Abstract
The RNA interference (RNAi) triggered by short/small interfering RNA (siRNA) was discovered in nematodes and found to function in most living organisms. RNAi has been widely used as a research tool to study gene functions and has shown great potential for the development of novel pest management strategies. RNAi is highly efficient and systemic in coleopterans but highly variable or inefficient in many other insects. Differences in double-stranded RNA (dsRNA) degradation, cellular uptake, inter- and intracellular transports, processing of dsRNA to siRNA, and RNA-induced silencing complex formation influence RNAi efficiency. The basic dsRNA delivery methods include microinjection, feeding, and soaking. To improve dsRNA delivery, various new technologies, including cationic liposome–assisted, nanoparticle-enabled, symbiont-mediated, and plant-mediated deliveries, have been developed. Major challenges to widespread use of RNAi in insect pest management include variable RNAi efficiency among insects, lack of reliable dsRNA delivery methods, off-target and nontarget effects, and potential development of resistance in insect populations.
Keywords: dsRNA delivery, RNAi efficiency, RNAi pathway, systemic RNAi, pest management
INTRODUCTION
RNA interference (RNAi) refers to highly conserved cellular mechanisms in which Argonaute (Ago) family proteins bind small RNAs to trigger the degradation of longer RNAs (i.e., targets) through the sequence complementarity between the small and target RNAs (71). RNAi mechanisms were first described in the nematode (Caenorhabditis elegans) by Andrew Z. Fire, Craig C.Mello, and their colleagues (24), who received the 2006 Nobel Prize in Physiology or Medicine. RNAi has been found in all eukaryotic organisms, including protozoans, invertebrates, vertebrates, fungi, algae, and plants (1), and is believed to have initially evolved as an antiviral innate immune response (19). There is compelling evidence to support a key role for RNAi in antiviral immunity in insects (51, 112). It has been hypothesized that RNAi efficiency among insects is correlated with the prevalence of viral infections in them (112).
Since its discovery, RNAi has rapidly emerged as a powerful reverse genetics tool for studying gene function, regulation, and interaction at the cellular and organismal levels (8, 78, 110). RNAi has also shown great potential in pest management (6, 91, 98). One breakthrough was the development of transgenic maize engineered to express double-stranded RNA (dsRNA) of an essential insect gene to protect maize roots from damage caused by feeding of the western corn rootworm (Diabrotica virgifera virgifera) larvae (6). The current rapid pace of target gene identification and the development of new dsRNA delivery systems will soon lead to an increase in the use of RNAi-based technologies for the management of insect pests (44, 154).
DIFFERENT RNA INTERFERENCE PATHWAYS IN INSECTS
RNA Interference Pathways and Mechanisms
In insects and other animals, three different classes of small RNAs have been known to trigger three corresponding RNAi pathways (150). These classes of the small RNAs include short/small interfering RNAs (siRNAs) that mediate the siRNA pathway, microRNAs (miRNAs) that mediate the miRNA pathway, and P-element induced wimpy testis (Piwi)-interacting RNAs (piRNAs) that mediate the piRNA pathway (29, 78) (Figure 1). The siRNAs are dsRNAs typically 19–21 bp in length, which are processed from long dsRNA molecules that originate either exogenously or endogenously (Figure 1a). Exogenous dsRNAs include viral RNAs and experimentally introduced dsRNAs, whereas the vast majority of endogenous dsRNAs are derived from repetitive or transposable elements (e.g., transposon RNAs), structured loci, and overlapping transcripts (9, 18, 20, 25). Accordingly, siRNAs can be divided into exogenous siRNAs (exo-siRNAs) and endogenous siRNAs (endo-siRNAs) (14). In contrast, miRNAs are single-stranded RNAs 21–24 nucleotides (nt) in length (14). They are derived from endogenous primary miRNAs (pri-miRNAs) that are transcribed by RNA polymerase II from miRNA genes and constitute a large family of noncoding RNAs (49) (Figure 1b). Both siRNAs and miRNAs are generated by the type III ribonuclease Dicer (Dcr). However, piRNAs are single-stranded RNAs that are generally approximately 23–36 nt in length (124). At least two mechanisms have been identified for biogenesis of piRNAs in the fruit fly (Drosophila melanogaster) germline nurse cells, which include the de novo pathway and the ping-pong cycle (Figure 1c). In insects and other arthropods, different RNAi pathways have been found to play crucial roles, including (a) defenses against viral infections and transposable elements (transposons) via the siRNA-mediated pathway (9, 75, 113), (b) regulations of gene expression via the miRNA-mediated pathway (59, 107, 136, 137), and (c) suppression of germline transposon expression via the piRNA-mediated pathway (29, 75, 78, 81, 106, 121).
Figure 1.
Mechanisms of different RNA interference (RNAi) pathways identified in insects. (a) In the short/small interfering RNA (siRNA)-mediated pathway, long double-stranded RNA (dsRNA) is cleaved by Dcr2 in association with R2D2 in the exo-siRNA pathway or Loqs in the endo-siRNA pathway (67). After a siRNA duplex is loaded into Ago2, an RNA-induced silencing complex (RISC) is assembled from Ago2 and other RISC-associated proteins (RP). The degradation of the sense strand of the siRNA in the RISC allows the antisense strand to guide the cleavage of complementary target RNA (e.g., mRNA) by Ago2 of the RISC (69). (b) In the microRNA (miRNA) pathway, the dsRNA-specific endoribonuclease (Drosha) and its partner (Pasha) trim the primary miRNA (pri-miRNA) into hairpins to form precursor miRNA (pre-miRNA). The pre-miRNA is then sliced by Dcr1 to yield a miRNA duplex. The duplex is unwound by RNA helicase to yield mature single-stranded miRNA, which is subsequently loaded into Ago1. After the RISC components are assembled, the active Ago1 enzyme within the RISC cleaves complementary target mRNA. (c) In the piwi-interacting RNA (piRNA)-mediated pathway, de novo piRNA production begins with the transcription of long single-stranded antisense RNA called the piRNA precursor by RNA polymerase II from particular genomic sites (piRNA clusters) that contain transposable elements (106). The piRNA precursor is then cut into primary piRNA by the endonuclease Zucchini (Zuc) (65). The resulting antisense piRNA is then loaded into the Piwi family proteins, Piwi and Aub (82). The Piwi/Aub complex cleaves a sense transposon transcript, which not only degrades the target, but also creates new sense piRNA. A different Piwi family protein, Ago3, then uses the new sense piRNA to guide the cleavage of the complementary antisense transposon transcript to generate more antisense piRNA, which can be loaded into the Piwi/Aub complex to cleave additional complementary sense transposon transcripts (82).
Core Machinery Genes and Their Variations in Insect RNA Interference Pathways
The core machinery genes of the three RNAi pathways were found to undergo various duplications and deletions in insects (23, 83). In the siRNA pathway, gene duplications have been reported for Ago2, Dcr2, and/or R2D2, which encodes the dsRNA-binding protein (dsRBP) R2D2 for at least some insect species (27, 52, 76, 120). The only exception is Blattodea; however, only the German cockroach (Blattella germanica) of this order has been examined (57). Specifically, both the common house mosquito (Culex pipiens) and the house fly (Musca domestica) were reported to have two Ago2 genes (12, 52), whereas the Tsetse fly (Glossina morsitans) has three Ago2 genes (76). The red flour beetle (Tribolium castaneum) has two Ago2 and two R2D2 or R2D2-like genes (120).
In the miRNA pathway, two Ago1 genes in the yellow fever mosquito (Aedes aegypti) and the Russian wheat aphid (Diuraphis noxia) and two Dcr1 genes in M. domestica have been reported (52, 76). However, the pea aphid (Acyrthosiphon pisum) appears to have two genes for each of Ago1, Loqs, which encodes the dsRBP Loquacious (Loqs), and Dcr1, and four genes of Pasha, which encode Pasha, a partner of the dsRNA-specific endoribonuclease Drosha (15, 23, 37, 76).
In the piRNA pathway, two or three Ago3 genes have been reported in A. pisum and G. morsitans (15, 23, 76). Two Aub/Piwi genes, which encode the Piwi family proteins, Aubergine (Aub) and Piwi, in the African malaria mosquito (Anopheles gambiae) (52, 76), and 2–3 Aub genes in the jewel wasp (Nasonia vitripennis), the Florida carpenter ant (Camponotus floridanus), and the Indian jumping ant (Harpegnathos saltator) have been reported (76). However, 5–8 Piwi genes have been reported in various insect species, including A.pisum, A. aegypti, A. gambiae, and D. noxia (23, 52, 76). Duplications of core RNAi-related genes may lead to subfunctionalization or neofunctionalization in RNAi pathways, which may increase RNAi efficiency in insects, whereas loss of these genes may decrease RNAi efficiency in certain insect lineages (18, 23).
CELLULAR UPTAKE AND SYSTEMIC RNA INTERFERENCE
Mechanisms of Double-Stranded RNA Cellular Uptake
The first identified cellular uptake mechanism represents a passive transport of dsRNA through an intestinal luminal transmembrane protein called systemic RNA interference defective 2 (Sid2), where it functions as an endocytic receptor for ingested dsRNA in C. elegans (70, 128). Although no Sid2 homologs have been identified in insects (18, 38, 120), it is possible that the proteins that function in transport of macromolecules may help in cellular uptake of ingested dsRNAs. In the D. melanogaster S2 cell line and the desert locust (Schistocerca gregaria), the endocytosis-mediated pathway was identified as a mechanism for dsRNA uptake (100, 131). In T. castaneum, studies using selective endocytosis inhibitors and the RNAi of RNAi strategies showed that clathrin-dependent endocytosis is a major mechanism of dsRNA cellular uptake (134). Clathrin-dependent endocytosis has now been identified or implicated as a mechanism for cellular uptake of dsRNA in other insects or insect cells (13, 54, 86, 130, 140). Also, macropinocytosis has been implicated in cellular uptake of dsRNA in the boll weevil (Anthonomus grandis) (26). Thus, the cellular uptake of dsRNA seems to be highly complex in insects not only because different species may have different mechanisms, but also because the same species may rely on more than one mechanism (13).
Intercellular Transport of Double-Stranded RNA and Systemic RNA Interference
The highly conserved transmembrane protein Sid1 has been known to function as a dsRNA-selective channel that enables passive bidirectional transport of dsRNA between cells in C. elegans (103, 127). However, most insect species do not possess Sid1 orthologous genes, but rather only Sid1-like genes (3, 13, 45, 62, 73, 135). Most studies on insect Sid1-like genes have mainly focused on cellular uptake of dsRNA, whereas the uptake of dsRNA into alimentary canal cells is mediated by the Sid2 gene in C. elegans (128). In T. castaneum, silencing three Sid1-like genes, which share high sequence identities with another C. elegans gene (Tag130), did not affect RNAi efficiency against other genes, suggesting that these genes do not play any significant role in either cellular uptake or intercellular transport of dsRNA (120). Nevertheless, research has shown that C. elegans Sid1 expressed in Drosophila S2 cells can enable the S2 cells to gain passive uptake of dsRNA from the culture medium (103). However, the migratory locust (Locusta migratoria) Sid1-like protein expressed in Drosophila S2 cells did not enhance dsRNA uptake (62), suggesting a functional difference between the C. elegans Sid1 and L. migratoria Sid1-like proteins.
Systemic RNAi refers to the intercellular spreading of RNAi silencing throughout the body or even into its progeny after dsRNA is delivered into a specific tissue or region. In C. elegans, systemic RNAi requires Sid1-mediated transport of dsRNA between cells (103, 128). Although systemic RNAi has been widely documented in different insect species (11, 36, 62, 73, 133), the mechanism(s) that mediate systemic RNAi in insects are largely unknown (18). However, nanotube-like structures have been found to mediate systemic RNAi in D. melanogaster by transporting dsRNA and components of the RNAi machinery between adjacent cells (39). In addition, exosome-like vesicles, which originate in hemocytes and participate in immune priming, have also been found to transport siRNAs and thereby to mediate the systemic RNAi–based adaptive antiviral response (116). However, these mechanisms and their roles need to be explored in other insects.
DELIVERY OF INTERFERING RNA TO INSECTS
Various methods have been tested or applied to deliver dsRNA in insect species (38, 78, 84, 141). Basic dsRNA delivery methods include microinjection, feeding, and soaking. In recent years, however, various technologies have been developed and incorporated into these basic methods, including cationic liposome–assisted, nanoparticle-enabled, symbiont-mediated, and plant-mediated deliveries.
Basic Delivery Methods
The most commonly used method for delivery of dsRNAs is microinjection, in which a small amount of dsRNA in an appropriate solution is directly injected into an insect embryo or insect body (38, 84, 141). The first successful dsRNA microinjection experiment was reported for silencing the frizzled and frizzled 2 genes in the wingless pathway by injecting their corresponding dsRNAs into D. melanogaster embryos in 1998 (40). Since then, microinjection has become a popular technique to deliver dsRNA into the embryos or bodies of various insect species, such as T. castaneum (58, 120), L. migratoria (53, 142, 145), and B. germanica (33, 68). The advantages of microinjection include the immediate and direct delivery of a known amount of dsRNA into the insect body at various developmental stages or even to specific body parts (102, 145). This method can also avoid the structural barriers, such as the integument, that prevent penetration of dsRNA and that are encountered by other delivery methods, such as topical applications and soaking. However, microinjection is more time-consuming and is sometimes challenging due to the small body size of the insects.
Another basic delivery method is to feed insects a diet containing synthetic or microorganism-expressed dsRNA (i.e., oral delivery) (141). The microorganisms that have been used to express dsRNAs for insect RNAi include Escherichia coli (119, 153) and Saccharomyces cerevisiae (77). Oral delivery of dsRNA is highly effective for RNAi in many coleopterans (6, 42, 84, 94). It is not laborious, is easy to perform, and is more applicable to insect pest management. However, oral delivery results in low RNAi efficiency in many other insect species. In L. migratoria, for example, RNAi efficiency is high if dsRNA is delivered by injection but extremely low if it is delivered by feeding (107). Such differential RNAi efficiencies between the two dsRNA delivery methods appear to be mainly due to dsRNA degradation by double-stranded ribonucleases (dsRNases) in the insect gut. In addition, supplementing dsRNA into the cell culture medium is a method widely applied to silence gene expressions in insect cell cultures. Applications of this deliverymethod have been extensively reviewed (129).
Cationic Liposome–Assisted Delivery
Cationic liposomes are lipid-based transfection reagents that can provide a simple and efficient means to transfer dsRNA into animal cells in a process known as lipofection (48). The interactions of positively charged cationic lipids and negatively charged dsRNA can form lipoplexes through electrostatic interactions (155). Cationic liposomes have been widely used to assist in the delivery of dsRNA in insect cell cultures (129). However, several studies have also reported an increase in RNAi efficiency after feeding or injecting the RNAi-recalcitrant arthropod species (e.g., D. melanogaster, Drosophila suzukii, Rhipicephalus haemaphysaloides) with the lipoplexes (114, 126, 148). However, different cationic liposomes have not been thoroughly evaluated in conjunction with basic delivery methods.
Nanoparticle-Enabled Delivery
The first proof-of-concept study on nanoparticle-enabled delivery used chitosan/dsRNA nanoparticles to silence two chitin synthase genes in A. gambiae larvae, for which the naked dsRNAs were ineffective with feeding as the delivery method (147). Chitosan is made up of highly deacetylated β-1,4-linked N-acetylglucosamine polymers and can form nanoparticles with dsRNA based on the electrostatic interactions between positively charged amino groups of chitosan and negatively charged phosphate groups of dsRNA. The increase in RNAi efficiency when chitosan/dsRNA nanoparticles were used is likely due to an increase in dsRNA stability and cellular uptake in insects (147). Li-Byarlay et al. (56) also developed a unique method to treat a large number of honey bees (Apis mellifera) quickly and noninvasively by coupling siRNA to perfluorocarbon-nanoparticles (PFC-NPs) for aerosol application. To date, chitosan and various other nanocarrier-based dsRNA nanoparticles have been used to improve RNAi efficiency in other insect species (21, 31, 93, 146). More recently, guanidine-containing polymers, nanocarrier/dsRNA/detergent formulation, and branched amphiphilic peptide bilayer conjugated gold nanoparticles have been reported to protect dsRNA against nucleolytic degradation (115), facilitate dsRNA penetration through the insect body wall (149), and likely improve the cellular uptake and endosomal escape of dsRNA (79), respectively.
Symbiont-Mediated Delivery
Many insect species frequently harbor culturable symbiotic bacteria in their guts. Whitten & Dyson (125) used an intriguing strategy to create RNase III–deficient, dsRNA-producing bacterial strains from the symbionts of the kissing bug (Rhodnius prolixus) and the western flower thrips (Frankliniella occidentalis). Once ingested by the host insect, the genetically manipulated symbiotic bacteria can colonize the insect, which allows constitutive synthesis of dsRNA to trigger the RNAi pathway. Furthermore, the dsRNA-producing symbionts can be horizontally transmitted to other individuals via ingestion of feces in R. prolixus. The conspecific spread of such dsRNA-producing symbiont bacteria through an insect colony represents sustained RNAi and can be used to study gene function and develop new RNAi-based pest management strategies.
Plant-Mediated Delivery
The first proof-of-concept study on plant-mediated delivery of dsRNA for insect pest management was reported by Baum and colleagues in 2007 (6); they developed transgenic maize expressing an insecticidal V-ATPase A dsRNA to protect maize roots from feeding damage by D. v. virgifera larvae. Similar studies have also been carried out to generate transgenic plants expressing dsRNAs targeting insect detoxification genes (e.g., cytochrome P450 genes). The target insects feeding on these plants demonstrated an increase in susceptibility to phytotoxins (66) or to spider predation (46). In plants, however, most of the expressed long dsRNA can be processed into siRNAs. This process can reduce RNAi efficiency because insect cells take up siRNAs much less efficiently than do long dsRNAs (11). To overcome this problem, researchers generated transgenic potato plants expressing long dsRNA in the chloroplasts (plastids), where long dsRNA is accumulated but is not processed into siRNA, as chloroplasts are devoid of the RNAi machinery. When the Colorado potato beetle (Leptinotarsa decemlineata) ingest transgenic potato plants, the long dsRNA expressed in the chloroplasts can be taken up by the insect gut cells to initiate the RNAi process (144).
MECHANISMS AFFECTING RNA INTERFERENCE EFFICIENCY
Variations in RNA Interference Efficiency Among Insect Groups
RNAi efficiency varies considerably among insects. RNAi is highly efficient and systemic in coleopterans, especially those belonging to the families Tenebrionidae (e.g., T. castaneum) and Chrysomelidae (e.g., L. decemlineata and D. v. virgifera). In general, RNAi functions well in beetles; injection or oral delivery of dsRNA has been shown to induce systemic and robust RNAi response in many economically important coleopterans (10, 47, 63, 89, 90, 96, 97). RNAi has been shown to function efficiently for knockdown of target gene expression after injection of dsRNA into hemimetabolous insects such as the milkweed bug (Oncopeltus fasciatus) (34), the brown marmorated stink bug (Halyomorpha halys) (74), the bed bug (Cimex lectularius) (151), B. germanica (17), S. gregaria, and L. migratoria (61, 133). RNAi efficiency is variable in insects belonging to the orders Lepidoptera, Diptera, Hymenoptera, and Hemiptera, which include major pests, disease vectors, and beneficial insects (15, 28, 72, 104, 117). Furthermore, RNAi efficiency varies among insects even within the same order. For example, in the order Hemiptera, RNAi has been reported to be effective in Cimex, Halyomorpha, and Oncopeltus but not highly effective in Acyrthosiphon.
Differences in Stability of Interfering RNA
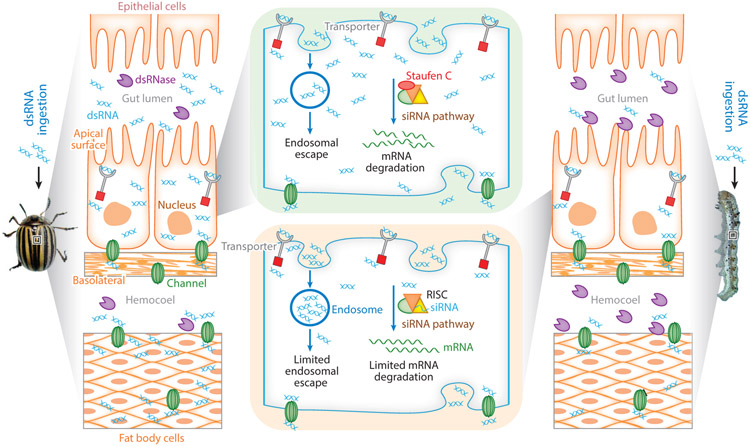
Nucleases, such as dsRNases produced by host cells, have been identified and shown to degrade dsRNA (2, 108, 132). In L. migratoria, injected, but not ingested, dsRNA triggers efficient RNAi (61). dsRNases produced in the alimentary canal and capable of degrading dsRNA have been identified in this and other insects (108). Degradation of dsRNA by nucleases is a major problem in hemipterans such as aphids (15). In L. migratoria and L. decemlineata, silencing of genes encoding dsRNases by injection of dsRNA targeting these genes improved RNAi triggered by orally delivered dsRNA (108, 109). Also, knockdown in expression of genes coding for dsRNases improved RNAi in other insects including aphid and whitefly (16, 60). Comparative studies on the degradation of dsRNA by nucleases in insects that showed variable RNAi efficiency revealed significant differences in their nuclease activity. In general, lepidopterans and hemipterans showed higher nuclease activity compared to that in other insects (105, 123). Comparison of dsRNA degradation by nucleases from L. decemlineata (highly efficient systemic RNAi) and the tobacco budworm (Heliothis virescens) (inefficient RNAi) showed that the dsRNA-degrading nucleases are much more active in H. virescens than in L. decemlineata (104) (Figure 2).
Figure 2.
Comparison of uptake, intracellular transport, and processing of dsRNA in coleopteran (RNAi-efficient) and lepidopteran (RNAi-inefficient) insects. In coleopterans (left), the ingested dsRNA is taken up by gut epithelial cells through clathrin-dependent endocytosis and other dsRNA transporters. The dsRNA is then released into the cytoplasm, and there is no major accumulation of dsRNA in endosomes (104). The dsRNA in the cytoplasm is processed to siRNA to trigger RNAi. Some of the dsRNA is exported to the hemocoel and taken by tissues such as fat body, where RNAi is induced. In contrast, in lepidopterans, some of the ingested dsRNA is degraded by dsRNases, and the rest is taken up by gut epithelial cells. Most of the dsRNA is trapped by the endosomes and is therefore not efficiently processed to siRNA, resulting in inefficient RNAi (138). Some of the dsRNA may also be exported to the hemocoel, but the dsRNases in the hemocoel degrade most of the dsRNA, resulting in inefficient transport into tissues such as fat body. In addition, an RNA binding protein, Staufen C, that is involved in processing dsRNA to siRNA for initiating RNAi, has so far been detected only in coleopterans and not in other insects (139). Abbreviations: dsRNA, double-stranded RNA; RISC, RNA-induced silencing complex; RNAi, RNA interference; siRNA, short/small interfering RNA.
Differences in Cellular Uptake and Intracellular Transport of Double-Stranded RNA
As discussed in the section titled Cellular Uptake and Systemic RNA Interference, Sid2 and Sid1 mediate gut cellular uptake and intercellular transport of dsRNA, respectively, in C. elegans. However, the proteins that function as dsRNA channels and transporters, similar to SID proteins of C. elegans, have not been identified in insects. Studies on insects showed that clathrin-dependent endocytosis and macropinocytosis might be involved in dsRNA uptake (100, 131, 134). Differences in the uptake of dsRNA among tissues in an insect have been reported. For example, in L. migratoria, the injected dsRNA is not taken up by the follicle cells and oocytes, and thus RNAi is not triggered in these tissues (95). The dsRNA taken up by cells through clathrin-dependent and other endocytic pathways is transported through endosomes to these cytoplasmic sites. In coleopterans, such as T. castaneum and L. decemlineata, where RNAi is efficient, the dsRNA is delivered to the cytoplasm, where it is converted to siRNA (104). In contrast, in lepidopterans, such as H. virescens and the fall armyworm (Spodoptera frugiperda), where RNAi does not work efficiently, the intracellular transport is inefficient, and most of the dsRNA is not able to escape endosomes and thus is not converted to siRNA (104, 138) (Figure 2).
Differences in Processing of Double-Stranded RNA to Short/Small Interfering RNA
Upon entering cells, dsRNA is transported into the cytoplasm and processed to siRNA by Dicers (43, 50, 64, 85, 92). The processing of dsRNA to siRNA varies among insects. For example, in cells and tissues of L. decemlineata where RNAi is efficient, the 32P-labeled dsRNA is processed to siRNA (104). In contrast, in cells and tissues of S. frugiperda, the 32P-labeled dsRNA is not efficiently processed to siRNA (104). Comparative analysis of processing of dsRNA to siRNA in insects belonging to suborder Heteroptera and orders Lepidoptera, Orthoptera, and Diptera showed that the dsRNA was processed to siRNA but not as efficiently as in coleopterans (105). Differences in the structure and activity of Dicers and their substrate specificity could contribute to differences in processing of dsRNA to siRNA among insects. However, further studies are needed to verify this hypothesis. In addition, recent studies showed that an RNA binding protein, Staufen C, that is involved in processing dsRNA to siRNA is detected only in coleopterans and not in other insects (139). The function of RNA binding proteins such as Staufen C also needs further investigation.
Differences in Argonaute Proteins
Recent studies on the evolution of Ago genes in insects suggest that these genes undergo frequent evolutionary expansions resulting in their functional divergence (23, 52). The number of Ago genes varies among insects. For example, four members of Ago genes coding for Ago1, Ago2, Ago3, and Piwi were identified in the silkworm (Bombyx mori) genome (122). In D. melanogaster, five genes coding for Ago1, Ago2, Ago3, Aub, and Piwi were identified (52, 111). Studies in D. melanogaster showed that Ago1 and Ago2 participate in siRNA and miRNA pathways, whereas Ago3, Aub, and Piwi interact with piRNAs (14, 30, 106). B. germanica seems to use both Ago1 and Ago2 in the miRNA pathway, but uses only Ago2 in the siRNA pathway (99). Interestingly, silencing of not only Ago2, but also Ago1 and Aub genes blocks dsRNA-triggered RNAi in the L. decemlineata cell line (140). In addition, Ago2 is duplicated in this insect, and both Ago2 genes are required for dsRNA-triggered RNAi (140). Enhancement of RNAi efficiency after expression of Ago2 in B. mori suggests that Ago2 may be one of the factors contributing to lower RNAi efficiency in lepidopterans (55).
Physiological Conditions Affecting RNA Interference Efficiency: Expression of Genes Coding for RNA Interference Pathway Components
Studies in D. melanogaster showed that feeding naked dsRNA did not trigger RNAi efficiently, but adding a transfection reagent such as Lipofectamine improved response to fed dsRNA, suggesting that the fruit fly lacks mechanisms for efficient uptake of dsRNA from the lumen (126). The physiological conditions of the target insect could influence RNAi efficiency. For example, the pH of the lumen, especially the highly alkaline pH observed in the lepidopterans, could be detrimental to successful RNA (22). In some lepidopterans, such as B. mori and the tobacco hornworm (Manduca sexta), the dsRNA triggers RNAi more efficiently during the nonfeeding wandering stage of the final instar larvae than during the feeding stages of the larvae. This may reflect changes in expression of genes involved in dsRNA degradation or RNAi response, as well as the physiological state of target tissues. For example, declines in food intake and digestion and thus in the titers of the dsRNA-degrading enzymes may enhance RNAi efficiency during the wandering stage.
OPPORTUNITIES AND CHALLENGES OF RNA INTERFERENCE
RNA Interference as a Powerful Research Tool
Before the discovery of RNAi, the analysis of gene function was limited to a few model insects such as Drosophila. RNAi has helped researchers perform large-scale functional genomics studies in many model insects, as well as in economically important non-model insects. This led to significant advances in our understanding of insect biology. Recent studies on genome and transcriptome sequencing and proteomic analyses identified thousands of genes, mRNAs, and proteins in hundreds of insect species. RNAi played a critical role in uncovering the function of the genes, RNA, and proteins identified by these studies (7). These advances facilitated an increase in our understanding of the developmental, physiological, and molecular processes that govern every aspect of insect life (32). Indeed, about 1,300 papers published during the past two decades studied RNAi or employed RNAi to investigate gene function in Drosophila. In addition, research in other model insects, such as A. aegypti, A. gambiae, B. mori, Locusta sp., Spodoptera sp., T. castaneum, the brown planthopper (Nilaparvata lugens), the cotton bollworm (Helicoverpa armigera), and the western honey bee (Apis mellifera), used RNAi methods to learn about the molecular mechanisms of physiological processes that are specific to each group of insects. RNAi methods also helped to identify targets for insecticide discovery and make significant advances in our understanding of the mode of action of insecticides and molecular mechanisms of insecticide resistance (4, 151, 152).
RNA Interference for Development of Novel Pest Management Methods
Soon after the discovery of RNAi and development of its methods for gene silencing, entomologists initiated research to apply this technology to pest management. The US Environmental Protection Agency and other regulatory agencies have approved commercial products based on RNAi technology to control D. v. virgifera. Products for management of L. decemlineata and canola flea beetles are under development. Successful knockdown of target gene expression in the insects feeding on plants expressing dsRNA has been demonstrated in H. armigera (80), L. decemlineata (144), M. sexta (88), N. lugens (143), the green peach aphid (Myzus persicae) (87), and the silverleaf whitefly (Bemisia tabaci) (118). Although several publications document successful knockdown in expression of genes by dsRNA in lepidopterans and hemipterans, whether the knockdown reaches the levels required for the successful development of commercial products needs further research. dsRNA targeting house-keeping genes in L. decemlineata produced in E. coli and delivered as a foliar application of heat-killed bacteria causes significant insect mortality (101, 153). This approach may be further developed for controlling this insect pest in the field.
Challenges in Widespread Use of RNA Interference Products
The major challenges to widespread use of RNAi to manage insect pests include the cost of dsRNA triggers, efficient delivery of dsRNA to the site of action, off-target and nontarget effects of dsRNA, and potential resistance development. Whyard et al. (126) fed in vitro synthesized dsRNA to insects and successfully silenced target genes; this suggests that it is possible to achieve silencing of target genes by feedings dsRNA. However, achieving insect control by feeding of dsRNA is challenging due to the lack of cost-effective methods for producing large quantities of dsRNA. One could produce large quantities of dsRNA using cell-free synthesis methods. Hunter et al. (35) successfully field-tested dsRNA produced using such methods to control Israeli acute paralysis virus in A. mellifera. Effective delivery of dsRNA to the site of action is another major hurdle faced by RNAi applications in agriculture and medicine. The half-life of naked siRNA in serum ranges from several minutes to about an hour (5). It is difficult to achieve efficient RNAi if the trigger is degraded rapidly in an organism unless a high dose of dsRNA or siRNA is applied.
Another significant roadblock to RNAi that is preventing aggressive investment in the development of dsRNA-based products for pest management is the concern that insects may evolve resistance to dsRNA-based products, as they do for conventional pesticides. Insects could use several different mechanisms to develop resistance to these products. These include mutations of target genes, mutations of RNAi core machinery genes, enhanced dsRNA degradation, increased dsRNA excretion, decreased dsRNA uptake and transport, and decreased dsRNA endosomal escape. Some of the resistance mechanisms could be mitigated more easily than the others. For example, target site mutations could be managed by employing a dsRNA that targets a different gene. However, if insects develop resistance to dsRNA products through mutations or change in expression of a critical gene required for RNAi mechanisms, management of resistance could become a difficult task. Recent studies on resistance development in D. v. virgifera to Snf7 dsRNA (41) and the L. decemlineata cell line to IAP dsRNA (139) showed that the resistance is not due to mutations in the dsRNA target site. Changes in the expression of proteins involved in uptake and intracellular transport of dsRNA are thought to be the major mechanisms for the resistance.
SUMMARY POINTS.
RNAi is triggered by the siRNA, miRNA, and piRNA pathways in insects.
Different cellular uptake mechanisms for dsRNA have been found or implicated, whereas the mechanisms of intercellular transport of dsRNA and systemic RNAi are largely unknown in insects.
Basic dsRNA delivery methods include microinjection, feeding, and soaking, but various new technologies have been developed and incorporated into the basic methods; these new technologies include cationic liposome–assisted, nanoparticle-enabled, symbiont-mediated, and plant-mediated deliveries.
Mechanisms affecting RNAi efficiency among insects include dsRNA degradation, cellular uptake and intracellular transport of dsRNA, efficiency in the processing of dsRNA to siRNA, the difference in RNA-induced silencing complex formation, and various physiological factors.
RNAi has provided not only powerful tools to study the developmental, physiological, and molecular processes that govern every aspect of insect life, but also great opportunities to develop new methods for insect pest management.
Major challenges to widespread use of RNAi for insect pest management include variable RNAi efficiency among insects, lack of reliable dsRNA delivery methods, off-target and nontarget effects, and potential resistance development.
FUTURE ISSUES.
What are the mechanisms governing cellular uptake, inter- and intracellular transport of dsRNA, and systemic RNAi?
What are the exact mechanisms causing significant differential RNAi responses among different taxonomic groups of insects?
How do duplication or deletion and upregulation or downregulation of the RNAi core machinery genes affect RNAi efficiency?
What are practical strategies to enhance RNAi efficiency by increasing the stability, cellular uptake, inter- and intracellular transport, and endosomal escape of dsRNA?
How can off-target and nontarget effects of dsRNA and potential development of resistance to RNAi be identified and managed?
ACKNOWLEDGMENTS
The authors acknowledge that only a fraction of more than 3,000 papers published on RNAi in insects could be included in this review due to space restrictions. We particularly thank Zhitao Yu for help in organizing the references. Relevant research in the Zhu laboratory is supported by the US Department of Agriculture (USDA) National Institute of Food and Agriculture (NIFA), including the Foundational Program (2014–67013–21714), Hatch (371324), and Hatch Multistate (372103) projects. Other support comes from a collaborative grant funded by the National Natural Science Foundation of China (NSFC31730074). Research in the Palli laboratory is supported by grants from the National Institutes of Health (GM070559–13 and 1R21AI131427–01), the National Science Foundation (Industry/University Cooperative Research Centers, the Center for Arthropod Management Technologies under grant IIP-1821936), and the USDA/NIFA (under Hatch Project 2351177000 and Agriculture and Food Research Initiative Competitive Grant 2019-67013-29351). This manuscript is contribution No. 19–208-J from the Kansas Agricultural Experiment Station, Kansas State University, Manhattan, Kansas.
Glossary
- RNA interference (RNAi)
small RNA–mediated downregulation of gene expression
- Argonaute (Ago)
the main siRNA-induced silencing complex component aiding in target recognition and cleavage during RNAi
- Short/small interfering RNA (siRNA)
double-stranded, approximately 19–21-base-pair RNA processed from long dsRNA by Dicer
- MicroRNA (miRNA)
single-stranded, approximately 21–24-nucleotide-long RNA processed from hairpin RNA precursors by Dicer
- P-element induced wimpy testis (Piwi)
highly conserved RNA-binding proteins
- Piwi-interacting RNA (piRNA)
single-stranded, approximately 23–36-nucleotide-long RNA processed mainly from transposons by Dicer-independent processes
- Dicer 2 (Dcr2)
RNase III nuclease family enzyme that cuts long dsRNA into siRNA
- R2D2
a dsRBP that forms the Dicer 2/R2D2 complex
- dsRNA-binding protein (dsRBP)
a class of proteins responsible for recognizing dsRNA
- Dicer 1 (Dcr1)
RNase III nuclease family enzyme that cuts precursor microRNA (pre-miRNA) to yield the miRNA duplex
- Aubergine (Aub)
a Piwi family protein that binds RNA
- Systemic RNA interference defective 2 (Sid2)
transmembrane protein involved in cellular uptake or cell-to-cell transport of dsRNA in C. elegans
- Systemic RNAi
a process in which local administration of dsRNA leads to RNAi responses in the whole body or even its progeny
- dsRNase
double-stranded ribonuclease capable of degrading dsRNA
- Nanocarrier
nanomaterial used as a transport module for other substances, such as RNA
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. 2003. RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev 67:657–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arimatsu Y, Kotani E, Sugimura Y, Furusawa T. 2007. Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol 37:176–83 [DOI] [PubMed] [Google Scholar]
- 3.Aronstein K, Pankiw T, Saldivar E. 2006. SID-1 is implicated in systemic gene silencing in the honey bee. J. Apic. Res 45:20–24 [Google Scholar]
- 4.Bai H, Palli SR. 2016. Identification of G protein-coupled receptors required for vitellogenin uptake into the oocytes of the red flour beetle, Tribolium castaneum. Sci. Rep 6:27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett DW, Davis ME. 2007. Effect of siRNA nuclease stability on the in vitro and in vivo kinetics of siRNA-mediated gene silencing. Biotechnol. Bioeng 97:909–21 [DOI] [PubMed] [Google Scholar]
- 6.Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, et al. 2007. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol 25:1322–26 [DOI] [PubMed] [Google Scholar]
- 7.Bellés X 2009. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol 55:111–28 [DOI] [PubMed] [Google Scholar]
- 8.Ben-Amar A, Daldoul S, Reustle GM, Krczal G, Mliki A. 2016. Reverse genetics and high throughput sequencing methodologies for plant functional genomics. Curr. Genom 17:460–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biryukova I, Ye T. 2015. Endogenous siRNAs and piRNAs derived from transposable elements and genes in the malaria vector mosquito Anopheles gambiae. BMC Genom. 16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodemann RR, Rahfeld P, Stock M, Kunert M, Wielsch N, et al. 2012. Precise RNAi-mediated silencing of metabolically active proteins in the defense secretions of juvenile leaf beetles. Proc. Biol. Sci 279:4126–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolognesi R, Ramaseshadri P, Anderson J, Bachman P, Clinton W, et al. 2012. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLOS ONE 7:e47534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell CL, Black WC, Hess AM, Foy BD. 2008. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genom 9:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappelle K, de Oliveira CFR, Van Eynde B, Christiaens O, Smagghe G. 2016. The involvement of clathrin-mediated endocytosis and two Sid-1-like transmembrane proteins in double-stranded RNA uptake in the Colorado potato beetle midgut. Insect Mol. Biol 25:315–23 [DOI] [PubMed] [Google Scholar]
- 14.Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christiaens O, Swevers L, Smagghe G. 2014. dsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 53:307–14 [DOI] [PubMed] [Google Scholar]
- 16.Chung SH, Jing X, Luo Y, Douglas AE. 2018. Targeting symbiosis-related insect genes by RNAi in the pea aphid–Buchnera symbiosis. Insect Biochem. Mol. Biol 95:55–63 [DOI] [PubMed] [Google Scholar]
- 17.Ciudad L, Piulachs MD, Bellés X. 2006. Systemic RNAi of the cockroach vitellogenin receptor results in a phenotype similar to that of the Drosophila yolkless mutant. FEBS J. 273:325–35 [DOI] [PubMed] [Google Scholar]
- 18.Cooper AM, Silver K, Zhang JZ, Park Y, Zhu KY. 2019. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci 75:18–28 [DOI] [PubMed] [Google Scholar]
- 19.Cullen BR. 2014. Viruses and RNA interference: issues and controversies. J. Virol 88:12934–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453:798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das S, Debnath N, Cui Y, Unrine J, Palli SR. 2015. Chitosan, carbon quantum dot, and silica nanoparticle mediated dsRNA delivery for gene silencing in Aedes aegypti: a comparative analysis. ACS Appl. Mater. Interfaces 7:19530–35 [DOI] [PubMed] [Google Scholar]
- 22.Dow JA. 1992. pH gradients in lepidopteran midgut. J. Exp. Biol 172:355–75 [DOI] [PubMed] [Google Scholar]
- 23.Dowling D, Pauli T, Donath A, Meusemann K, Podsiadlowski L, et al. 2016. Phylogenetic origin and diversification of RNAi pathway genes in insects. Genome Biol. Evol 8:3784–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–11 [DOI] [PubMed] [Google Scholar]
- 25.Ghildiyal M, Seitz H, Horwich MD, Li CJ, Du TT, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320:1077–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillet FX, Garcia RA, Macedo LLP, Albuquerque EVS, Silva MCM, Grossi-de-Sa MF. 2017. Investigating engineered ribonucleoprotein particles to improve oral RNAi delivery in crop insect pests. Front. Physiol 8:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo WC, Fu KY, Yang S, Li XX, Li GQ. 2015. Instar-dependent systemic RNA interference response in Leptinotarsa decemlineata larvae. Pestic. Biochem. Physiol 123:64–73 [DOI] [PubMed] [Google Scholar]
- 28.Guo X, Wang Y, Sinakevitch I, Lei H, Smith BH. 2018. Comparison of RNAi knockdown effect of tyramine receptor 1 induced by dsRNA and siRNA in brains of the honey bee, Apis mellifera. J. Insect Physiol 111:47–52 [DOI] [PubMed] [Google Scholar]
- 29.Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348:817–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartig JV, Tomari Y, Förstemann K. 2007. piRNAs—the ancient hunters of genome invaders. Genes Dev. 21:1707–13 [DOI] [PubMed] [Google Scholar]
- 31.He B, Chu Y, Yin M, Mullen K, An C, Shen J. 2013. Fluorescent nanoparticle delivered dsRNA toward genetic control of insect pests. Adv. Mater 25:4580–84 [DOI] [PubMed] [Google Scholar]
- 32.Heigwer F, Port F, Boutros M. 2018. RNA interference (RNAi) screening in Drosophila. Genetics 208:853–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang JH, Lee HJ. 2011. RNA interference unveils functions of the hypertrehalosemic hormone on cyclic fluctuation of hemolymph trehalose and oviposition in the virgin female Blattella germanica. J. Insect Physiol 57:858–64 [DOI] [PubMed] [Google Scholar]
- 34.Hughes CL, Kaufman TC. 2000. RNAi analysis of Deformed, proboscipedia and Sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the hemipteran head. Development 127:3683–94 [DOI] [PubMed] [Google Scholar]
- 35.Hunter W, Ellis J, Vanengelsdorp D, Hayes J, Westervelt D, et al. 2010. Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLOS Pathog. 6:e1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivashuta S, Zhang YJ, Wiggins BE, Ramaseshadri P, Segers GC, et al. 2015. Environmental RNAi in herbivorous insects. RNA 21:840–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaubert-Possamai S, Rispe C, Tanguy S, Gordon K, Walsh T, et al. 2010. Expansion of the miRNA pathway in the hemipteran insect Acyrthosiphon pisum. Mol. Biol. Evol 27:979–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joga MR, Zotti MJ, Smagghe G, Christiaens O. 2016. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front. Physiol 7:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlikow M, Goic B, Mongelli V, Salles A, Schmitt C, et al. 2016. Drosophila cells use nanotube-like structures to transfer dsRNA and RNAi machinery between cells. Sci. Rep 6:27085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennerdell JR, Carthew RW 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017–26 [DOI] [PubMed] [Google Scholar]
- 41.Khajuria C, Ivashuta S, Wiggins E, Flagel L, Moar W, et al. 2018. Development and characterization of the first dsRNA-resistant insect population from western corn rootworm, Diabrotica virgifera virgifera LeConte. PLOS ONE 13:e0197059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khajuria C, Velez AM, Rangasamy M, Wang HC, Fishilevich E, et al. 2015. Parental RNA interference of genes involved in embryonic development of the western corn rootworm, Diabrotica virgifera virgifera LeConte. Insect Biochem. Mol. Biol 63:54–62 [DOI] [PubMed] [Google Scholar]
- 43.Kim K, Lee YS, Harris D, Nakahara K, Carthew RW. 2006. The RNAi pathway initiated by Dicer-2 in Drosophila. Cold Spring Harb. Symp. Quant. Biol 71:39–44 [DOI] [PubMed] [Google Scholar]
- 44.Kim YH, Soumaila Issa M, Cooper AM, Zhu KY. 2015. RNA interference: applications and advances in insect toxicology and insect pest management. Pestic. Biochem. Physiol 120:109–17 [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi I, Tsukioka H, Komoto N, Uchino K, Sezutsu H, et al. 2012. SID-1 protein of Caenorhabditis elegans mediates uptake of dsRNA into Bombyx cells. Insect Biochem. Mol. Biol 42:148–54 [DOI] [PubMed] [Google Scholar]
- 46.Kumar P, Pandit SS, Steppuhn A, Baldwin IT. 2014. Natural history-driven, plant-mediated RNAi-based study reveals CYP6B46’s role in a nicotine-mediated antipredator herbivore defense. PNAS 111:1245–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laudani F, Strano CP, Edwards MG, Malacrino A, Campolo O, et al. 2017. RNAi-mediated gene silencing in Rhynchophorus ferrugineus (Oliver) (Coleoptera: Curculionidae). Open Life Sci. 12:214–22 [Google Scholar]
- 48.Lechanteur A, Sanna V, Duchemin A, Evrard B, Mottet D, Piel G. 2018. Cationic liposomes carrying siRNA: impact of lipid composition on physicochemical properties, cytotoxicity and endosomal escape. Nanomaterials 8:E270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee Y, Kim M, Han JJ, Yeom KH, Lee S, et al. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee YS, Nakahara K, Pham JW, Kim K, He Z, et al. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117:69–81 [DOI] [PubMed] [Google Scholar]
- 51.Leggewie M, Schnettler E. 2018. RNAi-mediated antiviral immunity in insects and their possible application. Curr. Opin. Virol 32:108–14 [DOI] [PubMed] [Google Scholar]
- 52.Lewis SH, Salmela H, Obbard DJ. 2016. Duplication and diversification of dipteran Argonaute genes, and the evolutionary divergence of Piwi and Aubergine. Genome Biol. Evol 8:507–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li D, Zhang JQ, Wang Y, Liu XJ, Ma EB, et al. 2015. Two chitinase 5 genes from Locusta migratoria: molecular characteristics and functional differentiation. Insect Biochem. Mol. Biol 58:46–54 [DOI] [PubMed] [Google Scholar]
- 54.Li XX, Dong XL, Zou C, Zhang HY. 2015. Endocytic pathway mediates refractoriness of insect Bactrocera dorsalis to RNA interference. Sci. Rep 5:8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z, Zeng B, Ling L, Xu J, You L, et al. 2015. Enhancement of larval RNAi efficiency by over-expressing Argonaute2 in Bombyx mori. Int. J. Biol. Sci 11:176–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li-Byarlay H, Li Y, Stroud H, Feng S, Newman TC, et al. 2013. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honeybee. PNAS 110:12750–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lozano J, Gomez-Orte E, Lee HJ, Bellés X. 2012. Super-induction of Dicer-2 expression by alien double-stranded RNAs: an evolutionary ancient response to viral infection? Dev. Genes Evol 222:229–35 [DOI] [PubMed] [Google Scholar]
- 58.Lu YH, Park Y, Gao XW, Zhang X, Yao JX, et al. 2012. Cholinergic and non-cholinergic functions of two acetylcholinesterase genes revealed by gene-silencing in Tribolium castaneum. Sci. Rep 2:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lucas KJ, Zhao B, Roy S, Gervaise AL, Raikhel AS. 2015.Mosquito-specific microRNA-1890 targets the juvenile hormone-regulated serine protease JHA15 in the female mosquito gut. RNA Biol. 12:1383–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo Y, Chen Q, Luan J, Chung SH, Van Eck J, et al. 2017. Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem. Mol. Biol 88:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo Y, Wang X, Wang X, Yu D, Chen B, Kang L. 2013. Differential responses of migratory locusts to systemic RNA interference via double-stranded RNA injection and feeding. Insect Mol. Biol 22:574–83 [DOI] [PubMed] [Google Scholar]
- 62.Luo Y, Wang X, Yu D, Kang L. 2012. The SID-1 double-stranded RNA transporter is not required for systemic RNAi in the migratory locust. RNA Biol. 9:663–71 [DOI] [PubMed] [Google Scholar]
- 63.Macedo LLP, Antonino de Souza JD Jr., Coelho RR, Fonseca FCA, Firmino AAP, et al. 2017. Knocking down chitin synthase 2 by RNAi is lethal to the cotton boll weevil. Biotechnol. Res. Innov 1:72–86 [Google Scholar]
- 64.Macrae IJ, Li F, Zhou K, Cande WZ, Doudna JA. 2006. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb. Symp. Quant. Biol 71:73–80 [DOI] [PubMed] [Google Scholar]
- 65.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, et al. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137:522–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, et al. 2007. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol 25:1307–13 [DOI] [PubMed] [Google Scholar]
- 67.Marques JT, Kim K, Wu PH, Alleyne TM, Jafari N, Carthew RW. 2010. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol 17:24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin D, Maestro O, Cruz J, Mane-Padros D, Bellés X. 2006. RNAi studies reveal a conserved role for RXR in molting in the cockroach Blattella germanica. J. Insect Physiol 52:410–16 [DOI] [PubMed] [Google Scholar]
- 69.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123:607–20 [DOI] [PubMed] [Google Scholar]
- 70.McEwan DL, Weisman AS, Huntert CP. 2012. Uptake of extracellular double-stranded RNA by SID-2. Mol. Cell 47:746–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meister G. 2013. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet 14:447–59 [DOI] [PubMed] [Google Scholar]
- 72.Miller SC, Brown SJ, Tomoyasu Y. 2008. Larval RNAi in Drosophila? Dev. Genes Evol 218:505–10 [DOI] [PubMed] [Google Scholar]
- 73.Miller SC, Miyata K, Brown SJ, Tomoyasu Y. 2012. Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLOS ONE 7:e47431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mogilicherla K, Howell JL, Palli SR. 2018. Improving RNAi in the brown marmorated stink bug: identification of target genes and reference genes for RT-qPCR. Sci. Rep 8:3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mondal M, Mansfield K, Flynt A. 2018. siRNAs and piRNAs collaborate for transposon control in the two-spotted spider mite. RNA 24:899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mongelli V, Saleh M-C. 2016. Bugs are not to be silenced: Small RNA pathways and antiviral responses in insects. Annu. Rev. Virol 3:573–89 [DOI] [PubMed] [Google Scholar]
- 77.Murphy KA, Tabuloc CA, Cervantes KR, Chiu JC. 2016. Ingestion of genetically modified yeast symbiont reduces fitness of an insect pest via RNA interference. Sci. Rep 6:22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nandety RS, Kuo YW, Nouri S, Falk BW. 2015. Emerging strategies for RNA interference (RNAi) applications in insects. Bioengineered 6:8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Natarajan P, Sukthankar P, Changstrom J, Holland CS, Barry S, et al. 2018. Synthesis and characterization of multifunctional branched amphiphilic peptide bilayer conjugated gold nanoparticles. ACS Omega 3:11071–83 [Google Scholar]
- 80.Ni M, Ma W, Wang X, Gao M, Dai Y, et al. 2017. Next-generation transgenic cotton: Pyramiding RNAi and Bt counters insect resistance. Plant Biotechnol. J 15:1204–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ninova M, Griffiths-Jones S, Ronshaugen M. 2017. Abundant expression of somatic transposon-derived piRNAs throughout Tribolium castaneum embryogenesis. Genome Biol. 18:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J. 2012. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol. Cell 47:954–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ortiz-Rivas B, Jaubert-Possamai S, Tanguy S, Gauthier JP, Tagu D, Claude R. 2012. Evolutionary study of duplications of the miRNA machinery in aphids associated with striking rate acceleration and changes in expression profiles. BMC Evol. Biol 12:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palli SR. 2014. RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr. Opin. Insect Sci 6:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. 2004. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117:83–94 [DOI] [PubMed] [Google Scholar]
- 86.Pinheiro DH, Velez AM, Fishilevich E, Wang HC, Carneiro NP, et al. 2018. Clathrin-dependent endocytosis is associated with RNAi response in the western corn rootworm, Diabrotica virgifera virgifera LeConte. PLOS ONE 13:e0201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA. 2011. Silencing of aphid genes by dsRNA feeding from plants. PLOS ONE 6:e25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poreddy S, Li J, Baldwin IT. 2017. Plant-mediated RNAi silences midgut-expressed genes in congeneric lepidopteran insects in nature. BMC Plant Biol. 17:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Powell ME, Bradish HM, Gatehouse JA, Fitches EC. 2017. Systemic RNAi in the small hive beetle (Aethina tumida Murray, Coleoptera: Nitidulidae), a serious pest of the European honey bee (Apis mellifera). Pest Manag. Sci 73:53–63 [DOI] [PubMed] [Google Scholar]
- 90.Prentice K, Christiaens O, Pertry I, Bailey A, Niblett C, et al. 2017. RNAi-based gene silencing through dsRNA injection or ingestion against the African sweet potato weevil Cylas puncticollis (Coleoptera: Brentidae). Pest Manag. Sci 73:44–52 [DOI] [PubMed] [Google Scholar]
- 91.Price DR, Gatehouse JA. 2008. RNAi-mediated crop protection against insects. Trends Biotechnol. 26:393–400 [DOI] [PubMed] [Google Scholar]
- 92.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21:5864–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramesh Kumar D, Saravana Kumar P, Gandhi MR, Al-Dhabi NA, Paulraj MG, Ignacimuthu S. 2016. Delivery of chitosan/dsRNA nanoparticles for silencing of wing development vestigial (vg) gene in Aedes aegypti mosquitoes. Int. J. Biol. Macromol 86:89–95 [DOI] [PubMed] [Google Scholar]
- 94.Rangasamy M, Siegfried BD. 2012. Validation of RNA interference in western corn rootworm Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae) adults. Pest Manag. Sci 68:587–91 [DOI] [PubMed] [Google Scholar]
- 95.Ren D, Cai Z, Song J, Wu Z, Zhou S. 2014. dsRNA uptake and persistence account for tissue-dependent susceptibility to RNA interference in the migratory locust, Locusta migratoria. Insect Mol. Biol 23:175–84 [DOI] [PubMed] [Google Scholar]
- 96.Rodrigues TB, Dhandapani RK, Duan JJ, Palli SR. 2017. RNA interference in the Asian longhorned beetle: identification of key RNAi genes and reference genes for RT-qPCR. Sci. Rep 7:8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodrigues TB, Rieske LK, Duan JJ, Mogilicherla K, Palli SR. 2017. Development of RNAi method for screening candidate genes to control emerald ash borer, Agrilus planipennis. Sci. Rep 7:7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rosa C, Kuo YW, Wuriyanghan H, Falk BW. 2018. RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol 56:581–610 [DOI] [PubMed] [Google Scholar]
- 99.Rubio M, Maestro JL, Piulachs M-D, Bellés X. 2018. Conserved association of Argonaute 1 and 2 proteins with miRNA and siRNA pathways throughout insect evolution, from cockroaches to flies. Biochim. Biophys. Acta Gene Regul. Mech 1861:554–60 [DOI] [PubMed] [Google Scholar]
- 100.Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, et al. 2006. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol 8:793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.San Miguel K, Scott JG. 2016. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci 72:801–9 [DOI] [PubMed] [Google Scholar]
- 102.Shi X, Zhang Y, Zhu KY, Ma E, Zhang J, et al. 2017. Comparison of silencing efficacy of the antenna-rich genes by different dsRNA delivery methods in Locusta migratoria. Chin. J. Appl. Entomol 54:780–90 [Google Scholar]
- 103.Shih JD, Hunter CP. 2011. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17:1057–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shukla JN, Kalsi M, Sethi A, Narva KE, Fishilevich E, et al. 2016. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 13:656–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh IK, Singh S, Mogilicherla K, Shukla JN, Palli SR. 2017. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep 7:17059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol 12:246–58 [DOI] [PubMed] [Google Scholar]
- 107.Song H, Fan Y, Zhang J, Cooper AM, Silver K, et al. 2019. Contributions of dsRNases to differential RNAi efficiencies between the injection and oral delivery of dsRNA in Locusta migratoria. Pest Manag. Sci 75:1707–17 [DOI] [PubMed] [Google Scholar]
- 108.Song HF, Zhang JQ, Li DQ, Cooper AMW, Silver K, et al. 2017. A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in migratory locust. Insect Biochem. Mol. Biol 86:68–80 [DOI] [PubMed] [Google Scholar]
- 109.Spit J, Philips A, Wynant N, Santos D, Plaetinck G, Broeck JV. 2017. Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol 81:103–16 [DOI] [PubMed] [Google Scholar]
- 110.Suzuki T, Nunes MA, Espana MU, Namin HH, Jin PY, et al. 2017. RNAi-based reverse genetics in the chelicerate model Tetranychus urticae: a comparative analysis of five methods for gene silencing. PLOS ONE 12:e0180654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, et al. 2014. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol 21:743–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Swevers L, Broeck JV, Smagghe G. 2013.The possible impact of persistent virus infection on the function of the RNAi machinery in insects: a hypothesis. Front. Physiol 4:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Swevers L, Liu J, Smagghe G. 2018. Defense mechanisms against viral infection in Drosophila: RNAi and non-RNAi. Viruses 10:E230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taning CNT, Christiaens O, Berkvens N, Casteels H, Maes M, Smagghe G. 2016. Oral RNAi to control Drosophila suzukii: laboratory testing against larval and adult stages. J. Pest Sci 89:803–14 [Google Scholar]
- 115.Taning CNT, Christiaens O, Li XX, Swevers L, Casteels H, et al. 2018. Engineered flock house virus for targeted gene suppression through RNAi in fruit flies (Drosophila melanogaster) in vitro and in vivo. Front. Physiol 9:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tassetto M, Kunitomi M, Andino R. 2017. Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell 169:314–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, et al. 2011. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol 57:231–45 [DOI] [PubMed] [Google Scholar]
- 118.Thakur N, Upadhyay SK, Verma PC, Chandrashekar K, Tuli R, Singh PK. 2014. Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase A gene. PLOS ONE 9:e87235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tian HG, Peng H, Yao Q, Chen HX, Xie Q, et al. 2009. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLOS ONE 4:e6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D, Bucher G. 2008. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 9:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Van Den Beek M, Da Silva B, Pouch J, Chaouche MEA, Carre C, Antoniewski C. 2018. Dual-layer transposon repression in heads of Drosophila melanogaster. RNA 24:1749–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang G, Jiang L, Zhu L, Cheng T, Niu W, et al. 2013. Characterization of Argonaute family members in the silkworm, Bombyx mori. Insect Sci. 20:78–91 [DOI] [PubMed] [Google Scholar]
- 123.Wang K, Peng Y, Pu J, Fu W, Wang J, Han Z.2016. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol 77:1–9 [DOI] [PubMed] [Google Scholar]
- 124.Wang W, Han BW, Tipping C, Ge DT, Zhang Z, et al. 2015. Slicing and binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol. Cell 59:819–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Whitten M, Dyson P 2017. Gene silencing in non-model insects: overcoming hurdles using symbiotic bacteria for trauma-free sustainable delivery of RNA interference. Bioessays 39(3):1600247. [DOI] [PubMed] [Google Scholar]
- 126.Whyard S, Singh AD, Wong S. 2009. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol 39:824–32 [DOI] [PubMed] [Google Scholar]
- 127.Winston WM, Molodowitch C, Hunter CP. 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295:2456–59 [DOI] [PubMed] [Google Scholar]
- 128.Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP. 2007. Caenorhabditis elegans SID-2 is required for environmental RNA interference. PNAS 104:10565–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu H, March J, Bentley W. 2016. Gene silencing in insect cells using RNAi. Methods Mol. Biol 1350:469–76 [DOI] [PubMed] [Google Scholar]
- 130.Wynant N, Santos D, Van Wielendaele P, Vanden Broeck J. 2014. Scavenger receptor-mediated endocytosis facilitates RNA interference in the desert locust, Schistocerca gregaria. Insect Mol. Biol 23:320–29 [DOI] [PubMed] [Google Scholar]
- 131.Wynant N, Santos D, Vanden Broeck J. 2014. Biological mechanisms determining the success of RNA interference in insects. Int. Rev. Cell Mol. Biol 312:139–67 [DOI] [PubMed] [Google Scholar]
- 132.Wynant N, Santos D, Verdonck R, Spit J, Van Wielendaele P, Vanden Broeck J. 2014. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol 46:1–8 [DOI] [PubMed] [Google Scholar]
- 133.Wynant N, Verlinden H, Breugelmans B, Simonet G, Vanden Broeck J. 2012. Tissue-dependence and sensitivity of the systemic RNA interference response in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol 42:911–17 [DOI] [PubMed] [Google Scholar]
- 134.Xiao D, Gao XW, Xu JP, Liang X, Li QQ, et al. 2015. Clathrin-dependent endocytosis plays a predominant role in cellular uptake of double-stranded RNA in the red flour beetle. Insect Biochem. Mol. Biol 60:68–77 [DOI] [PubMed] [Google Scholar]
- 135.Xu J, Nagata Y, Mon H, Li Z, Zhu L, et al. 2013. Soaking RNAi-mediated modification of Sf9 cells for baculovirus expression system by ectopic expression of Caenorhabditis elegans SID-1. Appl. Microbiol. Biotechnol 97:5921–31 [DOI] [PubMed] [Google Scholar]
- 136.Yang ML, Wei YY, Jiang F, Wang YL, Guo XJ, et al. 2014. MicroRNA-133 inhibits behavioral aggregation by controlling dopamine synthesis in locusts. PLOS Genet. 10:e1004206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ylla G, Fromm B, Piulachs MD, Bellés X. 2016. The microRNA toolkit of insects. Sci. Rep 6:37736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yoon JS, Gurusamy D, Palli SR. 2017. Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem Mol. Biol 90:53–60 [DOI] [PubMed] [Google Scholar]
- 139.Yoon JS, Mogilicherla K, Gurusamy D, Chen X, Chereddy S, Palli SR. 2018. Double-stranded RNA binding protein, Staufen, is required for the initiation of RNAi in coleopteran insects. PNAS 115:8334–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yoon JS, Shukla JN, Gong ZJ, Mogilicherla K, Palli SR. 2016. RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: identification of key contributors. Insect Biochem. Mol. Biol 78:78–88 [DOI] [PubMed] [Google Scholar]
- 141.Yu N, Christiaens O, Liu JS, Niu JZ, Cappelle K, et al. 2013. Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci. 20:4–14 [DOI] [PubMed] [Google Scholar]
- 142.Yu R, Liu W, Li D, Zhao X, Ding G, et al. 2016. Helicoidal organization of chitin in the cuticle of the migratory locust requires the function of the chitin deacetylase 2 enzyme (LmCDA2). J. Biol. Chem 291:24352–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zha WJ, Peng XX, Chen RZ, Du B, Zhu LL, He GC. 2011. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLOS ONE 6:e20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang J, Khan SA, Hasse C, Ruf S, Heckel DG, Bock R. 2015. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 347:991–94 [DOI] [PubMed] [Google Scholar]
- 145.Zhang TT, Liu WW, Li DQ, Gao L, Ma EB, et al. 2018. LmCht5–1 promotes pro-nymphal molting during locust embryonic development. Insect Biochem. Mol. Biol 101:124–30 [DOI] [PubMed] [Google Scholar]
- 146.Zhang X, Mysore K, Flannery E, Michel K, Severson DW, et al. 2015. Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. J. Vis. Exp 97:e52523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang X, Zhang J, Zhu KY. 2010. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol 19:683–93 [DOI] [PubMed] [Google Scholar]
- 148.Zhang YT, Cui J, Zhou YZ, Cao J, Gong HY, et al. 2018. Liposome mediated double-stranded RNA delivery to silence ribosomal protein P0 in the tick Rhipicephalus haemaphysaloides. Ticks Tick-Borne Dis. 9:638–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zheng Y, Hu Y, Yan S, Zhou H, Song D, et al. 2019. A polymer/detergent formulation improves dsRNA penetration through the body wall and RNAi-induced mortality in the soybean aphid Aphis glycines. Pest Manag. Sci 75:1993–99 [DOI] [PubMed] [Google Scholar]
- 150.Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. 2008. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol. Cell 32:592–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhu F, Gujar H, Gordon JR, Haynes KF, Potter MF, Palli SR. 2013. Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci. Rep 3:1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhu F, Parthasarathy R, Bai H, Woithe K, Kaussmann M, et al. 2010. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. PNAS 107:8557–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhu F, Xu J, Palli R, Ferguson J, Palli SR. 2011. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci 67:175–82 [DOI] [PubMed] [Google Scholar]
- 154.Zotti M, Dos Santos EA, Cagliari D, Christiaens O, Taning CNT, Smagghe G. 2018. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci 74:1239–50 [DOI] [PubMed] [Google Scholar]
- 155.Zuhorn IS, Engberts JBFN, Hoekstra D. 2007. Gene delivery by cationic lipid vectors: overcoming cellular barriers. Eur. Biophys. J 36:349–62 [DOI] [PubMed] [Google Scholar]