Abstract
Purpose
Germline loss-of-function variants in CTNNB1 cause Neurodevelopmental Disorder with Spastic Diplegia and Visual Defects (NEDSDV; OMIM: 615075) and are the most frequent, recurrent monogenic cause of cerebral palsy (CP). We investigated the range of clinical phenotypes due to disruptions of CTNNB1 to determine the association between NEDSDV and CP.
Methods
Genetic information from 404 individuals with collectively 392 pathogenic CTNNB1 variants were ascertained for the study. From these, detailed phenotypes for 52 previously unpublished individuals were collected and combined with 68 previously published individuals with comparable clinical information available. The functional effects of selected CTNNB1 missense variants were assessed by TOPFlash assay.
Results
The phenotypes associated with pathogenic CTNNB1 variants were similar. A diagnosis of CP was not significantly associated with any set of traits that defined a specific phenotypic subgroup, indicating that CP is not additional to NEDSDV. Two CTNNB1 missense variants were dominant negative regulators of WNT signalling, highlighting the utility of the TOPFlash assay to functionally assess variants.
Conclusions
NEDSDV is a clinically homogeneous disorder irrespective of initial clinical diagnoses, including CP, or entry points for genetic testing.
Introduction
Neurodevelopmental disorders (NDDs) are clinically diverse and predominantly genetic in origin. For NDDs such as epilepsy, intellectual disability (ID), vision, speech and movement disorders, early clinical genetic investigations have both financial and more importantly, clinical benefit.1 Until recently, cerebral palsy (CP) which is often comorbid with other NDDs, was under represented in clinical genomic research. Studies to date suggest at least one quarter of CP is monogenic2–4, however consideration of individuals for clinical genomic investigation was (and may still be) overlooked due to the pervasive view that CP is primarily a consequence of prenatal or perinatal ‘brain injury.’ One example that demonstrates this clinical ascertainment bias for genomic investigations is Neurodevelopmental Disorder with Spastic Diplegia and Visual Defects (NEDSDV; OMIM: 615075) is caused by heterozygous (typically de novo) loss-of-function variants of CTNNB1. In previous clinical reviews of NEDSDV, the most prominent contributors to the phenotype were impairments in (i) cognition, (ii) speech, (iii) movement due to abnormal muscle tones or delays in acquiring motor skills, (iv) morphology or physiology of the eye, (v) microcephaly, and (vi) mild craniofacial dysmorphic features.5,6 Germline loss-of-function variants of CTNNB1 have appeared in clinical sequencing cohorts where the basis for ascertainment included ID, developmental delay (DD) and autism spectrum disorders.7–9 Notably, CTNNB1 was also the most frequent recurrently affected gene (4% of all diagnoses) in a cohort of 1,345 individuals analysed retrospectively on the basis of a CP diagnosis.4 CTNNB1 variants have been detected in other CP sequencing cohorts, occasionally being used as grounds for change of clinical diagnosis.10,11 This led us to examine the breadth of phenotypic variation due to pathogenic and likely pathogenic (P/LP) germline CTNNB1 variants.
CTNNB1 encodes β-catenin, a member of the highly conserved Armadillo repeat protein family.12 β-catenin performs dual functions in cells: as a component of adherens junctions it links transmembrane cadherins to the actin cytoskeleton through α-catenin; and as an essential component of the WNT signalling pathway, it acts as a transcriptional co-activator in the nucleus.13 During brain development, the role of β-catenin in cell adhesions is essential for proper cell migration while the WNT signalling pathway regulates cell proliferation and cell fate determination.14–16
Here, we present phenotypes of 52 previously unpublished individuals with NEDSDV due to P/LP variants in CTNNB1 and compare them to 68 previously described individuals. We show that there is a common phenotype among all individuals with NEDSDV except for eye-related pathologies. A clinical diagnosis of CP was not significantly associated with any other set of traits that defined a specific phenotypic subgroup, indicating that CP is not due to an additional environmental or secondary genetic cause.
Materials and Methods
Inclusion criteria and collection of clinical data
Fields for clinical data in Supplementary Table 1 were selected based on the range of traits previously associated with P/LP CTNNB1 variants. For unpublished cases, the referring clinical team were required to specifically indicate presence or absence of a trait when known. Where data were unavailable, it was treated as missing rather than absence of the trait and the corresponding individual was excluded from calculations of proportions of that particular trait in the disease population. Individuals previously reported in sequencing studies in the literature or public clinical databases with four or less of the six known CTNNB1 traits described in the introduction were grouped with the unpublished individuals when new information was provided. Individuals previously published with more than four known CTNNB1 traits, were grouped with previously published even when additional information was collected. For published individuals, at least five out of the six known CTNNB1 traits were required for inclusion in the comparisons with the cohort of 52 unpublished individuals. Identification of unpublished and previously published individuals with CTNNB1 variants is summarized in Supplementary Figure 1.
Identification of 340 CTNNB1 variants in literature and public clinical genetic databases
Published literature indexed in PubMed and supplementary data from large sequencing studies were reviewed to identify CTNNB1 variants associated with neurodevelopmental phenotypes (see Supplementary Table 2 for references). ClinVar17 and DECIPHER18 were queried to identify additional CTNNB1 variants in NDDs (last accessed on April 30th, 2022) and are identified in Supplementary Table 3 by their respective accession numbers. All germline protein-truncating and canonical splice site variants in CTNNB1 were included irrespective of the depth of phenotypic information except for five protein-truncating variants that were implicated in cancers (Supplementary Table 4). Missense, in-frame, and splice region variants were included only when NDD phenotypes were present. All somatic variants, associated with cancers were excluded. Genomic and phenotypic information was combined when an individual was counted from a publication and was also in ClinVar or DECIPHER to exclude duplication. Three variants reported to ClinVar that were likely reported in published literature by the same group but with no specific link to the corresponding articles, were excluded from the list of published cases to avoid potential double-counting. Excluded individuals are listed with accession numbers in Supplementary Table 4. Variants which were likely double reported to ClinVar with a different submission identifier by a reporting laboratory and a testing laboratory were considered as one and both identifiers were noted in the Patient ID field in Supplementary Table 3. Structural variants impacting CTNNB1 only or CTNNB1 and the adjacent predicted dosage insensitive and loss-of-function tolerant gene, ULK4 were also counted into the collection of published CTNNB1 variants.
Identification of CTNNB1 variants not associated with NDDs
Predicted benign variants in CTNNB1 were obtained from gnomAD (v2.1.1)19.
Statistics
Statistical analysis was performed using R (version 4.0.4).
In-silico prediction of pathogenicity of missense CTNNB1 variants
Effects of CTNNB1 missense variants were predicted by VEST3, CADD, PROVEAN, DANN, Polyphen2, SIFT, Mutation Assessor, MetaSVM, and FATHMM using ANNOVAR (hg19 dbNSFP version 3.5a).20 Statistical significance of pathogenicity scores between different phenotypic groups was assessed for each predictive tool using two-tailed t-test assuming unequal variance.
Expression plasmids
A pcDNA 3.1 mammalian expression vector carrying wild-type CTNNB1 coding sequence with a C-terminal V5 tag was provided by Dr. Yoshitaka Sekido.21 From this vector, we substituted the V5 tag for a Myc tag by PCR-based cloning. Using overlap PCR, we generated four CTNNB1 missense variants identified in individuals with NDDs, c.1163T>C:p.Leu388Pro (rs1559474140), c.1723G>A:p.Gly575Arg (rs797044875), c.1271T>G:p.Leu424Arg (rs863224864), c.2128C>T:p.Arg710Cys (rs748653573), and two predicted benign variants from gnomAD database, c.860A>G:p.Asn287Ser (rs35288908; Allele frequency [AF] 6.02E-04 ) and c.1188A>C:p.Glu396Asp (rs751375496; AF 1.77E-05). Cloning strategies of these variants are summarized in Supplementary Table 5. Successful cloning of these variants was confirmed by Sanger sequencing. M50 Super 8x TOPFlash (Addgene plasmid # 12456; http://n2t.net/addgene:12456; RRID:Addgene_12456) and M51 Super 8x FOPFlash (Addgene plasmid # 12457; http://n2t.net/addgene:12457; RRID:Addgene_12457) were a gift from Randall Moon.22 Renilla luciferase vector, pRL-TK plasmid was obtained from Promega (Cat #E2241).
Cell culture and dual luciferase reporter assay
Culturing of HEK293T cells and dual luciferase reporter assay were performed as previously described.23 CTNNB1 constructs (200ng; wild type, mutant or 100ng of both) were co-transfected with TOPFlash or FOPFlash plasmid (200 ng per well) and pRL-TK plasmid (5ng per well) using lipofectamine 2000 (Invitrogen, Cat #11668019). A pcDNA3.1 vector lacking CTNNB1 coding sequence (empty vector) was used as a negative control.
Western blotting
Extraction of protein from HEK293T cells transfected with β-catenin expression constructs and luciferase reporter plasmids and western blotting were performed as previously described.23 Primary antibodies used in this study were anti-Myc tag 9E10 antibody (1:2000), anti-V5 tag antibody (1:2000, ThermoFisher Scientific, Cat #R960-25), anti-β-catenin antibody (1:1000, BD transduction laboratories, Cat#610153), and anti-β-actin antibody (1:2000, Sigma, Cat #A2228).
Results
Ascertainment of individuals with germline P/LP CTNNB1 variants
Fifty two individuals, comprising 28 females and 24 males with P/LP CTNNB1 variants were ascertained from the United States, Australia, and Europe using GeneMatcher24 and personal communications through the International Cerebral Palsy Genomics Consortium25 (Figure 1 and Supplementary Table 1). Three of these (Individual 6, 8, and 50) were previously published with limited or no clinical information10,26,27 and nine (Individual 13, 15, 18, 22, 23, 45, 48, 49, and 52) were previously reported through ClinVar17 or DECIPHER18 with no or limited clinical information, therefore we considered their phenotypes as unpublished (Supplementary Figure 1). At the time of ascertainment, the remainder (40/52) had not been reported in either the literature or variant databases.
Figure 1.
Graphical summary of 52 unpublished individuals with pathogenic variants in CTNNB1. (A) The division of males and females in the cohort. (B) The proportions of different types of variants. Colors represent different variant types: stop-gain variant (red), frameshift variant (blue), splice donor variant (purple), structural variant (yellow), and missense variant (orange). (C) Lollipop plot shows β-catenin structure at the bottom and blue boxes represent Armadillo repeat domains in β-catenin. Each dot represents a CTNNB1 variant identified in unpublished individuals. Recurrent variants identified in two or more unrelated individuals are labelled with the amino acid changes with the number of individuals in brackets. (D) Variants affecting splice donor sites were identified in two individuals. (E) Structural variants of a deletion and a balanced inversion in chromosome 3 were identified in two individuals. (F) Facial images of 10 individuals with CTNNB1 variants. Images were collected from unpublished individual 9, 10, 19 (at the age of four years old and nine years old), 28, 29, 30, 34 (at the age of six years old and 16 years and 9 months old), 40, 44, and previously published individual 180.
CTNNB1 variants were confirmed as de novo in 48 affected individuals while inheritance of the other four variants was unknown due to lack of parental samples. Fifty individuals carried single nucleotide variants (SNVs) in CTNNB1 comprising 27 stop-gain, 20 frameshift, two splice donor variants, and one missense variant. This missense variant, NM_001904.3(CTNNB1):p.Gly575Arg was recurrent,28,29 therefore, classified as pathogenic according to the American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) guidelines.30 These SNVs were distributed throughout the CTNNB1 gene with no apparent enrichment for variants in any domain (Figure 1C). All these SNVs were absent from the gnomAD database (v2.1.1) and therefore considered to be rare events.19 One of the remaining two individuals had a de novo deletion (ISCN 2016): arr[hg19] 3p22.1(41227620_43101021)x1), while the other had a de novo paracentric inversion of chromosome 3 NC_000003.11:g.16710965_41275270inv, with both of these structural variants impacting CTNNB1 (Figure 1E). Similar structural variants spanning CTNNB1 were absent in both gnomAD and the Database of Genomic Variants.19,31 Facial images of 10 individuals were provided with informed written consent. Thin upper lip vermillion was commonly identified in these individuals (Figure 1F). In line with previous reports of individuals with P/LP CTNNB1 variants,5,6 predicted loss-of-function variants including stop-gain, frameshift, canonical splice variants, and structural variants in CTNNB1 were predominant.
We compared these 52 individuals with those previously published in clinical reports or reviews, as well as the large number of individuals that have been reported in the ClinVar17 and DECIPHER18 clinical genetic databases without an associated publication. In total, we identified an additional 340 CTNNB1 variants in 352 individuals which were likely involved in NDDs (Supplementary Table 3). We selected 68 individuals from this group of 352 on the basis that they had sufficient clinical information available to make meaningful comparisons to our cohort of 52 new individuals (Supplementary Figure 1 and Supplementary Table 6). The cohort of 68 previously published individuals included nine individuals from three families with inherited CTNNB1 variants: two families with non-syndromic familial exudative vitreoretinopathy (FEVR) and one family with suspected parental germline mosaicism, otherwise CTNNB1 variants of published individuals were all de novo. This cohort comprised 35 females, 29 males, and four individuals of unreported sex.
Germline P/LP CTNNB1 variants delineate a homogeneous syndrome
Comparison of frequencies of previously reported CTNNB1-related traits between unpublished (n=52) and previously published individuals (n=68) identified significant differences in cognition (ID and / or DD), motor delay, and eye abnormalities (Fisher’s exact test, p< 0.05) (Supplementary Figure 2, Supplementary Table 7). Exclusion of the seven individuals with non-syndromic FEVR who were clinically distinct from the majority of individuals carrying CTNNB1 pathogenic variants from the cohort of previously published individuals was sufficient to ablate any significant differences in traits between unpublished and previously published individuals suggesting minimal ascertainment biases in the new cohort (Fisher’s exact test, p> 0.05; Supplementary Table 7).
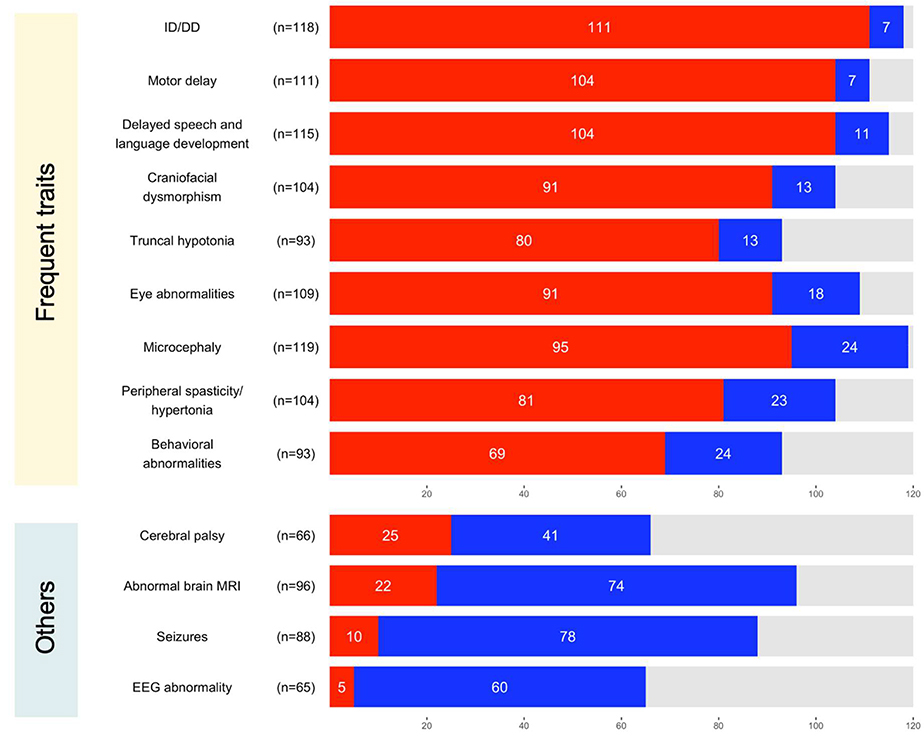
We combined data of 52 unpublished and 68 previously published individuals to delineate the common and rare traits associated with germline P/LP CTNNB1 variants. Frequencies of the nine most common neurological traits in a combined cohort of 120 individuals where the information of each trait was available, were as follows: 94.1% with ID/DD (111/118, 95% confidence interval (CI)= 89.8%–98.3%), 93.7% with motor delay (104/111, CI=89.2%–98.2%), 90.4% with delayed speech and language development (104/115, CI= 85.1%–95.8%), 87.5% with craniofacial dysmorphism (91/104, CI= 81.1%–93.9%), 86.0% with truncal hypotonia (80/93, CI= 79.0%–93.1%), 83.5% with mild to severe eye abnormalities (91/109, CI = 76.5%–90.5%), 79.8% with microcephaly (95/119, CI = 72.6%–87.0%), 77.9% with peripheral spasticity or hypertonia (81/104, CI = 69.9%–85.9%), and 74.2% with behavioral abnormalities (69/93, CI = 65.3%–83.1%). Neurological symptoms typically became apparent after two months of age and by 18 months at the latest. Onset of microcephaly was reported in 61/95 individuals with congenital onset (65.6%, n=40) more frequent than postnatal onset (34.4%, n=21). Available occipitofrontal circumference measurements of 73/120 individuals ranged from −8.18SD (standard deviations) to 0.50SD (mean= −3.16SD, median= −3.16SD). Brain morphology was unremarkable for 74/96 (77.1%) of individuals when examined by magnetic resonance imaging (MRI), despite the high frequency of microcephaly in this cohort. The frequency of seizures was low 11.4% (10/88, CI= 4.73%–18.0%). Seven out of the 10 individuals had febrile seizures or a history of seizures that were likely self-limiting in early childhood, suggesting P/LP CTNNB1 variants rarely cause epilepsy.
Motor and neurological phenotypes are homogenous irrespective of a clinical diagnosis of CP
One third (18/52) of unpublished individuals had a CP diagnosis, while in previously published cases, CP was reported in only 7/68 individuals but excluded in only 8/63. Records of gestational ages available from 43/52 of the unpublished individuals indicated most babies were born full term at an average of 39.3 (± 1.73) weeks; range 34– 42 weeks, therefore prematurity was not a factor associated with CP in this cohort. In previously published individuals with the records available (39/68), the average gestational age was 39.0 (± 2.94) weeks; range 24– 42 weeks. Comparison of movement phenotypes between individuals diagnosed with CP (n= 25) and others (n= 95) which include those where the CP diagnosis had been explicitly excluded (n=41, 33 unpublished and 8 previously published) and those who did not have specific mention of the diagnosis (n= 54, 1 unpublished and 53 previously published) found significantly increased frequency of peripheral spasticity/hypertonia in the group of individuals with CP (Fisher’s exact test, p< 0.05). However, the difference was not significant when we excluded seven individuals with non-syndromic FEVR who exhibited clinically distinct phenotypes from the majority of individuals with P/LP CTNNB1 variants from the analysis (Fisher’s exact test, p> 0.1). Regardless of inclusion or exclusion of individuals with non-syndromic FEVR, no significant difference in frequencies of any other traits, motor delay, truncal hypotonia, ID and / or DD, delayed speech and language development, craniofacial dysmorphism, eye abnormalities, microcephaly, behavioral abnormalities, and seizures was found between individuals diagnosed with CP and others (Fisher’s exact test, p> 0.05, Supplementary Table 8). Movement impairments of individuals with P/LP CTNNB1 variants were typically non-progressive. Slowly progressive spasticity in lower limbs was only reported in 4/68 previously published individuals (Supplementary Table 6). In summary, individuals with P/LP CTNNB1 variants were similarly affected irrespective of their CP diagnosis.
Diagnostic pathways for discovery of CTNNB1 genetic variants
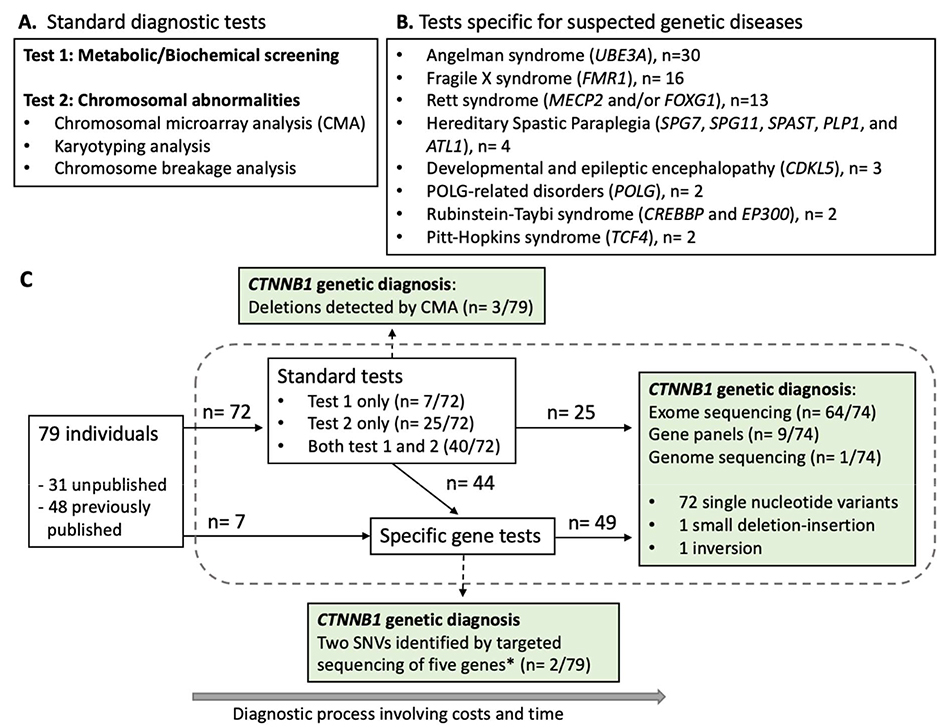
We summarized diagnostic pathways that 79 individuals followed prior to the discovery of their P/LP CTNNB1 variants (Figure 3). The information was newly collected from 31 unpublished and 2 previously published individuals and extracted from published information of 46 previously published individuals. Except for three deletions spanning CTNNB1 that were identified by chromosomal microarray analysis (CMA) and two published variants from a research cohort that were identified through targeted sequencing of five ID genes,8 P/LP CTNNB1 variants were mostly identified through exome sequencing (86.5%, 64/74). Prior to their CTNNB1 genetic diagnosis, all these individuals were assessed by standard diagnostic tests for abnormal metabolic / biochemical profiles and chromosomal abnormalities and / or tests specific for suspected genetic diseases (Figure 3 and Supplementary Table 9). In hindsight, early application of exome sequencing during testing process could have avoided unessential testing to deliver faster diagnosis to the majority of these individuals.
Figure 3.
Diagnostic pathways of 79 individuals prior to their CTNNB1 genetic diagnosis. (A) Standard diagnostic tests performed during diagnostic process. (B) A list of tests to assess suspected, specific genetic diseases that were performed in two or more individuals. (C) A graphical summary of diagnostic pathways of 79 individuals prior to their CTNNB1 genetic diagnosis. Seventy-four single nucleotide variants (SNVs) in CTNNB1 were identified through exome sequencing, targeted next-generation sequencing panels, or genome sequencing. These variants included 31 frameshift, 29 stop-gain, eight canonical splice site, and four missense variants. Two variants (*) were exceptionally identified by targeted sequencing of five intellectual disability genes including CTNNB1.
Sex bias
Sex was specified for 225 individuals that we identified with neurological impairments likely due to CTNNB1 variants, of whom 121 were female and 104 were male, thus, the frequency of predicted P/LP CTNNB1 variants does not appear to be biased towards a particular sex (Pearson’s Chi-squared test with Yates’ continuity correction, p= 0.48). Limiting our analysis to the combined cohort of 120 individuals with detailed clinical data (comprising 63 females, 53 males and four of unreported sex), we compared frequencies of each CTNNB1-related neurological trait between males and females. Behavioral abnormalities were more frequently reported in females regardless of inclusion or exclusion of individuals with non-syndromic FEVR (Fisher’s exact test, p<0.05, Supplementary Table 10). Truncal hypotonia was significantly frequent in males only when individuals with non-syndromic FEVR were excluded from the analysis (Fisher’s exact test, p<0.05, Supplementary Table 10).
Analysis of CTNNB1 variants
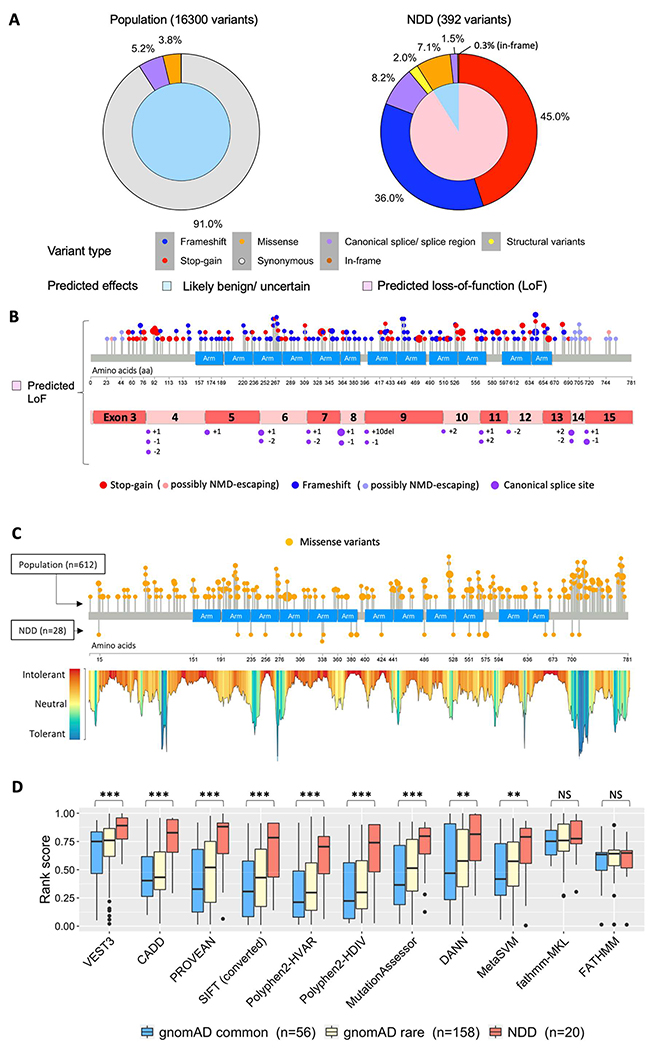
CTNNB1 variants implicated in NDD were compared to predicted benign variants in gnomAD.19 The majority of CTNNB1 variants in individuals with NDD phenotypes (91.1%, 357/392 variants) were predicted loss-of-function variants, predominantly stop-gain and frameshift variants that introduce premature termination codons in CTNNB1 mRNA (Figure 4A, Supplementary Table 11). These variants were expected to result in reduced expression of β-catenin due to nonsense-mediated mRNA decay (NMD) except for 20 variants which were predicted to escape from NMD due to their location in the last exon of CTNNB1, within 50 nucleotides upstream of the last exon-exon boundary, or proximal to the translation initiation codon32 (Figure 4B). The vast majority of variants in gnomAD, where P/LP CTNNB1 variants causing NDDs were expected to be depleted, were synonymous changes (91.0%, 14826/16300) and only two predicted loss-of-function variants in CTNNB1 were identified, each with an allele count of one (Supplementary Table 11). One of the two variants was a substitution at splice acceptor site of exon 14 (c.2077–2A>G) which likely alters normal splicing. The same variant was previously reported through ClinVar17 (variant ID: VCV000985127.1) in a male with DD, delayed speech and language development, muscular hypotonia, and several craniofacial traits (submission ID: SCV001444047.1). The second variant, c.−48–2A>G which was located at a splice donor site within the 5’ untranslated region had a low confidence loss-of-function annotation and was of uncertain significance. Locations of CTNNB1 canonical splice variants implicated in NDDs are shown in Figure 4B.
Figure 4.
Analysis of phenotypic outcome by CTNNB1 variants type. (A) Distinct patterns of CTNNB1 variants identified in the general population and neurodevelopmental disorders (NDD) with different predicted effects on CTNNB1. Inner pie charts show the ratio of variants with predicted effects of likely benign or uncertain (light blue) and loss of function (pink). Outer pie charts show the percentage of variants by type: synonymous (grey), missense (orange), frameshift (blue), stop-gain (red), splicing site (purple), in-frame insertions / deletions (brown), and structural variants mainly with deletions (yellow). Percentage labels of variant types are shown when the values are larger than 0.1%. (B) Variant plots showing distribution of stop-gain, frameshift, and canonical splice site variants of CTNNB1 identified in individuals with NDD. Lollipop plot shows μ-catenin structure at the bottom and blue box represents Armadillo repeat domains. Each dot represents an individual with stop-gain (red) or frameshift (blue) variant of CTNNB1. The larger size of a dot indicates multiple individuals with the same variant. Variants predicted to escape nonsense-mediated mRNA decay (NMD) are indicated with lighter blue (frameshift) and lighter red (stop-gain). CTNNB1 mRNA structure shows exons with canonical splice site variants (purple) likely affecting normal splicing of CTNNB1. Locations of these splice site variants in intron regions were noted with the number of nucleotides from the last nucleotide of an exon (+) or the first nucleotide of an exon (−). (C) Analysis of missense CTNNB1 variants identified in individuals with NDD compared to likely benign variants identified in the general population. Lollipop plots show distribution of missense variants of CTNNB1 (orange) identified in the general population (above) or individuals with NDD (below). Landscape of CTNNB1 variant tolerance generated using MetaDome is shown under the lollipop plot. (D) Summary of deleterious predictions of missense CTNNB1 variants using 11 predictive tools. Box plots show 1st quartile (bottom) to 3rd quartile (top) with each median value at the center. SIFT scores were calculated as 1-SIFT raw score. Student’s t-test was applied to assess the difference of NDD variants against population variants with allele frequency equal or greater than 1.0 × 10E-05 (gnomAD common). The significance marked with “***”=0.001,“**”=0.01,“*”=0.05, or “NS”=Not significant.
Missense variants accounted for 7.1%, (28/392) of CTNNB1 variants implicated in NDDs. Missense variants identified by clinical sequencing are typically classified as variants of uncertain significance (VUS) according to ACMG/AMP guidelines30 because their effects on β-catenin functions are largely unknown. Missense variants reported in the gnomAD database moderately clustered at the C terminus of β-catenin, as demonstrated by areas tolerant to genetic variation identified by MetaDome33 (Figure 4C). A VUS reported through ClinVar (p.Ile700Thr, rs2078481368, VCV001029547.1, SCV001522697.1) and a non-syndromic FEVR variant (p.Arg710Cys, rs748653573) were located within this variation tolerant region at the C terminus. The majority of NDD-associated missense variants were located in regions intolerant to genetic variation identified by MetaDome, supporting but not confirming the pathogenicity of these variants.
In-silico analyses of CTNNB1 missense variants
We investigated various in-silico tools for predicting pathogenicity of missense variants in CTNNB1. Score distributions of each prediction for variants identified in NDDs were compared to those for common variants in gnomAD (56 variants with allele frequencies equal or greater than 1.0E-05). NDD variants were scored significantly higher than common population variants by VEST3, CADD, PROVEAN, SIFT, Polyphen2, Mutation Assessor, DANN, and MetaSVM (Figure 4D). NDD variants were best distinguished from the common population variants by VEST3 (Student’s t-test, p= 7.72E-06), followed by CADD (Student’s t-test, p= 1.46E-05).
Functional investigation of CTNNB1 missense variants
We tested the functional impact of missense CTNNB1 variants identified in individuals with NDD phenotypes by TOPFlash dual-luciferase reporter assay in HEK293T cells. Transfection of a mutant β-catenin expression vector along with a luciferase reporter carrying TCF/LEF binding sites in the promoter region specifically assesses the impact of the mutant β-catenin on regulation of the WNT signalling pathway. We cloned four P/LP variants: p.Leu388Pro, p.Leu424Arg, p.Gly575Arg, and a non-syndromic FEVR variant, p.Arg710Cys, along with two predicted benign variants as controls, p.Asn287Ser and p.Glu396Asp from the gnomAD database. The variant, p.Leu388Pro was reported in a male exhibiting full CTNNB1-related neurological traits with an exception of abnormalities of the eye.5 The same variant reported in ClinVar17 was classified as VUS (VCV000560986.1, SCV000807393.1). The variant, p.Leu424Arg was identified in a male with CP, DD, microcephaly, and dysmorphic traits.34 The third variant, p.Gly575Arg was recurrently identified in six previously published individuals and one individual from this study (Supplementary Table 1 and 3). Neurological traits shared two or more among these seven individuals were DD, motor delay, truncal hypotonia, microcephaly, craniofacial dysmorphism, and eye abnormalities including FEVR, retinal detachment, and loss of vision.
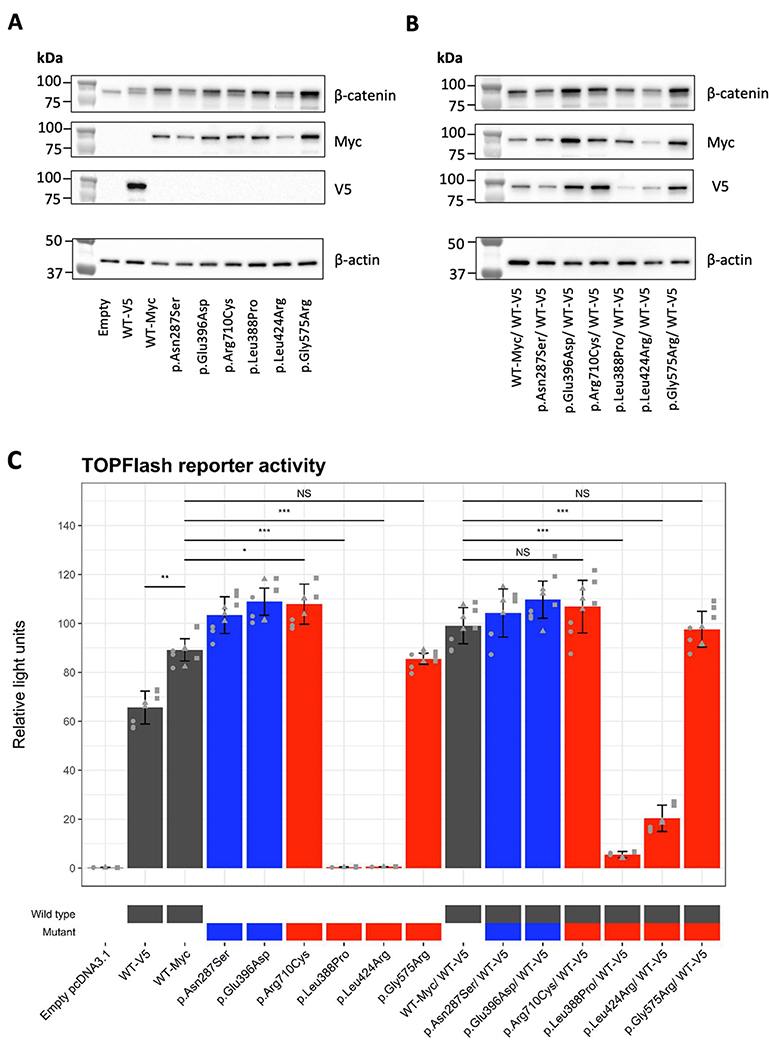
The abundance of some transfected β-catenin variant proteins was variable in comparison to wild type (Figure 5A and 5B), but relatively similar at the mRNA level (Supplementary Figure 3) suggesting some of these variants alter protein stability. We observed a significant difference in reporter activities with the addition of different epitope tags, Myc or V5 to the wild-type construct (Student’s t-test, p<0.001), therefore, we used Myc epitope tagged constructs for all comparisons between mutant and wild type β-catenin (Figure 5C).
Figure 5.
Functional assessment of missense CTNNB1 variants using TOPFlash assay. (A-B) Detection of Myc or V5-tagged wild-type (WT) β-catenin proteins and Myc-tagged mutant β-catenin proteins transfected into HEK293T cells by western blot. Expression constructs were transfected without co-transfection (A) and with co-transfection of a V5-tagged wild-type β-catenin (B). Molecular sizes of standard protein markers were indicated on the left of blots. Endogenous and exogenous β-catenin were detected with a β-catenin antibody (amino acid 571–781). Exogenous β-catenin was identified with a V5 antibody, and a Myc antibody. Endogenous levels of β-actin were detected to show equal loading by western blot. Full blots are available in Supplementary Figure 5. (C) Effects of missense CTNNB1 variants Wnt signaling as measured by the TOPFlash assay. Relative luciferase activity measured using the TOPFlash assay in HEK293T cells transfected with expression vectors for wild-type β-catenin or mutant β-catenin or an equal mix with wildtype β-catenin tagged with V5. Wild type, gnomAD variants, and pathogenic/likely pathogenic variants are highlighted in grey, blue, and red on the X-axis labels, respectively. Assay was performed in triplicate (shown with different shaped data points) with three technical replicate samples for each assay. Error bars indicate standard deviations between the three independent experiments. Student’s t-test was applied to assess the difference of relative light unit of pathogenic/likely pathogenic variants against that of wild-type β-catenin. The significance marked with “***”=0.001,“**”=0.01,“*”=0.05, or “NS”=Not significant.
TOPFlash activity was absent for two NDD variants, p.Leu388Pro and p.Leu424Arg compared to the activity of wild type β-catenin (Student’s t-test, p<0.001; Figure 5C). These two variants were dominant-negative and significantly repressed TOPFlash activity when co-expressed with wild type β-catenin (Figure 5C). In contrast, both of the predicted benign variants from gnomAD and the non-syndromic FEVR variant significantly increased TOPFlash activity compared to the wild type (Student’s t-test, p<0.05). Increases in TOPFlash activity were also observed when each of these three variants were co-transfected an equal amount of the expression construct of the wild type β-catenin; however, this was not statistically significant (Student’s t-test, p>0.05). Unexpectedly, TOPFlash activity was not altered by the recurrent p.Gly575Arg variant compared to wild type β-catenin in this assay (Student’s t-test, p=0.283). None of the constructs tested in this assay had an effect on, the negative control FOPFlash reporter which has non-functional TCF/LEF binding motifs (Supplementary Figure 4). In summary, the TOPFlash assay facilitated functional assessment of CTNNB1 missense variants. We were able to resolve p.Leu388Pro and p.Leu424Arg as likely dominant negative variants affecting the WNT signalling pathway which is greater than the effect of haploinsufficiency caused by the known pathogenic loss-of-function variants. The functional impact of p.Gly575Arg was not evident using this assay; however, given that this variant is recurrent, there is already sufficient evidence to determine that it is pathogenic.
Discussion
Prompted by multiple prior observations of individuals with P/LP CTNNB1 variants and clinical diagnosis of CP, we sought to identify if this diagnosis defines a specific phenotypic subgroup. Combined phenotypes from 120 individuals however revealed that P/LP CTNNB1 variants result in relatively consistent clinical traits in both males and females, suggesting that the CP diagnosis might reflect clinical ascertainment bias. These data support that genomic testing is beneficial for individuals with CP so they have clear and fast genetic diagnosis irrespective of initial clinical diagnosis. Our results show overwhelming evidence for heterozygous loss-of-function of CTNNB1 as the predominant disease mechanism. Analysis of missense variants however, showed that not all may affect WNT signalling which may influence the design of future targeted therapies.
Looking historically at the discovery of CTNNB1 variants implicated in NDDs, cohorts were recruited for clinical sequencing studies of ID, autism, epilepsy, DD, FEVR and CP. Though several reviews pointed towards a consistent syndrome it remained unclear if some genotype-phenotype relationships exist, or if these findings resulted from ascertainment biases. We assembled the majority of known individuals with CTNNB1 variants identified to date and our data overwhelmingly supports that P/LP CTNNB1 variants result in a syndrome with consistent neurological traits, except in the case of non-syndromic FEVR. In the case of CP, the prominent movement impairments in individuals with P/LP CTNNB1 variants have been well characterized as truncal hypotonia and usually non-progressive peripheral spasticity or hypertonia5,6 which is consistent with Surveillance of Cerebral Palsy in Europe guidelines for diagnosing CP.35,36 In previous studies, a genetic diagnosis of CTNNB1 variant was considered as grounds to remove a CP diagnosis11; however, our study now suggests that clinical CP diagnosis might also be appropriate for some individuals with P/LP CTNNB1 variants. Regardless of diagnostic clinical labels, it is most important that these individuals have early and equitable access to genomic testing. Over half of individuals in whom it possible to trace a diagnostic odyssey underwent a targeted single gene or gene-panel prior to a receiving a diagnosis from exome or genome sequencing, which highlights the importance of genomic analysis for identifying P/LP CTNNB1 variants (Figure 3).
We assessed the functional impact of missense CTNNB1 variants, including VUS, using TOPFlash dual-luciferase reporter assay. With this established assay, impact of these variants on μ-catenin mediated transactivation of WNT signalling pathway target genes can provide strong evidence for pathogenicity when loss-of-function or dominant negative effects are observed (PS3 in the ACMG/AMP guidelines). The TOPFlash assay does not account for all the functions of β-catenin, therefore, the result of the assay needs to be carefully interpreted. Negative results in the assay, as seen in one of the NDD variants; p.Gly575Arg assessed in this study, do not refute pathogenicity. The negative result of the p.Gly575Arg variant may be explained by cell-type specific impact of the variant on WNT signalling pathway. We note that delayed speech and language development, a trait frequently associated with P/LP CTNNB1 variants, was specifically ruled out in two out of seven individuals carrying the p.Gly575Arg variant and not mentioned as a trait affecting the remaining five individuals. Therefore, this variant may only affect a subset of CTNNB1 functions, manifesting as lack of speech delay.
CTNNB1 is currently associated with two neurological phenotypes: NEDSDV (MIM: 615075) and exudative vitreoretinopathy (EVR or FEVR, MIM: 617572) that is characterized by incomplete peripheral vascular development in the retina.37 Whether the pathogenic mechanisms of NEDSDV and FEVR overlap is unknown. Detailed ophthalmologic examination was not available for the majority of our unpublished individuals and reporting was variable in published individuals, therefore, we could not provide the exact frequency of FEVR in our cohort. Norrin induced Frizzled4/β-catenin signalling, a particular derivative of WNT signalling pathway likely attributes FEVR.38 Mouse models with knockout mutations in FEVR genes (Fzd4, Lrp5, Tspan12, and Ctnnb1) developed defects in retinal vasculature, suggesting that reduced activity of WNT signalling pathway leads to FEVR.37,39–41 However, studies of non-syndromic FEVR variants in CTNNB1 using the TOPFlash assay resulted in contradicting effects on the transcriptional activities.42 Further functional studies on these variants may be able to identify a specific cause of FEVR.
There are currently no established interventions or treatments for NEDSDV. Treatment with L-dopamine was used in one female with a stop-gain variant (p.Gln558*) of CTNNB1.43 The outcome was positive with improvements in her motor skills; however, a full scale randomised control trial is required to determine the benefits of L-dopamine treatment for individuals with P/LP CTNNB1 variants. Clinical homogeneity of individuals with P/LP CTNNB1 variants suggests there is minimal impact of individual-specific genetic or environmental factors on CTNNB1-related phenotypes, which would simplify modelling this disease for the purposes of identifying the potential interventions. The Batface (Bfc) mouse which has a heterozygous missense variant, p.Thr653Lys in Ctnnb1 was proposed as a potential model for NEDSDV based on the similar craniofacial features observed between the Bfc mouse and individuals carrying loss-of-function variants of CTNNB1.44 Molecular characterization of the Bfc variant in mice identified reduced interaction between β-catenin and N-cadherin at cell adhesions in hippocampus44 and surprisingly a gain of WNT signalling activity in embryos.45 Thus the Bfc mouse does not model CTNNB1 haploinsufficiency which is the typical effect of variants in patients. The effect of the Bfc variant and the p.Gly575Arg variant in the TOPFlash assay may suggest that dysregulation of the role of CTNNB1 in cell-cell adhesion may be the major contributor to phenotypes associated with NEDSDV. Heterozygous Ctnnb1 knockout mice also failed to recapitulate developmental abnormalities reported in individuals with P/LP CTNNB1 variants,46,47 possibly indicating that differences in developmental process between human and mice are critical to model this NDD. In our accumulated 392 CTNNB1 variants identified in NDDs, the most recurrent variant was p.Tyr333* (Supplementary table 13). Recently, an induced pluripotent stem cell (iPSC) line, which was capable of differentiating into all three germ layers, was established from a male individual heterozygous for the p.Tyr333* variant.48 This iPSC line or equivalent human cell models, are promising avenues elucidating the disease mechanism behind NEDSDV and potential identification of drugs capable of restoring normal development through stabilisation of CTNNB1.
Supplementary Material
Figure 2.
Neurological traits associated with pathogenic or likely pathogenic variants in CTNNB1. Traits that were frequently identified in a cohort of unpublished and previously published individuals are summarized at the top. Other relevant traits discussed in the present study are summarized at the bottom. Bar charts show the number of affected (red), unaffected (blue), and unknown (grey) individuals per trait. The number of individuals known for their affected status per trait is shown in brackets next to each trait. ID, intellectual disability; DD, developmental delay.
Acknowledgements
This work was supported by NHMRC Project Grant: 1099163 (A.H.M., C.L.v.E., M.A.C.), NHMRC Senior Fellowship App1155224 (J.G.), The Tenix Foundation and Commercial Motor Vehicles Association (J.G., A.H.M., C.L.v.E., M.A.C.). This work was partially funded by strategic action projects in health, from the Instituto de Salud Carlos III, awarded to Dr. María Palomares. Ref: PI19/01681. Marta Pacio Miguez is supported by a grant of the Raregenomics network, funded by the Dirección de General de Universidades e Investigación de la Comunidad de Madrid (S2017/ BMD-3721). We wish to acknowledge the Victorian Cerebral Palsy Register.
Footnotes
Conflicts of Interest
Two members of our authorship Dr Michelle Morrow and Dr Francisca Millan declare their employment with GeneDx as a conflict of interest in this study and there are no other conflicts to declare.
Ethics Declaration
This study was approved by the Women’s and Children’s Health Network Human Research Ethics Committee number 2020/HRE01273. Written informed consent was obtained for all individuals for whom new data are presented in this study. Individual level data in this study are de-identified. Copies of explicit informed written consent for patients providing photographs (Figure 1F) are archived with the corresponding author.
Data Availability Statement
Where not otherwise indicated (by e.g. ClinVar accession number) all source data for this paper and supplementary information is available from the corresponding author on reasonable request. Requests for potentially identifiable data are subject to approval by the Women’s and Children’s Health Network Human Research Ethics Committee.
References
- 1.Vissers LELM, van Nimwegen KJM, Schieving JH, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med. 2017;19(9):1055–1063. doi: 10.1038/gim.2017.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chopra M, Gable DL, Love- Nichols J, et al. Mendelian etiologies identified with whole exome sequencing in cerebral palsy. Ann Clin Transl Neurol. 2022;9(2):193–205. doi: 10.1002/acn3.51506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Eyk CL, Webber DL, Minoche AE, et al. Yield of clinically reportable genetic variants in unselected cerebral palsy by whole genome sequencing. Npj Genomic Med. 2021;6(1):74. doi: 10.1038/s41525-021-00238-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno-De-Luca A, Millan F, Pesacreta DR, et al. Molecular Diagnostic Yield of Exome Sequencing in Patients With Cerebral Palsy. JAMA. 2021;325(5):467. doi: 10.1001/jama.2020.26148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuechler A, Willemsen MH, Albrecht B, et al. De novo mutations in beta-catenin (CTNNB1) appear to be a frequent cause of intellectual disability: expanding the mutational and clinical spectrum. Hum Genet. 2015;134(1):97–109. doi: 10.1007/s00439-014-1498-1 [DOI] [PubMed] [Google Scholar]
- 6.Kharbanda M, Pilz DT, Tomkins S, et al. Clinical features associated with CTNNB1 de novo loss of function mutations in ten individuals. Eur J Med Genet. 2017;60(2):130–135. doi: 10.1016/j.ejmg.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542(7642):433–438. doi: 10.1038/nature21062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ligt J, Willemsen MH, van Bon BWM, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367(20):1921–1929. doi: 10.1056/NEJMoa1206524 [DOI] [PubMed] [Google Scholar]
- 9.O’Roak BJ, Vives L, Fu W, et al. Multiplex Targeted Sequencing Identifies Recurrently Mutated Genes in Autism Spectrum Disorders. Science. 2012;338(6114):1619–1622. doi: 10.1126/science.1227764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin SC, Lewis SA, Bakhtiari S, et al. Mutations disrupting neuritogenesis genes confer risk for cerebral palsy. Nat Genet. 2020;52(10):1046–1056. doi: 10.1038/s41588-020-0695-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takezawa Y, Kikuchi A, Haginoya K, et al. Genomic analysis identifies masqueraders of full-term cerebral palsy. Ann Clin Transl Neurol. 2018;5(5):538–551. doi: 10.1002/acn3.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butz S, Stappert J, Weissig H, Kemler R. Plakoglobin and beta-catenin: distinct but closely related. Science. 1992;257(5073):1142–1144. doi: 10.1126/science.257.5073.1142-a [DOI] [PubMed] [Google Scholar]
- 13.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin: β-Catenin: a life by, beyond, and against the Wnt canon. EMBO J. 2012;31(12):2714–2736. doi: 10.1038/emboj.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297(5580):365–369. doi: 10.1126/science.1074192 [DOI] [PubMed] [Google Scholar]
- 15.Backman M, Machon O, Mygland L, et al. Effects of canonical Wnt signaling on dorso-ventral specification of the mouse telencephalon. Dev Biol. 2005;279(1):155–168. doi: 10.1016/j.ydbio.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 16.Machon O, van den Bout CJ, Backman M, Kemler R, Krauss S. Role of β-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience. 2003;122(1):129–143. doi: 10.1016/S0306-4522(03)00519-0 [DOI] [PubMed] [Google Scholar]
- 17.Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firth HV, Richards SM, Bevan AP, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84(4):524–533. doi: 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164–e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Usami N, Sekido Y, Maeda O, et al. β-catenin inhibits cell growth of a malignant mesothelioma cell line, NCI-H28, with a 3p21.3 homozygous deletion. Oncogene. 2003;22(39):7922–7930. doi: 10.1038/sj.onc.1206533 [DOI] [PubMed] [Google Scholar]
- 22.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol CB. 2003;13(8):680–685. doi: 10.1016/s0960-9822(03)00240-9 [DOI] [PubMed] [Google Scholar]
- 23.Bennett MF, Hildebrand MS, Kayumi S, et al. Evidence for a Dual-Pathway, 2-Hit Genetic Model for Focal Cortical Dysplasia and Epilepsy. Neurol Genet. 2022;8(1):e0652. doi: 10.1212/NXG.0000000000000652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum Mutat. 2015;36(10):928–930. doi: 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLennan AH, Kruer MC, Baynam G, et al. Cerebral palsy and genomics: an international consortium. Dev Med Child Neurol. 2018;60(2):209–210. doi: 10.1111/dmcn.13643 [DOI] [PubMed] [Google Scholar]
- 26.Thevenon J, Duffourd Y, Masurel-Paulet A, et al. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test: Diagnostic odyssey in severe neurodevelopmental disorders. Clin Genet. 2016;89(6):700–707. doi: 10.1111/cge.12732 [DOI] [PubMed] [Google Scholar]
- 27.Stessman HAF, Xiong B, Coe BP, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49(4):515–526. doi: 10.1038/ng.3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossetti LZ, Bekheirnia MR, Lewis AM, et al. Missense variants in CTNNB1 can be associated with vitreoretinopathy—Seven new cases of CTNNB1 - associated neurodevelopmental disorder including a previously unreported retinal phenotype. Mol Genet Genomic Med. Published online December 22, 2020. doi: 10.1002/mgg3.1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monies D, Abouelhoda M, Assoum M, et al. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am J Hum Genet. 2019;104(6):1182–1201. doi: 10.1016/j.ajhg.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(D1):D986–D992. doi: 10.1093/nar/gkt958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Supek F, Lehner B, Lindeboom RGH. To NMD or Not To NMD: Nonsense-Mediated mRNA Decay in Cancer and Other Genetic Diseases. Trends Genet. 2021;37(7):657–668. doi: 10.1016/j.tig.2020.11.002 [DOI] [PubMed] [Google Scholar]
- 33.Wiel L, Baakman C, Gilissen D, Veltman JA, Vriend G, Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum Mutat. Published online May 22, 2019:humu.23798. doi: 10.1002/humu.23798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312(18):1880–1887. doi: 10.1001/jama.2014.14604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smithers-Sheedy H, Badawi N, Blair E, et al. What constitutes cerebral palsy in the twenty-first century? Dev Med Child Neurol. 2014;56(4):323–328. doi: 10.1111/dmcn.12262 [DOI] [PubMed] [Google Scholar]
- 36.Surveillance of Cerebral Palsy in Europe. Surveillance of cerebral palsy in Europe: a collaboration of cerebral palsy surveys and registers. Surveillance of Cerebral Palsy in Europe (SCPE). Dev Med Child Neurol. 2000;42(12):816–824. doi: 10.1017/s0012162200001511 [DOI] [PubMed] [Google Scholar]
- 37.Xu Q, Wang Y, Dabdoub A, et al. Vascular Development in the Retina and Inner Ear: Control by Norrin and Frizzled-4, a High-Affinity Ligand-Receptor Pair. Cell. 2004;116(6):883–895. doi: 10.1016/S0092-8674(04)00216-8 [DOI] [PubMed] [Google Scholar]
- 38.Gilmour DF. Familial exudative vitreoretinopathy and related retinopathies. Eye. 2015;29(1):1–14. doi: 10.1038/eye.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Junge HJ, Yang S, Burton JB, et al. TSPAN12 Regulates Retinal Vascular Development by Promoting Norrin- but Not Wnt-Induced FZD4/β-Catenin Signaling. Cell. 2009;139(2):299–311. doi: 10.1016/j.cell.2009.07.048 [DOI] [PubMed] [Google Scholar]
- 40.Xia CH, Liu H, Cheung D, et al. A model for familial exudative vitreoretinopathy caused by LPR5 mutations. Hum Mol Genet. 2008;17(11):1605–1612. doi: 10.1093/hmg/ddn047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y, Yang M, Zhao R, et al. Novel truncating variants in CTNNB1 cause familial exudative vitreoretinopathy. J Med Genet. Published online March 31, 2022:jmedgenet-2021–108259. doi: 10.1136/jmedgenet-2021-108259 [DOI] [PubMed] [Google Scholar]
- 42.Panagiotou ES, Sanjurjo Soriano C, Poulter JA, et al. Defects in the Cell Signaling Mediator β-Catenin Cause the Retinal Vascular Condition FEVR. Am J Hum Genet. 2017;100(6):960–968. doi: 10.1016/j.ajhg.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pipo-Deveza J, Fehlings D, Chitayat D, Yoon G, Sroka H, Tein I. Rationale for dopa-responsive CTNNB1/ß -catenin deficient dystonia: Letters: New Observations. Mov Disord. 2018;33(4):656–657. doi: 10.1002/mds.27320 [DOI] [PubMed] [Google Scholar]
- 44.Tucci V, Kleefstra T, Hardy A, et al. Dominant β-catenin mutations cause intellectual disability with recognizable syndromic features. J Clin Invest. 2014;124(4):1468–1482. doi: 10.1172/JCI70372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fossat N, Jones V, Khoo PL, et al. Stringent requirement of a proper level of canonical WNT signalling activity for head formation in mouse embryo. Development. 2011;138(4):667–676. doi: 10.1242/dev.052803 [DOI] [PubMed] [Google Scholar]
- 46.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Lack of beta-catenin affects mouse development at gastrulation. Dev Camb Engl. 1995;121(11):3529–3537. [DOI] [PubMed] [Google Scholar]
- 47.Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W. Requirement for β-Catenin in Anterior-Posterior Axis Formation in Mice. J Cell Biol. 2000;148(3):567–578. doi: 10.1083/jcb.148.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan R, Liu P, Li F, et al. Generation of a human induced pluripotent stem cell line (SBWCHi001-A) from a patient with NEDSDV carrying a pathogenic mutation in CTNNB1 gene. Stem Cell Res. 2020;49:102091. doi: 10.1016/j.scr.2020.102091 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Where not otherwise indicated (by e.g. ClinVar accession number) all source data for this paper and supplementary information is available from the corresponding author on reasonable request. Requests for potentially identifiable data are subject to approval by the Women’s and Children’s Health Network Human Research Ethics Committee.