Abstract
MicroRNAs (miRNAs) are a class of non‐coding single‐stranded RNA molecules with a length of approximately 18‐25 nt nucleotides that regulate gene expression post‐transcriptionally. MiR‐204‐5p originates from the sixth intron of the transient receptor potential cation channel subfamily M member 3 (TRPM3) gene. MiR‐204‐5p is frequently downregulated in various cancer types and is related to the clinicopathological characteristics and prognosis of cancer patients. So far, many studies have determined that miR‐204‐5p functions as a tumor suppressor for its extensive and powerful capacity to inhibit tumor proliferation, metastasis, autophagy, and chemoresistance in multiple cancer types. MiR‐204‐5p appears to be a promising prognostic biomarker and a therapeutic target for human cancers. This review summarized the latest advances on the role of miR‐204‐5p in human cancers.
Keywords: microRNA, miR‐204‐5p, tumor suppressor, cancer, gene regulation
MiR‐204‐5p regulates tumorigenesis and progression via various target genes in human cancers.
1. INTRODUCTION
The occurrence and development of cancer are dynamic and complex and involve a variety of genetic and epigenetic mechanisms. MicroRNAs (miRNAs) are a class of small endogenous regulatory RNAs (18‐25 nt) lacking protein‐coding abilities. MiRNAs have been widely identified in multiple species and play an important role in the development and diseases. 1 By binding to the 3'untranslated regions (3'UTRs) of their mRNA targets, miRNAs participate in the formation of RNA‐induced silencing complex and inhibit the expression of target mRNAs. Now, it is clear that the base‐pairing between miRNAs and their mRNA targets could induce mRNA degradation or translational repression. 2 In addition to the classical mechanism by which miRNAs inhibit mRNA expression, some non‐coding RNAs (ncRNAs), such as long non‐coding RNAs (lncRNAs) and circular RNAs (circRNAs), also could be bound and regulated by miRNAs. MiRNAs and their RNA targets form a regulatory network named the competitive endogenous RNA (ceRNA) network. 3
MiR‐204‐5p, previously named miR‐204 in the miRBase database before the release 19.0 (https://www.mirbase.org/), 4 originates from TRPM3 intron 6. Since miR‐204‐5p was discovered, numerous studies have revealed its key role in many essential physiological or pathological processes, especially tumorigenesis and progression. In normal tissues, miR‐204‐5p has been demonstrated to play a significant role in eye development, 5 , 6 lipogenesis, 7 and osteogenesis. 8 For instance, overexpression of miR‐204‐5p in the human retinal epithelium governs eye development. MiR‐204‐5p has also been shown to stimulate the development of human adipose mesenchymal stem cells into mature adipocytes. 9 Besides, by modulating the IL6/IL6R axis, miR‐204‐5p reduces inflammation, and chemokine production in renal tubular epithelial cells. 10
Increasing pieces of evidence have expounded that miR‐204‐5p functions as a key tumor suppressor in a variety of human tumors by regulating proliferation, stemness, metastasis, apoptosis, chemoresistance, and autophagy. Additionally, clinical analyses reveal that miR‐204‐5p expression is in relation to the prognosis and clinicopathological characteristics of cancer patients. In this review, we summarized the latest advances regarding the pivotal suppressive role of miR‐204‐5p in various tumor types and discussed its potential applications in cancer therapy.
2. ABNORMAL EXPRESSION OF MIR‐204‐5P IN HUMAN CANCERS
Generally, miR‐204‐5p plays a tumor inhibitory role and decreased expression of miR‐204‐5p promotes tumorigenesis and progression. Although a limited number of studies reported that miR‐204‐5p was overexpressed in prostate cancer (PCa), 11 breast cancer (BC), 12 and ovarian cancer (OC), 13 most studies showed that the expression of miR‐204‐5p is significantly decreased in a series of cancer types, including the abovementioned PCa, 14 BC, 15 , 16 , 17 and OC. 18
The expression of miR‐204‐5p could be regulated at transcriptional and post‐transcriptional levels. Firstly, DNA methylation has been shown to epigenetically silence miR‐204 in human cancers. 19 , 20 , 21 , 22 The miR‐204 coding sequence, located in the intron 6 of TRPM3, is produced from the same transcription unit as TRPM3 and has the same transcriptional regulatory motif. 23 , 24 It has been reported that the promoter of TRPM3/miR‐204 is hypermethylated in gliomas 21 and colorectal cancer (CRC), 22 leading to silenced miR‐204 expression. Secondly, many reports have shown that some ncRNAs, including lncRNAs and circRNAs, can bind and suppress the expression and/or activity of miR‐204‐5p in oncocytes. 25 , 26 , 27 , 28 , 29 , 30 Besides, some transcriptional factors (i.e., STAT3 and TFAP2A) have also been proved to regulate miR‐204 expression in tumors. 31 , 32 , 33 , 34 , 35
3. THE ROLE OF MIR‐204‐5P IN HUMAN CANCERS
3.1. Breast cancer
BC is the most commonly diagnosed cancer worldwide, surpassing lung cancer (LC) with a calculated 2.3 million new cases. 36 A growing number of studies showed that miR‐204‐5p is decreased in BC and exerts tumor‐suppressive functions. 15 , 16 , 17 , 25 , 37 , 38 , 39 , 40 These studies have identified many miR‐204‐5p targets by which miR‐204‐5p inhibits BC development and progression. For example, SOX4, a BC stem cell‐specific marker that promotes EMT, tumor growth, and metastasis, has been established as a key target of miR‐204 in BC cells. 41 In addition, miR‐204‐5p impairs the proliferation of BC cells via targeting TGFBR2, AKT, and PI3K. 37 MiR‐204‐5p can also inhibit the progression of BC by inhibiting ANGPT1, 38 AP1S3, 42 BDNF, 43 COX5A, 40 IL‐11, 44 PIK3CB, 16 PTEN, 37 and RRM2. 25 Interestingly, Findlay et al. declared that miR‐204‐5p is upgraded in BC and facilitates tumor cell invasion and migration by targeting PDEF. 12 However, in view of the very low case number of this study (n = 5), its conclusion of miR‐204‐5p downregulation in BC is subject to deliberation.
Recent studies showed that many lncRNAs are aberrantly expressed and play significant parts in human cancers. Some lncRNAs indirectly regulate gene expression through working as ceRNAs of miRNAs. For example, in triple‐negative breast cancer, lncRNA ARNILA acts as a ceRNA for miR‐204‐5p to promote the expression of its target genes, including SOX4, BCL2, RAB22A, SIRT1, and FOXA1. 39 Interestingly, although RUNX2 was also a verified target of miR‐204‐5p, ARNILA knockdown, or overexpression had no effect on its expression in triple‐negative breast cancer, 39 suggesting that the action of ARNILA on miR‐204‐5p is insufficient to concurrently inhibit all of its target genes. In addition, the expression of RUNX2 may be controlled by other regulatory mechanisms that counterbalance the impact of ARNILA on miR‐204. 39 Wang et al. revealed that MALAT1 induces EMT phenotype and promotes metastasis via regulating the miR‐204/ZEB2 axis in BC. 45 It has also been demonstrated that DSCAM‐AS1 25 could bind miR‐204‐5p and upregulate its target RRM2, thus promoting BC growth and metastasis. Furthermore, DGUOK‐AS1 also acts as a ceRNA of miR‐204‐5p and promotes BC progression and metastasis. 46
CircRNAs are endogenous ncRNAs with a covalently closed loop. Several circRNAs have been reported to regulate the targets of miR‐204‐5p via acting as a miR‐204‐5p sponge. For example, circPVT1 was reported to promote BC growth, EMT, and invasion via inhibiting miR‐204‐5p. 29
3.2. Colorectal cancer
CRC is the third most commonly diagnosed cancer and the second leading cause of cancer death. 36 We revealed that miR‐204‐5p is significantly downregulated in CRC, which was mediated by DNA hypermethylation of its promoter. 22 We previously reported that RAB22A expression is elevated in CRC and represents an independent survival risk factor. What is more, we revealed, for the first time, that RAB22A is a key functional target of miR‐204‐5p and mediates its tumor‐inhibitory functions in CRC. 22 We also elucidated that miR‐204‐5p inhibits CRC growth, metastasis, and chemoresistance by targeting CREB1. 28 In addition, we revealed that tumor‐associated macrophages‐secreted IL‐6 induces chemoresistance through regulating the IL‐6R/STAT3/miR‐204‐5p axis in CRC cells. 34 The tumor‐inhibitory effects of miR‐204‐5p in CRC have also been reported by other groups. 26 , 47
The methylation levels of TRPM3/miR‐204 promoter in CRC tissues were much higher than in corresponding non‐cancerous tissues, and the treatment with 5‐aza‐dC (a DNA methyltransferase inhibitor) restores miR‐204‐5p expression in CRC cells, 22 suggesting DNA methylation is a key way to regulate the expression and function of miR‐204‐5p in CRC. LncRNAs also regulate miR‐204‐5p in CRC. We demonstrated, for the first time, that lncRNA—UCA1 accelerates proliferation and induces 5‐fluorouracil (5‐FU) resistance in CRC through binding to miR‐204‐5p and then increasing the expression of its targets (CREB1, BCL2, and RAB22A). 28 Besides, Lu et al. showed that DSCAM‐AS1 promotes cell proliferation and migration by regulating the miR‐204‐5p/SOX4 axis in CRC. 47 Jia et al. demonstrated that PlncRNA‐1 promotes CRC cell proliferation as well as liver metastasis by regulating the miR‐204/Wnt/β‐catenin axis. 48 The abnormal overexpression of PCAT6 restrains miR‐204 expression, thus enhancing the activity of the HMGA2/PI3K axis, and finally induces the chemotherapeutic resistance of CRC cells to 5‐ FU. 26
3.3. Prostate cancer
PCa ranks the second most common cancer in men. 36 The main threat to PCa patients is the high rate of bone metastasis. PCa patients with bone metastasis showed significantly higher levels of miR‐204‐5p in tumor tissues and serum than those without bone metastasis. 14 Functionally, several studies have confirmed that miR‐204‐5p suppresses PCa growth and metastasis. For example, Wa et al. uncovered that miR‐204‐5p inhibits PCa bone metastasis by targeting multiple targets (MAP3K3, TAB3, and TRAF1) and then inactivating NF‐κB signaling. 14 In addition, miR‐204‐5p was also verified to promote apoptosis and chemosensitivity in PCa cells by downregulating BCL2 49 and SIRT1. 50 Moreover, Ding et al. demonstrated that the androgen receptor (AR)/miR‐204/XRN1 axis has dual regulatory effects on the growth of different PCa cells. 51 Interestingly, Turner et al. confirmed the overexpression of miR‐204‐5p in PCa tissues 11 and declared that miR‐204‐5p promotes PCa cell proliferation via suppressing PDEF. However, in this study, the case number is too small (n = 5) to prove the upregulation of miR‐204‐5p in PCa.
Chemoresistance is a key factor leading to tumor relapse and poor prognosis in human cancers, including PCa. We previously revealed that lncRNA—UCA1 promotes CRC tumorigenesis and chemoresistance by binding to miR‐204‐5p and restoring the expression of its target genes. 28 Similarly, Wang et al. reported that UCA1 modulates the sensitivity of PCa cells to docetaxel by regulating the miR‐204/Sirt1 axis. 52 Other groups also showed that UCA1 promotes tumor progression by acting as a ceRNA of miR‐204 and increasing the levels of ATF2 and CXCR4 in PCa. 53 , 54 In addition, lncRNA‐NEAT1 was found to promote docetaxel resistance by sponging miR‐204‐5p and then increasing ACSL4 expression in PCa. 55
3.4. Gastric cancer (GC)
GC is the fifth most commonly diagnosed cancer. 36 Many researchers have shown that miR‐204‐5p inhibits tumorigenesis and progression through regulating multiple targets in GC, 56 , 57 , 58 , 59 , 60 including CKS1B, CXCL1, CXCL12, CXCR4, ERBB3, GPRC5A, RAB22A, USP47, and ZNF52. For example, Our group showed that miR‐204‐5p suppresses GC growth by targeting RAB22A and USP47. 58 Extensive invasion and lymphatic metastasis is a key feature of advanced GC. Zhang et al. observed, for the first time, that the levels of miR‐204‐5p significantly decreased in tumor tissue and serum samples of GC patients, especially in those with lymphatic metastasis. 60 Further functional and mechanistic investigations revealed that miR‐204‐5p inhibits GC metastasis via impairing CXCL12 and CXCR4. 60 Besides, SOX4 61 and SIRT1 62 have also been reported as miR‐204‐5p key targets in regulating EMT, anoikis resistance and metastasis in GC.
Some lncRNAs, including BCYRN1, 63 DLX6‐AS1, 64 SNHG4, 65 and LINC01234 66 have been reported to be overexpressed in GC and inhibit the anti‐tumor function of miR‐204‐5p. For example, LINC01234 sponges miR‐204‐5p to upregulate CBFB expression, promoting GC tumorigenesis. 66 In addition, Liang et al. uncovered a positive regulatory loop of DLX6‐AS1/miR‐204‐5p/OCT1 that promotes GC progression. 64 CircRNAs also regulate miR‐204‐5p activity in GC. For instance, circSLAMF6 binds miR‐204‐5p and regulates the miR‐204‐5p/MYH9 axis in GC, thus facilitating cell migration invasion and glycolysis. 67
3.5. Lung cancer
LC remains the leading cause of cancer death worldwide. 36 JAK2/STAT3 pathway is a common signaling pathway with important regulatory functions in cell proliferation, differentiation, hemopoiesis, inflammation, and embryonic development. Aberrantly increased JAK2/STAT3 activity is frequently observed in a series of cancer types, including LC. Wang et al. demonstrated that miR‐204 suppresses cell migratory and invasive capacities in non‐small cell lung cancer (NSCLC) by inhibiting JAK2. 68 Later, Liu et al. elucidated that miR‐204 inhibits angiogenesis in LC by regulating the JAK2‐STAT3 pathway. 69 Interestingly, we previously revealed that STAT3 could transcriptionally repress miR‐204‐5p in CRC, indicating the involvement of a negative feedback loop. 34 In addition, miR‐204‐5p inhibits cell proliferation, migration, and invasion in NSCLC by downregulating NUAK1 70 and SIX1. 71 Lastly, lncRNA—NEAT1 acts as a ceRNA of miR‐204‐5p to enhance the expression of NUAK1, resulting in increased cell proliferation, migration, and invasion in NSCLC. 72 Similarly, lncRNA—MALAT1 functions as a sponge of miR‐204‐5p to enhance the expression of SLUG in lung adenocarcinoma, promoting EMT and metastasis. 73
3.6. Liver cancer
Liver cancer is the sixth most common cancer, and hepatocellular carcinoma (HCC) accounts for more than 90% of primary liver cancer. 36 MiR‐204‐5p also plays a critical role in regulating the development and progression of liver cancer. In HCC, multiple genes, including BCL2, 74 NUAK1, 75 SIR,T1 76 and SIX1, 77 have been identified as miR‐204‐5p targets, through which miR‐204‐5p exerts tumor‐suppressive functions.
Interestingly, miR‐204‐5p inhibits HCC cell proliferation by inhibiting HOTTIP, an oncogenic lncRNA. 78 In addition, NEAT1 also counteracts the tumor‐inhibitory activity of miR‐204 by acting as a miR‐204 sponge in HCC. 79 SNHG6 promotes cell cycle transition and tumorigenesis in HCC by suppressing miR‐204‐5p‐mediated inhibition of E2F1. 80
In intrahepatic cholangiocarcinoma, miR‐204‐5p facilitates chemotherapeutic drug‐triggered apoptosis and inhibits proliferation via downregulating BCL2 81 and SLUG. 82 It has been shown that MALAT1 interacts with miR‐204‐5p to increase CXCR4 expression, leading to enhanced cell proliferation and invasion in hilar cholangiocarcinoma. 83 Tu et al. reported that circ_0021205 sponges miR‐204‐5p and promotes RAB22A expression, thus promoting tumorigenesis in cholangiocarcinoma. 30
3.7. Glioma
Gliomas are the most common brain tumors. Of them, glioblastoma is a diffuse, highly invasive tumor with poor clinical outcomes. Due to the promoter hypermethylation, miR‐204 was significantly downregulated in glioma. 21 , 84 Ying et al. showed that restoring the expression of miR‐204 simultaneously suppressed stem cell‐like phenotypes and migration of glioma cells by targeting SOX4 and EphB2. 21 In addition, miR‐204 suppresses the development and progression of glioma by targeting ATF2, 85 BCL2, 86 ezrin, 87 FAP‐α, 88 CYP27A1, 89 RAB22A 84 or ZEB1. 90 Several lncRNAs, including XIST, 86 UCA1, 90 and HOXD‐AS1, 91 were also reported to bind and inhibit miR‐204‐5p in glioma.
3.8. Others
Apart from the abovementioned cancer types, miR‐204‐5p also plays an inhibitory role in other human tumors, including cervical cancer, 92 osteosarcoma, 93 pancreatic cancer, 94 , 95 and renal cell carcinoma. 96 , 97 In cervical cancer, miR‐204 inhibits tumor progression via regulating cell proliferation, apoptosis and autophagy. 35 , 98 In renal cell carcinoma, SNHG4 96 and HOTAIR 97 antagonized miR‐204‐5p to accelerate tumor proliferation and invasion. MiR‐204‐5p also inhibits tumorigenesis and progression in pancreatic cancer. For example, miR‐204 suppresses proliferation, migration, and invasion in pancreatic cancer by targeting MCL‐1 94 and RACGAP1 95 ; lncRNA ZEB2‐AS1 accelerates tumor growth and invasion by regulating the miR‐204/HMGB1 axis. 27
4. MIR‐204‐5P REGULATES TUMORIGENESIS AND PROGRESSION BY TARGETING MULTIPLE KEY SIGNALING PATHWAYS
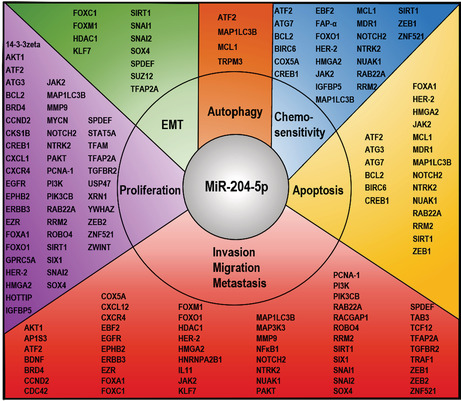
Sustaining proliferation signaling, activating invasion and metastasis, and resisting cell death are hallmarks of cancer cells. As mentioned above, as an essential tumor suppressor, miR‐204‐5p regulates cell proliferation, metastasis, invasion, autophagy, apoptosis, and chemoresistance by inhibiting dozens of target genes, demonstrating the extensive and universal functions of miR‐204‐5p (Tables 1 and 2 and Figure 1). For example, BCL2 has been identified as the target gene of miR‐204‐5p in different types of tumors, including CRC, 99 GC, 100 glioma, 86 neuroblastoma, 101 intrahepatic cholangiocarcinoma, 81 PCa 49 and HCC. 74 Next comes SOX4, which has been identified as a target of miR‐204‐5p in BC, 39 CRC, 47 GC, 61 glioma, 21 and oral squamous cell carcinoma. 102 When it comes to the number of miR‐204‐5p targets identified, to the best of our knowledge, at least 16 genes have been reported in GC. These studies indicate that miR‐204‐5p, as the core of a regulatory network, plays a tumor‐inhibitory role by regulating a large group of target genes in pan‐cancer.
TABLE 1.
Coding RNA target genes of miR‐204‐5p
| Target genes | Cancer types | Function | Reference |
|---|---|---|---|
| 14–3‐3zeta | OS | ‐proliferation | 112 |
| AKT1 | BC, ESCA | ‐proliferation and metastasis | 37, 103 |
| ANGPT1 | BC | ‐angiogenesis | 38 |
| AP1S3 | BC | ‐migration, and invasion | 42 |
| ATF2 | PCa, Glioma, CC, NSCLC | ‐proliferation, metastasis, autophagy, migration, and apoptosis | 53, 85, 98, 113 |
| ATG3 | NSCLC | ‐proliferation and apoptosis | 114 |
| ATG7 | OC | ‐apoptosis | 115 |
| BCL2 | PCa, HCC, ICC, CRC, GC, NB, Melanoma, RB, BC | ‐chemosensitivity, apoptosis, and proliferation | 49, 74, 81, 99, 100, 101 |
| BDNF | BC | ‐migration and invasion | 43 |
| BIRC6 | AML | ‐apoptosis | 116 |
| BRD4 | TSCC | ‐proliferation, migration, and invasion | 117 |
| CCND2 | RB | ‐proliferation and invasion | 118 |
| CDC42 | NPC | ‐invasion and metastasis | 31 |
| CKS1B | GC | ‐proliferation | 59 |
| COX5A | BC | ‐invasion, metastasis, and chemoresistance | 40 |
| CREB1 | CRC | ‐proliferation and apoptosis | 28 |
| CXCL1 | GC | ‐proliferation | 59 |
| CXCL12 | GC | ‐metastasis | 60 |
| CXCR4 | GC, OSCC, NPC | ‐metastasis and proliferation | 60, 119, 120 |
| E2F1 | HCC | ‐cell cycle | 80 |
| EBF2 | OS | ‐apoptosis and migration | 93 |
| EGFR | GC | ‐migration and proliferation | 121 |
| EPHB2 | Glioma, CC | ‐migration and stemness | 21, 122 |
| ERBB3 | GC | ‐invasion, proliferation, and metastasis | 57 |
| EZR | Glioma, GC | ‐proliferation, migration, and invasion | 87, 123 |
| FAP | Glioma | ‐chemoresistance | 88 |
| FOXA1 | BC | ‐proliferation, migration, invasion, and apoptosis | 124 |
| FOXC1 | EEC, LSCC | ‐metastasis, migration, invasion, and EMT | 125, 126 |
| FOXM1 | ESCA | ‐invasion and EMT | 127 |
| FOXO1 | BC | ‐different alterations of cellular activity | 128 |
| GPRC5A | GC | ‐proliferation | 59 |
| HDAC1 | HNSCC | ‐EMT | 129 |
| HER‐2 | GC | ‐proliferation, migration invasion, and apoptosis | 130 |
| HMGA2 | CRC, OSCC, THCA, ESCA | ‐chemosensitivity, proliferation, and metastasis | 131, 132, 133, 134 |
| HNRNPA2B1 | BC | ‐migration and invasion | 17 |
| HOTTIP | HCC | ‐proliferation | 78 |
| HOXA10 | AML | ‐regulation | 135 |
| IGFBP5 | PTC | ‐proliferation and apoptosis | 136 |
| IL11 | BC, ESCA | ‐metastasis and invasion | 44, 137 |
| JAK2 | NSCLC, LC, BC, HNSCC | ‐proliferation, invasion, and migration | 68, 69, 138, 139 |
| KHDRBS1 | BC | ‐self‐renewal | 140 |
| KLF7 | NSCLC | ‐migration, invasion, and EMT | 141 |
| MAP1LC3B | OC, ccRCC | ‐proliferation, chemosensitivity, and apoptosis | 115 |
| MAP3K3 | PCa | ‐invasion, migration, and metastasis | 14 |
| MCL1 | PC | ‐apoptosis and autophagy | 94 |
| MDR1 | OC | ‐apoptosis | 115 |
| MEIS1 | AML, Nephroblastoma | ‐tumorigenesis | 135, 142 |
| MET | OC | ‐cell infiltration | 143 |
| MMP9 | RB | ‐proliferation and invasion | 118 |
| MYCN | NB | ‐proliferation and tumorigenesis | 144 |
| NFκB1 | PCa | ‐invasion, migration, and metastasis | 14 |
| NOTCH2 | GBC | ‐proliferation, invasion, and apoptosis | 106 |
| NTRK2 | ESCA, NB | ‐proliferation, invasion, and chemosensitivity | 32, 101 |
| NUAK1 | NSCLC, HCC | ‐metastasis and chemosensitivity | 70, 75 |
| PAKT | BC | ‐proliferation and metastasis | 37 |
| PCNA‐1 | LC | ‐proliferation, migration, and invasion | 145 |
| PHOX2B | NB | ‐regulation | 146 |
| PI3K | BC, ESCA | ‐proliferation and metastasis | 37, 103 |
| PIK3CB | BC | ‐metastasis, proliferation, and migration | 16 |
| PTEN | BC | ‐regulation | 37 |
| PTPN11 | cSCC | ‐migration | 20 |
| RAB22A | GC, Glioma, RCC | ‐proliferation, invasion, and chemosensitivity | 58, 84, 147 |
| RACGAP1 | PDAC | ‐migration and invasion | 95 |
| ROBO4 | Bladder Cancer | ‐growth and metastasis | 148 |
| RRM2 | BC | ‐proliferation, metastasis, and apoptosis | 25 |
| RUNX2 | PCa | ‐regulation | 149 |
| SIRT1 | PCa, GC, HCC, RB | ‐proliferation, invasion, apoptosis, EMT, anoikis resistance, and chemosensitivity | 52, 62, 76, 150 |
| SIX1 | NSCLC, HCC, BC | ‐proliferation and invasion | 71, 77, 151 |
| SNAI1 | GC | ‐EMT, metastasis, and invasion | 152 |
| SNAI2 | ICC, OSCC, HNSCC | ‐metastasis, EMT, stemness, and self‐renewal | 82, 102, 129 |
| SOX4 | Glioma, BC, CRC, GC, OSCC, LC, T‐ALL, RCC | ‐stemness, proliferation, migration, invasion, metastasis, and EMT | 21, 39, 47, 61, 102, 153, 154, 155 |
| SPDEF | PCa, BC | ‐migration, invasion, metastasis, and EMT | 11, 12 |
| STAT3 | HNSCC | ‐regulation | 121 |
| STAT5A | B‐cell lymphoma | ‐proliferation | 156 |
| SUZ12 | HNSCC | ‐EMT | 121 |
| TAB3 | PCa | ‐invasion, migration, and metastasis | 14 |
| TCF12 | CC | ‐migration and invasion | 92 |
| TFAM | CRC | ‐proliferation | 157 |
| TFAP2A | CC | ‐proliferation, migration, invasion, and EMT | 35 |
| TGFBR2 | BC | ‐proliferation, migration, and angiogenesis | 38 |
| THBS1 | OC | ‐angiogenesis | 13 |
| TRAF1 | PCa | ‐invasion, migration, and metastasis | 14 |
| TRPM3 | ccRCC | ‐autophagy | 158 |
| USP47 | OC, GC | ‐proliferation | 18, 58 |
| XRN1 | PCa | ‐proliferation | 51 |
| YWHAZ | ESCA | ‐growth | 103 |
| ZEB1 | PCa, PC | ‐migration, invasion, chemosensitivity, and apoptosis | 159, 160 |
| ZEB2 | BC, HCC | ‐growth, migration, and invasion | 45, 161 |
| ZNF521 | GC | ‐apoptosis, proliferation, migration, and invasion | 56 |
| ZWINT | BC | ‐proliferation | 162 |
TABLE 2.
Non‐coding RNA targets of miR‐204‐5p
| Target genes | Cancer types | Function | Reference | |
|---|---|---|---|---|
| LncRNA | ARNILA | BC | ‐EMT, invasion, and metastasis | 39 |
| ATXN8OS | BC | ‐proliferation, viability, and invasion | 163 | |
| BANCR | Melanoma | ‐growth and invasion | 164 | |
| BCYRN1 | GC | ‐proliferation, migration, and invasion | 63 | |
| BRM | OC | ‐proliferation, migration, and invasion | 165 | |
| DGUOK‐AS1 | BC | ‐migration, angiogenesis, and metastasis | 46 | |
| DLX6‐AS1 | GC | ‐proliferation, migration, invasion, and EMT | 64 | |
| DNM3OS | Oral cancer | ‐viability and migration | 166 | |
| DSCAM‐AS1 | BC, CRC | ‐proliferation and apoptosis | 25, 47 | |
| HOTAIR | RCC, ESCA, CCA, | ‐invasion, migration, apoptosis, autophagy, and proliferation | 97, 190, 191 | |
| HOTTIP | HCC | ‐viability and proliferation | 78 | |
| HOXD‐AS1 | Glioma | ‐proliferation, migration, invasion, and cisplatin sensitivity | 91 | |
| KCNQ1OT1 | NSCLC, MSSCC | ‐proliferation, migration, and invasion | 114, 167 | |
| MALAT1 | BC, LC, HCCA, TC, HCC, GC, | ‐migration, invasion, EMT, autophagy, and proliferation | 45, 73, 83, 168, 169, 170 | |
| MIR100HG | LSCC | ‐proliferation, migration, and invasion | 171 | |
| NEAT1 | PCa, NSCLC, HCC, RB, NPC, | ‐proliferation, migration, invasion, apoptosis, EMT, radioresistance, sorafenib resistance, autophagy, and docetaxel resistance | 55, 72, 79, 172, 173 | |
| OIP5‐AS1 | LSCC | ‐proliferation, migration, invasion, and EMT | 174 | |
| PBB12 | OS | ‐proliferation and invasion | 175 | |
| PCAT6 | CRC | ‐chemoresistance | 26 | |
| PlncRNA‐1 | CRC | ‐proliferation and metastasis | 48 | |
| ROR | ESCA | ‐apoptosis | 176 | |
| SNHG1 | ESCA | ‐migration, invasion, and apoptosis | 177 | |
| SNHG4 | GC, RCC | ‐proliferation, metastasis, migration, invasion, and EMT | 65, 96, 178 | |
| SNHG6 | HCC | ‐cell cycle and proliferation | 80 | |
| LncRNA | UCA1 | CRC, PCa, Glioma, CC, AML, ESCA, PTC | ‐proliferation, invasion, docetaxel sensitivity, apoptosis, migration, and EMT | 28, 52, 53, 54, 90, 179, 180, 181, 182, 183 |
| XIST | Glioma, RB | ‐proliferation, autophagy, vincristine sensitivity, migration, and invasion and apoptosis | 86, 184 | |
| ZEB2‐AS1 | PC | ‐growth, cell cycle, and invasion | 27 | |
| LINC00518 | Melanoma | ‐metastasis | 185 | |
| LINC01234 | GC | ‐apoptosis and growth | 66 | |
| CircRNA | Circ0021205 | CCA | ‐proliferation, migration, and invasion | 30 |
| CircPVT1 | BC | ‐invasion and EMT | 29 | |
| CircSLAMF6 | GC | ‐glycolysis, migration, and invasion | 67 | |
| Circ0001971 | OSCC | ‐proliferation, migration, invasion, apoptosis, and chemosensitivity | 186 | |
| CircMTO1 | RCC | ‐proliferation, migration, invasion, and apoptosis | 187 | |
| CircNOP10 | GC | ‐proliferation, migration, and EMT | 188 | |
| Circ‐E2F3 | RB | ‐proliferation, migration, invasion, and apoptosis | 189 |
FIGURE 1.

MiR‐204‐5p regulates tumorigenesis and progression via various mRNA targets in human cancers. This figure summarizes miR‐204‐5p targets shown in Table 1 of this paper.
To better understand the role of miR‐204‐5p in human cancers, we applied the online tool Sangerbox (http://vip.sangerbox.com/) to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses using on all identified mRNA targets of miR‐204‐5p (Table 1). GO enrichment results showed these target genes mainly participated in the transcription regulation, cell proliferation, and apoptosis (Figure 2A). KEGG pathway analyses indicated that they were mainly enriched in Transcriptional Dysregulation in Cancer, PI3K/AKT and other cancer‐related pathways (Figure 2B). In addition, we constructed a PPI network by using the STRING database (https://cn.string‐db.org/) and Cytoscape 3.8.2 software. In the network, there were 84 nodes and 554 edges, with an average node degree of 13.2 and a local clustering coefficient of 0.539 (Figure 2C). In the PPI network, the core targets, including AKT1, CREB1, CDC42, SIRT1, PIK3CA, and MMP9, are key cancer‐related genes, which further indicate the important role of miR‐204‐5p in human cancers (Figure 2C).
FIGURE 2.

MiR‐204‐5p regulates multiple pathways in human cancers. (A and B) Gene ontology (GO) Analyses of miR‐204‐5p targets using the online tool Sangerbox (http://vip.sangerbox.com/). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of miR‐204‐5p targets using Sangerbox. (C) Protein–protein interaction (PPI) network was constructed using the mRNA targets of miR‐204‐5p. The web tool (https://cn.string‐db.org/) and the software Cytoscape 3.8.2 were applied to construct the PPI network.
Apoptosis is one of the most common phenotypes regulated by miR‐204‐5p and its target genes. BCL2 is a key apoptosis regulator and is frequently upregulated in cancer cells. As summarized above, BCL2, the most common target of miR‐204‐5p, was reported to be regulated by miR‐204‐5p in seven different types of tumors. In these cancer types, miR‐204‐5p enhances apoptosis and reverses chemoresistance by inhibiting BCL2 expression.
The other representative pathway enriched from miR‐204‐5p targets is PI3K/AKT which plays a critical role in cell proliferation, survival, migration, differentiation, angiogenesis, and metabolism. Due to its key role in these critical cellular processes, dysregulation of PI3K/AKT pathway is closely associated with many human diseases, especially cancers. Some members of this pathway, including the two most important factors of PI3K/AKT pathway AKT1 and PI3K, are reported to be direct targets of miR‐204‐5p in multiple cancer types. Aberrant activation of PI3K/AKT pathway due to miR‐204‐5p induces apoptosis inhibition, chemoresistance, migration, invasion, and angiogenesis in cancer cells. 37 , 103
5. POTENTIAL CLINICAL APPLICATIONS OF MIR‐204‐5P FOR CANCERS
Decreased expression of miR‐204‐5p in cancer tissues often predicts poor therapeutic effects and prognosis, strongly suggesting it as a cancer biomarker. MiR‐204‐5p has been reported as a prognostic factor in 20 types of malignancies in a TCGA‐based study. 104 Most studies indicate the protective role of overexpressed miR‐204‐5p. For example, in two independent melanoma cohorts, miR‐204 expression was associated with a better prognosis. 105 In addition, a low level of miR‐204 is related to poor prognosis in CRC, 22 GC, 100 BC 40 , and some other cancers. 31 , 106 , 107
In addition, accumulated data have confirmed the extensive anti‐cancer functions of miR‐204‐5p. Consequently, restoring the expression and tumor‐suppressive effects of miR‐204‐5p may be a promising strategy for cancer therapy. For example, drug resistance is a major obstruction to successful cancer treatment, and the strong chemotherapeutic sensitization effect of miR‐204‐5p highlight a new strategy to reverse drug resistance and improve chemotherapeutic efficacy. Many studies have confirmed that ectopic miR‐204‐5p expression increases the response of cancer cells to chemotherapeutic agents, including 5‐FU and oxaliplatin.
Due to the small molecular weight and high stability, miRNAs have been suggested as promising therapeutic molecules. Even though miRNAs have a great potential for cancer treatment, we should pay attention to it possible adverse reactions. Recently, a miR‐34a‐based clinical trial for cancer treatment was stopped by FDA on account of immune‐mediated toxicities, reflecting the importance of targeting the delivery system. 107 An ideal delivery system for miRNA‐based therapy should meet at least the following five requirements which we summarize as “three high and two low” characteristics: high affinity, high specificity, high stability, low cost, and low side effects.
Recently, different strategies, including aptamers, nanoparticles, and cell‐penetrating peptides, have been tried to deliver miRNAs. Zheng et al. developed PEGylated polymer nanoparticle for delivering miR‐204‐5p, which showed an obvious tumor‐inhibitory effect in a CRC xenograft model. 109 Fattore et al. confirmed the anti‐tumor efficiency of encapsulated miR‐204‐5p by lipid nanoparticles in melanoma. 110 Compared with these artificial materials, exosome appears to be a promising drug delivery carrier for its unique features, including low toxicity, immune compatibility, nanoscale size, and circulation stability in vivo. We showed that exosome‐encapsulated miR‐204‐5p significantly inhibits CRC growth and chemoresistance without obvious side effects. 191 In addition, due to the numerous miR‐205‐5p targets identified, when designing clinical trials, we should keep in mind that miR‐204‐5p may regulate different phenotypes in different cancer types by regulating different targets.
6. PERSPECTIVES
Due to their extensive regulatory functions for gene expression, miRNAs have been extensively studied in human diseases, especially cancers. 14 , 22 , 40 Several clinical trials have been performed to evaluate the value of miRNAs as cancer biomarkers or therapeutic targets. 40 , 109 , 110 As a pivotal tumor suppressor, miR‐204‐5p shows the latent clinical value for predicting cancer prognosis and therapeutic efficacy. However, multiple centers' clinical trials should be performed to evaluate the clinical importance of miR‐204‐5p before it is considered as a cancer biomarker.
Targets regulated by miR‐204‐5p formed a large network to affect cancer cell proliferation, metastasis, angiogenesis, apoptosis, and chemosensitivity, suggesting miR‐204‐5p as a candidate therapeutic molecule for cancers. As abovementioned, several groups have shown the broad perspective of miR‐204‐5p for cancer therapy. 109 , 110 , 111 However, to the best of our knowledge, there is still no clinical trial performed to evaluate the therapeutic efficacy of miR‐204‐5p in cancers. We hope that this review can promote further research on miR‐204‐5p to understand its biological role in human cancers and provide a theoretical foundation for the clinical application of miR‐204‐5p.
AUTHOR CONTRIBUTION
Zhaohui Huang, Fan Yang, and Zehua Bian designed and wrote the manuscript. Peiwen Xu collected and analyzed references. Fan Yang and Shengbai Sun participated in bioinformatic analyses.
FUNDING INFORMATION
This study was partially supported by grants from the Social Development Project of Jiangsu Province (BE2019632), the Six Talent Peaks Projects of Jiangsu Province (WSW‐WSW‐196), Wuxi Taihu Lake Talent Plan, and Wuxi Medical Key Discipline (ZDXK2021002).
CONFLICT OF INTEREST
No potential conflict of interest was reported by the author(s).
Yang F, Bian Z, Xu P, Sun S, Huang Z. MicroRNA‐204‐5p: A pivotal tumor suppressor. Cancer Med. 2023;12:3185‐3200. doi: 10.1002/cam4.5077
Fan Yang and Zehua Bian contributed equally to this work.
DATA AVAILABILITY STATEMENT
None.
REFERENCES
- 1. Johnson SJ, Cooper TA. Overlapping mechanisms of lncRNA and expanded microsatellite RNA. Wiley Interdiscip Rev RNA. 2021;12(1):e1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99‐110. [DOI] [PubMed] [Google Scholar]
- 3. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272‐283. [DOI] [PubMed] [Google Scholar]
- 4. Kozomara A, Birgaoanu M, Griffiths‐Jones S. MiRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155‐D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conte I, Carrella S, Avellino R, et al. MiR‐204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci USA. 2010;107(35):15491‐15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shaham O, Gueta K, Mor E, et al. Pax6 regulates gene expression in the vertebrate lens through miR‐204. PLoS Genet. 2013;9(3):e1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Divoux A, Xie H, Li JL, et al. MicroRNA‐196 regulates HOX gene expression in human gluteal adipose tissue. Obesity (Silver Spring). 2017;25(8):1375‐1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhai M, Zhu Y, Yang M, Mao C. Human mesenchymal stem cell derived exosomes enhance cell‐free bone regeneration by altering their miRNAs profiles. Adv Sci (Weinh). 2020;7(19):2001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He H, Chen K, Wang F, et al. MiR‐204‐5p promotes the adipogenic differentiation of human adipose‐derived mesenchymal stem cells by modulating DVL3 expression and suppressing Wnt/β‐catenin signaling. Int J Mol Med. 2015;35(6):1587‐1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li H, Wang J, Liu X, Cheng Q. MicroRNA‐204‐5p suppresses IL6‐mediated inflammatory response and chemokine generation in HK‐2 renal tubular epithelial cells by targeting IL6R. Biochem Cell Biol. 2019;97(2):109‐117. [DOI] [PubMed] [Google Scholar]
- 11. Turner DP, Findlay VJ, Moussa O, et al. Mechanisms and functional consequences of PDEF protein expression loss during prostate cancer progression. Prostate. 2011;71(16):1723‐1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Findlay VJ, Turner DP, Moussa O, Watson DK. MicroRNA‐mediated inhibition of prostate‐derived Ets factor messenger RNA translation affects prostate‐derived Ets factor regulatory networks in human breast cancer. Cancer Res. 2008;68(20):8499‐8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen X, Mangala LS, Mooberry L, et al. Identifying and targeting angiogenesis‐related microRNAs in ovarian cancer. Oncogene. 2019;38(33):6095‐6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wa Q, Huang S, Pan J, et al. MiR‐204‐5p represses bone metastasis via inactivating NF‐κB signaling in prostate cancer. Mol Ther Nucleic Acids. 2019;18:567‐579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W, Jin X, Zhang Q, Zhang G, Deng X, Ma L. Decreased expression of miR‐204 is associated with poor prognosis in patients with breast cancer. Int J Clin Exp Pathol. 2014;7(6):3287‐3292. [PMC free article] [PubMed] [Google Scholar]
- 16. Hong BS, Ryu HS, Kim N, et al. Tumor suppressor miRNA‐204‐5p regulates growth, metastasis, and immune microenvironment remodeling in breast cancer. Cancer Res. 2019;79(7):1520‐1534. [DOI] [PubMed] [Google Scholar]
- 17. Zhang L, Bai J, Hu Y, et al. MiR‐204 inhibits invasion and metastasis of breast cancer cells by targeted regulation of HNRNPA2B1. Nan Fang Yi Ke Da Xue Bao. 2020;40(6):869‐875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu L, Kolibaba H, Zhang S, et al. MicroRNA‐204‐5p inhibits ovarian cancer cell proliferation by down‐regulating USP47. Cell Transplant. 2019;28(1_suppl):51s‐58s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia F, Wang W, Jiang B, Chen Y, Li X. DNA methylation‐mediated silencing of miR‐204 is a potential prognostic marker for papillary thyroid carcinoma. Cancer Manag Res. 2019;11:1249‐1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toll A, Salgado R, Espinet B, et al. MiR‐204 silencing in intraepithelial to invasive cutaneous squamous cell carcinoma progression. Mol Cancer. 2016;15(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ying Z, Li Y, Wu J, et al. Loss of miR‐204 expression enhances glioma migration and stem cell‐like phenotype. Cancer Res. 2013;73(2):990‐999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin Y, Zhang B, Wang W, et al. MiR‐204‐5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20(23):6187‐6199. [DOI] [PubMed] [Google Scholar]
- 23. Harteneck C, Reiter B. TRP channels activated by extracellular hypo‐osmoticity in epithelia. Biochem Soc Trans. 2007;35(Pt 1):91‐95. [DOI] [PubMed] [Google Scholar]
- 24. Meloche J, Pflieger A, Vaillancourt M, et al. Role for DNA damage signaling in pulmonary arterial hypertension. Circulation. 2014;129(7):786‐797. [DOI] [PubMed] [Google Scholar]
- 25. Liang WH, Li N, Yuan ZQ, Qian XL, Wang ZH. DSCAM‐AS1 promotes tumor growth of breast cancer by reducing miR‐204‐5p and up‐regulating RRM2. Mol Carcinog. 2019;58(4):461‐473. [DOI] [PubMed] [Google Scholar]
- 26. Wu H, Zou Q, He H, et al. Long non‐coding RNA PCAT6 targets miR‐204 to modulate the chemoresistance of colorectal cancer cells to 5‐fluorouracil‐based treatment through HMGA2 signaling. Cancer Med. 2019;8(5):2484‐2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao H, Gong N, Ma Z, et al. LncRNA ZEB2‐AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR‐204/HMGB1 axis. Int J Biol Macromol. 2018;116:545‐551. [DOI] [PubMed] [Google Scholar]
- 28. Bian Z, Jin L, Zhang J, et al. LncRNA‐UCA1 enhances cell proliferation and 5‐fluorouracil resistance in colorectal cancer by inhibiting miR‐204‐5p. Sci Rep. 2016;6:23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bian Q, Circular RNA. PVT1 promotes the invasion and epithelial‐mesenchymal transition of breast cancer cells through serving as a competing endogenous RNA for miR‐204‐5p. Onco Targets Ther. 2019;12:11817‐11826. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Tu J, Chen W, Zheng L, et al. Circular RNA circ0021205 promotes cholangiocarcinoma progression through miR‐204‐5p/RAB22A axis. Front Cell Dev Biol. 2021;9:653207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma L, Deng X, Wu M, Zhang G, Huang J. Down‐regulation of miRNA‐204 by LMP‐1 enhances CDC42 activity and facilitates invasion of EBV‐associated nasopharyngeal carcinoma cells. FEBS Lett. 2014;588(9):1562‐1570. [DOI] [PubMed] [Google Scholar]
- 32. Bao W, Wang HH, Tian FJ, et al. A TrkB‐STAT3‐miR‐204‐5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer. 2013;12:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bishop JL, Thaper D, Zoubeidi A. The multifaceted roles of STAT3 signaling in the progression of prostate cancer. Cancers (Basel). 2014;6(2):829‐859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yin Y, Yao S, Hu Y, et al. The immune‐microenvironment confers chemoresistance of colorectal cancer through macrophage‐derived IL6. Clin Cancer Res. 2017;23(23):7375‐7387. [DOI] [PubMed] [Google Scholar]
- 35. Zhang P, Hou Q, Yue Q. MiR‐204‐5p/TFAP2A feedback loop positively regulates the proliferation, migration, invasion and EMT process in cervical cancer. Cancer Biomark. 2020;28(3):381‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 37. Fan X, Fang X, Liu G, Xiong Q, Li Z, Zhou W. MicroRNA‐204 inhibits the proliferation and metastasis of breast cancer cells by targeting PI3K/AKT pathway. J BUON. 2019;24(3):1054‐1059. [PubMed] [Google Scholar]
- 38. Flores‐Pérez A, Marchat LA, Rodríguez‐Cuevas S, et al. Dual targeting of ANGPT1 and TGFBR2 genes by miR‐204 controls angiogenesis in breast cancer. Sci Rep. 2016;6:34504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang F, Shen Y, Zhang W, et al. An androgen receptor negatively induced long non‐coding RNA ARNILA binding to miR‐204 promotes the invasion and metastasis of triple‐negative breast cancer. Cell Death Differ. 2018;25(12):2209‐2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeng J, Li G, Xia Y, et al. MiR‐204/COX5A axis contributes to invasion and chemotherapy resistance in estrogen receptor‐positive breast cancers. Cancer Lett. 2020;492:185‐196. [DOI] [PubMed] [Google Scholar]
- 41. Roukens MG, Frederiks CL, Seinstra D, et al. Regulation of a progenitor gene program by SOX4 is essential for mammary tumor proliferation. Oncogene. 2021;40(45):6343‐6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Toda H, Kurozumi S, Kijima Y, et al. Molecular pathogenesis of triple‐negative breast cancer based on microRNA expression signatures: antitumor miR‐204‐5p targets AP1S3. J Hum Genet. 2018;63(12):1197‐1210. [DOI] [PubMed] [Google Scholar]
- 43. Imam JS, Plyler JR, Bansal H, et al. Genomic loss of tumor suppressor miRNA‐204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS One. 2012;7(12):e52397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pollari S, Leivonen SK, Perälä M, Fey V, Käkönen SM, Kallioniemi O. Identification of microRNAs inhibiting TGF‐β‐induced IL‐11 production in bone metastatic breast cancer cells. PLoS One. 2012;7(5):e37361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y, Zhou Y, Yang Z, et al. MiR‐204/ZEB2 axis functions as key mediator for MALAT1‐induced epithelial‐mesenchymal transition in breast cancer. Tumour Biol. 2017;39(7):1010428317690998. [DOI] [PubMed] [Google Scholar]
- 46. Liang Y, Ye F, Wang Y, et al. DGUOK‐AS1 acts as a tumorpromoter through regulatingmiR‐204‐5p/IL‐11 axis in breast cancer. Mol Ther Nucleic Acids. 2021;26:1079‐1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu C, Xie T, Guo X, et al. LncRNA DSCAM‐AS1 promotes colon cancer cells proliferation and migration via regulating the miR‐204/SOX4 axis. Cancer Manag Res. 2020;12:4347‐4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jia GQ, Zhang MM, Wang K, Zhao GP, Pang MH, Chen ZY. Long non‐coding RNA PlncRNA‐1 promotes cell proliferation and hepatic metastasis in colorectal cancer. J Cell Biochem. 2018;119(8):7091‐7104. [DOI] [PubMed] [Google Scholar]
- 49. Lin YC, Lin JF, Tsai TF, Chou KY, Chen HE, Hwang TIS. Tumor suppressor miRNA‐204‐5p promotes apoptosis by targeting BCL2 in prostate cancer cells. Asian J Surg. 2017;40(5):396‐406. [DOI] [PubMed] [Google Scholar]
- 50. Shu Y, Ren L, Xie B, Liang Z, Chen J. MiR‐204 enhances mitochondrial apoptosis in doxorubicin‐treated prostate cancer cells by targeting SIRT1/p53 pathway. Oncotarget. 2017;8(57):97313‐97322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ding M, Lin B, Li T, et al. A dual yet opposite growth‐regulating function of miR‐204 and its target XRN1 in prostate adenocarcinoma cells and neuroendocrine‐like prostate cancer cells. Oncotarget. 2015;6(10):7686‐7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang X, Yang B, Ma B. The UCA1/miR‐204/Sirt1 axis modulates docetaxel sensitivity of prostate cancer cells. Cancer Chemother Pharmacol. 2016;78(5):1025‐1031. [DOI] [PubMed] [Google Scholar]
- 53. Zhang S, Dong X, Ji T, et al. Long non‐coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer. Am J Transl Res. 2017;9(2):366‐375. [PMC free article] [PubMed] [Google Scholar]
- 54. He C, Lu X, Yang F, et al. LncRNA UCA1 acts as a sponge of miR‐204 to up‐regulate CXCR4 expression and promote prostate cancer progression. Biosci Rep. 2019;39(5):BSR20181465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jiang X, Guo S, Zhang Y, et al. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR‐34a‐5p and miR‐204‐5p. Cell Signal. 2020;65:109422. [DOI] [PubMed] [Google Scholar]
- 56. Huan C, Xiaoxu C, Xifang R. Zinc finger protein 521, negatively regulated by microRNA‐204‐5p,promotes proliferation, motility and invasion of gastric cancer cells. Technol Cancer Res Treat. 2019;18:1533033819874783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi Y, Chen X, Xi B, et al. SNP rs3202538 in 3'UTR region of ErbB3 regulated by miR‐204 and miR‐211 promote gastric cancer development in Chinese population. Cancer Cell Int. 2017;17:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang B, Yin Y, Hu Y, et al. MicroRNA‐204‐5p inhibits gastric cancer cell proliferation by downregulating USP47 and RAB22A. Med Oncol. 2015;32(1):331. [DOI] [PubMed] [Google Scholar]
- 59. Shrestha S, Yang CD, Hong HC, et al. Integrated microRNA‐mRNA analysis reveals miR‐204 inhibits cell proliferation in gastric cancer by targeting CKS1B, CXCL1 and GPRC5A. Int J Mol Sci. 2017;19(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang J, Xing L, Xu H, et al. MiR‐204‐5p suppress lymph node metastasis via regulating CXCL12 and CXCR4 in gastric cancer. J Cancer. 2020;11(11):3199‐3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yuan X, Wang S, Liu M, et al. Histological and pathological assessment of miR‐204 and SOX4 levels in gastric cancer patients. Biomed Res Int. 2017;2017:6894675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang L, Wang X, Chen P. MiR‐204 down regulates SIRT1 and reverts SIRT1‐induced epithelial‐mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer. 2013;13:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhai H, Li Y. BCYRN1 is correlated with progression and prognosis in gastric cancer. Biosci Rep. 2019;39(11):BSR20190505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liang Y, Zhang CD, Zhang C, Dai DQ. DLX6‐AS1/miR‐204‐5p/OCT1 positive feedback loop promotes tumor progression and epithelial‐mesenchymal transition in gastric cancer. Gastric Cancer. 2020;23(2):212‐227. [DOI] [PubMed] [Google Scholar]
- 65. Cheng XB, Zhang T, Zhu HJ, et al. Knockdown of lncRNA SNHG4 suppresses gastric cancer cell proliferation and metastasis by targeting miR‐204‐5p. Neoplasma. 2021;68(3):546‐556. [DOI] [PubMed] [Google Scholar]
- 66. Chen X, Chen Z, Yu S, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR‐204‐5p in gastric cancer. Clin Cancer Res. 2018;24(8):2002‐2014. [DOI] [PubMed] [Google Scholar]
- 67. Fang X, Bai Y, Zhang L, Ding S. Silencing circSLAMF6 represses cell glycolysis, migration, and invasion by regulating the miR‐204‐5p/MYH9 axis in gastric cancer under hypoxia. Biosci Rep. 2020;40(6):BSR20201275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang P, Lv HY, Zhou DM, et al. MiR‐204 suppresses non‐small‐cell lung carcinoma (NSCLC) invasion and migration by targeting JAK2. Genet Mol Res. 2016;15(2):gmr6415. [DOI] [PubMed] [Google Scholar]
- 69. Liu X, Gao X, Zhang W, Zhu T, Bi W, Zhang Y. MicroRNA‐204 deregulation in lung adenocarcinoma controls the biological behaviors of endothelial cells potentially by modulating Janus kinase 2‐signal transducer and activator of transcription 3 pathway. IUBMB Life. 2018;70(1):81‐91. [DOI] [PubMed] [Google Scholar]
- 70. Shi L, Zhang B, Sun X, et al. MiR‐204 inhibits human NSCLC metastasis through suppression of NUAK1. Br J Cancer. 2014;111(12):2316‐2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xia Y, Zhu Y, Ma T, et al. MiR‐204 functions as a tumor suppressor by regulating SIX1 in NSCLC. FEBS Lett. 2014;588(20):3703‐3712. [DOI] [PubMed] [Google Scholar]
- 72. Zhao MM, Ge LY, Yang LF, et al. LncRNA NEAT1/miR‐204/NUAK1 axis is a potential therapeutic target for non‐small cell lung cancer. Cancer Manag Res. 2020;12:13357‐13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li J, Wang J, Chen Y, et al. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR‐204. Am J Cancer Res. 2016;6(5):1099‐1107. [PMC free article] [PubMed] [Google Scholar]
- 74. Li K, Xyu Q, Liu X, et al. Growth inhibition of human hepatocellular carcinoma by miRNA‐204 via down‐regulation of Bcl‐2 and Sirt1 expression. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2015;31(2):168‐172. [PubMed] [Google Scholar]
- 75. Yu Y, Wang Y, Xiao X, et al. MiR‐204 inhibits hepatocellular cancer drug resistance and metastasis through targeting NUAK1. Biochem Cell Biol. 2019;97(5):563‐570. [DOI] [PubMed] [Google Scholar]
- 76. Jiang G, Wen L, Zheng H, Jian Z, Deng W. MiR‐204‐5p targeting SIRT1 regulates hepatocellular carcinoma progression. Cell Biochem Funct. 2016;34(7):505‐510. [DOI] [PubMed] [Google Scholar]
- 77. Chu Y, Jiang M, Du F, et al. MiR‐204‐5p suppresses hepatocellular cancer proliferation by regulating homeoprotein SIX1 expression. FEBS Open Bio. 2018;8(2):189‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ge Y, Yan X, Jin Y, et al. MiRNA‐192 [corrected] and miRNA‐204 directly suppress lncRNA HOTTIP and interrupt GLS1‐mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 2015;11(12):e1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li X, Zhou Y, Yang L, et al. LncRNA NEAT1 promotes autophagy via regulating miR‐204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J Cell Physiol. 2020;235(4):3402‐3413. [DOI] [PubMed] [Google Scholar]
- 80. Chen K, Hou Y, Liao R, Li Y, Yang H, Gong J. LncRNA SNHG6 promotes G1/S‐phase transition in hepatocellular carcinoma by impairing miR‐204‐5p‐mediated inhibition of E2F1. Oncogene. 2021;40(18):3217‐3230. [DOI] [PubMed] [Google Scholar]
- 81. Chen L, Yan HX, Yang W, et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol. 2009;50(2):358‐369. [DOI] [PubMed] [Google Scholar]
- 82. Qiu YH, Wei YP, Shen NJ, et al. MiR‐204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell Physiol Biochem. 2013;32(5):1331‐1341. [DOI] [PubMed] [Google Scholar]
- 83. Tan X, Huang Z, Li X. Long non‐coding RNA MALAT1 interacts with miR‐204 to modulate human hilar cholangiocarcinoma proliferation, migration, and Invasion by targeting CXCR4. J Cell Biochem. 2017;118(11):3643‐3653. [DOI] [PubMed] [Google Scholar]
- 84. Xia Z, Liu F, Zhang J, Liu L. Decreased expression of miRNA‐204‐5p contributes to glioma progression and promotes glioma cell growth, migration and invasion. PLoS One. 2015;10(7):e0132399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Song S, Fajol A, Tu X, Ren B, Shi S. MiR‐204 suppresses the development and progression of human glioblastoma by targeting ATF2. Oncotarget. 2016;7(43):70058‐70065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shen J, Xiong J, Shao X, et al. Knockdown of the long noncoding RNA XIST suppresses glioma progression by upregulating miR‐204‐5p. J Cancer. 2020;11(15):4550‐4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mao J, Zhang M, Zhong M, Zhang Y, Lv K. MicroRNA‐204, a direct negative regulator of ezrin gene expression, inhibits glioma cell migration and invasion. Mol Cell Biochem. 2014;396(1–2):117‐128. [DOI] [PubMed] [Google Scholar]
- 88. Yang YN, Zhang XH, Wang YM, et al. MiR‐204 reverses temozolomide resistance and inhibits cancer initiating cells phenotypes by degrading FAP‐α in glioblastoma. Oncol Lett. 2018;15(5):7563‐7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Xin J, Zheng LM, Sun DK, Li XF, Xu P, Tian LQ. MiR‐204 functions as a tumor suppressor gene, at least partly by suppressing CYP27A1 in glioblastoma. Oncol Lett. 2018;16(2):1439‐1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liang C, Yang Y, Guan J, et al. LncRNA UCA1 sponges miR‐204‐5p to promote migration, invasion and epithelial‐mesenchymal transition of glioma cells via upregulation of ZEB1. Pathol Res Pract. 2018;214(9):1474‐1481. [DOI] [PubMed] [Google Scholar]
- 91. Zhou H, Ma Y, Zhong D, Yang L. Knockdown of lncRNA HOXD‐AS1 suppresses proliferation, migration and invasion and enhances cisplatin sensitivity of glioma cells by sponging miR‐204. Biomed Pharmacother. 2019;112:108633. [DOI] [PubMed] [Google Scholar]
- 92. Shu L, Zhang Z, Cai Y. MicroRNA‐204 inhibits cell migration and invasion in human cervical cancer by regulating transcription factor 12. Oncol Lett. 2018;15(1):161‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li M, Shen Y, Wang Q, Zhou X. MiR‐204‐5p promotes apoptosis and inhibits migration of osteosarcoma via targeting EBF2. Biochimie. 2019;158:224‐232. [DOI] [PubMed] [Google Scholar]
- 94. Chen Z, Sangwan V, Banerjee S, et al. MiR‐204 mediated loss of Myeloid cell leukemia‐1 results in pancreatic cancer cell death. Mol Cancer. 2013;12(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Khalid M, Idichi T, Seki N, et al. Gene regulation by antitumor miR‐204‐5p in pancreatic ductal adenocarcinoma: the clinical significance of direct RACGAP1 regulation. Cancers (Basel). 2019;11(3):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wu J, Liu T, Sun L, Zhang S, Dong G. Long noncoding RNA SNHG4 promotes renal cell carcinoma tumorigenesis and invasion by acting as ceRNA to sponge miR‐204‐5p and upregulate RUNX2. Cancer Cell Int. 2020;20:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ding J, Yeh CR, Sun Y, et al. Estrogen receptor β promotes renal cell carcinoma progression via regulating LncRNA HOTAIR‐miR‐138/200c/204/217 associated CeRNA network. Oncogene. 2018;37(37):5037‐5053. [DOI] [PubMed] [Google Scholar]
- 98. Li N, Guo X, Liu L, Wang L, Cheng R. Molecular mechanism of miR‐204 regulates proliferation, apoptosis and autophagy of cervical cancer cells by targeting ATF2. Artif Cells Nanomed Biotechnol. 2019;47(1):2529‐2535. [DOI] [PubMed] [Google Scholar]
- 99. Kuwano Y, Nishida K, Kajita K, et al. Transformer 2β and miR‐204 regulate apoptosis through competitive binding to 3' UTR of BCL2 mRNA. Cell Death Differ. 2015;22(5):815‐825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sacconi A, Biagioni F, Canu V, et al. MiR‐204 targets Bcl‐2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012;3(11):e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ryan J, Tivnan A, Fay J, et al. MicroRNA‐204 increases sensitivity of neuroblastoma cells to cisplatin and is associated with a favourable clinical outcome. Br J Cancer. 2012;107(6):967‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yu CC, Chen PN, Peng CY, Yu CH, Chou MY. Suppression of miR‐204 enables oral squamous cell carcinomas to promote cancer stemness, EMT traits, and lymph node metastasis. Oncotarget. 2016;7(15):20180‐20192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Shen Z, Chai T, Luo F, et al. Loss of miR‐204‐5p promotes tumor proliferation, migration, and invasion through targeting YWHAZ/PI3K/AKT pathway in esophageal squamous cell carcinoma. Onco Targets Ther. 2020;13:4679‐4690. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104. Ye ZH, Wen DY, Cai XY, et al. The protective value of miR‐204‐5p for prognosis and its potential gene network in various malignancies: a comprehensive exploration based on RNA‐seq high‐throughput data and bioinformatics. Oncotarget. 2017;8(62):104960‐104980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Galasso M, Morrison C, Minotti L, et al. Loss of miR‐204 expression is a key event in melanoma. Mol Cancer. 2018;17(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang B, Cui H, Sun Y, et al. Up‐regulation of miR‐204 inhibits proliferation, invasion and apoptosis of gallbladder cancer cells by targeting Notch2. Aging (Albany NY). 2021;13(2):2941‐2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Rajan C, Roshan VGD, Khan I, et al. MiRNA expression profiling and emergence of new prognostic signature for oral squamous cell carcinoma. Sci Rep. 2021;11(1):7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li WJ, Wang Y, Liu R, et al. MicroRNA‐34a: potent tumor suppressor, cancer stem cell Inhibitor, and potential anticancer therapeutic. Front Cell Dev Biol. 2021;9:640587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zheng B, Chen L, Pan CC, et al. Targeted delivery of miRNA‐204‐5p by PEGylated polymer nanoparticles for colon cancer therapy. Nanomedicine (Lond). 2018;13(7):769‐785. [DOI] [PubMed] [Google Scholar]
- 110. Fattore L, Campani V, Ruggiero CF, et al. In vitro biophysical and biological characterization of lipid nanoparticles co‐encapsulating oncosuppressors miR‐199b‐5p and miR‐204‐5p as potentiators of target therapy in metastatic melanoma. Int J Mol Sci. 2020;21(6):1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yao S, Yin Y, Jin G, et al. Exosome‐mediated delivery of miR‐204‐5p inhibits tumor growth and chemoresistance. Cancer Med. 2020;9(16):5989‐5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhao R, He H, Zhu Y, et al. MiR‐204/14‐3‐3ζ axis regulates osteosarcoma cell proliferation through SATA3 pathway. Pharmazie. 2017;72(10):593‐598. [DOI] [PubMed] [Google Scholar]
- 113. Zhang S, Gao L, Thakur A, et al. MiRNA‐204 suppresses human non‐small cell lung cancer by targeting ATF2. Tumour Biol. 2016;37(8):11177‐11186. [DOI] [PubMed] [Google Scholar]
- 114. Kang Y, Jia Y, Wang Q, et al. Long noncoding RNA KCNQ1OT1 promotes the progression of non‐small cell lung cancer via regulating miR‐204‐5p/ATG3 axis. Onco Targets Ther. 2019;12:10787‐10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tang J, Zhu J, Ye Y, et al. Inhibition LC3B can increase chemosensitivity of ovarian cancer cells. Cancer Cell Int. 2019;19:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang Z, Luo H, Fang Z, et al. MiR‐204 acts as a potential therapeutic target in acute myeloid leukemia by increasing BIRC6‐mediated apoptosis. BMB Rep. 2018;51(9):444‐449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zheng J, Zhang YW, Li TK, Bao Y, Zhang SX. Effect of miR‐204‐5p on the proliferation, migration, and invasion on tongue squamous cell carcinoma SCC25 cells by targeting bromodomain‐containing protein 4. Hua Xi Kou Qiang Yi Xue Za Zhi. 2020;38(2):185‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wu X, Zeng Y, Wu S, Zhong JX, Wang YZ, Xu JF. MiR‐204, down‐regulated in retinoblastoma, regulates proliferation and invasion of human retinoblastoma cells by targeting CyclinD2 and MMP‐9. FEBS Lett. 2015;589(5):645‐650. [DOI] [PubMed] [Google Scholar]
- 119. Wang X, Li F, Zhou X. MiR‐204‐5p regulates cell proliferation and metastasis through inhibiting CXCR4 expression in OSCC. Biomed Pharmacother. 2016;82:202‐207. [DOI] [PubMed] [Google Scholar]
- 120. Zong G, Han J, Yue Z, Liu Y, Cui Z, Shi L. Downregulation of miR‐204 facilitates the progression of nasopharyngeal carcinoma by targeting CXCR4 through NF‐κB signaling pathway. J BUON. 2020;25(2):1098‐1104. [PubMed] [Google Scholar]
- 121. Wang Y, Zhang H, Ge S, et al. Effects of miR‐138‐5p and miR‐204‐5p on the migration and proliferation of gastric cancer cells by targeting EGFR. Oncol Rep. 2018;39(6):2624‐2634. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 122. Duan S, Wu A, Chen Z, Yang Y, Liu L, Shu Q. MiR‐204 regulates cell proliferation and invasion by targeting EphB2 in human cervical cancer. Oncol Res. 2018;26(5):713‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lam EK, Wang X, Shin VY, et al. A microRNA contribution to aberrant Ras activation in gastric cancer. Am J Transl Res. 2011;3(2):209‐218. [PMC free article] [PubMed] [Google Scholar]
- 124. Shen SQ, Huang LS, Xiao XL, et al. MiR‐204 regulates the biological behavior of breast cancer MCF‐7 cells by directly targeting FOXA1. Oncol Rep. 2017;38(1):368‐376. [DOI] [PubMed] [Google Scholar]
- 125. Chung TK, Lau TS, Cheung TH, et al. Dysregulation of microRNA‐204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130(5):1036‐1045. [DOI] [PubMed] [Google Scholar]
- 126. Gao W, Wu Y, He X, et al. MicroRNA‐204‐5p inhibits invasion and metastasis of laryngeal squamous cell carcinoma by suppressing forkhead box C1. J Cancer. 2017;8(12):2356‐2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sun Y, Yu X, Bai Q. MiR‐204 inhibits invasion and epithelial‐mesenchymal transition by targeting FOXM1 in esophageal cancer. Int J Clin Exp Pathol. 2015;8(10):12775‐12783. [PMC free article] [PubMed] [Google Scholar]
- 128. Liang CY, Huang ZG, Tang ZQ, Xiao XL, Zeng JJ, Feng ZB. FOXO1 and hsa‐microRNA‐204‐5p affect the biologic behavior of MDA‐MB‐231 breast cancer cells. Int J Clin Exp Pathol. 2020;13(5):1146‐1158. [PMC free article] [PubMed] [Google Scholar]
- 129. Zhuang Z, Yu P, Xie N, et al. MicroRNA‐204‐5p is a tumor suppressor and potential therapeutic target in head and neck squamous cell carcinoma. Theranostics. 2020;10(3):1433‐1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yang S, Chen B, Zhang B, et al. MiR‐204‐5p promotes apoptosis and inhibits migration of gastric cancer cells by targeting HER‐2. Mol Med Rep. 2020;22(4):2645‐2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wu H, Liang Y, Shen L, Shen L. MicroRNA‐204 modulates colorectal cancer cell sensitivity in response to 5‐fluorouracil‐based treatment by targeting high mobility group protein A2. Biol Open. 2016;5(5):563‐570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wu ZY, Wang SM, Chen ZH, et al. MiR‐204 regulates HMGA2 expression and inhibits cell proliferation in human thyroid cancer. Cancer Biomark. 2015;15(5):535‐542. [DOI] [PubMed] [Google Scholar]
- 133. Zhang H, Wu X, Sui Z, et al. High‐mobility group AT‐hook 2 promotes growth and metastasis and is regulated by miR‐204‐5p in oesophageal squamous cell carcinoma. Eur J Clin Invest. 2021;51(8):e13563. [DOI] [PubMed] [Google Scholar]
- 134. Tsai SC, Huang SF, Chiang JH, et al. The differential regulation of microRNAs is associated with oral cancer. Oncol Rep. 2017;38(3):1613‐1620. [DOI] [PubMed] [Google Scholar]
- 135. Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105(10):3945‐3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Liu L, Wang J, Li X, et al. MiR‐204‐5p suppresses cell proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma. Biochem Biophys Res Commun. 2015;457(4):621‐626. [DOI] [PubMed] [Google Scholar]
- 137. Tang J, Li Z, Zhu Q, et al. MiR‐204‐5p regulates cell proliferation, invasion, and apoptosis by targeting IL‐11 in esophageal squamous cell carcinoma. J Cell Physiol. 2020;235(3):3043‐3055. [DOI] [PubMed] [Google Scholar]
- 138. Wang X, Qiu W, Zhang G, Xu S, Gao Q, Yang Z. MicroRNA‐204 targets JAK2 in breast cancer and induces cell apoptosis through the STAT3/BCl‐2/survivin pathway. Int J Clin Exp Pathol. 2015;8(5):5017‐5025. [PMC free article] [PubMed] [Google Scholar]
- 139. Wu Q, Zhao Y, Wang P. MiR‐204 inhibits angiogenesis and promotes sensitivity to cetuximab in head and neck squamous cell carcinoma cells by blocking JAK2‐STAT3 signaling. Biomed Pharmacother. 2018;99:278‐285. [DOI] [PubMed] [Google Scholar]
- 140. Wang L, Tian H, Yuan J, Wu H, Wu J, Zhu X. CONSORT: Sam68 is directly regulated by miR‐204 and promotes the self‐renewal potential of breast cancer cells by activating the Wnt/Beta‐Catenin signaling pathway. Medicine (Baltimore). 2015;94(49):e2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Liu XN, Zhang CB, Lin H, et al. MicroRNA‐204 shuttled by mesenchymal stem cell‐derived exosomes inhibits the migration and invasion of non‐small‐cell lung cancer cells via the KLF7/AKT/HIF‐1α axis. Neoplasma. 2021;68(4):719‐731. [DOI] [PubMed] [Google Scholar]
- 142. Koller K, Pichler M, Koch K, et al. Nephroblastomas show low expression of microR‐204 and high expression of its target, the oncogenic transcription factor MEIS1. Pediatr Dev Pathol. 2014;17(3):169‐175. [DOI] [PubMed] [Google Scholar]
- 143. Liu H, Li SR, Si Q. Regulation of miRNAs on c‐met protein expression in ovarian cancer and its implication. Eur Rev Med Pharmacol Sci. 2017;21(15):3353‐3359. [PubMed] [Google Scholar]
- 144. Ooi CY, Carter DR, Liu B, et al. Network modeling of microRNA‐mRNA interactions in neuroblastoma tumorigenesis identifies miR‐204 as a direct inhibitor of MYCN. Cancer Res. 2018;78(12):3122‐3134. [DOI] [PubMed] [Google Scholar]
- 145. Li P, Wang Q, Wang H. MicroRNA‐204 inhibits the proliferation, migration and invasion of human lung cancer cells by targeting PCNA‐1 and inhibits tumor growth in vivo. Int J Mol Med. 2019;43(3):1149‐1156. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146. Bachetti T, Di Zanni E, Ravazzolo R, et al. MiR‐204 mediates post‐transcriptional down‐regulation of PHOX2B gene expression in neuroblastoma cells. Biochim Biophys Acta. 2015;1849(8):1057‐1065. [DOI] [PubMed] [Google Scholar]
- 147. Xiong F, Liu K, Zhang F, et al. MiR‐204 inhibits the proliferation and invasion of renal cell carcinoma by inhibiting RAB22A expression. Oncol Rep. 2016;35(5):3000‐3008. [DOI] [PubMed] [Google Scholar]
- 148. Li Y, Chen R, Li Z, et al. MiR‐204 negatively regulates cell growth and metastasis by targeting ROBO4 in human bladder cancer. Onco Targets Ther. 2019;12:8515‐8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Todorova K, Metodiev MV, Metodieva G, Zasheva D, Mincheff M, Hayrabedyan S. MiR‐204 is dysregulated in metastatic prostate cancer in vitro. Mol Carcinog. 2016;55(2):131‐147. [DOI] [PubMed] [Google Scholar]
- 150. Ding J, Lu X. Expression of miR‐204 in pediatric retinoblastoma and its effects on proliferation and apoptosis of cancer cells. Oncol Lett. 2018;16(6):7152‐7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zeng J, Wei M, Shi R, et al. MiR‐204‐5p/Six1 feedback loop promotes epithelial‐mesenchymal transition in breast cancer. Tumour Biol. 2016;37(2):2729‐2735. [DOI] [PubMed] [Google Scholar]
- 152. Liu Z, Long J, Du R, et al. MiR‐204 regulates the EMT by targeting snai1 to suppress the invasion and migration of gastric cancer. Tumour Biol. 2016;37(6):8327‐8335. [DOI] [PubMed] [Google Scholar]
- 153. Hu WB, Wang L, Huang XR, Li F. MicroRNA‐204 targets SOX4 to inhibit metastasis of lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2019;23(4):1553‐1562. [DOI] [PubMed] [Google Scholar]
- 154. Yin JJ, Liang B, Zhan XR. MicroRNA‐204 inhibits cell proliferation in T‐cell acute lymphoblastic leukemia by down‐regulating SOX4. Int J Clin Exp Pathol. 2015;8(8):9189‐9195. [PMC free article] [PubMed] [Google Scholar]
- 155. Wu D, Pan H, Zhou Y, et al. Upregulation of microRNA‐204 inhibits cell proliferation, migration and invasion in human renal cell carcinoma cells by downregulating SOX4. Mol Med Rep. 2015;12(5):7059‐7064. [DOI] [PubMed] [Google Scholar]
- 156. Wu L, Li Y, Fan JM, et al. MicroRNA‐204 targets signal transducer and activator of transcription 5 expression and inhibits proliferation of B‐cell lymphoma cells. Mol Med Rep. 2015;11(6):4567‐4572. [DOI] [PubMed] [Google Scholar]
- 157. Wu K, He Y, Li G, Peng J. Expression and proliferative regulation of miR‐204 related to mitochondrial transcription factor A in colon cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 2015;18(10):1041‐1046. [PubMed] [Google Scholar]
- 158. Hall DP, Cost NG, Hegde S, et al. TRPM3 and miR‐204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell. 2014;26(5):738‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wu G, Wang J, Chen G, Zhao X. microRNA‐204 modulates chemosensitivity and apoptosis of prostate cancer cells by targeting zinc‐finger E‐box‐binding homeobox 1 (ZEB1). Am J Transl Res. 2017;9(8):3599‐3610. [PMC free article] [PubMed] [Google Scholar]
- 160. Zhang H, Li M, Xu X. MicroRNA‐204 attenuates the migration and invasion of pancreatic cancer cells by targeting ZEB1/EMT axis. Int J Clin Exp Pathol. 2018;11(7):3802‐3811. [PMC free article] [PubMed] [Google Scholar]
- 161. Hu B, Sun M, Liu J, Hong G, Lin Q. MicroRNA‐204 suppressed proliferation and motility capacity of human hepatocellular carcinoma via directly targeting zinc finger E‐box binding homeobox 2. Oncol Lett. 2017;13(5):3823‐3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Zhou G, Shen M, Zhang Z. ZW10 binding factor (ZWINT), a direct target of mir‐204, predicts poor survival and promotes proliferation in breast cancer. Med Sci Monit. 2020;26:e921659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Deng Z, Cai H, Lin L, et al. lncRNA ATXN8OS promotes breast cancer by sequestering miR‐204. Mol Med Rep. 2019;20(2):1057‐1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Cai B, Zheng Y, Ma S, et al. BANCR contributes to the growth and invasion of melanoma by functioning as a competing endogenous RNA to upregulate Notch2 expression by sponging miR‐204. Int J Oncol. 2017;51(6):1941‐1951. [DOI] [PubMed] [Google Scholar]
- 165. Xi J, Feng J, Zeng S. Long noncoding RNA lncBRM facilitates the proliferation, migration and invasion of ovarian cancer cells via upregulation of Sox4. Am J Cancer Res. 2017;7(11):2180‐2189. [PMC free article] [PubMed] [Google Scholar]
- 166. Fang X, Tang Z, Zhang H, Quan H. Long non‐coding RNA DNM3OS/miR‐204‐5p/HIP1 axis modulates oral cancer cell viability and migration. J Oral Pathol Med. 2020;49(9):865‐875. [DOI] [PubMed] [Google Scholar]
- 167. Sun Y, Xu C, Wu Q, Zhang L, Wang P. Long noncoding RNA KCNQ1OT1 promotes proliferation, migration, and invasion in maxillary sinus squamous cell carcinoma by regulating miR‐204/EphA7 axis. J Cell Biochem. 2020;121(4):2962‐2969. [DOI] [PubMed] [Google Scholar]
- 168. Ye M, Dong S, Hou H, Zhang T, Shen M. Oncogenic Role of Long Noncoding RNAMALAT1 in Thyroid Cancer Progression through Regulation of the miR‐204/IGF2BP2/m6A‐MYC Signaling. Mol Ther Nucleic Acids. 2021;23:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 169. Hou Z, Xu X, Zhou L, et al. The long non‐coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR‐204 and releasing SIRT1. Tumour Biol. 2017;39(7):1010428317718135. [DOI] [PubMed] [Google Scholar]
- 170. Shao G, Zhao Z, Zhao W, et al. Long non‐coding RNA MALAT1 activates autophagy and promotes cell proliferation by downregulating microRNA‐204 expression in gastric cancer. Oncol Lett. 2020;19(1):805‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Huang Y, Zhang C, Zhou Y. LncRNA MIR100HG promotes cancer cell proliferation, migration and invasion in laryngeal squamous cell carcinoma through the downregulation of miR‐204‐5p. Onco Targets Ther. 2019;12:2967‐2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Zhong W, Yang J, Li M, Li L, Li A. Long noncoding RNA NEAT1 promotes the growth of human retinoblastoma cells via regulation of miR‐204/CXCR4 axis. J Cell Physiol. 2019;234(7):11567‐11576. [DOI] [PubMed] [Google Scholar]
- 173. Lu Y, Li T, Wei G, et al. The long non‐coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR‐204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37(9):11733‐11741. [DOI] [PubMed] [Google Scholar]
- 174. Wang H, Qian J, Xia X, Ye B. Long non‐coding RNA OIP5‐AS1 serves as an oncogene in laryngeal squamous cell carcinoma by regulating miR‐204‐5p/ZEB1 axis. Naunyn Schmiedebergs Arch Pharmacol. 2020;393(11):2177‐2184. [DOI] [PubMed] [Google Scholar]
- 175. Ma T, Liu A, Xu D, Zhang T. Mechanisms underlying the promotion of osteosarcoma cell proliferation and invasion by lncRNA PBB12. Oncol Rep. 2020;43(2):736‐746. [DOI] [PubMed] [Google Scholar]
- 176. Gao H, Wang T, Zhang P, et al. Linc‐ROR regulates apoptosis in esophageal squamous cell carcinoma via modulation of p53 ubiquitination by targeting miR‐204‐5p/MDM2. J Cell Physiol. 2020;235(3):2325‐2335. [DOI] [PubMed] [Google Scholar]
- 177. Li HM, Yu YK, Liu Q, et al. LncRNA SNHG1 regulates the progression of esophageal squamous cell cancer by the miR‐204/HOXC8 axis. Onco Targets Ther. 2020;13:757‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Wang S, Zhu W, Qiu J, et al. LncRNA SNHG4 promotes cell proliferation, migration, invasion and the epithelial‐mesenchymal transition process via sponging miR‐204‐5p in gastric cancer. Mol Med Rep. 2021;23(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. He Q, Meng J, Liu S, et al. Long non‐coding RNA UCA1 upregulates KIF20A expression to promote cell proliferation and invasion via sponging miR‐204 in cervical cancer. Cell Cycle. 2020;19(19):2486‐2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Liang Y, Li E, Zhang H, et al. Silencing of lncRNA UCA1 curbs proliferation and accelerates apoptosis by repressing SIRT1 signals by targeting miR‐204 in pediatric AML. J Biochem Mol Toxicol. 2020;34(3):e22435. [DOI] [PubMed] [Google Scholar]
- 181. Jiao C, Song Z, Chen J, et al. LncRNA‐UCA1 enhances cell proliferation through functioning as a ceRNA of Sox4 in esophageal cancer. Oncol Rep. 2016;36(5):2960‐2966. [DOI] [PubMed] [Google Scholar]
- 182. Liu H, Li R, Guan L, Jiang T. Knockdown of lncRNA UCA1 inhibits proliferation and invasion of papillary thyroid carcinoma through regulating miR‐204/IGFBP5 axis. Onco Targets Ther. 2018;11:7197‐7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Li D, Cui C, Chen J, Hu Z, Wang Y, Hu D. Long non‐coding RNA UCA1 promotes papillary thyroid cancer cell proliferation via miR‐204‐mediated BRD4 activation. Mol Med Rep. 2018;18(3):3059‐3067. [DOI] [PubMed] [Google Scholar]
- 184. Yao L, Yang L, Song H, Liu TG, Yan H. Silencing of lncRNA XIST suppresses proliferation and autophagy and enhances vincristine sensitivity in retinoblastoma cells by sponging miR‐204‐5p. Eur Rev Med Pharmacol Sci. 2020;24(7):3526‐3537. [DOI] [PubMed] [Google Scholar]
- 185. Luan W, Ding Y, Ma S, Ruan H, Wang J, Lu F. Long noncoding RNA LINC00518 acts as a competing endogenous RNA to promote the metastasis of malignant melanoma via miR‐204‐5p/AP1S2 axis. Cell Death Dis. 2019;10(11):855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Tan X, Zhou C, Liang Y, Lai YF, Liang Y. Circ_0001971 regulates oral squamous cell carcinoma progression and chemosensitivity by targeting miR‐194/miR‐204 in vitro and in vivo. Eur Rev Med Pharmacol Sci. 2020;24(5):2470‐2481. [DOI] [PubMed] [Google Scholar]
- 187. Wang K, Lin X. Circular RNA circMTO1 suppressed the progression of renal cell carcinoma progression by sponging miR‐211 and miR‐204. Mamm Genome. 2022. doi: 10.1007/s00335-022-09944-1 [DOI] [PubMed] [Google Scholar]
- 188. Xu J, Wang X, Wang W, Zhang L, Huang P. Candidate oncogene circularNOP10 mediates gastric cancer progression by regulating miR‐204/SIRT1 pathway. J Gastrointest Oncol. 2021;12(4):1428‐1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Huang Y, Xue B, Pan J, Shen N. Circ‐E2F3 acts as a ceRNA for miR‐204‐5p to promote proliferation, metastasis and apoptosis inhibition in retinoblastoma by regulating ROCK1 expression. Exp Mol Pathol. 2021;120:104637. [DOI] [PubMed] [Google Scholar]
- 190. Wang AH, Tan P, Zhuang Y, Zhang XT, Yu ZB, Li LN. Down‐regulation of long non‐coding RNA HOTAIR inhibits invasion and migration of oesophageal cancer cells via up‐regulation of microRNA‐204. J Cell Mol Med. 2019;23(10):6595‐6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Lu M, Qin X, Zhou Y, et al. LncRNA HOTAIR suppresses cell apoptosis, autophagy and induces cell proliferation in cholangiocarcinoma by modulating the miR‐204‐5p/HMGB1 axis. Biomed Pharmacother. 2020;130:110566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
None.


