Abstract
The study of the size of cells and organelles has a long history, dating back to the 1600s when cells were defined. In particular, various methods have elucidated the size of the nucleus and the mitotic spindle in several species. However, little research has been conducted on oocyte size and organelles in mammals, and many questions remain to be answered. The appropriate size is essential to cell function properly. Oocytes have a very large cytoplasm, which is more than 100 times larger than that of general somatic cells in mammals. In this review, we discuss how oocytes acquire an enormous cytoplasmic size and the adverse effects of a large cytoplasmic size on cellular functions.
Keywords: Oocyte, Scaling, Size, Spindle, Spindle assembly checkpoint (SAC)
Introduction
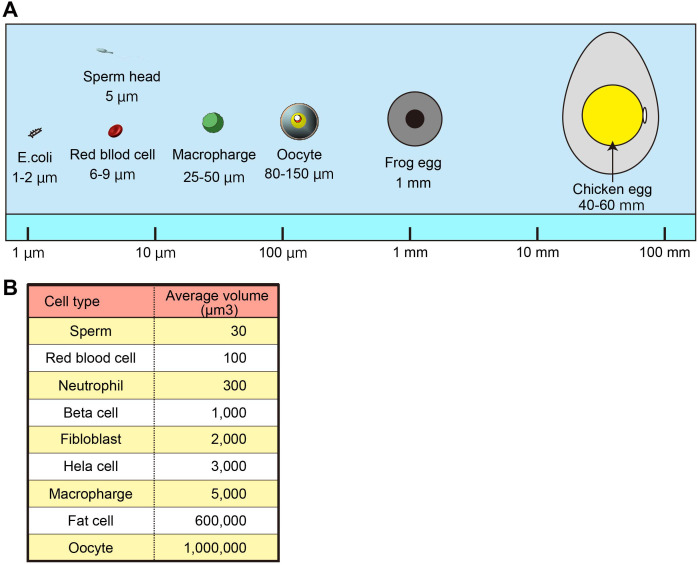
The cell size has been mentioned since Robert Hooke first described the “cell” in 1665. Although early cytologists have found that cell size is relatively constant within a species, in most mammals, oocytes are the largest single cells in the body. For example, the human body comprises 3.7 ± 0.8 × 1013 cells [1], distributed over 200 different cell types [2]. The shapes and sizes of cells depend highly on their function and span a wide range, as shown in Fig. 1. For example, red blood cells need to squeeze through narrow capillaries; their small size and biconcave disk shape make it possible and maximize the surface area to volume ratio contributing to carrying oxygen molecules [3]. In contrast, when transporting signals for long distances, neurons can reach lengths of over a meter with a width of only approximately 10 µm [4]. On the other hand, storage cells, such as fat cells and oocytes, have large volumes. In mammalian species, oocytes are typically spherical, with diameters of approximately 80 µm in mice, 120–130 µm in large animals, including humans, while a typical somatic cell has a diameter of approximately 10–20 µm.
Fig. 1.
Relative sizes of cells. A. Relative size of oocytes. B. The volume of the cells of various sizes in the body.
Oocytes have evolved special mechanisms for arresting the meiotic cell cycle. Oocytes are arrested at the diplotene stage of meiotic prophase I, in a state equivalent to the G2 phase of a mitotic division cycle, lasting from a few days to many years, depending on the species. During this long period, the primary oocytes synthesize and stock proteins, ribosomes, and mRNA necessary for early embryonic development. The oocyte diameter is enlarged from 30 µm to 80 µm in mice. The next phase of oocyte development is maturation. After gonadotropin stimulation, fully grown oocytes resume division I of meiosis and mature to metaphase II (MII). Nuclear envelope breakdown (NEBD) occurs to form spindles during metaphase I (MI) after the resumption of meiosis. The oocyte then undergoes asymmetric division to maintain a large cytoplasm and is arrested at MII until fertilization. After fertilization, division II of meiosis resumes, and the cytoplasm of the large secondary oocyte divides asymmetrically to produce the mature egg and the second small polar body. The oocytes maintain their large size by undergoing two asymmetrical meiotic divisions. Both polar bodies were small and eventually degenerated. After fertilization, the preimplantation embryo undergoes a series of reductive cell divisions without intervening in cell growth, progressing first to a 16–32 cell morula, followed by a 64–128 cell blastocyst stage.
Cells have long been said to maintain their appropriate size by coordinating cell division and growth [5]. Cells use many systems to maintain them; however, different sizes can make it difficult to achieve similar control. A cell type-specific balance between the sizes of various intracellular compartments can support specialized cellular functions. Nevertheless, it may also negatively affect functions involved in common cellular processes, such as cell division. In this review, we discuss the scaling mechanism of oocytes and the merits and demerits of oocytes having a large cytoplasm.
Size of Oocytes
Oocytes have special mechanisms for achieving a large size. In preparation for cell division, somatic cells with a diameter of 10–20 μm typically take approximately 24 h to double in mass. At this biosynthesis rate, somatic cells would take a long time to reach the oocyte size, which has a 1000-fold cytoplasmic volume. A simple strategy for rapid growth is to have additional cell gene copies. Thus, the oocyte delays the completion of the first meiosis and grows while containing a diploid chromosome set in duplicate. Therefore, twice as much DNA is available for RNA synthesis in the oocyte than in an average somatic cell in the G1 phase of the cell cycle. Indeed, oocytes with half a set of chromosomes without undergoing meiosis by knocking out Star8 are small [6]. Moreover, eight transcription factors that drive the primordial-to-primary-follicle transition can convert pluripotent stem cells into oocyte-like cells [7]. These directly induced oocyte-like cells (DIOLs) that do not enter meiosis also have a half-set of chromosomes and show a small size. However, DIOLs are not half the size of in vivo oocytes, suggesting that other mechanisms increase the size of oocytes other than the amount of the genome.
Subcellular Organelle Size in Oocytes
A cell type-specific balance between the sizes of the various intracellular compartments must be appropriately regulated to maintain cell physiology and division. Organelle scaling refers to the phenomenon in which the size and/or the number of organelles and other subcellular structures are generally positively correlated with cell size. Larger cells may contain proportionately larger or more abundant organelles.
Fully grown mammalian oocytes have a specific large nucleus called germinal vesicle (GV). The nucleus is one of the most studied organelles in scaling studies in many species, such as yeast [8, 9, 10], Xenopus [11, 12], worms [13], Arabidopsis [14], and mice [15]. However, there have been no reports of GV scaling. In general, nuclear size correlates with cytoplasmic size, and the nuclear-to-cytoplasmic ratio remains constant [16, 17]. However, the extent of size changes in the nucleus may be constrained by genome size [18]. Indeed, in fully grown mouse oocytes, the nuclear-cytoplasmic ratio is approximately 1:22, which is significantly smaller than that in 2-cell stage embryos (1:16). Therefore, it is thought that oocytes increase GV size through at least two scaling mechanisms. However, further detailed studies are required to confirm this hypothesis.
Fully grown oocytes and zygotes contain specific nucleoli, termed nucleolus precursor bodies (NPBs), which exhibit distinct structural and functional differences compared to nucleoli in somatic cells. In mouse zygotes, the volume of NPB material plays a major role in NPB scaling through a limiting component mechanism [19]. It is thought that NPBs are scaled by a similar mechanism in oocytes because oocytes from NPM2 heterozygous mice form half-sized NPBs [20]. NPM2 is an oocyte-specific nuclear protein, a component of oocytes and zygote NPBs in mice. Although NPM2 –/– mouse oocytes and embryos never form NPBs in the nuclei, NPM2 +/– mice can produce half the amount of protein compared to controls; thus, NPM2 +/– mice can form half the size of the NPBs.
Spindle Scaling in Oocytes
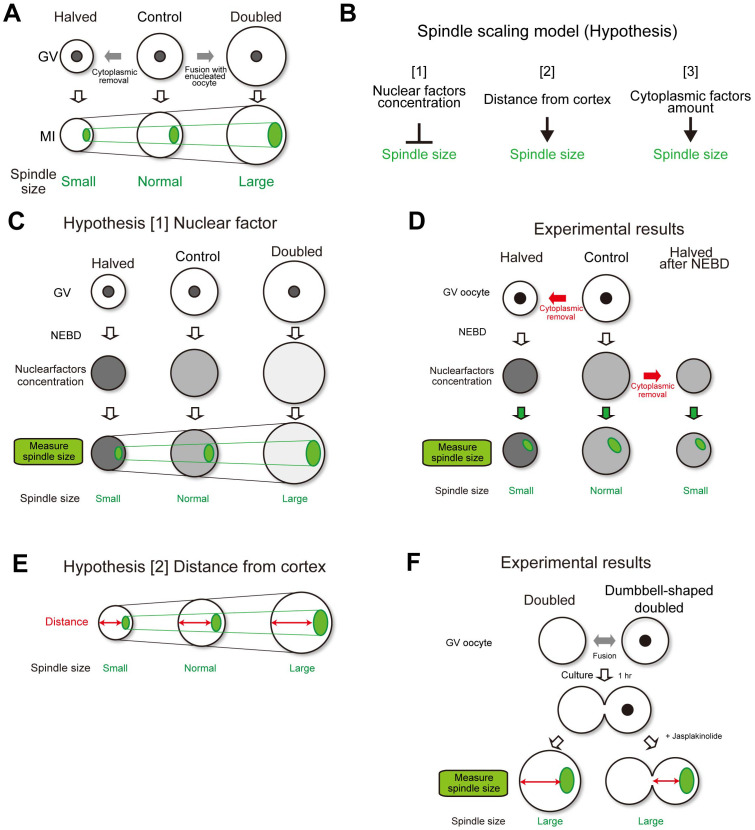
Spindle scaling is essential for properly assembling the spindle microtubule apparatus, which is crucial for faithful chromosome segregation. Spindle scaling has been studied for many decades for various species and methods. In particular, studies using non-mammalian embryos are progressing in vivo and in vitro. When Xenopus embryos undergo rapid cell division following fertilization, spindle size is dramatically changed by decreasing the cell size [16, 21]. In Caenorhabditis elegans embryos, spindle scale size depends on cell size [22, 23]. Moreover, the cell size-dependent spindle scaling is observed in in vitro experiments using encapsulated Xenopus egg extracts [24, 25]. Recent studies have suggested a strong relationship between cell size and spindle length during early embryonic development in mammalians [26,27,28,29]. However, the meiotic spindle shape and formation mechanism differ from that of mitotic spindle formation. In mouse oocytes, the meiosis I spindle lacks astral microtubule and centriole-based microtubule organizing centers [30], and the cortically localized spindle segregates meiotic chromosomes a short distance into the polar body, which is important for asymmetric cell division [30, 31]. These reports suggest that the spindle-scaling mechanism in meiosis I oocytes may differ from that in mitotic cells. Various mechanisms contribute to the regulation of spindle length, including the molecular gradients [32,33,34], density of kinetochore-microtubule attachments [35], balance of spindle forming forces [36], and limited availability of spindle building blocks in the cytoplasm [24, 25]. Our previous report showed an oocyte spindle scaling mechanism [37]. In that study, we generated mouse oocytes carrying half the normal cytoplasmic volume (halved oocytes) and oocytes carrying twice the normal cytoplasmic volume (doubled oocytes) using a micromanipulation technique. Live imaging analysis demonstrated that the spindle volume decreased by approximately half in the halved oocytes, whereas it increased by approximately 2-fold in the doubled oocytes (Fig. 2A).
Fig. 2.
Possible oocyte spindle scaling models. A. Oocyte spindle is scaled by cytoplasmic size. B. Hypotheses of spindle scaling models. C. Spindle scaling by cytoplasmic size is explained with a nuclear factor scaling model. D. In the nuclear factor scaling model, oocytes halved after NEBD should form normal-sized spindles. However, small-sized spindles were formed. E. Spindle scaling by cytoplasmic size is explained with a distance from the cortex scaling model. F. In the distance from the cortex scaling model, dumbbell-shaped doubled oocytes should form normal-sized spindles. However, large-sized spindles were formed.
There are three possible explanations for how the cytoplasm scales the spindles (Fig. 2B). The first hypothesis is that there are nuclear factors (Fig. 2C). Some reports have shown that accurate control of spindle length requires a balanced ratio between nuclear and cytoplasmic volumes [29, 38]. Because the halved and doubled oocytes have different sizes of cytoplasm but the same size of nucleus, the concentration of nuclear factors in the cytoplasm will differ after nuclear envelope breakdown. If nuclear factors have a negative effect on spindle size, this could explain why smaller oocytes have smaller spindles than control oocytes. To test this hypothesis, we removed half of the cytoplasmic volume from oocytes after NEBD. These oocytes carried half the amount of cytoplasm-derived factors and the same concentration of nucleus-derived factors as the control oocytes (Fig. 2D). In the oocytes that were halved after NEBD, the spindle size was significantly smaller than that in the controls but was not significantly different from that in oocytes that were halved before NEBD. Thus, the concentration of the nucleus-derived factors did not significantly influence spindle scaling.
The second hypothesis was based on the distance from the cortex (Fig. 2E). This hypothesis assumes that the oocyte senses the distance between the spindle and cell cortex to regulate spindle size, as observed in C. elegans embryos during anaphase [22]. In larger oocytes, the distance between the spindle and the distal cortex is longer than in control oocytes, which promotes the formation of a larger spindle. To test this hypothesis, dumbbell-shaped double oocytes were generated by interrupting cell rounding after cell fusion (Fig. 2F). The spindle, formed on one side of the dumbbell in the doubled oocytes, was significantly larger than that in the control oocytes but was not significantly different from that in the round doubled oocytes. This finding suggests that the geometry of the cell cortex did not significantly influence spindle scaling.
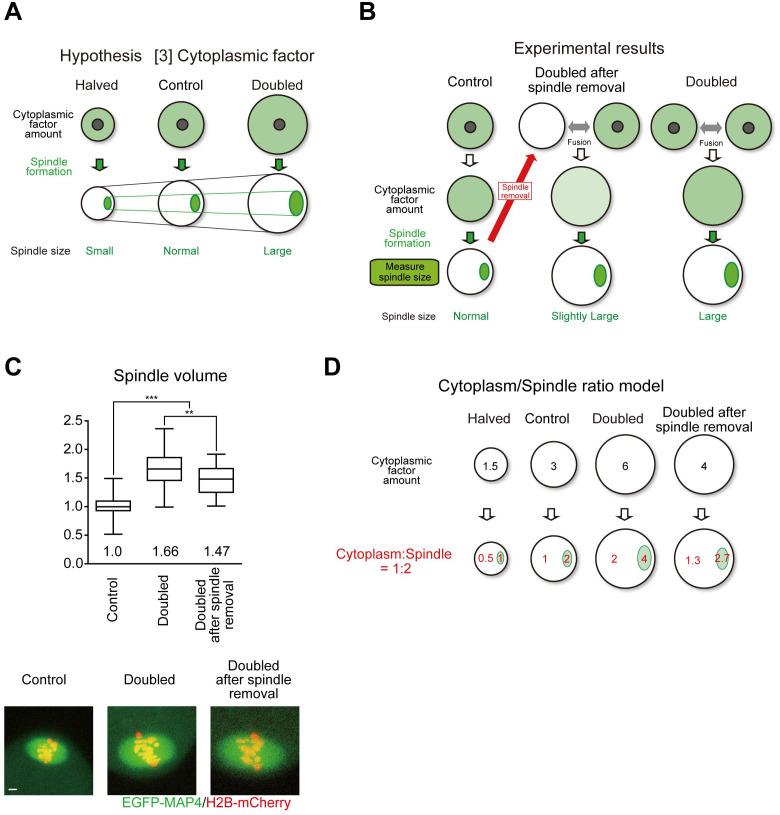
The third hypothesis was based on the presence of cytoplasmic factors (Fig. 3A). This hypothesis assumes that critical components of the spindle exist in the cytoplasm. The cytoplasm may provide finite amounts of spindle components, thus scaling the spindle through a limiting component mechanism [16, 39], as observed in in vitro cell-like compartment experiments using encapsulated Xenopus egg extracts [24, 25]. Larger oocytes have a higher number of critical components of the spindle than control oocytes, forming a larger spindle. In contrast, the number of components in smaller oocytes is limited, resulting in a smaller spindle size. According to this model, spindle components are initially diffused in the cytoplasm and assembled into spindles. Doubled oocytes have double the amount of components, forming a double-sized spindle. A key prediction of this model is that the cytoplasmic pool of spindle components is depleted in the spindle form. To test this hypothesis, we removed the spindle from the oocyte with a reduced number of spindle components and then fused this oocyte with an intact oocyte (unpublished data; Fig. 3B and C). In the oocytes that doubled after spindle removal, the spindle size was significantly smaller than that in the normally doubled oocytes but was still larger than that in the control. Combining this result with the exclusion of the first and second hypotheses, the amount of cytoplasm-derived factors plays a major role in the spindle scaling observed in our system, in agreement with spindle scaling through a limiting component mechanism. However, this mechanism cannot be fully explained. Based on these results, we are considering a model to test the limiting component mechanism with the cytoplasmic spindle ratio (Fig. 3D). In this model, not all spindle-forming factors in the cytoplasm move to the spindle, but are balanced between the cytoplasm and spindle (for example, 1:2). This model could explain all the data.
Fig. 3.
A reliable oocyte spindle scaling model. A. Spindle scaling by cytoplasmic size is explained with a cytoplasmic factor scaling model. B and C. In the cytoplasmic factor scaling model, doubled after spindle removal, oocytes should form normal-sized spindles, however, actually slightly large-sized spindles were formed. Spindle volume was determined at late metaphase (n = 92, 39, and 32 oocytes). Boxes show the 25th–75th percentiles and whiskers the 10th–90th percentiles. Two-tailed, unpaired Student’s t-tests were performed. ** P < 0.01. *** P < 0.001. Scale bars, 5 µm. D. Cytoplasm/Spindle ratio model.
What is the Scaling Factor of the Oocyte Spindle?
Although the scaling mechanism of the oocyte spindle was elucidated in our previous report, the spindle scaling factor remains unclear. For mitotic spindles, the molecular mechanisms of spindle length control have been broadly studied, and many spindle scaling factors have been determined.
The balance between microtubule polymerization and depolymerization is critical for controlling spindle length [28, 40]. Spindle size decreases or stops growing after inhibition of microtubule polymerization factors, such as TOG [41], EB1 [42], and CLASP [43]. The microtubule-depolymerizing kinesin KIF2A, a member of the kinesin-13 family, has been reported to play a role in spindle assembly during mitosis [44,45,46] and meiosis [47, 48]. KIF2A is released from importin α in smaller embryos, where it decreases the spindle size [49]. The microtubule-severing protein katanin is also a well-known spindle length-controlling factor in Xenopus mitotic and meiotic cells [50, 51]. RanGTP-dependent microtubule stabilization and nucleation factors, Cdk11 [52], CHD4 [53], ISWI [54], and MEL-28 [55] have been reported as regulators of spindle assembly. Other microtubule-related factors, such as TAC-1, which is a major regulator of microtubule length in C. elegans embryos [56], and GM130, which regulates microtubule organization in mouse oocytes [57], have also been reported. Many reports have shown that microtubule-associated proteins are involved in mitotic spindle size. Therefore, we tried to overexpress the microtubule-related proteins Cdk11, DKC1, ISWI, POP1, POP4, POP5, Cyclin-L1, Rpp38, Rpp30, TPX2, Ino80, RuvBL1, Rae1, RanBP2, CHD4, and chTOG in mouse oocytes. However, we could not find any proteins that significantly changed the meiotic spindle size (unpublished data).
There are several hypotheses to explain why microtubule-associated factors did not alter the oocyte spindle size. The first hypothesis is that the scaling factors differ among animal species. In a unique pig-mouse inter-species GV transfer model, the spindle size of meiotic progression is controlled by cytoplasmic components rather than cytoplasmic volume and GV materials [58]. A recent report showed that oocyte spindle shape was changed by pole organization [59]. In that report, when spindle pole organization was disrupted in mouse oocytes, the oocyte spindle shape resembled a mitotic spindle. Moreover, the authors showed that the spindle assembly system differed between mouse and human oocytes. The second hypothesis is that oocyte spindle scaling factors are not microtubule-associated factors. A study showed that methylation equilibrium by LCMT1-PME-1, an essential enzyme that regulates the methylation of the protein phosphatase 2A catalytic subunit, is critical for regulating mitotic spindle size [60]. In addition, importin α palmitoylation has recently been shown to play an important role in mitotic spindle size in Xenopus and human cells [61].
Oocyte Size Affects the Accuracy of Spindle Assembly Checkpoint (SAC)
The spindle assembly checkpoint (SAC), a mechanism that prevents chromosome segregation errors by delaying anaphase onset when kinetochores are not attached to microtubules [62, 63], exhibits several unique features in oocytes [64,65,66,67,68,69]. The low accuracy of SAC is one of the factors that makes oocytes more susceptible to error. While a single unattached kinetochore is sufficient to induce a checkpoint-dependent anaphase delay in normal mitotic cells [70], in oocytes, this delay is only triggered when multiple kinetochores are unattached [37, 64, 65, 67, 69, 71]. The low accuracy of SAC in oocytes may be due to its large cytoplasmic volume [72,73,74]. However, the influence of cytoplasmic volume on SAC response in oocytes remains unclear [75].
Our previous study showed that a large cytoplasm decreased SAC accuracy [37]. The timing of anaphase was significantly delayed in half of the oocytes and accelerated in double oocytes. When inhibitors such as Reversine, canceled the function of the SAC, the delay in anaphase timing observed in half of the oocytes was eliminated.
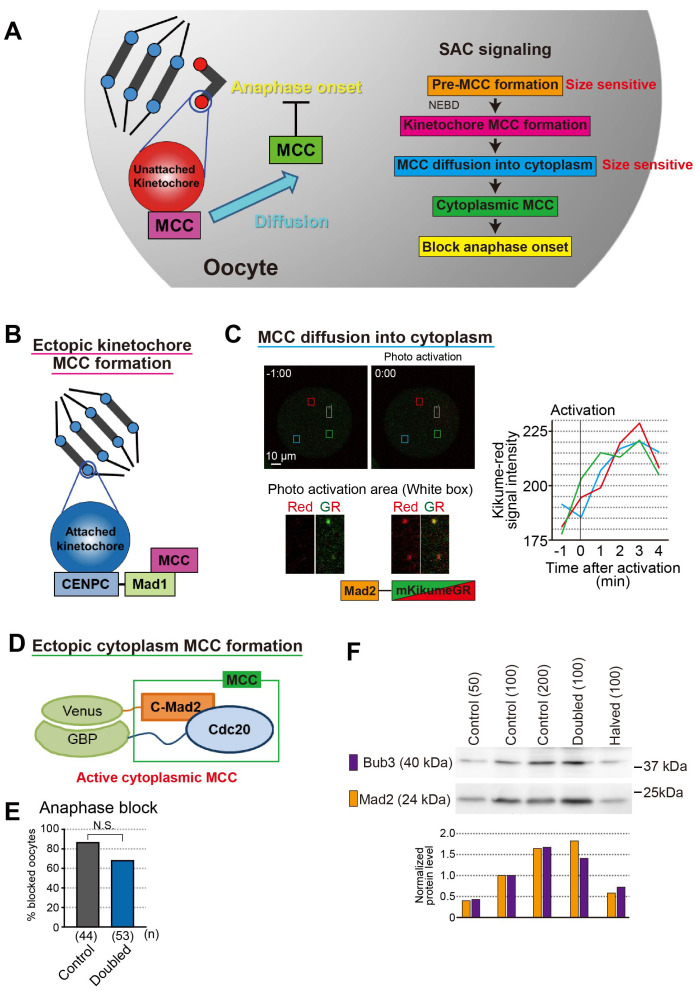
The SAC operates through several processes (Fig. 4A). SAC prevents the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase from recognizing cyclin B and securin by catalyzing the incorporation of the APC/C co-activator CDC20 into a complex called the mitotic checkpoint complex (MCC) [62, 63, 76]. In somatic cells, where on-kinetochore checkpoint activation is minimal, MCC is pre-formed at nuclear pores and then enriched on kinetochores after NEBD [77], suggesting that the anaphase timing onset may depend on the nuclear-to-cytoplasmic ratio prior to NEBD. The next hypothesis is that a larger cytoplasmic size dilutes the SAC signal generated on kinetochores during prometaphase [78]. The prevailing view is that a larger cytoplasmic volume may dilute the active checkpoint signal generated on the kinetochores, weakening the SAC response [78]. This hypothesis is consistent with the observation that Xenopus oocytes have no detectable SAC response and a much larger cytoplasmic volume than mouse oocytes [79]. Moreover, when mouse oocytes are bisected, a single chromosome is sufficient to support the SAC-dependent anaphase delay [80]. A previous study showed that in C. elegans embryos, the strength of SAC, which is activated on all kinetochores through microtubule disruption by nocodazole, depends on the kinetochore-to-cytoplasmic ratio [78]. To distinguish these hypotheses, we removed half of the cytoplasm from mouse oocytes after NEBD. In the oocytes halved after NEBD, which had an intact nuclear-to-cytoplasmic ratio prior to NEBD and a doubled kinetochore-to-cytoplasmic ratio after NEBD, anaphase onset was not significantly delayed compared with that in the control oocytes. The cytoplasmic dilution of checkpoint and MCC components, such as Mad1, Mad2, and BubR1 that are enriched in the nucleus may also contribute to lowering checkpoint accuracy [37], which indicates that the pre-MCC formation step is size sensitive.
Fig. 4.
Spindle assembly checkpoint in oocyte. A. SAC signal pathway. Pre-MCC formation and MCC diffusion into cytoplasm step size sensitive. B. Ectopic kinetochore MCC formation by tethering Mad1 to kinetochore. C. Kinetochore Mad2 diffusion rate was fast and spread uniformly. Mad2-kikumeGR was expressed in oocytes. After activation few kinetochore (white box area), kikume-red signal intensity was measured at three different points (red, blue, and green box areas). The graph showed signal intensities were increased within 3 minutes and increasing rates were same among three different area. D. Ectopic cytoplasm MCC formation by inducing dimerization of C-Mad2 and Cdc20 in the cytoplasm [86]. E. Anaphase block by ectopic cytoplasm MCC formation in control and doubled oocytes. Bar graph shows the percentages of metaphase-arrested oocytes 10.5 h after NEBD. Chi-square tests were performed. N.S., not significant. F. Bub3 and Mad2 protein levels in halved and doubled oocytes. Western blotting analysis was performed and analysis the protein level of Bub3 and Mad2. The protein levels of Bub3 and Mad2 depended on their cytoplasmic amount, suggesting that their concentrations in the cytoplasm were almost constant. The number of oocytes loaded is indicated in parentheses.
Next, we focused on kinetochore MCC formation. The formation of kinetochore C-Mad2, the active form of Mad2, is intact in oocytes of different cytoplasmic sizes oocytes [37]. Moreover, we ectopically activated checkpoint signaling on kinetochores by expressing Mad1, a scaffold protein for checkpoint activation that was fused to the constitutive kinetochore component CENP-C (Fig. 4B) [81]. In the control oocytes, the onset of anaphase was efficiently blocked. In contrast, forced MCC formation failed to block anaphase onset in doubled oocytes efficiently. These data suggest that the downstream events of kinetochore MCC formation are defective in double oocytes and responsiveness to on-kinetochore checkpoint activation depends on the nuclear-to-cytoplasmic ratio rather than on the kinetochore-to-cytoplasmic ratio.
The next step is the diffusion of MCC from the kinetochore to the cytoplasm. All MCC constituents cycle on and off the kinetochores with high turnover rates [82,83,84,85]. By observing the behavior of Mad2 fused with a photoactivated fluorescent protein (KikumeGR), we clarified whether cytoplasmic size affects the diffusion speed and diffusion range (Fig. 4C; unpublished data). The cytoplasmic diffusion rate of kinetochore MCCs was high and spread uniformly in the cytoplasm. However, regarding the diffusion, it possible to assume that the larger the cytoplasm, the more kinetochore MCC is diluted, and the MCC concentration in the cytoplasm decreases; thus, this step can be affected by the cytoplasm size.
Next, we focused on cytoplasmic MCC formation by inducing dimerization of C-Mad2 and Cdc20 [86] (Fig. 4D; unpublished data). This forced formation of cytoplasmic MCC blocked anaphase onset in double oocytes as efficiently as in control oocytes (Fig. 4E). These data suggest that the downstream event of cytoplasmic MCC formation was intact in double oocytes.
In summary, the large cytoplasm of oocytes dilutes nuclear factors, thereby increasing the number of unattached kinetochores required to trigger SAC arrest. MCC, which is pre-formed at the nuclear membrane and is critical for determining checkpoint accuracy [77], is a nuclear factor diluted by the cytoplasm. Further studies are needed to determine the critical molecules that limit SAC accuracy in oocytes. In the SAC molecular pathway, the Mad1-Mad2 complex is recruited to unattached kinetochores by upstream components of the SAC, including Mps1, Aurora B, and Bub1 kinases, and the Rod-Zw10-Zwilch complex [87, 88]. MCC is created dynamically at or near kinetochores [82,83,84,85]. Kinetochore MAD2 consists of a stable and a high-turnover pool [85]. Compelling evidence indicates that Mad1 binding shifts Mad2 from its “open” (O or N1) to “closed” (C or N2) conformation. This not only stabilizes the heterodimer, but also confers prion-like activity, which can induce similar conformation change in soluble O-Mad2 [88, 89]. Similar to C-Mad2, this pool can bind to Cdc20, a key activator of APC/C [90]. In conjunction with a second Cdc20 inhibitor, BubR1 and its cofactors Bub3, C-Mad2, and Cdc20 form one or more MCCs [91,92,93] that inhibit APC/C-mediated proteolysis of securin and cyclin B, thereby delaying sister-chromatid separation and mitotic exit [87, 88]. The concentrations of Mad2 and Bub3 before NEBD did not change in the different cytoplasmic oocytes (unpublished data; Fig. 4F). Therefore, the ratio of O-Mad2 to C-Mad2 may be important for size-dependent SAC activity.
Chromosome Segregation Error Due to Large Cytoplasmic Size of Oocytes
Weakness of the spindle checkpoint contributes to chromosome segregation errors when oocytes carry misaligned chromosomes. Misaligned chromosomes are generated through age-related defects, such as the precocious separation of bivalents into univalents and decreased function of kinetochores [94,95,96,97,98]. In human oocytes, chromosomal misalignment due to unstable spindle bipolarity does not block anaphase onset through the spindle checkpoint [99]. A recent study showed that the efficiency of spermatocyte injection in mice could be greatly improved by reducing the size of recipient oocytes [100]. In this study, spermatocyte chromosomes, which might be intrinsically more error-prone, were dramatically rescued from segregation error by reducing the cytoplasm. However, spermatocyte chromosomes lack the SAC activator MAD2, indicating that increased SAC activity due to cytoplasmic depletion did not reduce chromosome segregation errors. Alternatively, changes in the biochemical environment of the oocyte cytoplasm or spindle forces owing to changes in oocyte size may influence chromosome segregation.
Why do Oocytes have Large Cytoplasm?
The large cytoplasmic size of oocytes is linked to error-prone chromosomal segregation. Why do oocytes have a large cytoplasm? The most reasonable theory is that a large cytoplasmic volume plays a role in supporting embryogenesis. Fully grown oocytes have huge cytoplasmic volumes, thus creating ample space to stock all materials (e.g., maternal proteins and RNAs) needed for embryonic development after fertilization [101]. Indeed, a large cytoplasmic size is critical for post-fertilization development; decreasing the cytoplasmic volume significantly decreases the developmental potential after fertilization [37, 102]. Another hypothesis is that large cytoplasmic size is required to maintain the distance between the site of sperm fertilization and the meiotic MII spindle of oocytes. A study showed that sperm is released to the polar body side when sperm is microinjected near the MII spindle and that there is a mechanism by which sperm move away from the MII spindle after fertilization. [103]. In other words, if the cytoplasmic size is too small to keep away from the MII spindle, sperm may be accidentally released into the polar body. We hypothesized that the error-prone nature of mammalian oocytes before fertilization is a trade-off of the large oocyte volume, which is important for supporting fertilization and post-fertilization development.
Conclusion
In recent years, the development of an in vitro experimental system using egg extracts and microdevices has made it possible to study scaling without a special cellular environment, such as in early embryos, where the cytoplasmic size changes. The in vitro experimental systems have led to great progress in scaling spindle and nuclear sizes. In addition, by using micromanipulation methods that can artificially change the size of the oocyte, it is possible to study the enigmatic organelle scaling and function of the oocyte. There are many unexplained areas in oocyte-specific scaling mechanisms, which are expected to be clarified in the future.
Conflict of interests
The authors declare no competing or financial interests.
Acknowledgments
We thank Dr. T. Miyano for reviewing this manuscript. This work was supported by research grants JSPS KAKENHI 15K18823 to H.K. and 26650072/16H01226/16H06161 to T.S.K. This publication was supported by JSPS KAKENHI (22HP2009).
References
- 1.Naumova AV, Modo M, Moore A, Murry CE, Frank JA. Clinical imaging in regenerative medicine. Nat Biotechnol 2014; 32: 804–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan YS, Farhana A. Histology, Cell. StatPearls. Treasure Island (FL); 2022. [Google Scholar]
- 3.Fabry ME, Kaul DK, Raventos C, Baez S, Rieder R, Nagel RL. Some aspects of the pathophysiology of homozygous Hb CC erythrocytes. J Clin Invest 1981; 67: 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fletcher DA, Theriot JA. An introduction to cell motility for the physical scientist. Phys Biol 2004; 1: T1–T10. [DOI] [PubMed] [Google Scholar]
- 5.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol 2004; 14: R1014–R1027. [DOI] [PubMed] [Google Scholar]
- 6.Dokshin GA, Baltus AE, Eppig JJ, Page DC. Oocyte differentiation is genetically dissociable from meiosis in mice. Nat Genet 2013; 45: 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamazaki N, Kyogoku H, Araki H, Miura F, Horikawa C, Hamada N, Shimamoto S, Hikabe O, Nakashima K, Kitajima TS, Ito T, Leitch HG, Hayashi K. Reconstitution of the oocyte transcriptional network with transcription factors. Nature 2021; 589: 264–269. [DOI] [PubMed] [Google Scholar]
- 8.Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. The size of the nucleus increases as yeast cells grow. Mol Biol Cell 2007; 18: 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kume K, Cantwell H, Neumann FR, Jones AW, Snijders AP, Nurse P. A systematic genomic screen implicates nucleocytoplasmic transport and membrane growth in nuclear size control. PLoS Genet 2017; 13: e1006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol 2007; 179: 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara Y, Merten CA. Dynein-based accumulation of membranes regulates nuclear expansion in Xenopus laevis egg extracts. Dev Cell 2015; 33: 562–575. [DOI] [PubMed] [Google Scholar]
- 12.Levy DL, Heald R. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell 2010; 143: 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uppaluri S, Weber SC, Brangwynne CP. Hierarchical size scaling during multicellular growth and development. Cell Reports 2016; 17: 345–352. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Dittmer TA, Richards EJ. Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol 2013; 13: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsichlaki E, FitzHarris G. Nucleus downscaling in mouse embryos is regulated by cooperative developmental and geometric programs. Sci Rep 2016; 6: 28040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy DL, Heald R. Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol 2012; 28: 113–135. [DOI] [PubMed] [Google Scholar]
- 17.Webster A, Schuh M. Mechanisms of aneuploidy in human eggs. Trends Cell Biol 2017; 27: 55–68. [DOI] [PubMed] [Google Scholar]
- 18.Cavalier-Smith T. Skeletal DNA and the evolution of genome size. Annu Rev Biophys Bioeng 1982; 11: 273–302. [DOI] [PubMed] [Google Scholar]
- 19.Kyogoku H, Wakayama T, Kitajima TS, Miyano T. Single nucleolus precursor body formation in the pronucleus of mouse zygotes and SCNT embryos. PLoS One 2018; 13: e0202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, Eppig JJ, Matzuk MM. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science 2003; 300: 633–636. [DOI] [PubMed] [Google Scholar]
- 21.Wühr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, Mitchison TJ. Evidence for an upper limit to mitotic spindle length. Curr Biol 2008; 18: 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara Y, Kimura A. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr Biol 2009; 19: 1549–1554. [DOI] [PubMed] [Google Scholar]
- 23.Reber S, Goehring NW. Intracellular scaling mechanisms. Cold Spring Harb Perspect Biol 2015; 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science 2013; 342: 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 2013; 342: 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Courtois A, Schuh M, Ellenberg J, Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol 2012; 198: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamagata K, FitzHarris G. 4D imaging reveals a shift in chromosome segregation dynamics during mouse pre-implantation development. Cell Cycle 2013; 12: 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol 2010; 26: 21–57. [DOI] [PubMed] [Google Scholar]
- 29.Novakova L, Kovacovicova K, Dang-Nguyen TQ, Sodek M, Skultety M, Anger M. A balance between nuclear and cytoplasmic volumes controls spindle length. PLoS One 2016; 11: e0149535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ, Verlhac MH. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J Cell Biol 2007; 176: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verlhac MH, Lefebvre C, Guillaud P, Rassinier P, Maro B. Asymmetric division in mouse oocytes: with or without Mos. Curr Biol 2000; 10: 1303–1306. [DOI] [PubMed] [Google Scholar]
- 32.Bastiaens P, Caudron M, Niethammer P, Karsenti E. Gradients in the self-organization of the mitotic spindle. Trends Cell Biol 2006; 16: 125–134. [DOI] [PubMed] [Google Scholar]
- 33.Greenan G, Brangwynne CP, Jaensch S, Gharakhani J, Jülicher F, Hyman AA. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr Biol 2010; 20: 353–358. [DOI] [PubMed] [Google Scholar]
- 34.Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci 2008; 121: 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nannas NJ, O’Toole ET, Winey M, Murray AW. Chromosomal attachments set length and microtubule number in the Saccharomyces cerevisiae mitotic spindle. Mol Biol Cell 2014; 25: 4034–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol 2009; 19: R749–R761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyogoku H, Kitajima TS. Large cytoplasm is linked to the error-prone nature of oocytes. Dev Cell 2017; 41: 287–298.e4. [DOI] [PubMed] [Google Scholar]
- 38.Cui LB, Huang XY, Sun FZ. Nucleocytoplasmic ratio of fully grown germinal vesicle oocytes is essential for mouse meiotic chromosome segregation and alignment, spindle shape and early embryonic development. Hum Reprod 2005; 20: 2946–2953. [DOI] [PubMed] [Google Scholar]
- 39.Marshall WF. Subcellular size. Cold Spring Harb Perspect Biol 2015; 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alfaro-Aco R, Petry S. Building the microtubule cytoskeleton piece by piece. J Biol Chem 2015; 290: 17154–17162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheeseman LP, Harry EF, McAinsh AD, Prior IA, Royle SJ. Specific removal of TACC3-ch-TOG-clathrin at metaphase deregulates kinetochore fiber tension. J Cell Sci 2013; 126: 2102–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, Borisy GG, Steinmetz MO, Akhmanova A. Mammalian end binding proteins control persistent microtubule growth. J Cell Biol 2009; 184: 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Espiritu EB, Krueger LE, Ye A, Rose LS. CLASPs function redundantly to regulate astral microtubules in the C. elegans embryo. Dev Biol 2012; 368: 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uematsu K, Okumura F, Tonogai S, Joo-Okumura A, Alemayehu DH, Nishikimi A, Fukui Y, Nakatsukasa K, Kamura T. ASB7 regulates spindle dynamics and genome integrity by targeting DDA3 for proteasomal degradation. J Cell Biol 2016; 215: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsieh PC, Chiang ML, Chang JC, Yan YT, Wang FF, Chou YC. DDA3 stabilizes microtubules and suppresses neurite formation. J Cell Sci 2012; 125: 3402–3411. [DOI] [PubMed] [Google Scholar]
- 46.Kwon HJ, Park JE, Song H, Jang CY. DDA3 and Mdp3 modulate Kif2a recruitment onto the mitotic spindle to control minus-end spindle dynamics. J Cell Sci 2016; 129: 2719–2725. [DOI] [PubMed] [Google Scholar]
- 47.Yi ZY, Ma XS, Liang QX, Zhang T, Xu ZY, Meng TG, Ouyang YC, Hou Y, Schatten H, Sun QY, Quan S. Kif2a regulates spindle organization and cell cycle progression in meiotic oocytes. Sci Rep 2016; 6: 38574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen MH, Liu Y, Wang YL, Liu R, Xu BH, Zhang F, Li FP, Xu L, Lin YH, He SW, Liao BQ, Fu XP, Wang XX, Yang XJ, Wang HL. KIF2A regulates the spindle assembly and the metaphase I-anaphase I transition in mouse oocyte. Sci Rep 2016; 6: 39337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilbur JD, Heald R. Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife 2013; 2: e00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol 2006; 175: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loughlin R, Wilbur JD, McNally FJ, Nédélec FJ, Heald R. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 2011; 147: 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yokoyama H, Gruss OJ, Rybina S, Caudron M, Schelder M, Wilm M, Mattaj IW, Karsenti E. Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate. J Cell Biol 2008; 180: 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokoyama H, Nakos K, Santarella-Mellwig R, Rybina S, Krijgsveld J, Koffa MD, Mattaj IW. CHD4 is a RanGTP-dependent MAP that stabilizes microtubules and regulates bipolar spindle formation. Curr Biol 2013; 23: 2443–2451. [DOI] [PubMed] [Google Scholar]
- 54.Yokoyama H, Rybina S, Santarella-Mellwig R, Mattaj IW, Karsenti E. ISWI is a RanGTP-dependent MAP required for chromosome segregation. J Cell Biol 2009; 187: 813–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokoyama H, Koch B, Walczak R, Ciray-Duygu F, González-Sánchez JC, Devos DP, Mattaj IW, Gruss OJ. The nucleoporin MEL-28 promotes RanGTP-dependent γ-tubulin recruitment and microtubule nucleation in mitotic spindle formation. Nat Commun 2014; 5: 3270. [DOI] [PubMed] [Google Scholar]
- 56.Le Bot N, Tsai M-C, Andrews RK, Ahringer J. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr Biol 2003; 13: 1499–1505. [DOI] [PubMed] [Google Scholar]
- 57.Zhang CH, Wang ZB, Quan S, Huang X, Tong JS, Ma JY, Guo L, Wei YC, Ouyang YC, Hou Y, Xing FQ, Sun QY. GM130, a cis-Golgi protein, regulates meiotic spindle assembly and asymmetric division in mouse oocyte. Cell Cycle 2011; 10: 1861–1870. [DOI] [PubMed] [Google Scholar]
- 58.Wang ZW, Zhang GL, Schatten H, Carroll J, Sun QY. Cytoplasmic determination of meiotic spindle size revealed by a unique inter-species germinal vesicle transfer model. Sci Rep 2016; 6: 19827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.So C, Menelaou K, Uraji J, Harasimov K, Steyer AM, Seres KB, Bucevičius J, Lukinavičius G, Möbius W, Sibold C, Tandler-Schneider A, Eckel H, Moltrecht R, Blayney M, Elder K, Schuh M. Mechanism of spindle pole organization and instability in human oocytes. Science 2022; 375: eabj3944. [DOI] [PubMed] [Google Scholar]
- 60.Xia X, Gholkar A, Senese S, Torres JZ. A LCMT1-PME-1 methylation equilibrium controls mitotic spindle size. Cell Cycle 2015; 14: 1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brownlee C, Heald R. Importin alpha partitioning to the plasma membrane regulates intracellular scaling. Cell 2019; 176: 805–815.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.London N, Biggins S. Signalling dynamics in the spindle checkpoint response. Nat Rev Mol Cell Biol 2014; 15: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol 2015; 25: R1002–R1018. [DOI] [PubMed] [Google Scholar]
- 64.Gui L, Homer H. Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes. Development 2012; 139: 1941–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolano A, Brunet S, Silk AD, Cleveland DW, Verlhac MH. Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc Natl Acad Sci USA 2012; 109: E1858–E1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lane SIR, Jones KT. Non-canonical function of spindle assembly checkpoint proteins after APC activation reduces aneuploidy in mouse oocytes. Nat Commun 2014; 5: 3444. [DOI] [PubMed] [Google Scholar]
- 67.Lane SIR, Yun Y, Jones KT. Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development 2012; 139: 1947–1955. [DOI] [PubMed] [Google Scholar]
- 68.Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet 2012; 13: 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sebestova J, Danylevska A, Novakova L, Kubelka M, Anger M. Lack of response to unaligned chromosomes in mammalian female gametes. Cell Cycle 2012; 11: 3011–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol 1995; 130: 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagaoka SI, Hodges CA, Albertini DF, Hunt PA. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr Biol 2011; 21: 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gorbsky GJ. The spindle checkpoint and chromosome segregation in meiosis. FEBS J 2015; 282: 2471–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones KT, Lane SI. Molecular causes of aneuploidy in mammalian eggs. Development 2013; 140: 3719–3730. [DOI] [PubMed] [Google Scholar]
- 74.Verlhac MH, Terret ME. Oocyte maturation and development. F1000 Res 2016; 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerhold AR, Labbé JC, Maddox PS. Bigger isn’t always better: cell size and the spindle assembly checkpoint. Dev Cell 2016; 36: 244–246. [DOI] [PubMed] [Google Scholar]
- 76.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 2006; 7: 644–656. [DOI] [PubMed] [Google Scholar]
- 77.Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, Shah JV, Jallepalli PV. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 2014; 156: 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Galli M, Morgan DO. Cell size determines the strength of the spindle assembly checkpoint during embryonic development. Dev Cell 2016; 36: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shao H, Li R, Ma C, Chen E, Liu XJ. Xenopus oocyte meiosis lacks spindle assembly checkpoint control. J Cell Biol 2013; 201: 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoffmann S, Maro B, Kubiak JZ, Polanski Z. A single bivalent efficiently inhibits cyclin B1 degradation and polar body extrusion in mouse oocytes indicating robust SAC during female meiosis I. PLoS One 2011; 6: e27143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signaling from chromosome biorientation (vol 13, pg 475, 2011). Nat Cell Biol 2011; 13: 633–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol 2000; 150: 1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol 2004; 14: 953–964. [DOI] [PubMed] [Google Scholar]
- 84.Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol 2002; 158: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol 2004; 14: 942–952. [DOI] [PubMed] [Google Scholar]
- 86.Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature 2015; 517: 631–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 2013; 14: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 2007; 8: 379–393. [DOI] [PubMed] [Google Scholar]
- 89.Yu H. Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model. J Cell Biol 2006; 173: 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol 2011; 12: 427–438. [DOI] [PubMed] [Google Scholar]
- 91.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell 2002; 13: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sudakin V, Chan GKT, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 2001; 154: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell 2001; 1: 227–237. [DOI] [PubMed] [Google Scholar]
- 94.Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol 2010; 20: 1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, Jessberger R, Kirkwood TB, Höög C, Herbert M. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol 2010; 20: 1511–1521. [DOI] [PubMed] [Google Scholar]
- 96.Sakakibara Y, Hashimoto S, Nakaoka Y, Kouznetsova A, Höög C, Kitajima TS. Bivalent separation into univalents precedes age-related meiosis I errors in oocytes. Nat Commun 2015; 6: 7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shomper M, Lappa C, FitzHarris G. Kinetochore microtubule establishment is defective in oocytes from aged mice. Cell Cycle 2014; 13: 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zielinska AP, Holubcova Z, Blayney M, Elder K, Schuh M. Sister kinetochore splitting and precocious disintegration of bivalents could explain the maternal age effect. eLife 2015; 4: e11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holubcová Z, Blayney M, Elder K, Schuh M. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 2015; 348: 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ogonuki N, Kyogoku H, Hino T, Osawa Y, Fujiwara Y, Inoue K, Kunieda T, Mizuno S, Tateno H, Sugiyama F, Kitajima TS, Ogura A. Birth of mice from meiotically arrested spermatocytes following biparental meiosis in halved oocytes. EMBO Rep 2022; 23: e54992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alberts B, Johnson A, Lewis J, Raff M, Roberts K. Eggs. In: Molecular Biology of the Cell, Fourth Edition (Garland Science). 2002. [Google Scholar]
- 102.Wakayama T, Yanagimachi R. Fertilisability and developmental ability of mouse oocytes with reduced amounts of cytoplasm. Zygote 1998; 6: 341–346. [DOI] [PubMed] [Google Scholar]
- 103.Mori M, Yao T, Mishina T, Endoh H, Tanaka M, Yonezawa N, Shimamoto Y, Yonemura S, Yamagata K, Kitajima TS, Ikawa M. RanGTP and the actin cytoskeleton keep paternal and maternal chromosomes apart during fertilization. J Cell Biol 2021; 220: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]