Abstract
Background:
Tumor cells proliferation and apoptosis inhibition are the mechanisms through which the Colorectal Cancer (CRC) progression, metastasis and chemoresistance are promoted pathologically, offering clinical advantages for characterizing their molecular regulators.
Objectives:
In this study, to unravel the role of PIWIL2 as a potential CRC oncogenic regulator, we examined the effect of its overexpression on proliferation, apoptosis and colony formation of SW480 colon cancer cell line.
Material and Methods:
Established SW480-P (overexpression of PIWIL2) and SW480-control (SW480-empty vector) cell lines were cultured in DMEM containing 10% FBS with 1% penicillin-streptomycin. The total DNA and RNA was extracted for further experiments. Real-Time PCR and western blotting assay were performed to measure the differential expression of proliferation associated genes including the expression of cell cycle and anti-apoptotic genes as well as Ki-67 and PIWIL2 in both cell lines. Cell proliferation was determined using MTT assay, doubling time assay and the colony formation rate of transfected cells was measured with the 2D colony formation assay.
Results:
At the molecular level, PIWIL2 overexpression was associated with significant up-regulation of cyclin D1, STAT3, BCL2-L1, BCL2-L2 and Ki-67 genes. MTT and doubling time assay showed that PIWIL2 expression induced time-related effects on proliferation rate of SW480 cells. Moreover, SW480-P cells had markedly greater capacity to form colonies.
Conclusions:
PIWIL2 plays important roles to promote cancer cell proliferation and colonization via the cell cycle acceleration and inhibition of apoptosis, the mechanisms through which this gene seems to contribute to CRC development, metastasis and chemoresistance, hence potentially highlighting PIWIL2 targeted therapy as a valuable tool for CRC treatment.
Keywords: Apoptosis, Colorectal Cancer (CRC), PIWIL2, Proliferation
1. Background
Cellular proliferation is indispensably required for body development, from embryogenesis to tissue repair and growth ( 1 ). There are tightly controlled multiple steps in the normal process of cells proliferation. However, tumorigenesis is the result of uncontrolled proliferation that is the hallmark feature of cancers ( 2 ). In cancer cells, dysregulation of the cell cycle elements including cyclins, cyclin dependent kinases (Cdks), Cdk inhibitors (CKI) and growth factors promotes the cellular proliferation, while diminishing the proliferation inhibitors ( 3 ). Assessment of tumor cell proliferation have clinical advantages to predict tumor relapse, metastatic potential and survival in many types of cancers including Colorectal cancer (CRC) as the major cause of cancer related death worldwide ( 4 ). Several line of studies have reported the correlation between high proliferation, chemoresistance and invasive CRC phenotypes ( 5 - 7 ). CRC develops in a multi-step process which is based on uncontrolled cell proliferation and the progressive inhibition of apoptosis. When apoptosis is disrupted in CRC cells, these cells become immortal and accumulate further genetic and epigenetic alterations leading to aggressive tumorigenic behavior such as angiogenesis and metastasis ( 2 ). In addition, these cells become resistant to the common cancer treatments which is supposedly due to the apoptosis disruption in their cancer stem cells ( 8 ). Taking together, attempt to better understanding of the master regulators, controlling either cellular proliferation or apoptosis resistance, as well as their molecular targeting should provide effective therapeutic strategies to improve treatment response rates. P-element-induced wimpy testis (PIWI) proteins are one of the Argonaute (AGO) family members. PIWI gene subfamily is comprised of four members in the human genome including Piwil1/Hiwi, Piwil2/Hili, Piwil3, and Piwil4/Hiwi2. The PIWIL2 (piwi-like 2) gene is located on chromosome 8 (8p21) with 23 exons which coding 973 amino acids (110 kDa of MW). PIWIL2 harbors two conserved PAZ and PIWI domains, (whereas) the PAZ domain (PPD) is a central domain with RNA binding motif, which binding to the 3′ end of short RNAs, and the C-terminal PIWI domain acts as an RNase H catalytic region. PIWIL2, mainly functioning in germ cells, plays role in cell division, self-renewal, RNA silencing, translational regulation, chromatin remodeling and epigenetic modifications of GSCs. Although PIWIL2 expression is mainly confined to the germ cells but was wildly reported to be ectopically expressed in several cancers such as breast, lung, glioblastoma, cervical and prostate cancers ( 9 - 12 ). In addition, accumulating studies reveal that PIWIL2 expression was higher in colorectal carcinomas compared with adjacent normal colon mucosa, yet requiring further investigations to investigate the relation between PIWIL2 upregulation and the aggressive clinicopathological state of the colorectal carcinomas ( 13 , 14 ).
2. Objectives
In this study, we examined the effect of PIWIL2 expression on proliferation, apoptosis and colony formation of SW480 colon cancer cell line. Our results showed that PIWIL2 played an important role to promote cell proliferation and colony formation in CRC cells while inhibiting their apoptosis. PIWIL2 contributed to CRC proliferation through up-regulation of cyclin D1 resulting in the acceleration of cell cycle progression. Moreover, PIWIL2 contribution to apoptosis inhibition was undertaken via up-regulation of STAT3 and BCL2-L1 and BCL2-L2 genes involved in apoptotic pathway.
3. Materials and Methods
3.1. Cell Culture
SW480 was cultured in DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Brazil), at 37 °C in a humidified atmosphere of 95% air with 5% CO2. The stable SW480-PIWIL2 expressing (SW480-P) and SW480-empty vector (SW480-control) cell lines were generated by transfecting SW480 cell line using PCDNA3/PIWIL2 or PCDNA3/empty plasmid via electroporation with a NeonTM Transfection System (Thermo Fisher Scientific, USA) (supplementary Fig. 1). After 12h, infected cells were selected by G418 (500µg.mL-1), Sigma-Aldrich,USA) for 28 days. Survived cells were expanded for further analysis.
3.2. DNA Extraction and PCR Conditions
DNA was extracted from survived SW480-P and SW480-control stable cell lines by QIAamp DNA Mini Kit (Qiagen, Germany). DNA concentrations were measured by spectrophotometer (Nanodrop One, Thermo ScientificTM,USA). The ratio of absorbance at 260/280 nm were between 1.8 and 2.0. The PCR with CMV forward and PIWIL2 reverse primers was conducted as follows: after 5 min denaturation at 95 °C, 30 cycles of PCR were carried out by denaturation at 95 °C for 30 secs, annealing at 62 °C for 20 sec and elongation for 30 secs at 72 °C, followed by a final 5 min elongation at 72 ºC. The Primers were listed in supplementary Table 1.
3.3. RNA Extraction, cDNA Synthesis and Quantitative Real-Time PCR
Cells were grown to 70 % confluence on 6-well plates and subjected to total RNA isolation using High Pure RNA Tissue Kit (Roche, Germany) following the manufacturer’s protocol. Synthesis of cDNA with reverse transcriptase was performed by RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific, Inc). For Real-Time quantitative RT-PCR reactions using the Real Q Plus 2x Master Mix Green, without ROX (AMPLIQON, Denmark) was applied on Rotor-Gene 6000 HRM Real Time PCR Thermocycler (Corbett Life Science, Australia). Primers for candidate genes, used for qRT-PCR, were designated by Oligo 7 (version 7.56) software. Details of the primers were listed in Table 1 Calculation of relative mRNA expression was performed by the 2-∆∆Ct method. All the mRNA expression values were normalized to ß -actin as a reference gene.
Table 1.
The sequences of primers and the related PCR conditions.
| names | Forward primer | Reverse primer | Tm (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| PIWIL2 | CGAGGCTTGTCTGCTAATCTGG | CTTGTCCACTTCTCTGCTGATACC | 60 | 204 |
| STAT3 | GGAGAAGGACATCAGCGGTAAGAC | AGAGATAGACCAGTGGAGACACCAG | 60 | 149 |
| BCL2L2 | GGCCGCCTTGTAGCCTTCTTTGTCTTTGC | CCCGTATAGAGCTGTGAACTCCGCCCAG | 60 | 177 |
| BCL2L1 | ATATCAGAGCTTTGAACAGGTAGTG | TAGGTGGTCATTCAGGTAAGTGG | 60 | 178 |
| CCND1 | ATCAAGTGTGACCCGGACTGCCT | ACGTCGGTGGGTGTGCAAGC | 60 | 161 |
| KI67 | GGACTTTGGGTGCGACTTGAC | TTACAACTCTTCCACTGGGACGAT | 60 | 79 |
| β-actin | ACAGAGCCTCGCCTTTGCC | GATATCATCATCCATGGTGAGCTGG | 60 | 70 |
3.4. Western Blotting
Briefly, cells were lysed in a Lysis buffer with protease inhibitor cocktail and NP40 (1%) (Triton), and then clarified by centrifugation. Equal amounts of proteins were separated by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes and then blocked with 2% non-fat dry milk and washed with TBST buffer for 90 min at room temperature. Membranes were incubated with primary antibodies for B-actin (1:200 dilutions.), PIWIL2 (1:200 dilutions.) overnight before incubation with the corresponding HRP-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc.) and immunoreactive proteins were visualized with a chemiluminescence kit (Bio-Rad).
3.5. Colony Formation Assay
Single cells at a density of 200 cells/well were seeded in six well plate in triplicate wells and incubated at 37 °C for 2 weeks. Then, cells were fixed in methanol and stained with crystal violet. The plates were then photographed, and the number of colonies larger than 50 cells were counted. At least three independent experiments were carried out for each assay.
3.6. MTT Assay
The prompted proliferative effect of PIWIL2 on SW480 cells were determined by 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye. In brief, 2000 cells/well were seeded into 96-well plates and the proliferation was monitored for 4 days (0, 24, 48 and 72 h). On each day, the DMEM medium was removed and MTT reagent (Sigma-Aldrich, USA) was added to a well and incubated in the dark at 37 °C for 4 h. Then, the supernatant of the well was removed and 200 µL of Dimethyl sulfoxide (DMSO) at room temperature was added and the absorbance was measured at 580 nm.
3.7. Doubling Time Assay
To further investigate the promoting influence of PIWIL2 overexpression on the growth rate and division capacity of SW480-P cells, doubling time assay (time for a cell to double in number), to measure the cell cycle time of the cells was applied. Therefore, SW480-P and SW480-control cells were plated in triplicate wells (15000 cells per well) in 500 μL standard 10% FBS-containing medium in 24-well plates. Haemocytometer counting with trypan blue staining was applied every 24 h for 3 days (24, 48 and 72 h).
3.8. Statistical Analysis
Assessment of the statistical differences among the experimental and control groups was performed by Student’s t test, using statistical analysis software SPSS version 24.0 (SPSS Inc., Chicago, USA). The data were presented as mean ± standard error of the mean (SEM). P values < 0.05 were interpreted to indicate significant difference.
4. Results
4.1. Establishment of Stable SW480-P and SW480-Control Cell Lines
To verify successful overexpression of PIWIL2 in SW480-P cells, PIWIL2 overexpression was quantified by Real-Time PCR in SW480-P and SW480-control cells and normalized with β -actin housekeeping gene expressions. Substantial overexpression of PIWIL2 was measured in SW480-P cells compared to SW480-control cells (53.5 fold, P = 0.002) (supplementary Fig. 2A). In addition, western blotting assay was applied to detect the protein level of PIWIL2 in SW480-P and SW480-control cells. The results were consistent with the trend observed in the Real-Time PCR results (supplementary Fig. 2B). Further, to verify successful integration of PCDNA3/PIWIL2 and PCDNA3/empty plasmids into genomic DNA of SW480 cells to generate SW480-P and SW480-control stable cell lines, respectively, two PCRs with specific primers were performed in survived cells after antibiotic selection. A fragment harboring CMV promoter and PIWIL2 insertion was amplified only in SW480-P cell line, whereas no amplification was observed in SW480-control cell line confirming that only SW480-P cells were successfully transfected with the recombinant PCDNA3/PIWIL2 (supplementary Fig. 2C).
4.2. PIWIL2 Overexpression Promoted Cell Cycle Progression and Anti-Apoptotic Pathway
Furthermore, to study the mRNA expression pattern of cell cycle related protein and anti- apoptotic pathway in SW480-P and SW480-control cell lines, their mRNA levels were quantitatively determined by qRT-PCR. Cyclin D1 mRNA expression level showed a significant enhanced expression (1.87 fold, P = 0.004) and Ki-67 mRNA expression, as another known proliferative marker, was also increased (1.17 fold, P = 0.02) in SW480-P cells compared to SW480-control cells. Furthermore, the study revealed a statistically significant increase of mRNA levels of genes known to be involved in anti- apoptotic pathway such as the mRNA expression of STAT3, BCL2-L1 and BCL2-L2 with increase up to (1.67 fold, P = 0.02), (1.36 fold, P = 0.003) and (1.80 fold, P = 0.006) fold, respectively when compared with controls in Figure 1.
Figure 1.
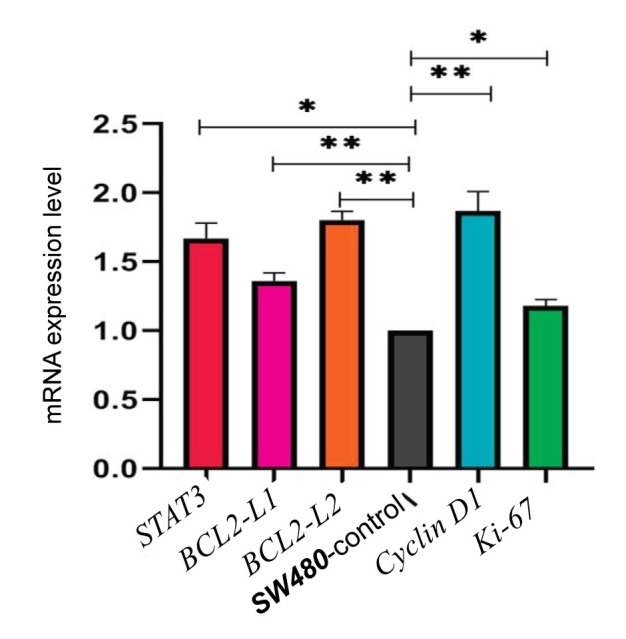
The effect of PIWIL2 expression on key regulators of proliferation related pathways. The control values have been normalized to 1 (black bar), and the data were expressed as fold change in treated cells. *P < 0.05, **P < 0.01.
4.3. PIWIL2 Promoted SW480 Cells Proliferation in a Time-Dependent Manner
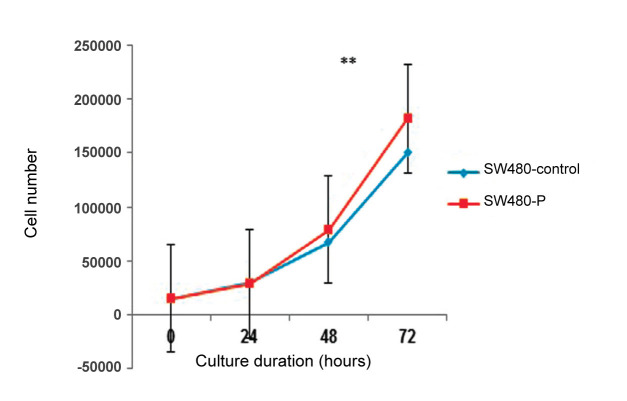
The time-dependent changes in SW480 cells proliferation were monitored using the MTT assay. As presented in Figure 2, both the SW480-P and SW480-control cell lines were shown to have similar growth during 24 and 48 h. After incubation for 72 h, a significant increase in the number of viable cells was observed in SW480-P cell lines as compared to SW480-control cells, reaching up to 1.13 fold (P = 0.007). This results suggested that PIWIL2 induced time-related effects on proliferation rate in SW480 cells.
Figure 2.
PIWIL2 overexpression induced time dependent growth of SW480 CRC cells. Cell lines after 0, 24, 48 and 72 h of incubation, respectively. Data were normalized to cells absorbance on day 1 and shown as fold changes. **P < 0.01. All experiments were performed in 9 repeats.
4.4. PIWIL2 Expression Stimulated Colony Formation of SW480 Cells
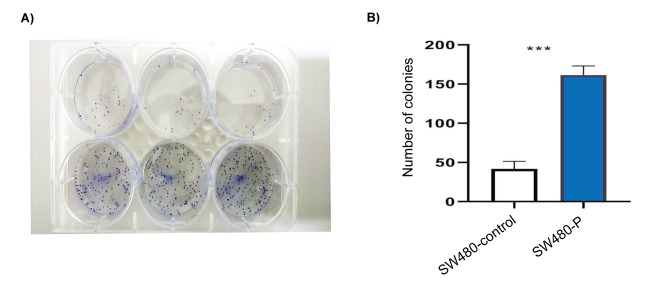
The role of PIWIL2 in the colony formation was assessed as shown in Figure 3A. In both cell lines, colonies were visible after two weeks but the number of colonies were significantly greater in SW480-P cells in 2D culture compared with vector control. Further, the PIWIL2 transfected cells formed largest clones than the control group. Statistical analysis indicated that expression of PIWIL2 promoted the colony formation of SW480 cells by 3.87 folds (P = 0.001) Figure 3B.
Figure 3.
PIWIL2 expression elevated colony formation capacity in PIWIL2 transfected cells. A) image of six-well plates with colonies. B) the bar graph was obtained by calculating the number of colonies from each cell line. **P < 0.01
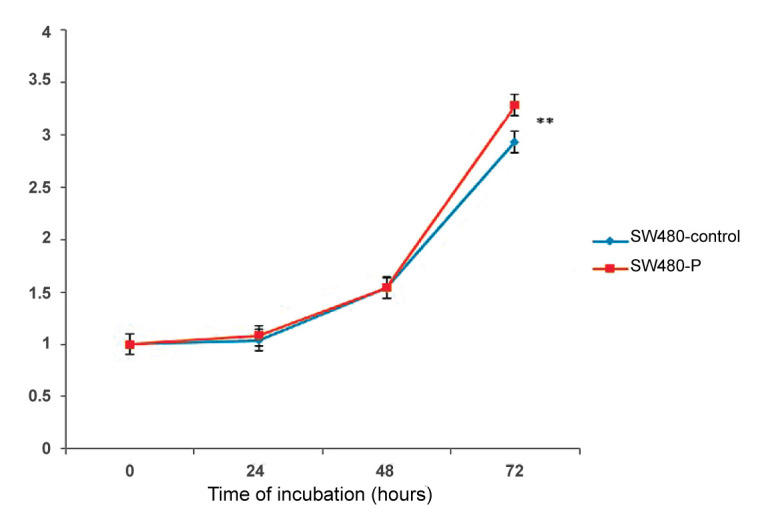
4.5. PIWIL2 Overexpression Reduced Cellular Doubling Time
Consistent with the MTT assay, doubling time experiment demonstrated higher growth rate for SW480-P cells by comparison with the SW480-control cells. As depicted in Figure 4, significant increase in cellular proliferation was observed in the SW480-P cells after 48-hour culture when the cells gained their optimal morphology and likely growth signals from each other. The doubling time calculation, undertaken during 72-hour monitoring of the cell counts, revealed nearly 2-hour decrease in doubling time of SW480-P cells in comparison to SW480-control cells (P = 0.01). According to our observations, doubling time of SW490-P and SW480-Control cells were measured as 19.23 h and 21.06 h, respectively; hence, signifying that PIWIL2 overexpression markedly led to the faster growth and proliferation activity of SW480-P cells.
Figure 4.
Doubling time assay. Counting the cells after 24, 48 and 72 h of culture, respectively. Each point indicated the average of counted cells in triplicate from SW480-P and SW480-control cells for the defined time. **P < 0.01.
5. Discussion
In the current study, we investigated the potential role of PIWIL2 in the proliferation of colorectal cancer cells, and demonstrated that PIWIL2 overexpression induced CRC cells proliferation through different mechanisms. In one mechanism, PIWIL2 expression reduced CRC cells apoptosis through up-regulation of STAT3 gene expression. Increasing expression level of STAT3 has been shown integrally involved in tumor initiation, progression and maintenance ( 15 - 17 ). STAT3 is also involved in angiogenesis, chemoresistance and inflammation, and its inhibition was reported to induce growth retardation and apoptosis ( 15 ). Furthermore, in this study, PIWIL2 expression was shown to link the up-regulation of STAT3 to expression promotion of downstream target genes such as BCL2-L1 (BCL-XL) and BCL2-L2. The BCL-2 family proteins, have pro- or anti-apoptotic effects, which have fundamental roles in promotion (BAX, BAK, BOK, BIK, BLK, HRK, BAD, BID, DIVA and EGL-1) or inhibition (BCL2, BCL-X, BCL-W, MCL-1 and CED-9) of apoptosis ( 12 , 18 ). BCL2-L1 and BCL2-L2 are two main anti-apoptotic members of BCL2 family proteins promoting cell survival ( 19 ). Lena Scherr et al. have demonstrated that among anti-apoptotic BCL-2 proteins, BCL-XL markedly increased in human CRC specimens. They generated a mouse model with defective BCL-XL in intestinal epithelial cells (Bcl-xLIEC-KO) and found marked increase in apoptosis of Bcl-xL-negative tumors ( 20 ). Similar results were obtained from other publications reporting up-regulation of BCL-XL associated with malignant behavior of colorectal cancer with worse clinical outcome ( 21 , 22 ). Our findings supported previous studies demonstrating PIWIL2 mediated anti-apoptotic state through the STAT3/BCL-XL signaling pathway ( 9 , 12 ). It seems that PIWIL2 drives some different pathways for repression of apoptosis in cancer cells. Recently, it has been reported that PIWIL2 suppresses the apoptosis in Esophageal squamous cell carcinoma (ESCC) by activating the classic IKK/IκB/NF-κB pathway ( 23 ). In another study, PIWIL2 was reported to prevent the Fas/FasL apoptotic pathway. Indeed, PIWIL2 binds to K8 and p38 and conformation of PIWIL2/K8/P38 complex results in K8 accumulation inhibiting Fas-mediated apoptosis ( 24 ). Another possible pathway or mechanism of action for PIWIL2 is supposed to be through its control over some cell cycle related proteins regulating the cell cycle progression of CRC cells. Our results suggested that PIWIL2 promoted the expression of cyclin D1, hence accelerating cell cycle progression and proliferation rate of CRC cells. Cyclin D1 is a key cell cycle related protein driving cell cycle progression through phosphorylation and subsequently inactivation of the retinoblastoma protein (pRB) ( 25 ). Cyclin D1 is essential to induce G1/S cell cycle transition as well as being a main downstream target of STAT3 ( 26 , 27 ). Several line of evidence showed correlation between cyclin D1 overexpression and tumorigenicity ( 25 , 28 ). In line with our observation, Qiu, et al. demonstrated that PIWIL2, via regulating ß-catenin/cyclin D1 pathway, contributes to entry of tumor cells from G1 into S-phase, thereby mediating tumor cell proliferation ( 29 ). Consistently, PIWIL2 ablation in glioma cells led to the cell cycle arrest of U87 cells in the G0/G1 phase indicating the involvement of PIWIL2 in the progression of the cell cycle of glioma cells ( 11 ). Interestingly, it seems that PIWIL2 participates in cancer proliferation via cell cycle progression mechanism through abnormal regulation of various cell cycle related proteins. By regulating CDK2 and cyclin A, PIWIL2 overexpression significantly increased cell proliferation in non-small cell lung cancer ( 10 ). Moreover, it has been shown that PIWIL2 mediated glioma G1/S phase transition by down-regulating p53 and p21 expression, while simultaneously up-regulating cyclin E and Cdk2 ( 11 ). Therefore, it is evident that aberrant expression of protein components of cell cycle control results in abnormal cell cycle progression influencing cell proliferation ( 29 ). As another line of evidence for PIWIL2 pro-survival activity, we investigated the expression of Ki-67 as a proliferation biomarker responding to the elevated expression level of PIWIL2-induded cyclin D1. Correlated with tumor cell proliferation, Ki-67 has been reported to be associated with metastasis and the clinical stage of tumors with predictive and prognostic potential ( 30 - 32 ). Ki-67 is a cell cycle associated antigen being expressed in all phases of cell cycle, except in G0 and early G1 phases, and its expression is robustly associated with proliferative cells ( 33 , 34 ). In line of our results, recently, it has been suggested that Ki-67 is necessary for normal G2/M transition and D-type cyclins are regulated mitosis by influencing Ki-67 expression as downstream target through E2F1 activation in HaCaT cells ( 35 ). In this study, concomitant increase in cyclin D1 and Ki-67 have been detected in SW480-P cells in comparison with SW480-control cells indicating that PIWIL2 may have boosting role for proliferation of CRC cells, an inference corroborated by the previous studies underlining PIWIL2 proliferative role in the other types of cancers ( 9 , 11 , 13 ).
Since PIWIL2 expression is ectopically limited to cancerous tissues where it works as a key mediator of oncogenic signaling, the detectable expression levels of PIWIL2 in tumors can serve as a biomarker for CRC treatment and prognosis ( 12 , 36 , 37 ). With respect to the capacity of PIWIL2 for driving cell proliferation while concomitantly inhibiting apoptosis, it’s nearly exclusive expression in cancerous tissues as well as lack of presence in normal tissues offers valuable advantage for its application in targeting therapy ( 36 , 38 ). Furthermore, several cohort and cross-sectional studies have been reported to elicit the prognostic and clinopathological implications of PIWIL2, many of which suggesting its correlation with poor prognosis and severe clinopathological state of the patients ( 39 , 40 ).
6. Conclusions
In conclusion, it seems that PIWIL2 by driving some proliferation associated cancer pathways contributes to the cell cycle regulation, apoptosis, proliferation and colonization in different kind of cancers. In this study, we investigated STAT3/BCL-XL and cyclin D1 pathway in CRC. Our results revealed that PIWIL2 induced CRC proliferation and colony formation through acceleration of cell cycle along with coincident suppression of apoptosis, the mechanisms by which PIWIL2 seemingly contributes to CRC development, metastasis and chemoresistance, all proving PIWIL2 as a valuable tool for target therapy of CRC.
Acknowledgements
This study was supported by research grants from National Institute of Genetic Engineering and Biotechnology (Grant no. 443) and Gilrayan co., Ltd. (Grant no. GRI-16-121) and Zistpooyesh knowledge-based company (ZPSP/19-10).
Conflict of Interest
The authors declare no conflict of interest.
Author contributions:
R.K.F. carried out the experiments; HR.S.L. acted as academic supervisor of this study, and designed the conceptual and experimental sketch of this study; R.K.F. provided the primary draft of the manuscript followed by significant modification by HR.S.L.; S.S. and M.G. contributed to the general experimental procedures. All the authors have approved the final version of the manuscript.
References
- 1.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Golias C, Charalabopoulos A, Charalabopoulos K. Cell proliferation and cell cycle control: a mini review. Int J Clin Pract. 2004;58(12):1134–1141. doi: 10.1111/j.1742-1241.2004.00284.x. [DOI] [PubMed] [Google Scholar]
- 4.Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14(10):101174. doi: 10.1016/j.tranon.2021.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pu J, Bai D, Yang X, Lu X, Xu L, Lu J. Adrenaline promotes cell proliferation and increases chemoresistance in colon cancer HT29 cells through induction of miR-155. Biochem Biophys Res Commun. 2012;428(2):210–215. doi: 10.1016/j.bbrc.2012.09.126. [DOI] [PubMed] [Google Scholar]
- 6.Soleimani A, Rahmani F, Ferns GA, Ryzhikov M, Avan A, Hassanian SM. Role of the NF-κB signaling pathway in the pathogenesis of colorectal cancer. Gene. 2020;726:144132. doi: 10.1016/j.gene.2019.144132. [DOI] [PubMed] [Google Scholar]
- 7.Jiang T, Tang H-m, Lu S, Yan D-w, Yang Y-x, Peng Z-h. Up-regulation of tripartite motif-containing 29 promotes cancer cell proliferation and predicts poor survival in colorectal cancer. Med Oncol. 2013;30(4):715. doi: 10.1007/s12032-013-0715-4. [DOI] [PubMed] [Google Scholar]
- 8.Serna N, Álamo P, Ramesh P, Vinokurova D, Sánchez-García L, Unzueta U, et al. Nanostructured toxins for the selective destruction of drug-resistant human CXCR4+ colorectal cancer stem cells. J Control Release. 2020;320:96–104. doi: 10.1016/j.jconrel.2020.01.019. [DOI] [PubMed] [Google Scholar]
- 9.Lee JH, Jung C, Javadian-Elyaderani P, Schweyer S, Schütte D, Shoukier M, et al. Pathways of proliferation and antiapoptosis driven in breast cancer stem cells by stem cell protein piwil2. Cancer Res. 2010;70(11):4569–4579. doi: 10.1158/0008-5472.CAN-09-2670. [DOI] [PubMed] [Google Scholar]
- 10.Qu X, Liu J, Zhong X, Li X, Zhang Q. PIWIL2 promotes progression of non-small cell lung cancer by inducing CDK2 and Cyclin A expression. J Transl Med. 2015;13(1):1–10. doi: 10.1186/s12967-015-0666-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Xu L, Bao Z, Xu P, Chang H, Wu J, et al. High expression of PIWIL2 promotes tumor cell proliferation, migration and predicts a poor prognosis in glioma. Oncol Rep. 2017;38(1):183–192. doi: 10.3892/or.2017.5647. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, Schütte D, Wulf G, Füzesi L, Radzun H-J, Schweyer S, Engel W, Nayernia K. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15(2):201–211. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- 13.Li D, Sun X, Yan D, Huang J, Luo Q, Tang H, Peng Z. Piwil2 modulates the proliferation and metastasis of colon cancer via regulation of matrix metallopeptidase 9 transcriptional activity. Exp Biol Med (Maywood) 2012;237(10):1231–1240. doi: 10.1258/ebm.2012.011380. [DOI] [PubMed] [Google Scholar]
- 14.Oh S-J, Kim S-M, Kim Y-O, Chang H-K. Clinicopathologic implications of PIWIL2 expression in colorectal cancer. Korean J Pathol. 2012;46(4):318. doi: 10.4132/KoreanJPathol.2012.46.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong A, Yang Z, Shen Y, Zhou J, Shen Q. Transcription factor STAT3 as a novel molecular target for cancer prevention. Cancers (Basel) 2014;6(2):926–957. doi: 10.3390/cancers6020926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garg M, Shanmugam MK, Bhardwaj V, Goel A, Gupta R, Sharma A, et al. The pleiotropic role of transcription factor STAT3 in oncogenesis and its targeting through natural products for cancer prevention and therapy. Med Res Rev. 2021;41(3):1291–1336. doi: 10.1002/med.21761. [DOI] [PubMed] [Google Scholar]
- 17.Lassmann S, Schuster I, Walch A, Göbel H, Jütting U, Makowiec F, Hopt U, Werner M. STAT3 mRNA and protein expression in colorectal cancer: effects on STAT3-inducible targets linked to cell survival and proliferation. J Clin Pathol. 2007;60(2):173–179. doi: 10.1136/jcp.2005.035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281(5381):1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 19.Opferman JT, Kothari A. Anti-apoptotic BCL-2 family members in development. Cell Death Differ. 2018;25(1):37–45. doi: 10.1038/cdd.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherr A-L, Gdynia G, Salou M, Radhakrishnan P, Duglova K, Heller A, et al. Bcl-x L is an oncogenic driver in colorectal cancer. Cell Death Dis. 2016;7(8):e2342–e2342. doi: 10.1038/cddis.2016.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y-L, Pang L-Q, Wu Y, Wang X-Y, Wang C-Q, Fan Y. Significance of Bcl-xL in human colon carcinoma. World J Gastroenterol. 2008;14(19):3069. doi: 10.3748/wjg.14.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biroccio A, Benassi B, D’Agnano I, D’Angelo C, Buglioni S, Mottolese M, et al. c-Myb and Bcl-x overexpression predicts poor prognosis in colorectal cancer: clinical and experimental findings. Am J Pathol. 2001;158(4):1289–1299. doi: 10.1016/S0002-9440(10)64080-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Huang L, Lu Y, Jiang W, Song Y, Qiu B, Tao D, Liu Y, Ma Y. PIWIL2 interacting with IKK to regulate autophagy and apoptosis in esophageal squamous cell carcinoma. Cell Death Differ. 2021;28(6):1941–1954. doi: 10.1038/s41418-020-00725-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang S, Zhao L, Lu Y, Wang M, Chen Y, Tao D, et al. Piwil2 inhibits keratin 8 degradation through promoting p38-induced phosphorylation to resist Fas-mediated apoptosis. Mol Cell Biol. 2014;34(21):3928–3938. doi: 10.1128/MCB.00745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K, Jiao X, Ashton A, Di Rocco A, Pestell TG, Sun Y, et al. The membrane-associated form of cyclin D1 enhances cellular invasion. Oncogenesis. 2020;9(9):1–13. doi: 10.1038/s41389-020-00266-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Q, Li Y, Li S, Jin L, Lai H, Wu Y, et al. LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nature communications. 2020;11(1):1–15. doi: 10.1038/s41467-020-19349-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin J-J, Yan L, Zhang J, Zhang W-D. STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J Exp Clin Cancer Res. 2019;38(1):1–16. doi: 10.1186/s13046-019-1206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drobnjak M, Osman I, Scher HI, Fazzari M, Cordon-Cardo C. Overexpression of cyclin D1 is associated with metastatic prostate cancer to bone. Clin Cancer Res. 2000;6(5):1891–1895. [PubMed] [Google Scholar]
- 29.Qiu B, Zeng J, Zhao X, Huang L, Ma T, Zhu Y, et al. PIWIL2 stabilizes β-catenin to promote cell cycle and proliferation in tumor cells. Biochem Biophys Res Commun. 2019;516(3):819–824. doi: 10.1016/j.bbrc.2019.06.136. [DOI] [PubMed] [Google Scholar]
- 30.Li LT, Jiang G, Chen Q, Zheng JN. Ki67 is a promising molecular target in the diagnosis of cancer. Mol Med Rep. 2015;11(3):1566–1572. doi: 10.3892/mmr.2014.2914. [DOI] [PubMed] [Google Scholar]
- 31.Warnakuiasuriya K, MacDonald D. Epithelial cell kinetics in oral leukoplakia. J Oral Pathol Med. 1995;24(4):165–169. doi: 10.1111/j.1600-0714.1995.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 32.Shah N, Trivedi T, Tankshali R, Goswami J, Shah J, Jetly D, et al. Molecular alterations in oral carcinogenesis: significant risk predictors in malignant transformation and tumor progression. Int J Biol Markers. 2007;22(2):132–143. doi: 10.1177/172460080702200207. [DOI] [PubMed] [Google Scholar]
- 33.Sawhney N, Hall PA. Ki67--structure, function, and new antibodies. J Pathol. 1992;168(2):161–162. doi: 10.1002/path.1711680202. [DOI] [PubMed] [Google Scholar]
- 34.Gerdes J, Lemke H, Baisch H, Wacker H-H, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133(4):1710–1715. [PubMed] [Google Scholar]
- 35.Belső N, Gubán B, Manczinger M, Kormos B, Bebes A, Németh I, et al. Differential role of D cyclins in the regulation of cell cycle by influencing Ki67 expression in HaCaT cells. Exp Cell Res. 2019;374(2):290–303. doi: 10.1016/j.yexcr.2018.11.030. [DOI] [PubMed] [Google Scholar]
- 36.Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R, et al. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics. 2018; 8(19):5213–5230. doi: 10.7150/thno.28001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H-L, Chen B-B, Cao X-G, Wang J, Hu X-F, Mu X-Q, et al. The clinical significances of the abnormal expressions of Piwil1 and Piwil2 in colonic adenoma and adenocarcinoma. Onco Targets Ther. 2015;8:1259–1264. doi: 10.2147/OTT.S77003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Xu L, Bao Z, Xu P, Chang H, Wu J, et al. High expression of PIWIL2 promotes tumor cell proliferation, migration and predicts a poor prognosis in glioma. Oncol Rep. 2017;38(1):183–192. doi: 10.3892/or.2017.5647. [DOI] [PubMed] [Google Scholar]
- 39.Wang S, Li F, Fan H, Xu J, Hu Z. Expression of PIWIL2 in oral cancer and leukoplakia: Prognostic implications and insights from tumors. Cancer Biomark. 2019;26(1):11–20. doi: 10.3233/CBM. [DOI] [PubMed] [Google Scholar]
- 40.Oh S-J, Kim S-M, Kim Y-O, Chang H-K. Clinicopathologic implications of PIWIL2 expression in colorectal cancer. Korean J Pathol. 2012;46(4):318– 323. doi: 10.4132/KoreanJPathol.2012.46.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]