Abstract
Background
HMGB1 is a highly conserved nuclear protein widely expressed in mammalian cells. This study aimed to comprehensively investigate the roles and mechanisms of HMGB1 in different tumors.
Methods
Original data on HMGB1 expression, localization, potential interacting proteins, genetics were obtained from The Cancer Genome Atlas, Genotype-Tissue Expression, Cancer Cell Line Encyclopedia, Human Protein Atlas, Compartmentalized Protein-Protein Interaction and cBioPortal databases. Then, correlation between HMGB1 expression levels and tumor stage, prognosis, potential pathways, tumor microenvironment, ESTIMATE score, immune-related genes, immune cell infiltration, microsatellite instability, tumor mutation burden, or anti-tumor drug resistance was investigated. The above results consistently indicated that high expression of HMGB1 protein may be related to clinical prognosis of HCC patients. Therefore, clinical tissues of HCC patients were selected to verify the differential expression of HMGB1 protein in HCC. The sensitivity of HMGB1-siRNA transfected HepG2 cells to sorafenib was assessed.
Results
HMGB1 was found to be differentially expressed in many tumors and normal tissues. HMGB1 was mainly located in the nucleus and might interact with proteins such as TLR2 and TLR4. Furthermore, HMGB1 expression was closely related to tumor stage, prognosis, tumor microenvironment, immune-related genes, immune cell infiltration, microsatellite instability, tumor mutation burden, and anti-tumor drug resistance and might be involved in different pathways of various tumors. Immunohistochemistry results further verified the differential expression of HMGB1 in HCC and paracancerous tissues. HMGB1-siRNA transfected HepG2 cells had a tendency to be more insensitive to sorafenib treatment compared to the control group.
Conclusions
HMGB1 was differentially expressed in most tumors and normal tissues, and was closely related to the clinical stage, prognosis, immune infiltration, tumor microenvironment, and drug resistance of tumors. Therefore, HMGB1 may serve as a novel biomarker for predicting tumor prognosis, efficacy of immune checkpoint inhibitors, and a potential target for anti-tumor therapy.
Keywords: high mobility group box 1, pan-cancer analysis, tumor, bioinformatics, prognosis
Introduction
In 2020, cancer was still a significant health burden, with 19.3 million new cases and 10 million deaths worldwide. The global tumor burden is expected to reach 28.4 million by 2040, an increase of 47% from 2020.1 Traditional treatments such as surgery, radiotherapy, chemotherapy, and targeted drugs have significantly improved the prognosis of cancer patients. However, many patients are diagnosed at an advanced stage and therefore have a poor prognosis.2 Recently, immune checkpoint inhibitors (ICIs) targeting CTLA4, PD-1, PD-L1, and other targets have been utilized in various tumors. Tumor mutation burden (TMB), microsatellite instability (MSI) and PD-L1 expression were the currently predictors of ICIs efficacy. However, only a subset of patients benefits from ICIs. Tumor microenvironment (TME) and immune infiltration are closely linked to tumor occurrence, progression, and drug efficacy. Therefore, while investigating a potential therapeutic target, it is insufficient only to analyze its correlation with tumor prognosis. There is also a need to further explore its correlation with immune infiltration and tumor microenvironment.
High mobility group box 1 (HMGB1) is a highly conserved nuclear protein widely distributed in various cell types. HMGB1 is mainly passively released by necrotic cells or actively secreted by monocyte-macrophages.3,4 As a non-histone DNA-binding protein in the nucleus, HMGB1 regulates gene transcription and plays an important role in inflammatory diseases as a damage-associated molecular pattern molecule after being released extracellularly.5,6 HMGB1 exerts different roles depending on its redox status and the receptors. HMGB1 can act alone or in complexes with other molecules, such as lipopolysaccharide, deoxyribonucleic acid (DNA), and histones. Known HMGB1 receptors include toll-like receptor (TLR) 4 and the receptor of advanced glycation endproducts (RAGE). HMGB1 can activate myeloid differentiation factor 2, being a part of a complex with TLR4, which induces the downstream signal transduction pathway and promotes inflammatory response.7 The binding of HMGB1 to RAGE activates nuclear factor-κB (NF-κB) and subsequently promotes the production of cytokines. However, HMGB1 may have different roles in different inflammatory diseases. For example, low levels of HMGB1 secreted by innate immune cells have pro-inflammatory effects in sepsis. The release of large doses of HMGB1 induces macrophage pyroptosis, immune tolerance, and immunosuppression.8 This phenomenon may be explained by HMGB1 binding to different receptors. HMGB1 is also involved in immune cell infiltration. Bourquin et al discovered that HMGB1-CXC chemokine ligand-12 complex promotes B cell migration from Peyer’s patches to other sites via CXC motif chemokine receptor 4.9 Rovere-Querini et al reported that necrotic cells activate dendritic cells, increase HMGB1 secretion, as well as stimulate T cell proliferation and Th1 cell differentiation through RAGE receptors.10,11 Beatriz Rendon-Mitchell et al found that IFN-γ promotes HMGB1 secretion by macrophages through TNF and Janus kinase 2 dependent mechanisms.12
HMGB1 has been found to mediate the progression of immune diseases such as systemic lupus erythematosus, Sjogren’s syndrome and tumors.13–15 In addition, HMGB1 is thought to be closely related to the occurrence, development, and drug sensitivities of various tumors. However, studies have recently found that HMGB1 plays contradictory roles in different tumors.16 On the one hand, HMGB1 overexpression in gastrointestinal stromal carcinoma was closely related to c-kit mutation.17 Animal studies showed that HMGB1 promoted the formation of hepatocellular carcinoma (HCC) in Mdr2 (-/-) mice through RAGE receptors.18 More importantly, HMGB1 was closely related to tumor invasion and metastasis. Under hypoxia, necrotic tumor cells could release HMGB1 into the tumor microenvironment and promote vascular endothelial cell proliferation and neovascularization through Myd88-dependent NF-κB pathway. Tsung et al identified that after HCC cells were stimulated, HMGB1 protein transferred from the nucleus to the cytoplasm and bound to mitochondrial DNA, which then activated TLR9 signal pathway and promoted tumor cell proliferation.19 Guo et al observed that HMGB1 promoted the expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in renal cell carcinoma, leading to increased angiogenesis.20 On the other hand, nuclear HMGB1 is involved in DNA damage repair and maintains genomic stability. Intracellular HMGB1 exerts its anti-tumor effect by promoting tumor apoptosis and inhibiting tumor proliferation by binding to the retinoblastoma protein p105.21 After chemotherapeutic drugs act on the tumor, HMGB1 released by necrotic cells can recruit more immune cells and lead to anti-tumor immune effects. Accordingly, HMGB1 silencing was observed to cause loss of response to paclitaxel in ovarian cancer.22
Therefore, HMGB1 has varying roles that depend on its location, binding receptor, and disease type, which warrants further exploration. In this study, the expression and mutation status of HMGB1 in 33 types of tumors in TGCA database were analyzed. The correlation of HMGB1 with tumor stage, prognosis, tumor microenvironment, immune infiltration, and drug resistance was also explored. The findings were expected to provide a potential therapeutic target for anti-tumor therapy.
Methods
Data Extraction and Processing
We performed the study using the following flow chart (Figure S1). RNA sequencing, gene mutation, and clinical data of 33 types of tumors and normal tissues were extracted from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases via UCSC Xena platform (https://xenabrowser.net/datapages/).23 Similarly, the expression data of HMGB1 in cancer cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE, https://sites.broadinstitute.org/ccle/) database. R (version 4.2.0) and RStudio software (version 2022.02.3) were used to process the data and visualization. All RNA expression data were presented as transcripts per million mapped reads (TPM). The abbreviations are listed in Table S1.
Genetics, Location, and Interaction Proteins of HMGB1
HMGB1 gene mutation were explored using cBioPortal database (https://www.cbioportal.org/).24 The information on immunofluorescence staining of HMGB1 protein in different cell lines was downloaded from Human Protein Atlas (HPA, https://www.proteinatlas.org/) database to analyze HMGB1 localization in cells. Proteins with potential for interaction with HMGB1 were downloaded from Compartmentalized Protein-Protein Interaction Database (ComPPI, https://comppi.linkgroup.hu/). The protein information was transformed by the biological DataBase network platform (bioDBnet, https://biodbnet-abcc.ncifcrf.gov/db/db2db.php). The data visualization was performed through the R packages of “ggplot2”, “igraph”, and “magrittr”.25
Prognostic Analysis
The correlation between HMGB1 expression and tumor prognosis was evaluated, including overall survival (OS), progression-free interval (PFI), disease-specific survival (DSS), and disease-free interval (DFI). R packages “survival” and “survminer” were used to analyze the effects of HMGB1 expression levels on the prognosis of different tumors. The optimal cut-off value was used to distinguish between the high and low expression subgroup of HMGB1. The Kaplan-Meier method and Log rank test were used to compare the prognosis between the two groups. The univariate Cox regression model was used to calculate the relationship between HMGB1 expression and tumor prognosis, and the results were expressed as hazard ratio and 95% confidence intervals.
Enrichment Analysis of HMGB1
The Gene Set Variation Analysis (GSVA) method was used to analyze the possible pathways of HMGB1 involved in different tumors based on HALLMARK gene sets of MSigDB database (http://www.gsea-msigdb.org/gsea/login.jsp). The results were analyzed using R packages “clusterProfiler” and “GSVA” and represented by a heat map.
Analysis of Tumor Microenvironment (TME)
The correlation between HMGB1 and TME was analyzed according to the method described by Liao et al.26 R package “ESTIMATE” was used to analyze the correlation between HMGB1 expression and TME scores in different tumors, including stromal score, immune score, ESTIMATE score (estimation of stromal and immune cells in malignant tumor tissues using expression data), and tumor purity score.27
Analysis of Tumor Immune Infiltration
The correlation between HMGB1 expression and immune cell infiltration in different tumors was analyzed by ImmuCellAI database (http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/). In addition, the correlation between HMGB1 expression and TMB, MSI, and immune checkpoints in different tumors was also evaluated.28 The correlation between HMGB1 expression and immune cells was calculated and demonstrated by a heat map using pan-cancer immune infiltration data downloaded from Tumor Immune Estimation Resource 2 database (TIMER2, http://timer.comp-genomics.org/).
Antitumor Drug Sensitivity Analysis
The relationship between HMGB1 expression and drug sensitivity of 192 anti-tumor drugs (represented as half maximal inhibitory concentration, IC50) was analyzed using The Genomics of Drug Sensitivity in Cancer platform (GDSC, https://www.cancerrxgene.org/). Correlation between HMGB1 expression and drug sensitivity was analyzed by Spearman method. The differences in IC50 of antitumor drugs in HMGB1 high and low-expression subgroups were further explored. The relationship between HMGB1 expression and drug efficacy in the real world was analyzed by downloading dataset GSE61676 from Gene expression omnibus (GEO) database.
Hepatocellular Carcinoma (HCC) Samples and Immunohistochemical Staining
The paraffin sections of HCC and matching pericarcinomatous tissues were purchased from Liaoding Company (cat: LD-LVC1805). The paraffin sections were dewaxed with xylene, rehydrated with ethanol, repaired by sodium citrate (Servicebio, cat: G1202), inactivated by hydrogen peroxide (Servicebio, cat: G0115) and blocked by bovine serum albumin (Servicebio, cat: G5001). Rabbit anti-human HMGB1 antibody (Abcam, cat: ab79823) was added and left at 4 degrees overnight. Next, goat anti-rabbit antibody (Servicebio, cat: GB23303) was added and incubated at room temperature for one hour. Then, the sections were stained with 3, 3’-diaminobenzidine tetrahydrochloride (Servicebio, cat: G1211) and hematoxylin, placed in alcohol and xylene in turn, and finally sealed with neutral gum. The nucleus was stained blue and the HMGB1 protein was brown, and finally scanned with microscope (CIC, XSP-C204). The staining intensity in immunohistochemical sections was scored according to the staining characteristics of the target cells: no staining was scored as 0, pale yellow as 1, brown as 2, and tan as 3; the positive rate of cells was 0–5%, 6% to 25%, 26% to 50%, 51% to 75%, and > 75% were recorded as 0, 1, 2, 3, and 4 points, respectively. The degree of staining (0–3 points) and the positive rate (0–4 points) were scored separately and then multiplied to obtain an immunoreactivity scoring system (IRS) score (0–12 points).29
Sensitivity of HepG2 Cells with Different Levels of HMGB1 Expression to Sorafenib
The following experiments were carried out to verify the sensitivity of hepatocellular carcinoma cells with different levels of HMGB1 expression to sorafenib. Human hepatocellular carcinomas cells (HepG2) were purchased from HyCyte company (cat: TCH-C196) and cultured with complete medium (HyCyte, cat: TCH-G196) in a humidified incubator at 37 degrees with 5% CO2. The siRNA for HMGB1 and negative control (NC) were designed and synthesized in RiboBio Company. According to the manufacturer’s instructions, NC-siRNA or HMGB1-siRNA were transfected into HepG2 cells using the transfection reagent (RiboBio, cat: siBDM2500). HMGB1 silencing efficiency was verified with Western blotting analysis. In brief, cells were washed 3 times with pre-cooled PBS, followed by treatment with RIPA lysis buffer (CWBIO, cat: CW2333S) containing protease inhibitor (CWBIO, cat: CW2200S) for 30 minutes, and finally the supernatant was collected by centrifugation. Protein quantification was performed using the BCA protein assay kit (CWBIO, cat: CW0014S). The protein samples were added to loading buffer (Fdbio, cat: FD002) and boiled at 98 degrees for 6 minutes. After electrophoresis in 10% sodium dodecyl sulfate-polyacrylamide gel (Beyotime, cat: P0012AC), the proteins and markers (SMDBIO, cat: PM2511) were transferred to polyvinylidene fluoride membrane (Millipore, cat: IPVH00010). Then, 5% skim milk blocking solution was applied to the membrane for 1h at room temperature and then incubated with primary antibodies against HMGB1 (Abcam, cat: ab228624) and β-actin (Abclonal, cat: AC038) overnight at 4 degrees. After washing three times with TBST, the membranes were incubated with goat anti-rabbit antibodies (Fdbio, cat: FDR007) for 1h at room temperature. Immunoblots were developed with chemiluminescent agent (GLPBIO, cat: GK10008) and gray value was quantified by ImageJ software. Cells in NC-siRNA and HMGB1-siRNA groups were exposed to sorafenib (APExBIO, cat: A3009) with different concentrations (0, 2.5umol/l, 5umol/l, 7.5umol/l and 10umol/l). At 72 hours, 10% cell counting kit-8 working reagent (DOJINDO, cat: CK04) was added to each well and incubated at 37 degrees for 2 h, and the optical density at 450nm (Synergy H1M, BioTek, USA) was detected to calculate the cell viability. The cell viability was equal to (optical density of experimental well - optical density of blank well) / (optical density of control well - optical density of blank well).
Statistical Analysis
Image arrangement was performed using Adobe Illustrator, Adobe Photoshop and GraphPad Prism software. Two-sided p < 0.05 was defined as the statistically significant difference. P < 0.05, 0.01, 0.001, and 0.0001 were presented as “*”, “**”, “***”, “****”, respectively. The difference expressed by “ns” was not statistically significant.
Results
The Differential Expression of HMGB1 in Various Tumors Compared with Normal Tissues
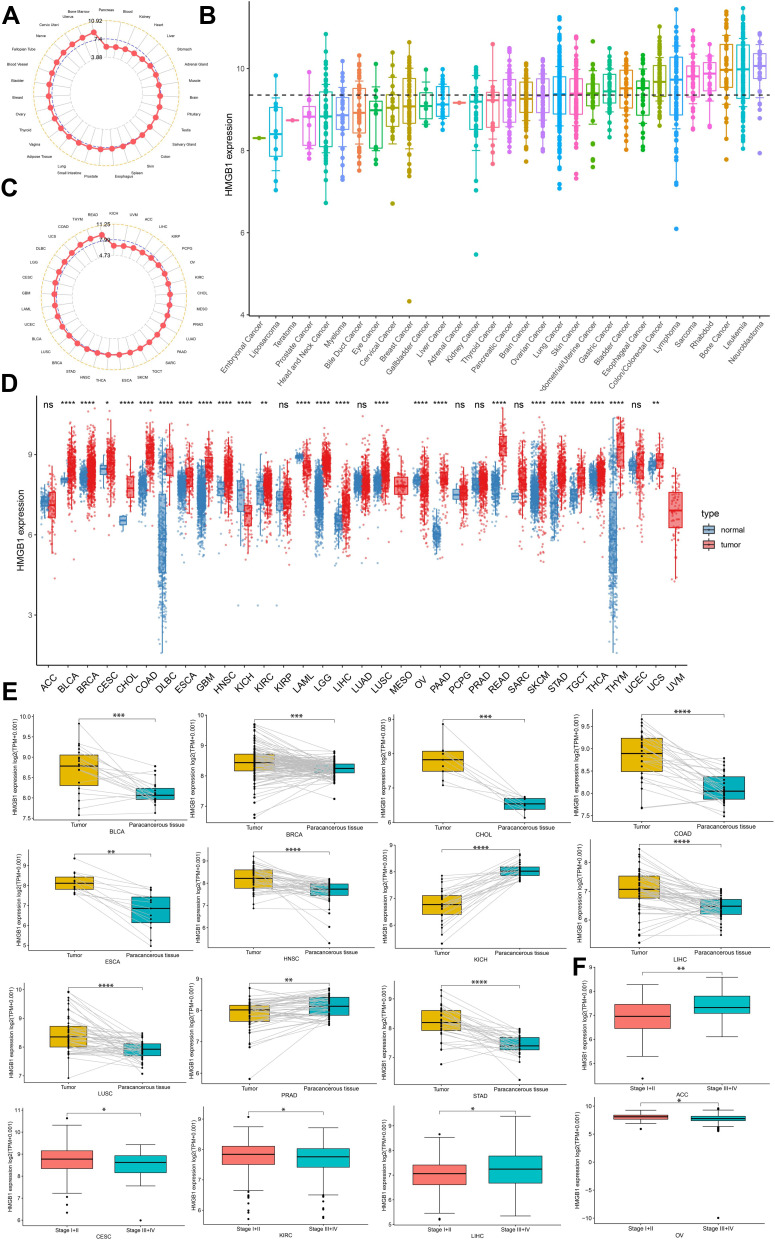
HMGB1 was widely expressed in normal tissues, with the highest expression levels noted in bone marrow, followed by uterus, and cervix uteri (Figure 1A). From the results of CCLE database, HMGB1 expression was the highest in the neuroblastoma, followed by leukemia and bone cancer cell lines (Figure 1B). Based on the results of HMGB1 expression in various tumors in TCGA database, HMGB1 expression was the highest in rectum adenocarcinoma (READ), followed by thymoma (THYM) and colon adenocarcinoma (COAD) (Figure 1C). Additionally, the results of differential expression of HMGB1 in various tumors and normal tissues revealed that, compared to normal tissues, the expression of HMGB1 was higher in 21 types of tumors, including bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), COAD, diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), brain lower-grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), READ, skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), THYM, uterine carcinosarcoma (UCS), and thyroid carcinoma (THCA). Conversely, HMGB1 expression in kidney chromophobe (KICH), acute myeloid leukemia (LAML), and ovarian serous cystadenocarcinoma (OV), was lower than in normal tissues, and there were no corresponding normal tissues in mesothelioma (MESO) and uveal melanoma (UVM) (Figure 1D). HMGB1 expression in BLCA, BRCA, CHOL, COAD, ESCA, HNSC, LIHC, LUSC, and STAD was higher than that in paracancerous tissues. However, HMGB1 expression in KICH and prostate adenocarcinoma (PRAD) was lower than that in paracancerous tissues (Figure 1E). Furthermore, HMGB1 was highly expressed in locally advanced and advanced adrenocortical carcinoma (ACC) and LIHC (stage III and IV) than in early-stage (stage I and II) tumors; the results were opposite in CESC, KIRC and OV (Figure 1F).
Figure 1.
HMGB1 expression levels in tumors, cancer cell lines, and normal tissues. HMGB1 expression levels in normal tissues (A), cancer cell lines (B), and tumors (C). (D) Comparison of HMGB1 expression levels between tumors and normal tissues. (E) Comparison of HMGB1 expression levels in tumors and matched paracancerous tissues. (F) Comparison of HMGB1 levels in different tumor stages. P > 0.05, < 0.05, < 0.01, < 0.001, and < 0.0001 were presented as “ns”, “*”, “**”, “***”, “****”, respectively.
Cellular Localization and Genetic Information of HMGB1
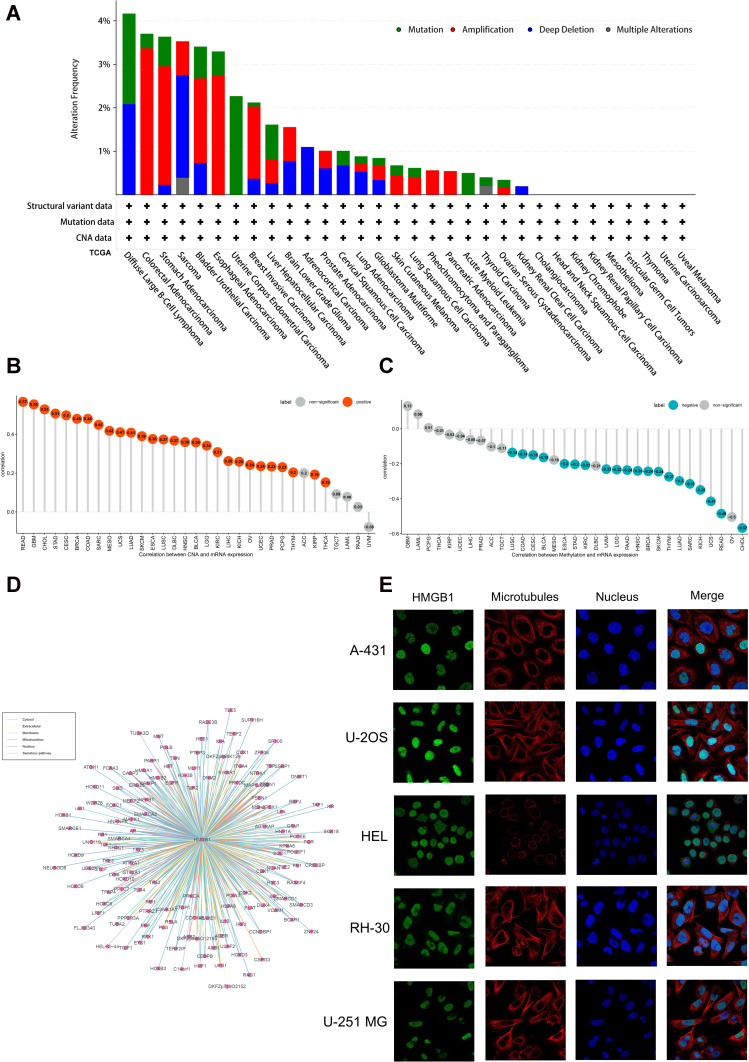
The cBioPortal tool was used to analyze genetic alterations of HMGB1 in different tumors in TCGA database. HMGB1 had the highest mutation frequency in uterine corpus endometrial carcinoma, the highest amplification frequency in colorectal adenocarcinoma, and more than 2% deep deletions in sarcoma (SARC) and DLBC (Figure 2A). As displayed in Figure 2B, the expression levels of HMGB1 were positively correlated with copy numbers in most tumors of TCGA database, except for ACC, TGCT, LAML, PAAD, and uveal melanoma (UVM) (Figure 2B). HMGB1 expression levels were negatively correlated with methylation levels in many tumors, including LUSC, COAD, CESC, BLCA, ESCA, STAD, KIRC, UVM, LGG, PAAD, HNSC, BRCA, SKCM, THYM, LUAD, SARC, KICH, UCS, READ, and CHOL (Figure 2C). Based on ComPPI data predictions, as many as 187 proteins had the potential to interact with HMGB1, including TLR2, TLR4, poly ADP-ribose polymerase (PARP), superoxide dismutase 1 (SOD1), heat shock protein member 8 (HSPA8), and tumor protein P53 (TP53), thus indicating that HMGB1 might be involved in multiple pathways of tumors (Figure 2D). HPA database provides the intracellular fluorescence staining of HMGB1 in five specific cell lines. The results indicated that HMGB1 protein was localized to the nucleus in A-431, U-2OS, HEL, RH-30, and U-251MG cell lines (Figure 2E).
Figure 2.
Genetic alterations, cellular localization, and interaction of HMGB1. (A) Mutation status of HMGB1 in different tumors. (B) Correlation between HMGB1 expression levels and copy number in different tumors. (C) Correlation between HMGB1 expression levels and methylation in different tumors. (D) Proteins that might interact with HMGB1 through ComPPI database. (E) Intracellular localization of HMGB1 protein in cell lines through HPA database.
Effects of HMGB1 Expression Levels on the Prognosis of Different Tumors
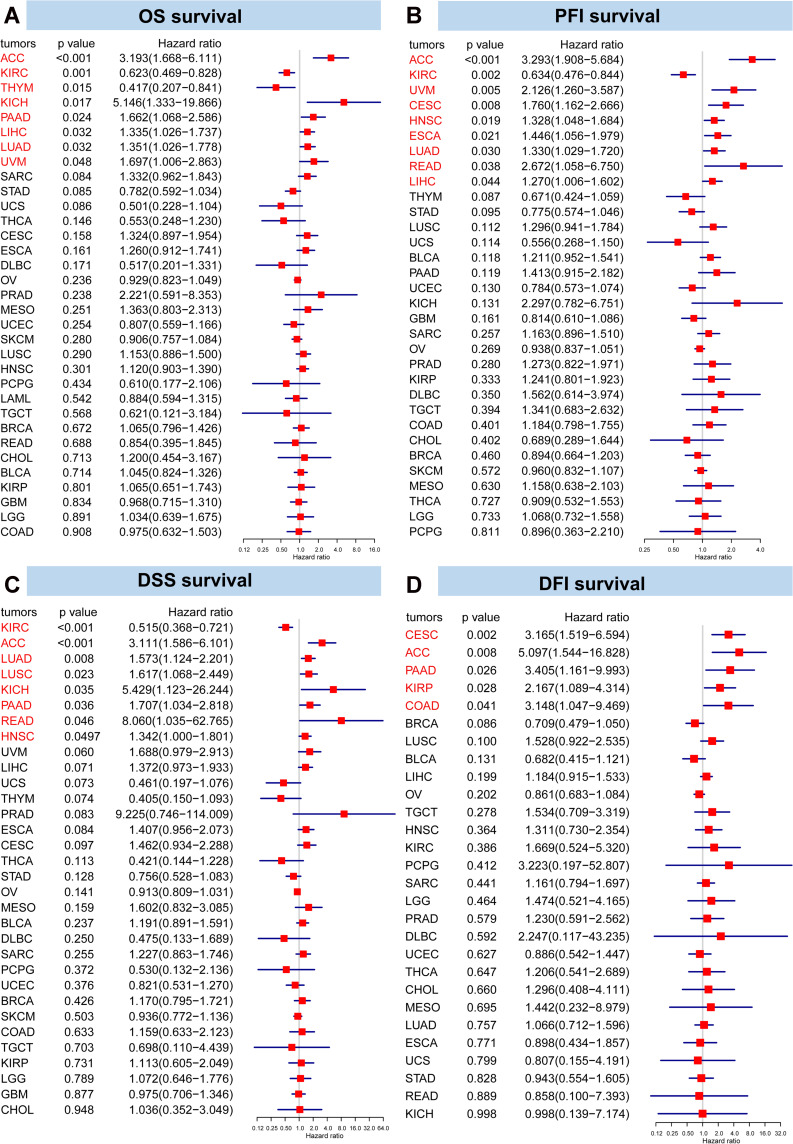
The expression levels of HMGB1 in tumors were divided into high and low expression levels according to the optimal cut-off values. In 15 types of tumors, including ACC, CESC, ESCA, HNSC, KICH, LGG, LIHC, LUAD, LUSC, MESO, PAAD, PRAD, SARC, SKCM, and UVM, HMGB1 high expression group had a lower OS than the low expression group, indicating a worse prognosis (Figure S2). However, among six types of tumors (ie, DLBC, KIRC, STAD, THCA, THYM, and UCS), OS in HMGB1 high expression group had a better OS than the low expression group, suggesting a better prognosis (Figure S2). A univariate Cox regression model was used to analyze the correlation between HMGB1 expression and OS. Interestingly, the results revealed that high HMGB1 expression was associated with a higher risk of death in ACC, KICH, PAAD, LIHC, LUAD, and UVM but lower risk of death in KIRC and THYM (Figure 3A). Eight kinds of tumors (ACC, PAAD, LIHC, LUAD, UVM, KICH, KIRC, and THYM) were significant in Kaplan-Meier and Cox analyses for OS outcome.
Figure 3.
Forest map of univariate Cox regression model for the impact of HMGB1 expression on prognosis in different tumors. The impacts of HMGB1 expression on (A) overall survival, (B) progression-free interval, (C) disease-specific survival, and (D) disease-free interval in different tumors.
Next, HMGB1 high expression subgroup of 19 types of tumors, including ACC, BLCA, CESC, COAD, DLBC, ESCA, HNSC, KICH, LIHC, LUAD, LUSC, MESO, PAAD, pheochromocytoma and paraganglioma (PCPG), READ, SARC, SKCM, TGCT, and UVM had worse PFI than HMGB1 low expression subgroup (Figure S3). Conversely, HMGB1 high expression subgroup of seven types of tumors, including BRCA, GBM, KIRC, STAD, THYM, USEC, and UCS, had better PFI than HMGB1 low expression subgroup (Figure S3). Furthermore, univariate Cox regression analysis confirmed that high HMGB1 expression was associated with a higher risk of progression of eight tumors, namely, ACC, UVM, CESC, HNSC, ESCA, LUAD, READ, and LIHC. However, high HMGB1 expression decreased the risk of progression of KIRC (Figure 3B). Nine kinds of tumors (ACC, UVM, CESC, HNSC, ESCA, LUAD, READ, LIHC, and KIRC) were significant in both Kaplan-Meier and Cox analyses for PFI outcome.
Similarly, DSS of 16 types of tumors in the high HMGB1 expression subgroup (ACC, CESC, ESCA, HNSC, KICH, LGG, LIHC, LUAD, LUSC, MESO, PAAD, PRAD, READ, SARC, SKCM, and UVM) was lower than the low HMGB1 expression subgroup, thus suggesting that the high HMGB1 expression subgroup had a higher risk of tumor-related death (Figure S4). The high expression subgroup of HMGB1 in six types of tumors (KIRC, DLBC, STAD, THCA, THYM, and UCS) had higher DSS than the low expression subgroup of HMGB1, indicating a lower risk of tumor-related death in the high HMGB1 expression subgroup (Figure S4). Based on the results of univariate Cox regression model, high HMGB1 expression was associated with worse DSS in seven types of tumors (ACC, LUAD, LUSC, KICH, PAAD, READ, and HNSC) but better DSS in KIRC (Figure 3C). Eight kinds of tumors (ACC, HNSC, KICH, LUAD, LUSC, PAAD, READ, and KIRC) were significant in Kaplan-Meier and Cox analyses for DSS outcome.
Next, the effects of HMGB1 expression on DFI of tumors were also analyzed. From the results, DFI of high HMGB1 expression subgroup for 12 types of tumors (ACC, CESC, CHOL, COAD, HNSC, kidney renal papillary cell carcinoma (KIRP), LIHC, LUSC, PAAD, PCPG, SARC, and TGCT) was lower than the low HMGB1 expression subgroup (Figure S5). Therefore, the high HMGB1 expression subgroup had a shorter disease-free survival time and a worse prognosis. On the contrary, the opposite findings were observed in BLCA, BRCA, and OV (Figure S5). A univariate Cox regression analysis further verified that high expression level of HMGB1 predicted worse DFI in CESC, ACC, PAAD, KIRP and COAD (Figure 3D). Five kinds of tumors (ACC, CESC, COAD, KIRP, and PAAD) were significant in both Kaplan-Meier and Cox analyses for DFI outcome.
Enrichment Analysis of HMGB1 in Different Tumors
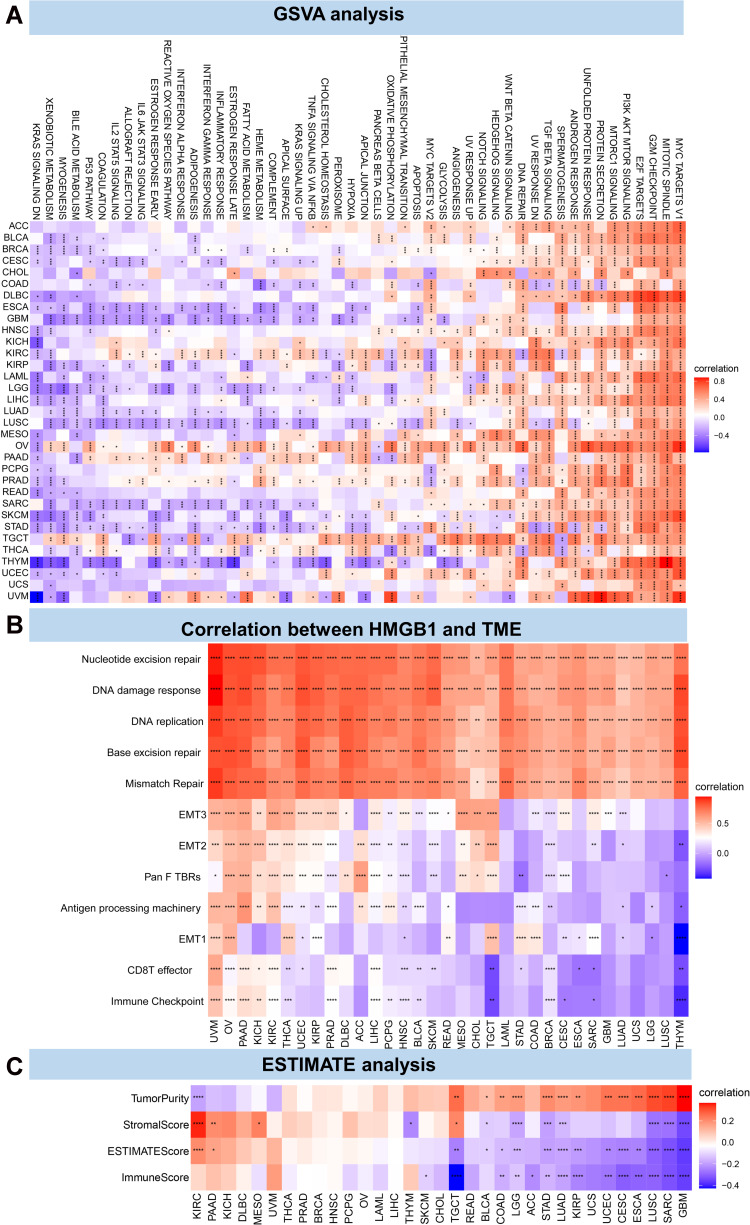
As depicted in Figure 4A, GSVA analysis of a total of 50 possible pathways in HALLMARK gene sets revealed that HMGB1 might be involved in the occurrence and development of different tumors through multiple pathways. The results demonstrated that HMGB1 had the highest scores in MYC targets V1, mitotic spindle, G2M checkpoint, E2F targets, PI3K AKT mTOR signaling, and other pathways, a finding worthy of further investigation.
Figure 4.
Correlation between HMGB1 expression and tumor microenvironment. (A) The correlation between HMGB1 expression and possible pathways in different tumors by GSVA method. (B) The correlation between HMGB1 expression and tumor microenvironment. (C) The correlation between HMGB1 expression and tumor microenvironment by ESTIMATE method. P > 0.05, < 0.05, < 0.01, < 0.001, and < 0.0001 were presented as “ns”, “*”, “**”, “***”, “****”, respectively.
HMGB1 Expression and TME
Next, we analyzed the correlation of HMGB1 with TME. As illustrated in Figure 4B, HMGB1 expression was associated with pathways such as nucleotide excision repair, DNA damage response, DNA repair, base excision repair, and mismatch repair in pan-cancer. The ESTIMATE method was used to evaluate the correlation between HMGB1 expression and tumor stromal cells, immune cell infiltration, and tumor purity in different tumors. Based on the results, the top three tumors most related to the immune score and HMGB1 expression were TGCT, GBM, and SARC, with KIRC, GBM, and SARC for stromal score; GBM, SARC, and LUSC for tumor purity; and GBM, SARC, and LUSC for ESTIMATE score, ie, the sum of stromal score and immune score (Figure 4C).
HMGB1 Expression and Tumor Immune Infiltration
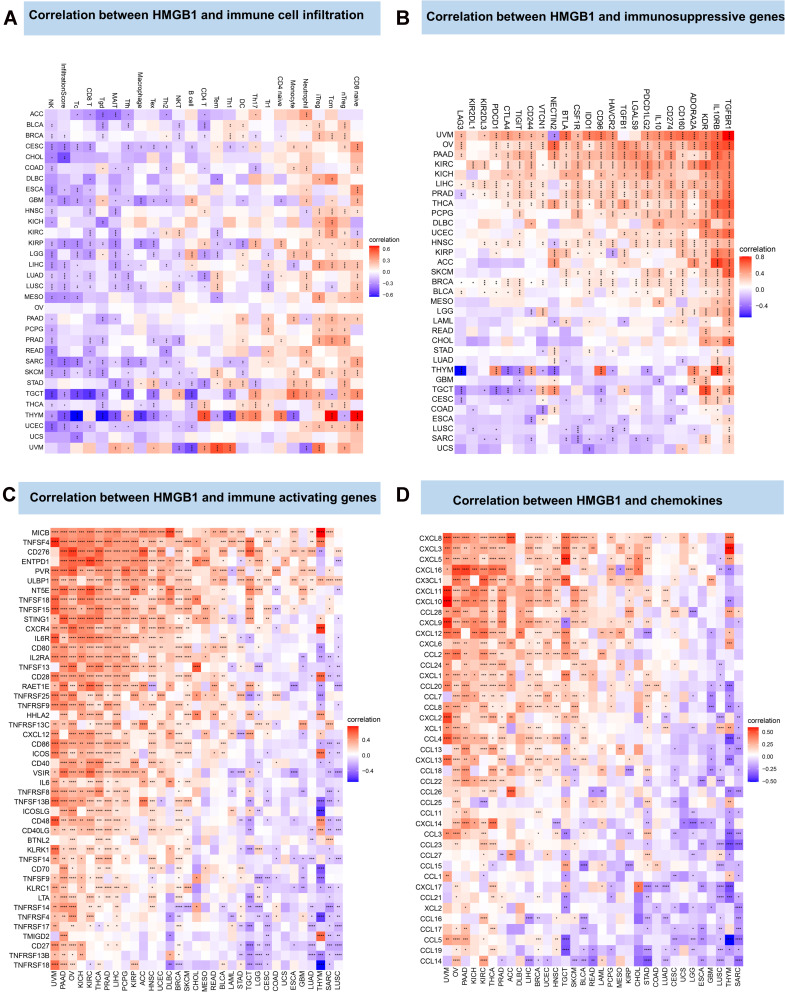
The CIBERSORT method was used to analyze the correlation between HMGB1 expression and tumor immune cell infiltration. Based on the findings, HMGB1 expression was related to various immune cell infiltration involving T cells, monocytes and neutrophils (Figure 5A). The association between HMGB1 gene and immune-related genes was further analyzed. As illustrated in Figure 5B, HMGB1 expression in most tumors was positively correlated with immunosuppressive genes, including transforming growth factor β receptor 1 (TGFBR1), interleukin-10 receptor subunit β (IL10RB), and kinase insert domain receptor (KDR). Notably, the expression levels of HMGB1 in 15 tumors (UVM, OV, PAAD, KIRC, KICH, LIHC, PRAD, THCA, PCPG, UCEC, HNSC, SKCM, BRCA, BLCA, and LAML) was positively correlated with PD-L1 (CD274) expression, suggesting a possible benefit from immune checkpoint inhibitors. As displayed in Figure 5C, in most tumors, HMGB1 expression was closely related to immune activation genes such as MICB, TNFSF4, and CD276. Similarly, HMGB1 expression was closely related to chemokines such as CXCL8 and CXCL3 (Figure 5D).
Figure 5.
Correlation between HMGB1 expression and tumor immune infiltration. (A) Correlation between HMGB1 expression and immune cell infiltration by CIBERSORT method. (B) Correlation between HMGB1 expression and immunosuppressive genes. (C) Correlation between HMGB1 expression and immune-activation genes. (D) Correlation between HMG1B expression and chemokines. P > 0.05, < 0.05, < 0.01, < 0.001, and < 0.0001 were presented as “ns”, “*”, “**”, “***”, “****”, respectively.
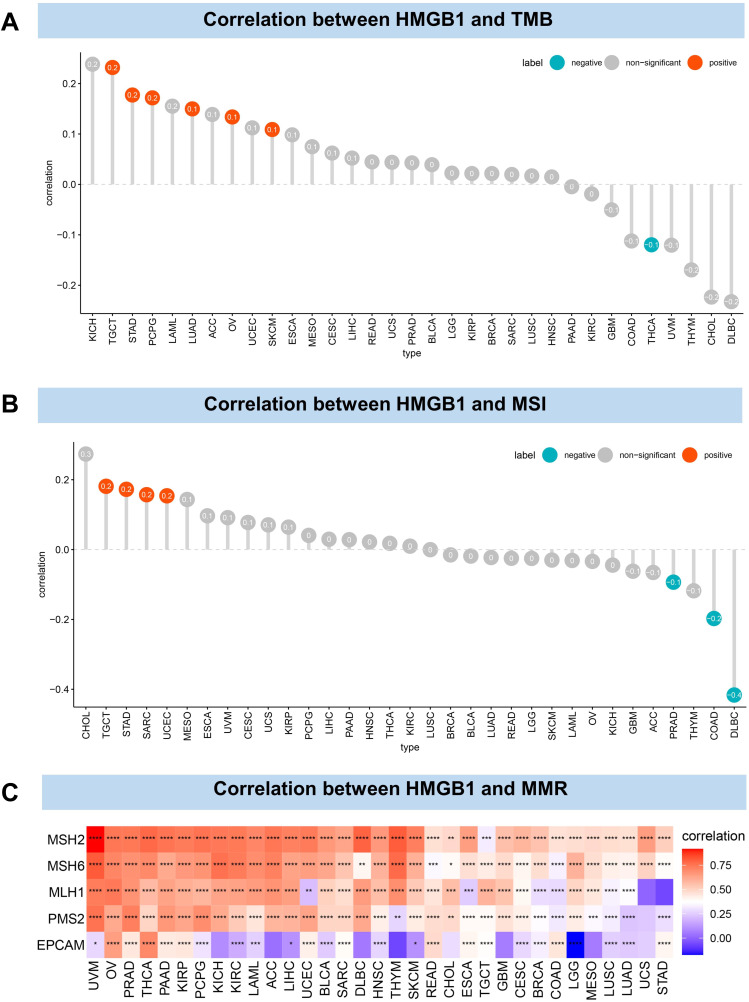
To predict the efficacy of immune checkpoint inhibitors, the relationship between HMGB1 expression and tumor mutations (TMB and MSI) was further analyzed. HMGB1 expression was positively correlated with TMB levels of TGCT, STAD, PCPG, LUAD, OV, and SKCM but negatively correlated with TMB levels in THCA (Figure 6A). In addition, the results revealed that HMGB1 expression was positively correlated with MSI levels of TGCT, STAD, SARC and UCEC but negatively correlated with the MSI levels of PRAD, COAD and DLBC (Figure 6B). The correlation between HMGB1 expression and mismatch repair (MMR) was further explored. The results revealed that HMGB1 expression was strongly correlated with MMR gene expression in most tumors, and the expression of MSH2, MSH6, and MLH1 were positively correlated with HMGB1 expression in most tumors (Figure 6C).
Figure 6.
Correlation between HMGB1 expression and tumor mutation. (A) Correlation between HMGB1 expression and tumor mutation burden. (B) Correlation between HMGB1 expression and microsatellite instability. (C) Correlation between HMGB1 expression and mismatch repair. P > 0.05, < 0.05, < 0.01, < 0.001, and < 0.0001 were presented as “ns”, “*”, “**”, “***”, “****”, respectively.
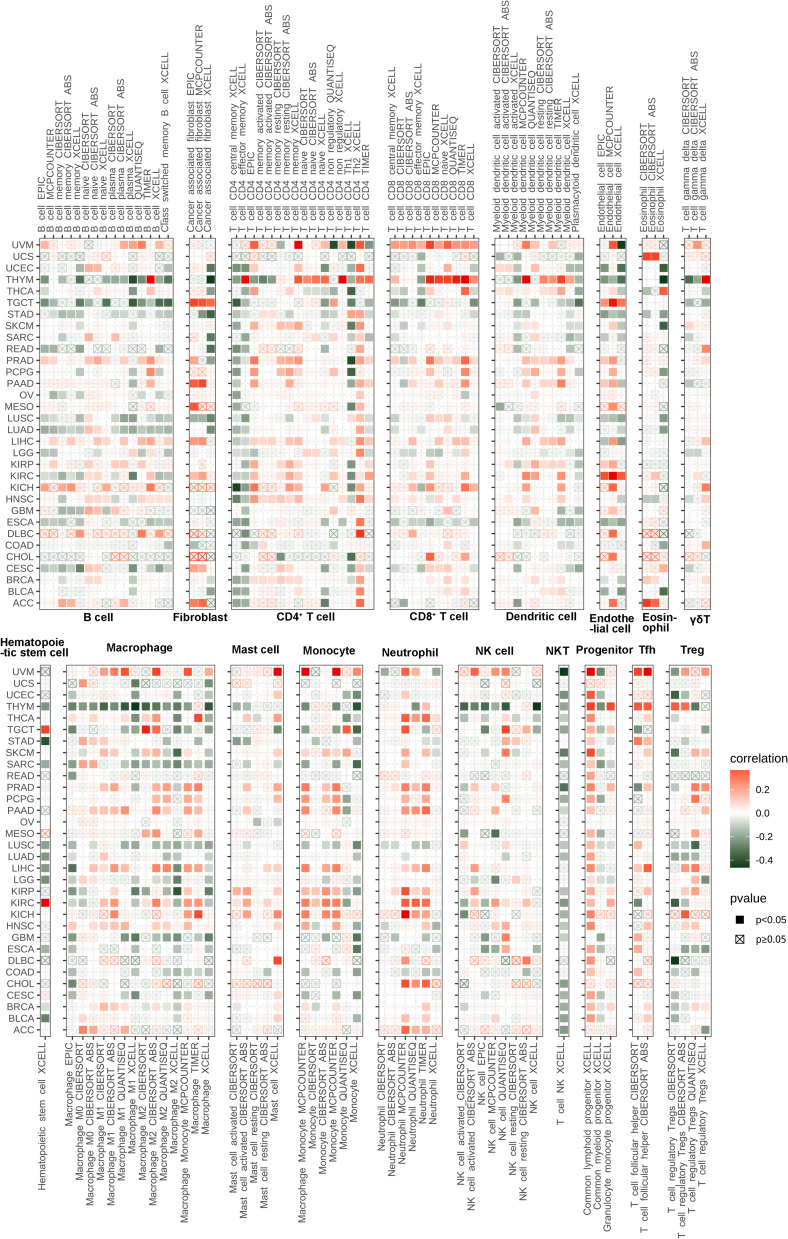
Based on pan-cancer immune infiltration data downloaded from TIMER2 database, the correlation between HMGB1expression and immune cell infiltration was calculated, including T cells, B cells, macrophages, natural killer cells, dendritic cell, neutrophils, and mast cells. The results demonstrated that HMGB1 expression was related to many kinds of immune cells, especially HMGB1 expression was positively correlated with fibroblasts in TGCT, naive CD4+ T cells in THYM, CD8+ T cells in UVM, endothelial cells in TGCT and KIRC, hematopoietic stem cells in TGCT and KIRC, M0 subtype macrophages in ACC and SARC, and follicular helper T cells in UVM, STAD, LIHC and THYM (Figure 7).
Figure 7.
Correlation between HMGB1 expression and immune cells infiltration by TIMER2 database.
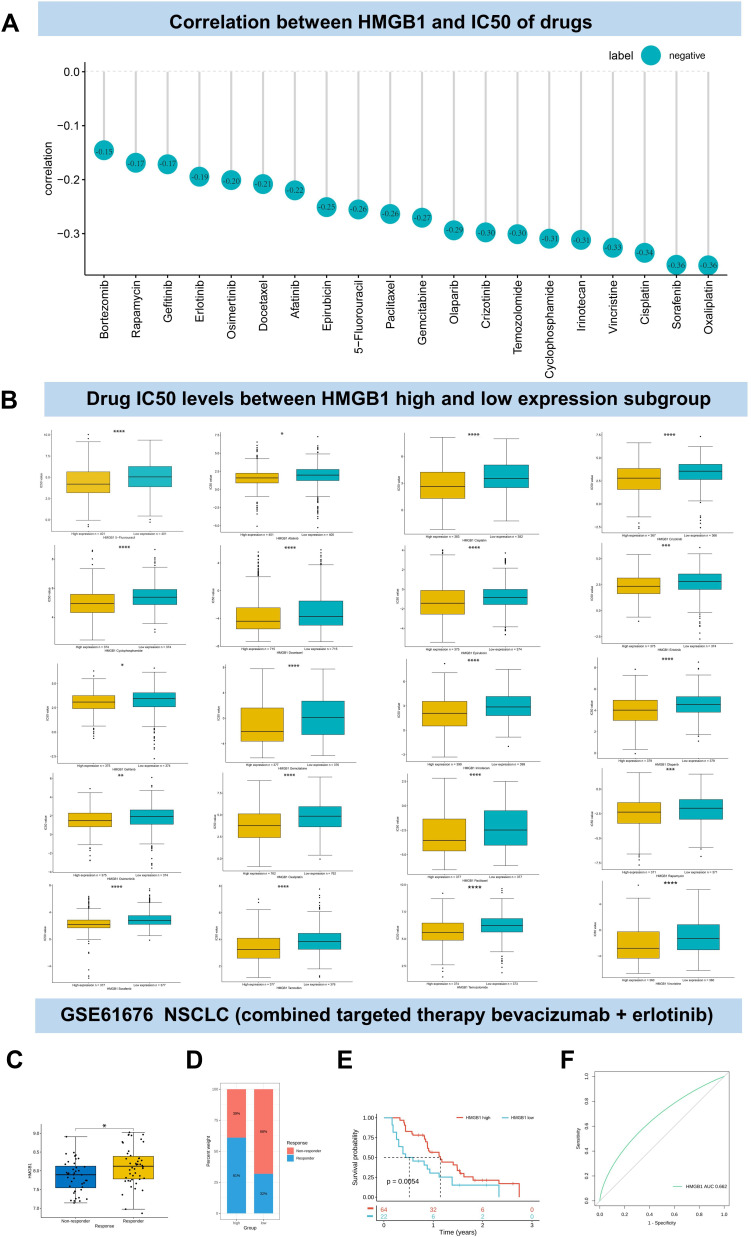
HMGB1 Expression and Anti-Tumor Drugs Sensitivity
The relationship between HMGB1 expression and drug sensitivity was analyzed via GDSC database (Table S2). The results indicated a significant correlation between HMGB1 expression and 169 anti-tumor drugs. HMGB1 expression levels were negatively correlated with the IC50 of 20 kinds of common chemotherapeutic drugs (cisplatin, docetaxel, 5-fluorouracil, paclitaxel, irinotecan, oxaliplatin, gemcitabine, temozolomide, epirubicin, cyclophosphamide, vincristine and rapamycin) and targeted drugs (gefitinib, olaparib, crizotinib, erlotinib, bortezomib, osimertinib, afatinib, and sorafenib) (Figure 8A). The levels of IC50 of the above drugs in HMGB1 high expression subgroup were lower than that in HMGB1 low expression subgroup (Figure 8B). Similarly, the relationship between HMGB1 and the combination of bevacizumab and erlotinib in the treatment of advanced non-small cell lung cancer (GSE61676) from GEO database showed that the high-level subgroup of HMGB1 had a higher proportion of response to the combination therapy and a better prognosis than the low-level subgroup. In other words, patients with high level of HMGB1 were more likely to benefit from bevacizumab and erlotinib therapy (Figure 8C–E). ROC analysis showed that the AUC of HMGB1 level predicting response to bevacizumab and erlotinib was 0.662 (Figure 8F).
Figure 8.
Association between HMGB1 expression and anti-tumor drug sensitivity. (A) Association between HMGB1 expression levels and IC50 of anti-tumor drugs. (B) Comparison of IC50 of anti-tumor drugs between HMGB1 high and low expression subgroup. (C) Comparison of HMGB1 levels in advanced non-small cell lung cancer patients with response and non-response to bevacizumab combined with erlotinib therapy. (D) Proportion of HMGB1 response to bevacizumab combined with erlotinib in high and low HMGB1 expression subgroups of advanced non-small cell lung cancer. (E) The survival probabilities of HMGB1-high and low expressing subgroups in patients with advanced non-small cell lung cancer receiving bevacizumab combined erlotinib therapy. (F) ROC analysis for HMGB1 to predict drug efficacy in patients with advanced non-small cell lung cancer receiving bevacizumab combined erlotinib therapy. P > 0.05, < 0.05, < 0.01, < 0.001, and < 0.0001 were presented as “ns”, “*”, “**”, “***”, “****”, respectively.
HMGB1 Expression and Hepatocellular Carcinoma (HCC)
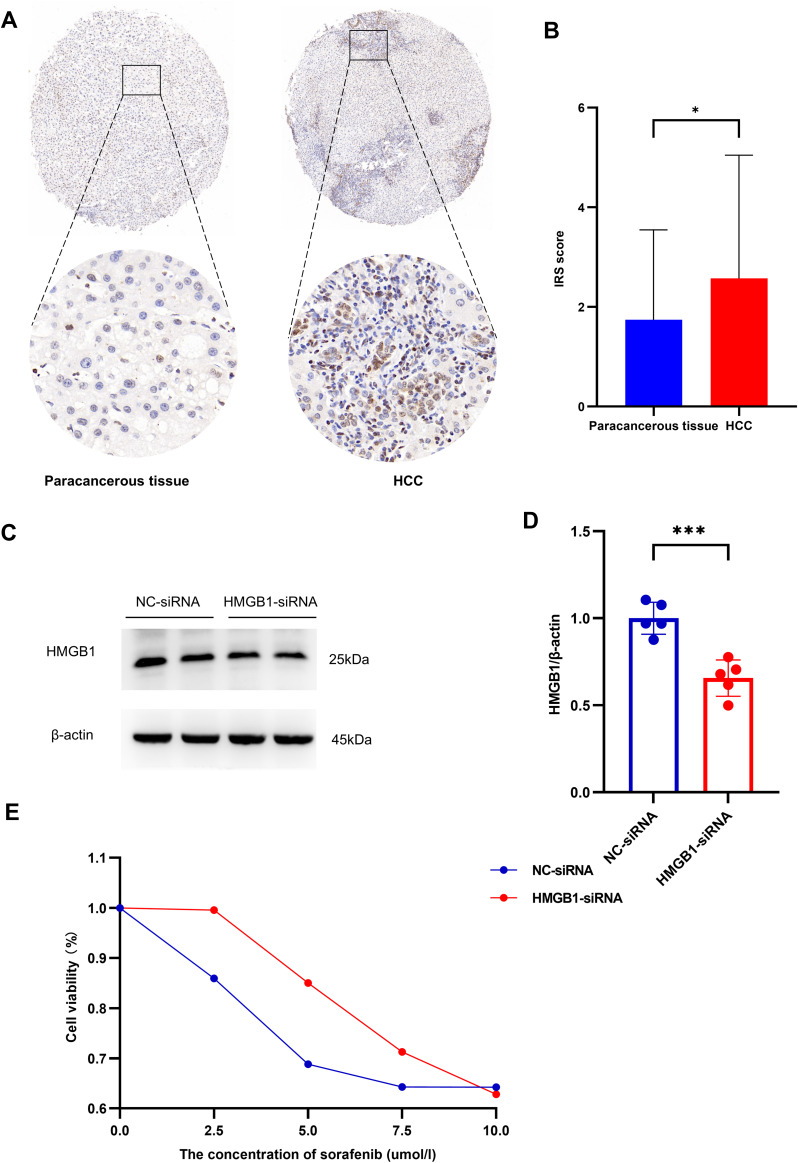
The above results consistently indicated that HMGB1 protein was highly expressed in HCC compared with normal tissues and correlated with the clinical prognosis of patients. Therefore, 54 pairs of paraffin sections of HCC patients and paracancerous tissues were collected for immunohistochemical staining IRS scores revealed that HMGB1 expression in HCC was higher than in paracancerous normal tissues (Figure 9A and B).
Figure 9.
Association between HMGB1 expression and hepatocellular carcinoma. (A) Immunohistochemical results showed HMGB1 expression in hepatocellular carcinoma and pericarcinomatous tissues. (B) Expression levels of HMGB1 in hepatocellular carcinoma and pericarcinomatous tissues. (C) Expression of HMGB1 in HepG2 cells after transfection with HMGB1-siRNA. (D) HMGB1/β-actin ratios of both NC-siRNA and HMGB1-siRNA groups in HepG2 cells. (E) Effect of silencing HMGB1 on cell viability in HepG2 cells. P < 0.05 and P < 0.01 were presented as “*” and “**”, respectively.
Based on the Western blotting results, HMGB1-siRNA was found to have the optimal HMGB1 silencing efficiency and was selected for the subsequent drug sensitivity experiments (9C and D). There was a trend of decreased response rate to sorafenib in the HMGB1-siRNA group compared to the NC-siRNA group (Figure 9E).
Discussion
In 1973, Johns et al extracted high mobility group (HMG) protein from calf thymus for the first time, and it was named such because of its rapid migration in polyacrylamide gel electrophoresis.30 The HMG family includes HMGB, HMGA, and HMGN. As one of the most abundant and widely used proteins, HMGB1 has been proven to play an important role in infectious diseases, tumors, autoimmune diseases, and other diseases.31 Even though it is known to be closely related to inflammation and immunity, the role and mechanism of HMGB1 molecule in tumors remain unclear. In this study, not only was HMGB1 differentially expressed in various tumors but also significantly correlated with both clinical prognosis and the immune microenvironment. Consequently, HMGB1 may be a potential target for tumor therapy.
First, our results revealed that HMGB1 existed widely in human organs, tumor tissues, and tumor cell lines. HMGB1 had different expression levels in tumors and normal tissues. The expression level of HMGB1 was closely related to the clinical stage and prognosis of different tumors, suggesting that HMGB1 might play different roles depending on tumor characteristics. These results were consistent with previous findings. Previous studies have demonstrated that HMGB1 is a highly conserved chromatin structural protein widely expressed in mammals.32 Mark I. MOSEVITSKY et al pointed out that HMGB1 was differentially expressed in different tissues of mammals, with high expression in lymphoid tissues and testis, but low expression in liver and brain.33 Several studies have confirmed that HMGB1 is highly expressed in various tumors than in normal tissues, including colon cancer, pancreatic cancer, esophageal cancers, breast cancer and melanoma.34,35
Next, cBioPortal database was used to explore the genetic information of HMGB1. The results revealed that the overall mutation frequencies of HMGB1 in various tumors did not exceed 5%. This confirmed that HMGB1 was a highly conserved nuclear protein, consistent with previous findings.36 As expected, in mostly tumors, HMGB1 expression level was positively correlated with HMGB1 gene copy numbers and negatively correlated with methylation. Studies have revealed that the translocation and release of HMGB1 protein are regulated by acetylation, methylation, glycosylation, phosphorylation, oxidation and other modifications.37 Methylation modification can change the conformation of HMGB1 protein in clear cell renal cell carcinoma, reduce its ability to bind to DNA, and promote its transformation from nucleus to cytoplasm.38
The results of intracellular localization of HMGB1 through HPA database demonstrated that HMGB1 was mainly localized in the nucleus in five tumor cell lines, namely, A-431, U-20S, HEL, RH-30, and U-251 MG. Previous studies have reported that HMGB1 is a position-dependent multifunctional protein.39 As an intranuclear protein, HMGB1 plays an important role in regulating gene transcription and maintaining the stability of DNA.40 HMGB1 is translocated from the nucleus to the cytoplasm and even secreted outside the cell in response to oxidative stress and hypoxia.4 The secretion and release of HMGB1 are regulated by many factors, including the mode of cell death and post-translational modification such as phosphorylation, acetylation, methylation, glycosylation, and oxidation.37,41 Using ComPPI platform, HMGB1 was predicted to interact with 187 proteins such as TLR2 and PARP. Previous studies found that HMGB1 could participate in various immune responses, including macrophage activation and B cell secretion of antibodies through TLR2/NF-κB signaling pathway.42 PARP led to poly-adenosine diphosphate-ribosylation of HMGB1, induced autophagy, and promoted resistance of tumor cells to chemotherapeutics.43 The above results suggested that HMGB1 might be involved in the occurrence and development of diseases through various pathways.
Afterward, the correlation of HMGB1 expression level with tumor prognosis was analyzed. Kaplan-Meier survival curves and univariate Cox regression analysis depicted that HMGB1 expression levels significantly affected the prognosis of various tumors, including OS, DFI, PFI, and DSS. Fachao Zhi et al found that HMGB1 induced cell proliferation and promoted the progression of colorectal cancer through the extracellular regulated protein kinases 1/2 pathway.44 Next, Zhongqiu Wang et al confirmed that HMGB1 promoted the proliferation and metastasis of pancreatic cancer cells through the Wnt pathway.45 The results of GSVA analysis demonstrated that HMGB1 expression was correlated with MYC targets, E2/F targets, G2/M checkpoint, and other pathways, indicating that HMGB1 might affect tumor cell proliferation by affecting the cell cycle.
Subsequentially, the correlation of HMGB1 expression with TME was analyzed. TME refers to the surrounding microenvironment in which tumor cells exist, including immune cells, stromal cells, and extracellular matrix. TME is a complex environment for tumor cells to survive and develop. Accordingly, it plays an important role in tumor progression, immune escape, and resistance of tumors to drugs. Innate immune cells such as macrophages, neutrophils, dendritic cells, natural killer cells, and adaptive immune cells such as T cells and B cells can both kill tumors and promote immune escape, significantly affecting tumor treatment and prognosis.46 Therefore, understanding tumor immune cell infiltration is critical for successful tumor therapy.47 The results revealed that HMGB1 expression was positively correlated with nucleotide excision repair, DNA damage response, DNA replication, base excision repair, and mismatch repair in 33 tumors, consistent with previous literature reports. Vasquez et al discovered that HMGB1 was involved in DNA replication, transcription, repair, and recombination.40 In addition, ESTIMATE method was used to evaluate the correlation between HMGB1 and stromal score, tumor purity, and immune score. The results demonstrated that the expression of HMGB1 in KIRC was positively correlated with tumor purity, stromal score, and ESTIMATE score. Additionally, HMGB1 was positively correlated with tumor purity for various tumors. Based on the above results, HMGB1 was closely related to TME, which might affect DNA replication and immune cell infiltration in tumors.
Next, four methods were used to further analyze the correlation of HMGB1 expression with immune infiltration. First, the results of ImmuCellAI database showed that the expression of HMGB1 in different tumors was related to various immune cell infiltration. TMB, MSI, and PD-L1 are common indicators to predict the efficacy of ICIs. TMB is the number of mutations per million base pairs, and elevated TMB may predict tumor sensitivity to ICIs.48 The correlation between HMGB1 and TMB depicted a correlation between high HMGB1 expression and increased TMB in TGTC, STAD, PCPG, LUAD, OV, and SKCM but decreased TMB in THCA.
MSI refers to a new microsatellite allele phenomenon in tumors. It occurs due to the change of microsatellite length caused by the insertion or deletion of short repetitive DNA sequences. It is mainly due to MMRdeficiency.49 MMR can cause tumors to lose the ability to repair DNA replication errors and accumulate mutations, resulting in high TMB and increased microsatellite instability. The more neoantigens recognized by immune cells, the better the efficacy of ICIs. Moreover, there was a positive correlation between HMGB1 and MSI in TGCT, STAD, SARC, and UCEC.
In addition, the correlation between HMGB1 expression and immune genes was determined, including immune-related genes, and MMR genes such as MSH2, MSH5, PMS2, MLH1, and EPCAM.50 A correlation was noted between HMGB1 and MMR gene expression in many types of tumors. Notably, HMGB1 was associated with immunosuppressive genes such as PD-1, PD-L1, CTLA4, TIGIT, and LAG3. The above results demonstrated that HMGB1 was closely related to TMB, MSI, MMR, and PD-L1, indicating that HMGB1 has the potential as a new biomarker to predict the effects of ICIs.
Moreover, the relationship between HMGB1 expression and immune infiltration was evaluated using TIMER2 database. The results revealed that HMGB1 expression was closely related to many types of cell infiltration, including T cells, B cells, mononuclear macrophages, NK cells, dendritic cells, fibroblasts, mast cells, neutrophils, and endothelial cells. The results of the above four methods all suggested that HMGB1 was closely related to tumor immune infiltration and significantly affected the tumor microenvironment. Yuhan Yang et al confirmed that HMGB1 mediated LPS-induced inflammation in colon cancer cells.51 In summary, HMGB1 seems to play different roles in different diseases due to differences in cell type, receptor, location, and expression level.16
The relationship between the expression level of HMGB1 and tumor response to common chemotherapeutic drugs and targeted drugs was analyzed by GDSC2 platform. IC50 refers to the drug concentration at which 50% of the cells are suppressed.52 The results showed that the subgroup with high HMGB1 expression was more sensitive to certain common antineoplastic drugs, and the IC50 was smaller. One possible reason is that HMGB1 promotes tumor cell proliferation and metastasis. That is to say, tumors with high HMGB1 expression proliferate actively and are more sensitive to drugs that interfere with the cell cycle, such as cisplatin, paclitaxel, docetaxel, 5-fluorouracil, and so on.53–56
The above results consistently indicated that the expression level of HMGB1 was higher in the tumor tissue than that of normal tissue of HCC patients and was related to clinical prognosis. Therefore, we focused on the role of HMGB1 in HCC. In 2020, there were 905,677 new cases (4.7% of all new tumors) and 830,180 deaths (8.3% of global tumor deaths) of liver cancer globally.1 In 2015, there were 466,100 new cases of liver cancer in China, with 422,100 related deaths mainly due to hepatitis B virus infection and alcoholic cirrhosis.57 Due to vague and unspecific early symptoms, most patients with HCC are diagnosed with locally advanced or advanced tumors with poor prognoses.58 Our data revealed that HMGB1 expression was higher in HCC than in normal tissues. High levels of HMGB1 expression were associated with later clinical stage and worse prognosis, including OS, DSS, PFI, and DFI. The immunohistochemical results of 54 pairs of HCC and paracancerous samples verified higher HMGB1 expression in HCC than that in distant paracancerous tissues. To further elucidate the role of HMGB1 in HCC, we transfected HepG2 cells with HMGB1-siRNA. The results showed that the cell viability of the HMGB1-siRNA group was higher than that of the NC-siRNA group with sorafenib treatment. The results showed that HMGB1 knockdown attenuated the sensitivity of HepG2 cells to sorafenib. Due to limitations of the experimental conditions, we were unable to obtain sorafenib-resistant HepG2 cells to further determine the effect of HMGB1 on the sensitivity of HepG2 cells to sorafenib and the IC50 of sorafenib. More experiments are needed in the future to confirm the effect of sorafenib on HepG2 cell proliferation. The results indicated that HMGB1 was involved in the proliferation of HepG2 cells. This was consistent with the results of previous studies. Accordingly, HMGB1 may be utilized as a new biomarker to predict prognosis and drug sensitivity in HCC.
We note that several studies have focused on the role of HMGB1 in different tumors. A study analyzed genetic variation of HMGB1 in different tumors in TCGA database. This study focused on the role of HMGB1 in THYM and found that HMGB1 was highly expressed in THYM, and high levels of HMGB1 were associated with higher OS.59 Another study analyzed the role of HMGB protein family in various tumors.60 The results showed that HMGBs were highly expressed in various tumors, including COAD, DLBC, LGG, PAAD, READ, STAD and THYM. High HMGB1 expression was associated with worse OS in ACC and KICH and higher OS in THYM. These were consistent with our findings (Figures 1D, 3A, and S2). The above two articles used the median value of HMGB1 expression level as the cutoff value to distinguish high expression and low expression of HMGB1, and we used the optimal cutoff value to divide high expression and low expression of HMGB1. We may provide a better method to analyze the relationship between HMGB1 expression and survival prognosis. Shoukai Yu et al used STRING tool to predict proteins that may interact with HMGB1, and we used ComPPI method instead. Differences in the analytical methods of previous articles and our study may be the reason for the inconsistency of some of the results.
This study had certain limitations. First, the results were derived from public database data that focused on the expression and role of HMGB1 in various tumors but ignored the role of HMGB1 in individualized therapy. Next, the immunohistochemical results only verified the high expression levels of HMGB1 in HCC. Due to limitations in experimental conditions, HMGB1 expression levels in other tumors and the mechanisms behind the effect of HMGB1 on tumor prognosis were unexplored. In the future, more human and animal studies are needed to further investigate the mechanism of HMGB1 in different tumors. More clinical studies should verify the relationship between HMGB1 and tumor prognosis.
Conclusions
Based on the pan-cancer analysis of HMGB1, this protein was differentially expressed in different tumors and corresponding normal tissues. In addition, HMGB1 expression was closely correlated with the clinical stage, prognosis, tumor microenvironment, immune-related genes and immune infiltration. Consequently, it may be a potential biomarker that predicts the prognosis of tumors.
Acknowledgments
We thank the patients and their families for their support of this study. We acknowledge TCGA, CCLE, GTEx, GDSC, HPA, GEO, ImmuCellAI, TIMER2 and ComPPI databases platforms and data contributors.
Funding Statement
This work was supported by Sanming Project of Medicine in Shenzhen (SZSM201911013), National Nature Science Foundation of China (82170690), the Shenzhen Science and Technology Innovation Committee of Guangdong Province of China (JCYJ20180307150634856 and JCYJ20210324123200003).
Data Sharing Statement
The data involved in the article can be downloaded from public databases. The raw data on hepatocellular carcinoma can be obtained by contacting the corresponding author.
Ethics Statement
This study complied with the Declaration of Helsinki. The collection of biological samples was approved by the Ethics Committee of Tong Ren Hospital (No. 2020-035-01). The study has obtained the informed consent of patients and their families.
Author Contributions
All authors made significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; participated in drafting the article or critically revising important intellectual content; agreed to submit the article to the current journal; gave final approval of the version to be published; and agreed to take responsibility for all aspects of the work.
Disclosure
All authors declare no conflict of interest in this work.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Soerjomataram I, Bray F. Planning for tomorrow: global cancer incidence and the role of prevention 2020–2070. Nat Rev Clin Oncol. 2021;18(10):663–672. doi: 10.1038/s41571-021-00514-z [DOI] [PubMed] [Google Scholar]
- 3.Yang H, Wang H, Andersson U. Targeting Inflammation Driven by HMGB1. Front Immunol. 2020;11:484. doi: 10.3389/fimmu.2020.00484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418(6894):191–195. doi: 10.1038/nature00858 [DOI] [PubMed] [Google Scholar]
- 5.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015 [DOI] [PubMed] [Google Scholar]
- 6.Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochem Soc Trans. 2001;29(Pt 4):395–401. doi: 10.1042/bst0290395 [DOI] [PubMed] [Google Scholar]
- 7.Yang H, Wang H, Ju Z, et al. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J Exp Med. 2015;212(1):5–14. doi: 10.1084/jem.20141318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grégoire M, Tadié JM, Uhel F, et al. Frontline science: HMGB1 induces neutrophil dysfunction in experimental sepsis and in patients who survive septic shock. J Leukoc Biol. 2017;101(6):1281–1287. doi: 10.1189/jlb.5HI0316-128RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spagnuolo L, Puddinu V, Boss N, et al. HMGB1 promotes CXCL12-dependent egress of murine B cells from Peyer’s patches in homeostasis. Eur J Immunol. 2021;51(8):1980–1991. doi: 10.1002/eji.202049120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumitriu IE, Baruah P, Valentinis B, et al. Release of high mobility group box 1 by dendritic cells controls T cell activation via the receptor for advanced glycation end products. J Immunol. 2005;174(12):7506–7515. doi: 10.4049/jimmunol.174.12.7506 [DOI] [PubMed] [Google Scholar]
- 11.Rovere-Querini P, Capobianco A, Scaffidi P, et al. HMGB1 is an endogenous immune adjuvant released by necrotic cells. EMBO Rep. 2004;5(8):825–830. doi: 10.1038/sj.embor.7400205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rendon-Mitchell B, Ochani M, Li J, et al. IFN-gamma induces high mobility group box 1 protein release partly through a TNF-dependent mechanism. J Immunol. 2003;170(7):3890–3897. doi: 10.4049/jimmunol.170.7.3890 [DOI] [PubMed] [Google Scholar]
- 13.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603 [DOI] [PubMed] [Google Scholar]
- 14.Kokkola R, Li J, Sundberg E, et al. Successful treatment of collagen-induced arthritis in mice and rats by targeting extracellular high mobility group box chromosomal protein 1 activity. Arthritis Rheum. 2003;48(7):2052–2058. doi: 10.1002/art.11161 [DOI] [PubMed] [Google Scholar]
- 15.Dupire G, Nicaise C, Gangji V, Soyfoo MS. Increased serum levels of high-mobility group box 1 (HMGB1) in primary Sjögren’s syndrome. Scand J Rheumatol. 2012;41(2):120–123. doi: 10.3109/03009742.2011.633099 [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Zhang Y. HMGB1 in inflammation and cancer. J Hematol Oncol. 2020;13(1):116. doi: 10.1186/s13045-020-00950-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi YR, Kim H, Kang HJ, et al. Overexpression of high mobility group box 1 in gastrointestinal stromal tumors with KIT mutation. Cancer Res. 2003;63(9):2188–2193. [PubMed] [Google Scholar]
- 18.Pusterla T, Nèmeth J, Stein I, et al. Receptor for advanced glycation endproducts (RAGE) is a key regulator of oval cell activation and inflammation-associated liver carcinogenesis in mice. Hepatology. 2013;58(1):363–373. doi: 10.1002/hep.26395 [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Yan W, Tohme S, et al. Hypoxia induced HMGB1 and mitochondrial DNA interactions mediate tumor growth in hepatocellular carcinoma through Toll-like receptor 9. J Hepatol. 2015;63(1):114–121. doi: 10.1016/j.jhep.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CZ, Zheng JJ, Bai YH, Xia P, Zhang HC, Guo Y. HMGB1/RAGE axis mediates the apoptosis, invasion, autophagy, and angiogenesis of the renal cell carcinoma. Onco Targets Ther. 2018;11:4501–4510. doi: 10.2147/OTT.S167197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao Y, Wang HC, Fan SJ. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta Pharmacol Sin. 2007;28(12):1957–1967. doi: 10.1111/j.1745-7254.2007.00669.x [DOI] [PubMed] [Google Scholar]
- 22.Cámara-Quílez M, Barreiro-Alonso A, Vizoso-Vázquez Á, et al. The HMGB1-2 ovarian cancer interactome. The role of HMGB proteins and their interacting partners MIEN1 and NOP53 in ovary cancer and drug-response. Cancers. 2020;12(9):2435. doi: 10.3390/cancers12092435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu Z, Peng J, Long X, et al. Sperm autoantigenic protein 17 predicts the prognosis and the immunotherapy response of cancers: a pan-cancer analysis. Front Immunol. 2022;13:844736. doi: 10.3389/fimmu.2022.844736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng D, Li M, Zhou R, et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol Res. 2019;7(5):737–750. doi: 10.1158/2326-6066.CIR-18-0436 [DOI] [PubMed] [Google Scholar]
- 27.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonneville R, Krook MA, Kautto EA, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017;2017. doi: 10.1200/PO.17.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo Z, Zhang X, Zhu H, et al. TELO2 induced progression of colorectal cancer by binding with RICTOR through mTORC2. Oncol Rep. 2021;45(2):523–534. doi: 10.3892/or.2020.7890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973;38(1):14–19. doi: 10.1111/j.1432-1033.1973.tb03026.x [DOI] [PubMed] [Google Scholar]
- 31.Kang R, Chen R, Zhang Q, et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40:1–116. doi: 10.1016/j.mam.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rrapaj E, Trisolini E, Bertero L, et al. Expression analysis of HMGB1 in histological samples of malignant pleural mesothelioma. Histopathology. 2018;72(6):1039–1050. doi: 10.1111/his.13470 [DOI] [PubMed] [Google Scholar]
- 33.Mosevitsky MI, Novitskaya VA, Iogannsen MG, Zabezhinsky MA. Tissue specificity of nucleo-cytoplasmic distribution of HMG1 and HMG2 proteins and their probable functions. Eur J Biochem. 1989;185(2):303–310. doi: 10.1111/j.1432-1033.1989.tb15116.x [DOI] [PubMed] [Google Scholar]
- 34.Sparvero LJ, Asafu-Adjei D, Kang R, et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J Transl Med. 2009;7:17. doi: 10.1186/1479-5876-7-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellerman JE, Brown CK, de Vera M, et al. Masquerader: high mobility group box-1 and cancer. Clin Cancer Res. 2007;13(10):2836–2848. doi: 10.1158/1078-0432.CCR-06-1953 [DOI] [PubMed] [Google Scholar]
- 36.Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162. doi: 10.1146/annurev-immunol-030409-101323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen R, Kang R, Tang D. The mechanism of HMGB1 secretion and release. Exp Mol Med. 2022;54(2):91–102. doi: 10.1038/s12276-022-00736-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu F, Zhao ZH, Ding ST, Wu HH, Lu JJ. High mobility group box 1 protein is methylated and transported to cytoplasm in clear cell renal cell carcinoma. Asian Pac J Cancer Prev. 2013;14(10):5789–5795. doi: 10.7314/APJCP.2013.14.10.5789 [DOI] [PubMed] [Google Scholar]
- 39.Chen G, Ward MF, Sama AE, Wang H. Extracellular HMGB1 as a proinflammatory cytokine. J Interferon Cytokine Res. 2004;24(6):329–333. doi: 10.1089/107999004323142187 [DOI] [PubMed] [Google Scholar]
- 40.Mandke P, Vasquez KM. Interactions of high mobility group box protein 1 (HMGB1) with nucleic acids: implications in DNA repair and immune responses. DNA Repair. 2019;83:102701. doi: 10.1016/j.dnarep.2019.102701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richard SA, Jiang Y, Xiang LH, et al. Post-translational modifications of high mobility group box 1 and cancer. Am J Transl Res. 2017;9(12):5181–5196. [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Li R, Peng Z, Hu B, Rao X, Li J. HMGB1 participates in LPS‑induced acute lung injury by activating the AIM2 inflammasome in macrophages and inducing polarization of M1 macrophages via TLR2, TLR4, and RAGE/NF‑κB signaling pathways. Int J Mol Med. 2020;45(1):61–80. doi: 10.3892/ijmm.2019.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pal Singh M, Pal Khaket T, Bajpai VK, et al. Morin hydrate sensitizes hepatoma cells and xenograft tumor towards cisplatin by downregulating PARP-1-HMGB1 mediated autophagy. Int J Mol Sci. 2020;21:21. doi: 10.3390/ijms21218253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan G, Huang C, Chen J, Zhi F. HMGB1 released from GSDME-mediated pyroptotic epithelial cells participates in the tumorigenesis of colitis-associated colorectal cancer through the ERK1/2 pathway. J Hematol Oncol. 2020;13(1):149. doi: 10.1186/s13045-020-00985-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu L, Ren S, Daniels MJ, et al. Exogenous HMGB1 promotes the proliferation and metastasis of pancreatic cancer cells. Front Med. 2021;8:756988. doi: 10.3389/fmed.2021.756988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043 [DOI] [PubMed] [Google Scholar]
- 47.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557–4566. doi: 10.1158/0008-5472.CAN-18-3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin A, Zhang J, Luo P. Crosstalk between the MSI status and tumor microenvironment in colorectal cancer. Front Immunol. 2020;11:2039. doi: 10.3389/fimmu.2020.02039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Velzen MJM, Derks S, van Grieken NCT, Haj Mohammad N, van Laarhoven HWM. MSI as a predictive factor for treatment outcome of gastroesophageal adenocarcinoma. Cancer Treat Rev. 2020;86:102024. doi: 10.1016/j.ctrv.2020.102024 [DOI] [PubMed] [Google Scholar]
- 50.Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49(3):151–157. doi: 10.1136/jmedgenet-2011-100714 [DOI] [PubMed] [Google Scholar]
- 51.Yang Y, Yang L, Jiang S, et al. HMGB1 mediates lipopolysaccharide-induced inflammation via interacting with GPX4 in colon cancer cells. Cancer Cell Int. 2020;20:205. doi: 10.1186/s12935-020-01289-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho K, Choi ES, Kim JH, Son JW, Kim E. Numerical learning of deep features from drug-exposed cell images to calculate IC50 without staining. Sci Rep. 2022;12(1):6610. doi: 10.1038/s41598-022-10643-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weaver BA, Bement W. How Taxol/paclitaxel kills cancer cells. Mol Biol Cell. 2014;25(18):2677–2681. doi: 10.1091/mbc.e14-04-0916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cortes JE, Pazdur R. Docetaxel. J Clin Oncol. 1995;13(10):2643–2655. doi: 10.1200/JCO.1995.13.10.2643 [DOI] [PubMed] [Google Scholar]
- 55.Fluorouracil. Drugs and Lactation Database (Lactmed). Bethesda (MD): National Library of Medicine (US); 2006. [Google Scholar]
- 56.Ghosh S. Cisplatin: the first metal based anticancer drug. Bioorg Chem. 2019;88:102925. doi: 10.1016/j.bioorg.2019.102925 [DOI] [PubMed] [Google Scholar]
- 57.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 58.Ren Z, Li A, Jiang J, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68(6):1014–1023. doi: 10.1136/gutjnl-2017-315084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu S, Qian L, Ma J. Genetic alterations, RNA expression profiling and DNA methylation of HMGB1 in malignancies. J Cell Mol Med. 2022;26(15):4322–4332. doi: 10.1111/jcmm.17454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin T, Zhang Y, Lin Z, Peng L. Roles of HMGBs in prognosis and immunotherapy: a pan-cancer analysis. Front Genet. 2021;12:764245. doi: 10.3389/fgene.2021.764245 [DOI] [PMC free article] [PubMed] [Google Scholar]