on MS-based blood biomarkers for Alzheimer’s disease. Neurol Ther. 2019;8(Suppl 2):113–127. doi: 10.1007/s40120-019-00165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneko N, Nakamura A, Washimi Y, Kato T, Sakurai T, Arahata Y, et al. Novel plasma biomarker surrogating cerebral amyloid deposition. Proc Jpn Acad Ser B Phys Biol Sci. 2014;90(9):353–364. doi: 10.2183/pjab.90.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rembach A, Faux NG, Watt AD, Pertile KK, Rumble RL, Trounson BO, et al. Changes in plasma amyloid beta in a longitudinal study of aging and Alzheimer’s disease. Alzheimers Dement J Alzheimers Assoc. 2014;10(1):53–61. doi: 10.1016/j.jalz.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Chen TB, Lai YH, Ke TL, Chen JP, Lee YJ, Lin SY, et al. Changes in plasma amyloid and tau in a longitudinal study of normal aging, mild cognitive impairment, and Alzheimer’s disease. Dement Geriatr Cogn Disord. 2019;48(3–4):180–195. doi: 10.1159/000505435. [DOI] [PubMed] [Google Scholar]
- 36.Sutphen CL, Jasielec MS, Shah AR, Macy EM, Xiong C, Vlassenko AG, et al. Longitudinal cerebrospinal fluid biomarker changes in preclinical Alzheimer disease during middle age. JAMA Neurol. 2015;72(9):1029–1042. doi: 10.1001/jamaneurol.2015.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansson O, Lehmann S, Otto M, Zetterberg H, Lewczuk P. Advantages and disadvantages of the use of the CSF amyloid β (Aβ) 42/40 ratio in the diagnosis of Alzheimer’s disease. Alzheimers Res Ther. 2019;11(1):34. doi: 10.1186/s13195-019-0485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Schindler SE, Bollinger JG, Ovod V, Mawuenyega KG, Weiner MW, et al. Validation of plasma amyloid-β 42/40 for detecting Alzheimer disease amyloid plaques. Neurology. 2022;98(7):e688–e699. doi: 10.1212/WNL.0000000000013211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Y, Kirmess KM, Meyer MR, Rabinovici GD, Gatsonis C, Siegel BA, et al. Assessment of a plasma amyloid probability score to estimate amyloid positron emission tomography findings among adults with cognitive impairment. JAMA Netw Open. 2022;5(4):e228392. doi: 10.1001/jamanetworkopen.2022.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zetterberg H. Blood-based biomarkers for Alzheimer’s disease-an update. J Neurosci Methods. 2019;319:2–6. doi: 10.1016/j.jneumeth.2018.10.025. [DOI] [PubMed] [Google Scholar]
- 41.Jack CR, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12(2):207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cullen NC, Leuzy A, Janelidze S, Palmqvist S, Svenningsson AL, Stomrud E, et al. Plasma biomarkers of Alzheimer’s disease improve prediction of cognitive decline in cognitively unimpaired elderly populations. Nat Commun. 2021;12(1):3555. doi: 10.1038/s41467-021-23746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hampel H, O’Bryant SE, Molinuevo JL, Zetterberg H, Masters CL, Lista S, et al. Blood-based biomarkers for Alzheimer disease: mapping the road to the clinic. Nat Rev Neurol. 2018;14(11):639–652. doi: 10.1038/s41582-018-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Details about the diagnosis in the other neurodegenerative diseases (NDD) and other neurological disorders (OND) groups. Table S2. Correlations between plasma Biomarkers with Simoa and IPMS-Shim and Demographic Features. Table S3. Characteristics of Study Participants According to CSF Aβ42/40 Status and AT or AT(N) Profiles. Table S4. Correlations between Plasma Amyloid Biomarkers and CSF/Plasma Amyloid Biomarkers. Table S5. Baseline Characteristics of the 29 Individuals with follow-up and repeated biomarkers assessment. Fig. S1. Performance of the plasma amyloid biomarkers to discriminate A+T+ from A-T- subjects. ROC analysis of the amyloid biomarkers. AUC is presented with 95% confidence interval (CI).
Data Availability Statement
Plasmaboost de-identifed data are available to qualified researchers upon approved request to the corresponding author.